Abstract
We have examined the antipneumococcal mechanisms of a series of novel fluoroquinolones that are identical to ciprofloxacin except for the addition of a benzenesulfonylamido group to the C-7 piperazinyl ring. A number of these derivatives displayed enhanced activity against Streptococcus pneumoniae strain 7785, including compound NSFQ-105, bearing a 4-(4-aminophenylsulfonyl)-1-piperazinyl group at C-7, which exhibited an MIC of 0.06 to 0.125 μg/ml compared with a ciprofloxacin MIC of 1 μg/ml. Several complementary approaches established that unlike the case for ciprofloxacin (which targets topoisomerase IV), the increased potency of NSFQ-105 was associated with a target preference for gyrase: (i) parC mutants of strain 7785 that were resistant to ciprofloxacin remained susceptible to NSFQ-105, whereas by contrast, mutants bearing a quinolone resistance mutation in gyrA were four- to eightfold more resistant to NSFQ-105 (MIC of 0.5 μg/ml) but susceptible to ciprofloxacin; (ii) NSFQ-105 selected first-step gyrA mutants (MICs of 0.5 μg/ml) encoding Ser-81-to-Phe or -Tyr mutations, whereas ciprofloxacin selects parC mutants; and (iii) NSFQ-105 was at least eightfold more effective than ciprofloxacin at inhibiting DNA supercoiling by S. pneumoniae gyrase in vitro but was fourfold less active against topoisomerase IV. These data show unequivocally that the C-7 substituent determines not only the potency but also the target preference of fluoroquinolones. The importance of the C-7 substituent in drug-enzyme contacts demonstrated here supports one key postulate of the Shen model of quinolone action.
The renewed interest in antibacterial fluoroquinolones derives from the recent development of agents active against gram-positive pathogens, particularly Streptococcus pneumoniae, the main cause of community-acquired pneumonia (4, 7, 29). Several new fluoroquinolones, e.g., clinafloxacin, gatifloxacin, gemifloxacin, and moxifloxacin, which are more potent in vitro than ciprofloxacin or levofloxacin, are at various stages of clinical development. With the likely increase in clinical use of antipneumococcal fluoroquinolones, attention has concentrated on their mechanisms of action and resistance in S. pneumoniae.
Genetic studies both with Escherichia coli and with gram-positive pathogens have shown that fluoroquinolones act through inhibition of the type II topoisomerases DNA gyrase and topoisomerase IV (12, 13, 17). Both enzymes function by making a transient double-stranded DNA break and passing through a second DNA duplex in an ATP-dependent reaction (19, 33). This topological maneuver allows DNA supercoiling by gyrase and decatenation of daughter chromosomes by topoisomerase IV, two essential activities in chromosomal DNA replication and segregation (1, 36). The DNA breakage-reunion and ATPase activities of these tetrameric enzyme complexes are provided by the respective GyrA and GyrB subunits of gyrase, encoded by the gyrA and gyrB genes; the ParC and ParE proteins (coded by parC and parE) fulfill similar functions in topoisomerase IV (17, 33). Fluoroquinolones interfere with enzymatic DNA resealing, leading to the generation of a double-stranded DNA break which is thought to be the cytotoxic lesion in vivo (12). Mutations giving rise to bacterial resistance to quinolones occur in limited areas of the GyrA/ParC and GyrB/ParE proteins termed the quinolone resistance-determining regions (QRDRs) (6, 21, 34, 35). In E. coli, quinolone resistance mutations arise first in gyrA or gyrB, suggesting that gyrase is the primary drug target (6, 34, 35). By contrast, in Staphylococcus aureus, parC or parE mutations are seen first, indicating that the primary target is topoisomerase IV (9, 10, 21, 22). Analysis of the QRDRs of quinolone-resistant S. pneumoniae mutants arising from stepwise drug challenge indicates that, depending on the drug used, either topoisomerase IV or gyrase mutations are selected (11, 25). Thus, one group of quinolones, whose prototype is ciprofloxacin, selects for parC or parE mutations, suggesting that these drugs act preferentially through topoisomerase IV in vivo (15, 16, 20, 23, 28, 32), whereas a second group, typified by sparfloxacin, selects gyrA mutants in the first step, consistent with DNA gyrase being the primary target in vivo (11, 25). Clinafloxacin selects gyrA mutants but appears to act through both enzymes (26).
