Abstract
Wild tomatoes (Solanum peruvianum) are important genomic resources for tomato research and breeding. Development of a foreign DNA-free clustered regularly interspaced short palindromic repeat (CRISPR)-Cas delivery system has potential to mitigate public concern about genetically modified organisms. Here, we established a DNA-free CRISPR-Cas9 genome editing system based on an optimized protoplast regeneration protocol of S. peruvianum, an important resource for tomato introgression breeding. We generated mutants for genes involved in small interfering RNAs biogenesis, RNA-DEPENDENT RNA POLYMERASE 6 (SpRDR6), and SUPPRESSOR OF GENE SILENCING 3 (SpSGS3); pathogen-related peptide precursors, PATHOGENESIS-RELATED PROTEIN-1 (SpPR-1) and PROSYSTEMIN (SpProSys); and fungal resistance (MILDEW RESISTANT LOCUS O, SpMlo1) using diploid or tetraploid protoplasts derived from in vitro-grown shoots. The ploidy level of these regenerants was not affected by PEG-Ca2+-mediated transfection, CRISPR reagents, or the target genes. By karyotyping and whole genome sequencing analysis, we confirmed that CRISPR-Cas9 editing did not introduce chromosomal changes or unintended genome editing sites. All mutated genes in both diploid and tetraploid regenerants were heritable in the next generation. spsgs3 null T0 regenerants and sprdr6 null T1 progeny had wiry, sterile phenotypes in both diploid and tetraploid lines. The sterility of the spsgs3 null mutant was partially rescued, and fruits were obtained by grafting to wild-type (WT) stock and pollination with WT pollen. The resulting seeds contained the mutated alleles. Tomato yellow leaf curl virus proliferated at higher levels in spsgs3 and sprdr6 mutants than in the WT. Therefore, this protoplast regeneration technique should greatly facilitate tomato polyploidization and enable the use of CRISPR-Cas for S. peruvianum domestication and tomato breeding.
A DNA-free CRISPR-Cas9 genome editing tool based on an optimized protoplast regeneration protocol of wild tomato creates stable and inheritable diploid and tetraploid regenerants.
Introduction
Tomato (Solanum lycopersicum) is an important vegetable crop, representing the sixth most economically important crop worldwide (http://www.fao.org/faostat/en/#data/QV). Wild tomato species are resistant to diverse biotic and abiotic stresses, and introgression lines are often used for tomato breeding (Schouten et al., 2019). Solanum peruvianum is considered the most variable tomato wild relative and represents an important gene pool for introducing early blight, leaf mold, fusarium wilt, Septoria leaf spot, root knot nematodes, bacterial canker, bud necrosis disease, and viruses resistance genes into modern tomato cultivars (Kaushal et al., 2020). The hallmarks of introgressions of resistance genes could be identified in the modern tomato genomes by functional characterization and whole-genome sequencing (WGS) analysis (Verlaan et al., 2013; Lin et al., 2014). Solanum peruvianum is a self-incompatible species and the existence of unilateral incompatibility has hitherto prevented its use as a female parent in the breeding (Hogenboom, 1972). De novo domestication of wild tomato was recently achieved within a short period by gene editing using clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein (Cas) (Li et al., 2018; Zsogon et al., 2018). Thus, CRISPR-Cas mutagenesis of wild tomato represents a strategy for tomato breeding and basic research.
Endopolyploidy, the occurrence of different ploidy levels within an organism, is widespread among plant taxa. It is often generated by endoreduplication where the complete genome is replicated without mitosis (Scholes and Paige, 2015). In tomato, cells with different ploidy are found in all organs (Smulders et al., 1994). Developmental stages such as young/old organs and growth conditions affect the ratio of ploidy level in cells. Furthermore, genome multiplication is a frequent occurrence during crop domestication. Many of the most economically important crops are polyploid, including potato (Solanum tuberosum), wheat (Triticum aestivum), and cotton (Gossypium hirsutum). Polyploidy conveys advantages in terms of genomic buffering, viability, and environmental robustness (Van de Peer et al., 2021). Triploids can also be used as seedless crops, such as watermelon (Citrullus lanatus) and bananas (Musa acuminata). Thus, CRISPR-Cas-edited tetraploid versions of crop species and their relatives represent important materials for crop breeding in the face of rapid climate change caused by global warming, among other challenges, as was recently demonstrated for tetraploid wild rice (Oryza alta) (Yu et al., 2021). Therefore, it is important to establish a gene editing platform for polyploid crops and related species.
The CRISPR-Cas system uses Agrobacterium tumefaciens-mediated stable transformation to deliver DNA encoding Cas protein and single guide RNA (sgRNA) into the nuclei of tomato cells. As an alternative approach, CRISPR ribonucleoprotein (RNP) or plasmids harboring the Cas and sgRNA sequences can be introduced directly into protoplasts using transient transfection, allowing recombinant DNA-free plants to be regenerated to circumvent concerns about genetically modified organisms (GMOs) (Woo et al., 2015; Andersson et al., 2018; Lin et al., 2018; Hsu et al., 2019, 2021a, 2021b; De Bruyn et al., 2020; Yu et al., 2021). This protocol is important for use with hybrids or plants with a long juvenile period and for vegetative propagation because the transgenes from stable transformation (selection markers and CRISPR reagent genes) cannot be removed from these crops by crossing. Also, the progeny will be different from their heterozygous parental lines due to segregation. The gene editing efficiency and specificity could be validated by targeted sequencing (Woo et al., 2015; Nekrasov et al., 2017) or WGS (Fossi et al., 2019; Hsu et al., 2021b). Nevertheless, previous analysis paid little attention to the overall chromosomal changes, especially in polyploid regenerants (Fossi et al., 2019). WGS and copy number analysis would help to elucidate the potential genome instability after different steps of cell manipulations.
The protoplast regeneration gene editing system has two other major advantages: (1) Gene-edited transformants derived from tissue-culture-based Agrobacterium-mediated transformation are often chimeric, especially in eudicots (Shimatani et al., 2017). If the transformant is an edited/wild-type (WT) chimera and the edited allele occurs only in somatic cells (and not germ cells), edited alleles cannot be passed on to the next generation (Zheng et al., 2020). In protoplast regeneration, there is a low incidence of chimerism, and all mutated alleles detected in the T0 generation can be transmitted to the next generation (Lin et al., 2018; Hsu et al., 2019, 2021a, 2021b). (2) The protoplast regeneration system can be used to introduce many CRISPR reagents and donor DNAs into plants for targeted insertion at the same time without the limitation of vector size (Hsu et al., 2019, 2021a). In addition, the second transfer step can be performed directly to obtain homozygous alleles in polyploids without self-fertilization which is very useful for hybrid, long juvenile period, and sterile plants (Hsu et al., 2019). However, the main bottleneck of this strategy is the difficulty of performing protoplast regeneration.
Here, we established a diploid/allotetraploid protoplast regeneration protocol for S. peruvianum, an important stress-resistant wild tomato, for use with CRISPR-Cas-mediated genome editing. We targeted several genes for editing, including RNA-DEPENDENT RNA POLYMERASE6 (SpRDR6) and SUPPRESSOR OF GENE SILENCING3 (SpSGS3), two key genes in the plant RNA silencing pathway (Mourrain et al., 2000) that mediate defense against tomato yellow leaf curl virus (TYLCV) (Verlaan et al., 2013); PATHOGENESIS-RELATED PROTEIN-1 (SpPR-1) encoding the cysteine-rich secretory proteins antigen 5 and pathogenesis-related 1 protein (CAP)-derived peptide 1 (CAPE1) precursor (Chen et al., 2014) and PROSYSTEMIN (SpProSys), two pathogen-resistance peptide precursors; and MILDEW RESISTANT LOCUS O (SpMlo1) (Nekrasov et al., 2017). Targeting of these genes which was performed using two types of CRISPR reagents, plasmids and RNPs, yielded diploid and tetraploid transgene-free lines. Stable genome structures of ten plants, including one explant derived from stem cutting, three diploid regenerants and six tetraploid of SpProSys or SpMlo1 RNP transfection regenerants were confirmed by WGS.
