Abstract
Since its first appearance, CRISPR–Cas9 has been developed extensively as a programmable genome-editing tool, opening a new era in plant genome engineering. However, CRISPR–Cas9 still has some drawbacks, such as limitations of the protospacer-adjacent motif (PAM) sequence, target specificity, and the large size of the cas9 gene. To combat invading bacterial phages and plasmid DNAs, bacteria and archaea have diverse and unexplored CRISPR–Cas systems, which have the potential to be developed as a useful genome editing tools. Recently, discovery and characterization of additional CRISPR–Cas systems have been reported. Among them, several CRISPR–Cas systems have been applied successfully to plant and human genome editing. For example, several groups have achieved genome editing using CRISPR–Cas type I-D and type I-E systems, which had never been applied for genome editing previously. In addition to higher specificity and recognition of different PAM sequences, recently developed CRISPR–Cas systems often provide unique characteristics that differ from well-known Cas proteins such as Cas9 and Cas12a. For example, type I CRISPR–Cas10 induces small indels and bi-directional long-range deletions ranging up to 7.2 kb in tomatoes (Solanum lycopersicum L.). Type IV CRISPR–Cas13 targets RNA, not double-strand DNA, enabling highly specific knockdown of target genes. In this article, we review the development of CRISPR–Cas systems, focusing especially on their application to plant genome engineering. Recent CRISPR–Cas tools are helping expand our plant genome engineering toolbox.
Recently discovered and characterized clustered regularly interspaced short palindromic repeats-CRISPR associated (CRISPR–Cas) systems allow additional applications to plant genome editing.
Introduction
The rapid progress of plant genome editing technologies has brought about a dramatic evolution in plant research and plant breeding. The most general and well-known genome editing tool is clustered regularly interspaced short palindromic repeats-CRISPR associated protein 9 (CRISPR–Cas9), which applies a programmable RNA-guided Cas9 endonuclease (Osakabe and Osakabe, 2015; Wang et al., 2016; Wada et al., 2020) to edit target sequences. Especially, CRISPR–Cas9 from Streptococcus pyogenes (SpyCas9) has been applied for genome editing in many organisms (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013; Osakabe and Osakabe, 2015; Wang et al., 2016; Jaganathan et al., 2018; Wada et al., 2020). The main drawbacks of CRISPR–Cas9 are the limitations of available protospacer-adjacent motif (PAM) sequences, target specificity, and its large size, which prevents delivery by virus-based vectors (Yang et al., 2021). To overcome these problems, engineering of SpyCas9 (Kleinstiver et al., 2015, 2016; Slaymaker et al., 2016; Chen et al., 2017; Hu et al., 2018; Lee et al., 2018; Nishimasu et al., 2018; Walton et al., 2020) and mining of Cas9 putative orthologs from many bacteria and archaea (Hou et al., 2013; Ran et al., 2015; Hirano et al., 2016; Müller et al., 2016; Kim et al., 2017) have progressed with surprising speed in recent years. These efforts have brought great success, such as the development of near-PAM-less engineered SpyCas9 variants (Walton et al., 2020), highly specific SpyCas9 variants (Kleinstiver et al., 2016; Slaymaker et al., 2016; Chen et al., 2017; Lee et al., 2018), and the smallest Cas9 yet discovered from Campylobacter jejuni (Kim et al., 2017). When fused with effector proteins, catalytically inactive Cas9 (dCas9) has also brought promising applications (Wang et al., 2016; Adli, 2018; Molla and Yang, 2019), including transcriptional control (Qi et al., 2013), epigenetic editing (Gjaltema and Rots, 2020), live cell imaging (Wu et al., 2019), and base editing (Komor et al., 2016; Nishida et al., 2016; Gaudeli et al., 2017; Molla and Yang, 2019). An additional strategy—prime editing—has also been developed to enable precise genome editing without inducing DNA double-strand breaks (Anzalone et al., 2019). Another CRISPR–Cas, CRISPR–Cas12a (previously known as Cpf1) has also been applied to genome editing in both mammalian and plant cells (Zetsche et al., 2015; Alok et al., 2020). More recently, even more CRISPR–Cas systems have been reported continuously and applied to genome editing (Murugan et al., 2017). Classification and general characteristics of each type of CRISPR–Cas systems have been summarized in Table 1 and Box 1. These CRISPR–Cas systems have opened a new era in CRISPR–Cas technology applications.
Table 1.
General characteristics of each type of CRISPR–Cas systema
| Class | Type | Subtype | Signature Gene | Target | crRNA | Representative Cas Proteins Related to Each Step |
||
|---|---|---|---|---|---|---|---|---|
| RNA Processing | Target Binding | Target Cleavage | ||||||
| I | Type I | 7 | cas3 | DNA | Single crRNA | Cas6 | Cas5, Cas7,Cas8, Cas11 | Cas3 |
| (db) | cas3 | DNA | Single crRNA | Cas6 | Cas5, Cas7 | Cas10 | ||
| I | Type III | 6 | cas10 | DNA/RNA | Single crRNA | Cas6 | Cas5, Cas7, Cas11, Cas10 | Cas10 |
| I | Type IV | 3 | csf1 c | – | – | Cas6 | Cas5, Cas7, Cas11, Csf1 | |
| II | Type II | 3 | cas9 | DNA | tracrRNA:crRNA | Cas9, RNase III | Cas9 | Cas9 |
| II | Type V | 10 | cas12 | DNA | Single crRNA/tracrRNA:crRNA | Cas12 | Cas12 | Cas12 |
| II | Type VI | 4 | cas13 | RNA | Single crRNA | Cas13 | Cas13 | Cas13 |
Classification is based on Makarova et al. (2020).
A cas3 gene is present as a signature gene in type I-D but cleavage is performed by Cas10, not Cas3.
Some type IV CRISPR–Cas systems lack a csf1 gene.
Box 1.
Classification of CRISPR–Cas systems
Bacteria and archaea have a variety of CRISPR–Cas systems that function naturally as an adaptive and heritable immune system (Makarova et al., 2020). In the current classification, CRISPR–Cas has been classified into 2 classes, 6 types, and 33 subtypes based on the gene organization of CRISPR–Cas loci, the presence of the signature cas gene, sequence similarity and phylogenetic analysis of conserved Cas proteins (Makarova et al., 2020). Class is defined by an organization of effector proteins. Class I CRISPR–Cas systems (types I, III, and IV) have a multi-effector complex termed CRISPR-associated complex for antiviral defense (Cascade), whereas Class II CRISPR–Cas systems (types II, V, and VI) has a single multi-domain effector Cas protein (Makarova et al., 2020; Figure 1; Table 1). Each type is represented by the presence of signature proteins: Cas3 in Type I, Cas9 in Type II, Cas10 in Type III, Csf1 in Type IV, Cas12 in Type V, and Cas13 in Type VI. Classification of subtype has been more complicated (for details, please refer to a review by Makarova et al., 2020). Recently, we identified that Cas10 is an important component in TiD (Osakabe et al., 2020, 2021). This diversification of CRISPR–Cas results from evolution in an intensive arms race between bacteria and their phage foes (Hampton et al., 2020). Each type and subtype has different characteristics in terms of recognition of polynucleotides (DNA or RNA), organization of cas genes, target recognition and cleavage mechanisms (Makarova et al., 2020; Table 1).
