Under water-deficit conditions, inactivation of OsWRKY5 – which acts as a direct negative regulator of OsMYB2 expression under normal growth conditions – increases ABA-induced drought tolerance in rice
Abstract
During crop cultivation, water-deficit conditions retard growth, thus reducing crop productivity. Therefore, uncovering the mechanisms behind drought tolerance is a critical task for crop improvement. Here, we show that the rice (Oryza sativa) WRKY transcription factor OsWRKY5 negatively regulates drought tolerance. We determined that OsWRKY5 was mainly expressed in developing leaves at the seedling and heading stages, and that its expression was reduced by drought stress and by treatment with NaCl, mannitol, and abscisic acid (ABA). Notably, the genome-edited loss-of-function alleles oswrky5-2 and oswrky5-3 conferred enhanced drought tolerance, measured as plant growth under water-deficit conditions. Conversely, the overexpression of OsWRKY5 in the activation-tagged line oswrky5-D resulted in higher susceptibility under the same conditions. The loss of OsWRKY5 activity increased sensitivity to ABA, thus promoting ABA-dependent stomatal closure. Transcriptome deep sequencing and reverse transcription quantitative polymerase chain reaction analyses demonstrated that the expression of abiotic stress-related genes including rice MYB2 (OsMYB2) was upregulated in oswrky5 knockout mutants and downregulated in oswrky5-D mutants. Moreover, dual-luciferase, yeast one-hybrid, and chromatin immunoprecipitation assays showed that OsWRKY5 directly binds to the W-box sequences in the promoter region of OsMYB2 and represses OsMYB2 expression, thus downregulating genes downstream of OsMYB2 in the ABA signaling pathways. Our results demonstrate that OsWRKY5 functions as a negative regulator of ABA-induced drought stress tolerance, strongly suggesting that inactivation of OsWRKY5 or manipulation of key OsWRKY5 targets could be useful to improve drought tolerance in rice cultivars.
Introduction
Crop plants are increasingly being confronted with water-deficient conditions due to global climate change, resulting in a reduction in crop productivity (Pandey and Shukla, 2015). Therefore, improving drought tolerance is an important strategy for stabilizing crop productivity, especially grain yield in cereal crops. Numerous studies have identified transcription factors (TFs) as positive regulators of drought tolerance mechanisms, since their overexpression in rice (Oryza sativa) plants resulted in drought-tolerant phenotypes. For instance, rice MYB DOMAIN PROTEIN 2 (OsMYB2), an R2R3-type MYB TF, mediates abscisic acid (ABA)-dependent pathways by upregulating ABA signaling genes, including LATE EMBRYOGENESIS ABUNDANT 3 (LEA3) and RESPONSIVE TO ABSCISIC ACID 16A (RAB16A). Transgenic plants overexpressing OsMYB2 thus exhibit enhanced tolerance to salt, cold, and dehydration stresses (Yang et al., 2012). In addition to this positive regulator, a handful of TFs have been characterized as negative regulators in response to drought stress (Jia et al., 2015; Wang et al., 2019a).
Members of many TF families participate in abiotic stress responses, including NAC [NAM (No apical meristem), ATAF (Arabidopsis transcription activation factor) and CUC (Cup-shaped cotyledon)], MYB (myeloblastosis), AP2/ERF (APETALA2/Ethylene Responsive Factor), WRKY, and bZIP (Basic leucine zipper) TFs (Atkinson and Urwin, 2012; Shaik and Ramakrishna, 2014; Wu et al., 2019). AQ3NAM, ATAF, CUC (NAC), MYB, APETALA2 (AP2)/ERF, WRKY, and Basic leucine zipper TFs (Atkinson et al., 2012; Shaik et al., 2014; Wu et al., 2019). Among these, the WRKY TF family is specific to plants and many WRKY TFs regulate abiotic stress tolerance (Eulgem et al., 2000). The Arabidopsis (Arabidopsis thaliana) and rice genomes encode 74 and 109 putative WRKY TFs, respectively (Eulgem and Somssich, 2007; Ross et al., 2007). At their N termini, WRKY proteins have one or two WRKY domains (containing the conserved motif WRKYGQK), followed by a zinc finger domain. These WRKY domains specifically bind to the W-box (TTGAC[T/C]) in the promoter regions of their target genes (Eulgem et al., 2000; Cai et al., 2008). In addition, WRKY proteins interact with other proteins, such as receptors, kinases, and other TFs, and are interwoven in transcriptional regulatory networks (Chen et al., 2019).
Studies with transgenic plants have shown that many WRKY TFs affect abiotic stress tolerance. For example, transgenic rice plants overexpressing the maize (Zea mays) WRKY gene ZmWRKY58 exhibited enhanced tolerance to drought and salt stresses (Cai et al., 2014). Among 12 drought-responsive wheat (Triticum aestivum) WRKY genes, the heterologous expression of TaWRKY1 and TaWRKY33 in Arabidopsis enhanced tolerance to drought and heat stresses (He et al., 2016). Arabidopsis WRKY46 (AtWRKY46) modulates endogenous auxin levels, thus promoting lateral root development under osmotic and salt stress conditions (Ding et al., 2015; Chen et al., 2017). Transgenic rice plants overexpressing OsWRKY47 exhibited a drought-tolerant phenotype, leading to higher grain yield even under drought stress (Raineri et al., 2015). The overexpression of GmWRKY12 enhanced tolerance to drought and salt stresses (Shi et al., 2018). However, the underlying molecular mechanisms by which other WRKY TFs affect abiotic stress tolerance are not yet fully understood.
Plants induce multiple responses to drought stress and of these responses, reducing transpirational water loss through stomatal closure is a key determinant of drought tolerance (Matsuda et al., 2016; Agurla et al., 2018). Indeed, a substantial body of work has focused on the regulation of stomatal movement. ABA is the major phytohormone that triggers stomatal closure (Bharath et al., 2021). ABA induces the expression of the rice NAC gene STRESS-RESPONSIVE NAC 1 (SNAC1) and the overexpression of SNAC1 increases ABA sensitivity and stomatal closure, leading to enhanced drought tolerance in transgenic rice lines (You et al., 2013). The overexpression of rice ABSCISIC ACID, STRESS AND RIPENING 5 (OsASR5) enhanced drought tolerance by regulating leaf water status and the transgenic plants were also hypersensitive to exogenous ABA treatment (Li et al., 2017). The overexpression of the harpin-encoding gene hrf1 increased endogenous ABA levels and promoted stomatal closure in rice (Zhang et al., 2011a). The transcription of rice CALCIUM-DEPENDENT PROTEIN KINASE 9 (OsCPK9) is induced by ABA, polyethylene glycol (PEG), and NaCl treatments, and OsCPK9 overexpression enhanced drought tolerance by increasing stomatal closure (Wei et al., 2014). In contrast to these positive regulators, rice SQUALENE SYNTHASE (OsSQS) functions as a negative regulator of drought stress responses since OsSQS silencing by RNA interference improves drought tolerance (Manavalan et al., 2012).
Some rice TFs participate in phytohormone-mediated pathways that regulate both leaf senescence and abiotic stress responses. For example, the NAC family TF OsNAP confers drought tolerance by regulating ABA signaling (Chen et al., 2014). The overexpression of OsNAP promotes leaf senescence by fine-tuning ABA biosynthesis and directly regulating senescence-associated genes (Liang et al., 2014). In addition, rice ETHYLENE RESPONSE FACTOR 101 (OsERF101) upregulates the expression of ABA-responsive genes such as RESPONSIVE TO DEHYDRATION 22, LEA3, and PEROXIDASE, thereby improving drought tolerance (Jin et al., 2018). OsERF101 promotes leaf senescence via jasmonic acid signaling involving OsNAP and OsMYC2 (Lim et al., 2020). Our previous study reported that OsWRKY5 promotes leaf senescence through upregulating ABA biosynthesis (Kim et al., 2019). Therefore, we speculated that OsWRKY5 is also involved in abiotic stress responses in rice. Indeed, OsWRKY5 expression in developing leaves is substantially downregulated upon dehydration, suggesting that OsWRKY5 acts as a negative regulator of drought tolerance mechanisms in rice. Therefore, in the present study, we focused on examining OsWRKY5 function and showed that mutation of OsWRKY5 by genome editing confers enhanced tolerance to drought stress, which is consistent with the direct or indirect downregulation of genes involved in drought stress tolerance. Our findings thus identify a potential biotechnological avenue for improving drought tolerance in rice and other cereal crops.
Results
Expression of OsWRKY5 in rice
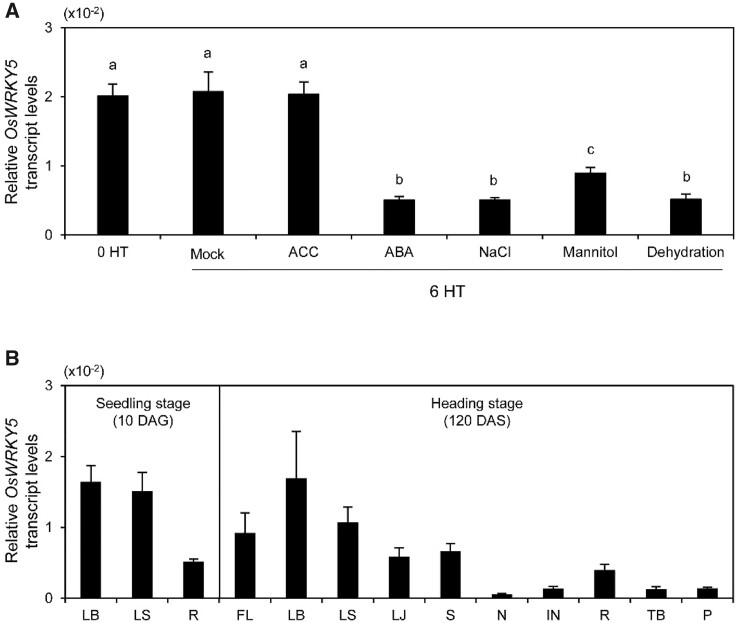
OsWRKY5 acts as a positive regulator of leaf senescence at the reproductive stage in rice (Kim et al., 2019). To evaluate whether phytohormone or stress treatments affect OsWRKY5 expression, we applied 1-aminocyclopropane-1-carboxylic acid (ACC), ABA, NaCl, mannitol, or imposed dehydration stress on 10-d-old wild-type (WT) seedlings for 6 h. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis indicated that, while OsWRKY5 was constitutively expressed in mock control and after treatment with ACC, OsWRKY5 transcript levels were significantly reduced by ABA, NaCl, mannitol, and dehydration treatments (Figure 1A), strongly suggesting that OsWRKY5 is responsive to abiotic stress and ABA signaling.
Figure 1.