Results
Protoplast regeneration in S. peruvianum
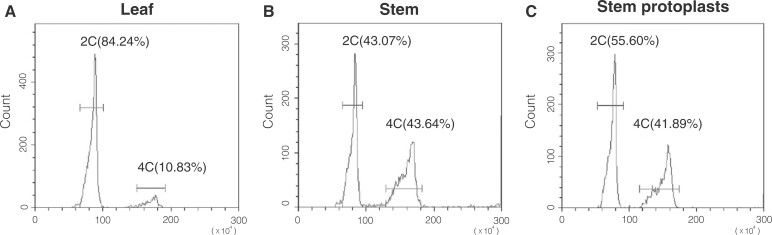
To obtain a high proportion of tetraploid protoplasts, we analyzed the genome sizes of different explants (leaves and stems) using flow cytometry to determine the proportion of tetraploid cells. In leaves, the ratio of diploid to tetraploid nuclei was 5:1 (Figure 1a), and in stems, the ratio was 1:1 (Figure 1b). The same ratio was detected in protoplasts derived from stems (Figure 1c). Therefore, since stems had a higher proportion of tetraploid cells, we used them in subsequent studies to increase the proportion of tetraploid regenerated plants.
Figure 1.
Flow cytometric analysis of the nuclear DNA contents of S. peruvianum tissues. The genome sizes of (a) leaves, (b) stems (c), and protoplasts derived from stems. X: fluorescence density; Y: count. Chicken erythrocyte nuclei (CEN: 2.5 Gb) were used as the calibration standard. The bar indicates the area used for counting nuclei. 2C: diploid; 4C: tetraploid. The number in brackets after the ploidy is the percentage of each different ploidy level versus the total counts.
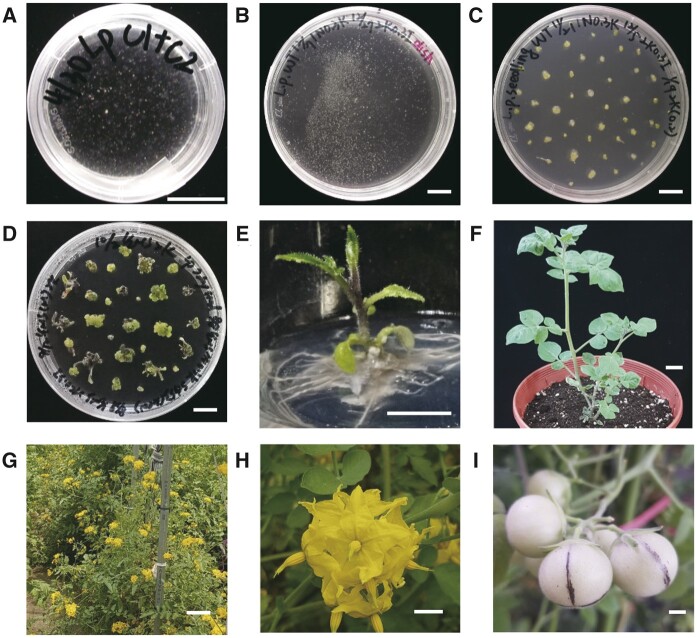
Using a method previously published for Nicotiana benthamiana (Lin et al., 2018), we successfully isolated S. peruvianum protoplasts from in vitro-grown shoots. We incubated the purified protoplasts in liquid medium consisting of half-strength Murashige and Skoog medium (1/2 MS), 0.4 M mannitol, 3% sucrose, 1 mg/L naphthaleneacetic acid (NAA), and 0.3 mg/L kinetin, pH 5.7, for 1 month in the dark, leading to the formation of fine, sand-like calli (Figure 2a). Next, we subcultured these calli in liquid medium containing 1/2 MS, 0.4 M mannitol, 3% sucrose, 2 mg/L kinetin, and 0.3 mg/L indole-3-acetic acid (IAA), pH 5.7, in the light (Figure 2b). After 1 month, these white calli turned green and were transferred to solid medium (1/2 MS, 0.2 M mannitol, 3% sucrose, and 2 mg/L kinetin; Figure 2c). We transferred the calli to fresh medium every month to induce the formation of small shoots (Figure 2d), which were incubated in medium without plant growth regulators until adventitious roots formed at the bottoms of the shoots (Figure 2e). Finally, we transferred the rooted plants to pots (Figure 2f) and grew them in the greenhouse (Figure 2g). The regenerated plants flowered (Figure 2h), fruited (Figure 2i), and produced seeds.
Figure 2.
Regeneration of S. peruvianum protoplasts. a, Protoplasts incubated in 1/2 MS medium supplemented with 3% sucrose, 0.4 M mannitol, 1 mg/L NAA, and 0.3 mg/L kinetin, pH 5.7 liquid medium for 1 month. b, Calli subcultured in 1/2 MS medium supplemented with 3% sucrose, 0.4 M mannitol, 2 mg/L kinetin, and 0.3 mg/L IAA, pH 5.7 liquid medium in the light. c, Calli subcultured in 1/2 MS medium supplemented with 3% sucrose, 0.4 M mannitol, 2 mg/L kinetin, pH 5.7, solid medium. d, Shoot bud formation after two subcultures in 2 mg/L kinetin solid medium. e, Adventitious root formation in plant growth regulator-free 1/2 MS solid medium supplemented with 3% sucrose. f, Regenerated plants after 1 month of growth in a pot. g, Regenerated plants grown in the field. h, Flowers of a regenerated plant. i, Fruits of a regenerated plant. Throughout, bars = 1 cm.
Optimized protoplast regeneration protocol
Compared with N. benthamiana (Lin et al., 2018), S. peruvianum protoplasts take longer to regenerate. According to our observations, the most important steps in the tomato regeneration process are those in liquid culture: callus induction in the dark (the first step) and callus proliferation in the light (the second step). Therefore, we tested several modifications to the composition of the culture medium to shorten the regeneration time. The results indicated that zeatin and 6-benzylaminopurine are the best hormonal treatments for the two liquid culture steps (Supplemental Figure S1), and zeatin is the best cytokinin for the third subculture step in solid medium (Supplemental Figure S2).
CRISPR-Cas9-targeted mutagenesis in S. peruvianum
We used this protoplast regeneration system to establish a method for CRISPR-Cas9-targeted gene mutagenesis of S. peruvianum. First, we used plasmids as CRISPR-Cas9 reagents for targeting mutagenesis of three important disease-resistance-related genes: SpSGS3, SpRDR6, and SpPR-1.
In the SpSGS3 experiment, we chose four target sites (Table 1), and the total efficiency of mutagenesis was 8.3%. Based on sequencing results, mutations occurred in all three target sites except GTAACAATGCTGGATCAGGC. Among these, GCGCAATTGAATGGTTTACA was targeted the most effectively, and mutations at this position were observed in all mutants (Supplemental Table S1). spsgs3#6 (2n), #11 (2n), and #13 (2n) are the null mutants and spsgs3#6 contains four mutated alleles. SpSGS3#7 (4n) also contains three mutated alleles and one non-mutated WT allele. A 68-bp insertion from the vector was detected in spsgs3#11 (2n).
Table 1.
CRISPR-Cas9 target sites and mutagenesis efficiencies
| Reagent | Target gene | Target site | Mutation (%) | |
|---|---|---|---|---|
| Plasmid | SpSGS3 | ATTCCCCCCAGGATAAAAGCGGG | GCGCAATTGAATGGTTTACAGGG | 8.3 (6/72) |
| GTTCCTCCTGCTCTGAAGAATGG | GTAACAATGCTGGATCAGGCCGG | |||
| SpRDR6 | TTAAAGCTGGGACCATTGCGAGG | TGCGAGGTCGAATTGAAACACGG | 13.2 (5/38) | |
| SpPR-1 | CCAGGAGAGAATCTTGCCAAGGG | CTCCGCCACCCACAATTCAGAGG | 13.9 (10/72) | |
| GGGCTCGTTGCAACAACGGATGG | TCTTGCAACTATGATCCTGTAGG | |||
| ACTATGATCCTGTAGGCAATTGG | GATCCTGTAGGCAATTGGGTCGG | |||
| TGTCCGACCCAATTGCCTACAGG | ||||
| RNP | SpProSys | TCATGGTGAAGTTTCACCTTTGG | GGAGGATCACGCTTTGATGGAGG | 45.8 (11/24) |
| SpMlo1 | GGTGTACCTGTGGTGGAGACTGG | GTACAAAGTTAATCAAGAATAGG | 63.6 (14/22) | |
Note: Underlined letters indicate the protoplast adjacent motif sequence.
