Abstract
Background
Emergence of new variant of SARS-CoV-2, namely omicron, has posed a global concern because of its high rate of transmissibility and mutations in its genome. Researchers worldwide are trying to understand the evolution and emergence of such variants to understand the mutational cascade events.
Methods
We have considered all omicron genomes (n = 302 genomes) available till 2nd December 2021 in the public repository of GISAID along with representatives of variants of concern (VOC), i.e., alpha, beta, gamma, delta, and omicron; variant of interest (VOI) mu and lambda; and variant under monitoring (VUM). Whole genome-based phylogeny and mutational analysis were performed to understand the evolution of SARS CoV-2 leading to emergence of omicron variant.
Results
Whole genome-based phylogeny depicted two phylogroups (PG-I and PG-II) forming variant specific clades except for gamma and VUM GH. Mutational analysis detected 18,261 mutations in the omicron variant, majority of which were non-synonymous mutations in spike (A67, T547K, D614G, H655Y, N679K, P681H, D796Y, N856K, Q954H), followed by RNA dependent RNA polymerase (rdrp) (A1892T, I189V, P314L, K38R, T492I, V57V), ORF6 (M19M) and nucleocapsid protein (RG203KR).
Conclusion
Delta and omicron have evolutionary diverged into distinct phylogroups and do not share a common ancestry. While, omicron shares common ancestry with VOI lambda and its evolution is mainly derived by the non-synonymous mutations.
Keywords: SARS-CoV-2, COVID-19, genome-wide, evolution, variants, VOC, VOI, VUM, SNP, mutation, non-synonymous, silent mutation, spike, RNA dependent RNA polymerase, NSP, UTR: Abbreviations: VOC, variant of concern; VOI, variant of interest; VUM, variant under monitoring; NSP, non-structural protein; UTR, untranslated region; rdrp, RNA dependent RNA polymerase
Abbreviations: VOC, Variant of concern; VOI, Variant of interest; VUM, Variant under monitoring; NSP, Non-structural protein; UTR, Untranslated region; rdrp, RNA dependent RNA polymerase
1. Introduction
Throughout the globe resurgence of COVID-19 cases has been linked to the emergence of new variants of concern (https://www.hopkinsmedicine.org/health/conditions-and-diseases/coronavirus/first-and-second-waves-of-coronavirus) (Thakur et al., 2021). Currently, the world is witnessing a new variant namely, omicron which was first reported in South Africa on 24th November 2021 from the specimen collected on 9th November 2021(https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states). On 26th November 2021, World Health Organisation (WHO) assigned omicron to the ‘variant of concern’ (VOC) category due to its ability to poses a higher risk of reinfection as compared to previously reported variants (https://www.who.int/news/item/26–11–2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern; https://www.who.int/news/item/28–11–2021-update-on-omicron). According to the 1st December 2021 update, omicron is reported in at least 23 countries from five out of six WHO regions, with most cases in Africa and Europe (https://www.cnbc.com/2021/12/01/who-says-omicron-has-been-found-in-23-countries-across-the-world.html).
There is a lot of uncertainty surrounding the omicron variant. For its risk assessment, scientists and researchers are investigating the intensity of its spread, extent of its infection, effectiveness of detection methods, therapeutics, and vaccine efficacy (Knoll & Wonodi, 2021; Lipsitch & Dean, 2020; Pegu et al., 2021). The onset of omicron is reported with mild diseases suggests its low or mild severity than its previous counterparts like delta (Ewen Callaway, 2021; E. Callaway & Ledford, 2021). It is known to have a very high mutation rate with more than 30 mutational changes in its spike protein (Ewen Callaway, 2021) (https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states)
Globally, high risk of reinfection with omicron variant and its ability to evade vaccine-induced immunity resulting in the emergence of new variants of SARS-CoV-2 (Pulliam et al., 2021). Since COVID-19 inception, researchers have been trying to investigate its origin and evolution (Bansal, Kumar, & Patil, 2021; Singh & Soojin, 2021; Tang et al., 2020). We are currently witnessing a global molecular arms race between SARS-CoV-2 and its preventive therapeutics based on diverse regimes such as DNA, RNA, protein or inactivated whole-virion, etc. (Andreadakis et al., 2020; Corey, Mascola, Fauci, & Collins, 2020; Sharma, Sultan, Ding, & Triggle, 2020). This global crisis can be addressed by a very rapid immunization program worldwide. Moreover, the real-time monitoring of evolutionary cascade of SARS-CoV-2 leading to novel variants is utmost. Earlier investigation of several VOC and VOI suggests some of the crucial mutations for viral survival and high infectivity in humans (Boehm et al., 2021; Kumar & Bansal, 2021; Schmidt et al., 2021). However, mutations giving rise to omicron and intra-omicron genomic diversity are not yet analyzed at a population level.
