Abstract
Mitochondria are small cellular constituents that generate cellular energy (ATP) by oxidative phosphorylation (OXPHOS). Dysfunction of these organelles is linked to a heterogeneous group of multisystemic disorders, including diabetes, cancer, ageing-related pathologies and rare mitochondrial diseases. With respect to the latter, mutations in subunit-encoding genes and assembly factors of the first OXPHOS complex (complex I) induce isolated complex I deficiency and Leigh syndrome. This syndrome is an early-onset, often fatal, encephalopathy with a variable clinical presentation and poor prognosis due to the lack of effective intervention strategies. Mutations in the nuclear DNA-encoded NDUFS4 gene, encoding the NADH:ubiquinone oxidoreductase subunit S4 (NDUFS4) of complex I, induce ‘mitochondrial complex I deficiency, nuclear type 1’ (MC1DN1) and Leigh syndrome in paediatric patients. A variety of (tissue-specific) Ndufs4 knockout mouse models were developed to study the Leigh syndrome pathomechanism and intervention testing.
Here, we review and discuss the role of complex I and NDUFS4 mutations in human mitochondrial disease, and review how the analysis of Ndufs4 knockout mouse models has generated new insights into the MC1ND1/Leigh syndrome pathomechanism and its therapeutic targeting.
Keywords: Leigh syndrome, mouse model, pathomechanism, intervention
The NDUFS4 gene encodes an accessory subunit of OXPHOS complex I and its mutation causes mitochondrial disease in children. Van de Wal et al. review how Ndufs4 knockout mouse models have provided new insights into the disease pathomechanism and potential intervention strategies for these disorders.
Mitochondria and oxidative phosphorylation
Mitochondria are constituents of virtually every eukaryotic cell. These organelles generate cellular energy in the form of adenosine triphosphate (ATP) and also play a central role in reactive oxygen species and redox metabolism, fatty acid oxidation, haem biosynthesis, apoptosis induction, heat generation and calcium (Ca2+) homeostasis.1-5 Structurally, mitochondria consist of a matrix compartment surrounded by the mitochondrial inner membrane (MIM) and mitochondrial outer membrane and an in-between intermembrane space. The MIM contains numerous folds (‘cristae’) that enlarge its surface area. Several nutrient-dependent pathways can deliver substrates for mitochondrial ATP generation by the MIM-embedded oxidative phosphorylation (OXPHOS) system.6–8 These pathways include: (i) the cytosolic glycolysis-mediated conversion of glucose, galactose or fructose into pyruvate, which subsequently enters the mitochondrion. There, pyruvate is converted into acetyl coenzyme A (acetyl-CoA) that is used as a substrate for the tricarboxylic acid (TCA) cycle to generate reduced nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FADH2); (ii) entry of glutamine into the TCA cycle; and (iii) entry of fatty acids into β-oxidation and the TCA cycle. In addition to OXPHOS, ATP is also generated by the glycolysis pathway in the cytosol and mitochondrial TCA cycle. Under physiological conditions, OXPHOS in the prime generator of ATP in most cells. However, during mitochondrial dysfunction increased glycolytic ATP production can compensate for loss of OXPHOS-mediated ATP generation. This compensation involves the conversion of glycolysis-generated pyruvate into lactate, which leaves the cell and acidifies the extracellular environment.9,10 The OXPHOS process requires the combined action of four electron transport chain (ETC) complexes (complex I–IV; CI–IV) and an ATP-producing fifth complex (CV or FoF1-ATPase) by a chemiosmotic coupling mechanism.11 ETC action sustains a matrix-directed proton-motive force by transporting electrons from NADH (via CI) and FADH2 (via CII) to molecular oxygen (O2; via CIV). This process further requires coenzyme Q10 and cytochrome-c, which mediate electron transport from CI/CII to CIII and from CIII to CIV, respectively. Of note, electrons can also enter the ETC via alternative coenzyme Q10-converging pathways.12,13 These are often tissue-specific and include: (i) the electron-transferring flavoprotein (ETF)-ubiquinone oxidoreductase; (ii) s, n-glycerophosphate dehydrogenase; and (iii) dihydroorotate dehydrogenase. At CI, CIII and CIV, protons (H+) are transported from the mitochondrial matrix to the intermembrane space, leading to a trans-MIM electrical potential (Δψ) and chemical proton gradient (ΔpH), which together constitute the proton-motive force.6 Matrix re-entry of H+ is then used at CV for ATP generation.14 In addition, the proton-motive force is also crucial to sustain other mitochondrial functions including ion/metabolite exchange and protein import.4,15,16
Complex I of the oxidative phosphorylation system
NADH:ubiquinone reductase (EC 1.6.5.3.) or complex I (CI) is the first and largest (∼1 MDa) OXPHOS complex, which couples H+ transport to electron transfer from NADH to coenzyme Q10.6,17–21 It was estimated that CI action is responsible for sustaining ∼40% of the total proton-motive force.22 CI is composed of 44 different subunits (Table 1), 14 of which are ‘core subunits’ that suffice to carry out CI catalytic function.22 In humans, seven hydrophobic core subunits are encoded by mitochondrial DNA (mtDNA): ND1, ND2, ND3, ND4, ND4L, ND5 and ND6, whereas the remaining core subunits are nuclear DNA (nDNA)-encoded: NDUFV1, NDUFV2, NDUFS1, NDUFS2, NDUFS3, NDUFS7 and NDUFS8. In addition to these core subunits, human CI also contains 30 nDNA-encoded ‘accessory’ (or ‘supernumerary’) subunits. Although the function of these subunits still is largely unknown, it is expected that they play a role in CI assembly, stabilization, functional regulation, prevention of electron escape and the formation of OXPHOS supercomplexes (see below). Currently, 14 assembly factors have been identified that mediate CI biogenesis: NDUFAF1, NDUFAF2, NDUFAF3, NDUFAF4, NDUFAF5, NDUFAF6, NDUFAF7, NDUFAF8, NUBPL, TIMMDC1, ECSIT, ACAD9, TMEM126B and FOXRED1.19,23 All of these are nDNA-encoded. Structurally, CI is L-shaped and consists of a hydrophobic MIM-embedded membrane arm and a matrix-protruding peripheral arm (Fig. 1). It appears that CI can exist in active and inactive states, which are associated with changes in CI structure.21,24 Functionally, CI consists of three modules: N, Q and P (Table 1), with the P module comprising two submodules (Pp and Pd). Electrons enter the N-module via an NDUFV1-bound flavin mononucleotide (FMN), after which the Q-module transports them to coenzyme Q10 via eight iron-sulphur (FeS) clusters (N1a/NDUFV2, N1b/NDUFS1, N2/NDUFS7, N3/NDUFV1, N4/NDUFS1, N5/NDUFS1, N6a/NDUFS8, N6b/NDUFS8). The P module mediates the trans-MIM proton pumping from the matrix to the intermembrane space.19,22,25 At a higher level of organization, CI can assemble into supercomplexes (also known as ‘respirasomes’) consisting of CI1CIII1, CI/CIII2/CIV, CI2CIII2CIV2, CI2CII2CIII2CIV2 or potentially even larger oligomers. It was hypothesized that formation of these supercomplexes mediates substrate channelling, prevents electrons from escaping and thereby reactive oxygen species production and/or serves protein stabilization purposes.26–34
Table 1.
Complex I subunits
| Number | LS-linkeda | Nameb | Modulec | Alternative names | Remarks |
|---|---|---|---|---|---|
| 1 | + | NDUFV1 | N | 51-kDa, Nqo1, NuoF | Contains FMN and N3 |
| 2 | + | NDUFV2 | N | 24-kDa, Nqo2, NuoE | Contains N1a |
| 3 | − | NDUFV3d | N | 10-kDa | |
| 4 | + | NDUFS1 | N | 75-kDa, Nqo3, NuoG | Contains N1b, N4, N5 |
| 5 | + | NDUFS6 | N | 13-kDa | Contains Zn2+ binding site |
| 6 | + | NDUFA12 | N | B17.2 | |
| 7 | + | NDUFS4 | N/Q? | AQDQ, 18-kDa | |
| 8 | + | NDUFA2 | N/Q? | B8 | Contains thioredoxin fold |
| 9 | + | NDUFS2 | Q | 49-kDa, Nqo4, NuoCD | |
| 10 | + | NDUFS3 | Q | 30-kDa, Nqo5, NuoCD | |
| 11 | + | NDUFS7 | Q | PSST, Nqo6, NuoB | Contains N2 |
| 12 | + | NDUFS8 | Q | TYKY, Nqo9, NuoI | Contains N6a and N6b |
| 13 | − | NDUFA5 | Q | B13 | |
| 14 | − | NDUFA6 | Q | B14 | LYR |
| 15 | − | NDUFA7 | Q | B14.5a | |
| 16 | + | NDUFA9 | Q | 39-kDa | Short-chain dehydrogenase/reductase fold with bound NAD(P)H |
| 17 | − | NDUFAB1 | Q & Pd | SDAP-a, SDAP-b | Acyl carrier protein; 2 copies present; contains phosphopantetheine cofactor |
| 18 | − | NDUFS5 | Pp | 15-kDa | Quadruple CX9C domain; double CHCH domain |
| 19 | + | NDUFA1 | Pp | MWFE | STMD |
| 20 | − | NDUFA3 | Pp | B9 | STMD |
| 21 | − | NDUFA8 | Pp | PGIV | Quadruple CX9C domain; double CHCH domain |
| 22 | + | NDUFA10 | Pp | 42-kDa | Nucleoside kinase family |
| 23 | + | NDUFA11 | Pp | B14.7 | |
| 24 | + | NDUFA13 | Pp | B16.6 | Identical to GRIM19, STMD |
| 25 | − | NDUFC1 | Pp | KFYI | STMD |
| 26 | + | NDUFC2 | Pp | B14.5b | |
| 27 | +/− | MT-ND1 | Pp | Nqo8, NuoH, ND1 | 8 TMHs |
| 28 | + | MT-ND2 | Pp | Nqo14, NuoN, ND2 | 11 TMHs |
| 29 | + | MT-ND3 | Pp | Nqo7, NuoA, ND3 | 3 TMHs |
| 30 | − | MT-ND4L | Pp | Nqo11, NuoK, ND4L | 3 TMHs |
| 31 | + | MT-ND6 | Pp | Nqo10, NuoJ, ND6 | 5 TMHs |
| 32 | − | NDUFB1 | Pd | MNLL | STMD |
| 33 | − | NDUFB2 | Pd | AGGG | STMD |
| 34 | − | NDUFB3 | Pd | B12 | STMD |
| 35 | − | NDUFB4 | Pd | B15 | STMD |
| 36 | − | NDUFB5 | Pd | SGDH | STMD |
| 37 | − | NDUFB6 | Pd | B17 | STMD |
| 38 | − | NDUFB7 | Pd | B18 | Double CX9C domain; double CHCH domain |
| 39 | + | NDUFB8 | Pd | ASHI | STMD |
| 40 | − | NDUFB9 | Pd | B22 | LYR |
| 41 | − | NDUFB10 | Pd | PDSW | |
| 42 | − | NDUFB11 | Pd | ESSS | STMD |
| 43 | + | MT-ND4 | Pd | Nqo7, NuoM, ND4 | 14 TMHs |
| 44 | + | MT-ND5 | Pd | Nqo12, NuoL, ND5 | 16 TMHs |
CHCH = coiled-coil-helix-coiled-coil-helix; FMN = flavin mononucleotide; LYR = member of mitochondrial LYR (LYRM) protein family; N = iron-sulphur cluster; O = for ovine CI; STMD = small single transmembrane domain; TMH = transmembrane helices. Adapted from Guerrero-Castillo et al.,19 Fiedorczuk and Sazanov,20 Adjobo-Hermans et al.,64 and Zhu et al.192
Linked to Leigh syndrome (LS; OMIM #256000; compiled using a literature search and information from: www.omim.org; +/− marks a potential link).
Human protein name according to HGNC (www.genenames.org). Core subunits are highlighted in bold. Subunits encoded by the mtDNA are in italics.
Functional modules: N (NADH binding and oxidation), Q (electron transfer to ubiquinone), P module (consisting of Pp and Pd submodules, proton pumping).
Represents the NDUFV3-10 subunit (10 kDa).
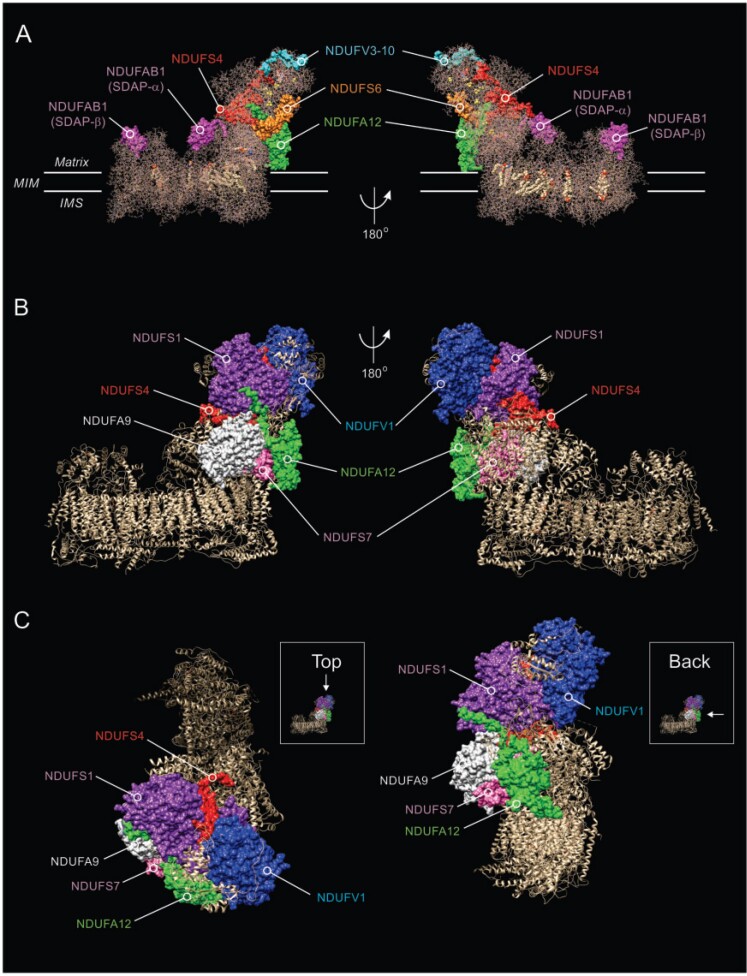
Figure 1.
Structure of CI and location of NDUFS4 and other subunits. (A) Side view of the cryogenic-electronic microscopy structure of CI in Ovis aries heart at 3.90 Å resolution (PDB accession number: 5LNK; www.rcsb.org), highlighting the position of the NDUFS4 protein (red) relative to the NDUFAB1, NDUFV3-10, NDUFS6 and NDUFA12 subunits.192 The two copies (α and β) of the NDUFAB1/SDAP accessory subunit are indicated. Yellow spheres mark iron-sulphur clusters. Transmembrane helices are depicted in the MIM-embedded part. (B) Same as A but now highlighting the position of the NDUFS4 protein relative to the NDUFV1, NDUFS1, NDUFA9, NDUFS7 and NDUFA12 subunits. (C) Same as B, but now depicting a view from the top and back of CI. The molecular graphics in this figure were created using the PyMOL Molecular Graphics System v.2.0 (Schrödinger-LLC, Mannheim, Germany).
