Abstract
Dynamic nuclear polarization (DNP) is a cutting-edge technique that markedly enhances the detection sensitivity of molecules using nuclear magnetic resonance (NMR)/magnetic resonance imaging (MRI). This methodology enables real-time imaging of dynamic metabolic status in vivo using MRI. To expand the targetable metabolic reactions, there is a demand for developing exogenous, i.e., artificially designed, DNP-NMR molecular probes; however, complying with the requirements of practical DNP-NMR molecular probes is challenging because of the lack of established design guidelines. Here, we report Ala-[1-13C]Gly-d2-NMe2 as a DNP-NMR molecular probe for in vivo detection of aminopeptidase N activity. We developed this probe rationally through precise structural investigation, calculation, biochemical assessment, and advanced molecular design to achieve rapid and detectable responses to enzyme activity in vivo. With the fabricated probe, we successfully detected enzymatic activity in vivo. This report presents a comprehensive approach for the development of artificially derived, practical DNP-NMR molecular probes through structure-guided molecular design.
In vivo detection of aminopeptidase N has been achieved by highly sensitive molecular probe for dynamic nuclear polarization MRI.
INTRODUCTION
Nuclear magnetic resonance imaging (NMR/MRI) is a powerful method for metabolic imaging. Metabolic reactions of endogenous molecules labeled with low-γ nuclei, such as 13C and 15N, can be observed with NMR. However, such an application in MRI is limited because of the extremely low sensitivity of low-γ nuclei compared with that of 1H, abundant tissue water (1–4). In 2003, Ardenkjær-Larsen et al. (5) reported dissolution dynamic nuclear polarization (dDNP) as a powerful method for overcoming this issue. Through dDNP, the NMR sensitivity of measured low-γ nuclei in a molecular probe is markedly enhanced by polarization transfer from electron spins to nuclear spins when a sample is subjected to the special conditions of cryogenic temperature, high magnetic field, and microwave irradiation. The hyperpolarized molecular probes can then be used for real-time MRI after quick dissolution (6). The NMR sensitivity of hyperpolarized molecules can be enhanced 104- to 106-fold in 13C and 15N nuclei. For instance, dDNP has been clinically applied to detect elevated lactate dehydrogenase activity in vivo using hyperpolarized [1-13C]pyruvate for diagnosing prostate cancer (7).
Although dDNP is useful for a variety of in vivo studies including in humans, only a limited number of molecular probes can be practically used in vivo (8–10). This situation is due to a number of physicochemical and biological requisites needed for the 13C- or 15N-labeled dynamic nuclear polarization (DNP)-NMR molecular probes. Because the hyperpolarized state of the nuclear spin decays exponentially depending on the spin-lattice relaxation value (T1), the labeled atom in the molecular probe should have a sufficiently long T1 for monitoring under physiological conditions (requisite 1). To detect enzymatic activity, target selectivity (requisite 2), rapid enzymatic reaction (requisite 3), and a large chemical shift difference between the substrate and the products (requisite 4) are also required to produce detectable signals and distinguish each component as a separate peak. Other fundamental properties include water solubility (requisite 5), biocompatibility (requisite 6), biostability (requisite 7), and high hyperpolarization efficiency (requisite 8).
Successful examples that meet the above requisites were found in naturally occurring endogenous small molecules labeled with 13C stable isotopes (8–10). These are sometimes further modified by deuteration and esterification to improve their physicochemical and pharmacokinetic properties. Some of these DNP-NMR molecular probes, including [1-13C]pyruvate, function in vivo and are used for biological and medical applications (11–16). Nonetheless, practical DNP-NMR molecular probes are still few. Although endogenous molecules have advantages in biocompatibility and in tracing their metabolic pathways, such molecules do not usually satisfy several of the above requisites. This situation leads to very limited enzymes and metabolic reactions that can be probed with DNP-NMR/MRI.
An alternative approach is the development of exogenous, i.e., artificially designed, molecular probes (8–10). This approach is effective for developing molecular probes to satisfy the requisites because molecular properties can be optimized through chemical structures and appropriate chemical modifications. However, there are no established guidelines for the design of such DNP-NMR molecular probes. Moreover, in terms of metabolic analysis, only a few artificially designed probes are useful in vivo (17, 18). Even if probe and metabolite signals can be detected in vivo, they are often barely visible, making these probes impractical for biological evaluation and other applications. Thus, developing DNP-NMR molecular probes applicable in vivo by rational molecular design is critical, and the establishment of necessary design guidelines is a great challenge in this field.
Aminopeptidase N (APN) (CD13) plays several important roles throughout the body. This zinc-containing enzyme hydrolyzes peptides at the N terminus by preferentially recognizing neutral residues such as alanine. APN has attracted attention because of its relevance in various physiological systems and diseases, including the immune system, pain sensation, and blood pressure regulation (19, 20). In particular, APN is highly expressed in a variety of tumors and is associated with malignancy, angiogenesis, and metastasis, making it an important therapeutic target and biomarker for cancer (21–26). The expression and localization of APN have been investigated using molecular probes that bind specifically to APN (24, 26); the direct detection of APN activity is also of interest because APN is a multifunctional enzyme that has roles other than that of a peptidase (19, 27). Although some luminescence and fluorescence probes for APN activity have been reported and partially applied for in vivo imaging, detection of APN activity has not yet been achieved because of the low transparency of light in deeper regions of the body (28–30).
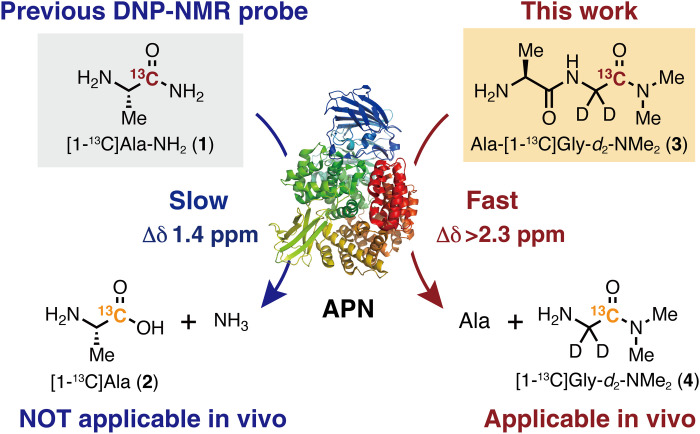
In 2016, we reported [1-13C]Ala-NH2 (1) as a DNP-NMR molecular probe (Fig. 1) that successfully detected APN activity in vitro (31). This molecule was designed to have APN specificity and prolonged T1. However, APN activity detection in vivo was not achieved because [1-13C]Ala-NH2 (1) did not satisfy several of the abovementioned requisites, such as rapid enzymatic reaction and large chemical shift change.
Fig. 1. Previous and new DNP-NMR probes for the detection of APN activity in vivo.
Here, we developed Ala-[1-13C]Gly-d2-NMe2 (3) as a practical DNP-NMR molecular probe for the detection of APN activity in vivo (Fig. 1) via structure-guided design to satisfy the requirements for in vivo application. Notably, slow and insufficient enzymatic conversion, which was a major disadvantage in previous probes, was overcome by focusing on the enzymatic parameter Km, which was previously overlooked. This probe enabled in vivo detection of APN activity in tumor xenograft mice; moreover, distinct parent 3 and corresponding product 4 signals were clearly observed in vivo in the magnetic resonance (MR) spectra (MRS), and chemical shift imaging (CSI) was successfully obtained. There are only a few DNP-NMR probes that provide large signals of the enzymatic products in vivo, and to the best of our knowledge, this is the first example of structure-guided design of an artificial DNP-NMR molecular probe that can provide such clear metabolic signals (8, 9). In this report, we demonstrate that artificial DNP-NMR molecular probes applicable in vivo can be developed by rational molecular design.
RESULTS
Design strategy for a practical DNP-NMR molecular probe that can detect APN activity in vivo
The previously reported probe [1-13C]Ala-NH2 (1) could not be used in vivo because of its (i) slow enzymatic reactions that led to an insufficient accumulation of the product and (ii) small chemical shift change [Δδ 1.4 parts per million (ppm)] that resulted in inseparable probe and product peaks (31). In particular, it was a serious issue that the metabolite is barely observed because of the weak product signals even in vitro. Thus, we envisioned that solving these problems, which are common issues in many other cases, would contribute to establishing design guidelines for DNP-NMR molecular probes (9).