In the present study, we aim to look for the mutational profile of under-monitoring variants reported till now to understand the emergence of a heavily mutated variant named omicron. Interestingly, whole genome-based phylogeny suggests two major phylogroups PG-I and PG-II. Further, mutational analysis depicted the key role of non-synonymous mutations in the evolution of novel variant. Such genome-wide mutational landscape is required for surveillance and vaccine development.
2. Results
2.1. Phylogenomics suggests common ancestry of omicron and lambda variants
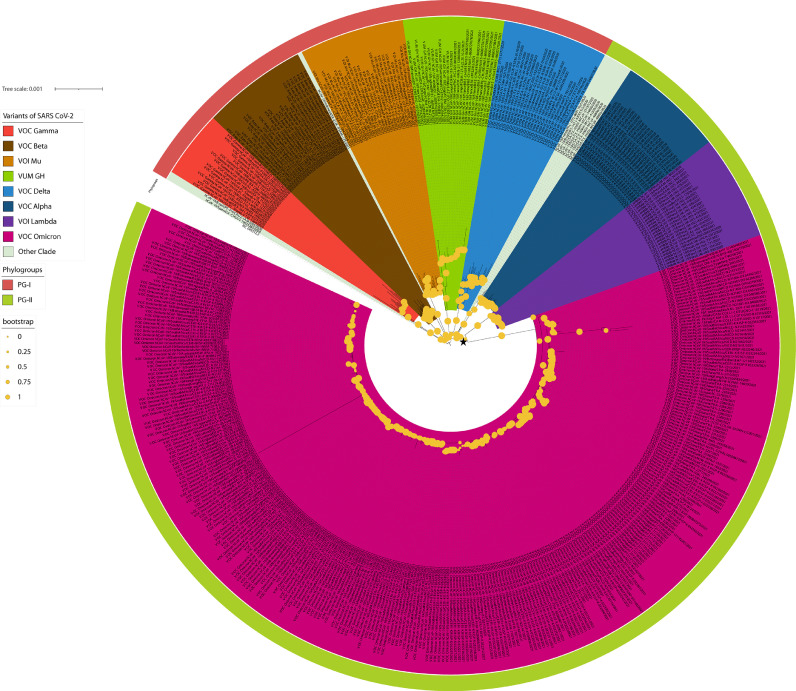
Whole genome-based phylogeny (n = 478 genomes) representing VOC (alpha, beta, gamma, delta, and omicron), VOI (mu and lambda) and VUM depicts two major phylogroups PG-I and PG-II (Fig. 1 and Table 1 ). Here, the reference strain of SARS-CoV-2 (Wuhan-Hu-1, NC_045512.2) is taken as an outgroup. PG-I has VOC: gamma, beta, and delta; VOI: mu and VUM: GH. Whereas, PG-II includes VOC: alpha, omicron and VOI: lambda. Interestingly, two VOCs, delta and omicron, belong to different phylogroups. Phylogeny depicted that omicron shares a common ancestry with VOI lambda represented by a black asterisk in Fig. 1. Interestingly, three isolates from Italy (EPI_ISL_6854346, EPI_ISL_6854347, and EPI_ISL_6854348) form a diversified sub-lineage among the omicron population. Additionally, EPI_ISL_6886594 from Germany is a diversified omicron strain.