Isolated complex I deficiency and Leigh syndrome
Mutations in nDNA- or mtDNA-encoded OXPHOS genes impair mitochondrial energy metabolism and result in multisystemic diseases, a heterogeneous group of severe, often fatal, pathologies affecting ∼1:5000 live births.6,35,36 However, mitochondrial dysfunction is not only observed in ‘rare’ mitochondrial syndromes, but also in more common diseases such as Parkinson’s disease, Alzheimer’s disease, age-related frailty, cancer, epilepsy, diabetes and obesity. In this sense, studying rare mitochondrial diseases is of great value to better understand mitochondrial dysfunction-related diseases with a higher incidence. We previously proposed to classify monogenic mitochondrial diseases as ‘primary’ and ‘secondary’ disorders.37 This classification is based on whether the disease arises from a mutation in a mitochondrial protein-encoding gene (primary disorder) or from an outside influence on mitochondria (secondary disorder). The latter include for instance viral infections, environmental toxins and off-target drug effects.38,39 In this review, we focus on isolated CI deficiencies due to nDNA mutations (OMIM #252010). The latter often induce Leigh syndrome (OMIM #256000),40,41 first described in 1951 by Denis Archibald Leigh.42 Currently, mutations in 24 CI subunit-encoding genes have been implicated in Leigh syndrome (Table 1). In addition, Leigh syndrome has been linked to mutations in various assembly factors of CI (NDUFAF1, NDUFAF2, NDUFAF3, NDUFAF4, NDUFAF5, NDUFAF6, NDUFAF8 and FOXRED1). Leigh syndrome is also referred to as subacute necrotizing encephalopathy and is characterized by an early-onset, often fatal, neurodegenerative disorder presenting with bilateral, symmetric lesions in the brainstem, midbrain, pons, thalamus, basal ganglia and cerebellum.43–48 Patients with Leigh syndrome are born without obvious clinical abnormalities and have a normal prenatal development.49 Most patients start developing symptoms within the first year of life and most die before the age of 3 years. Mutations are generally transferred to the offspring in an autosomal recessive manner,50 although X-linked inheritance (NDUFA1 mutation) has also been reported.51 Symptoms primarily develop in high-energy demanding tissues, such as the CNS, skeletal muscle and heart.37,49,52 Leigh syndrome presents with variable clinical symptoms such as progressive encephalopathy, failure to thrive, hypotonia, psychomotor retardation, breathing difficulties, recurrent vomiting, dysphagia, nystagmus, ataxia, neuropathy, hypertrophic cardiomyopathy, loss of vision, impaired hearing, seizures, lactic acidosis and eventually early death.43,44,46–48 Similar to most other mitochondrial diseases, the prognosis of CI deficiency-linked Leigh syndrome is poor, as currently no effective treatment strategies exist.40,54,55
The NDUFS4 subunit of complex I
In the context of mitochondrial disease, pathogenic mutations in the nDNA-encoded NDUFS4 gene (OMIM *602694) induce ‘mitochondrial complex I deficiency, nuclear type 1’ (MC1DN1; OMIM #252010) and Leigh syndrome. NDUFS4 is located on chromosome 5q11.2, contains five exons and encodes the accessory NADH-dehydrogenase subunit S4 (NDUFS4) of CI, also referred to as the ‘AQDQ’ or ‘18-kDa’ subunit.43,48,56,57 The NDUFS4 pre-protein is 175 amino acids long, with its ATG start codon located in exon one and the TGA stop codon in exon five, and contains a mitochondrial targeting sequence (MTS) of 42 amino acids. The latter is removed following mitochondrial import yielding a mature protein of 133 amino acids, which is highly similar between human and mouse (Fig. 2A–C). NDUFS4 contains two predicted phosphorylation sites (R-X1-2-S/T-X)58 for AMP-activated protein kinase A (PKA). These are located at position 36–38 (RTS/STS) in the MTS, and at position 171–173 (RVS) in the pre-protein (Fig. 2A and B). Evidence was provided that NDUFS4 exists in non-phosphorylated and phosphorylated forms and that PKA-mediated phosphorylation can occur at Ser173 of the RVS site.59 Of note, another study concluded that not NDUFS4 but another CI accessory subunit of 18 kDa (NDUFB11) was phosphorylated.60 It was proposed that NDUFS4 phosphorylation affects its mitochondrial import, MTS removal and subsequent incorporation in CI.57,59,61,62 In addition, PKA-mediated NDUFS4 phosphorylation might stimulate CI activity.61 Within ovine CI, the NDUFS4 subunit is located at the interface between the N- and Q-module (Fig. 1) and interacts with various other CI subunits within the N-module (NDUFV1, NDUFV2, NDUFV3-10, NDUFS1, NDUFA12) and Q-module (NDUFS3, NDUFS8, NDUFA6, NDUFA9). In case of bovine CI, the NDUFS4 protein also interacts with NDUFS2 but not with NDUFV2 (Table 2). Taken together, the current experimental data suggests that the NDUFS4 subunit is potentially PKA-phosphorylated and plays a key role in CI assembly and/or stabilization (likely involving N-to-Q-module attachment), thereby exerting control over CI levels and/or activity.63,64
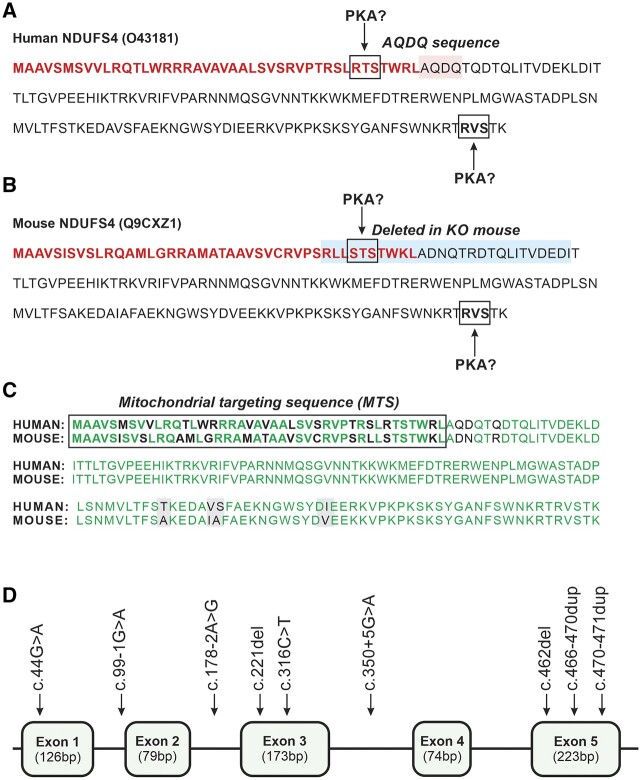
Figure 2.
Sequence of the human and mouse NDUFS4 and pathogenic NDUFS4 mutations. (A) Human (Homo sapiens) NDUFS4 pre-protein sequence (O43181 from UniProt: www.uniprot.org). The MTS is highlighted in bold red. The two PKA consensus phosphorylation sites in the MTS and NDUFS4 protein are highlighted by boxes. The AQDQ sequence (highlighted in pink) is also indicated. (B) Same as panel A but now for the mouse (Mus musculus) NDUFS4 protein sequence (Q9CXZ1). In the whole-body Ndufs4 knockout animal (Ndufs4−/−-WB), the last part of the MTS and the first 17 amino acids of NDUFS4 (highlighted in blue) were deleted.72 (C) Alignment of the pre-protein sequences in A and B. The MTS is highlighted in bold and identical amino acids are in green. The mature human and mouse NDUFS4 proteins differ by only four amino acids (highlighted in grey), rendering them 97% identical. (D) Schematic structure of NDUFS4 (NM_002495.2) consisting of five exons (not drawn to scale). The currently known mutations are highlighted (Table 2).
Table 2.
Interactions of NDUFS4 and NDUFA12 with other CI subununits
| Subunit | Modulea | Interacting withb | |||
|---|---|---|---|---|---|
| NDUFS4 | NDUFA12 | ||||
| Ovine | Bovine | Ovine | Bovine | ||
| NDUFV1/51-kDa | N | X | X | — | — |
| NDUFV2/24-kDa | N | X | — | — | — |
| NDUFV3-10/10-kDa | N | X | X | — | — |
| NDUFS1/75-kDa | N | X | X | X | X |
| NDUFS6/13-kDa | N | — | — | X | X |
| NDUFA12/B17.2 | N | X | X | NA | NA |
| NDUFS4/18-kDa | N/Q? | NA | NA | X | X |
| NDUFS2/49-kDa | Q | — | X | — | — |
| NDUFS3/30-kDa | Q | X | X | — | — |
| NDUFS7/PSST | Q | — | — | X | X |
| NDUFS8/TYKY | Q | X | X | X | X |
| NDUFA6/B14 | Q | X | X | — | — |
| NDUFA7/B14.5a | Q | — | — | X | X |
| NDUFA9/39-kDa | Q | X | X | — | — |
| MT-ND1/ND1 | Pp | — | — | X | X |
| NDUFA1/MWFE | Pp | — | — | — | X |
For each subunit the human (according to HGNC: www.genenames.org) and bovine names (kDa) are given. Core subunit are highlighted in bold, mtDNA-encoded subunits are highlighted in italics. NA = not appropriate.
Functional modules: N (NADH binding and oxidation), Q (electron transfer to ubiquinone), P module (consisting of Pp and Pd submodules, proton pumping).
NDUFS4 mutations and human mitochondrial disease
Pathogenic mutations in NDUFS4 linked to MC1DN1/Leigh syndrome are inherited in an autosomal recessive manner. Several of these mutations have been described (Table 3), occurring in intronic and exonic nDNA regions (Fig. 2D). The first reported mutation (c.466-470dup) induced a 5 bp duplication leading to a frameshift (K158fs) in a patient with symptoms resembling Leigh syndrome. This mutation resulted in an elongation of the mature protein by 14 amino acids and destruction of the RVS phosphorylation consensus site.56 Hereafter, various other NDUFS4 mutations associated with Leigh syndrome, Leigh-like syndrome and/or MC1DN1 were presented including: (i) a homozygous c.462del mutation disrupting the NDUFS4 reading frame44; (ii) introduction of a stop codon resulting in degradation of transcribed product and suppression of the truncated NDUFS4 subunit43,61,65; and (iii) nonsense mutations leading to premature truncation of NDUFS4.66 Analysis of 14 patients with pathogenic NDUFS4 mutations demonstrated that the age of death varied between 3.6 and 27 months.67 Compatible with the last study, analysis of a cohort of 22 patients (18 families) yielded an average age of disease onset of 4.5 ± 4.4 months and an average age of death of 10 ± 7.7 months.48 The last study further reported that NDUFS4 mutations were associated with diverse clinical features including: (i) CI deficiency in muscle (n = 18 patients) and fibroblasts (n = 12); (ii) lactate elevation in plasma (n = 16) and CSF (n = 11); (iii) lesions of the brainstem (n = 14) and basal ganglia (n = 9); (iv) cortical atrophy (n = 3); (v) hypotonia (n = 22); (vi) developmental arrest-regression (n = 11); (vii) absence of eye contact (n = 10); (viii) apnoeic episodes (n = 10); (ix) pyramidal signs (n = 6); (x) epilepsy/seizures (n = 4); (xi) movement disorder (n = 2); (xii) microcephaly (n = 1); (xiii) ocular abnormalities (n = 11); (xiv) hypertrophic cardiomyopathy (n = 5); (xv) feeding problems/failure to thrive (n = 8); and (xvi) ragged-red fibres and lipid accumulation in muscle biopsies (n = 4).
Table 3.
Pathogenic NDUFS4 gene mutations
| ClinVar accession | Variant | Variant type | Effect at protein level | Disease phenotype | Reference |
|---|---|---|---|---|---|
| VCV000006890 | c.44G>A | Single nucleotide (1 bp) | p.Trp15Ter (W15*). MTS mutation. NDUFS4 protein absent. | MC1DN1 | Petruzzella et al.66 |
| VCV000496165.3 | c.99-1G>A | Single nucleotide (1 bp) | Abnormal splicing. | Leigh syndrome; MC1DN1 | Bénit et al.193 |
| VCV000488559 | c.178-2A>G | Single nucleotide (1 bp) | Abnormal splicing. | Leigh syndrome | ClinVar only |
| VCV000006888 | c.291del | Deletion (1 bp) | p.Lys96_Trp97insTer (W96*). No full-length NDUFS4 protein. | MC1DN1 | Budde et al.61 |
| VCV000006889 | c.316C>T | Single nucleotide (1 bp) | p.Arg106Ter (R106*). NDUFS4 protein absent.64 | MC1DN1 | Budde et al.61 |
| VCV000930177 | c.350+5G>A | Single nucleotide (1 bp) | Abnormal splicing. Analysis of muscle and fibroblast cDNA from the patient showed reduced expression of NDUFS4 to 13% and 18% of control levels, respectively, and the presence of abnormal transcript species indicative of splicing abnormalities was detected in muscle. | MC1DN1 | González-Quintana et al.194 |
| VCV000040257 | c.462del | Deletion (1 bp) | p.Lys154fs (K154fs). Replacement of the last 22 amino acids with 34 novel amino acids. RVS phospho-site destroyed. A 60% reduction in NDUFS4 transcript levels due to nonsense-mediated mRNA decay. No full length NDUFS4 protein was detected in isolated mitochondria.46 | Leigh syndrome; MC1DN1 | Anderson et al.44 |
| VCV000006887.2 | c.466-470dup | Duplication (5 bp) | p.Lys158fs (K158fs). Mature NDUFS4 protein is 14 amino acids longer. RVS phospho-site destroyed. NDUFS4 protein absent.64 | Leigh syndrome; MC1DN1 | Van den Heuvel et al.56 |
| VCV000488560 | c.470-471del | Deletion (2 bp) | p.Lys156_Ser157insTer. | Leigh syndrome | ClinVar only |
The data in this table were compiled using ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/; only mutations marked as ‘Pathogenic’) and OMIM (www.omim.org).
