Highlights
-
•
CEACAM6 is overexpressed in most lung adenocarcinomas.
-
•
CEACAM6 is significantly expressed in lung cancer cells of non-smokers.
-
•
Lung adenocarcinoma patients overexpressing CEACAM6 have shorter overall survival.
-
•
Exogenous CEACAM5/6 interacts with cell membrane-bound CEACAM6 in lung cancers.
-
•
CEACAM6 homophilic interactions inhibit anoikis through Src-FAK activation.
Keywords: CEACAM6, Anoikis, Intercellular interaction, Lung adenocarcinoma, Src-FAK pathway
Abbreviations: CEACAM, carcinoembryonic antigen-related cell adhesion molecule; EGFR, epidermal growth factor receptor; ELISA, enzyme-linked immunosorbent assay; FAK, focal adhesion kinase; FBS, fetal bovine serum; IHC, immunohistochemical; NSCLC, non-small cell lung cancer; PBS, phosphate-buffered saline; poly-HEMA, poly 2-hydroxyethyl meth-acrylate; scRNA, single cell RNA; SF, serum-free; Src, steroid receptor coactivator; TCGA-LUAD, The Cancer Genome Atlas Lung Adenocarcinoma; TMA, tissue microarray; TME, tumor microenvironment
Abstract
Among carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family proteins, CEACAM6 has received less attention than CEACAM5 and its presence and role in lung cancer are largely unknown. The application of CellphoneDB on the single cell RNA sequencing dataset showed that the homophilic interactions among CEACAM6 molecules, which are overexpressed in lung cancer cells were highly significant. CEACAM6 was overexpressed in 80.1% of lung adenocarcinomas and its overexpression had a significant relationship with non-smoking history and activating EGFR mutations. The effect of CEACAM6 overexpression on patient prognosis was evaluated using TCGA-LUAD dataset; the CEACAM6 overexpression group showed a shorter overall survival than that of the control group when matched for stage, age, sex, and pack-years. Immunoblotting of cell culture soup and ELISA of human derived material suggested that the majority of CEACAM6 was present on the cancer cell surface and interacted with other cancer cells in the crowded tumor microenvironment. Treatment with CEACAM6 showed CEACAM6 homophilic interactions in the cell membrane and anoikis inhibition through the activation of the Src-FAK pathway. Inhibition of CEACAM6 or its homophilic interactions in the cancer cell membrane may provide another therapeutic strategy for lung cancer.
Introduction
After the discovery of increased cancer-specific proteins in primary and metastatic colorectal cancers more than 50 years ago, the discovery of carcinoembryonic antigen-related family proteins in various cancer tissues, embryos, or fetus tissues has ensued [1,2]. The human carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family is a family of proteins encoded from 12 independent genes located on chromosome 19q13.1–13.2. Except for CEACAM16, which has two N-terminal domains, all CEACAM family proteins are composed of one variable-like N-terminal domain and 0–6 C-terminal constant type 2 (C2)-like immunoglobulin domains [3], [4], [5]. These domains, along with the manner in which they are attached to the membrane, characterize each protein [6]. The CEACAM5, 6, 7, and 8 proteins are attached to the cell membrane via a glycosylphosphatidylinositol linkage whereas CEACAM1, 3, 4, 19, 20, and 21 are anchored through the transmembrane domain [7].
The diverse subtypes and structures of the CEACAM protein subfamily show a varied distribution in the organs and are involved in various cellular functions, such as survival and proliferation, suppression of anoikis, metastasis, and sometimes tumor suppression [8], [9], [10]. Cancer-related CEACAM family proteins are: CEACAM5, also known as CEA; CEACAM6, which is also expressed in granulocytes and monocytes; and CEACAM1, which is most widely distributed in non-malignant epithelial tissues including lymphoid and myeloid cells [7]. Their biological function is mediated by dimerization resulting from homophilic or heterophilic interactions of the N-glycosylated extracellular domain. CEACAM1 and CEACAM5 are homodimerized through N-terminal immunoglobulin-variable domains [11,12]. CEACAM6 generates homodimers as well as heterodimers with CEACAM1, 5, or 8 [13,14]. The cis- or trans-dimerization between CEACAM family proteins is presumed to play an important role in the interaction of cell clusters in the tumor microenvironment (TME) composed of various cell types [15].
High expression levels of CEACAM6 have been reported in several neoplastic diseases, such as pancreatic adenocarcinomas, colorectal polyps, and early adenocarcinomas. In a study that used immunohistochemical (IHC) staining of tissue microarray (TMA), the expression level of CEACAM6 was relatively higher than that of CEACAM5 in breast cancer, pancreatic cancer, colorectal cancer, and non-small cell lung cancer (NSCLC) [16]. CEACAM6 is an important molecule that inhibits anoikis, a cell-death that occurs due to the loss of cell adhesion [17]. A study that used pancreatic cancer cells found that the expression level of CEACAM6 had a positive correlation with the inhibition of anoikis [18].
The mechanism by which CEACAM6, that does not have an intracellular domain, inhibits anoikis is not well understood. Bonsor et al. reported the energetic stability of the homodimeric interaction of CEACAM6 and the heterodimeric interaction of CEACAM6 and CEACAM8 through X-ray crystallographic structure analysis and analytical ultracentrifugation [15]. In a study by Camacho-Leal et al., immunofluorescence staining showed co-localization between CEACAM6 and integrin, but activation of the signaling system through physical interaction between CEACAM6 and integrin has not been reported [19]. The EGFR pathway and/or the Src-FAK pathway, both are positive regulator of cell survival, proliferation and migration, have been reported to be activated in cells that are forced to overexpress CEACAM6 [20,21].
Although the role of CEACAM6 and its overexpression has been reported in a few cancers, its clinical implications in lung adenocarcinoma is unknown. Because CEACAM6 is located on the cell surface, it is expected to play an important biological role in cell-cell interactions, but the existence and function of CEACAM6 in the TME of lung cancer has been limitedly studied. In this study, using human derived specimens, we investigated the overexpression of CEACAM6 in lung adenocarcinoma, its presence in the blood, and its effects on anoikis and the Src-FAK signaling system.