Expression profiling of OsWRKY5. A, Relative OsWRKY5 mRNA levels in response to various abiotic stresses. WT seedlings were grown on half-strength MS phytoagar media for 10 d in a growth chamber under LD conditions (14-h light at 30°C/10-h dark at 25°C) and then incubated in half-strength MS liquid medium supplemented with 1-mM ACC, 100-µM ABA, 100-mM NaCl, or 100-mM mannitol for 6 h, or dehydrated by 6-h air drying. Seedlings incubated in half-strength MS liquid medium without treatment were used as mock control. WT leaves were harvested at 0 and 6 h of treatment (HT). B, OsWRKY5 is differentially expressed in various organs of WT plants (japonica cultivar ‘Dongjin’), including flag leaf (FL), leaf blade (LB), leaf sheath (LS), lamina joint (LJ), stem (S), node (N), internode (IN), root (R), tiller base (TB), and panicle (P). Seedling stage: WT seedlings were grown in a growth chamber for 10 DAG under LD conditions (14-h light at 30°C/10-h dark at 25°C). Heading stage: WT plants were grown in a paddy field for 120 DAS under NLD conditions (≥14-h light/day). Relative OsWRKY5 transcript levels were determined by RT-qPCR and normalized to OsUBIQUITIN5 (OsUbq5; Os01g22490). Relative expression was calculated using the ΔΔCT method. Different letters (A) indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (P < 0.05). Mean and standard deviation values were obtained from three biological replicates. These experiments were repeated three times with similar results.
To determine the expression pattern of OsWRKY5, we measured its transcript levels in various plant tissues (flag leaf, leaf blade, leaf sheath, lamina joint, stem, node, internode, root, tiller base, and panicle) of the WT (the japonica rice cultivar Dongjin) grown in a growth chamber for 10 d after germination (DAG, seedling stage) or for 120 d after sowing (DAS, heading stage) under natural long-day (NLD) conditions (≥14-h light/day in Suwon, South Korea, 37°N latitude). Consistent with a previous report (Kim et al., 2019), RT-qPCR analysis demonstrated that OsWRKY5 is preferentially expressed in leaves, including leaf blades, leaf sheaths, and flag leaf tissues (Figure 1B). These results indicate that OsWRKY5 may play a regulatory role in leaves during vegetative growth.
To determine the subcellular localization of OsWRKY5, we transiently expressed the Ubipro:OsWRKY5-sGFP construct in rice protoplasts isolated from rice suspension cultured cells. As expected for a TF, we detected green fluorescence in nuclei (Supplemental Figure S1A), indicating that OsWRKY5 localizes to the nucleus. Since homodimerization of TFs is commonly a prerequisite to regulate transcription (Funnell and Crossley, 2012), we tested whether OsWRKY5 could form homodimers using yeast two-hybrid assays. Indeed, in vivo plate assays showed that OsWRKY5 can form homodimers (Supplemental Figure S1B), consistent with OsWRKY5 functioning as a TF.
A loss-of-function mutation of OsWRKY5 induced by genome editing enhances drought tolerance in rice
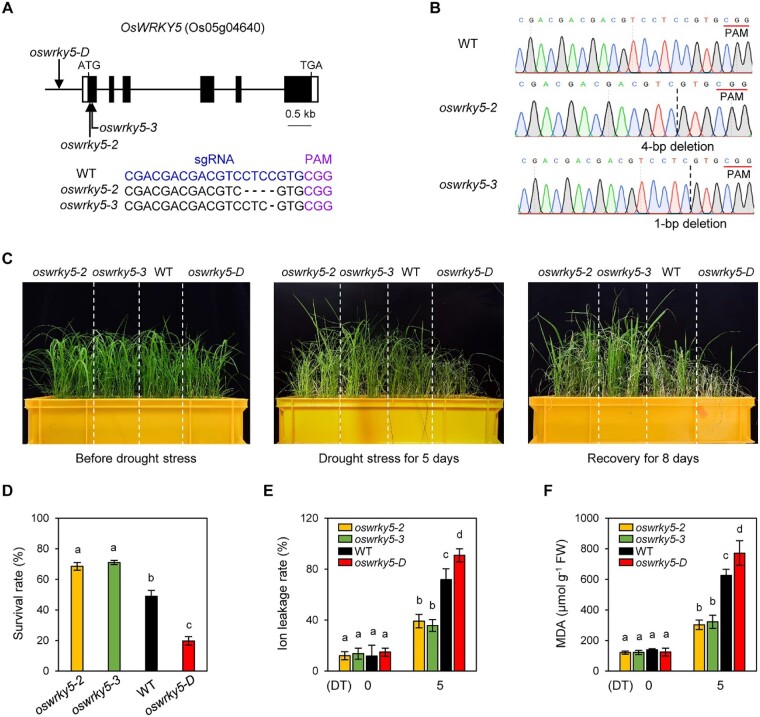
To examine the role of OsWRKY5 in drought stress responses, we used the enhancer-trapped T-DNA insertion mutant oswrky5-D described in our previous study (Kim et al., 2019), whereby OsWRKY5 expression is constitutive due to four copies of the 35S Cauliflower mosaic virus promoter inserted upstream of the gene. In addition, we employed clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9)-mediated genome editing to generate knockout mutants in the Dongjin background and isolated the alleles oswrky5-2 and oswrky5-3 with 4-bp and 1-bp deletions in the first exon, respectively (Figure 2, A and B). To assess drought tolerance of these mutants, we subjected 3-week-old WT, oswrky5-D, oswrky5-2, and oswrky5-3 plants grown in the same pot to progressive drought stress by withholding water for 5 d, followed by an 8-d recovery by watering with tap water (Figure 2C). We then scored the number of surviving rice plants after recovery. The oswrky5-2 and oswrky5-3 mutants exhibited ∼20% and ∼23% higher survival rates than the WT, respectively (Figure 2D).
Figure 2.
A loss-of-function mutation in OsWRKY5 enhances tolerance to drought stress. A, Schematic representation of the OsWRKY5 locus (Os05g04640) and its mutant alleles. Each arrow indicates the position of the T-DNA insertion (oswrky5-D, PFG_3A-15928) and the CRISPR-Cas9 edited site (oswrky5-2 and oswrky5-3). Black boxes represent exons and the two white boxes represent 5′-untranslated region and 3′-untranslated region. The nucleotides around the CRISPR-Cas9 edited site are compared in the WT, oswrky5-2, and oswrky5-3. Blue and purple nucleotides represent the sgRNA and PAM sequences, respectively. The edited nucleotides are shown as dashes. B, Sanger sequencing results showed a deletion of 4 bp and 1 bp in the genomic DNA of oswrky5-2 and oswrky5-3, respectively, compared to the WT. Red line represents the PAM sequence. Black dotted lines represent the predicted double-stranded break (DSB) site. C–F, WT, oswrky5-2, oswrky5-3, and oswrky5-D plants were grown in the greenhouse under NLD conditions for three weeks. C, Phenotypes of rice plants treated with drought stress for 5 d and recovered with water for 8 d. D, Survival rates measured after recovery. E, F, Ion leakage rate (E) and MDA contents (F) in rice leaves after 0 and 5 d of treatment (DT). Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (P < 0.05). Mean and standard deviation values were obtained from three biological replicates. These experiments were repeated three times with similar results. FW, fresh weight.
Under unfavorable circumstances such as water deficit or high salinity, a reduction of membrane integrity leads to an increase in ion leakage rates. In addition, lipid peroxidation due to excessive accumulation of reactive oxygen species (ROS) results in increased malondialdehyde (MDA) contents. Thus, both ion leakage and MDA contents serve as important indicators to evaluate abiotic stress tolerance. Although the ion leakage rate and MDA contents increased in all genotypes following a 5-d drought treatment (Figure 2, E and F), these indicators were significantly lower in oswrky5-2 and oswrky5-3 plants compared to the WT and oswrky5-D. These results suggest that the loss of OsWRKY5 function enhanced drought tolerance in rice.
Mannitol is widely used to induce osmotic stress in plants (Verslues et al., 2006). We therefore tested whether a loss of OsWRKY5 function might confer tolerance to mannitol treatment. We treated 3-week-old WT, oswrky5-2, and oswrky5-D plants grown in the same pot with 200-mM mannitol for 10 d, after which they were watered with tap water for 10 d to allow them to recover (Supplemental Figure S2A). The survival rate of oswrky5-2 was significantly higher than that of the WT and oswrky5-D (Supplemental Figure S2B). In addition, we assessed growth inhibition of rice seedlings grown in half-strength Murashige and Skoog (MS) phytoagar medium containing 100- or 150-mM mannitol for 10 d (Supplemental Figure S2C). Relative shoot elongation was less inhibited in oswrky5-2 compared with the WT or oswrky5-D (Supplemental Figure S2D). These results indicate that OsWRKY5 is closely associated with tolerance to drought and osmotic stresses.
oswrky5-2 mutants reduce water loss via stomatal closure under drought stress
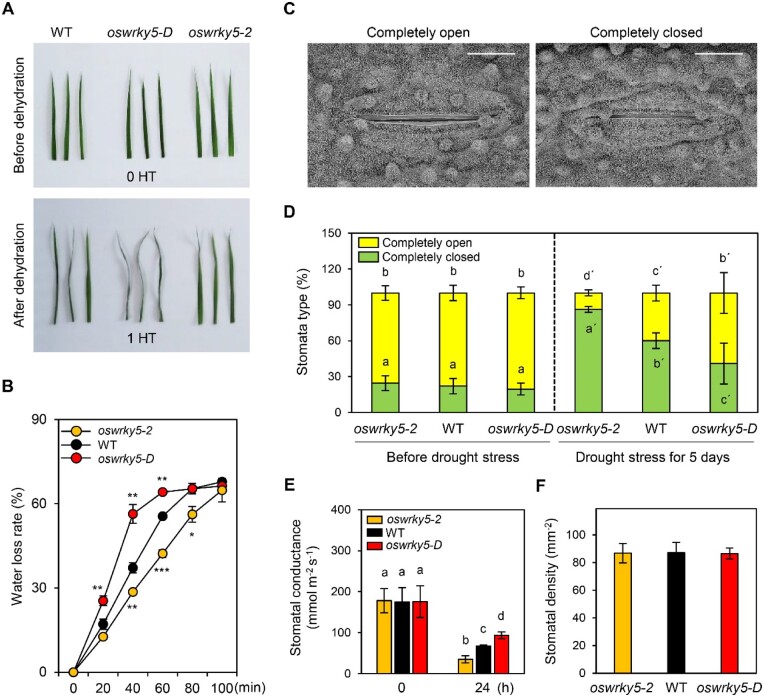
To examine the effect of OsWRKY5 under dehydration conditions, we dehydrated leaves that had been detached from 2-week-old seedlings by air-drying for 1 h. Unlike the leaves of oswrky5-D, which exhibited a severe rolling phenotype, oswrky5-2 leaves were only moderately rolled following dehydration (Figure 3A). In agreement with this leaf rolling phenotype, oswrky5-D leaves lost water faster than WT leaves, while oswrky5-2 leaves retained significantly more water than WT leaves (Figure 3B). These results were validated by using infrared thermal imaging to visualize the surface temperature of detached leaves: leaves with a higher transpiration rate appear cooler (blue) in this assay (You et al., 2013). Our infrared thermal analysis showed that the leaf surface temperature of the WT was lower due to more water loss than oswrky5-2 after 3.5-h dehydration, while that of oswrky5-D was lower than the WT after 2.5-h dehydration (Supplemental Figure S3).
