SUMMARY
Environmental light cycles entrain circadian feeding behaviors in animals that produce rhythms in exposure to foodborne bacteria. Here, we show that the intestinal microbiota generates diurnal rhythms in innate immunity that synchronize with feeding rhythms to anticipate microbial exposure. Rhythmic expression of antimicrobial proteins was driven by daily rhythms in epithelial attachment by segmented filamentous bacteria (SFB), members of the mouse intestinal microbiota. Rhythmic SFB attachment was driven by the circadian clock through control of feeding rhythms. Mechanistically, rhythmic SFB attachment activated an immunological circuit involving group 3 innate lymphoid cells. This circuit triggered oscillations in epithelial STAT3 expression and activation that produced rhythmic antimicrobial protein expression and caused resistance to Salmonella Typhimurium infection to vary across the day-night cycle. Thus, host feeding rhythms synchronize with the microbiota to promote rhythms in intestinal innate immunity that anticipate exogenous microbial exposure.
INTRODUCTION
Virtually all animals follow circadian cycles that are tied to the rising and setting of the sun (Wier et al., 2010; Stone et al., 2012; Partch et al., 2014; Patke et al., 2020). These rhythms in natural light entrain rhythmic behaviors such as eating and sleeping. Feeding exposes animals to microorganisms in the environment or associated with food, and thus feeding rhythms produce rhythmic exposure to foodborne pathogens. To protect against infection, the mammalian intestinal epithelium produces innate immune effectors including antimicrobial proteins (AMPs), which are secreted in large quantities and are energetically expensive to produce. This suggests that intestinal innate immunity might exhibit circadian rhythms that anticipate pathogen exposure upon host food intake.
Circadian clocks synchronize physiological processes with day-night light cycles, allowing anticipation of changes in the environment. In mammals, the circadian clock is a network of transcription factors that drives rhythmic gene expression over a ~24-hour cycle. Gene expression rhythms are generated by a transcriptional-translational feedback loop in which the transcriptional activators CLOCK and BMAL1 govern expression of the repressor proteins period (PER1–3) and cryptochrome (CRY1 and CRY2), which inhibit CLOCK/BMAL1-dependent transcription. (Mohawk et al., 2012; Takahashi et al. 2017). The transcription factors REV-ERBα/β and RORα fine tune the clock mechanism (Mohawk et al., 2012; Takahashi et al. 2017). Circadian clocks are in virtually all tissues and are synchronized to environmental light cycles through neuronal and hormonal signals from the central clock in the brain (Izumo et al., 2014; Mohawk et al., 2012).
Many biological processes in the intestine exhibit daily rhythms that are generated by the circadian clock. However, the intestine is unique in that many of its circadian rhythms also require the gut microbiota. For example, the microbiota coordinates with the circadian clock to generate rhythms in the expression of genes that govern lipid metabolism (Wang et al., 2017; Kuang et al., 2019).
Another way in which the circadian clock interacts with the microbiota is through regulation of host feeding behavior (Thaiss et al., 2014; 2016; Zarrinpar et al., 2014; Leone et al., 2015). Feeding behavior is regulated by sleep-wake cycles controlled by the central circadian clock (Mohawk et al., 2012). Rhythmic feeding drives daily oscillations in microbial community composition and abundance that generate host gene expression rhythms and impact host metabolism (Thaiss et al., 2014; 2016; Leone et al., 2015). Despite a growing understanding of how microbiota-clock interactions regulate metabolism, we know little about how these interactions regulate intestinal innate immunity.
Here we show that the gut microbiota and the circadian clock coordinate to generate diurnal rhythms in intestinal innate immunity. Clock-entrained host feeding rhythms generate rhythms in intestinal surface attachment of segmented filamentous bacteria (SFB), a member of the mouse gut microbiota. Rhythmic SFB attachment drives oscillations in the expression and activation of epithelial STAT3 (signal transducer and activator of transcription 3), which generates circadian oscillations in epithelial AMP expression. These innate immune rhythms cause resistance to Salmonella Typhimurium infection to vary across the day-night cycle. Thus, the microbiota generates rhythms in intestinal innate immunity that anticipate exposure to exogenous microorganisms.
RESULTS
Diurnal rhythms in antimicrobial protein expression depend on the microbiota
We looked for rhythmic expression of intestinal innate immune effectors by first examining regenerating islet-derived protein 3γ (REG3G). REG3G is secreted by the small intestinal epithelium and kills Gram-positive bacteria (Cash et al., 2006; Mukherjee et al., 2014), limiting bacterial colonization of the mucus layer (Vaishnava et al., 2011). Expression of both the Reg3g gene and its encoded protein (REG3G) was diurnally rhythmic in conventional mice, with lower expression at Zeitgeber time (ZT) 0 than at ZT12, where ZT0 is lights on and ZT12 is lights off (Figure 1A–1D, S1). REG3G expression was lower in germ-free mice at all time points and was not rhythmic (Figure 1A–1D). Thus, REG3G expression is diurnally rhythmic and these rhythms require the microbiota.
Figure 1. Diurnal rhythms in antimicrobial protein expression depend on the microbiota.
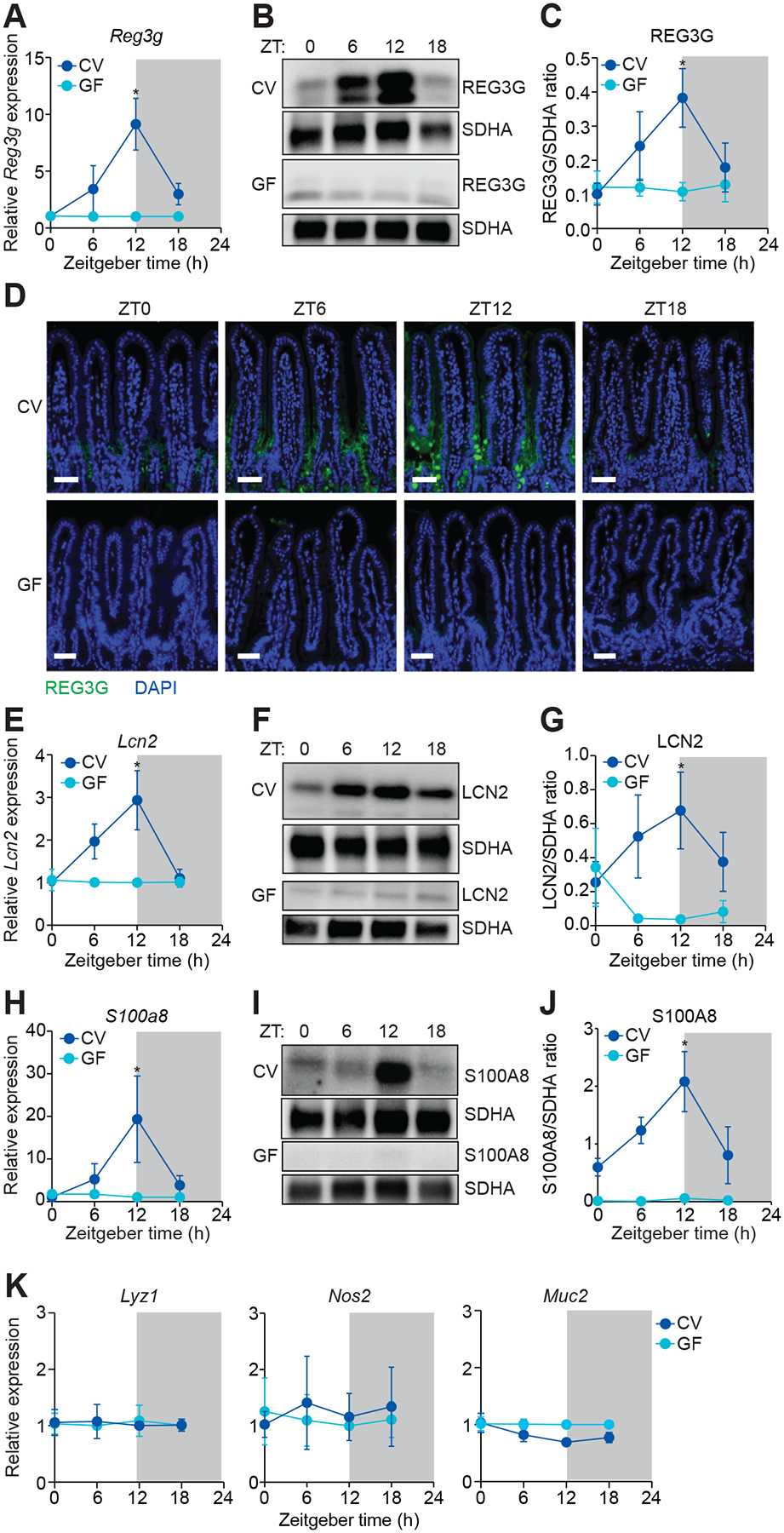
(A) Real-time quantitative PCR (Q-PCR) measurement of Reg3g transcript abundance in small intestines from conventional (CV) and germ-free (GF) mice across the day-night cycle. n=2–3 independent experiments.
(B and C) Representative immunoblot of small intestines from CV and GF mice, with detection of REG3G and succinate dehydrogenase subunit A (SDHA) as loading control (B). Band intensities were quantified by densitometry (C). n=3 independent experiments.
(D) Immunofluorescence detection of REG3G in the small intestines of CV and GF mice across the day-night cycle. Nuclei were stained with 4´,6-diamidino-2-phenylindole (DAPI). Scale bars, 50 μm.
(E) Q-PCR measurement of Lcn2 transcript abundance in small intestines from CV and GF mice across the day-night cycle. n=3 independent experiments.
(F and G) Representative immunoblot of small intestines from CV and GF mice, with detection of LCN2 and SDHA (control) (F). Band intensities were quantified by densitometry (G). n=3 independent experiments.
(H) Q-PCR measurement of S100a8 transcript abundance in small intestines from CV and GF mice across the day-night cycle. n=2–3 independent experiments.
(I and J) Representative immunoblot of small intestines from CV and GF mice, with detection of S100A8 and SDHA (control) (I). Band intensities were quantified by densitometry (J). n=3 independent experiments.
(K) Q-PCR analysis of Lyz1, Nos2, and Muc2 transcript abundance in small intestines from CV and GF mice across the day-night cycle. n=2–3 independent experiments.
ZT, Zeitgeber time. Means ± SEM are plotted; *p < 0.05 by Student’s t test. See also Figures S1 and S2 and Table S1.
Other epithelial AMPs also exhibited rhythmic expression that required the microbiota. These included lipocalin-2 (LCN2), an AMP that limits bacterial iron acquisition (Behnsen et al., 2014; Valeri and Raffatellu, 2016) (Figure 1E–1G), and S100A8, an AMP that limits bacterial calcium acquisition (Behnsen et al., 2014; Valeri and Raffatellu, 2016) (Figure 1H–1J). In contrast, expression of other innate immune genes was non-rhythmic. These included Lyz1, encoding lysozyme, and genes necessary for the generation of reactive nitrogen species (Nos2) and mucus (Muc2). Thus, rhythmic expression is characteristic of multiple AMPs, but is not a universal feature of intestinal innate immune effectors (Figures 1K, S1, and S2).