Fig. 1.
Maximum likelihood whole genome-based phylogeny of SARS-CoV-2 VOCs, VOIs and VUMs. Here, phylogroups (PG-I and PG-II) and clades (alpha, beta, gamma, delta, omicron, mu etc.) are marked with respective colors as indicated. Bootstrap values are represented by the radius of circle at the nodes. Common ancestry of omicron and lambda is marked by black star.
Table 1.
Metadata of the VOCs, VOIs and VUMs strains used in the present study.
 |
 |
 |
2.2. Very high non-synonymous mutations give rise to omicron
Mutation is driving the evolution and emergence of new variants of COVID-19 worldwide (Islam et al., 2021; Kumar & Bansal, 2021; Thakur et al., 2021). Availability of genomic resources have enabled the research community in tracking mutational events and linking them to new variants (Mercatelli & Giorgi, 2020; Rambaut et al., 2020). Analysis and routine surveillance from South Africa suggested omicron ability to evade immunity from prior infection as compared to other VOCs (Pulliam et al., 2021). In the present study, we intend to understand the evolution and emergence of omicron by its mutational landscape at population level.
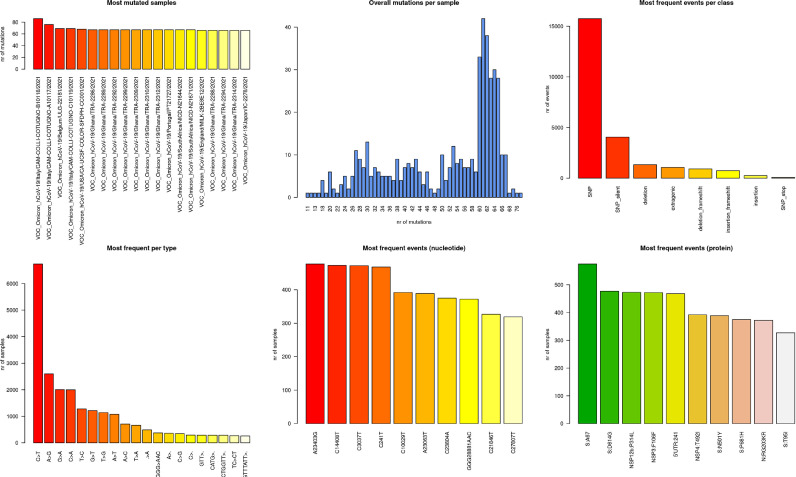
We have performed a mutational analysis with respect to the reference genome of SARS-CoV-2 (NC_045512.2) (Fig. 2 ). Total mutations detected in the dataset were 24,189, and omicron genomes constituted 18,261 mutations (supplementary table 1). For all the strains under study, we have calculated the total number of mutations detected (supplementary table 2). Average mutations per genome for the omicron variant were detected to be 60.5. For the limited genomes of VOCs, VOIs and VUMs, average mutations for GH, delta, mu, gamma, alpha, lambda and beta were 48, 39, 38.5, 37.8, 30.7, 27.4 and 24.2 respectively. This clearly depicts high number of mutations in the omicron variant as compared to other variants of SARS-CoV-2. Except for omicron, average mutations for other variants were calculated on the basis of limited genomes, which might not represent the true mutational events for them. Since, omicron is the recently emerged variant, aim of present study was to understand its mutational landscape at population level.
Fig. 2.
Mutational analysis of omicron. Six panel image displays the most mutated samples, overall mutations per samples, most frequent events per class of mutation category, changes of nucleotide per type, nucleotide wise most frequent events and protein level most frequent events for the genomes used in the study.
Interestingly, >97% (n = 17,703 mutations) of the mutations in omicron were in the coding region, and remaining 558 were detected in the extragenic region of the genome. Amongst the coding gene mutations, 2965 were indels while 14,738 were SNPs constituting non-synonymous (n = 11,995 mutations) and synonymous mutations (n = 2743 mutations). Single nucleotide transitions are shown to be major mutational types amongst the SARS-CoV-2 genomes (Kumar & Bansal, 2021; Mercatelli & Giorgi, 2020).
