Abstract
Malaria-causing parasites proliferate within erythrocytes through schizogony, forming multinucleated stages before cellularization. Nuclear multiplication does not follow a strict geometric 2n progression, and each proliferative cycle produces a variable number of progeny. Here, by tracking nuclei and DNA replication, we show that individual nuclei replicate their DNA at different times, despite residing in a shared cytoplasm. Extrapolating from experimental data using mathematical modeling, we provide strong indication that a limiting factor exists, which slows down the nuclear multiplication rate. Consistent with this prediction, our data show that temporally overlapping DNA replication events were significantly slower than partially overlapping or nonoverlapping events. Our findings suggest the existence of evolutionary pressure that selects for asynchronous DNA replication, balancing available resources with rapid pathogen proliferation.
In multinucleated malaria-causing parasites, asynchronous nuclear cycles balance limited resources with rapid proliferation.
INTRODUCTION
Eukaryotic cells usually proliferate by duplicating their genome and subsequently dividing into two daughter cells, each containing a single nucleus. Nevertheless, multinucleated cells can be readily found throughout the eukaryotic tree of life (1) and can be seen in several unicellular organisms (2), early embryos of insects (3), during plant seed development (4), or in osteoclasts and myotubes of vertebrates (5, 6). While some multinucleated cells, such as osteoclasts and myotubes, form by fusion of mononucleate cells, multinucleation can also result from repeated nuclear divisions without cell division, as seen in the early Drosophila embryo (3).
Seminal experiments fusing HeLa cells in different stages of the cell cycle yielded a synchronization of their nuclei (7), demonstrating that cell cycle control was mediated by diffusible cytoplasmic factors. In line with this mode of regulation, the nuclei of many multinucleated cells progress through cell cycle events synchronously (8). Yet, experiments with multinucleated mammalian cells, both fused and spontaneously arising, also reported asynchronous behavior of the nuclei (9, 10). Another example of asynchronous nuclear division cycles can be observed in the large multinucleated hyphae of the fungus Ashbya gossypii (11). Here, asynchrony is due to a combination of limited diffusion of cytoplasmic factors along the hypha and nucleus-intrinsic mechanisms, such as differences in transcriptional activity (12–14).
The unicellular parasite Plasmodium falciparum is the major contributor to global malaria-associated morbidity and mortality (15). During its complex life cycle, the parasite alternates between a female Anopheles mosquito host and a human host (Fig. 1A). In the mosquito host, a multinucleated cell occurs during the oocyst stage and in the human host during the liver and blood stages of infection. Within the liver and the blood, parasites proliferate via a process called schizogony. During schizogony, repeated rounds of nuclear division occur, forming a multinucleated cell before cellularization (16, 17). Through schizogony in the liver stage, a single parasite produces tens of thousands of daughter cells, called merozoites. Merozoites enter the blood stream again to invade erythrocytes. In this clinically relevant blood stage of infection, P. falciparum burden is directly related to disease severity (18, 19). Each blood-stage proliferative cycle takes roughly 48 hours and one parasite gives rise to approximately 20 ± 3 daughter merozoites (20–22). After invasion of an erythrocyte, merozoites quickly transform into a ring-stage parasite, which over time develops into a trophozoite and further into a schizont. The schizont stage is characterized by the presence of multiple nuclei, which actively divide while keeping their nuclear envelope intact (23, 24). Nuclear multiplication concludes with a relatively synchronous final round of division, which coincides with cellularization (17, 24). The daughter cells are subsequently released during egress and can then invade other erythrocytes, starting the proliferative cycle anew.
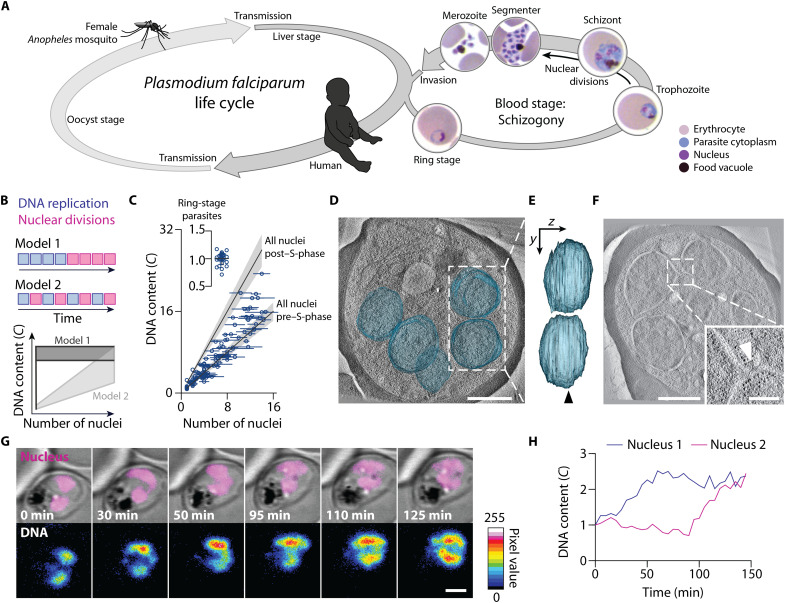
Fig. 1. P. falciparum proliferates through consecutive rounds of asynchronous DNA replications and nuclear divisions.
(A) Scheme of the P. falciparum life cycle, alternating between female Anopheles mosquitoes and humans. Multinucleation occurs in the oocyst, liver, and blood stage. Blood-stage development starts with the ring stage, followed by the trophozoite. Asynchronous nuclear divisions occur in the subsequent schizont stage. At the end of this stage, cellularization occurs and the segmenter is formed, containing nascent daughter cells called merozoites. Egress releases the merozoites that can then infect another erythrocyte. (B) Schematic and predictions of two models proposing the mode of P. falciparum proliferation in the blood stage of infection. (C) Gradual increase of the total DNA content and the number of nuclei of P. falciparum supports model 2. The DNA content was normalized to haploid ring-stage parasites (insert), defined as 1C. Horizontal bars, SD; gray lines, expected DNA contents of parasites with all nuclei before or after S-phase; gray bands, propagated error (SD) of ring-stage measurements. (D) Electron tomogram, overlayed with 3D-segmented inner nuclear membranes (blue); scale bar, 1 μm; see movie S1. (E) Side view of nuclear volumes showed no connection (90° rotation around the y axis); arrowhead, tomogram plane shown in (D). (F) Electron tomogram of connected nuclei; scale bar, 1 μm; inset highlights the connection (arrowhead); scale bar, 250 nm. (G) Time-lapse microscopy of a reporter parasite stained with the DNA dye 5-SiR-Hoechst showed asynchronous DNA replication in sister nuclei; scale bar, 2 μm; see movie S3. (H) Quantification of the DNA content of the nuclei shown in (G).
In a P. falciparum blood-stage schizont, odd numbers of nuclei are frequent and nuclear microtubule structures often differ between nuclei, indicating that nuclei divide asynchronously (23, 25–27). However, the dynamics of asynchronous nuclear division and its coordination with DNA replication are unknown. Here, we investigate nuclear multiplication in P. falciparum blood stages by combining different imaging modalities with computer simulations. We show how DNA replication and nuclear divisions are organized and provide data indicating that asynchronous nuclear cycles in P. falciparum blood stages balance limited resources with rapid parasite proliferation.
RESULTS
P. falciparum proliferates through consecutive rounds of DNA replication and nuclear division
Two distinct models have been proposed to describe the chronology of DNA replication and nuclear division events in a P. falciparum blood-stage schizont (Fig. 1B). Model 1 assumes several rounds of DNA replication in the trophozoite stage, which precede a phase of nuclear divisions in the schizont stage (28, 29) (Fig. 1B, top). This model predicts at first parasites with a single nucleus and varying DNA content and then parasites with varying number of nuclei and a relatively constant total DNA content (Fig. 1B, bottom). Model 2 proposes alternating rounds of DNA replication and nuclear divisions (26, 30, 31) (Fig. 1B, top), predicting a gradual increase of both number of separate nuclei per parasite and the total DNA content (Fig. 1B, bottom).