Rhythmic epithelial attachment of segmented filamentous bacteria drives diurnal rhythms in antimicrobial protein expression
We next sought to identify microbiota components that drive rhythmic AMP expression. Scanning electron microscopy (SEM) of the mouse small intestinal surface revealed numerous attaching bacteria with a segmented filamentous morphology (Figure S3A). SFB are Gram-positive members of the intestinal microbiota in rodents, non-human primates, and humans (Davis and Savage, 1974; Klaasen et al., 1993; Ley et al., 2008; Yin et al., 2013; Jonsson et al., 2020). SFB have a distinctive morphology characterized by long, segmented filaments, and have a unique ability to attach tightly to intestinal epithelial cells (Davis and Savage, 1974) (Figure S3A) and stimulate host gene expression (Ivanov et al., 2008; Ivanov et al., 2009; Sano et al., 2015; Atarashi et al., 2015).
To test whether SFB generate rhythms in REG3G expression, we studied mice from two vendors: Taconic Farms and Jackson Laboratory. SFB are present in Taconic but not Jackson mice (Ivanov et al., 2008) (Figure S3A and S3B). REG3G expression was rhythmic in Taconic but not Jackson mice (Figure 2A–2C), suggesting that SFB drive rhythmic REG3G expression in the intestinal epithelium. Indeed, SEM revealed rhythmic SFB attachment to the small intestinal surface in SFB+ (Taconic) mice (Figure 2D and S3C). Peak attachment occurred at ZT12 (Figure 2E), coinciding with peak REG3G expression (Figure 2C). In contrast, no bacteria were attached to the intestinal surface of SFB− (Jackson) mice across the day-night cycle (Figure 2D, 2E, and S3D). The overall abundance of SFB in the mucus was not rhythmic (Figure S3E), indicating that rhythmic SFB attachment was independent of SFB abundance. Co-housing of SFB+ and SFB− mice introduced SFB into the SFB− mice (Figure S3F) and led to rhythmic bacterial attachment and oscillating REG3G expression (Figure 2F–J). Finally, germ-free mice monocolonized with SFB (Shi et al., 2019) had more attached SFB at ZT12 than ZT0 (Figure 2K and 2L), coinciding with higher expression of REG3G, LCN2, and S100A8 (Figure 2M–P). SFB were not attached to the large intestines of SFB+ mice (Figure S3G), and REG3G expression was correspondingly lower in the large intestine (Figure S3H and S3I) than in the small intestine (Figure 1B and 1C). These findings suggest that rhythmic SFB attachment to the small intestinal epithelium drives diurnal rhythms in AMP expression.
Figure 2. Rhythmic epithelial attachment of segmented filamentous bacteria (SFB) drives diurnal rhythms in antimicrobial protein expression.
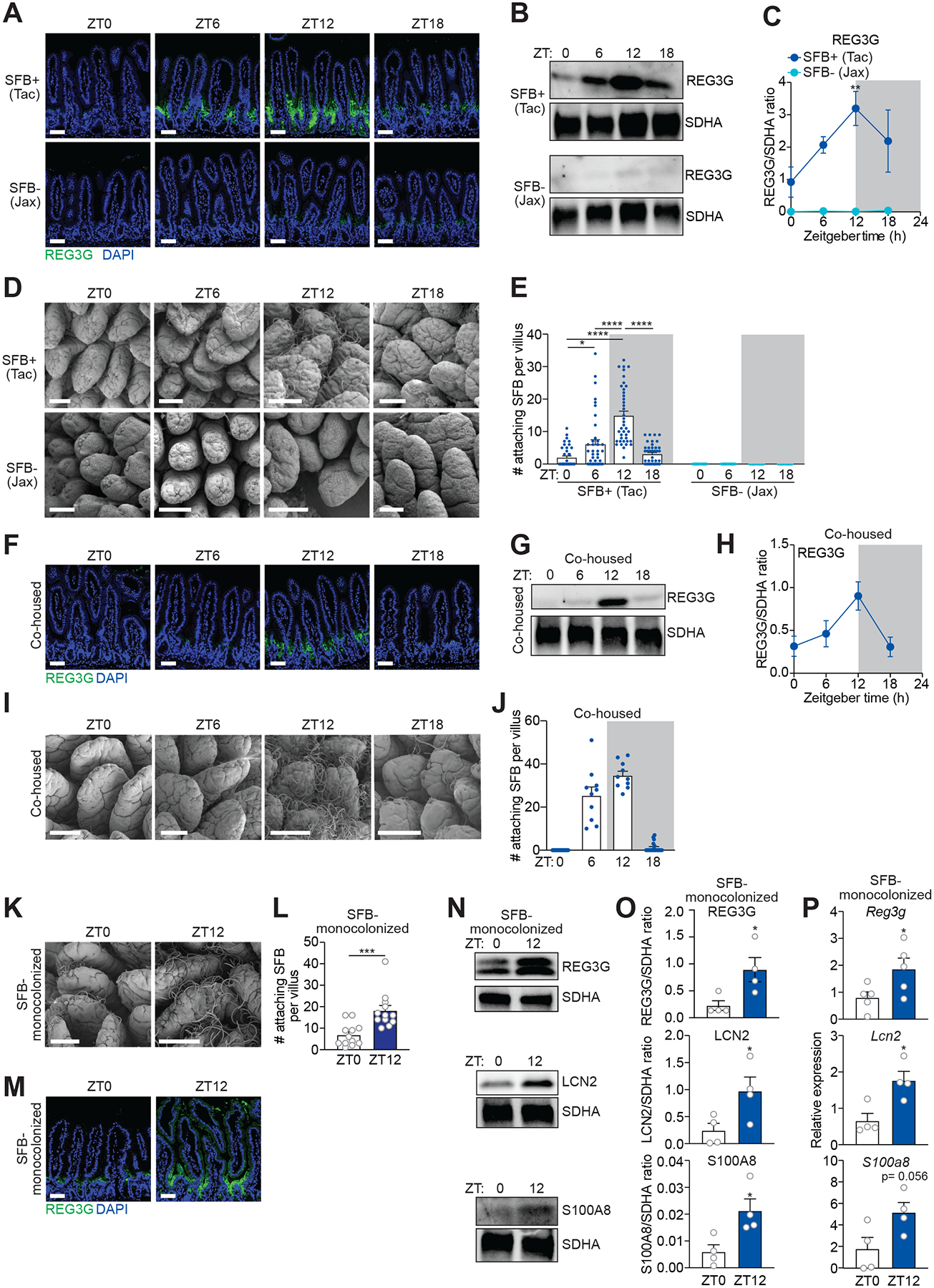
(A) Immunofluorescence detection of REG3G in the small intestines of SFB+ (Tac) and SFB− (Jax) mice. Nuclei were stained with DAPI.
(B and C) Representative immunoblot of small intestines from SFB+ (Tac) and SFB− (Jax) mice, with detection of REG3G and SDHA (control) (B). Band intensities were quantified by densitometry (C); n=3 independent experiments.
(D and E) Scanning electron microscopy of small intestinal villi from SFB+ (Tac) and SFB− (Jax) mice (D). Enumeration of attaching bacteria (E). The point of bacterial attachment was counted for 10 randomly selected villi across three visual fields per mouse; n=4 mice per group.
(F) Immunofluorescence detection of REG3G in the small intestines of SFB− (Jax) mice that were co-housed with SFB+ (Tac) mice for 14 days. Nuclei were stained with DAPI.
(G and H) Representative immunoblot of small intestines from co-housed mice, with detection of REG3G and SDHA (control) (G). Band intensities were quantified by densitometry (H).
(I and J) Scanning electron microscopy of intestinal villi from co-housed mice. Enumeration of attaching bacteria (J). The point of bacterial attachment was counted for five randomly selected villi across two visual fields per mouse; n=2 mice per group.
(K and L) Scanning electron microscopy of small intestinal villi from SFB-monocolonized mice (K). Enumeration of attaching bacteria (L). The point of bacterial attachment was counted for three randomly selected villi across one visual field per mouse; n=4 mice per group.
(M) Immunofluorescence detection of REG3G in the small intestines of SFB-monocolonized mice. Nuclei were stained with DAPI.
(N and O) Representative immunoblot of small intestines from SFB-monocolonized mice, with detection of REG3G, LCN2, S100A8, and SDHA (control) (N). Band intensities were quantified by densitometry (O). n=4 independent experiments.
(P) Q-PCR analysis of Reg3g, Lcn2, and S100A8 transcript abundance in small intestinal epithelial cells recovered by laser capture microdissection from mice monocolonized with SFB for four weeks. Tissues were collected at two timepoints across the day-night cycle. n=4–5 independent experiments.
ZT, Zeitgeber time; SFB, Segmented filamentous bacteria; Tac, Taconic; Jax, Jackson. Scale bars, 50 μm. Means ± SEM are plotted; *p < 0.05, **p < 0.01, ****p < 0.0001, ns, not significant by Student’s t test or one-way ANOVA. See also Figures S3 and S4 and Table S1.
We also considered whether REG3G expression might drive rhythmic SFB attachment. This could occur if REG3G kills SFB or bacteria that compete with SFB for epithelial attachment. Two of our findings argued against this idea. First, peak SFB attachment coincided with peak REG3G expression (Figure S4A). Second, SFB attachment remained diurnally rhythmic in Reg3g−/− mice (Figure S4B and S4C). These data indicate that REG3G does not drive rhythmic attachment of SFB, and instead support the idea that SFB attachment triggers REG3G rhythms.
REG3G is secreted into the small intestinal mucus layer where it restricts bacterial colonization of the mucus without markedly altering lumenal community composition (Vaishnava et al., 2011). Given the REG3G expression rhythms, we hypothesized that the densities of some mucus-associated bacteria would also oscillate with the day-night cycle. As predicted, the overall density of mucus-associated bacteria was inversely correlated with REG3G expression, with higher abundance at ZT0 than ZT12 in wild-type mice (Figure S4D). The rhythms in total bacterial density contrasted with SFB, which did not oscillate in overall abundance in the mucus (Figure S3E). In Reg3g−/− mice, the densities of mucus-associated bacteria were constant between ZT0 and ZT12 (Figure S4E), indicating that REG3G generates the rhythms in mucus-associated bacterial abundance. These rhythms were maintained in SFB+ but not SFB− mice (Figure S4F), consistent with our finding that SFB drive REG3G rhythms (Figure 2A–P). Thus, REG3G drives rhythms in the densities of mucus-associated bacteria in the small intestine.
An ILC3-STAT3 signaling relay drives diurnal rhythms in REG3G expression
REG3G expression requires signaling through Toll-like receptors (TLRs) and the TLR signaling adaptor MYD88 (Vaishnava et al., 2008; Vaishnava et al., 2011; Kinnebrew et al., 2012). Accordingly, REG3G expression was low at both ZT0 and ZT12 in Myd88−/− mice compared to wild-type mice (Figure 3A–3C). Although intestinal epithelial cell Myd88 was dispensable for rhythmic REG3G expression (Myd88ΔIEC mice; Figure 3A–3C), CD11c+ myeloid cell Myd88 was required (Myd88ΔDC mice; Figure 3A–3C).
Figure 3. An ILC3-STAT3 signaling relay drives diurnal rhythms in REG3G expression.