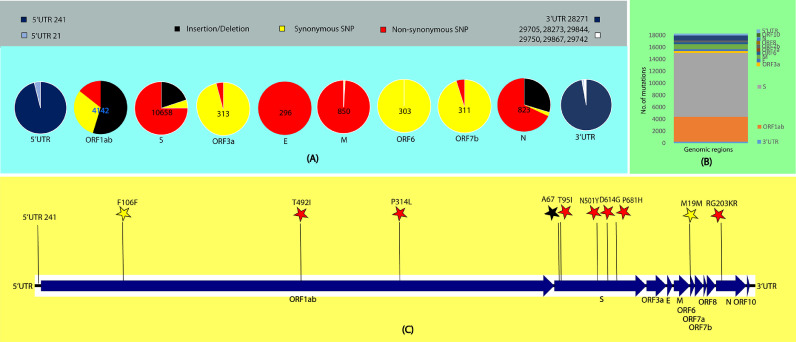
Interestingly, mutational events are highly skewed towards the spike protein, which constitutes ∼60% (n = 10,658) of the total mutations in the coding genomic region (n = 17,703) (Fig. 3 ). The majority of spike protein mutations encompass A67, T547K, D614G, H655Y, N679K, P681H, D796Y, N856K, Q954H, which are reported in all the omicron genomes analysed (Table 3). Count of mutations in the spike was followed by RNA dependent RNA polymerase (rdrp) (n = 4142) constituting A1892T, I189V, P314L, K38R, T492I, V57V in all omicron genomes analyzed (Fig. 3 and Table 3). Remaining 2903 mutations were detected in rest of the coding genomic region (Table 2 , 3 , and supplementary table 1), where M19M in ORF6, and RG203KR in nucleocapsid protein are amongst the most prevalent mutations in omicron (Fig. 3).
Fig. 3.
Mutational analysis of omicron (A) Number of mutations in the coding region is in the centre of the pie-chart representing indels (black), synonymous (yellow) and non-synonymous (red) SNPs. Type and number of mutations in the extergenic region is represented by pie charts blue, light blue and white as represented in the color legends. (B) Bar graph representing number of mutations in the genomic region of SARS-CoV-2. (C) Some of the top mutations (pl. refer Table 3 for all top mutations in omicron) among the omicron variant are represented by stars of black: indels, yellow: synonymous and red: non-synonymous mutations.
Table 3.
Top mutations (>185 in count) in omicron variant as compared to the reference sequence NC_045512.2.
| annotation | protein | variant | varclass | Count | Refpos | refvar | qvar | qpos | qlength |
|---|---|---|---|---|---|---|---|---|---|
| Spike | S | A67 | deletion_frameshift | 575 | 21,762 | C | . | 21,483 | 29,387 |
| Predicted phosphoesterase, papain-like proteinase | NSP3 | A1892T | SNP | 302 | 8393 | G | A | 8124 | 29,387 |
| Transmembrane protein | NSP6 | I189V | SNP | 302 | 11,537 | A | G | 11,259 | 29,387 |
| RNA-dependent RNA polymerase, post-ribosomal frameshift | NSP12b | P314L | SNP | 302 | 14,408 | C | T | 14,130 | 29,387 |
| Spike | S | T547K | SNP | 302 | 23,202 | C | A | 22,915 | 29,387 |
| Spike | S | D614G | SNP | 302 | 23,403 | A | G | 23,116 | 29,387 |
| Spike | S | H655Y | SNP | 302 | 23,525 | C | T | 23,238 | 29,387 |
| ORF6 protein | ORF6 | M19M | SNP_silent | 302 | 27,259 | A | C | 26,972 | 29,387 |
