Abstract
The Escherichia coli OmpF porin is a nonspecific channel involved in the membrane translocation of small hydrophilic molecules and especially in the passage of β-lactam antibiotics. In order to understand the dynamic of charged-compound uptake through bacterial porins, specific charges located in the E. coli OmpF channel were mutated. Substitutions G119D and G119E, inserting a protruding acidic side chain into the pore, decreased cephalosporin and colicin susceptibilities. Cefepime diffusion was drastically altered by these mutations. Conversely, substitutions R132A and R132D, changing a residue located in the positively charged cluster, increased the rate of cephalosporin uptake without modifying colicin sensitivity. Modelling approaches suggest that G119E generates a transverse hydrogen bond dividing the pore, while the two R132 substitutions stretch the channel size. These charge alterations located in the constriction area have differential effects on cephalosporin diffusion and substantially modify the profile of antibiotic susceptibility.
The outer membrane of gram-negative bacteria shelters them from external toxic compounds. In the membrane, porins are channel-forming proteins allowing diffusion of small hydrophilic solutes through this barrier (10, 18, 20). With bacterial resistance to various antibiotics due to the permeability barrier impairing chemotherapy (19), it is important to define the biochemical and biophysical parameters governing target access and intracellular drug concentration. In particular, since outer membrane porins are key to β-lactam penetration (19, 22), it is essential to understand the various possible interactions. The native Escherichia coli OmpF porin is a trimer, and the three-dimensional structure shows a monomeric β-barrel built of 16 antiparallel β-strands containing the pore (6). The longest loop, L3, is bent into the channel, forming a gate; in this constriction area, a positively charged cluster of amino acid residues protruding from the barrel wall faces the L3 negatively charged side chain residues. This generates a strong electrostatic field parallel to the membrane surface, and such an organization could facilitate the diffusion of molecules and modulate voltage gating (6, 12, 36). Several mutations have been selected on residues located in the channel; among them, G119D is a substitution located in L3 obtained from colicin N resistance screening after random mutagenesis (8). Structural and functional analyses of the G119D porin indicate that the mutation affects channel properties without causing large molecular alterations (11). To address the question of the effects of steric hindrance and charge movement in the flux through the pore lumen, mutant porins with site-specific mutations in positions 119 and 132 have been constructed: 119D and 119E, which are located in the negatively charged cluster, and 132A and 132D, which belong to the facing positive region. Using immunological probes directed against wild-type porin, we established the correct membrane insertion of the various modified molecules. The activities of antibacterial compounds and the kinetics of labeled antibiotic uptake demonstrated the role of amino acid residues in diffusion through the channel. Protein modelling addressed the substitution effect on the pore and illustrated the interaction between the porin lumen and cephalosporin.
(This work was presented in part at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, Calif., 24 to 27 September 1998.)
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and site-directed mutagenesis of the ompF gene.
The E. coli strains used in this work were AK101 (rnhA::cat derivative of JM101) (23)), DH5α mutS (DH5α mutS::Tn10) (27), and BZB1107 (ompF::Tn5) derived from the wild-type E. coli BE from the Biozentrum (Basel, Switzerland) collection (27). Enterobacter cloacae 201-RevM3, devoid of porin, was also used (17). Plasmids pLG361, encoding wild-type OmpF, and pBSK(+/−) Urnh, which can replicate only into AK101 cells due to its defective ori, have been described elsewhere (4, 8).
Bacteria were routinely grown in Luria-Bertani (LB) medium at 37°C with gentle shaking. If required, kanamycin (50 μg/ml), ampicillin (50 μg/ml), tetracycline (15 μg/ml), and chloramphenicol (60 μg/ml) (Sigma) were added.
The amino acid residues located in the constriction area of the E. coli ompF channel were replaced by the site-directed mutagenesis method as described by Ohmori (23). A 918-bp BglII-ClaI fragment of the ompF gene was excised from pLG361 and inserted into the BamHI and ClaI sites of the pBSK Urnh vector to give pBSK-ompF. This plasmid was used to transform E. coli AK101. pBSK-ompF single-stranded DNA was isolated using R408 helper phage (31). The mutations were created by the method of Ohmori (23) and confirmed by DNA sequencing. The plasmid was transformed into DH5α mutS. The BstYI-ClaI fragments carrying the various mutations were excised from pBSK-ompF and cloned into the pLG361 expression vector between the BglII and ClaI sites to give pVAV1, pVAV2, pVAV3, and pVAV4, encoding substitutions G119D, G119E, R132A, and R132D, respectively. These plasmids were used to transform BZB1107 or E. cloacae 201-RevM3 cells.