To test the predictions of both models, we stained the DNA and quantified the total DNA content as well as the apparent number of nuclei per parasite, assuming that every distinct DNA mass corresponds to an individual nucleus. We quantified the DNA content of haploid ring-stage parasites by integrating their fluorescence intensity and defined it as 1C (32). DNA content and number of nuclei per parasite were positively correlated, supporting model 2. In addition, the total DNA content did not exceed a value of 2C per nucleus of a given parasite (Fig. 1C and fig. S1A). These observations suggest that the DNA content of individual nuclei alternates between 1C and 2C. To exclude the possibility that membranous connections still existed between the nuclear envelopes that appeared separate in light microscopy, we recorded three-dimensional electron tomographic views of cell parts containing several entire nuclei (Fig. 1, D and E, and fig. S1, B and C). Although the nucleoplasms of some adjacent nuclei were only 75 nm apart (fig. S1, D and E), most nuclei appeared as separate compartments with clearly discernible nuclear envelopes and ribosomes filling the cytoplasmic gap. In only one of eight analyzed cells, we recorded a narrow bridge interconnecting two nuclei, which appeared to be completing nuclear division (Fig. 1F). Together, these data support a mode of P. falciparum blood-stage proliferation that consists of alternating rounds of DNA replication and nuclear divisions before cellularization. Although nuclear divisions lack synchronization (23, 25–27), it is unclear whether DNA replication in pairs of sister nuclei is synchronized. Using time-lapse live-cell microscopy of a cell line that expressed mCherry fused to a triple nuclear localization signal as a marker for nuclei (fig. S2, A to C), we quantified the DNA content using a live-cell–compatible DNA dye (32, 33). We found that the DNA content of sister nuclei increased at different times, indicating that DNA replications can occur asynchronously, i.e., onset and end of S-phases are desynchronized (Fig. 1, G and H, and fig. S2D).
A nuclear cycle sensor system
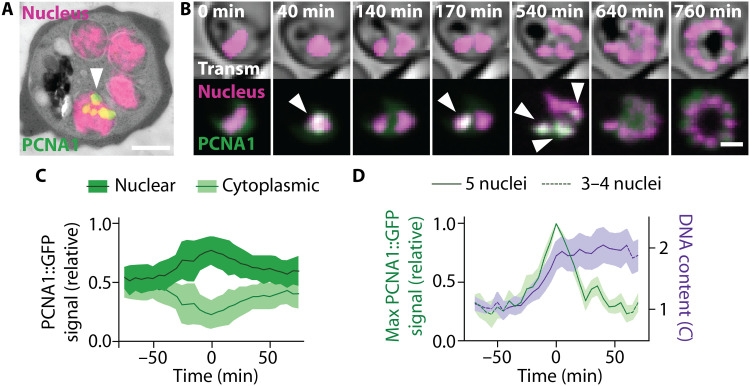
To understand how asynchronous DNA replications are orchestrated, we investigated the localization of the DNA replication machinery, using the P. falciparum proliferating cell nuclear antigen (PCNA) 1 as a proxy. PCNA is a critical cofactor of DNA polymerases and serves as a hub for many other components of the replication fork (34). As endogenous fusion of PCNA1 with green fluorescent protein (GFP) failed (fig. S3), we episomally expressed a PCNA1::GFP fusion protein in the background of our nuclear marker line (fig. S4A). Using correlative light and electron microscopy, we found that, in contrast to previous reports (35, 36), PCNA1::GFP localized unequally in nuclei of the same parasite, with only some nuclei showing distinct PCNA1::GFP foci (Fig. 2A and fig. S4B). In addition, time-lapse imaging revealed a dynamic localization and transient accumulation of PCNA1 in changing subsets of nuclei (Fig. 2B and fig. S4C). An increasing nuclear PCNA1::GFP signal was accompanied by a decreasing cytosolic signal and vice versa (Fig. 2, B and C, and fig. S4D), suggesting that nuclei access a common cytoplasmic pool of PCNA1. Moreover, nuclear accumulation of PCNA1::GFP coincided with a duplication of the DNA content in the same nuclei (Fig. 2D and fig. S4E). This allowed us to track individual DNA replication and nuclear division events over time in a given cell (Fig. 2B and fig. S4F). Hence, our parasite line expressing PCNA1::GFP can be considered a nuclear cycle sensor system analogous to the widely used FUCCI cell cycle sensor system for mammalian cells (37).
Fig. 2. Heterogeneous accumulation of PCNA1::GFP among nuclei permits development of a nuclear cycle sensor system.
(A) Correlative light and electron microscopy showed heterogeneous accumulation of PCNA1::GFP among P. falciparum nuclei; scale bar, 1 μm; arrowhead, PCNA1::GFP focus. (B) Time-lapse microscopy showed dynamic and transient accumulation of PCNA1::GFP; scale bar 2 μm; arrowheads, nuclear PCNA1::GFP accumulation; see movies S4 and S5. (C) Nuclear accumulation of PCNA1::GFP coincided with a depletion of the cytosolic pool; lines, average (n = 4); bands, SD. (D) Nuclear PCNA1::GFP accumulation caused a peak in the maximal pixel intensity, coinciding with DNA content duplication. DNA content was normalized to the average of 10 or all available values before the nuclear accumulation of PCNA1::GFP, defined as 1C; solid lines, average; bands, SD.
Single-cell dynamics of nuclear multiplication
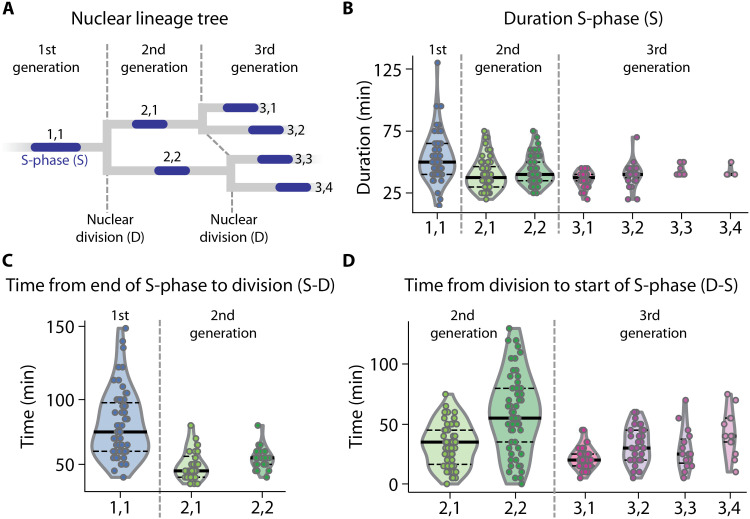
Using our PCNA1-based nuclear cycle sensor system, we next quantified the timing of individual events during nuclear multiplication over several generations of nuclei. We defined S-phase as the time interval between the onset and the end of visible PCNA1::GFP accumulation in a nucleus (Fig. 2, A and B). Completion of nuclear division was defined as the first time point where two separate nuclei could be observed. To enable the comparison of different events during nuclear multiplication, we numbered each event by the generation of nuclei and in the order of S-phase initiation. Hence, the initial S-phase in a parasite containing a single nucleus was labeled 1,1. Subsequent division of this nucleus resulted in two nuclei labeled 2,1 and 2,2, respectively, with 2,1 indicating the nucleus that entered S-phase first (Fig. 3A). This allowed us to draw nuclear lineage trees for individual parasites (Fig. 3A and fig. S5A). Notably, the timing of events and, thus, the resulting nuclear lineage trees varied markedly between individual parasites (fig. S5A). This cannot be explained by variable expression of PCNA1::GFP, which had no effect on DNA replication dynamics (fig. S5B). Investigating over 60 nuclear lineage trees showed that the first S-phase (S1,1) was on average significantly longer than the S-phase of the second-generation nuclei (S2,1; S2,2). The duration of S-phases then remained similarly long in the third generation of nuclei (S3,1 through S3,4) (Fig. 3B). Also, the time from the end of S-phase to nuclear division (S-D) was longer in the first generation than in the second generation (Fig. 3C). The following interval between nuclear division and S-phase (D-S) in nuclei of the second generation was longer than in the third generation (Fig. 3D). Hence, all initial phases of nuclear multiplication (i.e., duration of the first S-phase, time from the end of first S-phase to first nuclear division, and time from first division to the start of the second S-phases) were slower than the corresponding phases in the ensuing cycle. These data are consistent with the duration of different microtubule stages, which were recently reported for the initial and the subsequent nuclear divisions (27). We observed that the S-phases and the times from completed S-phase to nuclear division were similar in sister nuclei of the second and third generation. In contrast, the time from nuclear division to the start of the following S-phase varied markedly between sister nuclei (fig. S5C). Therefore, the interval between nuclear division and onset of S-phase predominantly introduces asynchrony during nuclear multiplication.
Fig. 3. Single-cell dynamics of nuclear multiplication.
(A) Nuclear lineage tree illustrating the three consecutive generations of nuclei quantified in (B) to (D). Dashed lines, nuclear divisions D demarcating generations and defined as the first time point where two separate daughter nuclei were observed; blue, S-phases S defined as the interval during which PCNA1::GFP accumulation was observed in a nucleus; nuclei are numbered by generation and in order of S-phase occurrence (e.g., 2,1: second generation, first S-phase). (B) S-phase durations of three generations of nuclei. S1,1 phases were longer than the pooled second-generation S-phases S2,1/2 (two-sided Mann-Whitney U test effect size f = 0.72, n1 = 54, n2 = 117, P = 3.2 × 10−6), and S2,1/2 was the same as S3,1/2/3/4 (f = 0.51, n1 = 117, n2 = 75, P = 0.85). (C) Time from end of S-phase to nuclear division (S-D) of two generations of nuclei. (S-D)1,1 was longer than (S-D)2,1/2 (f = 0.87, n1 = 63, n2 = 60, P = 8.1 × 10−13). (D) Time from nuclear division to start of S-phase (D-S) of two generations of nuclei. (D-S)2,1/2 was longer than (D-S)3,1/2/3/4 (f = 0.66, n1 = 119, n2 = 91, P = 4.2 × 10−5). Numbering of events in (B) to (D) as indicated in (A); each dot represents an event occurring in a single nucleus of a single parasite of 70 parasites analyzed; solid lines, median; horizontal dashed lines, quartiles.