| Predicted phosphoesterase, papain-like proteinase | NSP3 | K38R | SNP | 301 | 2832 | A | G | 2566 | 29,387 |
| Spike | S | N679K | SNP | 301 | 23,599 | T | G | 23,312 | 29,387 |
| Transmembrane protein | NSP4 | T492I | SNP | 301 | 10,029 | C | T | 9760 | 29,378 |
| Nucleocapsid protein | N | RG203K* | SNP | 301 | 28,881 | GGG | AAT | 28,806 | 29,693 |
| Growth-factor-like protein | NSP10 | V57V | SNP_silent | 300 | 13,195 | T | C | 12,917 | 29,387 |
| Spike | S | P681H | SNP | 300 | 23,604 | C | A | 23,317 | 29,387 |
| Spike | S | D796Y | SNP | 300 | 23,948 | G | T | 23,661 | 29,387 |
| Spike | S | N856K | SNP | 300 | 24,130 | C | A | 23,843 | 29,387 |
| Spike | S | Q954H | SNP | 300 | 24,424 | A | T | 24,137 | 29,387 |
| Nucleocapsid protein | N | RG203KR | SNP | 300 | 28,881 | GGG | AAC | 28,594 | 29,387 |
| RNA-dependent RNA polymerase, post-ribosomal frameshift | NSP12b | N591N | SNP_silent | 298 | 15,240 | C | T | 14,962 | 29,387 |
| Spike | S | T95I | SNP | 298 | 21,846 | C | T | 21,562 | 29,387 |
| Predicted phosphoesterase, papain-like proteinase | NSP3 | F106F | SNP_silent | 297 | 3037 | C | T | 2771 | 29,387 |
| Spike | S | G339D | SNP | 297 | 22,578 | G | A | 22,291 | 29,387 |
| ORF3a protein | ORF3a | T64T | SNP_silent | 297 | 25,584 | C | T | 25,297 | 29,387 |
| NA | 5′UTR | 241 | extragenic | 297 | 241 | C | T | 187 | 29,693 |
| 3C-like proteinase | NSP5 | P132H | SNP | 296 | 10,449 | C | A | 10,180 | 29,387 |
| 3′-to-5′ exonuclease | NSP14 | I42V | SNP | 296 | 18,163 | A | G | 17,885 | 29,387 |
| Envelope | E | T9I | SNP | 296 | 26,270 | C | T | 25,983 | 29,387 |
| ORF7b protein | ORF7b | L17L | SNP_silent | 296 | 27,807 | C | T | 27,520 | 29,387 |
| Spike | S | N969K | SNP | 294 | 24,469 | T | A | 24,182 | 29,387 |
| Predicted phosphoesterase, papain-like proteinase | NSP3 | A889A | SNP_silent | 293 | 5386 | T | G | 5120 | 29,387 |
| Spike | S | L981F | SNP | 292 | 24,503 | C | T | 24,216 | 29,387 |
| Spike | S | D1146D | SNP_silent | 292 | 25,000 | C | T | 24,713 | 29,387 |
| Membrane | M | A63T | SNP | 289 | 26,709 | G | A | 26,422 | 29,387 |
| Predicted phosphoesterase, papain-like proteinase | NSP3 | S1265 | deletion | 288 | 6513 | GTT | . | 6246 | 29,387 |
| Transmembrane protein | NSP6 | L105 | deletion | 287 | 11,286 | TGTCTGGTT | . | 11,016 | 29,387 |