We commenced this study by improving the enzymatic reaction rate of Ala-NH2 (5). In our previous work, Ala-NH2 (5) was found to have a lower affinity to rat APN compared with that of natural substrate Ala-Gly (7) [Km(Ala-NH2) = 9.7 mM versus Km(Ala-Gly) = 3.6 mM] and, instead, to have a higher turnover number [kcat(Ala-NH2) = 210 s−1 versus kcat(Ala-Gly) = 74 s−1] (31). Given that the typical blood concentration of the probe injected into mice is in the low millimolar range (13), the inefficiency of the enzymatic reaction was attributed to the large Km(Ala-NH2) and not to kcat(Ala-NH2). Therefore, improving Km, i.e., enhancing affinity to APN, is expected to be key.
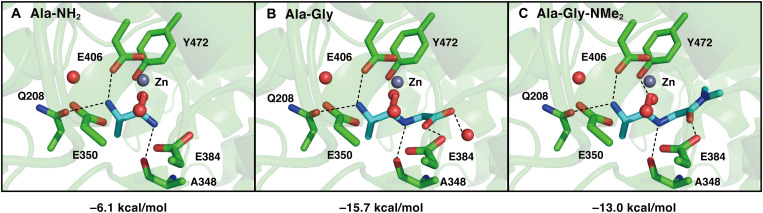
To improve affinity for APN, we calculated the binding energies of Ala-NH2 (5) and Ala-Gly (7) in the active pocket of APN (Fig. 2, A and B) using the reported cocrystal structure of porcine APN with polyalanine (Protein Data Bank ID: 4NAQ) (32). Porcine APN has high sequence identity and similar kinetic properties with human APN (hAPN) (32). As Km(Ala-NH2) is larger than Km(Ala-Gly), the calculated binding energy of Ala-Gly (7) was lower than that of Ala-NH2 (5), as protonated Glu384 donated a hydrogen bond to the substrate carbonyl oxygen site in the Ala-Gly-APN structure optimized using a quantum mechanical/molecular mechanical (QM/MM) approach. The corresponding hydrogen bond was absent in the QM/MM-optimized Ala-NH2-APN structure (Fig. 2A). Notably, the calculated binding energy was similar for these substrates when Glu384 was ionized and the hydrogen bond was absent (fig. S1). These results suggest that hydrogen bond formation between protonated Glu384 and the substrate (Fig. 2B) plays a important role in increasing substrate binding affinity to APN and decreasing Km. Accordingly, we concluded that the carbonyl oxygen of the second residue in the N terminus is crucial for increasing APN affinity.
Fig. 2. Substrate binding sites and binding energy.
QM/MM-optimized geometries of (A) Ala-NH2-APN, (B) Ala-Gly-APN, and (C) Ala-Gly-NMe2-APN. Red balls indicate water molecules. Dashed lines indicate electrostatic interactions. The calculated binding energies are described below the figures.
Exploration of molecular structure to achieve fast enzymatic reaction
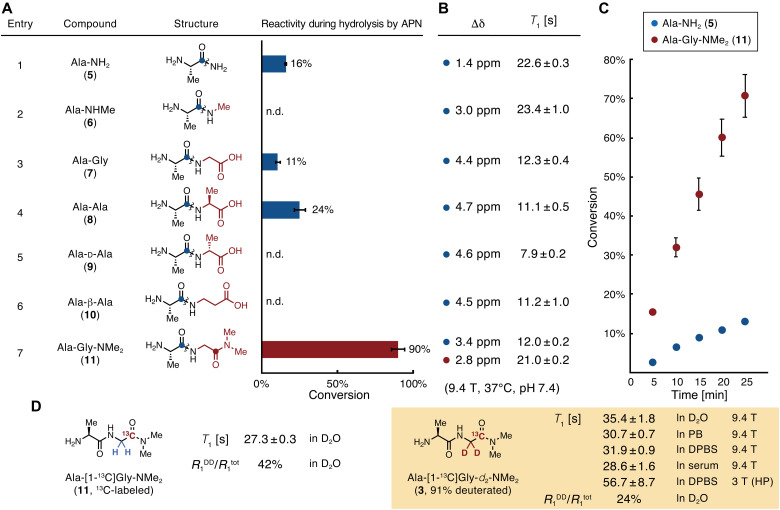
According to the suggestion above that carbonyl oxygen of the second residue from the N terminus is effective for affinity to APN, the molecular structures of the substrate were evaluated to improve the enzymatic reaction rate for hAPN. The efficiency of hydrolysis of the candidate substrates by APN was tested at a substrate concentration of 3.0 mM, assuming that the probe concentration is low millimolar when injected into mice (Fig. 3A). The conversion of Ala-NH2 (5) was 16% under the tested conditions (entry 1). Ala-NHMe (6), in which the carboxyl group was removed from the structure of Ala-Gly (7), showed low reactivity (entry 2); this was consistent with the theoretical calculation results. The conversion rates of Ala-NH2 (5) and Ala-Gly (7) were similar (entries 1 versus 3), probably because of the trade-off between their Km and kcat values. Moreover, Ala-Ala (8) was converted at a higher rate than Ala-Gly (7) (entry 4). Although the natural dipeptides Ala-Gly (7) and Ala-Ala (8) are promising candidates, their kcat to APN is reported to be low, and they may be cleaved by dipeptidyl peptidases and carboxyl peptidases in the body (31).
Fig. 3. Reaction efficiencies for APN, chemical shift changes, and spin-lattice relaxation times of APN probe candidates.
(A) A series of probe candidates and their conversion rates during enzymatic reactions with APN. Conditions: 3.0 mM compound in 100 mM phosphate buffer (pH 7.4), hAPN (9.7 nM), 37°C, 40 min, n = 3. Wavy lines indicate bonds that are cleaved by APN. (B) Magnetic parameters of the 13C atoms in probe candidates, highlighted as blue or red circles in (A). Δδ, change in 13C chemical shift between probe and product generated by hydrolysis by APN. Measurement conditions, 100 mM each of the probe and product in phosphate buffer [90 mM (pH 7.4)] containing 10% D2O, 37°C. T1, spin-lattice relaxation time. Measurement conditions, 500 mM compound in phosphate buffer [90 mM (pH 7.4)] containing 10% D2O, 37°C, 9.4 T. (C) Time course of Ala-NH2 (5) and Ala-Gly-NMe2 (11) conversion by APN. Conditions, 3.0 mM compound in phosphate buffer [100 mM (pH 7.4)], hAPN (9.7 nM), 37°C, n = 3. (D) T1 values and contributions of R1DD of Ala-[1-13C]Gly-NMe2 and Ala-[1-13C]Gly-d2-NMe2. Measurement conditions, 10 mM compound in D2O (In D2O), in phosphate buffer [90 mM (pH 7.4)] containing 10% D2O (In PB), in DPBS containing 10% D2O (In DPBS), or in human serum containing 10% D2O (In serum), 37°C, 9.4 T; 3 T (HP), ca. 1.4 mM compound in DPBS containing 3.4 mM glycerol, 32 μM OX063, and 227 μM EDTA, 37 °C, 3.0 T under a hyperpolarized state. R1DD, inverse of T1 relaxation time by dipole-dipole interaction; R1tot, the total value of the inverse of T1 relaxation times. HP, hyperpolarized state. Error bars indicate the SD. n.d. indicates that the product (Ala) was not detected.
Thus, we sought a structure similar to that of Ala-Gly (7) with a large kcat and resistance to nonexpected enzymatic digestions. Although Ala-d-Ala (9) and Ala-β-Ala (10), in which the C termini are unnatural amino acids, are expected to evade recognition by dipeptidyl peptidases and carboxyl peptidases, their reactions with APN were very slow (entries 5 and 6). On the basis of entries 3 and 4, we assumed that hydrophobicity at the C-terminal residue enhances reactivity. Thus, Ala-Gly-NMe2 (11) was designed to retain the hydrogen bond between the carbonyl oxygen and glutamate in the APN pocket, avoid enzymatic digestion by dipeptidyl peptidases and carboxyl peptidases, and exhibit increased hydrophobicity at the C terminus. Subsequently, this compound showed a much faster reaction with APN (entry 7) compared with that of the others. Figure 3C displays the conversion time course of Ala-Gly-NMe2 (11), which was much faster than that of Ala-NH2 (5).