Impact of NDUFS4 mutations in patient-derived cells
In patient-derived primary skin fibroblasts, NDUFS4 mutations typically result in reduced NDUFS4 mRNA levels and the absence of NDUFS4 protein, associated with reduced levels of the CI holocomplex and catalytic activity.44–46,53,64,68,69 In general, the expression/activity of OXPHOS complexes other than CI is not affected by NDUFS4 mutations and/or absence of the NDUFS4 protein.45,56,64,70 On native gels (i.e. using blue native polyacrylamide gel electrophoresis; BN–PAGE), NDUFS4-mutated patient fibroblasts exhibit complete absence of the fully assembled CI holocomplex. Instead, these cells display a catalytically-inactive CI subcomplex with a size of ∼830-kDa (‘CI-830’).44,45,68,69,71 This suggests that NDUFS4 mutations impair CI biogenesis and/or stability.68 Analysis of patient-derived cells further demonstrated that NDUFS4 mutations were associated with partial Δψ depolarization, increased reactive oxygen species and NAD(P)H levels, aberrations in cytosolic and mitochondrial calcium/ATP homeostasis and altered mitochondrial morphology.49,69
The Ndufs4 whole-body knockout mouse model
In human patients, most of the currently known pathogenic NDUFS4 mutations lead to absence of (full length) NDUFS4 protein (Table 3). These mutations are inherited in an autosomal recessive manner and thus present in every cell. Therefore, from a genetic point of view, whole-body Ndufs4 deletion is a valid strategy to create mouse models of MC1ND1/Leigh syndrome for pathomechanistic analysis. Next, we will primarily focus on the Ndufs4 whole-body knockout mouse model (Ndufs4−/−-WB), which was created by deleting exon 2 of NDUFS4 (Fig. 2B), resulting in a frameshift that prevented formation of NDUFS4 protein.72 Among Ndufs4 knockout models, Ndufs4−/−-WB mice are most extensively studied (Supplementary Table 1) and applied in intervention studies (Supplementary Table 2). However, to allow better interpretation of the Ndufs4−/−-WB model, results obtained with relevant (tissue-specific) Ndufs4 knockout mouse models will also be discussed. Mouse models focusing on the impact of Ndufs4 deletion on inflammation, bone resorption and immune cell fate are presented elsewhere.73,74
The phenotype of Ndufs4−/−-WB mice
Heterozygous Ndufs4+/−-WB mice exhibited no phenotype when compared to wild-type mice.72 In contrast, Ndufs4−/−-WB mice appeared smaller and displayed hair loss by postnatal Day (PD) 21, whereas hair grew back during the next hair-growth cycle. Detailed analysis revealed that Ndufs4−/−-WB mice display an infiltration of inflammatory monocytes and macrophages and elevated mRNA levels of inflammatory markers in their skin.73 The latter might explain the sudden hair loss and be linked to the systemic inflammation reported in Ndufs4−/−-WB mice.73 Until PD30, mice appeared healthy and displayed similar behaviour (e.g. grooming, feeding, socializing) as wild-type littermates. After PD30, mice became lethargic, hypothermic, blind and eventually developed severe ataxia, marked by uncoordinated gait/balance (also see below) and hindlimb clasping. Additionally, mice developed abnormal oxygen saturation, heart rate, breathing patterns and displayed elevated cerebral and serum lactate levels (see Supplementary Table 1 for further phenotypic information). These symptoms progressively worsened and mice died at ∼PD50.72,75–79 Neurologically, Ndufs4−/−-WB mice developed bilateral spongiform lesions in the vestibular nuclei, and neurodegeneration in various other brain regions including the olfactory bulb, optic chiasm, optic tract, superior colliculus, interpeduncular nucleus, lateral lemniscus, trapezoid body, cochlear nuclei, fastigial nucleus, inferior olivary complex, nodulus (X) and the uvula (IX) granular and molecular layer.75,77,80 In addition, progressive microgliosis was observed from PD26 (mid-stage disease) onwards.75,77 It is expected that the vestibular nuclei/fastigial nucleus lesions contribute to the breathing abnormalities observed in these mice.75Ndufs4−/−-WB mice also displayed (mild) glial lipid droplet accumulation in olfactory bulb/vestibular nuclei, periaqueductal grey, cerebellum, dorsal motor nucleus, vagus and abducens nuclei. Accumulation of lipid droplets in glial cells is strongly associated with neuroinflammation and progression of neurodegeneration in Drosophila mutants and Ndufs4−/−-WB mice.80 The last study also demonstrated that lipid droplet accumulation correlated with elevated reactive oxygen species levels, and that both phenomena were mitigated by antioxidants. This suggests that oxidative stress can induce lipid droplet accumulation during mitochondrial dysfunction. Therefore, reactive oxygen species-induced lipid peroxidation and ensuing neuroinflammation might be part of the pathomechanism in Ndufs4−/−-WB mice.
The behaviour of Ndufs4−/−-WB mice
Because of their severe and progressive phenotype, early death and small size, it is challenging to analyse Ndufs4−/−-WB knockout mice in behavioural studies. Mice suffered vision loss at PD21, as evidenced by an absent B wave in their electroretinogram and their failure to recognize a visual cliff.72 Until ∼PD30 normal locomotor activity during both day and night cycle was observed.72 Gait analysis performed at PD30, PD35 and PD40 revealed severe impairment.77 From PD35 onwards the mice: (i) failed to maintain balance on a ledge; (ii) failed in a negative geotaxis test; (iii) displayed a decline in locomotor activity in the open field test; (iv) exhibited deteriorating muscle strength on the wire grip hang test; and (v) were unable to remain on a rotating rod as long as wild-type littermates.72,77,81 Although we do not have information at the level of individual animals, the water intake of Ndufs4−/−-WB mice is ∼50% of that of wild-type mice and further decreases around PD32-36 (A.Q., unpublished observation). However, it was reported that food consumption of Ndufs4−/−-WB animals was within the normal range during day, night and fasting.72 This leaves open the question as to why Ndufs4−/−-WB animals have a smaller size. Between PD26-37 mice were housed in metabolic cages. Weight loss occurred from PD35–40 onwards, which coincides with the reduced water intake and ataxia induction. This might suggest that food intake is normal before this time window, but Ndufs4−/−-WB animals start to eat less and lose weight once the disease phenotype becomes more severe. Supporting this idea, conditional Ndufs4−/− knockout animals displayed reduced food intake coinciding with the appearance of symptoms.82
Sensitivity of Ndufs4−/−-WB mice to anaesthetics
It is well established that the use of anaesthetics in patients with mitochondrial disease requires careful consideration.83 In this sense, Ndufs4−/−-WB mice displayed resistance against ketamine sedation, but were hypersensitive to volatile anaesthetics, such as isoflurane and halothane.84–87 It is still incompletely understood why ketamine resistance occurs, but this phenomenon might be linked to its different mode-of-action when compared to volatile anaesthetics. It was suggested that ketamine action is mediated by increased neuronal activation and cortical synchronization rather than neuronal inactivation. Moreover, ketamine may induce the anaesthetized state via a different target than other anaesthetics.84 Glutamatergic neuron-specific Ndufs4 knockout animals (see below) also displayed hypersensitivity to volatile anaesthetics,86,88 whereas animals with GABAergic neuron- or cholinergic neuron-specific Ndufs4 loss (see below) did not display this phenomenon.82 The increased volatile anaesthetic sensitivity of Ndufs4−/−-WB mice may be due to volatile anaesthetic-induced CI inhibition89,90 and the key role played by CI function in maintaining neuronal activity.84 These findings suggest that excitatory glutamatergic transmission is the major contributor to the volatile anaesthetic hypersensitivity of Ndufs4−/−-WB mice.88 Mechanistically, it was reported that volatile anaesthetics rapidly depleted the blood levels of β-hydroxybutyrate (βHB) in neonatal mice.90 In this study volatile anaesthetic concentrations well below those required for anaesthesia were applied and depletion of β-HB was mediated by citrate accumulation, malonyl-CoA production by acetyl-CoA carboxylase and inhibition of fatty acid oxidation. Analysis of PD17 mice revealed significantly higher β-HB levels in Ndufs4−/−-WB neonates relative to their wild-type littermates.90 It was concluded that CI is not the direct target of volatile anaesthetic mediating the acute β-HB effect in neonates, but may contribute to the increased lactate observed in animals exposed to volatile anaesthetic.
Tissue analyses of Ndufs4−/−-WB mice
Comparison of residual CI activities in isolated mitochondria between seven tissues of wild-type, Ndufs4+/−-WB and Ndufs4−/−-WB mice revealed normal values for Ndufs4+/−-WB and reduced but non-zero values for Ndufs4−/−-WB animals.91 The last equalled: 44% of wild-type (heart), 29% (muscle), 26% (brain), 25% (kidney), 19% (liver), 17% (pancreas) and 9% (lung). No significant differences between wild-type and Ndufs4−/−-WB tissues were detected with respect to CII–CV activities.91 A differential reduction in CI activity was observed in various knockout brain regions: 14% of wild-type (olfactory bulb), 25% (brainstem), 28% (cerebellum) and 62% (anterior cortex).92,93 It was hypothesized that, for the affected brain regions, the inability of mitochondrial energy production to meet cellular demands is responsible for the observed neurodegeneration and disease progression in Ndufs4−/−-WB mice.92 In contrast to the findings of Calvaruso et al.,91 the original study reported no CI activity in liver of Ndufs4−/−-WB mice.72 As suggested in the latter paper, it is to be expected that differences in sample preparation will lead to different amounts of functional CI due to its destabilization by NDUFS4 absence. Using BN–PAGE, inactive CI-830 and active ∼200-kDa CI-subassemblies were observed in heart, muscle, brain and kidney. The smaller subassembly at least contained the NDUFV1 and NDUFV2 subunit, and therefore likely represents the N-module of CI.91 These data indicate that Ndufs4−/−-WB tissues display a reduced but not zero CI activity due to a destabilized CI holo-complex.91 This is compatible with a follow-up proteomics analysis, demonstrating that overall CI subunit levels were reduced by ∼50% in brain, liver, heart, kidney, diaphragm and skeletal muscle of Ndufs4−/−-WB mice.64 The latter study also demonstrated that overall CII–CV subunit levels were not greatly affected in these tissues. Analysis of the CI structure revealed that NDUFS4 and the CI accessory subunit NDUFA12 interact with each other and with CI subunits of the N- and Q-module (Table 2). Remarkably, absence of the NDUFS4 protein induced near complete absence of the NDUFA12 subunit and increased the protein level of NDUFA2 (an assembly factor of CI) in brain, liver, heart, kidney, diaphragm and skeletal muscle of Ndufs4−/−-WB animals.64 This finding was confirmed in Ndufs4−/−-WB-derived mouse embryonic fibroblasts (MEFs; see below), Ndufs4−/− brainstem glutamatergic neurons94 and NDUFS4-mutated Leigh syndrome patient cells. Compatible with previous data,91Ndufs4−/−-WB-derived MEFs displayed in situ CI activity, but BN–PAGE analysis revealed that NDUFAF2 attached to an inactive CI subcomplex (CI-830) and inactive assemblies of higher molecular weight. Interestingly, NDUFA12 absence did not reduce NDUFS4 levels but triggered NDUFAF2 association to active CI in NDUFA12-mutated Leigh syndrome patient cells.64 This strongly suggests that absence of NDUFS4 induces absence of NDUFA12 but not vice versa. Association of NDUFAF2 with active CI was not observed in BN–PAGE experiments with mutations in other CI subunit-encoding genes where NDUFAF2 was attached to CI-830 (NDUFS1, NDUFV1 mutation) or not detected (NDUFS7 mutation). Taken together (Fig. 3A), this evidence supports a model in which absence of NDUFS4 induces: (i) absence of NDUFA12; (ii) attachment NDUFAF2 to stabilize active CI in situ; (iii) a reduced stability of N- to Q-module attachment95; and (iv) a reduced, but not zero, level of active CI. In addition to NDUFAF2 binding, in situ CI stabilization might also involve MIM lipids and/or interaction with other ETC complexes.29,91,96–98 However, in multiple models (ovine, porcine, human) NDUFS4, NDUFA12 and their interacting subunits (Table 2) were not involved in ETC supercomplex interactions.29–32
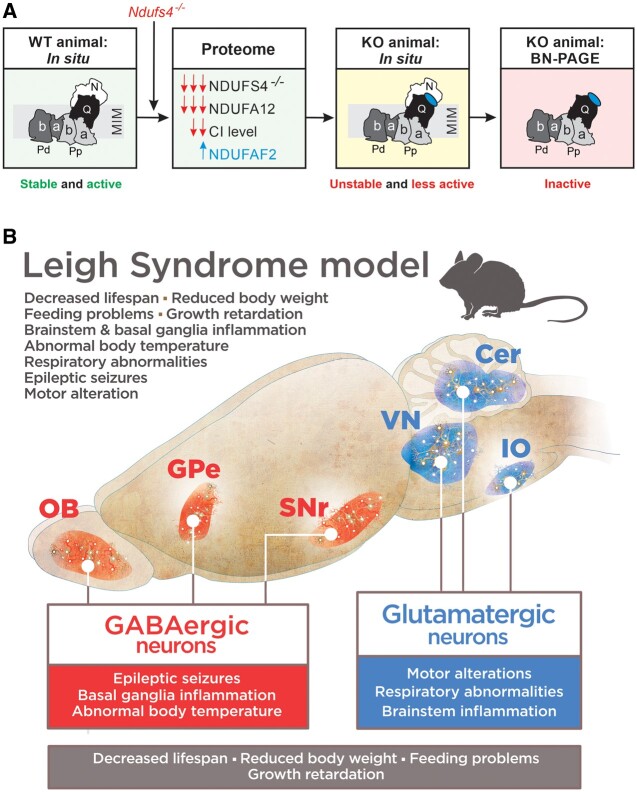
Figure 3.