In the SpRDR6 experiment, we selected two target sites (Table 1). Based on the sequencing results, both target sites could be mutated by CRISPR-Cas9, with a total mutation efficiency of 13.2%. TTAAAGCTGGGACCATTGCG gave the best results, as all five mutant plants contained mutations at this target site. The mutation TGCGAGGTCGAATTGAAACA was only identified in SpRDR6#38 (Supplemental Table S2). All regenerated mutants were heterozygous, and SpRDR6#38 had two mutated alleles and at least one WT allele.
In the SpPR-1 experiment, seven target sites were selected and used to construct two vectors. These two constructs, harboring sgRNAs targeting seven target sites, were co-transfected into protoplasts (Table 1). Among the 10 regenerated mutants, 4 contained fragment deletions, indicating that at least two cleavages had occurred. Except for TGTCCGATCCAGTTGCCTAC and CTATGATCCTGTAGGCAAC there were no mutations in the target sites; the five other sgRNAs caused mutations at the expected positions. The mature CAPE1 peptide is derived from the C-terminal end of tomato PR-1b. sppr-1#28, #31, and #52 were mutated only in the target sites located in CAPE1, all at ATCCTGTAGGCAACTGGAT, resulting in a 5-bp deletion. All SpPR-1 mutants were null mutants except for SpPR-1#72 (Supplemental Table S3).
In the experiments with N. tabacum (Lin et al., 2018) and SpSGS3 (Supplemental Table S1), the use of plasmid CRISPR reagent may still result in foreign DNA insertions. Therefore, RNP is used as a CRISPR reagent to achieve DNA-free gene editing. Here, we delivered two RNPs that target sites located in SpProSys to protoplasts and regenerated the transfected protoplasts into plants. Upon sequencing of the 24 regenerated SpProSys plants, 11 showed target mutagenesis (45.8%, Table 1). Prosystemin is a precursor of systemin, which is processed by phytaspase (Beloshistov et al., 2018). The target site GGAGGATCACGCTTTGATGG is at the C terminus of SpProSys, which is the position of systemin, and the mutations in lines #5, #16, and #19 occurred only at this site (Supplemental Table S4). Using two published SlMlo1 target sites (Nekrasov et al., 2017), we synthesized RNPs targeted to these sites in vitro and simultaneously delivered them into protoplasts. Of the regenerated calli and plants, 63.6% showed targeted mutagenesis (Table 1 and Supplemental Table S5).
Analysis of the genome sizes, phenotypes, and progeny of diploids and tetraploids
A higher proportion of tetraploid cells was observed in protoplasts derived from diploid stems compared to leaf tissue (Figure 1). In addition, during target gene genotyping, we observed that some mutants contained more than three alleles. For example, SpRDR6#38 contained three alleles (+1 bp, –7 bp, and WT, Supplemental Table S2), and its genome size was 4.40 ± 0.03 pg. Therefore, targeted mutant plants of tetraploids can be obtained using this method. We performed karyotype analysis of these regenerated plants (T0, sterile mutants) or their offspring (T1) to confirm the chromosome numbers (Figure 3). Except for a SpPR-1 tetraploid without targeting regenerant, we obtained diploid and tetraploid regenerated plants with or without targeting mutations derived from plasmid CRISR-Cas9 reagent-transfected protoplasts (Figure 3 and Supplemental Table S6). Similar results were obtained for SpProSys RNP transfection (Supplemental Figure S3 and Supplemental Table S7). The ploidy of the plants that were regenerated from transfected protoplasts is provided in Supplemental Tables S6 and S7. These results indicate that most tetraploid plants were derived from tetraploid protoplasts from the explants rather than by protoplast fusion caused by the presence of PEG-Ca2+ in the transfection medium.
Figure 3.
Karyotypes of S. peruvianum plants regenerated from protoplasts. Gray font: null mutant. Black font: heterozygous or WT. Underline: 4n. Bars = 5 μm.
In regenerated plants derived from SpSGS3 transfection, the tetraploids had a reduced seed set (Supplemental Figure S4a). The seeds of tetraploids were larger than those of diploids; this phenomenon was also observed in tetraploid regenerated plants derived from transfection with other CRISPR reagents (Supplemental Figure S4b). The tetraploid plants grew more slowly than the diploid plants (Supplemental Figure S4c). The leaf edges of tetraploid plants were more rounded than those of the diploid plants (Supplemental Figure S4c).
We subjected the offspring of SpSGS3#7 and #10 (Supplemental Figure S5); SpRDR6#6, #33, and #38 (Supplemental Figure S6); and sppr-1#52 and #61 (Supplemental Figure S7) to target gene sequencing. Except for sppr-1#52, which contained one mutant locus not present in the parent, all other offspring had the same mutated locus as the parent. These results demonstrate that these mutated loci can be transmitted to the next generation in diploids and tetraploids.
Stable genome structures in diploids and tetraploids
To further confirm the stability of genome structure in regenerants, we performed WGS of ten samples, including one diploid plant propagated by stem cutting (SpB), three diploids and six tetraploids derived from SpProSys or SpMlo1 RNP transfection (Supplemental Table S7). Taking into account the different genome sizes between diploid and tetraploid plants, each sample was sequenced to the anticipated 30x genome coverage. That is, 141–171 million pair-end reads were sequenced for diploid plants and at least 252–373 million pair-end reads were sequenced for tetraploid plants (Supplemental Table S7).
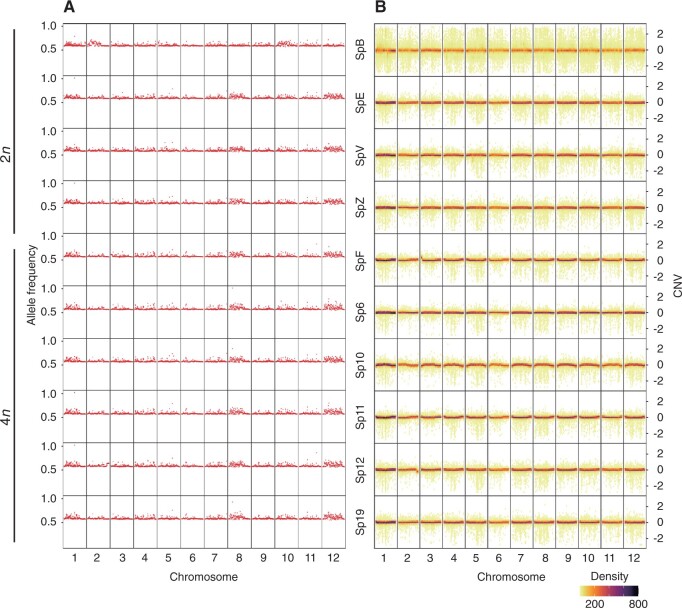
Multiple analysis strategies were used to study the genome structures. Despite the low mapping rate of both diploid and tetraploid samples at some chromosome locations, sequencing coverage analysis did not show inconsistent coverage changes between samples (Supplemental Figure S8). Deletion of large chromosomal segments, which were commonly seen in aneuploid cells (Musacchio and Salmon, 2007) cause allelic imbalance. By calculating heterozygous allele frequency of sequenced plants, we did not identify abnormal allele frequency variations or loss of heterozygosity (Figure 4a). A Bayesian approach to determine copy number variations (CNVs) along chromosomes and compared between uneven sequencing depth of samples did not identify abnormal copy number changes in sequenced plants (Figure 4b). Taking these findings together, we concluded that there is no abnormal chromosomal gain or loss in diploid and tetraploid plants. Neither the protoplast regeneration process nor the CRISPR reagents caused detectible chromosomal changes.
Figure 4.
