Abstract
Histone deacetylase enzymes participate in the regulation of many aspects of plant development. However, the genome-level targets of histone deacetylation during apple (Malus domestica) fruit development have not been resolved in detail, and the mechanisms of regulation of such a process are unknown. We previously showed that the complex of ethylene response factor 4 (MdERF4) and the TOPLESS co-repressor (MdTPL4; MdERF4–MdTPL4) is constitutively active during apple fruit development (Hu et al., 2020), but whether this transcriptional repression complex is coupled to chromatin modification is unknown. Here, we show that a histone deacetylase (MdHDA19) is recruited to the MdERF4–MdTPL4 complex, thereby impacting fruit ethylene biosynthesis. Transient suppression of MdHDA19 expression promoted fruit ripening and ethylene production. To identify potential downstream target genes regulated by MdHDA19, we conducted chromatin immunoprecipitation (ChIP) sequencing of H3K9 and ChIP-quantitative polymerase chain reaction assays. We found that MdHDA19 affects ethylene production by facilitating H3K9 deacetylation and forms a complex with MdERF4–MdTPL4 to directly repress MdACS3a expression by decreasing the degree of acetylation. We demonstrate that an early-maturing-specific acetylation H3K9ac peak in MdACS3a and expression of MdACS3a were specifically up-regulated in fruit of an early-maturing, but not a late-maturing, cultivar. We provide evidence that a C-to-G mutation in the ethylene-responsive element binding factor-associated amphiphilic repression motif of MdERF4 reduces the repression of MdACS3a by the MdERF4–MdTPL4–MdHDA19 complex. Taken together, our results reveal that the MdERF4–MdTPL–MdHDA19 repressor complex participates in the epigenetic regulation of apple fruit ripening.
One-sentence summary: Global changes in histone modifications that occur during apple fruit ripening.
Introduction
Fleshy fruit ripening is associated with numerous well-studied physiological and biochemical processes, which in many species include changes in color, soluble sugar accumulation, starch degradation, and coordinated changes in the levels of various hormones (Giovannoni et al., 2017). Notably, major increases in the rates of respiration and ethylene biosynthesis during ripening are features of fruit that are classified as climacteric, such as tomato (Solanum lycopersicum), apple (Malus domestica), and banana (Musa acuminata), whereas those that show no such increases are termed nonclimacteric and include strawberry (Fragaria), citrus (Citrus), and grape (Vitis vinifera; Klee and Giovannoni, 2011; Lü et al., 2018). In recent years, many aspects of the molecular mechanisms of climacteric fruit ripening involving internal gene regulation networks have been elucidated, such as those in apple, tomato, banana, and peach (Han et al., 2016; Lü et al., 2018). Ethylene plays a key regulatory role during the development and ripening of climacteric fruits, and studies have found that Ethylene Responsive Factor/APETALA2 (ERFs/AP2) transcription factors (TFs) are involved in ethylene signal transduction (Zhang et al., 2009; Xiao et al., 2013). In apple, which exhibits climacteric fruit ripening, ethylene response factor MdERF2 binds to MdERF3, thereby suppressing the binding of MdERF3 to the promoter of the 1-amino cyclopropane carboxylic acid synthase gene MdACS1. This gene encodes a key step in the ethylene biosynthesis pathway and the binding of MdERF2 to MdERF3 reduces ethylene production (Li et al., 2016).
Several studies suggest that some ERF proteins contain an ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif and function as suppressors of ripening. For example, in apple MdERF4 acts as an ethylene response repressor, and we reported that a mutation in the EAR motif of MdERF4 results in reduced binding to the MdERF3 promoter, thus accelerating fruit ripening (Hu et al., 2020). During kiwifruit (Actinidia deliciosa) ripening, the AdERF9 repressor, which also contains an EAR motif, substantially suppresses activity of the cell wall modification-associated gene AdXET5 (Yin et al., 2010), and in tomato, AP2 acts as a negative regulator of ripening that operates in a negative-feedback loop with COLORLESS NON-RIPENING (Chung et al., 2010). EAR motifs can recruit transcriptional repression complex members, such as the TOPLESS/TOPLESS-LIKE PROTEINS (TPL/TPRs) corepressors. Several studies have demonstrated that TPL/TPR proteins can interact with members of the histone deacetylase (HDAC) family and thereby epigenetically regulate gene expression (Long et al., 2006; Szemenyei et al., 2008; Wang et al., 2013; Tang et al., 2016).
Epigenetic modifications involving histone acetylation regulate gene expression by changing the state of the chromatin or recruiting protein complexes that affect transcriptional regulation (Strahl and Allis, 2000; Ma et al., 2013). Histone acetylation is one of the most well-studied modifications (Lawrence et al., 2016), and involves the removal of positively charged lysine (Lys) residues from histone tails, thereby decreasing nucleosome binding to negatively charged DNA, and leading to a more accessible chromatin structure, which is associated with higher levels of gene transcription (Verdone et al., 2006). Histone acetylation is a reversible, dynamic process involving gene activation by histone acetyltransferases and transcriptional inhibition by HDAC enzymes (Sridha and Wu, 2006; Ma et al., 2013). HDACs have been reported to play important roles in plant growth and development, including seed (Chhun et al., 2016) and flower development (Kang et al., 2015; Bollier et al., 2018), as well as stress responses (Luo et al., 2012) in Arabidopsis thaliana. In recent years, the involvement of HDACs in the regulation of fruit development has been studied in tomato (Guo et al., 2017, 2018), cucumber (Cucumis sativus; Zhang et al., 2020), pear (Pyrus communis L.; Li et al., 2020), and banana (Xiao et al., 2013; Han et al., 2016; Fu et al., 2018). However, much remains to be learnt about the scale and direct targets of HDAC-mediated gene repression in fruit development.
We previously reported that MdERF4 affects fruit ripening in apple by interacting with a MdTPL4 co-repressor to suppress ethylene production (Hu et al., 2020). We also provided evidence that allelic variation in MdACS1, MdACO1, and MdERF4 explained 72% of the total phenotypic variation in fruit ripening and softening within an apple cultivar panel. Here, we describe the identification of MdHDA19 as another factor with wide-ranging effects on fruit ripening through its interaction with MdERF4–MdTPL4 to directly repress the expression of MdACS3a through decreasing H3K9ac abundance. The data presented here provide mechanistic insights into the epigenetic regulation of climacteric fruit ripening.
Results
The MdERF4C/G mutant allele confers an early-maturing fruit phenotype by promoting ethylene production
Previously our studies showed that the fruit of progeny from a cross between “Zisai Pearl” and “Red Fuji” with early maturity were heterozygous for MdERF4C/G, while late maturing fruit were homozygous for the MdERF4C/C genotype (Hu et al., 2020), indicating that the MdERF4C/G allele may contribute to early-maturing phenotype. We selected two cultivars with different maturity, “Ralls Janet” and “Gala”, which have the same MdACO1 (MdACO1-1/2) and MdACS1 (MdACS1-2/2) alleles, but that show allelic variation in the MdERF4 sequence. MdERF4 in ‘Ralls Janet’ is the MdERF4C/C allele, which is associated with a late-maturing phenotype, while MdERF4 in “Gala” is the MdERF4C/G allele, which is associated with an early-maturing phenotype (Supplemental Figure S1). We observed that “Ralls Janet” takes substantially longer after bloom to reach full maturity and synthesizes relatively low levels of ethylene, while “Gala” showed peak ethylene production at an earlier time point (T4; Figure 1, A and B; Supplemental Figure S2).
Figure 1.
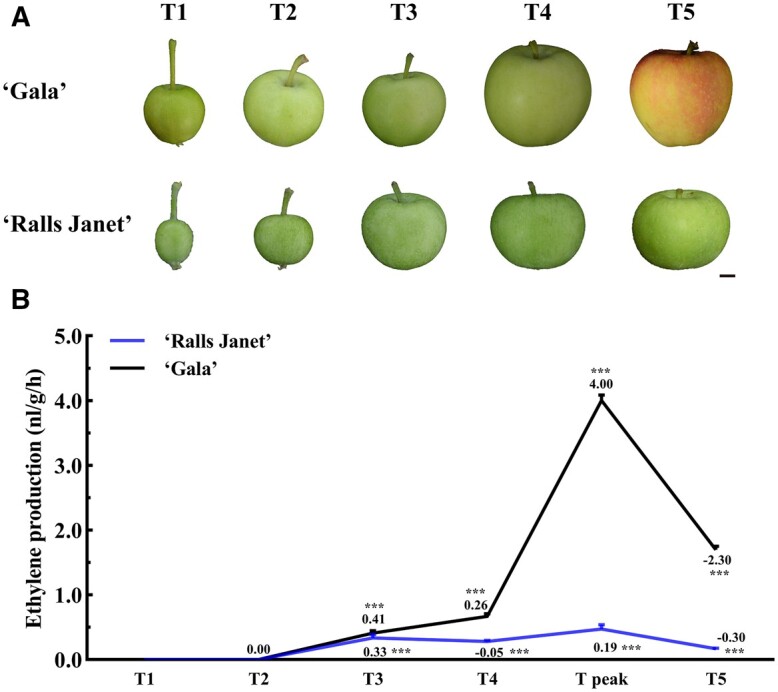
Phenotypes and ethylene production in different developmental timepoints of “Gala” and “Ralls Janet”. A, Phenotypes and B, Ethylene production at different developmental timepoints of “Ralls Janet” and “Gala” apple fruit (T1: timepoint 1; T2: timepoint 2 T3: timepoint 3; T4: timepoint 4; T5: timepoint 5; T peak: timepoint of peak ethylene production of “Gala”). Scale bar = 1 cm. Asterisks indicate a statistically significant difference (*P <0.05; **P <0.01; ***P <0.001), as determined by a one-way analysis of variance (ANOVA). Error bars indicate ±sd of three replicates. The images of (A) were digitally extracted for comparison.