Materials and methods
Study cases and ethical approvals. Three-hundred and eleven cases fulfilling the enrollment and exclusion criteria described below were recruited from the institutional tissue archive and assessed using TMA. The enrollment criteria included: (1) cases pathologically confirmed as lung adenocarcinoma from December 2010 to May 2018, (2) patients who underwent lung resection for treatment purposes, (3) cases where residual blocks were available for TMA production in the institutional tissue bank, and (4) cases in which EGFR mutation status was identified from tissue. Patients who received adjuvant or neoadjuvant chemotherapy were excluded. All hematoxylin and eosin stained slides from resected lung adenocarcinoma specimens were reviewed and representative areas were marked on them. Tissue cores (3 mm) were extracted from the matched formalin-fixed paraffin-embedded tumor blocks and placed into 6 × 5 recipient TMA blocks. To measure the concentration of CEACAM6 in the blood, the plasma of 29 patients with lung cancer of various stages and 11 patients with chronic obstructive pulmonary disease (COPD) (control group) were randomized from the institutional tissue archive and measured by ELISA. This study was approved by the Institutional Review Board of our institution (IRB # 4-2021-1206).
Estimation of intercellular interaction. Interactions between lung cancer cells from the scRNA analysis dataset of non-smoker lung adenocarcinomas were analyzed using CellPhoneDB v.2.0 [22]. The 10X chromium-based scRNA sequencing datasets were obtained from previously reported non-smoker lung cancers [23] (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA773987). We applied the Infercnv v1.10.1 package on the dataset to obtain the barcodes of epithelial cells inside the tumor tissue that showed significant copy number aberrations, which were then defined as lung cancer cells [24]. In parallel, the batch effect of the entire scRNA sequencing dataset was corrected by the integration function of the Seurat v4.0.4 package in R and made into one object [25]. Subsequently, the seurat object composed only of cancer cells was extracted using the barcodes obtained previously. A count_matrix file and annotation file were then generated, and CellphoneDB was used with the following options: threshold=0.333, result-precision=5, pvalue=0.001, and threads=12. Lastly, we applied the ‘secreted == True’ and ‘is_integrin==False’ options to secure meaningful interactions.
Immunoblotting. All cell lines used for in vitro experiments were purchased from the Korean Cell Line Bank (https://cellbank.snu.ac.kr/main/index.html). Immunoblotting was performed as described by Kim et al. [26] and the antibodies used for this study are listed in Supplementary Table 1. Cells treated with 50 ng/mL platelet-derived growth factor (R&D systems, Cat No: 200BB010) were used as positive controls for Src-FAK pathway activation analyses. The expression level of each protein was measured using ImageJ v1.53 g (http://rsbweb.nih.gov/ij/) and quantified relative to β-actin.
Immunohistochemistry and immunocytochemistry. IHC staining was performed according to the manufacturer's protocol using the LABS2 system (Dako, Carpentaria, CA, USA). CEACAM6 expression was evaluated using the H-score system, which captures both the intensity and the proportion of CEACAM6 expression and comprises of values between 0 and 300 [27]. For immunocytochemistry, cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS), incubated in either serum-free (SF) RPMI media or SF RPMI media containing 10 ng/mL of Recombinant Human CEACAM-6/CD66c Protein (R&D systems, Cat No: 3934-CM), and attached with His-Tag for 10 min. Staining was performed as described elsewhere [26] and imaging was taken with a confocal laser-scanning microscope (Zeiss, LSM780) with the ZEN 2.3 SP1 software (Zeiss) and stored as a CZI image format. Quantification of the colocalization between endogenous CEACAM6 and exogenous CEACAM5 or 6 was performed by Coloc 2 function of ImageJ [28].
Enzyme-linked immunosorbent assay. Plasma samples were analyzed using a Human CD66c/CEACAM6 (Sandwich ELISA) ELISA Kit (LSBio, Cat No: LS-F7305) for human CEACAM6 according to the manufacturer's instructions. The concentrations were calculated using the protein standards included in the kits.
Live-dead and anoikis assays. The effect of CEACAM6 on cell attachment and survival in an FBS-free culture environment was observed using the following protocol. Firstly, 5 × 105 cells were suspended in SF RPMI media supplemented with or without CEACAM6 at 10 ng/mL and plated on a 24-well Thermo Scientific™ Nunc™ Cell-Culture Treated Multidish Culture Plate® (Thermo Scientific™). After 16 h, cells were stained with the LIVE/DEAD™ Viability/Cytotoxicity Kit (Invitrogen™, Cat No: L3224) and observed with an EVO fluorescent microscope system (Leica) at 494 and 517 nm. Inhibition of anoikis by CEACAM6 was observed as described elsewhere [21,29]. Briefly, cells suspended either in 2 mL RPMI media supplemented with 5% FBS or SF RPMI media, were incubated in the poly-HEMA-coated (Sigma, Cat No: P3932) wells with or without addition of CEACAM6 for 16 h. Following the induction of anoikis, cells were stained with the LIVE/DEAD™ Viability/Cytotoxicity Kit and the image was captured with the EVO fluorescent microscope system. The anoikis fraction (percentage of cells undergoing anoikis) was calculated by using ImageJ (Green: Live, Red: Dead). All observations were repeated in triplicate and the mean ± SD reported.
Statistical analysis. Differences between groups were analyzed using the Student's t-test, multifactorial analysis of variance of initial measurements, and Mann-Whitney U test for nonparametric data (as appropriate) using R v4.1.0 (https://www.r-project.org/). Propensity score matching of TCGA-LUAD data was performed using MatchIt v4.3.0 [30] and survival analysis was performed using survival v3.2-13 and packHV v2.2 R packages. Multicollinearity was tested using the HH package version 3.1-43 and no interaction was inferred when an r value of less than 10 was shown. Statistical significance was considered when P < 0.05.