Stable genome structures in plants regenerated from stem cutting and protoplasts. a, Heterozygous allele frequency of WGS samples. The heterozygous allele frequency was attained by dividing the read depth of the heterozygous allele (labeled as 0/1 by GLnexus) by the total read depth of the variant. Heterozygous frequency is plotted using 10-kb chromosome window size on the X axis. A value of heterozygous allele frequency 0.5 indicates the frequency of the heterozygous genotype (0/1) from the DeepVariant is 0.5, regardless the ploidy level. b, CNV of WGS samples. CNV was predicted as 3 kb fragment size with minimum 10 fragments. Predicted CNV is plotted using 30 bins per chromosome on the X axis. Dot colors indicate the CNV density per bin. A value of zero on the Y axis indicates no copy number change was detected. Values above zero indicate copy number gain and below zero indicate copy number loss.
spsgs3 and sprdr6 diploid and tetraploid null mutants show wiry phenotypes
The regenerated plants containing a WT allele(s) produced flowers and fruits (Figure 2) with morphology and development similar to those of WT plants in the greenhouse. Biallelic spsgs3 mutants (carrying two distinct genome-edited alleles: spsgs3#11, Figure 5; spsgs3-6 and spsgs3-13; Supplemental Figure S9a) had a wiry leaf phenotype and abnormal flowers, which is similar to the previously reported sgs3 domesticated tomato mutants (Yifhar et al., 2012).
Figure 5.
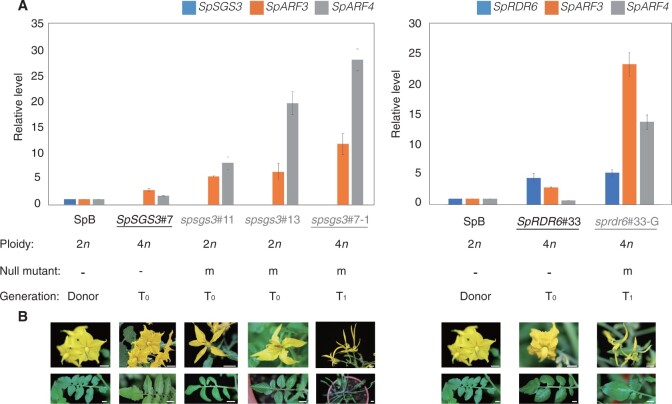
Gene expression and phenotypic profiles of S. peruvianum sgs3 and rdr6 mutants. a, RT-qPCR analysis of auxin response regulator genes (SpSGS3, SpARF3, SpARF4, and SpRDR6) in the WT and protoplast-derived regenerants (n = 3). T0: regenerated plants derived from protoplasts. T1: seedlings derived from T0 plants. Error bars represent the stand deviation. b, Phenotypes of spsgs3 and sprdr6 mutants. Bars = 1 cm.
Among the six progeny of SpSGS3#7, two progeny harbored the mutated alleles only (Supplemental Figure S5); these plants also showed a wiry phenotype (spsgs3#7-2; Supplemental Figure S9b). A similar phenomenon was also observed in the SpRDR6 regenerants. Although all SpRDR6 T0 plants were heterozygous and contained WT SpRDR6 alleles in their genomes, no wiry phenotypes were observed. The SpRDR6#33 and SpRDR6#38 offspring had wiry phenotypes (sprdr6#33-G, Figure 5b; sprdr6#38-16, Supplemental Figure S9b). The pollen of both null T0 and T1 mutated plants, including SpSGS3 and SpRDR6 mutants, was abnormal (Supplemental Figure S9c) and failed to produce seeds.
Because AUXIN RESPONSE FACTOR3 (ARF3) and ARF4 are the target genes of trans-acting secondary siRNA3 (TAS3), whose biogenesis requires RDR6 (Marin et al., 2010), we investigated the transcript levels of these genes in WT, spsgs3 and sprdr6 plants (Figure 5a). The spsgs3 null mutants (T0: spsgs3#11 and spsgs3#13; T1: spsgs3#7-1) lacked SpSGS3 expression. In contrast to the WT, the transcript levels of SpARF3 and SpARF4 were increased in the spsgs mutants, not only for null diploid mutants spsgs3#11 and spsgs3#13 but also for tetraploid mutant spsgs3#7-1 (Figure 5a). Similarly, the transcript levels of SpARF3 and SpARF4 were also increased in the SpRDR6 T1 mutant sprdr6#33-G (Figure 5a).
TYLCV proliferation
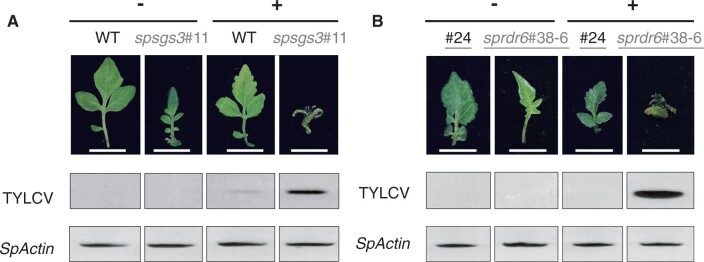
We evaluated the infectivity of TYLCV in the mutants by in vitro inoculation (Al Abdallat et al., 2010). After 8 weeks in vitro inoculation, plant growth was severely retarded (Supplemental Figure S10) and leaf morphology changed in the T1 diploid spsgs3#11 (Figure 6a) and the T2 tetraploid sprdr6#38-6 (Figure 6b). Compared to the WT, all of the null mutants (spsgs3/sprdr6 and diploid/tetraploid) showed higher levels of TYLCV accumulation (Figure 6, a and b, Supplemental Figure S10).
Figure 6.
Symptoms and TYLCV proliferation on in vitro-cultured S. peruvianum plants inoculated with the infectious TYLCV clone. a, Diploid WT and spsgs3#11 mutant. b, Tetraploid regenerant (#24) and sprdr6#38-6 mutant. TYLCV: PCR product of the TYLCV fragment. SpActin: PCR product of SpActin. Gray: null mutant. Black: Un-edited tetraploid regenerated plant (#24) or the WT. Underline: 4n. Bars = 1 cm.
Grafting rescued the fertility of the sgs3#11 null mutant
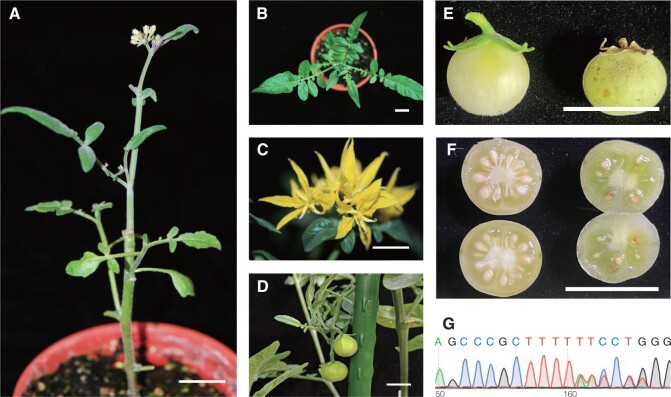
We used WT pollen for hybridization, which failed to pollinate the fruits of the spsgs3 and sprdr6 null mutants. Based on these results, these mutants could not produce the substrate(s) needed for the development of male or female reproductive organs. However, using grafting, the substrate(s) produced in WT stock was successfully transported to the spsgs3#11 scion (Figure 7a). Although there were no substantial differences in leaf (Figure 7b) or flower morphology (Figure 7c), spsgs3#11 failed to produce viable pollen (Supplemental Figure S9c) and the pollen viability of spsgs3#11 increased to 20% by grafting to the WT stock. Grafted spsgs3#11 produced fruits (Figure 7d), but non-grafted spsgs3#11 did not. The fruits from spsgs3#11 scions were smaller (Figure 7e) and contained fewer seeds than the WT (Figure 7f). Genotyping indicated that all of the progeny harbored spsgs3#11 mutated alleles (Figure 7g).
Figure 7.
Growth of a sterile spsgs3 #11 plant grafted with WT stock. a, Grafted plant. Leaves (b), flowers (c), and fruit (d) of spsgs3 #11 scion. Mature fruit (e) and seeds (f) of WT stock (left) and spsgs3 #11 scion (right). g, Results of Sanger sequencing of the seedling derived from spsgs3 #11 scion fruit, which is heterozygous, harboring spsgs3#11 mutated alleles mixed with the WT allele. Bars = 1 cm.