MdHDA19 forms a complex with MdERF4 and affects ethylene biosynthesis
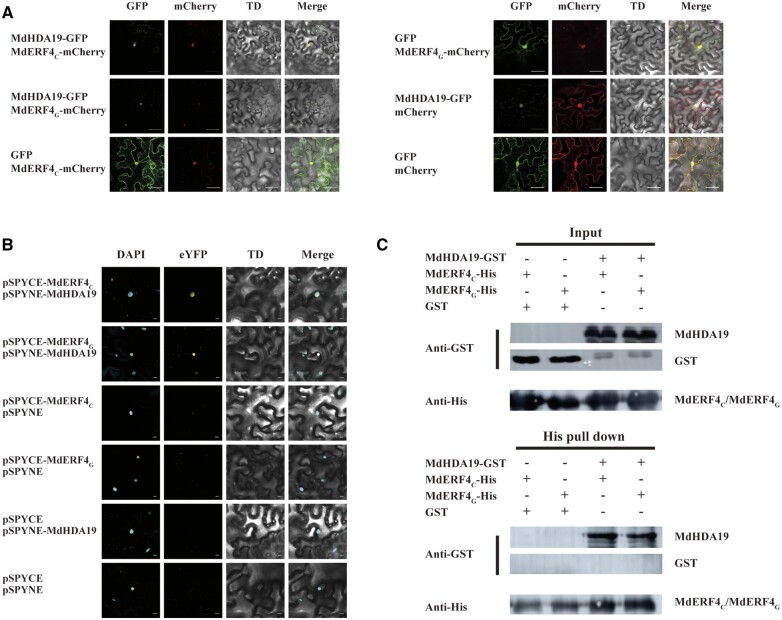
In plants, TFs containing EAR domains regulate downstream gene expression by recruiting TPL–HDA19 or SAP18–HDA19 protein complexes to modify deacetylation activity (Kagale and Rozwadowski, 2011). Since MdERF4 can form a complex with MdTPL4 (Hu et al., 2020), we hypothesized that MdERF4 might be involved in fruit development through recruitment of MdHDA19 to modify deacetylation. To test this idea, we first identified an apple homolog of A. thaliana AtHDA19 (AT4G38130) through a search of the apple genome sequence (http://www.rosaceae.org; Daccord et al., 2017). Phylogenetic analysis identified two putative MdHDA19 genes (MD10G1145400, MD05G1146000) in the apple genome that are expressed during early apple fruit development (Supplemental Figure S3). We then investigated whether MdHDA19 interacted with MdERF4 using bimolecular fluorescence complementation (BiFC) analysis. Specifically, we expressed allelic variants of MdERF4 with a C-terminal yellow fluorescent protein (YFP) fragment (MdERF4C-cYFP; pSPYCE-MdERF4C or MdERF4G-cYFP; pSPYCE-MdERF4G) together with a fusion of MdHDA19 with an N-terminal YFP fragment (nYFP-MdHDA19; pSPYNE-MdHDA19) in Nicotiana benthamiana leaves. We measured YFP fluorescence, as well as the fluorescence resulting from staining the transformed leaves with the nuclear dye, 4,6-diamidino-2-phenylindole (DAPI; Figure 2B). We found that MdHDA19 (MD10G1145400) interacted with both MdERF4C and MdERF4G (Figure 2), while the other gene (MD05G1146000) did not (Supplemental Figure S4). YFP was observed in the nuclei of leaves co-expressing pSPYCE-MdERF4C or pSPYCE-MdERF4G together with pSPYNE-MdHDA19 (MD10G1145400; Figure 2B), but not in the nuclei of the control leaves (transformed with pSPYCE-MdERF4C or pSPYCE-MdERF4G together with pSPYNE, pSPYCE, and pSPYNE-MdHDA19, pSPYCE, and pSPYNE; Figure 2A). We also performed protein colocalization studies by transiently expressing MdHDA19 tagged with green fluorescent protein (GFP) together with MdERF4 tagged with mCherry in N. benthamiana leaves. As shown in Figure 2A, when MdHDA19 was transiently co-expressed with MdERF4, they both localized to the nucleus. MdERF4C-His or MdERF4G-His and MdHDA19-glutathione S-transferase (GST) proteins were also expressed in Escherichia coli BL21 and purified for pull-down and detected by immunoblot with Anti GST-Tag Mouse Monoclonal Antibody. The co-immunodetection results suggested that MdHDA19 interacted with MdERF4C and MdERF4G in vitro (Figure 2C), taken together, we hypothesize that MdHDA19 can form a complex with MdERF4–MdTPL4 to regulate ethylene biosynthesis by modifying histone deacetylation.
Figure 2.
Interactions between MdHDA19 and MdERF4C or MdERF4G. A, Colocalization of MdHDA19 and MdERF4C or MdERF4G in N. benthamiana. MdHDA19 and MdERF4C or MdERF4G were transiently co-expressed in N. benthamiana. MdHDA19 was fused to GFP, whereas MdERF4C and MdERF4G were fused to mCherry (a red fluorescent protein). The cells were observed 3 d after A. tumefaciens infiltration. Scale bar = 50 μm. B, BiFC analysis of the association between MdHDA19 and MdERF4C or MdERF4G. Scale bar = 10 μm. There, DAPI is a fluorescent dye that can be used to label normal N. benthamiana nuclei. TD is transmitted light channel. pSPYCE with a C-terminal enhanced YFP fragment and pSPYNE with an N-terminal eYFP fragment, if there is interaction between the target proteins, the yellow fluorescence of eYFP will be generated under excitation light. C, Pull-down analysis of the interaction between MdHDA19 and MdERF4C or MdERF4G. There, GST (26 KDa) and His (6*His, 0.6KD) are tag–proteins.
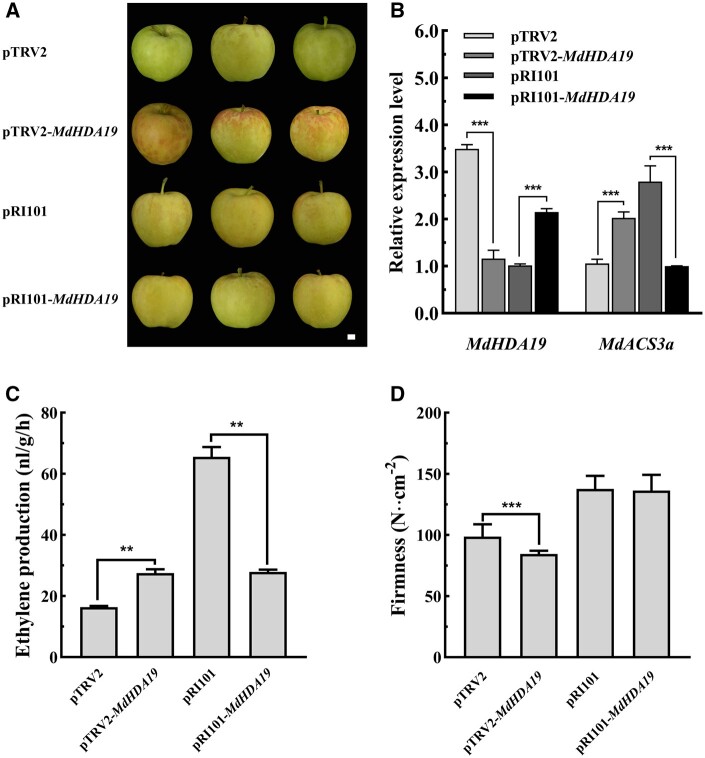
To further test the putative role of MdHDA19 in ethylene biosynthesis, we overexpressed or silenced MdHDA19 in “Gala” fruit harvested 20 d before fruit ripening, using Agrobacterium tumefaciens-mediated transient transformation (Figure 3, A and B). We observed that transient overexpression of MdHDA19 suppressed ethylene production (Figure 3C), while silencing of MdHDA19 promoted ethylene production and led to reduced fruit firmness (Figure 3, C and D). This result was similar to the outcome previously reported functional tests of MdERF4 and MdTPL4 (Hu et al., 2020), supporting our hypothesis that that MdHDA19 plays a regulatory role in ethylene biosynthesis.
Figure 3.
MdHDA19 functional analysis in apple fruit. A, Phenotypes of Gala fruit in which MdHDA19 was transiently overexpressed (pRI101 vector) or silenced (pTRV vector) using A. tumefaciens-mediated transformation (the images were digitally extracted for comparison). Scale bar = 1 cm. B, MdHDA19 and MdACS3a gene expression. C, Ethylene production in transformed fruit, in which MdHDA19 was transiently overexpressed or silenced (three biological replicates). D, Fruit firmness in transformed fruit, in which MdHDA19 was transiently overexpressed or silenced (three biological replicates and five technical replicates). Asterisks indicate a statistically significant difference (*P <0.05; **P <0.01; ***P <0.001), as determined by a one-way ANOVA. Error bars indicate ±sd.