Here, we review the application to plant genome editing of CRISPR–Cas systems other than the well-known CRISPR–Cas9 or -Cas12a. Several CRISPR–Cas systems show double-strand DNA (dsDNA) cleavage activity similar to Cas9, but have different unique and interesting functions to induce site-directed mutagenesis (Murugan et al., 2017). Most of these technologies have generally been applied and characterized in human cells; however, information gained from genome editing in humans can also be applied effectively to plant genetic engineering. Later in this review, we will discuss future developments and the application of alternative CRISPR–Cas systems to plant genome engineering.
Class I Type I-B, I-E, and I-F CRISPR–Cas
Class I Type I CRISPR–Cas is the most abundant CRISPR–Cas in bacteria and archaea (Makarova et al., 2020). However, its application to genome editing was not reported until recently. The structure and basic mechanisms of Type I-E CRISPR–Cas are the best characterized to date (Broun et al., 2008; Westra et al., 2012; Jackson et al., 2014; Mulepati et al., 2014; Zhao et al., 2014; Hayes et al., 2016; Xiao et al., 2017, 2018; Loeff et al., 2018). Type I-E CRISPR–Cas consists of Cas5e (CasD), Cas6e (CasE), Cas7e (CasC), Cas8e (Cse1 and CasA), Cas11e (Cse2 and CasB), Cas3e, and crispr RNA (crRNA; Westra et al., 2012; Figure 1B). Five Cas proteins (Cas5e, Cas6e, Cas7e, Cas8e, and Cas11e) form a CRISPR-associated complex for antiviral defense (Cascade) complex with crRNA, forming an R-loop structure between crRNA and the target DNA (Xiao et al., 2017). When formation of the R-loop structure is complete, a nuclease (Cas3e) is recruited to the target sequence, resulting in processive degradation of the target DNA strand (Xiao et al., 2018). Other Type I systems also have similar Cascade structures although with some differences. One of the expected advantages of Type I CRISPR–Cas in genome editing is the higher specificity than CRISPR–Cas9 as the Cascade complex generally recognizes a target sequence of 30 bp or longer (Young et al., 2019).
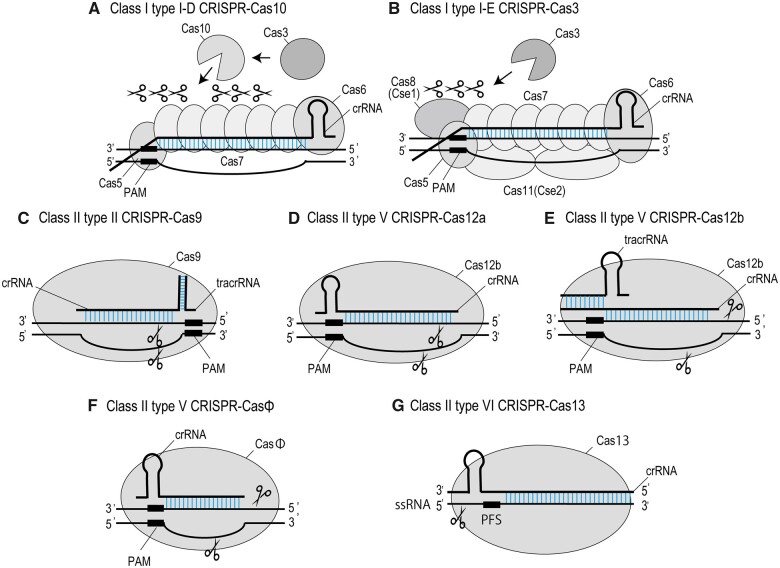
Figure 1.
Representative structures of CRISPR–Cas systems. Cas proteins with crRNAs are shown with double-strand DNAs in (A)–(F) and with single-strand RNA in (G). A, Class I type I-D CRISPR–Cas10. Cas10 and Cas3 bind to the Cascade complex containing Cas5, Cas6, Cas7, and crRNA, then Cas10 cleaves and digests dsDNA at target site. B, Class I type I-E CRISPR–Cas3. Cas3 is recruited to the Cascade complex containing Cas5, Cas6, Cas7, Cas8, Cas11, and crRNA, then digests dsDNA at target sites. C, Class II type II CRISPR–Cas9. Single Cas9 protein forms a complex with crRNA annealing to tracrRNA and cleaves dsDNAs at target sites, producing blunt ends. D, Class II type V CRISPR–Cas12a. Single Cas12a protein forms a complex with crRNA and cleaves dsDNAs at target site, producing cohesive ends. E, Class II type V CRISPR–Cas12b. Single Cas12b protein forms a complex with crRNA annealing to tracrRNA and cleaves dsDNAs at target site, producing cohesive ends. F, Class II type V CRISPR–CasΦ. Single CasΦ protein forms a complex with crRNA and cleaves dsDNA at target sites, producing cohesive ends. G, Class II type VI CRISPR–Cas13. Single Cas13 protein forms a complex with crRNA, and cleaves single strand RNA of target gene. PFS, protospacer flanking sequence.
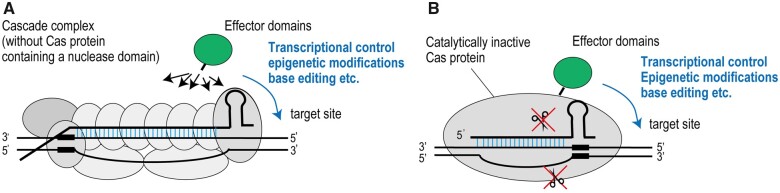
In 2019, several studies reported the successful application of Type I CRISPR–Cas to transcriptional control and gene editing in human cells (Cameron et al., 2019; Dolan et al., 2019; Morisaka et al., 2019; Pickar-Oliver et al., 2019; Chen et al., 2020). Representative structures of Class I Cascade-effectors and Class II inactive Cas-effectors used for transcriptional control are shown in Figure 2. Pickar-Oliver et al. (2019) modified gene expression of a target gene by tethering activation (human acetyltransferase p300) or repression (Krüppel-associated box, KRAB) domains to Escherichia coli type I-E Cascade (EcCascade) or Listeria monocytogenes Finland_1998 type I-B Cascade (LmoCascade) in human cells. They found that tethering of p300 to Cas8e, Cse2, Cas5e, or Cas6e could induce gene activation without abrogating complex formation. By tethering p300 or KRAB domains to Cas6e, gene expression could be modified with high efficiency and high specificity (Pickar-Oliver et al., 2019). Similarly, type I-F Cascade has also been applied for transcriptional control in human cells (Chen et al., 2020). Interestingly, Chen et al. (2020) demonstrated that tethering of transcription activation domain VP64-p65-Rta (VPR) to Csy3 (Cas7 equivalent) of Pseudomonas aeruginosa type I-F Cascade (PaeCascade) could activate expression of the target gene with high specificity and more efficiently than dCas9-VPR, dAsCas12a-VPR, or EcoCascade-VPR. Expression of PaeCascade containing PaeCsy3-VPR resulted in accumulation of six copies of Csy3-VPR at the target site (a Csy3 protein per every 6 nt of the crRNA). The extended crRNA also recruited more Csy3-VPR to target site, resulting in enhanced gene activation (Chen et al., 2020). Interestingly, expression of type I-B LmoCascade containing LmoCas7-VPR failed to induce gene activation (Pickar-Oliver et al., 2019), suggesting that optimization of linker sequence is required or that the effects of Cas7-VPR on transcriptional control differ depending on the CRISPR–Cas system used. In addition, type I-F PaeCascade needs fewer Cas proteins to function than the type I-E EcoCascade (four Cas proteins versus five Cas proteins). Requirement for fewer Cas components makes the vector system simpler, contributing to the development of easy-to-use genome manipulation tools.