A mathematical model of P. falciparum nuclear multiplication
To produce approximately 20 daughter cells (20–22), most nuclei should undergo four and some nuclei five rounds of S-phase and nuclear division. Because of the spatial proximity of nuclei (fig. S1, D and E), the S-phases and nuclear divisions that occurred during the latter half of schizogony could not be confidently analyzed by live-cell microscopy. Hence, we aimed to extrapolate the dynamics of the initial nuclear multiplication computationally. We modeled nuclear multiplication as a branching process, where nuclei stochastically progress through nuclear cycles. They consisted only of two phases: S-phase and the interval from the end of one S-phase to the start of the next in each daughter nucleus (Fig. 4A and fig. S6A). The model was parameterized using observed distributions of these two phases in each of the first two nuclear cycles, with the first cycle being slower than the second (Fig. 3, A to D, and table S1). In addition, we also determined the total replication time, i.e., the time from start of first S-phase to end of the last S-phase, and the time of egress in individual parasites (Fig. 4A and fig. S6B). As the first and second nuclear cycles showed no significant correlation (Fig. 4B), we did not include the inheritance of factors that facilitate nuclear multiplication in the model.
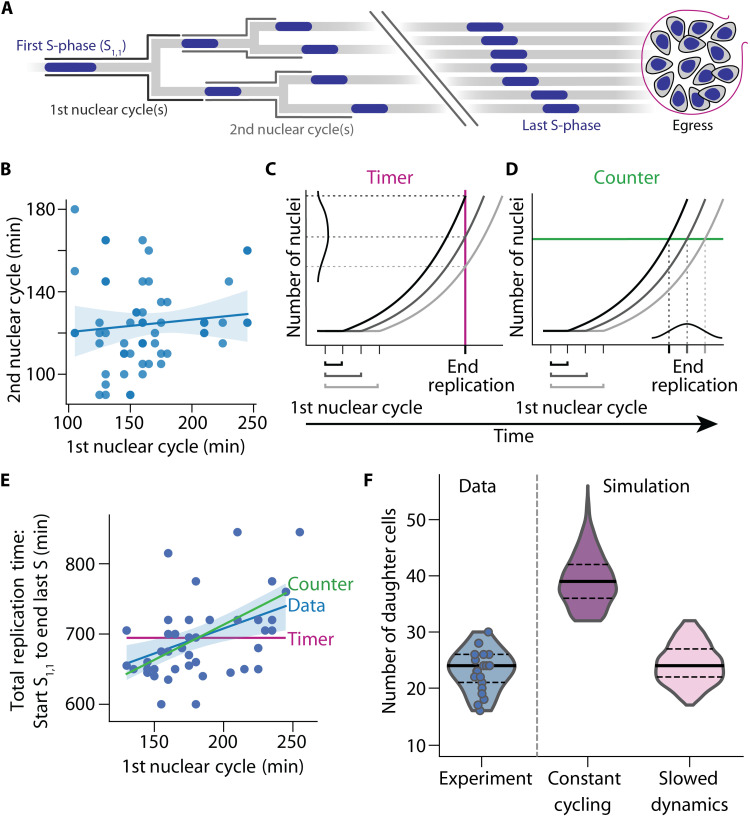
Fig. 4. Simulation of P. falciparum proliferation predicts slowing nuclear cycle dynamics.
(A) Nuclear lineage tree illustrating events of P. falciparum proliferation that were quantified in single parasites. S-phases depicted blue; nuclear cycles are defined as the total time from the start of an S-phase until the start of ensuing S-phases. Break indicates events that could not be individually resolved in the experiments. (B) The duration of the first and second nuclear cycles showed no correlation (Spearman’s ρ = 0.14, n = 58, P = 0.28); solid line, linear regression; band, bootstrapped 95% confidence interval. (C and D) Schematic illustrating how the duration of the first nuclear cycle affects the time needed to complete nuclear multiplication. (C) Timer mechanism with a set total time (red line) predicts no correlation. (D) Counter mechanism with a set total number of nuclei (green line) predicts a positive correlation. (E) Time-lapse imaging data showed a positive correlation between duration of first nuclear cycle and total time needed, i.e., time from start S1,1 to end of last S-phase (ρ = 0.42, n = 46, P = 0.0034), supporting a counter mechanism and contradicting a timer mechanism; blue solid line and band, linear regression and bootstrapped 95% confidence interval; red solid line, timer prediction; green solid line, counter prediction if all events were synchronous. (F) Mathematical model with slowing nuclear cycling dynamics (17% per cycle) fitted the experimental data best; solid lines, median; dashed lines, quartiles.
Because the observed nuclear lineage trees were very diverse (fig. S5A), we asked by what mechanism nuclear multiplication is stopped to achieve a well-defined end point that allows for cellularization. We examined two qualitatively different stopping mechanisms, which have been proposed in the context of cell proliferation (38) and nuclear multiplication in Plasmodium (22). The timer mechanism posits that growth of a system (e.g., increase in the number of nuclei) stops after a set time period. This set time is independent of the size of the system (e.g., the number of nuclei). If the initial nuclear cycle during a given time period is slow, less nuclei will be produced overall. Thus, the length of the first nuclear cycle should correlate with the number of nuclei but not with the overall duration of nuclear multiplication (Fig. 4C). Such a timer mechanism has been described for synchronous nuclear multiplication in the unicellular eukaryote Sphaeroforma arctica (39). Conversely, the counter mechanism stops growth after a certain system size has been reached (e.g., a certain number of nuclei in a schizont), regardless of the time needed (Fig. 4D). Longer first cycles lead to a delayed end of nuclear multiplication, as the required number of nuclei is reached later. The counter mechanism predicts that the length of the first nuclear cycle has an effect on the overall duration of nuclear multiplication. Hence, a counter positively correlates the time needed for the first nuclear cycle with the overall duration of nuclear multiplication.
To distinguish timer and counter mechanisms, we compared the duration of the first nuclear cycles to the total replication time, i.e., start of first S-phase to end of the last S-phase in single parasites (Fig. 4E). These two traits showed a significant positive correlation, which is incompatible with a timer but consistent with a counter. We also found a positive correlation between the first nuclear cycle and the overall time needed from the onset of the first S-phase to parasite egress, again supporting a counter but rejecting a timer (fig. S6E). A similar analysis using data from the second nuclear cycles also favored a counter (fig. S6, F and G). Consequently, we adopted a counter mechanism in our model by stopping the initiation of further S-phases after a set number of S-phases have been completed. At this point, a variable number of S-phases are still ongoing and these are allowed to conclude, producing a variable final number of nuclei. We adjusted this stopping criterion such that our simulations fitted the measured distribution of times from the start of the first S-phase to the end of last S-phase (fig. S6H). Without further adjustment, the simulations then reproduced the measured positive correlation between the first nuclear cycle and overall duration of nuclear multiplication and also the measured slope of their linear regression (Fig. 4E and fig. S6I). Thus, a counter mechanism alone is sufficient to explain the observed timing of nuclear multiplication.
To test the model, we asked how many progeny it generated overall and compared this to our experimentally determined values. When the cycling dynamics were kept unchanged from the third nuclear cycle onward, the simulations clearly overestimated the total progeny (Fig. 4F). This result implies that nuclear multiplication in P. falciparum must slow down overall after the second nuclear cycle. This overall slowdown could be accomplished in different ways, e.g., the replicative arrest of individual nuclei or the slowing of the cycling dynamics of all nuclei. Although we cannot exclude the arrest of nuclei, we did not readily observe this (movies S4 to S6). We therefore adapted the model by gradually slowing the cycling dynamics of all nuclei from the third cycle onward. A slowdown by 17% per cycle fitted our data best and recovered both the average and the variability of the number of progeny (Fig. 4F). In summary, nuclear multiplication during blood-stage schizogony initially accelerates from first to second cycle (Fig. 3, B to D) and then slows down.
A shared limiting factor can explain the dynamics of DNA replication in P. falciparum
As nuclear multiplication proceeds, the number of nuclei that share the same cellular resources increases. In addition, the probability that several nuclei are in the same stage of the nuclear cycle also increases, which can be immediately seen (Fig. 2B and movies S4 to S6). This suggest that the decrease in nuclear cycling speed could be explained by shared factors that become increasingly limiting as nuclei multiply.