Although several potent antipneumococcal fluoroquinolones belong to the DNA gyrase-targeting group in vivo, their structural dissimilarities make it difficult to discern the molecular determinant that governs target selection. However, we noted that novel benzenesulfonamide fluoroquinolones differing from ciprofloxacin or norfloxacin solely by the linkage of various benzenesulfonylamido groups to the 1-piperazinyl residue at C-7 of the parent drug were reported to have particularly high in vitro activity against gram-positive cocci such as S. aureus (2). Were these agents to target gyrase rather than topoisomerase IV, then it would be possible to establish an unequivocal structure-function relationship with important implications for drug design and clinical use. In this paper, we have examined the antibacterial mechanisms of benzenesulfonamide quinolones and their parent compounds by testing their activities against S. pneumoniae and its gyrA and parC mutants, by analyzing sulfanilyl fluoroquinolone-resistant mutants selected by stepwise challenge, and by determining the inhibitory activities of the various agents against purified S. pneumoniae gyrase and topoisomerase IV in vitro.
MATERIALS AND METHODS
Bacterial strains.
S. pneumoniae 7785, a quinolone-susceptible clinical isolate, and its quinolone-resistant isolates 1C1, 2C6, 2C7, 3C4, 1S1, 1S4, 2S1, and 2S4 have been described previously (23, 25).
Drugs and drug susceptibilities.
Ciprofloxacin hydrochloride and norfloxacin were provided by Bayer U.K., Newbury, United Kingdom, and Glaxo Wellcome, Stevenage, United Kingdom. NSFQ-105 and other benzenesulfonamide fluoroquinolones were synthesized as described previously (18; R. H. Manzo, M. R. Mazzieri, M. J. Nieto, and F. L. Alovero, 21 February 1997, Argentine patent application P970106669). MICs of S. pneumoniae strains were determined by two-fold dilution as described previously (23, 25, 26). Approximately 105 CFU of bacteria was spotted on the plates, which were examined after overnight aerobic incubation at 37°C.
Stepwise selection of NSFQ-105-resistant S. pneumoniae strains.
Mutants were selected by plating approximately 1010 CFU of strain 7785 or its drug-resistant derivatives on brain heart infusion plates containing 10% horse blood and NSFQ-105. Plates were incubated aerobically at 37°C for 24 to 48 h. Mutant frequencies were determined as described previously (23, 25).
PCR and restriction fragment length polymorphism analysis.
The conditions for bacterial growth and isolation of chromosomal DNA were as described previously (24). PCR was used to amplify DNA from the QRDRs of the gyrase and topoisomerase IV genes of NSFQ-105-resistant S. pneumoniae mutants. PCR primers and conditions have been reported previously, as has the restriction fragment length polymorphism analysis by HinfI digestion of PCR products (23–26).
DNA sequence analysis.
Generation of single-stranded S. pneumoniae QRDR DNA products by asymmetric PCR and their direct DNA sequencing by the chain termination method have been described previously (25, 30).
Inhibition of DNA gyrase and topoisomerase IV.
The purification of recombinant S. pneumoniae gyrase and topoisomerase IV proteins has been reported earlier, as have assay conditions for inhibition of DNA supercoiling by gyrase and kinetoplast DNA (kDNA) decatenation by topoisomerase IV (27).
RESULTS
Benzenesulfonamide derivatives of ciprofloxacin display enhanced activity against S. pneumoniae: evidence for an altered target preference in vivo.
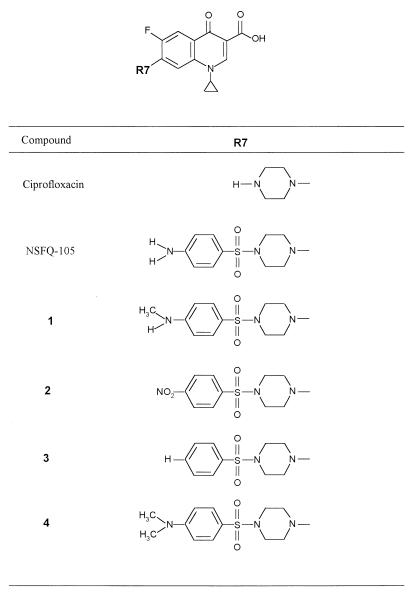
We have studied a series of benzenesulfonamide fluoroquinolones in which the 1-piperazinyl group of ciprofloxacin was replaced with various 4-(arylsulfonyl)-1-piperazinyl moieties, yielding NSFQ-105 and compounds 1 to 4 (Fig. 1) (2, 3). Except for the alteration at C-7, these compounds are structurally identical to ciprofloxacin. The activities of the different benzenesulfonamide fluoroquinolones against quinolone-susceptible S. pneumoniae isolate 7785 and its mutants carrying quinolone resistance mutations in parC, gyrA, and both gyrA and parC were determined (Table 1). As has been reported previously, the ciprofloxacin MIC for 7785 was 1 μg/ml, which is typical for wild-type S. pneumoniae (23). Interestingly, NSFQ-105, bearing a 4-(4-aminophenylsulfonyl)-1-piperazinyl group at the C-7 position, and its 4-aminomethylphenylsulfonyl and 4-nitrophenylsulfonyl homologues (compounds 1 and 2) were more active against strain 7785 than ciprofloxacin, with MICs of 0.06 to 0.125, 0.12, and 0.5 μg/ml, respectively (Table 1). Compounds 3 and 4, bearing 4-benzenesulfonyl or 4-dimethylaminobenzenesulfonyl additions to the piperazine moiety at C-7, were two- to fourfold less active against 7785 than ciprofloxacin. These results establish that modification of the C-7 group of ciprofloxacin alters antibacterial potency, with NSFQ-105 exhibiting an 8- to 16-fold increase in activity against S. pneumoniae 7785. Indeed, the NSFQ-105 MIC against strain 7785 compares favorably with MICs of 0.25 and 0.125 μg/ml determined for sparfloxacin and clinafloxacin, respectively (25, 26).
