Abstract
Introduction
The functional roles and mechanism of the XIST in osteoarthritis and the chondrogenic differentiation of BMSCs were clarified.
Methods
The expression levels of XIST, TAF15, FUT1 and YY1 were detected through quantitative RT-PCR. The protein expression of Sox9, ACAN, COL2A1 and FUT1 were detected by western blot and immunohistochemistry. The damage of cartilage tissue was detected by HE staining, and Safranin O-fast green. Alcian-Blue and Alizarin red S staining were performed to evaluate BMSCs chondrogenic differentiation. The relationship between XIST and TAF15, XIST and TAF15 were analyzed by RNA immunoprecipitation assay. Luciferase reporter assays and chromatin immunoprecipitation were performed to detect the interaction relationship between XIST and YY1. In addition, osteoarthritis mice were built to assess the function of XIST in vivo.
Results
The levels of XIST, TAF15 and FUT1 were upregulated in cartilage tissues from osteoarthritis patient. The level of XIST was decreased in BMSCs during chondrogenic differentiation. XIST overexpression inhibited the chondrogenic differentiation of BMSCs. Moreover, silencing of FUT1 reversed the effects of XIST overexpression on BMSCs chondrogenic differentiation. Mechanistically, in BMSCs, YY1 induced the expression of XIST in BMSCs, and XIST regulated FUT1 mRNA stability through targeting TAF15. Furthermore, silencing of XIST alleviated the symptoms of cartilage injury in OA mice.
Conclusion
Taken together, these results suggested that YY1 induced XIST was closely related to the chondrogenic differentiation of BMSCs and the progression of osteoarthritis by TAF15/FUT1 axis, and may be a new OA therapeutic target.
Keywords: Osteoarthritis, XIST, Chondrogenic differentiation, BMSCs, YY1
Abbreviations: OA, osteoarthritis; MSCs, mesenchymal stem cells; BMSCs, bone marrow-derived mesenchymal stem cells; TFs, transcription factors; YY1, ying yang 1; XIST, X inactive specific transcript; lncRNAs, long noncoding RNAs; TAF15, TATA-box-binding protein-associated factor 15; FUT1, fucosyltransferase 1; H&E, hematoxylin and eosin; RT, room temperature; qRT-PCR, quantitative real-time polymerase chain reaction; SOX9, sex-determining region Y (SRY)-box 9; ACAN, aggrecan; RIP, RNA immunoprecipitation; ChIP, chromatin immunoprecipitation
Highlights
-
•
XIST and TAF15 expression were upregulated in osteoarthritis.
-
•
Overexpression of XIST suppressed the chondrogenic differentiation of BMSCs.
-
•
XIST regulated BMSCs chondrogenic differentiation through enhancing FUT1 mRNA stability via TAF15.
-
•
XIST silencing relieved cartilage damage in OA mice.
1. Introduction
Osteoarthritis (OA) is a type of degenerative arthropathy with characteristics including cartilage erosion, osteophytes and subchondral sclerosis [1,2]. It is predicted that approximately 0.25 billion people suffer from OA in the world [3]. OA causes significant pain and stiffness in the joints and substantial societal burden [4]. The cause and development of OA is complex, including obesity, biomechanical, environmental, genetic, and aging [5]. The conventional treatment means for OA are pain relieving and symptom control [6]. However, the drugs using to relieve pain are often insufficient, and there are no effective therapies to prevent OA.
Recently, cell therapy, specifically with mesenchymal stem cells (MSCs), which could differentiate into chondrocytes and has unique immunoregulatory competence, gained increasing attention [7]. MSC-based therapy has been demonstrated to reduce cartilage degeneration, encourage pain reduction and inhibit OA progression [8,9]. Especially bone marrow-derived MSCs (BMSCs), one of the most common sources of MSCs, has been considered as an optimal approach for cartilage repair. Many studies have reported that BMSCs could repair osteochondral defects with regenerated cartilage both in animal and clinical studies [[10], [11], [12]]. For example, transplantation of BMSC in rabbit with defective articular cartilage could regenerate osteochondral tissue successfully [13]. Moreover, Orozco et al. confirmed that injection of autologous BMSCs relieved the clinical symptoms of chondral defects, and improve the knee function and cartilage quality of OA patients [14]. The differentiation and proliferation capacity of BMSCs were regulated by numerous factors, such as bioactive factors, culture conditions and gene editing. Therefore, understanding the mechanisms of regulation of BMSCs chondrogenic differentiation are essential for their therapeutic applications.
Transcription factors (TFs), which regulate genes transcription through binding DNA, have decisive roles in biological processes such as cell proliferation, differentiation and apoptosis [15]. More recently, the functional roles of TFs for cartilage development and regeneration are researched both in vitro and in vivo [16,17]. For example, transcription factor Sox9 is indispensable for chondrogenesis and the survival of chondrocyte [18]. Ying Yang 1 (YY1), one DNA-binding zinc finger TF, which could function as an activator or a repressor of many genes associated with cell survival, cell proliferation, DNA repair and autophagy [19]. Interestingly, study had demonstrated that YY1 is also an important regulator for MSC multipotency and differentiation [20]. For example, YY1 regulated p38-induced MSCs differentiation into osteoblasts [21]. Moreover, it was reported that YY1 regulated the expression of cartilage-specific gene (Chondromodulin-I) in MSCs [22]. However, the effects and mechanisms of YY1 on BMSCs chondrogenic differentiation remain unclear and need further investigation.
Long noncoding RNAs (lncRNAs) are non-coding transcripts >200 nt in length [23]. Many evidences have revealed the physical features, biological functions, and potential roles of lncRNAs in OA [24,25]. Moreover, lncRNAs are also participated in the chondrogenic differentiation by MSCs [26]. Notably, lncRNA X inactive specific transcript (XIST), which regulated progression of various types of cancer, was upregulated in cartilage from OA patients [27]. Furthermore, it was demonstrated that XIST promoted the proliferation and apoptosis of OA chondrocytes through the miR-211/CXCR4 signal pathway, or promote the degradation of extracellular matrix in OA by targeting miR-1277-5p [28,29]. However, the specific mechanism underlying how XIST regulated BMSCs chondrogenic differentiation has yet to be fully elucidated.