Consequences of Ndufs4 knockout. (A) Ndufs4 knockout induces absence of the NDUFS4 subunit of CI, near complete absence of the NDUFA12 subunit and increased levels of the CI-attached NDUFAF2 assembly factor. This results in an unstable CI holocomplex that is present at lower levels in situ and therefore displays a lower activity in Ndufs4−/− mice. On isolation, the unstable CI complex loses its N-module, resulting in an inactive ∼800 kDa subcomplex on BN–PAGE gels. Adapted from Adjobo-Hermans et al.64 (B) Genetic dissection of clinical signs in Ndufs4−/−-WB mice. Vglut2-expressing glutamatergic neurons mediate most of the phenotype of Ndufs4−/−-WB mice, such as motor and respiratory alterations, while GABAergic neurons are involved in basal ganglia inflammation, development of epilepsy and hypothermia. Conditional alteration in either population leads to reduced lifespan and decreased body weight. Cer = cerebellum; GPe = external globus pallidus; IO = inferior olive; KO = knockout; OB = olfactory bulb; SNr = substantia nigra pars reticulata; VN = vestibular nuclei; WT = wild-type. Adapted from Bolea et al.82
Skeletal muscle bioenergetics of Ndufs4−/−-WB mice
In soleus muscle of Ndufs4−/−-WB mice, ATP levels were slightly decreased and CI activity was reduced, whereas phosphocreatine, creatine and inorganic phosphate levels were normal.72 Compatible with these observations, the histology, physiology and metabolism of soleus muscle tissue appears to be minimally affected by Ndufs4 deletion. Enzymological analysis of isolated skeletal muscle mitochondria99 demonstrated a 79% reduction in maximal CI activity (Vmax), which was paralleled by a 45–72% increase in Vmax for CII, CIII, CIV and citrate synthase. Using integrated in silico and experimental analysis, the same study demonstrated that the maximal rates of mitochondrial pyruvate oxidation and ATP production were not significantly affected in muscle mitochondria of Ndufs4−/−-WB mice. Computer modelling further predicted that CI deficiency alters the concentration of intermediate metabolites, increases mitochondrial NADH/NAD+ ratio and stimulates the lower half of the TCA cycle, including CII.99 The computer model further predicted that CI deficiency only has a major metabolic impact when its activity decreases below 10% of normal levels, compatible with a biochemical threshold effect, and that mouse skeletal muscle mitochondria display a substantial CI overcapacity, minimizing the effect of CI dysfunction on mitochondrial metabolism. Both CI overcapacity and the biochemical threshold effects probably differ between tissues and therefore could be involved in the tissue-specific impact of Ndufs4 deletion.100–102
Metabolome analysis of Ndufs4−/−-WB mouse brain and skeletal muscle tissue
A disturbed redox balance is often associated with accumulation of pyruvate, lactate and alanine levels. Accordingly, pyruvate, lactate and glycolytic intermediates were increased in the whole brain of Ndufs4−/−-WB mice at PD30.103 Likewise, the ratios of alanine, leucine and isoleucine relative to glutamic acid, as well as pyruvate/lactate and pyruvate/acyl-carnitine ratios, were increased in lesion-prone brain regions (i.e. brainstem, cerebellum and olfactory bulb) of male Ndufs4−/−-WB mice at PD45–50. These alterations may be due to an increased NADH/NAD+ ratio, which controls the formation of amino acids and lactate, while the increased pyruvate/acyl-carnitine ratio suggests NAD+-dependent acetyl-CoA formation from pyruvate.93 Glutathione was increased in brainstem, olfactory bulb and lesion-resistant brain regions. Analysis of skeletal muscle revealed decreased levels of pyruvate, glycerol, alanine and lactate.104,105 In addition, Ndufs4−/−-WB skeletal muscle also displayed decreased levels of N,N-dimethylglycine, fatty acid acyl-carnitines (chain lengths of: C0, C3, C4, C5, C6, C8, C12, and C16) and 2-aminoadipate.105 Similar changes were observed in Ndufs4−/−-WB brains,93 suggesting that fatty acid β-oxidation and amino acid catabolism is altered. Furthermore, muscle creatinine and creatine levels were decreased as well as fumarate, malate and succinate. The last may indicate an increased use of succinate to fuel CII thereby bypassing CI. Proline, hydroproline, citrulline, glutamate and glutamine were decreased.105 Similar metabolic alterations were observed in lesion-prone brain regions of male Ndufs4−/−-WB mice.93 These metabolic alterations may indicate altered proline metabolism that has been suggested to play a central role in mTOR signalling.106,107 Branched-chain amino acids (BCAAs) were increased, while butyryl(C4)- and isovaleryl(C5)-carnitine were decreased in lesion-prone brain regions.93 BCAA accumulation may be a consequence of inhibition of the NAD+-dependent reaction catalysed by branched-chain α-keto acid dehydrogenase due to redox imbalance.108–112 In addition, glutamate, aspartic acid and α-hydroxyglutaric acid were also decreased in Ndufs4−/−-WB brains, which could point to a compensatory increase of α-ketoglutaric acid to the TCA cycle. These alterations are in line with disruptions of neuronal transport systems and the beneficial effects of mTOR complex 1 (mTORC1) inhibition that have been attributed to a restored glutamine/glutamate/α-ketoglutaric acid axis in presymptomatic Ndufs4−/−-WB mice.113 Pyruvate and alanine levels tended to be increased in all brain regions, lactate was only increased in lesion-prone brain regions. The olfactory bulb displayed higher levels of other glycolysis and pentose phosphate pathway-related intermediates, which were not observed in other brain regions. Last, metabolic changes were detected in dihydroxyacetone phosphate (DHAP) and glycerol-3-phosphate (G3P). These metabolites link glycolysis, lipid metabolism and OXPHOS. G3P was most severely decreased in lesion-prone brain regions, suggesting that G3P oxidation is involved in fuelling respiration in CI deficiency. Collectively, an adaptive increase in glutamate, α-hydroxyglutaric acid and G3P oxidation seems to drive energy-generation in lesion-resistant brain regions, while these mechanisms appear less effective in lesion-prone brain regions where CI activity is more severely reduced.93 In this context, analysis of brainstem tissue from Ndufs4−/−-WB mice suggested glycolysis impairment, as supported by increased levels of fructose-6-phosphate (F6P), G3P and DHAP, which are glycolysis intermediates upstream of the NADH-producing step.114 Suggestive of an increased brainstem NADH/NAD+ ratio, the latter study also demonstrated increased α-hydroxybutyrate (αHB) and lactate in the brainstem, reduced aspartate levels in the brainstem, and increased αHB plasma levels.
Analysis of Ndufs4−/−-WB mice-derived cell models
To gain further insight into the cellular consequences of Ndufs4 deletion, immortalized MEFs were generated from Ndufs4−/−-WB mice.115 Mitochondrial fractions isolated from these MEFs lacked NDUFS4 protein, displayed virtually no CI activity and reduced CII, CIII and CIV activity. Also, SCC activity (a combined measure of CII and CIII activity) was reduced whereas CV and CS activities were normal. Similar to tissues of Ndufs4−/−-WB mice, BN–PAGE analysis of mitochondrial fractions demonstrated the presence of CI-830 and ∼200-kDa CI-subassemblies, whereas the levels of fully assembled CII–CV were not affected in Ndufs4−/− MEFs. Using intact cells, it was observed that Ndufs4 knockout displayed a ∼50% reduced O2 consumption that was, however, sensitive to acute treatment with the CI inhibitor rotenone.115 The latter demonstrates that active CI (i.e. a complex in which the N-module is attached to the Q-module) is present in Ndufs4−/− MEFs. As stated previously, it is likely that differences in sample preparation lead to different amounts of functional CI due to its destabilization by NDUFS4 absence. This explains why no CI activity was detected in mitochondrial fractions of Ndufs4−/− MEFs, whereas rotenone effectively inhibited O2 consumption in intact Ndufs4−/− MEFs.115 Complementation of the Ndufs4−/− cells with the wild-type gene increased the levels of catalytically active CI and O2 consumption. At the intact cell level, Ndufs4−/− MEFs displayed an increase in the combined mitochondrial autofluorescence signal of NADH and NADPH, increased lactate release, an increased NADH/NAD+ ratio and slight Δψ hyperpolarization. In contrast, Ndufs4−/− MEFs displayed no detectable alterations in NADPH/NADP ratio, glucose consumption, protein expression of hexokinases (I and II), pyruvate dehydrogenase phosphorylation, total ATP content, free cytosolic ATP concentration, cell growth rate and levels of hydroethidium-oxidizing reactive oxygen species.115 These findings indicate that Ndufs4 deletion destabilizes CI and suggest that Ndufs4−/− MEFs display an increased mitochondrial NADH level and are slightly more glycolytic than wild-type cells. However, alterations in cell metabolism might be difficult to detect in the MEFs given their intrinsic bioenergetic properties (e.g. a relatively high glycolytic rate). Analysis of primary muscle- and skin-derived fibroblasts from Ndufs4−/−-WB mice revealed that these cells displayed no active CI on BN–PAGE, increased levels of hydroethidium-oxidizing reactive oxygen species and minor aberrations in mitochondrial morphology.116Ndufs4−/−-fibroblasts proliferated normally and displayed no obvious apoptotic signs.116 These findings are compatible with results obtained in patient-derived primary skin fibroblasts harbouring NDUFS4 mutations,49,64,68,69 and suggest a connection between CI deficiency, reactive oxygen species levels and (regulation of) mitochondrial structure in the fibroblast cell model.2,4,117,118 In summary, we conclude that the cellular consequences of Ndufs4 deletion (co)depend on the type of cell, cell metabolic state, culture conditions, external substrate (e.g. glucose) concentration and immortalization status.
Results with other whole-body Ndufs4 knockout mouse models
In general, results obtained with other whole-body knockout models (Supplementary Table 1) were highly similar (e.g. CRISPR–Cas9 Ndufs4−/− mice) but not always identical (e.g. Ndufs4fky/fky, Ndufs4GT/GT mice) to those from Ndufs4−/−-WB mice. For instance, Ndufs4fky/fky mice displayed a reduced CI activity in brain, heart, muscle, liver and kidney but presented with symptoms earlier than Ndufs4−/−-WB animals.95,119 Similar to the Ndufs4−/−-WB mouse model, NDUFS4 protein was not detectable in MEFs from Ndufs4fky/fky mice, and these cells displayed the CI-830 subassembly, reduced CI activity, reduced O2 consumption, but normal ATP content and lactate production.119,120 MEFs from Ndufs4fky/fky and Ndufs4+/fky (heterozygous) mice exhibited downregulation of genes involved in cellular function, transcriptional regulation, neural differentiation/signalling pathways and synaptic transmission. This suggests that these cells have variable gene expression patterns in early differentiation highlighting the effect of CI dysfunction on the cell’s differentiation potential.119 In addition to these genetic changes, Ndufs4 deletion induced shifts in acyl-carnitine and AA levels in Ndufs4fky/fky, MEFs, pointing towards a reverse TCA flux.95Ndufs4GT/GT mice displayed a milder phenotype than Ndufs4−/−-WB animals and their degree of CI deficiency was less than in Ndufs4−/−-WB and Ndufs4fky/fky mice.121,122 Relative to their wild-type littermates, Ndufs4GT/GT mice were hyperactive, which might indicate increased restlessness, a symptom that is also observed in patients with mitochondrial disease.123Ndufs4GT/GT mice displayed a normal maximal ATP production capacity and mitochondrial content in left hippocampus and, similar to Ndufs4−/−-WB and Ndufs4fky/fky mice, displayed a normal CII and CIII activity in left hippocampus and frontal cortex. Ndufs4GT/GT mice presented with elevated levels of acyl-carnitine C3, C4 and C12 in the frontal cortex,133 whereas no alterations were found in whole brain tissue.121 In addition, amino acid metabolism and TCA metabolites were also altered in brain tissue. These findings are compatible with a reverse TCA cycle flux in Ndufs4GT/GT mice due to altered brain bioenergetics.121 Individuals with mitochondrial disease often present with diabetes,124 and therefore, plasma corticosterone and glucose levels were measured in Ndufs4GT/GT mice. Relative to wild-type littermates, Ndufs4GT/GT mice presented with normal plasma glucose and insulin levels but lower baseline plasma cortisone levels. The last increased on chronic unpredictable stress.121 Given their minor CI deficiency and phenotype Ndufs4GT/GT mice are less suited as animal models of Leigh syndrome. However, these mice could be of particular value for pathomechanistic analysis of mild CI deficiencies.121
Results with neuron-specific Ndufs4 KO mouse models
Analysis of a neuron- and glia-specific model (NesKO mice) revealed a phenotype similar to that of Ndufs4−/−-WB animals (Supplementary Table 1) including failure to thrive, hypothermia, optic atrophy, cataracts, ptosis, seizures and breathing abnormalities.125 Furthermore, these animals exhibited severe progressive ataxia from ∼PD35 onwards, marked by an uncoordinated gait, reduced balance, hindlimb clasping and decreased rotarod performance.125,126 These results suggest that Ndufs4 deletion in NesKO mice induces glial activation, associated with increased oxidative stress, cytokine release and eventually necrotic, but not apoptotic, cell death due to ATP depletion. Animals with specific Ndufs4 knockout in vestibular nuclei (AAV-VN-KO mice) also presented with a similar phenotype as Ndufs4−/−-WB mice.75 The development of severe breathing abnormalities in AAV-VN-KO mice suggests that vestibular nuclei neurodegeneration might be responsible for these abnormalities in Ndufs4−/−-WB mice. Glutamatergic neuron-specific Ndufs4 knockout mice (Vglut2:Ndufs4cKO mice) developed several of the symptoms observed in Ndufs4−/−-WB mice, associated with a similar phenotype and leading to early death.82,113,127 However, Vglut2:Ndufs4cKO mice did have a longer lifespan (∼10 weeks)127 relative to Ndufs4−/−-WB mice.72,75 Although GABAergic neuron-specific loss of Ndufs4 (Gad2:Ndufs4cKO mice) resulted in failure to thrive, it was not associated with clinical symptoms compared to wild-type littermates.82 However, mice suffered from hypothermia as early as PD20–30, exhibited (spontaneous) seizures when being handled, and displayed an increase in seizure incidence from PD50 onwards. Gad2:Ndufs4cKO mice showed a different profile compared to Vglut2:Ndufs4cKO mice with increased microglial and astroglial activation in the external globus pallidus (GPe), basal ganglia and the substantia nigra pars reticulata (SNr). Ndufs4 deletion in cholinergic neurons (ChAT:Ndufs4cKO mice) did not induce development of fatal encephalopathy and animals remained clinically healthy throughout the study.82 Knockout of Ndufs4 in striatal medium spiny neurons (MSN Ndufs4−/− mice) did not affect body weight or life span up to 6 months after birth, and animals did not develop neuroinflammation.128 Finally, animals with dopaminergic neuron-specific Ndufs4 deletion (DA Ndufs4−/− mice) appeared healthy and were indistinguishable from wild-type littermates.129–131 In summary (Fig. 3B), the use of neuron-specific models suggests that Ndufs4 deletion affects glutamatergic neurons in the vestibular nuclei, cerebellum and inferior olive to induce motor alterations, respiratory (breathing) abnormalities and brainstem inflammation. In addition, it appears that malfunction of GABAergic neurons in the olfactory bulb, GPe and SNr probably underlies the epileptic seizures, basal ganglia inflammation and abnormal body temperature.
Results with heart- and/or skeletal muscle-specific Ndufs4 null mice
Next to the brain, the heart is one of the most severely affected organs in CI deficiencies due to its high dependency on aerobic metabolism.132,133 However, in sharp contrast to the Ndufs4−/−-WB model, heart-specific Ndufs4 null mice did not present with a pathological phenotype up to 1 year of age.134 Instead, mice developed hypertrophic cardiomyopathy marked by decreased left ventricular ejection fraction and increased left ventricular mass.135 Interestingly, hypertrophic cardiomyopathy was also observed in several patients (5 of 22) with NDUFS4 mutations.48 Animals with heart- and skeletal muscle-specific Ndufs4 knockout (Ckm-NLS-cre; Ndufs4loxP/loxPmice), developed an increased heart-to-body weight ratio. Although this may indicate cardiomyopathy, animals did not show signs of heart failure and remained clinically healthy up to at least 1 year of age.129 Taken together, analysis of the heart- and/or muscle-specific Ndufs4−/− models suggests that the hypertrophic cardiomyopathy in NDUFS4-mutated patients (see above) might be a secondary consequence of the CI deficiency, which develops at a later age.
Intervention studies in Ndufs4 mouse models
Intervention studies primarily have been carried out using the Ndufs4−/−-WB mouse model. These interventions involve: (i) reducing tissue oxygenation (carbon monoxide exposure, hypoxia, phlebotomy); (ii) small molecule treatment; (iii) injection of induced pluripotent stem cells (iPSCs); and (iv) genetic approaches (Supplementary Table 2).
Reduction of tissue oxygenation
As mitochondria are prime O2 consumers, defects in the OXPHOS system might increase tissue oxygenation. Supporting this idea, venous hyperoxia was observed in patients with mitochondrial disease and tissue oxygenation was used as a measure of impaired OXPHOS function.136 Whole-body O2 consumption decreased over time and appeared to correlate with disease phenotype in Ndufs4−/−-WB mice. More detailed examination revealed that the partial oxygen pressure (PO2) in brain tissue increased in an age-dependent manner in Ndufs4−/−-WB mice and correlated with disease severity.137 Different strategies were applied to reduce tissue oxygenation in these mice. Carbon monoxide (CO) exposure is an established approach to reduce tissue oxygenation, which has been linked to cytoprotective effects as well as oxidative stress induction.138,139 Although various positive effects were observed (Supplementary Table 2), chronic CO exposure induced hyperintense lesions in the caudoputamen region of the brain in Ndufs4−/−-WB mice, indicative of adverse effects.137 Similarly, hypoxic conditions prevented/mitigated part of the disease phenotype in Ndufs4−/−-WB mice (prolonged lifespan, no loss of body weight, improved motor function, prevention of hypothermia).140 However, beneficial effects were only induced by continuous hypoxia (11% O2 for 3 weeks starting from PD30) or by hypoxia applied during a late-stage of the disease (11% O2; Supplementary Table 2). In contrast, temporary or milder hypoxia conditions (17% O2 for 3 weeks) were ineffective.141,142 Decreasing the number of circulating red blood cells by phlebotomy is another approach to reduce brain oxygenation by decreasing oxygen delivery. Although phlebotomy increased the lifespan of Ndufs4−/−-WB mice (Supplementary Table 2) it only temporarily prevented development of vestibular nuclei lesions.137 Taken together, these studies provide evidence that a (partial) reduction in brain oxygenation and/or oxygen delivery ameliorates the disease phenotype of Ndufs4−/−-WB mice. Regarding the mechanism of this amelioration, it was proposed that simultaneous reduction of oxygen delivery and consumption may reverse disease progression by triggering adaptive programs and at the same time limiting oxygen toxicity.