FIG. 1.
Structures of fluoroquinolones used in this study.
TABLE 1.
Fluoroquinolone susceptibilities of S. pneumoniae mutants
| Strain | Mutation in QRDR of:
|
MIC (μg/ml) ofa:
|
||||||
|---|---|---|---|---|---|---|---|---|
| GyrA | ParC | CIP | NSFQ-105 | 1 | 2 | 3 | 4 | |
| 7785 | None | None | 1 | 0.06–0.125 | 0.12 | 0.5 | 4 | 2 |
| 2C6 | None | Ser-79→Tyr | 8 | 0.06–0.125 | 0.12 | 1 | 4 | 2 |
| 2C7 | None | Ser-79→Phe | 8 | 0.06–0.125 | 0.12 | 1 | 4 | 2 |
| 1S1 | Ser-81→Phe | None | 1–2 | 0.5 | 1 | 4 | >8 | >16 |
| 1S4 | Ser-81→Tyr | None | 1–2 | 0.5 | 0.5 | 2 | >8 | 16 |
| 3C4 | Ser-81→Tyr | Ser-79→Tyr | 64 | 1–2 | 2 | >4 | >8 | >16 |
| 2S1 | Ser-81→Phe | Ser-79→Tyr | 64 | 1–2 | 1–2 | >4 | >8 | >16 |
| 2S4 | Ser-81→Phe | Asp-83→Asn | 16 | 1 | 1–2 | >4 | >8 | >16 |
CIP, ciprofloxacin; NSFQ-105, 4-(4-aminophenylsulfonyl)-1-piperazinyl ciprofloxacin; 1 to 4, compounds 1 to 4, respectively.
Analysis of the responses of S. pneumoniae mutants bearing defined mutations in the gyrase or topoisomerase IV QRDR regions can provide useful information on the target specificity of the quinolone. Table 1 describes defined S. pneumoniae strains (originally derived by single-step or two-step drug selection) (26) and their drug responses. The data for ciprofloxacin are consistent with the view that topoisomerase IV is its primary target and gyrase is a secondary target in S. pneumoniae. Thus, the presence of a gyrA mutation changing Ser-81 in strains 1S1 and 1S4 did not significantly affect the ciprofloxacin MIC, whereas a parC mutation altering Ser-79 in mutants 2C6 and 2C7 increased the MIC to 8 μg/ml. Strains 3C4, 2S1, and 2S4, with mutations in both gyrA and parC, were highly resistant, with ciprofloxacin MICs of 16 to 64 μg/ml (Table 1). By contrast, the data for NSFQ-105 and compounds 1 to 4 suggest that in each case their primary in vivo target is gyrase and not topoisomerase IV. Thus, unlike the case for the response to ciprofloxacin, the presence of a parC mutation in strains 2C6 and 2C7 did not significantly affect the MIC of any of the benzenesulfonamide fluoroquinolones (Table 1). However, for each derivative, a gyrA mutation in strains 1S1 and 1S4 produced a four- to eightfold elevation in the MIC, indicating that modification at C-7 had switched the target preference of ciprofloxacin from topoisomerase IV to gyrase.
To confirm that a gyrA QRDR mutation alone is sufficient to confer resistance to benzenesulfonamide fluoroquinolones, we determined the complete nucleotide sequences of the parC and parE genes of strain 1S1 (X.-S. Pan and L. M. Fisher, unpublished work). The sequences were identical to those for the wild-type parent strain 7785. Therefore, changes in topoisomerase IV can be excluded in explaining the decreased susceptibility of 1S1 to benzenesulfonamide fluoroquinolones (and other fluoroquinolones examined previously, e.g., sparfloxacin [25]); this must accrue from the gyrA (Ser-81→Phe) change, a known quinolone resistance mutation. These studies reinforce the suggestion that gyrase, and not topoisomerase IV, is the target of NSFQ-105 and compounds 1 to 4.