Figure 3.
OsWRKY5 affects stomatal aperture. A, B, WT, oswrky5-2, and oswrky5-D plants were grown in the growth chamber under LD conditions for 2 weeks. Detached leaves were dehydrated by air-drying at room temperature (23°C, 40% relative humidity). A, Dehydration phenotypes of detached WT, oswrky5-D, and oswrky5-2 leaves after 1-h air-drying treatment (HT) on dry filter paper. B, Rate of water loss of rice leaves, measured at the indicated time points (n = 12). Asterisks above oswrky5-2 and below oswrky5-D data points indicate a statistically significant difference from the WT, as determined by Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001). C, Scanning electron microscopic images of stomatal opening in 3-week-old WT before drought stress. Scale bars = 10 µm. D–F, Three-week-old rice plants were exposed to drought stress for 5 d (D and F) or 24 h (E), as shown in Figure 2C. D, Stomata type (%) in the WT, oswrky5-2, and oswrky5-D leaves before and after drought stress (73, 71, and 74 stomata were evaluated in the WT, oswrky5-2, and oswrky5-D before drought stress; 78, 80, and 77 stomata were evaluated in the WT, oswrky5-2, and oswrky5-D after drought stress, respectively). E, Stomatal conductance in the WT, oswrky5-2, and oswrky5-D. F, Stomatal density of middle leaves of the WT, oswrky5-2, and oswrky5-D. Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (P < 0.05). Mean and standard deviation values were obtained from three biological replicates. These experiments were repeated three times with similar results.
Since the water loss rate mainly depends on stomatal opening, we investigated whether stomatal aperture was altered in rice leaves subjected to drought stress conditions. Scanning electron microscopy assays revealed that there was no difference in the percentage of closed stomata (around 20%) in the WT, oswrky5-2, and oswrky5-D plants before drought treatment, but that the percentage of closed stomata was significantly higher in oswrky5-2 (86%) relative to the WT (60%) and oswrky5-D (40%) after 5 d of water deficit (Figure 3, C and D). Next, we measured the stomatal conductance of drought-stressed leaves. We observed significantly lower stomatal conductance in oswrky5-2 compared to the WT and oswrky5-D, indicating that a loss of OsWRKY5 function leads to decreased gas and water exchange through stomata under drought stress (Figure 3E). All genotypes had similar numbers of stomata, indicating that OsWRKY5 might not affect stomatal differentiation or density (Figure 3F). Taken together, these results suggest that enhanced drought tolerance in oswrky5-2 is mainly due to much faster stomatal closure, which minimizes water loss at the whole-plant level.
OsWRKY5 inhibits ABA-induced stomatal closure
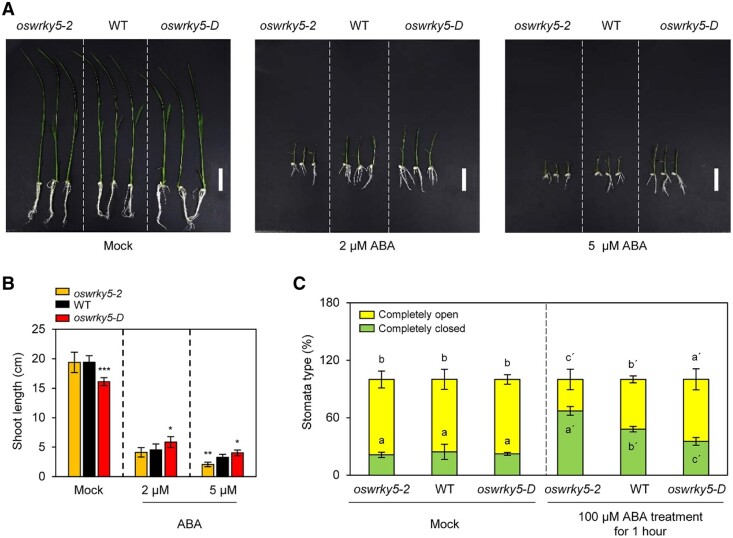
Downregulation of OsWRKY5 expression by ABA treatment suggested that OsWRKY5 is closely associated with ABA signaling (Figure 1A). To investigate whether changes in OsWRKY5 expression affect sensitivity to ABA, we characterized the rates of shoot growth in 10-d-old rice seedlings grown in half-strength MS phytoagar medium containing 2- or 5-µM ABA (Figure 4A). ABA treatment significantly inhibited shoot growth in oswrky5-2 compared to the WT and oswrky5-D (Figure 4B), indicating that the loss of OsWRKY5 function increases ABA sensitivity.
Figure 4.
The oswrky5-2 mutant shows increased ABA sensitivity. A and B, WT, oswrky5-2, and oswrky5-D seeds were germinated on half-strength MS phytoagar medium for 3 d and then grown on half-strength MS phytoagar medium containing 0-, 2-, or 5-µM ABA for 10 d under LD conditions. Seedlings grown on half-strength MS plates with 0-µM ABA were used as mock control. White bars = 3 cm. A, Growth phenotypes and (B) the shoot length of the WT, oswrky5-2, and oswrky5-D plants were measured after 10-d ABA treatment (n = 10). Asterisks above oswrky5-2 and oswrky5-D data points indicate a statistically significant difference from the WT, as determined by Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001). C, Effect of ABA on stomatal aperture. Detached leaves of 3-week-old WT, oswrky5-2, and oswrky5-D plants grown in the greenhouse under NLD conditions were incubated in 3-mM MES buffer (pH 5.8) containing 100-µM ABA for 1 h. Detached leaves floated on 3-mM MES buffer (pH 5.8) without phytohormones were used as mock control (79, 81, and 77 stomata were evaluated in the WT, oswrky5-2, and oswrky5-D under mock control; 72, 81, and 77 stomata were evaluated in the WT, oswrky5-2, and oswrky5-D under ABA treatment, respectively). Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (P < 0.05). Mean and standard deviation values were obtained from three biological replicates. These experiments were repeated three times with similar results.
As ABA activates downstream signaling pathways leading to stomatal closure (Munemasa et al., 2015), we analyzed ABA-induced stomatal closure in detached leaves that we floated on the ABA solution. In leaves that had been detached from 3-week-old plants on 100-µM ABA, the percentage of closed stomata was significantly higher in oswrky5-2 (64%) than in the WT (45%) or oswrky5-D (30%) leaves (Figure 4C). These results indicate that OsWRKY5 serves as a negative regulator of ABA-induced stomatal closure.
Identification of stress-related genes controlled by OsWRKY5
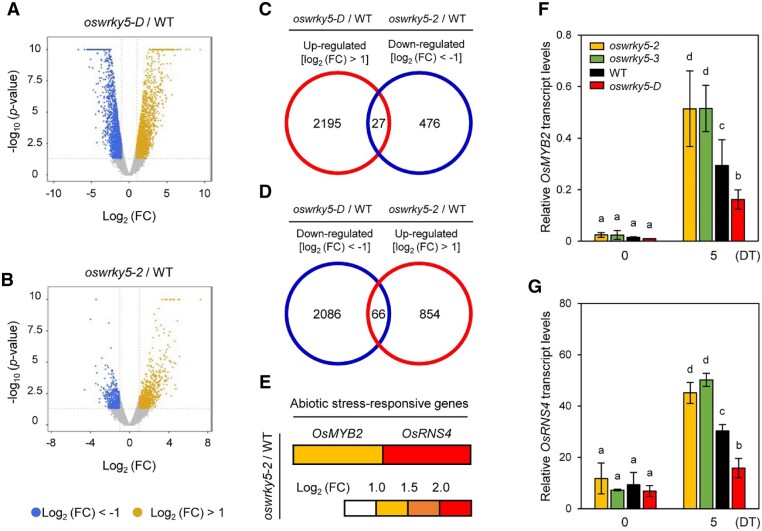
To understand the molecular mechanisms regulated by OsWRKY5 under drought stress, we compared the transcriptomes of leaves from drought-stressed 3-week-old WT, oswrky5-2, and oswrky5-D plants using transcriptome deep sequencing (RNA-seq). The threshold for significantly differentially expressed genes (DEGs) was set at a (log2 scale)-fold change (FC) value of more than 1 or less than −1 and adjusted P <0.05. Using these criteria, we identified 1,423 DEGs (920 upregulated and 503 downregulated) in oswrky5-2 compared to the WT, and 4,374 DEGs (2,222 upregulated and 2,152 downregulated) in oswrky5-D compared to the WT (Supplemental Data Sets S1 and S2). These DEGs altered by drought stress are shown in the volcano plots, which illustrate the asymmetry between upregulated (brown) and downregulated (blue) DEGs (Figure 5, A and B). Venn diagram analysis showed that 27 genes were upregulated in oswrky5-D and downregulated in oswrky5-2, and 66 genes were downregulated in oswrky5-D and upregulated in oswrky5-2 (Figure 5, C and D; Supplemental Data Sets S3 and S4).
Figure 5.
DEGs in the oswrky5-D and oswrky5-2 mutants. A and B, Volcano plots comparing the transcriptomes of oswrky5-D (A) and oswrky5-2 (B) mutants with the WT. X-axis and Y-axis represent log2 FC and −log10(P-value), respectively. The blue and brown dots represent downregulated DEGs with log2(FC) less than −1 and upregulated DEGs with log2(FC) >1, respectively. The gray dots represent no significant difference in transcriptomes. C and D, Venn diagrams of the DEGs upregulated in oswrky5-D and downregulated in oswrky5-2 (C), and downregulated in oswrky5-D and upregulated in oswrky5-2 (D). E, The abiotic stress-responsive genes OsMYB2 and OsRNS4 were upregulated in oswrky5-2 compared to the WT. F, G, Altered expression of OsMYB2 (F) and OsRNS4 (G) in the WT, oswrky5-2, oswrky5-3, and oswrky5-D in response to drought stress. Total RNA was isolated from the leaves of drought-stressed WT, oswrky5-2, oswrky5-3, and oswrky5-D plants at 0 and 5 d of treatment (DT) as shown in Figure 2C. The relative transcript levels of abiotic stress-responsive genes (OsMYB2 and OsRNS4) were determined by RT-qPCR and normalized to OsUbq5. Relative expression was calculated using the ΔΔCT method. Mean and standard deviation values were obtained from three biological replicates. Different letters indicate significantly different values according to a one-way ANOVA and Duncan’s least significant range test (P < 0.05). These experiments were repeated three times with similar results.