| Spike | S | I68 | deletion_frameshift | 287 | 21,767 | CATG | . | 21,486 | 29,387 |
| Spike | S | E484A | SNP | 284 | 23,013 | A | C | 22,726 | 29,387 |
| Spike | S | S477N | SNP | 283 | 22,992 | G | A | 22,705 | 29,387 |
| Spike | S | T478K | SNP | 283 | 22,995 | C | A | 22,708 | 29,387 |
| Spike | S | Q493R | SNP | 282 | 23,040 | A | G | 22,753 | 29,387 |
| Spike | S | Q498R | SNP | 281 | 23,055 | A | G | 22,768 | 29,387 |
| Spike | S | N501Y | SNP | 281 | 23,063 | A | T | 22,776 | 29,387 |
| Spike | S | G496S | SNP | 280 | 23,048 | G | A | 22,761 | 29,387 |
| Spike | S | Y505H | SNP | 277 | 23,075 | T | C | 22,788 | 29,387 |
| Membrane | M | D3G | SNP | 275 | 26,530 | A | G | 26,243 | 29,387 |
| Membrane | M | Q19E | SNP | 272 | 26,577 | C | G | 26,290 | 29,387 |
| Spike | S | S371L | SNP | 270 | 22,673 | TC | CT | 22,386 | 29,387 |
| Spike | S | S373P | SNP | 270 | 22,679 | T | C | 22,392 | 29,387 |
| Spike | S | G142 | deletion | 260 | 21,987 | GTGTTTATT | . | 21,702 | 29,387 |
| Spike | S | S375F | SNP | 260 | 22,686 | C | T | 22,399 | 29,387 |
| ORF7b protein | ORF7b | E3* | SNP_stop | 253 | 27,762 | G | T | 27,687 | 29,752 |
| Spike | S | I210 | insertion_frameshift | 243 | 22,193 | . | T | 21,901 | 29,387 |
| Spike | S | R214 | insertion_frameshift | 243 | 22,203 | . | A | 21,916 | 29,387 |
| Spike | S | R214R | SNP_silent | 243 | 22,204 | T | A | 21,917 | 29,387 |
| Nucleocapsid protein | N | E31 | deletion | 243 | 28,362 | GAGAACGCA | . | 28,074 | 29,378 |
| Spike | S | L212* | SNP_stop | 243 | 22,197 | T | G | 22,118 | 29,749 |
| Spike | S | N211K | SNP | 242 | 22,195 | T | G | 21,903 | 29,387 |
| Spike | S | L212C | SNP | 242 | 22,197 | TA | GC | 21,905 | 29,387 |
| Spike | S | S214 | insertion | 242 | 22,201 | . | AGC | 21,910 | 29,387 |
| Spike | S | V213 | insertion_frameshift | 242 | 22,202 | . | A | 21,914 | 29,387 |
| NA | 3′UTR | 28,271 | extragenic | 242 | 28,271 | A | T | 27,984 | 29,378 |
| Nucleocapsid protein | N | P13L | SNP | 241 | 28,311 | C | T | 28,024 | 29,378 |
| Spike | S | N764K | SNP | 234 | 23,854 | C | A | 23,567 | 29,387 |
| Spike | S | G446S | SNP | 203 | 22,898 | G | A | 22,611 | 29,387 |
| Spike | S | N440K | SNP | 199 | 22,882 | T | G | 22,595 | 29,387 |
| Spike | S | K417N | SNP | 183 | 22,813 | G | T | 22,526 | 29,387 |
Table 2.
Genomic region wise mutational count of the omicron isolates by taking NC_045512.2 as a reference.