Results
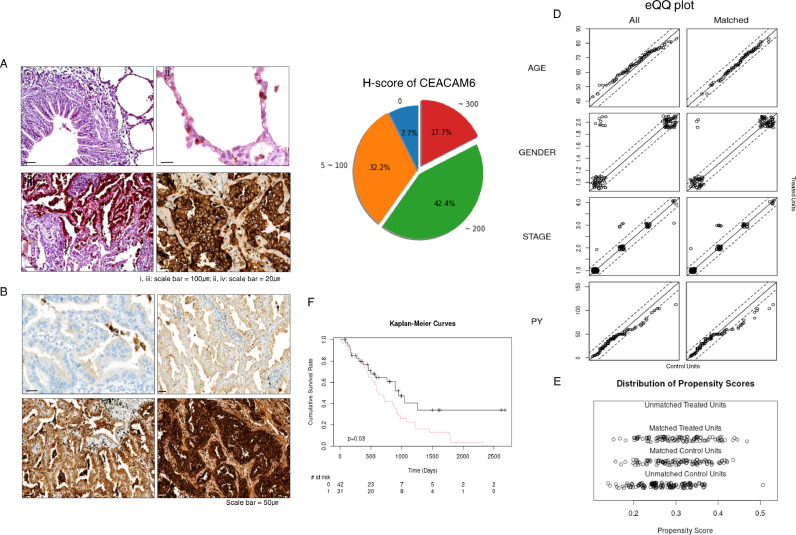
CEACAM6 is frequently overexpressed in lung adenocarcinomas. In a previous single-cell transcriptome study using non-smoker lung cancer cells, CEACAM6 was one of the most differentially expressed genes overexpressed in lung cancer cells when compared to the adjacent normal-appearing pulmonary epithelial cells [23]. To identify the candidate molecules involved in inter-cancer-cell interactions, we analyzed that same dataset using CellPhoneDB v.2.0 [22,23]. Table 1 shows a list of interacting molecules that were significant ligand-receptor pairs in lung cancer cell of non-smokers (A full list of interactions is shown in the supplementary Table 2). Among them, CEACAM6 was one of the molecules specifically and significantly overexpressed in lung cancer cells compared to that in normal-appearing adjacent lung epithelial cells [23]. We further investigated CEACAM6 overexpression in 311 lung adenocarcinoma samples and found that 60.1% showed moderate to high expression levels, 32.2% showed low expression levels, and only 7.7% of the samples showed no expression (Fig. 1A–C). Table 2 shows the basic demographic and molecular pathological characteristics of the samples used.
Table 1.
List of interacting molecules within lung cancer cell of non-smokers.
| Interacting pair | Secreted | Annotation_strategy | Significant_means* | P-values** |
| CD74 : APP | No | Curated | 10.1916 | <0.001 |
| CEACAM6 : CEACAM6 | No | Curated | 9.9630 | <0.001 |
| CD74 : MIF | Yes | I2D, InnateDB-All | 9.8360 | <0.001 |
| CD74 : COPA | Yes | IMEx, IntAct | 9.4922 | <0.001 |
| ICAM1 : AREG | Yes | InnateDB-All | 7.5263 | <0.001 |
| EGFR : AREG | Yes | Curated | 6.6550 | <0.001 |
| CEACAM5 : CEACAM6 | No | Curated | 6.4427 | <0.001 |
| CD55 : ADGRE5 | Yes | Curated | 6.3808 | <0.001 |
| CXCL2 : DPP4 | Yes | Curated | 3.1502 | <0.001 |
| CADM1 : CADM1 | No | Curated | 2.4790 | <0.001 |
| LGALS9 : CD47 | Yes | InnateDB-All | 2.2030 | <0.001 |
| EFNA1 : EPHA2 | Yes | Curated | 1.9217 | <0.001 |
| CD44 : HBEGF | Yes | I2D, InnateDB-All | 1.9061 | <0.001 |
| EGFR : GRN | Yes | InnateDB-All | 1.8111 | <0.001 |
| EGFR : MIF | Yes | IMEx, InnateDB-All | 1.7214 | <0.001 |
*Significant_means denotes the significant mean calculation for all the interacting partners. If P < 0.05, the value will be the mean; alternatively, the value is set to 022.
** P values for all the interacting partners: p-value refers to the enrichment of the interacting ligand-receptor pair in each of the interacting pairs of cell types22.
Fig. 1.
CEACAM6 is overexpressed in the majority of lung adenocarcinomas and is associated with poor prognosis. (A) Representative image of CEACAM6 immunohistochemical staining (IHC) of (ⅰ) bronchial epithelium, (ⅱ) alveolar structure, (ⅲ) transitional area from normal structure to lung cancer, and (ⅳ) lung cancer showing strong expression in the cell membrane. (B) Representative IHC staining of lung adenocarcinoma showing (ⅰ) negative, (ⅱ) mild, (ⅲ) moderate, and (ⅳ) strong CEACAM6 expression. (C) Pie chart showing the frequency of H-score for CEACAM6 expression in lung adenocarcinoma. The number outside the circle denotes the H-score of CEACAM6. 0; no, 5–100; low, ∼ 200; moderate, and ∼ 300; strong expression. (D) Empirical quantile-quantile (eQQ) plots before and after propensity score matching for age, gender, stage, and pack-year (PY). (E) Distribution of propensity scores before and after matching for age, gender, stage and PY. “Treated” refers to the top one third of TCGA-LUAD cases overexpressing CEACAM6. (F) Cumulative survival rate of TCGA-LUAD cases according to the CEACAM6 expression obtained by the Kaplan-Meier estimator.
Table 2.