Small molecule treatment
Intervention studies in Ndufs4−/−-WB mice have been carried out with several classes of small molecules targeting: redox metabolism, mitochondrial biogenesis, energy metabolism, the mechanistic target of rapamycin (mTOR), toll-like receptor 4 (TLR4), benzodiazepine receptors and cyclic-AMP (cAMP) homeostasis. Below, we summarize the impact of these interventions, whereas additional details are provided in Supplementary Table 2.
Redox active compounds
Idebenone is an antioxidant that rescues vision impairment in CI-deficiency-linked Leber’s hereditary optic neuropathy.143 However, idebenone treatment did not reverse visual impairment in Ndufs4−/−-WB mice. N-acetyl cysteine amide (AD4) is a blood–brain barrier penetrating antioxidant that reduces oxidative stress.144,145 AD4 delayed disease onset and reduced the severity of the disease phenotype, marked by improved motor function at PD30.80 The reactive oxygen species scavenger KH176 (sonlicromanol) is based on the vitamin E-derivative trolox.117 KH176 diminished cellular damage caused by oxidative stress and improved residual enzyme activity of different OXPHOS complexes without inducing drug toxicity in Leigh syndrome patient-derived fibroblasts.146 Although KH176 displayed various positive effects, including improved motor function and normalization of lipid peroxidation, this molecule did not improve disease onset/severity and brain pathology, and did not restore residual CI activity.147,148 Individually, KH176 and clofibrate (see below) displayed positive effects in Ndufs4−/−-WB mice. However, combined treatment with these molecules did not prolong lifespan or improve motor function.148 NAD+ levels were reduced and NADH/NAD+ ratios were increased in brain tissue of Ndufs4−/−-WB mice, suggesting that NAD(H) redox imbalance might be involved in the pathomechanism. Compatible with this hypothesis and reduced CI-mediated NAD+ formation from NADH, galactose-induced death of Leigh syndrome patient primary skin fibroblasts with isolated CI deficiency was rescued by NAD+ increasing interventions.149 However, administration of the NAD+ precursor nicotinamide mononucleotide (NMN) prolonged lifespan, but did not ameliorate the clinical phenotype of Ndufs4−/−-WB mice.150 P7C3 is an aminopropyl carbazole that was first identified by an in vivo screen in search of chemicals that enhanced neuron formation in the hippocampus of adult mice.151 It is thought that P7C3 activates nicotinamide phosphoribosyltransferase (NAMPT; a key enzyme in the NAD+ salvage pathway), thereby increasing intracellular NAD+ levels. P7C3 moderately prolonged lifespan, but did not increase NAD+ levels in Ndufs4−/−-WB mice brains or alter NAMPT protein levels.150
Stimulating mitochondrial biogenesis
Stimulation of peroxisome proliferator-activated receptors (PPARs) and PPARG (gamma) coactivator 1α (PGC-1α) enhances fatty acid β-oxidation and mitochondrial biogenesis.152,153 Treatment with the PPARA (alpha) stimulator clofibrate prolonged lifespan and motor function of Ndufs4−/−-WB mice without inducing adverse hepatic effects.148 Similar to clofibrate, the PPARA stimulator fenofibrate prolonged lifespan and partially improved motor function in Ndufs4−/−-WB mice.154
Manipulation of energy metabolism
Dimethyl α-ketoglutarate (DMKG), a cell-permeable form of α-ketoglutarate, was administered to Ndufs4−/−-WB mice to increase brain α-ketoglutarate levels. DMKG was investigated since the beneficial effects of NMN (see above) were attributed to elevated brain levels of α-ketoglutarate. Moreover, DMKG increased cellular NAD+ levels and rescued galactose-induced death of primary skin fibroblasts of Leigh syndrome patients with isolated CI deficiency.149 DMKG prolonged lifespan and delayed onset of hind limb clasping of Ndufs4−/−-WB mice.150 In certain models, tetracyclines induced mitochondrial proteotoxic stress impacting on mitochondrial dynamics and function.155 Nevertheless, tetracyclines such as doxycycline were identified as potent small molecules to rescue against cell death in low-glucose culture conditions in mitochondrial disease cell models. It was found that doxycycline-mediated suppression of mitochondrial translation ameliorates the neuroinflammatory profile of Ndufs4−/−-WB mice.156
Inhibition of mTOR
Rapamycin (also known as sirolimus) and the macrocyclic lactone tacrolimus (also known as FK-506) display powerful immunosuppressant properties.157 Both rapamycin and tacrolimus can bind to FK506-binding protein 12 (FKBP12), thereby inhibiting mTORC1, which is composed of several proteins including mTOR itself.158 mTORC1 positively regulates cell growth and proliferation by promoting protein and lipid biosynthesis and inhibits catabolic processes such as autophagy.159,160 Tacrolimus was also described to inhibit calcineurin.161 Elevated mTOR levels have not been reported in Ndufs4−/−-WB mice; however, the mTOR kinase target ribosomal protein S6 (rpS6) was increased in whole brain homogenates from PD >45 mice.103 In contrast, elevated rpS6 phosphorylation was not detected in presymptomatic Ndufs4−/−-WB mice,113 suggesting that the mTOR pathway may be involved in the disease mechanism. Rapamycin delayed disease onset, prevented the development of neurological symptoms (ataxia, uncoordinated balance and hindlimb clasping) and prolonged the lifespan of Ndufs4−/−-WB mice.103,162,163 In contrast, tacrolimus did not affect disease onset or progression, suggesting that the beneficial effects of rapamycin might not be solely due to immunosuppression nor off-target disruption of calcineurin.103 Similarly, modulation of the mTOR pathway using the poly(ADP-ribose)polymerase-1 (PARP1) inhibitor PJ34, delayed disease onset but did not extend the lifespan of Ndufs4−/−-WB mice.164 Evidence was provided that PARP1 inhibition modulates the mTOR pathway by decreasing the phosphorylation of AMPK, Raptor, s6 kinase, Rictor and Akt. In this context, PJ34-mediated PARP1 inhibition prevented reactive oxygen species-stimulated phosphorylation of these proteins.165 Evidence was provided that rapamycin reduces neuroinflammation and glial activation through inhibition of protein kinase C (PKC).166 Treatment with pan-PKC inhibitors (GO6983, GF109203X) or the PKC-β-specific inhibitor ruboxistaurin prevented hair loss, prolonged survival and delayed the onset of neurological symptoms (hindlimb clasping) in Ndufs4−/−-WB mice. Only ruboxistaurin prevented skin inflammation and reduced glial fibrillary acidic protein (GFAP) levels and the NF-κB inflammatory response in brain. These findings suggest that inhibition of PKC-β and NF-κB-mediated inflammation increases survival of Ndufs4−/−-WB mice.166 Importantly, rapamycin can display severe adverse effects,167 especially in young children, which limits its therapeutic potential in paediatric Leigh syndrome patients.103 In this context, encapsulated rapamycin did have similar efficacy as intraperitoneal injections, but did not result in toxicity in treated mice162 potentially increasing its suitability for patient applications. In addition to encapsulated rapamycin, it was reported that the FDA-approved immunosuppressant everolimus is better tolerated by mitochondrial disease patients, although not all patients were responsive to this drug.168 Collectively, mTOR inhibition may prove to be a suitable strategy to augment the severe phenotype of patients with Leigh syndrome. However, the toxicity and off-target effects of mTOR inhibitors should be closely monitored when using these compounds in clinical trials.
TLR4 inhibition, benzodiazepine-agonist treatment and cAMP homeostasis
The TLR4 inhibitor TAK-242, a drug suppressing inflammatory mediators by binding to TLR4, rescued hair loss in Ndufs4−/−-WB mice, suggesting that systemic inflammation in these mice is mediated by TLR4.73 Similar to rapamycin, the benzodiazepine-agonist zolpidem, rescued visual function, prevented starburst amacrine cell degeneration and innate immune and inflammatory responses that occurred at time of vision loss.169 Papaverine is an opium alkaloid antispasmodic drug that elevates cAMP levels by inhibiting phosphodiesterase.170 Papaverine also restored visual function and prevented starburst amacrine cell degeneration and innate immune/inflammatory responses that occurred at time of vision loss.169
Injection of human induced pluripotent stem cell-derived mesenchymal stem cells
Mitochondria can be uni- and bi-directionally transferred between cells via cell–cell contacts (‘tunnelling nanotubes’).171 Using co-cultures of mesenchymal stem cells (MSCs) and CI-deficient mouse or human fibroblasts, it was demonstrated that mitochondria were transported from MSCs to CI-deficient cells.172 Although reactive oxygen species levels were reduced in CI-deficient cells, mitochondrial transfer efficiency was low and (partial) restoration of CI assembly/activity was not detected. Intravitreal injection of iPSC-MSCs into retinal ganglion cells (RGCs) of 3-week-old Ndufs4−/−-WB mice resulted in transfer of mitochondria to neurons. The latter was associated with rescue of retinal function, prevention of RGC loss and prevention of abnormal RGC activation. RGCs that received mitochondria displayed extended longevity and normalized levels of pro-inflammatory cytokines. These results demonstrate that donation of healthy mitochondria reduces retinal degeneration, likely by improving retinal mitochondrial bioenergetics.173 Although certainly promising, the therapeutic impact of iPSC-MSC delivery still faces several major challenges including tumorigenicity, immune rejection and genetic instability.174
Genetic approaches
Adeno-associated virus vectors
Various adeno-associated virus (AAV) vectors were used to deliver human NDUFS4 (hNDUFS4) in Ndufs4−/−-WB mice. Delivery of AAV-PHP.B-hNDUFS4 (PD26–28) extended lifespan and reduced disease severity (normalization of growth rate, improvement of motor coordination, preventing development of failure to thrive, epileptic seizures and cardiac abnormalities).175,176 This vector, in contrast to other tissues, did not transduce brain in newborn mice (PD1). The latter possibly relates to inefficient AAV transport mechanisms in neonates. Bilateral delivery of an AAV serotype 1 construct (AAV1-Ndufs4-IRES-GFP; i.e. containing the mouse Ndufs4 gene) into the vestibular nuclei at PD21 extended lifespan and delayed disease progression.75 Intravenous injection of the AAV2/9-hNDUFS4 construct resulted in AAV2/9-hNDUFS4 being present in skeletal muscle, heart and liver. However, this vector was absent in the brain and did not ameliorate clinical symptoms.177 Intracerebroventricular injection in newborn mice increased hNDUFS4 levels in the brain but did not improve the clinical phenotype. Combined intravenous and intracerebroventricular injections in newborn mice were associated with detectable levels of hNDUFS4 in brain, muscle and heart. This resulted in near complete restoration of CI activity, extended lifespan, increased body weight and improved motor coordination.177 These results suggest that AAV2/9-hNDUFS4 cannot pass the blood–brain barrier, and further strengthen the hypothesis that the Ndufs4−/−-WB phenotype is brain-specific. Recently, intravenous injection of AAV-EF1α-GPD1 was applied to express glycerol-3-phosphate dehydrogenase 1 (GPD1) in the brain of Ndufs4−/−-WB mice in a neuron- and glia-specific manner.114 GPD1 is a cytosolic protein, which catalyses the reversible conversion of DHAP and NADH to G3P and NAD+. Together with the mitochondrial protein glycerol-3-phosphate dehydrogenase, it also forms a G3P shuttle that facilitates the transfer of reducing equivalents from the cytosol to mitochondria. It was found that GDP1 expression normalized several of the metabolic aberrations in the brainstem of Ndufs4−/−-WB mice, ameliorated neuroinflammation, partially prevented motor function decline and reduction in body temperature and extended lifespan to PD84.144 These results provide evidence that stimulation of G3P biosynthesis regenerates cytosolic NAD+ to mitigate the phenotype in Ndufs4−/−-WB mice. Taken together, although AAV vector delivery might be a promising strategy for mitochondrial disease patients with NDUFS4 mutations, blood–brain barrier permeability differs between mice and human. In this sense, the ability of AAV vectors to penetrate the blood–brain barrier needs to be determined, or vector delivery should be performed directly in the affected brain region, to be applicable in a clinical setting.176
Overexpression of specific proteins
Metallothioneins are small intracellular proteins that are induced in fibroblasts from mitochondrial disease patients with isolated CI deficiency178 and potentially protect against oxidative stress. However, Mt1 overexpression in Ndufs4−/−-WB mice (confirmed in quadriceps muscle and brain) did not prevent protein oxidation, oxidative stress or inflammation and did not rescue the disease phenotype.81 This is compatible with other antioxidant-based therapies (see above), which did not rescue disease onset and severity. Evidence was provided that overexpression of the MIM-fusion and cristae-shaping protein optic atrophy 1 (OPA1) normalized aberrant mitochondrial cristae morphology in the forebrain and cerebellum of Ndufs4−/−-WB mice.179 OPA1 overexpression was demonstrated in various tissues (brain, skeletal muscle, heart) and paralleled by improved rotarod performance at 5 weeks and a moderate increase in lifespan. However, no phenotypic improvement was observed at 7 weeks with respect to disease progression and death of the animals. Furthermore, the therapeutic potential of OPA1 overexpression may be limited by its antiapoptotic properties.180 Small molecule mTOR inhibition ameliorated the disease phenotype of Ndufs4−/−-WB mice (see above). In this context, it was investigated whether the serine/threonine kinase S6K1, which is downstream of mTORC1 and involved in ribosomal protein synthesis and autophagy induction, plays a role in disease progression.181 Whole-body and liver-specific S6k1 knockout prolonged the lifespan of Ndufs4−/−-WB mice and delayed onset of hindlimb clasping. In contrast, fat- or brain-specific S6k1 showed no beneficial effects, suggesting that the mitigating effects of rapamycin-induced mTOR inhibition are partially mediated by active S6K1, although whole-body or liver-specific S6K1 knockout was not as efficient as a high dose (8 mg/kg) of rapamycin.103 In yeast and other models, the single-subunit NADH-dehydrogenase Ndi1 functions as a non-proton pumping alternative enzyme that replaces CI and oxidizes intramitochondrial NADH.182,183 This suggests that Ndi1 expression might compensate for reduced CI-mediated NADH oxidation in Ndufs4−/−-WB mice. Introduction of Ndi1 in neuron- and glia-specific Ndufs4 knockout mice (NesKO mice) did not prevent motor function and breathing abnormalities but prolonged lifespan to a median of >1 year.126 Importantly, in contrast to NesKO mice, Ndi1-expressing mice did not develop seizures, suggesting that seizures are a major cause of death in NesKO mice. These findings demonstrate that normalization of NADH oxidation partially prevents disease progression in NesKO mice, but does not fully restore neuronal function.126
Summary and conclusions
Development of Ndufs4−/− mouse models for MC1DN1/Leigh syndrome delivered novel pathomechanistic insights and allowed in vivo evaluation of potential intervention strategies. Mechanistically, Ndufs4 deletion induces the combined absence of two CI subunits (NDUFS4 and NDUFA12), paralleled by increased levels of the CI assembly factor NDUFAF2. As a consequence, the CI holocomplex becomes destabilized and, though still (partially) active in situ, dissociates during isolation prior to BN–PAGE analysis (Fig. 3A). Comprehensive analysis of the Ndufs4−/−-WB model demonstrated that these mice develop a fatal encephalopathy, which closely resembles the phenotype of human MC1DN1/Leigh syndrome patients with NDUFS4 mutations (Fig. 3B). Ndufs4−/−-WB mice primarily presented with neurological symptoms that were similar to those of brain/neuron-specific Ndufs4 knockout animals (Fig. 3B). This strongly suggests that whole-body Ndufs4 deletion and joint absence of NDUFS4/NDUFA12 induces brain-specific pathology. The last might be due to tissue-specific differences in CI expression levels, CI-dependent energy demand and/or biochemical threshold effects. Of note, the Ndufs4 mouse model(s) fail(s) to recapitulate the whole spectrum of manifestations encountered in different cases of Leigh syndrome.184,185 Moreover, it was reported that spontaneous mutations in mouse (sub)strains due to excessive breeding programs can affect animal metabolism and study outcomes. This difference in genetic background might explain why homozygous Ndufs4−/− knock-in mice were embryonically lethal, whereas Ndufs4−/−-WB mice were healthy until ∼5 weeks of age. Therefore, mouse (sub)strain selection should be scrutinized to minimize genotypic and phenotypic differences, potentially leading to misinterpretation of study outcomes.186,187 In this context, it is likely that the genetic background also impacts on the detailed disease outcome and phenotype in human MC1DN1/Leigh syndrome patients. Most of the interventions discussed in this review partially rescue the phenotype in Ndufs4−/− mice, which is promising. Although various clinical trials are starting or ongoing, there is no evidence yet supporting the effectiveness of these interventions in human mitochondrial disease and MC1DN1/Leigh syndrome patients.188–190
Funding
M.v.d.W. was supported by a PhD grant to W.J.H.K. and C.v.K. from the Radboudumc. M.R.W. was supported by the Polish National Science Centre (Grant: UMO-2014/15/B/NZ1/00490). A.Q. was supported by the European Research Council (Starting grant NEUROMITO, ERC-2014-StG-638106), MINECO Proyectos I + D de Excelencia (SAF2017-88108-R) and ‘la Caixa’ Foundation (ID 100010434), under the agreement LCF/PR/HR20/52400018.
