Abstract
Ethambutol (EMB) is a central component of drug regimens used worldwide for the treatment of tuberculosis. To gain insight into the molecular genetic basis of EMB resistance, approximately 2 Mb of five chromosomal regions with 12 genes in 75 epidemiologically unassociated EMB-resistant and 33 EMB-susceptible Mycobacterium tuberculosis strains isolated from human patients were sequenced. Seventy-six percent of EMB-resistant organisms had an amino acid replacement or other molecular change not found in EMB-susceptible strains. Thirty-eight (51%) EMB-resistant isolates had a resistance-associated mutation in only 1 of the 12 genes sequenced. Nineteen EMB-resistant isolates had resistance-associated nucleotide changes that conferred amino acid replacements or upstream potential regulatory region mutations in two or more genes. Most isolates (68%) with resistance-associated mutations in a single gene had nucleotide changes in embB, a gene encoding an arabinosyltransferase involved in cell wall biosynthesis. The majority of these mutations resulted in amino acid replacements at position 306 or 406 of EmbB. Resistance-associated mutations were also identified in several genes recently shown to be upregulated in response to exposure of M. tuberculosis to EMB in vitro, including genes in the iniA operon. Approximately one-fourth of the organisms studied lacked mutations inferred to participate in EMB resistance, a result indicating that one or more genes that mediate resistance to this drug remain to be discovered. Taken together, the results indicate that there are multiple molecular pathways to the EMB resistance phenotype.
Ethambutol [EMB; (S, S)-2,2′-(ethylenediimino)di-1-butanol] is used worldwide as one of the primary antituberculosis agents. The mechanism of action and the molecular genetic basis of resistance to EMB are not fully understood. Only the dextro isomer of EMB is biologically active, an observation consistent with the idea that the drug binds to a specific cellular target (7, 37). Several studies have implicated membrane-associated arabinosyltransferases as targets for EMB (1, 5, 20, 22). These enzymes are well conserved in mycobacteria and are involved in the biosynthesis of arabinan, a component of arabinogalactan present in cell walls (6, 8, 17, 33, 34, 39). Inhibition of arabinan synthesis leads to accumulation of mycolic acids and eventually to cell death.
Three contiguous genes encoding arabinosyltransferases and designated embC, embA, and embB have been identified in Mycobacterium tuberculosis (35). The proteins encoded by these genes are about 65% identical to each other. Previous studies based on limited sequencing of the 10-kb region containing the embCAB genes have identified mutations that result in replacement of amino acid residues and are found only in EMB-resistant organisms cultured from humans. The most commonly affected amino acid was Met306 of EmbB. For example, Sreevatsan et al. (31) identified five distinct mutant codons that resulted in replacement of wild-type Met306 with Ile, Leu, or Val, a result suggesting that EmbB is one of the targets of EMB. Mutations located at the corresponding codon of the embB gene in Mycobacterium smegmatis were also present in EMB-resistant strains (19). Gene transfer experiments showed that mutations in embB conferred EMB resistance in M. smegmatis (1, 5, 19, 35). Telenti et al. (35) postulated that EmbB amino acid 306 is located in a cytoplasmic loop that forms an EMB resistance-determining region (ERDR), and Alcaide et al. (1) showed that amino acids in this region are well conserved among EmbB proteins made by M. tuberculosis, Mycobacterium leprae, M. smegmatis, and many other mycobacterial species. In addition, it was reported that high-level natural resistance to EMB was associated with a variant amino acid motif in the ERDRs of M. leprae, Mycobacterium abscessus, and Mycobacterium chelonae (1). Transfer of a variant M. abscessus embB allele to M. smegmatis resulted in a 500-fold increase in the EMB MIC, a result that provides additional support for the idea that the ERDR participates in resistance to this drug.
Although analysis of EMB-resistant clinical isolates of M. tuberculosis identified embB amino acid-conferring mutations in approximately 50 to 70% isolates with resistance-associated polymorphisms, few resistant organisms have been sequenced in their entirety for the embCAB genes. Importantly, the cause of EMB resistance in isolates that lack mutations in the ERDR of EmbB is unknown (27). In addition, there have been no studies of resistance-associated polymorphisms in genes recently found to be upregulated in vitro in response to EMB exposure. Identification of additional mutations that occur in EMB-resistant organisms will assist in providing an understanding the mechanisms of resistance to this primary antituberculosis agent and provide a starting point for biochemical and other studies of the variant molecules. In the present analysis, we sequenced all genes implicated in EMB resistance in a sample of 75 EMB-resistant isolates from diverse localities that have arisen independently in the course of therapy.
MATERIALS AND METHODS
Bacterial isolates, MIC determination, and DNA extraction.
Seventy-five epidemiologically unrelated EMB-resistant and 33 EMB-susceptible isolates of M. tuberculosis cultured from patients with pulmonary and extrapulmonary tuberculosis were studied. The resistant isolates were obtained from diverse geographic localities in the New York City region (n = 49 isolates), the former Soviet Union (n = 22 isolates), and Texas and Colorado (n = 2 each). EMB-susceptible organisms were obtained from these same geographic areas. The EMB-resistant and EMB-susceptible isolates were chosen to represent all three principal genetic groups of M. tuberculosis. In addition, on the basis of IS6110 profiling and genetic group designation, the chromosomal genotypes of the susceptible control organisms were judged to be closely similar to those of many of the EMB-resistant organisms.
All isolates were initially classified as EMB resistant or susceptible in routine diagnostic laboratories by the BACTEC radiorespiratory method (7.5 μg/ml) or by agar diffusion with Middlebrook 7H10 medium (5 μg/ml) (15). To determine EMB MICs, susceptibility testing was performed by agar diffusion with Middlebrook 7H10 medium and EMB at the following concentrations: 0, 1, 5, 10, 20, 30, 40, and 50 μg/ml (25). Discrepancies between phenotypic susceptibility results and genotypic data were resolved by redetermination of the EMB MICs and reanalysis of the genotypic data. Genomic DNA was isolated from bacteria grown on Lowenstein-Jensen media. All bacterial growth and DNA extraction procedures were conducted in a biosafety level 3 facility.
IS6110 profiling.
Epidemiological independence was assessed for the 75 resistant and 33 susceptible isolates by IS6110 profiling (36). The hybridizing DNA fragments were visualized by enhanced chemiluminescence, and the band patterns were compared by computer-assisted image analysis. The number of hybridizing bands ranged from 1 to 20. The majority of organisms had unique IS6110 profiles, a result indicating epidemiological independence. Fourteen of the 75 isolates shared five IS6110 patterns designated C, KY, P, and W187 (n = 2 isolates each) and W148 (n = 6 isolates). Although these organisms shared IS6110 profiles, drug resistance was thought to have arisen independently in these organisms on the basis of available epidemiological information, including recovery of the isolates from patients living in communities located thousands of miles apart.
Principal genetic group assignment.
All isolates were assigned to one of three principal genetic groups on the basis of polymorphisms present in gyrA codon 95 and katG codon 463 (32).
PCR amplification and sequencing strategy.