In this study, XIST was found to be upregulated in OA cartilage and BMSCs during chondrogenic differentiation. XIST overexpression suppressed the chondrogenic differentiation of BMSCs. Mechanistic studies revealed that YY1-induced XIST regulates the chondrogenic differentiation of BMSCs by binding with TATA-box-binding protein-associated factor 15 (TAF15) to stabilizing fucosyltransferase 1 (FUT1) expression. Therefore, our study identified a new YY1/XIST/TAF15/FUT1 axis in the differentiation of BMSCs into chondrocytes, thereby providing a novel avenue for OA treatment.
2. Materials and methods
2.1. Clinical samples
All procedures were approved by the Medical Ethics Committee of Jiangxi Provincial People's Hospital Affiliated to Nanchang University. Informed consent was obtained from all patients. Cartilage tissues were obtained from 22 OA patients (11 females and 11 males, 43–72 years) who underwent knee replacement and 22 patients without OA (10 females and 12 males, 36–58 years) who underwent the amputation at the Jiangxi Provincial People's Hospital Affiliated to Nanchang University and were preserved in - 80 °C. OA was diagnosed using the World Health Organization parameters.
2.2. Cell culture and differentiation
Human BMSCs were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured using DMEM medium supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA), and 1% penicillin-streptomycin (Invitrogen) at 37 °C in 5% CO2. Medium was changed every three days.
For chondrogenic differentiation, 1 × 106 BMSCs were seeded in 12-well plate and induced using StemPro® Chondrogenesis Differentiation kit (Thermo Scientific, Waltham, MA USA) for 0, 7 and 14 days. Medium was changed every 4 days.
2.3. Cell transfection
SiRNA-XIST, siRNA-TAF15, siRNA-FUT1, siRNA-YY1, XIST overexpression plasmid pcDNA3.1-XIST, YY1 overexpression lentivirus and their respective controls were designed and synthesized through GenePharma (Shanghai, China). Cells were transfected with Invitrogen Lipofectamine™3000 reagent (Thermo Fisher Scientific, Inc.), following the manufacturer's recommendations. 24 h after transfection, the cells were collected for subsequent experimentation.
2.4. Hematoxylin and eosin (H&E) and Alcian blue staining
Cartilage tissues from OA patients or OA mice were fixed by 4% formaldehyde, decalcified with 0.2 M EDTA for two weeks. Dehydration and paraffin embedding were conducted conventionally. Paraffin sections (5 μm) were then dewaxed and rehydrated. The sections were stained with an H&E staining kit (Beyotime, Shanghai, China) or 1% Alcian Blue 8GX (Abcam; cat# ab145250) at room temperature (RT). After washing, stained sections were observed under light microscope (Nikon Corporation).
2.5. Alizarin red S staining
BMSCs were fixed by 70% ethanol, stained with 2% Alizarin red staining reagent (Sigma) at RT for 20 min. After washing, the signals were visualized using a light microscope (Zeiss, Germany).
2.6. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cartilage tissues and cultured BMSCs through TRIzol Reagent (Invitrogen; cat#10296028). The concentration of RNA was measured with a NanoDrop 2000 instrument (Thermo Scientific). Then, the first-strand cDNA was synthesized by a TaqMan Reverse Transcription Kit (Takara, Dalian, China). The quantitative real-time PCR (qRT-PCR) was performed with SYBR®-Green PCR kit (Thermo Scientific) on an ABI 7500HT real-time PCR system as follows: 95 °C for 5 min, 40 cycles of 30 s of 94 °C and 30 s of 60 °C, 72 °C for 10 min. 2−ΔΔCq method was used to analyze the data. Primer sequences were used as follows:
XIST R: 5′-CGATCTGTAAGTCCACCA-3′,
XIST F: 5′-CAGACGTGTGCTCTTC-3′.
TAF15 R: 5′-GCCAAGTTAACTGCTTCGTGG-3′,
TAF15 F: 5′-AGGTGACTTCTTCAGCGAGC-3′,
FUT1 R: 5′-GGACACAGGATCGACAGG-3′,
FUT1 F: 5′-AAAGCGGACTGTGGATCT-3′,
YY1 R: 5′-AAAGGGCTTCTCTCCAGT-3′,
YY1 F: 5′-TCTCAGATCCCAAACAACT-3′,
β-actin R: 5′-GTCCACCGCAAATGCTTCTA-3′,
β-actin F: 5′-TGCTGTCACCTTCACCGTTC-3′.
2.7. Western blot assay
Total protein was isolated from cartilage tissue or cultured BMSCs using RIPA lysis buffer (Beyotime, Shanghai, China). The concentration was detected with NanoDrop 2000c spectrophotometer. The samples (30 μg/lane) were electrophoresed with 10% SDS-PAGE and transferred to PVDF membrane (Millipore). Membrane was blocked by 5% non-fat milk for 2 h at RT and incubated with respective primary antibodies against FUT1 (1:500; Abcam; cat#ab121411), SOX9 (1;500; Santa Cruz; cat#20095), aggrecan (ACAN) (1:200; Abcam; cat#ab3778) and COL2A1 (1:200; Abcam; cat# ab188570) overnight at 4 °C. Finally, samples were incubated with secondary HRP antibodies for 2 h at RT and were detected with chemiluminescence kit (Thermo Fisher Scientific) and band intensity was analyzed by ImageJ software v1.8.0.
2.8. RNA immunoprecipitation (RIP) assay
RIP assay was performed with a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) and primary antibodies against TAF15 (Abcam; cat#ab134916) or IgG (Abcam; cat#ab205718). BMSCs were lysed in RIP buffer and incubated with magnetic beads conjugated with anti TAF15 or IgG antibody overnight at 4 °C following incubation with Proteinase K (Abcam; cat#ab281339) at 60 °C for 1 h. Finally, the immunoprecipitated RNAs were isolated and measured through qRT-PCR.
2.9. Luciferase reporter assay
Wild type (WT) XIST, Site-1 mutant type (MUT) XIST, Site-2 MUT XIST, and Site-3 MUT XIST were purchased from Hanbio Biotechnology Co., Ltd., and cloned into the luciferase reporter plasmid (psi CHECK2; Promega Corporation). BMSCs were co-transfected with WT XIST or MUT XIST combined with YY1 vector or NC vector using Lipofectamine™3000 reagent (Thermo). After 48 h, the luciferase activity was detected with the Dual Luciferase Assay System (Promega Corporation).