MdHDA19 affects ethylene production by facilitating H3K9 deacetylation
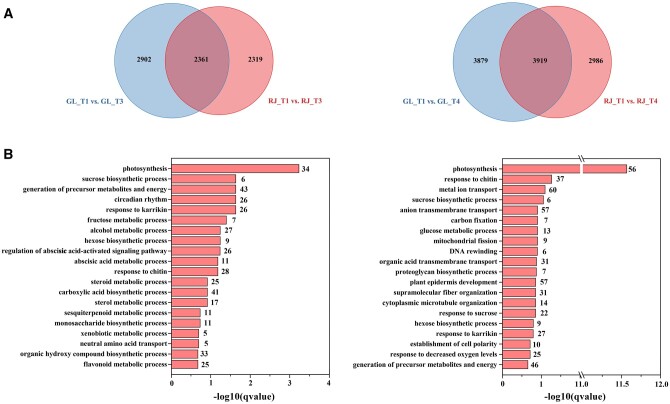
Histone acetylation is a marker for gene transcriptional activation status. HDAs play an important role in controlling gene expression by changing histone acetylation levels. Previous studies have shown that HDA proteins regulate biological processes through deacetylation of the ninth lysine of the histone H3 protein (H3K9ac; Ueda and Seki, 2020). To elucidate the direct targets of MdHDA19 and a potential mechanism of MdERF4–MdTPL4–MdHDA19-mediated repression of apple fruit development, we performed RNA sequencing (RNA-seq) analysis and histone modification chromatin immunoprecipitation sequencing (ChIP-seq) analysis of H3K9ac spanning fruit development. We selected a set of “Gala”-specific hyperacetylation genes which were also identified by RNA-seq analysis, as being differentially expressed between “Gala” and “Ralls Janet” fruits and potential candidates involved in fruit ripening. Fruit flesh from four developmental timepoints (T1–T4) of “Ralls Janet” and “Gala” (Figure 1) were used for transcriptome analysis. We detected 46,558 transcripts in fruit (Supplemental Table S1), and identified the differentially expressed genes (DEGs) during fruit development (Supplemental Figure S5A). Comparisons were made between four timepoints of “Ralls Janet” and “Gala” (Supplemental Figure S5B), and 2,902 and 3,879 “Gala” T3-specific and “Gala” T4-specific gene sets were identified, respectively (Figure 4A). Gene ontology term analysis of these genes revealed an enrichment of the terms “sugar biosynthetic and metabolic processes”, “lipid metabolism”, “terpene metabolic process”, and “flavonoid metabolic process” (Figure 4B). A Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) pathway enrichment analysis revealed an enrichment in the terms “tyrosine and tryptophan biosynthesis”, “carotenoid biosynthesis”, and “hormone biosynthesis” for the “Gala” T3-specific and “Gala” T4-specific DEGs (Supplemental Figure S5C). These biological processes are all often associated with fleshy fruit ripening, suggesting that the expression of many ripening-related genes was substantially upregulated in “Gala” T3 and T4 fruit, leading to earlier maturation. In “Gala”, ethylene production also started to increase at T3 and reached its peak after T4 (Figure 1B; Supplemental Figure S2).
Figure 4.
DEG analysis between “Gala” and “Ralls Janet”. A, Venn diagrams showing the DEGs between “Gala” and “Ralls Janet” at T3 and T4 by RNA-seq analysis. GL, “Gala”; RJ, “Ralls Janet”; T1, T3, T4 refer to timepoints 1, 3, and 4, respectively. B, The enriched gene ontology terms of the unique DEGs identified from pairwise comparisons of T3 “Gala” versus “Ralls Janet” (left) and T4 of “Gala” versus “Ralls Janet” (right). The numbers of DEGs in each category are indicated at the end of each bar. The fold enrichment values −log10 (q value) are showed.
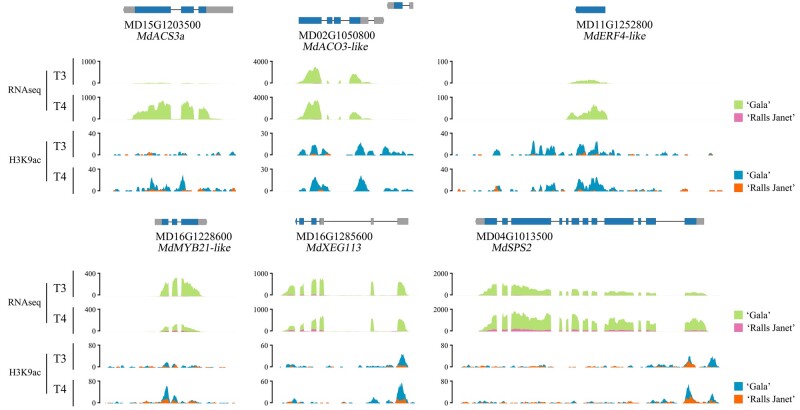
Based on these results, we selected fruit flesh from T3 and T4 of “Gala” and “Ralls Janet”, for the analysis of histone modification of H3K9ac using ChIP-seq. We identified 970 and 1,343 “Gala”-specific hyper-H3K9ac acetylation peaks at T3 and T4, respectively, which were associated with 761 genes in total (Supplemental Table S2). This “Gala”-specific hyperacetylation set of genes included several encoding ethylene biosynthetic enzymes and signaling components, anthocyanin biosynthetic regulatory TF, and sucrose biosynthetic enzymes, which were also identified by RNA-seq analysis as being differentially expressed between “Gala” and “Ralls Janet” fruits at T3 and T4 (Supplemental Table S3). These included MdACS3a (MD15G1203500), MdACO3-like (MD02G1050800), MdERF4-like (MD11G1252800), MdMYB21-like (MD16G1228600), MdSPS2 (SUCROSE PHOSPHATE SYNTHASE, MD04G1013500), and MdXEG113 gene (XYLOGLUCANASE, MD16G1285600; Figure 5; Supplemental Table S3).
Figure 5.
Genome browser screenshot of merged data sets (RNA-seq and H3K9ac ChIP-seq showing representative ripening-related genes and changes in their H3K9ac levels at T3 and T4 in “Gala” and “Ralls Janet” fruit. “Gala” and “Ralls Janet” RNA-seq and H3K9ac ChIP-seq tracks are overlaid.
The relative genes MdACS3a, MdACO3-like, MdMYB21-like, MdXEG113, and MdSPS2 were verified by reverse transcription quantitative polymerase chain reaction (RT-qPCR) to verify the accuracy of transcriptome data (Supplemental Figure S6). The results showed that the expression of MdACS3a, MdACO3-like, MdMYB21-like, MdXEG113, and MdSPS2 was consistent with the data of transcriptome. The relative expression of these five genes as determined by RT-qPCR closely correlated with their TPM values, with R2 ranging from 0.755 (MD16G1285600) to 0.998 (MD15G1203500) at a P <0.01 significance level (Supplemental Table S4).
MdHDA19 can bind to the gene body of MdACS3a to modify deacetylation
Previous studies have shown that MdACS3a is important for ethylene biosynthesis, and hence the timing of the ethylene peak (Dougherty et al., 2016). There are two natural mutant alleles of MdACS3a: one is the functional null allele MdACS3a-G289 (G289 to V289, resulting in a functionally inactive enzyme) and another, a transcriptionally null allele Mdacs3a which is nondetectable mRNA. The combinations of Mdacs3a and MdACS3a-G289V alleles, regardless of whether they are homozygous or heterozygous, are highly associated with lower ethylene production. (Dougherty et al., 2016). The allelic genotypes of MdACS3a in both “Gala” and “Ralls Janet” were MdACS3a/MdACS3a-G289V and the background of MdACS1 and MdACO1 was the same (Wang et al., 2009).
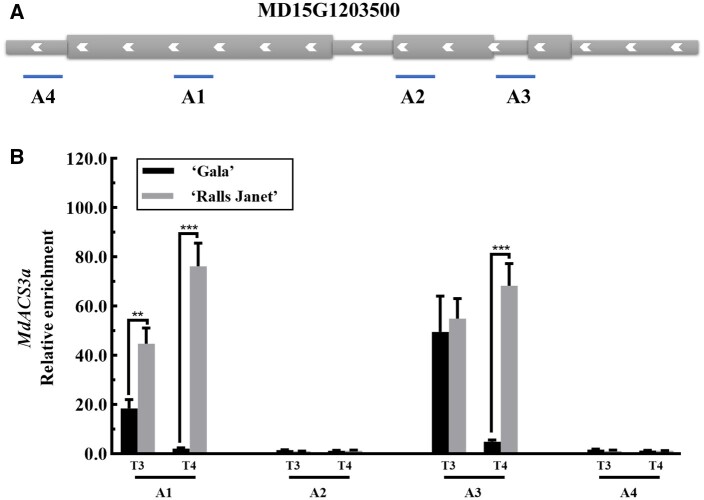
Our ChIP-seq and RNA-seq analyses indicated that the “Gala”-specific H3K9ac acetylation peak at the MdACS3a locus was enriched and MdACS3a expression was upregulated in “Gala” T4 but not in “Ralls Janet”. Accordingly, we hypothesized that the earlier ethylene peak in “Gala” fruit reflects a regulatory effect of MdERF4–MdTPL4–MdHDA19 in modifying acetylation at the MdACS3a locus. To test whether MdHDA19 binds to MdACS3a, we used ChIP-qPCR analysis of fruit flesh samples from the two cultivars at unripe and ripe stages (Figure 6). The presence of MdHDA19 substantially enhanced the PCR-based detection of the MdACS3a region (A1 and A3, these sites were selected by the H3K9ac ChIP-seq results; Figure 6B), indicating that MdHDA19 binds to the MdACS3a in vivo. We also observed that the relative enrichment of MdACS3a acetylation in “Gala” fruit was lower than in “Ralls Janet” fruit, and this was particularly evident in the ripe stage (Figures 5 and 6, B). To investigate the effect of MdHDA19 on MdACS3a further, we analyzed the expression of MdACS3a in apple fruit with silenced or overexpressed MdHDA19 expression. Compared with the control, the MdHDA19 silenced fruit (pTRV-MdHDA19) showed significantly higher MdACS3a expression (Figure 3B). Conversely, the expression of MdACS3a was significantly lower when MdHDA19 was overexpressed (Figure 3B). Meanwhile, we analyzed the expression of MdACS3a in apple calli with silenced or overexpressed MdHDA19 expression. Compared with the control, the MdHDA19 silenced calli (pRI101-RNAi-MdHDA19) showed significantly higher MdACS3a expression (Figure 7A; Supplemental Figure S7A). Conversely, the expression of MdACS3a was significantly lower when MdHDA19 was overexpressed (Figure 7A; Supplemental Figure S7A), which further indicated that MdHDA19 regulates the expression of MdACS3a through its deacetylation.
