Abstract
The alveolar bone is a unique osseous tissue due to the presence of the teeth and the proximity of commensal oral microbes. Commensal microbe effects on alveolar bone homeostasis have been attributed to the oral microbiota, yet the impact of commensal gut microbes is unknown. Study purpose was to elucidate whether commensal gut microbes regulate osteoimmune mechanisms and skeletal homeostasis in alveolar bone. Male C57BL/6T germfree (GF) littermate mice were maintained as GF or monoassociated with segmented filamentous bacteria (SFB), a commensal gut bacterium. SFB has been shown to elicit broad immune response effects, including the induction of TH17/IL17A immunity, which impacts the development and homeostasis of host tissues. SFB colonized the gut, but not oral cavity, and increased IL17A levels in the ileum and serum. SFB had catabolic effects on alveolar bone and non-oral skeletal sites, which was attributed to enhanced osteoclastogenesis. The alveolar bone marrow of SFB vs. GF mice had increased dendritic cells, activated helper T-cells, TH1 cells, TH17 cells, and upregulated Tnf. Primary osteoblast cultures from SFB and GF mice were stimulated with vehicle-control, IL17A, or TNF to elucidate osteoblast-derived signaling factors contributing to the pro-osteoclastic phenotype in SFB mice. Treatment of RAW264.7 osteoclastic cells with supernatants from vehicle-stimulated SFB vs. GF osteoblasts recapitulated the osteoclast phenotype found in vivo. Supernatants from TNF-stimulated osteoblasts normalized RAW264.7 osteoclast endpoints across SFB and GF cultures, which was dependent on the induction of CXCL1 and CCL2. This report reveals that commensal gut microbes have the capacity to regulate osteoimmune processes in alveolar bone. Outcomes from this investigation challenge the current paradigm that alveolar bone health and homeostasis is strictly regulated by oral microbes.
Keywords: Gastrointestinal Microbiome, Alveolar Bone Loss, Osteoclasts, Immunomodulation
INTRODUCTION
Commensal microbes colonize the host to establish a relationship in which one derives benefit without obvious harm or benefit to the other1. Collections of microbes colonize distinct anatomical sites to form microbiota communities (i.e., oral microbiota, gut microbiota, skin microbiota)1-3. The commensal microbiota critically regulates host immunity3-6, which has indirect effects on the physiology and homeostasis of host tissues6-10. Commensal microbiota effects on host immunity has recently been shown to impact osteoimmunology11-14, which is the process by which immune cells regulate bone cells15. Commensal microbiota regulation of osteoimmune mechanisms critically influence the turnover and homeostasis of the skeleton in both health and disease11-15.
Alveolar bone formation and homeostasis is dependent on the eruption and maintenance of the teeth. The teeth are a mineralized transmucosal organ that is vigorously colonized by oral microbes. The alveolar bone is distinct from all other bone tissues, due to the teeth acting as a scaffold for oral microbe biofilms that form in close proximity to alveolar bone16,17. Commensal oral microbes elicit local immune responses in barrier epithelial and gingival connective tissues, which modulate osteoimmune signaling and coupled osteoclast-osteoblast actions at the subjacent alveolar bone12,17.
Gnotobiology is the science of studying animals with a defined microbiota. Germfree (GF) animals are associated (colonized) with specific microorganisms to elucidate host-microbe interactions. Early investigators monoassociated GF rats with different commensal oral bacteria to delineate that commensal oral microbes critically regulate alveolar bone homeostasis18-20. Due to the proximity of the oral microbiota to alveolar bone, current theory supports the notion that alveolar bone homeostasis is strictly regulated by oral microbes17. Recognizing that commensal gut microbes modulate systemic immunity3-6, the purpose of the current report was to delineate whether commensal gut microbes have the capacity to regulate osteoimmune mechanisms and skeletal homeostasis in the alveolar bone complex.
We innovatively utilized the segmented filamentous bacteria (SFB)-monoassociated mouse model, which is devoid of oral microbes, to discern whether commensal gut microbes impart osteoimmune response effects in alveolar bone. SFB is a commensal gut bacterium that resides in the gastrointestinal tracts of a wide variety of species, including rodents and humans21-26. SFB colonizes the terminal ileum of mice post weaning and maintains its presence in the intestinal tract throughout post-natal development and young adulthood27-30. SFB adheres to intestinal epithelial cells to elicit broad innate and adaptive immune response effects, including the induction of TH17/IL17A immunity31-38. SFB promotion of TH17/IL17A immunity confers protection against pathogenic microbes, both locally in the gut31,39,40 and at distant anatomic sites41. SFB actions on systemic immunity influences host tissue homeostasis at extraintestinal sites, including the skeleton42-45. Two recent osteoimmunology reports relied on the presence of SFB in the gut to discern that specific commensal gut microbes regulate osteoimmune processes at non-oral skeletal sites44,45.
To our knowledge, this is the first known report to employ a gnotobiotic animal model to determine whether commensal gut microbes have the capacity to regulate osteoimmune mechanisms in the alveolar bone complex. In line with prior work which showed that SFB enhanced both IL17A and TNF45-47, we demonstrated that SFB osteoimmune actions are linked to the induction of IL17A and TNF. Based on knowledge that IL17A and TNF can promote osteoblasts to secrete pro-osteoclastic factors48-53, we established an in vitro osteoblast-osteoclast coupling assay targeting IL17A and TNF. This model system provides initial evidence demonstrating that commensal gut microbe immunomodulatory actions influence osteoimmune crosstalk. Notably, this work discerns that commensal gut microbes have the ability to regulate osteoimmune processes and skeletal homeostasis in the alveolar bone complex. Findings from this study challenge the current paradigm that alveolar bone health and homeostasis is strictly regulated by oral microbes.
MATERIALS AND METHODS
Mice.
GF C57BL/6T mice were acquired from Taconic Biosciences (Rensselaer, NY), bred, and maintained in sterile isolators at Medical University of South Carolina (MUSC) Gnotobiotic Animal Core. Male GF littermates were maintained as GF or monoassociated with SFB at age 5-weeks. SFB-monoassociated mice were generated as previously described44. Animals were euthanized at age 11-weeks and tissues were harvested for analyses. Animal protocols were approved by the MUSC Animal Protocols Review Board and were done in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals.
Micro-CT.
Femurs and maxillae were fixed in 10% phosphate-buffered-formalin for 24 hours at room temperature and then stored in 70% ethanol. Specimens were scanned with Bruker Skyscan 1176 (Bruker, Kontich, Belgium) with the following acquisition parameters: aluminum 0.5mm filter, rotation step of 0.3°, average of 2 frames at 4000x2612 dpi, X-ray tube potential at 50kVp, X-ray intensity of 497μA, 65ms integration time, and isotropic voxel size of 9μm3. Calibrated three-dimensional images were reconstructed.
Trabecular bone volume per tissue volume was analyzed in femurs and maxillae via AnalyzePro 12.0 Bone Microarchitecture Analysis software (Analyze Direct, Seattle, WA). For femurs, transverse CT slices were evaluated beginning 450μm distal to the proximal growth plate and extending 1500μm distally. A fixed threshold of 250 Hounsfield Units was utilized to discern mineralized tissue. Data are reported in accordance with standardized nomenclature54, as previously described13,44,55. For maxillae, trabecular bone volume per tissue volume analysis was performed in the maxillary 1st molar trifurcation, within a defined volume of interest (VOI). The VOI was created by linearly morphing a cylinder within the trifurcation, excluding the periodontal ligament lining the mesiobuccal, distobuccal, and lingual roots and the endocortical surface of the buccal and lingual cortical plates. The height of the cylinder was set so that the superior aspect of the cylinder was positioned apical to the fornix of the trifurcation, and the inferior aspect of the cylinder was positioned at the apex of the distobuccal root. Fixed threshold of 250 Hounsfield units was used to discriminate mineralized tissue.
Reconstructed maxillae images were also used to assess linear alveolar bone loss. Linear alveolar bone loss was assessed at the maxillary first molar using AnalyzePro Analysis software (Analyze Direct, Seattle, WA). Linear alveolar bone loss was evaluated by measuring the linear distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) at the mesiobuccal line angle, distobuccal line angle, and mid-lingual aspect of the maxillary first molar (Supp Fig 1). The CEJ to ABC linear measurements were performed to evaluate alveolar bone loss. CEJ to ABC linear distance is used both clinically and experimentally to accurately assess alveolar bone loss16,56. The maxillary first molar was oriented in the axial plane such that the mid buccal-lingual aspect of the mesiobuccal and distobuccal roots were aligned to a horizontal plane at 0°. The first molar was oriented in the sagittal plane such that the CEJ at the mesial and distal aspect of the tooth were aligned to a horizontal plane at 0°. Within the axial plane, the coronal height of contour was determined at the mesiobuccal line angle, distobuccal line angle, and mid-lingual aspect of the molar. These landmarks served as the midpoint for carrying out CEJ to ABC linear measurements. The initial measurement was performed at the mid-point, two subsequent measurements were carried out 45μm and 90μm mesial to the midpoint, and two additional measurements were performed 45μm and 90μm distal to the midpoint.
Histomorphometry.
Maxillae and femurs were fixed in 10% phosphate-buffered-formalin for 24 hours at room temperature. Specimens were then decalcified in 14% EDTA for 3 weeks at room temperature, processed for paraffin-embedding, and 5μm serial sagittal sections were cut using an Accu-Cut SRM 200 Sakura microtome (Sakura Finetek Europe B.V, Zoeterwoude, NL). Serial sagittal sections were cut through the maxillary first molar and distal femur and stained with tartrate-resistant acid phosphatase (TRAP). Maxillae were counterstained with aniline blue. Histomorphometric analysis of osteoclast cellular outcomes were performed in TRAP-stained sections where TRAP+ multinucleated (≥3 nuclei) cells lining bone were scored as osteoclasts. The region of interest for histomorphometric analysis was limited to the furcal alveolar bone within the maxillary first molar trifurcation, excluding the periodontal ligament. The region of interest in the distal femur was limited to the secondary spongiosa, beginning 250μm proximal to the growth plate and extending 1000μm proximally, and 50μm from endocortical surfaces. Images were taken at 400X for maxillary sections and 200X for femoral sections, acquired by Nikon’s Inverted Microscope Eclipse TS100-F (Nikon Instruments, Inc., Melville, NY), and analyzed using NIH ImageJ software, version 1.51j8, https://imagej.nih.gov/ij/ (NIH, Bethesda, MD). Data are reported in accordance with standardized nomenclature57, as previously described13,44,55.
In situ Immunofluorescence.
