Abstract
The age-related cognitive decline of normal aging is exacerbated in neurodegenerative diseases including Alzheimer’s disease (AD). However, it remains unclear whether age-related cognitive regulators in AD pathologies contribute to life span. Here, we show that C/EBPβ, an Aβ and inflammatory cytokine–activated transcription factor that promotes AD pathologies via activating asparagine endopeptidase (AEP), mediates longevity in a gene dose–dependent manner in neuronal C/EBPβ transgenic mice. C/EBPβ selectively triggers inhibitory GABAnergic neuronal degeneration by repressing FOXOs and up-regulating AEP, leading to aberrant neural excitation and cognitive dysfunction. Overexpression of CEBP-2 or LGMN-1 (AEP) in Caenorhabditis elegans neurons but not muscle stimulates neural excitation and shortens life span. CEBP-2 or LGMN-1 reduces daf-2 mutant–elongated life span and diminishes daf-16–induced longevity. C/EBPβ and AEP are lower in humans with extended longevity and inversely correlated with REST/FOXO1. These findings demonstrate a conserved mechanism of aging that couples pathological cognitive decline to life span by the neuronal C/EBPβ/AEP pathway.
Neuronal C/EBPβ/AEP triggers GABAnergic neuronal degeneration, leading to aberrant neural excitation and short life span.
INTRODUCTION
The nervous system plays a critical role in the regulation of aging. In the nematode Caenorhabditis elegans, deletion of specific sensory or neurosecretory neurons alters life span (1–3), and life-span extension from reduced insulin/insulin-like growth factor (IGF)–like signaling can be reversed by restoring function specifically in neurons (4). Ablation in the Drosophila brain of specific neurosecretory cells that produce insulin-like peptides extends life span (5). In C. elegans, neural excitation increases with age and inhibition of excitation increases longevity, for which the transcription factors REST (RE1 silencing transcription factor)/FOXO (Forkhead Box O) are indispensable, and they are up-regulated in humans with extended longevity and they repress excitation-related genes (6). Reduction of insulin/IGF-1 signaling is a strongly conserved mechanism of life-span extension in worms, flies, and mammals. An IGF signaling–regulated mechanism that protects from β-amyloid (Aβ) toxicity is conserved from worms to mammals. In worms, reduced insulin signaling ameliorates Aβ aggregation and cytotoxicity (7). Similarly, knockout of Irs2 or the IGF-1 receptor (IGF-1R) reduces cognitive impairment, neurodegeneration, and premature mortality in mouse models of Alzheimer’s disease (AD) (8, 9). IGF-1R and insulin receptor (IR) signalings are compromised in human AD neurons, suggestive of resistance to IGF-1R/IR signaling (10). FOXO transcription factors function downstream of insulin/IGF signaling and play a conserved role in longevity and cellular homeostasis and cognitive performance (11). They integrate signals emanating from nutrient deprivation and stress stimuli to coordinate programs of genes involved in cellular metabolism and resistance to oxidative stress for maintaining organelle and protein homeostasis (12). FOXOs are also implicated in protection against neurodegenerative conditions. In C. elegans, FOXO/DAF-16 (abnormal DAuer Formation-16) activity is required for reduced toxicity of Aβ (7) and SOD1 (superoxide dismutase 1) (13) aggregates in low insulin signaling conditions (daf-2 mutants), suggesting a protective role in neurodegenerative diseases.
CCAAT/enhancer binding protein β (C/EBPβ), an Aβ and inflammatory cytokine–activated transcription factor, primarily regulates various genes in memory formation, neuroprotection, axon growth, and neurogenesis in neurons, and it modulates proinflammatory programs in astrocytes and microglia. Lipopolysaccharide, brain injury, and learning paradigms elevate C/EBPβ expression in glial and hippocampal neurons (14). Apart from its role in neuropathies, C/EBPβ is a well-known transcription factor in regulating life span, nutrition metabolism, energy homeostasis, and adipose tissue differentiation (15, 16). Accumulating evidence suggests that C/EBPβ may play a role in health span and life-span determination. For instance, DNA methylation changes are associated with aging. Notably, access of the C/EBPβ transcription factor to cognate binding sites is regulated by DNA demethylation and that impaired demethylation of C/EBPβ sites can lead to premature aging (17). Moreover, transgenic (Tg) mice replacing Cebpa by Cebpb gene display an increased median life span of 20% and show reduced fat storage and increased mitochondrial biogenesis in white adipose tissue (18, 19), suggesting that C/EBPβ acts as a pro-longevity factor. Cebpb is a top hit in screens identifying candidate transcriptional regulators of aging-associated genes (20).
The expression of C/EBPβ progressively increases in the central nervous system (CNS) during aging (21) and various neurodegenerative diseases including AD (22). C/EBPβ expresses several isoforms. Noticeably, reduced expression of C/EBPβ-LIP (liver-enriched inhibitory protein) (the shortest isoform) extends health and life span in female but not in male mice (15). Moreover, C/EBPβ up-regulates AEP (asparagine endopeptidase; also known as legumain; LGMN, gene name) expression in the brain in an age-dependent manner. Age is the most important known risk factor for AD; we have shown that C/EBPβ/AEP signaling tempospatially mediates AD pathologies (23). Furthermore, C/EBPβ is age-dependently up-regulated in hippocampal neurons, implicated in neurogenesis and excitotoxicity (24, 25). GABAnergic (Gamma-Aminobutyric Acid-nergic) neurons are the most important negative regulator in excitatory and inhibitory balance in aging and AD (26, 27). To better understand the mechanism how neuronal C/EBPβ/AEP signaling contributes to life span, we generated neuronal specific Thy1-human C/EBPβ Tg mice and neuronal specific cebp-2 and lgmn-1 overexpression worms. Our study demonstrates a conserved mechanism of aging that couples pathological cognitive decline to reduced life span by the neuronal C/EBPβ/AEP pathway.
RESULTS
Neuronal C/EBPβ overexpression in Thy1-C/EBPβ Tg mice results in short life span and behavioral impairment
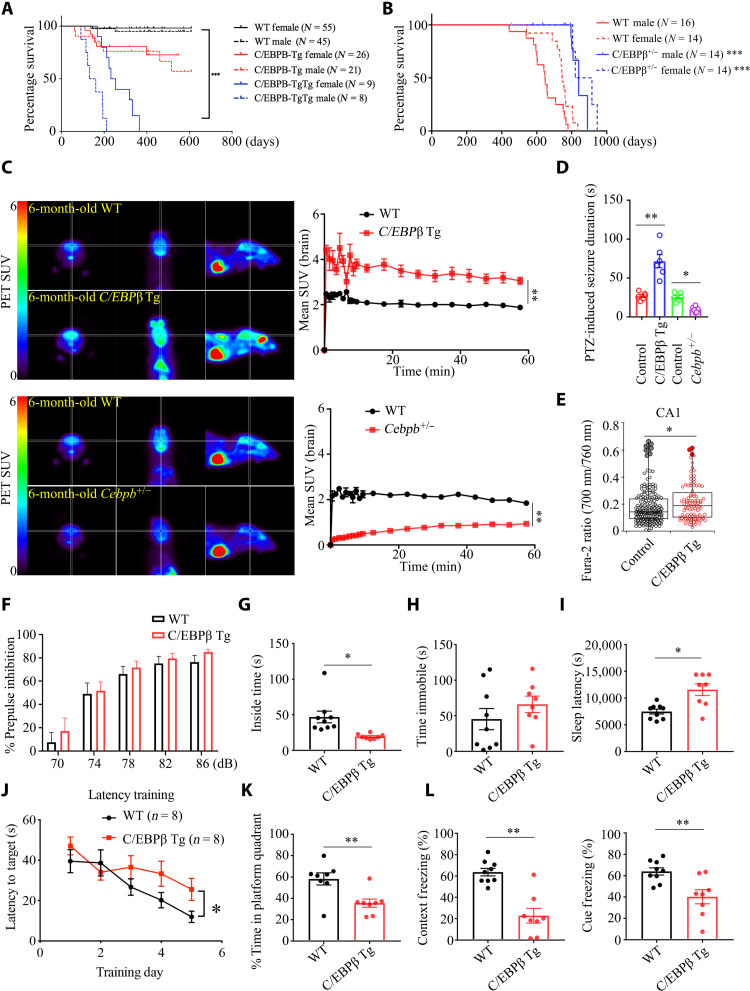
To interrogate the biological roles of neuronal C/EBPβ, we generated neuronal specific Thy1-human C/EBPβ Tg mice (fig. S1, A to C). Human C/EBPβ was gene dose–dependently expressed in the brains of Thy1-C/EBPβ single Tg or double Tg/Tg mice for the total C/EBPβ and the LAP (liver activator protein)/LIP ratio (fig. S1D). Neuronal C/EBPβ in the hippocampus was further confirmed by immunofluorescent (IF) costaining with NeuN (fig. S1E). Unexpectedly, these mice displayed a gene dose–dependent shortening of life span (Fig. 1A). In contrast, C/EBPβ+/− mice exhibited longer life span than their wild-type (WT) littermates (Fig. 1B). Because neural excitation increases with age and inhibition of excitation increases longevity (6), we wondered whether Thy1-C/EBPβ Tg and C/EBPβ+/− mice life span correlates with neural excitation status. Positron emission tomography–computed tomography (PET-CT) scanning of fluorodeoxyglucose (18F-FDG) uptake showed that Thy1-C/EBPβ Tg mice displayed much stronger signals than WT, whereas C/EBPβ+/− mice exhibited significantly weaker glucose uptake activities (Fig. 1C), indicating that neural excitation is highly increased in C/EBPβ Tg mice and pronouncedly reduced in C/EBPβ+/− mice. Abnormal neural excitation is frequently associated with seizures. An acute model of seizure induced by systemic injection of pentylenetetrazole (PTZ), a γ-aminobutyric acid type A (GABAA) receptor antagonist, showed that seizure was much longer in C/EBPβ Tg mice than in WT controls, and it was substantially diminished in C/EBPβ+/− mice (Fig. 1D). As another indication of elevated excitability of neurons, we conducted intracellular calcium ([Ca2+]i) imaging through two-photon microscopy in acute hippocampal brain slices loaded with a ratiometric fluorescence Ca2+ indicator, Fura-2. [Ca2+]i measured in the cell bodies of CA1 pyramidal cells from Thy1-C/EBPβ Tg mice was significantly higher than from WT mice (Fig. 1E). By contrast, Fura-2 excitation ratio in the dentate gyrus (DG) and CA3 remained comparable (fig. S1F). DG and CA3 might be less susceptible to degeneration of GABAnergic cells as those regions in the mouse hippocampus contain fewer interneurons than CA1 (28, 29). The brain plays a central role in the regulation of aging and longevity. Various behavioral tests demonstrated that Thy1-C/EBPβ mice displayed anxiety and sleep disorders; however, the prepulse inhibition (PPI) assay showed no schizophrenia phenotype, and forced swim test (FST) revealed no depression in C/EBPβ Tg mice (Fig. 1, F to I, and fig. S1H). The I/O (input/output) function of PPI demonstrated that C/EBPβ Tg mice displayed increased hearing sensitivity as compared to WT mice (fig. S1G). MWM (Morris water maze) and fear condition tests supported that Thy1-C/EBPβ Tg mice revealed impaired cognitive functions (Fig. 1, J to L). Together, these behavioral tests suggest that neuronal C/EBPβ up-regulation facilitates aging in the brain.
Fig. 1. Neuronal C/EBPβ overexpression elicits short life span, associated with behavioral impairments in Thy1-C/EBPβ Tg mice, related to fig. S1.
(A) Human C/EBPβ overexpression in neurons shortens life span. C/EBPβ Tg strain of origin: C57BL/6; P < 0.0001, log-rank test; ***P < 0.001. (B) C/EBPβ knockdown extends life span. C/EBPβ+/− strain of origin: 129S1/Sv-Oca2+ Tyr+ Kitl+; P < 0.0001, log-rank test; ***P < 0.001. (C) Left: Images from PET-CT scanning of 18F-FDG uptake in 6-month-old Thy1-C/EBPβ Tg, C/EBPβ+/−, and age-matched WT littermates. Right: Standardized uptake value (SUV) at increasing time intervals after injection of 18F-FDG. Means ± SEM; n = 3 mice per group; **P < 0.01; Mann-Whitney U test. (D) Seizure duration after administration of PTZ (35 mg kg−1). Control (WT), n = 6; C/EBPβ Tg, n = 6; C/EBPβ+/−, n = 6 mice. *P < 0.05 and **P < 0.01; Mann-Whitney U test. (E) Elevated [Ca2+]i in the C/EBPβ-overexpressing hippocampal CA1. The box plots indicate first, median, and third quartiles as well as outliers. N = 196 (WT control) and 95 cells (C/EBPβ Tg) pooled from six animals each; *P = 0.043, t(289) = 2.04; two-tailed Student’s t test. (F) PPI. Means ± SEM; n = 8 mice per group; unpaired t test with Welch’s correction. (G) Open field test. Means ± SEM; n = 8 mice per group; *P < 0.05; unpaired t test with Welch’s correction. (H) FST. Means ± SEM; n = 8 mice per group; unpaired t test with Welch’s correction. (I) Sleep latency test. Means ± SEM; n = 8 mice per group; *P < 0.05; unpaired t test with Welch’s correction. (J and K) MWM analysis of cognitive functions. Means ± SEM; n = 8 mice per group; *P < 0.05 and **P < 0.01; unpaired t test with Welch’s correction. (L) Fear condition tests. Means ± SEM; n = 8 mice per group; *P < 0.05, and **P < 0.01; unpaired t test with Welch’s correction.