Figure 6.
ChIP-qPCR assay. A, Schematic representation of the MdACS3a (MD15G1203500) gene showing the location of the A1, A2, A3, and A4 sites, these sites were selected by the H3K9ac ChIP-seq results of “Gala” T4 fruit flesh. B, Binding of MdHDA19 to MdACS3a in the A1, A2, A3, and A4 areas. Fruit flesh DNA samples from T3 and T4 fruit were immunoprecipitated with an MdHDA19 antibody or an IgG control. Asterisks indicate a statistically significant difference (**P <0.01; ***P <0.001), as determined by a one-way ANOVA. Error bars indicate ±sd of three technical replicates.
Figure 7.
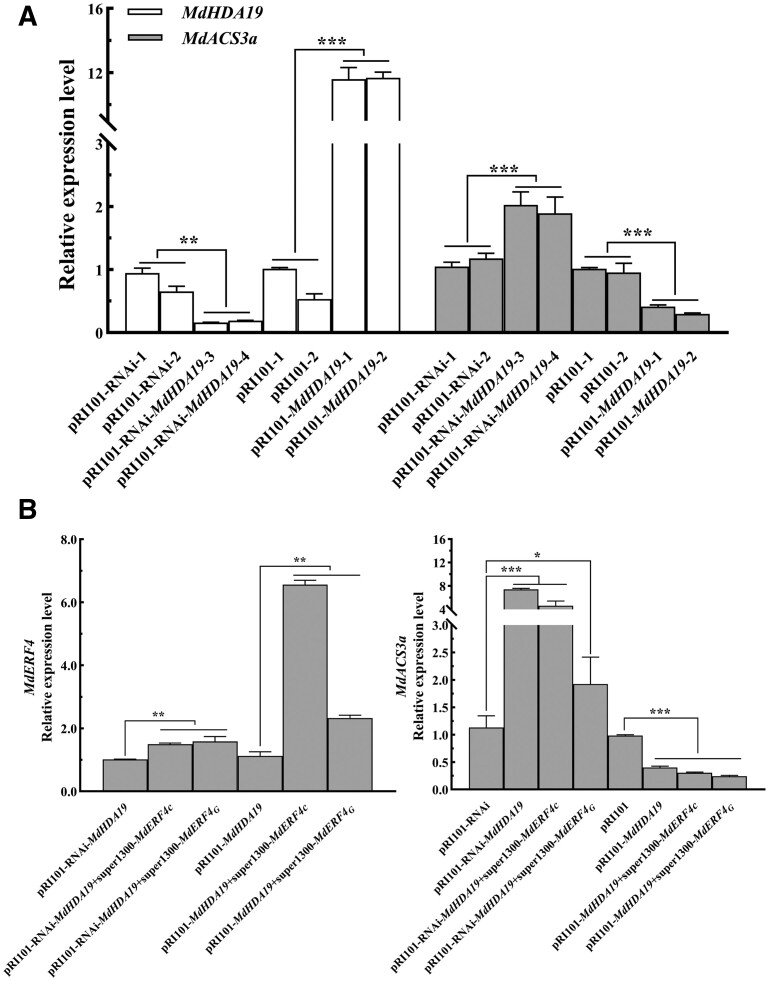
Identification and analysis from transgenic apple “Orin” calli. A, Gene relative expression levels of MdHDA19 and MdACS3a in apple “Orin” calli (pRI101-MdHDA19 and pRI101-RNAi-MdHDA19). B, C, Gene relative expression levels of MdERF4 and MdACS3a in apple “Orin” calli (pRI101-MdHDA19+super1300-MdERF4C/G and pRI101-RNAi-MdHDA19+super1300-MdERF4C/G). Asterisks indicate a statistically significant difference (*P <0.05; **P <0.01; ***P <0.001), as determined by a one-way ANOVA. Error bars indicate ±sd (n = 3).
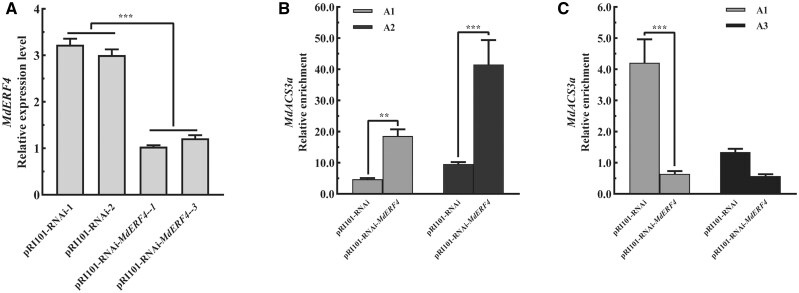
To verify that MdERF4 and MdHDA19 can function together to regulate histone 3 deacetylation of MdACS3a, we evaluated MdACS3a acetylation in the MdERF4-suppressed calli (Figure 8A; Supplemental Figure S7C). ChIP-qPCR analysis was performed using an H3K9ac antibody, where we observed that the relative enrichment of MdACS3a acetylation in MdERF4-suppressed calli was higher than in the control (Figure 8B). Furthermore, the relative enrichment of MdACS3a deacetylation in MdERF4-suppressed calli was lower than in the control (Figure 8C). We further overexpressed MdERF4 in the MdHDA19-overexpressing or -suppressed calli (Figure 7B; Supplemental Figure S7B) and found, by RT-qPCR (quantitative real-time PCR) analysis, that MdERF4 overexpression led to lower expression of MdACS3a in OE-MdHDA19 calli, and overexpressed MdERF4 can lead to limited downregulation of MdASC3a in the MdHDA19 silenced calli compared with the control (pRI101-RNAi) (Figure 7C). These results further indicated that MdERF4 regulates the expression of MdACS3a together with MdHDA19.
Figure 8.
Identification and ChIP-qPCR analysis of MdERF4 from transgenic apple “Orin” calli. A, Gene relative expression levels of MdERF4 in apple “Orin” calli. B, H3K9ac ChIP-qPCR analysis of MdACS3a gene in transgenic “Orin” calli. C, MdHDA19 ChIP-qPCR analysis of MdACS3a gene in transgenic “Orin” calli. A1 and A3 were binding sites of MdHDA19 to MdACS3a and A1 and A2 were binding sites of H3K9ac to MdACS3a (A1–A4 binding sites were shown in Figure 6). Asterisks indicate a statistically significant difference (*P <0.05; **P <0.01; ***P <0.001), as determined by a one-way ANOVA. Error bars indicate ±sd (n = 3).
To further investigate whether MdACS3a contributes to fruit maturity, we investigated the role of MdACS3a by transient expression (Supplemental Figure S8, A and B). We observed that MdACS3a-overexpression promoted ethylene production, resulting in fruit softening, while ethylene production decreased and firmness was higher in MdACS3a-silenced fruit compared to the control (Supplemental Figure S8, C and D).