| Genomic region | Mutational count | Annotation |
|---|---|---|
| 5′UTR | 309 | 5′ Untranslated region |
| NSP1 | 5 | RNA dependent RNA polymerase |
| NSP2 | 31 | |
| NSP3 | 1572 | |
| NSP4 | 325 | |
| NSP5 | 317 | |
| NSP6 | 595 | |
| NSP7 | 0 | |
| NSP8 | 2 | |
| NSP9 | 9 | |
| NSP10 | 301 | |
| NSP11 | 0 | |
| NSP12a | 0 | |
| NSP12b | 632 | |
| NSP13 | 14 | |
| NSP14 | 319 | |
| NSP15 | 6 | |
| NSP16 | 14 | |
| S | 10,658 | Spike |
| ORF3a | 313 | ORF3a protein |
| E | 296 | Envelope |
| M | 850 | Membrane |
| ORF6 | 303 | ORF6 protein |
| ORF7a | 2 | ORF7a protein |
| ORF7b | 311 | ORF7b protein |
| ORF8 | 4 | ORF8 protein |
| N | 823 | Nucleocapsid protein |
| ORF10 | 1 | ORF10 protein |
| 3′UTR | 249 | 3′ Untranslated region |
2.3. Low intra-sequence diversity amongst omicron variant
Intra-strain diversity among the omicron variant strains reported worldwide will be crucial in understanding the genome dynamics and rapid evolution of SARS-CoV-2. We performed the mutational analysis on the current dataset using omicron (OL677199) isolated from Canada on 23rd November 2021 as the reference genome (supplementary table 3). Most of the strains (n = 298), irrespective of their geographic origin, had less than ten mutations depicting low intra-strain diversity among omicron strains. We found omicron variants had >55 mutations when compared with other VOCs and VOIs. However, four of the isolates two from Europe (Italy) (EPI_ISL_6854347 (n = 23 mutations) and EPI_ISL_6854346 (n = 14 mutations) and two from South Africa (EPI_ISL_6699742 (n = 12 mutations) and EPI_ISL_6774091 (n = 11 mutations) were most diversified among the omicron genomes.
3. Methods
3.1. Identification and procurement of SARS-CoV-2 genome from the public repository
We have considered all the available genomes of omicron variant available in public domain until 6 pm Indian Standard Time (IST) on 2nd December 2021 from GISAID (n = 302 genomes). A total of 25 strains from each variant of concern, namely alpha (B.1.1.7), beta (B.1.351), gamma (P.1) and delta (B.1.617.2) and variant of interest, namely lambda (C.37) and mu (B.1.621). We have also considered 25 strains from variant under monitoring, namely GH (B.1.640). These all strains are from their respective earlier reports in the public domain. Pangolin COVID-19 lineage assigner webserver (https://pangolin.cog-uk.io/) was used to truly demarcate the strains of across variants. The investigation suggested that 9 out of 25 strains does not belong to gamma (P.1) and 1 out of 25 strains doesn't belong to VUM GH (B.1.640) and were wrongly classified earlier. A detailed list of all the strains used in the study is provided in Table 1.
3.2. Phylogenetic analysis
A total of 477 high-quality genomes, including the major variants spread across the globe were taken into consideration. Multiple sequence alignment was performed for all the genomes using MAFFT v7.467 (Nakamura, Yamada, Tomii, & Katoh, 2018) followed by phylogenetic tree construction using fasttree v2.1.8 with double precision (Price, Dehal, & Arkin, 2010) with gamma time reversal method. Visualization of the obtained phylogenetic tree was performed using iTol v6 (Letunic & Bork, 2019). Different variants were marked in accordance with different colors as mentioned in the legends.
3.3. Mutational analysis
Mutational analysis of all the strains (n=477) in the study was performed with two different reference genomes. First with NC_045512.2 (Wuhan-Hu-1) strain (reference SARS CoV-2 strain) and another with first reported strain of omicron variant (OL677199.1) (https://www.ncbi.nlm.nih.gov/nuccore/OL677199) using nucmer v3.1 (Delcher, Phillippy, Carlton, & Salzberg, 2002). We have used a well-documented R script described earlier (Mercatelli & Giorgi, 2020). Here, we have used gff3 annotation and reference genome file to extract genomic coordinate of SARS-CoV-2 proteins. R library package seqinr (https://cran.r-project.org/web/packages/seqinr/index.html) and biostring package (https://bioconductor.org/packages/release/bioc/html/Biostrings.html) of bioconductor was implemented to obtain the list of all the mutational events. Mutational events were calculated with respect to two different references (Reference SARS CoV-2 strain: NC_045512.2) (https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2) and omicron (OL677199.1) (https://www.ncbi.nlm.nih.gov/nuccore/OL677199) separately. Further, the average mutations for a variant were calculated by adding up the mutations in each variant and dividing them by the total number of genomes of the variant used in the present study.
Funding Information
Nil
Author contribution statement
Both the authors’ KB and SK have contributed equally to the data curation, analysis, and writing of the manuscript.