2.10. Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using an anti-YY1 antibody (Santa-Cruz; cat#sc-1703) as previously described [30]. In brief, chromatin collected from BMSCs was cross-linked using 1.0% formaldehyde and fragmented to about 200–1000 bp through sonication. Then 20 μg of chromatin was precleared with protein G-agarose beads (Santa Cruz) for 2 h at 4 °C, followed by immunoprecipitation with 3 μg of anti-YY1 antibody overnight at 4 °C. Precipitated samples were eluted and reverse cross-linked, and the ChIP DNA was analyzed using q-PCR. Four PCR primers were used: 5′- AGGTAGCGTTTGCTTTCTCACCCA -3′ (forward) and 5′-CAGCAATGCCAAGGGTAAACGGAA -3′ (reverse) (XIST promoter region); 5′- AGGGCTGCTCAGAAGTCTATCTCGGGGGCTC -3′ (forward) and 5′-GAGCCCCCGAGATAGACTTCTGAGCAGCCCT -3′ (reverse) (XIST promoter site1 region);
5′- CATTTAGGTCGTACAGGAACTCAAGTTCTTGGTGCGG -3′ (forward) and 5′- CGCACCAAGAACTTGAGTTCCTGTACGACCTAAATG -3′ (reverse) (XIST promoter site2 region); 5′- A ATGCTCTTGAATGTGTCTAAGTCATGTGACCTGCCC -3′ (forward) and 5′- GGGCAGGTCACATGACTTAGACACATTCAAGAGCAT -3′ (reverse) (XIST promoter site3 region).
2.11. Osteoarthritis (OA) mice and intraarticular injection
All animal experimental protocols were approved by the Institutional Animal Use and Care Committee of Jiangxi Provincial People's Hospital Affiliated to Nanchang University. Ten-week-old male C57BL/6 J mice were purchased from Jackson Labs, housed and maintained according to the IACUC guideline. Surgery to destabilize the medial meniscus (DMM) was performed on the right knee to induce OA as previously described [31]. For sham group, the right knee was opened without intervention of the menisco-tibial ligament. Ten days later, si-NC or si-XIST was injected into the knee joint of OA mice using a 33G needle and a micro-syringe (Hamilton). Mice were sacrificed 8-weeks post-surgery and the right Knee joints were harvested. Articular cartilage was collected from the medial tibial plateau under a dissecting microscope.
2.12. Safranin O-fast green and IHC
Mice were euthanized, and knee joints were dissected and processed for Safranin O-fast green and immunohistochemical staining as described [32]. Briefly, knee joints were fixed using 4% PFA, decalcified in 0.2 M EDTA, paraffin embedded and 5 μm sections were cut followed by deparaffinized using xylene and hydrated through using a graded series of ethanol. For Safranin O-fast green, sections were stained with 0.02% Fast Green for 5 min (followed by 3 dips in 1% acetic acid) and then 0.1% Safranin-O for 10 min. For immunohistochemistry (IHC), after antigen unmasking, sections were washed using PBS, treated by hydrogen peroxide and blocked with 10% goat serum for 30 min at RT. After incubating with primary antibodies against SOX9 (1;1000; Santa Cruz; cat#20095), ACAN (1:2 = 1000; Abcam; cat#ab3778) and COL2A1 (1:200; Abcam; cat# ab188570). After washing, sections were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at RT. Finally, sections were stained with DAB substrate kit (Pierce, #34002). Images were captured using an Olympus VS120 Scanning microscope.
2.13. Statistical analyses
Prism software version 6.0 was used for the data analyses. All data were expressed as mean ± standard deviation (SD). Student's t-test or one-way analysis of variance (ANOVA) analysis was carried out to calculate the differences between groups, P < 0.05 was considered statistically significant.
3. Results
3.1. The expression levels of XIST, TAF15 and FUT1 were upregulated in OA tissues
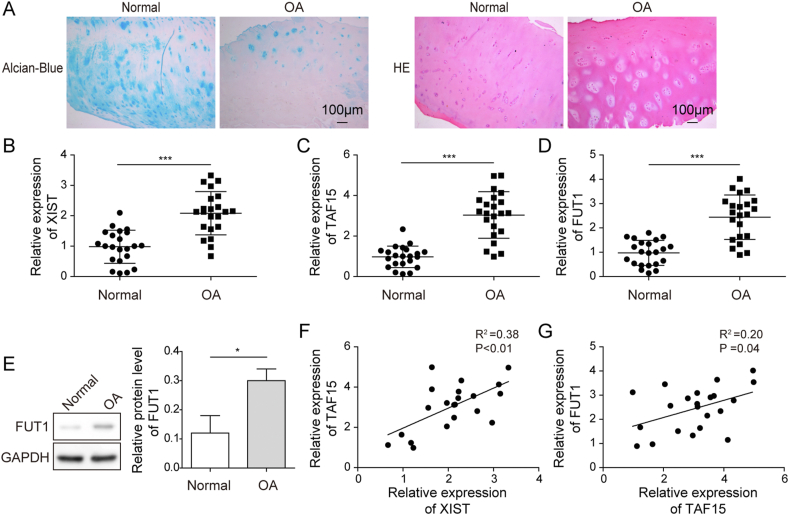
The H&E/Alcian-Blue staining was performed and the cartilage tissue of the knee joints of OA patients showed appearance of defects, structural breakage and damage compared with the normal knee cartilage (Fig. 1A). Moreover, the levels of XIST, TAF15 and FUT1 in cartilages of 22 OA and 22 normal subjects were measured with qRT-PCR. The expression levels of XIST, TAF15 and FUT1 were higher in OA group compared with normal group (Fig. 1B–D). As shown in Fig. 1E, the protein level of FUT1 was increased in OA group. Further Pearson correlation analysis demonstrated positive correlation between XIST and TAF15, TAF15 and FUT1 (Fig. 1F,G). These data hinted that XIST, TAF15 and FUT1 might be involved in the progression of OA.
Fig. 1.
The dysregulation of XIST, TAF15 and FUT1 in OA tissues. (A) Representative images of normal and OA cartilage sections for HE staining and Alcian Blue staining (magnification, ×100). (B–D) The RNA expression levels of XIST (B), TAF15 (C) and FUT1 (D) in normal and OA cartilage were measured by qRT-PCR. (E) The levels of FUT1 in normal and OA cartilage were measured by western blot. (F and G) Pearson correlation analysis about XIST and TAF15 (F), TAF15 and FUT1 (G) in the OA cartilage.