Of 93 genes from the intersection of the Venn diagram, 80 genes were classified by the agriGO Classification System (http://bioinfo.cau.edu.cn/agriGO; Du et al., 2010; Tian et al., 2017) for gene ontology (GO) analysis. The biological process and molecular function included response to stimuli (GO:0050896), metabolic process (GO:0008152), cellular process (GO:0009987), biological regulation (GO:0065007), binding (GO:0005488), and catalytic activity (GO:0003824; Supplemental Figure S4 and Supplemental Data Set S5). These results indicate that OsWRKY5 is involved in a diverse range of biological processes and molecular functions, including those related to abiotic stress responses.
Interestingly, among 80 genes subjected to GO analysis, we found that OsMYB2 and rice S-LIKE RIBONUCLEASE 4 (OsRNS4), belonging to the binding (GO:0005488) category, were upregulated in oswrky5-2 and downregulated in oswrky5-D (Figure 5E; Supplemental Data Set S3). These genes were reported to function in the response to abiotic stresses such as drought and high salinity (Yang et al., 2012; Zheng et al., 2014). RT-qPCR analysis showed that their expression patterns coincided in the drought-stressed leaves of the WT, oswrky5-2, oswrky5-3, and oswrky5-D plants (Figure 5, F and G). OsRNS4 is known to function in high salinity stress responses and therefore we did not analyze it further (Zheng et al., 2014). In contrast, OsMYB2 is involved in abiotic stress responses and ABA signaling and its overexpression confers drought tolerance by upregulating the expression of a number of stress- and ABA-responsive genes (OsLEA3, OsRAB16A, and DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN [OsDREB2A]; Yang et al., 2012). Therefore, we speculated that OsWRKY5 participates in OsMYB2-mediated regulatory pathways. To test this, we examined the expression of downstream genes of OsMYB2 in the WT, oswrky5-2, oswrky5-3, and oswrky5-D. RT-qPCR analysis showed that transcript levels of these downstream genes were significantly upregulated in oswrky5-2 and oswrky5-3 and downregulated in oswrky5-D compared to the WT under drought stress (Supplemental Figure S5). These results suggest that OsWRKY5 represses the OsMYB2-regulated pathway in response to drought stress.
OsWRKY5 directly binds to the promoter region of OsMYB2
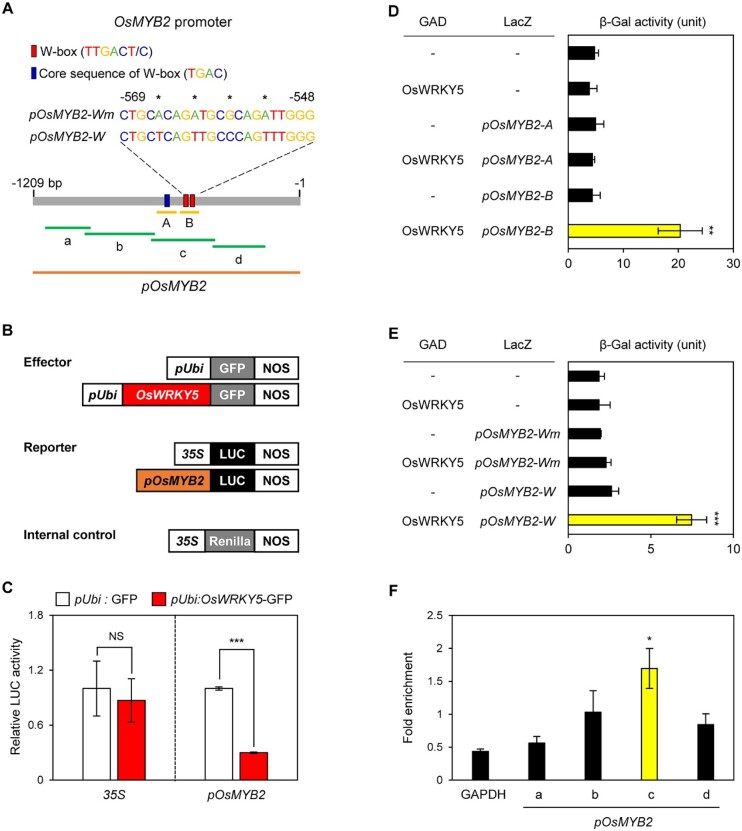
Under drought conditions, the expression of OsMYB2 was closely associated with that of OsWRKY5 (Figure 5, E and F). WRKY TFs regulate the transcription of their target genes by recognizing the consensus cis-element W-box, defined as 5′-TTGAC(T/C)-3′, with an invariant TGAC core sequence (Eulgem et al., 2000). We searched for W-box and TGAC core sequences upstream of the translation initiation sites of OsMYB2 (Figure 6A). The OsMYB2 promoter region contains W-box sequences and a single TGAC core sequence (Figure 6A).
Figure 6.
OsWRKY5 downregulates the transcription of OsMYB2. A, The blue and red boxes in the promoter of OsMYB2 (gray horizontal line) represent the positions of the core sequence (TGAC) of the W-box and W-boxes (TTGACT/C), respectively. The position of fragments used for the Y1H (uppercase letters A–B with yellow horizontal lines) assay, ChIP (lowercase letters a–d with green horizontal lines) assay, and transient LUC reporter (orange horizontal line) assay, respectively. Asterisks above nucleotides indicate substituted positions in the mutated W-box (pOsMYB2-Wm) construct from W-box sequences (pOsMYB2-W). B, Effector, reporter, and internal control constructs used in the transient LUC reporter assay. pUbi, Ubiquitin promoter; 35S, 35S promoter; LUC, luciferase. Each construct also contains the NOS terminator. C, The activation of the OsMYB2 promoter by OsWRKY5-GFP expression in a protoplast transient assay. The 35S promoter was used as a negative control. D and E, The binding activity of OsWRKY5 to the promoter regions of OsMYB2 [pOsMYB2-A and pOsMYB2-B, uppercase letters with yellow horizontal lines (D); pOsMYB2-W and pOsMYB2-Wm (E)] was examined by Y1H assays. Empty bait and prey plasmids (−) were used for the negative controls. The relative β-galactosidase (β-Gal) activity was obtained by normalizing to the level of each negative control. F, OsWRKY5 binding affinity to the promoter regions of OsMYB2 in planta examined by ChIP assays. OsWRKY5-GFP was transiently expressed in protoplasts isolated from 10-d-old WT rice seedlings. Fold enrichment of the promoter fragments (lowercase letters a–d with green horizontal lines) was measured by immunoprecipitation with an anti-GFP antibody (see “Materials and methods”). The OsGAPDH gene (Os04g40950) encoding glyceraldehyde 3-phosphate dehydrogenase was used as the negative control. The mean and sd values were obtained from three biological replicates (C and F). The mean and sd values were obtained from more than five independent colonies (D and E). Asterisks above bars (C–F) indicate a statistically significant difference compared to the negative controls, as determined by Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001). These experiments were repeated twice with similar results.
To determine whether OsWRKY5 might directly bind to the promoter regions of OsMYB2, we conducted dual-luciferase (LUC), yeast one-hybrid (Y1H), and chromatin immunoprecipitation (ChIP) assays. First, we performed LUC assays to determine whether OsWRKY5 represses OsMYB2 transcription. To this end, the OsMYB2 promoter (pOsMYB2) was fused with the LUC reporter gene (Figure 6B). LUC activity in rice protoplasts harboring the pOsMYB2:LUC plasmid was significantly decreased when it was co-transfected with the Ubi:OsWRKY5-GFP, compared to co-transfection with Ubi:GFP (Figure 6C). We further tested binding of OsWRKY5 by Y1H assays, revealing that OsWRKY5 directly binds to the OsMYB2-B promoter region, which includes the W-boxes, but does not bind to the OsMYB2 TGAC core sequence (Figure 6D).
To examine whether OsWRKY5 specifically binds to the W-box of OsMYB2, we performed further Y1H assays using a mutated W-box (pOsMYB2-Wm) in which nucleotides were substituted from pyrimidine to purine (Figure 6A). While OsWRKY5 exactly binds to the WT W-boxes (pOsMYB2-W), it did not recognize pOsMYB2-Wm (Figure 6E). ChIP assays further confirmed that OsWRKY5 binds to the amplicon-c region of the OsMYB2 promoter in planta, but it did not bind to other regions (Figure 6F). These results suggest that OsWRKY5 functions as a direct negative regulator of OsMYB2 transcription through binding to the repeated W-boxes of the OsMYB2 promoter region.
Genome editing of OsWRKY5 improves grain yield under drought stress
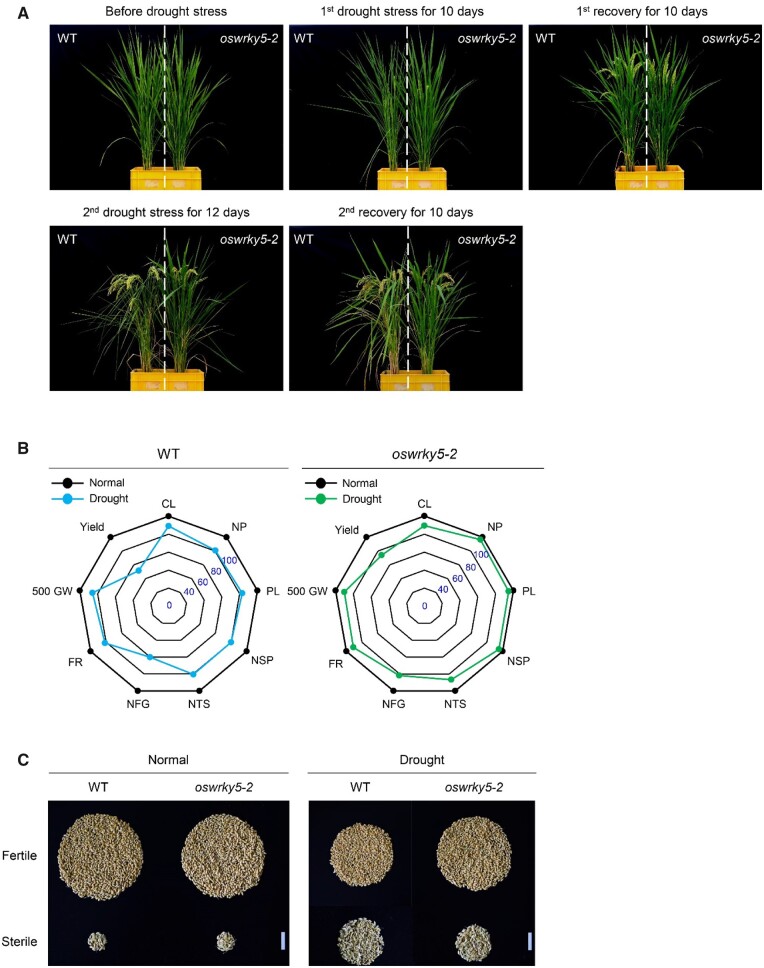
To determine whether the loss of OsWRKY5 function might affect grain yield under water-deficient conditions, we transplanted WT and oswrky5-2 plants after growth for 100 DAS in a paddy field under NLD conditions (≥14-h light/day in Suwon, South Korea, 37°N latitude) to the same plastic boxes filled with paddy soil. We then evaluated agronomic traits after two sequential drought stress exposures and observed a drought-tolerant phenotype in oswrky5-2 plants compared to the WT (Figure 7A). However, yield components such as number of panicles per plant, 500-grain weight and filling rate were not significantly different between oswrky5-2 and WT plants grown under normal growth conditions (Table 1).