The 12 genes analyzed for nucleotide sequence variation are listed in Table 1. A GeneAmp System 9700 thermocycler (Perkin-Elmer Corp., Foster City, Calif.) was used for all DNA amplifications. For each nucleotide polymorphism identified in the EMB-resistant isolates, we sequenced the relevant gene region in at least five EMB-susceptible isolates selected to be genetically similar to the resistant organism on the basis of principal genetic group assignment and the IS6110 profile. This sampling strategy was used to maximize the likelihood of identifying the polymorphic nucleotide in the susceptible organisms. The PCR primers and conditions used to sequence the 12 genes are listed in Table 2. DNA sequencing reactions were performed with the BigDye terminator cycle sequencing kit with AmpliTaq DNA polymerase FS (Applied Biosystems, Inc., Foster City, Calif.). Sequence data generated with an ABI 377 automated instrument were assembled and edited electronically with ALIGN, EDITSEQ, and MEGALIGN programs (DNASTAR, Madison, Wis.) and were compared with the corresponding sequence found in the published H37Rv sequence (10).
TABLE 1.
Genes analyzed for nucleotide sequence diversity in EMB-resistant M. tuberculosis
| Gene | Product or function | Gene size (bp) | Nucleotides sequenced (positions) | No. of nucleotide sites with nonsynonymous substitutions | No. of nucleotide sites with synonymous substitutions | GenBank accession no. |
|---|---|---|---|---|---|---|
| embC | Arabinosyltransferase | 3,285 | −128 to 3285 | 4 | 2 | Z80343 |
| embA | Arabinosyltransferase | 3,285 | −85 to 3285 | 9 | 10 | Z80343 |
| embB | Arabinosyltransferase | 3,297 | 1 to 3384 | 12 | 7 | Z80343 |
| embR | Putative regulator | 1,167 | −295 to 1479 | 2 | 2 | Z77137 |
| Rv3124 | Putative regulator | 870 | −430 to 948 | 2 | 0 | Z95150 |
| Rv0340 | EMB induced | 540 | −231 to 619 | 1 | 3 | Z97991 |
| iniB | INHa and EMB induced | 1,440 | −192 to 1440 | 1 | 0 | Z95324 |
| iniA | INH and EMB induced | 1,923 | −36 to 1923 | 2 | 0 | Z95324 |
| iniC | INH and EMB induced | 1,482 | 1 to 1566 | 2 | 2 | Z95324 |
| rmlD | EMB induced | 915 | −112 to 915 | 2 | 2 | Z92771 |
| wbbL | EMB induced | 906 | −10 to 906 | 0 | 0 | Z92771 |
| rmlA2 | EMB induced | 1,080 | −1 to 1157 | 1 | 1 | Z92771 |
INH, isoniazid
TABLE 2.
PCR primers and conditions used to amplify the designated M. tuberculosis gene regions
| Gene region | Forward primer | Reverse primer | Size (bp) | PCR conditionsa
|
||
|---|---|---|---|---|---|---|
| D (s) | A (temp [°C], length [s]) | E (s) | ||||
| embC | 5′-CCCAACCAGCCCAATGTTC-3′ | 5′-GGCGGTGTCCAGGATGTG-3′ | 880 | 40 | 64.5, 30 | 40 |
| 5′-GCTGCACATCCTGGACAC-3′ | 5′-ACGACATTGCCACCGATAC-3′ | 914 | 40 | 60.0, 30 | 40 | |
| 5′-GTATCGGTGGCAATGTCGT-3′ | 5′-CGGGATGGCGGACAGTGGT-3′ | 1176 | 40 | 60.0, 30 | 45 | |
| 5′-ACCACTGTCCGCCATCCCG-3′ | 5′-GACGACGGCTGCTAGGCGTG-3′ | 635 | 30 | 67.0, 30 | 30 | |
| embA | 5′-GTGACTCGCAGCGGGCTGTG-3′ | 5′-CGGTGAACACAGCGACCCGG-3′ | 1223 | 35 | 68.0, 30 | 40 |
| 5′-TGGACCGGCTCAGCAGGGG-3′ | 5′-TCAGGTTGGCCTTGGCGGTG-3′ | 1500 | 40 | 67.0, 30 | 45 | |
| embA-embB | 5′-CTGGTGGTCGCGGTGATCAT-3′ | 5′-AATTGGCGTCCTTGCCTT-3′ | 1542 | 40 | 61.5, 30 | 50 |
| embB | 5′-GGTGCGCGCCATGCCACC-3′ | 5′-GGTCTGGCAGGCGCATCC-3′ | 803 | 35 | 68.0, 30 | 35 |
| 5′-GGATGCGCCTGCCAGACC-3′ | 5′-AGATGACGCCCATCAGCC-3′ | 730 | 30 | 63.0, 30 | 30 | |
| 5′-GGCTGATGGGCGTCATCT-3′ | 5′-GGACCAGCCGTTGGAGTAGGTC-3′ | 497 | 30 | 63.0, 30 | 40 | |
| 5′-CCCGACCTACTCCAACGGC-3′ | 5′-TGGTGCATACCGAGCAGCAT-3′ | 1195 | 40 | 66.0, 30 | 40 | |
| embR | 5′-CGATCACCACAGCGGGCAGC-3′ | 5′-GTTCGAATGTCAGAGCCTCG-3′ | 896 | 30 | 62.0, 30 | 30 |
| 5′-CGAGGCTCTGACATTCGAAC-3′ | 5′-GCCGACACTATCAACAACGG-3′ | 898 | 30 | 62.0, 30 | 30 | |
| Rv3124 | 5′-GAAACCCGGAGTGGTTCA-3′ | 5′-TCCGGTCTGTGTGACGGAG-3′ | 1,378 | 40 | 61.5, 30 | 40 |
| rmlD | 5′-TACGAACCGTACGAACCAC-3′ | 5′-CGATCGAAGTTGAGTTCGC-3′ | 998 | 40 | 59.0, 30 | 40 |
| wbbL-rmlA2 | 5′-GCGAACTCAACTTCGATCG-3′ | 5′-GACAGCAGATGGGTGAGGAA-3′ | 1,096 | 40 | 60.0, 30 | 40 |
| rmlA2 | 5′-TTCCTCACCCATCTGCTGTC-3′ | 5′-GCGGATCTCGGCGATAAC-3′ | 1,045 | 40 | 62.0, 30 | 40 |
| Rv0340 | 5′-ATGCGTCGTATGCTTGG-3′ | 5′-CCAAACACCTATCGGGATC-3′ | 850 | 40 | 58.5, 30 | 40 |
| iniB | 5′-ATAAGTTCCGGACCGGCG-3′ | 5′-CGACAGATGAGGCATAGCAG-3′ | 1,053 | 40 | 56.0, 30 | 45 |
| iniB-iniA | 5′-TTGAACGGCGCTGCTATG-3′ | 5′-GTGCTGATGTCATCGACGG-3′ | 1,070 | 40 | 62.0, 30 | 45 |
| iniA | 5′-CAACCGCAGCGGTTGACAT-3′ | 5′-CCGCATGCCGATAATCATT-3′ | 1,079 | 40 | 61.0, 30 | 45 |
| iniA-iniC | 5′-GGAATCGAAACCGCTGCG-3′ | 5′-CCAGCCCACCGATCTGTTTGA-3′ | 1,090 | 40 | 64.0, 30 | 45 |
| iniC | 5′-TCCTGTTGCGCACCCTGAAC-3′ | 5′-AACATGTTCCACCCGGTGGC-3′ | 1,040 | 40 | 65.0, 30 | 45 |
D, length of denaturation at 94°C; A, primer annealing conditions; E, length of extension at 72°C. All PCRs were 25 cycles, were preceded by a denaturation step at 94°C for 3 min, and included a final extension step at 72°C for 7 min.
RESULTS
Overall nucleotide variation.