In situ immunofluorescence analysis was carried out in paraffin-embedded sagittal sections of the maxillary first molar and sagittal sections of the distal femur. Specimens were deparaffinized with xylenes, rehydrated with graded ethanols, and washed with saline. Antigen retrieval was achieved in 10mM sodium citrate buffer (pH 6.0). Samples were permeabilized in 1x PBS containing 0.2% Triton X-100 and blocked in 10% goat serum for one hour at room temperature. A 1:100 dilution of anti-osterix monoclonal antibody (clone sc #343396 Santa Cruz Biotechnology, Dallas, TX, USA) overnight at 4°C. Sections were washed in 1X PBS and then incubated with a 1:2000 rhodamine-goat anti-mouse (Santa Cruz Biotechnology) for one hour at room temperature (protected from light). Samples were washed in 1X PBS and mounted via ProLong Diamond Antifade Mountant with DAPI (Life Technologies, Carlsbad, CA, USA). Images were acquired at 200X with a Keyence BZ-X810 fluorescence microscope. Images were stitched and overlaid with the Keyence BZ-X Analysis Software (Keyence, Osaka, Japan). Analysis of bone-lining osterix+ osteoblasts were determined in the maxillary first molar furcation alveolar bone. In the distal femur, analysis was limited to the secondary spongiosa, beginning 250μm proximal to the growth plate, extending proximally 1000μm, and 50μm from endocortical surfaces. Images were scored using ImageJ software version 1.52a.
Bone Marrow Cultures.
Femurs were isolated from each animal, and marrow was flushed using α-MEM media (Gibco, Fisher Scientific) + 20% Hyclone FBS + 1% 2mM glutamine + 100 U/ml penicillin + 100 mg/ml streptomycin (PSG). Bone marrow cells were disassociated, counted, and plated at 3x106 cells/cm2 in a 60 mm dish as described previously13,44,55. Twelve hours later, the non-adherent hematopoietic progenitor cells (HPCs) were collected for osteoclast-precursor (OCP) assays. Fresh α-MEM media + 20% FBS + 1% PSG was added back to the cultures. Forty-eight hours later, adherent bone marrow mesenchymal stem cells (BMMSCs) were isolated for the osteoblast stimulation assay. Importantly, marrow cells were not combined from animals for initial bone marrow cultures or subsequent osteoclast-precursor / osteoblast assays; n-values reported for in vitro assays are representative of biological replicates.
In vitro Osteoclast-Precursor (OCP) Assays.
Non-adherent hematopoietic progenitor cells (HPCs) were collected from whole marrow cultures, washed, and labeled with CD11b microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany). AutoMACS Sorter (Miltenyi Biotec) was used to separate CD11b− HPCs, as previously described13,44,55. CD11b− HPCs were plated at 1.5x105 cells/cm2 in culture media (α-MEM media + 10% Hyclone FBS + 1% PSG) for assays; plated in 12-well plates for gene expression assays and 96-well plates for the TRAP stain assay. CD11b− HPCs were plated in culture media supplemented with 10ng/mL CSF1 (R&D Systems; Minneapolis, MN), to enrich for CD11b− osteoclast-precursor (OCP) cells having high osteoclast potential13,44,55. Thirty-six hours later, the culture media was changed. Gene expression assay: CD11b− OCP cells were stimulated with fresh control (25ng/mL CSF1; R&D Systems) or treatment (25ng/mL CSF1 + 50ng/mL RANKL; R&D Systems) media for 4-days; media refreshed every other day. Day-4 CD11b− OCP cells were isolated for qRT-PCR gene expression analysis; performed in duplicate (technical replicate) cultures13,44,55. Data are representative of two separate experiments. TRAP stain assay: CD11b− OCP cells were stimulated with fresh control (25ng/mL CSF1; R&D Systems) or treatment (25ng/mL CSF1 + 50ng/mL RANKL; R&D Systems) media for 6-days; media refreshed every other day. Day-6 cultures were stained by the TRAP method, as described previously13,44,55. Cytomorphometric analysis of TRAP+ osteoclast cells was carried out in triplicate (technical replicate) cultures; five fields of view were analyzed per technical replicate culture at 100X. TRAP+ cells with ≥3 nuclei were scored as an osteoclast13,44,55. Analysis was performed using NIH ImageJ software, version 1.51j8, https://imagej.nih.gov/ij/ (NIH, Bethesda, MD). Data are representative of two separate experiments.
In vitro Osteoblast Stimulation Assay.
BMMSCs were plated in culture media (α-MEM media + 10% Hyclone FBS + 1% PSG) at 2.0x104 cells/cm2 in 12-well plates. Upon reaching confluency, cultures were stimulated with osteogenic media (α-MEM media + 10% Hyclone FBS + 1% PSG + 50ug/ml ascorbic acid) for 5 days, media changed every other day. Cells were then cultured in serum-deprived osteogenic media (α-MEM media + 0.3% Hyclone FBS + 1% PSG + 50ug/ml ascorbic acid) overnight. On Day-6, cultures were stimulated for 2 hours with serum-deprived osteogenic media supplemented with either vehicle-control, IL17A (200ng/mL, R&D Systems), or TNF (2ng/mL, R&D Systems), as described previously50,51. After the 2-hour stimulation, supernatants were aspirated, and cells were washed to remove recombinant cytokines. Fresh serum-deprived osteogenic media was added back to cultures for 6 hours. Cell culture supernatants were then collected for ELISA protein assays and the in vitro osteoblast-osteoclast coupling assay, and cells were isolated for qRT-PCR gene expression analysis. In vitro osteoblast stimulation assay was performed in triplicate (technical replicate) cultures. Data are representative of two separate experiments.
In vitro Osteoblast-Osteoclast Coupling Assay.
RAW264.7 murine macrophage cells were expanded in culture media (α-MEM media + 10% Hyclone FBS + 1% PSG). RAW264.7 cells were passed and plated at 1.5x105 cells/cm2 in 96-well plates for the osteoblast-osteoclast coupling assay. RAW264.7 cells were plated in 100uL culture media for 4-hours and then supplemented with 100uL of culture media containing 100ng/ml RANKL (R&D Systems); final concentration of RANKL at 50ng/ml. The cultures were not disturbed for three days, which allowed for the RAW264.7 cells to initiate fusing into small multinucleated cells.
On day-3, the culture media was changed in order to treat the RAW264.7 cells with supernatants derived from the in vitro osteoblast stimulation assay. The old media was aspirated and 100uL of fresh culture media was added back to the cultures. 50uL of osteoblast culture supernatant was immediately added to each culture, followed by 50uL of fresh culture media containing 50ng/ml RANKL; final concentration of RANKL at 25ng/ml. Neutralizing antibodies αCXCL1 (500pg/mL; R&D Systems) and αCCL2 (2000pg/mL; R&D Systems) were added into cultures with supernatants derived from the in vitro osteoblast stimulation assay on day-3. Concentrations of αCXCL1 and αCCL2 neutralizing antibodies were based on outcomes from osteoblast supernatant ELISA assays.
Forty-eight hours later at day-5 of the culture system, the RAW264.7 cells were fixed and stained by the TRAP method. TRAP stain assay was carried out in triplicate (technical replicate) cultures. Images were acquired and technical replicate cultures analyzed at 200X. TRAP+ cell with ≥3 nuclei was scored an osteoclast for cytomorphometric analysis. Analysis was performed using NIH ImageJ software, version 1.51j8, https://imagej.nih.gov/ij/ (NIH, Bethesda, MD).
Prior to executing the investigations with osteoblast culture supernatants from experimental GF and SFB mice, optimization studies were performed with RAW264.7 cells and osteoblast culture supernatants derived from 11-12 week-old male specific-pathogen-free mice. Optimization studies discerned that culturing the RAW264.7 cells in 25ng/ml RANKL from day-3 to day-5 of the culture system supported the formation of small TRAP+ osteoclastic cells. Supplementing the cultures with 50uL of osteoblast culture supernatant promoted the formation of larger TRAP+ osteoclastic cells throughout the culture well.
qRT-PCR for 16S rDNA.
Distal ileum contents and maxillary gingiva were harvested at euthanasia. Previous studies characterizing SFB colonization along the murine gastrointestinal tract have shown that SFB predominates in the distal/terminal ileum 58,59. Therefore, the most distal 20mm of the ileum was isolated for qRT-PCR assays assessing bacterial 16S rDNA and murine mRNA. Ileal contents were utilized for bacterial 16S rDNA analysis and ileal tissue was used for murine mRNA analyses. Bacterial DNA was isolated from distal ileum contents and maxillary gingiva using the Qiagen DNeasy Powersoil Pro Kit, per manufacturer’s protocols (Qiagen, Hilden, Germany). qRT-PCR protocol was executed on the StepOnePlus System (Applied Biosystems) for 30 cycles55,60. Universal 16S rDNA target gene was tested using the forward (Fwd) primer: 5’- ACT CCT ACG GGA GGC AGC AGT-3’, and reverse (Rev) primer: 5’- ATT ACC GCG GCT GGC-3’44,55,60. SFB was assessed via primers: Fwd: 5’- GAC GCT GAG GCA TGA GAG CAT-3’, Rev: 5’- GAC GGC ACG GAT TGT TAT TCA-3’, as described previously44. SFB rDNA from ileum and gingiva specimens of GF and SFB mice were normalized to a bacterial DNA standard (ZymoBIOMICS, Irvine, CA) as previously described44. Relative quantification of data was carried out by the 2−ΔCT method61,62. All reactions were performed in duplicate.
Quantitative Real-Time PCR (qRT-PCR) for mRNA.
Mandible bone marrow was flushed with TRIzol Reagent (Invitrogen, Carlsbad, CA). Mandibular mucogingival tissues were collected at euthanasia and homogenized in TRIzol. Distal ileum tissue was flash frozen, pulverized, and homogenized in TRIzol. Cell cultures were washed twice with 1X PBS, and TRIzol was directly applied to lyse cells. RNA was isolated by the TRIzol method following manufacturer’s instructions, as previously described13,44,55. Total RNA was quantified via NanoDrop 1000 (Thermo Scientific, Waltham, MA). cDNA was produced using Taqman Random Hexamers and Reverse Transcription Reagents (Applied Biosystems, Foster City, CA), according to the manufacturer’s protocol. cDNA was amplified using TaqMan gene expression primers/probes, Universal PCR Master Mix, and the StepOnePlus System (Applied Biosystems). Primers used included Tnfrsf11a (Rank), Dcstamp, Tnfsf11 (Rankl), Tnfrsf11b (Opg), Il17a, Tnf, Ccl2, Cxcl1, Cxcl5, Gapdh, and Rn18s; all primers were purchased from Thermo Scientific. Gapdh was used as an endogenous control for ileum, mandible marrow, and cell culture specimens, and Rn18s was utilized as the endogenous control for mucogingival samples. Relative quantification of data was performed via the comparative CT method (2−ΔΔCT)62,63, as previously described13,44,55. All reactions were performed in duplicate.
Flow Cytometric Analysis.