3.2. XIST overexpression suppressed the chondrogenic differentiation of BMSCs
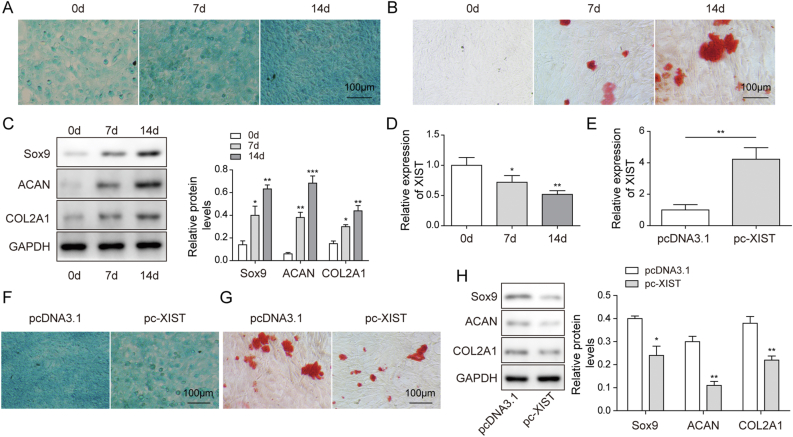
First, the chondrogenic differentiation of BMSCs were identified by alcian blue staining, Alizarin red S staining and western blot on days 0, 7 and 14. The results indicated that the glycosaminoglycan deposition (Fig. 2A) and mineralized nodule formation (Fig. 2B), as well as the protein levels of Sox9, ACAN and Col2A1 (as chondrogenic differentiation markers) (Fig. 2C) increased from day 7 in a time-dependent manner, suggesting successful chondrogenic induction. In addition, chondrogenic induction decreased XIST level in BMSCs in a time-dependent manner (Fig. 2D). To study the effects of XIST on chondrogenic differentiation, BMSCs were transfected with pc-XIST to overexpress XIST (Fig. 2E). The staining intensity of Alcian blue and Alizarin red S were decreased in pc-XIST group compared with pc-DNA3.1 group at 14 days (Fig. 2F,G). Besides, the protein expression of Sox9, ACAN and Col2A1 in BMSCs were all downregulated by XIST overexpression at 14 days (Fig. 2H). These results confirmed that XIST overexpression inhibited the chondrogenic differentiation of BMSCs.
Fig. 2.
XIST overexpression suppressed the chondrogenic differentiation of BMSCs. (A and B) Chondrogenic differentiation of BMSCs was evaluated with Alcian blue staining (A) and Alizarin red S staining (B) at 0, 7 and 14 days after chondrogenic induction. (C) The protein expression of Sox9, ACAN and COL2A1 in BMSCs were determined western blot at 0, 7 and 14 days. (D) The expression level of XIST in BMSCs was determined by qRT-PCR at 0, 7 and 14 days. (E) The transfection efficacy of pcDNA3.1 and pc- XIST in BMSCs was measured by qRT-PCR. (F–H) BMSCs were transfected with pcDNA3.1 or pc-XIST for 24 h, and undergo chondrogenic induction for 14 days. The acidic mucopolysaccharide (F), mineralized nodule formation (G) and protein expression of Sox9, ACAN and COL2A1 (H) in BMSCs were measured by Alcian blue staining, Alizarin red S staining and western blot, respectively.
3.3. XIST regulated FUT1 mRNA stability via TAF15 in BMSCs
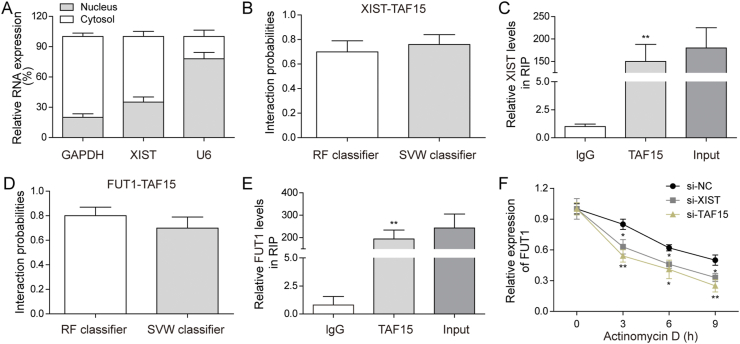
The underlying mechanisms of the effects of XIST on BMSCs chondrogenic differentiation was furtherly explored. Because differential localization of lncRNAs may have various mechanisms of function (Kopp and Mendell 2018), we measured the cellular sub-localization of XIST in BMSCs through qRT-PCR and found that the localization of XIST higher in cytoplasm than in nucleus (Fig. 3A). XIST-protein interactions were predicted by RPIseq (http://pridb.gdcb.iastate.edu/RPISeq/). The result showed that XIST may interact with TAF15 by RF (P = 0.7) and SVM (P = 0.76) (Fig. 3B). Next, RIP assay was carried out to confirm the combination between XIST and TAF15. XIST was significantly higher in TAF15 group compared with IgG group in BMSCs (Fig. 3C). Moreover, TAF15 was predicted to combine with FUT1 by RF (P = 0.8) and SVM (P = 0.7) (Fig. 3D). And the results of RIP assay showed that FUT1 was significantly higher in TAF15 group compared with IgG group in BMSCs (Fig. 3E). Moreover, the FUT1 mRNA levels and the half-life of the FUT1 mRNA were significantly reduce in XIST knockdown or TAF15 knockdown BMSCs (Fig. 3F). These results indicated that XIST regulated FUT1 expression by increasing FUT1 mRNA stability via bombinating with TAF15.
Fig. 3.
XIST regulated FUT1 mRNA stability by targeting TAF15 in BMSCs. (A) The levels of XIST in cytosol and nucleus were measured by qRT-PCR. (B) The interaction probability between XIST and TAF15 predicted by RPIseq based on RF and SVM classifiers. (C) RIP assay was performed to verify the association between XIST and TAF15 in BMSCs. (D) The interaction probability between FUT1 and TAF15 predicted by RPIseq based on RF and SVM classifiers. (E) RIP assay was performed to verify the association between FUT1 and TAF15 in BMSCs. (F) qRT-PCR was performed to measure FUT1 mRNA level in XIST silencing, TAF15 silencing or control BMSCs treated with Actinomycin D for the indicated time.
