Abstract
Migraine is three times more prevalent in people with bipolar disorder or depression. The relationship between schizophrenia and migraine is less certain although glutamatergic and serotonergic neurotransmission are implicated in both. A shared genetic basis to migraine and mental disorders has been suggested but previous studies have reported weak or non-significant genetic correlations and five shared risk loci. Using the largest samples to date and novel statistical tools, we aimed to determine the extent to which migraine’s polygenic architecture overlaps with bipolar disorder, depression and schizophrenia beyond genetic correlation, and to identify shared genetic loci.
Summary statistics from genome-wide association studies were acquired from large-scale consortia for migraine (n cases = 59 674; n controls = 316 078), bipolar disorder (n cases = 20 352; n controls = 31 358), depression (n cases = 170 756; n controls = 328 443) and schizophrenia (n cases = 40 675, n controls = 64 643). We applied the bivariate causal mixture model to estimate the number of disorder-influencing variants shared between migraine and each mental disorder, and the conditional/conjunctional false discovery rate method to identify shared loci. Loci were functionally characterized to provide biological insights.
Univariate MiXeR analysis revealed that migraine was substantially less polygenic (2.8 K disorder-influencing variants) compared to mental disorders (8100–12 300 disorder-influencing variants). Bivariate analysis estimated that 800 (SD = 300), 2100 (SD = 100) and 2300 (SD = 300) variants were shared between bipolar disorder, depression and schizophrenia, respectively. There was also extensive overlap with intelligence (1800, SD = 300) and educational attainment (2100, SD = 300) but not height (1000, SD = 100). We next identified 14 loci jointly associated with migraine and depression and 36 loci jointly associated with migraine and schizophrenia, with evidence of consistent genetic effects in independent samples. No loci were associated with migraine and bipolar disorder. Functional annotation mapped 37 and 298 genes to migraine and each of depression and schizophrenia, respectively, including several novel putative migraine genes such as L3MBTL2, CACNB2 and SLC9B1. Gene-set analysis identified several putative gene sets enriched with mapped genes including transmembrane transport in migraine and schizophrenia.
Most migraine-influencing variants were predicted to influence depression and schizophrenia, although a minority of mental disorder-influencing variants were shared with migraine due to the difference in polygenicity. Similar overlap with other brain-related phenotypes suggests this represents a pool of ‘pleiotropic’ variants that influence vulnerability to diverse brain-related disorders and traits. We also identified specific loci shared between migraine and each of depression and schizophrenia, implicating shared molecular mechanisms and highlighting candidate migraine genes for experimental validation.
Keywords: migraine, schizophrenia, bipolar disorder, depression, genetics
Migraine and mental disorders are heritable, but most of their genetic determinants are unknown. In this post-GWAS analysis, Bahrami et al. show that most common genetic variants that influence migraine also influence schizophrenia and depression, and identify 50 jointly associated genetic loci and 335 putative shared causal genes.
Introduction
People with migraine are three times more likely to suffer from bipolar disorder or depression than the general population and vice versa.1-5 The clinical overlap between schizophrenia and migraine is less well delineated. Epidemiological studies report positive,6 negative7 or no association,8 but are limited by small or non-representative samples. Nonetheless, several neurobiological systems are implicated in both disorders, including serotonergic9 and glutamatergic dysfunction.10 It has been hypothesized that a shared genetic basis contributes to the phenotypic and patho-aetiological overlap observed between migraine and mental disorders.11
Although the aetiologies of all four disorders remain elusive,12–15 twin studies have revealed strong genetic influences, with broad-sense heritabilities of ∼40% for migraine and between 40% and 80% for mental disorders.16–19 Genome-wide association studies (GWAS), which test for association between a disorder and millions of common single nucleotide polymorphisms (SNPs), have since demonstrated that approximately half of the total heritability for each disorder is due to common genetic variants.20–23 GWAS have also identified 38, 30, 102 and 176 genetic risk loci for migraine, bipolar disorder, depression and schizophrenia, respectively, with the latest migraine GWAS implicating a predominance of genes expressed in vascular tissues.20–23 However, thousands of variants are predicted to influence their risk with small effects, the majority of which have not been discovered.24 The discovery of disorder-influencing variants (also referred to as ‘causal variants’) is further complicated by the tendency for physically approximate alleles to be inherited together, termed linkage disequilibrium (LD). SNP effect sizes are therefore highly correlated across blocks of SNPs, effectively concealing disorder-influencing variants among large numbers of correlated SNPs.
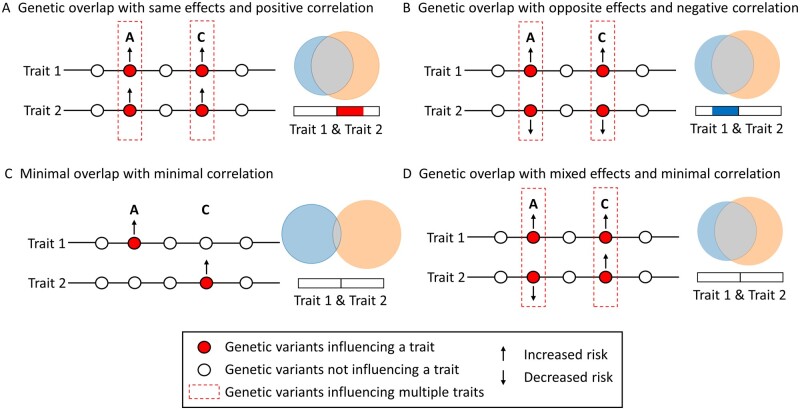
Traditional methods investigating genetic overlap include genetic correlation and polygenic risk scores, which return a single estimate of genome-wide genetic overlap. A significant positive genetic correlation of 0.32 has been reported between migraine and depression but not bipolar disorder or schizophrenia.25 This implies that increased genetic risk of migraine increases risk of depression and vice versa, possibly contributing to their comorbidity, but not for bipolar disorder or schizophrenia. An increased polygenic risk score for schizophrenia was also associated with reduced risk of migraine, although this study was limited by small sample sizes.10 An important limitation to genetic correlation and polygenic risk scores, however, is that they require a predominant correlation of effect sizes across all SNPs between two traits, which may not be the case with brain-related traits where genetic effects with a mixture of same and opposite effect directions have been reported in several cross-GWAS analyses (Fig. 1).27–29 In contrast, the bivariate causal mixture model (MiXeR) provides a ‘bird’s eye view’ of genetic overlap by first inferring the total number of disorder-influencing variants beyond the confounding effect of LD. This is a key advance on standard frameworks such as linkage disequilibrium score regression (LDSR), which are based on the infinitesimal model that assumes that all variants have an infinitesimally small effect on all phenotypes.30 Bivariate analysis next estimates the total number of shared and unique variants influencing two traits regardless of effect direction.31 This has enabled the discovery of extensive genetic overlap between brain-related traits with non-significant genetic correlation and a mixture of variants with same and opposite effect directions.26,29,32,33 While this indicates that genetic risk for one disorder does not correlate with risk for the second at the genome-wide level, this is nevertheless biologically relevant since it may suggest shared molecular mechanisms.34 It is important to note, however, that MiXeR only estimates the number of disorder-influencing variants, without identifying genomic loci hosting those variants.
Figure 1.
Conceptual figure illustrating MiXeR’s ability to characterise genetic overlap beyond genetic correlation adapted from Smeland et al.26 Four scenarios of genetic overlap (A–D) with corresponding MiXeR Venn diagrams and genetic correlations. Genetic overlap with a preponderance of shared variants with the same (A) or opposite (B) effect directions results in either positive (A, red bar) or negative (B, blue bar) genetic correlation. However, since genetic correlation provides an estimate of the correlation of effect sizes between two traits, it is unable to discriminate between scenarios C and D, both of which return minimal genetic correlations. In contrast, since MiXeR estimates the number of shared variants regardless of effect direction, MiXeR successfully estimates more extensive genetic overlap in D compared to C, represented by the larger shared component (grey).
