Abstract
Sphingosine-1-phosphate (S1P) regulates immune cell trafficking, angiogenesis, and vascular function via its five receptors. Inherited mutations in S1P receptor 2 (S1PR2) occur in individuals with hearing loss, and acquired mutations in S1PR2 and Gα13 occur in a malignant lymphoma. Here, we present the cryo–electron microscopy structure of S1P-bound S1PR2 coupled to the heterotrimeric G13. Interaction between S1PR2 intracellular loop 2 (ICL2) and transmembrane helix 4 confines ICL2 to engage the α5 helix of Gα13. Transforming growth factor–α shedding assays and cell migration assays support the key roles of the residues in S1PR2-Gα13 complex assembly. The structure illuminates the mechanism of receptor disruption by disease-associated mutations. Unexpectedly, we showed that FTY720-P, an agonist of the other four S1PRs, can trigger G13 activation via S1PR2. S1PR2F274I variant can increase the activity of G13 considerably with FTY720-P and S1P, thus revealing a basis for S1PR drug selectivity.
The study reveals S1PR2-G13 interaction details and their physiological roles and how S1PR2 responds to immune modulator FTY720.
INTRODUCTION
The diversity of lipids ensures the structural rigidity and flexibility of cellular membranes. Lipids and their metabolites also serve as extracellular ligands for more than 50 G protein (heterotrimeric guanine nucleotide–binding protein)–coupled receptors (GPCRs), enabling the transduction of signals from the cell surface into the intracellular space (1–3). Sphingosine-1-phosphate (S1P), a signaling sphingolipid, functions as a regulator of the human immune and vascular systems through binding G protein–coupled S1P receptors (S1PR1 to S1PR5) (fig. S1) (4–7). S1P can act to increase or decrease vascular permeability, cell growth, and cell migration depending on which receptor is engaged, and it plays a crucial role in promoting lymphocyte egress from lymphoid organs by engaging S1PR1. Several S1PRs modulators—including Fingolimod (FTY720), Siponimod, and Ozanimod—have been approved for treating the autoimmune disease multiple sclerosis (8–10).
Intriguingly, the five S1PRs share high sequence homology but have been shown to bind different G proteins (11, 12). G proteins are classified into four families according to their α subunit: Gi, Gs, Gq/11, and G12/13. S1PR1 and S1PR5 couple preferentially to Gi/o, while S1PR2 and S1PR3 can couple to Gi/o, Gq/11, and G12/13 (11–13). In the case of S1PR2, in vivo studies in lymphocytes have shown a strict dependence on Gα13 (14, 15), and endothelial S1PR2 also appears to signal dominantly via Gα12 or Gα13 (16). Inherited missense mutations in S1PR2 have been identified in patients with hearing impairment, and mice lacking S1PR2 are deaf due to functional defects in auditory and vestibular systems (17, 18). Acquired mutations in GNA13 (encoding Gα13) and S1PR2 were found in germinal center B cell–like diffuse large B cell lymphoma (GCB-DLBCL) by deep sequencing studies (15, 19–21). Deficiency in S1PR2 or Gα13 caused a loss of germinal center B cell confinement and was sufficient to cause lymphomagenesis (14, 15), demonstrating the important role of Gα13 in inhibiting the growth and dissemination of germinal center B cells (15).
Previous structural studies on S1PRs have provided important insights into receptor-ligand interactions and G protein coupling. The crystal structure of S1PR1 bound to the sphingosine-mimic antagonist ML056 presented the inactive state of S1PRs (22), and comparison to the crystal structure of S1P-bound S1PR3 revealed transmembrane-domain residues that may regulate G protein selectivity (23). Recently, the structures of S1PR1, S1PR3, and S1PR5 coupled to Gi in the presence of different agonists provided a molecular basis for S1PR-mediated Gi activation (24, 25). However, the molecular details of how S1PRs recruit heterotrimeric G13 protein remain unknown. Notably, G13 plays an important role in regulating cell migration and actin cytoskeletal remodeling (26), and there is no available structure of any GPCR coupled to G12/13 to date. Because Gα12 and Gα13 share high sequence homology and, sometimes, function redundantly in signal transmission (26), structural studies on the S1PR2-G13 complex will provide molecular insights into the G12/13 activation mechanism that may apply to other GPCRs, as well as the molecular basis for G12/13 protein coupling.
RESULTS
Structure of S1PR2-G13 complex
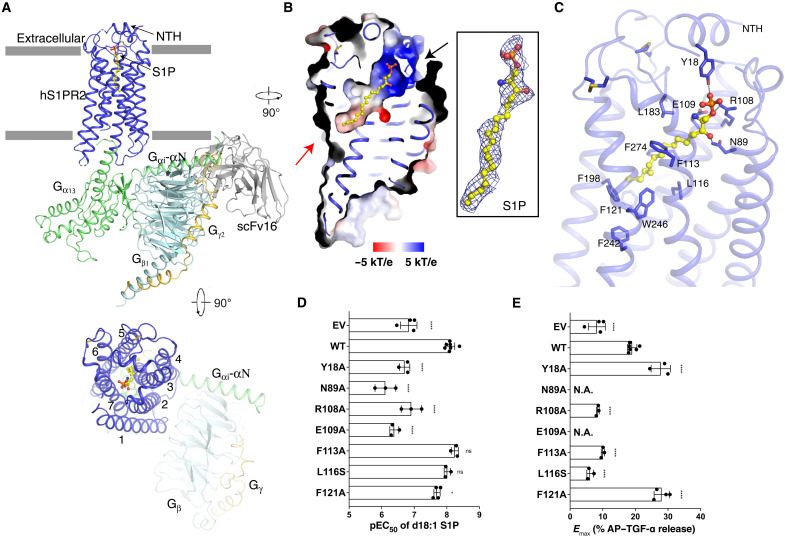
We purified human S1PR2 in the presence of S1P and incubated with Gα13β1γ2 heterotrimer. However, the complex was unstable in solution during purification. We then substituted the αN helix of wild-type Gα13 with the αN helix of Gαi (referred here as “G13iN”) according to previous studies (27, 28). To stabilize the complex, we added scFv16, which binds to G13iN-αN and Gβ (29). The purified complex was concentrated for cryo–electron microscopy (cryo-EM) studies (fig. S2A). We obtained 13 million particles from ~33,000 cryo-EM images. The particles exhibit severe dissociation upon grid preparation, resulting in only 5% of the particles in complex for final three-dimensional (3D) reconstruction (fig. S2B and table S1). The overall structure was determined at 3.2 Å resolution by 640,483 particles. The structure revealed well-defined features for the canonical seven transmembrane helices (7-TMs) of S1PR2, the G13iN Ras-like domain, the Gβ and Gγ subunits, and scFv16 (Fig. 1A and figs. S2, C and D, and S3). The density of the S1P ligand was observed in the cavity created by the transmembrane region (Fig. 1B).
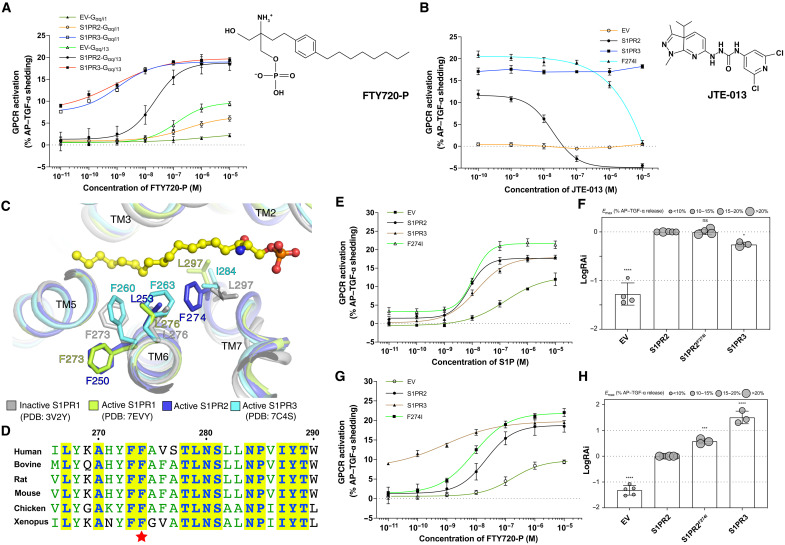
Fig. 1. Overall structure of S1PR2-G13 complex and the S1P binding site.
(A) Ribbon representation of the structure viewed from the side of the membrane (top) and the extracellular space (bottom). NTH, N-terminal helix. (B) Electrostatic surface representation of S1PR2 and the cryo-EM map of d18:1 S1P ligand (yellow sticks) at 5σ level. The putative S1P entrances are indicated by black arrow (the outer leaflet) and red arrow (the lipid bilayer). (C) Interaction details of S1P with residues in the binding site indicated. (D and E) Comparison of pEC50 values and Emax of each variant. The representative dose-response curves from same-day experiment and membrane expression of S1PRs are shown in fig. S4A. The not applicable (N.A.) value means curve not reaching Emax. Data are means ± SD (n = 3 to 7 independent experiments). *P < 0.05 and ****P < 0.0001, mutant versus wild-type (WT) values according to one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison test. ns, not significant. AP–TGF-α, alkaline phosphatase–fused TGF-α. EV, empty vector.