Neuronal overexpression of C/EBPβ selectively triggers GABAnergic degeneration, leading to neuroexcitation
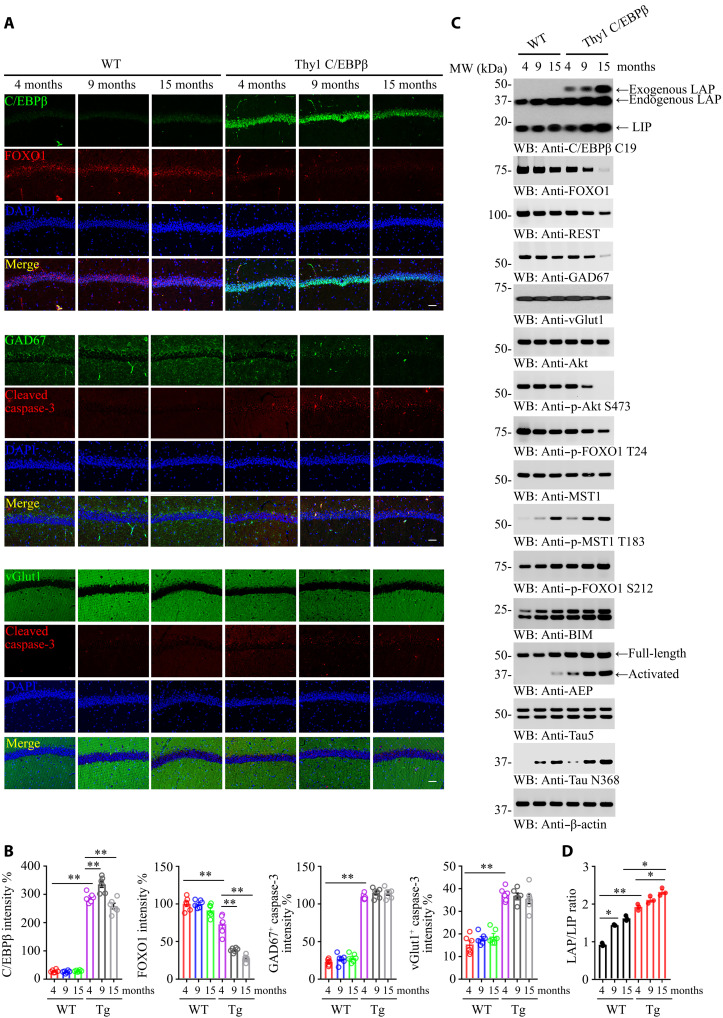
The Tg mice exhibited prominent neural excitation and seizures, indicating that the inhibitory GABAnergic neurons might be impaired. IF costaining showed that GAD67 (67 KDa glutamic acid decarboxylase)-positive GABAnergic neurons were reduced in an age-dependent manner in Tg mice in comparison with WT mice, coupled with apoptotic caspase-3 escalation; in contrast, vGluT1-positive glutamatergic neurons undergoing apoptosis were significantly fewer than apoptotic GABAnergic neurons (Fig. 2A, middle and bottom, and 2B). Noticeably, neuronal FOXO1 immunoreactivity was pronouncedly attenuated in Thy1-C/EBPβ Tg mice versus WT mice during aging. It was barely detectable in Tg mice at 9 and 15 months old (Fig. 2A, top, and 2B). Both FOXO1 and REST, a transcription factor for FOXOs, were decreased in Tg mice in an age-dependent way. Although GAD67 levels were diminished during aging in both WT and Tg mice, vGluT1 displayed a reversed pattern (Fig. 2C, top-fifth). FOXOs can be phosphorylated by Akt and Mst1 on T24 and S212 residues, respectively. Akt-phosphorylated FOXO1 is sequestered in the cytoplasm and blocks the apoptotic genes similar to BIM (bcl-2 interacting mediator of cell death) transcription, promoting cell survival (30), whereas Mst1-phosphorylated FOXO1 translocates into the nucleus, promoting apoptosis (31). Phosphorylated Akt (p-Akt) was repressed in Tg mice during aging, leading to p-FOXO1 T24 reduction. By contrast, p-Mst1 was elevated in Tg mice as compared to WT mice, resulting in more robust p-FOXO1 S212 in Tg mice than WT mice (Fig. 2C, 6th to 11th panels). C/EBPβ consists of LAP and LIP, which function oppositely, such that LAP acts as a transcriptional activator and LIP acts as a transcriptional inhibitor. Compared with WT, the LAP/LIP ratio of C/EBPβ was higher in the C/EBPβ Tg group and increased in an age-dependent manner (Fig. 2C, first panel, and Fig. 2D). Consequently, the downstream target BIM, a proapoptotic effector, was escalated in an age-dependent manner with Tg stronger than WT controls. As a positive control, AEP, a well-characterized downstream target of C/EBPβ, was progressively augmented during aging, with Tg mice much stronger than WT mice. Subsequently, active AEP cleaved Tau N368 that is prone to aggregation and is neurotoxic (Fig. 2C, 12th bottom panel).
Fig. 2. Neuronal C/EBPβ represses FOXO1 during aging and promotes GABAnergic neuronal degeneration in mice.
(A) Neuronal C/EBPβ inhibits FOXO1 expression and selectively triggers GABAnergic neuronal apoptosis. IF analysis of C/EBPβ, FOXO1, GAD67, vGlut1, and cleaved caspase-3 was analyzed in neurons from the different ages of mice hippocampus. Confocal immunofluorescence microscopy was performed in mice hippocampus. Scale bars, 40 μm. The image shown is representative of immunofluorescence labeling performed in six individuals. (B) Quantification of (A). Data are represented as means ± SEM; n = 6 per group. **P < 0.01; two-way analysis of variance (ANOVA) and Sidak’s multiple comparisons test. (C) C/EBPβ overexpression in the neurons represses FOXO1 and GAD67 in the mouse brain during aging. The cerebral cortex frozen tissues from different ages of WT and Thy1-C/EBPβ Tg mice were used for WB detection with various indicated antibodies. (D) The LAP/LIP isoform ratios were calculated from quantification by immunoblots of (C). Data are represented as means ± SEM; n = 3 per group. *P < 0.05 and **P < 0.01; two-way ANOVA and Bonferroni’s post hoc test.
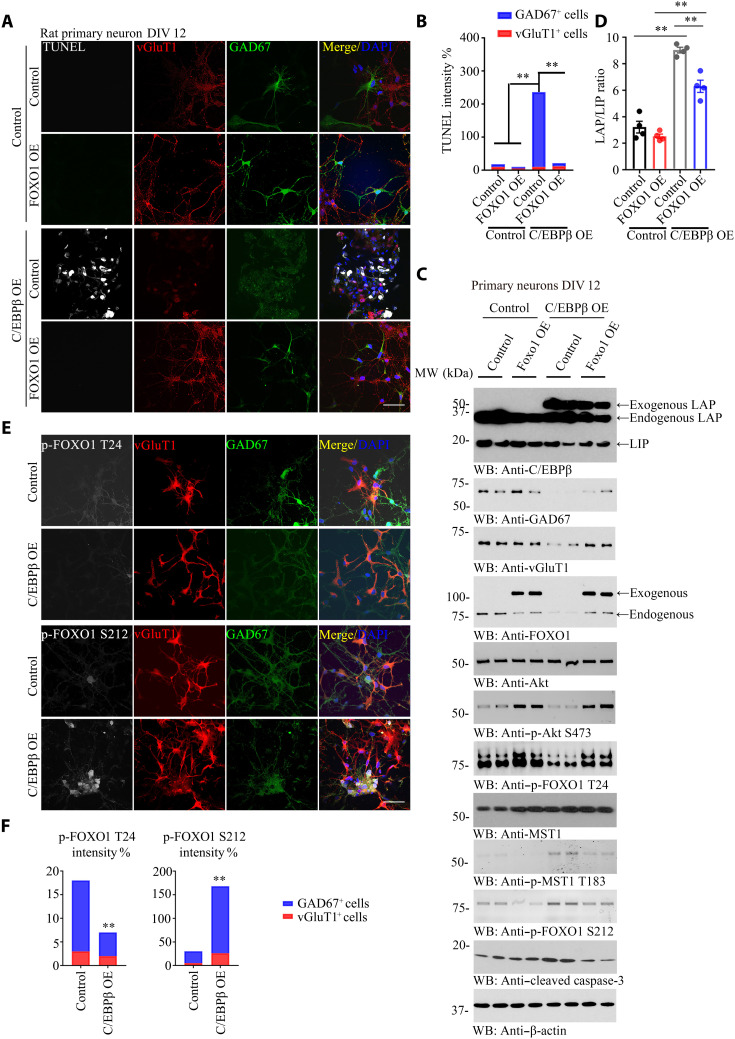
To explore why GABAnergic neurons are more vulnerable than glutamatergic neurons in Thy1-C/EBPβ Tg mice, we prepared primary neuronal cultures from embryonic day 18 (E18) rats [days in vitro (DIV) 12] and infected the neurons with control virus or virus expressing C/EBPβ in the presence or absence of virus overexpressing FOXO1. C/EBPβ overexpression selectively elicited prominent apoptosis in GABAnergic but not glutamatergic neurons, and this effect was suppressed by FOXO1 overexpression (Fig. 3, A and B). C/EBPβ overexpression completely eradicated GAD67 and diminished vGluT1 levels. Notably, overexpressed C/EBPβ eliminated FOXO1 as compared to control. On the other hand, FOXO1 overexpression also attenuated C/EBPβ levels (Fig. 3C, top to fourth panels), indicating that C/EBPβ and FOXO1 mutually repress each other. The LAP/LIP ratios of C/EBPβ were higher in C/EBPβ overexpression groups and suppressed by FOXO1 overexpression (Fig. 3C, first panel, and Fig. 3D). C/EBPβ overexpression decreased p-Akt S473, resulting in p-FOXO1 T24 reduction. On the contrary, p-Mst1 T183 was elevated in C/EBPβ-overexpressed neurons, leading to p-FOXO1 S212 increase and caspase-3 activation. Notably, these effects were alleviated by FOXO1 overexpression. For instance, caspase-3 activation by C/EBPβ was suppressed by FOXO1 overexpression, accompanied with GAD67 and vGluT1 level augmentation (Fig. 3C). C/EBPβ overexpression repressed p-FOXO1 T24 and elevated p-FOXO1 S212, and these effects were much more robust in GABAnergic neurons than glutamatergic neurons (Fig. 3, E and F). Moreover, C/EBPβ potently suppressed both GAD67 and vGluT1 signals, and FOXO1 significantly restored both (fig. S2, A, B, and D). C/EBPβ overexpression selectively induced AEP escalation in GABAnergic neurons, which was repressed by FOXO1 (fig. S2, C and D). In alignment with AEP expression, C/EBPβ induced Tau up-regulation and Tau N368 cleavage by AEP, associated with p-Tau AT8 increase, which were inhibited by FOXO1 (fig. S2E).
Fig. 3. Neuronal C/EBPβ selectively triggers GABAnergic neuronal degeneration, related to figs. S2 to S4.
(A) C/EBPβ overexpression promotes GABAnergic neuronal apoptosis. Rat primary neurons (DIV 12) were infected with FOXO1 or C/EBPβ lentivirus for 72 hours. Neurons were fixed and permeabilized; after terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining, cells were incubated with vGlut1 and GAD67 antibodies. vGlut1 marked the glutamatergic neurons, and GAD67 marked GABAnergic neurons. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Control, vector virus. Scale bar, 50 μm. OE, overexpression. (B) Quantification of TUNEL-positive neurons induced by C/EBPβ overexpression, and the percentage of glutamatergic or GABAnergic neuron also showed. (Means ± SEM; n = 6; **P < 0.01; unpaired t test with Welch’s correction). (C) Immunoblotting analysis using C/EBPβ-overexpressed neurons. C/EBPβ overexpression inhibited Akt activation and FOXO1 T24 phosphorylation, and it promoted MST1 activation and FOXO1 S212 phosphorylation. The treated neurons from (A) were used for Western blotting (WT) and detected with various indicated antibodies. MW, molecular weight. (D) The LAP/LIP isoform ratios were calculated from quantification by immunoblots of (C). Data are represented as means ± SEM; n = 4 per group. **P < 0.01; two-way ANOVA and Bonferroni’s post hoc test. (E) C/EBPβ regulates p-FOXO1 status. C/EBPβ overexpression inhibited p-FOXO T24 and promoted p-FOXO1 S212 in GABAnergic other than glutamatergic neurons, leading to p-FOXO1 S212–induced GABAnergic neuronal degeneration. Rat primary neurons (DIV 12) were infected with control virus or C/EBPβ lentivirus for 72 hours. Neurons were fixed and permeabilized, and cells were incubated with p-FOXO1, vGlut1, and GAD67 antibodies. The nuclei were stained with DAPI. Control, vector virus. Scale bar, 50 μm. (F) Quantification of p-FOXO1 T24 or p-FOXO1 S212–positive neurons induced by C/EBPβ overexpression, and the percentage of glutamatergic or GABAnergic neurons also showed. (Means ± SEM; n = 6; **P < 0.01; unpaired t test with Welch’s correction).
On the other hand, depletion of FOXO1 diminished GAD67 and vGluT1, whereas eradication of C/EBPβ strongly increased both. Nonetheless, these effects were blunted when FOXO1 was also deleted (fig. S3, A, B, and D). In particular, knocking down FOXO1 selectively escalated AEP in GABAnergic neurons, which was completely abolished when C/EBPβ was deleted (fig. S3, C and D). Again, C/EBPβ knockdown elicited total Tau and Tau N368 reduction, leading to lessened AT8 activities (fig. S3E). FOXO1 knockdown selectively induced apoptosis in GABAnergic but not glutamatergic neurons, which was antagonized by deletion of C/EBPβ (fig. S4, A and B). These biochemical effects were validated by IB (immunoblotting) and IF analysis (fig. S4, C to F). The LAP/LIP ratios of C/EBPβ were higher in C/EBPβ knockdown groups and reversed by FOXO1 deletion (fig. S4C, first panel, and fig. S4D). Hence, GAD67-positive GABAnergic neurons were selectively lost in C/EBPβ-overexpressed neurons, whereas this effect was substantially decreased in excitatory vGluT1-positive glutamatergic neurons in the hippocampus, resulting in neuronal excitation in Thy1-C/EBPβ Tg mice.