Antibiotic and colicin susceptibility tests.
For the determination of MICs, approximately 106 cells were inoculated into 1 ml of Mueller-Hinton broth containing twofold serial dilutions of each antibiotic, and the results were read after 18 h at 37°C (5).
Colicin N was purified from strain BZB1019(pCHAP4) (28). It was assayed with cells grown in LB medium. One hundred microliters of cell suspension at an optical density of 0.5 at 600 nm was added to various dilutions (10−1 to 10−6) of colicin N (1.0 mg/ml) in phosphate buffer (150 mM NaCl, 3 mM KCl, 1 mM KH2PO4, 10 mM Na2HPO4, pH 7) and incubated for 20 min at 37°C. The cell suspension was then diluted with 15 volumes of fresh LB medium. The percentage of surviving cells with or without bacteriocin treatment was monitored after 2 h at 37°C by determining the ratio of the optical densities at 600 nm (11).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and porin immunocharacterization.
Exponential-phase bacteria in LB broth were pelleted and solubilized in loading buffer (160 mM Tris, 0.8 M sucrose, 0.01% bromophenol blue, 3% sodium dodecyl sulfate, 1% 2-mercaptoethanol) at 96°C. Samples (an amount corresponding to 0.02 optical density unit at 600 nm) were loaded on sodium dodecyl sulfate-polyacrylamide gels (10% polyacrylamide, 0.1% sodium dodecyl sulfate) as described previously (8) and then electrotransferred to nitrocellulose membranes. An initial saturating step with TBS (50 mM Tris-HCl, 150 mM NaCl, pH 8) containing 10% (wt/vol) skim milk was carried out overnight at 4°C. The nitrocellulose membranes were then incubated in TBS containing 10% skim milk and 0.2% Triton X-100 for 2 h at room temperature in the presence of polyclonal antibodies directed against denatured OmpF (8). After four washings in the same buffer, detection was performed with alkaline phosphatase-conjugated AffinitiPure goat anti-rabbit immunoglobulin G antibodies (Jackson ImmunoResearch, West Grove, Pa.) as described previously (5).
For immunodotting detection, equivalent amounts of bacteria expressing the various OmpF porins from exponential cell cultures were loaded on nitrocellulose. Immunodetections were carried out in TBS containing 10% skim milk with two monoclonal antibodies (MoF 18 and 19) directed against cell surface-exposed monomeric epitopes located on the subunit, with a monoclonal antibody (MoF 21) specific to a cell surface-exposed trimeric epitope, and with polyclonal antibodies directed against cell envelope (15). After four washings in the same buffer, detection was carried out with alkaline phosphatase-conjugated AffinitiPure anti-mouse antibodies (Jackson ImmunoResearch) (8, 15).
Measurement of cefepime uptake.
E. cloacae 201-RevM3 (17), an isolate devoid of porins, was selected as the recipient strain for the various constructs and wild-type OmpF. Exponential-phase bacteria in nutrient broth were removed by centrifugation, and pellets were resuspended in sodium phosphate buffer (50 mM, pH 7), supplemented with 5 mM MgCl2, to a density of 3 × 106 CFU/ml. Fifty microliters, containing 50 nM 14C-labeled cefepime (a gift from Bristol-Myers Squibb, Syracuse, N.Y.) mixed with unlabeled cefepime (final specific activity, 25.4 μCi/mg), was added to a 450-μl cell suspension at 37°C in a shaking water bath. At set intervals, samples were mixed with 7% cold trichloroacetic acid as described by Lee et al. (13). After 10 min on ice, samples were filtered through GF/C filters (Whatman LTD, Maidstone, England), washed twice, and dried. The radioactivity was measured in a Packard scintillation spectrophotometer.
Protein modelling.