Demographic and molecular pathological characteristics of the study population used for CEACAM6 immunohistochemistry.
| Parameters | Number of cases | 311 |
| Age (median [range], years) | 66 years | 26–86 |
| Gender (n, (%)) | Male | 165 (53.1%) |
| Female | 146 (46.9%) | |
| Stage | I | 189 |
| II | 43 | |
| III | 24 | |
| IV | 2 | |
| N.A. | 53 | |
| Location of primary Tumor* | RUL | 101 |
| LUL | 74 | |
| RLL | 48 | |
| LLL | 47 | |
| RML | 15 | |
| LUL_LLL | 8 | |
| RUL_RML | 10 | |
| RUL_RLL | 5 | |
| RML_RLL | 3 | |
| Smoking | Non-smoker | 233 |
| Ex-smoker | 48 | |
| Current smoker | 30 | |
| Predominant pattern | Acinar | 211 |
| Solid | 39 | |
| Papillary | 34 | |
| Lepidic | 20 | |
| Micropapillary | 7 | |
| EGFR | Wild type | 116 |
| L858R | 96 | |
| E19D | 85 | |
| **Rare mutation | 10 | |
| †Compound mutations | 4 |
* RUL: right upper lobe, RML: right middle lobe, RLL: right lower lobe, LUL: left upper lobe, LLL: left lower lobe.
** Rare mutations include E20ins, G719A, L861Q, V786M, G719X, G719A.
† Compound mutations include S768I + V774M, G719C + S768I, E19D + T790M, E19Del + L858R.
CEACAM6 is more strongly expressed in lung cancer cells of non-smokers than in that of smokers. Demographic, pathological, and molecular factors related to CEACAM6 overexpression in these samples were investigated through regression analysis. Interestingly, non- and ex-smokers and the presence of EGFR activation mutations were significantly associated with overexpression of CEACAM6 (P = 0.0013, P = 0.0228, and P = 0.0152, respectively; linear regression). Further evaluation by multivariate analysis revealed a significant relationship between CEACAM6 overexpression and ex- and non-smokers (Table 3). To clarify the relationship between smoking history and EGFR mutations, a multicollinearity test was performed; the VIF score was 2.2 for ex-smokers, 2.3 for non-smokers, and 1.1 for EGFR mutants, suggesting that multicollinearity does not exist between these parameters.
Table 3.
Univariate and multivariate analysis predicting CEACAM6 overexpression in lung adenocarcinoma.
| Univariate | Multivariate | ||||
| OR (95% CI) | P-value* | OR (95% CI) | P-value* | ||
| Age | 1.3747 (0.5096–3.7082) | 0.5285 | – | – | |
| Gender | Male | Reference | – | – | |
| Female | 3.8889 X 103 (0–2.8085 X 1012) | 0.4259 | – | – | |
| Stage | I | Reference | – | – | |
| II | 0.2484(0–3.4530 X 1012) | 0.9278 | – | – | |
| III | 0.0011(0–7.7045 X 1013) | 0.7289 | – | – | |
| IV | 0 (0–8.0545 X 1025) | 0.2962 | – | – | |
| Smoking | Current smoker | Reference | Reference | ||
| Ex-smoker | 6.3226 X 1020 (8.1209 X 102–4.9225 X 1038) | 0.0228 | 6.5863 X 1019 (9.2860 X 10–4.6716 X 1037) | 0.0297 | |
| Non-smoker | 3.4659 X 1024 (4.2514 X 109–2.8255 X 1039) | 0.0013 | 5.1067 X 1021 (3.6028 X 106–7.2383 X 1036) | 0.0051 | |
| EGFR mutation | Wild | Reference | reference | ||
| Mutant | 1.7641 X 1011 (1.5211 X 102–2.0460 X 1020) | 0.0152 | 5.3840 X 108 (0.31110–9.3170 X 1017) | 0.0639 |
* P-value was obtained from linear regression.
Lung adenocarcinoma patients overexpressing CEACAM6 have a poor prognosis. The effect of CEACAM6 overexpression on clinical outcome was further evaluated using TCGA-LUAD dataset from the Genomic Data Commons data portal. (https://portal.gdc.cancer.gov/). First, the top third TCGA-LUAD cases overexpressing CEACAM6 were extracted based on the RNA-seq by expectation maximization1 value using OncoLnc (http://www.oncolnc.org/)2. In parallel, we selected cases in TCGA-LUAD dataset for which stage, age, sex, pack-year, and survival data were available on the clinical dataset as on October 1, 2021. Using these data, survival analysis was performed by 1:1 matching of the propensity scores for stage, age, sex, and pack-year of the CEACAM6 overexpression group and control group3 (Fig. 1D and E). The group overexpressing CEACAM6 and the matched control dataset are shown in Supplementary Table 3. When the effect of CEACAM6 overexpression on prognosis was evaluated using the TGCA-LUAD dataset, patients in the CEACAM6 overexpression group showed a significantly shorter overall survival than those in the control group when matched for stage, age, sex, and smoking history (Fig. 1F, P = 0.030, LogRank test).