Overexpression of C/EBPβ in neurons represses REST and FOXO1 expression
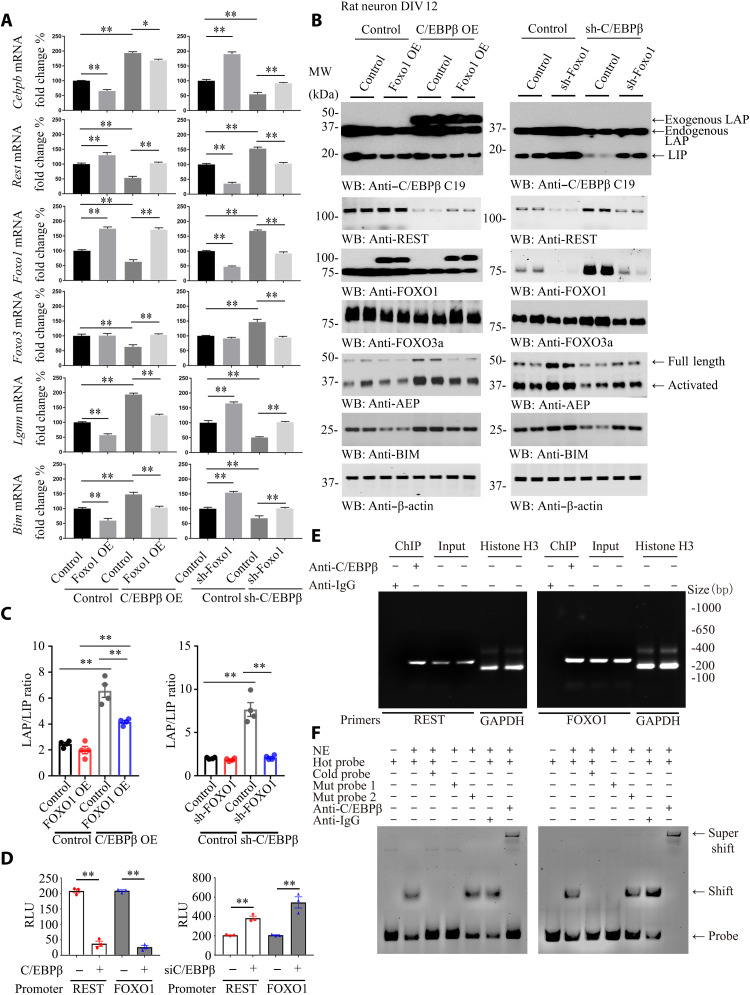
To assess whether C/EBPβ directly mediates FOXOs and REST expression, we conducted quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis using primary neuronal cultures infected with virus expressing C/EBPβ or sh (short hairpin RNA) of C/EBPβ in the presence of virus expressing FOXO1 or its short hairpin RNA (shRNA) and found that C/EBPβ overexpression suppressed FOXO1, FOXO3, and REST mRNA levels, resulting in escalation of Bim and LGMN (AEP) mRNAs. Because C/EBPβ and FOXO1 mutually repress each other, overexpression of FOXO1 led to CEBPβ mRNA reduction, and, consequently, both Bim and LGMN mRNAs were also blunted. Accordingly, C/EBPβ overexpression–modulated gene expression patterns were reversed (Fig. 4A, left). On the other hand, deletion of C/EBPβ triggered FOXO1, FOXO3, and REST mRNA escalation, associated with both Bim and LGMN mRNAs being reduced. As expected, depletion of FOXO1 stimulated C/EBPβ mRNA up-regulation, resulting in Bim and LGMN transcriptional augmentation. The C/EBPβ eradication–triggered genes were significantly overturned by sh-FOXO1 (Fig. 4A, right). IB analysis showed that the protein levels mirrored their mRNA concentration oscillations (Fig. 4B). The LAP/LIP ratios of C/EBPβ were higher in C/EBPβ overexpression groups and reversed by FOXO1 overexpression. Moreover, the LAP/LIP ratios of C/EBPβ were higher in C/EBPβ knockdown groups and decreased by FOXO1 depletion (Fig. 4B, first panel, and Fig. 4C). Although both C/EBPβ OE (overexpression) and shRNA treatment lead to LAP/LIP ratio escalation, the LIP isoform is more selectively eradicated by sh-C/EBPβ.
Fig. 4. Neuronal C/EBPβ represses REST and FOXO1 expression in the brain, related to figs. S5 to S7.
(A and B) Rat primary neurons (DIV 12) were infected with lentivirus expressing FOXO1 or C/EBPβ, sh-FOXO1, or sh-C/EBPβ for 72 hours. Neurons were harvested for real-time PCR (A) and WB (B). Control, vector virus. (Means ± SEM; *P < 0.05 and **P < 0.01; n = 3; two-way ANOVA and Bonferroni’s post hoc test). (C) The LAP/LIP isoform ratios were calculated from quantification by immunoblots of (B). Data are represented as means ± SEM; n = 4 per group. **P < 0.01; two-way ANOVA and Bonferroni’s post hoc test. (D) Luciferase assay. Human REST and FOXO1 promoter luciferase plasmids were cotransfected into SH-SY5Y cells with C/EBPβ–green fluorescent protein (GFP) overexpression plasmid for 48 hours. The luciferase activities were calculated by using the Luciferase Reporter Assay System, which indicated the promoter activities. (Means ± SEM; **P < 0.01; n = 3; one-way ANOVA). RLU, relative light units. (E) Chromatin immunoprecipitation (ChIP) assay. ChIP assay was performed to detect the binding sites of C/EBPβ on the human REST and FOXO1 promoters. The DNA-protein cross-linking ChIP samples were immunoprecipitated with anti-C/EBPβ antibody or immunoglobulin G (IgG). After reversing cross-links, PCR was performed by using primer pairs at −1409 to −1189 (REST) or −1587 to −1357 (FOXO1) of the PCR assays that also detected each input sample. Genomic DNA samples were pulled down with anti–histone H3 and normalized by glyceraldehyde phosphate dehydrogenase (GAPDH) primers as positive control. The equal amount of input was confirmed with REST and FOXO1. (F) EMSA. Nuclear extract proteins (NE) were isolated from SH-SY5Y cells transfected with C/EBPβ-GFP for 48 hours. EMSA was used to detect the C/EBPβ binding ability on sites −1250 to −1260 (REST) or −1504 to −1495 (FOXO1) promoter [ATGTTGTAATA (REST) or TTTACTTAAC (FOXO1)], mutation probe 1 [GCGTTGTAATA (REST) or TTTACTTGGC (FOXO1)] or mutation probe 2 [(ATTTTGTAATA (REST) or TTTACTTAAT (FOXO1)], and supershift. Data are representative of three independent experiments.
Luciferase assay showed that C/EBPβ overexpression repressed both REST and FOXO1 promoter activities. By contrast, deletion of C/EBPβ increased both promoter activities (Fig. 4D), suggesting that C/EBPβ might bind to their promoters and act as a transcriptional repressor. ChIP (chromatin immunoprecipitation) assay revealed that anti-C/EBPβ but not control immunoglobulin G (IgG) specifically pulled down DNA containing REST and FOXO1 promoters, although equal amounts of fragmented chromatin were used (Fig. 4E). EMSA (electrophoretic mobility shift assay) supported that C/EBPβ specifically bound to these promoters, which was further validated by supershift using anti-C/EBPβ (Fig. 4F). Thus, C/EBPβ directly binds to REST and FOXO1 promoters and acts as a transcriptional repressor.
Single-cell transcriptome (GSE67835) analysis from human brains revealed that both CEBPB and LGMN mRNAs were expressed in various brain cell types. However, LGMN was highly expressed in neurons, whereas CEBPB was expressed at a low level. Conversely, REST and FOXO1 were both expressed in endothelial cells and astrocytes. Whereas FOXO1 were demonstrable in both microglia and neurons, REST was barely expressed in microglia and neurons (fig. S5A). Quantification revealed that CEBPB was elevated in the brain during aging and peaked in individuals who were 60 to 84 years old and declined in those >85 years old. Nevertheless, FOXO1 mRNA escalated in an age-dependent manner. However, the ratios for FOXO3 and REST mRNAs were not significantly changed in the brain during aging (fig. S5B). To gain insight into changes in gene expression profiles in the brain associated with human longevity, we analyzed RNA sequencing data from aged individuals with intact cognitive functions in two cohorts: ROSMAP (Religious Orders Study and Memory and Aging Project) and CMC (Common Mind Consortium). There is no correlation between down-regulated genes in either cohort and levels of CEBPB mRNA levels. However, CEBPB was significantly associated with up-regulated genes in ROSMAP but insignificant in CMC (fig. S6, A and B). Nevertheless, there was a borderline significant association between CEBPB with REST and FOXO1 in ROSMAP but not in the CMC cohort (fig. S6C). Stratification of different age groups showed that CEBPB was significantly associated with up-regulated genes in ROSMAP with extended longevity (fig. S6, D and E). The most significant changes associated with extended longevity groups occurred with LGMN. Down-regulated expression of genes in individuals with extended longevity (≥85 versus ≤80 years old) in the ROSMAP (n = 117 individuals) and CMC (n = 155 individuals) cohorts were positively correlated with LGMN levels, whereas up-regulated genes were inversely correlated with LGMN (fig. S7, A and B). There is significant association with down-regulated REST and FOXO1 expression in both ROSMAP and CMC cohorts related to levels of LGMN mRNA (fig. S7C). Stratification of different age groups (≥85 or ≤80 years old) also revealed the positive correlation between down-regulated genes with LGMN levels and reverse correlation with up-regulated genes (fig. S7, D and E). Overall, the association between LGMN expression and down-regulated genes including REST and FOXO1 is tightly related, but CEBPB displays no correlation, which fits our data that CEBPB has little effect on glutamatergic neurons where we saw the greatest age-related transcriptional changes.
Neuronal overexpression of C/EBPβ or AEP shortens the life span of C. elegans and is associated with neural excitation
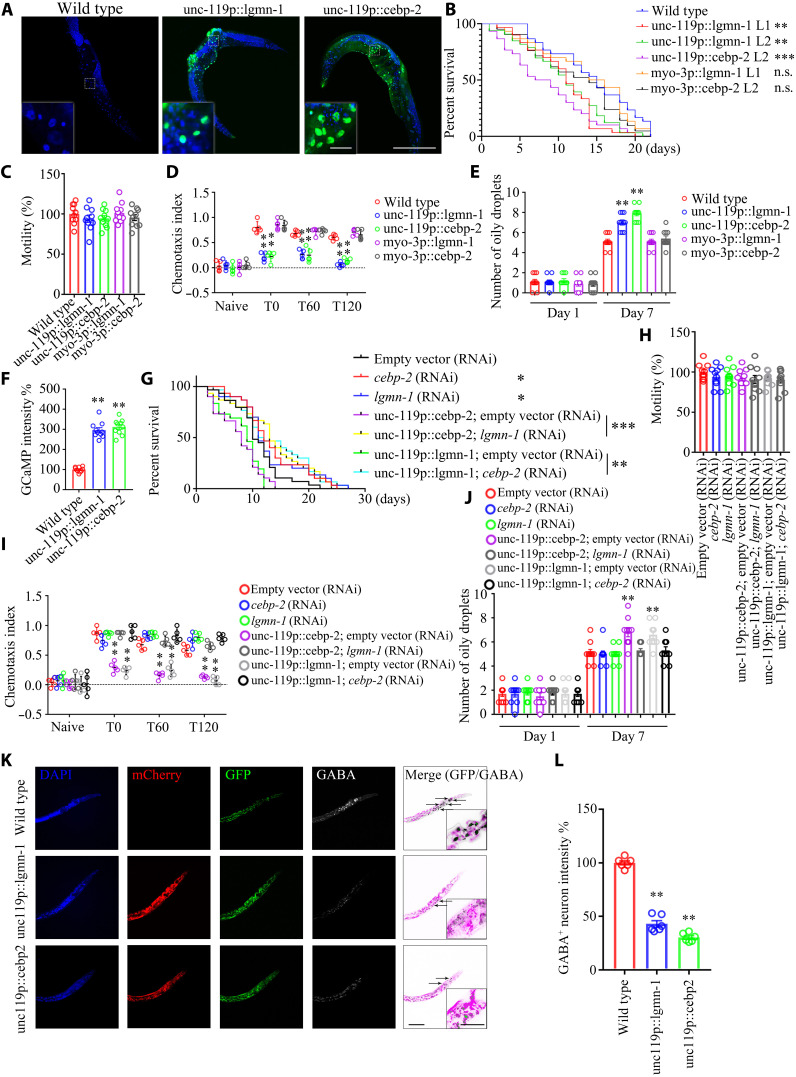
To explore the neural regulation of longevity, we used C. elegans, a well-established model system for studying aging. The C. elegans genome contains orthologs of mammalian CEBPB and LGMN, namely, cebp-2 and lgmn-1. Transgenic nematodes with a WT background were created that expressed either cebp-2 or lgmn-1 under the control of the pan-neuronal promoter from unc-119, or the body wall muscle cell promoter from myo-3. In three independent transgenic lines, the arrays containing unc-119p::lgmn-1 and unc-119p::cebp-2 were widely expressed in neurons in the head and the nerve ring around the pharynx (Fig. 5A). The myo-3p::lgmn-1 and myo-3p::cebp-2 transgenic lines showed life spans comparable to N2 WT worms. The unc-119p::lgmn-1 lines 1 and 2 (L1 and L2) displayed significantly shorter life span than N2 WT nematodes, and unc-119p::cebp-2 exhibited the shortest life span (Fig. 5B). However, all of the transgenic strains demonstrated whole animal locomotion similar to N2 worms (Fig. 5C). In the positive butanone association chemotaxis (CTX) assay, a measure of learning and memory, myo-3p::lgmn-1 and myo-3p::cebp-2 lines, presented CTX index (CI) values similar to that of N2; by contrast, unc-119p::lgmn-1 and unc-119p::cebp-2 showed significantly impaired learning and memory compared with N2 worms from T0 (time 0 min.) to T120 duration (Fig. 5D). At day 7, neuronal cebp-2 or lgmn-1 overexpression led to significantly more oily droplets than N2 nematodes or muscle overexpression strains (Fig. 5E), suggesting that neuronal cebp-2 or lgmn-1 overexpression accelerates tissue aging together with reducing cognitive functions and life span. We monitored neural excitation in C. elegans by GCaMP (green fluorescent protein-calmodulin fusion protein) calcium imaging in the neurons (32). In WT worms, we observed rapid, transient pulses of GCaMP fluorescence indicative of neuronal excitation. Noticeably, calcium influx into neurons increased in unc-119p::lgmn-1 and unc-119p::cebp-2 strains (Fig. 5F), fitting with a previous report that elevated calcium influx correlates with shorter life span (6). To further explore the effect of neuronal cebp-2 or lgmn-1 on life span, we suppressed their neuronal expression by using RNA interference (RNAi). When hypersensitive neuronal RNAi strain TU3335 worms were treated with RNAi against cebp-2 or lgmn-1, there was a demonstrable increase in life span. However, the increase of life span upon knockdown of these genes may not reflect a neuronal specific effect on longevity. Therefore, to determine whether neuronal cebp-2 or lgmn-1 specifically regulates life span, we asked whether decreasing their expression in neuronal cebp-2 or lgmn-1 overexpressed lines increases life span. To do this, we crossed in our neuronal cebp-2 or lgmn-1 overexpression transgenes into the TU3335 background and then performed RNAi. Notably, RNAi knockdown of lgmn-1 in neuronal overexpressed cebp-2 or RNAi knockdown of cebp-2 from neuronal overexpressed lgmn-1 strongly extended life span as compared to their vector controls, and these genetic manipulations did not affect the worms’ motility (Fig. 5, G and H). As indicated above, CTX assays showed that neuronal overexpression of cebp-2 or lgmn-1 resulted in defective cognitive functions, which were rescued by RNAi knockdown of lgmn-1 and cebp-2, respectively (Fig. 5I). Consistent with impaired learning and memory, the oily droplet assay at day 7 also revealed that overexpression of cebp-2 or lgmn-1 exhibited aging phenotypes, which were alleviated by RNAi against lgmn-1 or cebp-2, respectively (Fig. 5J). Most individual neurons in worms can be identified on the basis of their unique position, connectivity, and morphology. IF costaining with anti-GABA showed that overexpression of neuronal cebp-2 or lgmn-1 selectively decreased the number of neurons that produce GABA or decreased GABA expression in these neurons (Fig. 5, K and L).