Identifying genetic overlap at the individual locus level is therefore essential to obtain biological insights. Previously, cross-GWAS analysis identified three SNPs associated with migraine and depression, two of which mapped to the ANKDD1B and KCNK5 genes, respectively.35 However, standard cross-GWAS techniques rely on family-wise error rate-based methods, requiring massive correction for multiple testing which limits the statistical power to discover shared genetic loci.36 In contrast, the conditional/conjunctional false discovery rate method (cond/conjFDR) uses a Bayesian approach that leverages information from two GWAS to identify shared loci without the same loss in statistical power, thus increasing the yield in comparison to standard approaches.32,36
Despite their comorbidity, there have been few attempts to investigate genetic overlap between migraine and bipolar disorder, depression and schizophrenia, which possess overlapping symptom profiles, large proportions of overlapping common genetic variants,26,31 and moderate-to-strong positive genetic correlation (rg = 0.3–0.7).25 Understanding how migraine’s genetic determinants relate to this clinico-genetic spectrum may offer insights into the landscape of shared genetic architecture across brain-related traits and highlight putative shared aetiological processes, with potential applications to personalized treatment. It can also form the basis for follow-up experimental studies to determine the biological significance of the overlapping regions.
We therefore applied MiXeR and cond/conjFDR to GWAS summary statistics to (i) describe the shared genetic architecture of migraine and mental disorders beyond genetic correlations; (ii) identify and characterize specific shared genomic loci; and (iii) identify novel risk loci for migraine.
Materials and methods
Samples
We acquired summary statistics for migraine, comprising 59 674 cases and 316 078 controls, including self-reported migraine21 and a replication sample comprising 18 682 cases and 362 593 controls.37,38 The bipolar disorder sample comprised 20 352 cases and 31 358 controls,20 the depression sample 170 756 cases and 328 443 controls,39,40 and the schizophrenia sample 40 675 cases and 64 643 controls.41 Studies were published between June 2016 and May 2019 and selected to maximize available sample size. For further details, including data availability and additional replication samples, see the Supplementary material. Full genotyping procedures are detailed in the original publications. The Regional Committee for Medical and Health Research Ethics South-East Norway evaluated the protocol and found that no additional ethical approval was required because no individual data were used.
Data analysis
We applied MiXeR v.1.3 to quantify polygenic overlap between migraine and mental disorders.31 We constructed a univariate mixture model to estimate the number of disorder-influencing SNPs based on the distributions of the primary GWAS test statistics (i.e. SNPs underlying the association with the disorder beyond LD, also termed ‘casual SNPs’). Next, a bivariate gaussian mixture model was constructed to quantify the additive effect of four components: (i) SNPs not influencing either phenotype; SNPs uniquely influencing either the (ii) primary; or (iii) secondary phenotype (unique disorder-influencing SNPs); and (iv) SNPs influencing both phenotypes (shared disorder-influencing SNPs). Results were presented as Venn diagrams displaying the proportion of unique and shared SNPs. Model fit was based on likelihood maximization of signed test statistics (GWAS z-scores), the Akaike Information Criterion (AIC) and predicted versus observed conditional quantile–quantile (Q-Q) plots (Supplementary material). We also calculated LD score regression based genetic correlations.42
We next applied cond/conjFDR, which leverages polygenic overlap between two traits to boost statistical power to identify novel loci associated with a single trait (condFDR) and loci jointly associated with two traits (conjFDR).36 Cross-trait enrichment of SNP associations between migraine and each mental disorder was visualized using conditional Q–Q plots. The condFDR value of each SNP was computed for migraine conditional on mental disorders and vice versa. CondFDR represents the probability that a SNP is not associated with the primary trait given that the P-values in the primary and conditional trait are as small or smaller than the observed P-values. The conjFDR value for each SNP was next calculated as the maximum of the two condFDR values (i.e. migraine conditional on bipolar disorder and vice versa). This represents a conservative estimate of the FDR for the association between each SNP with both traits. SNPs with a cond/conjFDR < 0.05 were assigned statistical significance.36 Further details are provided in Supplementary material and previous publications.36,43
Genomic loci definition
Because of the confounding effect of LD and in line with previous literature, significantly associated SNPs were clumped into discrete ‘genomic loci’ defined by their cond/conjFDR value, LD structure (r2-value) and physical proximity.44 Each locus comprised: a lead SNP—the most significantly associated SNP; independent significant SNPs—significant SNPs (cond/conjFDR < 0.05) which are not in LD with each other (r2 < 0.6) but physically approximate (<250 kb apart); and candidate SNPs—in LD with independent significant SNPs (r2 ≥ 0.6) and with evidence of association with the primary trait/traits (cond/conjFDR < 0.1). In line with previous literature, the candidate SNP cond/conjFDR thresholds were lowered to maximize the probability that putative disorder-influencing SNPs were captured within each locus.34 Novel loci were identified by cross-referencing candidate SNPs against the NHGRI-EBI catalogue and minimum and maximum base pair positions with loci reported in original GWAS publications and secondary GWAS analyses of migraine and mental disorders.20,21,28,39,41,45–47
Replication of conditional FDR and conjunctional FDR significant loci in independent samples
Since the probability of replicating individual loci at genome-wide significance is low due to weak genetic effects, we first tested for en masse sign concordance of effect direction in primary and replication samples, in line with previous literature.48–50 For condFDR analyses, there was sign concordance if the lead SNP had concordant effect directions in the primary and replication migraine samples. For conjFDR, there was sign concordance if the lead SNP possessed concordant effect directions for both phenotypes. If lead SNPs were missing in the independent datasets for conjFDR analyses, we replaced them with the next most significant candidate SNP which was present in the independent datasets. We employed a one-sided exact binomial test of significance under the null hypothesis that sign concordance was randomly distributed.48–50 We also identified lead SNPs (or the next most significant candidate SNPs) that were nominally significant at P < 0.05 in each independent sample.48,50
Functional annotation
Candidate SNPs were functionally annotated to characterize their biological significance (Supplementary material), identify SNPs with pathogenic potential [Combined Annotation Dependent Depletion (CADD) scores >12.37]51 and highlight putative causal genes. Genes were mapped using three strategies: (i) positional mapping—SNP mapped to all genes within 10 kb distance; (ii) expression quantitative trait locus (eQTL) mapping—SNPs mapped to genes whose expression has been associated with the SNPs allelic variation by one of three eQTL resources (Supplementary material); and (iii) chromatin interaction mapping—SNPs mapped to genes with which they are predicted to physically interact by 3D modelling of chromatin structure.44
On each set of mapped genes, we performed gene-set enrichment analysis within the Gene Ontology classification system, corrected for multiple testing using Benjamini–Hochberg FDR.44 The Gene Ontology resource is a comprehensive database of computationally or biologically derived gene sets.52,53 We also constructed spatiotemporal heat maps of gene-expression levels across 11 brain tissues at 11 developmental time points in the R package ‘cerebroViz’ using BrainSpan RNA sequencing data.54–56 Expression across brain tissues was clustered using unsupervised hierarchical cluster analysis. We tested for overrepresentation of mapped genes within pathways derived from 12 public resources, collated by ConsensusPathDB and corrected for multiple testing using the q-value.57
Data availability
Data supporting the findings of this study are openly available from an online repository or are available on request from study authors. For further details, refer to the Supplementary material. All code is freely available at https://github.com/precimed and https://github.com/bulik/ldsc. Analyses were conducted in Python v.3.5, MATLAB R2020b and R v.3.6.3. Locus definition, functional annotation and gene-set analysis were performed using FUMA (https://fuma.ctglab.nl/).44
Results
Quantifying polygenic overlap
Univariate MiXeR estimated that mental disorders were 2.9–4.4 times more polygenic relative to migraine, with 8100, 12 300 and 9700 variants estimated to influence bipolar disorder, depression and schizophrenia, respectively, compared to 2800 in migraine.