Competing interests
W.J.H.K. acts as an adhoc scientific adviser of the SME Khondrion B.V. (Nijmegen, The Netherlands). This company was not involved in the writing of the manuscript, nor in the decision to submit the manuscript for publication.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Abbreviations
- AAV
adeno-associated virus
- BN–PAGE
blue native polyacrylamide gelelectrophoresis
- CI–V
complex I–V
- ETC
electron transport chain
- OXPHOS
oxidative phosphorylation
- MC1DN1
mitochondrial complex I deficiency, nuclear type 1
- MEFs
mouse embryonic fibroblasts
- MIM
mitochondrial inner membrane
- MTS
mitochondrial targeting sequence
- Ndufs4 −/−-WB
whole-body Ndufs4 knockout
- TCA
tricarboxylic acid
References
- 1. Mailloux RJ, McBride SL, Harper ME.. Unearthing the secrets of mitochondrial ROS and glutathione in bioenergetics. Trends Biochem Sci. 2013;38(12):592–602. [DOI] [PubMed] [Google Scholar]
- 2. Willems PHGM, Rossignol R, Dieteren CE, Murphy MP, Koopman WJH.. Redox homeostasis and mitochondrial dynamics. Cell Metab. 2015;22(2):207–218. [DOI] [PubMed] [Google Scholar]
- 3. Raffaello A, Mammucari C, Gherardi G, Rizzuto R.. Calcium at the center of cell signaling: Interplay between endoplasmic reticulum, mitochondria, and lysosomes. Trends Biochem Sci. 2016;41(12):1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bulthuis EP, Adjobo-Hermans MJW, Willems PHGM, Koopman WJH.. Mitochondrial morphofunction in mammalian cells. Antioxid Redox Signal. 2019;30(18):2066–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pfanner N, Warscheid B, Wiedemann N.. Mitochondrial proteins: From biogenesis to functional networks. Nat Rev Mol Cell Biol. 2019;20(5):267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smeitink JAM, van den Heuvel L, DiMauro S.. The genetics and pathology of oxidative phosphorylation. Nat Rev Genet. 2001;2(5):342–352. [DOI] [PubMed] [Google Scholar]
- 7. Vander Heiden MG, Cantley LC, Thompson CB.. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koopman WJH, Beyrath J, Fung CW, et al. Mitochondrial disorders in children: Toward development of small-molecule treatment strategies. EMBO Mol Med. 2016;8(4):311–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liemburg-Apers DC, Schirris TJ, Russel FG, Willems PHGM, Koopman WJH.. Mitoenergetic dysfunction triggers a rapid compensatory increase in steady-state glucose flux. Biophys J. 2015;109(7):1372–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Teixeira J, Basit F, Swarts HG, et al. Extracellular acidification induces ROS- and mPTP-mediated death in HEK293 cells. Redox Biol. 2018;15:394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. [DOI] [PubMed] [Google Scholar]
- 12. Koopman WJH, Nijtmans LG, Dieteren CE, et al. Mammalian mitochondrial complex I: Biogenesis, regulation, and reactive oxygen species generation. Antioxid Redox Signal. 2010;12(12):1431–1470. [DOI] [PubMed] [Google Scholar]
- 13. Dard L, Blanchard W, Hubert C, Lacombe D, Rossignol R.. Mitochondrial functions and rare diseases. Mol Aspects Med. 2020;71:100842. [DOI] [PubMed] [Google Scholar]
- 14. Walker JE. The ATP synthase: The understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41(1):1–16. [DOI] [PubMed] [Google Scholar]
- 15. Kamer KJ, Mootha VK.. The molecular era of the mitochondrial calcium uniporter. Nat Rev Mol Cell Biol. 2015;16(9):545–553. [DOI] [PubMed] [Google Scholar]
- 16. Jackson TD, Palmer CS, Stojanovski D.. Mitochondrial diseases caused by dysfunctional mitochondrial protein import. Biochem Soc Trans. 2018;46(5):1225–1238. [DOI] [PubMed] [Google Scholar]
- 17. Stroud DA, Surgenor EE, Formosa LE, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538(7623):123–126. [DOI] [PubMed] [Google Scholar]
- 18. Wirth C, Brandt U, Hunte C, Zickermann V.. Structure and function of mitochondrial complex I. Biochim Biophys Acta. 2016;1857(7):902–914. [DOI] [PubMed] [Google Scholar]
- 19. Guerrero-Castillo S, Baertling F, Kownatzki D, et al. The assembly pathway of mitochondrial respiratory chain complex I. Cell Metab. 2017;25(1):128–139. [DOI] [PubMed] [Google Scholar]
- 20. Fiedorczuk K, Sazanov LA.. Mammalian mitochondrial complex I structure and disease-causing mutations. Trends Cell Biol. 2018;28(10):835–867. [DOI] [PubMed] [Google Scholar]
- 21. Agip AA, Blaza JN, Fedor JG, Hirst J.. Mammalian respiratory complex I through the lens of cryo-EM. Annu Rev Biophys. 2019;48:165–184. [DOI] [PubMed] [Google Scholar]
- 22. Pagniez-Mammeri H, Loublier S, Legrand A, Bénit P, Rustin P, Slama A.. Mitochondrial complex I deficiency of nuclear origin I. Structural genes. Mol Genet Metab. 2012;105(2):163–172. [DOI] [PubMed] [Google Scholar]
- 23. Formosa LE, Dibley MG, Stroud DA, Ryan MT.. Building a complex complex: Assembly of mitochondrial respiratory chain complex I. Semin Cell Dev Biol. 2018;76:154–162. [DOI] [PubMed] [Google Scholar]
- 24. Grivennikova VG, Gladyshev GV, Vinogradov AD.. Deactivation of mitochondrial NADH: Ubiquinone oxidoreductase (respiratory complex I): Extrinsically affecting factors. Biochim Biophys Acta. 2020;1861(8):148207. [DOI] [PubMed] [Google Scholar]
- 25. Brandt U. Energy converting NADH: Quinone oxidoreductase (complex I). Annu Rev Biochem. 2006;75:69–92. [DOI] [PubMed] [Google Scholar]
- 26. Schägger H, Pfeiffer K.. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schägger H, de Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U.. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J Biol Chem. 2004;279(35):36349–36353. [DOI] [PubMed] [Google Scholar]
- 28. Letts JA, Fiedorczuk K, Sazanov LA.. The architecture of respiratory supercomplexes. Nature. 2016;537(7622):644–648. [DOI] [PubMed] [Google Scholar]
- 29. Wu M, Gu J, Guo R, Huang Y, Yang M.. Structure of mammalian respiratory supercomplex I1III2IV1. Cell. 2016;167(6):1598–1609.e10. [DOI] [PubMed] [Google Scholar]
- 30. Gu J, Wu M, Guo R, et al. The architecture of the mammalian respirasome. Nature. 2016;537(7622):639–643. [DOI] [PubMed] [Google Scholar]
- 31. Guo R, Zong S, Wu M, Wu M, Gu J, Yang M.. Architecture of human mitochondrial respiratory megacomplex I2III2IV2. Cell. 2017;170(6):1247–1257.e12. [DOI] [PubMed] [Google Scholar]
- 32. Letts JA, Sazanov LA.. Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nat Struct Mol Biol. 2017;24(10):800–808. [DOI] [PubMed] [Google Scholar]
- 33. Fedor JG, Hirst J.. Mitochondrial supercomplexes do not enhance catalysis by quinone channeling. Cell Metab. 2018;28(3):525–531.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vercellino I, Sazanov LA.. Structure and assembly of the mammalian supercomplex CIII2CIV. Nature. 2021;598(7880):364–367. [DOI] [PubMed] [Google Scholar]
- 35. Lightowlers RN, Taylor RW, Turnbull DM.. Mutations causing mitochondrial disease: What is new and what challenges remain? Science. 2015;349(6255):1494–1499. [DOI] [PubMed] [Google Scholar]
- 36. Gorman GS, Chinnery PF, DiMauro S, et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. [DOI] [PubMed] [Google Scholar]
- 37. Koopman WJH, Willems PHGM, Smeitink JAM.. Monogenic mitochondrial disorders. N Engl J Med. 2012;366(12):1132–1141. [DOI] [PubMed] [Google Scholar]
- 38. Dykens JA, Will Y.. Drug-induced mitochondrial dysfunction. John Wiley & Sons; 2008. [Google Scholar]
- 39. Schirris TJJ, Renkema GH, Ritschel T, et al. Statin-induced myopathy is associated with mitochondrial complex III inhibition. Cell Metab. 2015;22(3):399–407. [DOI] [PubMed] [Google Scholar]
- 40. Baertling F, Rodenburg RJ, Schaper J, et al. A guide to diagnosis and treatment of Leigh syndrome. J Neurol Neurosurg Psychiatry. 2014;85(3):257–265. [DOI] [PubMed] [Google Scholar]
- 41. Lake NJ, Compton AG, Rahman S, Thorburn DR.. Leigh syndrome: One disorder, more than 75 monogenic causes. Ann Neurol. 2016;79(2):190–203. [DOI] [PubMed] [Google Scholar]
- 42. Leigh D. Subacute necrotizing encephalomyelopathy in an infant. J Neurol Neurosurg Psychiatry. 1951;14(3):216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Budde SM, van den Heuvel LP, Smeets RJ, et al. Clinical heterogeneity in patients with mutations in the NDUFS4 gene of mitochondrial complex I. J Inherit Metab Dis. 2003;26(8):813–815. [DOI] [PubMed] [Google Scholar]
- 44. Anderson SL, Chung WK, Frezzo J, et al. A novel mutation in NDUFS4 causes Leigh syndrome in an Ashkenazi Jewish family. J Inherit Metab Dis. 2008;31(S2):461- S467. [DOI] [PubMed] [Google Scholar]
- 45. Assouline Z, Jambou M, Rio M, et al. A constant and similar assembly defect of mitochondrial respiratory chain complex I allows rapid identification of NDUFS4 mutations in patients with Leigh syndrome. Biochim Biophys Acta. 2012;1822(6):1062–1069. [DOI] [PubMed] [Google Scholar]
- 46. Assereto S, Robbiano A, Di Rocco M, et al. Functional characterization of the c.462delA mutation in the NDUFS4 subunit gene of mitochondrial complex I. Clin Genet. 2014;86(1):99–101. [DOI] [PubMed] [Google Scholar]
- 47. Lamont RE, Beaulieu CL, Bernier FP, et al. A novel NDUFS4 frameshift mutation causes Leigh disease in the Hutterite population. Am J Med Genet A. 2017;173:595–600. [DOI] [PubMed] [Google Scholar]
- 48. Ortigoza-Escobar JD, Oyarzabal A, Montero R, et al. Ndufs4 related Leigh syndrome: A case report and review of the literature. Mitochondrion. 2016;28:73–78. [DOI] [PubMed] [Google Scholar]
- 49. Distelmaier F, Koopman WJH, van den Heuvel LP, et al. Mitochondrial complex I deficiency: From organelle dysfunction to clinical disease. Brain. 2009;132(Pt 4):833–842. [DOI] [PubMed] [Google Scholar]
- 50. Koene S, Willems PHGM, Roestenberg P, Koopman WJH, Smeitink JAM.. Mouse models for nuclear DNA-encoded mitochondrial complex I deficiency. J Inherit Metab Dis. 2011;34(2):293–307. [DOI] [PubMed] [Google Scholar]
- 51. Fernandez-Moreira D, Ugalde C, Smeets R, et al. X-linked NDUFA1 gene mutations associated with mitochondrial encephalomyopathy. Ann Neurol. 2007;61(1):73–83. [DOI] [PubMed] [Google Scholar]
- 52. Koopman WJH, Distelmaier F, Smeitink JAM, Willems PHGM.. OXPHOS mutations and neurodegeneration. EMBO J. 2013;32(1):9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lombardo B, Ceglia C, Tarsitano M, Pierucci I, Salvatore F, Pastore L.. Identification of a deletion in the NDUFS4 gene using array-comparative genomic hybridization in a patient with suspected mitochondrial respiratory disease. Gene. 2014;535(2):376–379. [DOI] [PubMed] [Google Scholar]
- 54. Alston LA, Rocha MC, Lax NZ, Turnbull DM, Taylor RW.. The genetics and pathology of mitochondrial disease. J Pathol. 2017;241(2):236–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schubert Baldo M, Vilarinho L.. Molecular basis of Leigh syndrome: A current look. Orphanet J Rare Dis. 2020;15(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Van den Heuvel LP, Ruitenbeek W, Smeets R, et al. Demonstration of a new pathogenic mutation in human complex I deficiency: A 5-bp duplication in the nuclear gene encoding the 18-kD (AQDQ) subunit. Am J Hum Genet. 1998;62(2):262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. De Rasmo D, Panelli D, Sardanelli AM, Papa S.. cAMP-dependent protein kinase regulates the mitochondrial import of the nuclear encoded NDUFS4 subunit of complex I. Cell Signal. 2008;20(5):989–997. [DOI] [PubMed] [Google Scholar]
- 58. Kennelly PJ, Krebs EG.. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J Biol Chem. 1991;266(24):15555–15558. [PubMed] [Google Scholar]
- 59. De Rasmo D, Palmisano G, Scacco S, et al. Phosphorylation pattern of the NDUFS4 subunit of complex I of the mammalian respiratory chain. Mitochondrion. 2010;10(5):464–471. [DOI] [PubMed] [Google Scholar]
- 60. Chen R, Fearnley IM, Peak-Chew SY, Walker JE.. The phosphorylation of subunits of complex I from bovine heart mitochondria. J Biol Chem. 2004;279(25):26036–26045. [DOI] [PubMed] [Google Scholar]
- 61. Budde SM, van den Heuvel LP, Janssen AJ, et al. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem Biophys Res Commun. 2000;275(1):63–68. [DOI] [PubMed] [Google Scholar]
- 62. Papa S, Rasmo DD, Technikova-Dobrova Z, et al. Respiratory chain complex I, a main regulatory target of the cAMP/PKA pathway is defective in different human diseases. FEBS Lett. 2012;586(5):568–577. [DOI] [PubMed] [Google Scholar]
- 63. Kahlhöfer F, Kmita K, Wittig I, Zwicker K, Zickermann V.. Accessory subunit NUYM (NDUFS4) is required for stability of the electron input module and activity of mitochondrial complex I. Biochim Biophys Acta. 2017;1858(2):175–181. [DOI] [PubMed] [Google Scholar]
- 64. Adjobo-Hermans MJW, de Haas R, Willems PHGM, et al. NDUFS4 deletion triggers loss of NDUFA12 in Ndufs4-/- mice and Leigh syndrome patients: A stabilizing role for NDUFAF2. Biochim Biophys Acta. 2020;1861(8):148213. [DOI] [PubMed] [Google Scholar]
- 65. Scacco S, Petruzzella V, Budde S, et al. Pathological mutations of the human NDUFS4 gene of the 18-kDa (AQDQ) subunit of complex I affect the expression of the protein and the assembly and function of the complex. J Biol Chem. 2003;278(45):44161–44167. [DOI] [PubMed] [Google Scholar]
- 66. Petruzzella V, Vergari R, Puzziferri I, et al. A nonsense mutation in the NDUFS4 gene encoding the 18 kDa (AQDQ) subunit of complex I abolishes assembly and activity of the complex in a patient with Leigh-like syndrome. Hum Mol Genet. 2001;10(5):529–535. [DOI] [PubMed] [Google Scholar]
- 67. Koene S, Rodenburg RJ, van der Knaap MS, et al. Natural disease course and genotype-phenotype correlations in Complex I deficiency caused by nuclear gene defects: What we learned from 130 cases. J Inherit Metab Dis. 2012;35(5):737- 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ugalde C, Janssen RJ, van den Heuvel LP, Smeitink JAM, Nijtmans LGJ.. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum Mol Genet. 2004;13(6):659–667. [DOI] [PubMed] [Google Scholar]