3.4. FUT1 silencing reversed the inhibitory effects of XIST overexpression on the chondrogenic differentiation of BMSCs
The mRNA and protein level of FUT1 were downregulated in BMSCs with a time-dependent manner during chondrogenic induction (Fig. 4A,B). To verify whether XIST regulated BMSCs chondrogenic differentiation through FUT1, pc-XIST, si-FUT1, pc-XIST+si-FUT1 were transfected into BMSCs. Alcian blue staining indicated that knockdown of FUT1 increased the staining intensity, and the inhibitory effect of pc-XIST on glycosaminoglycan deposition was rescued by si-FUT1 (Fig. 4C). Moreover, transfection with si-FUT1 upregulated the mineralized nodule formation and abolish the pc-XIST-induced descending of calcium nodules in BMSCs chondrogenic differentiation (Fig. 4D). In addition, western blot showed that the expression of Sox9, ACAN and Col2A1 were up-regulated in si-FUT1 transfected BMSCs, and XIST overexpression induced downregulation of Sox9, ACAN and Col2A1 was reversed by si-FUT1 (Fig. 4E). Taken together, XIST negatively regulated chondrogenic differentiation of BMSCs through regulating FUT1.
Fig. 4.
FUT1 knockdown reversed the inhibitory effects of XIST overexpression on chondrogenic differentiation of BMSCs. (A–B) The mRNA (A) and protein level (B) of FUT1 in BMSCs was determined by qRT-PCR and western blot after 0, 7 and 14 days of chondrogenic culture. BMSCs were transfected with pc-XIST or si-FUT1 alone or co-transfected with pc-XIST and si-FUT1 for 24 h, and undergo chondrogenic induction for 14 days. The acidic mucopolysaccharide (C), mineralized nodule formation (D) and protein levels of Sox9, ACAN and COL2A1 (E) in BMSCs were measured by Alcian blue staining, Alizarin red S staining or western blot, respectively.
3.5. YY1 induced XIST upregulation in BMSCs
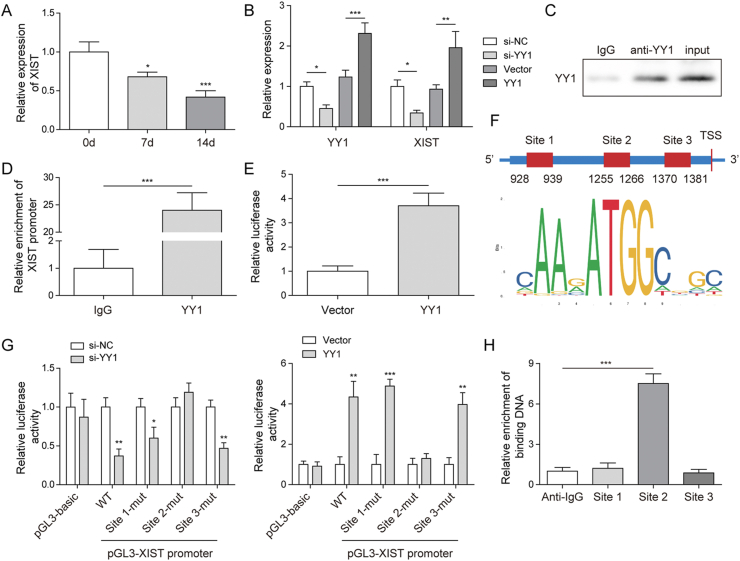
To study how XIST was downregulated in BMSCs during chondrogenic differentiation, the upstream of XIST was explored. We found YY1 expression was decreased in BMSCs after chondrogenic induction (Fig. 5A). Then YY1 was knockdown or overexpressed in BMSCs. The qRT-PCR results showed that the expression of XIST and YY1 were reduced by YY1 silencing and were upregulated by YY1 overexpression (Fig. 5B). Next, chIP assay verified that YY1 could target to the XIST promoter (Fig. 5C,D). The luciferase activity of XIST was increased by YY1 overexpression (Fig. 5E). The binding motif of YY1 and YY1 binding sites on XIST promoter were predicted by JASPAR database (http://jaspar.genereg.net/) and shown in Fig. 5F. Subsequently, the luciferase activity in XIST promoter-WT, Site1-Mut and Site3-Mut group were reduced by si-YY1 and evaluated by YY1 overexpression but were not changed in pGL3-basic and Site2-Mut groups (Fig. 5G). Also, ChIP assay also confirmed that only Site2 region was responsive to YY1-mediated transcription of XIST (Fig. 5H). Taken together, YY1 induced XIST transcription and increased its expression.
Fig. 5.
The upregulation of XIST induced by YY1. (A) The level of YY1 in BMSCs during chondrogenic differentiation was determined by qRT-PCR. (B) BMSCs were transfected with si-YY1, YY1 vector or their NC for 24 h, the levels of YY1 and XIST were determined by qRT-PCR. (C–D) ChIP assay demonstrated the targeted relationship between YY1 and XIST promoter. (E) Luciferase reporter assay was performed and showed that YY1 could bind with XIST promoter. (F) The potential binding region of YY1 on XIST promoter predicted by JASPAR database. (G) Luciferase reporter assay was performed to detect the targeted site between YY1 and XIST promoter. (H) ChIP assay demonstrated that YY1 binds the site 2 region of XIST promoter.