Figure 2.
Representative structure of Cas proteins with effector proteins. Class I Cascade complex fused to an effector domain (A) and Class II catalytically inactive Cas protein fused to an effector domain (B) are shown. These engineered Cas proteins have been applied for transcriptional control, epigenetic modification, base editing, etc. at target sites. Effector domains can be fused to any Cas proteins of Cascade complex (A) whereas an effector domain can be fused to one Cas protein (B).
For genome editing, Cameron et al. (2019) fused a dimerization-dependent, nonspecific FokI nuclease domain to Cas8e and found that the FokI-Cascade from Pseudomonas sp. S-6–2 (PseCascade) using a paired gRNA showed higher genome editing efficiency (up to 50%) than other FokI-Cascade complexes such as FokI-EcCascade. Specificity of the FokI-Cascade was analyzed by GUIDE-seq. While more than 250 off-targets were recovered and their read counts ranged from 11 to 41,733 reads/421,646 total reads in Cas9-treated samples, at most 2 off-target sites were detected, and their reads were only 20 reads/10,757 total reads and 35 reads/10,757 total reads, respectively in any of the FokI-PseCascade paired gRNAs-treated samples. These results indicate the high specificity of FokI-Cascade.
Genome editing using full components of type I-E CRISPR–Cas (including Cas3e) was reported by three groups in 2019 (Dolan et al., 2019; Morisaka et al., 2019; Pickar-Oliver et al., 2019). Dolan et al. (2019) achieved genome editing using Thermobifida fusca type I-E CRISPR–Cas. They purified the TfCascade and Cas3 complex and delivered them as a ribonucleoprotein (RNP) into human embryonic stem cells, resulting in successful induction of mutations at target sites. Interestingly, type I-E CRISPR–Cas induced long-range genomic deletions of up to 100 kb at target sites—a characteristic unique to type I-E CRISPR–Cas and different from the mutation patterns induced by Cas9. The deletions were unidirectional, toward the region upstream of the PAM sequence. Cameron et al. (2019) has also successfully induced mutations using a full PseCascade–Cas3 complex in human cells. Their strategy involved plasmid DNA-based delivery of the full PseCascade–Cas3 complex into human HEK293 cells. The mutation patterns were similar to those reported by Dolan et al. (2019). Morisaka et al. (2019) also delivered EcCascade–Cas3 into human HEK293T cells by using plasmid DNA, and achieved genome editing in human cells. A comprehensive analysis of mutation patterns demonstrated that type I-E CRISPR–Cas can induce unidirectional long deletions with high specificity. In addition, they indicated that type I-E CRISPR–Cas can knock out target genes efficiently and can also be used for knock-in of a DNA fragment via the homology-directed repair (HDR) pathway. By achieving exon skipping of the dystrophin gene in patient-induced pluripotent stem cells, they demonstrated the potential of type I-E CRISPR–Cas for future therapeutic applications (Morisaka et al., 2019).
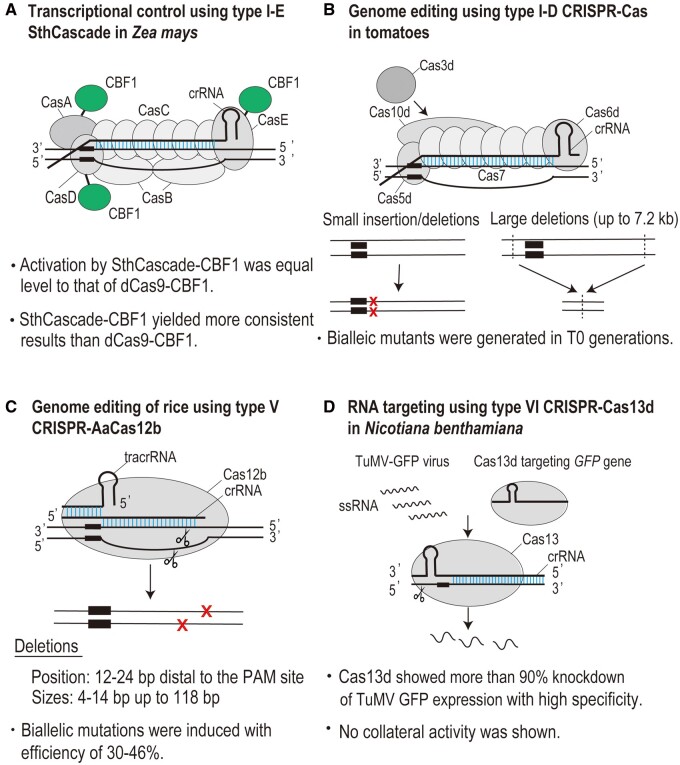
On the other hand, the application of type I-E CRISPR–Cas to plant genome engineering has been limited to date. The only application of type I-E CRISPR–Cas in plant cells has been in transcriptional control in Zea mays (Young et al., 2019; Figure 3A). Young et al. (2019) optimized and engineered type I-E CRISPR–Cas from Streptococcus thermophilus for gene activation in corns (Z. mays L.). They fused a C-terminal acidic plant transcriptional activation domain from Arabidopsis (Arabidopsis thaliana) C-REPEAT BINDING FACTOR/DRE BINDING FACTOR 1 (CBF1) to the 3′-end of Cas8e, Cas5e, and Cas6e, respectively. Each gene was expressed by the Zm ubiquitin promoter-intron-5′-untranslated region and potato (Solanum tuberosum L.) proteinase inhibitor (PinII) terminator (Ter). The repeat-spacer-repeat sequence was expressed by a polymerase III promoter and Ter from a Z. mays U6 gene. Using these vectors, Young and coworkers successfully activated the expression of target genes on the reporter plasmid and also on endogenous chromosomes. Interestingly, they indicated that fusion of CBF1 into multiple Cas proteins gave synergistic effects for gene activation. As a target of endogenous gene activation, they selected a transcription factor r gene and c1 gene, which are involved in the production of anthocyanin. By simultaneously overexpressing the c1 gene, targeting the r gene by SthCascade-CBF1 produced a near equivalent anthocyanin signal to targeting by dCas9-CBF1. SthCascade yielded more consistent chromosomal activation than dCas9-CBF1. These results suggest the advantage of using the multi-effector complex of type I CRISPR–Cas for gene manipulation, leading to the accumulation of multiple activation domains at target sites. Young et al. (2019) did not address specificity of gene activation in their study, but from the study of Chen et al. (2020) it would be expected that the StCascade has higher specificity than dCas9. A disadvantage of using the multi-effector complex is the requirement for simultaneous or sequential introduction of multiple genes. The RNP-based delivery system is one of the promising strategies to overcome this problem. It would be also interesting to see if unidirectional long-range deletions can be induced in plant cells using type I-E CRISPR–Cas.
Figure 3.
Application of recently developed CRISPR–Cas tools to plant genome engineering. (A) Type I-E SthCascade-CBF1 for targeted gene activation in Z. mays (Young et al., 2019). B, TiD for targeted mutagenesis in tomatoes (Osakabe et al., 2020). C, Type V CRISPR–AaCas12b for targeted mutagenesis in rice (Ming et al., 2020). D, RNA targeting using type VI CRISPR–Cas13d in N. benthamiana (Mahas and Mahfouz, 2018). Sth, S. thermophilus DGCC7710.