The protein modelling was carried out by Synt:em (Nimes, France). The frame structures used for modelling were wild-type OmpF (6) and the mutants R42C, R82C, G119D, D113G, and deletion 109-114 (11, 14, 26, 29). These structures were analyzed with the COMPOSER program (3) to search the structurally conserved area (24, 30). The flexible part was analyzed by AMBER treatment with the SYBYL 6.4 program (24).
RESULTS
Folding and stability of OmpF mutants.
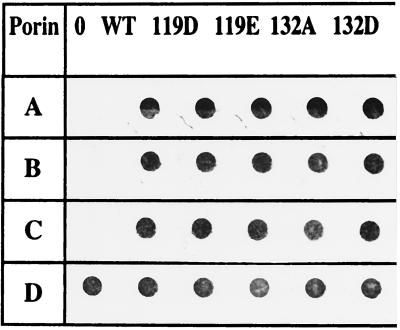
The folding and membrane locations of the mutated porins were checked by immunodotting of bacteria containing the various plasmids (Fig. 1). The signals obtained with monoclonal antibodies recognizing monomeric epitopes (Fig. 1, rows A and B) indicated similar expression and antigenic accessibility of the various modified porins at the cell surface compared to wild-type OmpF. In addition, the probe specific to a cell surface-exposed trimeric epitope clearly indicated the correct folding of modified porins in the outer membrane (Fig. 1, row C). The amounts of the OmpF mutants were roughly equal to that of the wild-type porin, and the monomers issued from these constructs had an electrophoretic mobility different from that of wild-type OmpF (data not shown). It has been previously reported that some mutated or chimeric porins have reduced thermal stability, especially when the modification or the fusion site mapped on residues neighboring the channel constriction area or in the L2 loop (7, 8, 25). The 119E trimer was strongly heat sensitive; the trimeric form was observed only at 50°C and was completely dissociated at 55°C. Only a 5°C shift distinguished the 119E and 119D trimer stabilities (Table 1). In contrast, 132D and to a greater extent 132A were markedly less affected by the temperature, suggesting that no perturbation occurred during trimer arrangement.
FIG. 1.
Folding and locations of the various constructs. Immunodetections were carried out with two monoclonal antibodies directed against epitopes located on the monomeric form (rows A and B), with a monoclonal antibody specific to a trimeric epitope (C), and with polyclonal antibodies directed against cell envelope (D). Lanes: 0, E. coli BZB1107, devoid of porin; WT, BZB1107 (pLG361), encoding the wild-type OmpF; 119D, 119E, 132A, and 132D, BZB1107 expressing the various mutants.
TABLE 1.
Porin thermostabilitya
| OmpF porin | Heat stabilityb at:
|
||||
|---|---|---|---|---|---|
| 50°C | 55°C | 60°C | 65°C | 70°C | |
| Wild type | T | T | T | T | T |
| 119D | T | T | M | M | M |
| 119E | T | M | M | M | M |
| 132A | T | T | T | T | M |
| 132D | T | T | T | M | M |
BZB1107 cells expressing wild-type or mutated OmpF porins were solubilized at various temperatures and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunodetection was carried out with polyclonal antiserum directed against OmpF porin (8).
T, trimer detected; M, monomer detected.
Colicin and antibiotic susceptibilities.
Resistance of the G119D mutant to colicin N, which requires only OmpF to bind and enter the bacterial cell, has been previously reported (8, 11). Consequently, it was of interest to determine the levels of colicin N sensitivity for the various porins. Substitutions located at position 119 conferred greater colicin resistance (Table 2). Moreover, the colicin sensitivity of the bacterial cells expressing the 119E substitution was decreased by a factor of 10 relative to that for 119D. Mutations 132A and 132D did not significantly modify the colicin N susceptibility of producing cells (Table 2).
TABLE 2.