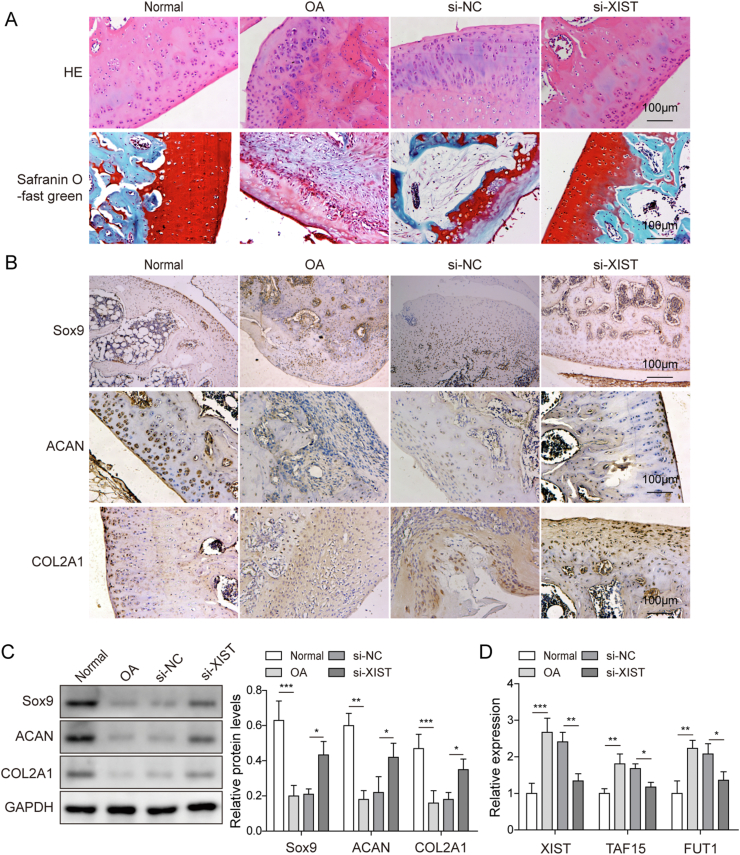
3.6. XIST silencing suppressed the cartilage damage in the mice model of osteoarthritis
To investigate XIST function in OA in vivo, si-XIST or si-NC were injected into mice with ACLT-induced osteoarthritis through knee joint cavity. Safranin-O staining and HE staining of knee joints showed severe articular cartilage lesions and proteoglycan loss in OA group than sham group, and the damage induced by ACLT were decreased by XIST silencing (Fig. 6A). The results of immunohistochemistry staining and western blot demonstrated that the levels of Sox9, ACAN and COL2A1 were lower in the knee joints in OA group, while this downregulation was restored by XIST knockdown (Fig. 6B,C). Moreover, the increased expression of XIST, TAF15 and FUT1 in knee joints induced by osteoarthritis was inhibited after XIST silencing (Fig. 6D). These data confirmed that knockdown of XIST suppressed the development of osteoarthritis in mice.
Fig. 6.
Silence of XIST alleviated the symptoms of cartilage injury in OA mice. Mice were randomly divided into four groups: Sham, OA, OA+si-NC and OA+si-XIST groups. (A) Representative images of cartilage sections after Safranin O-fast and HE staining. (B) Sox9, ACAN and COL2A1 expression in cartilage were assessed by IHC. (C) The protein levels of Sox9, ACAN and COL2A1 in cartilage were measured by western blot. (D) qRT-PCR analysis of XIST, TAF15 and FUT1 RNA levels in cartilage.
4. Discussion
OA is considered as chronic joint disorders, and usually result in gradual cartilage degeneration and disability in the world [33,34]. Risk factors, such as aging, obesity, genetic architecture, injury and inflammation, may contribute to the incidence of OA [35]. The pathophysiology of OA was very complicated and limited knowledge, which makes the limited symptomatic and disease-modifying treatments [36]. Cell-based strategies using chondrocytes or multipotent stem cells include BMSCs may be the effective therapies for OA to improve functional repair cartilage tissue [37]. Clarifying the underlying molecular mechanisms that regulate BMSCs chondrogenic differentiation is important for its application in cartilage regeneration. In this study, we demonstrated that overexpression of XIST inhibited BMSCs chondrogenic differentiation through TAF15/FUT1 axis, and silencing of XIST alleviated the cartilage injury in OA mice model, which will provide a reliable and effective target for OA.
As important post-transcriptional regulators, lncRNAs play vital crucial roles in many biological processes [38]. Many studies demonstrated the many lncRNAs could regulate the chondrogenesis and cartilage homeostasis, and play essential roles in the progression of OA [24,39]. For example, lncRNA-GAS5 ameliorated the articular cartilage injury in arthritis mice through downregulating miR-103 level [40]. Recently, several researches had reported that XIST was associated with OA. For example, XIST promoted the degradation of extracellular matrix and collagen in OA [29]. Our study found that XIST was upregulated in cartilage tissues of OA patient. Furthermore, XIST silencing upregulated the expression of chondrogenic differentiation markers, thereby partially attenuated cartilage destruction of OA mice.
To develop BMSCs-based cartilage reconstruction therapy for OA treatment, it is necessary to identify factors regulating chondrogenic differentiation of BMSCs. Accumulating evidences demonstrated that lncRNAs were implicated in chondrogenic differentiation [41]. For example, lncRNA ADAMTS9-AS2 promoted the human MSCs chondrogenic differentiation by sponging miR-942-5p [42]. Previous study showed that XIST inhibited BMSCs osteogenic differentiation through miR-19a-3p/Hoxa5 pathway [43]. XIST expression was downregulated over chondrogenic differentiation of BMSCs. Moreover, overexpression of XIST repressed BMSCs chondrogenic differentiation. LncRNAs exert their functions by regulating the expression of target mRNAs. Through RPISeq software and RIP assay, we discovered that XIST combined with TAF15, thereby regulated the stability of FUT1 mRNA in BMSCs. FUT1 is important in chondrocyte growth and apoptosis, and is associated with OA [44]. For example, Li et al. reported that FUT1 knockdown increased cell apoptosis and ECM degradation in chondrocytes [45]. Consistent with previous report, we found that FUT1 was upregulated in OA cartilage tissue. More importantly, knockdown of FUT1 attenuated the inhibitory effects of XIST overexpression on BMSCs chondrogenic differentiation. These findings revealed that XIST has a vital role in BMSCs chondrogenic differentiation through binding with TAF15 to stabilizing FUT1 expression.
Many studies have reported the importance of YY1 in neuronal development and differentiation [46,21]. It was reported that YY1 promoted p38-mediated osteogenic differentiation of MSCs by upregulating p38 expression [21]. Moreover, YY1 regulated the expression of chondromodulin-I, which is pivotal for regulation of chondrocyte differentiation and cartilage homeostasis, suggesting that YY1 is also important for chondrogenesis. Previous reports showed that YY1 could bind directly the promoter to upregulate of XIST expression [47,48]. Here we identify that YY1 could bind XIST regulatory region and control XIST expression in BMSCs. Moreover, YY1 was downregulated in BMSCs during chondrogenic differentiation. Overall, YY1 may regulate chondrogenic differentiation of BMSCs by binding XIST.
5. Conclusions
In the present study, we highlighted that YY1 induced XIST inhibited the chondrogenic differentiation of BMSCs by binding with TAF15 to stabilizing FUT1 expression. Importantly, it was shown that knockdown of XIST alleviated the cartilage injury in OA mice. Thus, XIST would be an effective target for BMSC-based OA cartilage regeneration.
Funding
This work was supported by Analysis of YY1-activated IncRNA XIST regulating cartilage differentiation through RBP pathway, Health Commission of Jiangxi Provincial (No.202130032).