The TMs generate an amphiphilic pocket with a positively charged head in the outer leaflet and hydrophobic region in the middle of the receptor to accommodate the S1P ligand (Fig. 1B). Several polar residues are responsible for binding the sphingosine head and phosphate group, and several hydrophobic residues contact the alkyl tail of S1P (Fig. 1C). The N-terminal helix (NTH) functions as a lid to retain the ligand in the cavity (Fig. 1C). We used the established transforming growth factor–α (TGF-α) shedding assay that reports on heterotrimeric G protein signaling to validate S1P-mediated S1PR2 coupling (11, 30). The human S1PR2 (wild-type or point mutant variants), Gαq/13 (a chimeric Gq with Gα13-α5 helix), and alkaline phosphatase–fused TGF-α (AP–TGF-α) were cotransfected into human embryonic kidney (HEK) 293 ΔGq/11/12/13 cells to measure G protein signal. We obtained a potency [median effective concentration (EC50)] and an amplitude of ligand-induced response (Emax) from the sigmoidal concentration-response curve of each variant. The results show that S1P can activate S1PR2 and trigger Gαq/13 protein signaling (fig. S4A). Mutations on the key residues that accommodate the sphingosine head (Y18NTH, N892.60, R1083.28, and E1093.29, superscripts refer to Ballesteros-Weinstein number) interfere with the EC50 and/or Emax of S1P activity considerably (Fig. 1, C to E, and fig. S4A), while mutations on the residues that contact the alkyl tail (F1133.33 and L1163.36) attenuate the Emax of S1PR2 (Fig. 1E and fig. S4A).
Structural comparisons reveal activation mechanism
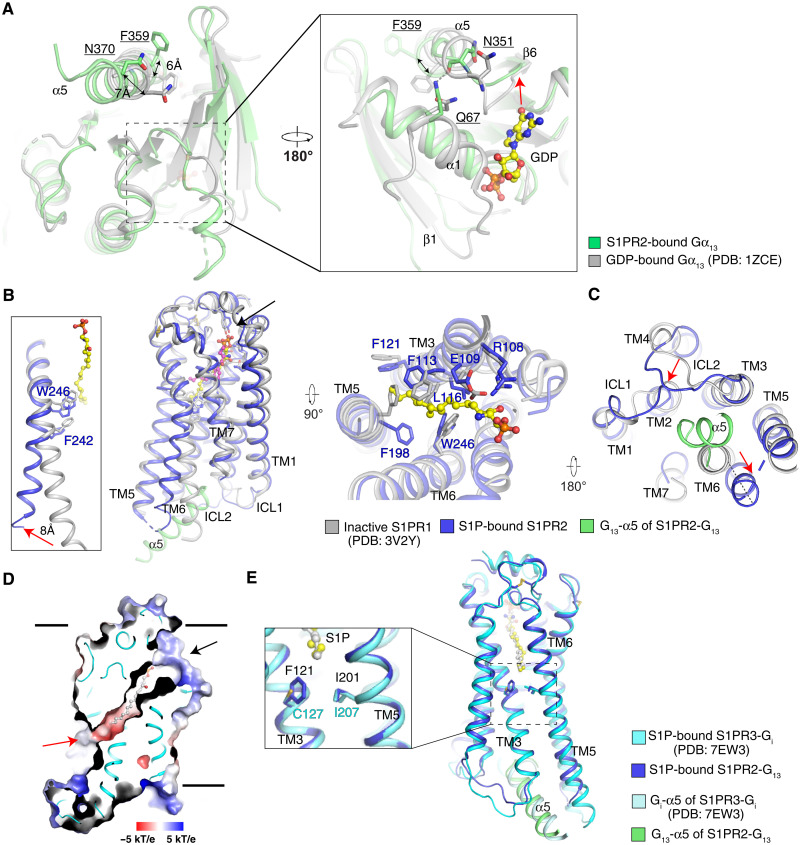
Comparison of our G13 structure with a guanosine diphosphate (GDP)–bound G13 structure provides molecular insights into the structural rearrangements associated with S1PR2-induced G13 activation (28). The Gα13-α5 undergoes a notable conformational change with a translation of more than 6 Å and a ~45° rotation upon nucleotide release (Fig. 2A). The conformational change of α5 accompanies the concerted movements of the β6-α5 loop and the α1 helix (Fig. 2A). This reorganization is consistent with the findings in other GPCR-engaged Gi, Gs, and Gq proteins (27, 31, 32). The shift of the α5 induces a movement of the β6-α5 loop of more than 5 Å. This movement enhances the hydrogen bond between Q67 and the backbone carbonyl of N351 and relieves the interactions between the β6-α5 loop and the guanine base of GDP to further trigger GDP dissociation from G13 (Fig. 2A). The nucleotide-free G13 protein may, in turn, allosterically enhance S1P-binding affinity as suggested by studies on the β2-adrenergic receptor (33).
Fig. 2. Structural comparisons reveal conformational changes associated with activation and differences in ligand access.
(A) Structural comparison with GDP-bound Gα13 with S1PR2-coupled Gα13. The movement of β6-α5 loop is indicated by a red arrow. (B) Superimposition of TMs of inactive S1PR1- and S1P-bound S1PR2 viewed from the side of the membrane (left) and extracellular space (right). The antagonist ML056 of S1PR1 is shown as magenta sticks, and the S1P is shown as yellow sticks. The residues that engage the ligands are shown in sticks. The movements of TM6 in the G13-bound S1PR2 compared with the inactive S1PR1 are shown with red arrow. The putative S1P entrance in the outer leaflet is indicated by black arrow. (C) Superimposition as in (B) but viewed from the side of the cytosol. The movements of TMs are indicated by red arrows. (D) Electrostatic surface representation of S1PR3. The putative S1P entrances are indicated by black arrow (the outer leaflet) and red arrow (the lipid bilayer). The S1P is shown as gray sticks. (E) Superimposition of TMs of S1P-bound S1PR2 and S1PR3 viewed from the side of the membrane. The comparison of the putative S1P entrance is zoomed.
Structural comparison of our active S1PR2 structure with the inactive S1PR1 state (22) suggests that S1P induces the conformational changes of several hydrophobic residues, including F1133.33, F1213.41, F1985.47, and W2466.48 in the center of S1PR2 (Fig. 2B and fig. S5A). The shift of W2466.48 further forces F2426.44 toward the edge of the receptor to trigger the notable movement of TM6 that is characteristic of receptor activation (Fig. 2B) (34). This conformational change creates enough room in the cytosolic side of S1PR2 to accommodate the Gα13-α5 (Fig. 2C). The structural analysis reveals that the alkyl tail of S1P stimulates the movement of the PIF motif (fig. S5B), and Y2095.58 and Y2887.53 of the NPXXY motif have a hydrophilic interaction in the active S1PR2 (fig. S5, C and D), resembling the activation of other class A GPCRs (34, 35).
The smoothened (SMO) receptor structures showed a cholesterol entrance in the middle of its transmembrane domain (36, 37), suggesting that lipid substrates may access the receptor from deep within the lipid bilayer. In contrast, the structures of S1PR1 and S1PR3 showed that a cavity opens to the outer leaflet, suggesting an entrance of S1P between TM1 and TM7 close to the membrane surface (Fig. 2, B and D) (22, 23, 38). The S1P-bound S1PR3 also revealed an opening to the middle of the lipid bilayer, implying that S1P may access the receptor through a deep bilayer entrance (Fig. 2D) (23); however, the analogous cavity is closed in S1PR2 (Fig. 1B). F1213.41 of S1PR2, which provides hydrophobic contacts with the alkyl tail of S1P, may impede the access of S1P via the 7-TMs deep within the bilayer (Fig. 2E). The TGF-α shedding assay shows that the F121A mutation leads to a slight decrease of the EC50 of S1P but considerably enhances the Emax, which implies that reducing the steric hindrance of the putative lipid entrance via the 7-TMs deep in the bilayer may accelerate the access of S1P into the receptor (Fig. 1, D and F, and fig. S4A). S1P may enter via the membrane, perhaps after delivery by one of the two best defined carriers (e.g., apolipoprotein M+ high-density lipoprotein and albumin (39)].
The receptor and Gα13 interface
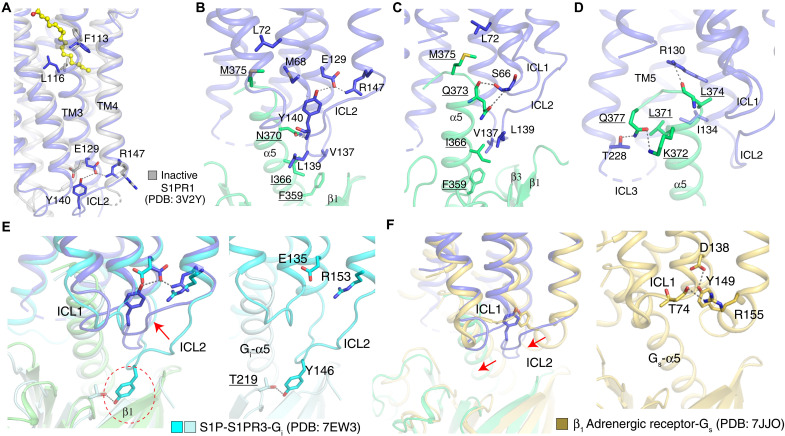
In the active conformation, E1293.49 of the DRY motif forms an intramolecular bond with Y140ICL2 and a salt bridge with R1474.41 (Fig. 3, A and B). These interactions likely stabilize Gα13-N370 through a hydrophilic bond between N370 and the main chain amino group of Y140ICL2, keeping V137ICL2 and L139ICL2 buried in the hydrophobic groove generated by the β1, β3, β2-β3 loop, and α5 of Gα13 (Fig. 3, B and C). Our previous work showed that mutant R147C loses germinal center growth suppressive activity (15), thus providing physiological evidence to support our observation. This finding is also supported by a previous study that demonstrated that the intracellular loop 2 (ICL2) plays a key role in the G protein selectivity (40). The Q373 of Gα13-α5 binds S662.37 through hydrophilic interactions to confine the ICL1 (Fig. 3C). Moreover, L371, L374, M375, and Q377 contact M682.39, L722.43, R1303.50, I1343.54, V2165.65, T2286.30, and L2326.34, thus building the interaction network between Gα13 and S1PR2 (Fig. 3D).