Mandible bone marrow cells were isolated, washed, and counted as previously described13,44,55. Live Cell Analysis: Cells were treated with FcR-block (Miltenyi Biotec) and stained for cell surface markers. CD4+ Naïve / Activated T-cells: Naïve/activated helper T-cells were gated on CD3+ cells, followed by gating for helper T-cells (CD4+CD8−) or cytotoxic T-cells (CD4−CD8+). CD3+CD4+CD8− (Q1) were subsequently gated on naïve (CD62L+CD69−) or activated (CD62L−CD69+) surface markers; anti-CD3-PE-Vio770 (Miltenyi Biotec; clone REA641), anti-CD4-VioBlue (Miltenyi Biotec; clone REA604), anti-CD8-PE (Miltenyi Biotec; clone REA601), anti-CD62L-FITC (Miltenyi Biotec; clone REA828), anti-CD69-APC (Miltenyi Biotec; cloneH1.2F3). Dendritic cells (DCs): Dendritic cells were negatively gated on B220- cells, followed by examining cell surface markers MHC II+CD11c+ (Q2); anti-CD11c-PE-Vio770 (Miltenyi Biotec; clone REA754), anti-MHC II-FITC (Miltenyi Biotec; clone REA528), anti-B220-VioBlue (Miltenyi Biotec; clone REA755). Dead cells were excluded from analysis via propidium iodide viability dye (Miltenyi Biotec). CD4+ Helper T-cell Subset Analysis: Cells were treated with FcR-block (Miltenyi Biotec) and stained for cell surface markers. Cells were then treated with fixation/permeabilization buffer (eBioscience) to label intracellular transcription factors. TREG cells: TREG cells were gated on CD3+CD4+ (Q2), followed by positive gating for CD25 and transcription factor FoxP3; anti-CD3-APC-Vio770 (Miltenyi Biotec; clone REA641), anti-CD4-FITC (Miltenyi Biotec; clone REA604), anti-CD25-PE-Vio770 (Miltenyi Biotec; clone 7D4), and anti-FoxP3-PE (Miltenyi Biotec; clone REA788). TH1 cells: TH1 cells were gated on CD3+CD4+ (Q2), followed by positive gating for CD183 and transcription factor T-bet; anti-CD3-PE-Vio770 (Miltenyi Biotec; clone REA641), anti-CD4-FITC (Miltenyi Biotec; clone REA604), anti-CD183-PE (Miltenyi Biotec; clone CXCR3-173), anti-T-bet-APC (Miltenyi Biotec; clone REA102). TH2 cells: TH2 cells were gated on CD3+ cells, followed by positive gating of TH2 surface markers CD4+CD184+. CD3+CD4+CD184+ (Q2) were gated on transcription factors GATA3+IRF4−;anti-CD3-APC-Vio770 (Miltenyi Biotec; clone REA641), anti-CD4-FITC (Miltenyi Biotec; clone REA604), anti-CD184-PE-Vio770 (Miltenyi Biotec; clone REA107), anti-GATA3-PE (Miltenyi Biotec; clone REA174), anti-IRF4-APC (Miltenyi Biotec; clone REA201). TH17 cells: TH17 cells were gated on CD3+ cells, followed by positive gating of TH17 surface markers CD4+CD196+. CD3+CD4+CD196+ (Q2) were gated on transcription factors RORγt+AHR−; anti-CD3-APC-Vio770 (Miltenyi Biotec; clone REA641), anti-CD4-FITC (Miltenyi Biotec; clone REA604), anti-RORγT-APC (Miltenyi Biotec; clone REA278), anti-AHR-PE-Vio770 (eBioscience; clone 4MEJJ). Dead cells were excluded from analysis via e450 viability dye (Invitrogen). A minimum of 5,000 gated cells were analyzed per specimen. Data were acquired by MACSQuant System (Miltenyi Biotec) and analyzed by FlowJo 11.0 software (TreeStar, Ashland, OR).
Enzyme-Linked Immunosorbent Assays (ELISA).
Whole blood was collected via cardiac puncture at euthanasia, serum was isolated, and stored at −80°C. IL17A (Quantikine, R&D Systems), TNF (Quantikine, R&D Systems), P1NP (Immunodiagnostics Systems Inc, Gaithersburg, MD, USA), and TRAP5b (Immunodiagnostics Systems Inc) were assessed in serum isolates. CXCL1 (Quantikine, R&D Systems), CCL2 (Quantikine, R&D Systems), and CXCL5 (Quantikine, R&D Systems) was evaluated in osteoblast culture supernatants. ELISA kits were used following manufacturer’s protocols. All reactions were performed in triplicate.
Statistical Analysis.
Power analysis was performed in consultation with the Biostatistical Unit of the Medical University of South Carolina Bioinformatics Core and was established on the authors’ previous studies utilizing gnotobiotic murine models. Unpaired t-tests or 2-way ANOVA with Tukey Post Hoc tests were performed using GraphPad Prism 8.0 (La Jolla, CA, USA). Data are presented as mean +/− SEM. Significance is designated as p<0.05.
RESULTS
SFB does not colonize or induce immune responses in oral mucogingival tissues.
SFB inoculation was strategically performed in 5-week-old weanling GF mice to generate SFB-monoassociated mice. GF and SFB mice were intentionally euthanized at age 11-weeks (Fig 1A). The rationale for the experimental timeline was based on prior investigations in the SFB-monoassociated mouse model29,32,64 and murine alveolar bone development65-67. When young GF mice are inoculated with SFB, host immune response effects are typically induced within 1-2 weeks and persist for at least 2 months29,32,64. Different from non-oral skeletal sites, alveolar bone formation is dependent on the development and eruption of the teeth67,68. Teeth are typically fully erupted and alveolar bone formation is complete in mice by age 5 weeks65-67.
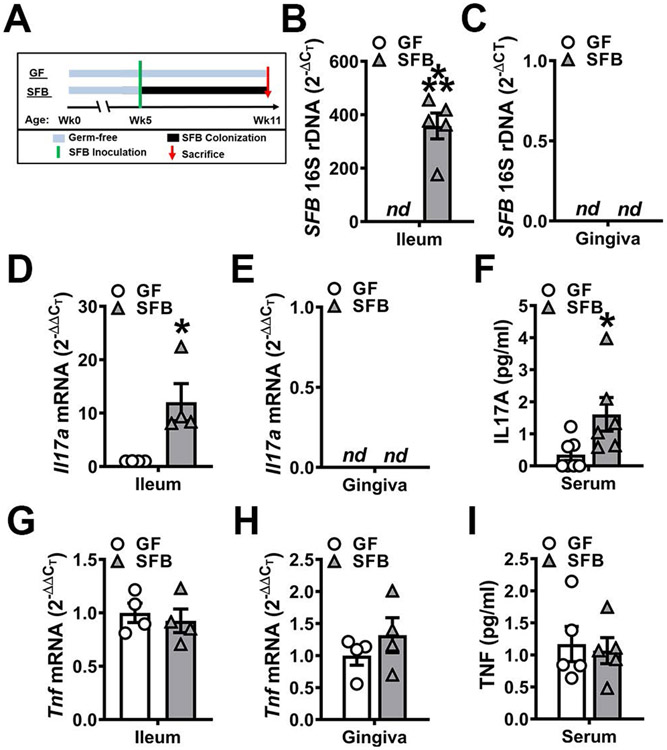
Figure 1. SFB does not colonize or induce immune responses in oral mucogingival tissues.
A. Experimental timeline. Eleven-week-old male GF and SFB mice were euthanized, and specimens harvested for analysis. B-C. 16S rDNA analysis for SFB colonization; relative quantification of rDNA was performed via the CT method (2−ΔCT). B. 16S rDNA analysis of SFB expression in distal ileum (n=5/gp). C. 16S rDNA analysis for SFB colonization in gingiva (n=5/gp). D-E. Il17a mRNA qRT-PCR analysis; relative quantification of mRNA was performed via the 2−ΔΔCT method. D. Il17a mRNA expression in ileums of GF vs. SFB mice (n=4/gp). E. Il17a mRNA expression in gingiva of GF vs. SFB mice (n=4/gp). F. Serum ELISA analysis of IL17A levels (n=6/gp). G-H. Tnf mRNA qRT-PCR analysis; relative quantification of mRNA was performed via the 2−ΔΔCT method. G. Tnf mRNA expression in ileums of GF vs. SFB mice (n=4/gp). H. Tnf mRNA expression in gingiva of GF vs. SFB mice (n=4/gp). I. Serum ELISA analysis of TNF levels (n=5/gp). Unpaired t-test; data are presented as mean ± SEM, *p<0.05, **p<0.01, ***p<0.001.
16S rDNA analysis demonstrated the presence of SFB in the ileum (Fig 1B), but not in the oral cavity (Fig 1C), of SFB-monoassociated mice. SFB induction of TH17/IL17A-mediated immunity within the gut was validated by increased Il17a expression in the distal ileum of SFB vs. GF mice (Fig 1D). Il17a expression was not detected in barrier mucogingival tissues (Fig 1E), which corroborates prior findings that SFB does not colonize the oral cavity or induce TH17/IL17A immunity in the gingiva69. Serum IL17A levels were enhanced in SFB vs. GF mice (Fig 1F). SFB interactions with the host have been reported upregulate TNF expression in intestinal cells45-47. Therefore, alterations in Tnf / TNF were analyzed in mucosal barrier tissues and the circulation (Fig 1G-I). There were no differences in Tnf expression in the ileum (Fig 1G) or mucogingival tissues (Fig 1H). Furthermore, circulating TNF levels were similar in GF and SFB mice (Fig 1I).
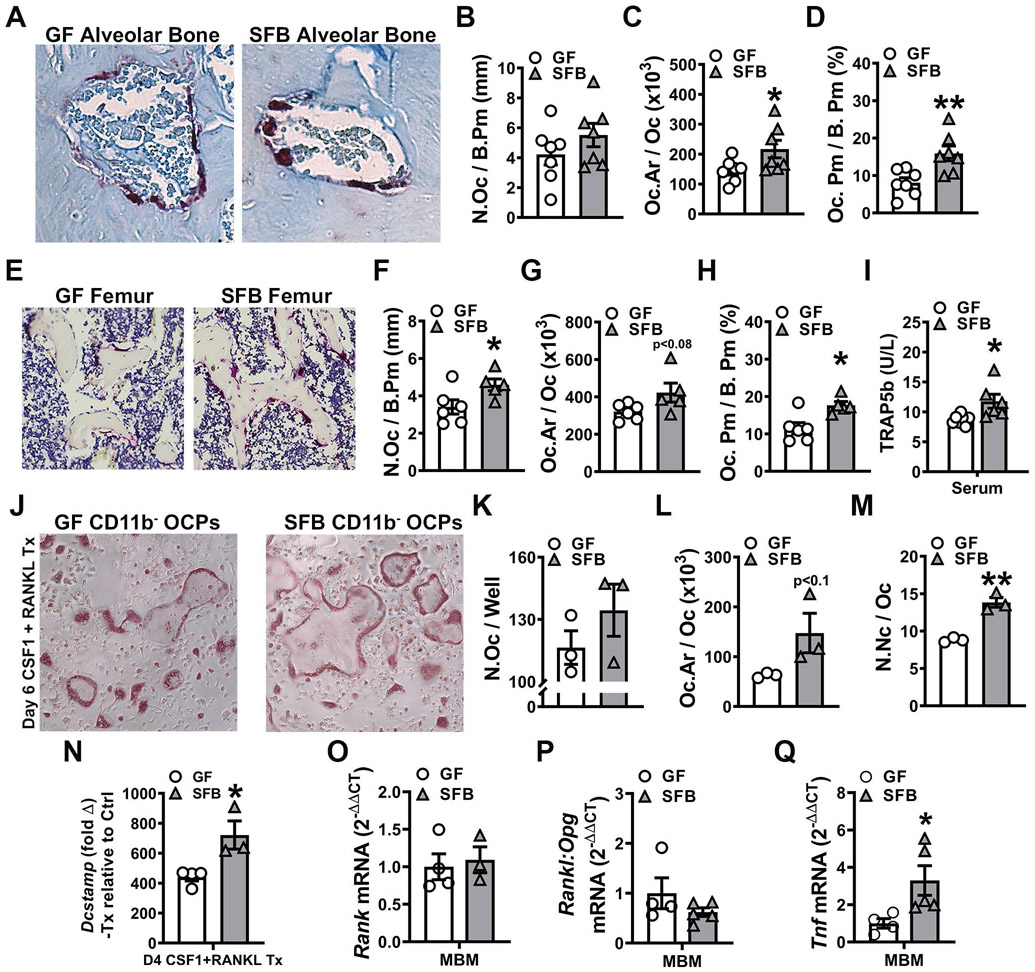
SFB has catabolic effects on alveolar bone.