Colicin and antibiotic susceptibilities
| OmpF porin | Colicin N sensitivity (dilution)a | MIC (μg/ml) ofb:
|
|||||
|---|---|---|---|---|---|---|---|
| CFO | CTX | FAM | FEP | MOX | OFX | ||
| Wild type | 10−5 | 2 | 0.06 | 0.5 | 0.06 | 0.12 | 0.25 |
| 119D | 10−1 | 16 | 0.5 | 4 | 1 | 4 | 0.25 |
| 119E | Rc | 16 | 0.5 | 4 | 1 | 8 | 0.25 |
| 132A | 10−5 | 2 | 0.06 | 0.5 | 0.06 | 1 | 0.25 |
| 132D | 10−5 | 2 | 0.06 | 0.5 | 0.06 | 0.25 | 0.25 |
Maximal dilution of colicin suspension which caused cell death in the direct assay (8).
MICs were determined in Mueller-Hinton broth. Values are means of three independent determinations. CFO, cephaloridine; CTX, cefotaxime; FAM, cefamandole; FEP, cefepime; MOX, moxalactam; OFX, ofloxacin.
R, resistant with the maximal colicin concentration used in the test.
A panel of antibiotics was used to test susceptibilities of strains expressing the various mutants. The strongest modifications, reflected by the increase of MICs, affected cephem activities (Table 2). Mutations located at residue 119 conferred the greater resistance profile. This resistance concerned all of the tested cephalosporins whatever their global charge, with some of the tested molecules being monoanionic, zwitterionic, or dianionic (21, 24). Mutation 132A or 132D did not significantly change the cephalosporin sensitivity, except for the dianionic moxalactam with 132A.
Cefepime diffusion.
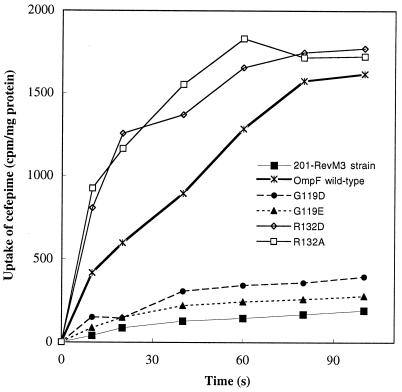
We analyzed the uptake of radiolabeled cefepime by using E. cloacae cells (Fig. 2). When the wild-type OmpF was synthesized, a linear rate was observed during the first 60 s and the steady-state level was reached at about 1.5 to 2 min. This cefepime accumulation reflected the functional porin, while no significant uptake was obtained in the absence of porin. Diffusion of radiolabeled cefepime was drastically impaired for cells expressing mutations 119D or 119E, indicating a strong alteration of channel properties (Fig. 2). Interestingly, the extension of the side chain, induced by the D→E transition, did not markedly change the level of cefepime diffusion. In contrast, when bacteria expressed the 132D or 132A substitution, the initial rate of cefepime uptake was appreciably increased without modification of the steady-state level.
FIG. 2.
Cefepime diffusion through the various porins. Values are means of three independent determinations. E. cloacae isolate 201-RevM3, which is devoid of porins, and 201-RevM3 expressing wild-type OmpF or the 119D, 119E, 132D, or 132A mutant were used.
Protein modelling.
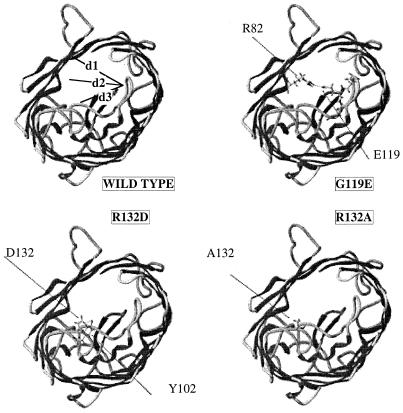
In the case of the 119E mutant, and taking into account the energetic hindrances in this area, the longer side chain of glutamic acid relative to aspartic acid generated a hydrogen bond with R82 (Fig. 3). This transverse bond could definitively separate the channel into two smaller compartments, as previously observed for the three-dimensional structure of the 119D mutant (11). Interestingly, three distances between residues located in the constriction area, d1 (between residues 42 and 119), d2 (between residues 82 and 119), and d3 (between residues 119 and 132), were lower than those of wild-type porin: 50, 55, and 50%, respectively, with the 119D substitution and 45, 39, and 35%, respectively, with the 119E substitution. With the substitutions located at R132, no significant modification in d1 or d2 was observed, while a noticeable increase in d3 (110 to 127%) compared with that in wild-type OmpF was obtained. These data suggested that 132A and 132D, in addition to the charge modification, stretch the channel, generating an increase of the cavity.