Figure 7.
Genome editing of OsWRKY5 improves grain yield under drought stress conditions. A, Phenotypes of oswrky5-2 mutants under drought stress at the heading stage. WT and oswrky5-2 plants grown in paddy soil for 100 ds after sowing were treated with a first drought stress by withholding water for 10 d, followed by a first recovery for 10 d, and then a second drought stress for 12 d. Agronomic traits of rice plants were measured after a second recovery for 10 d. B, Spider plots of yield components of the WT and oswrky5-2 under normal and drought conditions. Each point represents the mean value (n = 8) for each trait listed in Table 1. Each value is presented as a percentage relative to the value for the same genotype under control conditions. For example, relative yield=100× (yield under drought stress treatment)/(yield under normal growth conditions). CL, culm length; NP, number of panicles per plant; PL, panicle length; NSP, number of spikelets per panicle; NTS, number of total spikelets; NFG, number of filled grains; FR, filling rate; 500 GW, 500-grain weight. C, Photographs of the relative abundances of fertile and sterile spikelets in a single WT or oswrky5-2 plants grown under normal and drought conditions, respectively. Scale bars = 3 cm.
Table 1.
Agronomic traits of oswrky5-2 mutants under normal and drought conditions
| Treatment | Genotype and P-value | Culm length (cm) | No. of panicles (/plant) | Panicle length (cm) | No. of spikelets (/panicle) | No. of total spikelets (/plant) | No. of filled grains | Filling rate (%) | 500-grain weight (g) | Yield (g/plant) |
|---|---|---|---|---|---|---|---|---|---|---|
| Normal | WT | 104.83 | 13.13 | 21.70 | 122.12 | 1,625.83 | 1,516.66 | 92.66 | 13.38 | 40.59 |
| oswrky5-2 | 100.83** | 14.00 | 21.62 | 115.10 | 1,601.16 | 1,440.50 | 91.15 | 13.42 | 38.66 | |
| P-value | 0.002 | 0.380 | 0.800 | 0.240 | 0.700 | 0.200 | 0.140 | 0.780 | 0.190 | |
| Drought | WT | 93.21 | 10.60 | 18.00 | 97.77 | 1311.58 | 911.95 | 75.49 | 11.47 | 20.92 |
| oswrky5-2 | 90.16* | 13.50** | 20.48 | 109.88** | 1,387.33 | 1,179.50** | 83.02** | 12.10* | 28.54** | |
| P-value | 0.011 | 0.005 | 0.060 | 0.008 | 0.090 | 0.006 | 0.002 | 0.020 | 0.002 |
Each value represents the mean (n = 8) for the WT and oswrky5-2 (*P < 0.05 and ** P < 0.01, Student’s t test).
In contrast, under drought stress, the yield components of oswrky5-2 were significantly higher than those of the WT (Figure 7B and Table 1). In contrast to oswrky5-2, OsWRKY5 overexpression in the oswrky5-D background increased drought susceptibility and reduced total grain yield compared to the WT (Supplemental Figure S6 and Supplemental Table S1). Thus, genome editing of OsWRKY5 provides a breeding goal for enhancing drought tolerance not only at the vegetative growth stage but also to reduce the formation of sterile spikelets at the ripening stage under drought stress (Figure 7C).
Discussion
Land plants have evolved complex regulatory mechanisms to respond to abiotic stresses. These mechanisms involve transcriptional activators and repressors that modulate the expression of their downstream genes typically associated with tolerance. For example, the expression of transcriptional activators is generally upregulated by abiotic stress inducers (mannitol, NaCl, or dehydration) and stress-induced phytohormones (ABA; OsERF101, Jin et al., 2018; OsMYB2, Yang et al., 2012; OsNAC2, Shen et al., 2017; NAC-LIKE, ACTIVATED BY AP3/P1 [OsNAP], Chen et al., 2014). Finally, stress-induced TFs activate the molecular pathways that confer tolerance to relevant abiotic stresses. The expression of regulatory TFs may also be repressed in response to a given stress: for example, transcript levels of the cotton (Gossypium hirsutum) WRKY TF GhWRKY68 are reduced in response to PEG treatment. The overexpression of GhWRKY68 in transgenic Nicotiana benthamiana repressed the expression of ROS-scavenging and ABA-responsive genes under drought and salt stresses, leading to increased susceptibility to salt stress (Jia et al., 2015). Similarly, transgenic Arabidopsis plants heterologously overexpressing GhWRKY33 also exhibited reduced tolerance of drought stress (Wang et al., 2019a). WRKY TFs in land plants are thought to be good candidates for factors that may confer tolerance to distinct biotic and abiotic stresses. However, thus far, there have been few considerations of the potential uses of rice WRKY TFs for improving plant tolerance to abiotic stresses.
In this study, we established that targeted genome editing of OsWRKY5 leads to improved tolerance to drought stress. Furthermore, similar to the expression pattern of the cotton gene GhWRKY68 under abiotic stress conditions, OsWRKY5 expression significantly decreased upon dehydration and mannitol treatments. Based on these findings, we speculated that OsWRKY5 might specifically function as a transcriptional repressor of drought tolerance under normal growth conditions, likely by repressing drought stress-activated pathways. Through RNA-seq analysis and subsequent RT-qPCR analysis, we found that the OsWRKY5 gain-of-function allele oswrky5-D led to decreased expression of OsMYB2 and OsRNS4, which positively regulate abiotic stress tolerance. LUC and binding assays confirmed the possible role of OsWRKY5 in directly repressing OsMYB2-regulated pathways.
Drought stress raises ABA levels, resulting in the upregulation of a number of TFs that then activate drought tolerance mechanisms (Shinozaki and Yamaguchi-Shinozaki, 2007; Joshi et al., 2016). In contrast to the general trend for the upregulation of TF gene expression in response to ABA, we showed here that OsWRKY5 expression was significantly downregulated by ABA treatment. OsWRKY5 repressed the expression of OsMYB2 by directly binding to the W-box sequences within its promoter region. Accordingly, a loss-of-function mutation in OsWRKY5 led to the upregulation of genes downstream of OsMYB2, such as OsLEA3, OsRAB16A, and OsDREB2A, which are responsible for conferring drought tolerance and participate in ABA signaling. Thus, ABA attenuates the repressor activity of OsWRKY5 in relation to its downstream regulators, thereby enhancing tolerance to drought stress.
Controlling stomatal closure is a crucial step in preventing water loss under water-deficient conditions. Therefore, plant leaves have developed various regulatory mechanisms to modulate stomatal movement; these mechanisms are governed by phytohormones, environmental signals, and the activity of ion channels (Daszkowska-Golec and Szarejko, 2013). In rice, drought-responsive genes regulate stomatal aperture by differentially participating in ABA signaling pathways. Transgenic rice plants overexpressing SNAC1, OsSDIR1, hrf1, OsCPK9, or OsASR5 are more sensitive to ABA treatment (compared with the WT), leading to ABA-dependent stomatal closure (You et al., 2013; Gao et al., 2011; Zhang et al., 2011a; Wei et al., 2014; Li et al., 2017). In this study, ABA treatment significantly inhibited the growth of oswrky5-2 shoots compared to the WT (Figure 4, A and B), indicating that the loss of OsWRKY5 function increases plant sensitivity to ABA. Indeed, oswrky5-2 mutants showed stomatal closure in response to ABA treatment (Figure 4C). These observations demonstrate that OsWRKY5 participates in ABA-dependent stomatal movement.
OsWRKY5 might also be involved in the salt stress regulatory pathway that is mediated by the S-like ribonuclease OsRNS4; OsRNS4 expression increases in response to salt or ABA treatment and its overexpression results in enhanced tolerance to salt stress (Zheng et al., 2014). Considering that OsWRKY5 transcript levels significantly decreased in response to salt treatment, OsWRKY5 might participate in the regulatory pathways related not only to drought stress, but also to high salinity stress. Although we did not pursue the OsWRKY5–OsRNS4 interaction in this study, it will be worth investigating the molecular functions of OsWRKY5 in response to salt stress in future studies.
Some WRKY TFs are involved in coping with both abiotic and biotic stresses (Phukan et al., 2016). For example, OsWRKY11 integrates plant responses to pathogens and abiotic stresses by positively modulating the expression of biotic and abiotic stress-related genes (Lee et al., 2018). The heterologous expression of GhWRKY25 in N. benthamiana reduced plant tolerance to drought stress but enhanced tolerance to salt stress (Liu et al., 2016). Transgenic N. benthamiana plants overexpressing GhWRKY27a were more susceptible to drought stress and Rhizoctonia solani infection compared with the WT (Yan et al., 2015). In this study, we identified biotic stress-related genes whose expression was upregulated in oswrky5-2 and downregulated in oswrky5-D in drought stress (Supplemental Dat Set S3). In rice, the role of biotic stress-related genes has been widely studied. For example, A rice XYLANASE-INHIBITING PROTEIN-type (XIP) gene, OsHI-XIP, enhances resistance to herbivores (Xin et al., 2014). In addition, rice PLANT AND FUNGI ATYPICAL-DUAL SPECIFICITY PHOSPHATASE 2 (OsPFA-DSP2) negatively regulates the pathogen response in transgenic rice plants (He et al., 2012). Rice RECEPTOR-LIKE CYTOPLASMIC KINASE 5 (OsRLCK5) interacts with GLUTAREDOXIN 20 (OsGRX20), to regulate levels of ROS, which serve as secondary messengers in signal transduction, leading to the activation of pathogenesis-related (PR) genes and an immune response (Wang et al., 2021). Taken together, our findings suggest the potential role of OsWRKY5 not only in drought stress but also in biotic stress resistance.