Fig. 5. Neuronal overexpression of C/EBPβ or AEP shortens the life span in C. elegans, associated with neural excitation, related to figs. S8 and S9.
(A) GFP signal from neurons in transgenic lines. Scale bars, 200 μm and 20 μm (inset). (B) Neuronal but not muscle overexpression of lgmn-1 or cebp-2 reduces the life span of worm. (n = 30 worms per group; P < 0.0001, log-rank test; **P < 0.01 and ***P < 0.001 versus WT). n.s., not significant. (C) Motility assay. Shown are mean motility scores for the first 30 s. (n = 10 worms per group). (D) CTX assay. Data were analyzed by two-way ANOVA and Sidak’s multiple comparisons test (n = 14 to 20 worms per experiment from five independent experiments). Color circles represent the individual CIs. (E) Oily droplet characterization. Data are represented as means ± SEM (day 7; two-way ANOVA and Sidak’s multiple comparisons test; **P < 0.01). (F) Neural excitation assay. Quantification of GCaMP fluorescence changes in adult day 2 worms: n = 10 worms per group. **P < 0.01 versus WT. (G) Knocking down lgmn-1 or cebp-2 by RNAi reverses life-span reduction by neuronal overexpression lgmn-1 or cebp-2. n = 30 worms per group. P < 0.0001, log-rank test; *P < 0.05, **P < 0.01, and ***P < 0.001. (H) Motility assay. n = 10 worms per group. (I) CTX assay. Data were analyzed by two-way ANOVA and Sidak’s multiple comparisons test (n = 14 to 20 worms per experiment from five independent experiments). (J) Oily droplet assay. Data are presented as means ± SEM (day 7; two-way ANOVA and Sidak’s multiple comparisons test; **P < 0.01). (K) Neuronal overexpression of lgmn-1 or cebp-2 induces GABAnergic neuronal degeneration in worms. Scale bars, 40 μm and 10 μm (inset). (L) Quantification of GABA+ neurons’ fluorescence intensity in day 2 worms. Data are represented as means ± SEM; n = 10 worms per group. **P < 0.01 versus WT; Mann-Whitney U test with multiple testing correction by Holm’s method.
Because knocking down lgmn-1 extended the life span of the strain that overexpresses cebp-2 in neurons, we wondered whether biochemical inhibition of LGMN (legumain)-1 with its small molecular inhibitor compound #11 (C11) exerts a similar effect (33). Notably, treatment with C11 rescued the short life spans of both unc-119p::lgmn-1 and unc-119p::cebp-2 transgenic lines, which were comparable to the life spans of N2 worms treated with C11. All of the strains exhibited similar motility regardless of C11 treatment or not, indicating that C11 does not affect whole animal locomotion (fig. S8, A and B). CI and oily droplet analysis revealed that C11 significantly restored cognitive functions and delayed the age-associated phenotypes (fig. S8, C and D). Protein sequence alignment shows that the enzymatic domains of LGMN/AEP/legumain proteins are well conserved among multiple species (fig. S8E). As a result, C11 potently inhibited AEP enzymatic activity from worms (fig. S8F). In addition, lgmn-1 or cebp-2 RNAi treatment significantly decreased AEP activities in unc-119p::lgmn-1 and unc-119p::cebp-2 transgenic lines (fig. S8G). Thus, the effects of cebp-2 and lgmn-1 on life span are bidirectional. Life span is extended by reducing these genes from neurons and shortened by increasing neuronal expression, and cebp-2 and lgmn-1 mutually regulate each other’s role in age-associated cognitive decline and longevity.
daf-2, the gene that encodes the IR, regulates longevity in C. elegans. Decreased DAF-2 signaling causes an increase in life span. The forkhead transcription factor DAF-16 is the essential downstream target of DAF-2–insulin/IGF-like signaling that regulates life span in C. elegans. To further explore the effects of C/EBPβ/AEP orthologs on the DAF-2–DAF-16 pathway, we crossed transgenes neuronally overexpressing cebp-2 or lgmn-1 into a daf-2 mutant or into a line in which DAF-16 was overexpressed. Noticeably, neuronal overexpression of cebp-2 or lgmn-1 significantly shortened the life span of daf-2(e1370) without interfering with locomotion (fig. S9, A and C). Overexpressing cebp-2 or lgmn-1 repressed both daf-16 and spr-4 (encodes one of the two REST orthologs in worms) (fig. S9B). daf-2 mutants in which cebp-2 or lgmn-1 were overexpressed exhibited age-associated parameter and cognitive disorders as compared to daf-2(e1370) itself; in addition, compared with WT, daf-2 mutant reduces the accumulation of oily droplets on day 7 (fig. S9, D and E).
To examine the effect of overexpressing DAF-16 in neurons, we crossed our neuronally overexpressing lgmn-1 or cebp-2 lines into a strain that contained the integrated transgene muIs131 [unc-119p::GFP::daf-16 + rol-6(su1006)], which overexpresses daf-16 in all neurons. Neuronal overexpression of lgmn-1 or cebp-2 (purple or yellow lines in fig. S9F) shortened the life span of daf-16 overexpression worms (blue line in fig. S9F). Again, neuronal cebp-2 or lgmn-1 overexpression significantly suppressed both spr-4 and daf-16 expression (fig. S9G). Neuronal overexpression of lgmn-1 or cebp-2 (purple or gray bars in fig. S9H) substantially increased the accumulation of oily droplets compared to daf-16 overexpression worms (green bar in fig. S9F). Therefore, overexpression of lgmn-1 or cebp-2 in neurons in C. elegans accelerated the appearance of physical markers of aging as compared to control nematodes. Together, our results suggest that neuronal cebp-2 or lgmn-1 specifically shortens life span and promotes senescence. Both spr-4 and daf-16 mRNAs in various DAF-2 mutant and DAF-16 transgenic lines are robustly repressed when neural cebp-2 or lgmn-1 is overexpressed.
Insulin regulates the expression of C/EBPβ in various peripheral tissues (34, 35). C/EBPβ also implicates in mediating IGF-1 and human IR expression via binding their promoters (36–38). C/EBPβ in liver and pancreas negatively regulates insulin levels (39). Accordingly, we analyzed the relationship of C/EBPβ and the insulin signaling in neurons. Under control conditions, insulin temporally activated p-IR/p-IRS and downstream effectors p-Akt and p–mitogen-activated protein kinase (MAPK). Consequently, p-FOXO1 T24 echoed upstream p-Akt signals. Insulin reduced both endogenous and overexpressed C/EBPβ and p-C/EBPβ signals in a time-dependent manner. Compared with control groups, the LAP/LIP ratios were higher in C/EBPβ overexpression groups but lower in C/EBPβ knockdown groups. The LAP/LIP ratios were repressed by insulin treatment in both C/EBPβ overexpression and knockdown groups (fig. S10A, first panel, and fig. S10B). Notably, overexpression of C/EBPβ diminished p-IR and p-IRS triggered by insulin, whereas downstream p-Akt and p-MAPK remained intact. As expected, C/EBPβ suppressed FOXO1 levels, resulting in reduced p-FOXO1 T24 activities (fig. S10A). Insulin time-dependently repressed Cebpb and increased Foxo1 mRNA levels in both control and C/EBPβ-overexpressed neurons. When C/EBPβ was depleted by its shRNA, the remnant Cebpb mRNA transcription was also temporally suppressed by insulin, whereas Foxo1 mRNA remained unaltered and IRS-1 mRNA concentrations were escalated. Nevertheless, Insr mRNA levels were unchanged, no matter whether C/EBPβ was modified or insulin stimulation (fig. S10C). p-IR signals were not affected by C/EBPβ in GABAnergic neurons, but, in contrast, p-FOXO1 T24 was antagonized by C/EBPβ overexpression. Conversely, p-IR activities were inhibited by C/EBPβ overexpression in glutamatergic neurons, whereas p-FOXO1 T24 were constant regardless of C/EBPβ levels (fig. S10, D and E, and fig. S11, A to D). Hence, neuronal C/EBPβ up-regulation displays distinctive effects on insulin-triggered p-IR and p-FOXO1 T24/FOXO1 activities in GABAnergic and glutamatergic neurons.
C/EBPβ mediates insulin signaling in neurons, modulating FOXO1-induced longevity in humans
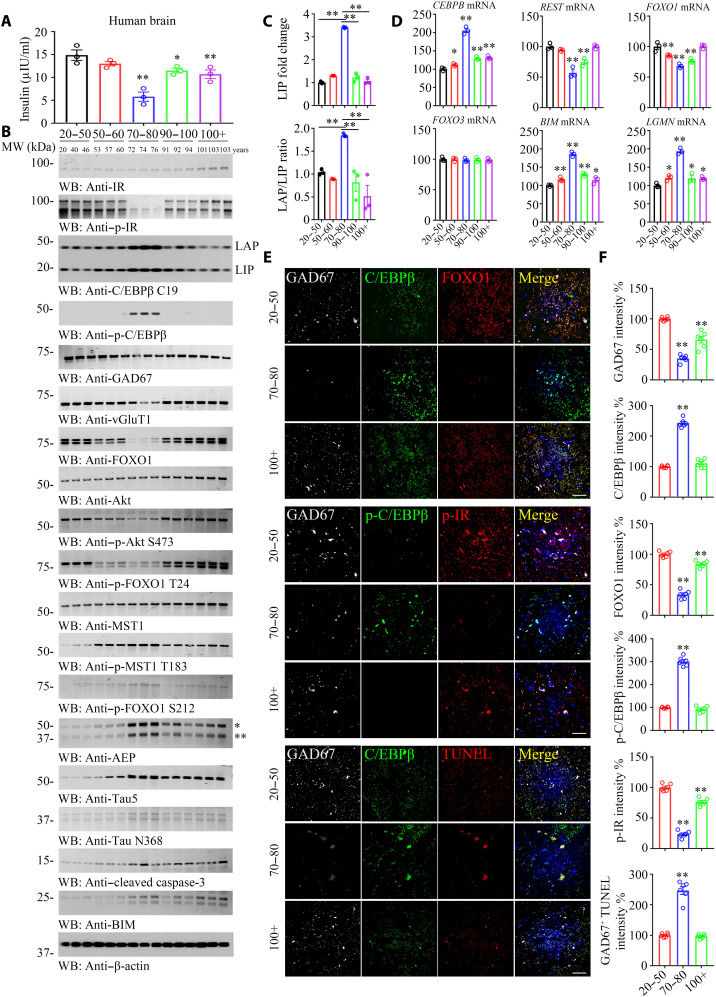
Life-span regulation by insulin-like metabolic control is analogous to mammalian longevity enhancement induced by caloric restriction, suggesting a general link between metabolism and longevity. To gain further insight into the correlation between C/EBPβ and insulin signaling and life span, we monitored insulin concentrations and performed Western blotting (WB) with human brains from different age groups. Notably, the brain insulin concentrations exhibited a concave curve with the lowest levels in the seventh decade and the highest levels in the younger and older age groups (Fig. 6A). Noticeably, p-IR signals were also lowest in the 70 to 80 years old groups compared to other age groups, mirroring the ligand insulin concentrations, whereas total human IR levels remained comparable among the groups. By sharp contrast, p-C/EBPβ activities displayed a convex curve, with the 70 to 80 years old group being the highest, although total C/EBPβ levels were slightly higher compared to other age groups (Fig. 6B). Compared with the 20 to 50 years old group, both C/EBPβ isoforms were highly augmented, and the LAP/LIP ratio of C/EBPβ was highest in the 70 to 80 years old group. After 70 to 80 years of age, both LAP and LIP isoforms evidently decreased in humans with longevity (Fig. 6B, third panel, and Fig. 6C). Consequently, the total levels of FOXO1, GAD67, and vGluT1 were lower within this group. Remarkably, p-Akt S473 and its downstream p-FOXO1 T24 signals were progressively lower from 50 to 80 years old, with the 70 to 80 years of age group being the lowest. By contrast, p-Mst1 and p-FOXO1 S212 activities were gradually higher within this duration and lower thereafter. As a downstream target of p-C/EBPβ, AEP also presented a parabolic curve pattern. Total Tau started to rise from 70 years old and plateaued within the 70- to 80-year-old period and slowly lowered from 90 to 100+ years old. AEP-cleaved Tau N368 oscillated with upstream active AEP levels. BIM correlated with p-FOXO S212 activities (Fig. 6B). qRT-PCR demonstrated the inverse correlation between cebpb and Rest/Foxo1 mRNA levels, while both Bim and LGMN mRNA concentrations mirrored cebpb signals. By contrast, Foxo3 mRNA remained constant from young to aged groups (Fig. 6D). C/EBPβ fluorescent intensities inversely coupled to GAD67 and FOXO1 activities. Moreover, p-C/EBPβ contrariwise correlated to p-IR signals in GABAnergic neurons. Terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining showed that GABAnergic neuronal apoptosis was greatest in the 70 to 80 years old group (Fig. 6, E and F, and fig. S12), fitting with the findings of augmented proapoptotic Bim and active caspase-3 amount in IB. We made the similar observations in vGluT1-positive glutamatergic neurons. However, glutamatergic neuronal apoptotic activities were much weaker than GABAnergic neurons (fig. S12, A to C and F). Again, both AEP and Tau N368 fluorescent activities were also the highest in 70 to 80 years old group in both GABAnergic and glutamatergic neurons (fig. S12, D, E, and G). Therefore, neuronal C/EBPβ escalation appears to make GABAnergic neurons being more vulnerable and prone to degeneration than glutamatergic neurons during aging.