Approximately 1.6 Mb of the genes of the 75 EMB-resistant M. tuberculosis isolates were sequenced. Only 29 sites with polymorphic synonymous (silent, not resulting in amino acid replacement) substitutions were identified, a result confirming that unselected nucleotide variation in structural genes is very limited in M. tuberculosis (32). Inasmuch as these 29 silent nucleotide changes would not result in amino acid replacements, they are unlikely to participate in drug resistance. Hence, they will not be considered further when describing the sequencing results for the 12 genes studied. (A listing of the 29 silent nucleotide polymorphisms is available by request from S.V.R.). Thirty-eight (51%) of the 75 EMB-resistant M. tuberculosis isolates studied had an EMB resistance-associated mutation in only 1 of the 12 genes sequenced (Table 3).
TABLE 3.
EMB resistance-associated mutations in M. tuberculosis
| Gene or gene region | No. of isolates with mutations
|
% of isolates with single-locus mutation | MIC rangea (μg/ml) | |
|---|---|---|---|---|
| Single locusb | Multiple loci | |||
| embC | 0 | 2 | 0 | |
| embC-embA intergenic region | 0 | 8 | 0 | |
| embA | 4 | 5 | 5.3 | 10–30 |
| embB | 26 | 15 | 34.7 | 10–50 |
| embR | 2 | 0 | 2.7 | 20–40 |
| Rv3124 | 0 | 3 | 0 | |
| Rv0340 | 0 | 1 | 0 | |
| iniB | 1 | 1 | 1.3 | 30 |
| iniA | 2 | 5 | 2.7 | >50 |
| iniC | 0 | 2 | 0 | |
| rmlD | 3 | 0 | 4.0 | >50 |
| wbbL | 0 | 0 | 0 | |
| rmlA2 | 0 | 1 | 0 | |
| Total | 38 | 43 | 50.7 | |
The MIC range shown is for the isolates with single-locus mutations. The MICs for isolates with multiple-locus mutations are shown in Table 4.
The four embA mutations were Ala462Val (n = 2) and Gly321Ser and Asp833Ala (n = 1 each). The embB mutations were Met306Val (n = 7), Met306Ile (n = 4), Gln497Arg (n = 3), Gly406Ala and Gly406Cys (n = 2 each), and Ser297Ala, Met306Leu, Asp328Gly, Phe330Val, Gly406Asp, Asp959Ala, Met1000Arg, and Asp1024Asn (n = 1 each). The two embR mutations were Gln379Arg and a change at position −137, an A insertion (n = 1 each). The iniB mutation was A47T. Two isolates had Ser501Trp changes in iniA. The rmlD changes were T284K (n = 1) and a change at position −71, G→T (n = 2). Nineteen isolates had EMB-resistance associated mutations in two or more genes.
Polymorphisms in the embCAB operon.
Twenty-five codons in the embCAB operon had mutations that would result in amino acid replacements. The analysis also identified three polymorphic nucleotide sites and one nucleotide insertion located in the embC-embA intergenic region. To determine if these mutations occur among EMB-resistant organisms, we next sequenced the analogous gene region containing each polymorphic nucleotide in at least five genetically related EMB-susceptible organisms. The susceptible organisms characterized had the same principal genetic group as the resistant organisms and IS6110 profiles similar to those for the resistant organisms (the profiles generally differed by one hybridizing IS6110 band). Four of the 25 variant codons were also identified in the EMB-susceptible bacteria, which means that 21 of the variable amino acid sites were uniquely represented in the EMB-resistant strains (Fig. 1). The four variable codons found in both susceptible and resistant organisms (embC codon 270, embC codon 981, embA codon 206, and embB codon 378) were each confined to strains of the same principal genetic group, including many with related IS6110 profiles. The sequencing results support the idea that these four variant amino acids do not confer EMB resistance but, rather, are changes that have arisen in the course of evolution and that have been maintained by clonally related organisms. All nucleotide polymorphisms located in the embC-embA intergenic region occurred only in EMB-resistant organisms, suggesting that these changes are resistance associated rather than surrogate markers of resistant strains.
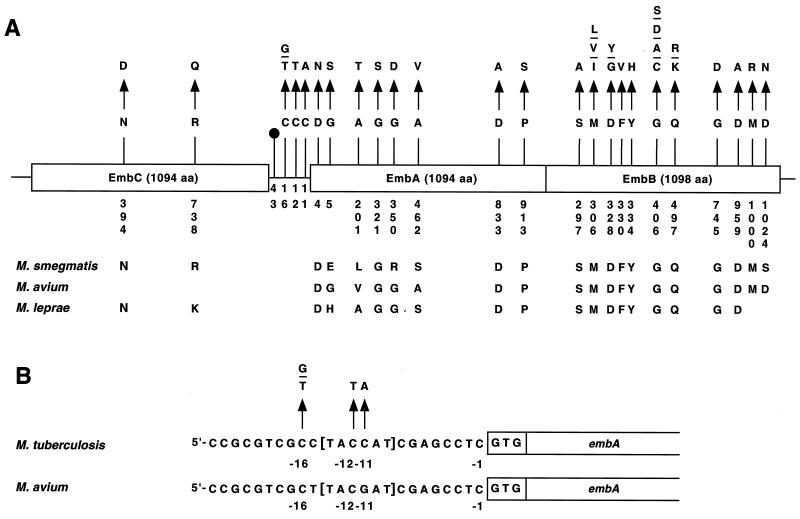
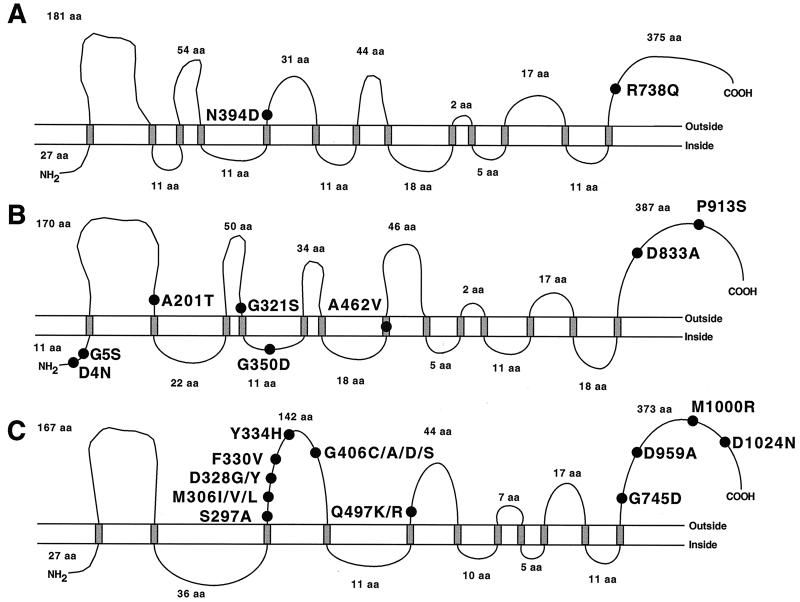
FIG. 1.
Schematic representation of EMB resistance-related polymorphisms in the embCAB genes. (A) Overview of the mutations identified. Variant amino acid residues and nucleotides are numbered vertically. The corresponding amino acid residues found in M. smegmatis, M. avium, and M. leprae are shown. The symbol at position −43 denotes a dinucleotide deletion of a guanine and a cytosine. (B) Expanded view of the embC-embA intergenic region. The nucleotide substitutions identified in the region containing a putative TATA box (marked in brackets) are shown. The single-letter amino acid (aa) abbreviations are used. A, alanine; C, cysteine; D, aspartic acid; E, glutamic acid; F, phenylalanine; G, glycine; H, histidine; I, isoleucine; K, lysine; L, leucine; M, methionine; N, asparagine; P, proline; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; Y, tyrosine.