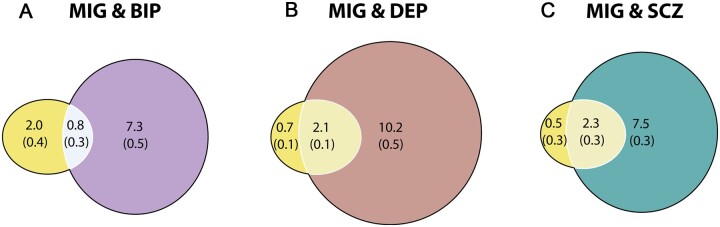
Bivariate MiXeR analysis revealed substantial polygenic overlap of migraine-influencing variants with each of depression and schizophrenia, and less overlap with bipolar disorder (Fig. 2). Of the 2800 migraine-influencing variants, 2100 (SD = 100) and 2300 (SD = 300) were also predicted to influence depression and schizophrenia, respectively, compared to 800 (SD = 0.3) predicted to influence bipolar disorder. This is despite minimal negative genetic correlation between migraine and each of schizophrenia [rg = −0.077, standard error (SE) = 0.027, P = 0.0046] and bipolar disorder (rg = −0.035, SE = 0.037, P = 0.35) and moderate positive genetic correlation between migraine and depression (rg = 0.33, SE = 0.030 P = 8.31 × 10−28), replicating previous findings.25 The difference in polygenicity between migraine (2800) and mental disorders (8100–12 300) limits the overlap between migraine and each mental disorder, demonstrated by the difference in the number of migraine-specific variants (500–2000) and mental disorder-specific variants (7300–10 200).
Figure 2.
Total number of shared and unique variants estimated to influence migraine and mental disorders. Results from the MiXeR analysis for migraine (MIG) and each of (A) bipolar disorder (BIP), (B) depression (DEP) and (C) schizophrenia (SCZ). Venn diagrams representing the unique and shared variants associated with migraine and each of bipolar disorder, depression and schizophrenia. Polygenic overlap is represented in a lighter shade, migraine in yellow, bipolar disorder in purple, depression in brown and schizophrenia in green. The numbers indicate the estimated number of variants in thousands per component with standard deviations in parentheses. The size of the circle reflects the extent of polygenicity for each disorder.
The AIC demonstrated adequate power for all analyses, but model fit was suboptimal for depression and bipolar disorder (Supplementary material, Supplementary Table 1 and Supplementary Fig. 1). These findings must therefore be interpreted with caution.
Identifying shared loci
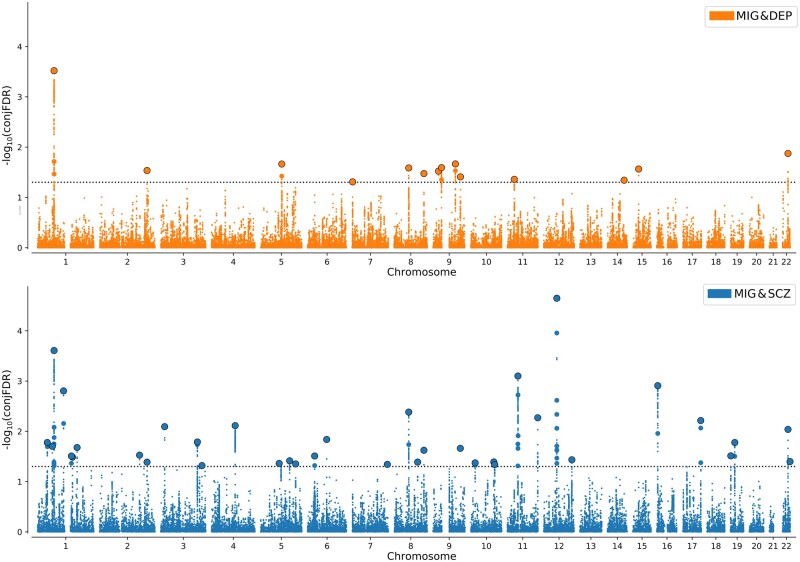
ConjFDR analysis identified 14 loci jointly associated with depression and migraine and 36 loci associated with schizophrenia and migraine (Table 1, Fig. 3, Supplementary Tables 2 and 3 and Supplementary Fig. 2). Five loci were identified across both analyses, four of which had the same lead SNP. Furthermore, 71% of lead SNPs (10/14) had the same effect direction on depression and migraine compared to 42% for schizophrenia and migraine (15/36), consistent with the moderate positive genetic correlation between depression and migraine and minimal negative genetic correlation between schizophrenia and migraine. There were no loci jointly associated with bipolar disorder and migraine.
Table 1.
Top 10 most strongly associated loci with migraine and each of depression (depression) and schizophrenia (schizophrenia)
| Chr | Min-max base pairs | Lead SNPs | ConjFDR | Psych | Migraine | Overlapping | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect size (OR) | P-value | Novel | Effect size (Beta) | P-value | Novel | |||||
| Migraine and depression | ||||||||||
| 1 | 73 458 846–74 108 971 | rs11210247 | <0.001 | 1.06 | 6.62 × 10−9 | No | −0.04 | 1.69 × 10−7 | No | Yes |
| 2 | 208 017 033–208 088 987 | rs7592120 | 0.03 | 0.94 | 4.46 × 10−8 | No | −0.03 | 4.78 × 10−5 | Yes | Yes |
| 5 | 92 362 700–92 538 853 | rs10514370 | 0.02 | 0.96 | 9.39 × 10−6 | Yes | −0.03 | 2.95 × 10−5 | Yes | No |
| 8 | 64 496 159–64 624 581 | rs1217091 | 0.03 | 0.95 | 1.23 × 10−5 | Yes | 0.04 | 3.93 × 10−5 | Yes | Yes |
| 8 | 131 030 628–131 361 477 | rs143725649 | 0.03 | 1.04 | 5.85 × 10−5 | Yes | −0.04 | 6.08 × 10−5 | Yes | No |
| 9 | 23 736 400–23 737 627 | rs10119773 | 0.03 | 0.95 | 7.70 × 10−8 | No | −0.03 | 5.02 × 10−5 | Yes | No |
| 9 | 37 045 825–37 406 391 | rs10973193 | 0.03 | 1.08 | 4.96 × 10−8 | No | 0.04 | 3.85 × 10−5 | Yes | No |
| 9 | 98 191 712–98 314 415 | rs10512249 | 0.02 | 1.07 | 3.28 × 10−5 | No | 0.05 | 1.48 × 10−5 | Yes | No |
| 15 | 47 615 220–47 685 504 | rs281264 | 0.03 | 0.96 | 2.25 × 10−5 | No | −0.03 | 4.26 × 10−5 | Yes | No |
| 22 | 41 408 754–41 713 111 | rs71327107 | 0.01 | 0.95 | 4.02 × 10−6 | No | −0.04 | 1.45 × 10−5 | Yes | Yes |
| Migraine and schizophrenia | ||||||||||
| 1 | 73 305 593–74 161 292 | rs11210247 | <0.001 | 1.07 | 1.92 × 10−11 | No | −0.04 | 1.69 × 10−7 | No | Yes |
| 1 | 115 677 183–115 763 649 | rs12134493 | 0.002 | 1.08 | 6.29 × 10−7 | Yes | 0.11 | 1.01 × 10−22 | No | No |
| 3 | 16 947 451–16 972 211 | rs11128810 | 0.008 | 1.04 | 8.66 × 10−6 | No | −0.04 | 3.39 × 10−6 | No | No |
| 4 | 103 618 023–103 975 060 | rs6810668 | 0.008 | 1.04 | 8.04 × 10−6 | No | −0.03 | 1.04 × 10−5 | Yes | No |
| 8 | 64 496 159–64 842 662 | rs1217112 | 0.004 | 1.06 | 2.94 × 10−6 | No | −0.04 | 4.60 × 10−6 | Yes | Yes |
| 11 | 46 257 757–47 371 598 | rs7932866 | <0.001 | 0.92 | 1.06 × 10−10 | No | 0.05 | 6.36 × 10−7 | Yes | No |
| 12 | 57 331 741–57 527 283 | rs324015 | <0.001 | 0.93 | 1.42 × 10−10 | No | 0.05 | 1.26 × 10−8 | No | No |
| 16 | 4 447 771–4 596 447 | rs4786505 | 0.001 | 0.95 | 2.85 × 10−7 | No | −0.04 | 1.07 × 10−6 | Yes | No |
| 17 | 78 442 650–78 738 796 | rs112821038 | 0.006 | 0.93 | 5.51 × 10−6 | No | 0.06 | 4.76 × 10−6 | No | No |
| 22 | 41 408 754–41 713 111 | rs71327107 | 0.009 | 0.93 | 3.71 × 10−10 | No | −0.04 | 1.45 × 10−5 | Yes | Yes |
For the complete list of loci please refer to Supplementary Tables 2 and 3. Chromosome (Chr), minimum and maximum base pairs, lead SNPs and conjFDR statistic are presented from each conjFDR analysis. Effect sizes [either odds ratio (OR) for depression and schizophrenia or beta for migraine] and P-values are presented from the original mental disorder (psych) and migraine GWAS. The novelty of each locus for migraine, depression and schizophrenia in relation to previous GWAS and conjFDR studies is indicated by Yes (novel) or No (not novel). ‘Overlapping’ indicates loci that were physically overlapping across both migraine and depression, and migraine and schizophrenia analyses.