Fig. 3. Interaction details between S1PR2 and Gα13.
(A) The structural comparison of the inactive S1PR1 (gray) with S1PR2. (B to D) The interactions between S1PR2 (blue) and Gα13 (green). The hydrophilic interaction is indicated by the dashed line, and the residues of Gα13 are underlined. (E) Comparison of ICL2 in S1PR2-G13 complex and S1PR3-Gi complex. The putative steric clash between Y145 of S1PR3 and β1 of G13 is indicated by a red dashed circle. The interaction detail between S1PR3 and Gi is shown on the right. (F) Comparison of ICL2 in S1PR2-G13 complex and β1-Gs complex. The interaction detail between β1 and Gs is shown on the right. The interactions between the residues are indicated by dashed lines. The structural differences between the receptors are indicated by arrows.
The intramolecular interaction between the E1293.49 of the DRY motif and Y140ICL2 in S1PR2 facilitates the coupling of the receptor to Gα13-α5; however, the conserved tyrosine in the S1PR3-Gi complex did not bind to E1353.49 of the DRY motif (Fig. 3E) (25). Instead, it interacts with the T219 of Gαi to aid the interactions between the receptor and Gαi. While superimposing the S1PR3-Gi complex onto the S1PR2-G13 complex, the Y146ICL2 of S1PR3 would be predicted to clash with the β1 of G13 (Fig. 3E). However, our study along with prevous studies (11–13) showed that S1PR3 is also able to engage G13 (fig. S6A). Because Y140ICL2 is only conserved between S1PR2 and S1PR3 (fig. S1), it is tempting to speculate that the conserved tyrosine in S1PR3 may adopt a similar conformation as that in the S1PR2-G13 complex when S1PR3 couples to G13.
Although the overall arrangement of S1PR2-G13 is similar to other GPCR–G protein complexes (including M1-Gq, μR-Gi, and β1-Gs complexes) (27, 31, 41), several distinct features are apparent (figs. S7 and S8). Superimposing the structure of S1PR2 with M1 in its Gq-bound complex shows a different orientation of the G protein relative to the receptor (fig. S7A). The C terminus of M1 inserts into the cavity between Gα and Gβ, but the C-terminal tail of S1PR2 is not resolved in the cryo-EM map (fig. S7B). The ICL2 of M1, μR-Gi, and β1 forms a helix, while ICL2 of S1PR2 resembles a loop and faces inward toward the α5 (figs. S7C and S8, A and B). The extensive contacts between the ICL2 of S1PR2 and Gα13-α5 induce a ~10° rotation of α5 toward TM5 and TM6, which is more than in the other complexes (figs. S7C and S8, A and B). In S1PR2-G13 and M1-Gq complexes, the Gln residue in α1 has a hydrophilic interaction with the β6-α5 loop to assist the disassociation of the nucleotide and retain the conformation of α5 for recruiting the receptor (fig. S7D). The structures of the active aminergic receptors reveal that the conserved Tyr in ICL2 also forms a hydrogen bond with Asp3.49 of the DRY motif (Fig. 3F) (32, 41). The ICL2 of the β1 receptor does not contact with Gs-α5; in contrast, the flexibility of S1PR2-ICL2 allows Y140ICL2 to form the hydrogen bond with N370 of Gα13-α5 (Fig. 3B). Furthermore, unlike R1474.41 of S1PR2, which forms a hydrophilic network with E1293.49 and Y140ICL2 (Fig. 3B), R1554.41 of the β1 receptor binds the backbone carbonyl of T74ICL1, thus keeping ICL2 away from Gs-α5 (Fig. 3F).
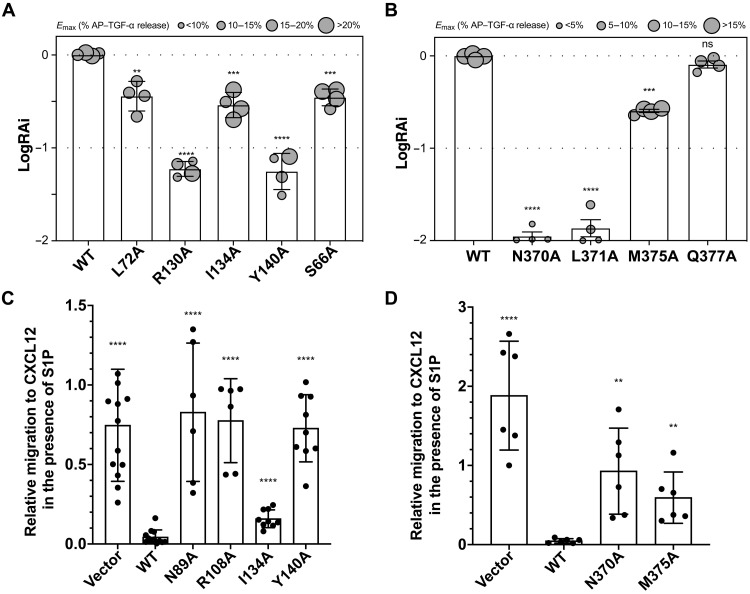
To validate this interface, we performed a substantial mutagenesis study using TGF-α shedding assays. The Emax/EC50 value of each variant was normalized by the value of the wild-type protein to generate a relative intrinsic activity (RAi). Then, LogRAi is ranged from −2 to 0 and used as an indicator of G protein coupling (11). Either S1PR2 (S662.37, L722.43, R1303.50, I1343.54, and Y140ICL2) or Gα13 (N370, L371, and M375) variant presents less intrinsic activity than the wild-type complex (Fig. 4, A and B, and fig. S4, B and C). Although the LogRAi of Gα13Q377A did not decrease, the mutation affects the Emax and EC50, weakening the activation of G protein signaling via S1PR2 (Fig. 4B and fig. S4C).
Fig. 4. The physiological importance of S1PR2 and Gα13 interface.
(A and B) Relative intrinsic activity (RAi) of each varient of S1PR2 (A) and Gα13 (B). G protein coupling is scored as logarithmic values of RAi, which is defined as a relative Emax/EC50 value normalized by the wild-type value. Symbol size is proportional to Emax. Each LogRAi plot denotes a single measurement from a dose-response experiment. The representative dose-response curves from same-day experiment and protein expression are shown in fig. S4 (B and C). Data are means ± SD (n = 4 independent experiments). (C) Cell migration inhibition assay with S1PR2 wild-type, S1PR2 variant, or control (vector) expressing WEHI-231 cells. Membrane expression of S1PRs is shown in fig. S9. Data are means ± SD (pooled from three to four independent experiments). (D) Cell migration inhibition assay with Gα13 wild-type, Gα13 variant, or control (vector) expressing M12 cells. Data are means ± SD (pooled from three independent experiments). **P < 0.01, ***P < 0.001, and ****P < 0.0001; mutant versus wild-type values according to one-way ANOVA with Dunnett’s multiple comparison test.
To further validate the physiological role of the S1PR2-G13 complex, we tested four S1PR2 mutations (S1PR2N89A, S1PR2R108A, S1PR2I134A, and S1PR2Y140A) for their influence on S1P-mediated migration inhibition of WEHI-231 B lymphoma cells (Fig. 4C and fig. S9). Three of the mutants caused an almost complete loss of S1P-mediated migration inhibition, while the S1PR2I134A mutant had only a mild effect, findings in close accord with the TGF-α shedding assay (Fig. 4A). We also tested two G13 mutations (G13-N370A and G13-M375A) in the migration inhibition assay by introducing them into the M12 B lymphoma line that expresses S1PR2 but lacks endogenous G13 function. Both mutations led to a severe defect in the ability of S1P to mediate migration inhibition (Fig. 4D). These data further support the conclusions from the TGF-α shedding assays regarding the physiological relevance of the structural observations. Thus, our findings reveal the molecular basis of how S1PR2 couples to G13 protein.
Unique drug selectivity of S1PR2
Fingolimod/FTY720 is used for multiple sclerosis treatment. FTY720 can be converted to FTY720 (S)-phosphate (FTY720-P) by sphingosine kinase 2 in vivo (42), which then stimulates the internalization of S1PR1 on lymphocytes, inhibiting their migration to S1P and egress from lymphoid tissues (43, 44). Previous studies showed that FTY720-P also binds and activates S1PR3, S1PR4, and S1PR5 (44, 45). In particular, FTY720-P acts as a biased agonist to trigger Gi and G12/13 signaling (Fig. 5A) but not Gq signaling via S1PR3, causing off-target effects (23, 46). Early studies indicated that FTY720-P did not engage S1PR2 (44, 45), and this is still a widely held view (5, 6), although cell signaling studies have suggested that S1PR2 can be activated by FTY720-P for coupling G12/13 (47) and β-arrestin (48). Thus, there is still some uncertainty in the possible effect of FTY720-P on S1PR2.
Fig. 5. FTY720-P triggers G13 activation via S1PR2.