(i) Polymorphisms in the embC gene.
The embC gene had two EMB resistance-associated nonsynonymous (resulting in an amino acid replacement) mutations located in codon 394 (AAC→GAC, Asn→Asp) and codon 738 (CGG→CAG, Arg→Gln) (Fig. 1). Each of these variant amino acids was identified in one strain. Although the Asn394Asp and Arg738Gln replacements were present only in EMB-resistant organisms in our sample, these two strains also had other amino acid replacements exclusively found in resistant organisms. Hence, it is unclear if the EmbC amino acid replacements alone would confer EMB resistance. We note that Asn394 was conserved in M. smegmatis and M. leprae and Arg738 was conserved in M. smegmatis (Fig. 1).
(ii) Polymorphisms in the embC-embA intergenic region.
Eight isolates had resistance-associated nucleotide changes located in the embC-embA intergenic region (Fig. 1). All isolates with these mutations also had resistance-associated amino acid replacements in EmbA (n = 1 isolate) or EmbB (n = 7 isolates), suggesting that the intergenic polymorphisms are secondary or compensatory changes. The observation that the mutations that occurred at positions −11 (C→A), −12 (C→T), and −16 (C→T or G) are all located in or immediately adjacent to a 6-bp putative TATA box (TACCAT) (Fig. 1) is consistent with this idea (3).
(iii) Polymorphisms in the embA gene.
Eight EMB resistance-associated amino acid replacements were identified in EmbA (Fig. 1). Four of these eight amino acids were conserved in M. smegmatis, M. leprae, and M. avium, and the other four amino acids were conserved in at least one of these three mycobacterial species. Amino acid changes at positions 462 (Ala→Val) and 913 (Pro→Ser) were present in two isolates each, and the other six amino acid replacements were identified in a total of five isolates.
(iv) Polymorphisms in the embB gene.
Our sequence analysis of the embB gene identified 41 isolates with EMB resistance-associated nucleotide substitutions in 11 distinct codons (Fig. 1). Four codons (codons 306, 328, 406, and 497) had mutations that would result in two, three, or four different amino acid replacements. For example, Met306 was replaced by Ile, Leu, and Val, and Gly406 was altered to Ala, Asp, Cys, and Ser (Fig. 1). Twenty-six isolates had resistance-associated mutations only in EmbB. The 15 isolates with EmbB amino acid replacements plus a resistance-associated change in one or more additional genes usually had a putative regulatory mutation in the embC-embA intergenic region (n = 7 isolates) or a resistance-associated mutation that was identified only in organisms with an EmbB amino acid replacement (n = 8 isolates).
(v) Summary.
To summarize, 49 of the 75 (65%) isolates studied had EMB resistance-associated mutations identified in the embCAB operon. Twenty-one distinct codons had mutations that were exclusively represented among EMB-resistant organisms. Thirty of the 49 isolates had resistance-associated mutations in only 1 of the 12 genes sequenced, whereas 19 isolates with an embCAB mutation had a resistance-associated mutation in one or more genes. All amino acids with resistance-associated replacements were conserved in M. smegmatis, M. leprae, or M. avium, and 14 of the affected amino acids were conserved in the homologous genes present in all of these species. Interestingly, no amino acid replacements were identified between positions 498 and 737 in all the Emb proteins, an observation suggesting that amino acid changes in these residues are selected against.
Polymorphisms in embR.
The embC gene is absent from M. avium, and a gene designated embR is found in its place immediately upstream of the embAB genes (5). Belanger et al. (5) used an M. avium EMB-sensitive cell-free assay for arabinan biosynthesis to show that overexpression of EmbAB was associated with high-level EMB-resistant arabinosyltransferase activity. EmbR modulated the level of this arabinosyltransferase activity in vitro (5). This observation and the relatedness of EmbR to transcriptional activators such as ToxR, which regulates cholera toxin production, led to the hypothesis that EmbR regulated expression of embAB genes in M. avium. However, the role of EmbR in M. tuberculosis physiology is unknown, and no information regarding embR mutations in EMB-resistant organisms is available. In contrast to M. avium, an embR homologue is located 2 Mb from the embCAB locus in M. tuberculosis rather than immediately upstream of the embAB genes (10).
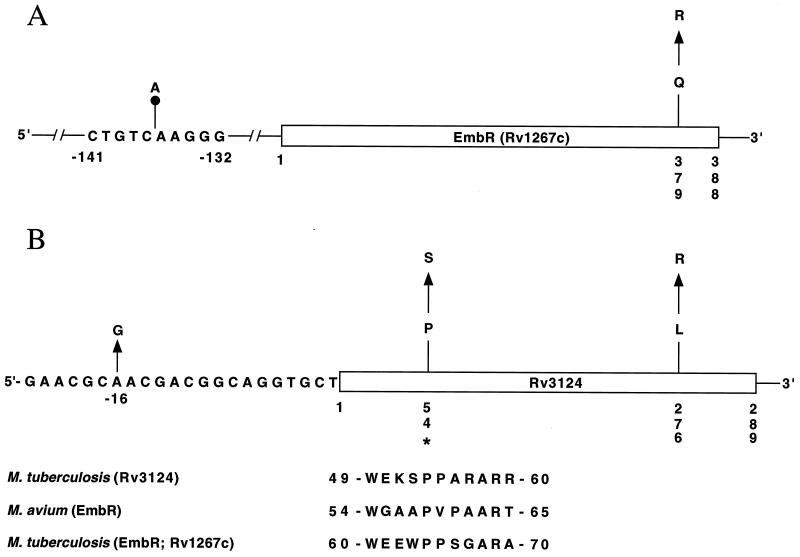
We sequenced embR and its upstream putative regulatory region in the 75 EMB-resistant isolates. Two nonsynonymous nucleotide substitutions resulting in Cys110Tyr and Gln379Arg replacements and two upstream regulatory region changes were identified. However, sequence analysis of the homologous gene regions in EMB-susceptible strains revealed that only the Gln379Arg replacement and an adenine insertion at position −137 upstream of the EmbR start codon were exclusively represented in EMB-resistant isolates (Fig. 2A). Each of these two changes was identified in one isolate, and these were the only resistance-associated mutations identified in the organisms.
FIG. 2.
(A) Schematic representation of EMB resistance-associated mutations identified in the embR gene. Variant amino acids and nucleotides are numbered vertically. The single-letter amino acid designations used are Q (glutamine) and R (arginine). (B) Schematic representation of EMB resistance-associated nucleotide and nonsynonymous mutations identified in the Rv3124 gene. Variant amino acids are numbered vertically. The asterisk indicates an identical amino acid residue present in M. avium EmbR and the corresponding homologous regions from M. tuberculosis Rv3124 and EmbR. The single-letter amino acid (aa) designations used are L (leucine), P (proline), R (arginine), and S (serine).
Polymorphisms in Rv3124.
The M. tuberculosis H37Rv genome also contains a gene designated Rv3124 that encodes a protein of unknown function that is 55% identical to M. tuberculosis EmbR (10). This gene and upstream region were sequenced in the 75 EMB-resistant isolates, and mutations were identified in three strains. One organism each had a nonsynonymous substitution resulting in a Pro54Ser or Leu276Arg replacement (Fig. 2B). In addition, one organism had an upstream nucleotide substitution (adenine→guanine) located at position −16 relative to the start codon. These polymorphisms were not present in the EMB-susceptible isolates characterized. However, all three isolates with the Rv3124 mutations had EMB resistance-associated changes in other genes including embC (n = 1 isolate) and embB (n = 2 isolates).