Figure 3.
Chromosomal distribution of genetic loci jointly associated with migraine and mental disorders. Manhattan plots showing the −log10 transformed conjunctional FDR values for (A) depression (DEP) and migraine (MIG) (orange) and (B) schizophrenia (SCZ) and migraine (blue) for each SNP (y-axis) against chromosomal position (x-axis). The dotted line represents the conjFDR threshold for significant association <0.05. Black outlined circles represent independent lead SNPs.
The boost in power derived from condFDR analysis provided a total of 138 unique novel loci associated with migraine conditional on bipolar disorder (n = 56), depression (n = 84) and schizophrenia (n = 119). Forty-nine were identified across all three analyses (Supplementary material, Supplementary Tables 4–9 and Supplementary Figs 3 and 4).
Consistency of genetic effects in independent samples
For 176 unique loci associated with migraine across condFDR and conjFDR analyses, 153 lead SNPs showed sign concordance (P = 1.30 × 10−27) and 76 (43%) had nominal P < 0.05 in the independent migraine datasets. Among the conjFDR loci, 11 of 14 loci jointly associated with migraine and depression had lead SNPs or next most significant candidate SNPs with concordant effects in primary and independent samples for both migraine and depression (P = 0.0292). Three of 14 had nominally significant P-values in the independent depression sample and one in the independent migraine sample, none of which were nominally significant in both samples. For schizophrenia and migraine, three loci had neither lead SNPs nor secondary lead SNPs present in the schizophrenia replication sample, leaving 33 conjFDR lead SNPs. Twenty-seven of 33 lead SNPs had concordant effect directions in the primary and independent samples in both migraine and schizophrenia (P = 0.00016). Nineteen lead SNPs were nominally significant in the independent migraine cohort and eight in the independent schizophrenia sample. Five lead SNPs were nominally significant in both independent samples. The consistency of associations in independent datasets is comparable to that of other GWAS on complex polygenic phenotypes,48,50 supporting the validity of these findings. Complete condFDR replication analyses are presented in the Supplementary material.
Genetic overlap between migraine and other brain and non-brain-related phenotypes
We performed post hoc analyses to test the specificity of the observed overlaps. Compared to schizophrenia and depression, there was similar MiXeR estimated polygenic overlap between migraine and intelligence (1800, SD = 300) and educational attainment (2100, SD = 300). In contrast, there were fewer shared variants with coronary artery disease (1300, SD = 100) and more unique migraine variants (1500, SD 200), although the shared variants represented a large proportion coronary artery disease-influencing variants (1300/1400 variants). Migraine shared the fewest variants with height (1000K, SD = 100) (Supplementary Fig. 5). Other neurological and non-neurological disorders were not suitable for MiXeR analysis due to small sample sizes and the inability of MiXeR to accurately model less polygenic phenotypes. However, post hoc conjFDR analysis identified multiple shared loci between migraine and educational attainment (34), intelligence (29), Parkinson’s disease (2), multiple sclerosis (4) and prostate cancer (2), alongside previously identified loci between migraine and coronary artery disease (3).47 Interestingly, 15 loci associated with either depression and migraine and/or schizophrenia and migraine were physically overlapping with loci associated with migraine and educational attainment (5), intelligence (8) and Parkinson’s disease (1), but not multiple sclerosis, prostate cancer or coronary-artery disease (Supplementary material and Supplementary Figs 6 and 7).
Biological insights from shared loci
We mapped 298 and 37 protein-coding genes to candidate SNPs jointly associated with migraine and each of schizophrenia and depression, respectively (Supplementary Tables 10 and 11). Among genes mapped to depression and migraine loci, PAX5, a transcription factor involved in neurogenesis,58ELAVL2, a regulator of neurodevelopmental and synaptic genes,59 and L3MBTL2, a transcriptional repressor, were all mapped to probable pathogenic SNPs from novel migraine loci, and have all previously been linked to both depression and schizophrenia.58,60–63 Notably, L3MBTL2 was mapped to a locus that was nominally significant in the depression replication sample, and was mapped by both positional and eQTL mapping, with evidence of differential expression in the anterior cingulate cortex and frontal cortex. With regards to schizophrenia and migraine, several genes have previously been linked to both disorders, including LRP1, ASTN2, HPSE2 and IGSF9B. Among the genes mapped to novel migraine loci, CSNK1G2 was mapped by chromatin interaction mapping within foetal and adult cortex. CSNK1G2 is a casein kinase that has previously been implicated in schizophrenia.64 It is also a paralogue of CSNK1D, a monogenic cause of familial advanced sleep phase syndrome with migraines and a component of the circadian clock.65 Additionally, KLF10 was positionally mapped to a probable pathogenic lead SNP (CADD score >12.37). KLF10 is a transcriptional repressor that also plays a role in the circadian clock.66 There were also several genes coding transmembrane ion channel proteins including the calcium channel subunit CACNB2, which has been strongly linked to schizophrenia,67 and the sodium/hydrogen exchanger SLC9B1 which was mapped using all three strategies, including eQTL mapping to brain tissue in two independent datasets. Both CACNB2 and SLC9B1 were mapped to loci that were nominally significant in both migraine and schizophrenia replication samples. Further functional annotation analyses are described in the Supplementary material and Supplementary Fig. 6.
We conducted gene-set analyses on each set of mapped genes. While there were no significantly enriched depression and migraine gene sets, 27 gene sets were enriched with mapped genes for schizophrenia and migraine with a predominance of gene sets related to transmembrane transport (e.g. ‘GO_TRANSPORTER_ACTIVITY’), mitochondria (e.g. ‘GO_MITOCHONDRIA’) and neuron structure (e.g. ‘GO_SOMATODENDRITIC_COMPARTMENT’) gene sets (Supplementary Table 12).
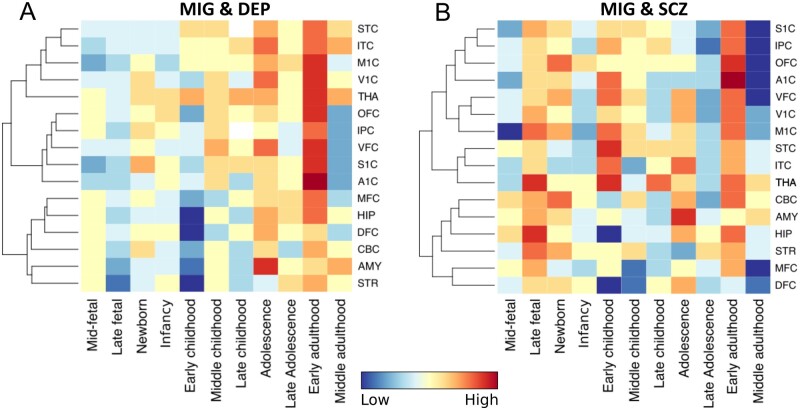
We also present spatiotemporal gene-expression analyses of mapped genes using RNA sequencing data (Fig. 4).
Figure 4.