The mutation of the EAR motif affects the repression ability of the MdERF4–MdTPL4–MdHDA19 complex to MdACS3a
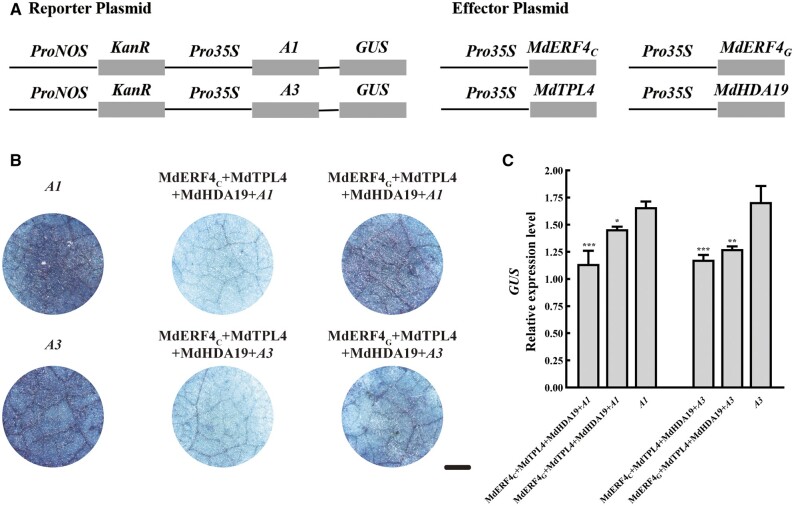
Our previous studies showed that a mutation (C–G) in an EAR motif in MdERF4 resulted in a decrease in its transcriptional repressor activity and in its ability to recruit the co-repressor, MdTPL4 (Hu et al., 2020). To investigate whether this mutation affected the ability of the MdERF4–MdTPL4–MdHDA19 complex to repress expression of the MdHDA19-binding-site-containing reporter, we performed a β-glucuronidase (GUS) activity assay in N. benthamiana leaves to compare the repressive ability of the different MdERF4 alleles (MdERF4C and MdERF4G). The Pro35S: MdTPL4/Pro35S: MdERF4C/Pro35S: MdHDA19 plasmids were co-transformed into N. benthamiana leaves with the MdACS3a: GUS plasmid (A1 or A3, these sites were selected by the H3K9ac ChIP-seq and MdHDA19 ChIP-qPCR results). GUS activity was significantly lower when the Pro35S: MdTPL4/Pro35S: MdERF4C/Pro35S: MdHDA19 plasmids were co-transformed with A1 or A3: GUS than after transformation with A1 or A3: GUS alone (Figure 9). A GUS transactivation assay verified that the GUS activity was significantly lower when the Pro35S: MdTPL4/Pro35S: MdERF4C/Pro35S: MdHDA19 plasmids were co-transformed with A1 or A3: GUS, than following a Pro35S: MdTPL4/Pro35S: MdERF4G/Pro35S: MdHDA19 co-transformation (Figure 9). Taken together, these results indicated that the EAR motif mutation (C–G) leads to reduced repression of MdACS3a expression.
Figure 9.
The MdERF4–MdTPL4–MdHDA19 complex repression ability analysis. A, Reporter vector containing the A1 or A3 alone or together with the effector vectors of MdERF4, MdHDA19, and MdTPL4 were infiltrated into N. benthamiana leaves to analyze GUS activity. A1 and A3 were binding sites of MdHDA19 to MdACS3a (A1–A4 binding sites were shown in Figure 6). B, Phenotypes of GUS staining. Scale bar = 0.5 mm. C, GUS relative expression levels using RT-qPCR. GUS expression was measured relative to that of KanaR, Error bars represent ±sd of measurements from three independent repeat experiments. Asterisks indicate a statistically significant difference (*P <0.05; **P <0.01; ***P <0.001), as determined by a one-way ANOVA.
Discussion
Plant hormones and TFs have a central function in controlling the development and ripening of both climacteric and nonclimacteric fruits (Adams-Phillips et al., 2004; Giovannoni et al., 2017; Lü et al., 2018), and there is increasing evidence that epigenetic regulators also play critical and coordinated roles (Manning et al., 2006; Lang et al., 2017; Lü et al., 2018). In tomato, 32 HAT family members and 15 HDAC family members were identified based on whole genome sequencing (Cigliano et al., 2013). For example, tomato HDAC proteins have been shown to accumulate during fruit development (Cigliano et al., 2013), and MaHDA6 may be involved in banana ripening through binding to the MaERF11/15 promoters (Fu et al., 2018). Moreover, EAR motif-containing proteins have been proposed to form transcriptional repression complexes by recruiting other co-repressors, such as HDAC proteins (Kagale and Rozwadowski, 2011), and it has been suggested that MaERF11 may recruit MaHDA1 to transcriptionally repress its MaACO1 target (Han et al., 2016). However, global changes in histone modifications that occur during apple fruit development are not well documented. Here, the use of members of the Malus genus with natural variation in the EAR motif allowed us to demonstrate that such variation contributes to differences in H3K9ac histone modification, and that these account for variation in ethylene peaks.
It has been reported that a recent whole-genome duplication (WGD) shaped the genome of the domesticated apple (Velasco et al., 2010), and there is strong collinearity between large segments of apple chromosomes 3 and 11. Our results show the two collinear MdERF4 alleles, MD03G1231800 and MD11G1252800, which are located on chromosome 11, and MD11G1252800 was not expressed in apple fruit (Supplemental Figure S9). The advent of genome-scale sequencing has resulted in several puzzles regarding duplicate gene evolution, including their high frequency, since if null mutations are more common than those that are beneficial it might be expected that most duplicate genes would be lost (Conant et al., 2014). Interestingly, here we uncovered differences in MdERF4–MdTPL4–MdHDA19-mediated acetylation in MD11G1252800, which is consistent with gene copy dosage and quantitative genetics being tightly regulated by epigenetic paths following WGD. Our results therefore indicate that MdERF4 may regulate and maintain the dosage balance of its own gene copy number through acetylation modification.
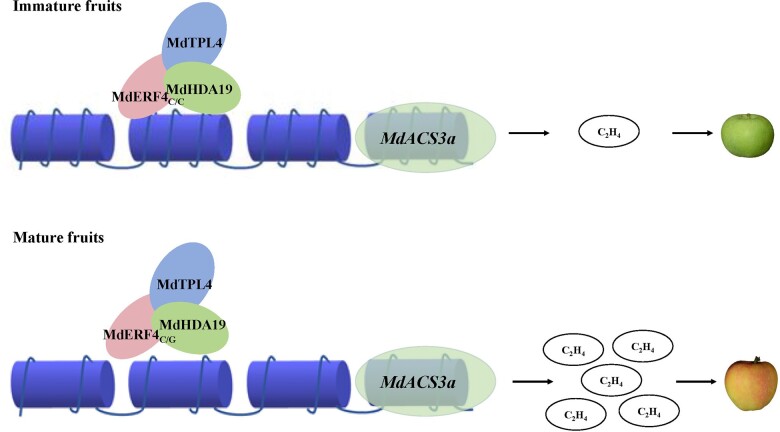
Based on our results and those of previous studies (Hu et al., 2020), we propose that MdERF4 regulates apple fruit ripening in two ways. First, as a TF it regulates the transcription of targets by binding them directly, and the mutation in the MdERF4 EAR motif reduces its ability to interact with MdTPL4, which in turn leads to reduced repression of MdERF3 expression and increased ethylene production (Hu et al., 2020). Second, MdERF4 recruits MdHDA19 and thus promotes epigenetic modification of targets. In immature fruit, MdERF4 and the co-suppressor MdTPL4 recruit MdHDA19 to form a deacetylation complex to regulate ripening-related genes, such as MdACS3a, and the expression of these genes will then be suppressed. During ripening, the deacetylation of MdHDA19 in the complex is weakened, thus promoting the expression of ripening-related genes, and accelerating fruit ripening. These two functions have complementary actions during apple fruit development and jointly regulate ripening.
In Figure 10, we propose a model of an acetylation regulatory module that affects apple fruit ripening. MdHDA19 acts via MdERF4–MdTPL4 to directly repress the expression of MdACS3a through decreasing H3K9ac abundance. A C–G mutation in the MdERF4 EAR motif reduces the deacetylation activity of MdERF4–MdTPL4–MdHDA19, which in turn results in lower transcriptional suppression, and increased MdACS3a expression in early-maturing apple cultivars. Our study reveals an association between MdERF4 allelic variation and histone deacetylase proteins in the epigenetic regulation of apple fruit development.
Figure 10.
A model of the regulatory acetylation module, MdERF4–MdTPL4–MdHDA19, involved in apple fruit ripening. A C–G mutation in MdERF4 EAR motif reduces the deacetylation activity of the module, which will facilitate the histone acetylation and more accessible chromatin on the promoters of ripening genes, such as MdACS3a in early-maturing apple cultivars.
Materials and methods
Plant materials
Apple (Malus domestica cv. “Gala” and M. domestica cv. “Ralls Janet”) trees were grown in the Northern suburb farm (Beijing, China). “Gala” and “Ralls Janet” fruits were harvested as follows: The sampling time of “Gala” was as follows: 30 d (timepoint 1; T1), 50 d (timepoint 2; T2), 70 d (timepoint 3;T3), 90 d (timepoint 4; T4), 100 d (timepoint of peak ethylene production of “Gala”; T peak), 110 d (timepoint 5; T5) after flower blossom; the sampling time of “Ralls Janet” was 30 d (timepoint 1; T1), 50 d (timepoint 2; T2), 70 d (timepoint 3;T3), 90 d (timepoint 4; T4), 100 d (timepoint of peak ethylene production of “Gala”; T peak), 110 d (timepoint 5; T5), 130 d (timepoint 6; T6), 150 d (timepoint 7; T7), 170 d (timepoint 8; T8), 190 d (timepoint 9; T9) after flower blossom (Supplemental Table S5). At each of the above sampling points, ethylene production was determined (Hu et al., 2020).
“Gala” fruit at 90–110 d after flower blossom and “Golden Delicious” (Malus domestica cv. “Golden Delicious”) fruit at 120 d after flower blossom were used for A. tumefaciens infection.
Apple “Orin” (Malus domestica cv. “Orin”) calli were used for A. tumefaciens transformation. Nicotiana benthamiana plants were used for A. tumefaciens infection, and grown in a growth chamber at 25°C and a cycle of 14 h of light 10 h of dark.