FIG. 3.
Representation of the various porins. Molecules are viewed from the periplasmic space, and only residues 82, 102, 119, and 132 are indicated for clarity. d1, d2, and d3 illustrate the intervals between residues 119 to 42, 119 to 82, and 119 to 132, respectively, which are used to evaluate the induced deviation.
DISCUSSION
Various OmpF site-specific mutants were analyzed to investigate the role of charged residues located in the porin constriction area in the diffusion rate of hydrophilic compounds. In 119E substitution, compared to 119D, the conservation of negative charge is associated with a CH2 extension of the side chain favoring a transverse hydrogen bond with R82. The previous X-ray data indicated that 119D generates only local effects (11). The crystal structure of the colicin N fragment suggests a model for its translocation (35). The results support the role of the L3 loop in conjunction with the colicin translocation domain during bacteriocin uptake through the membrane. The redistribution of charged side chains due to the hydrogen bond 119E-R82 inhibited the penetration of colicin into the periplasm. Moreover, the presence of negatively charged residues in the porin channel was sufficient per se to decrease cephalosporin sensitivity. The initial rate of cefepime diffusion was seriously affected, with reductions of flux of 8- to 10-fold for 119D and 119E. Of significant importance is that the zwitterionic cefepime will presumably form one or several salt bridges during diffusion. Taking into account the modelling of mutations at position 119 and the distribution of charges inside the channel (12, 26, 33, 36), a strong deviation of the bulky cefepime, which is a very large molecule relative to pore diameter (1), probably occurred through the newly orientated electrostatic field.
The 132A substitution eliminated the hydrogen bond with Y102 residue and released this position from the positively charged cluster, while the 132D substitution preserved a hydrogen bond with Y102. In two cases, no significant modification in the colicin activity was observed. These two substitutions speeded up the diffusion of cefepime. The previous studies on OmpF mutants selected for larger pore size, which reported the modification of electrical properties of mutant porin channels (2, 14, 29), are worth mentioning. Substitution R42C or R132P, removing the charged side chain, had a more cation-selective pore and a decreased critical voltage. The increase of cefepime uptake with mutants 132A and 132D indicated that the guanidium group of R132 plays an important role in the cephalosporin orientation through the native eyelet. In the case of PhoE, the substitution removing the R residue in position 37 or 75 (equivalent to positions 42 and 82 in OmpF, respectively), distally located to R132, increases the rate of uptake of cephaloridine (34). However, this is a small zwitterionic cephalosporin compared to cefepime used in this study (1, 20, 21, 37).
This concept is especially important with the increasing bacterial resistance conferred by modification of membrane permeability (19). A recently isolated clinical strain of E. aerogenes (16) producing an altered porin had characteristics similar to those of the in vitro-designed mutants described here. In addition, Gill et al. (9) reported the existence of discrete mutations in the Neisseria gonorrhoeae porin that increased the negative charge in the putative gonococcal equivalent of E. coli loop 3. These substitutions were associated with a noteworthy β-lactam resistance found in clinical isolates. Recently, molecular dynamic studies of the OmpF pore that focused on the constriction zone supported an active role for the ionic environment of the charged residues located inside the channel (32, 33). In this context, the location of our selected substitutions is essential: the effects reported here illustrate the putative role of the opposite charges in the pore eyelet as orientation-determining regions for charged compounds such as cephalosporins.
ACKNOWLEDGMENTS
We thank D. Fourel, V. Géli, B. I. Holland, and F. Pattus for the generous gifts of colicins and plasmids. We gratefully acknowledge H. Bénédetti, J.-M. Bolla, and J. Chevalier for helpful advice. We thank Bristol-Myers Squibb for its generous gift of radiolabeled cefepime.
This work was supported by the Institut National de la Santé et de la Recherche Médicale, the Fondation pour la Recherche Médicale, the Université de la Méditerranée, and the Région PACA and Marseille-Métropole.