Polymorphisms in the ini chromosomal region.
The ini chromosomal region has five genes designated Rv0339c, Rv0340, iniB, iniA, and iniC (Fig. 3). Three of these genes (iniB, iniA, and iniC) are organized as an operon. The Rv0340 gene is located upstream of the iniBAC operon and is transcribed in the same orientation. The Rv0339c gene is upstream of Rv0340 but is transcribed in the opposite direction. The ini genes were originally identified on the basis of being induced by isoniazid and EMB treatment in vitro (2). Hence, we elected to sequence Rv0340 and the iniBAC genes in all 75 EMB-resistant isolates.
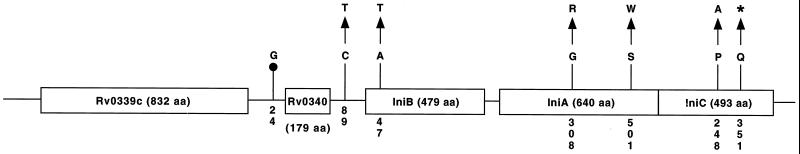
FIG. 3.
Schematic representation of EMB resistance-associated nucleotide and nonsynonymous mutations identified in the ini locus. The symbol upstream of Rv0340 denotes deletion of a guanine nucleotide. The asterisk indicates a termination mutation. Variant amino acids and nucleotides are numbered vertically. The single-letter amino acid (aa) designations used are A (alanine), G (glycine), P (proline), Q (glutamine), R (arginine), S (serine), T (threonine), and W (tryptophan).
Four EMB-resistant isolates had mutations in the Rv0340 gene (Fig. 3). Three isolates had a Thr143Met replacement, and one isolate had a deletion of a guanine residue located at position −24 upstream of the start codon. All four strains with these polymorphisms also had other EMB resistance-associated mutations including changes in embC (n = 1 isolate) and embB (n = 3 isolates). Sequence analysis of the Rv0340 gene in EMB-susceptible organisms identified the Thr143Met polymorphism in two organisms. The three EMB-resistant and two EMB-susceptible organisms with this change were all members of principal genetic group 1 and had related IS6110 profiles. Taken together, the results indicate that this polymorphism does not confer EMB resistance. In contrast, the guanine deletion at position −24 was not identified in EMB-susceptible control organisms.
The iniB gene would encode a protein with weak homology to alanine-glycine-rich cell wall structural proteins (2). Two EMB-resistant isolates had polymorphisms in the iniB gene (Ala47Thr) or upstream region (C→T at position −89) that were exclusively represented in EMB-resistant organisms.
iniA would encode a protein with a phosphopantetheine attachment site motif that is characteristic of acyl-carrier proteins. Eight isolates had EMB resistance-associated amino acid replacements in IniA, including one organism with a Gly308Arg polymorphism and seven strains with Ser501Trp changes. Although these amino acid replacements were not identified in the EMB-susceptible control organisms that we characterized, five of the seven strains with iniA mutations also had EMB resistance-associated polymorphisms in other genes studied. We note that computer modeling suggests that the Ser501Trp polymorphism is located in the middle of a transmembrane domain of IniA (30).
The iniC gene would encode a protein that is 34% identical to IniA (2). Two isolates had EMB resistance-associated changes in the iniC gene. Both of these organisms also had other resistance-associated polymorphisms.
Polymorphisms in the rmlD chromosomal region.
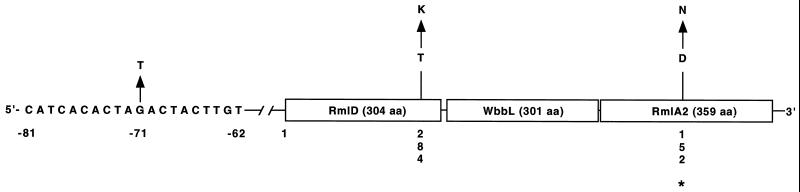
The rmlD chromosomal region consists of three genes designated rmlD, wbbL, and rmlA2 that are transcribed in the same direction. On the basis of amino acid homology, the proteins encoded by these genes may be enzymes that participate in the modification of rhamnose residues prior to incorporation into the M. tuberculosis cell wall and that are suspected on the basis of biochemical analyses to participate in EMB interactions (20, 21, 23; C. E. Barry III, personal communication). Sequence analysis of these three genes in the 75 EMB-resistant organisms identified four nucleotide polymorphisms, including two and one nonsynonymous substitutions in rmlD (Ser257Pro and Thr284Lys) and rmlA2 (Asp152Asn), respectively (Fig. 4). Two isolates also had a guanine→thymidine change located at position −71 upstream of the start codon in rmlD. These two isolates did not have EMB resistance-associated changes in the other 11 genes sequenced, and the MICs for the isolates were greater than 50 μg/ml. For one other isolate the MIC was >50 μg/ml, and it had a Thr284Lys substitution in RmlD and lacked other EMB-resistance associated changes.
FIG. 4.
Schematic representation of nucleotide and nonsynonymous changes identified in the rml locus. Variant amino acids and nucleotides are numbered vertically. Asterisk indicates identical residue present in a variety of gram-positive, and gram-negative bacteria, and archaebacteria. The single-letter amino acid (aa) designations used are D (aspartic acid), K (lysine), N (asparagine), and T (threonine).
Relationship of mutations and EMB MICs.
Several reports have noted a strong correlation between MICs and distinct resistance-associated mutations for several antituberculosis drugs (27, 31). For example, Sreevatsan et al. (31) reported that the MICs for EMB-resistant organisms with EmbB Met306Ile replacements were generally lower (20 μg/ml) than those for strains with the Met306Leu and Met306Val substitutions (40 μg/ml). Analysis of the MICs for the 75 EMB-resistant strains confirmed this observation for the amino acid polymorphisms for EmbB at codon 306. We also found that EMB MICs were generally ≥30 μg/ml for organisms with amino acid replacements in EmbB at codon 406 and EmbB at codon 497. There was no apparent difference in the EMB MICs for organisms with mutations in the other genes analyzed. There was also no simple correlation between MICs for strains with single or multiple resistance-associated mutations (Table 4).
TABLE 4.
EMB-resistant M. tuberculosis isolates with mutations in two or more genes
| Isolate | MIC (μg/ml) | Mutationa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| embC | embA | embB | Rv3124 | Rv0340 | iniB | iniA | iniC | rmlA2 | ||
| TN2050 | >50 | Q497K | 89 UPS, C→T | |||||||
| TN2946 | 40 | G745D | G308R | |||||||
| TN4864 | 30 | 12 UPS, C→T | Q497K | |||||||
| TN5480 | 20 | M306I | D152N | |||||||
| TN5534 | 40 | R738Q | 16 UPS, C→T | M306I | P54S | |||||
| TN6980 | 40 | D4N | G406D | |||||||
| TN7289 | 50 | N394D | G350D | L276R | 24 UPS, G del | |||||
| P913S | ||||||||||
| TN8199 | 20 | 43 UPS, CG del | M306I | |||||||
| TN8560 | 30 | 12 UPS, C→T | Q497R | S501W | ||||||
| TN8561 | 30 | 43 UPS, CG del | G406S | |||||||
| TN8564 | >10 | 16 UPS, C→G | S501W | |||||||
| TN8565 | 5 | S501W | P248A | |||||||
| TN8568 | 30 | M306I | S501W | |||||||
| TN8575 | 30 | 12 UPS, C→T | M1000R | |||||||
| TN8708 | 10 | M306I | 16 UPS, A→G | |||||||
| HN56 | 40 | P913S | Q351STOP | |||||||
| NHN203 | 30 | A201T | D328Y | |||||||
| Y334H | ||||||||||
| TN718 | 20 | 11 UPS, C→A | M306I | |||||||
| G406A | ||||||||||
| TN1618 | 20 | G5S | M306I | |||||||
The single-letter amino acid designations used are as follows: A, alanine; D, aspartic acid; G, glycine; H, histidine; I, isoleucine; K, lysine; L, leucine; M, methionine; N, asparagine; P, proline; Q, glutamine; R, arginine; S, serine; T, threonine; W, tryptophan; and Y, tyrosine. The single-letter nucleotide designations used are as follows: A, adenine; C, cytosine; G, guanine; and T, thymidine. The other abbreviations are as follows: UPS, nucleotide position upstream of the start codon; del, deletion of the nucleotide bases; STOP, termination mutation.