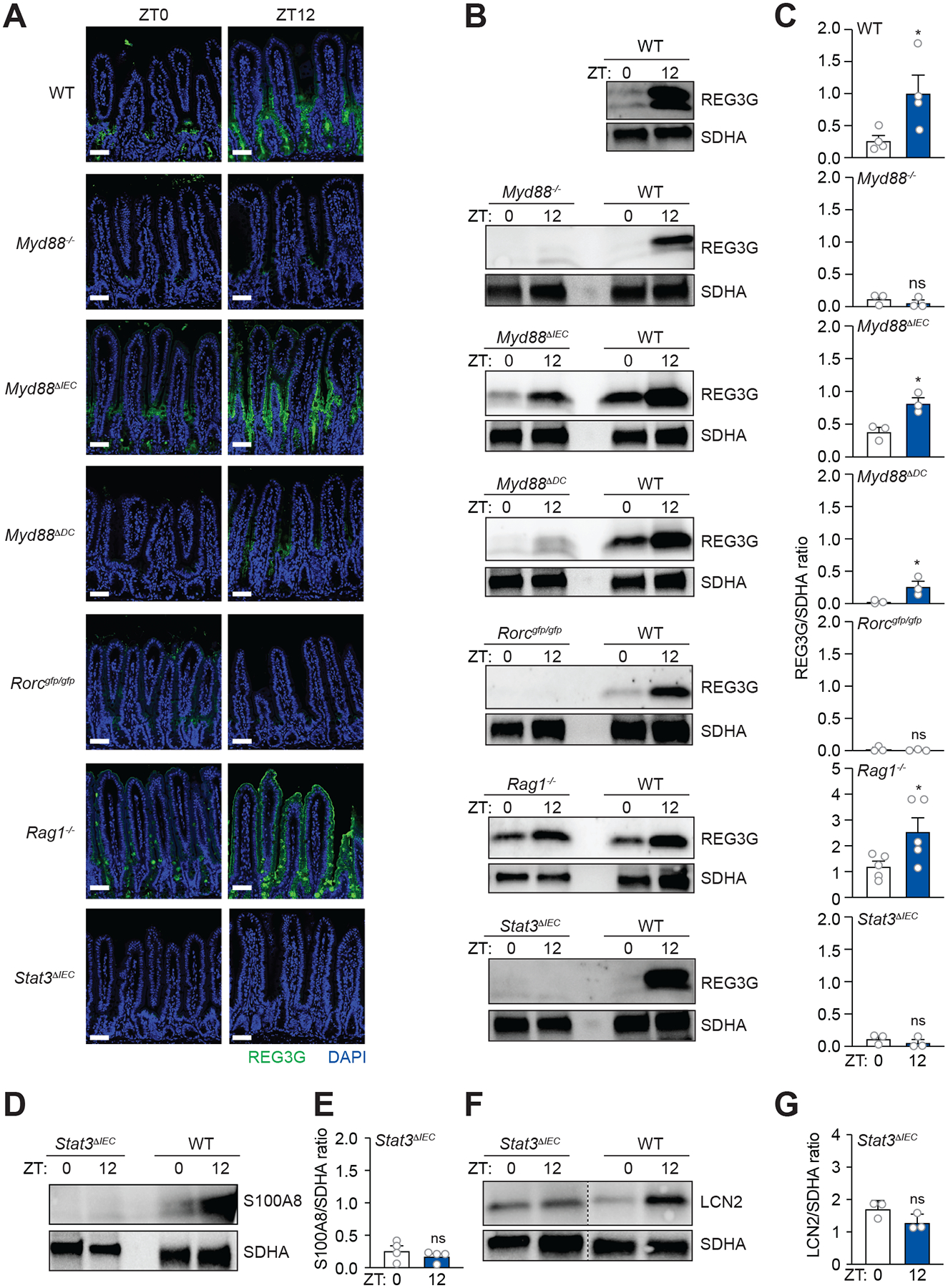
(A) Immunofluorescence detection of REG3G in the small intestines of wild-type (WT), Myd88−/−, Myd88ΔIEC, Myd88ΔDC, Rorcgfp/gfp, Rag1−/−, and Stat3ΔIEC mice. Scale bars, 50 μm.
(B and C) Representative immunoblot of small intestines from WT, Myd88−/−, Myd88ΔIEC, Myd88ΔDC, Rorcgfp/gfp, Rag1−/−, and Stat3ΔIEC mice, with detection of REG3G and SDHA (control) (B). Band intensities were quantified by densitometry (C).
(D and E) Representative immunoblot of small intestines from Stat3ΔIEC mice, with detection of S100A8 and SDHA (control) (E). Band intensities were quantified by densitometry (F).
(F and G) Representative immunoblot of small intestines from Stat3ΔIEC mice, with detection of LCN2 and SDHA (control) (G). Band intensities were quantified by densitometry (H).
ZT, Zeitgeber time. All results are representative of at least three independent experiments. Means ± SEM are plotted; *p < 0.05, **p < 0.01, ns, not significant by Student’s t test. See also Figures S5 and S6.
An immune cell signaling circuit, involving CD11c+ myeloid cells and group 3 innate lymphoid cells (ILC3), drives expression of epithelial genes including Reg3g (Sanos et al., 2009; Sano et al., 2015). In this circuit, myeloid cell TLR-MYD88 signaling triggers IL-23 production, which elicits IL-22 production by ILC3. IL-22 activates epithelial cell STAT3 to drive Reg3g expression (Sano et al., 2015) (Figure S5A). Epithelial attachment by SFB triggers the ILC3-STAT3 relay and stimulates expression of genes including Reg3g (Sano et al., 2015; Atarashi et al., 2015).
The requirement for CD11c+ myeloid cell Myd88 suggested that SFB might generate REG3G rhythms through the ILC3-STAT3 pathway. To test whether ILC3 are required, we studied mice lacking Rorc (Rorcgfp/gfp), which encodes the RAR-related orphan receptor γ (RORγt). RORγt is a transcription factor that governs the development of IL-22 producing cells, including ILC3, T helper 17 (TH17) cells, and γδ T cells (Sano et al., 2015). REG3G expression in Rorcgfp/gfp mice was low at both ZT0 and ZT12, supporting a role for IL-22-producing cells in driving REG3G rhythms (Figure 3A–3C). However, REG3G expression was low at ZT0 and high at ZT12 in mice lacking Recombination-activating gene 1 (Rag1) (Mombaerts et al., 1992), which lack mature T and B cells but retain ILC3 (Figure 3A–3C). These findings rule out a requirement for TH17 and γδ T cells and suggest that ILC3 drive rhythmic REG3G expression.
To test whether STAT3 was required for rhythmic REG3G expression, we generated mice lacking epithelial cell Stat3 (Stat3ΔIEC; Figure S5B). REG3G was low at ZT0 and ZT12 in Stat3ΔIEC mice (Figure 3A–3C), indicating that STAT3 is required for REG3G rhythms. Similarly, expression of S100A8 (Figure 3D and E) and LCN2 (Figure 3F and G) was low at both ZT0 and ZT12 in Stat3ΔIEC mice, indicating that epithelial STAT3 is required for the rhythmic expression of multiple AMPs. In contrast, non-rhythmic lysozyme expression was maintained in mice with genetic disruptions of the ILC3-STAT3 pathway, indicating that this pathway is not required for expression of all epithelial AMPs (Figure S5C). Further, SFB attachment was retained in mice with genetic disruptions of the ILC3-STAT3 pathway (Figure S6A and S6B), indicating that the lack of rhythmic REG3G expression was not due to an absence of attaching SFB. Thus, the ILC3-STAT3 pathway drives the rhythmic expression of select epithelial AMPs.
SFB drive diurnal rhythms in STAT3 expression and activation
We next determined whether intestinal IL-23 and IL-22 were rhythmically produced. IL-23 and IL-22 levels were higher at ZT12 than ZT0 (Figure 4B and 4C). Further, rhythmic IL-22 expression was maintained in Rag1 mice and abrogated in Rorcgfp/gfp mice (Figure 4C), corresponding with rhythmic REG3G expression (Figure 3A–C). Thus, there is rhythmic expression of the cytokines required for the ILC3-STAT3 circuit.
Figure 4. SFB drive diurnal rhythms in STAT3 expression and activation.
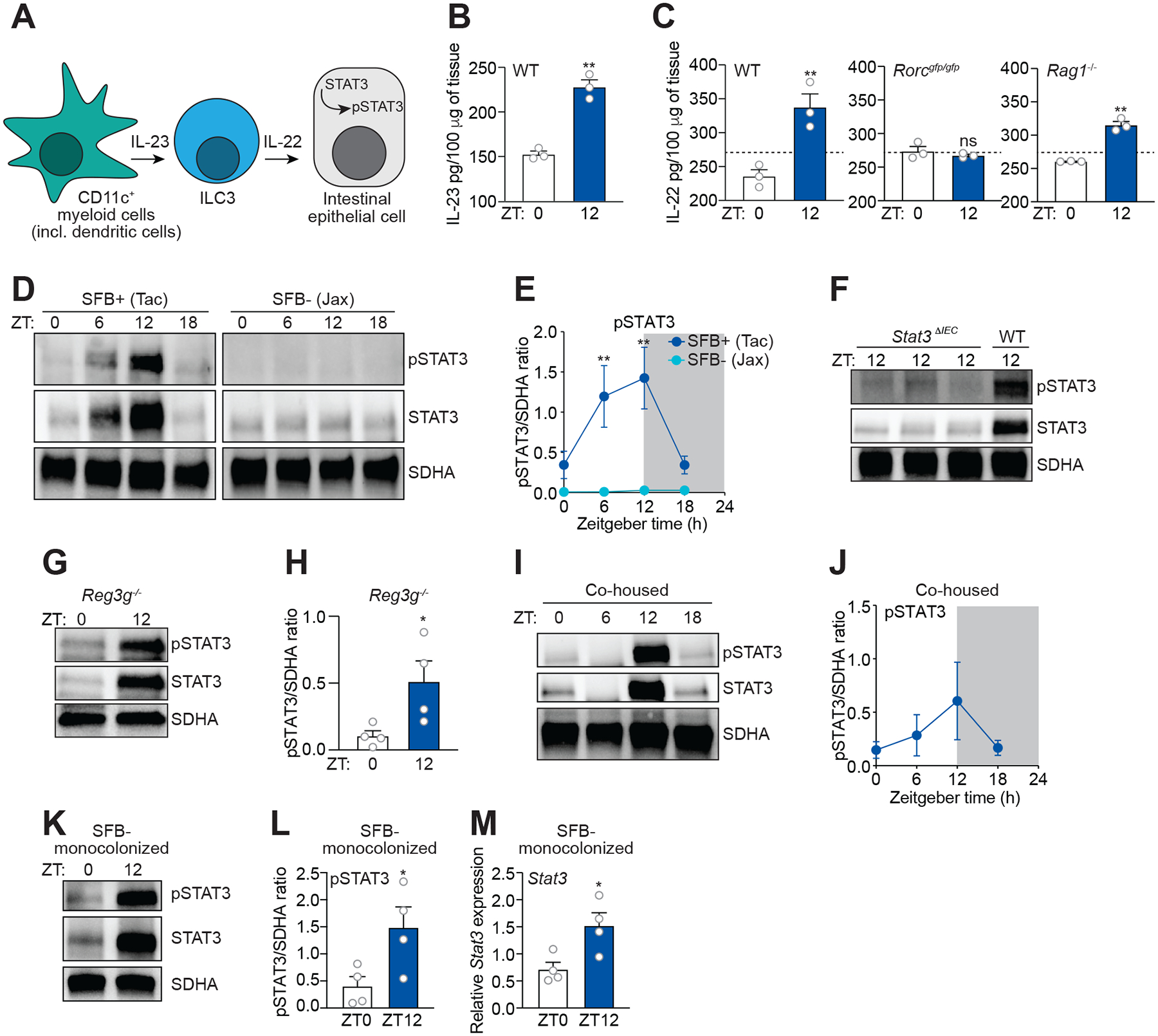
(A) Schematic of the small intestinal ILC3-STAT3 pathway.
(B) Measurement of IL-23 in mouse small intestine by enzyme-linked immunosorbent assay (ELISA). n=3 mice per group.