Class I type I-D CRISPR–Cas
Recently, we characterized type I-D (TiD) CRISPR–Cas loci from Microcystis aeruginosa, and successfully developed a genome editing tool, which we named TiD, based on this system (Osakabe et al., 2020, 2021; Figure1A; Figure 3B). Several groups have reported in vitro DNA binding and cleavage capability of several type I-D systems from bacteria such as Sulfolobus islandicus and Synechocystis sp. PCC 6803 (Manav et al., 2020, McBride et al., 2020, Lin et al., 2020); however, its applications to genome editing in eukaryotic cells have not been reported. TiD consists of five Cas proteins (Cas 3d, 5d, 6d, 7d, and 10d) and a crRNA that recognizes a 35-nt or 36-nt target sequence. Among TiD Cas proteins, Cas10d is a unique protein not found in other type I CRISPR–Cas systems. Generally, Cas3 has a histidine–aspartate (HD) nuclease domain and functions as a nuclease. However, in the TiD system, it is Cas10d that instead has the HD nuclease domain. Cas10 is known as a signature protein of the type III CRISPR–Cas family, but the Cas10d in TiD has highly diverged in comparison with its type III counterparts. The Cas10d HD domain is rather similar to the Cas3d HD domains of type I-B, -C, -E, and -F. Therefore, although TiD is a unique system that possesses both types I and III signature genes, it is assigned to type I. The presence of an HD domain in Cas10d raised the hypothesis that Cas10d plays a role as a nuclease. Hence, we analyzed ssDNA nuclease activity in vitro and the results indeed indicated that Cas10d, but not Cas3d, has ssDNA nuclease activity (Osakabe et al., 2020). Both these Cas proteins also showed ATPase activity, suggesting that they function as a helicase to unwind the dsDNA. TiD recognizes 5′-GTH-3′ as a PAM together with the following 35- or 36-nt target sequences. Like other type I-based systems, recognition of a longer target sequence (35 or 36 nt) than that of Cas9 (20 nt) suggests that TiD has higher specificity than CRISPR–Cas9. Using TiD, we have successfully induced mutations at target genes in human cells (Osakabe et al., 2021). Interestingly, our results showed that the mutation patterns produced by TiD in both human cells and plants were different from those induced by known Cas proteins such as Cas9, Cas12a, and Cas3: TiD introduced not only small insertion/deletion but also long-range deletions (ranging from 2.5 kb to 18.5 kb), and its direction was not uni-directional, but bi-directional (Osakabe et al., 2020, 2021). Although the mutation patterns induced by genome editing would depend on the host organisms and the presence of active repair pathways, TiD would be a tool that can induce both small indels and long deletions, whereas majority of mutations induced by CRISPR–Cas9 and Cas12 are small indels (they also induce long deletions in some cases), and type I-E CRISPR–Cas can induce only unidirectional long deletions. These differences can be attributed to the function of a unique nuclease protein, Cas10d.
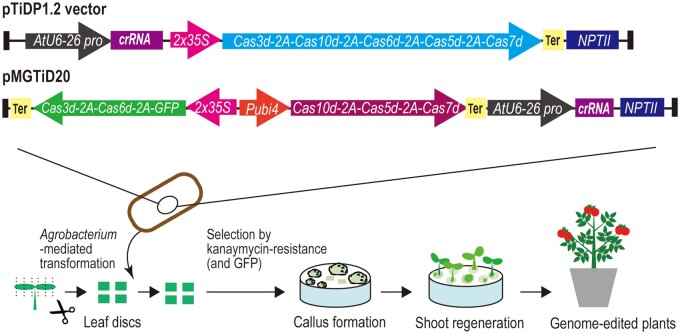
We optimized TiD for genome editing in plant cells by using plant-cell specific-promoters (CaMV35S, Parsley Ubiquitin 4-2) for expression of codon-optimized cas genes and the AtU6-26 promoter for expression of crRNA (Figure 4). Using plant-optimized TiD vectors targeting tomato INDOLE-3-ACETIC ACID9 (SlIAA9, an important factor for parthenocarpy) and RIPENING INHIBITOR (an important factor for fruit ripening) genes, we introduced mutations successfully into tomato plants (S. lycopersicum L., cv. Micro-Tom and Ailsa Craig) (Osakabe et al., 2020; Figure 4. As with the mutation patterns induced in human cells, short indels and long deletions were detected in callus and shoots. Small indels were induced in 64% of transgenic calli. Long-range deletions were also detected in transgenic calli and shoots. We detected up to 7.2 kb of bi-directional long-range deletions around the target sequence. Sequence analysis of regenerated shoots indicated that the transgenic shoots included 100% mutated DNA sequence, revealing that TiD could induce bi-allelic mutations in the T0 generation. SlIAA9 knockout phenotypes (seedless fruit, changes in leaf morphology) were also observed.
Figure 4.
Genome editing by TiD. Scheme for generations of genome-edited tomatoes by TiD is shown. All-in one vector (pTiDP1.2 vector) and two cassette vector (pMGiD20) were used for expression of cas genes and crRNA. Several cas genes and the GFP gene were linked by 2A sequences to enable co-expression of these genes in a single expression cassette. 2x35S, 2x cauliflower mosaic virus 35S promoter, Pubi4: Parsley ubiquitin 4 promoter, NPT, neomycin phosphotransferase.
To identify the basic characteristics of TiD as a genome editing tool, we searched on-target sites and off-target candidate sequences with 0–5 mismatches against Arabidopsis, rice (O. sativa L.) and tomato whole genome for TiD (Osakabe et al., 2020). The results indicated that more on-target sites for TiD exist in tomato and Arabidopsis than for Cas9, but there are fewer in rice. For off-target sites, TiD has clearly fewer off-target candidate sites than Cas9 in these plant genomes, suggesting an advantage for TiD as a highly specific genome editing tool in plant cells. Off-target mutations, including short indels and long deletions, were also not detected from SlIAA9 knockout plants, and the on-target mutations were transmitted to the next generation (Osakabe et al., 2020). These results show that TiD is a useful and unique alternative genome editing tool for both human and plant genome editing. Although further improvements and research is needed, such as identifying regulatory control mechanisms, long-range deletions would allow TiD to be used for chromosome engineering, further expanding what is possible in plant genome engineering.