Structural investigation of kinetic parameters
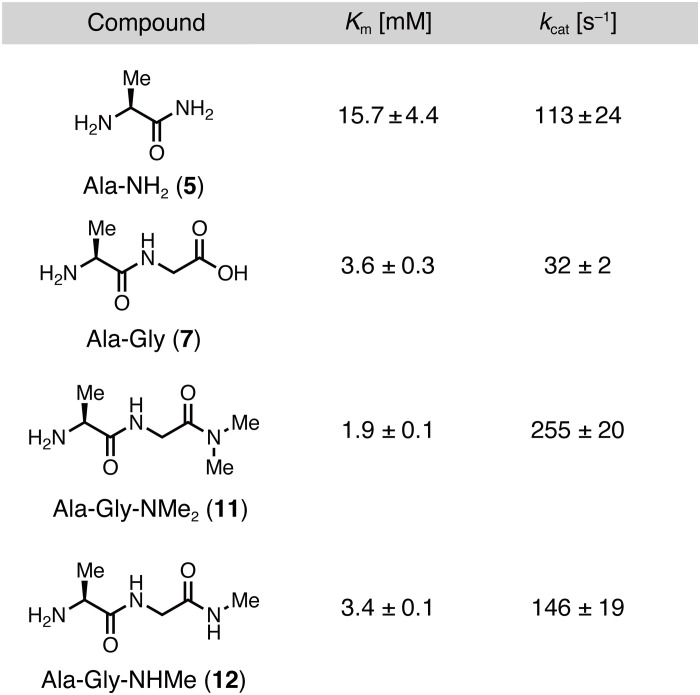
To confirm the validity of our enzymatic kinetic parameter-based strategy and to elucidate the mechanism by which Ala-Gly-NMe2 (11) shows a high hydrolysis rate by APN, we determined the Km and kcat of Ala-NH2 (5), Ala-Gly (7), Ala-Gly-NMe2 (11), and Ala-Gly-NHMe (12) for hAPN (Fig. 4). Consistent with previous results on rat APN (31), Km(Ala-Gly) was lower than Km(Ala-NH2), and kcat(Ala-Gly) was lower than kcat(Ala-NH2), indicating that the Gly moiety contributes to the increased APN affinity and decreased catalytic turnover number. Moreover, Ala-Gly-NMe2 (11) showed slightly lower Km than that of Ala-Gly (7), suggesting that the Gly moiety in the second residue contributes to the increased APN affinity. These findings are consistent with the calculated binding energies (Fig. 2C and fig. S1). Furthermore, Ala-Gly-NMe2 (11) showed a higher kcat than that of Ala-Gly (7), indicating that the dimethylamide group increases kcat. This is supported by the fact that Ala-Gly-NHMe (12), in which the terminal amide group is modified with only one methyl group, has a lower kcat than that of Ala-Gly-NMe2 (11). Consequently, we conclude that the Gly moiety increased affinity to APN, while the dimethylamide group increased kcat of Ala-Gly-NMe2 (11).
Fig. 4. Structural effect of Ala-Gly-NMe2 on the rapid APN enzymatic reactions.
Kinetic parameters (Km and kcat) of Ala-Gly-NMe2 and its analogs for hAPN.
Structural investigation of molecular magnetic properties
For DNP-NMR molecular probes, satisfying the criteria for molecular magnetic properties in addition to enzymatic reaction properties is also essential. In particular, T1, which determines the hyperpolarization lifetime, and the magnitude of the chemical shift change (Δδ) are important for obtaining separate probe and product peaks for a sufficient period of time. In this structural investigation, on the basis of our experimental knowledge, we tentatively set the criteria for these two parameters under physiological conditions as T1 > 20 s and Δδ > 2.0 ppm.
We thus focused on the quaternary carbon atom of the carbonyl group, which has a relatively large T1 value (Fig. 3B) (8). While Ala-NH2 (5) has a T1 value of 22.6 s [500 mM in 90 mM phosphate buffer containing 10% D2O (pH 7.4), 37°C, 9.4 T], its Δδ value is small (1.4 ppm; entry 1). The introduction of a methyl group into the amide moiety increased Δδ at physiological pH (entries 1 versus 2). This trend of increased Δδ was observed for Ala-Gly (7), Ala-Ala (8), Ala-d-Ala (9), and Ala-β-Ala (10), but the T1 values were considerably decreased (entries 3 to 6).
The carbonyl carbon of the N-terminal Ala residue in Ala-Gly-NMe2 (11) exhibited a large Δδ value (3.4 ppm), but the T1 value (12.0 s) was small (entry 7; carbon is highlighted with a blue circle). However, this molecule has another carbonyl carbon in the C-terminal moiety (entry 7; carbon is highlighted with a red circle), which was found to satisfy our tentative criteria (T1 = 21.0 s, Δδ = 2.8 ppm). Considering that Ala-Gly-NMe2 (11) is converted by APN to two products, Ala and Gly-NMe2, and that Ala is readily incorporated into further metabolic pathways (22, 23), the Gly-NMe2 moiety would be a better tracer for monitoring APN activity. A similar trend in T1 values was observed in Dulbecco’s phosphate-buffered saline (DPBS) and in human serum at 3.0 T used for animal experiments (table S1). Therefore, Ala-[1-13C]Gly-NMe2 was chosen as a potential molecular probe candidate.
Ala-[1-13C]Gly-d2-NMe2 as a DNP-NMR probe for APN
Ala-[1-13C]Gly-d2-NMe2 (3) was lastly obtained as a molecular probe. In Ala-[1-13C]Gly-d2-NMe2 (3), the Gly moiety α-position was deuterated because the adjacent 1H nucleus to the measured 13C causes T1 relaxation due to dipole-dipole interactions (Fig. 3D) (33–35). Nondeuterated Ala-[1-13C]Gly-NMe2 had a T1 value of 27.3 s in D2O (10 mM, 37°C, 9.4 T), whereas that of Ala-[1-13C]Gly-d2-NMe2 (3) (deuterated efficiency: 91%) was 35.4 s with an elongation of about 8 s. The ratio of the dipole-dipole relaxation (R1DD) to total relaxation (R1tot) decreased from 42 to 24%, indicating that the dipole-dipole relaxation was effectively attenuated. Considering that the dipole-dipole interaction is inversely proportional to the sixth power of the distance (35), the remaining dipole-dipole relaxation is thought to originate mainly from the 1H of the dimethylamide group. Although it is possible to further deuterate the dimethylamide group, we considered that Ala-[1-13C]Gly-d2-NMe2 (3) would be the best choice for the imaging probe in terms of synthetic cost as it already has a sufficiently long T1. The details and effects of other relaxation pathways are summarized in fig. S2. Under aqueous conditions [10 mM in 90 mM phosphate buffer containing 10% D2O (pH 7.4), 37°C, 9.4 T], the T1 of the 13C nucleus in Ala-[1-13C]Gly-d2-NMe2 (3) was 30.7 ± 0.7 s and that of [1-13C]Gly-d2-NMe2 (4), the corresponding product, was 38.3 ± 2.2 s. Under the biological conditions such as DPBS and human serum, T1 values of Ala-[1-13C]Gly-d2-NMe2 (3) were hardly decreased (31.9 ± 0.9 s in DPBS and 28.6 ± 1.6 s in human serum, 37°C, 9.4 T).
Synthesis of Ala-[1-13C]Gly-d2-NMe2
Ala-[1-13C]Gly-d2-NMe2 (3) was synthesized in six steps starting from [1-13C]Gly as a 13C source at a relatively low cost (figs. S3 and S4). [1-13C]Gly-d2-NMe2 (4) was prepared by deuteration of the α-position of [1-13C]Gly catalyzed by ruthenium on carbon (36), benzyloxycarbonyl (Cbz) protection, condensation with dimethylamine, and deprotection. Ala-[1-13C]Gly-d2-NMe2 (3) was obtained via coupling with Cbz-Ala and [1-13C]Gly-d2-NMe2 (4), followed by removal of the Cbz group. The total yield was 43% from [1-13C]Gly, with 91% deuterated efficiency at the α-position of the [1-13C]Gly moiety. This synthetic protocol can be applied to gram-scale synthesis (total yield: 63%).