(C) Measurement of IL-22 in the small intestines of wild-type (WT), Rorcgfp/gfp, and Rag1−/− mice by ELISA. n=3 mice per group.
(D and E) Representative immunoblot of small intestines from SFB+ (Tac) and SFB− (Jax) mice, with detection of STAT3, pSTAT3, and SDHA (control) (D). Band intensities were quantified by densitometry (E). n=3 independent experiments.
(F) Immunoblot of WT and Stat3ΔIEC mice, with detection of STAT3, pSTAT3, and SDHA (control). Representative of three independent experiments.
(G and H) STAT3 and pSTAT3 rhythms are maintained in Reg3−/− mice. (G) Representative immunoblot of small intestines from Reg3g−/− mice, with detection of STAT3, pSTAT3, and SDHA (control). (H) Band intensities were quantified by densitometry. n=4 independent experiments.
(I and J) Representative immunoblot of small intestines from SFB− (Jax) mice that were co-housed with SFB+ (Tac) mice for 14 days with detection of STAT3, pSTAT3, and SDHA (control) (F). Band intensities were quantified by densitometry (G). n=3 independent experiments.
(K and L) Representative immunoblot of small intestines from SFB-monocolonized mice, with detection of STAT3, pSTAT3, and SDHA (control) (H). Band intensities were quantified by densitometry (I). n=4 independent experiments.
(M) Q-PCR measurement of Stat3 transcript abundance in small intestinal epithelial cells recovered by laser capture microdissection from mice monocolonized with SFB for four weeks. Tissues were collected at two timepoints across the day-night cycle. n=4 independent experiments.
ZT, Zeitgeber time; SFB, Segmented filamentous bacteria; Tac, Taconic; Jax, Jackson. Means ± SEM are plotted; **p < 0.01, ***p < 0.001 by Student’s t test. See also Figure S6 and Table S1.
Because IL-22 stimulates the phosphorylation and thus activation of epithelial cell STAT3 (Sano et al., 2015), we asked whether STAT3 activation is diurnally rhythmic. In SFB+ mice, both expression and activation of STAT3 were rhythmic with peak expression and activation at ZT12, coinciding with peak expression of REG3G, LCN2, and S100A8 (Figure 4D and 4E). Further, the rhythmic expression and activation of STAT3 were attenuated in SFB− mice (Figure 4D and 4E), indicating that SFB are required for STAT3 rhythms. The STAT3 rhythms were localized to epithelial cells, as little STAT3 was detected in the intestines of Stat3ΔIEC mice at ZT12 as compared to wild-type mice (Figure 4F). Rhythms in STAT3 expression and activation were maintained in Reg3g−/− mice (Figure 4G and 4H), indicating that SFB drive rhythms in REG3G expression. Rhythmic STAT3 expression and activation were acquired by SFB− mice when co-caged with SFB+ mice (Figure 4I and 4J). Finally, in SFB-monocolonized animals, STAT3 expression and activation were higher at ZT12 than ZT0, and Stat3 expression exhibited a similar pattern (Figure 4K–M). Thus, rhythmic attachment of SFB drives diurnal rhythms in STAT3 expression and activation.
The circadian clock regulates diurnal rhythms in SFB attachment that drive rhythmic STAT3 and REG3G expression
We previously found that signals from the microbiota integrate with the host circadian clock through the ILC3-STAT3 pathway. Specifically, epithelial pSTAT3 represses expression of the circadian clock transcription factor REV-ERBα, which governs a rhythmic lipid metabolic program in intestinal epithelial cells (Wang et al. 2017). However, the mice in that study were devoid of SFB and therefore lacked pSTAT3 rhythms (Wang et al., 2017). Based on these earlier findings, we hypothesized that REV-ERBα might be necessary for REG3G rhythms in SFB+ animals. However, SFB attachment remained rhythmic in Rev-erbα−/− mice, coinciding with rhythms in both pSTAT3 and REG3G (Figure 5A–5G). Thus, REV-ERBα is not required for REG3G rhythms, indicating that these rhythms are generated through a different mechanism.
Figure 5. The circadian clock regulates diurnal rhythms in SFB attachment that drive rhythmic STAT3 and REG3G expression.
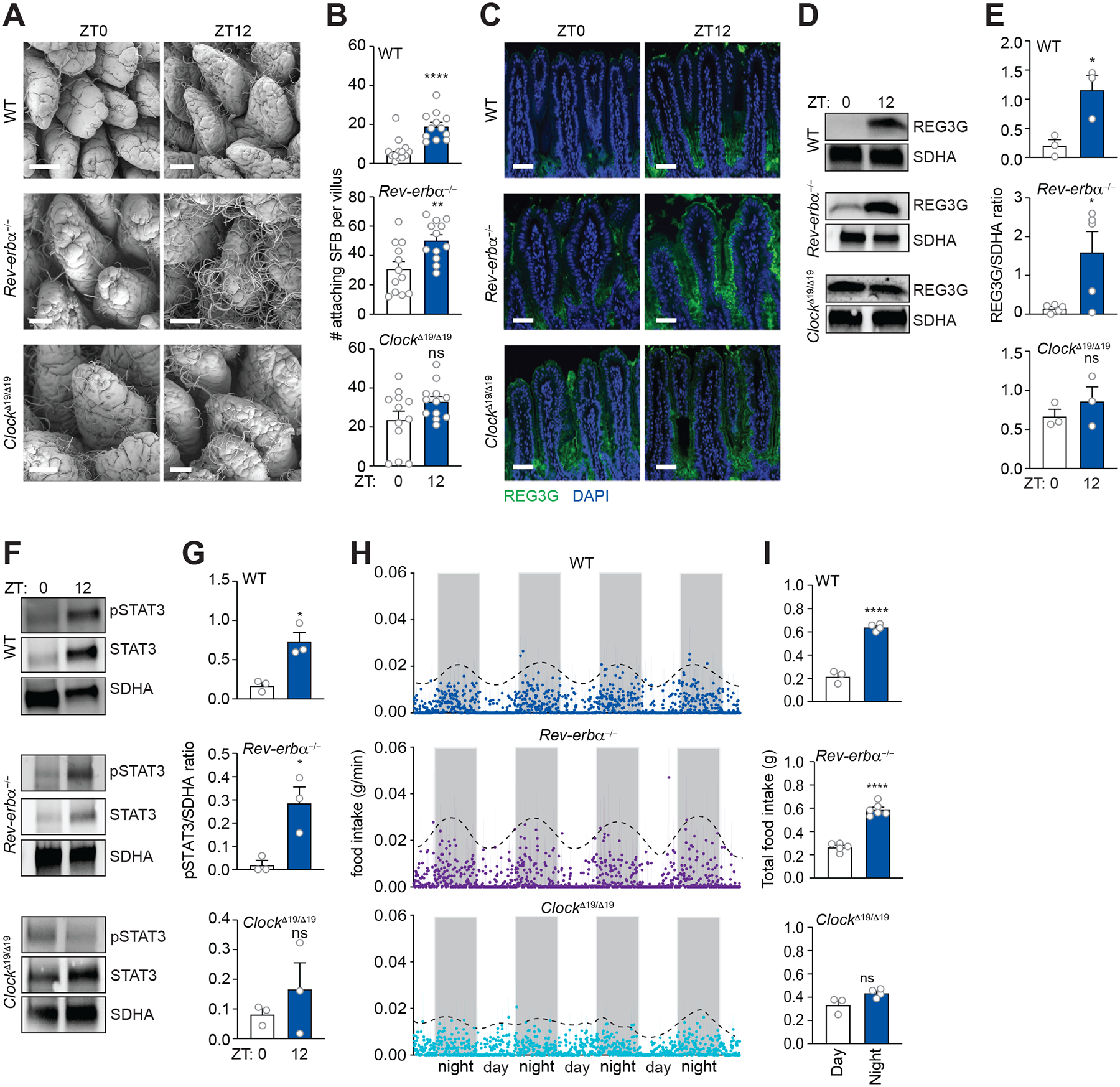
(A and B) Scanning electron microscopy of intestinal epithelium from WT, Rev-erbα−/−, and Clock Δ19/Δ19 mice (A). Scale bars, 50 μm. Enumeration of attaching bacteria (B). The point of bacterial attachment was counted for 4 randomly selected villi across two visual fields per mouse. n=3 mice per group.
(C) Immunofluorescence detection of REG3G in the small intestines of WT, Rev-erbα−/−, and Clock Δ19/Δ19 mice. Nuclei were stained with DAPI. Scale bars, 50 μm.
(D and E) Representative immunoblot of small intestines from WT, Rev-erbα−/−, and Clock Δ19/Δ19 mice, with detection of REG3G and SDHA (control) (D). Band intensities were quantified by densitometry (E); n=3 independent experiments.
(F and G) Representative immunoblot of small intestines from WT, Rev-erbα−/−, and Clock Δ19/Δ19 mice, with detection of STAT3, pSTAT3 and SDHA (control). Band intensities were quantified by densitometry (G); n=3 independent experiments.
(H and I) Measurement of food intake rate in WT, Rev-erbα−/−, and Clock Δ19/Δ19 mice (H). Total food intake during day and night (I); Each data point represents one mouse. n=3–6 mice per group.
ZT, Zeitgeber time. Means ± SEM are plotted; *p < 0.05, ****p < 0.0001; ns, not significant by Student’s t test.
Although REV-ERBα is a component of the molecular clock, deletion of Rev-erbα is not sufficient to inhibit some rhythms generated by the BMAL1/CLOCK complex (Preitner et al., 2002). This is due to the existence of the related circadian clock transcription factor REV-ERBβ (Dumas et al., 1994). Therefore, we tested whether the circadian clock drives REG3G rhythms by examining REG3G expression in ClockΔ19/Δ19 mice. ClockΔ19/Δ19 mice harbor a dominant-negative CLOCK allele that inhibits the function of the BMAL1/CLOCK complex (King et al., 1997). ClockΔ19/Δ19 mice had attenuated rhythms in SFB attachment, pSTAT3 levels, and REG3G expression as compared to wild-type mice (Figure 5A–5G). Thus, the circadian clock is required for rhythmic REG3G expression.
The circadian clock entrains host feeding rhythms that regulate rhythmic SFB attachment
We next assessed how the circadian clock regulates REG3G expression. The circadian clock entrains rhythms in the numbers, composition, and function of the gut microbiota, largely by regulating the timing of host feeding (Zarrinpar et al., 2014; Thaiss et al., 2014; 2016; Leone et al., 2015; Liang et al., 2015). We therefore considered whether clock-generated feeding rhythms might drive rhythmic attachment of SFB. Mice are nocturnal, and thus wild-type mice ate rhythmically with maximum feeding at night, (Figure 5H, 5I, and S7). Similarly, Rev-erbα−/− mice ate rhythmically with maximum feeding at night (Figure 5H and 5I). By contrast, and consistent with prior findings (King et al., 1997), the ClockΔ19/Δ19 mice lacked pronounced feeding rhythms (Figure 5H and 5I).
To determine whether the disrupted feeding rhythms in the ClockΔ19/Δ19 mice caused the attenuated REG3G rhythms, we restricted feeding to either day or night for five days. Total food intake was similar under the restricted feeding regimens, arguing against a role for altered caloric intake on REG3G rhythm generation (Figure 6A and 6B). SFB attachment was maximal at ZT6 and ZT12 and minimal at ZT0 and ZT18 in night-fed mice, similar to mice fed ad libitum (Figure 6C and 6D). In contrast, peak attachment of SFB occurred at ZT0 and ZT18 in day-fed mice, corresponding to a phase shift of 12 hours compared to night-fed mice (Figure 6C and 6D), and rhythms in REG3G and pSTAT3 correlated with SFB attachment (Figure 6E–6G).