Micro-CT analysis was carried out to elucidate SFB-induced tissue level alterations in the skeleton (Fig 2). Alveolar bone loss was examined at the maxillary first molar by measuring the linear distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) (Fig 2A,B) and by analyzing the trabecular bone volume fraction in the trifurcation (Fig 2C,D). The presence of SFB exacerbated linear alveolar bone loss at the mesiobuccal, distobuccal, and mid-lingual line angles (Fig 2A,B; Supp Fig 1) and blunted the trabecular bone volume fraction in the furcal alveolar bone (Fig 2C,D).
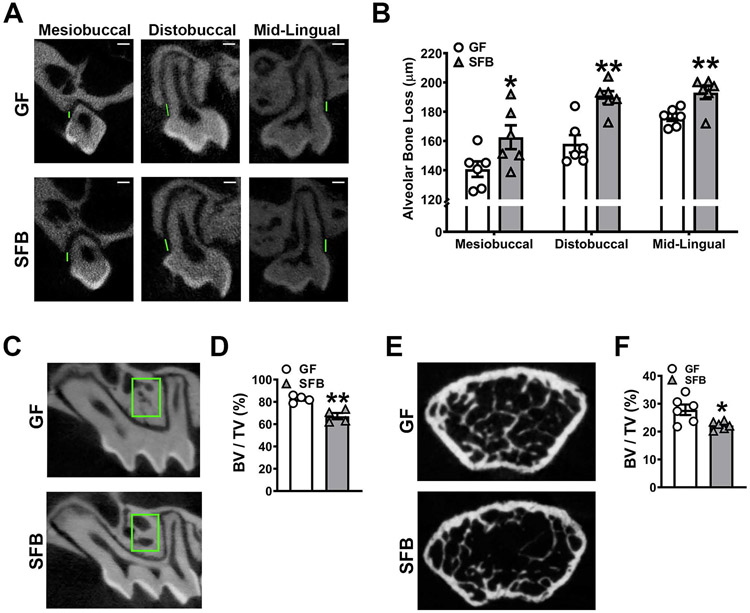
Figure 2. SFB has catabolic effects on alveolar bone.
A-B. Alveolar bone loss was measured by calculating the linear distance between the cementoenamel junction (CEJ) and alveolar bone crest (ABC) at the maxillary first molar in reconstructed micro-CT images. A. Representative micro-CT images displaying CEJ-ABC linear distance (green line) at the mesiobuccal line angle, distobuccal line angle, and mid-lingual aspect of the maxillary first molar; scale bar (white line) = 200μm. B. Quantitative measures of CEJ-ABC linear distance at the mesiobuccal line angle, distobuccal line angle, and mid-lingual aspect of the maxillary first molar (n=6/gp). C. Representative micro-CT images of trabecular bone in the maxillary first molar trifurcation, region of interest outlined (green box). D. Micro-CT quantitative analysis of trabecular bone volume per tissue volume (BV/TV) in the maxillary first molar furcal alveolar bone (n=4/gp). E. Representative micro-CT reconstructed cross-sectional images of trabecular bone in the distal femur, extending 50μm proximally from where analysis began. F. Micro-CT quantitative analysis of BV/TV in the distal femur (n=6/gp). Unpaired t-test; data are presented as mean ± SEM; *p<0.05, **p<0.01.
Micro-CT morphometric analysis of the femur showed that SFB-induced bone loss is not specific to oral skeletal sites. Trabecular bone volume fraction was diminished by 18.6% in the maxillary first molar trifurcation (Fig 2C,D) and by 20% in the distal femur (Fig 2E,F) of SFB vs. GF mice. The reduced trabecular bone volume outcomes found in the distal femur of male SFB vs. GF mice reported herein (Fig 2E,F) is similar to outcomes found in the proximal tibia of female SFB vs. GF mice44.
SFB upregulates innate / adaptive immune response effects in alveolar bone.
SFB induces TH17/IL17A-mediated immunity in the lamina propria through the presentation of antigens by intestinal dendritic cells33. Furthermore, SFB has been shown to induce IL17A-dependent protection against intestinal pathogens via extraintestinal effects on bone marrow dendritic cells39. Recent osteoimmunology studies have discerned that SFB upregulates CD4+ helper T-cell subsets in the bone marrow at non-oral skeletal sites44,45. However, it is unknown whether SFB influences helper T-cell-mediated immunity in the alveolar bone complex. Therefore, flow cytometric analysis of mandibular bone marrow cells was performed to evaluate dendritic cells, activated and naïve CD4+ T-cells, and CD4+ helper T-cell subsets (Fig 3).
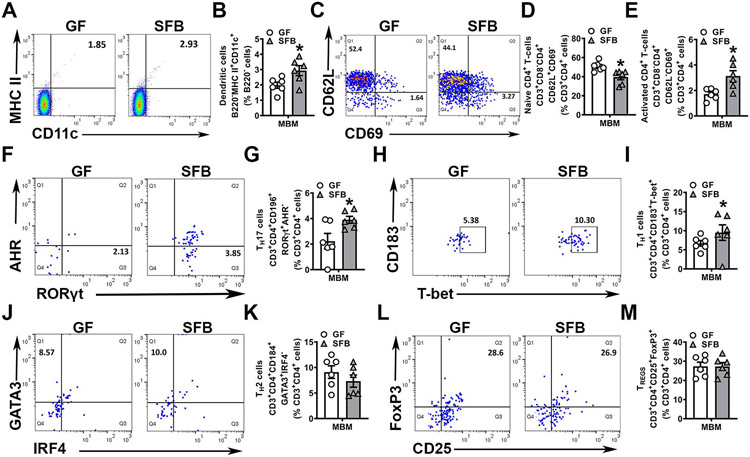
Figure 3. SFB upregulates immune response effects in alveolar bone marrow.
A-H. Mandible bone marrow (MBM) cells were isolated and stained for flow cytometric analysis (n=6/gp); cell percentages are expressed relative to the appropriate gated population. A. Representative gating of B220−MHC II+CD11c+ dendritic cells (DCs). B. Frequency of B220−MHC II+CD11c+ DCs, gated on % B220− cells. C. Representative gating of CD4+ naïve and activated T-cells. D. Frequency of CD3+CD8−CD4+CD62L+CD69− naïve T-cells, gated on % CD3+CD4+ cells. E. Frequency of CD3+CD8−CD4+CD62L−CD69+ activated T-cells, gated on % CD3+CD4+ cells. F. Representative gating of CD3+CD4+RORγt+AHR− TH17 cells. G. Frequency of CD3+CD4+RORγt+AHR− TH17 cells, gated on % CD3+CD4+ cells. H. Representative gating of CD3+CD4+CD183+T-bet+ TH1 cells. I. Frequency of CD3+CD4+CD183+T-bet+ TH1 cells, gated on % CD3+CD4+ cells. J. Representative gating of CD3+CD4+CD184+GATA3+ IRF4− TH2 cells K. Frequency of CD3+CD4+CD184+GATA3+ IRF4− TH2 cells, gated on % CD3+CD4+ CD184+ cells. L. Representative gating of CD3+CD4+CD25+FoxP3+ TREG cells. M. Frequency of CD3+CD4+CD25+FoxP3+ TREG cells, gated on % CD3+CD4+ cells. Unpaired t-test; data are presented as mean ± SEM, *p<0.05, **p<0.01.
The frequency of dendritic cells in the mandibular bone marrow was elevated in SFB vs. GF mice (Fig 3A,B), which supports the notion that SFB-derived circulating mediators alter bone marrow dendritic cells39. In line with the expanded dendritic cell population found in SFB mice (Fig 3A,B), naïve CD4+ T-cells were decreased (Fig 3C,D) and activated CD4+ T-cells were increased (Fig 3C,E) in the mandible bone marrow of SFB vs. GF mice. CD4+ helper T-cell subset analysis revealed increased TH17 cells (Fig 3F,G) and TH1 cells (Fig 3H,I), but no differences in TH2 cells (Fig 3J,K) or TREG cells (Fig 3L,M), in the mandible bone marrow of SFB vs. GF mice. Research has discerned that activated CD4+ T-cells support bone-resorbing osteoclasts15,70,71 and TH17 cells have pro-osteoclastic actions that drive alveolar bone loss72-74. Considering that SFB colonization increased activated CD4+ T-cells (Fig 3C,E) and TH17 cells (Fig 3F,G) in mandible bone marrow, the reduced alveolar bone phenotype found in SFB mice (Fig 2A-D) may be attributed to alterations in osteoclastogenesis.
SFB colonization does not alter osteoblastogenesis.