DISCUSSION
EMB has been used as a primary drug for the treatment of tuberculosis since the 1960s (13, 15, 16). Several hypotheses have been advanced to explain the mechanism of action of EMB. Most early studies pointed toward a detrimental alteration of the mycobacterial cell wall structure, although pleiotropic effects have been described (4, 9, 12, 14, 18, 26, 29, 34). Biochemical evidence has implicated arabinosyltransferases as EMB targets, and this concept has been supported by recent molecular genetic data obtained from study of M. avium, M. smegmatis, and M. tuberculosis (1, 5, 20, 39). Telenti et al. (35) reported that mutations in the M. tuberculosis and M. smegmatis embCAB operon were associated with EMB resistance. Sreevatsan et al. (31) confirmed that certain mutations in the embCAB operon were exclusively associated with EMB-resistant M. tuberculosis. In particular, amino acid replacements at position 306 of EmbB were shown to be abundantly represented among EMB-resistant, but not EMB-susceptible, organisms. However, knowledge of the spectrum of mutations that may participate in EMB resistance is relatively limited for three reasons. First, the embCAB operon is approximately 10 kb in length, a size that hinders complete sequence analysis. For example, only 7.5 kb of this 10-kb chromosomal region has been sequenced for 19 M. tuberculosis strains (27, 31). Second, recently several additional chromosomal loci that encode proteins that may participate in the response of M. tuberculosis to EMB treatment have been identified (2). The identification of these genes raises the possibility that mutations in them confer EMB resistance. However, thus far sequence variation in these genes has not been studied in EMB-resistant and -susceptible organisms. This strategy has been fruitful in the early stages of investigating the molecular genetic basis of resistance to several other antituberculosis drugs, including rifampin, isoniazid, streptomycin, and pyrazinamide (27). In addition, information of this type is required to assist in the development of rapid genetic molecular methods to identify drug-resistant M. tuberculosis isolates. Third, although some information has been reported, there is a need for additional data bearing on the association between EMB resistance-associated mutations and MICs of this drug. Analysis of multiple candidate genes is also important because Telenti et al. (35) have provided evidence that EMB resistance is selected in a stepwise fashion that could involve several genes. To address these areas in which knowledge is lacking, we sequenced all 12 genes that may participate in EMB resistance in a sample of 75 organisms recovered from diverse geographic localities and judged to have acquired EMB resistance independently.
Low rate of silent nucleotide substitutions.
Our data confirmed the rarity of synonymous nucleotide substitutions in structural genes in M. tuberculosis isolates recovered from intercontinental sources. We identified only 29 polymorphic synonymous sites in the ∼1.6 Mb sequenced in the 75 EMB-resistant organisms. The restricted occurrence of silent nucleotide substitutions in M. tuberculosis is consistent with the hypothesis that the species is evolutionarily new, perhaps having arisen as recently as 15,000 to 20,000 years ago (32).
embCAB resistance-associated mutations are common.
Previous molecular genetic studies provided evidence that the arabinosyltransferases encoded by the embCAB operon are targets for EMB and that amino acid replacements in these proteins confer EMB resistance (1, 5, 19, 31, 35). Our analysis provided additional data that support this idea. We identified 21 distinct codons that had mutations that confer amino acid replacements that were present in EMB-resistant organisms but not related EMB-susceptible control strains. Importantly, four embB codons (codons 306, 328, 406, and 497) had multiple mutations that would result in two, three, or four different amino acid replacements in the EmbB protein. Multiple amino acid replacements in EmbB306 were reported previously, but the identification of distinct mutations that repeatedly alter amino acid residues Asp328, Gly406, and Glu497 is new (31). The occurrence of multiple mutations that affect the same codon and produce different amino acid replacements is a hallmark of positive Darwinian selection by antibiotic pressure. The amino acid polymorphisms found at position 406 of EmbB were unanticipated because previous studies of embB sequence variation in EMB-resistant organisms did not identify mutations in this codon. The reason for the lack of identification of codon 406 changes in previous studies is unclear, although it may relate to the relatively few strains sequenced completely for the embB gene (31). In addition, we note that the PCR primers used by several investigators to identify codon 306 polymorphisms would not amplify the embB codon 406 region (1, 31, 35).
Another unexpected finding from sequencing of the complete embCAB operon was the identification of five distinct resistance-associated nucleotide changes in the embC-embA intergenic region. Previous studies had not identified EMB resistance-associated mutations in this region of the operon. Four of the five nucleotides with mutations were located at positions −11, −12, and −16 relative to the embA start codon. Inspection of this area of the intergenic region found that these nucleotide changes fall within or adjacent to a putative TATA box, an observation suggesting that these mutations may be in a regulatory region (3, 11, 24). Consistent with this idea is the fact that all EMB-resistant organisms with these embC-embA intergenic changes also had resistance-associated amino acid replacements in EmbA or EmbB. On the presumption that the EmbA and EmbB polymorphisms confer EMB resistance, we speculate that the intergenic region changes are compensatory or secondary mutations. Under this hypothesis these mutations result in altered regulation of expression of EmbA and/or EmbB. Compensatory mutations selected by exposure to toxic drugs also have been identified in isoniazid-resistant organisms in the upstream regulatory region of the two-gene operon containing mabA and inhA and in the regulatory region of the ahpC gene encoding alkylhydroperoxide reductase (27). Potentially at variance with the compensatory change hypothesis is the report by Telenti et al. (35) that no transcription initiation site was identified in the wild-type embC-embA intergenic region by primer extension analysis. However, it is possible that the mutations that we identified create a new transcriptional start site in the mutant organisms where none existed previously in the wild-type strains. Studies are under way to determine molecular support for this compensatory mutation hypothesis.
Of note, considerably fewer distinct EMB resistance-associated amino acid replacements were identified in EmbC (n = 2) compared to the number identified in EmbA (n = 8) and EmbB (n = 18). One possibility that may account for this observation is that EMB treatment results in less selective pressure on EmbC relative to that on EmbA and EmbB. This could occur because the expression level of EmbC is less than those of the other two Emb proteins or because EmbC interacts with EMB in a fundamentally different way compared to the way in which it interacts with EmbA and EmbB. On the presumption that M. avium is evolutionarily older than M. tuberculosis, it is reasonable to speculate that the embC gene was added to the genome after embA and embB were already present, presumably by a gene duplication event. Under this hypothesis, the region immediately upstream of embA in M. tuberculosis is analogous to the regulatory region upstream of embA in M. avium. The identities of 35 of 47 nucleotides upstream of embA in M. avium and M. tuberculosis and the lack of identity of nucleotides upstream of embC with embA of M. avium supports this idea. If this thesis is correct, it suggests that the region upstream of embA in M. tuberculosis may retain the capability to be transcriptionally regulated under appropriate conditions. The occurrence of EMB resistance-associated mutations in the embC-embA intergenic region and the dominance of EmbA and EmbB amino acid replacements in resistant organisms support this idea.