Drought stress, particularly at the reproductive stage, severely limits rice yield (Panda et al., 2021). The loss-of-function in OsWRKY5 minimized drought stress damage though ABA-mediated stomatal closure; it might have allowed more spikelets to develop and flower normally, thereby increasing grain yield (Figure 7B and Table 1). In addition to the yield components, both oswrky5-2 and oswrky5-D plants were shorter in height under normal and drought conditions compared to the WT (Table 1; Supplemental Table 1). These phenomena could be due to changes in the expression of growth-related genes classified into GO terms including nitrogen compound metabolic process (GO:0006807), carbohydrate metabolic process (GO:0005975), and phosphorus metabolic process (GO:0006793; Supplemental Figure S4). Among these genes, rice HOMEODOMAIN-LEUCINE ZIPPER TRANSCRIPTION FACTOR 12 (OsHOX12) belongs to the nitrogen compound metabolic process (GO:0006807). The overexpression of OsHOX12 in rice reduced panicle length and caused a dwarf phenotype (Shao et al., 2018). Protein phosphatases regulate protein phosphorylation, which affects critical roles in plant growth and biotic stress responses (Singh et al., 2010). OsPFA-DSP2, which encodes a protein phosphatase, was included in the GO term phosphorus metabolic process (GO:0006793). The balance of phosphorus (P) and nitrogen (N) affects plant growth rate and functional traits, as well as resource allocation to plant biomass (Luo et al., 2016). Thus, it will be worth investigating the molecular functions of OsWRKY5 in P- and N-related signaling to affect plant height in the future. Another potential explanation for the reduction in height of oswrky5 mutants is that the expression of OsWRKY5 in the rice organs relevant to plant height (including internodes and stem) might have caused the reduction in the plant height under normal and drought conditions (Figure 1B). Finally, the use of constitutive promoters to express TFs often causes unnecessary effects leading to unfavorable growth abnormalities (Jeong et al., 2012). Therefore, additional study will be required to untangle the effects of OsWRKY5 on biotic stress resistance, plant architecture, and other aspects of plant growth and development.
In conclusion, OsWRKY5 has an important biological function in drought tolerance, acting as a transcriptional repressor that downregulates many genes related to ABA-dependent stomatal movement and abiotic stress-related genes (Figure 8; Supplemental Figure S5). OsWRKY5 downregulates the expression of drought tolerance genes by directly binding to the OsMYB2 promoter region. In addition, genome editing of OsWRKY5 improves drought tolerance and limits the reduction in total grain yield that occurs under low-water conditions. Therefore, this study suggests that genome editing of OsWRKY5 and/or manipulation of its downstream targets by CRISPR/Cas9 might provide insight into the function of WRKY genes and be a useful strategy for crop biotechnology.
Figure 8.
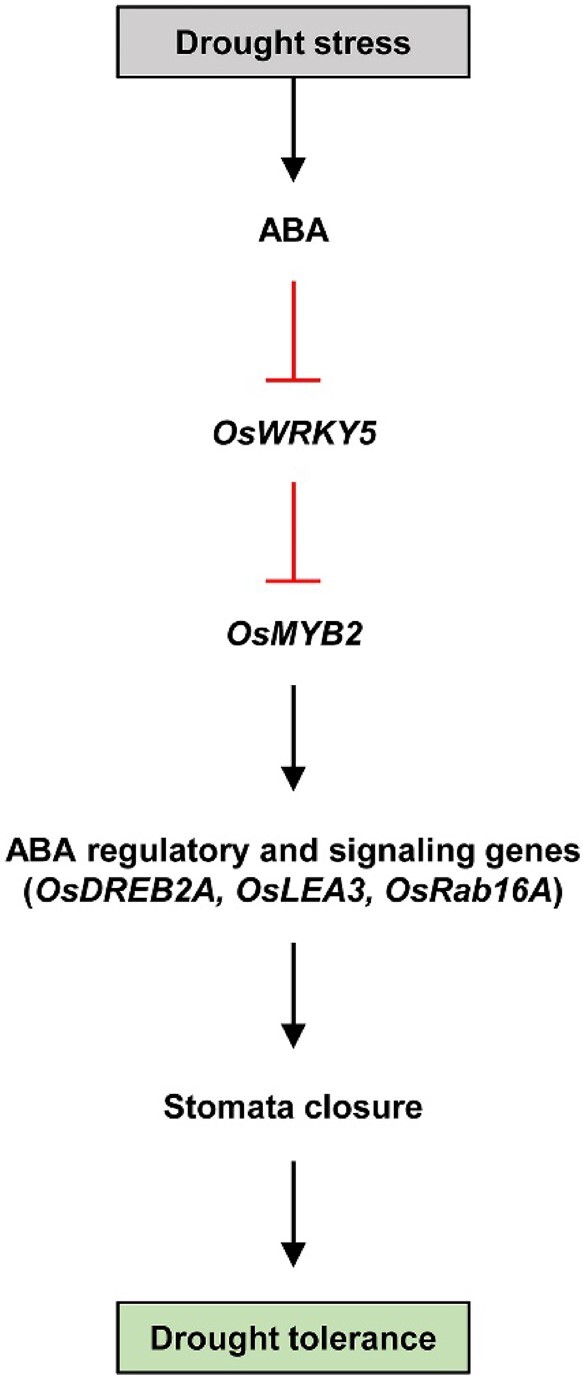
Proposed model of the negative roles of OsWRKY5 in drought tolerance. Solid lines represent direct regulation of downstream genes, respectively. Arrows indicate upregulation, and lines ending with bars represent downregulation. Red lines represent our findings in this study.
Materials and methods
Plant materials and growth conditions
The japonica rice (O. sativa) cultivar Dongjin (WT, parental line) and the activation-tagged oswrky5-D T-DNA insertion line (PFG_3A-15928) were obtained from Crop Biotech Institute at Kyung Hee University, Korea (Jeong et al., 2002; Kim et al., 2019). We generated the genome-edited rice mutants (oswrky5-2 and oswrky5-3) using the CRISPR/Cas9 system. For drought stress treatment, rice plants were grown in pots filled with paddy soil for 3 weeks under long-day (LD) conditions (16-h light at 30°C/8-h dark at 25°C) in the glasshouse. Drought stress was imposed by withholding water for 5 d, followed by 8 d of rewatering. To analyze the expression pattern of OsWRKY5 in response to abiotic stress or phytohormones, 10-d-old WT seedlings grown on half-strength MS phytoagar medium under LD conditions were transferred to half-strength MS liquid medium supplemented with 1-mM ACC, 100-μM ABA, 100-mM NaCl, or 100-mM mannitol, and were dehydrated by air-drying.
For post-germination assays, WT, oswrky5-2, and oswrky5-D seeds were first germinated on half-strength MS phytoagar medium for 3 d and then transferred to half-strength MS phytoagar medium containing 0-, 2-, or 5-µM ABA for 10 d under LD conditions. Seedlings grown on half-strength MS plates with 0-µM ABA were used as the mock control. The shoot length was measured after 10 d of ABA treatment.
Measurements of agronomic traits
To measure the agronomic traits of the WT, oswrky5-2, and oswrky5-D, the plants that were grown for 100-d after sowing in a paddy field under NLD conditions (>14-h sunlight/day in Suwon, South Korea, 37°N latitude) were transplanted to the same plastic boxes filled with the paddy soil. Drought stress was imposed when inflorescence meristems started to appear. When the WT plants showed visual symptoms of drought stress such as leaf rolling, they were irrigated and the same drought treatment was repeated. After two sequential drought stress exposures, the plants were grown with irrigation until harvesting. The control plants were irrigated normally without stress. The yield components of eight plants per each line were measured.
Plasmid constructs and rice transformation for genome editing of OsWRKY5
For CRISPR/Cas9-mediated editing of OsWRKY5, we introduced a custom-designed sgRNA (5′-CGACGACGACGTCCTCCGTG-3′) targeting the OsWRKY5 coding region into the pOs-sgRNA vector, followed by Gateway recombination to mobilize the sgRNA into the destination vector pH-Ubi-Cas9-7 (Miao et al., 2013). The resulting construct was introduced into Agrobacterium (Agrobacterium tumefaciens) strain LBA4404 using the freeze–thaw method (Weigel and Glazebrook, 2006). Agrobacterium-mediated transformation of embryogenic calli generated from the japonica rice cultivar Dongjin was performed as previously described (Jeon et al., 2000). To determine whether genome editing of OsWRKY5 was successful, genomic DNA extracted from regenerated transgenic lines was amplified by PCR with the primers listed in Supplemental Table S2, followed by sequencing of the PCR products (Macrogen Inc., Seoul, Korea).
Measurement of water loss, ion leakage rates, and MDA contents
To measure water loss, fully expanded second leaves were detached from the WT, oswrky5-2, and oswrky5-D seedlings and were placed on Whatman filter paper (Grade 1) on a laboratory bench (at 23 ± 1°C and 30%–40% humidity). The fresh weight of detached leaves was measured every 20 min for 100 min. Water loss was expressed as a percentage of initial fresh weight. Ion leakage was determined by the concentration of electrolytes (or ions) leaking from normal or drought stressed rice leaf discs (1 cm2). Three leaf discs for each treatment were immersed in 6 mL of 0.4-M mannitol at room temperature with gentle shaking for 3 h. The initial conductivity of the solution was measured with a conductivity meter (CON 6 METER, LaMotte Co., USA). Total conductivity was determined after incubation at 85°C for 20 min. The ion leakage rate was expressed as the percentage of initial conductivity, divided by total conductivity. The MDA assay was conducted according to the method proposed by (Chao and Hsueh, 2019). To measure MDA, 100 mg of normal or drought-stressed rice leaves was homogenized in 0.1% (w/v) trichloroacetic acid (TCA) followed by centrifugation at 15,000g for 10 min at 4°C. Four volumes of 0.5% (w/v) thiobarbituric acid in 20% (w/v) TCA were added to one volume of supernatant; the mixture was incubated at 95°C for 25 min. The reaction was terminated by incubating the mixture on ice for 15 min, after which the absorbance was measured spectrophotometrically at 532 nm and 600 nm. The amount of MDA was calculated using an extinction coefficient of 155 mM−1 cm−1 as described previously (Wang et al., 2019b).
Subcellular localization of OsWRKY5
To determine the localization of OsWRKY5, the full-length OsWRKY5 cDNA was cloned into the pGA3651 vector containing the superfolder green fluorescent protein (sGFP) gene. The plasmids were transformed into rice protoplasts isolated from rice suspension cultured cells (Toriyama and Hinata, 1985) using PEG-mediated protoplast transformation (Yoo et al., 2007). Protoplasts were then transferred into multiwell plates and cultured in the dark at room temperature for 6–16 h. After incubation, green fluorescence signals from transfected protoplasts were observed using a Carl Zeiss Axioskop 2 confocal microscope and the image acquisition software ZEN blue edition (Carl Zeiss, Oberkochen, Germany). A sGFP fluorescence was detected with 488 nm excitation and 505–530 nm emission wavelengths.