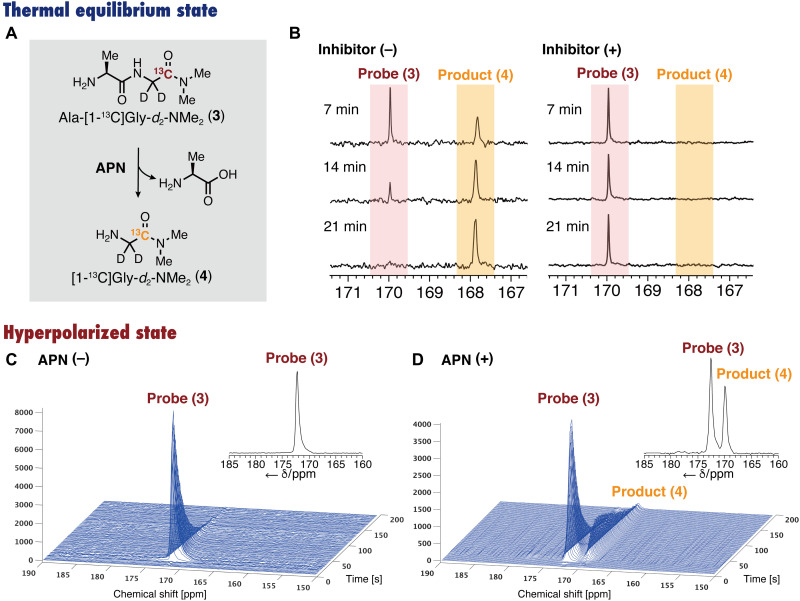
Enzymatic reaction under thermal equilibrium state
The ability of Ala-[1-13C]Gly-d2-NMe2 (3) to detect APN activity was evaluated under a thermal equilibrium state. Probe 3 was added to mouse kidney homogenate, and the conversion was monitored using 13C NMR at 37°C (Fig. 5, A and B). The 13C NMR signal of probe 3 (δ 170.0 ppm) gradually decreased, and an increase in the new signal (δ 167.7 ppm) was observed. The newly generated peak was assigned to the corresponding product [1-13C]Gly-d2-NMe2 (4) by comparison with an authentic sample. During this experiment, no peaks other than those of 3, 4, and dimethyl sulfoxide (DMSO; an internal standard) were observed, indicating that no side reactions occurred (fig. S5). In the presence of phebestin, an APN inhibitor (37), 4 was not generated, and the peaks of 3 did not change, suggesting that Ala-[1-13C]Gly-d2-NMe2 (3) can detect APN activity by selectively reacting with APN (fig. S6). Similar conversion and inhibition by phebestin were observed also in homogenate of mouse xenograft tumor tissue (MIA PaCa-2), although APN activity was lower than that in kidney homogenate (figs. S7 and S8).
Fig. 5. Detection of APN activity with hyperpolarized Ala-[1-13C]Gly-d2-NMe2 in vitro.
(A) Scheme for the reaction of Ala-[1-13C]Gly-d2-NMe2 (3) by APN to produce Ala and [1-13C]Gly-d2-NMe2 (4). (B) Thermally equilibrated 13C NMR spectra of Ala-[1-13C]Gly-d2-NMe2 (3) after incubation with mouse kidney homogenate in the absence (left) and presence (right) of phebestin, an APN inhibitor. (C and D) Hyperpolarization studies in vitro. (C) Stacked hyperpolarized 13C NMR spectra of 3 (10 mM in DPBS solution), acquired every 1 s (flip angle, 10°). (D) Detection of APN activity (250 nM; rat) in vitro using the hyperpolarized probe 3. Inset spectrum: Sum of individual scans (scans 1 to 200).
Hyperpolarization experiments in vitro
Next, we performed in vitro hyperpolarization experiments of Ala-[1-13C]Gly-d2-NMe2 (3) (Fig. 5, C and D). The aqueous solution of 4.4 M probe 3 containing 19 mM OX063 and 23% glycerol was hyperpolarized with HyperSense (Oxford Instruments, UK). The hyperpolarized sample was rapidly dissolved with hot DPBS containing 0.68 mM EDTA, and the 13C MRS (3.0 T) was then acquired at room temperature (Fig. 5C). The 13C NMR signal was observed over 160 s. The apparent T1 at 3.0 T was found to be 26 s by curve fitting. The corrected T1, taking into account the magnetization loss due to successive flip angles, was determined to be 43.7 ± 1.1 s. When this measurement was performed at 37°C, the corrected T1 was elongated to 56.7 ± 8.7 s (Fig. 3D and table S1), indicating that this probe has sufficiently long enough T1 for biological use at 3.0 T.
We further performed APN detection experiments. The hyperpolarized solution of probe 3 was added to a DPBS solution containing 250 nM APN (Fig. 5D). The peak of Ala-[1-13C]Gly-d2-NMe2 (3) appeared at δ 172.6 ppm. With a few seconds of delay, a distinct [1-13C]Gly-d2-NMe2 (4) peak appeared at δ 170.0 ppm. This indicates the successful sensitive detection of APN activity in vitro using the hyperpolarized probe 3.
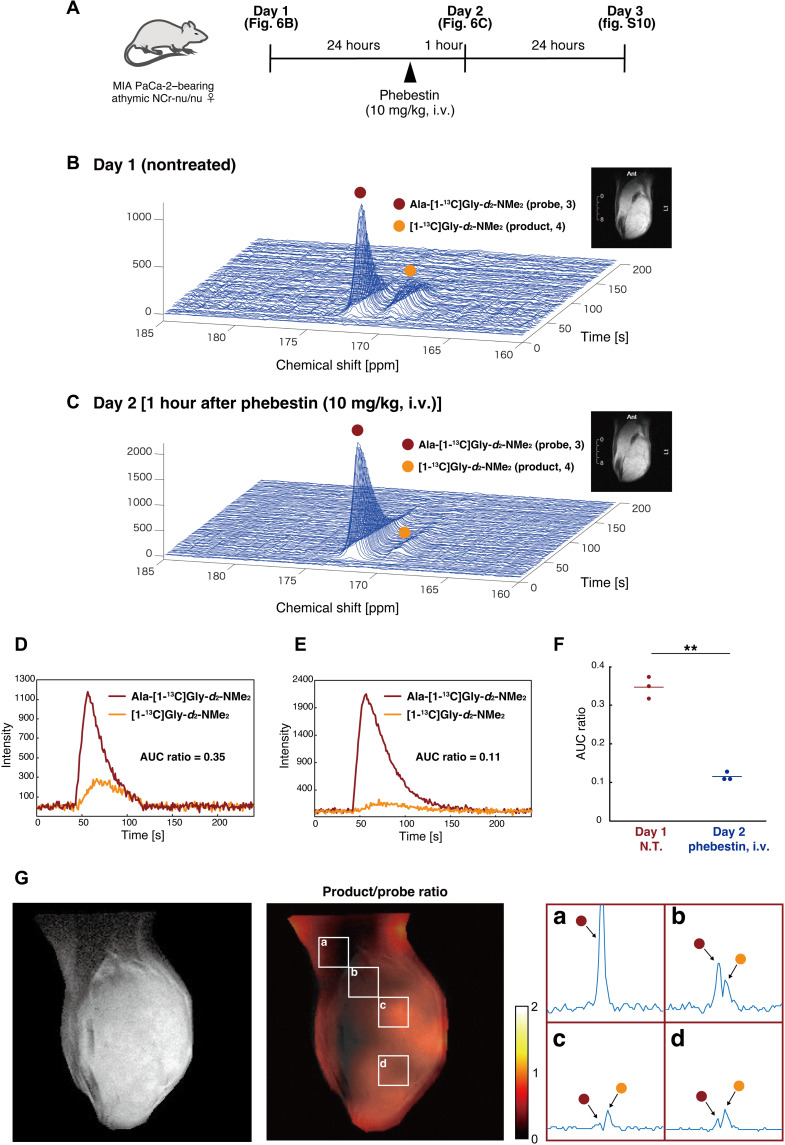
In vivo detection of APN activity
With Ala-[1-13C]Gly-d2-NMe2 (3), a potential hyperpolarized MR probe for APN, in hand, we then assessed the performance of Ala-[1-13C]Gly-d2-NMe2 (3) in direct detection of APN activity in vivo (Fig. 6). The neat sample of Ala-[1-13C]Gly-d2-NMe2 (3) (ca. 5 M) containing 19 mM OX063 was hyperpolarized in the polarizer. This sample was successfully hyperpolarized for about 1.5 to 2.5 hours without any additional glassing agents. General glassing agents such as DMSO and glycerol affect metabolic systems; this property, applicable to hyperpolarization at high concentrations without the use of glassing agents, is a distinct advantage for practical hyperpolarization. The hyperpolarized sample was rapidly dissolved with DPBS containing 0.68 mM EDTA for subsequent in vivo use.