On the basis of computer modeling, the Emb proteins were predicted to be integral membrane proteins that consist of multiple α-helical segments spanning the bacterial membrane. There were reported to be 12 transmembrane domains and a carboxy-terminal globular region of approximately 375 amino acids with a predicted noncytoplasmic location. It was reported that the ERDR of EmbB that contains amino acid residue 306 maps to a region predicted to be located in a cytoplasmic loop. Telenti et al. (35) modeled EmbB by the method described by Rost and Sander (28) which uses multiple amino acid sequence alignments as input to neural networks in predicting protein secondary structure. The method had an overall accuracy of 72% in a multiple cross-validation test with 126 unique proteins. We used the method described by Sonnhammer et al. (30) to analyze the potential locations of all EMB resistance-associated variant amino acids identified in EmbC, EmbA, and EmbB (Fig. 5). Only 1 of the 21 variant amino acid positions (EmbA462) was predicted to be located in a transmembrane region. Interestingly, 17 of the other 20 amino acids with EMB resistance-associated replacements were predicted to be located exterior to the cell membrane. Moreover, the region of EmbB that contains the majority of resistance-associated mutations identified in our study and previous analyses (EmbB297 to EmbB406), including the ERDR, was predicted to be on a single segment of EmbB located outside the cell membrane.
FIG. 5.
Schematic representation of EMB resistance-associated amino acid replacements identified in the EmbCAB proteins, modeled by the method described by Sonnhammer et al. (30). (A) EmbC; (B), EmbA; (C), EmbB. The single-letter amino acid (aa) designations used are A (alanine), C (cysteine), D (aspartic acid), F (phenylalanine), G (glycine), H (histidine), I (isoleucine), K (lysine), L (leucine), M (methionine), N (asparagine), P (proline), Q (glutamine), R (arginine), S (serine), T (threonine), V (valine), and Y (tyrosine).
The modeling method used by Sonnhammer et al. (30) is based on a hidden Markov model (HMM) with an architecture that corresponds closely to the biology of the protein. This modeling method correctly predicted the entire topology for 77% of 160 proteins in a validation test. The neural network model used by Telenti et al. (35) predicted 12 transmembrane helices in EmbB, whereas the HMM method predicted only 11 transmembrane helices. Both modeling methods predicted seven of the transmembrane helices located toward the carboxy end, and the orientations of the cytoplasmic and extracytoplasmic loops were identical in both models. However, the two models diverged at the fourth transmembrane helix predicted by the neural network method; this helix is not predicted by the HMM method. The lack of prediction of this helix results in a reversal of the orientation of the major loop containing the majority of amino acid replacements identified in resistant organisms. Instead of a cytoplasm orientation predicted by the method of Rost and Sander (28), this loop is now predicted to be located on the extracytoplasmic side of the membrane. The prediction of loops is generally based on a “positive-inside rule” because positively charged amino acids are commonly located on the cytoplasmic side of the membrane. These positively charged amino acids are usually found in relatively short loops and function to guide the orientation of helices by preventing translocation across the membrane. Longer loops that contain positively charged amino acid residues are transferred across the membrane by a different mechanism. The HMM uses algorithms to predict short loops and long loops separately for the noncytoplasmic loops, an approach that corresponds to the biology of the two known membrane insertion mechanisms. Clearly, modeling alone is not sufficient to determine the true orientations of the amino acids in proteins. However, the model predicted by the method of Sonnhammer et al. (30) has the potential advantage of orienting the loop containing the majority of altered amino acids on the extracytoplasmic side of the membrane, a location that may permit ready protein-drug interaction. Studies are under way to investigate this issue more fully.
Mutations in putative regulatory genes.
Our analysis identified several EMB resistance-associated polymorphisms in embR and Rv3124, two genes that encode proteins that are homologous with EmbR in M. avium. EmbR is related to known transcriptional activators such as ToxR, a protein that regulates cholera toxin production in Vibrio cholerae (5). Belanger et al. (5) reported that EmbR in M. avium modulated the level of arabinosyltransferase activity in vitro, a result suggesting a role for EmbR in regulating transcription of embA and/or embB. Although no data that bear on the possibility that EmbR or the protein encoded by Rv3124 participate in regulation of the embCAB genes are available, the occurrence of mutations in these putative regulatory genes that are exclusively found in EMB-resistant organisms suggests that this may be the case. The mutant alleles identified in our study provide an important resource for future studies to address this issue.
Implications for rapid identification of EMB-resistant M. tuberculosis.
The identification of missense and other mutations in the 12 genes that were sequenced and that are exclusively found in EMB-resistant organisms has practical implications for tuberculosis diagnostics. Many EMB-resistant organisms could be identified rapidly by molecular methods that interrogate the region encompassing embB codon 306 to embB codon 406. Contrary to the situation with rifampin and pyrazinamide resistance, in which greater than 90% of resistant organisms have sequence alterations in a relatively short gene or gene region, the technical problems associated with the development of a rapid method for the detection of mutations in at least 12 genes at a reasonable cost are unusually daunting (27). Presumably, DNA array technology could be used for this purpose. However, even if technical difficulties could be overcome, our data suggest that only about 75% of EMB-resistant organisms could be rapidly detected. The best strategy at present for molecular diagnostics is selective targeting of the region of embB encoding roughly amino acids 300 to 500.
EMB-resistant organisms lacking mutations in the 12 genes studied.
Although we sequenced in entirety all genes that have thus far been found to potentially participate in EMB resistance, our analysis found that 24% of isolates in this sample lacked a resistance-associated mutation. The EMB MICs for the 18 strains that lacked mutations ranged from 10 to >50 μg/ml. This range of MICs suggests that phenotypic resistance in these organisms is unlikely to be due to the same mutation(s) in all 18 strains. These 18 EMB-resistant strains will be useful in the search for additional genetic loci that participate in EMB resistance. Isolation and characterization of all genes that participate in EMB resistance is critical to the development of a comprehensive understanding of the mechanism of action of this drug and potentially to the development of new therapeutics.
Summary.
The identification of amino acid replacements and other molecular changes exclusively among EMB-resistant M. tuberculosis isolates does not prove that they confer or otherwise participate in resistance to this drug. Additional molecular genetic, biochemical, and enzymatic studies are required to prove that the mutations that we identified participate in the response of M. tuberculosis to EMB treatment. Nevertheless, the sequence data provided by analysis of all 12 genes potentially involved in EMB resistance is an important first step toward gaining additional insight into the molecular genetics of resistance to this drug in M. tuberculosis.
ACKNOWLEDGMENTS
This research was supported by Public Health Service grant AI-37004 to J.M.M.
We thank C. E. Barry III for helpful discussions.