NSFQ-105 selects gyrA mutants of S. pneumoniae.
In previous work, we have shown that stepwise challenge of S. pneumoniae 7785 with ciprofloxacin selects for quinolone resistance mutations in the parC QRDR before those in gyrA (23). This observation complements studies of defined mutants in suggesting that ciprofloxacin acts through topoisomerase IV in vivo. To examine the situation for the benzenesulfonamide fluoroquinolones, we chose to generate and characterize stepwise-selected mutants of S. pneumoniae 7785 by using NSFQ-105, the most potent of the derivatives (Table 1). Approximately 1010 CFU of strain 7785 was challenged with NSFQ-105 at 0.125 μg/ml, i.e., one to two times the MIC. After 24 to 48 h of growth on plates, mutants appeared at a frequency of 10−6, consistent with selection of strains carrying a single mutation; mutants with double mutations would be expected at a frequency of 10−14 to 10−16. Eight of these first-step mutants were characterized further in terms of drug susceptibility and the presence of resistance mutations. The NSFQ-105 MIC for all eight mutants was 0.5 μg/ml, and there was a <2-fold change in ciprofloxacin MIC. The NSFQ-105 MIC for the mutants was the same as that for gyrA strain 1S1 (Table 1), which lacks topoisomerase IV mutations. The result suggested that a single gyrA change could be responsible for the resistance of first-step mutants. PCR was employed to amplify the gyrase and topoisomerase IV QRDR regions of the mutants. The 382-bp gyrA QRDR and 366-bp parC QRDR PCR products were isolated using genomic DNAs from the eight first-step mutants as templates. In all eight strains, the gyrA PCR product did not undergo cleavage with HinfI at an internal site overlapping codon 81 (codon 83 in E. coli gyrA), indicating that this codon carried a mutation. By contrast, all eight parC products gave the wild-type HinfI digestion pattern yielding 183-, 127-, and 56-bp fragments and suggesting the absence of a parC mutation affecting hot spot Ser-79. DNA sequence analysis of PCR products from three mutants, NSF12, NSF18, and NSF22, confirmed the presence of mutations in gyrA leading to alteration of Ser-81 to Phe or Tyr at the protein level, alterations known to confer resistance to quinolones in transformation studies (Table 2). The parC, gyrB, and parE QRDR sequences in each of the strains were wild type. Thus, NSFQ-105 selects gyrA mutants in the first step.
TABLE 2.
Characterization of S. pneumoniae mutants selected stepwise for resistance to the sulfanilyl fluoroquinolone NSFQ-105
| Strain | MIC (μg/ml) ofa:
|
Mutation in QRDR ofb:
|
||||
|---|---|---|---|---|---|---|
| CIP | NSFQ-105 | GyrA | ParC | GyrB | ParE | |
| 7785 | 1–2 | 0.06–0.125 | ||||
| NSF12c | 2 | 0.5 | Ser-81→Tyr | None | None | None |
| NSF18c | 2 | 0.5 | Ser-81→Tyr | None | None | None |
| NSF22c | 2 | 0.5 | Ser-81→Phe | None | None | None |
CIP, ciprofloxacin; NSFQ-105, 4-(4-aminophenylsulfonyl)-1-piperazinyl derivative of CIP.
GyrA mutations resulted from the following nucleotide changes: Ser-81→Phe, TCC to TTC; Ser-80→Tyr, TCC to TAC.
Derived from strain 7785 by selection with NSFQ-105 at 0.125 μg/ml.
Second-step mutants were selected at a frequency of ∼10−5 from NSF18 and NSF22 by challenge with NSFQ-105 at 0.5 μg/ml. (No mutants were recovered using NSFQ-105 at 1 μg/ml.) Mutants NSF18.11, NSF18.12, NSF18.13, NSF22.1, and NSF22.4 were chosen for study; the NSFQ-105 MICs for these mutants were 2 to 4, 2, 1, 2 to 4, and 1 to 2 μg/ml, respectively, and ciprofloxacin MICs were 4, 4, 2 to 4, 2, and 4 μg/ml, respectively. By DNA sequence analysis, none of these second-step mutants had acquired additional mutations in the gyrA, parC, gyrB, and parE QRDRs (data not shown). Although it is possible that the second-step mutants have topoisomerase mutations lying outside the QRDRs, it seems more likely that the increased quinolone resistance of strains arises from another mechanism such as altered efflux. This aspect was not investigated further. However, despite the absence of identifiable new topoisomerase IV mutations in second-step mutants, the increases in MIC for the parC gyrA double mutants relative to the single gyrA mutants shown in Table 1 indicate that the benzenesulfonamido derivatives do have in vivo activity against topoisomerase IV.