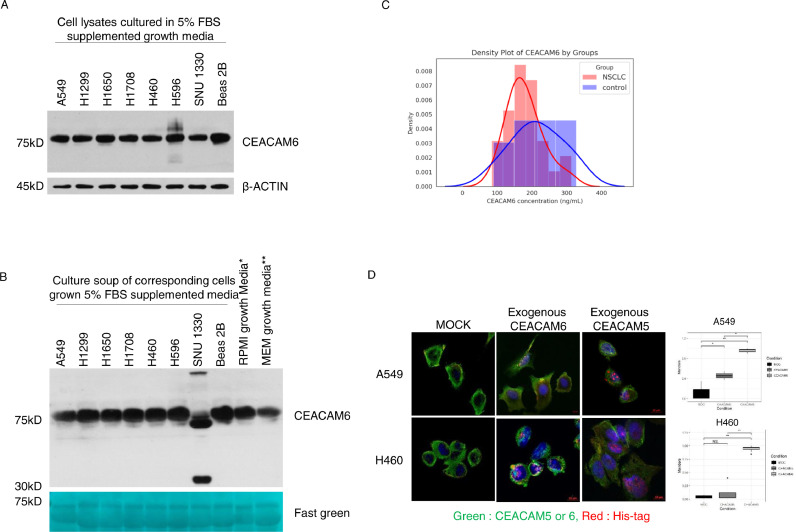
Cell surface-attached CEACAM6, not secreted form, mediates its role in TME. To further evaluate the role of CEACAM6 in the lung cancer TME, we first performed immunoblotting using lysates and culture media that were obtained during lung cancer cell culture. Immunoblotting of the cells grown in fetal bovine serum (FBS)-supplemented growth media showed strong expression of CEACAM6 whereas that of the cells maintained in FBS-free media showed no or very weak expression (Fig. 2A, Supplementary Fig. 1A). These findings suggest that the expression of CEACAM6 in lung cancer cells is strongly influenced by the presence of FBS in the culture environment. Therefore, to detect the presence of CEACAM6 in the cell culture environment, culture soups with NSCLC cells grown in FBS-supplemented media and FBS-supplemented fresh media were blotted for CEACAM6 (Fig. 2B). The results showed strong expression of CEACAM6 at 75 kDa, which is corresponding to the size of the glycosylated form, in culture soup of all lung cancer cells and in FBS-supplemented fresh media. Interestingly, new bands from cancer cell origin were observed only in one culture soup out of total eight cell lines. In other words, in the culture soup of SNU1330 cells, which harbor homozygous deletions in exon 19 of EGFR, new bands of approximately 37 kDa and 210 kDa were shown. To detect the CEACAM6 secreted from lung cancer cells—which may be masked by added FBS—we immunoblotted (1) culture soups with NSCLC cells grown in FBS-free media, (2) FBS-free culture media, and (3) FBS-supplemented growth media. CEACAM6 was observed only in FBS-supplemented media (Supplementary Fig. 1B). These findings suggested that CEACAM6 was present in large amounts in the cell culture environment, which was related to the expression of CEACAM6 in lung cancer cells and not secreted to the media in the most of the cells.
Fig. 2.
Exogenous CEACAM5/6 interacts with membrane-bound CEACAM6 in lung cancer cells. (A) Immunoblotting for CEACAM6 using the lysate of various lung cancer cells maintained in 5% fetal bovine serum (FBS) supplemented media. (B) Immunoblotting of culture soup of the indicated lung cancer cells incubated in growth media supplemented with 5% FBS. (C) Kernel density plot of plasma CEACAM6 concentrations from 29 lung adenocarcinomas and 11 control patients. (D) Interaction and co-localization of endogenous CEACAM6 and exogenous His-Tagged CEACAM5/6 in lung cancer cells Co-localization results are shown in yellow. (E) Histogram showing the quantification of colocalization using Mander's colocalization index. *p < 0.05, **p < 0.01, ***p < 0.001 (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
To investigate the presence and concentration of circulating CEACAM6 in the blood of lung cancer patients, a pilot test with immunoblotting and enzyme-linked immunosorbent assay (ELISA) was conducted using plasma from patients with COPD as a control group. The average plasma concentration of CEACAM6 in 29 lung cancer patients with various stages was 171.1 ng/mL (149.2–203.6 ng/mL), whereas that of the 11 control patients with COPD was 211.1 ng/mL (176.3–263.4 ng/mL). No significant difference in plasma CEACAM6 was observed between the two groups (Fig. 2C). Besides, no significant relationships were observed between CEACAM6 plasma concentration and parameters such as stage, tumor size, and other clinical measures (data not shown). Collectively, these results suggest that most CEACAM6 interacts with other cancer cells in the crowded TME as a cell surface attached form.
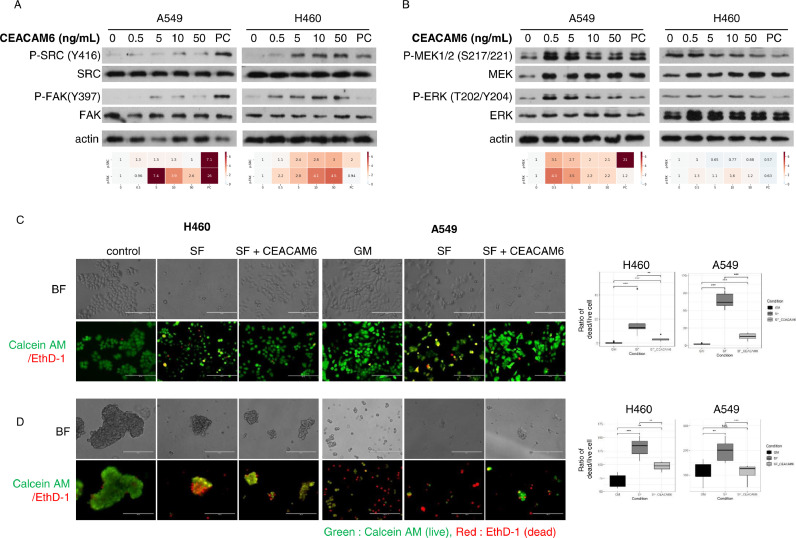
Homophilic interactions of CEACAM6 inhibit anoikis through Src-FAK pathway activation. To simulate the interaction between CEACAM6 on the surface of lung cancer cells and CEACAM5 or CEACAM6 in the TME, synthetic CEACAM5 or CEACAM6 attached with 6X His-Tags were treated in SF conditions, stained with His antibodies, and observed with a confocal microscope (Fig. 2D). Most of the signals observed on the cell surface were derived from the co-localization of His-tagged CEACAM6 and endogenous CEACAM6, whereas the co-localization of CEACAM5 and CEACAM6 was barely observed (Fig. 2D and E). Previous reports indicated that intracellular overexpression of CEACAM6 or modulation of N-glycosylation induce the activation of signaling systems which are involved in cell fates [20,21]. The effect of CEACAM6 on these signaling systems was observed by gradually increasing the concentration of CEACAM6 treatment in the cells (Fig. 3A and B). Among the signaling pathways evaluated, the activation of the Src-FAK signaling system was observed in a CEACAM6 dose-dependent manner, which lead to the phosphorylation of MAPK/extracellular signal-regulated kinase 1/2 (MEK1/2) and extracellular signal-regulated kinase (ERK). On the other hand, there was no effect on EGFR phosphorylation (Supplementary Fig. 2). As a comparison, cells were treated in the same manner with CEACAM5, but no effect on Src-FAK signaling was observed (Supplementary Fig. 3). These findings indicate that CEACAM6, not CEACAM5, interacts with itself in a trans- manner and activates the Src-FAK pathway, which has a positive effect on cancer cell fate. The effect of the homophilic interaction between exogenous CEACAM6 and its cell membrane-bound form on cell survival was further evaluated using poly 2-hydroxyethyl meth-acrylate (poly-HEMA) coated culture plate. Unlike previous studies that overexpressed CEACAM6 by introducing its gene [31] or silenced CEACAM6 using small interfering RNA [18], we investigated the biologic effect of CEACAM6 by adding exogenous CEACAM6 to SF culture media (Fig. 3C and D). When CEACAM6 was added to FBS-free media while culturing cells in a poly-HEMA coated culture plate, anoikis was significantly reduced in the CEACAM6-treated group compared to that in the control group. These findings suggest that CEACAM6 in the TME may play an important role in the inhibition of anoikis through the activation of the Src-FAK signaling system.