Fig. 6. Detection of APN activity with hyperpolarized Ala-[1-13C]Gly-d2-NMe2 in vivo.
(A) Schematic representation of the experimental outline. i.v., intravenously. (B and C) Time series of 13C NMR spectra acquired in vivo with the coil placed over the leg tumor after intravenous injection of hyperpolarized probe 3. Inset: an example of the anatomical images from the observed area (1H MRI). (D and E) Time course plots of 13C signals of Ala-[1-13C]Gly-d2-NMe2 (3) (red line) and [1-13C]Gly-d2-NMe2 (4) (orange line) in (B) and (C). (F) Dot plot of the AUC ratio on days 1 and 2 (n = 3 and P = 0.009). The experiments were performed using 40 μl (n = 1) and 80 μl (n = 2) of the probe solution for hyperpolarization. AUC, area under the curve. N.T., nontreated. G) CSI of a mouse leg bearing xenograft tumor (MIA PaCa-2). Left: Anatomical image from the observed area (T2-weighted 1H MRI). Middle: Image of the product (4) to the probe (3) ratio. Right: Expanded spectra in representative voxels a to d. The voxel size is 2.8 mm × 2.8 mm × 10 mm. The left peak is Ala-[1-13C]Gly-d2-NMe2 (3; red circle), and the right peak is [1-13C]Gly-d2-NMe2 (4; orange circle). 13C spectra are with denoising process. The image was acquired at 18 to 26 s after injection of the hyperpolarized probe.
The hyperpolarized probe 3 was intravenously injected into mice bearing tumor xenografts (MIA PaCa-2), followed by acquisition of 13C MRS (3.0 T) from the tumor regions. A distinct peak derived from Ala-[1-13C]Gly-d2-NMe2 (3) was observed immediately after injection. Furthermore, the NMR signal of 4, the corresponding product by APN, was detected in a time-dependent manner (Fig. 6B). The parent and product signals were observed as distinct and separate peaks of in vivo DNP-MRS. The area under the curve (AUC) ratio, which is the ratio of the integrated peak intensities of probe 3 and product 4, was 0.35 (Fig. 6D), suggesting a sufficient conversion of the exogenous hyperpolarized MR molecular probe 3 by APN in vivo, which had not been achieved by the conventional hyperpolarized MR probe for APN (31).
Next, we examined whether hyperpolarized probe 3 can be used to directly assess the in vivo efficacy of inhibitors. Phebestin is an APN inhibitor that suppresses its activity in vivo. The day after the first injection of hyperpolarized probe 3, phebestin was administered to the same mouse, and the hyperpolarized probe 3 was again injected to monitor APN activity (Fig. 6, A and C). The AUC ratio decreased to 0.11, indicating that APN activity was suppressed by phebestin (Fig. 6, C and E). One day later, the APN activity of the same mouse was monitored without preadministration of phebestin, which confirmed the recovery of APN activity (fig. S10). A series of animal experiments showed a significant difference in the AUC ratio between nontreated and inhibitor-treated mice (Fig. 6F). These results indicate that probe 3 and product 4 do not affect APN activity and that probe 3 reacts with APN with high selectivity in vivo. Notably, no abnormalities were observed in mice when hyperpolarized probe 3 was injected.
Last, we performed CSI with the developed probe 3 (Fig. 6G and figs. S11 and S12). The hyperpolarized solution was intravenously injected to tumor xenograft mice, followed by acquisition of MR signals from each voxel in the tumor-bearing leg region. MRS with different probe-to-product ratios were obtained in each voxel. An increase in the signal ratio of the product was particularly observed in tumor regions (Fig. 6G, voxels b to d). On the other hand, a strong hyperpolarized signal of the probe only was detected in the muscle regions where large blood vessels exist (Fig. 6G, voxel a). These results suggest that the present probe is capable of site-specifically monitoring APN activities in tumors, and it can be promising as a bioimaging tool for tumor diagnosis based on APN activity and evaluation of cancer therapeutics by APN inhibition. This is the first example of a DNP-NMR probe that allows observation of APN activity in vivo.
DISCUSSION
We have successfully developed a practical DNP-NMR molecular probe to detect APN activity in vivo by structure-guided molecular design. This probe fulfills all major requisites typically found with [1-13C]pyruvate, which is widely used in preclinical and clinical research. Here, in vivo APN activity and the inhibitory effect by phebestin were clearly observed with the developed probe 3, and imaging of APN activity was successful in the tumor region, validating its practicality for in vivo biological assessment. Because of its robust performance, the Ala-[1-13C]Gly-d2-NMe2 (3) probe could serve as a powerful tool for the detection of malignancy and evaluation of treatment response, because APN, also known as CD13, is a key biomarker for malignancy, angiogenesis, and metastasis (21–26). In addition to its application as a tumor biomarker, considering the multifunctional role of APN (19, 20), the DNP-NMR probe is expected to contribute to elucidating the pathogenesis of various APN-related diseases and not just cancer.
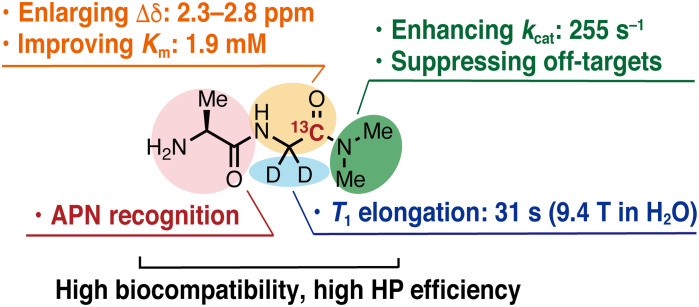
The realization of this probe is highly important from the perspective of rational design of exogenous DNP-NMR probes. Although a variety of DNP-NMR molecular probes have been reported to date, there are few examples of DNP-NMR molecular probes that provide distinct hyperpolarized NMR peaks for the probe and product in vivo. To the best of our knowledge, Ala-[1-13C]Gly-d2-NMe2 (3) is the first example of structure-guided design of an artificial DNP-NMR molecular probe that provides unambiguously distinct MRS of enzymatic conversion in vivo. This probe enabled such observation by satisfying the eight requisites for hyperpolarized MR molecular probes to function in vivo, including (i) long lifetime of hyperpolarized state, (ii) high selectivity for APN, (iii) fast enzymatic reactions at concentrations of in vivo DNP-MRI, (iv) large chemical shift change, (v) water solubility, (vi) biocompatibility, (vii) stability in blood (no change during the measurement time of DNP-MRI, a few minutes in serum; fig. S9), and (viii) high hyperpolarization efficiency. These properties are achieved by the elaborate molecular structure design in which each part plays its own role, as shown in Fig. 7. In particular, a fast enzymatic reaction (requisite 3) was achieved by focusing on the enzymatic kinetic parameter Km. The practical Km of this probe was achieved through a rational process involving calculations of the binding affinity between substrates and the target protein, explorations of appropriate molecular structures, and biochemical assessments. This report demonstrates the importance of Km and provides a set of guidelines for developing DNP-NMR molecular probes that detect enzymatic activity in vivo.
Fig. 7. Fine-tuned molecular structure of Ala-[1-13C]Gly-d2-NMe2 as a DNP-NMR probe that can detect APN activity in vivo.