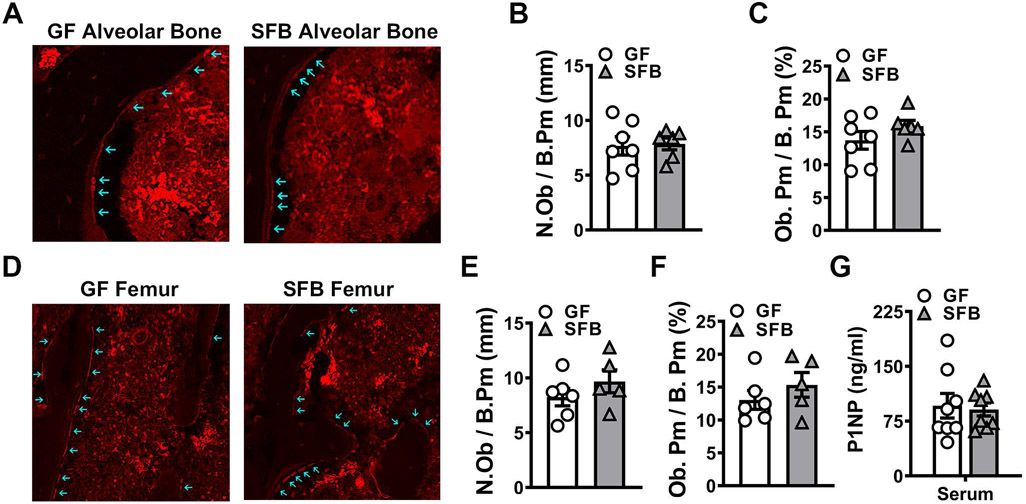
Skeletal bone loss occurs when the actions of bone resorbing osteoclasts exceed those of bone-forming osteoblasts 15,17. Osteoblastic cell alterations were evaluated by in situ immunofluorescence analysis of osterix+ bone-lining osteoblast cells in the maxillary alveolar bone (Fig 4A-C) and distal femur trabecular bone (Fig 4D-F). SFB colonization did not affect osteoblast cell numbers or interface with alveolar bone in the maxillary first molar furcation (Fig 4A-C) or the distal femur (Fig 4D-F). Serum P1NP, a marker for osteoblast-mediated bone formation, was not different in SFB vs. GF mice (Fig 4G), which suggests that SFB colonization does not alter osteoblast function across skeletal sites.
Figure 4. SFB colonization does not alter osteoblastogenesis.
A-C. Immunofluorescent analysis of osteoblasts in the maxillary first molar furcal alveolar bone (n=6-7/gp). Osterix+ cuboidal cells lining bone were designated as osteoblasts (red, osterix – Rhodamine). A. Representative images of osterix+ osteoblasts lining the maxillary first molar furcal alveolar bone; arrows indicate labeled osteoblasts (200X). B. N.Ob/B.Pm = osteoblast number per bone perimeter. C. Ob.Pm/B.Pm = osteoblast perimeter per bone perimeter. D-F. Immunofluorescent analysis of osteoblasts in the secondary spongiosa of distal femur (n=5-6/gp). Osterix+ cuboidal cells lining bone were designated as osteoblasts (red, osterix – Rhodamine). D. Representative images of osterix+ osteoblasts lining the secondary spongiosa trabecular bone of distal femur; arrows indicate labeled osteoblasts (200X). E. N.Ob/B.Pm. F. Ob.Pm/B.Pm. G. Serum ELISA analysis of P1NP levels (n=8/gp). Unpaired t-test; data are reported as mean ± SEM.
SFB colonization enhances osteoclastogenesis.
To elucidate whether the presence of SFB impacted osteoclastogenesis in the alveolar bone complex, histomorphometric analysis of TRAP+ osteoclastic cells lining alveolar bone was performed in the furcation of the maxillary first molar (Fig 5A-D). Whereas osteoclast numbers were similar (Fig 5A,B), the osteoclast size was larger (Fig 5A,C) in the alveolar bone of SFB vs. GF mice. The larger osteoclast size (Fig 5A,C) contributed to an increased osteoclast perimeter per bone perimeter (Fig 5A,D) at the alveolar bone surface of SFB mice. TRAP+ osteoclastic cell analysis in the femoral trabecular bone demonstrated that SFB actions promoting osteoclastogenesis are not specific to alveolar bone (Fig 5E-H). SFB mice had increased osteoclast numbers (Fig 5E,F) and a trend towards larger osteoclast size (Fig 5E,G), which contributed to an enhanced osteoclast perimeter per bone perimeter (Fig 5E,H), in the distal femur. Serum TRAP5B levels were higher in SFB vs. GF mice (Fig 5I), which supports that SFB colonization increases osteoclastogenesis across skeletal sites.
Figure 5. SFB colonization enhances osteoclastogenesis.
A-D. Maxillae harvested for histomorphometric analyses; sagittal sections cut and stained (n=7/gp). Histomorphometric analyses of TRAP+ osteoclast cellular endpoints were performed in the maxillary first molar furcal alveolar bone. TRAP+ cells lining bone with ≥3 nuclei were designated an osteoclast. A. Representative images of TRAP furcal alveolar bone (400X). B. N.Oc/B.Pm = osteoclast number per bone perimeter. C. Oc.Ar/Oc = average osteoclast area. D. Oc.Pm/B.Pm = osteoclast perimeter per bone perimeter. E-H. Femurs were harvested for histomorphometric analyses; sagittal sections cut and stained (n=5-6/gp). Histomorphometric analyses of TRAP+ osteoclast cellular endpoints were performed in the secondary spongiosa of the distal femur. TRAP+ cells lining bone with ≥3 nuclei were designated an osteoclast. E. Representative images of TRAP distal femur (200X). F. N.Oc/B.Pm. G. Oc.Ar/Oc. H. Oc.Pm/B.Pm. I. Serum ELISA analysis of TRAP5b levels (n=6-7/gp). J-M. Day-6 OCP culture TRAP stain assay (n=3/gp). J. Representative images (100X) of CD11b− OCP cultures stimulated with treatment (CSF1 & RANKL) media for 6 days. K. N.Oc/Well = number of osteoclasts per well. L. Oc.Ar/Oc = average osteoclast area. M. N.Nc/Oc = nuclei number per osteoclast. N. qRT-PCR gene expression studies were carried out in Day-4 CD11b− OCP cultures to detect early transcription level alterations in RANKL-stimulated osteoclast differentiation (n=3-4/gp). Relative quantification of Dcstamp mRNA was performed by the 2−ΔΔCT method; data expressed as treatment (CSF1 and RANKL) fold change relative to control (CSF1). O-Q. Mandible bone marrow (MBM) qRT-PCR analysis of (O) Tnfrsf11a (Rank) mRNA, (P) Tnfsf11(Rankl):Tnfrsf11b(Opg) ratio and (Q) Tnf mRNA (n=3-5/gp). Relative quantification of mRNA was performed via the 2−ΔΔCT method. Unpaired t-test; data are presented as mean ± SEM, *p<0.05, **p<0.01.
Osteoclast-precursor (OCP) cell in vitro differentiation assays were employed to further investigate SFB effects on osteoclastogenesis (Fig 5J-N). Magnetic cell sorting was used to enrich for marrow CD11b− HPCs, which are monocyte progenitor cells. CD11b− HPCs were primed in culture with CSF1 to drive the cells down the OCP lineage75,76. CD11b− OCP cultures were stimulated with control media (25ng/mL CSF1) or treatment media (25ng/mL CSF1 + 50ng/mL RANKL) for 4 or 6 days. Day-6 in the CD11b− OCP culture system is when terminal osteoclast maturation occurs44,55. Therefore, day-6 OCP cultures were TRAP stained for osteoclast cytomorphometric analysis (Fig 5J-M). CD11b− OCPs were isolated from day-4 OCP cultures for qRT-PCR analysis to target osteoclastic genes (Fig 5N) implicated in cell level alterations found in day-6 OCP cultures. While osteoclast numbers were not different (Fig 5J,K), osteoclast size (Fig 5J,L) and number of nuclei per osteoclast (Fig 5J,M) were increased in day-6 CD11b− OCP cultures from SFB vs. GF mice. Treatment over control analysis was carried out in RANKL+CSF1 treatment cultures relative to CSF1 control cultures to assess alterations in Dcstamp, a RANKL-induced transmembrane protein critical for osteoclast fusion77,78. RANKL treatment promoted a greater induction of Dcstamp in SFB vs. GF CD11b− OCP cultures (Fig 5N), which delineates that SFB immunomodulation supports RANKL-induced osteoclast fusion.
In vivo gene expression studies were performed in mandible bone marrow to delineate immunoregulatory mechanisms mediating SFB actions supporting osteoclastogenesis. Rank and the Rankl:Opg ratio were evaluated to assess alterations in the RANKL signaling axis (Fig 5O,P). When investigating alterations in RANKL signaling at the RANK receptor, the RANKL/OPG ratio must be evaluated to account for changes in the endogenous decoy receptor, osteoprotegerin (OPG)75,76. Rank (Tnfrsf11a) levels (Fig 5O) and the Rankl (Tnfsf11) / Opg (Tnfrsf11b) ratio (Fig 5P) were similar in the mandible bone marrow of GF and SFB mice. This data suggests that SFB’s pro-osteoclastic actions are not meditated by alterations in RANKL signaling at the RANK receptor. Appreciating that paracrine signaling effects in the bone marrow modulate RANKL-induced osteoclastogenesis, Tnf levels were assessed in the mandible bone marrow of GF and SFB mice. TNF is a proinflammatory cytokine that potently enhances RANKL-mediated osteoclastogenesis70,71,79. The rationale for assessing alterations in Tnf within the alveolar bone marrow was based on knowledge that SFB colonization upregulates TNF in the bone marrow at non-oral skeletal sites45. Tnf was greater than 3-fold higher in the mandible bone marrow of SFB vs. GF mice (Fig 5Q).
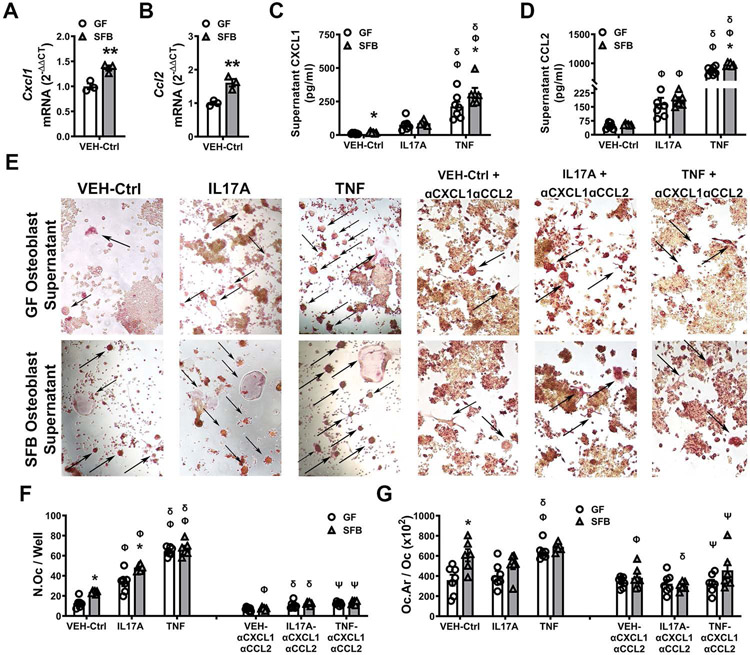
SFB induces osteoblast-derived signaling factors that support osteoclastogenesis.