Figure 6. The circadian clock entrains host feeding rhythms that regulate rhythmic SFB attachment.
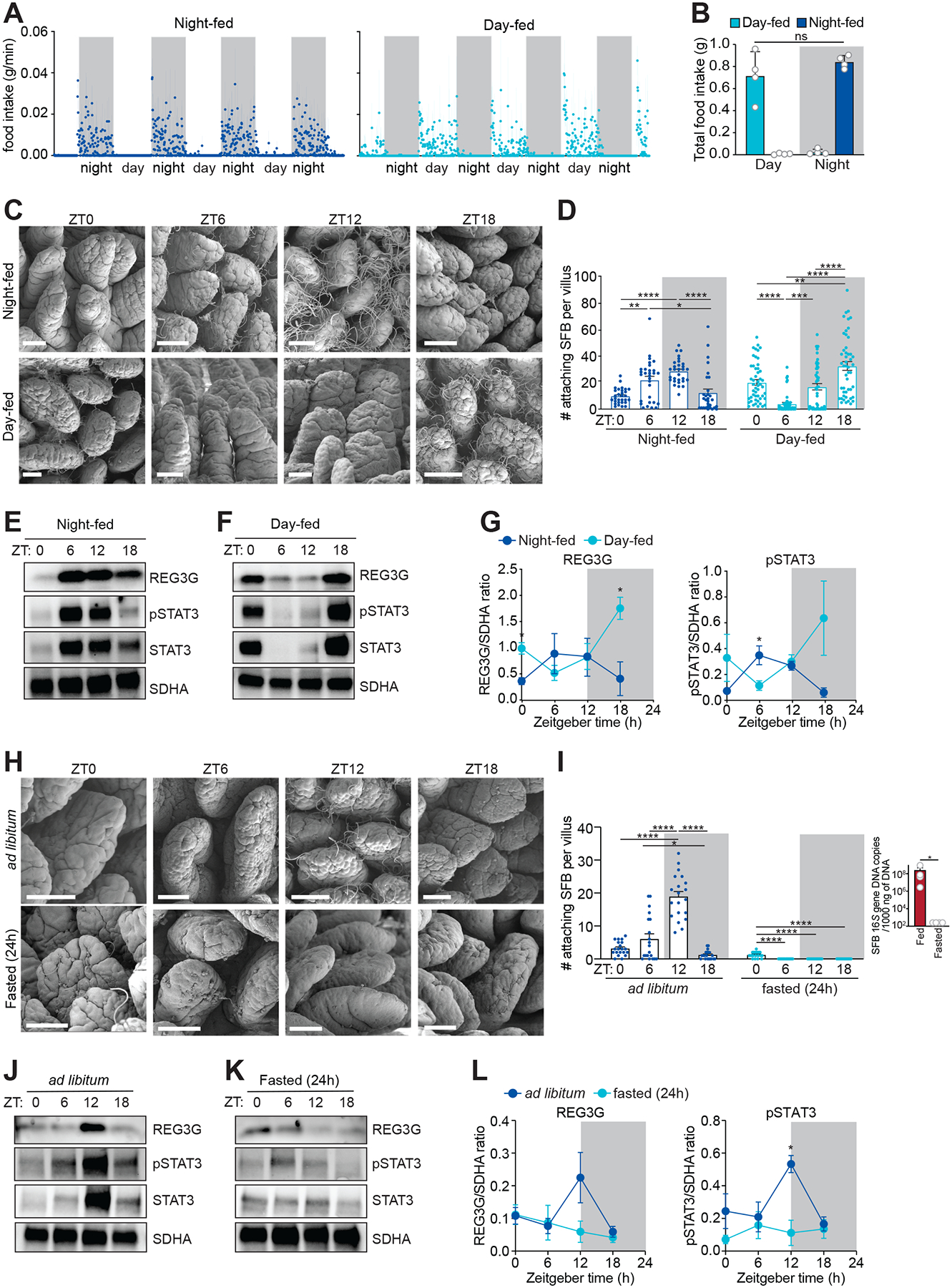
(A and B) Measurement of food intake rate in day- or night-fed mice (A). Total food intake during day and night (B). Each data point represents one mouse. n=4 mice per group.
(C and D) Scanning electron microscopy of intestinal epithelium from day- or night-fed mice (C). Scale bars, 50 μm. Enumeration of attaching bacteria (D). Attaching bacteria were counted as described in Figure 2E. n=3–5 mice per group.
(E-G) Representative immunoblots of small intestines from night-fed (E) or day-fed (F) mice, with detection of REG3G, STAT3, pSTAT3, and SDHA (control). Band intensities were quantified by densitometry (G). n=4 independent experiments.
(H and I) Scanning electron microscopy of intestinal epithelium from ad libitum fed or fasted (24 h) mice (H). Scale bars, 50 μm. Enumeration of attaching bacteria (I). The point of bacterial attachment was counted for 5 randomly selected villi across two visual fields per mouse. n=3–5 mice per group. Overall SFB abundance (I, right panel) was measured by Q-PCR analysis of 16S rRNA gene copy number in the ileum.
(J and K) Representative immunoblot of small intestines from ad libitum fed or fasted (24 h) mice, with detection of REG3G, STAT3, pSTAT3, and SDHA (control) (F). Band intensities were quantified by densitometry (G). n=4 independent experiments.
ZT, Zeitgeber time. Means ± SEM are plotted. *p < 0.05, **p < 0.01, ****p < 0.0001, ns, not significant by Student’s t test or one-way ANOVA. See also Figure S7 and Table S1.
To further assess whether SFB attachment was driven by feeding, we fasted mice for 24 hours. Numbers of attaching SFB were markedly reduced in fasted mice, but this was due to an overall depletion of SFB (Figure 6I). Accordingly, REG3G and pSTAT3 rhythms were lost in fasted mice (Figure 6H–6L). These data support the idea that feeding promotes rhythmic epithelial attachment of SFB, which drives rhythms in STAT3, pSTAT3, and REG3G. These findings also indicate that feeding is necessary for the retention of SFB in the intestine.
SFB acts through STAT3 to cause diurnal variation in resistance to Salmonella infection
Our finding of diurnal rhythms in intestinal innate immunity suggested that infection resistance might vary across the day-night cycle. To test this idea we conducted oral infection experiments with the enteric pathogen Salmonella Typhimurium. We chose S. Typhimurium as our model organism for two reasons. First, S. Typhimurium is a natural mouse pathogen that colonizes the intestine in the absence of antibiotics that would deplete SFB and thus suppress rhythms in innate immunity. Second, STAT3-dependent AMPs impact intestinal growth and survival of S. Typhimurium. For example, S. Typhimurium are resistant to LCN2, and thus have a competitive advantage over resident intestinal commensal bacteria when expression of STAT3-dependent AMPs is high (Raffatellu et al., 2009; Behnsen et al., 2014).
We tested whether S. Typhimurium infection varies with inoculation time. S. Typhimurium burdens were higher in SFB+ mice inoculated at ZT12 than ZT0 (Figure 7B). This accords with the competitive advantage of S. Typhimurium in the presence of STAT3-dependent AMPs such as LCN2, as STAT3 activation is highest at ZT12 and lowest at ZT0 (Figure 4A and 4B). In contrast, bacterial burdens were similar across the day-night cycle in SFB− mice (Figure 7C). Further, streptomycin treatment reduced SFB abundance in SFB+ mice (Figure 7D) and abolished the time-dependent variation in S. Typhimurium burden (Figure 7E and 7F), suggesting that SFB caused the time-dependent variation in resistance. Finally, Stat3ΔIEC mice lacked time-dependent differences in bacterial burden when infected at ZT0 versus ZT12 (Figure 7G), consistent with the fact that SFB promotes rhythmic AMP expression through STAT3 (Figure 3) (Coorens et al., 2019). Thus, SFB acts through STAT3 to drive diurnal variation in resistance to S. Typhimurium infection.
Figure 7. SFB acts through STAT3 to cause diurnal variation in resistance to Salmonella infection.
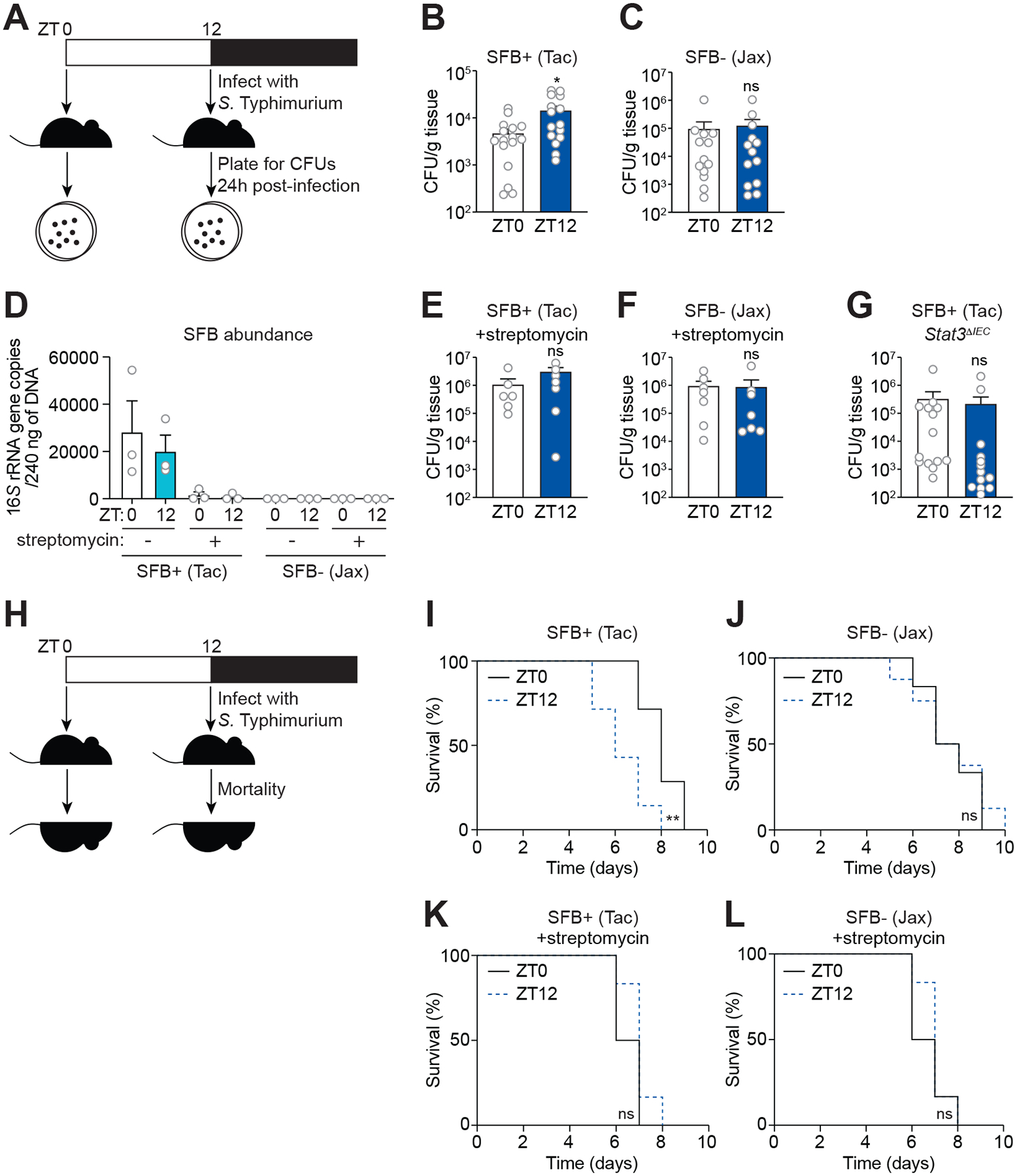
(A) Oral infection of mice with S. Typhimurium for determination of bacterial burden.