Structural investigation to achieve a long T1 (requisite 1) is also an important part of this research. The total relaxation rate (R1tot = 1/T1) can be expressed as the sum of each relaxation factor, including dipole-dipole (R1DD), chemical shift anisotropy (R1CSA), spin rotation (R1SR), scalar coupling (R1SC), and all other residual relaxations (R1res) (34, 35). The carbonyl 13C of the Gly moiety in Ala-Gly-NMe2 (11) exhibited a longer T1 than that of the amide carbons in the Ala moiety (Fig. 3B). This result could be attributed to the absence of nearby exchangeable amide protons, resulting in reduced R1DD and R1SC. In addition, deuteration of the α-position of the Gly moiety eliminated R1DD between the 1H nuclei and the measured 13C, further extending T1 (Fig. 3D). Ala-[1-13C]Gly-d2-NMe2 (3) exhibited a longer T1 at 3.0 T than that at 9.4 T. This probably occurred because the T1 value of carbonyl 13C is strongly affected by R1CSA, which depends on the magnetic field strength (B0) (8). In carbonyl 13C, R1CSA is usually much larger than other B0-dependent factors (8). Under such conditions, R1tot is proportional to the square of the magnetic field (B02) (38). The measured R1tot values at 9.4, 11.7, and 14.1 T were in proportion to B02 (fig. S13). Extrapolating this plot, T1 of probe 3 in D2O at 3.0 T was predicted to be 62.5 s, which is longer than T1 measured in DPBS at 3.0 T (56.7 s). The shorter experimentally determined T1 may have occurred because of paramagnetic relaxation from the coexisting radicals. A longer T1 could be achieved by removing radicals after hyperpolarization, as is the case in clinical studies, or by adapting radical-free hyperpolarization methods.
We found a unique and useful property for this probe in the hyperpolarization process. This sample was successfully hyperpolarized without the use of any glassing agents. Because of the concern that common glassing agents, such as DMSO and glycerol, may perturb metabolic systems, methods of hyperpolarization without glassing agents or with alternative glassing agents that are biologically inert have been sought (39). The property of being hyperpolarized at high concentrations without glassing agents is advantageous for practical hyperpolarization. Although it is difficult to explain why this probe has this unique property, the dimethylamide group is considered one of the most important factors. Ala-Gly (7) readily forms a solid at room temperature. In contrast, Ala-Gly-NMe2 (11) and Ala-[1-13C]Gly-d2-NMe2 (3) are sticky and hygroscopic, allowing the preparation of solutions with extremely high concentrations and viscosities, which may prevent ice formation, crystallization, and phase separation. If this property could also be implemented by molecular design, then it would be a breakthrough in the development of DNP-NMR molecular probes.
The possibility of preclinical and clinical applications of the newly developed probe requires further experiments. However, this probe exhibits promising properties. First, the probe retained a long T1 under biological conditions, such as DPBS and human serum (Fig. 3D). Second, the probe was stable in blood within the measurement time of DNP-NMR. The probe was not digested within 20 min in the mouse serum (fig. S9). Third, the T1 at 3.0 T was 56.7 s (Fig. 3D), which is a promising length for clinical application. If further elongation in T1 is required, then one approach is to deuterate the dimethylamide group to reduce R1DD. The 14N-to-15N substitution of the nitrogen atom next to the 13C carbonyl is another approach to obtain enhanced signals. The scalar relaxation of 13C by the neighboring 14N quadrupole is not negligible at Earth’s magnetic field, resulting in hyperpolarization loss during the sample transfer process. Substitution of 14N with 15N can suppress the relaxation, as reported previously (40). Alternatively, taking the 14N-containing hyperpolarized probe in a transport magnet maintained at a higher field is another option. It should be noted that further careful and detailed studies, including those related to toxicity to the human body, are necessary.
Until now, it has been considered that the molecular design of DNP-NMR is interesting but impractical. However, this report successfully demonstrates that rational molecular design is essential for the development of practical DNP-NMR molecular probes applicable in vivo. We expect that this research will be followed by the design and development of more sophisticated molecular probes, which would further improve hyperpolarized MR for in vivo imaging.
MATERIALS AND METHODS
Enzymatic reaction with APN
The solution of the APN substrate (6.0 mM) in phosphate buffer [50.0 μl and 100 mM (pH 7.4)] was preincubated at 37°C for 10 min. hAPN (19.4 nM) in phosphate buffer [50.0 μl and 100 mM (pH 7.4)] preincubated at 37°C for 10 min was then added to the solution. The resulting solution (100 μl, 3.0 mM APN substrate, 9.7 nM hAPN) was incubated at 37°C for 40 min. The reaction was quenched by the addition of inhibitor solution (20.0 μl, 25 mM EDTA, 5.0 mM 1,10-phenanthroline in H2O). After mixing, the solution was immediately chilled on ice, followed by the addition of 400 μl of D2O containing 1,4-dioxane (an internal standard for chemical shifts). The conversion of the compound was determined using 1H NMR spectroscopy.
Chemical shift and T1 measurements
Chemical shift changes were measured using a JEOL ECS400 (9.4 T; JOEL Ltd., Tokyo, Japan). Measurement conditions included 100 mM each of the probe candidate and product in phosphate buffer containing 10% D2O [90 mM (pH 7.4), adjusted with 1 M NaOH aq.] at 37°C. T1 measurements under a thermal equilibrium state were conducted with a JEOL ECS400 (9.4 T) using the inversion recovery method. Representative measurement conditions were as follows: 500 mM for nonlabeled or 10 mM for 13C-enriched compound in phosphate buffer containing 10% D2O [90 mM (pH 7.4), adjusted with 35 weight % (wt %) DCl in D2O and 40 wt % NaOD in D2O], 37°C, and 9.4 T. T1 measurements under a hyperpolarized state (Fig. 3D) were conducted with a 3-T MR Solutions animal scanner (MR Solutions Ltd., Guildford, UK).
DNP-NMR/MRI measurements in vitro
A 4.4 M solution of probe 3 containing 23% glycerol and 19 mM OX063 (GE Healthcare, Chicago, IL) was prepared (pH adjusted to 7 with 10 M HCl aq.). The sample was submerged in liquid helium in a DNP polarizer magnet (3.35 T; HyperSense, Oxford Instruments, Oxford, UK). Hyperpolarization was achieved using microwave irradiation at 94 GHz and 100 mW for approximately 1.5 hours under 2.8 mbar at 1.45 K. Then, hyperpolarized samples were dissolved in DPBS containing 0.68 mM EDTA. DNP-NMR measurements were performed using a 3-T MR Solutions animal scanner (MR Solutions Ltd., Guildford, UK) in the absence or presence of 250 nM rat APN (164599; Merck, Darmstadt, Germany). The apparent T1 was obtained by curve fitting to signal decay profile, and the corrected T1 was estimated by considering the magnetization loss due to successive flip angles. The details are described in the Supplementary Materials.
DNP-NMR/MRI measurements in vivo
All animal experiments were carried out in compliance with the Guide for the Care and Use of Laboratory Animal Resources, and experimental protocols were approved by the Animal Care and Use Committee of the National Cancer Institute (NCI-CCR-ACUC). A solution of probe 3 (4 to 5 M) containing 19 mM OX063 was prepared (pH was adjusted to 7 with 10 M HCl aq.) and hyperpolarized as described above. The hyperpolarized solution was injected into the tail vein of athymic nude mice bearing a tumor xenograft (MIA PaCa-2) in the hind leg. Dynamic 13C MRS were acquired using the 3-T MR Solutions animal scanner (MR Solutions Ltd., UK) with a 17-mm-diameter home-built 13C solenoid coil with a saddle 1H coil. 13C two-dimensional CSIs were acquired with a field of view of 28 × 28 mm2 in a 10-mm axial slice through the tumor, a matrix size of 10 × 10, a spectral width of 3.33 kHz, a repetition time of 85 ms, and 250 ms of Gaussian excitation pulse with a flip angle of 10°. MRS and CSI data were analyzed on MATLAB (MathWorks, Natick, MA, USA) using previously reported denoising methods (41).
Acknowledgments
We thank H. Nonaka (Kyoto University) for the fruitful discussion and advice. We thank A. Senoo, S. Nagatoishi, and K. Tsumoto (The University of Tokyo) for the fruitful discussion and support with the protein expression experiments. We thank C. Gadisetti and J. R. Brender for the fruitful discussions and inputs regarding the CSI processing. The expression vector pEFs (42) was provided by A. Yamashita (Yokohama City University). We thank K. Ideta (Evaluation Center of Materials Properties and Function, Institute for Materials Chemistry and Engineering, Kyushu University) for support with the NMR measurements.