Osteoimmunology research has discerned that IL17A- and TNF-expressing T-cells in the bone marrow are increased due to inflammatory conditions in the gut80 and the presence of SFB45. The current report has shown that SFB colonization in the gut upregulates circulating IL17A levels (Fig 1E) and increases the frequency of TH17 cells in alveolar bone marrow (Fig 3F,G). Furthermore, activated CD4+ T-cells (Fig 3C,E) and Tnf (Fig 5Q) were enhanced in the alveolar bone marrow of SFB vs. GF mice. IL17A and TNF signaling at osteoblastic cells promotes the synthesis of pro-osteoclastic chemokines/cytokines that support bone osteolysis48-53. Therefore, we established an in vitro osteoblast-osteoclast coupling assay targeting IL17A and TNF signaling effects on primary osteoblastic cells to elucidate whether SFB pro-osteoclastic actions are mediated through osteoblast-osteoclast crosstalk.
First passage confluent bone marrow mesenchymal stem cells (BMMSCs) from GF and SFB mice were cultured in osteogenic media to drive the cells down the osteoblastic lineage. Osteoblast cultures were then subjected to a single treatment with vehicle-control (VEH-Ctrl), IL17A, or TNF. Two hours later, the supernatants were aspirated, and cultures were washed to remove the recombinant cytokines. Fresh media was added back to the osteoblast cultures to allow for the secretion of pro-osteoclastic factors. Six hours later, the culture supernatants were collected for the in vitro osteoblast-osteoclast coupling assay and ELISA protein analysis. Osteoblast cells were isolated for qRT-PCR gene expression analysis.
To begin elucidating the molecular underpinnings of SFB actions on osteoblast-osteoclast crosstalk, qRT-PCR gene expression studies were performed in osteoblast cell lysates (Fig 6A,B; Supp Fig 2A-E). The pro-osteoclastic factors, CXCL1, CCL2, and CXCL5 were purposely targeted, since they are IL17A and TNF responsive genes in osteoblasts50,51,81,82. Cxcl1 (Fig 6A) and Ccl2 (Fig 6B) mRNA were increased, whereas there were no differences in Cxcl5 expression (Supp Fig 2A), in VEH-Ctrl-treated osteoblasts from SFB vs. GF mice. IL17A- and TNF-induced fold change increases in Cxcl1 (Supp Fig 2B), Ccl2 (Supp Fig 2C), and Cxcl5 (Supp Fig 2D) were consistent in osteoblasts derived from SFB vs. GF mice. qRT-PCR studies were also performed in SFB vs. GF osteoblast culture lysates, which ruled out differences in the Rankl:Opg axis (Supp Fig 2E).
Figure 6. SFB induces osteoblast-derived signaling factors that support osteoclastogenesis.
Osteoblast (OB) cultures were stimulated with vehicle-control (VEH-Ctrl), IL-17A, or TNF. OB cells were isolated for qRT-PCR gene expression analysis. OB supernatants were collected for ELISA protein analysis and the in vitro osteoblast-osteoclast coupling assay. A-B. RNA was isolated from VEH-Ctrl-stimulated OBs for qRT-PCR analysis of (A) Cxcl1 and (B) Ccl2 mRNA (n=3/gp); relative quantification of mRNA was performed via the 2−ΔΔCT method. Unpaired t-test; data are presented as mean ± SEM, **p<0.01. C-D. OB supernatant ELISA analysis of (C) CXCL1 and (D) CCL2 levels (n=6-7/gp). E-G. Osteoblast-osteoclast coupling assay. RANKL-induced RAW264.7 osteoclast cells were stimulated with supernatants from VEH-Ctrl-, IL17A-, or TNF-treated OBs +/− αCXCL1 and αCCL2 neutralizing antibodies on day-3. TRAP stain performed on day-5 of the culture system; TRAP+ cells with ≥ 3 nuclei were designated an osteoclast (n=6-7/gp). E. Representative images (200X) of TRAP+ RAW264.7 osteoclast cells stimulated with supernatants from VEH-Ctrl-, IL17A-, or TNF-treated OBs +/− αCXCL1 and αCCL2 neutralizing antibodies. F. N.Oc/Well = number of osteoclasts per well. G. Oc.Ar/Oc = average osteoclast area. Two-way ANOVA with Tukey post hoc test; data are presented as mean ± SEM. *p<0.05 between treatment-matched GF vs. SFB groups; Φp<0.05 vs. VEH-Ctrl (group-matched); δp<0.05 vs. IL17A treatment (group-matched); Ψp<0.05 vs. TNF treatment (group-matched).
Appreciating that protein levels of osteoblast-derived cytokines critically regulate osteoblast-osteoclast crosstalk, supernatants from osteoblast cultures treated with VEH-Ctrl, IL17A, and TNF were assessed for differences in CXCL1 (Fig 6C, Supp Table 1), CCL2 (Fig 6D, Supp Table 2), and CXCL5 (Supp Fig 2F). CXCL1 (Fig 6C) was increased in VEH-Ctrl-treated osteoblast culture supernatants from SFB vs. GF mice, while there no differences in unstimulated levels of CCL2 (Fig 6D) and CXCL5 (Supp Fig 2F). Compared to VEH-Ctrl treatment, IL17A treatment induced SFB and GF osteoblasts to secrete higher levels of CCL2 (Fig 6D). However, the upregulated CCL2 levels were similar in IL17A-treated osteoblast cultures from SFB vs. GF mice (Fig 6D). Relative to VEH-Ctrl or IL17A treatment, TNF treatment induced SFB and GF osteoblasts to secrete substantially higher levels of CXCL1 (Fig 6C) and CCL2 (Fig 6D). The increased levels of CXCL1 (Fig 6C) and CCL2 (Fig 6D) were greater in TNF-treated osteoblast cultures from SFB vs. GF mice. IL17A treatment and TNF treatment did not modulate CXCL5 levels in SFB or GF osteoblast cultures (Supp Fig 2F).
The osteoblast-osteoclast coupling assay was performed by stimulating RANKL-induced RAW264.7 osteoclastic cells with osteoblast culture supernatants from GF and SFB mice (Fig 6E-G; Supp Table 3,4). Based on our findings that secreted levels of CXCL1 (Fig 6C) and CCL2 (Fig 6D) were different in SFB vs. GF osteoblast culture supernatants, blocking antibodies were employed to elucidate the role of these factors in SFB immunomodulation of osteoblast-osteoclast crosstalk. Supernatants from VEH-Ctrl-treated SFB osteoblasts compared to VEH-Ctrl-treated GF osteoblasts induced the formation of larger numbers of RAW264.7 osteoclast cells (Fig 6F), which were greater in size (Fig 6G). The addition of αCXCL1 + αCCL2 neutralizing antibodies reduced the osteoclast numbers and size in cultures stimulated with VEH-Ctrl-treated SFB osteoblast supernatants, which normalized osteoclast outcomes driven by VEH-Ctrl-treated osteoblast supernatants (Fig 6F,G). These outcomes imply that CXCL1 and CCL2 support SFB actions promoting basal levels of osteoclastogenesis. Relative to supernatants from VEH-Ctrl-treated osteoblasts, supernatants from IL17A-treated osteoblasts increased osteoclast cell numbers, and this increase was greater in osteoblasts from SFB vs. GF mice (Fig 6F,G). The αCXCL1 + αCCL2 neutralizing antibodies attenuated the enhanced osteoclast cell numbers driven by the IL17A-treated osteoblast supernatants (Fig 6F,G). Supernatants from TNF-treated GF osteoblasts vs. VEH-Ctrl-treated and IL17A-treated GF osteoblasts induced greater numbers of osteoclastic cells (Fig 6F), which were larger in size (Fig 6G). Notably, osteoclast outcomes were similar in RAW264.7 osteoclast cells stimulated with supernatants from TNF-treated GF osteoblasts and TNF-treated SFB osteoblasts (Fig 6B-D). The presence of αCXCL1 + αCCL2 neutralizing antibodies blocked the increased osteoclast cell numbers and size driven by the TNF-treated GF osteoblast supernatants. These findings support the notion that TNF signaling at osteoblastic cells promotes SFB-induced pro-osteoclastic actions through the induction of CXCL1 and CCL2.
DISCUSSION
Four decades ago, researchers monoassociated GF rats with different commensal oral bacteria to show that commensal oral microbes critically regulate alveolar bone homeostasis18-20. Extensive subsequent research delineated that oral microbes stimulate immune responses in barrier gingival tissues, which lead to paracrine signaling effects that impact osteoimmune processes in the proximal alveolar bone17,56,83. The current report innovatively utilized a gnotobiotic murine model colonized by a commensal gut bacterium to show that commensal gut microbe effects on systemic immunity influence alveolar bone homeostasis.
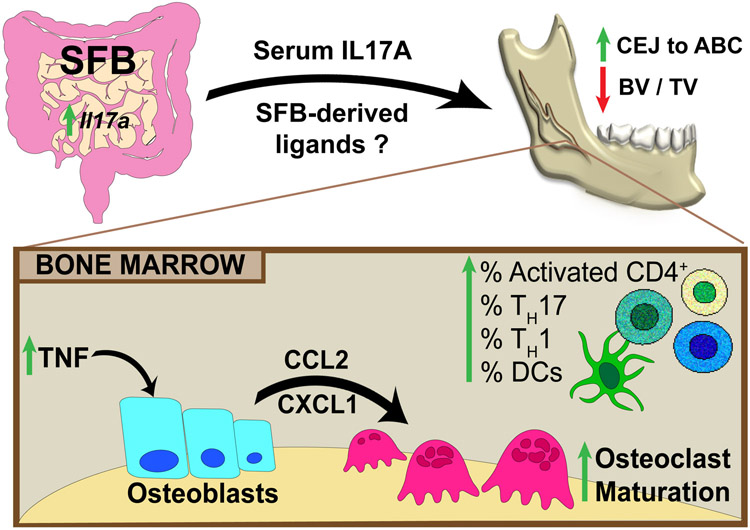
SFB was strategically chosen for the gnotobiotic murine model since the purpose was to associate mice with a commensal gut bacterium that does not colonize the oral cavity69. SFB was also selected based on the intent to utilize a commensal gut bacterium that modulates systemic immunity31-38. SFB regulation of systemic immunity has been shown to exacerbate inflammatory autoimmune conditions afflicting the central nervous system84, lungs85, and skeleton42,43. SFB driven TH17/IL17A immune responses have been linked to changes in innate immune cells residing in the bone marrow39,86, and SFB has been shown to expand marrow TH17 cells44,45. Osteoimmunology studies have recently delineated that SFB immunomodulatory actions impact the development44 and homeostasis45 of osseous tissues at non-oral skeletal sites. Herein, we employed the SFB-monoassociated mouse model to show that commensal gut microbes have the capacity to regulate osteoimmune processes and skeletal homeostasis in the alveolar bone complex (Fig 7).
Figure 7. Schematic of SFB immunomodulatory actions on skeletal health.
SFB colonization of the gut has immunomodulatory actions, which impairs trabecular bone microarchitecture at oral and non-oral skeletal sites. SFB elevates IL17A in the ileum and serum and potentially increases bacterial antigens/ligands in circulation, which have immunoregulatory effects on alveolar bone. Importantly, SFB increased TNF expression in the alveolar bone marrow microenvironment. TNF signaling at osteoblastic cells induced CXCL1 and CCL2 secretion which enhanced osteoclastogenesis. Therefore, commensal gut microbes have the capacity to regulate osteoimmune responses and skeletal homeostasis in alveolar bone.