(B and C) Bacterial burdens in the ileum of wild-type SFB+ (Tac) mice (B) and SFB− (Jax) mice (C) infected with S. Typhimurium (108 CFU per animal). Each data point represents one mouse. n=13–16 mice per group.
(D) Q-PCR analysis of SFB abundance from fecal material derived from SFB+ (Tac) mice and SFB− (Jackson) mice at two timepoints (ZT0 and ZT12) before and after treatment with streptomycin.
(E and F) Bacterial burdens in the ileum of wild-type SFB+ (Tac) mice (E), SFB− (Jax) mice (F) infected with S. Typhimurium (108 CFU per animal) 24 hours after streptomycin administration. Each data point represents one mouse. n=6–7 mice per group.
(G) Bacterial burdens in the ileum of Stat3ΔIEC mice infected with S. Typhimurium (108 CFU per animal). Each data point represents one mouse. n=13–14 mice per group.
(H) Oral infection of mice with S. Typhimurium for determination of lethal morbidity rates.
(I and J) Lethal morbidity in wild-type SFB+ (Tac) mice (E) and SFB− (Jax) mice (F) infected with S. Typhimurium (107 CFU per animal). n=6–8 mice per group.
(K and L) Lethal morbidity in wild-type SFB+ (Tac) mice (E) and SFB− (Jax) mice (F) infected with S. Typhimurium (107 CFU per animal) 24 h after streptomycin administration. n=6–7 mice per group.
ZT, Zeitgeber time; SFB, segmented filamentous bacteria; Tac, Taconic; Jax, Jackson. Means ± SEM are plotted. *p < 0.05, **p < 0.01, ns, not significant by Student’s t test (Figures 7B–G) or log-rank (Mantel-Cox) test (Figures 7I–L). See also Table S1.
We next examined whether mortality rates varied with the timing of S. Typhimurium infection. SFB+ mice infected with a lethal dose of S. Typhimurium at ZT12 exhibited higher lethal morbidity than mice infected at ZT0 (Figure 7I). In contrast, SFB− mice had similar rates of lethal morbidity following infection at the two timepoints (Figure 7J). Upon streptomycin treatment, the lethal morbidity rates of both SFB+ and SFB− mice were similar following infection at ZT0 and ZT12 (Figure 7K and 7L). Thus, SFB drive daily variation in resistance to S. Typhimurium infection.
DISCUSSION
The circadian clock coordinates physiological processes with day-night light cycles, allowing animals to anticipate changes in their environment. In this study, we show that the circadian clock coordinates intestinal innate immunity with host feeding rhythms, thus anticipating exposure to foodborne bacteria. The mechanism involves circadian clock entrainment of host feeding rhythms, which generate oscillating epithelial attachment of SFB. Rhythmic SFB attachment stimulates a myeloid cell-ILC3 immunological circuit that drives rhythms in epithelial STAT3 expression and activation. The STAT3 rhythms cause oscillating expression of AMPs, including REG3G, LCN2, and S100A8. In this way, host feeding rhythms are synchronized with rhythms in intestinal innate immunity.
Circadian rhythms in intestinal innate immunity generate diurnal variation in resistance to infection
Rhythms in AMP expression cause daily fluctuations in innate immunity to enteric infection. When we infected mice orally with S. Typhimurium at ZT12, the intestinal bacterial burdens and mortality rates were higher than when mice were infected at ZT0. The time-dependent difference in bacterial burden was dependent on STAT3, and the peak bacterial burden at ZT12 coincided with peak expression of the STAT3-dependent AMP LCN2. This is consistent with the fact that S. Typhimurium is resistant to LCN2, and thus has a competitive advantage over indigenous gut bacteria when LCN2 expression is high (Behnsen et al., 2014; Raffatellu et al., 2009). Our results thus indicate that rhythms in STAT3-controlled innate immunity cause infection resistance to vary over the day-night light cycle. Further, daily variation in infection susceptibility depended on SFB, revealing that the microbiota produces the daily oscillations in infection resistance.
A prior study also reported that resistance to oral S. Typhimurium infection varies with inoculation timing (Bellet et al., 2013). However, in this study resistance was minimal with inoculation at ZT4, at the beginning of the light phase, and was maximal with inoculation at ZT16, at the beginning of the dark phase. This contrasts with our finding of maximal resistance following inoculation at the beginning of the light phase (ZT0) and minimal resistance following inoculation at the beginning of the dark phase (ZT12). The discordant findings may be due to the fact that the mice in the Bellet et al. study were pretreated with streptomycin (Bellet et al., 2013). Since we found that streptomycin treatment markedly reduces SFB colonization (Figure 7D), this treatment likely abolished the natural innate immune rhythms that arise from SFB attachment. In contrast, we infected SFB+ mice that were not antibiotic-treated and thus retained SFB-dependent rhythms in AMP expression. The difference in the findings suggests that antibiotic treatment alters SFB-dependent rhythms in infection resistance, perhaps by altering AMP expression.
A recent report from Talbot et al. (2020) identified a distinct pathway by which feeding impacts Reg3g expression in mice. This study showed that feeding regulates IL-22 production and hence Reg3g expression through a signaling pathway involving enteric neurons and ILC3. Following feeding that was restricted to a daily 12-hour window, there was reduced IL-22 production by ILC3, lowered Reg3g expression, and increased SFB filament length. After a 12-hour fast, Reg3g expression was elevated.
In contrast to the Talbot et al. (2020) study, which used a restricted feeding regimen, our study analyzed Reg3g expression during natural diurnal rhythms in ad libitum feeding. Reg3g expression was maximal following the inactive phase of mice (light) and was minimal following the active phase (dark)(Figure 1A–D) in accordance with the patterns of rhythmic SFB attachment (Figure 2A–P). Night-restricted feeding produced results similar to those of ad libitum feeding, while day-restricted feeding altered SFB attachment such that it peaked on the opposite phase of the day-night cycle with concordant changes in peak pSTAT3 and REG3G levels (Figure 6A–D). Thus, our findings about the timing of REG3G expression accord with those of Talbot et al. (2020).
Our data support a model in which SFB drive a distinct signaling pathway, involving ILC3-STAT3, that synchronizes feeding rhythms with AMP expression. Our finding that SFB drive Reg3g rhythms through epithelial attachment distinguishes this pathway from the ILC3-neuronal pathway, where SFB morphology changes in response to pathway activation and reduction in Reg3g expression. Our conclusion that SFB drives, rather than responds to, Reg3g rhythms is supported by our finding that rhythms in SFB attachment are maintained in Reg3g−/− mice (Figure S4B and S4C).
SFB synchronize innate immunity with host feeding
The circadian clock uses light cues to synchronize the expression of metabolic pathways with food intake, enabling the efficient use of energetic resources (Mohawk et al., 2012). Our findings reveal that food intake is also synchronized with the expression of innate immune effectors. The mechanism involves circadian clock-driven rhythms of host food intake acting through the microbiota to promote daily oscillations in innate immunity. This coordination limits the energetic cost of AMP expression to times at which exposure to exogenous microorganisms is likely to be highest. Indeed, feeding rhythms and attaching small intestinal bacteria also promote diurnal rhythms in epithelial cell expression of major histocompatibility complex class II, suggesting that rhythms in other aspects of intestinal immunity are generated through similar mechanisms (Tuganbaev et al., 2020).
Interestingly, S. Typhimurium is a gastrointestinal pathogen that has co-opted this system to attain a competitive advantage. Given that all foodborne pathogens must contend with the same coordinated antimicrobial response, there are likely daily oscillations in host susceptibility to infection with other bacterial pathogens.
We found that SFB attach to the intestinal epithelium coincident with feeding and thus mediate the rhythmic coupling of feeding and immunity in mice. This raises the question of whether there are adherent microbes in the human gut that could also synchronize feeding and innate immunity. Two findings support this idea. First, SFB are a rare but detectable component of the gut microbiota of humans (Chen et al., 2018; Jonsson et al., 2020). Second, other members of the human microbiota adhere to the intestinal epithelium and thus shape immune responses in a manner similar to that of SFB (Yin et al., 2013; Atarashi et al., 2015).
A remaining question concerns the mechanisms by which SFB attachment activates the ILC3-STAT3 pathway. One possible mechanism involves direct transfer of SFB antigens into epithelial cells. When SFB adhere to intestinal epithelial cells, they trigger endocytosis that results in epithelial cell uptake of SFB antigens. These antigens activate antigen-specific TH17 cells in the lamina propria (Ladinsky et al., 2019). For SFB to activate the ILC3-STAT3 pathway, this might involve further activation of CD11c+ myeloid cells through their close interactions with epithelial cells. A second possible mechanism involves direct recognition of SFB-derived molecular patterns by TLRs on intestinal CD11c+ myeloid cells. Although SFB do not readily penetrate the epithelial barrier, myeloid cells could contact SFB molecular patterns through the diffusion of soluble molecules across the intestinal barrier or by capture of bacterial molecules from the luminal surface of epithelial cells (Rescigno et al., 2001; Chieppa et al. 2006).
Another important question is how host feeding behavior promotes SFB attachment to the intestinal epithelium. One possibility is that SFB attach to the epithelium to acquire nutrients from intestinal epithelial cells. This idea is suggested by the SFB genome sequence, which indicates that SFB lack many of the metabolic pathways normally found in free-living bacteria (Sczesnak et al., 2013). Thus, it seems likely that SFB depend on host cells for key nutrients and may acquire them through close contact with the epithelial surface. In this way, nutrient availability in host cells, driven by host feeding, could trigger SFB attachment through mechanisms that are still unclear.
Therapeutic implications
Chronic sleep disruption is tied to increased susceptibility to infection in humans. For example, people who work at night show elevated susceptibility to bacterial and viral infection compared to people who work during the day (Knutsson and Bøggild, 2010; Mohren et al., 2002). Our findings suggest that altered feeding behavior could be a causative factor. By identifying mechanisms by which the circadian clock drives diurnal rhythms in microbiota function, we can begin to illuminate how rhythmic functions of the microbiota impact host immunity. These studies could lead to the development of timed therapeutic interventions and vaccinations.