REFERENCES
- 1.Bellido F, Pechère J C, Hancock R E W. Reevaluation of the factors involved in the efficacy of new β-lactams against Enterobacter aerogenes. Antimicrob Agents Chemother. 1991;35:73–78. doi: 10.1128/aac.35.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson S A, Occi J L L, Sampson B A. Mutations that alter the pore function of the OmpF porin of Escherichia coli K-12. J Mol Biol. 1988;203:961–970. doi: 10.1016/0022-2836(88)90121-0. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein F, Koetzle T F, Williams G, Meyer E F, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. The protein data bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 4.Bouveret E, Rigal A, Lazdunski C, Benedetti H. Distinct regions of the colicin A translocation domain are involved in the interaction with Tol A and Tol B proteins upon import into Escherichia coli. Mol Microbiol. 1998;27:143–157. doi: 10.1046/j.1365-2958.1998.00667.x. [DOI] [PubMed] [Google Scholar]
- 5.Charrel R N, Pagès J-M, De Micco P, Malléa M. Prevalence of outer membrane porin alteration in β-lactam-antibiotic-resistant Enterobacter aerogenes. Antimicrob Agents Chemother. 1996;40:2854–2858. doi: 10.1128/aac.40.12.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J A, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 7.Fourel D, Bernadac A, Pagès J-M. Involvement of exposed polypeptide loops in trimeric and membrane insertion of Escherichia coli OmpF porin. Eur J Biochem. 1994;222:625–630. doi: 10.1111/j.1432-1033.1994.tb18905.x. [DOI] [PubMed] [Google Scholar]
- 8.Fourel D, Mizushima S, Bernadac A, Pagès J-M. Specific regions of Escherichia coli OmpF protein involved in antigenic and colicin receptor sites and in stable trimerization. J Bacteriol. 1993;175:2754–2757. doi: 10.1128/jb.175.9.2754-2757.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gill M J, Simjee S, Al-Hattawi K, Robertson B D, Easmon C S F, Ison C A. Gonococcal resistance to β-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob Agents Chemother. 1998;42:2799–2803. doi: 10.1128/aac.42.11.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 11.Jeanteur D, Schirmer T, Fourel D, Simonet V, Rummel G, Rosenbusch J P, Pagès J-M. Structural and functional alterations of a colicin resistant mutant of OmpF from E. coli. Proc Natl Acad Sci USA. 1994;91:10675–10679. doi: 10.1073/pnas.91.22.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karshikoff A, Cowan S W, Spassov V, Ladenstein R, Schirmer T. Electrostatic properties of two porin channels from E. coli. J Mol Biol. 1994;240:372–384. doi: 10.1006/jmbi.1994.1451. [DOI] [PubMed] [Google Scholar]
- 13.Lee E H, Nicolas M H, Kitzis M D, Pialoux G, Collatz E, Gutmann L. Association of two resistance mechanisms in a clinical isolate of Enterobacter cloacae with high-level resistance to imipenem. Antimicrob Agents Chemother. 1991;35:1093–1098. doi: 10.1128/aac.35.6.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lou K L, Saint N, Prilipov A, Rummel G, Benson S A, Rosenbusch J P, Schirmer T. Structural and functional characterization of OmpF porin mutants selected for larger pore size. Crystallographic analysis. J Biol Chem. 1996;271:20669–20675. [PubMed] [Google Scholar]
- 15.Lupi N, Bourgois A, Bernadac A, Laboucarié S, Pagès J-M. Immunological analysis of porin polymorphism in Escherichia coli B and K-12. Mol Immunol. 1989;26:1027–1036. doi: 10.1016/0161-5890(89)90067-9. [DOI] [PubMed] [Google Scholar]
- 16.Malléa M, Chevalier J, Bornet C, Eyraud A, Davin-Regli A, Bollet C, Pagès J-M. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology. 1998;144:3003–3009. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- 17.Malléa M, Simonet V, Eun-Hee L, Collatz E, Gervier R, Gutmann L, Pagès J-M. Biological and immunological comparisons of Enterobacter cloacae and Escherichia coli porins. FEMS Microbiol Lett. 1995;129:273–280. doi: 10.1111/j.1574-6968.1995.tb07592.x. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido H. Porins and specific diffusion channels in bacterial outer membranes. J Biol Chem. 1994;269:3905–3908. [PubMed] [Google Scholar]