Y1H assays
The OsWRKY5 coding sequence was subcloned into the EcoRI and BamHI sites of the pGADT7 vector (Clontech) as prey. Fragments of the promoter regions of OsMYB2 (pOsMYB2-A, pOsMYB2-B, pOsMYB2-W, and pOsMYB2-Wm) were amplified by genomic PCR and cloned into the pLacZi vector (Clontech) as baits. The resulting plasmids or empty vectors were transformed into yeast strain YM4271 using the PEG/LiAc method, and yeast cells were incubated on SD/–His/–Leu liquid medium. β-Galactosidase activity was determined based on the absorbance of chloramphenicol red, a hydrolysis product of chlorophenol red-β-d-galactopyranoside, at 595 nm using a ultra violet-visible spectroscopy (UV/VIS) spectrophotometer (BioTek Instruments, USA) according to the Yeast Protocol Handbook (Clontech). Primers used in this study are listed in Supplemental Table S2.
Yeast two-hybrid assays
Yeast two-hybrid (Y2H) assays were performed using the Matchmaker Y2H system (Clontech). The OsWRKY5 coding sequence was amplified from Dongjin first-strand cDNA by PCR. Full-length OsWRKY5 was then cloned into the EcoRI and BamHI sites of the Y2H vectors pGBKT7 and pGADT7 (Clontech). The pair of constructs were co-transformed into yeast strain AH109; positive colonies were spotted onto selective medium (SD–Leu–Trp or SD–Leu–Trp–His–Ade) to test for homodimer formation. Plates were incubated at 30°C for 3 d before taking photographs. Primers used in this study in Supplemental Table S2.
Physiological measurements of guard cells
To measure the status of stomatal closure upon ABA treatment or water deficit, we used fully expanded second leaves of 3-week-old rice plants. Stomatal apertures were examined after leaves were treated with 100-μM ABA for 1 h or exposed to drought stress for 5 d. The samples were first fixed in 2.5% glutaraldehyde in 0.1-M phosphate buffer, pH 7.2, then rinsed three times in distilled water, dehydrated gradually in an ethanol series (30%, 50%, 70%, 80%, and 95%), and rinsed three times in 100% ethanol. The samples were finally dried using a critical point dryer (EM CPD300, Leica, Wetzlar, Germany). Stomatal aperture pictures were obtained using a Zeiss SUPRA 55VP scanning electron microscope (Carl Zeiss, Oberkochen, Germany). To measure stomatal conductance, a portable porometer (AP4, Delta-T Devices, Burwell, England) was used in auto mode every 30 s for 5 min after drought stress treatment for 24 h. Stomatal density (number of stomata per unit area) was counted from three random areas on each leaf under a light microscope (Olympus BX-50, Tokyo, Japan). Stomatal imprints were made by coating the abaxial surface with instant glue (Kusumi, 2013). Infrared thermal images of dehydrated leaves were taken with an infrared thermal camera (ThermaCAM T420, FLIR, USA) at the indicated time points.
RT-qPCR
Total RNA was extracted from plant tissue using the MG Total RNA Extraction Kit (MGmed, Seoul, Korea), according to the manufacturer’s instructions. First-strand cDNA was synthesized with 2 μg of total RNA in a 25-μL volume using M-MLV reverse transcriptase, 5× reaction buffer, 10-mM dNTPs, and 101-mM oligo(dT)15 primer (Promega corp., Wisconsin, USA). The synthesized first-strand cDNAs were then diluted with 75-μL water. Each 20-μL qPCR amplification consisted of 2-μL diluted first-strand cDNA mixture, 10-μL 2X GoTaq PCR Mix (Roche), 7-μL nuclease-free water, and 1 μL of 10-pM primers. RT-qPCR amplification was performed with the gene-specific primers listed in Supplemental Table S2 using a LightCycler 2.0 instrument (Roche Diagnostics) with the following program: 95°C for 2 min, followed by 50 cycles at 95°C for 5 s, 59°C for 15 s, and 72°C for 10 s. Relative transcript levels were normalized to OsUbq5 (Os01g22490) according to the ΔΔCT method (Livak and Schmittgen, 2001).
Transcriptome deep sequencing (RNA-Seq) and GO analysis
The 3-week-old WT, oswrky5-D, and oswrky5-2 plants grown under LD conditions were treated with drought stress by withholding water for 5 d (Figure 2C). Total RNA was extracted from the wilted second leaves of 3-week-old WT, oswrky5-D, and oswrky5-2 plants with the MG total RNA Extraction kit (MGmed, Seoul, Korea) according to the manufacturer’s instructions. The cDNA libraries were prepared using the TruSeq Stranded mRNA Library Prep Kit (Illumina) and sequenced (Macrogen Inc., Seoul, Korea) using an Illumina NovaSeq 6000 platform (Illumina). Raw sequence reads were trimmed to remove low-quality bases (Q < 20), short sequence reads (length <20), and adapter sequence using Trimmomatic v0.38 (Bolger et al., 2014). The trimmed reads were then mapped onto the reference rice genome from RAP-DB (https://rapdb.dna.affrc.go.jp). Transcript abundance was calculated as Fragments Per Kilobase of transcript per Million mapped reads values. The GO analysis was performed using the agriGO classification system (http://bioinfo.cau.edu.cn/agriGO; Du et al., 2010; Tian et al., 2017).
ChIP
For ChIP assays, the Ubipro:GFP and Ubipro:OsWRKY5-GFP constructs were transfected into rice protoplasts as previously described (Zhang et al., 2011b). Protoplasts were then subjected to crosslinking with 1% formaldehyde in W5 solution (154-mM NaCl, 125-mM CaCl2, 5-mM KCl, and 2-mM MES [pH 5.8]) for 30 min under vacuum. Nuclei were isolated and lysed, and chromatin complexes were purified and sonicated, as described previously (Saleh et al., 2008). DNA was sonicated using a VCX500 ultrasonic processor (Sonics, Connecticut, USA). Anti-GFP polyclonal antibody, ChIP grade (ab290; Abcam, Cambridge, UK) and protein A agarose beads (Merck Millipore) were used for immunoprecipitation. DNA recovered from agarose beads was purified using the DNeasy Plant Mini Kit (Qiagen, Weinbunt, Germany). qPCR analysis was conducted on a LightCycler 2.0 instrument (Roche Diagnostics, Basel, Switzerland) using the following conditions: 95°C for 5 s, 59°C for 15 s and 72°C for 10 s. OsGAPDH (Os04g40950) was used as a negative control. Gene-specific primers are listed in Supplemental Table S2.
Luciferase assays in protoplasts
To construct the plasmids containing the LUC reporter gene under the control of various promoters, a fragment of the OsMYB2 promoter region (−1,209 to −1 bp) was cloned into the pJD301 vector, which contains the LUC reporter gene at the C-terminus. For the effector plasmids, the cDNA of OsWRKY5 was cloned upstream of a sequence encoding one copy of a GFP epitope tag in the pGA3651 vector (Kim et al., 2009). The reporter (2 μg), effector plasmids (4 μg), and internal control (1 μg) plasmids were co-transfected into 5 × 104 rice protoplasts by PEG-mediated transfection (Yoo et al., 2007). Transfected protoplasts were then suspended in protoplast culture medium (0.4-mM mannitol, 4-mM MES buffer, and 15-mM MgCl2, pH 5.8) and kept in darkness for 12 h. The LUC activity in each cell lysate was determined using the LUC reporter assay system kit (PR-E1910, Promega, USA).
Statistical analysis
For Student’s t test, asterisks indicate the P-values (*<0.05; **<0.01; ***<0.001). For analysis of variance (ANOVA) test, different letters indicate significant differences at P < 0.05 based on Duncan’s least significant range test.
Accession numbers
The sequence data of this article can be found in the Rice Genome Annotation Project Database and Resource (http://rice.plantbiology.msu.edu) under the following accession numbers: OsWRKY5 (Os05g04640); OsUbq5 (Os01g22490); OsGAPDH (Os04g40950); OsMYB2 (Os03g20090); OsRAB16A (Os11g26790); OsLEA3 (Os05g46480); OsDREB2A (Os01g07120); OsRNS4 (Os09g36680).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Subcellular localization and homodimerization of OsWRKY5.
Supplemental Figure S2. The oswrky5-2 mutant exhibits tolerance to mannitol-induced osmotic stress.
Supplemental Figure S3. Infrared thermal images of the WT, oswrky5-2, and oswrky5-D leaves.
Supplemental Figure S4. AgriGO analysis for biological process and molecular function of DEGs from intersection of genes (n = 80) related to Figure 5.
Supplemental Figure S5. Altered expression of drought-inducible genes in the WT, oswrky5-2, oswrky5-3, and oswrky5-D in response to drought stress.
Supplemental Figure S6. Overexpression of OsWRKY5 reduces grain yield under normal and drought conditions.
Supplemental Table S1. Agronomic traits of oswrky5-D plants under normal and drought conditions.
Supplemental Table S2. Primer sequences used in this study.
Supplemental Data Set S1. List of the up- or downregulated genes in oswrky5-2/WT in drought conditions.
Supplemental Data Set S2. List of the up- or downregulated genes in oswrky5-D/WT in drought conditions.
Supplemental Data Set S3. List of the intersection of the genes (n = 66) upregulated in oswrky5-2/WT and downregulated in oswrky5-D/WT under drought conditions.
Supplemental Data Set S4. List of the intersection of the genes (n = 27) downregulated in oswrky5-2/WT and upregulated in oswrky5-D/WT under drought conditions.
Supplemental Data Set S5. AgriGO analysis of intersection of genes (n = 80) related to oswrky5-2/WT and oswrky5-D/WT related to Figure 5.
Supplementary Material
Acknowledgments
We thank Dr. Jong-Seong Jeon for providing pOs-sgRNA and pH-Ubi-Cas9-7 vectors. The contents of this study are solely the responsibility of the authors.
Funding
This work was supported by the National Research Foundation (NRF) of Korea funded by the Ministry of Education (NRF-2017R1A2B3003310 to N.-C.P.) and the New Breeding Technologies Development Program (PJ01492704 to K.K.), Rural Development Administration, Republic of Korea.
Conflict of interest statement. None declared.
Contributor Information
Chaemyeong Lim, Department of Agriculture, Forestry and Bioresources, Plant Genomics and Breeding Institute, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul 08826, Republic of Korea.
Kiyoon Kang, Division of Life Sciences, Incheon National University, Incheon 22012, Republic of Korea.