Limitations of the study
Our study illuminates how host feeding rhythms generate rhythms in intestinal innate immunity, allowing anticipation of exposure to foodborne and environmental bacteria. However, key questions remain. First, our study used a single infection model, S. Typhimurium. Thus, a remaining question is whether other intestinal pathogens also show daily variations in resistance due to these same innate immune rhythms. However, exploring other infection models is likely to be challenging as many mouse models of intestinal bacterial infection require prior antibiotic treatment, which depletes the SFB that are required for rhythmic AMP expression. A second key question is whether AMP expression is also rhythmic in the human intestine. Although SFB are present in the human gut microbiota (Chen et al., 2018; Jonsson et al., 2020), it is not yet clear whether they exhibit rhythmic attachment to the human intestinal epithelium, or whether other epithelial-adherent members of the human microbiota show daily attachment rhythms that could drive rhythmic AMP expression (Yin et al., 2013; Atarashi et al., 2015). Answering this question would begin to address whether the human microbiota, like the mouse microbiota, synchronizes feeding rhythms with innate immune rhythms in order to anticipate pathogen exposure.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact Lora Hooper (lora.hooper@utsouthwestern.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
16S rRNA gene sequencing data (Figure S3) have been deposited in the Sequence Read Archive (SRA) with BioProject ID PRJNA733561 and are publicly available.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Wild-type C57BL/6, Reg3g−/− (Vaishnava et al., 2011), Myd88−/− (Adachi et al., 1998), Myd88ΔIEC (Vaishnava et al., 2011), Myd88ΔDC (Wang et al., 2017), Stat3ΔIEC (Wang et al., 2017), Rag1−/− (Mombaerts et al., 1992), Rorcgfp/gfp (Eberl et al., 2004), Stat3ΔIEC, Nr1d1 −/− (Rev-erbα-deficient) (Preitner et al. 2002), and Clock Δ19/19 mice (Vitaterna et al. 1994; King et al., 1997) were bred and maintained in the SPF barrier facility at the University of Texas Southwestern Medical Center. Stat3ΔIEC mice were generated as previously described in Wang et al. (2017) by crossing Stat3fl/fl mice (Jackson Laboratory; Moh et al., 2007) with a mouse expressing Cre recombinase under the control of the intestinal epithelial cell-specific Villin promoter (Jackson Laboratory; Madison et al., 2002). Myd88 ΔIEC mice were generated as previously described (Ismail et al., 2011) by crossing Myd88fl/fl mice (Hou et al., 2008) with Villin-Cre mice. Myd88ΔDC mice were generated as previously described (Hou et al., 2008; Wang et al., 2017) by crossing Myd88fl/fl mice with Cd11c-Cre mice (Jackson Laboratories). Germ-free C57BL/6 mice were bred and maintained in isolators at the University of Texas Southwestern Medical Center. C57BL/6 SFB+ mice were purchased from Taconic Farms and C57BL/6 SFB− mice were purchased from Jackson Laboratory, and used immediately. 8–18-week-old mice were used for all experiments, and both male and female mice were analyzed. All experiments were performed using protocols approved by IACUC at the University of Texas Southwestern Medical Center.
Bacterial strains
Salmonella enterica Serovar Typhimurium (IR715) was cultured in Luria broth at 37°C. Segmented filamentous bacteria were acquired from Andrew Gewirtz at Georgia State University and were used directly in germ-free mouse colonization experiments.
METHOD DETAILS
Bacterial infections and inoculations
Male mice were gavaged with 108 CFU S. Typhimurium cultured in Luria broth. 24 hours post-infection, mice were euthanized and the distal small intestines (ileum) were homogenized and quantified by dilution plating on selective LB agar (50 μg/ml nalidixic acid). Colonies were counted after 24 hours at 37°C. In some cases, mice were pre-treated intragastrically with 20 mg of streptomycin sulfate 24 hours prior to oral gavage with 108 CFU S. Typhimurium. For the survival curves, female mice were gavaged with 107 CFU S. Typhiumurium. Lethal morbidity was monitored in accordance with IACUC protocols at UT Southwestern.
For SFB monocolonization experiments, fecal material from SFB-monocolonized mice (derived from Andrew Gewirtz’s lab at the Georgia State University) was diluted in 1 ml PBS and gently vortexed for 10 minutes. The suspension was centrifuged at 2,000 rpm for 2 minutes, and 100 μl of the supernatant was used to gavage GF mice. After four weeks, tissues were harvested at ZT0 and ZT12. 16S rRNA gene sequencing of fecal material confirmed that these animals harbored >95% SFB.
Immunoblot
Proteins were extracted from mouse ileum by homogenizing in T-PER Tissue Protein Extraction Reagent (Thermo Fischer, 78510) supplemented with a protease inhibitor cocktail (cOmplete ULTRA Tablets, Sigma-Aldrich; 5892953001) and a phosphatase inhibitor (PhosSTOP, Sigma-Aldrich; 4906845001). 40 μg of total protein was loaded onto 4–20% gradient SDS-PAGE and transferred to a PVDF membrane. Membranes were blocked with 5% nonfat dry milk in PBS with 0.1% Tween-20. For detection with anti-STAT3 antibodies, membranes were blocked with 5% BSA in PBS with 0.1% Tween-20. Membranes were incubated at room temperature for one hour with the following primary antibodies: anti-REG3G antiserum raised against recombinant REG3G (Cash et al., 2006) and anti-succinate dehydrogenase (Abcam; ab14715), and at 4°C overnight with the following antibodies: anti-STAT3 (Cell Signaling; 4904S), anti-phospho-STAT3 (Tyr705) (Abcam; ab76315), and anti-lipocalin-2 (R&D systems; AF1857). After washing with PBS with 0.1% Tween, membranes were incubated with HRP-conjugated secondary antibodies. Membranes were visualized using a Bio-Rad ChemiDoc™ Touch system, and band density was quantified using Bio-Rad Image Lab Software 5.2.1.
Scanning electron microscopy
Mouse small intestines (ileum) were fixed with 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer overnight at 4°C. After three rinses in 0.1 M sodium cacodylate buffer, the tissues were post-fixed with 2% osmium tetroxide in 0.1 M sodium cacodylate buffer for 2 hours. The tissue samples were rinsed with water and dehydrated with increasing concentrations of ethanol, followed by increasing concentrations of hexamethyldisilazane in ethanol. Samples were air dried under the hood, mounted on SEM stubs and sputter coated with gold palladium in a Cressington 108 auto sputter coater. Images were acquired on a Field-Emission Scanning Electron Microscope (Zeiss Sigma) at 1.5–3kV accelerating voltage in the UT Southwestern Electron Microscopy Core Facility. The point of bacterial attachment was counted for 4–10 random villi across two visual fields per mouse.
Immunofluorescence microscopy
The terminal ileum was flushed with 5 ml of cold PBS, fixed in Bouin’s fixative and embedded in paraffin. Paraffin-embedded sections were washed twice in xylene followed by re-hydration in decreasing concentrations of ethanol. Antigen retrieval was performed by boiling in 0.1 M trisodium citrate and washing in PBS. Slides were blocked with 1% BSA, 1% Triton X-100 in PBS and then incubated with the following primary antibodies: anti-REG3G antiserum raised against recombinant REG3G (Cash et al., 2006) and anti-Lysozyme (Dako A0099) at 4°C overnight at 1:250 dilutions. Secondary antibodies AlexaFluor ® 488/647 (ThermoFisher) were diluted 1:400 and applied to slides for 1 hour at room temperature in a humidified chamber, in the dark. Slides were washed and incubated with Hoechst dye (Thermo Scientific, 62249) at a 1:200 dilution for 15 minutes at room temperature. Slides were washed and mounted with Fluoromount-G ® (Southern Biotechnology, 0100–01) and imaged on a Zeiss AxioImager M1 microscope or a Keyence Fluorescence Microscope BZ-X800. In some cases, VECTASHIELD® antifade mounting medium with DAPI (Vector labs, H-1200–10) was used instead of Hoechst dye.
DNA extraction for 16S rRNA gene sequencing
For analysis of mucus-associated bacteria, the terminal ileum was flushed with 5 ml of cold PBS and placed at −20°C until DNA extraction. For DNA extraction, samples were thawed and homogenized, lysates were transferred into a sterile Eppendorf tube, and DNA was extracted with phenol:chloroform. The DNA was precipitated with sodium acetate and ethanol and resuspended with Elution Buffer (Qiagen, 1014609).
Laser capture microdissection
For laser capture microdissection, a 4 cm portion of the distal ileum was washed with 10 ml of cold PBS and snap frozen in optimum cutting temperature (OCT) compound (Fisher scientific, 23-730-571). A 7 μm frozen section was cut on a Leica CM1950 cryostat and immediately fixed in 70% ethanol. The section was then stained with methyl green 1% (R&D Systems, 4800-30-18) and then with eosin Y solution (Sigma-Aldrich, HT110116–500ML). Stained sections were used for laser capture microdissection of intestinal epithelial cells using an Arcturus PixCell IIe system, and 8,000–10,000 pulses were obtained per sample. RNA was extracted from the captured intestinal epithelial cells using the PicoPure RNA Isolation Kit (Arcturus, KIT0202) following the manufacturer’s protocol. Purified RNA was stored at −80°C until use.
Whole tissue RNA purification
Whole tissue from the terminal ileum was washed with 20 ml of ice-cold PBS. After washing, the tissue was submerged in 5 ml of RNAlater. 50 mg of tissue was homogenized in 900 μl of QIAzol reagent solution using a TissueRuptor, and the RNeasy Plus Universal Mini kit (Qiagen, 73404) was used to extract and purify RNA.
Quantitative real-time PCR (Q-PCR)
cDNA was constructed using the M-MLV Reverse Transcriptase kit (Invitrogen, 28025–013) according to the manufacturer’s protocol. Q-PCR reactions were performed using the Platinum SYBR green kit (Life Technologies, 11733046) on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems). Relative expression values were determined using the comparative Ct (ΔΔCt) method, and transcript abundances were normalized to Actb transcript abundance. Primer sequences are listed in Table S1.
Enzyme-linked immunosorbent assay (ELISA)
Proteins were extracted from mouse ileum by homogenizing in T-PER Tissue Protein Extraction Reagent (Thermo Fischer, 78510), supplemented with a protease inhibitor cocktail (cOmplete ULTRA Tablets, Sigma-Aldrich; 5892953001) and a phosphatase inhibitor (PhosSTOP, Sigma-Aldrich; 4906845001). Homogenized tissues were diluted to a final protein concentration of 3 mg/ml in Reagent solution and 300 μg of tissue was assayed per sample. Each sample was assessed in triplicate. IL-22 and IL-23 abundance were measured using the Mouse IL-22 DuoSet ELISA (R&D Systems, DY582) and the Mouse IL-23 DuoSet ELISA (R&D Sytems, DY1887) kit, respectively. Plates were read on a SpectraMax plate reader.
16S Q-PCR analysis
For measurement of the abundance of tissue-associated bacteria, tissue from mouse ileum was homogenized in PBS. DNA was purified by phenol:chloroform extraction. Next, 2 μg of DNA from each sample was used for amplification with primers specific for the universal 16S rRNA or SFB gene sequence (listed in Table S1). The target sequence was amplified with the HotStarTaq® Master Mix Kit (Qiagen, 203443) to enrich for bacterial DNA. PCR reactions were diluted 1:10 into water and analyzed by Q-PCR using the Platinum SYBR green kit (Life Technologies, 11733046). For quantification of bacterial abundance in feces, fecal pellets were collected directly from the mouse, homogenized in PBS, and the DNA was purified by phenol:chloroform extraction. However, only 1 μg of DNA was amplified from each sample using primers specific for the SFB 16S rRNA gene sequence. PCR reactions were analyzed using standard curves generated with template controls designed for each primer set that were run in tandem with the experimental samples. Primer sequences are listed in Table S1.
16S rRNA gene sequencing
The hypervariable regions V3 and V4 of the bacterial 16S rRNA gene were captured using Illumina Nextera protocol (Part # 15044223 Rev. B). A single amplicon of about 460 bp was amplified using the 16S Forward Primer and 16S Reverse Primer (listed in Table S1) as described in the Illumina protocol. The PCR product was cleaned using Agencourt AmpureXP beads from Beckman Counter Genomics. Illumina adapter and barcode sequences were ligated to the amplicon in order to attach them to MiSeqDx flow cell and for multiplexing. Quality and quantity of each sequencing library were assessed using Bioanalyzer and picogreen measurements, respectively. About 6 pM of the pooled libraries were loaded onto a MiSeqDX flow cell and sequenced using PE300 (Paired end 300 bp) v3 kit. Raw fastq files were demultiplexed based on unique barcodes and assessed for quality. Samples with more than 50K QC pass sequencing reads were used for downstream 16S Operational Taxonomic Unit (OTU) analysis.