- 19.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 20.Nikaido H. Outer membrane. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 29–47. [Google Scholar]
- 21.Nikaido H, Liu W, Rosenberg E Y. Outer membrane permeability and β-lactamase stability of dipolar ionic cephalosporins containing methoxyimino substituents. Antimicrob Agents Chemother. 1990;34:337–342. doi: 10.1128/aac.34.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikaido H, Normark S. Sensitivity of Escherichia coli to various β-lactams is determined by the interplay of outer membrane permeability and degradation by periplasmic β-lactamases: a quantitative predictive treatment. Mol Microbiol. 1987;1:29–36. doi: 10.1111/j.1365-2958.1987.tb00523.x. [DOI] [PubMed] [Google Scholar]
- 23.Ohmori H. A new method for strand discrimination in sequence-directed mutagenesis. Nucleic Acids Res. 1994;22:884–885. doi: 10.1093/nar/22.5.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearlman D A, Case D A, Caldwell J W, Ross W S, Cheatham T E, Ferguson D M, Seibel G L, Chandra Singh U, Keiner P K, Kollman P A. AMBER 4.1. San Franscisco: University of California; 1995. [Google Scholar]
- 25.Phale P S, Philippsen A, Kiefhaber T, Koebnik R, Phale V P, Schirmer T, Rosenbusch J P. Stability of trimeric OmpF porin: the contribution of the latching loop L2. Biochemistry. 1998;37:15663–15670. doi: 10.1021/bi981215c. [DOI] [PubMed] [Google Scholar]
- 26.Phale P S, Schirmer T, Prilipov A, Hardmeyer A, Rosenbusch J P. Voltage gating of E. coli porin channels: role of the constriction loop. Proc Natl Acad Sci USA. 1997;94:6741–6745. doi: 10.1073/pnas.94.13.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prilipov A, Phale P S, Van Gelder P, Rosenbusch J P, Koebnik R. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol Lett. 1998;163:65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
- 28.Pugsley A P. Nucleotide sequencing of the structural gene for colicin N reveals homology between the catalytic, C-terminal domains of colicins A and N. Mol Microbiol. 1987;1:317–325. doi: 10.1111/j.1365-2958.1987.tb01938.x. [DOI] [PubMed] [Google Scholar]
- 29.Saint N, Lou K L, Widmer C, Luckey M, Schirmer T, Rosenbusch J P. Structural and functional characterization of OmpF porin mutants selected for larger pore size. Functional characterization. J Biol Chem. 1996;271:20676–20680. [PubMed] [Google Scholar]
- 30.Sali A, Blundell T. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1995;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Suenaga A, Komeiji Y, Uebayasi M, Meguro T, Saito M, Yamato I. Computational observation of an ion permeation through a channel protein. Biosci Rep. 1998;18:39–48. doi: 10.1023/a:1022292801256. [DOI] [PubMed] [Google Scholar]
- 33.Tieleman D P, Berendsen H J C. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys J. 1998;74:2786–2801. doi: 10.1016/S0006-3495(98)77986-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Gelder P, Saint N, Phale P, Eppens E F, Prilipov A, van Boxtel R, Rosenbusch J P, Tommassen J. Voltage sensing in the PhoE and OmpF outer membrane porins of Escherichia coli: role of charged residues. J Mol Biol. 1997;269:468–472. doi: 10.1006/jmbi.1997.1063. [DOI] [PubMed] [Google Scholar]
- 35.Vetter I R, Parker M W, Tucker A D, Lakey J H, Pattus F, Tsernoglou D. Crystal structure of a colicin N fragment suggests a model for toxicity. Structure. 1998;6:863–874. doi: 10.1016/s0969-2126(98)00088-4. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe M, Rosenbusch J P, Schirmer T, Karplus M. Computer simulations of the OmpF porin from the outer membrane of Escherichia coli. Biophys J. 1997;72:2094–2102. doi: 10.1016/S0006-3495(97)78852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshimura F, Nikaido H. Diffusion of β-lactam antibiotics through the porin channels of E. coli K-12. Antimicrob Agents Chemother. 1985;27:84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]