Fig. 6. C/EBPβ mediates insulin signaling in neurons, modulating FOXO1 expression in human brains, related to figs. S10 to S13.
(A) Brain insulin concentration decreased along with aging. Brain tissues from different age groups are detected by human insulin enzyme-linked immunosorbent assay kit. (B to D) p-C/EBPβ and p-IR are inversely coupled in the human brains from 70 to 80 years of age group. C/EBPβ and p-C/EBPβ/AEP displayed a parabolic curve during aging while reduced in long-lived individuals. A series of human cerebral cortex frozen tissues of different ages were used for WB detection with various indicated antibodies (B) and real-time PCR (D). Data are represented as means ± SEM; n = 3 per group. *P < 0.05 and **P < 0.01 versus the 20 to 50 group; two-way ANOVA and Bonferroni’s post hoc test. The LAP/LIP isoform ratios were calculated from quantification by immunoblots of (B) (C). Data are represented as means ± SEM; n = 3 per group. **P < 0.01; two-way ANOVA and Bonferroni’s post hoc test. (E) IF costaining on human brain sections. Colocalization of C/EBPβ, FOXO1, p-C/EBPβ, p-IR, and TUNEL in GAD67+ neurons from different ages’ human cerebral cortexes. Confocal immunofluorescence microscopy was performed in human cerebral cortex. Scale bars, 40 μm. The image shown is representative of immunofluorescence labeling performed in three individuals. (F) Quantification of (E). Data are represented as means ± SEM; n = 6 per group. **P < 0.01 versus the 20 to 50 group; Mann-Whitney U test with multiple testing correction by Holm’s method.
C/EBPβ/AEP pathway plays an essential role in driving AD pathogenesis (23, 40). IF costaining showed that, as the disease progressed from Braak I to VI stages, C/EBPβ was steadily higher and FOXO1 was gradually lower. Moreover, AEP and its truncated Tau N368 echoed the upstream C/EBPβ activities in both GABAnergic and glutamatergic neurons. GAD67 signals were progressively lower, but, by contrast, vGluT1 levels remained relatively constant, indicating selective loss of GABAnergic neurons in human AD brains. In alignment with this finding, GAD67-positive neurons displayed approximately threefold higher TUNEL signals as compared to vGluT1-positive neurons (fig. S13). Hence, C/EBPβ/AEP is progressively escalated from Braak I to VI during aging, and FOXO1 was gradually lower, conceivably leading to GABAnergic neuronal degeneration.
DISCUSSION
In this study, we found an evolutionarily conserved signaling link between the C/EBPβ/AEP pathway and the REST/FOXO transcription factors that mediates neural excitation and shortens life span in mice and nematodes. Identification of the REST/FOXO proteins as major repression targets of C/EBPβ suggests that this essential transcription factor plays crucial roles in diverse biological processes including learning and memory and longevity during aging. The transcription factor REST appears to be up-regulated in humans with extended longevity and represses excitation-related genes. REST-deficient mice exhibit increased cortical activity and neuronal excitability during aging (6). Similarly, loss-of-function mutations in the C. elegans REST ortholog genes, spr-3 and spr-4, elevate neural excitation and reduce the life span of long-lived daf-2 mutants (6). Our results suggest that DAF-16 acts downstream of neuronal CEBP-2 (CCAAT/enhancer-binding protein 2) or LGMN-1, because neuronal overexpression of CEBP-2 or LGMN-1 reduces the life span of long-lived animals in which DAF-16 has been overexpressed in neurons. Notably, neuronal C/EBPβ represses both FOXO and REST and selectively triggers GABAnergic neuronal loss, resulting in neural excitation and short life span. Deletion of C/EBPβ rescues the longevity in both mice and worms (Figs. 1 and 5). Although C/EBPβ that dictates a plethora of important gene expression is escalated in neurons in an age-dependent manner, conceivably, there is the caveat for neural overexpression as an experimental approach recapitulating aging. Numerous other factors, which are independent of neural C/EBPβ, contribute to the aging process. In alignment with this view, the gene expression profiles in the brains that are associated with human longevity reveal that there is no strong, consistent association between CEBPB expression and REST or FOXO1. This is in general agreement with the lack of enrichment of CEBPB gene sets listed in the ENCODE (The Encyclopedia of DNA Elements) ChIP sequencing supplemental tables from the report (6). By contrast, LGMN displays a tight correlation of down-regulated genes and inverse association with up-regulated genes, in addition to REST and FOXO1 (figs. S6 and S7).
Reddy et al. (41) identified CEBP-2 as the ortholog of human C/EBPγ. In this study, C/EBPγ was used as a template to BLAST (Basic Local Alignment Search Tool) the C. elegans genome. However, when CEBP-2 (NP_871835.1) is blasted with C/EBPβ (NP_005185.2) and C/EBPγ (NP_001797.1), respectively, it shares 48% amino acid identity with human C/EBPβ and 46% amino acid identity with human C/EBPγ. There are 6 amino acids where C/EBPβ but not C/EBPγ is the same as CEBP-2, and 10 amino acids where C/EBPγ but not C/EBPβ is the same as CEBP-2 (fig. S14). In addition, RNAi of cebp-2 in C. elegans reduced Oil Red O fat staining (42), the same as we observed (Fig. 5). Moreover, C/EBPβ but not C/EBPγ has a role in adipocyte differentiation (43). Thus, these findings support that C/EBPβ but not C/EBPγ isoform might be more appropriate to act as a mammalian ortholog of C. elegans cebp-2.
The mTORC1 (mammalian target of rapamycin complex 1) pathway integrates intracellular nutrient and energy availability with growth factor and/or hormonal signals. Inhibition of the mTORC1 pathway in response to CR (calorie restriction) is a key event that mediates the health- and life-span–extending effect of CR. mTORC1 controls the translation of the Cebpb mRNA into its two functionally different protein isoforms, LAP (transcriptional activator) and LIP (transcriptional inhibitor), adjusting metabolic gene regulation downstream of C/EBPβ to energy/nutrient availability (44). mTORC1 stimulates the expression of LIP through the second AUG translation initiation site. Hence, inhibiting mTORC1 activity results in lower LIP expression and thereby in reduced inhibitory C/EBPβ function, leading to C/EBPβ transcriptional activity increase (16). In Cebpb∆uORF mice, LIP expression is abrogated. Consequently, the C/EBPβ transcriptional function is increased (C/EBPβ super-mice). Markedly, female C/EBPβ super-mice show increased median life span by 20% (15). Furthermore, a mouse model that only expresses the LIP isoform but lacks LAP isoform expression displays an earlier onset of aging-associated tumorigenesis and a reduction in life span (45).
In addition, the CebpbΔuORF expressed LAP throughout the body, but Thy1-C/EBPβ mice only expressed C/EBPβ with LAP much more than LIP in the neurons. In alignment with our current observations, recent studies support that C/EBPβ also is involved in aging. For instance, impaired DNA demethylation of C/EBP sites causes premature aging and enhanced methylation can affect aging, implying that C/EBP proteins play an unexpected role in this process. Specifically, aberrant DNA methylation affects aging via C/EBPβ (16, 17). Moreover, reduced expression of C/EBPβ-LIP extends health and life span in mice. Müller et al. (15) showed that LIP levels increase upon aging in WT mice and restriction of LIP expression sustains health and fitness in female but not male mice. This study indicates that LIP somehow shortens the life span of female mice (15). In our current study, we demonstrate that LAP neuronal overexpression shortens the life span in both male and female Thy1-C/EBPβ Tg mice and C/EBPβ knockdown increases life span via manipulating REST/FOXOs in the brain. Thus, both LAP and LIP affect the aging process, although these two isoforms may have opposite transcriptional functions, e.g., LAP activates some gene expression, whereas LIP inhibits others. The different effects on aging from LAP and LIP might be due to the tissue specificity.
A key conclusion of our study is that the C/EBPβ/AEP-REST/FOXO signaling link is well conserved. We find that extended longevity and cognitive preservation in humans is associated with coordinate down-regulation of both C/EBPβ and AEP. In the model system of C. elegans, an increase in the activity of neurons, a normal aspect of aging, is observed in worms with neuronal overexpression of cebp-2 or lgmn-1 (Fig. 5). Nematodes in which lgmn-1 or cebp-2 is overexpressed in neurons demonstrate early onset in the development of age-associated oily droplets and are defective in learning and memory versus control nematodes (Fig. 5 and figs. S8 and S9). Loss-of-function mutations in daf-2 extends life span through activation of DAF-16 (46). Here, we show that neural overexpression of cebp-2 or lgmn-1 shortens the life span in both daf-2 mutant and DAF-16 overexpressing worms by repressing daf-16 transcription. It is well established that loss of function of daf-2 elongates life span via reduced phosphorylation and consequently increased nuclear translocation of DAF-16 (47). In the current study, we demonstrate that increased levels of CEBP-2 decrease life span via repressing daf-16 mRNA transcription, leading to DAF-16 deficiency in both DAF-2 mutant and DAF-16 overexpressed strains (figs. S8 and S9). The characterization of the C. elegans orthologs CEBP-2 and LGMN-1 broadens this pathway’s function beyond the control of AD pathologies to the regulation of life span. A previous study shows that C/EBPβ plays a predominantly inhibitory role on transcription in the normal CNS, because deletion of C/EBPβ up-regulates mRNA levels (48). In alignment with these observations, our finding reveals that C/EBPβ represses both REST and FOXO expression in neurons and triggers GABAnergic neuronal loss, leading to neuroexcitation and short life span. Notably, C/EBPβ, p-C/EBPβ, and FOXO1 protein levels exhibit the inverse correlation in 70- to 80-year-old human brains. Now, it remains unclear whether FOXO1 down-regulates C/EBPβ in centenarian human brains or not. Because C/EBPβ and FOXO1 mutually repress each other (Fig. 4), conceivably, the escalated FOXO1/REST may suppress C/EBPβ in the long-lived human brains, leading to a parabolic expression curve (Fig. 6). Noticeably, C/EBPβ binds to FOXO1 and antagonizes its transcriptional functions (49). Moreover, both FOXO1 and C/EBPβ can be polyubiquitinated by ubiquitin E3 ligase COP1 (constitutive photomorphogenic protein 1). Most recently, it has been reported the COP1 triggers C/EBPβ degradation in microglia, attenuating Tau pathology spreading in AD (50).
Neurodegenerative diseases target specific neuronal populations. GABAnergic interneuron dysfunction, in particular, is found in a range of neurological and psychiatric disorders, including AD (51). Loss of GABA, the primary inhibitory neurotransmitter in the brain, is a key component of AD. Loss of GABA and GABAnergic interneurons in patients with AD may be responsible for network hyperactivity manifesting as seizures (52). Substantial evidence shows that loss of GABAnergic tone leads to seizures and 10 to 22% of patients with AD exhibit seizures (53), as do hAPPFAD mice, and the onset of these seizures precedes cognitive decline (54). Accordingly, we show that GABAnergic but not glutamatergic neurons appear selectively and progressively degenerated from Braak I to VI stages in human AD brains, coupled with gradually higher of C/EBPβ/AEP activities (fig. S13). This is consistent with our previous reports that C/EBPβ/AEP signaling drives AD pathogenesis (23, 40). Note that both C/EBPβ LAP and LIP were up-regulated by Aβ in 12-month-old 5xFAD as compared to WT littermates (fig. S1I).
To investigate in-depth the roles of C/EBPβ/AEP pathway in brain aging and longevity control and to understand the mechanisms that underlie age-associated brain pathophysiologies will provide insight into the molecular pathologies of neurodegenerative diseases and normal aging. Together, our study supports that the well-conserved C/EBPβ/AEP pathway in neurons from worms to mice to human acts as a master regulator in modulating cognitive decline during aging, exacerbated by pathological stresses in AD, and dictating life span.
MATERIALS AND METHODS
Animals
For the generation of Thy1-human C/EBPβ Tg mouse, mouse genomic fragments containing homology arms (HAs) were amplified from the bacterial artificial chromosome clone by using high-fidelity Taq and were sequentially assembled into a targeting vector together with recombination sites and selection markers. After confirming correctly targeted embryonic stem clones via Southern blotting, we selected some clones for blastocyst microinjection, followed by founder production. Founders were confirmed as germline-transmitted via crossbreeding with WT. In the end, male and female F1 heterozygous mutant mice were confirmed as the final deliverables for this project. The genotypes of Tg mice were validated by PCR. (See the details of transgenic gene from file S1). The C/EBPβ knockout mice on a C57BL/6J background were generated as reported (55). 5XFAD mice on a C57BL/6J background were obtained from The Jackson Laboratory (stock no. 006554). The animals were assigned to different experimental groups based on the litter and gender in a way that every experimental group had similar number of sibling males and females. Animal care and handling were performed according to the National Institutes of Health (NIH) animal care guidelines and the Declaration of Helsinki and Emory Medical School guidelines. The protocol was reviewed and approved by the Emory Institutional Animal Care and Use Committee.