SFB-monoassociated mice had a reduced skeletal phenotype in femur and alveolar bone. These findings show that commensal gut microbe effects on skeletal tissue homeostasis are not limited to non-oral sites, but also extend to the alveolar bone complex. Study outcomes demonstrating that SFB colonized and induced immune response effects in intestinal mucosal tissues, but not oral mucogingival tissues, supports the notion that commensal gut microbe effects on alveolar bone are mediated through alterations in systemic immunity. The presence of SFB in the gut promoted an enhanced osteoclast phenotype in alveolar bone, which appears to be secondary to the increased fusion of pre-osteoclast / osteoclast cells. Candidate immune mediators supporting SFB pro-osteoclastic effects were identified, including upregulated circulating IL17A levels and increased TH17 cells, activated CD4+ T-cells, and Tnf in alveolar bone marrow.
Inflammatory changes in the gut80 and the presence of SFB45 increase IL17A and TNF expressing T-cells in bone marrow that drive osteoclast mediated bone destruction at non-oral skeletal sites. IL17A and TNF signaling at osteoblastic cells upregulates the synthesis of pro-osteoclastic chemokines/cytokines that promotes bone osteolysis48-53. Therefore, we speculated that SFB immunoregulatory actions supporting osteoclastogenesis in alveolar bone are mediated by IL17A and TNF paracrine signaling effects at osteoblastic cells. The osteoblast-osteoclast coupling assay discerned that osteoblast-derived CXCL1 and CCL2 support SFB actions promoting osteoclastogenesis. Supernatants from TNF-treated osteoblasts normalized osteoclast outcomes in GF and SPF mice, and this effect was dependent on CXCL1 and CCL2.
CXCL1 and CCL2 are IL17A and TNF responsive genes in osteoblasts50,51,81,82. Mechanical and ligand stimulation of osteoblastic cells induces CXCL1- and CCL2-mediated paracrine signaling that supports monocyte recruitment and osteoclast fusion / maturation53,87-90. The commensal gut microbiota has been shown to elicit Ccl2 expression in BMMSCs91, and we demonstrated that a commensal gut microbe promotes Cxcl1 and Ccl2 in BMMSCs. SFB-derived antigens and ligands stimulate immunity locally in the gut38,47. However, it is unclear whether these microbial signaling mediators pass into the bloodstream to signal to BMMSC / osteoblastic cells in the bone marrow environment.
The current report provides initial evidence that commensal gut microbes regulate osteoimmune processes and skeletal homeostasis in the alveolar bone complex. Indirect support for the concept that the gut microbiota influences alveolar bone homeostasis is provided by clinical oral manifestations of inflammatory bowel diseases. Inflammatory bowel conditions are linked to dysbiotic shifts in the indigenous gut microbiota92,93. Therefore, progressive alveolar bone loss observed in patients afflicted by inflammatory bowel diseases94-96 may be related to changes in the gut microbiota. Further support for the concept that the gut microbiota affects alveolar bone homeostasis is lent by experimental periodontitis investigations. Probiotic bacteria strains naturally found in the gut have been shown to protect against periodontitis-driven alveolar bone loss17,97-100. Enteral administration of the probiotics via the oral route prevented alveolar bone destruction, which was associated with improved intestinal tissue morphology97, reduced intestinal inflammation98,99, and symbiotic effects on the gut microbiota99. Ongoing research is needed to discern how alterations in the commensal gut microbiota influence osteoimmune mechanisms regulating alveolar bone health and homeostasis.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. Subramanya Pandruvada and Dr. Hong Yu for sharing RAW264.7 cells for experimentation.
FUNDING STATEMENT
NIH/NIDCR K08DE025337, American Society for Bone and Mineral Research Rising Star Award, NIH/NIDCR T32DE017551. NIH/NIGMS P20GM130457, NIH/NIGMS P20GM121342, NIH/NIDDK P30DK123704, NIH/NIDCR R01DE029637, and NIH/NIDCR R01DE023783.
Abbreviations:
- GF
Germfree
- SFB
segmented filamentous bacteria
- CEJ
cementoenamel junction
- ABC
alveolar bone crest
- TRAP
tartrate-resistant acid phosphatase
- HPC
hematopoietic progenitor cell
- OCP
osteoclast-precursor cell
- BMMSC
bone marrow mesenchymal stem cell
- DC
dendritic cell
- VEH-Ctrl
vehicle-control
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
DATA AVAILABILITY
The datasets used and/or analyzed during the current study are available from the corresponding author upon request.
References
- 1.Microbial Ecology in States of Health and Disease: Workshop Summary. Forum on Microbial Health; Board on Global Health; Institute of Medicine., (National Academies Press (US): Washington (DC), 2014). [PubMed] [Google Scholar]
- 2.Belkaid Y & Harrison OJ Homeostatic Immunity and the Microbiota. Immunity 46, 562–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belkaid Y & Naik S Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol 14, 646–653 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanov II & Honda K Intestinal commensal microbes as immune modulators. Cell Host Microbe 12, 496–508 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thaiss CA, Zmora N, Levy M & Elinav E The microbiome and innate immunity. Nature 535, 65–74 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Brestoff JR & Artis D Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 14, 676–684 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blander JM, Longman RS, Iliev ID, Sonnenberg GF & Artis D Regulation of inflammation by microbiota interactions with the host. Nat Immunol 18, 851–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sommer F & Backhed F The gut microbiota--masters of host development and physiology. Nat Rev Microbiol 11, 227–238 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Lackey DE & Olefsky JM Regulation of metabolism by the innate immune system. Nat Rev Endocrinol 12, 15–28 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Clemente JC, Ursell LK, Parfrey LW & Knight R The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjogren K, Engdahl C, Henning P, Lerner UH, Tremaroli V, Lagerquist MK et al. The gut microbiota regulates bone mass in mice. J Bone Miner Res 27, 1357–1367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irie K, Novince CM & Darveau RP Impact of the Oral Commensal Flora on Alveolar Bone Homeostasis. J Dent Res 93, 801–806 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novince CM, Whittow CR, Aartun JD, Hathaway JD, Poulides N, Chavez MB et al. Commensal Gut Microbiota Immunomodulatory Actions in Bone Marrow and Liver have Catabolic Effects on Skeletal Homeostasis in Health. Sci Rep 7, 5747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li JY, Chassaing B, Tyagi AM, Vaccaro C, Luo T, Adams J et al. Sex steroid deficiency-associated bone loss is microbiota dependent and prevented by probiotics. J Clin Invest 126, 2049–2063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh MC, Takegahara N, Kim H & Choi Y Updating osteoimmunology: regulation of bone cells by innate and adaptive immunity. Nat Rev Rheumatol 14, 146–156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novince CM & Kirkwood KL Alveolar Bone Homeostasis in Health and Disease. In: Bilezikian JP (ed). Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism Ch. 121, 933–940 (Wiley-Blackwell: Ames, Iowa, 2019). [Google Scholar]
- 17.Hathaway-Schrader JD & Novince CM Maintaining homeostatic control of periodontal bone tissue. Periodontol 2000 86, 157–187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garant PR Light and electron microscopic observations of osteoclastic alveolar bone resorption in rats monoinfected with Actinomyces naeslundii. J Periodontol 47, 717–723 (1976). [DOI] [PubMed] [Google Scholar]
- 19.Behling UH, Sallay C, Sanavi F, Pham PH & Nowotny A Humoral immunity and reduced periodontal bone loss in Eikenella corrodens-monoassociated rats. Infect Immun 33, 801–805 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burckhardt JJ, Gaegauf-Zollinger R, Schmid R & Guggenheim B Alveolar bone loss in rats after immunization with Actinomyces viscosus. Infect Immun 31, 971–977 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaasen HL, Koopman JP, Poelma FG & Beynen AC Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev 8, 165–180 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG & Beynen AC Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab Anim 27, 141–150 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Snel J, Heinen PP, Blok HJ, Carman RJ, Duncan AJ, Allen PC et al. Comparison of 16S rRNA sequences of segmented filamentous bacteria isolated from mice, rats, and chickens and proposal of "Candidatus Arthromitus". Int J Syst Bacteriol 45, 780–782 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Yin Y, Wang Y, Zhu L, Liu W, Liao N, Jiang M et al. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J 7, 615–621 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen B, Chen H, Shu X, Yin Y, Li J, Qin J et al. Presence of Segmented Filamentous Bacteria in Human Children and Its Potential Role in the Modulation of Human Gut Immunity. Front Microbiol 9, 1403 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Wang L, Wang X, Wang X, Liu H & Yin Y Distribution and Strain Diversity of Immunoregulating Segmented Filamentous Bacteria in Human Intestinal Lavage Samples. Microb Ecol 79, 1021–1033 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Klaasen HLK, J.P.; Beynen AC Effects of age, strain and social hierarchy on colonization of autochthonous, segmented, filamentous bacteria in the ileum of mice. . Microecology and Therapy 20, 17–20 (1990b). [Google Scholar]
- 28.Snel J, Hermsen CC, Smits HJ, Bos NA, Eling WM, Cebra JJ et al. Interactions between gut-associated lymphoid tissue and colonization levels of indigenous, segmented, filamentous bacteria in the small intestine of mice. Can J Microbiol 44, 1177–1182 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Talham GL, Jiang HQ, Bos NA & Cebra JJ Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immun 67, 1992–2000 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang HQ, Bos NA & Cebra JJ Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect Immun 69, 3611–3617 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 40, 594–607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panea C, Farkas AM, Goto Y, Abdollahi-Roodsaz S, Lee C, Koscso B et al. Intestinal Monocyte-Derived Macrophages Control Commensal-Specific Th17 Responses. Cell Rep 12, 1314–1324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flannigan KL, Ngo VL, Geem D, Harusato A, Hirota SA, Parkos CA et al. IL-17A-mediated neutrophil recruitment limits expansion of segmented filamentous bacteria. Mucosal Immunol 10, 673–684 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ladinsky MS, Araujo LP, Zhang X, Veltri J, Galan-Diez M, Soualhi S et al. Endocytosis of commensal antigens by intestinal epithelial cells regulates mucosal T cell homeostasis. Science 363 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgess SL, Buonomo E, Carey M, Cowardin C, Naylor C, Noor Z et al. Bone marrow dendritic cells from mice with an altered microbiota provide interleukin 17A-dependent protection against Entamoeba histolytica colitis. MBio 5, e01817 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Z, Zou J, Zhang Z, Zhao X, Noriega J, Zhang B et al. Segmented Filamentous Bacteria Prevent and Cure Rotavirus Infection. Cell 179, 644–658.e613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gauguet S, D'Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R et al. Intestinal Microbiota of Mice Influences Resistance to Staphylococcus aureus Pneumonia. Infect Immun 83, 4003–4014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chappert P, Bouladoux N, Naik S & Schwartz RH Specific gut commensal flora locally alters T cell tuning to endogenous ligands. Immunity 38, 1198–1210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hathaway-Schrader JD, Poulides NA, Carson MD, Kirkpatrick JE, Warner AJ, Swanson BA et al. Specific Commensal Bacterium Critically Regulates Gut Microbiota Osteoimmunomodulatory Actions During Normal Postpubertal Skeletal Growth and Maturation. JBMR Plus 4, e10338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu M, Malik Tyagi A, Li JY, Adams J, Denning TL, Weitzmann MN et al. PTH induces bone loss via microbial-dependent expansion of intestinal TNF(+) T cells and Th17 cells. Nat Commun 11, 468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schnupf P, Gaboriau-Routhiau V, Gros M, Friedman R, Moya-Nilges M, Nigro G et al. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520, 99–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Yin Y, Chen X, Zhao Y, Wu Y, Li Y et al. Induction of Intestinal Th17 Cells by Flagellins From Segmented Filamentous Bacteria. Front Immunol 10, 2750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson BM, Mundy GR & Chambers TJ Tumor necrosis factors alpha and beta induce osteoblastic cells to stimulate osteoclastic bone resorption. J Immunol 138, 775–779 (1987). [PubMed] [Google Scholar]