Spatiotemporal gene expression of all mapped genes. Dendrogram and heat map showing spatiotemporal gene expression of all mapped genes for (A) migraine (MIG) and depression (DEP) and (B) migraine and schizophrenia (SCZ) using RNA sequencing data from BrainSpan over 11 developmental periods (columns) and 16 brain regions (rows). Global expression of mapped genes for depression and migraine was low from the mid-foetal stage to early childhood, increasing from middle childhood to early adulthood with highest expression during early adulthood. Expression of mapped genes for schizophrenia and migraine was more distributed, with highest expression during the late foetal stage, early childhood and early adulthood. Highest expression of mapped genes for both migraine and depression and migraine and schizophrenia occurred in the primary auditory cortex (A1C) during early adulthood, during which there was also high expression in the primary sensory (S1C), primary motor (M1C), primary visual (V1C), inferior temporal (ITC), ventrolateral prefrontal (VFC), orbitofrontal cortices (OFC) and thalamus (THA) for both sets of genes. There was also high expression in the thalamus at several developmental stages from late foetal to early adulthood for schizophrenia and migraine, while expression in the thalamus increased steadily from the newborn stage to early adulthood for depression and migraine. Brain regions were clustered using unsupervised hierarchical cluster analysis. Gene expression is indicated from high (red) to low (blue). AMY = amygdala; CBC = cerebellum; DFC = dorsolateral prefrontal cortex; HIP = hippocampus; IPC = inferior parietal cortex; MFC = medial prefrontal cortex; STC = superior temporal cortex; STR = striatum. Refer to the Supplementary material for more details.
Finally, 31 pathways were over-represented with mapped genes for depression and migraine, most of which implicated cell-signalling (e.g. ‘Signalling by Hedgehog’). Thirty-five pathways were identified for schizophrenia and migraine, with a predominance of signalling (e.g. ‘Hedgehog, off state’), immune (e.g. ‘Type II interferon signalling’) and CNS related pathways (e.g. ‘Twelve loci associated with ADHD’) (Supplementary Tables 13 and 14).
Discussion
The present study revealed that most migraine-influencing variants also influenced depression and schizophrenia, although this represented a minority of depression and schizophrenia-influencing variants due to their greater polygenicity. This was alongside moderate positive and weak negative genetic correlation, respectively. There was smaller overlap and weak positive genetic correlation between bipolar disorder and migraine. Taken with evidence of similar extensive overlap between migraine and intelligence and educational attainment but not height, these findings indicate the presence of a pool of ‘pleiotropic’ genetic variants that effect general vulnerability to brain-related disorders and traits, as suggested for other mental disorders.26 We next used conjFDR to identify 14 genomic loci jointly associated with depression and migraine and 36 with schizophrenia and migraine, 15 of which were overlapping with loci associated with other brain-related disorders and traits. No loci were associated with bipolar disorder and migraine. CondFDR analysis additionally identified 138 novel loci associated with migraine, of which 119 had sign concordance and 56 were nominally significant in an independent sample, supporting the validity of these findings. We mapped 37 genes to candidate SNPs associated with depression and migraine and 298 genes to SNPs associated with schizophrenia and migraine, including several putative novel migraine genes. These findings have implications for how the genetic risk of complex polygenic disorders is conceptualized, offer insights into the genetic basis of the comorbidity between depression and migraine, and highlight several putative migraine genes for experimental validation with potential for drug discovery and personalized treatment.
First, these findings demonstrate widespread genetic overlap of migraine-influencing variants with depression and schizophrenia beyond previously reported genetic correlations.25 While this may be surprising given their clinical differences, these findings contribute to a growing body of evidence indicating extensive polygenic overlap across a range of complex, brain-related phenotypes, with mixed directions of effect and minimal genetic correlation.26 A proportion of this overlap is likely driven by non-specific factors consistent with the omnigenic model and illustrated by the polygenic overlap between migraine and height.68 However, approximately double the number of migraine-influencing variants were shared with schizophrenia, depression, intelligence and educational attainment. Such extensive overlap contrasts with a ‘traditional’ conceptualization of genetic risk which assumes a list of disorder-specific variants with limited overlap across disorders and traits. While extensive overlap is increasingly recognized across mental disorders, our findings indicate this also encompasses migraine. In addition to the overlapping loci identified across migraine, mental disorders, educational attainment, intelligence and Parkinson’s disease, recent evidence has also revealed genetic overlap between Parkinson’s disease and schizophrenia.34 Taken together, this is suggestive of a subset of ‘pleiotropic’ variants associated with a general vulnerability to brain disorders and influence several brain-related traits. This may also encompass brain structure, which has recently been associated with several hundreds of loci with distributed genetic effects.69 If there are few specific genetic variants, the specificity of genetic risk for a given disorder could instead be determined by the differing effect sizes and directions of the variants on each phenotype and disorder-specific gene × gene or gene × environment interactions.26 This also places greater emphasis on identifying disease-specific and rare variants, which may be of high relevance for understanding pathological processes and improving diagnosis and treatment.
Our findings also contribute to ongoing debate over the aetiology of migraine.70 Migraine was previously thought to be a vascular disorder, supported by the association with cardiovascular events and GWAS findings showing association with vascular genes.21,70,71 An alternative theory implicates a primary neurological origin, related to inappropriate activation in the hypothalamus, midbrain ventral tegmentum and noradrenergic and serotonergic nuclei.71 These regions are proposed to modulate cerebral blood flow leading to secondary alterations in vasculature. Our finding that ∼75% of genetic risk for migraine overlaps with both schizophrenia and depression, alongside the identification of multiple shared genetic risk loci, could support the latter theory. Vascular dysfunction is not prominent in schizophrenia or depression, while dysfunction of neurotransmitter-based systems, such as serotonin72,73 and glutamate,74,75 as well as cortical excitability are implicated in all three disorders.76,77 This is supported by the finding that transmembrane transporter gene sets were enriched with genes mapping to schizophrenia and migraine loci, and that several putative causal genes coded for ion channel subunits. However, previous studies have also reported genetic overlap between cardiovascular disease phenotypes and both migraine and mental disorders. Interestingly, our post hoc analysis found that most coronary-artery influencing variants were shared with migraine, but in addition there was a large number of unique migraine variants.47,78 It is therefore tempting to speculate that migraine possesses a spectrum of genetic overlap with psychiatric, neurological and cardiovascular disorders, possibly indicating distinct ‘neurological’ and ‘vascular’ components of genetic risk. Larger sample sizes and improved statistical modelling will enable more complete cross-disorder comparisons.
When comparing migraine to mental disorders, there was a substantial difference in polygenicity, which may reflect greater neurobiological heterogeneity. For example, the quality and laterality of migraine pain may be more directly linked to specific underlying neurobiological mechanisms than anhedonia or low energy in depression, which may represent multiple phenomena with similar phenotypic expression. This could mean that migraine maps to a more distinct underlying neurobiological substrate, whereas mental disorders encompass several, overlapping neurobiological processes. This interpretation is supported by the finding of less genetic correlation between neurological disorders than between mental disorders.25 Moreover, MiXeR has previously demonstrated extensive overlap across bipolar disorder, depression and schizophrenia, and almost complete overlap with autism spectrum disorder.26 It is therefore possible that the subset of migraine-influencing variants shared with mental disorders represent a specific neurobiological subset of genetic risk in mental disorders. The shared loci identified by conjFDR, which have the strongest joint effects, provide some insights into the neurobiological mechanisms that this subset may relate to. However, the biological significance of this will gain clarity once a greater proportion of shared variants are discovered.
The smaller overlap and lack of genetic correlation between bipolar disorder and migraine, alongside the failure of conjFDR to identify any shared loci, is surprising given the evidence of increased rates of bipolar disorder among migraine patients and vice versa.1,3 It is important to note that the suboptimal MiXeR model fit means that this result should be interpreted with caution, particularly considering the extensive overlap observed between migraine and each of intelligence and educational attainment. Furthermore, the primary reason for the absence of conjunctional loci is likely to be sample size (bipolar disorder n = 51 720 versus schizophrenia n = 105 318 and depression n = 375 752). However, this may also relate to the fact that migraine is more strongly associated with bipolar-II than bipolar-I.79,80 Bipolar-II, a less severe form of bipolar disorder, is underrepresented in the current cohort (n = 3421) despite an equivalent prevalence to bipolar-I.81 There may therefore be a genetic basis to the association with bipolar-II patients that is not captured given the current sample make-up. Additionally, bipolar-I patients with migraine have been hypothesized to represent a distinct neurobiological subset, since they differ significantly from those without migraine in several key clinical features.81 A follow-up analysis of bipolar subtypes would be of great interest.