C. elegans strains
Nematode strains were grown on NGM (nematode growth media) plates using standard methods and were maintained at 20°C unless otherwise noted. The following strains were obtained from the Caenorhabditis Genetics Center: N2 (WT, Bristol) (Brenner, Genetics, 77, 71-94, 1974); OH15265 [otIs672 (rab-3::NLS::GCaMP6s + arrd-4:NLS:::GCaMP6s)]; hypersensitive neuronal RNAi strain TU3335[lin-15B(n774)X; uIs57 [unc-119p::YFP + unc-119p::sid-1 + mec-6p::mec-6]]; CF2093 [daf-16(mu86) I; daf-2(e1370) III; muIs131 [unc-119p::GFP::daf-16 + rol-6(su1006)]]; daf-2 (e1370)ts, maintained at 15°C; and WB141 [pat-6(st561) IV; and zpEx99 [pat-6::GFP + rol-6(su1006)]]. The following strains were made during the course of this study: GB320 to GB322 (three independent lines) [sfEx64-66 [unc-119p::HA::lgmn-1 + sur-5p::sur-5::GFP]]; GB323 to GB325 (three independent lines) [sfEx67-69 [myo-3p::HA::lgmn-1 + sur-5p::sur-5::GFP]]; GB326 [sfEx70 [unc-119p::myc::cebp-2 + sur-5p::sur-5::GFP]]; GB327 [sfEx71 [myo-3p::myc::cebp-2 + sur-5p::sur-5::GFP]]; GB328 [lin-15B(n744) X; uIs57; sfEx64 [unc-119p::HA::lgmn-1 + sur-5p::sur-5::GFP]]; GB329 [lin-15B(n744) X; uIs57; sfEx70 [unc-119p::myc::cebp-2 + sur-5p::sur-5::GFP]]; GB330 [otIs672; sfEx72 [unc-119p::HA::lgmn-1 + sur-5p::mCherry]]; GB331 [otIs672; sfEx73 [unc-119p::myc::cebp-2 + sur-5p::mCherry]]; GB332 [muIs131 [unc-119p::GFP::daf-16 + rol-6(su1006)]]; GB333 [daf-2(e1370); sfEx64 [unc-119p::HA::lgmn-1 + sur-5p::sur-5::GFP]]; GB334 [daf-2(e1370); sfEx70 [unc-119p::myc::cebp-2 + sur-5p::sur-5::GFP]]; GB335 [muIs131 [unc-119p::GFP::daf-16 + rol-6(su1006)]]; sfEx64 [unc-119p::HA::lgmn-1 + sur-5p::sur-5::GFP]]; GB336 [muIs131 [unc-119p::GFP::daf-16 + rol-6(su1006)]; and sfEx70 [unc-119p::myc::cebp-2 + sur-5p::sur-5::GFP]].
Beginning with CF2093, we crossed out the daf-2(e1370) and daf-16(mu86) mutations to generate strain GB332, noted above. To follow the progress of these crosses and identify the mutations by PCR and sequencing, we used the followed primers: daf-2_e1370-1: atgattcatcaatgcgtactc, and daf-2_e1370-2: ggaagctaatattatgaagaac; daf-16_mu86-1: acatagacgatttcgaaaagttc; daf-16_mu86-2: aacacgagacgacgatccagg; and daf-16_mu86-3: ctcttggaagatttcatcagg. To create GB328 and GB329 in which the neuronal overexpression of lgmn-1 and cebp-2 occurs in the TU3335 background (for neuronal RNAi), we followed the presence of the lin-15B(n744) mutation in TU3335 by using PCR and sequencing with primers Lin-15b_n744-1: ctacattgacaatgaaacattc and Lin-15b_n744-2: tcggtggaggtgctgcacgag, and we also followed the presence of uIs57 by observing neuronal-YFP.
Transgenic nematode lines
The following primers were used to generate segments inserted into plasmids for microinjection to create transgenics: Unc-119p-2: cgcgctagcctgaaaattttggggattatggg; Unc-119p-3: aattgttttgtgccaagcttcagtaaaag; T28H10.3-1: gcggatatcatgagaccacttgctcttttaatttg; T28H10.3-2: cgcgtcgacttacaagaactccttctccaaacg.
We used pPD49.83, which is a heat shock promoter expression vector (provided by A. Fire, Stanford University), and substituted the heat shock promoter with the unc-119 promoter to drive expression in all neurons to create a different plasmid called pPD-u119. To create this unc-119 promoter, we PCR-amplified from genomic DNA [2180 base pairs (bp)] using primers Unc-119p-2 and Unc-119p-3. To create a plasmid that would direct expression of lgmn-1 in all neurons, pPD-u119-HA-lgmn-1, we first PCR-amplified cDNA from the RB2 cDNA library (provided by R. Barstead, Oklahoma Medical Research Foundation) for lgmn-1 with primers T28H10.3-1 and T28H10.3-2 and inserted the fragment into pKS-HA8(Nhex2) to generate the plasmid pKS-HA-lgmn-1, and then the Nhe I fragment was excised and ligated into pPD-u119. To create a plasmid that would direct expression of lgmn-1 in body wall muscle cells, the same Nhe I fragment from pKS-HA-lgmn-1 was ligated into pPD95.86 (myo-3 promoter plasmid). To generate a plasmid that would express cebp-2 in all neurons, we first ordered chemical synthesis of a full-length cDNA for cebp-2 and cloned it into pBS-myc, and then the Spe I fragment of pBS-myc-cebp-2 was excised and ligated into pPD-u119. To generate a plasmid that would express cebp-2 in body wall muscle cells, the same Spe I fragment of pBS-myc-cebp-2 was excised and ligated into pPD95.86 (myo-3 promoter plasmid). These various plasmids were supplied to SunyBiotech Corporation for microinjection and generation of the transgenic lines described above, using as host strain N2 WT for extrachromosomal arrays sfEx64 to sfEx71 (and using the transformation marker sur-5p::sur-5::GFP) and using as host strain OH15265 for extrachromosomal arrays sfEx72 and sfEx73 (and using transformation marker sur-5p::mCherry).
Human samples
Postmortem brain samples were dissected from paraffin-embedded or frozen brains of AD cases and nondemented controls from the Rush Alzheimer’s Disease Center and the Emory Alzheimer’s Disease Research Center. The former samples were all from the Religious Orders Study (ROS) (56): three ages 70 to 80 and three ages 100+. ROS was approved by the Institutional Review Board of Rush University Medical Center. All participants signed an informed consent, Anatomic Gift Act, and repository consent to allow their resources to be shared. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. AD was diagnosed by the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association criteria (57). See the information in table S1 for details.
Primary cultured neurons
Briefly, primary cortical neurons were isolated from E18 Sprague-Dawley rats. Brain tissues were dissected, dissociated, and incubated with 5 ml of D-Hanks containing 0.25% trypsin for 5 min, centrifuged at 1000g for 5 min after addition of 4 ml of the neuronal plating medium containing Dulbecco’s modified Eagle’s medium/F12 with 10% fetal bovine serum. Then, the cells were resuspended, about 5 × 105 cells were plated onto each well of 12-well plates for WB, and 1 × 105 cells were plated onto each glass coverslip for cell imaging. The neurons were then put into a humidified incubator with 5% CO2 at 37°C. The medium was changed to neurobasal medium supplemented with 2% B27 (maintenance medium) after 2 to 4 hours. Neurons at 11 to 13 DIV were treated with viruses or drugs. LV (lentivirus)-C/EBPβ (titer: 5 × 109 IU/ml), LV–sh-C/EBPβ (titer: 3 × 109 IU/ml), and LV-vector (titer: 5 × 109 IU/ml). The virus was packaged by the Viral Vector Core (VVC) of Emory University. AAV2 (adeno-associated viruses 2)-FOXO1 (titer: 0.5 × 1013 GC (genome copies)/ml) and AAV2–sh-FOXO1 (titer: 0.5 × 1013 GC/ml) were purchased from GeneCopoeia (AA02-Rn23466-AV01-200 and AA02-RSH055426-AV03-200). A total of 1.5 μl of virus was added to 1 ml of culture medium and was applied to primary neurons.
Luciferase assay
SH-SY5Y cells were seeded in 12-well plates and transfected with REST and FOXO1 Gaussia luciferase plasmid (HPRM46237 and HPRM44029, GeneCopoeia), treated with/without overexpressing plasmids or small interfering RNA. Forty-eight hours later, the cells were harvested in passive lysis buffer and analyzed using the Secrete-Pair Gaussia Luciferase Assay Kit (LF061, GeneCopoeia) on a microplate reader. The experiments were performed in triplicate.
Chromatin immunoprecipitation
SH-SY5Y cells were fixed with 1% formaldehyde for 10 min, and cross-linking was quenched with 2 M glycine for 5 min. After nuclei isolation, the chromatin was sonicated using a Covaris S220 sonicator to obtain the desired DNA fragment size (~500 bp). The sonicated chromatin was precleaned by two rounds of centrifugation with maximum speed at 4°C. A total of 5 μl of anti-C/EBPβ (C19, Santa Cruz Biotechnology) or 5 μl of anti-IgG (ab133470, Abcam) was used to precipitate the chromatin fragments that contain DNA-protein cross-linking ChIP samples, which was collected with 50 μl of Dynabeads protein A (Invitrogen, 10001D). The enrichment of specific DNA sequences was examined by PCR using primer pairs of the different species ApoE promoters. PCR assay also detected each input sample and internal reference β-actin. The following primers were used: hREST-1409, ACTGTGTAGAGTGTTCCTTG (forward); hREST-1189, CCTTAGTGTGGCGTTCAA (reverse); hFOXO1-1587, CACAGCAAGTAGCCTCAA (forward); hFOXO1-1357, TCCGTCCACTAAGTCCAG (reverse).
Electrophoretic mobility shift assay
Nuclear proteins of SH-SY5Y were extracted by NE-PER Nuclear and Cytoplasmic Extraction Reagents (Life Technologies). Double-stranded oligonucleotide probe for REST/FOXO1 promoter or its mutations was labeled with biotin. Unlabeled probes were used as cold competitors. EMSA was performed according to the manufacturer’s protocol of the LightShift Chemiluminescent EMSA Kit (Life Technologies).
Real-time PCR
RNAs from cells and tissues were isolated with TRIzol. After reverse transcription with SuperScriptIII reverse transcriptase, real-time PCR reactions were performed using the ABI 7500-Fast Real-Time PCR System. Gene-specific primers and the TaqMan Universal Master Mix Kit were designed and bought from Taqman. All kits and reagents were purchased from Life Technologies. The 2−𝚫𝚫Ct method was used for the relative quantification of gene expression. For each data point, at least two duplicated wells were used. Glyceraldehyde phosphate dehydrogenase was used for housekeeping gene control.
Insulin enzyme-linked immunosorbent assay
Brain samples were rapidly weighed and homogenized at maximum speed for 1 min in nine volumes of medium containing 10 mM Mops (pH 7.6), 120 mM NaCl, 1 mM EDTA, 0.1 mM benzethonium chloride, 1 mM benzamidine, and 0.1% Trasylol. The supernatant was collected by centrifuging at 2000 rpm for 20 min at 4°C and then detected by insulin enzyme-linked immunosorbent assay kit (RAB0327, Sigma-Aldrich).
Western blotting
The tissues or cells were washed with ice-cold phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay buffer [20 mM tris-HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 2.5 mM sodium pyrophosphate, 1% NP-40, 1% sodium deoxycholate, 1 mM Na3VO4, and 1 mM β-glycerophosphate] with protease inhibitor cocktail for 20 min on ice. The supernatant was collected by centrifuging at 14,000 rpm for 20 min at 4°C. Then, the protein extract was diluted to 5 mg/ml. After electrophoresis, the samples were incubated overnight at 4°C with the recommended amount of antibody, followed by detection of immunoblotting. See details of the antibodies from table S2.
Immunofluorescence
Transfected/treated cells or mice brain slices or human tissues were fixed and incubated for 24 to 48 hours at 4°C with primary antibodies followed by 1 hour at 37°C with Alexa Fluor 568–, Alexa Fluor 488–, or Alexa Fluor 647–conjugated secondary antibodies (Invitrogen). 4′,6-Diamidino-2-phenylindole (1 μg/ml) (Sigma-Aldrich) was used for the nuclei staining. See details of the antibodies in table S2. Images were acquired through a confocal microscope (Olympus FV1000). To quantify the fluorescent intensities, we split multicolor images into the single channels, converted the single-channel color images to 8-bit gray scale, and then copied the image to create a binary image. Next, a region of interest around the object was drawn and highlighted. The background was subtracted with rolling ball to create a binary version of the image with only two pixel intensities: black = 0 and white = 255. After setting measurements and the “Redirect to” line to the name of the copy of the image, the image fluorescent intensities on the binary image were analyzed.
C. elegans life-span analysis
Each experiment was carried out over several plates such that, for an experiment with n = 30, three plates containing 10 nematodes were used. During life-span analysis, C. elegans were observed daily for movements. If no movement was detected, then nematodes were prodded gently with a toothpick and examined for pharyngeal pumping to determine whether alive. Worms that escaped from the plates or exploded were censored. Statistical analysis (log-rank, Mantel-Cox) was carried out using OASIS 2 (Online Application for Survival Analysis 2) statistical software (https://sbi.postech.ac.kr/oasis2/).
Appetitive olfactory learning in response to butanone
The butanone learning assay was performed as previously reported (58) in a climate-controlled room at 20° ± 0.5°C and 35 to 55% relative humidity. Briefly, synchronized 1-day-old hermaphrodites were collected into a 15-ml conical tube. After three washes, half of the population was immediately tested for naive CTX toward butanone, while the rest was kept for 1 hour in the tube. Next, these starved worms were transferred to 100 mm NGM conditioning plates seeded with 1 ml of OP50 Escherichia coli bacteria, and 2 ml of 10% butanone (diluted in 100% ethanol) was pipetted on the lid. After 1 hour, a proportion of the conditioned worms was tested for CTX toward butanone (T0), whereas the rest of the population was placed on 100-mm NGM hold plates seeded with 1-ml OP50 for 60 or 120 min. At the end of each interval, the CTX toward butanone for the specific resting group was quantified (T60 and T120). CTX was tested on 100-mm unseeded NGM plates where worms were pipetted at the origin spot. Worms were given a choice between two equidistant spots containing 1 μl of NaN3 + 10% butanone and 1 μl of NaN3 + ethanol, respectively. After 1 hour, worms were counted, and the CI was calculated as follows: CI = (no. of worms at the butanone spot − no. of worms at the EtOH spot)/(total no. of worms − no. of worms at the origin spot).