Funding: This research was supported by MEXT Q-LEAP (grant number JPMXS0120330644 to Y.T. and S.S.); JST CREST (grant numbers JPMJCR13L4 to S.S. and JPMJCR1656 to H.I.); JSPS KAKENHI (grant numbers JP19H00919 to S.S.; JP20K15396 to Y.S.; JP19KK0369 to Y.T.; JP18H05155, JP18H01937, JP20H03217, and JP20H05090 to H.I.; and JP18H01186 and JP16H06560 to K.S.); the Interdisciplinary Computational Science Program in CCS, University of Tsukuba (to K.S.); QST President’s Strategic Grant (Exploratory Research) (to Y.T.); and the Intramural Research Program of the National Cancer Institute, NIH (to R.I., T.S., K.Y., and M.C.K.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and the mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
Author contributions: S.S. conceived and designed the project. Y.S. and H.Y. synthesized and assessed the compounds under a thermal equilibrium state with the help of Y.T. Y.S., H.Y., and A.E. performed protein expression and purification. I.T., Y.K., R.I., T.S., and Y.T. performed experiments under a hyperpolarized state with the help of N.O., K.Y., and M.C.K. K.H., K.S., and H.I. performed the calculation study. Y.S., H.I., and S.S. wrote the manuscript, which was edited by all coauthors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S13
Table S1
References
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.Howe F. A., Opstad K. S., 1H MR spectroscopy of brain tumours and masses. NMR Biomed. 16, 123–131 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Ross B., Lin A., Harris K., Bhattacharya P., Scweinsburg B., Clinical experience with 13C MRS in vivo. NMR Biomed. 16, 358–369 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Gilles R. J., Morse D. L., In vivo magnetic resonance spectroscopy in cancer. Annu. Rev. Biomed. Eng. 7, 287–326 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Kohut S. J., Kaufman M. J., Magnetic resonance spectroscopy studies of substance use disorders: Current landscape and potential future directions. Pharmacol. Biochem. Behav. 200, 173090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardenkjær-Larsen J. H., Fridlund B., Gram A., Hansson G., Hansson L., Lerche M. H., Servin R., Thaning M., Golman K., Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 100, 10158–10163 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golman K., in ‘t Zandt R., Thaning M., Real-time metabolic imaging. Proc. Natl. Acad. Sci. U.S.A. 103, 11270–11275 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson S. J., Kurhanewicz J., Vigneron D. B., Larson P. E. Z., Harzstark A. L., Ferrone M., van Criekinge M., Chang J. W., Bok R., Park I., Reed G., Carvajal L., Small E. J., Munster P., Weinberg V. K., Ardenkjær-Larsen J. H., Chen A. P., Hurd R. E., Odegardstuen L. I., Robb F. J., Tropp J., Murray J. A., Metabolic imaging of patients with prostate cancer using hyperpolarized [1-13C]pyruvate. Sci. Transl. Med. 5, 198ra108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keshari K. R., Wilson D. M., Chemistry and biochemistry of 13C hyperpolarized magnetic resonance using dynamic nuclear polarization. Chem. Soc. Rev. 43, 1627–1659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo Y., Nonaka H., Takakusagi Y., Sando S., Design of nuclear magnetic resonance molecular probes for hyperpolarized bioimaging. Angew. Chem. Int. Ed. 60, 14779–14799 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Mu C., Korenchan D. E., Wang S., Wilson D. M., Flavell R. R., Tumor microenvironment biosensors for hyperpolarized carbon-13 magnetic resonance spectroscopy. Mol. Imaging Biol. 23, 323–334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart N. J., Matsumoto S., Biomedical applications of the dynamic nuclear polarization and parahydrogen induced polarization techniques for hyperpolarized 13C MR imaging. Magn. Reson. Med. Sci. 20, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meier S., Jensen P. R., Karlsson M., Lerche M. H., Hyperpolarized NMR probes for biological assays. Sensors 14, 1576–1597 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comment A., Dissolution DNP for in vivo preclinical studies. J. Magn. Reson. 264, 39–48 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Kurhanewicz J., Vigneron D. B., Ardenkjær-Larsen J. H., Bankson J. A., Brindle K., Cunningham C. H., Gallagher F. A., Keshari K. R., Kjær A., Laustsen C., Mankoff D. A., Merritt M. E., Nelson S. J., Pauly J. M., Lee P., Ronen S., Tyler D. J., Rajan S. S., Spielman D. M., Wald L., Zhang X. L., Malloy C. R., Rizi R., Hyperpolarized 13C MRI: Path to clinical translation in oncology. Neoplasia 21, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z. J., Ohliger M. A., Larson P. E. Z., Gordon J. W., Bok R. A., Slater J., Villanueva-Meyer J. E., Hess C. P., Kurhanewicz J., Vigneron D. B., Hyperpolarized 13C MRI: State of the art and future directions. Radiology 291, 273–284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Page L. M., Guglielmetti C., Taglang C., Chaumeil M. M., Imaging brain metabolism using hyperpolarized 13C magnetic resonance spectroscopy. Trends Neurosci. 43, 343–354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishihara T., Yoshihara H. A. I., Nonaka H., Takakusagi Y., Hyodo F., Ichikawa K., Can E., Bastiaansen J. A. M., Takado Y., Comment A., Sando S., Direct monitoring of γ-glutamyl transpeptidase activity in vivo using a hyperpolarized 13C-labeled molecular probe. Angew. Chem. Int. Ed. 55, 10626–10629 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Jensen P. R., Serra S. C., Miragoli L., Karlsson M., Cabella C., Poggi L., Venturi L., Tedoldi F., Lerche M. H., Hyperpolarized [1,3-13C2] ethyl acetoacetate is a novel diagnostic metabolic marker of liver cancer. Int. J. Cancer 136, E117–E126 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Mina-Osorio P., The moonlighting enzyme CD13: Old and new functions to target. Trends Mol. Med. 14, 361–371 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu C. Y., Amin M. A., Fox D. A., CD13/aminopeptidase N is a potential therapeutic target for inflammatory disorders. J. Immunol. 204, 3–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amin S. A., Adhikari N., Jha T., Design of aminopeptidase N inhibitors as anti-cancer agents. J. Med. Chem. 61, 6468–6490 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Dixon J., Kaklamanis L., Turley H., Hickson I. D., Leek R. D., Harris A. L., Gatter K. C., Expression of aminopeptidase-n (CD 13) in normal tissues and malignant neoplasms of epithelial and lymphoid origin. J. Clin. Pathol. 47, 43–47 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzman-Rojas L., Rangel R., Salameh A., Edwards J. K., Dondossola E., Kim Y. G., Saghatelian A., Giordano R. J., Kolonin M. G., Staquicini F. I., Koivunen E., Sidman R. L., Arap W., Pasqualini R., Cooperative effects of aminopeptidase N (CD13) expressed by nonmalignant and cancer cells within the tumor microenvironment. Proc. Natl. Acad. Sci. U.S.A. 109, 1637–1642 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K., Ma W. H., Li G. Q., Wang J., Yang W. D., Yap L. P., Hughes L. D., Park R., Conti P. S., Synthesis and evaluation of 64Cu-labeled monomeric and dimeric NGR peptides for MicroPET imaging of CD13 receptor expression. Mol. Pharm. 10, 417–427 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Bhagwat S. V., Lahdenranta J., Giordano R., Arap W., Pasqualini R., Shapiro L. H., CD13/APN is activated by angiogenic signals and is essential for capillary tube formation. Blood 97, 652–659 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shao Y. H., Liang W. S., Kang F., Yang W. D., Ma X. W., Li G. Y., Zong S., Chen K., Wang J., A direct comparison of tumor angiogenesis with 68Ga-labeled NGR and RGD peptides in HT-1080 tumor xenografts using microPET imaging. Amino Acids 46, 2355–2364 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Delmas B., Gelfi J., Lharidon R., Vogel L. K., Sjostrom H., Noren O., Laude H., Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature 357, 417–420 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J., Chen L. Z., Wu W. X., Zhang W., Ma Z., Cheng Y. N., Du L. P., Li M. Y., Discovery of bioluminogenic probes for aminopeptidase N imaging. Anal. Chem. 86, 2747–2751 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Chen L. Z., Sun W., Li J., Liu Z. Z., Ma Z., Zhang W., Du L. P., Xu W. F., Fang H., Li M. Y., The first ratiometric fluorescent probes for aminopeptidase N cell imaging. Org. Biomol. Chem. 11, 378–382 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Chen L. Z., Sun W., Li W. H., Li J., Du L. P., Xu W. F., Fang H., Li M. Y., The first ratiometric fluorescent probe for aminopeptidase N. Anal. Methods 4, 2661–2663 (2012). [Google Scholar]