The extent of MiXeR estimated polygenic overlap between depression and migraine is striking despite phenotypic correlation and previously reported positive genetic correlation.25 Furthermore, our identification of 14 shared loci, 10 of which had concordant effects and 10 of which had P-values <0.05 in an independent sample, provides detail to their shared genetic basis beyond genetic correlation.35 A single conjunctional lead SNP also met genome-wide significance in the combined GWAS of Yang et al.,35 but missed genome-wide significance in the original migraine GWAS and hence was not reported as shared. Among the 37 mapped genes for depression and migraine, we highlight PAX5,58ELAVL259 and L3MBTL2 that mapped to probable pathogenic SNPs, and so may be of high interest for in vitro and in vivo validation and have potential for drug discovery. In contrast, the extensive overlap with schizophrenia and the 36 shared loci represent entirely novel findings given the dearth of previous genetics studies.10 The prominence of ion channel subunit genes and gene sets related to transmembrane transport was particularly striking. Ion channel dysfunction has been suggested to play a key role in migraine pathophysiology, while GWAS have previously implicated calcium channels in schizophrenia.82,83 These findings suggest possible mechanistic convergence between these distinct brain-related disorders, although further experimental studies are required to validate this.
While gene-set and pathway analyses provided insights into putative shared mechanisms, our spatiotemporal gene-expression analysis identified brain regions and developmental time periods during which these processes may impact disorder risk. Interestingly, global expression was highest during early adulthood for both sets of shared genes, while there was an additional period of high gene expression during the late foetal stage for migraine and schizophrenia. This widespread increase in expression during early adulthood broadly maps on to the clinical manifestation of all three disorders, with prevalence increasing from adolescence through to early adulthood.84 There is also extensive evidence implicating disturbances of early neurodevelopment in schizophrenia, including changes in gene expression.85,86 In contrast, few studies have explored the relationship between neurodevelopment and migraine risk.87 Evidence of increased rates of migraine in autism spectrum disorder, a common neurodevelopmental disorder, however, suggest this may be of relevance.88 The fact that gene expression during this time period was highest in the thalamus and the hippocampus, two subcortical structures implicated in the precipitation of migraines,71 is particularly intriguing, possibly indicating that altered expression of migraine risk genes during early development within these core structures may result in a predisposition to migraine later in life.
There were limitations to the current study. First, due to available sample sizes, we were unable to include multi-ancestral data for migraine or depression. Efforts to boost trans-ancestral samples and the development of statistical techniques to translate findings across ancestral groups are crucially important. Second, a subset of migraine and depression diagnoses were by self-report. Given that 20% of self-report migraines do not meet diagnostic criteria,89 it is possible that the genetic overlap is driven by a non-specific mental trait related to self-reporting rather than a specific migraine-signal. Similarly, the depression phenotype included self-report diagnoses and a ‘broad definition’ of depression.40 Nonetheless, this is weighed against the extra power for discovery offered by studies with self-report measures. These analyses should be repeated once larger samples of strictly defined depression and migraine are achieved. Third, although MiXeR explicitly models genetic effects beyond the confounding influence of LD, it is possible that uncontrolled environmental confounders could influence our findings. This limitation is inherited from the primary GWAS, and LDSR intercept estimates and genomic inflation factors indicated minimal effects of non-genetic confounders in all four studies.20,21,23,41 Despite this, further in vitro studies are required to confirm the causative effects of the genetic associations identified. Fourth, the comorbidity between migraine and bipolar disorder and depression may confound our findings since the rate of migraine among the cases is likely to be higher than controls, and vice versa. However, if this was a significant confounder, we would expect to see more substantial overlap with bipolar disorder and less overlap with schizophrenia, which does not have the same evidence of migraine comorbidity as bipolar disorder and depression. Nonetheless, individual level data with deeper phenotyping are required to confirm these findings, similar to other studies of genetic overlap.25,27,90–92 Finally, while functionally annotating candidate SNPs reduces the probability of missing causal variants, this approach increases the number of false positives to the gene-mapping, gene-set enrichment and gene-expression analyses. Further work will be required to refine functional annotation once larger primary GWASs are achieved.
To conclude, we have found substantial polygenic overlap of migraine-influencing variants with both schizophrenia and depression, and moderate overlap with bipolar disorder. These findings have implications for how the genetic risk of complex polygenic brain disorders is conceptualized, indicate the presence of a subset of highly pleiotropic genetic variants that influence diverse brain-related disorders and traits and provide biological insights into the shared variants with strongest effects on migraine, depression and schizophrenia.
Supplementary Material
Acknowledgements
We thank the research participants, employees and investigators of the Schizophrenia, Depression, and Bipolar Disorder Working Groups of the Psychiatric Genomics Consortium, 23andMe, the International Headache Genetics Consortium (IHGC), the UK Biobank, the Social Sciences Genetic Association Consortium, the FinnGen Study and the Genetic Investigation of Anthropomorphic Traits consortium for making this research possible. The Trøndelag Health Study (HUNT) is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology NTNU), Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health. The genotype quality control and imputation has been conducted by the KG Jebsen Center for Genetic Epidemiology, Department of Public Health and Nursing, Faculty of Medicine and Health Sciences, NTNU. This research has been conducted using the UK Biobank Resource under Application Number 22627.
Funding
We gratefully acknowledge support from the American National Institutes of Health (NS057198, EB00790); the Research Council of Norway (RCN) (#229129, #213837, #223273, #283798); the South-East Norway Regional Health Authority (#2017–112, #2020–034); KG Jebsen Stiftelsen; Norwegian Health Association (#22731), EU Joint Programme – Neurodegenerative Disease Research (JPND): RCN (#311993, #PMI-AD) and the EU’s Horizon2020 Programme (#847776, CoMorMent). The genotyping of the HUNT Study was financed by the National Institute of health (NIH), University of Michigan, The RCN, and Central Norway Regional Health Authority and the Faculty of Medicine and Health Sciences, NTNU. The funding bodies had no role in the design or conduct of the study; the collection, management, analysis or interpretation of the data; the preparation, review or approval of the manuscript; nor the decision to submit the manuscript for publication.
Competing interests
O.A.A. has received speaker’s honorarium from Sunovion, and Lundbeck and is a consultant for Healthlytix. A.M.D. is a founder of and holds equity interest in CorTechs Labs and serves on its scientific advisory board. He is also a member of the Scientific Advisory Board of Healthlytix and receives research funding from General Electric Healthcare (GEHC). The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. All other authors have no conflicts of interest to declare.
Abbreviations
- cond/conjFDR
conditional/conjunctional false discovery rate
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- MiXeR
bivariate causal mixture model
- SNP
single nucleotide polymorphism
Appendix I
Full details of the HUNT All-In Headache members are provided in the Supplementary material.
HUNT all-in headache
Amy E. Martinsen, Anne Heidi Skogholt, Ben Brumpton, Cristen J. Willer, Erling Tronvik, Espen Saxhaug Kristoffersen, John-Anker Zwart, Jonas Bille Nielsen, Knut Hagen, Kristian Bernhard Nilsen, Kristian Hveem, Lars Jacob Stovner, Lars G. Fritsche, Laurent F. Thomas, Linda M. Pedersen, Maiken E. Gabrielsen, Marianne Bakke Johnsen, Marie Udnesseter Lie, Oddgeir Holmen, Sigrid Børte, Synne Øien Stensland, Wei Zhou.