Discussion
It remains challenging to introduce desired alleles from multiple wild species into a single cultivar. In the history of the development of tomato disease resistant cultivars, the root knot nematode resistance gene Mi-1 was introduced from S. peruvianum and the TYLCV resistance gene Ty-1 1 was introduced from Solanum chilense. These two introgressed fragments are located at a nearby genomic location (26.6–27.7 Mb versus 30.9–32.5 Mb) on chromosome 6 and remain intact under severe recombination suppression (Lin et al., 2014). It was difficult to combine both genes into a single cultivar likely due to chromosomal inversions or close to the centromeric location. Development of CRISPR-Cas editing in wild tomatoes would allow the introduction of the desired alleles from wild species without the limitation of recombination frequency or the cost of potential linkage drag.
In addition to its use in wild tomatoes, CRISPR is also utilized in commercial varieties of S. lycopersicum. Dozens of studies using this technique in tomato have been published, most involving breeding trials for traits such as quality (fruit architecture, color, metabolism, and postharvest), anti-stress (biotic and abiotic stress), and domestication (Li et al., 2018; Zsogon et al., 2018). These studies were performed using several CRISPR platforms established in tomato, including (1) Cas9 (Brooks et al., 2014) and Cas12a (Bernabe-Orts et al., 2019), to generate DNA double-strand breaks that are preferentially repaired by non-homologous end joining to introduce target mutations; (2) precise modification of plant genomes using DNA repair templates via homologous recombination (Cermak et al., 2015); and (3) the cytidine base editor, an inactive Cas9 fusion with cytidine deaminase, which converts cytosine to uracil without cutting DNA and introducing mutations (Shimatani et al., 2017). Therefore, CRISPR is emerging as a powerful tool for tomato breeding.
For commercial breeding, however, it is desirable to produce DNA-free plants to avoid concerns about GMOs. Although there are several reports demonstrating successful DNA-free genome editing via biolistic methods in many crops (Svitashev et al., 2016; Liang et al., 2018; Banakar et al., 2020), protoplast regeneration systems have higher efficiency. Many studies have been performed using RNPs and plasmids to achieve DNA-free genome editing (Woo et al., 2015; Andersson et al., 2018; Lin et al., 2018; Hsu et al., 2019, 2021a, 2021b; De Bruyn et al., 2020; Yu et al., 2021). These reports indicate that it is possible to establish protoplast regeneration platforms for tomato and various target crops/plants.
Tomato and related species have been important materials in the development of protoplast isolation and regeneration techniques. Tomato protoplasts were isolated by enzymatic digestion, and this landmark achievement allowed sufficient amounts of protoplasts to be obtained for further application (Cocking, 1960). Solanum peruvianum was the first tomato-related species for which a protoplast regeneration system was reported, and such systems have subsequently been achieved in many tomato and wild tomato species (Kut and Evans, 1982). In this study, we combined these techniques to achieve DNA-free genome editing of a wild tomato. This method could be applied to other tomato-related species to facilitate breeding.
Although protoplast regeneration was first reported 50 years ago (Takebe et al., 1971), it still represents a major bottleneck in DNA-free genome editing. The shoot regeneration capacity in Lycopersicon is highly genotype dependent (Peres et al., 2001). Understanding how a cell is regenerated into a complete plant is an important topic of scientific and agricultural research (Sugimoto et al., 2019), but information about this process is still limited. Such knowledge could be applied to develop efficient tissue culture, gene transformation, and genome editing systems, tools that are important for de novo plant domestication (Li et al., 2018; Zsogon et al., 2018; Maher et al., 2020; Yu et al., 2021). In this study, we assessed the effects of plant growth regulators in the medium on protoplast regeneration. In addition to the chemical approach, several genes encoding morphogenic regulators have been identified and used to improve the efficiency of plant regeneration. It is possible to control the expression of these genes to establish a non-tissue-culture regeneration system for gene editing (Maher et al., 2020).
In addition to their roles in the domestication of wild species, polyploid crops have other benefits, including larger plants (Chung et al., 2017), and higher yields (Chen et al., 2018). In addition, triploid crop cultivars of species such as bananas and watermelons can produce commercially desirable seedless fruits. Most previous methods for chromosome multiplication have used colchicine. This procedure is complicated and inefficient, producing regenerated plants with mixed cell populations of various ploidy levels (Cola et al., 2014). Similar to haploid culture, in this report, using isolated protoplasts from polyploid cells in explants for regeneration and gene editing, we were able to obtain edited polyploid regenerated wild tomatoes without colchicine treatment. This phenomenon has also been reported in other plant species. In witloof chicory plants (Cichorium intybus var. foliosum) generated from CRISPR/Cas-edited protoplasts, 77.2% diploid and 21.5% tetraploid plants were produced and the remaining 1.3% consisted of haploids, hexaploids, and mixoploids (De Bruyn et al., 2020). Therefore, explants containing high proportions of polyploidized cells could be widely used for protoplast regeneration for crop polyploidization. However, in this study, we found no substantial enlargement in the leaves or flowers of tetraploid versus diploid lines, similar to the pattern reported for tetraploid tomatoes (Nilsson, 1950).
In addition to technological difficulties, the presumed mutagenicity of protoplast regeneration is another reason why researchers are reluctant to use this system as a gene editing platform. Indeed, WGS has revealed widespread genome instability in potatoes regenerated from protoplasts (Fossi et al., 2019), which has increased the concerns about this technology. The original purpose of protoplast regeneration was to use protoplast fusion to improve hybridization or as a platform for mutagenesis. Since only successful cases of mutation or fusion have been reported, and most such experiments have not been compared with other tissue culture methods, many researchers have the impression that protoplast regeneration readily leads to mutagenesis. In fact, other tissue culture technologies, including multiple shoot proliferation (Lin et al., 2007) and somatic embryogenesis (Lin et al., 2007), can also cause mutations. Although this study involved the use of PEG-Ca2+ in the transfection process, which could promote cell fusion, non-transfected tetraploid regenerated plants were also obtained. Based on our finding that the proportion of tetraploid regenerated plants was similar to that of shoot explants, we believe that the formation of polyploid regenerated plants was primarily due to the presence of polyploid cells in the explants. In addition to protoplast regeneration, there are also opportunities to obtain polyploid plants using other tissue culture technologies (Chung et al., 2017). In an Agrobacterium-mediated transformation experiment in tomato, the rate of tetraploid transgenic plants ranged from 24.5% to 80% and depended on both the genotype and the transformation procedure (Ellul et al., 2003). In Arabidopsis thaliana T-DNA insertion mutagenesis, large-scale genomic rearrangements have occurred (Pucker et al., 2021). Therefore, we believe that protoplast regeneration is an excellent tool for gene editing as well as other transgenic platforms.
Unlike the previous report of widespread genome changes in the autotetraploid potato (Fossi et al., 2019), the WGS analysis in this study does not identify aneuploidy and abnormal chromosomal changes in either diploid or tetraploid regenerants. Chromosomes in the autotetraploid genome, such as cultivated potato, were derived from the merging of two different chromosome sets (Van de Peer et al., 2021). On the other hand, tetraploid plants in this study, which were derived from chromosome doubling, contained the two identical sets of chromosomes. As the tissue culture steps caused a certain level of cell stresses, pairing of non-homologous chromosomes in the autotetraploid genomes (Fossi et al., 2019) likely has a higher probability of incorrect chromosome pairing than in the allotetraploid genomes (Hsu et al., 2019; Yu et al., 2021). Incorrect chromosome segregation during mitosis in the autotetraploid cells likely has a higher probability of evading the spindle-assembly checkpoint (Musacchio and Salmon, 2007). Furthermore, by analyzing changes in the allele frequency and CNV, we confirmed that the CRISPR-Cas9 editing did not introduce large scale chromosomal changes and unintended genome editing sites (Hsu et al., 2021b).
In this study, all tetraploid and diploid spsgs3 and sprdr6 null mutants had wiry phenotypes, similar to other microRNA biogenesis null mutants in tomatoes (Yifhar et al., 2012; Brooks et al., 2014). sgs3 and rdr6 null mutants show various phenotypes in different species. N. benthamiana spsgs3 and sprdr6 mutants have a wiry flower morphology and sterile phenotype, but their leaves are similar to those of the WT (Hsu et al., 2021a). The Arabidopsis sgs3 mutant shows no substantial phenotype (Adenot et al., 2006). Therefore, we would like to discover ways to improve the fertility of these mutants.