Oil Red O staining
Oil Red O staining was performed as previously described (59). After fixation, worms were resuspended and dehydrated in 60% isopropanol. Approximately 250 μl of 60% Oil Red O stain (catalog no. O9755, Sigma-Aldrich, St. Louis, MO, USA) was added to each sample overnight at room temperature.
C. elegans RNAi
Feeding RNAi was performed on NGM plates supplemented with ampicillin (100 μg/ml) and 2 mM isopropyl-β-d-thiogalactopyranoside and E. coli strain HT115 (DE3) cells that express double-stranded RNA by L4440 vector (60) for portions of the cDNAs for lgmn-1 and cebp-2. Worms were transferred to fresh plates every day.
C. elegans treatments
Worms were treated as follows with the C11 (inhibitor of AEP) in the following way. Worms were synchronously grown to early adult stage and placed in NGM plates containing 10 μM C11. Worms were transferred to fresh plates every day.
Thrashing motility assays
Worms were synchronously grown to early adult stage and placed in individual wells of a 96-well microtiter plate containing 50 μl of M9. After a 10-min exposure period to M9, thrashes were counted at 21°C for 30 s. A single thrash was defined as a complete change in the direction of bending at the mid body. During manual thrash counts, the data for a particular worm were dropped if the worm remained still for >10 s or if the worm was visibly damaged.
Confocal imaging
GCaMP imaging in C. elegans
GCaMP imaging was performed in lines OH15265 [otIs672 (rab-3::NLS::GCaMP6s + arrd-4:NLS:::GCaMP6s)], GB330 [otIs672; sfEx72 [unc-119p::HA::lgmn-1 + sur-5p::mCherry]], and GB331 [otIs672; sfEx73 [unc-119p::myc::cebp-2 + sur-5p::mCherry]], which expresses green fluorescent protein (GFP) in all the neurons. Worms were removed from plates and mounted on 7.5% agarose pads in liquid NGM, mixed 1:1 with 0.05 μm of polystyrene beads (Polysciences Inc., catalog no. 08691). A coverslip was very gently applied, and worms were imaged for a maximum of 30 min after mounting. Imaging was recorded on the confocal microscope (FV1000).
GABA staining
GABA staining was performed as previously described (61). Young adult hermaphrodites were fixed and washed three times in PBS/0.5% Triton X-100. Then, the worms were incubated for 1 hour at 4°C in a freshly made solution of PBS and NaBH4 (1 mg/ml) (Sigma-Aldrich, 71321) to quench the autofluorescence due to the glutaraldehyde. Samples were blocked for 30 min at room temperature with 0.2% gelatin from fish (Sigma-Aldrich). Anti-GABA antibodies (Abcam, ab17413) were used at a 1:250 dilution. For triple labeling, anti-GFP (sc-9996, Santa Cruz Biotechnology) and anti-mCherry (ab167453, Abcam) were used at a 1:1000 dilution. Incubations were done overnight at 4°C. Secondary antibodies included Alexa Fluor 488–labeled goat anti-mouse (A10680, Thermo Fisher Scientific), Alexa Fluor 594–labeled donkey anti-rabbit (A21207, Thermo Fisher Scientific), and Cy5 (cyanine dye 5)-labeled goat anti–guinea pig (GTX26567, GeneTex).
AEP activity assay
C. elegans tissue homogenates (20 μg) and 20 μM AEP substrate Z-Ala-Ala-Asn-AMC (4-methyl-7-coumarylamide) (Bachem) were incubated with 200-μl reaction buffer, which contained 60 mM Na2HPO4, 20 mM citric acid, 1 mM EDTA, 1 mM dithiothreitol, and 0.1% CHAPS (pH 5.5) at 37°C. Then, the AMC released fluorescence by substrate and cleavage was measured in a fluorescence plate reader in kinetic mode at 460 nm.
Mice life-span studies
One-month-old male or female Thy1-C/EBPβ Tg or C/EBPβ+/− mice and their littermates were obtained and grouped. Mice were housed four to five per cage. After that, no further operations were performed on these mice except for checking their cages. Some of the mice were moved from their original cages during the course of the study to minimize single cage housing stress. The mice activity and status were observed every day; each mouse’s date of death was recorded.
Calcium imaging
Mice were decapitated after deep anesthesia with inhalation of saturated isoflurane. The forebrain was obtained and immediately placed in an ice-cold artificial cerebrospinal fluid (ACSF) containing the following: 125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, 20 mM d(+)-glucose, 2 mM CaCl2, and 1.3 mM MgCl2. The solution was bubbled with 5% CO2 balanced with 95% O2 to obtain pH 7.40. Coronal sections (250 to 350 μm thickness) containing the hippocampus were obtained using a vibratome sectioning system. Fura-2 of the cell membrane permeable acetoxymethyl (AM) ester form (F1221; Invitrogen, Carlsbad, CA) was first dissolved in dimethyl sulfoxide supplemented with the surfactant Pluronic F-127 [20% (w/v); P3000MP, Invitrogen] at 10 mM and then diluted into oxygenated ACSF at a final concentration of 50 μM. After slicing, hippocampal slices were exposed to the Fura-2 AM solution for an hour and washed in oxygenated ACSF for at least an hour at room temperature. The slices were kept in the dark at room temperature afterwards until imaging experiments for up to 5 hours.
The slices loaded with Fura-2 were then imaged with a two-photon microscope (Scientifica HyperScope, Brambleside, UK) equipped with galvo mirrors and a tunable ultrafast laser (Spectra-Physics InSight X3) through 16× 0.8 numerical aperture water-immersion objective lens (Nikon CFI75 LWD 16X W) and a green emission filter (525 ± 25 nm bandpass) (62, 63). Images were obtained at ~1 Hz and averaged over 10 frames from at least three different locations in DG, CA3, and CA1 regions from at least three slices per animal at two excitation wavelengths of 700 and 760 nm. The regions of interest were set in somata in an automated fashion by thresholding raw fluorescence images at 760 nm. Fura-2–excitation ratio, 700 nm/760 nm, which directly reflected [Ca2+]i levels, was calculated after background subtraction from off-focus regions in the same field using a scientific and engineering software (IgorPro 8, WaveMetrics, Lake Oswego, OR). Imaging experiments and analysis were performed blindly to the genotypes of animals.
Small animal PET
Inhalation anesthesia with isoflurane was used, and the eyes of the animals were protected with dexpanthenol eye ointment. Anesthesia was initiated 15 min ahead of experiments by placing the animal in a cage ventilated with oxygen (3.5 liter/min) containing 3% isoflurane. Throughout the experiments, anesthesia was maintained by adjusting the isoflurane content (0.6 to 2%) to ensure a respiratory rate in the range 80 to 100/min. For intravenous injection, we inserted a tailored catheter into the lateral tail vein and administered a slow bolus injection of 50 to 200 μl of tracer solution. The radioactivity in the syringe was measured immediately before and after injection with a Capintec CRC 15R (Capintec Inc., 6 Arrow Road Ramsey, NJ, USA) dose calibrator. After tracer injection, we flushed the catheter with 50 to 100 μl of isotonic sodium chloride solution. At 60 min after administration, the animals were euthanized, and brains were extracted. Brains were chilled and placed on a plastic tray and then imaged for 60 min in three-dimensional list mode on a Siemens Inveon system (axial field of view of 12.7 cm with a bore diameter of 12 cm, approximately 1.4-mm full width at half-maximum spatial resolution; Siemens Healthcare, Erlangen, Germany). Individual brain PET images were registered to a T2-weighted magnetic resonance imaging mouse atlas (USC Laboratory of NeuroImaging). Data are reported as the standardized update value [% ID (injected dose)/g × body weight]. All data are expressed as means ± SEM from three or more independent experiments, and the level of significance between two groups was assessed with Student’s t test.
Seizure induction and analysis
Mice were provoked with a convulsive dose (35 mg/kg, i.p.) of the GABAA receptor antagonist PTZ (catalog no. 18682, Cayman Chemical) to measure seizure susceptibility and evaluate seizure thresholds. Following PTZ administration, latency to generalized tonic-clonic seizures (GTCs), number of seizures, and total duration of GTCs were recorded for each mouse. Mice without seizures were assigned a time of 20 min at the end of the PTZ challenge observation period.
Open field
The open field apparatus is a rectangular arena (96.5 cm in diameter) with opaque gray plexiglass walls (28 cm high). Mice will be placed individually in the center of the inner circle and allowed to roam freely about the apparatus for 5 min. The amount of time spent in the middle of the open field and the total distance traveled in the open field will be measured via an overhead tracking system (TopScan, CleverSys).
Prepulse inhibition
PPI is used to measure the deficit in the normal inhibition of the startle reflex by a prestimulus observed in schizophrenia patient and will be assessed in a sound-attenuated chamber (Med Associates). Mice will be allowed to habituate to the chamber, the 70-dB background white noise, and to the prepulses (20 ms of white noise at 75, 80, or 85 dB) and auditory-evoked startle stimuli (120 dB, 20 ms) for 5 min. In the PPI test, mice will be subjected to 12 startle trials (120 dB, 20 ms) and 12 prepulse/startle trials (20 ms of white noise at 75, 80, or 85 dB at 100-ms intervals and 20 ms of 120-dB startle stimulus). Each of the trial types will be presented in a pseudo-random fashion such that each trial will be presented 12 times and no two consecutive trials will be identical. Mouse movement will be measured by a piezoelectric accelerometer during 100 ms after startle stimulus onset for 100 ms. PPI (%) will be calculated according to the formula: [100 − (startle amplitude on prepulse-pulse trials/startle amplitude on pulse alone trials) × 100]. At the end of the session, subjects will be removed from the sound-attenuated chamber and will be placed in their home cage.
Forced swim test
Mice will be forced to swim and videotaped for 6 min in 20 cm of fresh 25°C water in a 4-liter beaker (18 cm in diameter). In this test, struggling, swimming, and immobile floating behavior is typically observed, with immobility considered as a “depression-like” phenotype. A mouse will be considered immobile when it is only making movements necessary to remain floating. Subtle movements of the feet, tail, or head required to maintain the eyes, ears, and nose above the surface of the water will be excluded as immobility. The videotapes will be scored for latency to immobility and total time spent immobile. The mice will then be removed from the water, dried, and returned to their home cage. The cage will be placed half-on and half-off a heating pad until the mice dry off.
Sleep latency
Mice will be placed in plexiglass activity cages (with cobb bedding) for 4 hours, and video will be recorded. Latency to sleep will be noted for each subject.
Morris water maze
The water maze is a round, water-filled tank (132 cm in diameter), which was surrounded by extra-maze visual cues that was kept at the same position during the whole training time. The platform was placed in the northwest quadrant of the maze. Water was made with the right amount of fat-free milk powder, filled to cover the platform by 1 cm at 22°C. Each mouse was given 4 trials/day; the maximum trial length was 60 s for five consecutive days with a 15-min intertrial interval. If mice did not reach the platform in time, then they were manually guided to stay on the platform for another 10 s. After 5 days of task acquisition, a probe trial was presented. The platform was removed and the percentage of time spent in the quadrant was measured over 60 s. MazeScan (CleverSys) was used for analyzing all trial latency and swim speed.
Contextual fear conditioning
Mice were placed in a test chamber (7″ W, 7″ D 3 12″ H, Coulbourn) composed of plexiglass with a metal shock grid floor. After wiping with 70% alcohol, we allowed the mice to explore the enclosure for 3 min, following with three conditioned stimulus (CS)–unconditioned stimulus (US) pairings (tone: 2000 Hz, 85 db, 20 s; foot shock: 0.5 mA, 2 s) with a 1-min intertrial interval. One minute following the last CS-US presentation, mice were removed from the chamber. Twenty-four hours later, the mice were presented with a context test, during which subjects were placed in the same chamber used during conditioning on day 1; no shocks were given during the context test. The amount of freezing was recorded. On day 3, a cue test was performed, during which subjects were exposed to the CS in a novel compartment. Animals were allowed to explore the novel context for 2 min; the 85-db tone was presented later. The freezing behavior in the 6 min was recorded via a camera and the software provided by Coulbourn.
Quantification and statistical analysis
All data are expressed as means ± SEM from three or more independent experiments and analyzed using GraphPad Prism statistical software (GraphPad Software). All of the statistical details of experiments can be found in the figure legends for each experiment, including the statistical tests used, number of mice in animal experiments (represented as n, unless otherwise stated), number of wells in cell culture experiments (represented as n, unless otherwise stated), and definition of center (means). Sample size was determined by Power and Precision (Biostat). The level of significance between two groups was assessed with unpaired t test with Welch’s correction. For more than two groups, one-way analysis of variance (ANOVA) and Bonferroni’s multiple comparisons test were applied. The two-way ANOVA and Bonferroni’s post hoc test compared the differences between groups that have been split on two independent factors. A value of P < 0.05 was considered to be statistically significant.
Acknowledgments
We thank ADRC at Emory University for human patients with AD and healthy control samples (P30AG066511) and the Rush Alzheimer’s Disease Center for ROS samples (P30AG10161 and R01AG15819; ROS resources can be requested at www.radc.rush.edu).
Funding: This study was supported in part by the Rodent Behavioral Core (RBC), which is subsidized by the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the VVC of the Emory Neuroscience NINDS Core Facilities (P30NS055077). Further support was provided by Georgia Clinical and Translational Science Alliance of the National Institutes of Health under award number UL1TR002378. Some of the C. elegans strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by National Institutes of Health grant R01 AG065517 (to K.Y.), National Institutes of Health grant S10 OD021773 (to K.B.), National Natural Science Foundation of China (NSFC) no. 31771114 (to X.-C.W.), National Science Foundation (NSF) CBET-1512826 (to K.B.), National Institutes of Health grant P30AG010161 (to D.A.B.), National Institutes of Health grant P30AG072975 (to D.A.B.), National Institutes of Health grant R01AG015819 (to D.A.B.), National Institutes of Health grant R01AG017917 (to D.A.B.), and National Institutes of Health grant U01AG61356 [De Jager (Broad Institute), Bennett, MPI].