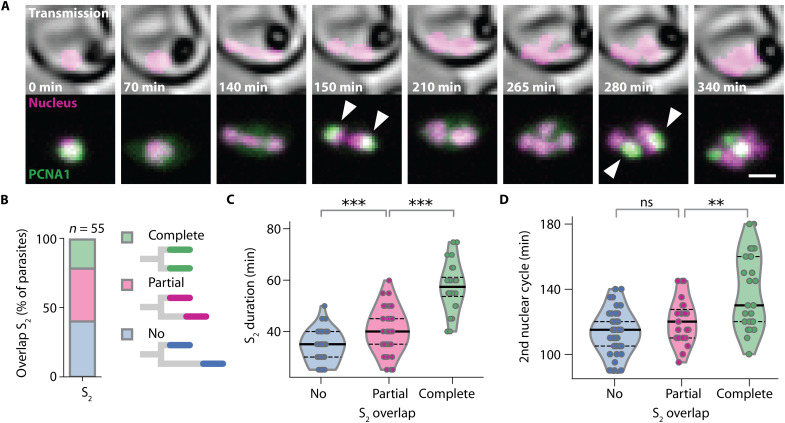
If nuclei share a limited resource that is needed for multiplication, then simultaneously multiplying nuclei should experience a stronger limitation than nuclei that multiply sequentially. To test this prediction, we compared pairs of sister nuclei, where S-phases showed a varying degree of temporal overlap (Fig. 5, A and B). While the intervals between S-phases did not differ significantly, partially overlapping S-phases were significantly longer than nonoverlapping S-phases, and completely overlapping S-phases were again markedly longer (Fig. 5C and fig. S7, A to D). This increase supports the notion that the speed of DNA replication is affected by a shared limiting factor. Next, we tested whether the increased duration of S-phases also translated into a longer nuclear cycle, and we found that partially overlapping S-phases had no effect. By contrast, a sizeable fraction of sister nuclei with synchronous S-phases displayed a prolonged nuclear cycle (Fig. 5D and fig. S7E), suggesting that delays caused by synchronous S-phases cannot be fully compensated.
Fig. 5. Asynchronous nuclear cycles facilitate rapid parasite proliferation.
(A) Time-lapse microscopy of a parasite with synchronous DNA replication events (arrowheads); scale bar, 2 μm; see movie S6. (B) Fraction of parasites with completely, partially, and not overlapping S2,1 and S2,2. (C) Durations of S-phases increased with the degree of temporal overlap; solid lines, median; dashed lines, quartiles; two-sided Mann-Whitney U test, no versus partial overlap, f = 0.28, n1 = 44, n2 = 42, P = 2.7 × 10−4; partial versus complete overlap, f = 0.10, n1 = 42, n2 = 24, P = 5.9 × 10−8. (D) Nuclear cycles containing synchronous S-phases were longer; solid lines, median; dashed lines, quartiles; no versus partial overlap, f = 0.36, n1 = 41, n2 = 23, P = 0.07; partial versus complete overlap, f = 0 .27, n1 = 23, n2 = 26, P = 0.0067. ns, not significant; **P < 0.01; ***P < 0.001.
DISCUSSION
The present work was made possible by a novel PCNA1-based nuclear cycle sensor system, which allowed us to investigate asynchronous nuclear multiplication in P. falciparum. Our results show that this parasite proliferates through alternating, consecutive rounds of DNA replication and nuclear division. Although nuclei reside in close proximity in a shared cytoplasm, DNA replications and nuclear divisions occur asynchronously (Figs. 1 to 3 and fig. S2).
Notably, both genetically identical nuclei and parasites show large variation in nuclear cycling dynamics (Fig. 3 and figs. S2D, S4F, and S5A). Our data show that variability in nuclear lineages is mostly introduced during the time between nuclear division and the subsequent S-phase, an interval sharing similarities with the G1 phase of the canonical cell cycle (Fig. 3D and fig. S5C). In mammalian and yeast cells, differences in cell cycle timing of genetically identical cells cultured in the same environment are predominantly introduced during the G1 phase (40–42). Similarly, in the multinucleated cells of A. gossypii, nuclear asynchrony is introduced in G1, causing a variable start of S-phase (43). To date, the molecular details of P. falciparum cell cycle regulation are not well understood, in part because the parasite displays a very divergent repertoire of cell cycle–related proteins (44). Many canonical proteins could not be identified by sequence homology and, for example, no G1-, S-, or M-phase cyclins have been found (45). However, the nuclear kinase CRK4 is known to be essential for S-phase initiation for each generation of nuclei (46), and hence, variability in CRK4 activity may be involved in generating asynchrony during nuclear multiplication.
In A. gossypii, asynchrony is usually associated with cytoplasmic domains that are established by spatial separation of the nuclei (12, 47, 48). Yet, in a dynactin mutant, nuclei cluster closely together and still maintained autonomy (47, 49). This indicates that nucleus-intrinsic factors are sufficient to establish asynchrony. In P. falciparum, the very close spatial proximity of nuclei (fig. S1, D and E) and the fast, heterogeneous, and transient nuclear accumulation of PCNA1::GFP suggest that nucleus-intrinsic factors are pivotal for nuclear autonomy. Nucleus-intrinsic differences could be achieved by physical asymmetry during nuclear division, resulting in the uneven distribution of factors that are necessary for the progression of the nuclear cycle, which has been proposed for asynchronous nuclear divisions in P. falciparum (26, 50, 51). In addition, differential nucleocytoplasmic transport may account for nucleus-intrinsic differences. The nuclei of terminally differentiated myotubes show an unequal nuclear import (52), and when myotubes are induced to reenter the cell cycle, their nuclei undergo asynchronous DNA replication (53).
P. falciparum blood-stage parasites can produce variable numbers of daughter cells (Fig. 4F) (20–22). Our results indicate that nuclear multiplication and, consequently, the number of daughter cells are regulated by a counter mechanism (Fig. 4E and fig. S6, E to G). Although several P. falciparum proteins have been implicated in the regulation of parasite progeny number, e.g., the kinases PK7 and CRK5 (54, 55), it is yet unclear how the counter operates at the molecular level.
Nuclear multiplication in P. falciparum is likely also influenced by nucleus-extrinsic factors, such as competition for a shared resource. Synchronous S-phases, which occurred in approximately a quarter of cells with two nuclei, significantly slowed down the nuclear cycles (Fig. 5). This suggests the existence of a shared factor that is required for DNA replication. Moreover, slow initial cycles translated in slower overall completion of nuclear multiplication (Fig. 4E), which suggests that asynchronous nuclear cycles may allow the parasite to exploit scarce resources more steadily, thereby shortening nuclear cycles and facilitating a faster completion of schizogony. Scarcity of nutrients led to a higher frequency of asynchrony in multinucleated human cells in tissue culture (10). In addition, P. falciparum cultured in serum of caloric restricted mice had fewer daughter cells per schizont (56), and P. falciparum grows better in blood from diabetic patients with elevated glucose levels (57). However, it remains unclear how altered nutrient availability affects the nuclear multiplication dynamics and the relative frequency of synchronous events. Overall, our findings support the notion that asynchrony has evolved to enable P. falciparum blood-stage parasites to balance available resources with rapid proliferation, which is the main driver of virulence and critical for life cycle progression.
MATERIALS AND METHODS
P. falciparum cell culture
The P. falciparum strain 3D7 is laboratory-adapted and derived from the isolate NF54 by limiting dilution. NF54 originated from a case of airport malaria in The Netherlands (58). P. falciparum 3D7 parasites were routinely cultured in human O+ peripheral blood erythrocytes in RPMI 1640 (GlutaMAX, Gibco, Thermo Fisher Scientific) medium supplemented with 0.5% AlbuMAX II Lipid-Rich BSA (Gibco, Thermo Fisher Scientific), 0.2 mM hypoxanthine (c.c.pro GmbH), 25 mM Hepes (pH 7.3; Sigma-Aldrich, Merck), and gentamicin sulfate (12.5 μg/ml; Carl Roth) (complete RPMI) at a 4% hematocrit and a temperature of 37°C in 90% relative humidity, 3% CO2, and 5% oxygen (59).
If needed, parasites were synchronized by incubating 400 μl of packed erythrocytes with 10 ml of prewarmed 5% sorbitol solution (w/v) for 10 min at 37°C (60). Parasites were washed once with complete RPMI before return to culture.
Parasitemia was counted using either a Zeiss Axiostar Plus (Carl Zeiss Microscopy GmbH) or Nikon Eclipse E100 (Nikon Corporation) with 100× oil immersion objective on thin blood smears fixed with 100% methanol (Honeywell) for 10 s and stained with Hemacolor rapid staining of blood smear solutions (Sigma-Aldrich, Merck). For genomic DNA extraction, parasite DNA was isolated from 200 μl of washed erythrocyte pellet with a parasitemia of 1 to 3% using the DNeasy Blood & Tissue Kit (Qiagen).
