Abstract
Lysimachia mauritiana Lam. (Primulaceae) is known as a medicinal plant with anti-tumor and anti-viral effects. In this study, we reported the first complete chloroplast genome sequence of L. mauritiana. The total length of the complete chloroplast genome was 152,691 bp, comprising a small-single copy region (SSC) of 17,928 bp, a large-single copy region of 83,811 bp, and a pair of inverted repeat regions of 25,476 bp. It contained 114 genes comprising 80 protein-coding genes, four rRNA, and 30 tRNA genes. The phylogenetic analysis showed that a chloroplast genome of L. mauritiana was a sister to a monophyletic clade, including L. christinae, L. congestiflora, L. hemsleyana. Our study will help to provide basic information for further studies on phylogenetics and population genetics.
Keywords: Lysimachia mauritiana, Primulaceae, chloroplast genome, phylogenetic analysis
Lysimachia mauritiana (Lamarck, 1792) is a medicinal plant belonging to the genus Lysimachia (Primulaceae). The plant extract of this species has known as a good source with anti-tumor and anti-viral effects (Yasukawa and Takido 1987; Bae and Song 2017). The plant is a biennial herb distributing coastlines in East Asia, the Philippines, Micronesia, Polynesia, and the Indian Ocean islands (Iwatsuki et al. 1993; Hu and Kelso 1996; Kono et al. 2013). Until recently, the chloroplast genome of this species has not been reported. In this study, we determined the complete chloroplast genome of L. mauritiana and investigated its phylogenetic position in the family Primulaceae.
Fresh leaves of L. mauritiana was collected from Byeonsan-myeon, Buan-gun, Jeollabuk-do in Korea (N 35°39′22", E 126°29′41") and the voucher specimen (voucher number KRIB0088806, Jin-Hyub Paik, jpaik@kribb.re.kr) was deposited in International Biological Material Research Center (IBMRC), Korea Research Institute of Bioscience & Biotechnology (KRIBB). L. mauritiana is not an endangered or protected species, and we did not collected from private or protected area that required permission. Ethics approval for this study was obtained from the Institutional Bioethics Committee of KRIBB.
The total genomic DNA of this plant was extracted using modified CTAB methods (Healey et al. 2014) and sequenced by Illumina HiseqXten platform (San Diego, CA). The raw data were filtered using Trimmomatic v.0.36 (Bolger et al. 2014), and the filtered data were assembled using the chloroplast genome of L. coreana (NC_026197.1) as the reference with Geneious Prime (v.2021.1.1; Kearse et al. 2012). Finally, the chloroplast genome was annotated using Geneious Prime (v.2021.1.1; Kearse et al. 2012) and GeSeq (Tillich et al. 2017) based on the chloroplast genome of L. coreana (NC_026197.1), and the complete chloroplast genome of L. mauritiana was submitted to GenBank (accession number: OK491633).
The complete chloroplast genome of L. mauritiana determined in this study has 152,691 bp in length, which contains a large-single copy region (LSC) of 83,811 bp, a small-single copy region (SSC) of 17,928 bp, and a pair of inverted repeat regions (IRs) of 25,476 bp. The chloroplast genome includes a total of 114 genes comprising 80 protein-coding genes, four rRNA, and 30 tRNA genes.
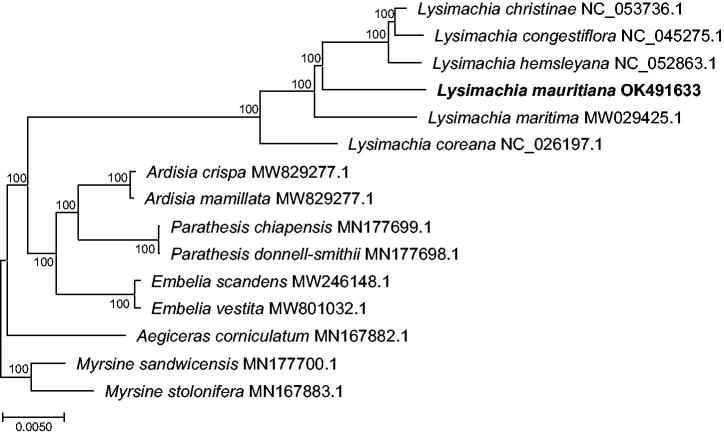
The phylogenetic analysis was performed with 14 accessions of Primulaceae deposited on Genbank, including a chloroplast genome of L. mauritiana determined in this study and an outgroup taxa from the genus Myrsine (MN167883.1 and MN177700.1). The sequences of these chloroplast genomes were aligned using MAFFT alignment v.7 (Katoh and Standley 2013; http://mafft.cbrc.jp/alignment/server/index.html). We selected TVM + F + I + G4 as the best model using ModelFinder module (Kalyaanamoorthy et al. 2017) in IQ-tree v.1.6.3 (Nguyen et al. 2015), and the maximum likelihood phylogenetic tree was reconstructed using in IQ-tree v.1.6.3 (Nguyen et al., 2015) with 1000 bootstrap replicates.
A Maximum-likelihood tree (Figure 1) shows that each genus is a highly supported monophyletic group (bootstrap support value = 100%). A chloroplast genome of L. mauritiana is a sister to a monophyletic clade including L. christinae, L. congestiflora, and L. hemsleyana with a high bootstrap value (bootstrap support value = 100%). Our study will provide basic information to understand the evolution and diversification of L. mauritiana.
Figure 1.
A Maximum-likelihood tree using 15 chloroplast genomes from Primulaceae, including a chloroplast genome of Lysimachia mauritiana determined in this study and an outgroup taxon (Myrsine stolonifera). The numbers near the nodes indicate bootstrap values.
Author contributions
Conceptualization: YL, SC, JP; Resources: NY, JK, SC, JP; Performed the experiments: YL, JK; Formal analysis: YL, NY, JK; Writing: YL; All authors reviewed the manuscript.
Funding Statement
This work was supported by the KRIBB Initiative Program [KGM4582221] of the Republic of Korea.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. OK491633. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA791132, SRR17284494, and SAMN24255752 respectively.
References
- Bae S, Song YJ.. 2017. Inhibition of varicella‑zoster virus replication by an ethanol extract of Lysimachia mauritiana. Mol Med Rep. 15(6):3847–3851. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healey A, Furtado A, Cooper T, Henry RJ.. 2014. Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods. 10(1):21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CM, Kelso S.. 1996. Primulaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 15. Myrsinaceae through Loganiaceae. Beijing (China)/St. Louis (MI): Science Press/Missouri Botanical Garden Press; p. 39–189. [Google Scholar]
- Iwatsuki K, Yamazaki T, Boufford DE, Ohba H.. 1993. Flora of Japan. Vol IIIa. Tokyo: Kodansha Ltd. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono Y, Hoshi Y, Setoguchi H, Yokota M, Oginuma K.. 2013. Widespread cytotypic variation, cytogeography and dynamic analysis of Lysimachia mauritiana (Primulaceae) on Takarajima Island in the Ryukyu Archipelago of Japan. Chromosom Bot. 8(3):47–52. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa K, Takido M.. 1987. A flavonol glycoside from Lysimachia mauritiana. Phytochemistry. 26(4):1224–1226. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. OK491633. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA791132, SRR17284494, and SAMN24255752 respectively.