NSFQ-105 is a more potent catalytic inhibitor of S. pneumoniae gyrase than is ciprofloxacin, but ciprofloxacin is more active against topoisomerase IV.
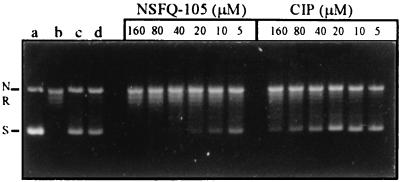
To study the role of target interactions in the mechanism of sulfanilyl fluoroquinolones, we examined the inhibitory effects of NSFQ-105 and ciprofloxacin against purified S. pneumoniae gyrase and topoisomerase IV. For gyrase, NSQF-105 showed a dose-dependent inhibition with an IC50 (the drug concentration that inhibits supercoiling by 50%) of 5 to 10 μM (Fig. 2). Ciprofloxacin was less active, with an IC50 of 80 μM (Fig. 2).
FIG. 2.
Inhibitory effects of NSFQ-105 and ciprofloxacin on the DNA-supercoiling activity of S. pneumoniae DNA gyrase. Relaxed pBR322 DNA (0.4 μg) was incubated with S. pneumoniae GyrA (1 U), GyrB (1 U), and 1.4 mM ATP in the absence or presence of NSFQ-105 or ciprofloxacin (CIP). DNA was analyzed by electrophoresis in 1% agarose. The concentrations of NSFQ-105 and ciprofloxacin included in the reaction mixtures are indicated. Lanes a and b, supercoiled and relaxed pBR322 DNA controls, respectively; lane c, relaxed DNA plus DNA gyrase in the absence of added drug or dimethyl sulfoxide (DMSO); lane d, as lane c but with 1% DMSO. All other gyrase reaction mixtures contained 1% DMSO. N, R, and S, nicked, relaxed, and supercoiled pBR322 DNA, respectively.
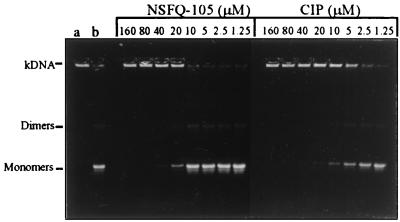
Figure 3 compares the inhibitory activities of NSFQ-105 and ciprofloxacin on the decatenation of kDNA by S. pneumoniae topoisomerase IV. kDNA comprises a large network of interlocked DNA circles that does not move from the wells. The DNA strand passage activity of topoisomerase IV leads to minicircle release and their migration into the gel on electrophoresis. NSFQ-105 showed a dose-dependent response, with an IC50 of 10 to 20 μM (Fig. 3). This value can be compared with an IC50 for ciprofloxacin of 5 μM. Clearly, NSFQ-105 is a less effective inhibitor of topoisomerase IV than ciprofloxacin, the inverse behavior to that seen for gyrase. Table 3 collects the various IC50 and MIC data for NSFQ-105 and ciprofloxacin. It can be seen that the 10- to 20-fold reduction in MIC for NSFQ-105 (expressed in micromolar) over that of ciprofloxacin parallels the greater inhibitory activity of NSFQ-105 against DNA gyrase.
FIG. 3.
Ciprofloxacin is more potent than NSQF-105 as an inhibitor of DNA decatenation by S. pneumoniae topoisomerase IV. kDNA (0.4 μg) was incubated with ParC (1 U), ParE (1 U), and 1.4 mM ATP in the absence or presence of the indicated concentrations of NSFQ-105 and ciprofloxacin (CIP). DNA samples were analyzed by agarose gel electrophoresis. Lane a, kDNA; lane b, kDNA plus ParC, ParE, and ATP in the absence of drug. Monomers, released minicircles; dimers, catenated dimeric minicircles, which are intermediates in the enzyme reaction.
TABLE 3.