Fig. 3.
Exogenous CEACAM6 activates Src-FAK signaling and inhibits anoikis. (A) The dose-dependent effect of CEACAM6 on Src-FAK. (B) MEK-ERK phosphorylation. Top: Immunoblots. Bottom: Relative expression ratio heat maps of phospho- to total-form proteins. Increasing color darkness indicates increasing relative expression ratios. (C) Representative fluorescent images (left) and histogram (right) of the CEACAM6 effect on the fate of lung cancer cells grown in serum-free (SF) media and SF media with CEACAM6. (D) Representative fluorescence images (left) and histograms (right) of lung cancer cells cultured under the same conditions as (C) on poly-HEMA-coated plates. Images of the cells cultured in 5% fetal bovine serum supplemented medium were used as controls. BF: bright field. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
CEACAM6 has received less attention than CEACAM5 but has recently gained increasing interest as it is often overexpressed in early-stage NSCLC [23,32]. Studies on the clinical implications of CEACAM6 overexpression or its effect on cellular signaling systems in lung cancer are very limited. This study showed that CEACAM6 is frequently overexpressed in lung adenocarcinoma, adversely affects patient prognosis, and inhibits anoikis by activation of Src-FAK signaling through homophilic interactions.
Unlike CEACAM1, 3, 4, 19, 20, and 21, which have a bona fide transmembrane domain, CEACAM5 and 6 are loosely attached to the membrane with a glycosylphosphatidylinositol anchor composed of 26 hydrophobic amino acids. Therefore, it was hypothesized that CEACAM6, like CEACAM5, could be easily released into and detected in the blood circulation, but this was not observed in our pilot study using plasma. One possible reason is that CEACAM5, which has a total of six C2-like domains, can be more easily detached from the cell surface by phosphatidylinositol-specific phospholipase C or physical interactions compared to CEACAM6, which has only two C2-like domains.
A clinical study using the anti-CEACAM6 monoclonal antibody, Tinurilimab (NTC03596372; Bay 1834942), is in progress based on the rationale that CEACAM6 is an immune checkpoint regulator suppressing the activity of effector T-cells against tumors (https://clinicaltrials.gov/ct2/show/NCT03596372) [33]. The researchers showed that when CEACAM6 was overexpressed or attached to cancer cells using beads, it inhibited the phosphorylation of zeta-chain-associated protein kinase 70 to suppress T-cell receptor-mediated T-cell responses. Furthermore, the Tinurilimab showed an increased tumor cell killing effect in the tumor-cell/T-cell co-culture system [34,35]. Our findings provide additional evidence that CEACAM6 is a possible target candidate for the treatment of lung cancer.
However, this study had some limitations. First, the dataset used to evaluate CEACAM6 overexpression in lung adenocarcinoma tissues, the dataset used to analyze the effect of overexpression on lung cancer patient prognosis, and the blood samples used to identify the presence of circulating CEACAM6 were different from each other. In the TMA dataset obtained from the patients who underwent lung resection at the research institution, information on cancer-related events such as recurrence and cancer-death were not included; therefore, TCGA-LUAD dataset was used to analyze the effect on prognosis. In addition, since blood samples were obtained from other datasets, the inability to compare the CEACAM6 expression levels and the blood concentration levels was a limitation. Second, a significant proportion of the lung cancer samples were from non-smokers and carried EGFR- tyrosine kinase inhibitors (TKI) sensitizing mutations. Our results were obtained from a dataset with high frequencies of non-smokers, women, and EGFR-TKI mutations which may have biased the conclusion that the majority of lung cancers show CEACAM6 overexpression. Since overexpression is also shown in single cell (scRNA) sequencing data [32] and another IHC study [36] with various cancer tissues, it would be easy to generalize the fact that CEACAM6 is overexpressed in most lung cancers. Third, CEACAM5/6 used for in vitro experimentation was not in glycosylated form. Glycosylation, a post-translational modification related to protein folding, localization, and stability, has a great influence on protein interaction and intracellular signal transduction [37]. Chiang et al. showed that when N-glycosylation of CEACAM6 on the cell surface was quenched with swainsonine and N-Acetyl-Glucosaminyltransferase 5 short hairpin RNA, its interaction with EGFR and EGFR signaling decreased [20]. It may be necessary to use the N-glycosylated form of CEACAM6 for a more precise evaluation of the binding or signaling effects. Finally, the identification of motifs involved in the homophilic interaction of CEACAM6 and the R&D of the tools controlling interaction were left for further research tasks. If the antibodies or chemicals, which block the interaction of CEACAM6 demonstrate, inhibition of the FAK-Src pathway and restoration of anoikis, it would be additional evidence supporting their clinical development. Along with the recent report by Pinkert et al., their humanized monoclonal antibody selectively blocking CEACAM6 would be of great help in solving this problem [35].