Measurement of fruit flesh firmness and ethylene production
Fruit firmness was measured using a texture analyzer as in Hu et al. (2020; TA. XT; Stable Micro Systems, Surrey, UK). The TA settings were as follows: pre-test speed 1.0 mm·s–1, test speed of 1.0 mm·s–1 and post-test speed 10.00 mm·s–1. The probe (diameter 2 mm) was pressed into the fruit to a depth of 10 mm. Three fruit were used for each treatment with five measurements per fruit. Ethylene production was measured as previously published (Hu et al., 2020). At each sampling point, three fruits without no apparent physical damage were selected and sealed in a 0.5-L gas collection chamber and placed at room temperature for 3 h. A sampling needle was used to extract 1 mL of gas for determination of ethylene production, and each sample was tested three times. Ethylene levels were determined using a gas chromatograph (6890N, Agilent Technologies, Santa Clara, CA, USA).
Total RNA isolation and cDNA synthesis
RNA from the fruit flesh was extracted using the cetyltrimethylammonium bromide (CTAB) method, and cDNA synthesis was performed as previously described (Han et al., 2018).
Cloning
The MdERF4, MdHDA19, MdHDA19-chr05, MdTPL4, and MdACS3a coding sequences (CDSs) were downloaded from the apple genome database (http://www.rosaceae.org) and amplified by PCR from fruit flesh cDNA using gene-specific primers (Supplemental Table S6). The amplicons were cloned into the pTOPO-Blunt vector (Aidlab, Beijing, China; code no. CV1702) and sequenced (Sangon, Beijing, China). The PCR reactions were performed in a final volume of 50 μL with 400-ng cDNA, 25-μL KOD buffer, 1-μL KOD FX, 10-μL dNTP, 9-μL H2O (TOYOBO, Japan; code no. KFX101), 1.5-μL 10-mM forward and reverse primers (Sangon, Beijing, China). Thermal cycling (Bio-Rad, Hercules, CA, USA) started with a denaturation step at 94°C for 2 min followed by 35 cycles of denaturation at 98°C for 10 s, annealing at 56°C for 30 s and extension at 68°C for 1 kb·min–1; and finishing with a final extension at 72°C for 10 min.
Genotype identification
The leaves DNA concentrations of “Gala” and “Ralls Janet” with CTAB methods were diluted to 100–200 ng·μL–1 Sequencing of MdACS1, MdACO1, MdERF4 were performed with specific primers (Sunako et al., 1999; Costa et al., 2005; Hu et al., 2020; Supplemental Table S6).
Phylogenetic analysis of MdHDA19 and MdERF4
Download the amino acid sequences of related website The Arabidopsis Information Resource (TAIR) (TAIR-Home Page [arabidopsis.org]), National Center for Biotechnology Information [nih.gov], Genome Database for Rosaceae (GDR) (GDR [rosaceae.org]). The format is FASTA, analysis software MEGA7 (Kumar et al., 2016) for phylogenetic analysis. The related parameters are: adjacency method (Saitou and Nei, 1987), bootstrap test (1,000 repeat; Felsenstein, 1985) for cluster analysis. Poisson’s correlation analysis (Zuckerkandl and Pauling, 1965) calculates the evolutionary distance.
RT-qPCR assay
First-strand cDNA was synthesized using the TRUEscript RT Kit for qPCR (Aidlab, Beijing, China; code no. PC5402). RT-qPCR was performed using the ABI QuantStudio 6 Flex system (Applied Biosystems Inc., Foster City, CA, USA) with a SensiFAST SYBR Lo-ROX Kit (Bioline, London, England, code no. BIO-94005). For RT-qPCR reactions, the thermo cycler protocol was as follows: pre-denaturation, 1 min at 95°C, 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 45 s. ACTIN (M. domestica) (MD12G1140800) was used as the internal reference, and the relative expression levels were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). MdERF4, MdHDA19, MdACS3a, MdACS3a, MdACO3-like, MdMYB21-like, MdSPS2, MdXEG113, and MdActin primers are listed in Supplemental Table S6.
Protein expression and purification
The MdHDA19 CDS was amplified by PCR, and the amplicons digested with EcoRI and XhoI and ligated into the corresponding sites of the pGEX-6p-1 vector to produce the MdHDA19-GST plasmid. Escherichia coli BL21 cells were transformed with the plasmids and cultures expressing the plasmid were induced with 0.4-mM isopropyl β-d-1-thiogalactopyranoside for 16 h at 16°C. The recombinant GST-tagged MdHDA19 protein was purified using glutathione-Sepharose resin (Cwbio, Beijing, China; code no. CW0190) according to the manufacturer’s specifications. MdERF4C-His, MdERF4G-His recombinant proteins were similarly generated using the His-Tagged Protein Purification Kit (Cwbio, Beijing, China; code no. CW0894) as previously described (Hu et al., 2020). The primers used are listed in Supplemental Table S6.
Preparation of MdHDA19 polyclonal antibody
MdHDA19-anti-(CFLRNITPETQQDQL) polyclonal antisera were generated using PolyExpressTM methods (GenScript Biotech Corporation, NanJing, China), with the antigen being the respective synthesized peptide. Express affinity-purified peptides were used to generate polyclonal antibodies in rabbit (AgResearch, Hamilton, New Zealand).
Protein co-localization analysis
The MdERF4C and MdERF4G CDSs were individually cloned into the pBI121-mCherry vector using EcoRI restriction enzymes, the MdHDA19 coding sequence was cloned into the pRI101-eGFP vector using BamHI restriction enzymes. Protein co-localization analysis was conducted in N. benthamiana leaves with mCherry and eGFP. Confocal images were taken with an FV3000 microscope (Olympus, System Version: 2.3.1.163.) 3 d after Agro-infiltration. All images correspond to single optical slices of leaf epidermal cells. The experimental setup follows: Excitation DM Name: DM405/488/561; C.A.; 138 µm; Bits/Pixel: 12 bits; Channel 1 Name: SD1; Dye Name: eGFP; Emission Wavelength: 510 nm; PMT Voltage: 780 V; Laser Wavelength: 488 nm; Laser Transmissivity: 22.1%; Detection Wavelength: 500–540 nm; Channel 2 Name: SD2; Dye Nam: mCherry; Emission Wavelength: 610 nm; PMT Voltage: 591 V; Laser Wavelength: 561 nm; Laser Transmissivity: 17.1%; Detection Wavelength: 570–670 nm; Channel 3 Name: TD; PMT Voltage: 175 V; Laser Wavelength: 561 nm; Laser Transmissivity: 17.1%, respectively. Primers are listed in Supplemental Table S6.
BiFC assay
The MdERF4C and MdERF4G CDSs were individually cloned into the pSPYCE155 vector using BamHI and XhoI restriction enzymes, the MdHDA19 coding sequence was cloned into the pSPYNE173 vector using XhoI and KpnI restriction enzymes, and the MdHDA19-chr05-coding sequence was cloned into the pSPYNE173 vector using BamHI restriction enzymes (vector provided by Professor Junping Gao, China Agricultural University; Aparicio and Pallas, 2017; Han et al., 2018; Hu et al., 2020). BiFC assays were conducted in N. benthamiana leaves with YFP visualization as previously described (Aparicio and Pallas, 2017; Han et al., 2018). Confocal images were taken with an FV3000 microscope (Olympus) 3 d after Agro-infiltration. All images correspond to single optical slices of leaf epidermal cells. Excitation and emission wavelengths were 407–457 nm for DAPI, and 514 and 527 nm for enhanced YFP, respectively. Primers are listed in Supplemental Table S6.
Pull-down assay
The putative interactions between MdHDA19 and MdERF4C or MdERF4G were investigated using a pull-down assay. The respective recombinant proteins were fractionated on a sodium dodecyl sulphate (SDS)–polyacrylamide gel using a Tricine–SDS–polyacrylamide gel electrophoresis Gel Kit (Cwbio, code no. CW2384) and polyvinylidene fluoride membranes (Millipore, code no. HATF00010). The membranes were then incubated with either an anti-His-tag mouse monoclonal antibody (Cwbio, code no. CW0286M), diluted 1:3,000 in TBST (Tris, Buffer, Solution, Tween) buffer (10-mM Tris-base, pH 8.0, 150-mM NaCl, 3 g/100-mL bovine serum albumin, 50-µL/100 mL Tween-20), or with an anti-GST (Abcam, code no. ab92) antibody diluted 1:5,000 in TBST. The secondary antibody was a Goat Anti-Rat IgG H&L (HRP; Abcam, code no. ab205719), which was diluted 1:10,000 in TBST buffer. The eECL Western Blot Kit (Cwbio, code no. CW0049S) was used for visualization using a chemiluminescence apparatus (Amershan Imager 680, GE, USA).
Agrobacterium-mediated transient transformation
For overexpression of MdHDA19 and MdACS3a, the CDSs were amplified by PCR using apple fruit as described above and the amplicons were ligated into the pRI101 or super1300-GFP vectors using the One Step Seamless Cloning kit (Aidlab, Beijing, China; code no. CV1901) to generate the pRI101-MdHDA19 and super1300-MdACS3a constructs, respectively (Li et al., 2016; Hu et al., 2020). To silence MdHDA19 or MdACS3a expression using virus-induced gene silencing, partial MdHDA19 and MdACS3a CDSs were ligated into the pTRV vector to generate the pTRV-MdHDA19 and pTRV-MdACS3a constructs, respectively (Xie et al., 2012; Hu et al., 2020). The recombinant plasmids were transformed into A. tumefaciens strain GV3101 Stable Chemically Competent Cell (Weidi Biotechnology, Beijing, China, AC1001; Xie et al., 2012; Hu et al., 2020) and the resulting A. tumefaciens lines containing the pRI101-MdHDA19 or pTRV-MdHDA19 constructs were transformed into “Gala” fruit (Tian et al., 2015; Hu et al., 2020). The super1300-MdACS3a and pTRV-MdACS3a constructs were transformed into ‘Golden Delicious’ fruit using vacuum infiltration (Tian et al., 2015). qPCR was performed as described above. Primers listed in Supplemental Table S6.