REFERENCES
- 1.Alcaide F, Pfyffer G E, Telenti A. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob Agents Chemother. 1997;41:2270–2273. doi: 10.1128/aac.41.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland D, Kramnik I, Weisbrod T T, Otsubo L, Cerny R, Miller L P, Jacobs W R, Jr, Bloom B R. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1998;95:13227–13232. doi: 10.1073/pnas.95.22.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashyam M D, Kaushal D, DasGupta S K, Tyagi A K. A study of mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beggs W H, Andrews F A. Nonspecific ionic inhibition of ethambutol binding by Mycobacterium smegmatis. Antimicrob Agents Chemother. 1973;4:115–119. doi: 10.1128/aac.4.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belanger A E, Besra G S, Ford M E, Mikusová K, Belisle J T, Brennan P J, Inamine J M. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc Natl Acad Sci USA. 1996;93:11919–11924. doi: 10.1073/pnas.93.21.11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besra G S, Khoo K-H, McNeil M R, Dell A, Morris H R, Brennan P J. A new interpretation of the structure of the mycolyl-arabinogalactan complex of Mycobacterium tuberculosis as revealed through characterization of oligoglycosylalditol fragments by fast-atom bombardment mass spectrometry and 1H nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:4257–4266. doi: 10.1021/bi00013a015. [DOI] [PubMed] [Google Scholar]
- 7.Blessington B, Beiraghi A. Study of the sterochemistry of ethambutol using chiral liquid chromatography and synthesis. J Chromatogr. 1990;522:195–203. [Google Scholar]
- 8.Brennan P J, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:39–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 9.Cheema S, Astora S, Khuller G K. Ethambutol induced leakage of phospholipids in Mycobacterium smegmatis. Int Res Commun Syst Med Sci. 1985;13:843–844. [Google Scholar]
- 10.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Dellagostin O A, Esposito G, Eales L-J, Dale J W, McFadden J. Activity of mycobacterial promoters during intracellular and extracellular growth. Microbiology. 1995;141:1785–1792. doi: 10.1099/13500872-141-8-1785. [DOI] [PubMed] [Google Scholar]
- 12.Deng L, Milkusová K, Robuck K G, Scherman M, Brennan P J, McNeil M R. Recognition of multiple effects of ethambutol on metabolism of mycobacterial cell envelope. Antimicrob Agents Chemother. 1995;39:694–701. doi: 10.1128/AAC.39.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes M, Kuck N A, Peets E A. Mode of action of ethambutol. J Bacteriol. 1962;84:1099–1103. doi: 10.1128/jb.84.5.1099-1103.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes M, Kuck N A, Peets E A. Effect of ethambutol on nucleic acid metabolism in Mycobacterium smegmatis and its reversal by polyamines and divalent cations. J Bacteriol. 1965;89:1299–1305. doi: 10.1128/jb.89.5.1299-1305.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heifets L B, Iseman M D, Lindholm-Levy P J. Ethambutol MICs and MBCs for Mycobacterium avium complex and Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1986;30:927–932. doi: 10.1128/aac.30.6.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwainsky H. EMB: mode of action, biotransformation and pharmacokinetics. In: Bartman K, editor. Antituberculosis drugs. Berlin, Germany: Springer-Verlag; 1988. pp. 533–540. [Google Scholar]
- 17.Khoo K-H, Douglas E, Azadi P, Inamine J M, Besra G S, Mikusová K, Brennan P J, Chatterjee D. Truncated structural variants of lipoarabinomannan in ethambutol drug-resistant strains of Mycobacterium smegmatis. Inhibition of arabinan biosynthesis by ethambutol. J Biol Chem. 1996;271:28682–28690. doi: 10.1074/jbc.271.45.28682. [DOI] [PubMed] [Google Scholar]
- 18.Kilburn J O, Takayama K, Armstrong E L, Greenberg J. Effects of ethambutol on phospholipid metabolism in Mycobacterium smegmatis. Antimicrob Agents Chemother. 1981;19:346–348. doi: 10.1128/aac.19.2.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lety M A, Nair S, Berche P, Escuyer V. A single point mutation in the embB gene is responsible for resistance to ethambutol in Mycobacterium smegmatis. Antimicrob Agents Chemother. 1997;41:2629–2633. doi: 10.1128/aac.41.12.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maddry J A, Suling W J, Reynolds R C. Glycosyltransferases as targets for inhibition of cell wall synthesis in M. tuberculosis and M. avium. Res Microbiol. 1996;147:106–112. doi: 10.1016/0923-2508(96)80211-7. [DOI] [PubMed] [Google Scholar]
- 21.McNeil M, Daffe M, Brennan P J. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J Biol Chem. 1990;265:18200–18206. [PubMed] [Google Scholar]
- 22.Mikusová K, Slayden R A, Besra G S, Brennan P J. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob Agents Chemother. 1995;39:2484–2489. doi: 10.1128/aac.39.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikusová K, Mikus M, Besra G S, Hancock I, Brennan P J. Biosynthesis of the linkage region of the mycobacterial cell wall. J Biol Chem. 1996;271:7820–7828. doi: 10.1074/jbc.271.13.7820. [DOI] [PubMed] [Google Scholar]
- 24.Mulder M A, Zappe H, Steyn L M. Mycobacterial promoters. Tuberc Lung Dis. 1997;78:211–223. doi: 10.1016/s0962-8479(97)90001-0. [DOI] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. Antimycobacterial susceptibility testing. Proposed standard M24-P. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 26.Paulin L G, Brander E E, Pîsî H J. Specific inhibition of spermidine synthesis in Mycobacteria spp. by the dextro isomer of ethambutol. Antimicrob Agents Chemother. 1985;28:157–159. doi: 10.1128/aac.28.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramaswamy S, Musser J M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 28.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 29.Silve G, Valero-Guillen P, Quemard A, Dupont M-A, Daffe M, Lanelle G. Ethambutol inhibition of glucose metabolism in mycobacteria: a possible target of the drug. Antimicrob Agents Chemother. 1993;37:1536–1538. doi: 10.1128/aac.37.7.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonnhammer E L L, Heijne G V, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of Sixth International Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 31.Sreevatsan S, Stockbauer K E, Pan X, Kreiswirth B N, Moghazeh S L, Jacobs W R, Jr, Telenti A, Musser J M. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother. 1997;41:1677–1681. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takayama K, Armstrong E L, Kunugi K A, Kilburn J O. Inhibition by ethambutol of mycolic acid transfer into the cell wall of Mycobacterium smegmatis. Antimicrob Agents Chemother. 1979;16:240–242. doi: 10.1128/aac.16.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takayama K, Kilburn J O. Inhibition of synthesis of arabinogalactan by ethambutol in Mycobacterium smegmatis. Antimicrob Agents Chemother. 1989;33:1493–1499. doi: 10.1128/aac.33.9.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Telenti A, Philipp W J, Sreevatsan S, Bernasconi C, Stockbauer K E, Wieles B, Musser J M, Jacobs W R., Jr The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nature Med. 1997;3:567–570. doi: 10.1038/nm0597-567. [DOI] [PubMed] [Google Scholar]
- 36.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson R G, Sheperd R G, Thomas J P, Baughn C. Stereospecificity of a new type of synthetic antituberculosis agent. J Am Chem Soc. 1961;83:2212–2213. [Google Scholar]
- 38.Winder F G. Mode of action of the antimycobacterial agents and associated aspects of the molecular biology of the mycobacteria. In: Ratledge C, Standford J, editors. The biology of the mycobacteria. Vol. 1. London, United Kingdom: Academic Press; 1982. pp. 354–442. [Google Scholar]
- 39.Wolucka B A, McNeil M R, de Hoffmann E, Chojnacki T, Brennan P J. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J Biol Chem. 1994;269:23328–23335. [PubMed] [Google Scholar]