16S gene sequencing analysis pipeline
Taxonomic classification and OTU abundance analysis were done using the CLC Bio microbial genomics module (https://www.qiagenbioinformatics.com/plugins/clc-microbial-genomics-module/). Individual sample reads were annotated using the Greengene database and taxonomic features were determined. Alpha and beta diversity analysis were done to measure the within- and between-sample diversity, respectively. Abundance data were used for numeric Principal Component Analysis (PCA) in SVS, Golden Helix Software. Raw fastq files from this study were submitted to the Sequence Read Archive and are also available on request.
Analysis of feeding behavior
Food intake, feeding times, energy expenditure, and locomotor activity were monitored using a combined indirect calorimetry system (Labmaster, TSE Systems GmbH, Germany). Experimental mice were individually housed in a light (12 hours on/12 hours off, 7 am-7 pm) and temperature (22.5–23.5°C) controlled environment and acclimated in the home cage for five days before data collection. Mice were analyzed in the metabolic chambers for four days and were provided with food and water ad libitum. O2 consumption and CO2 production were measured by indirect calorimetry to determine energy expenditure. Locomotor activity was measured using a multidimensional infrared light beam detection system. Food and water intake were recorded continuously using lid-mounted sensors.
For the 24-hour fasting experiment, 16 male mice were ordered from Taconic Farms. For each time point assessed, a group of four co-caged animals were split into two cages with two animals each. One cage was supplied with food while food was withdrawn from the other cage. Water was made freely available to all animals. This procedure was performed every six hours across the day-night cycle (ZT0, 6, 12, and 18) to ensure that each cage was fasted for a total of 24 hours. After 24 hours, tissue was harvested from each group of mice. Prior to separation, each cage was tested for the presence of SFB by Q-PCR. This experiment was performed twice, with a total of four mice assessed per timepoint for each condition.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of experiments can be found in the figure legends, including how significance was defined and the statistical methods used. Data represent mean ± standard error of the mean. Numbers of experiments noted in figure legends reflect independent experiments that were performed on different days. No method was used to predetermine sample size. Blinding was not performed for these experiments. Formal randomization techniques were not used; however, mice were allocated to experiments randomly and samples processed in an arbitrary order. All statistical analyses were performed with GraphPad Prism software. To assess the statistical significance of a difference between two groups of mice, we used two-tailed Student’s t tests. To assess the statistical significance of differences between more than two groups, we used one-way ANOVAs. For survival analysis, the log-rank (Mantel-Cox) test was used. The only mice excluded from experiments were mice that died during the course of experimentation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Brian Hassell, Madison Spence, and Nicole Salinas for assistance with mouse experiments, and Phoebe Doss, Robyn Leidel, Rebecca Jackson, and Dr. Anza Darehshouri for assistance with electron microscopy. We thank Dr. Andrew Gewirtz (Georgia State University) for providing the SFB used in the monocolonization experiments. Part of the Graphical Abstract was created at Biorender.com. The Zeiss Sigma scanning electron microscope was purchased with the NIH Shared Instrumentation program grant 1S10OD020103-01 to Dr. Katherine Luby-Phelps. This work was supported by NIH grant R01 DK070855 (L.V.H.), a Welch Foundation (Grant I-1874 to L.V.H.), and the Walter M. and Helen D. Bader Center for Research on Arthritis and Autoimmune Diseases (L.V.H.). J.F.B. was supported by NIH Grant T32 AI005284 and a Howard Hughes Medical Institute Hanna Gray Fellowship. L.V.H. and J.S.T. are Investigators of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, and Akira S (1998). Targeted disruption of the MyD88 gene results in loss of IL-1-and IL-18-mediated function. Immunity 9, 143–150. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen J, Jellbauer S, Wong CP, Edwards RA, George MD, Ouyang W, and Raffatellu M (2014). The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40, 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, Edwards RA, Sahar S, Dandekar S, Baldi P, et al. (2013). Circadian clock regulates the host response to Salmonella. Proc. Natl. Acad. Sci. U.S.A 110, 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash HL, Whitham CV, Behrendt CL, and Hooper LV (2006). Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Chen H, Shu X, Yin Y, Li J, Qin J, Chen L, Peng K, Xu F, Gu W, et al. (2018). Presence of segmented filamentous bacteria in human children and its potential role in the modulation of human gut immunity. Front. Microbiol 9, 1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieppa M, Rescigno M, Huang AYC, and Germain RN (2006). Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med 203, 2841–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coorens M, Rao A, Gräfe SK, Unelius D, Lindforss U, Agerberth B, Mjösberg J, and Bergman P (2019). Innate lymphoid cell type 3-derived interleukin-22 boosts lipocalin-2 production in intestinal epithelial cells via synergy between STAT3 and NF-κB. J. Biol. Chem 294, 6027–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CP and Savage DC (1974). Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect. Immun 10, 948–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas B, Harding HP, Choi HS, Lehmann KA, Chung M, Lazar MA, and Moore DD (1994). A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol. Endocrinol 8, 996–1005. [DOI] [PubMed] [Google Scholar]
- Eberl G, and Littman DR (2004). Thymic origin of intestinal ab T cells revealed by fate mapping of RORγt+ cells. Science 305, 248–251. [DOI] [PubMed] [Google Scholar]
- Hou B, Saudan P, Ott G, Wheeler ML, Ji M, Kuzmich L, Lee LM, Coffman RL, Bachmann MF, and DeFranco AL (2011). Selective utilization of Toll-like receptor and MyD88 signaling in B cells for enhancement of the antiviral germinal center response. Immunity 34, 375–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL, Ruhn K Hou B, DeFranco AL, Yarovinsky F, and Hooper LV (2011) gd intraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. Proc. Natl. Acad. Sci. USA 108, 8743–8748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, de Llanos Frutos R, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, and Littman DR (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo M, Pejchal M, Schook AC, Lange RP, Walisser JA, Sato TR, Wang X, Bradfield CA, and Takahashi JS (2014). Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. Elife 3, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson H, Hugerth LW, Sundh J, Lundin E, and Andersson AF (2020). Genome sequence of segmented filamentous bacteria present in the human intestine. Commun. Biol 3, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. (1997). Positional cloning of the mouse circadian clock gene. Cell 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, Hohl TM, Flavell RA, Littman DR, and Pamer EG (2012). Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 36, 276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaasen HL, Koopman JP, van den Brink ME, Bakker MH, Poelma FG, and Beynen AC (1993). Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab Anim 27, 141–150. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, and Glöckner FO (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41, e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsson A and Bøggild H (2010). Gastrointestinal disorders among shift workers. Scand. J. Work Environ. Health 36, 85–95. [DOI] [PubMed] [Google Scholar]
- Kuang Z, Wang Y, Yu X, Ruhn KA, Kubo M, and Hooper LV (2019). The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 357, 912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan-Diez M, Soualhi S, Lee C, Irie K, Pinker EY, Narushima S, et al. (2019). Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363, eaat4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, et al. (2015). Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe 17, 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, and Gordon JI (2008). Evolution of mammals and their gut microbes. Science 322, 1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Bushman FD, and FitzGerald GA (2015). Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. U.S.A 112, 10479–10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, and Gumucio DL (2002). Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem 277, 33275–33283. [DOI] [PubMed] [Google Scholar]
- Moh A, Iwamoto Y, Chai GX, Zhang SS, Kano A, Yang DD, Zhang W, Wang J, Jacoby JJ, Gao B, Flavell RA, and Fu XY (2007). Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab Invest 87, 1018–28 [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, and Takahashi JS (2012). Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci 35, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohren DCL, Jansen NWH, Kant IJ, Galama J, van den Brandt PA, and Swaen GMH (2002). Prevalence of common infections among employees in different work schedules. J. Occup. Environ. Med 44, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, and Papaioannou VE (1992). RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, Rollins D, Propheter DC, Rizo J, Grabe M, Jiang Q-X, and Hooper LV (2014). Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature 505, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patke A, Young MW, Axelrod S (2020) Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol 21, 67–84. [DOI] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS (2014) Molecular architecture of the mammalian circadian clock. Trends Cell. Biol 24, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U 3 and Schibler U (2002). The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260. [DOI] [PubMed] [Google Scholar]
- Propheter DC, Chara AL, Harris TA, Ruhn KA, and Hooper LV (2017) Resistin-like molecule β is a bactericidal protein that promotes spatial segregation of the microbiota and the colonic epithelium. Proc. Natl. Acad. Sci. USA 114, 11027–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio S-P, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, et al. (2009). Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A, Lepage P, Nolte A, Hellmig S, Schreiber S, and Ott SJ (2010). Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. J. Med. Microbiol 59, 1114–1122. [DOI] [PubMed] [Google Scholar]
- Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, and Ricciardi-Castagnoli P (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol 2, 361–367. [DOI] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. (2010). Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol 11, 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee J-Y, Ziel JW, Miraldi ER, Domingos AI, et al. (2015). An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 163, 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, and Diefenbach A (2008). RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol 10, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczesnak A, Segata N, Qin X, Geyers D, Petrosino JF, Huttenhower C, Littman DR, and Ivanov II (2013) The genome of TH17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe 10, 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Zou J, Zhang Z, Zhao X, Noriega J, Zhang B, Zhao C, Ingle H, Bittinger K, Mattei LM, Pruijssers AJ, Plemper RK, Nice TJ, Baldridge MT, Dermody TS, Chassaing B, and Gewirtz AT (2019) Cell 179, 644–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EF, Fulton BO, Ayres JS, Pham LN, Ziauddin J, and Shirasu-Hiza MM (2012) The circadian clock protein timeless regulates phagocytosis of bacteria in Drosophila. PLoS Pathog 8, e1002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet 18, 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J, Hahn P, Kroehling L, Nguyen H, Li D, and Littman DR (2020). Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature 579, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. (2014). Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 159, 514–529. [DOI] [PubMed] [Google Scholar]
- Thaiss CA, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, et al. (2016). Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510. [DOI] [PubMed] [Google Scholar]
- Tuganbaev T, Mor U, Bashiardes S, Liwinski T, Nobs SP, Leshem A, Dori-Bachash M, Thaiss CA, Pinker EY, Ratiner K, et al. (2020). Diet diurnally regulates small intestinal microbiome-epithelial-immune homeostasis and enteritis. Cell 182, 1441–1459. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, and Hooper LV (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U.S.A 105, 20858–20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, and Hooper LV (2011). The antibacterial lectin RegIIIg promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeri M, and Raffatellu M (2016). Cytokines IL-17 and IL-22 in the host response to infection. Pathog. Dis 74, ftw111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez EM, Nguyen H, Heasley KT, Saechao CH, Gil LM, Rogers AWL, Miller BM, Rolston MR, Lopez CA, Litvak Y, et al. (2019). Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat. Microbiol 4, 1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, and Takahashi JS (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, and Hooper LV (2017). The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 357, 912–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiassé RP, Schaefer AL, Koroleva I, Splinter-Bondurant S, Brown B, Manzella L, Snir E, et al. (2010) Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc. Natl. Acad. Sci. U.S.A 107, 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang Y, Zhu L Liu W, Liao N, Jiang M, Zhu B, Yu HD, Xiang C, and Wang X (2013) Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J 7, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Chaix A, Yooseph S, and Panda S (2014). Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 20, 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene sequencing data (Figure S3) have been deposited in the Sequence Read Archive (SRA) with BioProject ID PRJNA733561 and are publicly available.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