Author contributions: Conceptualization: K.Y. Methodology: K.Y., Y.X., H.Q., C.J.M., K.B., D.D., G.M.B., and D.A.B. Investigation: Y.X., H.Q., Z.-H.W., P.L., C.J.M., X.L., K.B., and D.D. Project administration: K.Y. and Y.X. Writing—Original draft: K.Y., Y.X., H.Q., and G.M.B. Writing—Review and editing: K.Y., Y.X., K.B., S.P.Y., D.D., D.A.B., X.-C.W., B.A.Y., H.Q., and G.M.B. Investigation: Y.X., H.Q., Z.-H.W., P.L., C.J.M., X.L., K.B., D.D., and K.X.Y.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Some frozen human brain tissue and slices come from the prefrontal cortex can be provided by Rush University Medical Center/Rush Alzheimer’s Disease Center pending scientific review and a completed material transfer agreement. Requests for the materials should be submitted to D.A.B.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Tables S1 and S2
Other Supplementary Material for this manuscript includes the following:
Supporting file S1
REFERENCES AND NOTES
- 1.Apfeld J., Kenyon C., Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402, 804–809 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Alcedo J., Kenyon C., Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41, 45–55 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Bishop N. A., Guarente L., Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447, 545–549 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Wolkow C. A., Kimura K. D., Lee M. S., Ruvkun G., Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290, 147–150 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Broughton S. J., Piper M. D. W., Ikeya T., Bass T. M., Jacobson J., Driege Y., Martinez P., Hafen E., Withers D. J., Leevers S. J., Partridge L., Longer lifespan, altered metabolism, and stress resistance in drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. U.S.A. 102, 3105–3110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zullo J. M., Drake D., Aron L., O’Hern P., Dhamne S. C., Davidsohn N., Mao C. A., Klein W. H., Rotenberg A., Bennett D. A., Church G. M., Colaiácovo M. P., Yankner B. A., Regulation of lifespan by neural excitation and REST. Nature 574, 359–364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen E., Bieschke J., Perciavalle R. M., Kelly J. W., Dillin A., Opposing activities protect against age-onset proteotoxicity. Science 313, 1604–1610 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Freude S., Hettich M. M., Schumann C., Stöhr O., Koch L., Köhler C., Udelhoven M., Leeser U., Müller M., Kubota N., Kadowaki T., Krone W., Schroder H., Brüning J. C., Schubert M., Neuronal IGF-1 resistance reduces Abeta accumulation and protects against premature death in a model of Alzheimer’s disease. FASEB J. 23, 3315–3324 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Cohen E., Paulsson J. F., Blinder P., Burstyn-Cohen T., du D., Estepa G., Adame A., Pham H. M., Holzenberger M., Kelly J. W., Masliah E., Dillin A., Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell 139, 1157–1169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moloney A. M., Griffin R. J., Timmons S., O’Connor R., Ravid R., O’Neill C., Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer’s disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiol. Aging 31, 224–243 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Kim S. Y., Webb A. E., Neuronal functions of FOXO/DAF-16. Nutr Healthy Aging 4, 113–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb A. E., Brunet A., FOXO transcription factors: Key regulators of cellular quality control. Trends Biochem. Sci. 39, 159–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boccitto M., Lamitina T., Kalb R. G., Daf-2 signaling modifies mutant SOD1 toxicity in C. elegans. PLoS One 7, e33494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pulido-Salgado M., Vidal-Taboada J. M., Saura J., C/EBPβ and C/EBPδ transcription factors: Basic biology and roles in the CNS. Prog. Neurobiol. 132, 1–33 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Müller C., Zidek L. M., Ackermann T., de Jong T., Liu P., Kliche V., Zaini M. A., Kortman G., Harkema L., Verbeek D. S., Tuckermann J. P., von Maltzahn J., de Bruin A., Guryev V., Wang Z.-Q., Calkhoven C. F., Reduced expression of C/EBPβ-LIP extends health and lifespan in mice. eLife 7, e34985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niehrs C., Calkhoven C. F., Emerging role of C/EBPβ and epigenetic DNA methylation in ageing. Trends Genet. 36, 71–80 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Schäfer A., Mekker B., Mallick M., Vastolo V., Karaulanov E., Sebastian D., von der Lippen C., Epe B., Downes D. J., Scholz C., Niehrs C., Impaired DNA demethylation of C/EBP sites causes premature aging. Genes Dev. 32, 742–762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S. S., Chen J. F., Johnson P. F., Muppala V., Lee Y. H., C/EBPbeta, when expressed from the C/ebpalpha gene locus, can functionally replace C/EBPalpha in liver but not in adipose tissue. Mol. Cell. Biol. 20, 7292–7299 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu C. H., Lin W. D., Huang S. Y., Lee Y. H., Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 18, 1970–1975 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shavlakadze T., Morris M., Fang J., Wang S. X., Zhu J., Zhou W., Tse H. W., Mondragon-Gonzalez R., Roma G., Glass D. J., Age-related gene expression signature in rats demonstrate early, late, and linear transcriptional changes from multiple tissues. Cell Rep. 28, 3263–3273.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Akar C. A., Feinstein D. L., Modulation of inducible nitric oxide synthase expression by sumoylation. J. Neuroinflammation 6, 12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strohmeyer R., Shelton J., Lougheed C., Breitkopf T., CCAAT-enhancer binding protein-β expression and elevation in Alzheimer’s disease and microglial cell cultures. PLoS One 9, e86617 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z. H., Gong K., Liu X., Zhang Z., Sun X., Wei Z. Z., Yu S. P., Manfredsson F. P., Sandoval I. M., Johnson P. F., Jia J., Wang J. Z., Ye K., C/EBPβ regulates delta-secretase expression and mediates pathogenesis in mouse models of Alzheimer’s disease. Nat. Commun. 9, 1784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortes-Canteli M., Aguilar-Morante D., Sanz-SanCristobal M., Megias D., Santos A., Perez-Castillo A., Role of C/EBPβ transcription factor in adult hippocampal neurogenesis. PLoS One 6, e24842 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortes-Canteli M., Luna-Medina R., Sanz-SanCristobal M., Alvarez-Barrientos A., Santos A., Perez-Castillo A., CCAAT/enhancer binding protein beta deficiency provides cerebral protection following excitotoxic injury. J. Cell Sci. 121, 1224–1234 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Mills D. J., The aging gabaergic system and its nutritional support. J Nutr Metab 2021, 6655064 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bi D., Wen L., Wu Z., Shen Y., GABAergic dysfunction in excitatory and inhibitory (E/I) imbalance drives the pathogenesis of Alzheimer’s disease. Alzheimers Dement. 16, 1312–1329 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Jinno S., Aika Y., Fukuda T., Kosaka T., Quantitative analysis of GABAergic neurons in the mouse hippocampus, with optical disector using confocal laser scanning microscope. Brain Res. 814, 55–70 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Pelkey K. A., Chittajallu R., Craig M. T., Tricoire L., Wester J. C., McBain C., Hippocampal GABAergic inhibitory interneurons. Physiol. Rev. 97, 1619–1747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E., Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Lehtinen M. K., Yuan Z., Boag P. R., Yang Y., Villén J., Becker E. B. E., DiBacco S., de la Iglesia N., Gygi S., Blackwell T. K., Bonni A., A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125, 987–1001 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Hilliard M. A., Apicella A. J., Kerr R., Suzuki H., Bazzicalupo P., Schafer W. R., In vivo imaging of C. elegans ASH neurons: Cellular response and adaptation to chemical repellents. EMBO J. 24, 63–72 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z., Obianyo O., Dall E., du Y., Fu H., Liu X., Kang S. S., Song M., Yu S. P., Cabrele C., Schubert M., Li X., Wang J. Z., Brandstetter H., Ye K., Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer’s disease. Nat. Commun. 8, 14740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosch F., Sabater J., Valera A., Insulin inhibits liver expression of the CCAAT/enhancer-binding protein beta. Diabetes 44, 267–271 (1995). [DOI] [PubMed] [Google Scholar]
- 35.MacDougald O. A., Cornelius P., Liu R., Lane M. D., Insulin regulates transcription of the CCAAT/enhancer binding protein (C/EBP) alpha, beta, and delta genes in fully-differentiated 3T3-L1 adipocytes. J. Biol. Chem. 270, 647–654 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Foti D., Iuliano R., Chiefari E., Brunetti A., A nucleoprotein complex containing Sp1, C/EBP beta, and HMGI-Y controls human insulin receptor gene transcription. Mol. Cell. Biol. 23, 2720–2732 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu M., Seufert J., Habener J. F., Pancreatic beta-cell-specific repression of insulin gene transcription by CCAAT/enhancer-binding protein beta. Inhibitory interactions with basic helix-loop-helix transcription factor E47. J. Biol. Chem. 272, 28349–28359 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Tang Y., Xiong K., Shen M., Mu Y., Li K., Liu H., CCAAT-enhancer binding protein (C/EBP) β regulates insulin-like growth factor (IGF) 1 expression in porcine liver during prenatal and postnatal development. Mol. Cell. Biochem. 401, 209–218 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Matsuda T., Kido Y., Asahara S. I., Kaisho T., Tanaka T., Hashimoto N., Shigeyama Y., Takeda A., Inoue T., Shibutani Y., Koyanagi M., Hosooka T., Matsumoto M., Inoue H., Uchida T., Koike M., Uchiyama Y., Akira S., Kasuga M., Ablation of C/EBPbeta alleviates ER stress and pancreatic beta cell failure through the GRP78 chaperone in mice. J. Clin. Invest. 120, 115–126 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Z., Liu X., Cheng L., Ye K., Delta-secretase triggers Alzheimer’s disease pathologies in wild-type hAPP/hMAPT double transgenic mice. Cell Death Dis. 11, 1058 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reddy K. C., Dunbar T. L., Nargund A. M., Haynes C. M., Troemel E. R., The C. elegans CCAAT-enhancer-binding protein gamma is required for surveillance immunity. Cell Rep. 14, 1581–1589 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu X. Y., Hu J. P., Wu M. M., Wang L. S., Fang N. Y., CCAAT/enhancer-binding protein CEBP-2 controls fat consumption and fatty acid desaturation in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 468, 312–318 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Darlington G. J., Ross S. E., MacDougald O. A., The role of C/EBP genes in adipocyte differentiation. J. Biol. Chem. 273, 30057–30060 (1998). [DOI] [PubMed] [Google Scholar]
- 44.Zidek L. M., Ackermann T., Hartleben G., Eichwald S., Kortman G., Kiehntopf M., Leutz A., Sonenberg N., Wang Z. Q., Maltzahn J., Müller C., Calkhoven C. F., Deficiency in mTORC1-controlledC/EBPβ-mRNAtranslation improves metabolic health in mice. EMBO Rep. 16, 1022–1036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bégay V., Smink J. J., Loddenkemper C., Zimmermann K., Rudolph C., Scheller M., Steinemann D., Leser U., Schlegelberger B., Stein H., Leutz A., Deregulation of the endogenous C/EBPβ LIP isoform predisposes to tumorigenesis. J. Mol. Med. (Berl) 93, 39–49 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Lin K., Dorman J. B., Rodan A., Kenyon C., daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319–1322 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Lin K., Hsin H., Libina N., Kenyon C., Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 28, 139–145 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Kfoury N., Kapatos G., Identification of neuronal target genes for CCAAT/enhancer binding proteins. Mol. Cell. Neurosci. 40, 313–327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zatara G., Hertz R., Shaked M., Mayorek N., Morad E., Grad E., Cahan A., Danenberg H. D., Unterman T. G., Bar-Tana J., Suppression of FoxO1 activity by long-chain fatty acyl analogs. Diabetes 60, 1872–1881 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ndoja A., Reja R., Lee S.-H., Webster J. D., Ngu H., Rose C. M., Kirkpatrick D. S., Modrusan Z., Chen Y.-J. J., Dugger D. L., Gandham V., Xie L., Newton K., Dixit V. M., Ubiquitin ligase COP1 suppresses neuroinflammation by degrading c/EBPbeta in microglia. Cell 182, 1156–1169.e2 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Ramamoorthi K., Lin Y., The contribution of GABAergic dysfunction to neurodevelopmental disorders. Trends Mol. Med. 17, 452–462 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Najm R., Jones E. A., Huang Y., Apolipoprotein E4, inhibitory network dysfunction, and Alzheimer’s disease. Mol Neurodegener 14, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palop J. J., Mucke L., Epilepsy and cognitive impairments in Alzheimer disease. Arch. Neurol. 66, 435–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DiFrancesco J. C., Tremolizzo L., Polonia V., Giussani G., Bianchi E., Franchi C., Nobili A., Appollonio I., Beghi E., Ferrarese C., Adult-onset epilepsy in presymptomatic Alzheimer’s disease: A retrospective study. J. Alzheimers Dis. 60, 1267–1274 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Sterneck E., Tessarollo L., Johnson P. F., An essential role for C/EBPbeta in female reproduction. Genes Dev. 11, 2153–2162 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bennett D. A., Buchman A. S., Boyle P. A., Barnes L. L., Wilson R. S., Schneider J. A., Religious orders study and rush memory and aging project. J. Alzheimers Dis. 64, S161–S189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E. M., Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 34, 939–944 (1984). [DOI] [PubMed] [Google Scholar]
- 58.Kauffman A., Parsons L., Stein G., Wills A., Kaletsky R., Murphy C., C. elegans positive butanone learning, short-term, and long-term associative memory assays. J. Vis. Exp., 2490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soukas A. A., Kane E. A., Carr C. E., Melo J. A., Ruvkun G., Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23, 496–511 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Timmons L., Court D. L., Fire A., Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Gendrel M., Atlas E. G., Hobert O., A cellular and regulatory map of the GABAergic nervous system of C. elegans. eLife 5, e17686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eilers J., Plant T. D., Marandi N., Konnerth A., GABA-mediated Ca2+ signalling in developing rat cerebellar Purkinje neurones. J. Physiol. 536, 429–437 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu C., Zipfel W. R., Multiphoton excitation of fluorescent probes. Cold Spring Harb. Protoc. 2015, 250–258 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Tables S1 and S2
Supporting file S1