CRediT authorship contribution statement
Kanika Bansal: Data curation, Formal analysis, Writing – original draft. Sanjeet Kumar: Data curation, Formal analysis, Writing – original draft.
Declaration of Competing Interest
The author declares no competing interest.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Authors acknowledge the support and motivation from Dr. Prabhu B.Patil – CSIR-Institute of Microbial Technology, Chandigarh. We are also thankful to Dr. Santosh Kumar Sethi for his kind support during the process of study. We also acknowledge GISAID initiative for extensive curation and availability of genomic resource in public domain.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.198765.
Appendix. Supplementary materials
References
- Andreadakis Z., Kumar A., Román R.G., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(5):305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Bansal K., Kumar S., Patil P.B. bioRxiv; 2021. Codon usage pattern reveals SARS-CoV-2 as a monomorphic pathogen of hybrid origin with role of silent mutations in rapid evolutionary success. 2020.2010.2012.335521. [DOI] [Google Scholar]
- Boehm E., Kronig I., Neher R.A., Eckerle I., Vetter P., Kaiser L. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin. Microbiol. Infect. 2021 doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Heavily mutated coronavirus variant puts scientists on alert. Nature. 2021;25 doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- Callaway E., Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021 doi: 10.1038/d41586-021-03614-z. [DOI] [PubMed] [Google Scholar]
- Corey L., Mascola J.R., Fauci A.S., Collins F.S. A strategic approach to COVID-19 vaccine R&D. Science. 2020;368(6494):948–950. doi: 10.1126/science.abc5312. [DOI] [PubMed] [Google Scholar]
- Delcher A.L., Phillippy A., Carlton J., Salzberg S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30(11):2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam O.K., Al-Emran H.M., Hasan M.S., Anwar A., Jahid M.I.K., Hossain M.A. Emergence of European and North American mutant variants of SARS-CoV-2 in South-East Asia. Transboundary Emerg. Dis. 2021;68(2):824–832. doi: 10.1111/tbed.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll M.D., Wonodi C. Oxford–AstraZeneca COVID-19 vaccine efficacy. Lancet North Am. Ed. 2021;397(10269):72–74. doi: 10.1016/S0140-6736(20)32623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Bansal K. Virus research; 2021. Cross-sectional genomic perspective of epidemic waves of SARS-CoV-2: a pan India study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Dean N.E. Understanding COVID-19 vaccine efficacy. Science. 2020;370(6518):763–765. doi: 10.1126/science.abe5938. [DOI] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Yamada K.D., Tomii K., Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34(14):2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A., O’Connell S.E., Schmidt S.D., O’Dell S., Talana C.A., Lai L., Corbett K.S. Durability of mRNA-1273 vaccine–induced antibodies against SARS-CoV-2 variants. Science. 2021;373(6561):1372–1377. doi: 10.1126/science.abj4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam J.R.C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Moultrie H. medRxiv; 2021. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. 2021.2011.2011.21266068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Volz E. Virological; 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. [Google Scholar]
- Schmidt M., Arshad M., Bernhart S.H., Hakobyan S., Arakelyan A., Loeffler-Wirth H., Binder H. The evolving faces of the SARS-CoV-2 genome. Viruses. 2021;13(9):1764. doi: 10.3390/v13091764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma O., Sultan A.A., Ding H., Triggle C.R. A Review of the Progress and Challenges of Developing a Vaccine for COVID-19. Front. Immunol. 2020;11:2413. doi: 10.3389/fimmu.2020.585354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D., Soojin V.Y. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021;53(4):537–547. doi: 10.1038/s12276-021-00604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Wu C., Li X., Song Y., Yao X., Wu X., Qian Z. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7(6):1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur V., Bhola S., Thakur P., Patel S.K.S., Kulshrestha S., Ratho R.K., Kumar P. Infection; 2021. Waves and variants of SARS-CoV-2: understanding the causes and effect of the COVID-19 catastrophe; pp. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.