Class II type V-B CRISPR–Cas12b (C2c1)
CRISPR–Cas type V features an RNA-guided effector protein, Cas12, which contains RuvC domain. Distinct architectures and the diverged RuvC sequences of Cas12 proteins suggest functional diversity (Yan et al., 2019). Identified Cas12 proteins indicate a range of functional activities, including targeting and collateral cleavage of single-strand RNA (ssRNA) and DNA, as well as dsDNA nicking and cleavage (Yan et al., 2019). CRISPR–Cas12a is a well-studied tool and we refer the reader to some excellent detailed reviews of its application (Zetsche et al., 2015; Alok et al., 2020) rather than covering this topic here. On the other hand, CRISPR–Cas12b (formerly known as C2c1) is an alternative type V CRISPR–Cas system with a dual-RNA-guided endonuclease, meaning that it requires two kinds of RNA (crRNA and trans-activating crRNA [tracrRNA]) for its nuclease function, whereas Cas12a requires only crRNA (Shmakov et al., 2015; Table 1; Figure 1, D and E). Cas12b has a conserved RuvC nuclease domain and Nuc domain that has no similarity with Cas12a. CRISPR–Cas12b is an attractive tool because Cas12b produces a long staggered end distal to the PAM, and it is smaller than Cas9 (Teng et al., 2018). However, its optimal temperature for DNA cleavage of Cas12b is generally higher than 40°C, suggesting that it is not suitable for genome editing in mammalian and plant cells. In 2018, Teng et al. (2018) identified a Cas12b from Alicyclobacillus acidiphilus (AaCas12b) that can maintain optimal nuclease activity at 31–59°C. Using AaCas12b, they successfully induced mutations at target genes in human and mouse cells (Teng et al. 2018). Mutation efficiency by AaCas12b in human cells was 21.5%. Multiplex genome editing using four gRNAs was also achieved, with a mutation efficiency of 3%–20% depending on the target site. Gene activation using a nuclease-deficient mutant of AaCa12b was also possible, although activation was not as strong as when using dSpyCas9. AaCas12b did not induce mutations at 88 predicted off-target sites, while SpyCas9 induced 3 out of 82 predicted off-target sites, indicating that AaCas12b has higher specificity than SpyCas9. Cas12b putative orthologs were also identified and their interchangeability between Cas12b effectors and dual-RNAs derived from other CRISPR–Cas12b systems revealed (Teng et al., 2019).
Strecker et al. (2019a, 2019b) performed protein engineering of Cas12b to change the optimal DNA cleavage temperature to 37°C. They identified mesophilic Bacillus hisashii Cas12b and produced a gain-of-function mutant, BhCas12b v4, that exhibited increased dsDNA cleavage activity and reduced nickase activity to the nontarget DNA strand at 37°C. BhCas12b v4 induced prominent larger deletions of 5–15 bp at target sites. Mutation efficiency was comparable to, or a little lower than, that of SpyCas9, depending on the target site. Also, no off-target cleavages were detected in BhCas12b v4-treated samples while SpyCas9-treated samples included off-target mutations, indicating the higher specificity of BhCas12b v4 when compared with SpyCas9 (similar to AaCas12b).
For the application of Cas12b to plant genome editing, Ming et al. (2020) compared Cas12b proteins from various bacteria in monocot rice (O. sativa): Cas12b from Alicyclobacillus acidoterrestris (Aac), A. acidiphilus (Aa), Bacillus thermoamylovorans (Bth), and B. hisashii. Comparison of genome editing efficiency suggested that AaCas12b is a more efficient genome editing tool than other Cas12b proteins in rice. AaCas12b and AacCas12b recognized VTTV (V: A, C, G) PAMs, preferentially ATTV and GTTG PAM. In particular, AaCas12b induced mutations with high efficiency (>50%) at ATTA, ATTC, and GTTG PAM in rice protoplasts, although the mutation efficiency was very different depending on the target site. AaCas12b generated biallelic mutants with an efficiency of 30%–46%. The deletions occurred at about 12–24 bp distal to the PAM site, and they were larger than those induced by Cas9 (Figure 3C). Moreover, inactivated AaCas12b variants with transcriptional repression/activation domains have controlled the gene expression of targeted genes successfully. These results reveal Cas12b as a promising tool for genome editing in rice.
Because cotton (Gossypium hirsutum) is resistant to high temperature, it raised the possibility that heat-inducible AaCas12b could be used for genome editing in cotton. Wang et al. (2020) assessed this possibility, testing various temperature conditions and durations during the callus induction stage after Agrobacterium tumefaciens-infection of cotton. They found that exposure of explants to 45°C for 4 d resulted in highest genome editing efficiency (17.4%) with little adverse effect on cotton cell culture. AaCas12b induced deletions of 1–16 bp, with the majority ranging from 9 to 14 bp. This is larger than the average size of deletions induced by SpyCas9, which is consistent with results in human cells (Teng et al., 2018). Since cotton is an allotetraploid plant, derived from the ancestral hybridization of two diploid genome (A and D), multiple copies exist in almost all genes. Interestingly, mutations were induced in the GhCLA gene in the Dt sub-genome more efficiently than in the GhCLA gene in the At sub-genome. This suggests that differences in chromatin structure affect genome editing efficiency by Cas12b in cotton (Wang et al. 2020). Off-target mutations were not detected in genome-edited cotton.
The third application of CRISPR–Cas12b in plants is genome editing of the dicot Arabidopsis (Wu et al., 2020). Plant codon-optimized BvCas12b and BhCas12b v4 were first tested in Arabidopsis protoplasts. Both BvCas12b and BhCas12b v4 proteins induced mutations, with efficiencies ranging from 1.0% to 1.7% with BvCas12b and from 1.0% to 1.5% using BhCas12b v4. Deletions of 5–13 bp (larger than generally induced by Cas9) were induced most frequently at FLS2 gene mutations by BvCas12b and BhCas12b version 4. Summarizing the results obtained at four target loci, editing efficiency differed depending on the target site, reaching a maximum of 4.3%, and was lower than that of SpyCas9. Wu et al. (2020) also showed that multiplex genome editing can be induced successfully using BvCas12b and BhCas12b v4, creating a deletion of approximately 1 kb between two target sites. No off-target effects were detected. They also introduced BvCas12b or BhCas12b v4 and gRNA into Arabidopsis plants by a floral dip method. The PDS3 gene, mutation of which gives an albino phenotype, was chosen as a target gene. Several transgenic T1 plants selected by hygromycin resistance following Agrobacterium infection in mature plants showed an albino phenotype, indicating that CRISPR–Cas12b can be applied for genome editing in the dicot plant Arabidopsis.
Class II type V CRISPR–CasΦ
CRISPR–CasΦ is a recently identified hypercompact CRISPR–Cas discovered in bacteriophage genomes (Pausch et al. 2020; Figure 1F). CasΦ is very small (–70 kDa), about half the size of SpyCas9 and Cas12a. CasΦ has a RuvC domain at its C-terminus with remote homology to the RuvC domain of the TnpB nuclease superfamily that is considered an ancestor of type V CRISPR–Cas proteins (Pausch et al. 2020). Unlike other CRISPR–Cas systems such as SpyCas9 or Cas12a, the RuvC domain of CasΦ catalyzes both crRNA processing and target DNA cleavage. CRISPR–CasΦ recognizes 5′-TBN-3′ (B: G, T, and C) as a PAM and cleaves dsDNA, ssDNA but not ssRNA. tracrRNA was not required for DNA cleavage activity. CasΦ generates staggered 5′-overhangs of 8–12 nt at cleavage sites. Similar to other type V CRISPR–Cas systems, trans ssDNA cleavage activity was observed upon the activation of CRISPR–Cas by binding to target sites in vitro.
CRISPR–CasΦ has been applied successfully to genome editing in human cells with efficiency ranging from 10% to 30% (in the case of CasΦ-2 protein; Pausch et al. 2020). CRISPR–CasΦ has also been delivered into Arabidopsis protoplasts as RNPs targeting the PDS3 gene (Pausch et al. 2020). Deletions of around 8–10 bp were induced at target sites with an efficiency of 0.85%. Although the efficiency is not high at this stage, it should be noted that this system is only at an early stage of testing, and further optimization will be needed to utilize this CRISPR–CasΦ in genetic engineering. Off-target effects have not yet been investigated. Specificity should also be addressed for further application of CRISPR–CasΦ. The biggest advantage of CRISPR–CasΦ is the small size of the CasΦ protein, which means it can be packaged in virus-based vectors. Such vectors offer high expression of transgenes without the need to integrate the transgenes into host genomic DNA, but there is a limit to the size of DNA that can be packaged. Thus, CRISPR–CasΦ would be useful for genome editing via virus-based vectors.