Ethical approval
All procedures were approved by the Medical Ethics Committee of Jiangxi Provincial People's Hospital Affiliated to Nanchang University. Informed consent was obtained from all patients. All animal experimental protocols were approved by the Institutional Animal Use and Care Committee of Jiangxi Provincial People's Hospital Affiliated to Nanchang University.
Consent for publication
The informed consent was obtained from study participants.
Availability of data and material
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contribution
JYH: Conceptualization; Writing-original draft; Methodology; Formal analysis;
MC: Supervision; Validation;
JLY: Data curation;
CHP: Resources;
JC: Investigation;
BL: Software;
XYZ: Visualization;
QF: Funding acquisition; Project administration; Writing-review & editing.
All authors have read and approved the final version of this manuscript to be published.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgements
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
References
- 1.Katz J.N., Arant K.R., Loeser R.F. Diagnosis and treatment of hip and knee osteoarthritis: a review. JAMA. 2021;325(6):568–578. doi: 10.1001/jama.2020.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspden R.M., Scheven B.A., Hutchison J.D. Osteoarthritis as a systemic disorder including stromal cell differentiation and lipid metabolism. Lancet. 2001;357(9262):1118–1120. doi: 10.1016/S0140-6736(00)04264-1. [DOI] [PubMed] [Google Scholar]
- 3.Vos T., Flaxman A.D., Naghavi M., Lozano R., Michaud C., Ezzati M., et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu J., Ji M.L., Zhang X.J., Shi P.L., Wu H., Wang C., et al. MicroRNA-218-5p as a potential target for the treatment of human osteoarthritis. Mol Ther. 2017;25(12):2676–2688. doi: 10.1016/j.ymthe.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thysen S., Luyten F.P., Lories R.J. Targets, models and challenges in osteoarthritis research. Dis Model Mech. 2015;8(1):17–30. doi: 10.1242/dmm.016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le L.T., Swingler T.E., Crowe N., Vincent T.L., Barter M.J., Donell S.T., et al. The microRNA-29 family in cartilage homeostasis and osteoarthritis. J Mol Med (Berl) 2016;94(5):583–596. doi: 10.1007/s00109-015-1374-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitney K.E., Liebowitz A., Bolia I.K., Chahla J., Ravuri S., Evans T.A., et al. Current perspectives on biological approaches for osteoarthritis. Ann N Y Acad Sci. 2017;1410(1):26–43. doi: 10.1111/nyas.13554. [DOI] [PubMed] [Google Scholar]
- 8.Ryu J.S., Jeong E.J., Kim J.Y., Park S.J., Ju W.S., Kim C.H., et al. Application of mesenchymal stem cells in inflammatory and fibrotic diseases. Int J Mol Sci. 2020;21(21) doi: 10.3390/ijms21218366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kangari P., Talaei-Khozani T., Razeghian-Jahromi I., Razmkhah M. Mesenchymal stem cells: amazing remedies for bone and cartilage defects. Stem Cell Res Ther. 2020;11(1):492. doi: 10.1186/s13287-020-02001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang R., Ma J., Han J., Zhang W., Ma J. Mesenchymal stem cell related therapies for cartilage lesions and osteoarthritis. Am J Transl Res. 2019;11(10):6275–6289. [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou G., Liu W., Cui L., Wang X., Liu T., Cao Y. Repair of porcine articular osteochondral defects in non-weightbearing areas with autologous bone marrow stromal cells. Tissue Eng. 2006;12(11):3209–3221. doi: 10.1089/ten.2006.12.3209. [DOI] [PubMed] [Google Scholar]
- 12.Nukavarapu S.P., Dorcemus D.L. Osteochondral tissue engineering: current strategies and challenges. Biotechnol Adv. 2013;31(5):706–721. doi: 10.1016/j.biotechadv.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Yamasaki S., Mera H., Itokazu M., Hashimoto Y., Wakitani S. Cartilage repair with autologous bone marrow mesenchymal stem cell transplantation: review of preclinical and clinical studies. Cartilage. 2014;5(4):196–202. doi: 10.1177/1947603514534681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orozco L., Munar A., Soler R., Alberca M., Soler F., Huguet M., et al. Treatment of knee osteoarthritis with autologous mesenchymal stem cells: a pilot study. Transplantation. 2013;95(12):1535–1541. doi: 10.1097/TP.0b013e318291a2da. [DOI] [PubMed] [Google Scholar]
- 15.Latchman D.S. Transcription factors: an overview. Int J Biochem Cell Biol. 1997;29(12):1305–1312. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzaki T., Alvarez-Garcia O., Mokuda S., Nagira K., Olmer M., Gamini R., et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med. 2018;10(428) doi: 10.1126/scitranslmed.aan0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neefjes M., van Caam A.P.M., van der Kraan P.M. Transcription factors in cartilage homeostasis and osteoarthritis. Biology (Basel) 2020;9(9) doi: 10.3390/biology9090290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dy P., Wang W., Bhattaram P., Wang Q., Wang L., Ballock R.T., et al. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell. 2012;22(3):597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarvagalla S., Kolapalli S.P., Vallabhapurapu S. The two sides of YY1 in cancer: a friend and a foe. Front Oncol. 2019;9:1230. doi: 10.3389/fonc.2019.01230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawamura K., Higuchi T., Fujiwara S. YAF2-mediated YY1-Sirtuin6 interactions responsible formitochondrial down-regulation in aging tunicates. Mol Cell Biol. 2021 doi: 10.1128/MCB.00047-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y.H., Chung C.C., Liu Y.C., Lai W.C., Lin Z.S., Chen T.M., et al. YY1 and HDAC9c transcriptionally regulate p38-mediated mesenchymal stem cell differentiation into osteoblasts. Am J Cancer Res. 2018;8(3):514–525. [PMC free article] [PubMed] [Google Scholar]