- 69. Iuso A, Scacco S, Piccoli C, et al. Dysfunctions of cellular oxidative metabolism in patients with mutations in the NDUFS1 and NDUFS4 genes of complex I. J Biol Chem. 2006;281(15):10374–10380. [DOI] [PubMed] [Google Scholar]
- 70. Willems PHGM, Smeitink JA, Koopman WJH.. Mitochondrial dynamics in human NADH: Ubiquinone oxidoreductase deficiency. Int J Biochem Cell Biol. 2009;41(10):1773–1782. [DOI] [PubMed] [Google Scholar]
- 71. Leshinsky-Silver E, Lebre AS, Minai L, et al. NDUFS4 mutations cause Leigh syndrome with predominant brainstem involvement. Mol Genet Metab. 2009;97(3):185–189. [DOI] [PubMed] [Google Scholar]
- 72. Kruse SE, Watt WC, Marcinek DJ, Kapur RP, Schenkman KA, Palmiter RD.. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab. 2008;7(4):312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Jin Z, Wei W, Yang M, Du Y, Wan Y.. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 2014;20(3):483–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bertaux A, Cabon L, Brunelle-Navas M-N, Bouchet S, Nemazanyy I, Susin SA.. Mitochondrial OXPHOS influences immune cell fate: Lessons from hematopoietic AIF-deficient and NDUFS4-deficient mouse models. Cell Death Dis. 2018;9(6):581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Quintana A, Zanella S, Koch H, et al. Fatal breathing dysfunction in a mouse model of Leigh syndrome. J Clin Invest. 2012;122(7):2359–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yu AK, Song L, Murray KD, et al. Mitochondrial complex I deficiency leads to inflammation and retinal ganglion cell death in the Ndufs4 mouse. Hum Mol Genet. 2015;24(10):2848–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. De Haas R, Russel FG, Smeitink JAM.. Gait analysis in a mouse model resembling Leigh disease. Behav Brain Res. 2016;296:191–198. [DOI] [PubMed] [Google Scholar]
- 78. Song L, Yu A, Murray K, Cortopassi G.. Bipolar cell reduction precedes retinal ganglion neuron loss in a complex 1 knockout mouse model. Brain Res. 2017;1657:232–244. [DOI] [PubMed] [Google Scholar]
- 79. Takahashi Y, Kioka H, Shintani Y, et al. Detection of increased intracerebral lactate in a mouse model of Leigh syndrome using proton MR spectroscopy. Magn Reson Imaging. 2019;58:38–43. [DOI] [PubMed] [Google Scholar]
- 80. Liu L, Zhang K, Sandoval H, et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160(1-2):177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Miller HC, Louw R, Mereis M, et al. Metallothionein 1 overexpression does not protect against mitochondrial disease pathology in Ndufs4 knockout mice. Mol Neurobiol. 2021;58(1):243–262. [DOI] [PubMed] [Google Scholar]
- 82. Bolea I, Gella A, Sanz E, et al. Defined neuronal populations drive fatal phenotype in a mouse model of Leigh syndrome. eLife. 2019;8:e47163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Niezgoda J, Morgan PG.. Anesthetic considerations in patients with mitochondrial defects. Paediatr Anaesth. 2013;23(9):785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Quintana A, Morgan PG, Kruse SE, Palmiter RD, Sedensky MM.. Altered anesthetic sensitivity of mice lacking Ndufs4, a subunit of mitochondrial complex I. PLoS One. 2012;7(8):e42904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Roelofs S, Manjeri GR, Willems PHGM, Scheffer GJ, Smeitink JAM, Driessen JJ.. Isoflurane anesthetic hypersensitivity and progressive respiratory depression in a mouse model with isolated mitochondrial complex I deficiency. J Anesth. 2014;28(6):807–814. [DOI] [PubMed] [Google Scholar]
- 86. Ramadasan-Nair R, Hui J, Zimin PI, Itsara LS, Morgan PG, Sedensky MM.. Regional knockdown of NDUFS4 implicates a thalamocortical circuit mediating anesthetic sensitivity. PLoS One. 2017;12(11):e0188087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zimin PI, Woods CB, Kayser EB, Ramirez JM, Morgan PG, Sedensky MM.. Isoflurane disrupts excitatory neurotransmitter dynamics via inhibition of mitochondrial complex I. Br J Anaesth. 2018;120(5):1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zimin PI, Woods CB, Quintana A, Ramirez JM, Morgan PG, Sedensky MM.. Glutamatergic neurotransmission links sensitivity to volatile anesthetics with mitochondrial function. Curr Biol. 2016;26(16):2194–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hanley PJ, Ray J, Brandt U, Daut J.. Halothane, isoflurane and sevoflurane inhibit NADH: Ubiquinone oxidoreductase (complex I) of cardiac mitochondria. J Physiol. 2002;544(3):687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stokes J, Freed A, Bornstein R, et al. Mechanisms underlying neonate-specific metabolic effects of volatile anesthetics. eLife. 2021;10:e65400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Calvaruso MA, Willems PHGM, van den Brand M, et al. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum Mol Genet. 2012;21(1):115–120. [DOI] [PubMed] [Google Scholar]
- 92. Kayser EB, Sedensky MM, Morgan PG.. Region-specific defects of respiratory capacities in the Ndufs4(KO) mouse brain. PLoS One. 2016;11(1):e0148219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Terburgh K, Coetzer J, Lindeque JZ, van der Westhuizen FH, Louw R.. Aberrant BCAA and glutamate metabolism linked to regional neurodegeneration in a mouse model of Leigh syndrome. Biochim Biophys Acta. 2021;1867(5):166082. [DOI] [PubMed] [Google Scholar]
- 94. Gella A, Prada-Dacasa P, Carrascal M, et al. Mitochondrial proteome of affected glutamatergic neurons in a mouse model of Leigh syndrome. Front Cell Dev Biol. 2020;8:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Leong DW, Komen JC, Hewitt CA, et al. Proteomic and metabolomic analyses of mitochondrial complex I-deficient mouse model generated by spontaneous B2 short interspersed nuclear element (SINE) insertion into NADH dehydrogenase (ubiquinone) Fe-S protein 4 (Ndufs4) gene. J Biol Chem. 2012;287(24):20652–20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Acín-Pérez R, Bayona-Bafaluy MP, Fernández-Silva P, et al. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol Cell. 2004;13(6):805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lobo-Jarne T, Pérez-Pérez R, Fontanesi F, et al. Multiple pathways coordinate assembly of human mitochondrial complex IV and stabilization of respiratory supercomplexes. EMBO J. 2020;39(14):e103912- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Protasoni M, Pérez-Pérez R, Lobo-Jarne T, et al. Respiratory supercomplexes act as a platform for complex III-mediated maturation of human mitochondrial complexes I and IV. EMBO J. 2020;39(3):e102817- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Alam MT, Manjeri GR, Rodenburg RJ, et al. Skeletal muscle mitochondria of NDUFS4-/- mice display normal maximal pyruvate oxidation and ATP production. Biochim Biophys Acta. 2015;1847(6-7):526–533. [DOI] [PubMed] [Google Scholar]
- 100. Rossignol R, Malgat M, Mazat JP, Letellier T.. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. J Biol Chem. 1999;274(47):33426–33432. [DOI] [PubMed] [Google Scholar]
- 101. Mazat JP, Rossignol R, Malgat M, Rocher C, Faustin B, Letellier T.. What do mitochondrial diseases teach us about normal mitochondrial functions…that we already knew: Threshold expression of mitochondrial defects. Biochim Biophys Acta. 2001;1504(1):20–30. [DOI] [PubMed] [Google Scholar]
- 102. Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T.. Mitochondrial threshold effects. Biochem J. 2003;370(Pt 3):751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Johnson SC, Yanos ME, Kayser EB, et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342(6165):1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Mason S, Terburgh K, Louw R.. Miniaturized 1H-NMR method for analyzing limited-quantity samples applied to a mouse model of Leigh disease. Metabolomics. 2018;14(6):74. [DOI] [PubMed] [Google Scholar]
- 105. Terburgh K, Lindeque Z, Mason S, van der Westhuizen F, Louw R.. Metabolomics of Ndufs4-/- skeletal muscle: Adaptive mechanisms converge at the ubiquinone-cycle. Biochim Biophys Acta. 2019;1865(1):98–106. [DOI] [PubMed] [Google Scholar]
- 106. Sahu N, Dela Cruz D, Gao M, et al. Proline starvation induces unresolved ER stress and hinders mTORC1-dependent tumorigenesis. Cell Metab. 2016;24(5):753–761. [DOI] [PubMed] [Google Scholar]
- 107. Karna E, Szoka L, Huynh TYL, Palka JA.. Proline-dependent regulation of collagen metabolism. Cell Mol Life Sci. 2020;77(10):1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Falk MJ, Zhang Z, Rosenjack JR, et al. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol Genet Metab. 2008;93(4):388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Clarke C, Xiao R, Place E, et al. Mitochondrial respiratory chain disease discrimination by retrospective cohort analysis of blood metabolites. Mol Genet Metab. 2013;110(1-2):145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Vergano SS, Rao M, McCormack S, et al. In vivo metabolic flux profiling with stable isotopes discriminates sites and quantifies effects of mitochondrial dysfunction in C. elegans. Mol Genet Metab. 2014;111(3):331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Esterhuizen K, van der Westhuizen FH, Louw R.. Metabolomics of mitochondrial disease. Mitochondrion. 2017;35:97–110. [DOI] [PubMed] [Google Scholar]
- 112. Lozoya OA, Martinez-Reyes I, Wang T, et al. Mitochondrial nicotinamide adenine dinucleotide reduced (NADH) oxidation links the tricarboxylic acid (TCA) cycle with methionine metabolism and nuclear DNA methylation. PLoS Biol. 2018;16(4):e2005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Johnson SC, Kayser EB, Bornstein R, et al. Regional metabolic signatures in the Ndufs4(KO) mouse brain implicate defective glutamate/α-ketoglutarate metabolism in mitochondrial disease. Mol Genet Metab. 2020;130(2):118–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Liu S, Fu S, Wang G, et al. Glycerol-3-phosphate biosynthesis regenerates cytosolic NAD+ to alleviate mitochondrial disease. Cell Metab. 2021;33(10):1974–1987. [DOI] [PubMed] [Google Scholar]
- 115. Valsecchi F, Monge C, Forkink M, et al. Metabolic consequences of NDUFS4 gene deletion in immortalized mouse embryonic fibroblasts. Biochim Biophys Acta. 2012;1817(10):1925–1936. [DOI] [PubMed] [Google Scholar]
- 116. Valsecchi F, Grefte S, Roestenberg P, et al. Primary fibroblasts of NDUFS4-/- mice display increased ROS levels and aberrant mitochondrial morphology. Mitochondrion. 2013;13(5):436–443. [DOI] [PubMed] [Google Scholar]
- 117. Distelmaier F, Valsecchi F, Forkink M, et al. Trolox-sensitive reactive oxygen species regulate mitochondrial morphology, oxidative phosphorylation and cytosolic calcium handling in healthy cells. Antioxid Redox Signal. 2012;17(12):1657–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Willems PHGM, Wanschers BF, Esseling J, et al. BOLA1 is an aerobic protein that prevents mitochondrial morphology changes induced by glutathione depletion. Antioxid Redox Signal. 2013;18(2):129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Johnson J, Lee W, Frazier AE, et al. Deletion of the Complex I subunit NDUFS4 adversely modulates cellular differentiation. Stem Cells Dev. 2016;25(3):239–250. [DOI] [PubMed] [Google Scholar]
- 120. Bird MJ, Wijeyeratne XW, Komen JC, et al. Neuronal and astrocyte dysfunction diverges from embryonic fibroblasts in the Ndufs4fky/fky mouse. Biosci Rep. 2014;34(6):e00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Emmerzaal TL, Preston G, Geenen B, et al. Impaired mitochondrial complex I function as a candidate driver in the biological stress response and a concomitant stress-induced brain metabolic reprogramming in male mice. Transl Psychiatry. 2020;10(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Emmerzaal TL, Jacobs L, Geenen B, et al. Chronic fluoxetine or ketamine treatment differentially affects brain energy homeostasis which is not exacerbated in mice with trait suboptimal mitochondrial function. Eur J Neurosci. 2021;53(9):2986–3001. [DOI] [PubMed] [Google Scholar]
- 123. Martikainen MH, Ng YS, Gorman GS, et al. Clinical, genetic, and radiological features of extrapyramidal movement disorders in mitochondrial disease. JAMA Neurol. 2016;73(6):668–674. [DOI] [PubMed] [Google Scholar]
- 124. Schaefer AM, Walker M, Turnbull DM, Taylor RW.. Endocrine disorders in mitochondrial disease. Mol Cell Endocrinol. 2013;379(1-2):2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Quintana A, Kruse SE, Kapur RP, Sanz E, Palmiter RD.. Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc Natl Acad Sci U S A. 2010;107(24):10996–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. McElroy GS, Reczek CR, Reyfman PA, Mithal DS, Horbinski CM, Chandel NS.. NAD+ regeneration rescues lifespan, but not ataxia, in a mouse model of brain mitochondrial complex I dysfunction. Cell Metab. 2020;32(2):301–308.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang L, Klingeborn M, Travis AM, et al. Progressive optic atrophy in a retinal ganglion cell-specific mouse model of complex I deficiency. Sci Rep. 2020;10(1):16326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chen B, Hui J, Montgomery KS, et al. Loss of mitochondrial Ndufs4 in striatal medium spiny neurons mediates progressive motor impairment in a mouse model of Leigh Syndrome. Front Mol Neurosci. 2017;10:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Sterky FH, Hoffman AF, Milenkovic D, et al. Altered dopamine metabolism and increased vulnerability to MPTP in mice with partial deficiency of mitochondrial complex I in dopamine neurons. Hum Mol Genet. 2012;21(5):1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kim HW, Choi WS, Sorscher N, et al. Genetic reduction of mitochondrial complex I function does not lead to loss of dopamine neurons in vivo. Neurobiol Aging. 2015;36(9):2617–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Choi WS, Kim HW, Tronche F, Palmiter RD, Storm DR, Xia Z.. Conditional deletion of Ndufs4 in dopaminergic neurons promotes Parkinson's disease-like non-motor symptoms without loss of dopamine neurons. Sci Rep. 2017;7:44989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Di Lisa F, Menabò R, Canton M, Petronilli V.. The role of mitochondria in the salvage and the injury of the ischemic myocardium. Biochim Biophys Acta. 1998;1366(1-2):69–78. [DOI] [PubMed] [Google Scholar]