Collectively, CEACAM6 is overexpressed in most lung adenocarcinomas and adversely affects patient prognosis. CEACAM6 on the surface of lung cancer cells inhibits anoikis through the activation of Src-FAK signaling through homophilic trans-interactions with adjacent cancer cells. These findings suggest that CEACAM6 contributes to a positive survival feedback loop between cancer cells in a crowded TME.
CRediT authorship contribution statement
Eun Young Kim: Data curation, Formal analysis, Methodology, Writing – review & editing, Writing – original draft. Yoon Jin Cha: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Sukin Jeong: Investigation, Writing – review & editing. Yoon Soo Chang: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This study was supported by the National Research Foundation of the Republic of Korea (Grant No. NRF-2020R1A2B5B01001883) awarded to YS Chang.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101402.
Appendix. Supplementary materials
References
- 1.Gold P., Freedman S.O. Specific carcinoembryonic antigens of the human digestive system. J. Exp. Med. 1965;122:467–481. doi: 10.1084/jem.122.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gold P., Freedman S.O. Demonstration of tumor-specific antigens in human colonic carcinomata by immunological tolerance and absorption techniques. J. Exp. Med. 1965;121:439–462. doi: 10.1084/jem.121.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zebhauser R., Kammerer R., Eisenried A., McLellan A., Moore T., Zimmermann W. Identification of a novel group of evolutionarily conserved members within the rapidly diverging murine Cea family. Genomics. 2005;86:566–580. doi: 10.1016/j.ygeno.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Thompson J.A., Pande H., Paxton R.J., Shively L., Padma A., Simmer R.L., Todd C.W., Riggs A.D., Shively J.E. Molecular cloning of a gene belonging to the carcinoembryonic antigen gene family and discussion of a domain model. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2965–2969. doi: 10.1073/pnas.84.9.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauchemin N., Benchimol S., Cournoyer D., Fuks A., Stanners C.P. Isolation and characterization of full-length functional cDNA clones for human carcinoembryonic antigen. Mol. Cell. Biol. 1987;7:3221–3230. doi: 10.1128/mcb.7.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmermann W., Ortlieb B., Friedrich R., von Kleist S. Isolation and characterization of cDNA clones encoding the human carcinoembryonic antigen reveal a highly conserved repeating structure. Proc. Natl. Acad. Sci. U. S. A. 1987;84:2960–2964. doi: 10.1073/pnas.84.9.2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beauchemin N., Arabzadeh A. Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) in cancer progression and metastasis. Cancer Metastasis Rev. 2013;32:643–671. doi: 10.1007/s10555-013-9444-6. [DOI] [PubMed] [Google Scholar]
- 8.Tchoupa A.K., Schuhmacher T., Hauck C.R. Signaling by epithelial members of the CEACAM family - mucosal docking sites for pathogenic bacteria. Cell Commun. Signal. 2014;12:27. doi: 10.1186/1478-811X-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray-Owen S.D., Blumberg R.S. CEACAM1: contact-dependent control of immunity. Nat. Rev. Immunol. 2006;6:433–446. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 10.Singer B.B., Scheffrahn I., Kammerer R., Suttorp N., Ergun S., Slevogt H. Deregulation of the CEACAM expression pattern causes undifferentiated cell growth in human lung adenocarcinoma cells. PLoS One. 2010;5:e8747. doi: 10.1371/journal.pone.0008747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fedarovich A., Tomberg J., Nicholas R.A., Davies C. Structure of the N-terminal domain of human CEACAM1: binding target of the opacity proteins during invasion of Neisseria meningitidis and N. gonorrhoeae. Acta. Crystallogr. D Biol. Crystallogr. 2006;62:971–979. doi: 10.1107/S0907444906020737. [DOI] [PubMed] [Google Scholar]
- 12.Korotkova N., Yang Y., Trong I.Le, Cota E., Demeler B., Marchant J., Thomas W.E., Stenkamp R.E., Moseley S.L., Matthews S. Binding of Dr adhesins of Escherichia coli to carcinoembryonic antigen triggers receptor dissociation. Mol. Microbiol. 2008;67:420–434. doi: 10.1111/j.1365-2958.2007.06054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oikawa S., Inuzuka C., Kuroki M., Arakawa F., Matsuoka Y., Kosaki G., Nakazato H. A specific heterotypic cell adhesion activity between members of carcinoembryonic antigen family, W272 and NCA, is mediated by N-domains. J. Biol. Chem. 1991;266:7995–8001. [PubMed] [Google Scholar]
- 14.Skubitz K.M., Skubitz A.P. Interdependency of CEACAM-1, -3, -6, and -8 induced human neutrophil adhesion to endothelial cells. J. Transl. Med. 2008;6:78. doi: 10.1186/1479-5876-6-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonsor D.A., Günther S., Beadenkopf R., Beckett D., Sundberg E.J. Diverse oligomeric states of CEACAM IgV domains. Proc. Natl. Acad. Sci. 2015;112:13561–13566. doi: 10.1073/pnas.1509511112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumenthal R.D., Leon E., Hansen H.J., Goldenberg D.M. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer. 2007;7:2. doi: 10.1186/1471-2407-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paoli P., Giannoni E., Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta. 2013;1833:3481–3498. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Duxbury M.S., Ito H., Zinner M.J., Ashley S.W., Whang E.E. CEACAM6 gene silencing impairs anoikis resistance and in vivo metastatic ability of pancreatic adenocarcinoma cells. Oncogene. 2004;23:465–473. doi: 10.1038/sj.onc.1207036. [DOI] [PubMed] [Google Scholar]