- 22.Aoyama T., Okamoto T., Fukiage K., Otsuka S., Furu M., Ito K., et al. Histone modifiers, YY1 and p300, regulate the expression of cartilage-specific gene, chondromodulin-I, in mesenchymal stem cells. J Biol Chem. 2010;285(39):29842–29850. doi: 10.1074/jbc.M110.116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H., Yang S., Shao R. Long non-coding XIST raises methylation of TIMP-3 promoter to regulate collagen degradation in osteoarthritic chondrocytes after tibial plateau fracture. Arthritis Res Ther. 2019;21(1):271. doi: 10.1186/s13075-019-2033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H., Peng G., Ning X., Wang J., Yang H., Deng J. Emerging roles of long noncoding RNA in chondrogenesis, osteogenesis, and osteoarthritis. Am J Transl Res. 2019;11(1):16–30. [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Sun Y., Liu J., Yang B., Wang T., Zhang Z., et al. Roles of long noncoding RNA in osteoarthritis (Review) Int J Mol Med. 2021;48(1) doi: 10.3892/ijmm.2021.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., Li Z., Li Z., Yu B., Wang Y. Long noncoding RNAs expression signatures in chondrogenic differentiation of human bone marrow mesenchymal stem cells. Biochem Biophys Res Commun. 2015;456(1):459–464. doi: 10.1016/j.bbrc.2014.11.106. [DOI] [PubMed] [Google Scholar]
- 27.Fu M., Huang G., Zhang Z., Liu J., Zhang Z., Huang Z., et al. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis Cartilage. 2015;23(3):423–432. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Li L., Lv G., Wang B., Kuang L. The role of lncRNA XIST/miR-211 axis in modulating the proliferation and apoptosis of osteoarthritis chondrocytes through CXCR4 and MAPK signaling. Biochem Biophys Res Commun. 2018;503(4):2555–2562. doi: 10.1016/j.bbrc.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 29.Wang T., Liu Y., Wang Y., Huang X., Zhao W., Zhao Z. Long non-coding RNA XIST promotes extracellular matrix degradation by functioning as a competing endogenous RNA of miR-1277-5p in osteoarthritis. Int J Mol Med. 2019;44(2):630–642. doi: 10.3892/ijmm.2019.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L., Wang J.F., Fan J., Rao Y.S., Liu F., Yan Y.E., et al. Nicotine suppressed fetal adrenal StAR expression via YY1 mediated-histone deacetylation modification mechanism. Int J Mol Sci. 2016;17(9) doi: 10.3390/ijms17091477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glasson S.S., Blanchet T.J., Morris E.A. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15(9):1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y., Cossman J., Jayasuriya C.T., Li X., Guan Y., Fonseca V., et al. Deficient mechanical activation of anabolic transcripts and post-traumatic cartilage degeneration in matrilin-1 knockout mice. PLoS One. 2016;11(6) doi: 10.1371/journal.pone.0156676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh J.A., Noorbaloochi S., MacDonald R., Maxwell L.J. Chondroitin for osteoarthritis. Cochrane Database Syst Rev. 2015;1:CD005614. doi: 10.1002/14651858.CD005614.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neogi T. The epidemiology and impact of pain in osteoarthritis. Osteoarthritis Cartilage. 2013;21(9):1145–1153. doi: 10.1016/j.joca.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieberthal J., Sambamurthy N., Scanzello C.R. Inflammation in joint injury and post-traumatic osteoarthritis. Osteoarthritis Cartilage. 2015;23(11):1825–1834. doi: 10.1016/j.joca.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuyinu E.L., Narayanan G., Nair L.S., Laurencin C.T. Animal models of osteoarthritis: classification, update, and measurement of outcomes. J Orthop Surg Res. 2016;11:19. doi: 10.1186/s13018-016-0346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makris E.A., Gomoll A.H., Malizos K.N., Hu J.C., Athanasiou K.A. Repair and tissue engineering techniques for articular cartilage. Nat Rev Rheumatol. 2015;11(1):21–34. doi: 10.1038/nrrheum.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang S., Xie Z., Wang P., Li J., Wang S., Liu W., et al. LncRNA-OG promotes the osteogenic differentiation of bone marrow-derived mesenchymal stem cells under the regulation of hnRNPK. Stem Cell. 2019;37(2):270–283. doi: 10.1002/stem.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silva A.M., Moura S.R., Teixeira J.H., Barbosa M.A., Santos S.G., Almeida M.I. Long noncoding RNAs: a missing link in osteoporosis. Bone Res. 2019;7:10. doi: 10.1038/s41413-019-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H., He C., Liu Y., Li X., Zhang C., Qin Q., et al. LncRNA-GAS5 inhibits expression of miR 103 and ameliorates the articular cartilage in adjuvant-induced arthritis in obese mice. Dose Response. 2020;18(4) doi: 10.1177/1559325820942718. 1559325820942718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H., Cao Y., Zhang J., Liang Y., Su X., Zhang C., et al. DLX5 and HOXC8 enhance the chondrogenic differentiation potential of stem cells from apical papilla via LINC01013. Stem Cell Res Ther. 2020;11(1):271. doi: 10.1186/s13287-020-01791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang M.J., Zhao J.Y., Xu J.J., Li J., Zhuang Y.F., Zhang X.L. lncRNA ADAMTS9-AS2 controls human mesenchymal stem cell chondrogenic differentiation and functions as a ceRNA. Mol Ther Nucleic Acids. 2019;18:533–545. doi: 10.1016/j.omtn.2019.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S., Li Y., Zhi S., Ding Z., Huang Y., Wang W., et al. lncRNA xist regulates osteoblast differentiation by sponging miR-19a-3p in aging-induced osteoporosis. Aging Dis. 2020;11(5):1058–1068. doi: 10.14336/AD.2019.0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ballarino M., Jobert L., Dembele D., de la Grange P., Auboeuf D., Tora L. TAF15 is important for cellular proliferation and regulates the expression of a subset of cell cycle genes through miRNAs. Oncogene. 2013;32(39):4646–4655. doi: 10.1038/onc.2012.490. [DOI] [PubMed] [Google Scholar]
- 45.Li Z., Wang J., Yang J. TUG1 knockdown promoted viability and inhibited apoptosis and cartilage ECM degradation in chondrocytes via the miR-17-5p/FUT1 pathway in osteoarthritis. Exp Ther Med. 2020;20(6):154. doi: 10.3892/etm.2020.9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen L., Borgs L., Vandenbosch R., Mangin J.M., Beukelaers P., Moonen G., et al. The Yin and Yang of cell cycle progression and differentiation in the oligodendroglial lineage. Ment Retard Dev Disabil Res Rev. 2006;12(2):85–96. doi: 10.1002/mrdd.20103. [DOI] [PubMed] [Google Scholar]
- 47.Makhlouf M., Ouimette J.F., Oldfield A., Navarro P., Neuillet D., Rougeulle C. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat Commun. 2014;5:4878. doi: 10.1038/ncomms5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Syrett C.M., Sindhava V., Hodawadekar S., Myles A., Liang G., Zhang Y., et al. Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. 2017;13(10) doi: 10.1371/journal.pgen.1007050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.