(A) Dose-response curves of S1PR2 and S1PR3 for the TGFα shedding assay using FTY720-P. The chemical structure of FTY720-P is shown. Data are means ± SD (n = 3). The membrane expression of S1PRs is shown in figs. S4A and S6C. EV, empty vector. (B) JTE-013 reduces the FTY720-P–induced activation of S1PR2 but not S1PR3. The chemical structure of JTE-013 is shown. Data are means ± SD (n = 3). (C) Structural comparisons among S1PRs imply the unique roles of residue F274 of S1PR2. The residues are shown in sticks. (D) The sequence alignment of S1PR2-TM7 from different species. The conserved residues are highlighted. The residue F274 is indicated by a star. (E and G) Mutagenesis analysis of S1PR2F274I for S1P (E) and FTY720-P (G). The representative dose-response curves from same-day experiment are shown. Data are means ± SD (n = 3). (F and H) RAi of each varient for S1P (F) and FTY720-P (H). Each LogRAi plot denotes a single measurement from a dose-response experiment. Data are means ± SD (n = 3 to 5 independent experiments). *P < 0.05, ***P < 0.001, and ****P < 0.0001; mutant versus wild-type values according to one-way ANOVA with Dunnett’s multiple comparison test.
Our TGF-α shedding assays show that FTY720-P, like S1P, can trigger strong Gq/13 activation via S1PR2 (Fig. 5A and fig. S6A). The shedding assays also reveal that S1P and FTY720-P can weakly activate Gq/i via S1PR2, although only near the micromolar range (Fig. 5A and fig. S6, A and B). In contrast, FTY720-P and S1P can trigger the activation of both Gq/i and Gq/13 via S1PR3 equally (fig. S6, A to D). Notably, the FTY720-P–mediated S1PR2 activation can be abolished by the highly S1PR2 selective inhibitor JTE-013 (49). However, JTE-013 cannot interfere with the FTY720-P–mediated S1PR3 activation, demonstrating that FTY720-P can specifically trigger S1PR2 activation (Fig. 5B). These findings extend the evidence for selective coupling of S1PR2 to Gα13 versus Gαi while also showing that S1PR3 signals via Gα13 and Gαi with similar efficiency (fig. S6D). Dual signaling via Gαi and Gα12/13 has been linked to enhancements in directed cell migration (50) and might contribute to the efficacy of S1PR3 as a promigratory receptor (51, 52). By contrast, the near selective Gα13 coupling of S1PR2 may be critical for its ability to antagonize S1PR1-mediated Gαi responses and thereby promote lymphocyte retention in tissues (53).
To uncover the molecular details of FTY720-P selectivity in S1PRs, we compared the structures of S1PRs. F2747.39 of S1PR2 contacts L2536.55 to force F2506.52 to face the edge of S1PR2 and thereby adopt a unique conformation compared to that of inactive S1PR1 and active S1PR3 (Fig. 5C). Notably, F274 is not conserved in the other S1PRs (fig. S1), but it is widely conserved in S1PR2 across different species, implying that this residue may play a unique role in the ligand recognition of S1PR2 (Fig. 5D). The shedding assay shows that the S1PR2F274I variant presents an increased efficacy for S1P-mediated Gq/13 activation (Fig. 5, E and F) and stronger potency and efficacy for FTY720-P–mediated Gq/13 activation (Fig. 5, G and H), because it may reduce the steric hindrance of F2506.52 with L2536.55 to facilitate ligand engagement and the structural rearrangement of TM6. F274I mutation leads to a reduced response of S1PR2 to its inhibitor JTE-013 (Fig. 5B).
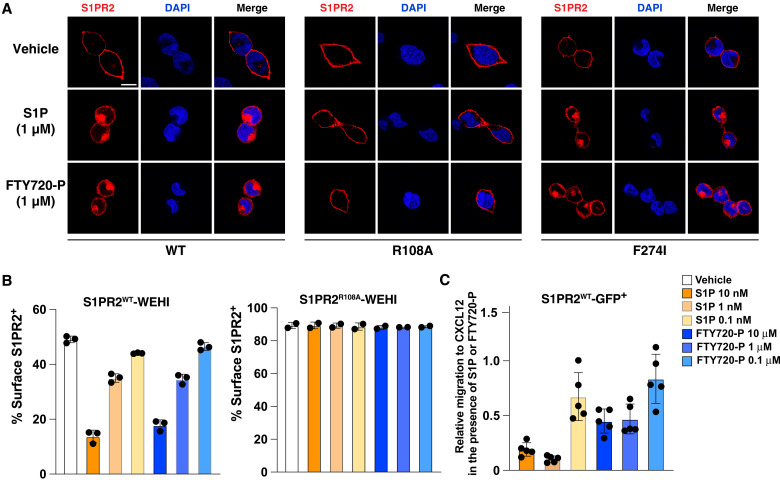
As further tests of the ability of FTY720-P to engage S1PR2, we conducted S1PR2 internalization and migration inhibition experiments. Immunofluorescence microscopy of S1PR2-expressing HEK293 cells and flow cytometric anlaysis of S1PR2-expressing WEHI-231 cells showed that FTY720-P caused internalization of S1PR2 with similar efficacy to S1P when used at 1 μM (Fig. 6, A and B). Moreover, FTY720-P was active in inhibiting migration of S1PR2-expressing WEHI-231 cells to CXC motif chemokine ligand 12 (CXCL12) (Fig. 6C). Together, these findings confirm that FTY720-P can act as an agonist of S1PR2, and they show the essential role and unique feature of S1PR2-F2747.39 in FTY720-P selectivity.
Fig. 6. The S1PR2 internalization assays.
(A) The immunofluorescence staining of HEK293 cells that express S1PR2 or its variants after vehicle or 1 μM ligand treatment. Scale bar, 10 μm. DAPI, 4′,6-diamidino-2-phenylindole. (B) Internalization of S1PR2WT or S1PR2R108A in WEHI-231 cells in response to the indicated amounts of S1P or FTY720-P treatment. Graphs show percent cells with detectable S1PR2 surface staining determined using flow cytometry. (C) Cell migration inhibition assay with S1PR2WT WEHI-231 cells in response to S1P or FTY720-P at the indicated concentrations. Data are means ± SD (pooled from three independent experiments).
Disease-associated S1PR2 variants
The physiological importance of the S1PR2-G13 complex in lymphoma has been demonstrated (15, 54, 55). We have mapped a series of GCB-DLBCL–associated mutations (15, 19–21, 56–59) into the complex structure (fig. S10). Mutations in S1PR2 in either the S1P binding site (including Y18, N89, R108, E109, and L116) or the Gα13 interface (including L72, I134,Y140, and R147) cause a substantial reduction in G protein signaling (Figs. 1, D and E, and 4, A and C) (15). Moreover, mutations of Gα13 on the receptor interface (including N370, L371, M375, and Q377) also attenuate G protein signaling (Fig. 4, B and D). These findings support that GCB-DLBCL–associated mutations, which may not be predicted to disrupt protein expression, are likely to reduce or abolish ligand binding or the S1P-mediated activation of G13, thereby contributing to tumorigenesis.
Three S1PR2 variants have been associated with hearing loss in humans: R108P, R108Q, and Y140C (17, 18). The R108A variant interferes with ligand binding directly, reducing the S1P ligand potency (Fig. 1D). Given that R108 interacts with the phosphate group of S1P, mutation to Pro and Gln would weaken the S1P engagement. A change of Y140 to Cys would have a similar adverse impact on bonding to E129 of the DRY motif as a change to Ala, thereby abolishing interaction with Gα13-α5 (Fig. 4, A and C). A mouse S1PR2T289R variant (also T289 in human) was identified as the stonedeaf mutation (60). Residue T289 is located in the C terminus of TM7 (fig. S10, A and C) and close to the NPXXY motif. This variant likely interferes with the cytosolic side of the receptor that recruits the Gα13-α5 or affects conformational rearrangement of the NPXXY motif for receptor activation. The genetic findings along with our structural analysis suggest the S1PR2-G13 signaling complex may contribute to the biogenesis of the auditory system.
DISCUSSION
Here, we report the cryo-EM structure of S1P-bound S1PR2 coupled to heterotrimeric G13 protein. The structure adds to the existing evidence that lipid ligands of GPCRs access their binding pockets through an opening within the lipid bilayer. These findings suggest that S1P must partition into the plasma membrane of the S1PR-expressing cell before engaging the receptor. A key distinction from other GPCR structures is the presence of an intramolecular hydrogen bond between the DRY motif and a tyrosine in ICL2 that allows insertion of Gα13-α5. Notably, this tyrosine is conserved in S1PR3, the only other S1PR also able to couple G13. This structure explains how multiple S1PR2 and Gα13 variants arising in GCB-DLBCL lead to receptor dysfunction and thus deregulated migration and growth of germinal center B cells. It also illuminates the disruptive properties of hearing loss-associated S1PR2 variants.
The therapeutic agent FTY720 has been used to treat autoimmune disease in more than 300,000 people (10). Low activity of the drug on S1PR2 is thought to be crucial in avoiding unwanted effects on the immune, vascular, and auditory systems. At the recommended daily dose for treatment of multiple sclerosis of 0.5 mg, the steady-state blood FTY720-P concentration is estimated to reach ~1 ng/ml (~2.6 nM) (61). The drug target, S1PR1, is functionally antagonized by low-nanomolar concentrations of FTY720-P (44), while 100- to 1000-fold greater amounts are needed to have strong functional effects on S1PR2 [this study and (48)]. Thus, at the approved dosing, the drug is unlikely to interfere with S1PR2 function. In rodent studies, however, it is common to use a dose of 1 mg/kg. A linear extrapolation from 0.5-mg daily dosing in a 70-kg human would suggest that an FTY720-P concentration of 360 nM may be reached in rodents treated with FTY720 (1 mg/kg) daily. The possible influence of FY720-P agonism of S1PR2 needs to be considered when interpreting in vivo effects of FTY720 treatment of rodents in this dose range. The suggestion in early studies that FTY720-P does not engage S1PR2 may have been a consequence of the low signal-to-noise ratio in binding assays, making it challenging to detect weak interactions. By revealing conserved features of the S1PR2 binding pocket that interfere with FTY720-P signaling, this work enables improved structure-guided drug design efforts for next-generation S1PR1 targeting therapeutics. S1PR2 signaling via Gα12/13 and Rho in endothelial cells reduces junctional integrity and promotes vascular permeability (16). In models of multiple sclerosis, acute lung injury, and endotoxemia, S1PR2 signaling contributes to endothelial leakiness and disease (16, 62–64). In myeloid cells, S1PR2 activation suppresses macrophage migration and promotes their retention in pathological lesions (65, 66). In toxin-induced liver and lung damage, S1PR2 signaling impedes tissue regeneration and increases fibrosis (67, 68). Development of new classes of S1PR2 antagonists as potential therapeutics to treat these conditions will be aided by this S1PR2-Gα13 structure.