Generation of transgenic P. falciparum
All primer and plasmid sequences are available upon request. The plasmid for visualization of parasite nucleoplasm was generated by restriction digest–mediated removal of the FKBP-rapamycin binding domain (FRB) sequence from the plasmid p3xNLS-FRB-mCherry-hsp86-BSD gifted by T. Spielmann (61). The resulting plasmid p3xNLS-mCherry-hsp86-BSD expresses mCherry coupled to three nuclear localization sequences (NLSs) under the constitutive hsp86 promoter. Furthermore, the plasmid contains a BSD-resistance cassette.
PCNA1::GFP was expressed from the plasmid pARL_PCNA1-GS-eGFP under the control of the P. falciparum CRT promoter. In addition, it contains a WR99210 resistance cassette (62).
Plasmids were amplified by transformation into 50 μl of competent 5-alpha F′Iq Competent Escherichia coli cells (New England Biolabs GmbH), overnight culture at 37°C, and extraction via the GenElute HP Plasmid Miniprep Kit (Sigma-Aldrich, Merck) or NucleoBond Xtra Midi Kit (Macherey-Nagel) depending on required purity and yield. Monoclonal colonies were verified by Sanger sequencing (Eurofins Genomics).
Transgenic parasites were created by electroporation (310 kV, 950 μF; Gene Pulser II, Bio-Rad) of 50 to 100 μg of purified DNA resuspended in 30 μl of TE buffer [10 mM tris (pH 8.0) and 1 mM EDTA (pH 8.0)] and 370 μl of CytoMix [120 mM KCl, 0.15 M CaCl2, 2 mM EGTA, 5 mM MgCl2, 10 mM K2HPO4/KH2PO4, and 25 mM Hepes (pH 7.6)] into 200 μl of packed erythrocytes of a ring-stage 3D7 culture with a parasitemia of at least 4% (63). The presence of episomes was maintained by continuous selection with blasticidin (BSD) (InvivoGen) at 5 μg/ml and/or WR99210 (Jacobus Pharmaceutical Company) at 2.5 μM, respectively.
Endogenous tagging of PCNA1 with GFP was attempted by CRISPR-Cas9–mediated homology repair. The plasmid pDC2-cam-coCas9-U6-hDHFR was a gift from M. Lee (64). Briefly, this plasmid expresses a guide RNA (gRNA) sequence from a U6 cassette together with a Cas9 nuclease under the P. falciparum calmodulin promoter as well as a WR99210 resistance cassette for selection in P. falciparum. gRNA binding sites were selected using the Protospacer Workbench software (65). The selection of gRNA sequences was based on distance to the C terminus of PCNA1 and the activity prediction scores (66). gRNAs were ordered as unmodified oligonucleotides (Thermo Fisher Scientific), annealed, and phosphorylated before ligation in the pDC2-cam-coCas9-U6-hDHFR plasmid linearized by Bbs I restriction digest (New England Biolabs GmbH). Template regions for double-crossover homology repair were cloned into the same plasmid backbone. The final plasmids were assembled via Gibson Assembly (NEBuilder HiFi DNA Assembly, New England Biolabs GmbH) using the polymerase chain reaction (PCR)–amplified homology and GFP sequences and the plasmid containing the selected guides, which was linearized by Eco RI and Aat II restriction digest (New England Biolabs GmbH).
Transfection was carried out as described above, but selection with WR99210 was discontinued 12 days after transfection. The presence of GFP fluorescence was investigated in live parasites under a coverslip on a glass slide using a 60× oil immersion objective on a Zeiss Axio Observer (Carl Zeiss Microscopy GmbH) equipped with a Prime BSI sCMOS camera (Teledyne Photometrics). Diagnostic PCR was carried out on isolated genomic DNA from the transfected pool.
Sample preparation for correlative light and electron tomography and serial section tomography
Red blood cells infected with P. falciparum 3D7 expressing 3xNLS-mCherry and PCNA1::GFP in multinucleated stages were purified via varioMACS (Miltenyi Biotec GmbH) magnetic purification, washed in phosphate-buffered saline (PBS) (Gibco, Thermo Fisher Scientific), and resuspended in incomplete RPMI [0.2 mM hypoxanthine, 25 mM Hepes (pH 7.3), and gentamicin (12.5 μg/ml)]. The pellet was pipetted into 0.2-mm-deep aluminum carriers (Engineering Office M. Wohlwend GmbH) and cryo-immobilized by high-pressure freezing using Leica EM ICE (Leica Microsystems).
Freeze substitution was done using a freeze substitution device (EM-AFS2, Leica Microsystems), the freeze substitution solution contained 0.3% (w/v) uranyl acetate dissolved in anhydrous acetone, and the samples were substituted at −90°C for 14 hours. The temperature was then increased at a rate of 5°C/hour to −45°C, followed by 2-hour incubation at −45°C. The samples were rinsed with ethanol for 1 hour, followed by LR Gold (London Resin Company) infiltration at −25°C in 2-hour steps with 25, 50, and 75% LR Gold in ethanol. The samples were then left in 100% LR Gold for 12 and 4 hours before the onset of polymerization. Ultraviolet (UV) polymerization was applied for 24 hours at −25°C, and the temperature was increased to 20°C at a rate of 5°C per hour. The samples were left exposed to UV at room temperature for 24 hours.
Correlative light and electron microscopy
Sectioning was done on a Leica UC6 ultramicrotome (Leica Microsystems), and sections were collected on formvar-coated finder grids (Plano). Sections (200 nm thick) on grids were placed in PBS and sandwiched between two cover glasses, and fluorescence signal was imaged on a Zeiss observer Z1 fluorescence microscope (Zeiss). The identical cells were imaged on a JEOL JEM-1400 electron microscope (JEOL) operating at 80 kV and equipped with a 4K TemCam F416 camera (Tietz Video and Image Processing Systems GmbH). In the final step, the correlation of fluorescence and electron micrographs, based on morphological features, was done using the eC-CLEM software (67).
Serial section tomography
Electron tomography
The resin block was sectioned in 200-nm sections using a Leica UC7 ultramicrotome (Leica Microsystems). Ribbons of serial sections were placed onto formvar-coated slot copper grids and imaged with a Tecnai F30 transmission electron microscope (FEI) operating at 300 keV. Tilt series were recorded at the angle ranging from −60° to 60° with 2° increments using SerialEM software (68). Digital images were recorded on a fast Gatan OneView 4k camera with a nominal magnification of 12,000.
Image processing and analysis
Three-dimensional (3D) tomograms were generated from the tilt series using weighted back projection and combined into serial tomograms using the IMOD software package (69). Schizont nuclei were segmented using AMIRA software (Thermo Fischer Scientific). Minimal distance between adjacent nuclei was measured with the line tool in ImageJ by manually connecting the edge of the nucleoplasm of neighboring nuclei.
Live P. falciparum labeling and imaging
For live-cell imaging, parasites were seeded on sterile glass-bottom 35-mm μ- or eight-well dishes (ibidi GmbH) as described by Grüring and Spielmann (70) with minor modifications. Briefly, the bottom of the dish was coated with concanavalin A (5 mg/ml; Sigma-Aldrich) and rinsed with PBS. Resuspended parasite culture was washed twice with incomplete RPMI and left to settle on the dish for 10 min at 37°C before unattached cells were washed off using incomplete RPMI until a single-cell layer remained. Cells were left to recover at standard culturing conditions in complete RPMI for at least 8 hours before medium was exchanged to phenol red–free complete RPMI imaging medium [RPMI 1640 l-glutamine (PAN-Biotech), 0.5% AlbuMAX, 0.2 mM hypoxanthine, 25 mM Hepes (pH 7.3), and gentamicin (12.5 μg/ml)] that had been equilibrated to incubator gas conditions for at least 6 hours. Dishes were completely filled, closed, and sealed tightly using parafilm before imaging was started.
For staining of parasite DNA, phenol red–free complete RPMI imaging medium was supplemented with 5-SiR-Hoechst provided by G. Lukinavičius und J. Bucevičius (33). 5-SiR-Hoechst was added at 250 nM in imaging medium to parasites 2 hours before imaging.
Routine long-time live-cell imaging was carried out on a PerkinElmer UltraVIEW VoX microscope equipped with Yokogawa CSU-X1 spinning disk head and Nikon TiE microscope body. An Apo total internal reflection fluorescence 60×/1.49 numerical aperture (NA) oil immersion objective and Hamamatsu C9100-23B electron-multiplying charge-coupled device (EM-CCD) camera were used. Live-cell imaging was performed at 36.5°C. Images were acquired at multiple positions using an automated stage and the Perfect Focus System (PFS) for focus stabilization with a time resolution of 5 min per stack. Multichannel images were acquired sequentially using solid-state lasers with excitation at 488 and 561 nm and matching emission filters in addition to differential interference contrast (DIC) images; 8-μm stacks were acquired with a z-spacing of 500 nm.