Grafting is a traditional agricultural tool that is used to control flowering, improve fruit quality, and increase resistance to biotic and abiotic stress (Haroldsen et al., 2012). In N. benthamiana, gene silencing was transmitted with 100% efficiency in a unidirectional manner from silenced stocks to non-silenced scions expressing the corresponding transgene (Palauqui et al., 1997). In this study, a mutant of SpSGS3, an RNA silencing-related gene, was used as a scion and grafted onto RNA-silenced normal WT rootstock. The fertility of spsgs3#11 scions was rescued, and they produced seeds with mutated alleles. In Arabidopsis, more than 3,000 mobile genes have been identified. The mRNA from these genes could be transported long distance, including SGS3 mRNA (Thieme et al., 2015). In addition to mRNA, organellar DNA, proteins, and plant growth regulators can also move across graft unions (Haroldsen et al., 2012). Whether these mobile substances were also involved in rescuing the fertility of the spsgs3#11 scions or whether grafting with WT plants could rescue other sterile mutants of mobile RNA requires further investigation.
Conclusions
To obtain tetraploid S. peruvianum DNA-free genome-edited plants, we used in vitro-grown shoots, which contain high proportions of tetraploid cells, as explants for protoplast isolation and regeneration. The medium components were optimized, and genome-edited regenerants were obtained within 6 months. This is the first study in S. peruvianum describing the use of both RNP and plasmid CRISPR reagents for DNA-free genome editing, yielding a targeted mutagenesis efficiency of 60% without the need for marker gene selection. Diploid and tetraploid heritable mutants were obtained for all pathogen-related genes targeted in this study, including SpSGS3, SpRDR6, SpPR-1, SpProSys, and SpMlo1, and the expected phenotypes were obtained. In comparative WGS analysis, protoplast derived CRISPR-Cas9 edited plants, either diploid or tetraploid, showed stable genome structure. The proliferation of TYLCV, an important viral disease of tomato, was increased in spsgs3 and sprdr6 null mutants. The reproductive growth defect of the SpSGS3 mutant was successfully rescued by grafting with WT stock. The protocols and materials described in this study will be useful for tomato breeding.
Materials and methods
Plant materials
Sterile S. peruvianum plantlets (seeds obtained from the National Plant Genetic Resources Center, Taiwan Agricultural Research Institute with US National Plant Germplasm System Accession number: PI 126441, https://npgsweb.ars-grin.gov/gringlobal/accessiondetail?id=1133100) were propagated by cutting and growing them in 0.5 MS medium supplemented with 30 mg/L sucrose and 1% (v/v) agar, pH 5.7. The plantlets were incubated in a 26°C culture room (12 h light/12 h in dark, light intensity of 75 µmol m−2 s−1). The plantlets were cut and subcultured in fresh medium monthly.
Protoplast isolation and transfection
Protoplast isolation and transfection of S. peruvianum were performed following our previously published method with minor modifications (Hsu et al., 2019). Protoplasts were isolated from the stems and petioles of in vitro-grown plantlets. Five or more stems (∼5 cm/each, total 0.2–0.25 g) were used to isolate roughly 1 × 106 protoplasts. These materials were place in a 6-cm glass Petri dish with 10 mL digestion solution (1/4 MS liquid medium containing 1% [v/v] cellulose and 0.5% [v/v] macerozyme, 3% [v/v] sucrose, and 0.4 M mannitol, pH 5.7) and cut into 0.5-cm-wide strips longitudinally. The material was incubated at room temperature in the dark overnight. The digested solution was diluted in 10 mL W5 (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, and 5 mM glucose) solution and filtered through a 40-µm nylon mesh. The sample was centrifuged at low speed (360× g) for 3 min to collect the protoplasts. The protoplasts were purified in 20% (v/v) sucrose solution and washed three times with W5 solution. The protoplasts were transferred to transfection buffer (1/2 MS medium supplemented with 3% sucrose, 0.4 M mannitol, 1 mg/L NAA, and 0.3 mg/L kinetin, pH 5.7) and adjusted to a concentration of 3 × 105 cells/mL.
The protoplasts were transfected with plasmids by PEG-Ca2+-mediated transfection (Woo et al., 2015; Lin et al., 2018). A 400-µL sample (1.2 × 105 protoplasts) was combined with 40 µL of CRISPR reagent (DNA: 20–40 µg; RNP: 10 µg) and mixed carefully. The same volume (440 µL) of PEG-Ca2+ solution was added to the sample, mixed, and incubated for 30 min. To end the reaction, 3 mL of W5 was added and the sample was mixed well. Transfected protoplasts were collected by centrifugation at 360× g for 3 min. The protoplasts were washed with 3 mL of W5 and centrifuged at 360× g for 3 min. The target sites are shown in Table 1.
CRISPR/Cas reagents
The SpCas9 vector for dicot transformation (pYLCRISPR/Cas9P35S-N) (Ma et al., 2015) was isolated using a Plasmid Midi-prep kit (Bio-Genesis). Preparation of Cas9 protein and sgRNA and Cas9 RNP nucleofection were performed according to Huang et al. (2020). Cas9 RNP complexes were assembled immediately before nucleofection by mixing equal volumes of 40 μM Cas9 protein and 88.3 μM sgRNA at a molar ratio of 1:2.2 and incubating at 37°C for 10 min.
Protoplast regeneration
Pooled protoplast DNA was used as a template to amplify the target genes for validation by sequencing. The putatively edited protoplasts were transferred to 5-cm-diameter Petri dishes containing 3 mL 1/2 MS liquid medium supplemented with 3% (v/v) sucrose, 0.4 M mannitol, 1 mg/L NAA, and 0.3 mg/L kinetin for plant regeneration. Calli formed from the protoplasts after 1 month of incubation in the dark. The calli were subcultured in a 9-cm-diameter Petri dish containing fresh medium with cytokinin for 3–4 weeks in the light. Calli that had turned green were transferred to solid medium containing the same plant growth regulators. The explants were subcultured every 4 weeks until shoots formed after several subcultures. The shoots were subcultured in solid rooting medium (HB1: 3 g/L Hyponex No. 1, 2 g/L tryptone, 20 g/L sucrose, 1 g/L activated charcoal, and 10 g/L Agar, pH 5.2) for adventitious roots formation.
Analysis of the genotypes of regenerated plants
Two pairs of primers were designed to amplify the sgRNA-targeted DNA region for each target gene. The PCR conditions were 94°C for 5 min, 35 cycles of denaturing (94°C for 30 s), annealing (55°C for 30 s), and polymerization (72°C for 30 s), followed by an extension reaction at 72°C for 3 min. The PCR product was sequenced by Sanger sequencing to confirm mutagenesis. The multiple sequences derived from mutated regenerated plants were bioinformatically separated using Poly Peak Parser (http://yosttools.genetics.utah.edu/PolyPeakParser/; (Hill et al., 2014)) or further confirmed by sequential T/A cloning and sequencing. The primer sequences are listed in Supplemental Table S8.
Estimation of genome size
Fresh leaves were finely chopped with a new razor blade in 250 µL isolation buffer (200 mM Tris, 4 mM MgCl2-6H2O, and 0.5% Triton X-100) and mixed well (Dolezel et al., 2007). The mixture was filtered through a 40-μm nylon mesh, and the filtered suspensions were incubated with a DNA fluorochrome (50 μg/mL propidium iodide containing RNase A). The samples were analyzed using a MoFlo XDP Cell Sorter (Beckman Coulter Life Sciences) and an Attune NxT Flow Cytometer (Thermo Fisher Scientific). Chicken erythrocyte (BioSure) was used as an internal reference.