Inhibitory effects of fluoroquinolones on S. pneumoniae gyrase and topoisomerase IV and on growth of S. pneumoniae 7785
| Quinolone | IC50 (μM) for:
|
MIC (μM) | |
|---|---|---|---|
| DNA supercoiling by DNA gyrase | kDNA decatenation by topoisomerase IV | ||
| NSFQ-105 | 5–10 | 20 | 0.13–0.26 |
| Ciprofloxacin | 80 | 5 | 2.7 |
DISCUSSION
We have shown that addition of a benzenesulfonylamido group to the C-7 piperazinyl ring of ciprofloxacin markedly affects potency in S. pneumoniae and changes the target specificity of the quinolone from topoisomerase IV to gyrase. The latter conclusion is based on the responses of defined gyrA or parC S. pneumoniae mutants to sulfanilyl agents such as NSFQ-105 (Table 1), the selection of first-step gyrA mutants by NSFQ-105 (Table 2), and the differential activity of NSFQ-105 against purified S. pneumoniae gyrase over topoisomerase IV in catalytic inhibition assays (Fig. 2 and 3 and Table 3). In further support, we have recently shown in DNA cleavage assays that NSFQ-105 was fourfold more efficient than ciprofloxacin in stimulating DNA breakage by S. pneumoniae gyrase, whereas ciprofloxacin was twice as active as NSFQ-105 in promoting DNA cleavage by topoisomerase IV (F. L. Alovero, X.-S. Pan, and L. M. Fisher, unpublished work). Although the nature of the C-7 substituent is known to influence quinolone activity in bacteria (5), our results provide the first direct evidence that it is also a key factor in target selection by quinolones. Moreover, we identify addition of the benzenesulfonylamido group as a particular chemical modification that allows manipulation of target preference in S. pneumoniae. The increased antipneumococcal potency and altered target specificity of the benzenesulfonamide fluoroquinolones have important mechanistic and clinical implications.
Previous studies have shown that NSQF-105 and compounds 1 and 2 display significantly enhanced activity compared with ciprofloxacin against S. aureus (2, 3). In exploring possible causes of this enhanced potency, it was noted that sulfanilyl fluoroquinolones could be viewed as hybrid drugs incorporating a quinolone and a sulfonamide moiety (3). It is known that the sulfa drugs act by competitive inhibition of p-aminobenzoic acid utilization by dihydropteroate synthase, an important step in the production of dihydrofolate. However, by a variety of approaches, it was shown that a sulfonamide mechanism makes little or no contribution to sulfanilyl fluoroquinolone action in S. aureus (3). Similarly, our genetic data, both using a panel of defined mutants and from analysis of stepwise-selected mutants, are not consistent with a sulfonamide mechanism in S. pneumoniae. In particular, the selection of first-step mutants carrying gyrA mutations indicates that gyrase is the primary drug target. Moreover, the enzymatic studies show that sulfanilyl fluoroquinolones do inhibit gyrase and topoisomerase IV at drug concentrations in a range broadly similar to those effective with ciprofloxacin (Table 3). Thus, it appears that in S. pneumoniae, as in S. aureus, sulfanilyl fluoroquinolones act by a fluoroquinolone mechanism.
Two general factors that contribute to antibacterial potency of fluoroquinolones are the kinetics of drug uptake and the ability to inhibit gyrase or topoisomerase IV. Previous studies have associated the enhanced activity of sulfanilyl fluoroquinolones against S. aureus with more favorable kinetics of uptake rather than improved topoisomerase inhibition (3). Ciprofloxacin and norfloxacin are zwitterionic quinolones, but their structural modification to yield the corresponding benzenesulfonamide derivatives produces compounds with only one ionizable group in the biological pH range (3). This charge difference was suggested to account for the better uptake into S. aureus for the sulfanilyl compounds. A role for target-mediated differences was discounted, because DNA supercoiling by E. coli gyrase was inhibited similarly by ciprofloxacin and its sulfanilyl homologues. However, given that topoisomerase IV is a primary target and gyrase is a secondary target for ciprofloxacin in S. aureus (9, 10, 22), the influence of drug targeting on drug activity may be more complex than suggested by experiments with E. coli gyrase. Indeed, in the case of S. pneumoniae, where we have access to the relevant target enzymes, NSFQ-105 and ciprofloxacin have differential activities against gyrase and topoisomerase IV in vitro (Fig. 2 and 3). It is interesting to speculate on whether altered target specificity also contributes to enhanced potency in S. pneumoniae and S. aureus. Because we have focused exclusively on the determinants of quinolone target specificity, we do not know whether enhanced drug uptake influences sulfanilyl fluoroquinolone activity in S. pneumoniae.
NSFQ-105 belongs to an expanding group of new quinolones that select gyrA QRDR mutants of S. pneumoniae in the first step. These agents include the archetype, sparfloxacin, and gatifloxacin (11, 25). A second group of quinolones comprising ciprofloxacin, norfloxacin, levofloxacin, and trovafloxacin select parC QRDR changes in S. pneumoniae before gyrA QRDR changes (11, 15, 16, 20, 28, 32). There appears to be little cross-resistance between the drug classes when first-step gyrA or parC mutants are tested, consistent with the interpretation that the drugs act through different targets in vivo (11, 25). Unfortunately, attempts to understand the structure-function relationships governing target preferences in vivo have been obfuscated by the multiple structural differences even between quinolones of the same class. Previously, we have suggested that the presence of a C-8 substituent is important in directing the quinolone to act through gyrase in vivo rather than through topoisomerase IV (26). Although a role for C-8 is not excluded, the results reported here indicate unequivocally that a simple modification at C-7 of ciprofloxacin is sufficient to switch the drug from the topoisomerase IV-targeting class of agents to one that acts through gyrase.