Based on the foundation of our observation that silencing of MdHDA19 promoted ethylene production and fruit ripening, to make the phenotype more obvious, MdHDA19-overexpressing fruit and their controls were collected two weeks after transient transformation, and MdHDA19-suppressed fruit and the corresponding controls were collected 10 d after transient transformation.
Generation of transgenic apple “Orin” calli
SalI and BamHI were selected to connect the sense and antisense chains of MdHDA19 (284 bp) to generate the vector pRI101-RNAi-MdHDA19. XbaI and XmaI were selected to connect the sense and antisense chains of MdERF4 (220 bp) to generate the vector pRI101-RNAi-MdERF4. The CDS of MdHDA19 was cloned into the binary pRI101 vector to generate vector pRI101-MdHDA19. The CDS of MdERF4 was cloned into the binary super1300 vector to generate vector super1300-MdERF4C and super1300-MdERF4G. Agrobacterium-mediated transformation of apple “Orin” calli was carried out as described (Li et al., 2021). Primers listed in Supplemental Table S6.
RNA-seq analysis
Total 24 RNA samples were extracted from “Gala” and “Ralls Janet” fruit flesh at 30, 50, 70, and 90 d after flower blossom using the CTAB method (Han et al., 2018). RNA-seq analysis was performed (Berry Genomics, Beijing, China) with each sampling point analyzed in triplicate. The workflow was as follows: Nanodrop/Gel electrophoresis technology was used for sample detection. Illumina TruSeq RNA sequencing libraries were constructed and sequenced by using NovaSeq6000 platform in the PE150 sequencing mode. Each sample produced ≥6 Gb of data. After generation of the FASTQ file, raw reads were filtered using Fastp (version 0.20.1) with the default parameters. Next, the cleaned reads were mapped to the apple reference genome (GDDH13 version 1.1) using Hisat2 (v2.2.0) with the default parameters. Transcripts Per Kilobase of exon model per Million mapped reads value of each gene was calculated using featureCounts of Rsubread (version 2.0.1) with the default parameters. DEGs were called using DESeq2 (1.16.1). For pairwise DEGs, fold-change ≥ 2 and false-discovery rate (FDR) ≤0.01 were required.
ChIP-seq and ChIP-qPCR
Apple fruit tissue or pRI101-RNAi-MdERF4 transgenic apple calli was fixed with formaldehyde (4-g paraformaldehyde with 100-mL PBS buffer, pH 7.0) for 15 min under vacuum, then nuclei were extracted and purified as previously described (Lü et al., 2018). The nuclei extract was sonicated to 300–500 bp using a Covaris M220 (Covaris, Inc.), and fragments were pre-cleared with Dynabeads protein A/G (Invitrogen) for 1 h before incubation for 6 h with new Dynabeads A/G with antibodies. The beads were then washed twice with low salt (150-mM NaCl in TE) and high salt (300-mM LiCl in TE) buffer.
The H3K9ac ChIP-qPCR was performed using an H3K9ac antibody (Abcam, ab10812), and MdHDA19 ChIP-qPCR was performed with an anti-MdHDA19 rabbit serum (Genscript, Nanjing, China). For ChIP-qPCR, the washed beads were directly reverse crosslinked and the DNA fragments were purified as qPCR templates. We used the IgG data as control to calculate the enrichment values. Two biological replicates for transgenic apple “Orin” calli and three biological replicates for apple fruit tissue were analyzed.
The histone modification ChIP-seq was performed using an H3K9ac antibody (Abcam, ab10812). For ChIP-seq, the washed magnetic beads were treated with a Tn5 transposase at 37°C for 30 min, and then reverse crosslinked for 8 h. The purified DNA was amplified using Illumina type N50x and N70x index primers, and sequenced by using illumina HiSeq X-ten platform in PE150 mode. The H3K9ac ChIP-seq datasets were mapped to the unmasked apple genome using Bwa (version 0.7.17) with the default parameters. Unmapped reads were then filtered using SAMTools view (version 1.7) with options -b -F 3852 -f 2 -q 30 and -q 30, and duplicated reads were removed using SAMTools rmdup. The clean reads were supplied to MACS2 (version 2.2.7.1, -f BED -keep-dup all -B -q 0.01) for peak calling. Differentially enriched peaks between two histone ChIP-seq data sets were called using MAnorm (fold-change >1.2 and FDR ≤0.05). Peaks were then associated to genes if they were located within region of the gene in genome annotation file.
GUS activity analysis
The CDS of MdERF4, MdHDA19, MdTPL4 were individually cloned into the binary pRI101 vector to generate effector constructs pRI101-MdERF4, pRI101-MdTPL4, and pRI101-MdHDA19 (Li et al., 2016). Reporter constructs were generated using the sequences of MdACS3a, A1, or A3, which was binding sites of MdHDA19 to MdACS3a, cloned upstream of the GUS reporter gene in the binary vector pBI121 (Yang et al., 2020). For the transient expression assay, the reporter vector and the effector vector were transformed into A. tumefaciens GV3101 and co-infiltrated into N. tabacum leaves as in Yin et al. (2010). After culture for 3 d, RNA was extracted for GUS activity analysis, GUS expression was measured relative to that of KanR. Each treatment was repeated three times. Primers are listed in Supplemental Table S6.
Statistical analysis
In this article, the software GraphPad Prism 8.0.2 was used for statistical analysis (GraphPad Software, Inc., La Jolla, CA, USA), to obtain mean ± sd (n = 3). The significance of the differences between different groups was determined through a one-way or two-way analysis of variance with a significance level of 0.001, 0.01, and 0.05 (*P <0.05; **P <0.01; ***P <0.001).
Accession numbers
Sequence data from this article can be found in the apple genome database (http://www.rosaceae.org). MdHDA19 (MD10G1145400), MdHDA19-chr05 (MD05G1146000) MdERF4 (MD03G1231800), MdERF4-like (MD11G1252800), MdACS3a (MD15G1203500), MdACO3-like (MD02G1050800), MdMYB21-like (MD16G1228600), MdSPS2 (MD04G1013500), MdXEG113 (MD16G1285600), MdActin (MD12G1140800).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . The MdACS1, MdACO1, and MdERF4 genotypes in “Gala” and “Ralls Janet”.
Supplemental Figure S2 Phenotypes and ethylene production in different developmental timepoints of “Gala” and “Ralls Janet”.
Supplemental Figure S3 . Homologous sequence analysis of MdHDA19.
Supplemental Figure S4 . BiFC analysis of the association between MdHDA19-chr05 (MD05G1146000) and MdERF4C or MdERF4G.
Supplemental Figure S5 . Analysis of DEGs in different fruit developmental timepoints.
Supplemental Figure S6 . Relative expression levels of MdACS3a, MdACO3-like, MdMYB21-like, MdXEG113, and MdSPS2 in “Gala” and “Ralls Janet” at different development timepoints.
Supplemental Figure S7 . Identification of genomic amplification in transgenic apple “Orin” calli.
Supplemental Figure S8 . Gene functional analysis of MdACS3a in apple fruit.
Supplemental Figure S9 . Homologous sequence analysis of MdERF4.
Supplemental Table S1 . Normalized expression (TPM) of “Gala” and “Ralls Janet” fruit in different developmental timepoints.
Supplemental Table S2 . H3K9ac ChIP-seq data generated from “Gala” and “Ralls Janet” fruit.
Supplemental Table S3 . Hyper-H3K9ac adjacent genes overlap with DEGs of “Gala” and “Ralls Janet” fruit at T3 and T4.
Supplemental Table S4 . The correlation analysis of TPM and gene expression level in “Gala” and “Ralls Janet” fruit different timepoints.
Supplemental Table S5 . The sampling time of “Gala” and “Ralls Janet” days after flower blossom.
Supplemental Table S6 . Primers used for cloning, RT-qPCR, protein purification, BiFC, and ChIP-qPCR.
Supplementary Material
Acknowledgments
We thank PlantScribe (www.plantscribe.com) for editing this manuscript.
Contributor Information
Yanan Hu, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
Zhenyun Han, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China; Institute of Crop Sciences, the Chinese Academy of Agricultural Sciences, Beijing 100081, China.
Ting Wang, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
Hua Li, College of Horticulture, FAFU-UCR Joint Center for Horticultural Biology and Metabolomics, Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University, Fuzhou, 35002, China.
Qiqi Li, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
Shuai Wang, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
Ji Tian, Plant Science and Technology College, Beijing University of Agriculture, Beijing, 102206, China.
Yi Wang, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
Xinzhong Zhang, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
Xuefeng Xu, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
Zhenhai Han, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
Peitao Lü, College of Horticulture, FAFU-UCR Joint Center for Horticultural Biology and Metabolomics, Haixia Institute of Science and Technology, Fujian Agriculture and Forestry University, Fuzhou, 35002, China.