Class II type VI CRISPR–Cas13
Type VI CRISPR–Cas is a unique system that can recognize and cleave single-stranded RNA, but not double-stranded DNA by a signature protein, Cas13 (Shmakov et al., 2015, 2017; Figure 1G). Cas13 has two structurally distinct higher eukaryote and prokaryote nucleotide-binding (HEPN) domains, by which Cas13 can target and process precrRNA into mature and functional crRNAs. Cas13 proteins have been divided into four subtypes (Cas13a, b, c, and d; Shmakov et al., 2017). In 2016, using experiments in vitro and in E. coli, Abudayyeh et al. (2016) characterized a Cas13a protein (formerly named C2c2) from Leptotrichia shahii (LshCas13a) as an RNA-guided RNA-targeting effector. They demonstrated that Cas13a has a collateral RNase activity, which cleaves RNAs nonspecifically once activated, in vitro and in bacteria cells. Abudayyeh et al. (2017) first demonstrated that Leptrotrichia wadei Cas13a (LwCas13a) can knockdown targeted RNA with higher specificity than RNAi in human cells. No collateral RNase activity was detected in human cells. Other than LshCas13a and LwCas13a, various Cas13 putative orthologs have been identified and applied to RNA editing (Shmakov et al., 2015; Cox et al., 2017; Yan et al., 2018; Konerman et al., 2018; Mahas and Mahfouz, 2018; Xu et al., 2021). Cas13 proteins have also been engineered by inactivating HEPN domains and by fusing effector domains for several applications such as live cell imaging of RNA, base editing, and nucleic acid detection (Abudayyeh et al., 2017; Cox et al., 2017; Gootenberg et al., 2017).
RNA targeting by CRISPR–Cas13 is a promising approach for plant research (Wolter and Puchta, 2018). Abudayyeh et al. (2017) successfully targeted three different genes (EPSPS, HCT, and PDS genes) in rice protoplasts using LwaCas13a. More than 50% knockdown was achieved with seven out of nine gRNAs, showing that LwaCas13a can efficiently disrupt the cytoplasmic RNA pool in plants. In addition, CRISPR–Cas13 technology has offered an approach to combat plant RNA viruses. RNA viruses are the most common type of plant virus, and many plant DNA viruses have an RNA intermediate form in their life cycle (Roossinck 2003). Using codon-optimized LshCas13a, Aman et al. (2018a, 2018b) demonstrated that CRISPR–Cas13a can interfere with Turnip mosaic virus (TuMV) in Nicotiana benthamiana and Arabidopsis. A reduction of up to 50% of green fluorescent protein (GFP) signals derived from GFP-TuMV was observed, and the reduction level differed depending on the RNA target, suggesting that RNA accessibility affects Cas13a activity. The effectiveness of CRISPR–LshCas13a to combat virus infection was also shown in a monocot plant, rice, using Southern rice black-streaked dwarf virus and Rice Stripe Mosaic Virus (Zhang et al. 2019). Furthermore, Mahas and Mahfouz (2018) identified a Cas13 variant showing the most efficient RNA virus interference activity in planta in N. benthamiana (Figure 3D). They identified CasRX (Cas13d from Ruminococcus flavefaciens) as the most robust and specific Cas13 variant in plant cells, as is also the case in mammalian cells (Konermann et al., 2018). CasRX significantly reduced GFP signal expressed from targeted virus with no collateral activity in plant cells. Simultaneous targeting of two different RNA viruses was also achieved with high specificity. RNA knockdown and editing by CRISPR–Cas13 would open new approaches for plant research and provide new tools to combat plant viruses.
Future perspectives
In this review, we have summarized the current CRISPR–Cas toolbox, which has expanded dramatically with recently identified CRISPR–Cas systems. The diversity of CRISPR–Cas is surprising; for example, the recently identified CasΦ derived from bacteriophage genomes is only half the size of Cas9 (Pausch et al., 2020). These recently discovered CRISPR–Cas systems have different unique characteristics compared with generally used Cas9 and Cas12a for plant genome editing (summarized in Table 2). However, the discovery of additional CRISPR–Cas tools raises some questions (see “Outstanding Questions”). For example, the applicability of several of the more recent CRISPR–Cas systems to plant genome editing has not yet been validated in plant cells. For example, the transposon-associated CRISPR–Cas system, which can insert DNA fragments at the target site, has been validated only in E. coli cells (Klompe et al., 2019; Strecker et al., 2019a, 2019b). Importantly, unlike the preceding Cas9 and Cas12a technologies, these recently identified CRISPR–Cas systems have not yet been engineered extensively, suggesting that there is still room for improvement of these tools to facilitate their application to plant genome editing.
Table 2.
Summary of recently developed CRISPR–Cas tools that have been applied for genome editing and gene manipulation in plant cells
| CRISPR–Cas System | Origin | Target Cells | Purpose | PAM/PFS | Brief Description | References |
|---|---|---|---|---|---|---|
| Type I-E CRISPR–Cas3 | S. thermophilus | Z. mays | Gene activation | AA | Gene activation was achieved at a similar level as dCas9-CBF1 in Z. mays. | Young et al. (2019) |
| Type I-D CRISPR–Cas10 (TiD) | M. aeruginosa | S. lycopersicum L. | Genome editing | GTT, GTC | TiD induced small indels and large deletions (∼7.5 kb) at target sites in tomatoes. | Osakabe et al. (2020) |
| Type V-B CRISPR–Cas12b |
A. acidoterrestris, A. acidiphilus, and B. thermoamylovorans B. hisashii |
O. sativa | Genome editing | VTTV (V: A, C, G) | Targeted mutagenesis in monocot rice was achieved with high specificity using AaCas12b, which was more efficient than other Cas12b. Deletions (4–14 bp) were larger than those induced by Cas9. | Ming et al. (2020) |
| Type V-B CRISPR–Cas12b | A. acidoterrestris | G. hirsutum | Genome editing | TTN | Genome editing of cottons, which is resistant to high temperature, were achieved using AacCas12b. | Wang et al. (2020) |
| Type V-B CRISPR–Cas12b | Bacillus sp. V3-13 and B. hisashii | A. thaliana | Genome editing | ATTA, ATTG | Genome editing of dicot Arabidopsis was achieved using BvCas12b and BhCas12b v4. | Wu et al. 2020 |
| Type V CRISPR–CasΦ | Bacteriophage genomes | A. thaliana | Genome editing | TBN (B: G, T, and C) | Small CasΦ has induced mutations into target genes in Arabidopsis protoplasts. | Pausch et al. (2020) |
| Type VI CRISPR–Cas13a | L. wadei | O. sativa | RNA targeting | no | More than 50% knockdown of target gene was achieved by LwaCas13 in rice. | Abudayyeh et al. (2017) |
| Type VI CRISPR–Cas13a | L. shahii | N. benthamiana and Arabidopsis | RNA targeting | A, U, and C | LshCas13a produced interference against TuMV expressing GFP in N. benthamiana and Arabidopsis. | Aman et al. (2018a, 2018b) |
| Type VI CRISPR–Cas13a | L. shahii | O. sativa, N. benthamiana | RNA targeting | No descriptions | LshCas13a inhibited virus infection in monocot and dicot plants. | Zhang et al. (2019) |
| Type VI CRISPR–Cas13d | R. flavefaciens | N. benthamiana | RNA targeting | No descriptions | CasRx indicated robust interference efficiencies with high specificity in N. benthamiana. | Mahas and Mahfouz (2018) |
In the field of plant genome editing, the development of new tools is not sufficient to achieve efficient generation of genome-edited plants. There are plant-specific problems with plant genome editing: it generally requires tissue culture, it has low HDR efficiency, and it often takes time to establish genome-edited homozygous lines, especially when optimization in nonmodel plants and/or in polyploid plants is necessary. Codon and vector optimization would also be needed depending on the plant species used. For polyploid plants, multiple copies of target genes have to be mutated to establish knockout plants. In addition, establishment of transgene-free genome-edited plants (null-segregant) is also preferable to avoid off-target risks, and to alleviate concerns about genome-edited plants (Wada et al., 2020). Recently, strategies to overcome some of these conventional problems have been reported: plant gene editing through de novo induction of meristems (Maher et al., 2020), efficient gene targeting in Arabidopsis (Miki et al., 2018), and genome editing during haploid induction (Kelliher et al., 2019), etc. Combined with these technological developments, plant genome engineering will become faster, more efficient, and more precise.