Live-cell DNA quantification in parasites containing one nucleus was performed at 37°C on a PerkinElmer UltraVIEW VoX microscope equipped with Yokogawa CSU-X1 spinning disk head and Nikon TiE microscope body. A Plan Apo VC 100×/1.4 NA oil immersion objective and Hamamatsu C9100-23B EM-CCD camera were used in combination with an additional zoom of 1.5×. Live-cell imaging was performed at 36.5°C. Images were acquired at multiple positions using an automated stage and the PFS for focus stabilization with a time resolution of 5 min per stack. Multichannel images were acquired sequentially using solid-state lasers with excitation at 488, 561, and 640 nm and matching emission filters in addition to DIC images; 6-μm stacks were acquired with a z-spacing of 500 nm.
Additional live-cell DNA quantification in parasites with one or two nuclei was done on a point laser scanning Leica SP8 microscope using HC PL APO CS2 63×/1.4-NA oil immersion objective. Images were acquired at multiple positions using an automated stage and the Adaptive Focus Control for focus stabilization with a time resolution of 5 min per stack. Multichannel images were acquired sequentially using a photomultiplier tube (PMT) detector with a gain of 800 to 1000 V, a laser excitation at 561 nm, and a spectral detection window set between 570 and 620 nm for mCherry imaging and using a HyD detector in the standard mode with laser excitation at 633 nm and spectral detection window set between 640 and 700 nm for 5-SiR-Hoechst. In parasites with one nucleus, additional images were acquired using a HyD detector in the standard mode with laser excitation at 488 nm and spectral detection window set between 490 and 550 nm for GFP imaging. Bright-field images were obtained from a transmitted light PMT detector. A 2.25- to 4.5-μm stack was acquired with a sampling of 60 to 100 nm in the xy axis and 0.75 to 1 μm in the z axis with a scanning speed of 400 to 700 Hz.
For DNA quantification in fixed cells, P. falciparum–infected erythrocytes were seeded in eight-well dishes (ibidi GmbH) in the same manner as for live-cell imaging. After a minimum of 4-hour recovery time in complete RPMI, cells were fixed using prewarmed 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 20 min at 37°C before washing with PBS. For staining of DNA, 5-SiR-Hoechst at a concentration of 250 nM was added to PBS 30 min before imaging.
Point laser scanning confocal microscopy was performed on a Leica SP8 microscope using an HC PL APO CS2 63×/1.4 NA oil immersion objective. Images were acquired using HyD detectors in the standard mode with the laser excitation at 633 nm and spectral detection window set between 640 and 700 nm for 5-SiR-Hoechst imaging. Bright-field images were obtained from a transmitted light PMT detector. A 6-μm stack was acquired with a sampling of 29 nm in the xy axis and 132 nm in the z axis with a scanning speed of 400 Hz. Subsequently, images were processed with the Lightning algorithm in the adaptive mode using default settings.
Image analysis
Image analysis was done using the Fiji distribution of ImageJ (71). Quantitative results were exported as comma-separated value (csv) files, analyzed, and plotted using Microsoft Excel, GraphPad Prism version 5.0.0 for Windows, or Python with Matplotlib 3.2.2, NumPy 1.19.2, pandas 1.2.0, and SciPy 1.6.0.
DNA quantification in fixed cells
Nuclei were counted by three independent researchers in Lightning algorithm–processed 3D stacks. For DNA intensity measurements, 5-SiR-Hoechst signal in the total area occupied by nuclei was measured in individual parasites. Briefly, a threshold (300 and 65536) was applied to Lightning algorithm–processed 3D stacks, and the resulting mask was used to measure raw integrated density of the 5-SiR-Hoechst channel within segmented areas of unprocessed images. Signal intensities were normalized to the mean 5-SiR-Hoechst signal intensity in nuclei of ring-stage parasites.
Live-cell DNA quantification in parasites with two nuclei
For quantification of DNA in live-cell microscopy, 5-SiR-Hoechst signal intensities in individual nuclei, segmented via the mCherry signal, were measured over time. Areas with individual nuclei were manually separated. A median filter with a radius of two pixels was applied to the mCherry signal of individual nuclei, and a threshold (20, 255-35, 255) depending on the fluorescence intensity of mCherry in the cell was applied to create masks of individual nuclei. 5-SiR-Hoechst signal intensity was then determined in the individual nuclei via measurement of raw integrated density within nuclear masks. Signal intensities in individual nuclei were normalized to the first time point of the time course or to the time point in which individual nuclei were first observed.
Live-cell DNA quantification in parasites with one nucleus
For parasites that had one nucleus and expressed PCNA1::GFP, the 5-SiR-Hoechst signal intensity in the nucleus was measured over time and correlated to the maximal PCNA1::GFP signal. Data acquired on the PerkinElmer spinning disk microscope were processed as follows: Background values for each channel were determined by the mean signal in an area not containing a parasite. The mCherry signal was used to determine the nucleus area. A background subtraction of 2020, as well as a median filter with a radius of one pixel, was applied to the mCherry signal of individual cells, followed by a threshold (170, 65,536) to create masks of individual nuclei. 5-SiR-Hoechst signal intensity was then determined in the individual nuclei via measurement of raw integrated density after background subtraction of 2300. Signal intensities were normalized to the average of the first 10 values of the time course or to the average of all available values from the start of the time course to the start of nuclear PCNA1::GFP accumulation if both time points were less than 10 time points apart from each other. In addition, maximal GFP signal intensity values were measured from an average intensity z-projection of the GFP channel and normalized to minimal and maximal values for each cell. Data from individual cells were aligned to the time point at which the highest maximal GFP signal intensity occurred.
Data acquired on the Leica Sp8 confocal laser scanning microscope were processed as follows: The mCherry signal was used to determine the nucleus area. A median filter with a radius of one pixel was applied to the mCherry signal of individual cells, followed by a threshold (15, 255) to create masks of individual nuclei. 5-SiR-Hoechst signal intensity was then determined in the individual nuclei via measurement of raw integrated density after background subtraction of 10. The area of the nucleus was determined via measurement of the area of the mCherry mask, and 5-SiR-Hoechst signal intensity was divided by the nuclear area to avoid total signal variations due to changing z-focus during imaging. Signal intensities per area were normalized to the average of the first 10 values of the time course or to the average of all available values from the start of the time course to the start of nuclear PCNA1::GFP accumulation if both time points were less than 10 time points apart from each other. In addition, maximal GFP signal intensity values were measured from an average intensity z-projection of the GFP channel and normalized to minimal and maximal values for each cell. Data from individual cells were aligned to the time point at which the highest maximal GFP signal intensity occurred.
Analysis of long-term live-cell imaging
Cells were manually evaluated for parasite survival and health and grouped into different categories depending on which events of schizogony were clearly observable, i.e., onset of S-phase, number of nuclei at start of imaging, parasite egress, and start of first S-phase and parasite egress in the same cell. Individual parasites were manually analyzed for start and end of S-phase using nuclear PCNA1::GFP accumulation as proxy, and for time points of nuclear division, i.e., earliest observable occurrence of completely separate nuclei.
Determination of GFP fluorescence intensity before first nuclear PCNA1::GFP accumulation
Time points for determination of GFP fluorescence intensity before start of nuclear PCNA1::GFP accumulation were manually selected. Background intensity was measured by determining maximum fluorescence in an area not occupied by the parasite and subtracted from the GFP channel. Subsequently, the area containing the parasite and raw integrated density of the GFP channel within this area was measured.
Analysis of fluorescence intensity over time
To analyze fluorescence levels over time, representative parasites were chosen where both onset of first S-phase and egress were clearly visible. Raw integrated density was measured in the mCherry and GFP channel of average intensity z-projections and normalized to maximal and minimal values per cell. Data from individual cells were aligned by the time point of parasite egress.
Visualization of nuclear PCNA1::GFP accumulation over time
To visualize occurrence of PCNA1::GFP accumulation over time, normalized maximal GFP pixel intensity values over time were plotted as a heatmap. Maximal GFP pixel values per time point were measured from average intensity z-projections and normalized to maximal and minimal values per cell. Cells were aligned by occurrence of first PCNA1::GFP nuclear accumulation and ordered by occurrence of second PCNA1::GFP accumulation. The heatmap was created in ImageJ by renormalizing values to the eight-bit value range (0 to 255) importing values as text image.
Determination of GFP fluorescence within nucleus and cytoplasm
Four representative parasites were chosen to analyze levels of PCNA1::GFP in the nucleus and in the cytoplasm over time. Fifty consecutive time points were chosen manually that showed parasites with one nucleus and an increasing and decreasing nuclear PCNA1::GFP signal. Background was determined for each parasite by measuring mean and maximum GFP values in an area not containing a parasite and set at . xbackground was subtracted from the GFP channel in each plane of the 3D stack at every time point. The area of the nucleus was identified via automatic thresholding in the mCherry channel to create a nuclear mask. Subsequently, raw integrated density of the GFP channel was determined per time point in the total parasite area as well as in the nucleus. Values for the cytoplasm were calculated by subtracting values for the nucleus from the values of the total parasite. Data from individual cells were aligned by occurrence of highest nuclear PCNA1::GFP signal.