MATERIALS AND METHODS
Protein expression, purification, and assembly of hS1PR2-G13-scFv16 complex
The full-length cDNA of wild-type human S1PR2 (UniProt: O95136) was cloned into pEG-BacMam vector with a C-terminal FLAG tag. The protein was expressed using baculovirus-mediated transduction of mammalian HEK293S GnTI− cells [American Type Culture Collection (ATCC)]. Baculovirus was generated in Sf9 cells (ATCC), and P2 virus was used to infect HEK293S GnTI− cells at 37°C. At 8 hours after infection, sodium butyrate at a final concentration of 10 mM was added to the culture. After further incubation for 60 hours at 30°C, cells were harvested and resuspended in buffer A [20 mM Hepes (pH 7.5) and 150 mM NaCl] supplemented with 1 mM phenylmethylsulfonyl fluoride, leupeptin (10 μg/ml), and 5 μM S1P and then homogenized by sonication. The lysate was cleared by centrifugation at 3000g for 5 min at 4°C. The resulting supernatant was supplemented with 1% L-MNG (lauryl maltose neopentyl glycol) /0.1% CHS (cholesteryl hemisuccinate) and incubated for 1 hour at 4°C. Insoluble components were removed by centrifugation at 40,000g, 4°C for 30 min, and the clarified lysate was then loaded onto a disposable gravity column (Bio-Rad) containing anti-FLAG M2 affinity agarose resin (Sigma-Aldrich). After washing two times with 10 column volumes (CVs) of buffer B [20 mM Hepes (pH 7.5), 150 mM NaCl, 0.01% L-MNG/0.001% CHS, and 5 μM S1P], the protein was eluted with 4 CVs of buffer B supplemented with 3× FLAG peptide (0.1 mg/ml; ApexBio). The eluted protein was further purified by size exclusion chromatography using a Superose 6 Increase 10/300 GL column (Cytiva) preequilibrated with buffer C [20 mM Hepes (pH 7.5), 150 mM NaCl, 0.001% L-MNG/0.0001% CHS, and 0.0025% GDN (glyco-diosgenin)] with 5 μM S1P. The peak fractions were collected for complex assembly.
For expression of chimeric G13iN, a DNA fragment encoding amino acid residues 1 to 31 of human Gαi1 was fused to the N terminus of human Gα13 (residues 47 to 377) and cloned into the expression vector pFastBac1 without any tag. The cDNAs of wild-type human β1γ2 subunits were cloned into pFastBac Dual vector with a 6×His tag inserted at the N terminus of the β1 subunit. The expression and purification of heterotrimeric G13iN protein were performed on the basis of a published method that was used for purifying heterotrimeric Gi (69). scFv16 was expressed and purified as previously described (31).
To assemble the S1PR2-G13-scFv16 complex, purified S1PR2 was mixed with the G13 heterotrimer at a 1:1.2 molar ratio. This mixture was incubated on ice for 1 hour, followed by the addition of apyrase to catalyze the hydrolysis of unbound GDP on ice for 1 hour. Then, scFv16 was added at a 1.5:1 molar ratio (scFv16: S1PR2) followed by 30-min incubation on ice. The mixture was diluted 10-fold by buffer C containing 5 μM S1P. To remove excess G13 and scFv16 proteins, the mixture was purified by anti-FLAG M2 antibody affinity chromatography in the presence of 5 μM S1P. The complex was eluted in buffer C supplemented with 5 μM S1P and 3×FLAG peptide (0.1 mg/ml). After concentration, the complex was further purified by size exclusion chromatography (Superose 6 Increase 10/300 GL) in buffer C with 5 μM S1P. Peak fractions were concentrated to ~10 to 12 mg/ml for cryo-EM studies.
Cryo-EM sample preparation and data acquisition
The freshly purified hS1PR2-G13-scFv16 complex was added to Quantifoil R1.2/1.3 400-mesh Au holey carbon grids (Quantifoil), blotted using a Vitrobot Mark IV (FEI), and frozen in liquid ethane. The grids were imaged in a 300-keV Titan Krios (FEI) with a Gatan K3 Summit direct electron detector. Data were collected in super-resolution mode at a pixel size of 0.421 Å with a dose rate of 23 electrons per physical pixel per second. Images were recorded for 1.8-s exposures in 60 subframes to give a total dose of 60 electrons/Å2.
Imaging processing and 3D reconstruction
A total of four datasets with 6696, 9876, 11,259, and 5349 micrographs, respectively, were collected. For each dataset, dark subtracted images were normalized by gain reference and binned twofold that resulted in the original pixel size of 0.842 Å. Drift correction was performed using the program MotionCor2 (70). The contrast transfer function (CTF) was estimated using CTFFIND4 (71). For the first dataset, about 2000 particles were manually picked and classified by 2D classification in RELION-3. The 2D class averages were used as the templates for autopicking in RELION-3 (72). After autopicking in RELION, the particles were extracted with a pixel size of 2.526 Å. The extracted particles were imported into cryoSPARC (73) for 2D classification followed by ab initio reconstruction with a class number of 5. The resulting ab initio maps and the cryo-EM map of SMO-Gi complex (EMD-22120) (37) were used as models in the following parallel heterogeneous refinements. The particles from heterogeneous refinements were combined, and duplicated particles were removed. Then, the resulting particles were transformed into RELION format, and one round of 3D classification was performed. The particles from the best class were recentered and reextracted with a pixel size of 0.842 Å and imported into cryoSPARC for further parallel heterogeneous refinements with various class number (K = 3, 4, 5, and 6). The resulting particles were combined, deduplicated, transformed into RELION format, and subjected to another round of 3D classification followed by 3D refinement and Bayesian polishing, which gave rise to a reconstruction at the resolution of 4.41 Å. For the other three datasets, a similar procedure was applied with the exception that 2D references were made by projection of 3D map generated from the first dataset. The resulting 123,752, 242,485, 335,498, and 150,558 particles from all four datasets were then combined and imported into cryoSPARC for further 2D classification and heterogeneous refinement with a class number of 2. A final number of 640,483 particles were selected from the better class and subjected to nonuniform refinement and local CTF refinement, which gave rise to a final reconstruction at the resolution of 3.19 Å using the 0.143 cutoff criteria. Local resolutions were estimated using cryoSPARC.
Model construction and refinement
The cryo-EM structure of the CB1-Gi [Protein Data Bank (PDB): 6N4B] (74) and the crystal structure of chimeric Gαi/13 (PDB: 1ZCB) (28) together with a homology model of S1PR2 were used as initial models and manually docked into cryo-EM density map with Chimera (75). The resulting model was subjected to iterative rounds of manual adjustment and rebuilding in Coot (76) and refinement using Phenix real-space refine (77). The model of the ligand S1P was manually built in Coot. MolProbity (78) was used to validate the geometries of the model. Structural figures were generated using PyMOL (www.pymol.org) and Chimera.
TGF-α shedding assay
The cDNAs encoding human S1PR2 and S1PR3 were cloned into the pCAGGS expression vector followed by a FLAG epitope tag at their C terminus. Point mutations of S1PR2 were introduced by two-step overlapping polymerase chain reaction. HEK293 ΔGq/11/12/13 cell line (79) was a gift from A. Inoue at the Tohoku University, Japan. The cells were maintained in Dulbecco’s modified Eagle’s medium-low glucose (DMEM-LG; Sigma-Aldrich) supplemented with heat-inactivated fetal calf serum and penicillin-streptomycin (100 U/ml and 100 μg/ml, Corning) at 37°C in a 5% CO2 incubator.
The TGF-α shedding assay was performed as described previously (11, 23). HEK293 ΔGq/11/12/13 cells were seeded in a 12-well plate at a density of 1 × 105 cells per well in 1 ml of DMEM-LG supplemented with 10% charcoal-stripped fetal bovine serum (Gibco) and penicillin-streptomycin and cultured at 37°C with 5% CO2 for 24 hours. Transient transfection was performed with a mixture of AP–TGF-α–encoding plasmid (250 ng per well, hereafter), S1PR2- or S1PR3-encoding plasmid (100 ng), and Gα-encoding plasmid using 1.2 μl of FuGene 6 transfection reagent (Promega). After 24 hours, the transfected cells were detached with 0.05% Trypsin-EDTA solution (Gibco) followed by adding complete DMEM to inactivate trypsin. The cells were palleted by centrifugation at 200g for 5 min and resuspended in 4 ml of Hanks’ balanced salt solution (HBSS) with calcium and magnesium (Gibco) containing 5 mM Hepes (pH 7.4). After incubation at room temperature for 10 min, cells were palleted by centrifugation and resuspended in 2.5 ml of Hepes-containing HBSS. The cell suspension was seeded into a 96-well culture plate with lid at a volume of 90 μl per well and incubated at 37°C in a 5% CO2 incubator for 30 min. The ligand S1P (Tocris) or FTY720 (S)-phosphate (FTY720-P, Echelon) was serially diluted in HBSS supplemented with 5 mM Hepes (pH 7.4) and 0.01% fatty acid–free bovine serum albumin (BSA; GoldBio) to prepare 10× stock solutions. The cells of each well were treated with 10 μl of ligand stock solution, where each concentration was tested in triplicate, and further incubated for 1 hour. Then, the plates were centrifuged at 400g for 5 min, and 80 μl of conditioned medium (CM) was transferred to a new 96-well culture plate. For each well of both cell plate and CM plate, 80 μl of AP reaction solution [120 mM tris-HCl (pH 9.5), 40 mM NaCl, 10 mM MgCl2, and 10 mM p-nitrophenyl phosphate (Thermo Fisher Scientific)] was added, and the absorbance at 405 nm (A) was measured using a microplate reader (Synergy Neo2, BioTek). After incubation at room temperature for 2 hours, the absorbance at 405 nm (A′) was measured again to get the AP activity (∆A = A′ − A). S1P- or FTY720-P–induced AP–TGF-α release was calculated as described previously (30). AP–TGF-α release signal over ligand concentration was fitted to a four-parameter logistic sigmoidal curve using Prism 9 software (GraphPad), from which EC50 and Emax values were obtained. For JTE-013 inhibition experiment, JTE-013 was serially diluted in HBSS with 5 mM Hepes (pH 7.4) and 0.01% fatty acid–free BSA in the presence or absence of 1 μM FTY720-P.