- 31.Hata R., Nonaka H., Takakusagi Y., Ichikawa K., Sando S., Design of a hyperpolarized molecular probe for detection of aminopeptidase N activity. Angew. Chem. Int. Ed. 55, 1765–1768 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Chen L., Lin Y. L., Peng G. Q., Li F., Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. U.S.A. 109, 17966–17971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonaka H., Hata R., Doura T., Nishihara T., Kumagai K., Akakabe M., Tsuda M., Ichikawa K., Sando S., A platform for designing hyperpolarized magnetic resonance chemical probes. Nat. Commun. 4, 2441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imakura Y., Nonaka H., Takakusagi Y., Ichikawa K., Maptue N. R., Funk A. M., Khemtong C., Sando S., Rational design of [13C,D14] tert-butylbenzene as a scaffold structure for designing long-lived hyperpolarized 13C probes. Chem. Asian J. 13, 280–283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berker E. D., Shoup R. R., Farrar T. C., 13C NMR spectroscopy: Relaxation times of 13C and methods for sensitivity enhancement. Pure Appl. Chem. 32, 51–66 (1972). [Google Scholar]
- 36.Taglang C., Korenchan D. E., von Morze C., Yu J., Najac C., Wang S. N., Blecha J. E., Subramaniam S., Bok R., VanBrocklin H. F., Vigneron D. B., Ronen S. M., Sriram R., Kurhanewicz J., Wilson D. M., Flavell R. R., Late-stage deuteration of 13C-enriched substrates for T1 prolongation in hyperpolarized 13C MRI. Chem. Commun. 54, 5233–5236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagai M., Kojima F., Naganawa H., Hamada M., Aoyagi T., Takeuchi T., Phebestin, a new inhibitor of aminopeptidase N, produced by Streptomyces sp. MJ716-m3. J. Antibiot. 50, 82–84 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Blicharski J. S., Nuclear magnetic relaxation by anisotropy of the chemical shift. Z. Naturforsch. 27A, 1456–1458 (1972). [Google Scholar]
- 39.Brender J. R., Kishimoto S., Eaton G. R., Eaton S. S., Saida Y., Mitchell J., Krishna M. C., Trehalose as an alternative to glycerol as a glassing agent for in vivo DNP MRI. Magn. Reson. Med. 85, 42–48 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho A., Eskandari R., Granlund K. L., Keshari K. R., Hyperpolarized [6-13C,15N3]-arginine as a probe for in vivo arginase activity. ACS Chem. Biol. 14, 665–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brender J. R., Kishimoto S., Merkle H., Reed G., Hurd R. E., Chen A. P., Ardenkjaer-Larsen J. H., Munasinghe J., Saito K., Seki T., Oshima N., Yamamoto K., Choyke P. L., Mitchell J., Krishna M. C., Dynamic imaging of glucose and lactate metabolism by 13C-MRS without hyperpolarization. Sci. Rep. 9, 3410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López-Perrote A., Castaño R., Melero R., Zamarro T., Kurosawa H., Ohnishi T., Uchiyama A., Aoyagi K., Buchwald G., Kataoka N., Yamashita A., Llorca O., Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res. 44, 1909–1923 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong A. H. M., Zhou D. X., Rini J. M., The x-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J. Biol. Chem. 287, 36804–36813 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alger T. D., Hamill W. D. Jr., Pugmire R. J., Grant D. M., Sllcox G. D., Solum M., Carbon-13 spin-lattice relaxation in condensed aromatic compounds. J. Phys. Chem. 84, 632–636 (1980). [Google Scholar]
- 45.Wong T. C., Ang T. T., Guziec F. S. Jr., Moustakis C. A., The chemical-shift anisotropy mechanism in 77Se spin–lattice relaxation. measurement of 77Se T1 at several magnetic fields. J. Magn. Reson. 57, 463–470 (1984). [Google Scholar]
- 46.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M., CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983). [Google Scholar]
- 47.Bashford D., Karplus M., pKa’s of ionizable groups in proteins: Atomic detail from a continuum electrostatic model. Biochemistry 29, 10219–10225 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Nozaki Y., Tanford C., Acid-base titrations in concentrated guanidine hydrochloride. Dissociation constants of the guamidinium ion and of some amino acids. J. Am. Chem. Soc. 89, 736–742 (1967). [DOI] [PubMed] [Google Scholar]
- 49.Tanokura M., 1H nuclear magnetic resonance titration curves and microenvironments of aromatic residues in bovine pancreatic ribonuclease A. J. Biochem. 94, 51–62 (1983). [DOI] [PubMed] [Google Scholar]
- 50.Tanokura M., 1H-NMR study on the tautomerism of the imidazole ring of histidine residues: I. Microscopic pK values and molar ratios of tautomers in histidine-containing peptides. BBA-Protein Struc. M. 742, 576–585 (1983). [DOI] [PubMed] [Google Scholar]
- 51.Tanokura M., 1H-NMR study on the tautomerism of the imidazole ring of histidine residues: II. Microenvironments of histidine-12 and histidine-119 of bovine pancreatic ribonuclease a. BBA-Protein Struc. M. 742, 586–596 (1983). [DOI] [PubMed] [Google Scholar]
- 52.Rabenstein B., Knapp E.-W., Calculated pH-dependent population and protonation of carbon-monoxy-myoglobin conformers. Biophys. J. 80, 1141–1150 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.QSite, version 5.8, Schrödinger, LLC, New York, NY, 2012.
- 54.Jorgensen W. L., Maxwell D. S., Tirado-Rives J., Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 (1996). [Google Scholar]
- 55.Molecular Operating Environment (MOE), 2018.0101; (Chemical Computing Group Inc. Montreal, Canada, 2018).
- 56.Eguchi A., Nakakido M., Nagatoishi S., Kuroda D., Tsumoto K., Nagamune T., Kawahara M., An epitope-directed antibody affinity maturation system utilizing mammalian cell survival as readout. Biotechnol. Bioeng. 116, 1742–1751 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto S., Kishimoto S., Saito K., Takakusagi Y., Munasinghe J. P., Devasahayam N., Hart C. P., Gillies R. J., Mitchell J. B., Krishna M. C., Metabolic and physiologic imaging biomarkers of the tumor microenvironment predict treatment outcome with radiation or a hypoxia-activated prodrug in mice. Cancer Res. 78, 3783–3792 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oshima N., Ishida R., Kishimoto S., Beebe K., Brender J. R., Yamamoto K., Urban D., Rai G., Johnson M. S., Benavides G., Squadrito G. L., Crooks D., Jackson J., Joshi A., Mott B. T., Shrimp J. H., Moses M. A., Lee M.-J., Yuno A., Lee T. D., Hu X., Anderson T., Kusewitt D., Hethaway H. H., Jadhav A., Picard D., Trepel J. B., Mitchell J. B., Stott G. M., Moore W., Simeonov A., Sklar L. A., Norenberg J. P., Linehan W. M., Maloney D. J., Dang C. V., Waterson A. G., Hall M., Darley-Usmar V. M., Krishna M. C., Neckers L. M., Dynamic imaging of LDH inhibition in tumors reveals rapid In Vivo metabolic rewiring and vulnerability to combination therapy. Cell Rep. 30, 1798–1810.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiavazza E., Kubala E., Gringeri C. V., Düwel S., Durst M., Schulte R. F., Menzel M. I., Earth’s magnetic field enabled scalar coupling relaxation of 13C nuclei bound to fast-relaxing quadrupolar 14N in amide groups. J. Magn. Reson. 227, 35–38 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S13
Table S1
References
Data S1