In the near future, we will be able to choose the most suitable genome editing strategy from a toolbox full of various kinds of unique tools, depending on the experimental purposes, the nature of the target gene, and the plant species. New tools will also expand what is possible, ranging from precise modifications of plant genomes, gene expressions, and epigenomes to chromosomal manipulation. For example, Schmidt et al. (2020) achieved a targeted inversion of a 1.1-Mb heterochromatic knob (hk4S) in Arabidopsis Col-0 chromosome 4, which resulted in the restoration of crossover between the short arms of Col-0 and Ler-1 chromosome 4. Translocations between different chromosomes at targeted sites have also been achieved using CRISPR–Cas9 by Beying et al. (2020). These achievements suggest that manipulation of genetic linkages by chromosome engineering for plant breeding is now possible. Newly developed technologies are set to further accelerate expansion of the possibilities of plant genome editing.
ADVANCES.
Type I CRISPR–Cas systems, which recognize longer sequences than Cas9 or Cas12a, have been applied for genome editing and transcriptional control.
Type I CRISPR–Cas3 has been applied to transcriptional control and genome editing in human cells but not yet in plant cells.
Type I CRISPR–Cas10 induces bi-directional long-range deletions and small indels in human and plant cells.
Type V CRISPR–Cas12b has been applied to genome editing in monocot and dicot plants with high specificity at target sites, inducing deletions larger than those induced by Cas9.
Type IV CRISPR–Cas13, which can target RNA, has been used for knockdown of target gene expression and has also provided RNA virus interference activity in plants.
OUTSTANDING QUESTIONS.
Can recently discovered CRISPR–Cas systems be developed as useful plant genome editing tools?
How can modified CRISPR–Cas systems be combined with other technologies to facilitate plant genome engineering?
What do recently developed CRISPR–Cas tools bring to plant genome engineering?
Acknowledgments
We apologize to the researchers whose works were not cited due to space constraints.
Funding
Research in the author’s laboratories was funded by New Energy and Industrial Technology Development Organization (NEDO) to Y.O., Japan Science and Technology Agency (JST) Adaptable and Seamless Technology transfer Program through Target-driven R&D (A-STEP) to K.O., and JST Core Research for Evolutional Science and Technology (CREST) to K.O.
Conflict of interest statement. The authors have no conflicts of interest to declare.
Contributor Information
Naoki Wada, Graduate School of Technology, Industrial and Social Sciences, Tokushima University, Tokushima, Japan.
Keishi Osakabe, Graduate School of Technology, Industrial and Social Sciences, Tokushima University, Tokushima, Japan.
Yuriko Osakabe, School of Life Science and Technology, Tokyo Institute of Technology, Yokohama 226-8502, Kanagawa, Japan.
N.W., K.O., and Y.O. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Yuriko Osakabe (osakabe.y.ab@m.titech.ac.jp).
References
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017) RNA targeting with CRISPR-Cas13. Nature 550: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353: aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M (2018) The CRISPR tool kit for genome editing and beyond. Nat Commun 9: 1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alok A, Sandhya D, Jogam P, Rodrigues V, Bhati KK, Sharma H, Kumar J (2020) The rise of the CRISPR/Cpf1 system for efficient genome editing in plants. Front Plant Sci 11: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S, Mahfouz M (2018a) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman R, Mahas A, Butt H, Aljedaani F, Mahfouz M (2018b) Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis. Viruses 10: 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen P J, Wilson C, Newby GA, Raguram A, Liu DR (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576: 149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beying N, Schmidt C, Pacher M, Houben A, Puchta H (2020) CRISPR-Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat Plants 6: 638–645 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron P, Coons MM, Klompe SE, Lied AM, Smith SC, Vidal B, Donohoue P D, Rotstein T, Kohrs BW, Nyer DB, et al. (2019) Harnessing type I CRISPR-Cas systems for genome engineering in human cells. Nat Biotechnol 37: 1471–1477 [DOI] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA (2017) Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550: 407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu J, Zhi S, Zheng Q, Ma W, Huang J, Liu Y, Liu D, Liang P, Songyang Z (2020) Repurposing type I-F CRISPR-Cas system as a transcriptional activation tool in human cells. Nat Commun 11: 3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017) RNA editing with CRISPR-Cas13. Science 358: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan AE, Hou Z, Xiao Y, Gramelspacher MJ, Heo J, Howden SE, Freddolino P L, Ke A, Zhang Y (2019) Introducing a spectrum of long-range genomic deletions in human embryonic stem cells using type I CRISPR-Cas. Mol Cell 74: 936–950.e935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu D R (2017) Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551: 464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjaltema RAF, Rots MG (2020) Advances of epigenetic editing. Curr Opin Chem Biol 57: 75–81 [DOI] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton HG, Watson BNJ, Fineran PC (2020) The arms race between bacteria and their phage foes. Nature 577: 327–336 [DOI] [PubMed] [Google Scholar]
- Hayes RP, Xiao Y, Ding F, van Erp PB, Rajashankar K, Bailey S, Wiedenheft B, Ke A (2016) Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature 530: 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, et al. (2016) Structure and engineering of Francisella novicida Cas9. Cell 164: 950–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA (2013) Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA 110: 15644–15649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, et al. (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B (2014) Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345: 1473–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaganathan D, Ramasamy K., Sellamuthu G, Jayabalan S, Venkataraman G (2018) CRISPR for crop improvement: an update review. Front Plant Sci 9: 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Su X, Tang G, Chen Z, Carter J, Wittich PE, Dong S, Green J, Burch E, et al. (2019) One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol 37: 287–292 [DOI] [PubMed] [Google Scholar]
- Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, et al. (2017) In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun 8: 14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales AP, Li Z, Peterson RT, Yeh JR, et al. (2015) Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH (2019) Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 571: 219–225 [DOI] [PubMed] [Google Scholar]
- Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533: 420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD (2018) Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173: 665–676.e614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, Lee K, Jung I, Kim D, Kim S, et al. (2018) Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun 9: 3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Fuglsang A, Kjeldsen AL, Sun K, Bhoobalan-Chitty Y, Peng X (2020) DNA targeting by subtype I-D CRISPR–Cas shows type I and type III features. Nucleic Acids Res 48: 10470–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeff L, Brouns SJJ, Joo C (2018) Repetitive DNA reeling by the Cascade-Cas3 complex in nucleotide unwinding steps. Mol Cell 70: 385–394.e383 [DOI] [PubMed] [Google Scholar]