Contributor Information
HUNT All-In Headache:
Amy E Martinsen, Anne Heidi Skogholt, Ben Brumpton, Cristen J Willer, Erling Tronvik, Espen Saxhaug Kristoffersen, John-Anker Zwart, Jonas Bille Nielsen, Knut Hagen, Kristian Bernhard Nilsen, Kristian Hveem, Lars Jacob Stovner, Lars G Fritsche, Laurent F Thomas, Linda M Pedersen, Maiken E Gabrielsen, Marianne Bakke Johnsen, Marie Udnesseter Lie, Oddgeir Holmen, Sigrid Børte, Synne Øien Stensland, and Wei Zhou
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Breslau N, Lipton RB, Stewart WF, Schultz LR, Welch KMA.. Comorbidity of migraine and depression: Investigating potential etiology and prognosis. Neurology. 2003;60(8):1308–1312. [DOI] [PubMed] [Google Scholar]
- 2. Minen MT, De Dhaem OB, Van Diest AK, et al. Migraine and its psychiatric comorbidities. J Neurol Neurosurg Psychiatry. 2016;87(7):741–749. [DOI] [PubMed] [Google Scholar]
- 3. Fornaro M, Stubbs B.. A meta-analysis investigating the prevalence and moderators of migraines among people with bipolar disorder. J Affect Disord. 2015;178:88–97. [DOI] [PubMed] [Google Scholar]
- 4. Leo RJ, Singh J.. Migraine headache and bipolar disorder comorbidity: A systematic review of the literature and clinical implications. Scand J Pain. 2016;11(1):136–145. [DOI] [PubMed] [Google Scholar]
- 5. Louter MA, Pelzer N, De Boer I, et al. Prevalence of lifetime depression in a large hemiplegic migraine cohort. Neurology. 2016;87(22):2370–2374. [DOI] [PubMed] [Google Scholar]
- 6. Birgenheir DG, Ilgen MA, Bohnert ASB, et al. Pain conditions among veterans with schizophrenia or bipolar disorder. Gen Hosp Psychiatry. 2013;35(5):480–484. [DOI] [PubMed] [Google Scholar]
- 7. Baptista T, Uzcátegui E, Arapé Y, et al. Migraine life-time prevalence in mental disorders: concurrent comparisons with first-degree relatives and the general population. Invest Clin. 2012;53(1):38–51. [PubMed] [Google Scholar]
- 8. Connaughton J, Wand B.. Prevalence, characteristics and management of headache experienced by people with schizophrenia and schizoaffective disorder: cross sectional cohort study. Australas Psychiatry. 2017;25(4):381–384. [DOI] [PubMed] [Google Scholar]
- 9. Gasparini CF, Smith RA, Griffiths LR.. Genetic and biochemical changes of the serotonergic system in migraine pathobiology. J Headache Pain. 2017;18(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van der Auwera S, Teumer A, Hertel J, et al. The inverse link between genetic risk for schizophrenia and migraine through NMDA (N-methyl-D-aspartate) receptor activation via D-serine. Eur Neuropsychopharmacol. 2016;26(9):1507–1515. [DOI] [PubMed] [Google Scholar]
- 11. Baskin SM, Smitherman TA.. Migraine and psychiatric disorders: Comorbidities, mechanisms, and clinical applications. Neurol Sci. 2009;30(S1):61–65. [DOI] [PubMed] [Google Scholar]
- 12. Charles A. The pathophysiology of migraine: Implications for clinical management. Lancet Neurol. 2018;17(2):174–182. [DOI] [PubMed] [Google Scholar]
- 13. Grande I, Berk M, Birmaher B, Vieta E.. Bipolar disorder. Lancet. 2016;387(10027):1561–1572. [DOI] [PubMed] [Google Scholar]
- 14. Malhi GS, Mann JJ.. Depression. Lancet. 2018;392(10161):2299–2312. [DOI] [PubMed] [Google Scholar]
- 15. Owen MJ, Sawa A, Mortensen PB.. Schizophrenia. Lancet. 2016;388(10039):86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mulder EJ, Van Baal C, Gaist D, et al. Genetic and environmental influences on migraine: A twin study across six countries. Twin Res Hum Genet. 2003;6(5):422–431. [DOI] [PubMed] [Google Scholar]
- 17. Hilker R, Helenius D, Fagerlund B, et al. Heritability of schizophrenia and schizophrenia spectrum based on the Nationwide Danish Twin Register. Biol Psychiatry. 2018;83(6):492–498. [DOI] [PubMed] [Google Scholar]
- 18. Johansson V, Kuja-Halkola R, Cannon TD, Hultman CM, Hedman AM.. A population-based heritability estimate of bipolar disorder – In a Swedish twin sample. Psychiatry Res. 2019;278:180–187. [DOI] [PubMed] [Google Scholar]
- 19. Kendler KS, Ohlsson H, Lichtenstein P, Sundquist J, Sundquist K.. The genetic epidemiology of treated major depression in Sweden. Am J Psychiatry. 2018;175(11):1137–1144. [DOI] [PubMed] [Google Scholar]
- 20. Stahl EA, Breen G, Forstner AJ, et al. ; eQTLGen Consortium. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gormley P, Anttila V, Winsvold BS, et al. ; International Headache Genetics Consortium. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat Genet. 2016;48(8):856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lam M, Chen CY, Li Z, et al. ; Genetic REsearch on schizophreniA neTwork-China and the Netherlands (GREAT-CN). Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat Genet. 2019;51(12):1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Howard DM, Adams MJ, Clarke TK, et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holland D, Frei O, Desikan R, et al. Beyond SNP heritability: Polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLOS Genet. 2020;16(5):e1008612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anttila V, Bulik-Sullivan B, Finucane HK, et al. ; The Brainstorm Consortium. Analysis of shared heritability in common disorders of the brain. Science. 2018;360(6395):eaap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smeland OB, Frei O, Dale AM, Andreassen OA.. The polygenic architecture of schizophrenia – rethinking pathogenesis and nosology. Nat Rev Neurol. 2020;16(7):366–379. [DOI] [PubMed] [Google Scholar]
- 27. Lee PH, Anttila V, Won H, et al. Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell. 2019;179(7):1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smeland OB, Bahrami S, Frei O, et al. Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. 2020;25(4):844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bahrami S, Shadrin A, Frei O, et al. Genetic loci shared between major depression and intelligence with mixed directions of effect. Nat Hum Behav. 2021;5:795–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fisher RA. The genetical theory of natural selection. Рипол Классик; 1958.