WGS
Leaves of S. peruvianum regenerates were harvested and genomic DNA was extracted using two independent protocols. A nuclei isolation protocol (Sikorskaite et al., 2013) was used on the WT (SpB) sample to recover higher quality and quantity of DNA samples. Briefly, nuclei were extracted by 36 mM sodium bisulfite, 0.35 M Sorbitol, 0.1 M Tris-base, 5 mM EDTA, 2 M NaCl, 2% (w/v) CTAB, and 2 mL 5% (v/v) N-lauroylsarcosine sodium salt. The genomic DNA was then extracted by chloroform–isoamyl alcohol (24:1), ethanol precipitation, and further cleaned up by DNeasy Blood & Tissue Kit (69504, Qiagen) and AMPure (Beckman Coulter Life Sciences). The other nine samples used the chloroform–isoamyl alcohol (24:1) for DNA extraction, followed with Zymo Genomic DNA Clean & Concentrator-25 (D4064, Zymo), and Zymo OneStep PCR Inhibitor Removal Kit (D6030, Zymo) to obtain high quality genomic DNA. DNA integrity was checked using the D1000 Screen Tape on the Agilent TapeStation 4150 System with DIN value >8. Genomic DNA were sheared using a Covaris E220 sonicator (Covaris) and paired‐end sequencing libraries were constructed by the NEBNext Ultra DNA Library Prep Kit II for Illumina (E7370S, NEB). DNA libraries were validated again on the Agilent TapeStation 4150, and were quantified by qPCR (E7630, NEB). The 2 × 150 bp paired-end sequencing with average insert size of 700 bp was performed by Welgene Biotech on an Illumina NovaSeq 6000 platform.
WGS data analysis
Since there was no assembled S. peruvianum genome, high quality Illumina reads were mapped to the S. lycopersicum Heinz 1706 reference genome (SL4.0) (Hosmani et al., 2019) by the GPU-based NVIDIA Clara Parabricks package (NVIDIA). To determine the variant frequency, we used the deep learning-based Google DeepVariant (Yun et al., 2021) with “WGS model” to identify variants. All samples were then combined by GLnexus (Yun et al., 2021) to perform “joint genotype calling” using “DeepVariant” model to combine samples. We then calculated the heterozygous allele frequency by dividing the read depth of the heterozygous allele (labeled as 0/1 by GLnexus) over the total read depth of the variant. A large chromosomal region with heterozygous allele frequency lower than 0.5 indicated either a chromosome region with low recombination rate or deletion of the chromosome fragments. To determine CNVs between samples, we used the cn.mops pipeline (Klambauer et al., 2012) to analyze mapped Illumina reads. To minimize the effects of repetitive sequence regions, we set the segment size to 3,000 bp and minimum number of segments as 10 to identify high confidence CNVs.
Reverse transcription quantitative PCR
Expression of four genes was analyzed using reverse transcription quantitative PCR (RT-qPCR). These genes were: SpSGS3, SpARF3, SpARF4, and SpRDR6. RT-qPCR analysis was performed on three biological replicates and three replicates in a CFX Connect instrument (Bio-Red, Hercules, CA, USA). RT-qPCR was carried out in 96-well optical reaction plates using the iQ SYBR Green Supermix (Bio-Rad). The reference gene Actin and gene-specific primers for the RT-qPCR are listed in Supplemental Table S8.
Data availability statement
The Illumina sequencing reads generated for this study have been deposited at NCBI under BioProject PRJNA768623.
Accession numbers
Sequence data from this article can be found in Supplemental Table S9.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Effects of cytokinins on callus induction (first subculture) and callus proliferation (second subculture).
Supplemental Figure S2. Effects of cytokinins on callus in solid medium (third subculture).
Supplemental Figure S3. Flow cytometric analysis of the nuclear DNA contents of tetraploid plants regenerated from SpProSys RNP-transfected protoplasts.
Supplemental Figure S4. Phenotypes of diploid and tetraploid plants regenerated from protoplasts transfected with CRISPR reagents.
Supplemental Figure S5 . Progeny analysis of SpSGS3.
Supplemental Figure S6. Progeny analysis of SpRDR6.
Supplemental Figure S7. Progeny analysis of SpPR-1.
Supplemental Figure S8. Illumina sequencing coverage for the tomato SL4.0 genome assembly.
Supplemental Figure S9. Phenotypes of the spsgs3 and sprdr6 null mutants.
Supplemental Figure S10. Symptoms and TYLCV proliferation on in vitro-cultured S. peruvianum plants inoculated with the infectious TYLCV clone.
Supplemental Table S1. SpSGS3 gene sequences of the SpSGS3 mutants.
Supplemental Table S2. SpRDR6 gene sequences of the SpRDR6 mutants.
Supplemental Table S3. SpPR-1 gene sequences of the SpPR-1 mutants.
Supplemental Table S4. SpProSys gene sequences of the SpProSys mutants.
Supplemental Table S5. SpMlo1 gene sequences of the SpMlo1 mutants.
Supplemental Table S6 . Karyotypes of plants regenerated from protoplasts transfected with CRISPR reagents.
Supplemental Table S7. Overview of Illumina WGS sequencing, mapping rate, and SRA number.
Supplemental Table S8. Primers used in these studies.
Supplemental Table S9 . Locus ID, orthologs in S. lycopersicum and the cDNA sequence used in this study.
Supplementary Material
Acknowledgments
We thank Te-Chang Hsu and the AS-BCST Bioinformatics Core for the computational support and Miranda Loney and Plant Editors for English editing. Experiments and data analysis were performed in part using the confocal microscope at the Division of Instrument Service of Academia Sinica with the assistance of Shu-Chen Shen. We also thank the IPMB Flow Cytometry Analysis and Sorting Service of Academia Sinica for flow cytometry analysis. Finally, we thank Ruei-Shiuan Wang for genomic DNA preparation, Yu-Jung Cheng for tissue culture, Wei-Fong Hung for genotyping, Jheng-Yang Ou for figure preparation, and Song-Bin Chang for karyotyping.
Funding
This research was supported by the Innovative Translational Agricultural Research Program (AS-KPQ-107-ITAR-10; AS-KPQ-108-ITAR-10; AS-KPQ-109-ITAR-10; AS-KPQ-110-ITAR-03) and Academia Sinica Institutional funding to Y.-C.L. and C.-S.L., and the Ministry of Science and Technology (105-2313-B-001-007-MY3; 108-2313-B-001-011-; 109-2313-B-001-011-), Taiwan, to C.-S.L. These funding bodies played no role in the design of the study, collection, analysis, or interpretation of data or in writing the manuscript.
Conflict of interest statement. The authors declare no potential conflict of interest.
Contributor Information
Choun-Sea Lin, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan.
Chen-Tran Hsu, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan.
Yu-Hsuan Yuan, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan.
Po-Xing Zheng, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan; Biotechnology Research Center in Southern Taiwan, Academia Sinica, Tainan 711, Taiwan.
Fu-Hui Wu, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan.
Qiao-Wei Cheng, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan.
Yu-Lin Wu, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan; Biotechnology Research Center in Southern Taiwan, Academia Sinica, Tainan 711, Taiwan.
Ting-Li Wu, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan; Biotechnology Research Center in Southern Taiwan, Academia Sinica, Tainan 711, Taiwan.
Steven Lin, Institute of Biochemistry, Academia Sinica, Taipei 115, Taiwan.
Jin-Jun Yue, Research Institute of Subtropical Forestry, Chinese Academy of Forestry, Hangzhou 311, China.
Ying-Huey Cheng, Plant Pathology Division, Taiwan Agricultural Research Institute, Taichung 413, Taiwan.
Shu-I Lin, Department of Horticulture and Landscape Architecture, National Taiwan University, Taipei 106, Taiwan.
Ming-Che Shih, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan.
Jen Sheen, Department of Molecular Biology and Center for Computational and Integrative Biology, Massachusetts General Hospital, and Department of Genetics, Harvard Medical School, Boston, Massachusetts 02114, USA.
Yao-Cheng Lin, Agricultural Biotechnology Research Center, Academia Sinica, Taipei 115, Taiwan; Biotechnology Research Center in Southern Taiwan, Academia Sinica, Tainan 711, Taiwan.
C.-S.L., Y.-C.L., J.S., and M.-C.S. conceived and designed the experiments. C.-T.H. and Y.-H.Y. performed the CRISPR-Cas9 experiments. C.-T.H., Y.-H.Y., Q.-W.C., J.-J.Y., and F.-H.W. conducted the protoplast regeneration, cell biology, molecular biology, and targeted mutagenesis experiments. S.L. conducted SpCas9 purification. Y.-L.W. performed WGS library preparation and qPCR analysis. P.-X.Z., T.-L.W., and Y.-C.L. performed the bioinformatics analysis. Y.-H.C., C.-T.H., C.-S.L., Q.-W.C., and F.-H.W. performed the virus-related analysis. C.-T.H. performed cell biology. C.-T.H. and S.-I.L. performed the grafting. J.S., M.-C.S., Y.-C.L., and C.-S.L. wrote the manuscript with input from all co-authors. All authors read and approved the final manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is Yao-Cheng Lin (yalin@sinica.edu.tw).