Migration inhibition transwell bioassays
S1PR2 and GNA13 wild-type and point mutant retroviral constructs were made by inserting the human open reading frame into the MSCV2.2 retroviral vector followed by an internal ribosome entry site and Thy1.1 or green fluorescent protein (GFP) as an expression marker. For S1PR2 constructs, an OX56 (rat CD43-derived) epitope was included at the N terminus to track surface expression levels of each receptor using the OX56 antibody (80). The retrovirus-encoding S1PR2 or GNA13 wild-type or point mutants were produced using the Platinum-E packaging cell line. Approximately 1 × 106 WEHI-231 or M12 cells were placed in a six-well plate along with the retroviral supernatant, and the cells were centrifuged at 1340g (2400 rpm) for 2 hours at room temperature. The viral supernatant was aspirated, and the cells were resuspended in growth medium and returned to culture. This spinfection was repeated with fresh retrovirus for a second time 24 hours later. Forty-eight hours after the second spinfection, the cells were analyzed on flow cytometry for the percentage of GFP- or Thy1.1-expressing cells. Cells were cultured to confluency in T75 flasks before use in transwell bioassays.
For transwell bioassays, transduced WEHI-231 or M12 cells were washed twice in prewarmed migration medium (RPMI 1640 containing 0.5% fatty acid–free BSA, 10 mM Hepes, and 50 IU of penicillin/streptomycin). The cells were resuspended in migration medium at 2.5 × 106 cells/ml and incubated for 20 min in a 37°C water bath. Recombinant human CXCL12 (PeproTech) was diluted to 100 ng/ml in migration medium. S1P (d18:1) (Avanti Polar Lipids) was diluted to 1 nM in CXCL12-containing migration medium. Alternatively, FTY720-P was diluted to 0.1 μM in an equivalent way. Six hundred microliters of each of these mixtures was added to a 24-well tissue culture plate. Transwell filters (6-mm insert, 5-μm pore size; Corning) were placed on top of each well, and 100 μl of transduced WEHI-231 or M12 cells (2.5 × 105 cells) was added to the transwell insert. The cells were allowed to migrate for 3 hours, after which the cells in the bottom well were counted by flow cytometry. To assess migration inhibition, the proportion of S1PR2-GFP+ or G13-Thy1.1+ cells that migrated to CXCL12 with S1P was divided by the proportion of S1PR2-GFP+ or G13-Thy1.1+ cells that migrated to CXCL12 alone.
Internalization assay in WEHI-231 cells
WEHI-231 cells expressing OX56-S1PR2-GFP were prepared as for the cell migration assays, and then, 40 μl of medium containing 2 × 105 cells was placed in each well of a round bottom 96-well plate. Appropriately diluted S1P or FTY720-P (160 μl) prepared in migration medium was added, and the plate was incubated at 37°C for 60 min. The fraction of GFP+ cells that were stained for OX56 was then determined by flow cytometry. Gates used for OX56 staining were set on the basis of GFP− cells.
Antibodies used for analyzing Gαq/13 expression level
Anti-Gαq mouse monoclonal antibody (Santa Cruz Biotechnology, catalog no. sc-136181) was used for detecting the expression level of Gαq/13 protein and its mutants. Anti–β-tubulin mouse monoclonal antibody (Cell Signaling Technology, catalog no. 86298S) was used for normalizing the amount of loaded sample. Anti-mouse immunoglobulin G (IgG) secondary antibody conjugated with horseradish peroxidase was from Cell Signaling Technology (catalog no. 7076S). Similar results were obtained in three biologically independent experiments.
Immunofluorescence staining
HEK293 ΔGq/11/12/13 cells were seeded on poly-d-lysine–coated coverslips in 24-well plates and transfected with plasmids of wild-type or mutant S1PR2 (or S1PR3) as used in the above TGF-α shedding assay. At 24 hours after transfection, the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, permeabilized with 100% methanol (prechilled at −20°C) for 5 min, blocked with 2% BSA in PBS for 1 hour, and labeled with anti-FLAG mouse antibody (2 μg/ml; Sigma-Aldrich, catalog no. F1804-50UG) for 3 hours at room temperature. Donkey anti-mouse IgG secondary antibody conjugated with Alexa Fluor 568 (Thermo Fisher Scientific, catalog no. A10037) was used at a concentration of 2 μg/ml in blocking buffer for 1 hour at room temperature for detection of FLAG-tagged S1PR2 or S1PR3. The coverslips were mounted onto glass slides with anti-fade reagent with 4′,6-diamidino-2-phenylindole (Thermo Fisher Scientific, catalog no. P36931). Fluorescence images were acquired using a Zeiss LSM 800 microscope system with ZEN imaging software (Zeiss). For internalization analysis, transfected HEK293 cells were serum-starved for 1 hour before S1P or FTY720-P treatment. After treatment for 1 hour, the cells were subjected to fixation, immunostaining, and imaging as described above. Similar results were obtained in three biologically independent experiments.
Acknowledgments
The data were collected at the UT Southwestern Medical Center Cryo-EM Facility (funded, in part, by the CPRIT Core Facility Support Award RP170644). We highly appreciate A. Inoue for sharing the materials of TGF-α shedding assay. We thank L. Beatty, L. Esparza, Y. Qin, and Y. Xu for technical support; F. Wolfreys for comments on the manuscript; and P. Schmiege for editing the manuscript.
Funding: This work was supported by NIH grants P01HL020948 and R01GM135343 (to X.L.) and R01AI045073 to J.G.C. J.G.C. is an investigator of Howard Hughes Medical Institute. X.L. is a Damon Runyon-Rachleff Innovator supported by the Damon Runyon Cancer Research Foundation (DRR-53S-19) and a Rita C. and William P. Clements Jr. Scholar in Biomedical Research at UT Southwestern Medical Center.