Mathematical modeling of P. falciparum nuclear multiplication
We constructed a mathematical model of nuclear multiplication to systematically explore the temporal evolution of nuclear multiplication during schizogony. We modeled nuclear multiplication as a branching process and parameterized this model with the observed nuclear multiplication dynamics (Fig. 4, fig. S6, and Supplementary Text).
Acknowledgments
We thank E. Egan for critically reading the manuscript and G. Lukinavičius and J. Bucevičius for providing the 5-SiR-Hoechst dye. We thank the Infectious Diseases Imaging Platform (IDIP) at the Center for Integrative Infectious Disease Research, Heidelberg, Germany, for the microscopy support and the Electron Microscopy Core Facilities at the European Molecular Biology Laboratory and Heidelberg University, Heidelberg, Germany. The Plasmodium database PlasmoDB facilitated this work.
Funding: This study was supported by German Research Foundation project number 240245660-SFB 1129 (to C.F., T.H., F.F., U.S.S., and M.G.), German Research Foundation project number 349355339 (to J.G.), Baden-Württemberg Foundation grant 1.16101.17 (to M.G.), Fundação para a Ciência e Tecnologia (FCT, Portugal) grant PD/BD/128002/2016 (to M.M.), and Research Training Group Mathematical Modeling for the Quantitative Biosciences (to P.B.).
Author contributions: Conceptualization: S.K., P.B., N.B.B., U.S.S., and M.G. Methodology: S.K., P.B., N.B.B., and M.G. Software: S.K., P.B., and N.B.B. Investigation: S.K., P.B., J.K., M.M., C.F., V.S., D.K., and M.C. Formal analysis: S.K., J.K., A.K., and P.B. Resources: V.L. and J.G. Writing—original draft: S.K., P.B., N.B.B., and M.G. Writing—review and editing: N.B.B., F.F., J.G., U.S.S., and M.G. Funding acquisition: T.H., F.F., U.S.S., and M.G. Supervision: F.F., T.H., U.S.S., and M.G.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S7
Table S1
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S6
REFERENCES AND NOTES
- 1.Adl S. M., Bass D., Lane C. E., Lukeš J., Schoch C. L., Smirnov A., Agatha S., Berney C., Brown M. W., Burki F., Cárdenas P., Čepička I., Chistyakova L., Campo J., Dunthorn M., Edvardsen B., Eglit Y., Guillou L., Hampl V., Heiss A. A., Hoppenrath M., James T. Y., Karnkowska A., Karpov S., Kim E., Kolisko M., Kudryavtsev A., Lahr D. J. G., Lara E., Gall L. L., Lynn D. H., Mann D. G., Massana R., Mitchell E. A. D., Morrow C., Park J. S., Pawlowski J. W., Powell M. J., Richter D. J., Rueckert S., Shadwick L., Shimano S., Spiegel F. W., Torruella G., Youssef N., Zlatogursky V., Zhang Q., Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchison-Field L. M. Y., Vargas-Muñiz J. M., Stormo B. M., Vogt E. J. D., Dierdonck S. V., Pelletier J. F., Ehrlich C., Lew D. J., Field C. M., Gladfelter A. S., Unconventional cell division cycles from marine-derived yeasts. Curr. Biol. 29, 3439–3456.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrell J. A., O’Farrell P. H., From egg to gastrula: How the cell cycle is remodeled during the Drosophila mid-blastula transition. Annu. Rev. Genet. 48, 1–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger F., Endosperm development. Curr. Opin. Plant Biol. 2, 28–32 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Boyle W. J., Simonet W. S., Lacey D. L., Osteoclast differentiation and activation. Nature 423, 337–342 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Chal J., Pourquié O., Making muscle: Skeletal myogenesis in vivo and in vitro. Development 144, 2104–2122 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Rao P. N., Johnson R. T., Mammalian cell fusion: Studies on the regulation of DNA synthesis and mitosis. Nature 225, 159–164 (1970). [DOI] [PubMed] [Google Scholar]
- 8.Johnson R. T., Rao P. N., Nucleo-cytoplasmic interactions in the achievement of nuclear synchrony in DNA synthesis and mitosis in multinucleate cells. Biol. Rev. 46, 97–155 (1971). [DOI] [PubMed] [Google Scholar]
- 9.Westerveld A., Freeke M. A., Cell cycle of multinucleate cells after cell fusion. Exp. Cell Res. 65, 140–144 (1971). [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S., Paweletz N., Ghosh I., Mitotic asynchrony of multinucleate cells in tissue culture. Chromosoma 65, 293–300 (1978). [DOI] [PubMed] [Google Scholar]
- 11.Gladfelter A. S., Hungerbuehler A. K., Philippsen P., Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172, 347–362 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C., Zhang H., Baker A. E., Occhipinti P., Borsuk M. E., Gladfelter A. S., Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev. Cell. 25, 572–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson C. A., Eser U., Korndorf T., Borsuk M. E., Skotheim J. M., Gladfelter A. S., Nuclear repulsion enables division autonomy in a single cytoplasm. Curr. Biol. 23, 1999–2010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H., Elbaum-Garfinkle S., Langdon E. M., Taylor N., Occhipinti P., Bridges A. A., Brangwynne C. P., Gladfelter A. S., RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization, World Malaria Report 2020: 20 Years of Global Progress and Challenges (World Health Organization, 2020). [Google Scholar]
- 16.Burda P.-C., Schaffner M., Kaiser G., Roques M., Zuber B., Heussler V. T., A Plasmodium plasma membrane reporter reveals membrane dynamics by live-cell microscopy. Sci. Rep. 7, 9740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudlaff R. M., Kraemer S., Marshman J., Dvorin J. D., Three-dimensional ultrastructure of Plasmodium falciparum throughout cytokinesis. PLOS Pathog. 16, e1008587 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dondorp A. M., Desakorn V., Pongtavornpinyo W., Sahassananda D., Silamut K., Chotivanich K., Newton P. N., Pitisuttithum P., Smithyman A. M., White N. J., Day N. P. J., Estimation of the total parasite biomass in acute falciparum malaria from plasma PfHRP2. PLOS Med. 2, e204 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowman A. F., Healer J., Marapana D., Marsh K., Malaria: Biology and disease. Cell 167, 610–624 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Mancio-Silva L., Lopez-Rubio J. J., Claes A., Scherf A., Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum. Nat. Commun. 4, 1530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrade C. M., Fleckenstein H., Thomson-Luque R., Doumbo S., Lima N. F., Anderson C., Hibbert J., Hopp C. S., Tran T. M., Li S., Niangaly M., Cisse H., Doumtabe D., Skinner J., Sturdevant D., Ricklefs S., Virtaneva K., Asghar M., Homann M. V., Turner L., Martins J., Allman E. L., N’Dri M.-E., Winkler V., Llinás M., Lavazec C., Martens C., Färnert A., Kayentao K., Ongoiba A., Lavstsen T., Osório N. S., Otto T. D., Recker M., Traore B., Crompton P. D., Portugal S., Increased circulation time of Plasmodium falciparum underlies persistent asymptomatic infection in the dry season. Nat. Med. 26, 1929–1940 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Simon C. S., Stürmer V. S., Guizetti J., How many is enough?—Challenges of multinucleated cell division in malaria parasites. Front. Cell. Infect. Microbiol. 11, 658616 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Read M., Sherwin T., Holloway S. P., Gull K., Hyde J. E., Microtubular organization visualized by immunofluorescence microscopy during erythrocytic schizogony in Plasmodium falciparum and investigation of post-translational modifications of parasite tubulin. Parasitology 106, 223–232 (1993). [DOI] [PubMed] [Google Scholar]
- 24.Francia M. E., Striepen B., Cell division in apicomplexan parasites. Nat. Rev. Microbiol. 12, 125–136 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Reininger L., Wilkes J. M., Bourgade H., Miranda-Saavedra D., Doerig C., An essential Aurora-related kinase transiently associates with spindle pole bodies during Plasmodium falciparum erythrocytic schizogony. Mol.Microbiol. 79, 205–221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnot D. E., Ronander E., Bengtsson D. C., The progression of the intra-erythrocytic cell cycle of Plasmodium falciparum and the role of the centriolar plaques in asynchronous mitotic division during schizogony. Int. J. Parasitol. 41, 71–80 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Simon C. S., Funaya C., Bauer J., Voβ Y., Machado M., Penning A., Klaschka D., Cyrklaff M., Kim J., Ganter M., Guizetti J., An extended DNA-free intranuclear compartment organizes centrosome microtubules in malaria parasites. Life Sci. Alliance 4, e202101199 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnot D. E., Gull K., The Plasmodium cell cycle: Facts and questions. Ann. Trop. Med. Parasit. 92, 361–365 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Gupta A., Mehra P., Dhar S. K., Plasmodium falciparum origin recognition complex subunit 5: Functional characterization and role in DNA replication foci formation. Mol Microbiol. 69, 646–665 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janse C. J., van der Klooster P. F. J., van der Kaay H. J., van der Ploeg M., Overdulve J. P., DNA synthesis in Plasmodium berghei during asexual and sexual development. Mol. Biochem. Parasit. 20, 173–182 (1986). [DOI] [PubMed] [Google Scholar]