- 31. Frei O, Holland D, Smeland OB, et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat Commun. 2019;10(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smeland OB, Frei O, Fan CC, Shadrin A, Dale AM, Andreassen OA.. The emerging pattern of shared polygenic architecture of psychiatric disorders, conceptual and methodological challenges. Psychiatr Genet. 2019;29(5):152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Watanabe K, Stringer S, Frei O, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51(9):1339–1348. [DOI] [PubMed] [Google Scholar]
- 34. Smeland OB, Shadrin A, Bahrami S, et al. Genome-wide association analysis of Parkinson’s disease and schizophrenia reveals shared genetic architecture and identifies novel risk loci. Biol Psychiatry. 2021;89(3):227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang Y, Zhao H, Boomsma DI, et al. ; International Headache Genetics Consortium. Molecular genetic overlap between migraine and major depressive disorder. Eur J Hum Genet. 2018;26(8):1202–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smeland OB, Frei O, Shadrin A, et al. Discovery of shared genomic loci using the conditional false discovery rate approach. Hum Genet. 2020;139(1):85- 10. [DOI] [PubMed] [Google Scholar]
- 37. FinnGen. FinnGen Documentation of R4 release. Published 2020. Accessed 27 January 2021. https://finngen.gitbook.io/documentation/
- 38. Krokstad S, Langhammer A, Hveem K, et al. Cohort profile: The HUNT study, Norway. Int J Epidemiol. 2013;42(4):968–977. [DOI] [PubMed] [Google Scholar]
- 39. Wray NR, Ripke S, Mattheisen M, et al. ; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Howard DM, Adams MJ, Shirali M, et al. ; 23andMe Research Team. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat Commun. 2018;9(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pardiñas AF, Holmans P, Pocklington AJ, et al. ; CRESTAR Consortium. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Andreassen OA, Thompson WK, Schork AJ, et al. ; Bipolar Disorder and Schizophrenia Working Groups. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 2013;9(4):e1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watanabe K, Taskesen E, van Bochoven A, Posthuma D.. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turley P, Walters RK, Maghzian O, et al. ; Social Science Genetic Association Consortium. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O’Connell KS, Shadrin A, Smeland OB, et al. Identification of genetic loci shared between attention-deficit/hyperactivity disorder, intelligence, and educational attainment. Biol Psychiatry. 2020;87(12):1052–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winsvold BS, Bettella F, Witoelar A, et al. ; The International Headache Genetics Consortium. Shared genetic risk between migraine and coronary artery disease: A genome-wide analysis of common variants. PLoS One. 2017;12(9):e0185663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Savage JE, Jansen PR, Stringer S, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50(7):912–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ripke S, Walters JTR, Donovan MC. Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. medRxiv. [Preprint] doi: 10.1101/2020.09.12.20192922. [DOI]
- 50. Lee JJ, Wedow R, Okbay A, et al. ; Social Science Genetic Association Consortium. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50(8):1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M.. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carbon S, Douglass E, Good BM, et al. ; The Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miller JA, Ding S-L, Sunkin SM, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508(7495):199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. BrainSpan. BrainSpan atlas of the developing human brain. Published 2010. Accessed 16 October 2020. http://www.brainspan.org/
- 56. Bahl E, Koomar T, Michaelson JJ.. cerebroViz: an R package for anatomical visualization of spatiotemporal brain data. Bioinformatics. 2017;33(5):762–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Herwig R, Hardt C, Lienhard M, Kamburov A.. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc. 2016;11(10):1889–1907. [DOI] [PubMed] [Google Scholar]
- 58. Benes FM, Lim B, Matzilevich D, Walsh JP, Subburaju S, Minns M.. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc Natl Acad Sci USA. 2007;104(24):10164–10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Berto S, Usui N, Konopka G, Fogel BL.. ELAVL2-regulated transcriptional and splicing networks in human neurons link neurodevelopment and autism. Hum Mol Genet. 2016;25(12):2451–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Amare AT, Schubert KO, Tekola-Ayele F, et al. Association of the polygenic scores for personality traits and response to selective serotonin reuptake inhibitors in patients with major depressive disorder. Front Psychiatry. 2018;9:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamada K, Iwayama Y, Hattori E, et al. Genome-wide association study of schizophrenia in Japanese population. PLoS One. 2011;6(6):e20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hyde CL, Nagle MW, Tian C, et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet. 2016;48(9):1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bianchin MM, De Castro LA, Cunha DRMF, Mota SM, Bianchini T, Bizzi JWJ.. Allelic variants of PAX5 and MEF2C-AS2 genes are associated with depression in temporal lobe epilepsy (4185). Neurology. 2020;94(Suppl 15). [Google Scholar]
- 64. Wu C, Pan W.. Integration of methylation QTL and enhancer-target gene maps with schizophrenia GWAS summary results identifies novel genes. Bioinformatics. 2019;35(19):3576–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sutherland HG, Albury CL, Griffiths LR.. Advances in genetics of migraine. J Headache Pain. 2019;20(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guillaumond F, Gréchez-Cassiau A, Subramaniam M, et al. Krüppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol. 2010;30(12):3059–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Juraeva D, Haenisch B, Zapatka M, et al. ; GROUP Investigators. Integrated pathway-based approach identifies association between genomic regions at CTCF and CACNB2 and schizophrenia. PLoS Genet. 2014;10(6):e1004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Boyle EA, Li YI, Pritchard JK.. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169(7):1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van der Meer D, Frei O, Kaufmann T, et al. Understanding the genetic determinants of the brain with MOSTest. Nat Commun. 2020;11(1):3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rothrock JF. ‘Outside-In’ vs ‘Inside-Out’: revisiting migraine’s vascular hypothesis. Headache J Head Face Pain. 2008;48(9):1409–1410. [DOI] [PubMed] [Google Scholar]
- 71. Dodick DW. Migraine. Lancet. 2018;391(10127):1315–1330. [DOI] [PubMed] [Google Scholar]
- 72. Gouzoulis-Mayfrank E, Heekeren K, Neukirch A, et al. Inhibition of return in the human 5HT2A agonist and NMDA antagonist model of psychosis. Neuropsychopharmacology. 2006;31(2):431–441. [DOI] [PubMed] [Google Scholar]
- 73. Fakhoury M. Revisiting the serotonin hypothesis: Implications for major depressive disorders. Mol Neurobiol. 2016;53(5):2778–2786. [DOI] [PubMed] [Google Scholar]
- 74. Poels EMP, Kegeles LS, Kantrowitz JT, et al. Imaging glutamate in schizophrenia: review of findings and implications for drug discovery. Mol Psychiatry. 2014;19(1):20–29. [DOI] [PubMed] [Google Scholar]
- 75. A. P. The involvement of glutamate in the pathophysiology of depression. Drug News Perspect. 2005;18(4):262–268. [DOI] [PubMed] [Google Scholar]
- 76. Lissemore JI, Bhandari A, Mulsant BH, et al. Reduced GABAergic cortical inhibition in aging and depression. Neuropsychopharmacology. 2018;43(11):2277–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Carment L, Dupin L, Guedj L, et al. Impaired attentional modulation of sensorimotor control and cortical excitability in schizophrenia. Brain. 2019;142(7):2149–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Andreassen OA, Djurovic S, Thompson WK, et al. ; Psychiatric Genomics Consortium Schizophrenia Working Group. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am J Hum Genet. 2013;92(2):197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Low NCP, Du Fort GG, Cervantes P.. Prevalence, clinical correlates, and treatment of migraine in bipolar disorder. Headache J Head Face Pain. 2003;43(9):940–949. [DOI] [PubMed] [Google Scholar]
- 80. Fasmer OB. The prevalence of migraine in patients with bipolar and unipolar affective disorders. Cephalalgia. 2001;21(9):894–899. [DOI] [PubMed] [Google Scholar]
- 81. Pini S, de Queiroz V, Pagnin D, et al. Prevalence and burden of bipolar disorders in European countries. Eur Neuropsychopharmacol. 2005;15(4):425–434. [DOI] [PubMed] [Google Scholar]
- 82. Nanou E, Catterall WA.. Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron. 2018;98(3):466–481. [DOI] [PubMed] [Google Scholar]
- 83. Yan J, Dussor G.. Ion channels and migraine. Headache. 2014;54(4):619–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Victor T, Hu X, Campbell J, Buse D, Lipton R.. Migraine prevalence by age and sex in the United States: A life-span study. Cephalalgia. 2010;30(9):1065–1072. [DOI] [PubMed] [Google Scholar]
- 85. Gulsuner S, Walsh T, Watts AC, et al. XSpatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154(3):518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jia P, Chen X, Fanous AH, Zhao Z.. Convergent roles of de novo mutations and common variants in schizophrenia in tissue-specific and spatiotemporal co-expression network. Transl Psychiatry. 2018;8(1):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Børte S, Winsvold BS, Stensland SØ, Småstuen MC, Zwart JA.. The effect of foetal growth restriction on the development of migraine and tension-type headache in adulthood. The HUNT Study. PLoS One. 2017;12(4):e0175908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Vetri L. Autism and migraine: An unexplored association? Brain Sci. 2020;10(9):615–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lipton RB, Stewart WF, Liberman JN.. Self-awareness of migraine: Interpreting the labels that headache sufferers apply to their headaches. Neurology. 2002;58(9 Suppl 6):S21–S26. [DOI] [PubMed] [Google Scholar]
- 90. Lee PH, Feng YCA, Smoller JW.. Pleiotropy and cross-disorder genetics among psychiatric disorders. Biol Psychiatry. 2021;89(1):20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lo MT, Hinds DA, Tung JY, et al. Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders. Nat Genet. 2017;49(1):152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wu Y, Cao H, Baranova A. et al. Multi-trait analysis for genome-wide association study of five psychiatric disorders. Transl Psychiatry 2020;10(1):209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are openly available from an online repository or are available on request from study authors. For further details, refer to the Supplementary material. All code is freely available at https://github.com/precimed and https://github.com/bulik/ldsc. Analyses were conducted in Python v.3.5, MATLAB R2020b and R v.3.6.3. Locus definition, functional annotation and gene-set analysis were performed using FUMA (https://fuma.ctglab.nl/).44