These findings are not restricted to ciprofloxacin. In a less detailed analysis, we have also examined C-7 benzenesulfonamide derivatives of norfloxacin akin to those of ciprofloxacin shown in Fig. 1. When we tested these derivatives against the panel of S. pneumoniae strains with defined resistance mutations (Table 1), we found that gyrA mutations increased resistance to NSFQ-104 (the norfloxacin derivative equivalent to NSFQ-105 [2]), whereas a parC mutation had little effect (data not shown). The opposite was seen for norfloxacin. These observations suggest that NSFQ-104 targets gyrase in S. pneumoniae, unlike norfloxacin, which is known to act through topoisomerase IV (11). Thus, it appears that the C-7 substituent is an important factor governing target selection by quinolones in vivo. It remains to be established whether the C-7 substituent is the primary determinant of differential targeting or whether there is a complex series of structure-activity relationships, only one of which relates to substituents at C-7.
In the absence of a crystal structure for a cleavable complex involving gyrase or topoisomerase IV, it is not presently possible to define in detail the molecular interactions that determine the target preferences of NSFQ-105 and ciprofloxacin in S. pneumoniae. However, our results can be considered in the context of a molecular model for the quinolone-gyrase-DNA complex proposed by Shen et al. (31). According to this model, four quinolone molecules are envisaged to bind to the single-stranded DNA regions opened up in the gyrase-DNA complex by covalent attachment of the two GyrA subunits to each complementary DNA strand at sites staggered by 4 bp. Two quinolones are envisaged to lie above the other pair of drug molecules, making hydrophobic interactions with each other through substituents on N-1, C-2, and C-8. Binding to DNA strands is suggested to involve a hydrogen bonding domain on the drug comprising the C-3 carboxyl group, the ketone at C-4, and C-5 and C-6, the latter carrying the fluorine substituent in ciprofloxacin. Furthermore, Shen et al. postulated that the substituent on C-7 is involved in drug-enzyme interactions. This proposal was based on the finding that quinolones with different C-7 substituents exhibited different inhibitory activities against DNA gyrase in vitro. However, many of the quinolones tested in the study also differed in their substituents at other positions in addition to C-7. Therefore, aside from concluding that bulky C-7 substituents were tolerated, only a tentative suggestion could be made regarding a role in drug-enzyme interactions (31). Other studies have reported that addition of methyl groups to the piperazine ring at C-7 of a quinolone, or its replacement with a hydroxyphenyl group, dramatically enhanced the inhibitory activity of fluoroquinolones against mammalian topoisomerase II (8, 14). These results suggest that C-7 substituents interact with topoisomerase II. However, our observations provide the first direct support for the idea that the fluoroquinolone C-7 substituent makes critical enzyme contacts that determine bacterial target recognition in vivo and in vitro. Data from high-resolution X-ray structure analysis of drug complexes with S. pneumoniae gyrase and topoisomerase IV will be essential in elucidating the nature of these key drug-enzyme contacts.
Finally, irrespective of the molecular mechanisms involved, it is remarkable that the 4-aminobenzenesulfonamide group in NSFQ-105 (or the 4-N-methylaminobenzenesulfonamide group in compound 1) converts ciprofloxacin into a drug with a potency against strain 7785 in vitro that rivals that of many antipneumococcal fluoroquinolones currently in clinical use or under clinical evaluation. It remains to be established whether there is scope for engineering C-7 modifications of fluoroquinolones that not only improve activity against S. pneumoniae and other gram-positive pathogens but also, by switching in vivo specificity, overcome resistance of first-step mutants likely to arise from conventional quinolone usage. Further studies of benzenesulfonamide derivatives will be important in exploring these interesting questions.
ACKNOWLEDGMENTS
Fabiana Alovero was supported by funds from the FOMEC Project 247 (Facultad de Ciencias Quimicas, Universidad Nacional de Cordoba, Cordoba, Argentina).
We thank M. R. Mazzieri and M. J. Nieto for synthesis of benzenesulfonamide derivatives.