- Mahas A, Mahfouz M (2018) Engineering virus resistance via CRISPR-Cas systems. Curr Opin Virol 32: 1–8 [DOI] [PubMed] [Google Scholar]
- Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF (2020) Plant gene editing through de novo induction of meristems. Nat Biotechnol 38: 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, et al. (2020) Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18: 67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manav MC, Van LB, Lin J, Fuglsang A, Peng X, Brodersen DE. (2020) Structural basis for inhibition of an archaeal CRISPR–Cas type I-D large subunit by an anti-CRISPR protein. Nat Commun 11: 5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride TM, Schwartz EA, Kumar A, Taylor DW, Fineran PC, Fagerlund RD. (2020) Diverse CRISPR-Cas complexes require independent translation of small and large subunits from a single gene. Mol Cell 80: 971–979.e7 [DOI] [PubMed] [Google Scholar]
- Miki D, Zhang W, Zeng W, Feng Z, Zhu JK (2018) CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 9: 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming M, Ren Q, Pan C, He Y, Zhang Y, Liu S, Zhong Z, Wang J, Malzahn AA, Wu J, Zheng X, Qi Y (2020) CRISPR-Cas12b enables efficient plant genome engineering. Nat Plants 6: 202–208 [DOI] [PubMed] [Google Scholar]
- Molla KA, Yang Y (2019) CRISPR/Cas-mediated base editing: technical considerations and practical applications. Trends Biotechnol 37: 1121–1142 [DOI] [PubMed] [Google Scholar]
- Morisaka H, Yoshimi K, Okuzaki Y, Gee P, Kunihiro Y, Sonpho E, Xu H, Sasakawa N, Naito Y, Nakada S, et al. (2019) CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat Commun 10: 5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Héroux A, Bailey S (2014) Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science 345: 1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG (2017) The revolution continues: newly discovered systems expand the CRISPR-Cas toolkit. Mol Cell 68: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Lee CM, Gasiunas G, Davis TH, Cradick TJ, Siksnys V, Bao G, Cathomen T, Mussolino C (2016) Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol Ther 24: 636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, Mochizuki M, Miyabe A, Araki M, Hara KY, et al. (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353: aaf8729. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, et al. (2018) Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361: 1259–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Wada N, Miyaji T, Murakami E, Marui K, Ueta R, Hashimoto R, Abe-Hara C, Kong B, Yano K, et al. (2020) Genome editing in plants using CRISPR type I-D nuclease. Commun Biol 3: 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Wada N, Murakami E, Miyashita N, Osakabe Y (2021) Genome editing in mammalian cells using the CRISPR type I-D nuclease. Nucleic Acids Res 49: 6347–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y, Osakabe K (2015) Genome editing with engineered nucleases in plants. Plant Cell Physiol 56: 389–400 [DOI] [PubMed] [Google Scholar]
- Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, Doudna JA (2020) CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369: 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickar-Oliver A, Black JB, Lewis MM, Mutchnick KJ, Klann TS, Gilcrest KA, Sitton MJ, Nelson CE, Barrera A, Bartelt LC, et al. (2019) Targeted transcriptional modulation with type I CRISPR-Cas systems in human cells. Nat Biotechnol 37: 1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ., Zetsche B, Shalem O, Wu X, Makarova KS, et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roossinck MJ (2003) Plant RNA virus evolution. Curr Opin Microbiol 6: 406–409 [DOI] [PubMed] [Google Scholar]
- Schmidt C, Fransz P, Rönspies M, Dreissig S, Fuchs J, Heckmann S, Houben A, Puchta H (2020) Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering. Nat Commun 11: 4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. (2015) Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell 60: 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, et al. (2017) Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol 15: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Jones S, Koopal B, Schmid-Burgk J, Zetsche B, Gao L, Makarova K. S, Koonin EV, Zhang F (2019a) Engineering of CRISPR-Cas12b for human genome editing. Nat Commun 10: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, Zhang F (2019b) RNA-guided DNA insertion with CRISPR-associated transposases. Science 365: 48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Cui T, Feng G, Guo L, Xu K, Gao Q, Li T, Li J, Zhou Q, Li W (2018) Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov 4: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Cui T, Gao Q, Guo L, Zhou Q, Li W (2019) Artificial sgRNAs engineered for genome editing with new Cas12b orthologs. Cell Discov 5: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada N, Ueta R, Osakabe Y, Osakabe K (2020) Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol 20: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton RT, Christie KA, Whittaker MN, Kleinstiver BP (2020) Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368: 290–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, La Russa M, Qi LS (2016) CRISPR/Cas9 in Genome Editing and Beyond. Annu Rev Biochem 85: 227–264 [DOI] [PubMed] [Google Scholar]
- Wang Q, Alariqi M, Wang F, Li B, Ding X, Rui H, Li Y, Xu Z, Qin L, Sun L, et al. (2020) The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol J 18: 2436–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter F, Puchta H (2018) The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J 94: 767–775 [DOI] [PubMed] [Google Scholar]
- Wu F, Qiao X, Zhao Y, Zhang Z, Gao Y, Shi L, Du H, Wang L, Zhang YJ, Zhang Y, et al. (2020). Targeted mutagenesis in Arabidopsis thaliana using CRISPR-Cas12b/C2c1. J Integr Plant Biol 62: 1653–1658 [DOI] [PubMed] [Google Scholar]
- Wu X, Mao S, Ying Y, Krueger CJ, Chen AK (2019) Progress and challenges for live-cell imaging of genomic loci using CRISPR-based platforms. Genom Proteom Bioinform 17: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Luo M, Dolan AE, Liao M, Ke A (2018) Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 361: eaat0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Luo M, Hayes RP, Kim J, Ng S, Ding F, Liao M, Ke A (2017) Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell 170: 48–60.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Zhou Y, Xiao Q, He B, Geng G, Wang Z, Cao B, Dong X, Bai W, Wang Y, et al. (2021) Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat Methods 18: 499–506 [DOI] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA (2018) Cas13d Is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell 70: 327–339.e325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, Garrity AJ, Chong S, Makarova KS, Koonin EV, et al. (2019) Functionally diverse type V CRISPR-Cas systems. Science 363: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xu J, GS, Lai L (2021) CRISPR/Cas: advances, limitations, and applications for precision cancer research. Front Med (Lausanne) 8: 649896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JK, Gasior SL, Jones S, Wang L, Navarro P, Vickroy B, Barrangou R (2019) The repurposing of type I-E CRISPR-Cascade for gene activation in plants. Commun Biol 2: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhao Y, Ye J, Cao X, Xu C, Chen B, An H, Jiao Y, Zhang F, Yang X, et al. (2019) Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol J 17: 1185–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y (2014) Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature 515: 147–150 [DOI] [PubMed] [Google Scholar]