- 31.Leete T. H., Rubin H., Malaria and the cell cycle. Parasitol. Today 12, 442–444 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Swift H. H., The desoxyribose nucleic acid content of animal nuclei. Physiol. Zool. 23, 169–198 (1950). [DOI] [PubMed] [Google Scholar]
- 33.Bucevičius J., Keller-Findeisen J., Gilat T., Hell S. W., Lukinavičius G., Rhodamine–Hoechst positional isomers for highly efficient staining of heterochromatin. Chem. Sci. 10, 1962–1970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choe K. N., Moldovan G.-L., Forging ahead through darkness: PCNA. Mol. Cell 65, 380–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitra P., Banu K., Deshmukh A. S., Subbarao N., Dhar S. K., Functional dissection of proliferating-cell nuclear antigens (1 and 2) in human malarial parasite Plasmodium falciparum: Possible involvement in DNA replication and DNA damage response. Biochem. J. 470, 115–129 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Banu K., Mitra P., Subbarao N., Dhar S. K., Role of tyrosine residue (Y213) in nuclear retention of PCNA1 in human malaria parasite Plasmodium falciparum. FEMS Microbiol. Lett. 365, (2018). [DOI] [PubMed] [Google Scholar]
- 37.Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., Imamura T., Ogawa M., Masai H., Miyawaki A., Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Facchetti G., Chang F., Howard M., Controlling cell size through sizer mechanisms. Curr. Opin. Syst. Biol. 5, 86–92 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ondracka A., Dudin O., Ruiz-Trillo I., Decoupling of nuclear division cycles and cell size during the coenocytic growth of the ichthyosporean Sphaeroforma arctica. Curr. Biol. 28, 1964–1969.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spudich J. L., Koshland D. E., Non-genetic individuality: Chance in the single cell. Nature 262, 467–471 (1976). [DOI] [PubMed] [Google Scholar]
- 41.Fox T. O., Pardee A. B., Animal cells: Noncorrelation of length of G1 phase with size after mitosis. Science 167, 80–82 (1970). [DOI] [PubMed] [Google Scholar]
- 42.Zetterberg A., Larsson O., Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc. Natl. Acad. Sci. U.S.A. 82, 5365–5369 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nair D. R., D’Ausilio C. A., Occhipinti P., Borsuk M. E., Gladfelter A. S., A conserved G1 regulatory circuit promotes asynchronous behavior of nuclei sharing a common cytoplasm. Cell Cycle. 9, 3795–3803 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews H., Duffy C. W., Merrick C. J., Checks and balances? Parasit. Vectors 11, 216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roques M., Wall R. J., Douglass A. P., Ramaprasad A., Ferguson D. J. P., Kaindama M. L., Brusini L., Joshi N., Rchiad Z., Brady D., Guttery D. S., Wheatley S. P., Yamano H., Holder A. A., Pain A., Wickstead B., Tewari R., Plasmodium P-type cyclin CYC3 modulates endomitotic growth during oocyst development in mosquitoes. PLOS Pathog. 11, e1005273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganter M., Goldberg J. M., Dvorin J. D., Paulo J. A., King J. G., Tripathi A. K., Paul A. S., Yang J., Coppens I., Jiang R. H. Y., Elsworth B., Baker D. A., Dinglasan R. R., Gygi S. P., Duraisingh M. T., Plasmodium falciparum CRK4 directs continuous rounds of DNA replication during schizogony. Nat. Microbiol. 2, 17017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dundon S. E. R., Chang S.-S., Kumar A., Occhipinti P., Shroff H., Roper M., Gladfelter A. S., Clustered nuclei maintain autonomy and nucleocytoplasmic ratio control in a syncytium. Mol. Biol. Cell 27, 2000–2007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerbich T. M., McLaughlin G. A., Cassidy K., Gerber S., Adalsteinsson D., Gladfelter A. S., Phosphoregulation provides specificity to biomolecular condensates in the cell cycle and cell polarity. J. Cell Biol. 219, e201910021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grava S., Keller M., Voegeli S., Seger S., Lang C., Philippsen P., Clustering of nuclei in multinucleated hyphae is prevented by dynein-driven bidirectional nuclear movements and microtubule growth control in Ashbya gossypii. Eukaryot. Cell 10, 902–915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gladfelter A. S., Nuclear anarchy: Asynchronous mitosis in multinucleated fungal hyphae. Curr. Opin. Microbiol. 9, 547–552 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Mazanka E., Weiss E. L., Sequential counteracting kinases restrict an asymmetric gene expression program to early G1. Mol. Biol. Cell 21, 2809–2820 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cutler A. A., Jackson J. B., Corbett A. H., Pavlath G. K., Non-equivalence of nuclear import among nuclei in multinucleated skeletal muscle cells. J. Cell Sci. 131, jcs207670 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cardoso M. C., Leonhardt H., Nadal-Ginard B., Reversal of terminal differentiation and control of DNA replication: Cyclin A and cdk2 specifically localize at subnuclear sites of DNA replication. Cell 74, 979–992 (1993). [DOI] [PubMed] [Google Scholar]
- 54.Dorin-Semblat D., Sicard A., Doerig C., Ranford-Cartwright L., Doerig C., Disruption of the PfPK7 gene impairs schizogony and sporogony in the human malaria parasite Plasmodium falciparum. Eukaryot. Cell 7, 279–285 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dorin-Semblat D., Carvalho T. G., Nivez M.-P., Halbert J., Poullet P., Semblat J.-P., Goldring D., Chakrabarti D., Mehra P., Dhar S., Paing M. M., Goldberg D. E., McMillan P. J., Tilley L., Doerig C., An atypical cyclin-dependent kinase controls Plasmodium falciparum proliferation rate. Kinome 1, 4–16 (2013). [Google Scholar]
- 56.Mancio-Silva L., Slavic K., Ruivo M. T. G., Grosso A. R., Modrzynska K. K., Vera I. M., Sales-Dias J., Gomes A. R., MacPherson C. R., Crozet P., Adamo M., Baena-Gonzalez E., Tewari R., Llinás M., Billker O., Mota M. M., Nutrient sensing modulates malaria parasite virulence. Nature 547, 213–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ch’ng J.-H., Moll K., Wyss K., Hammar U., Rydén M., Kämpe O., Färnert A., Wahlgren M., Enhanced virulence of Plasmodium falciparum in blood of diabetic patients. PLOS ONE 16, e0249666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ponnudurai T., Leeuwenberg A. D., Meuwissen J. H., Chloroquine sensitivity of isolates of Plasmodium falciparum adapted to in vitro culture. Trop. Geogr. Med. 33, 50–54 (1981). [PubMed] [Google Scholar]
- 59.Trager W., Jensen J., Human malaria parasites in continuous culture. Science 193, 673–675 (1976). [DOI] [PubMed] [Google Scholar]
- 60.Lambros C., Vanderberg J. P., Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65, 418–420 (1979). [PubMed] [Google Scholar]
- 61.Birnbaum J., Flemming S., Reichard N., Soares A. B., Mesén-Ramírez P., Jonscher E., Bergmann B., Spielmann T., A genetic system to study Plasmodium falciparum protein function. Nat. Methods 14, 450–456 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Fidock D. A., Wellems T. E., Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U.S.A. 94, 10931–10936 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Y., Sifri C. D., Lei H. H., Su X. Z., Wellems T. E., Transfection of Plasmodium falciparum within human red blood cells. Proc. Natl. Acad. Sci. U.S.A. 92, 973–977 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim M. Y.-X., LaMonte G., Lee M. C. S., Reimer C., Tan B. H., Corey V., Tjahjadi B. F., Chua A., Nachon M., Wintjens R., Gedeck P., Malleret B., Renia L., Bonamy G. M. C., Ho P. C.-L., Yeung B. K. S., Chow E. D., Lim L., Fidock D. A., Diagana T. T., Winzeler E. A., Bifani P., UDP-galactose and acetyl-CoA transporters as Plasmodium multidrug resistance genes. Nat. Microbiol. 1, 16166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacPherson C. R., Scherf A., Flexible guide-RNA design for CRISPR applications using Protospacer Workbench. Nat. Biotechnol. 33, 805–806 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B. L., Xavier R. J., Root D. E., Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paul-Gilloteaux P., Heiligenstein X., Belle M., Domart M.-C., Larijani B., Collinson L., Raposo G., Salamero J., eC-CLEM: Flexible multidimensional registration software for correlative microscopies. Nat. Methods. 14, 102–103 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Kremer J. R., Mastronarde D. N., McIntosh J. R., Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- 70.Grüring C., Spielmann T., Imaging of live malaria blood stage parasites. Methods Enzymol. 506, 81–92 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S7
Table S1
Movies S1 to S6