Ting Wu, State Key Laboratory of Agrobiotechnology, College of Horticulture, China Agricultural University, Beijing, 100193, China.
T.Wu. conceived and designed the research. P.L. conceived the original screening. Y.H., Z.Y.H., P.L., T.Wa., Q.L., and S.W. conducted the experiments. P.L. and H.L. analyzed the data. Y.W., X.Z., X.X., and Z.H.H. contributed reagents and/or analytical tools. T.Wu. and P.L. wrote the manuscript. J.T. gave advice and edited the manuscript. All authors read and approved the final manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Ting Wu (wuting@cau.edu.cn) and Peitao Lü (ptlv@fafu.edu.cn).
Funding
The work was supported by the National Key R&D Program of China (2018YFD1000200), the National Natural Science Foundation of China (Nos. 31872941, 32072543), the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects (CEFF-PXM2019_014207_000032), the 111 Project (B17043), and the Construction of Plateau Discipline of the Fujian Province (102/71201801104).
Conflict of interest statement. The authors declare that they have no conflict of interest.
References
- Adams-Phillips L, Barry C, Giovannoni J (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci 9: 331–338 [DOI] [PubMed] [Google Scholar]
- Aparicio F, Pallas V (2017) The coat protein of Alfalfa mosaic virus interacts and interferes with the transcriptional activity of the bHLH transcription factor ILR3 promoting salicylic acid-dependent defence signalling response. Mol Plant Pathol 18: 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollier N, Sicard A, Leblond J, Latrasse D, Gonzalez N, Gevaudant F, Benhamed M, Raynaud C, Lenhard M, Chevalier C, et al. (2018) At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. Plant Cell 30: 83–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun T, Chong SY, Park BS, Wong ECC, Yin JL, Kim M, Chua NH (2016) HSI2 repressor recruits MED13 and HDA6 to down-regulate seed maturation gene expression directly during Arabidopsis early seedling growth. Plant Cell Physiol 57: 1689–1706 [DOI] [PubMed] [Google Scholar]
- Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Cigliano RA, Sanseverino W, Cremona G, Ercolano MR, Conicella C, Consiglio FM (2013) Genome-wide analysis of histone modifiers in tomato: gaining an insight into their developmental roles. BMC Genomics 14: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Birchler JA, Pires JC (2014) Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr Opin Plant Biol 19: 91–98 [DOI] [PubMed] [Google Scholar]
- Costa F, Stella S, Van de Weg WE, Guerra W, Cecchinel M, Dalla Via J, Koller B, Sansavini S (2005) Role of the genes Md-ACO1 and Md-ACS1 in ethylene production and shelf life of apple (Malus domestica Borkh). Euphytica 141: 181–190 [Google Scholar]
- Daccord N, Celton J M, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R, et al. (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Dougherty L, Zhu Y, Xu K (2016) Assessing the allelotypic effect of two aminocyclopropane carboxylic acid synthase-encoding genes MdACS1 and MdACS3a on fruit ethylene production and softening in Malus. Hortic Res 3: 16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fu CC, Han YC, Guo YF, Kuang JF, Chen JY, Shan W, Lu WJ (2018) Differential expression of histone deacetylases during banana ripening and identification of MaHDA6 in regulating ripening-associated genes. Postharvest Biol Technol 141: 24–32 [Google Scholar]
- Giovannoni J, Cuong N, Ampofo B, Zhong S, Fei Z (2017) The epigenome and transcriptional dynamics of fruit ripening. Annu Rev Plant Biol 68: 61–84 [DOI] [PubMed] [Google Scholar]
- Guo JE, Hu Z, Yu X, Li A, Li F, Wang Y, Tian S, Chen G (2018) A histone deacetylase gene, SlHDA3, acts as a negative regulator of fruit ripening and carotenoid accumulation. Plant Cell Rep 37: 125–135 [DOI] [PubMed] [Google Scholar]
- Guo JE, Hu Z, Zhu M, Li F, Zhu Z, Lu Y, Chen G (2017) The tomato histone deacetylase SlHDA1 contributes to the repression of fruit ripening and carotenoid accumulation. Sci Rep 7: 7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YC, Kuang JF, Chen JY, Liu XC, Xiao YY, Fu CC, Wang JN, Wu KQ, Lu WJ (2016) Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol 171: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Hu Y, Lv Y, Rose JKC, Sun Y, Shen F, Wang Y, Zhang X, Xu X, Wu T, et al. (2018) Natural variation underlies differences in ETHYLENE RESPONSE FACTOR17 activity in fruit peel degreening. Plant Physiol 176: 2292–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Han Z, Sun Y, Wang S, Wang T, Wang Y, Xu K, Zhang X, Xu X, Han Z, et al. (2020) ERF4 affects fruit firmness through TPL4 by reducing ethylene production. Plant J 103: 937–950 [DOI] [PubMed] [Google Scholar]
- Kagale S, Rozwadowski K (2011) EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Jin HS, Noh YS, Noh B (2015) Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol 206: 281–294 [DOI] [PubMed] [Google Scholar]
- Kenneth Manning Klee HJ, Giovannoni JJ (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J, Zhang Y, Handa AK, Zhu JK (2017) Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci USA 114: E4511–E4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Daujat S, Schneider R (2016) Lateral thinking: how histone modifications regulate gene expression. Trends Genet 32: 42–56 [DOI] [PubMed] [Google Scholar]
- Li D, Sun Q, Zhang G, Zhai L, Li K, Feng Y, Wu T, Zhang X, Xu X, Wang Y, et al. (2021) MxMPK6-2-bHLH104 interaction is involved in reactive oxygen species signaling in response to iron deficiency in apple rootstock. J Exp Bot 72: 1919–1932 [DOI] [PubMed] [Google Scholar]
- Li T, Jiang ZY, Zhang LC, Tan DM, Wei Y, Yuan H, Li TL, Wang AD (2016) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J 88: 735–748 [DOI] [PubMed] [Google Scholar]
- Li X, Guo W, Li J, Yue P, Bu H, Jiang J, Liu W, Xu Y, Yuan H, Li T, et al. (2020) Histone acetylation at the promoter for the transcription factor PuWRKY31 affects sucrose accumulation in pear fruit. Plant Physiol 182: 2035–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Lü P, Yu S, Zhu N, Chen YR, Zhou B, Pan Y, Tzeng D, Fabi JP, Argyris J, Garcia-Mas J, et al. (2018) Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat Plants 4: 784–791 [DOI] [PubMed] [Google Scholar]
- Luo M, Yu CW, Chen FF, Zhao L, Tian G, Liu X, Cui Y, Yang JY, Wu K (2012) Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX Genes in Arabidopsis. PLoS Genet 8: e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Lv S, Zhang C, Yang C (2013) Histone deacetylases and their functions in plants. Plant Cell Rep 32: 465–478 [DOI] [PubMed] [Google Scholar]
- Manning K, Tör M, Poole M, Hong Y, Thompson A, King G, Giovannoni J, Seymour G (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sridha S, Wu KQ (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46: 124–133 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Sunako R, Sakuraba W, Senda M, Akada S, Ishikawa R, Niizeki M, Harada T (1999) An allele of the ripening-specific 1-aminocyclopropane-1-carboxylic acid synthase gene (ACS1) in apple fruit with a long storage life. Plant Physiol 119: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tang N, Ma SQ, Zong W, Yang N, Lv Y, Yan C, Guo ZL, Li J, Li X, Xiang Y, et al. (2016) MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell 28: 2161–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Han ZY, Zhang J, Hu Y, Song T, Yao Y (2015) The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in crabapples. Sci Rep 5: 12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda M, Seki M (2020) Histone modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiol 182: 15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010). The genome of the domesticated apple (Malus x domestica Borkh.). Nature Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Verdone L, Agricola E, Caserta M, Di Mauro E (2006) Histone acetylation in gene regulation. Brief Funct Genomic Proteomic 5: 209–221 [DOI] [PubMed] [Google Scholar]
- Wang A, Yamakake J, Kudo H, Wakasa Y, Hatsuyama Y, Igarashi M, Kasai A, Li T, Harada T (2009) Null mutation of the MdACS3 gene, coding for a ripening-specific 1-aminocyclopropane-1-carboxylate synthase, leads to long shelf life in apple fruit. Plant Physiol 151: 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YY, Chen JY, Kuang JF, Shan W, Xie H, Jiang YM, Lu WJ (2013) Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J Exp Bot 64: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, Yao YX, You CX, Zhang XS, Hao YJ (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35: 1884–1897 [DOI] [PubMed] [Google Scholar]
- Yang JJ, Zhan RL, Jin YR, Song JY, Li DX, An GY, Li MJ (2020) Functional analysis of the promoter of the MdFRK2 gene encoding a high affinity fructokinase in apple (Malus x domestica). Sci Hortic 265: 109088 [Google Scholar]
- Yin X-R, Allan AC, Chen KS, Ferguson IB (2010) Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol 153: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang B, Wang S, Lin T, Yang L, Zhao Z, Zhang Z, Huang S, Yang X (2020) Genome-wide target mapping shows histone deacetylase complex 1 regulates cell proliferation in cucumber fruit. Plant Physiol 182: 167–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang H, Quan R, Wang XC, Huang R (2009) Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol 150: 365–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In V Bryson, HJ Vogel, eds, Evolving Genes and Proteins. Academic Press, New York: , pp 97–166 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.