- 133. Halestrap AP, Clarke SJ, Javadov SA.. Mitochondrial permeability transition pore opening during myocardial reperfusion - a target for cardioprotection. Cardiovasc Res. 2004;61(3):372–385. [DOI] [PubMed] [Google Scholar]
- 134. Karamanlidis G, Lee CF, Garcia-Menendez L, et al. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metab. 2013;18(2):239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chouchani ET, Methner C, Buonincontri G, et al. Complex I deficiency due to selective loss of Ndufs4 in the mouse heart results in severe hypertrophic cardiomyopathy. PLoS One. 2014;9(4):e94157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Taivassalo T, Abbott A, Wyrick P, Haller RG.. Venous oxygen levels during aerobic forearm exercise: An index of impaired oxidative metabolism in mitochondrial myopathy. Ann. Neurol. 2002;51(1):38–44. [DOI] [PubMed] [Google Scholar]
- 137. Jain IH, Zazzeron L, Goldberger O, et al. Leigh syndrome mouse model can be rescued by interventions that normalize brain hyperoxia, but not HIF activation. Cell Metab. 2019;30(4):824–832.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Chen RJ, Lee YH, Chen TH, et al. Carbon monoxide-triggered health effects: The important role of the inflammasome and its possible crosstalk with autophagy and exosomes. Arch Toxicol. 2021;95(4):1141–1159. [DOI] [PubMed] [Google Scholar]
- 139. Chenoweth JA, Albertson TE, Greer MR.. Carbon monoxide poisoning. Crit Care Clin. 2021;37(3):657–672. [DOI] [PubMed] [Google Scholar]
- 140. Jain IH, Zazzeron L, Goli R, et al. Hypoxia as a therapy for mitochondrial disease. Science. 2016;352(6281):54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ferrari M, Jain IH, Goldberger O, et al. Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc Natl Acad Sci U S A. 2017;114(21):E4241–E4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Grange RMH, Sharma R, Shah H, et al. Hypoxia ameliorates brain hyperoxia and NAD+ deficiency in a murine model of Leigh syndrome. Mol Genet Metab. 2021;133(1):83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Klopstock T, Yu-Wai-Man P, Dimitriadis K, et al. A randomized placebo-controlled trial of idebenone in Leber's hereditary optic neuropathy. Brain. 2011;134(Pt 9):2677–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Amer J, Atlas D, Fibach E.. N-acetylcysteine amide (AD4) attenuates oxidative stress in 7beta-thalassemia blood cells. Biochim Biophys Acta. 2008;1780(2):249–255. [DOI] [PubMed] [Google Scholar]
- 145. Atlas D. Emerging therapeutic opportunities of novel thiol-amides, NAC-amide (AD4/NACA) and thioredoxin mimetics (TXM-Peptides) for neurodegenerative-related disorders. Free Rad Biol Med. 2021;176:120–141. [DOI] [PubMed] [Google Scholar]
- 146. Beyrath J, Pellegrini M, Renkema H, et al. KH176 safeguards mitochondrial diseased cells from redox stress-induced cell death by interacting with the thioredoxin system/peroxiredoxin enzyme machinery. Sci Rep. 2018;8(1):6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. De Haas R, Das D, Garanto A, et al. Therapeutic effects of the mitochondrial ROS-redox modulator KH176 in a mammalian model of Leigh Disease. Sci Rep. 2017;7(1):11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Frambach SJCM, van de Wal MAE, van den Broek PHH, et al. Effects of clofibrate and KH176 on life span and motor function in mitochondrial complex I-deficient mice. Biochim Biophys Acta. 2020;1866(6):165727. [DOI] [PubMed] [Google Scholar]
- 149. Iannetti EF, Smeitink JAM, Willems PHGM, Beyrath J, Koopman WJH.. Rescue from galactose-induced death of Leigh Syndrome patient cells by pyruvate and NAD +. Cell Death Dis. 2018;9(11):1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Lee CF, Caudal A, Abell L, Nagana Gowda GA, Tian R.. Targeting NAD+ metabolism as interventions for mitochondrial disease. Sci Rep. 2019;9(1):3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Pieper AA, Xie S, Capota E, et al. Discovery of a proneurogenic, neuroprotective chemical. Cell. 2010;142(1):39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Kleiner S, Nguyen-Tran V, Baré O, Huang X, Spiegelman B, Wu Z.. PPARδ agonism activates fatty acid oxidation via PGC-1α but does not increase mitochondrial gene expression and function. J Biol Chem. 2009;284(28):18624–18633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. D'Errico I, Salvatore L, Murzilli S, et al. Peroxisome proliferator-activated receptor-gamma coactivator 1-alpha (PGC1α) is a metabolic regulator of intestinal epithelial cell fate. Proc Natl Acad Sci U S A. 2011;108(16):6603–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Schirris TJJ, Rossell S, de Haas R, et al. Stimulation of cholesterol biosynthesis in mitochondrial complex I-deficiency lowers reductive stress and improves motor function and survival in mice. Biochim Biophys Acta. 2021;1867(4):166062. [DOI] [PubMed] [Google Scholar]
- 155. Moullan N, Mouchiroud L, Wang X, et al. Tetracyclines disturb mitochondrial function across eukaryotic models: A call for caution in biomedical research. Cell Rep. 2015;10(10):1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Perry EA, Bennett CF, Luo C, et al. Tetracyclines promote survival and fitness in mitochondrial disease models. Nat Metab. 2021;3(1):33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Chang JY, Sehgal SN, Bansbach CC.. FK506 and rapamycin: Novel pharmacological probes of the immune response. Trends Pharmacol Sci. 1991;12(6):218–223. [DOI] [PubMed] [Google Scholar]
- 158. Fruman DA, Wood MA, Gjertson CK, Katz HR, Burakoff SJ, Bierer BE.. FK506 binding protein 12 mediates sensitivity to both FK506 and rapamycin in murine mast cells. Eur J Immunol. 1995;25(2):563–571. [DOI] [PubMed] [Google Scholar]
- 159. Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20(7):1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Laplante M, Sabatini DM.. mTOR signaling at a glance. J Cell Sci. 2009;122(Pt 20):3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Taylor AL, Watson CJ, Bradley JA.. Immunosuppressive agents in solid organ transplantation: Mechanisms of action and therapeutic efficacy. Crit Rev Oncol Hematol. 2005;56(1):23–46. [DOI] [PubMed] [Google Scholar]
- 162. Johnson SC, Yanos ME, Bitto A, et al. Dose-dependent effects of mTOR inhibition on weight and mitochondrial disease in mice. Front Genet. 2015;6:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Felici R, Buonvicino D, Muzzi M, et al. Post onset, oral rapamycin treatment delays development of mitochondrial encephalopathy only at supramaximal doses. Neuropharmacology. 2017;117:74–84. [DOI] [PubMed] [Google Scholar]
- 164. Felici R, Cavone L, Lapucci A, Guasti D, Bani D, Chiarugi A.. PARP inhibition delays progression of mitochondrial encephalopathy in mice. Neurotherapeutics. 2014;11(3):651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ethier C, Tardif M, Arul L, Poirier GG.. PARP-1 modulation of mTOR signaling in response to a DNA alkylating agent. PLoS One. 2012;7(10):e47978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Martin-Perez M, Grillo AS, Ito TK, et al. PKC downregulation upon rapamycin treatment attenuates mitochondrial disease. Nat Metab. 2020;2(12):1472–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Li J, Kim SG, Blenis J.. Rapamycin: One drug, many effects. Cell Metab. 2014;19(3):373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Sage-Schwaede A, Engelstad K, Salazar R, et al. Exploring mTOR inhibition as treatment for mitochondrial disease. Ann Clin Transl Neurol. 2019;6(9):1877–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Yu AK, Datta S, McMackin MZ, Cortopassi GA.. Rescue of cell death and inflammation of a mouse model of complex I-mediated vision loss by repurposed drug molecules. Hum Mol Genet. 2017;26(24):4929–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Triner L, Vulliemoz Y, Schwartz I, Nahas GG.. Cyclic phosphodiesterase activity and the action of papaverine. Biochem Biophys Res Commun. 1970;40(1):64–69. [DOI] [PubMed] [Google Scholar]
- 171. Zampieri LX, Silva-Almeida C, Rondeau JD, Sonveaux P.. Mitochondrial transfer in cancer: A comprehensive review. Int J Mol Sci. 2021;22(6):3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Melcher M, Danhauser K, Seibt A, et al. Modulation of oxidative phosphorylation and redox homeostasis in mitochondrial NDUFS4 deficiency via mesenchymal stem cells. Stem Cell Res Ther. 2017;8(1):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Jiang D, Xiong G, Feng H, et al. Donation of mitochondria by iPSC-derived mesenchymal stem cells protects retinal ganglion cells against mitochondrial complex I defect-induced degeneration. Theranostics. 2019;9(8):2395–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Doss MZ, Sachinidis A.. Current challenges of iPSC-based disease modeling and therapeutic implications. Cells. 2019;8(5):403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Reynaud-Dulaurier R, Benegiamo G, Marrocco E, et al. Gene replacement therapy provides benefit in an adult mouse model of Leigh syndrome. Brain. 2020;143(6):1686–1696. [DOI] [PubMed] [Google Scholar]
- 176. Silva-Pinheiro P, Cerutti R, Luna-Sanchez M, Zeviani M, Viscomi C.. A single intravenous injection of AAV-PHP.B-hNDUFS4 ameliorates the phenotype of Ndufs4-/- mice. Mol Ther Methods Clin Dev. 2020;17:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Di Meo I, Marchet S, Lamperti C, Zeviani M, Viscomi C.. AAV9-based gene therapy partially ameliorates the clinical phenotype of a mouse model of Leigh syndrome. Gene Ther. 2017;24(10):661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Van der Westhuizen FH, van den Heuvel LP, Smeets R, et al. Human mitochondrial complex I deficiency: Investigating transcriptional responses by microarray. Neuropediatrics. 2003;34(1):14–22. [DOI] [PubMed] [Google Scholar]
- 179. Civiletto G, Varanita T, Cerutti R, et al. Opa1 overexpression ameliorates the phenotype of two mitochondrial disease mouse models. Cell Metab. 2015;21(6):845–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Varanita T, Soriano ME, Romanello V, et al. The OPA1-dependent mitochondrial cristae remodeling pathway controls atrophic, apoptotic, and ischemic tissue damage. Cell Metab. 2015;21(6):834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ito TK, Lu C, Khan J, et al. Hepatic S6K1 partially regulates lifespan of mice with mitochondrial complex I deficiency. Front Genet. 2017;8:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Sanz A, Soikkeli M, Portero-Otín M, et al. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc Natl Acad Sci U S A. 2010;107(20):9105–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Catania A, Iuso A, Bouchereau J, et al. Arabidopsis thaliana alternative dehydrogenases: A potential therapy for mitochondrial complex I deficiency? Perspectives and pitfalls. Orphanet J Rare Dis. 2019;14(1):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. El-Desouky S, Taalab YM, El-Gamal M, Mohamed W, Salama M.. Animal model for Leigh syndrome. Methods Mol Biol. 2019;2011:451–464. [DOI] [PubMed] [Google Scholar]
- 185. Finsterer J. Early-age Ndufs4 knockout mice are an inappropriate animal model of Leigh syndrome. Radiol Phys Technol. 2019;12(2):230–231. [DOI] [PubMed] [Google Scholar]
- 186. Goldstein JM, Wagers AJ.. What's in a (sub)strain? Stem Cell Rep. 2018;11(2):303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Enríquez JA. Mind your mouse strain. Nature Metab. 2019;1(1):5–7. [DOI] [PubMed] [Google Scholar]
- 188. Weissig V. Drug development for the therapy of mitochondrial diseases. Tr Mol Med. 2020;26(1):40–54. [DOI] [PubMed] [Google Scholar]
- 189. Khayat D, Kurtz TL, Stacpoole PW.. The changing landscape of clinical trials for mitochondrial diseases: 2011 to present. Mitochondrion. 2020;50:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Tinker RJ, Lim AZ, Stefanetti RJ, McFarland R.. Current and emerging clinical treatment of mitochondrial disease. Mol Diag Ther. 2021;25(2):181–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Fiedorczuk K, Letts JA, Degliesposti G, Kaszuba K, Skehel M, Sazanov LA.. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538(7625):406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Zhu J, Vinothkumar KR, Hirst J.. Structure of mammalian respiratory complex I. Nature. 2016;536(7616):354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Bénit P, Steffann J, Lebon S, et al. Genotyping microsatellite DNA markers at putative disease loci in inbred/multiplex families with respiratory chain complex I deficiency allows rapid identification of a novel nonsense mutation (IVS1nt -1) in the NDUFS4 gene in Leigh syndrome. Hum Genet. 2003;112(5-6):563–566. [DOI] [PubMed] [Google Scholar]
- 194. González-Quintana A, Trujillo-Tiebas MJ, Fernández-Perrone AL, et al. Uniparental isodisomy as a cause of mitochondrial complex I respiratory chain disorder due to a novel splicing NDUFS4 mutation. Mol Genet Metab. 2020;131(3):341–348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.