REFERENCES
- 1.Adams D E, Shekhtman E M, Zechiedrich E L, Schmid M B, Cozzarelli N R. The role of topoisomerase IV in partitioning DNA replicons and the structure of catenated intermediates in DNA replication. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 2.Allemandi D A, Alovero F L, Manzo R H. In-vitro activity of new sulfanilyl fluoroquinolones against Staphylococcus aureus. J Antimicrob Chemother. 1994;34:261–265. doi: 10.1093/jac/34.2.261. [DOI] [PubMed] [Google Scholar]
- 3.Alovero F, Nieto M, Mazzieri M R, Then R, Manzo R H. Mode of action of sulfanilyl fluoroquinolones. Antimicrob Agents Chemother. 1998;42:1495–1498. doi: 10.1128/aac.42.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartlett J G, Grundy L M. Community-acquired pneumonia. N Engl J Med. 1995;333:1618–1624. doi: 10.1056/NEJM199512143332408. [DOI] [PubMed] [Google Scholar]
- 5.Bryskier A, Chantot J-F. Classification and structure-activity relationships of fluoroquinolones. Drugs. 1995;49(Suppl. 2):16–28. doi: 10.2165/00003495-199500492-00005. [DOI] [PubMed] [Google Scholar]
- 6.Cullen M E, Wyke A W, Kuroda R, Fisher L M. Cloning and characterization of a DNA gyrase A gene from Escherichia coli that confers clinical resistance to 4-quinolones. Antimicrob Agents Chemother. 1989;33:886–894. doi: 10.1128/aac.33.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eliopoulos G M. In vitro activity of fluoroquinolones against gram-positive bacteria. Drugs. 1995;49(Suppl. 2):48–57. doi: 10.2165/00003495-199500492-00009. [DOI] [PubMed] [Google Scholar]
- 8.Elsea S H, McGuirk P R, Gootz T D, Moynihan M, Osheroff N. Drug features that contribute to the activity of quinolones against mammalian topoisomerase II and cultured cells: correlation between enhancement of enzyme-mediated DNA cleavage in vitro and cytotoxic potential. Antimicrob Agents Chemother. 1993;37:2179–2186. doi: 10.1128/aac.37.10.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrero L, Cameron B, Manse B, Lagneux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target for quinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda H, Hiramatsu K. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:410–412. doi: 10.1128/aac.43.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gellert M, Mizuuchi K, O'Dea M H, Itoh T, Tomizawa J-I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellert M, Mizuuchi K, O'Dea M H, Nash H A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gootz T D, McGuirk P R, Moynihan M S, Haskell S L. Placement of alkyl substituents on the C-7 piperazine ring of fluoroquinolones: dramatic effects on mammalian topoisomerase II and DNA gyrase. Antimicrob Agents Chemother. 1994;38:130–133. doi: 10.1128/aac.38.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gootz T D, Zaniewski R, Haskell S, Schmieder B, Tankovic J, Girard D, Courvalin P, Polzer R J. Activity of the new fluoroquinolone trovafloxacin (CP-99,219) against DNA gyrase and topoisomerase IV mutants of Streptococcus pneumoniae selected in vitro. Antimicrob Agents Chemother. 1996;40:2691–2697. doi: 10.1128/aac.40.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janoir C, Keller V, Kitzis M-D, Moreau N J, Gutmann L. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob Agents Chemother. 1996;40:2760–2764. doi: 10.1128/aac.40.12.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato J, Nishimura Y, Imamura R, Niki H, Higara S, Suzuki H. New topoisomerase essential for chromosomal segregation in E. coli. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 18.Manzo, R. H., D. A. Allemandi, and J. D. Perez. 7 March 1995. U.S. patent 5,395,936. (Argentine patent application 322,011.)
- 19.Mizuuchi K, Fisher L M, O'Dea M H, Gellert M. DNA gyrase action involves the introduction of transient double strand breaks into DNA. Proc Natl Acad Sci USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz R, De la Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of quinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura S. Mechanisms of quinolone resistance. J Infect Chemother. 1997;3:128–138. [Google Scholar]
- 22.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that gyrase is the primary target and topoisomerase IV is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan X-S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan X-S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan X-S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan X-S, Fisher L M. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan X-S, Fisher L M. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: overexpression, purification, and differential inhibition by fluoroquinolones. Antimicrob Agents Chemother. 1999;43:1129–1136. doi: 10.1128/aac.43.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perichon B, Tankovic J, Courvalin P. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piddock L J V. New quinolones and gram-positive bacteria. Antimicrob Agents Chemother. 1994;38:163–169. doi: 10.1128/aac.38.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen L L, Mitscher L A, Sharma P N, O'Donnell T J, Chu W T, Cooper C S, Rosen T, Pernet A G. Mechanism of inhibition of DNA gyrase by quinolone antibacterials: a co-operative drug-DNA binding model. Biochemistry. 1989;28:3886–3894. doi: 10.1021/bi00435a039. [DOI] [PubMed] [Google Scholar]
- 32.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in the gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2502–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zechiedrich E L, Cozzarelli N R. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]