- 49.Kotake S, Udagawa N, Takahashi N, Matsuzaki K, Itoh K, Ishiyama S et al. IL-17 in synovial fluids from patients with rheumatoid arthritis is a potent stimulator of osteoclastogenesis. J Clin Invest 103, 1345–1352 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruddy MJ, Shen F, Smith JB, Sharma A & Gaffen SL Interleukin-17 regulates expression of the CXC chemokine LIX/CXCL5 in osteoblasts: implications for inflammation and neutrophil recruitment. J Leukoc Biol 76, 135–144 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Shen F, Ruddy MJ, Plamondon P & Gaffen SL Cytokines link osteoblasts and inflammation: microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J Leukoc Biol 77, 388–399 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Souza PP & Lerner UH The role of cytokines in inflammatory bone loss. Immunol Invest 42, 555–622 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Brylka LJ & Schinke T Chemokines in Physiological and Pathological Bone Remodeling. Front Immunol 10, 2182 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ & Muller R Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 25, 1468–1486 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Hathaway-Schrader JD, Steinkamp HM, Chavez MB, Poulides NA, Kirkpatrick JE, Chew ME et al. Antibiotic Perturbation of Gut Microbiota Dysregulates Osteoimmune Cross Talk in Postpubertal Skeletal Development. Am J Pathol 189, 370–390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kinane DF, Stathopoulou PG & Papapanou PN Periodontal diseases. Nat Rev Dis Primers 3, 17038 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 28, 2–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schnupf P, Gaboriau-Routhiau V & Cerf-Bensussan N Host interactions with Segmented Filamentous Bacteria: an unusual trade-off that drives the post-natal maturation of the gut immune system. Semin Immunol 25, 342–351 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Farkas AM, Panea C, Goto Y, Nakato G, Galan-Diez M, Narushima S et al. Induction of Th17 cells by segmented filamentous bacteria in the murine intestine. J Immunol Methods 421, 104–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bacchetti De Gregoris T, Aldred N, Clare AS & Burgess JG Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods 86, 351–356 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Gaboriau-Routhiau V, Lecuyer E & Cerf-Bensussan N Role of microbiota in postnatal maturation of intestinal T-cell responses. Curr Opin Gastroenterol 27, 502–508 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Livak KJ & Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Schmittgen TD & Livak KJ Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Umesaki Y, Okada Y, Matsumoto S, Imaoka A & Setoyama H Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiol Immunol 39, 555–562 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Page RC & Schroeder HE Periodontitis in man and other animals: a comparative review. (Karger: New York, 1982). [Google Scholar]
- 66.Tucker A & Sharpe P The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet 5, 499–508 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Treuting PM, Morton TH & Vogel P 7 - Oral Cavity and Teeth. In: Treuting Piper M., Dintzis Suzanne M., & Montine Kathleen S. (eds). Comparative Anatomy and Histology (Second Edition) 115–133 (Academic Press: San Diego, 2018). [Google Scholar]
- 68.Cho MI & Garant PR Development and general structure of the periodontium. Periodontol 2000 24, 9–27 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Dutzan N, Abusleme L, Bridgeman H, Greenwell-Wild T, Zangerle-Murray T, Fife ME et al. On-going Mechanical Damage from Mastication Drives Homeostatic Th17 Cell Responses at the Oral Barrier. Immunity 46, 133–147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weitzmann MN & Ofotokun I Physiological and pathophysiological bone turnover - role of the immune system. Nat Rev Endocrinol 12, 518–532 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Redlich K & Smolen JS Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat Rev Drug Discov 11, 234–250 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Monasterio G, Fernandez B, Castillo F, Rojas C, Cafferata EA, Rojas L et al. Capsular-defective Porphyromonas gingivalis mutant strains induce less alveolar bone resorption than W50 wild-type strain due to a decreased Th1/Th17 immune response and less osteoclast activity. J Periodontol 90, 522–534 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Dutzan N, Kajikawa T, Abusleme L, Greenwell-Wild T, Zuazo CE, Ikeuchi T et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci Transl Med 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsukasaki M, Komatsu N, Nagashima K, Nitta T, Pluemsakunthai W, Shukunami C et al. Host defense against oral microbiota by bone-damaging T cells. Nat Commun 9, 701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin TJ & Sims NA RANKL/OPG; Critical role in bone physiology. Rev Endocr Metab Disord 16, 131–139 (2015). [DOI] [PubMed] [Google Scholar]
- 76.Boyce BF & Xing L Functions of RANKL/RANK/OPG in bone modeling and remodeling. Arch Biochem Biophys 473, 139–146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kukita T, Wada N, Kukita A, Kakimoto T, Sandra F, Toh K et al. RANKL-induced DC-STAMP is essential for osteoclastogenesis. J Exp Med 200, 941–946 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N et al. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med 202, 345–351 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mbalaviele G, Novack DV, Schett G & Teitelbaum SL Inflammatory osteolysis: a conspiracy against bone. J Clin Invest 127, 2030–2039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ciucci T, Ibanez L, Boucoiran A, Birgy-Barelli E, Pene J, Abou-Ezzi G et al. Bone marrow Th17 TNFalpha cells induce osteoclast differentiation, and link bone destruction to IBD. Gut 64, 1072–1081 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Graves DT, Jiang Y & Valente AJ The expression of monocyte chemoattractant protein-1 and other chemokines by osteoblasts. Front Biosci 4, D571–580 (1999). [DOI] [PubMed] [Google Scholar]
- 82.Di Benedetto G, Lempereur L, Valle D, Greco EA, Bernardini R, Lenzi A et al. Redundant modulatory effects of proinflammatory cytokines in human osteoblastic cells in vitro. Clin Exp Rheumatol 36, 959–969 (2018). [PubMed] [Google Scholar]
- 83.Curtis MA, Zenobia C & Darveau RP The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe 10, 302–306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee YK, Menezes JS, Umesaki Y & Mazmanian SK Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 108 Suppl 1, 4615–4622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bradley CP, Teng F, Felix KM, Sano T, Naskar D, Block KE et al. Segmented Filamentous Bacteria Provoke Lung Autoimmunity by Inducing Gut-Lung Axis Th17 Cells Expressing Dual TCRs. Cell Host Microbe 22, 697–704.e694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu C, Lee SK, Zhang D & Frenette PS The Gut Microbiome Regulates Psychological-Stress-Induced Inflammation. Immunity (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rahimi P, Wang CY, Stashenko P, Lee SK, Lorenzo JA & Graves DT Monocyte chemoattractant protein-1 expression and monocyte recruitment in osseous inflammation in the mouse. Endocrinology 136, 2752–2759 (1995). [DOI] [PubMed] [Google Scholar]
- 88.Li X, Qin L, Bergenstock M, Bevelock LM, Novack DV & Partridge NC Parathyroid hormone stimulates osteoblastic expression of MCP-1 to recruit and increase the fusion of pre/osteoclasts. J Biol Chem 282, 33098–33106 (2007). [DOI] [PubMed] [Google Scholar]
- 89.Cambré I, Gaublomme D, Burssens A, Jacques P, Schryvers N, De Muynck A et al. Mechanical strain determines the site-specific localization of inflammation and tissue damage in arthritis. Nat Commun 9, 4613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Onan D, Allan EH, Quinn JM, Gooi JH, Pompolo S, Sims NA et al. The chemokine Cxcl1 is a novel target gene of parathyroid hormone (PTH)/PTH-related protein in committed osteoblasts. Endocrinology 150, 2244–2253 (2009). [DOI] [PubMed] [Google Scholar]
- 91.Xiao E, He L, Wu Q, Li J, He Y, Zhao L et al. Microbiota regulates bone marrow mesenchymal stem cell lineage differentiation and immunomodulation. Stem Cell Res Ther 8, 213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sommer F, Anderson JM, Bharti R, Raes J & Rosenstiel P The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol 15, 630–638 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y & Andoh A Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol 11, 1–10 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Lamster I, Sonis S, Hannigan A & Kolodkin A An association between Crohn's disease, periodontal disease and enhanced neutrophil function. J Periodontol 49, 475–479 (1978). [DOI] [PubMed] [Google Scholar]
- 95.Ozmeric N, Bissada N & da Silva APB The Association between Inflammatory Bowel Disease and Periodontal Conditions: Is There a Common Bacterial Etiology? J Int Acad Periodontol 20, 40–51 (2018). [PubMed] [Google Scholar]
- 96.She YY, Kong XB, Ge YP, Liu ZY, Chen JY, Jiang JW et al. Periodontitis and inflammatory bowel disease: a meta-analysis. BMC Oral Health 20, 67 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Messora MR, Oliveira LF, Foureaux RC, Taba M Jr., Zangeronimo MG, Furlaneto FA et al. Probiotic therapy reduces periodontal tissue destruction and improves the intestinal morphology in rats with ligature-induced periodontitis. J Periodontol 84, 1818–1826 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Kobayashi R, Kobayashi T, Sakai F, Hosoya T, Yamamoto M & Kurita-Ochiai T Oral administration of Lactobacillus gasseri SBT2055 is effective in preventing Porphyromonas gingivalis-accelerated periodontal disease. Sci Rep 7, 545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gatej SM, Bright R, Weyrich LS, Marino V, Christophersen CT, Gibson RJ et al. Probiotic Lactobacillus Rhamnosus GG Protects Against P. Gingivalis And F. Nucleatum Gut Dysbiosis. J Int Acad Periodontol 22, 18–27 (2020). [PubMed] [Google Scholar]
- 100.Gatej S, Gully N, Gibson R & Bartold PM Probiotics and Periodontitis - A Literature Review. J Int Acad Periodontol 19, 42–50 (2017). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon request.