- 19.Camacho-Leal P., Zhai A.B., Stanners C.P. A co-clustering model involving alpha5beta1 integrin for the biological effects of GPI-anchored human carcinoembryonic antigen (CEA) J. Cell. Physiol. 2007;211:791–802. doi: 10.1002/jcp.20989. [DOI] [PubMed] [Google Scholar]
- 20.Chiang W.F., Cheng T.M., Chang C.C., Pan S.H., Changou C.A., Chang T.H., Lee K.H., Wu S.Y., Chen Y.F., Chuang K.H., Shieh D.B., Chen Y.L., Tu C.C., Tsui W.L., Wu M.H. Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) promotes EGF receptor signaling of oral squamous cell carcinoma metastasis via the complex N-glycosylation. Oncogene. 2018;37:116–127. doi: 10.1038/onc.2017.303. [DOI] [PubMed] [Google Scholar]
- 21.Duxbury M.S., Ito H., Ashley S.W., Whang E.E. CEACAM6 cross-linking induces caveolin-1-dependent, Src-mediated focal adhesion kinase phosphorylation in BxPC3 pancreatic adenocarcinoma cells. J. Biol. Chem. 2004;279:23176–23182. doi: 10.1074/jbc.M402051200. [DOI] [PubMed] [Google Scholar]
- 22.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 23.Kim E.Y., Cha Y.J., Lee S.H., Jeong S., Choi Y.J., Moon D.H., Lee S., Chang Y.S. Early lung carcinogenesis and tumor microenvironment observed by single-cell transcriptome analysis. Transl. Oncol. 2022;15 doi: 10.1016/j.tranon.2021.101277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., Deschler D.G., Varvares M.A., Mylvaganam R., Rozenblatt-Rosen O., Rocco J.W., Faquin W.C., Lin D.T., Regev A., Bernstein B.E. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624. doi: 10.1016/j.cell.2017.10.044. e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E.P., Jain J., Srivastava A., Stuart T., Fleming L.M., Yeung B., Rogers A.J., McElrath J.M., Blish C.A., Gottardo R., Smibert P., Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587. doi: 10.1016/j.cell.2021.04.048. e3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E.Y., Lee J.G., Lee J.M., Kim A., Yoo H.C., Kim K., Lee M., Lee C., Han G., Han J.M., Chang Y.S. Therapeutic effects of the novel Leucyl-tRNA synthetase inhibitor BC-LI-0186 in non-small cell lung cancer. Ther. Adv. Med. Oncol. 2019;11 doi: 10.1177/1758835919846798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyerholz D.K., Beck A.P. Principles and approaches for reproducible scoring of tissue stains in research. Lab. Investig. 2018;98:844–855. doi: 10.1038/s41374-018-0057-0. [DOI] [PubMed] [Google Scholar]
- 28.Dunn K.W., Kamocka M.M., McDonald J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ordoñez C., Screaton R.A., Ilantzis C., Stanners C.P. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res. 2000;60:3419–3424. [PubMed] [Google Scholar]
- 30.Austin P.C. An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivar. Behav. Res. 2011;46:399–424. doi: 10.1080/00273171.2011.568786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duxbury M.S., Ito H., Benoit E., Zinner M.J., Ashley S.W., Whang E.E. Overexpression of CEACAM6 promotes insulin-like growth factor I-induced pancreatic adenocarcinoma cellular invasiveness. Oncogene. 2004;23:5834–5842. doi: 10.1038/sj.onc.1207775. [DOI] [PubMed] [Google Scholar]
- 32.Sinjab A., Han G., Treekitkarnmongkol W., Hara K., Brennan P.M., Dang M., Hao D., Wang R., Dai E., Dejima H., Zhang J., Bogatenkova E., Sanchez-Espiridion B., Chang K., Little D.R., Bazzi S., Tran L.M., Krysan K., Behrens C., Duose D.Y., Parra E.R., Raso M.G., Solis L.M., Fukuoka J., Zhang J., Sepesi B., Cascone T., Byers L.A., Gibbons D.L., Chen J., Moghaddam S.J., Ostrin E.J., Rosen D., Heymach J.V., Scheet P., Dubinett S.M., Fujimoto J., Wistuba I.I., Stevenson C.S., Spira A., Wang L., Kadara H. Resolving the spatial and cellular architecture of lung adenocarcinoma by multiregion single-cell sequencing. Cancer Discov. 2021;11:2506–2523. doi: 10.1158/2159-8290.CD-20-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willuda J., Boehm H.H., Pinkert J., Trautwein M., Doecke W.-.D., Ahsen O.v., Ziegelbauer K., Offringa R., Kreft B., Beckhove P. Increased T cell- activation resulting from the combination of the anti-CEACAM6 function-blocking antibody BAY 1834942 with checkpoint inhibitors targeting either PD-1/PD-L1 or TIM-3. Proceedings of the AACR Annual Meeting; Atlanta, GA; 2019. AACR2019. [Google Scholar]
- 34.Willuda J., Trautwein M., Pinkert J., Doecke W.-.D., Boehm H.-.H., Wessel F., Ge Y., Gutierrez E.M., Weiske J., Freiberg C., Gritzan U., Glueck J., Zopf D., Golfier S., Ahsen O.v., Zierz R., Wittemer-Rump S., Apeler H., Karl Z., Offringa R., Kreft B., Philipp B. Proceedings of the AACR Annual Meeting; Chicago, IL; AACR; 2018. AACR Annual Meeting2018. [Google Scholar]
- 35.Pinkert J., Boehm H.H., Trautwein M., Doecke W.D., Wessel F., Ge Y., Gutierrez E.M., Carretero R., Freiberg C., Gritzan U., Luetke-Eversloh M., Golfier S., Von Ahsen O., Volpin V., Sorrentino A., Rathinasamy A., Xydia M., Lohmayer R., Sax J., Nur-Menevse A., Hussein A., Stamova S., Beckmann G., Glueck J.M., Schoenfeld D., Weiske J., Zopf D., Offringa R., Kreft B., Beckhove P., Willuda J. T cell-mediated elimination of cancer cells by blocking CEACAM6-CEACAM1 interaction. Oncoimmunology. 2022;11 doi: 10.1080/2162402X.2021.2008110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blumenthal R.D., Leon E., Hansen H.J., Goldenberg D.M. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer. 2007;7:2. doi: 10.1186/1471-2407-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taniguchi N., Kizuka Y. Glycans and cancer: role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv. Cancer Res. 2015;126:11–51. doi: 10.1016/bs.acr.2014.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.