Yejin Shim, Department of Agriculture, Forestry and Bioresources, Plant Genomics and Breeding Institute, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul 08826, Republic of Korea.
Soo-Cheul Yoo, Department of Plant Life and Environmental Science, Hankyong National University, Anseong 17579, Republic of Korea.
Nam-Chon Paek, Department of Agriculture, Forestry and Bioresources, Plant Genomics and Breeding Institute, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul 08826, Republic of Korea.
K.K. and N.-C.P. conceived the project and designed the experiments; C.L. and Y.S. carried out all of the experiments; C.L., K.K. and S.-C.Y. analyzed the data; C.L., K.K., and N.-C.P. wrote the article; and all authors approved the article for publication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Nam-Chon Paek (ncpaek@snu.ac.kr).
References
- Agurla S, Gahir S, Munemasa S, Murata Y, Raghavendra AS (2018) Mechanism of stomatal closure in plants exposed to drought and cold stress. In Iwaya-Inoue M, Sakurai M, Uemura M, eds, Survival Strategies in Extreme Cold and Desiccation. Advances in Experimental Medicine and Biology, Vol 1081. Springer, Singapore, pp 215–232 [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523–3543 [DOI] [PubMed] [Google Scholar]
- Bharath P, Gahir S, Raghavendra AS (2021) Abscisic acid-induced stomatal closure: an important component of plant defense against abiotic and biotic stress. Front Plant Sci 12: 615114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Qiu D, Yuan T, Ding X, Li H, Duan L, Xu C, Li X, Wang S (2008) Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ 31: 86–96 [DOI] [PubMed] [Google Scholar]
- Cai R, Zhao Y, Wang Y, Lin Y, Peng X, Li Q, Chang Y, Jiang H, Xiang Y, Cheng B (2014) Overexpression of a maize WRKY58 gene enhances drought and salt tolerance in transgenic rice. Plant Cell Tiss Organ Cult 119: 565–577 [Google Scholar]
- Chao Y-Y, Hsueh I-E (2019) Insights into physiological mechanisms of salt stress tolerance in Djulis (Chenopodium formosanum Koidz.) sprouts. J Plant Biol 62: 263–273 [Google Scholar]
- Chen J, Nolan T, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y (2017) Arabidopsis WRKY46, WRKY54 and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought response. Plant Cell 29: 1425–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li C, Wang H, Guo Z (2019) WRKY transcription factors: evolution, binding, and action. Phytopathol Res 1: 13 [Google Scholar]
- Chen X, Wang Y, Lv B, Li J, Luo L, Lu S, Zhang X, Ma H, Ming F (2014) The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol 55: 604–619 [DOI] [PubMed] [Google Scholar]
- Daszkowska-Golec A, Szarejko I (2013) Open or close the gate – stomata action under the control of phytohormones in drought stress conditions. Front Plant Sci 4: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ (2015) Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J 84: 56–69 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Funnell APW, Crossley M (2012) Homo- and heterodimerization in transcriptional regulation. In Matthews JM, ed, Protein Dimerization and Oligomerization in Biology. Springer, New York, pp 105–121 [DOI] [PubMed] [Google Scholar]
- Gao T, Wu Y, Zhang Y, Liu L, Ning Y, Wang D, Tong H, Chen S, Chu C, Xie Q (2011) OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Mol Biol 76: 145–156 [DOI] [PubMed] [Google Scholar]
- He H, Su J, Shu S, Zhang Y, Ao Y, Liu B, Feng D, Wang J, Wang H (2012) Two Homologous Putative Protein Tyrosine Phosphatases, OsP FA-DSP2 and AtP FA-DSP4, Negatively Regulate the Pathogen Response in Transgenic Plants. PLOS ONE 7: e34995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G-H, Xu J-Y, Wang Y-X, Liu J-M, Li P-S, Chen M, Ma Y-Z, Xu Z-S (2016) Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol 16: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J-S, Lee S, Jung K-H, Jun S-H, Jeong D-H, Lee J, Kim C, Jang S, Lee S, Yang K, et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22: 561–570 [DOI] [PubMed] [Google Scholar]
- Jeong D-H, An S, Kang H-G, Moon S, Han J-J, Park S, Lee HS, An K, An G (2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Redillas MCFR, Jang G, Jung H, Bang SW, Choi YD, Ha S-H, Reuzeau C, Kim J-K (2012) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J 11: 101–114 [DOI] [PubMed] [Google Scholar]
- Jia H, Wang C, Wang F, Liu S, Li G, Guo X (2015) GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PLoS ONE 10: e0120646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Pan W, Zheng X, Cheng X, Liu M, Ma H, Ge X (2018) OsERF101, an ERF family transcription factor, regulates drought stress response in reproductive tissues. Plant Mol Biol 98: 51–65 [DOI] [PubMed] [Google Scholar]
- Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL (2016) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 7: 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Kang K, Kim S-H, An G, Paek N-C (2019) OsWRKY5 promotes rice leaf senescence via senescence-associated NAC and abscisic acid biosynthesis pathway. IJMS 20: 4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-R, Lee D-Y, Yang J-I, Moon S, An G (2009) Cloning vectors for rice. J Plant Biol 52: 73–78 [Google Scholar]
- Kusumi K (2013) Measuring stomatal density in rice. Bio-protoc 3: e753 [Google Scholar]
- Lee H, Cha J, Choi C, Choi N, Ji H-S, Park SR, Lee S, Hwang D-J (2018) Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Y, Yin Z, Jiang J, Zhang M, Guo X, Ye Z, Zhao Y, Xiong H, Zhang Z, et al. (2017) OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol J 15: 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, et al. (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111: 10013–10018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Kang K, Shim Y, Sakuraba Y, An G, Paek N-C (2020) Rice ETHYLENE RESPONSE FACTOR 101 promotes leaf senescence through jasmonic acid-mediated regulation of OsNAP and OsMYC2. Front Plant Sci 11: 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Song Y, Xing F, Wang N, Wen F, Zhu C (2016) GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 253: 1265–1281 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Luo X, Mazer SJ, Guo H, Zhang N, Weiner J, Hu S (2016) Nitrogen:phosphorous supply ratio and allometry in five alpine plant species. Ecol Evol 6: 8881–8892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan LP, Chen X, Clarke J, Salmeron J, Nguyen HT (2012) RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J Exp Bot 63: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Takano S, Sato M, Furukawa K, Nagasawa H, Yoshikawa S, Kasuga J, Tokuji Y, Yazaki K, Nakazono M, et al. (2016) Rice stomatal closure requires guard cell plasma membrane ATP-binding cassette transporter RCN1/OsABCG5. Mol Plant 9: 417–427 [DOI] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu L-J (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23: 1233–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D, Mishra SS, Behera PK (2021) Drought tolerance in rice: focus on recent mechanisms and approaches. Rice Sci 28: 119–132 [Google Scholar]
- Pandey V, Shukla A (2015) Acclimation and tolerance strategies of rice under drought stress. Rice Sci 22: 147–161 [Google Scholar]
- Phukan UJ, Jeena GS, Shukla RK (2016) WRKY transcription factors: molecular regulation and stress responses in plants. Front Plant Sci 7: 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raineri J, Wang S, Peleg Z, Blumwald E, Chan RL (2015) The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol Biol 88: 401–413 [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ (2007) The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol 49: 827–842 [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Shaik R, Ramakrishna W (2014) Machine learning approaches distinguish multiple stress conditions using stress-responsive genes and identify candidate genes for broad resistance in rice. Plant Physiol 164: 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Haider I, Xiong L, Zhu X, Hussain RMF, Övernäs E, Meijer AH, Zhang G, Wang M, Bouwmeester HJ, et al. (2018) Functional analysis of the HD-Zip transcription factor genes Oshox12 and Oshox14 in rice. PLoS ONE 13: e0199248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Lv B, Luo L, He J, Mao C, Xi D, Ming F (2017) The NAC-type transcription factor OsNAC2 regulates ABA-dependent genes and abiotic stress tolerance in rice. Sci Rep 7: 40641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W-Y, Du Y-T, Ma J, Min D-H, Jin L-G, Chen J, Chen M, Zhou Y-B, Ma Y-Z, Xu Z-S, et al. (2018) The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. IJMS 19: 4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK (2010) Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45: W122–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama K, Hinata K (1985) Cell suspension and protoplast culture in rice. Plant Sci 41: 179–183 [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu J-K (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Wang A, Shu X, Jing X, Jiao C, Chen L, Zhang J, Ma L, Jiang Y, Yamamoto N, Li S, et al. (2021) Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome‐wide association study. Plant Biotechnol J 19: 1553–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N-N, Xu S-W, Sun Y-L, Liu D, Zhou L, Li Y, Li X-B (2019a) The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci Rep 9: 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu H, Yu F, Hu B, Jia Y, Sha H, Zhao H (2019b) Differential activity of the antioxidant defence system and alterations in the accumulation of osmolyte and reactive oxygen species under drought stress and recovery in rice (Oryza sativa L.) tillering. Sci Rep 9: 8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S, Hu W, Deng X, Zhang Y, Liu X, Zhao X, Luo Q, Jin Z, Li Y, Zhou S, et al. (2014) A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol 14: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2006) Transformation of Agrobacterium using the freeze-thaw method. Cold Spring Harbor Protocols 2006: pdb.prot4666. [DOI] [PubMed] [Google Scholar]
- Wu T, Zhang M, Zhang H, Huang K, Chen M, Chen C, Yang X, Li Z, Chen H, Ma Z, et al. (2019) Identification and characterization of EDT1 conferring drought tolerance in rice. J Plant Biol 62: 39–47 [Google Scholar]
- Xin Z, Wang Q, Yu Z, Hu L, Li J, Xiang C, Wang B, Lou Y (2014) Overexpression of a Xylanase inhibitor gene, OsHI-XIP, enhances resistance in rice to herbivores. Plant Mol Biol Rep 32: 465–475 [Google Scholar]
- Yan Y, Jia H, Wang F, Wang C, Liu S, Guo X (2015) Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to Rhizoctonia solani infection in transgenic Nicotiana benthamiana. Front Physiol 6: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Dai X, Zhang W-H (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63: 2541–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- You J, Zong W, Li X, Ning J, Hu H, Li X, Xiao J, Xiong L (2013) The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J Exp Bot 64: 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xiao S, Li W, Feng W, Li J, Wu Z, Gao X, Liu F, Shao M (2011a) Overexpression of a Harpin-encoding gene hrf1 in rice enhances drought tolerance. J Exp Bot 62: 4229–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011b) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Wang Y, He Y, Zhou J, Li Y, Liu Q, Xie X (2014) Overexpression of an S-like ribonuclease gene, OsRNS4, confers enhanced tolerance to high salinity and hyposensitivity to phytochrome-mediated light signals in rice. Plant Sci 214: 99–1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.