Author contributions: H.C. purified the protein for cryo-EM study, carried out cryo-EM work, and performed TGF-α shedding and internalization assays. K.C. performed cell migration and internalization assays. J.G.C. and X.L. conceived and supervised the project. All authors contributed to conceptualization of the research, analyzed the data, and contributed to manuscript preparation.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The 3D cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under the accession number EMD-25712. Atomic coordinates for the atomic model have been deposited in the Protein Data Bank under the accession number 7T6B. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Table S1
REFERENCES AND NOTES
- 1.Lu E., Cyster J. G., G-protein coupled receptors and ligands that organize humoral immune responses. Immunol. Rev. 289, 158–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizuno H., Kihara Y., Druggable lipid GPCRs: Past, present, and prospects. Adv. Exp. Med. Biol. 1274, 223–258 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Qi X., Li X., Mechanistic insights into the generation and transduction of hedgehog signaling. Trends Biochem. Sci. 45, 397–410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spiegel S., Milstien S., Sphingosine-1-phosphate: An enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4, 397–407 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Cartier A., Hla T., Sphingosine 1-phosphate: Lipid signaling in pathology and therapy. Science 366, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosen H., Stevens R. C., Hanson M., Roberts E., Oldstone M. B. A., Sphingosine-1-phosphate and its receptors: Structure, signaling, and influence. Annu. Rev. Biochem. 82, 637–662 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Proia R. L., Hla T., Emerging biology of sphingosine-1-phosphate: Its role in pathogenesis and therapy. J. Clin. Invest. 125, 1379–1387 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kappos L., Radue E. W., O’Connor P., Polman C., Hohlfeld R., Calabresi P., Selmaj K., Agoropoulou C., Leyk M., Zhang-Auberson L., Burtin P.; FREEDOMS Study Group , A placebo-controlled trial of oral fingolimod in relapsing multiple sclerosis. N. Engl. J. Med. 362, 387–401 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Brinkmann V., Billich A., Baumruker T., Heining P., Schmouder R., Francis G., Aradhye S., Burtin P., Fingolimod (FTY720): Discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Discov. 9, 883–897 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Chun J., Giovannoni G., Hunter S. F., Sphingosine 1-phosphate receptor modulator therapy for multiple sclerosis: Differential downstream receptor signalling and clinical profile effects. Drugs 81, 207–231 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue A., Raimondi F., Kadji F. M. N., Singh G., Kishi T., Uwamizu A., Ono Y., Shinjo Y., Ishida S., Arang N., Kawakami K., Gutkind J. S., Aoki J., Russell R. B., Illuminating G-protein-coupling selectivity of GPCRs. Cell 177, 1933–1947.e25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugimoto N., Takuwa N., Okamoto H., Sakurada S., Takuwa Y., Inhibitory and stimulatory regulation of Rac and cell motility by the G12/13-Rho and Gi pathways integrated downstream of a single G protein-coupled sphingosine-1-phosphate receptor isoform. Mol. Cell. Biol. 23, 1534–1545 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Windh R. T., Lee M.-J., Hla T., An S., Barr A. J., Manning D. R., Differential coupling of the sphingosine 1-phosphate receptors Edg-1, Edg-3, and H218/Edg-5 to the Gi, Gq, and G12 families of heterotrimeric G proteins. J. Biol. Chem. 274, 27351–27358 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Green J. A., Suzuki K., Cho B., Willison L. D., Palmer D., Allen C. D. C., Schmidt T. H., Xu Y., Proia R. L., Coughlin S. R., Cyster J. G., The sphingosine 1-phosphate receptor S1P2 maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat. Immunol. 12, 672–680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muppidi J. R., Schmitz R., Green J. A., Xiao W., Larsen A. B., Braun S. E., An J., Xu Y., Rosenwald A., Ott G., Gascoyne R. D., Rimsza L. M., Campo E., Jaffe E. S., Delabie J., Smeland E. B., Braziel R. M., Tubbs R. R., Cook J. R., Weisenburger D. D., Chan W. C., Vaidehi N., Staudt L. M., Cyster J. G., Loss of signalling via Gα13 in germinal centre B-cell-derived lymphoma. Nature 516, 254–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takuwa Y., Okamoto Y., Yoshioka K., Takuwa N., Sphingosine-1-phosphate signaling in physiology and diseases. Biofactors 38, 329–337 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Santos-Cortez R. L., Faridi R., Rehman A. U., Lee K., Ansar M., Wang X., Morell R. J., Isaacson R., Belyantseva I. A., Dai H., Acharya A., Qaiser T. A., Muhammad D., Ali R. A., Shams S., Hassan M. J., Shahzad S., Raza S. I., Bashir Z. E., Smith J. D., Nickerson D. A., Bamshad M. J.; University of Washington Center for Mendelian Genomics, Riazuddin S., Ahmad W., Friedman T. B., Leal S. M., Autosomal-recessive hearing impairment due to rare missense variants within S1PR2. Am. J. Hum. Genet. 98, 331–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofrichter M. A. H., Mojarad M., Doll J., Grimm C., Eslahi A., Hosseini N. S., Rajati M., Müller T., Dittrich M., Maroofian R., Haaf T., Vona B., The conserved p.Arg108 residue in S1PR2 (DFNB68) is fundamental for proper hearing: Evidence from a consanguineous Iranian family. BMC Med. Genet. 19, 81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin R. D., Mendez-Lago M., Mungall A. J., Goya R., Mungall K. L., Corbett R. D., Johnson N. A., Severson T. M., Chiu R., Field M., Jackman S., Krzywinski M., Scott D. W., Trinh D. L., Tamura-Wells J., Li S., Firme M. R., Rogic S., Griffith M., Chan S., Yakovenko O., Meyer I. M., Zhao E. Y., Smailus D., Moksa M., Chittaranjan S., Rimsza L., Brooks-Wilson A., Spinelli J. J., Ben-Neriah S., Meissner B., Woolcock B., Boyle M., McDonald H., Tam A., Zhao Y., Delaney A., Zeng T., Tse K., Butterfield Y., Birol I., Holt R., Schein J., Horsman D. E., Moore R., Jones S. J. M., Connors J. M., Hirst M., Gascoyne R. D., Marra M. A., Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohr J. G., Stojanov P., Lawrence M. S., Auclair D., Chapuy B., Sougnez C., Cruz-Gordillo P., Knoechel B., Asmann Y. W., Slager S. L., Novak A. J., Dogan A., Ansell S. M., Link B. K., Zou L., Gould J., Saksena G., Stransky N., Rangel-Escareno C., Fernandez-Lopez J. C., Hidalgo-Miranda A., Melendez-Zajgla J., Hernandez-Lemus E., Schwarz-Cruz y Celis A., Imaz-Rosshandler I., Ojesina A. I., Jung J., Pedamallu C. S., Lander E. S., Habermann T. M., Cerhan J. R., Shipp M. A., Getz G., Golub T. R., Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 3879–3884 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morin R. D., Mungall K., Pleasance E., Mungall A. J., Goya R., Huff R. D., Scott D. W., Ding J., Roth A., Chiu R., Corbett R. D., Chan F. C., Mendez-Lago M., Trinh D. L., Bolger-Munro M., Taylor G., Hadj Khodabakhshi A., Ben-Neriah S., Pon J., Meissner B., Woolcock B., Farnoud N., Rogic S., Lim E. L., Johnson N. A., Shah S., Jones S., Steidl C., Holt R., Birol I., Moore R., Connors J. M., Gascoyne R. D., Marra M. A., Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122, 1256–1265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanson M. A., Roth C. B., Jo E., Griffith M. T., Scott F. L., Reinhart G., Desale H., Clemons B., Cahalan S. M., Schuerer S. C., Sanna M. G., Han G. W., Kuhn P., Rosen H., Stevens R. C., Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda S., Shiimura Y., Asada H., Hirata K., Luo F., Nango E., Tanaka N., Toyomoto M., Inoue A., Aoki J., Iwata S., Hagiwara M., Endogenous agonist–bound S1PR3 structure reveals determinants of G protein–subtype bias. Sci. Adv. 7, eabf5325 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y., Jia G., Wu C., Wang W., Cheng L., Li Q., Li Z., Luo K., Yang S., Yan W., Su Z., Shao Z., Structures of signaling complexes of lipid receptors S1PR1 and S1PR5 reveal mechanisms of activation and drug recognition. Cell Res. 31, 1263–1274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao C., Cheng L., Wang W., Wang H., Luo Y., Feng Y., Wang X., Fu H., Cai Y., Yang S., Fu P., Yan W., Shao Z., Structural insights into sphingosine-1-phosphate recognition and ligand selectivity of S1PR3–Gi signaling complexes. Cell Res. 32, 218–221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Worzfeld T., Wettschureck N., Offermanns S., G12/G13-mediated signalling in mammalian physiology and disease. Trends Pharmacol. Sci. 29, 582–589 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Maeda S., Qu Q., Robertson M. J., Skiniotis G., Kobilka B. K., Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes. Science 364, 552–557 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreutz B., Yau D. M., Nance M. R., Tanabe S., Tesmer J. J. G., Kozasa T., A new approach to producing functional Gα subunits yields the activated and deactivated structures of Gα12/13 proteins. Biochemistry 45, 167–174 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maeda S., Koehl A., Matile H., Hu H., Hilger D., Schertler G. F. X., Manglik A., Skiniotis G., Dawson R. J. P., Kobilka B. K., Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun. 9, 3712 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue A., Ishiguro J., Kitamura H., Arima N., Okutani M., Shuto A., Higashiyama S., Ohwada T., Arai H., Makide K., Aoki J., TGFα shedding assay: An accurate and versatile method for detecting GPCR activation. Nat. Methods 9, 1021–1029 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Koehl A., Hu H., Maeda S., Zhang Y., Qu Q., Paggi J. M., Latorraca N. R., Hilger D., Dawson R., Matile H., Schertler G. F. X., Granier S., Weis W. I., Dror R. O., Manglik A., Skiniotis G., Kobilka B. K., Structure of the μ-opioid receptor-Gi protein complex. Nature 558, 547–552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rasmussen S. G., De Vree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T. A., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K., Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeVree B. T., Mahoney J. P., Vélez-Ruiz G. A., Rasmussen S. G. F., Kuszak A. J., Edwald E., Fung J. J., Manglik A., Masureel M., du Y., Matt R. A., Pardon E., Steyaert J., Kobilka B. K., Sunahara R. K., Allosteric coupling from G protein to the agonist-binding pocket in GPCRs. Nature 535, 182–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weis W. I., Kobilka B. K., The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem. 87, 897–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Q., Yang D., Wu M., Guo Y., Guo W., Zhong L., Cai X., Dai A., Jang W., Shakhnovich E. I., Liu Z.-J., Stevens R. C., Lambert N. A., Babu M. M., Wang M.-W., Zhao S., Common activation mechanism of class A GPCRs. eLife 8, e50279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deshpande I., Liang J., Hedeen D., Roberts K. J., Zhang Y., Ha B., Latorraca N. R., Faust B., Dror R. O., Beachy P. A., Myers B. R., Manglik A., Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature 571, 284–288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi X., Friedberg L., De Bose-Boyd R., Long T., Li X., Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling. Nat. Chem. Biol. 16, 1368–1375 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katritch V., Cherezov V., Stevens R. C., Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 53, 531–556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obinata H., Hla T., Sphingosine 1-phosphate and inflammation. Int. Immunol. 31, 617–625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flock T., Hauser A. S., Lund N., Gloriam D. E., Balaji S., Babu M. M., Selectivity determinants of GPCR-G-protein binding. Nature 545, 317–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su M., Zhu L., Zhang Y., Paknejad N., Dey R., Huang J., Lee M.-Y., Williams D., Jordan K. D., Eng E. T., Ernst O. P., Meyerson J. R., Hite R. K., Walz T., Liu W., Huang X.-Y., Structural basis of the activation of heterotrimeric Gs-protein by isoproterenol-bound β1-adrenergic receptor. Mol. Cell 80, 59–71.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paugh S. W., Payne S. G., Barbour S. E., Milstien S., Spiegel S., The immunosuppressant FTY720 is phosphorylated by sphingosine kinase type 2. FEBS Lett. 554, 189–193 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Chiba K., FTY720, a new class of immunomodulator, inhibits lymphocyte egress from secondary lymphoid tissues and thymus by agonistic activity at sphingosine 1-phosphate receptors. Pharmacol. Ther. 108, 308–319 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Mandala S., Hajdu R., Bergstrom J., Quackenbush E., Xie J., Milligan J., Thornton R., Shei G.-J., Card D., Keohane C.-A., Rosenbach M., Hale J., Lynch C. L., Rupprecht K., Parsons W., Rosen H., Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science 296, 346–349 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Brinkmann V., Davis M. D., Heise C. E., Albert R., Cottens S., Hof R., Bruns C., Prieschl E., Baumruker T., Hiestand P., Foster C. A., Zollinger M., Lynch K. R., The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 277, 21453–21457 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Sensken S. C., Stäubert C., Keul P., Levkau B., Schöneberg T., Gräler M. H., Selective activation of G alpha i mediated signalling of S1P3 by FTY720-phosphate. Cell. Signal. 20, 1125–1133 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Sobel K., Monnier L., Menyhart K., Bolinger M., Studer R., Nayler O., Gatfield J., FTY720 Phosphate activates sphingosine-1-phosphate receptor 2 and selectively couples to Gα12/13/Rho/ROCK to induce myofibroblast contraction. Mol. Pharmacol. 87, 916–927 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Wetter J. A., Revankar C., Hanson B. J., Utilization of the TangoTM β-Arrestin recruitment technology for cell-based EDG receptor assay development and interrogation. J. Biomol. Screen. 14, 1134–1141 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Osada M., Yatomi Y., Ohmori T., Ikeda H., Ozaki Y., Enhancement of sphingosine 1-phosphate-induced migration of vascular endothelial cells and smooth muscle cells by an EDG-5 antagonist. Biochem. Biophys. Res. Commun. 299, 483–487 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Xu J., Wang F., van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., Bourne H. R., Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell 114, 201–214 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Cinamon G., Matloubian M., Lesneski M. J., Xu Y., Low C., Lu T., Proia R. L., Cyster J. G., Sphingosine 1-phosphate receptor 1 promotes B cell localization in the splenic marginal zone. Nat. Immunol. 5, 713–720 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Keul P., Lucke S., von Wnuck Lipinski K., Bode C., Gräler M., Heusch G., Levkau B., Sphingosine-1-phosphate receptor 3 promotes recruitment of monocyte/macrophages in inflammation and atherosclerosis. Circ. Res. 108, 314–323 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Laidlaw B. J., Gray E. E., Zhang Y., Ramirez-Valle F., Cyster J. G., Sphingosine-1-phosphate receptor 2 restrains egress of γδ T cells from the skin. J. Exp. Med. 216, 1487–1496 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Basso K., Dalla-Favera R., Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 15, 172–184 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Healy J. A., Nugent A., Rempel R. E., Moffitt A. B., Davis N. S., Jiang X., Shingleton J. R., Zhang J., Love C., Datta J., McKinney M. E., Tzeng T. J., Wettschureck N., Offermanns S., Walzer K. A., Chi J. T., Rasheed S. A. K., Casey P. J., Lossos I. S., Dave S. S., GNA13 loss in germinal center B cells leads to impaired apoptosis and promotes lymphoma in vivo. Blood 127, 2723–2731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitz R., Wright G. W., Huang D. W., Johnson C. A., Phelan J. D., Wang J. Q., Roulland S., Kasbekar M., Young R. M., Shaffer A. L., Hodson D. J., Xiao W., Yu X., Yang Y., Zhao H., Xu W., Liu X., Zhou B., du W., Chan W. C., Jaffe E. S., Gascoyne R. D., Connors J. M., Campo E., Lopez-Guillermo A., Rosenwald A., Ott G., Delabie J., Rimsza L. M., Tay Kuang Wei K., Zelenetz A. D., Leonard J. P., Bartlett N. L., Tran B., Shetty J., Zhao Y., Soppet D. R., Pittaluga S., Wilson W. H., Staudt L. M., Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 378, 1396–1407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chapuy B., Stewart C., Dunford A. J., Kim J., Kamburov A., Redd R. A., Lawrence M. S., Roemer M. G. M., Li A. J., Ziepert M., Staiger A. M., Wala J. A., Ducar M. D., Leshchiner I., Rheinbay E., Taylor-Weiner A., Coughlin C. A., Hess J. M., Pedamallu C. S., Livitz D., Rosebrock D., Rosenberg M., Tracy A. A., Horn H., van Hummelen P., Feldman A. L., Link B. K., Novak A. J., Cerhan J. R., Habermann T. M., Siebert R., Rosenwald A., Thorner A. R., Meyerson M. L., Golub T. R., Beroukhim R., Wulf G. G., Ott G., Rodig S. J., Monti S., Neuberg D. S., Loeffler M., Pfreundschuh M., Trümper L., Getz G., Shipp M. A., Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 24, 679–690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sha C., Barrans S., Cucco F., Bentley M. A., Care M. A., Cummin T., Kennedy H., Thompson J. S., Uddin R., Worrillow L., Chalkley R., van Hoppe M., Ahmed S., Maishman T., Caddy J., Schuh A., Mamot C., Burton C., Tooze R., Davies A., du M. Q., Johnson P. W. M., Westhead D. R., Molecular high-grade B-cell lymphoma: Defining a poor-risk group that requires different approaches to therapy. J. Clin. Oncol. 37, 202–212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye X., Ren W., Liu D., Li X., Li W., Wang X., Meng F. L., Yeap L. S., Hou Y., Zhu S., Casellas R., Zhang H., Wu K., Pan-Hammarström Q., Genome-wide mutational signatures revealed distinct developmental paths for human B cell lymphomas. J. Exp. Med. 218, e20200573 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ingham N. J., Carlisle F., Pearson S., Lewis M. A., Buniello A., Chen J., Isaacson R. L., Pass J., White J. K., Dawson S. J., Steel K. P., S1PR2 variants associated with auditory function in humans and endocochlear potential decline in mouse. Sci. Rep. 6, 28964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Y. P., Mizugishi K., Bektas M., Sandhoff R., Proia R. L., Sphingosine kinase 1/S1P receptor signaling axis controls glial proliferation in mice with Sandhoff disease. Hum. Mol. Genet. 17, 2257–2264 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cruz-Orengo L., Daniels B. P., Dorsey D., Basak S. A., Grajales-Reyes J. G., McCandless E. E., Piccio L., Schmidt R. E., Cross A. H., Crosby S. D., Klein R. S., Enhanced sphingosine-1-phosphate receptor 2 expression underlies female CNS autoimmunity susceptibility. J. Clin. Invest. 124, 2571–2584 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skoura A., Sanchez T., Claffey K., Mandala S. M., Proia R. L., Hla T., Essential role of sphingosine 1-phosphate receptor 2 in pathological angiogenesis of the mouse retina. J. Clin. Invest. 117, 2506–2516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanchez T., Skoura A., Wu M. T., Casserly B., Harrington E. O., Hla T., Induction of vascular permeability by the sphingosine-1-phosphate receptor-2 (S1P2R) and its downstream effectors ROCK and PTEN. Arterioscler. Thromb. Vasc. Biol. 27, 1312–1318 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Skoura A., Michaud J., Im D. S., Thangada S., Xiong Y., Smith J. D., Hla T., Sphingosine-1-phosphate receptor-2 function in myeloid cells regulates vascular inflammation and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 31, 81–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Michaud J., Im D. S., Hla T., Inhibitory role of sphingosine 1-phosphate receptor 2 in macrophage recruitment during inflammation. J. Immunol. 184, 1475–1483 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ikeda H., Watanabe N., Ishii I., Shimosawa T., Kume Y., Tomiya T., Inoue Y., Nishikawa T., Ohtomo N., Tanoue Y., Iitsuka S., Fujita R., Omata M., Chun J., Yatomi Y., Sphingosine 1-phosphate regulates regeneration and fibrosis after liver injury via sphingosine 1-phosphate receptor 2. J. Lipid Res. 50, 556–564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao J., Okamoto Y., Asano Y., Ishimaru K., Aki S., Yoshioka K., Takuwa N., Wada T., Inagaki Y., Takahashi C., Nishiuchi T., Takuwa Y., Sphingosine-1-phosphate receptor-2 facilitates pulmonary fibrosis through potentiating IL-13 pathway in macrophages. PLOS ONE 13, e0197604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dror R. O., Mildorf T. J., Hilger D., Manglik A., Borhani D. W., Arlow D. H., Philippsen A., Villanueva N., Yang Z., Lerch M. T., Hubbell W. L., Kobilka B. K., Sunahara R. K., Shaw D. E., Structural basis for nucleotide exchange in heterotrimeric G proteins. Science 348, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng S. Q., Palovcak E., Armache J. P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Krishna Kumar K., Shalev-Benami M., Robertson M. J., Hu H., Banister S. D., Hollingsworth S. A., Latorraca N. R., Kato H. E., Hilger D., Maeda S., Weis W. I., Farrens D. L., Dror R. O., Malhotra S. V., Kobilka B. K., Skiniotis G., Structure of a signaling cannabinoid receptor 1-G protein complex. Cell 176, 448–458.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., Coy A. J. M., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Devost D., Sleno R., Pétrin D., Zhang A., Shinjo Y., Okde R., Aoki J., Inoue A., Hébert T. E., Conformational profiling of the AT1 angiotensin II receptor reflects biased agonism, G protein coupling, and cellular context. J. Biol. Chem. 292, 5443–5456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu E., Wolfreys F. D., Muppidi J. R., Xu Y., Cyster J. G., S-Geranylgeranyl-L-glutathione is a ligand for human B cell-confinement receptor P2RY8. Nature 567, 244–248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Table S1