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H (2006) DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol 16: 927–932 [DOI] [PubMed] [Google Scholar]
- Al Abdallat AM, Al Debei HS, Asmar H, Misbeh S, Quraan A, Kvarnheden A (2010) An efficient in vitro-inoculation method for Tomato yellow leaf curl virus. Virol J 7: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Turesson H, Olsson N, Falt AS, Ohlsson P, Gonzalez MN, Samuelsson M, Hofvander P (2018) Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol Plant 164: 378–384 [DOI] [PubMed] [Google Scholar]
- Banakar R, Schubert M, Collingwood M, Vakulskas C, Eggenberger AL, Wang K (2020) Comparison of CRISPR-Cas9/Cas12a ribonucleoprotein complexes for genome editing efficiency in the rice phytoene desaturase (OsPDS) gene. Rice (NY) 13: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloshistov RE, Dreizler K, Galiullina RA, Tuzhikov AI, Serebryakova MV, Reichardt S, Shaw J, Taliansky ME, Pfannstiel J, Chichkova NV, et al. (2018) Phytaspase-mediated precursor processing and maturation of the wound hormone systemin. New Phytol 218: 1167–1178 [DOI] [PubMed] [Google Scholar]
- Bernabe-Orts JM, Casas-Rodrigo I, Minguet EG, Landolfi V, Garcia-Carpintero V, Gianoglio S, Vazquez-Vilar M, Granell A, Orzaez D (2019) Assessment of Cas12a-mediated gene editing efficiency in plants. Plant Biotechnol J 17: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C, Nekrasov V, Lippman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166: 1292–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EG, Tsai KL, Chung HH, Chen JT (2018) Chromosome doubling-enhanced biomass and dihydrotanshinone I production in Salvia miltiorrhiza, a traditional Chinese medicinal plant. Molecules 23: 3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Lee CY, Cheng KT, Chang WH, Huang RN, Nam HG, Chen YR (2014) Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26: 4135–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HH, Shi SK, Huang B, Chen JT (2017) Enhanced agronomic traits and medicinal constituents of autotetraploids in Anoectochilus formosanus hayata, a top-grade medicinal orchid. Molecules 22: 1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocking EC (1960) A method for the isolation of plant protoplasts and vacuoles. Nature 187: 962–963 [Google Scholar]
- Cola GPA, Marques AM, Damasceno S, Carvalho CR, Clarindo WR (2014) In Vitro polyploidization in Solanum lycopersicum Mill. ‘Santa Cruz Kada Gigante’. Cytologia 79: 351–358 [Google Scholar]
- De Bruyn C, Ruttink T, Eeckhaut T, Jacobs T, De Keyser E, Goossens A, Van Laere K (2020) Establishment of CRISPR/Cas9 genome editing in witloof (Cichorium intybus var. foliosum). Fron Genome Ed 2: 604876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2: 2233–2244 [DOI] [PubMed] [Google Scholar]
- Ellul P, Garcia-Sogo B, Pineda B, Rios G, Roig LA, Moreno V (2003) The ploidy level of transgenic plants in Agrobacterium-mediated transformation of tomato cotyledons (Lycopersicon esculentum Mill.) is genotype and procedure dependent. Theor Appl Genet 106: 231–238 [DOI] [PubMed] [Google Scholar]
- Fossi M, Amundson K, Kuppu S, Britt A, Comai L (2019) Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol 180: 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haroldsen VM, Szczerba MW, Aktas H, Lopez-Baltazar J, Odias MJ, Chi-Ham CL, Labavitch JM, Bennett AB, Powell AL (2012) Mobility of transgenic nucleic acids and proteins within grafted rootstocks for agricultural improvement. Front Plant Sci 3: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JT, Demarest BL, Bisgrove BW, Su YC, Smith M, Yost HJ (2014) Poly peak parser: method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev Dyn 243: 1632–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenboom NG (1972) Breaking breeding barriers in Lycopersicon. 4. Breakdown of unilateral incompatibility between L. peruvianum (L.) Mill. and L. esculentum Mill. Euphytica 21: 397–404 [Google Scholar]
- Hosmani PS, Flores-Gonzalez M, van de Geest H, Maumus F, Bakker LV, Schijlen E, van Haarst J, Cordewener J, Sanchez-Perez G, Peters S, et al. (2019) An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv: 767764. https://doi.org/10.1101/767764
- Hsu CT, Cheng YJ, Yuan YH, Hung WF, Cheng QW, Wu FH, Lee LY, Gelvin SB, Lin CS (2019) Application of Cas12a and nCas9-activation-induced cytidine deaminase for genome editing and as a non-sexual strategy to generate homozygous/multiplex edited plants in the allotetraploid genome of tobacco. Plant Mol Biol 101: 355–371 [DOI] [PubMed] [Google Scholar]
- Hsu CT, Lee WC, Cheng YJ, Yuan YH, Wu FH, Lin CS (2021a) Genome editing and protoplast regeneration to study plant–pathogen interactions in the model plant Nicotiana benthamiana. Front Genome Ed 2: 627803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CT, Yuan YH, Lin YC, Lin S, Cheng QW, Wu FH, Sheen J, Shih MC, Lin CS (2021b) Efficient and economical targeted insertion in plant genomes via protoplast regeneration. CRISPR J 4: 752–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Shih HA, Lai MC, Chang YJ, Lin S (2020) Enhanced NK-92 Cytotoxicity by CRISP R Genome Engineering Using Cas9 Ribonucleoproteins. Front Immunol 11: 1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal A, Sadashiva AT, Ravishankar KV, Singh TH, Prasanna HC, Rai AK, Jatav VK (2020) A rapid disease resistance breeding in tomato (Solanum lycopersicum L.). In Gosal SS, Wani SH, eds, Accelerated Plant Breeding, Vegetable Crops, Vol. 2, Springer International Publishing, Cham, Switzerland, pp 17–55 [Google Scholar]
- Klambauer G, Schwarzbauer K, Mayr A, Clevert D-A, Mitterecker A, Bodenhofer U, Hochreiter S (2012) cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res 40: e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kut SA, Evans DA (1982) Plant regeneration from cultured leaf explants of eight wild tomato species and two related Solanum species. In Vitro 18: 593–598 [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C (2018) Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol 36: 1160–1163 [DOI] [PubMed] [Google Scholar]
- Liang Z, Chen K, Zhang Y, Liu J, Yin K, Qiu JL, Gao C (2018) Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat Protoc 13: 413–430 [DOI] [PubMed] [Google Scholar]
- Lin CS, Kalpana K, Chang WC, Lin NS (2007) Improving multiple shoot proliferation in bamboo mosaic virus-free Bambusa oldhamii Munro propagation by liquid culture. HortScience 42: 1243–1246 [Google Scholar]
- Lin CS, Hsu CT, Yang LH, Lee LY, Fu JY, Cheng QW, Wu FH, Hsiao HC, Zhang Y, Zhang R, et al. (2018) Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol J 16: 1295–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Zhu G, Zhang J, Xu X, Yu Q, Zheng Z, Zhang Z, Lun Y, Li S, Wang X, et al. (2014) Genomic analyses provide insights into the history of tomato breeding. Nat Genet 46: 1220–1226 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF (2020) Plant gene editing through de novo induction of meristems. Nat Biotechnol 38: 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S (2017) Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep 7: 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E (1950) Some experiments with tetraploid tomatoes. Hereditas 36: 181–204 [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16: 4738–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres LEP, Morgante PG, Vecchi C, Kraus JE, van Sluys M-A (2001) Shoot regeneration capacity from roots and transgenic hairy roots of tomato cultivars and wild related species. Plant Cell Tissue Organ Cult 65: 37–44 [Google Scholar]
- Pucker B, Kleinbolting N, Weisshaar B (2021) Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis. BMC Genomics 22: 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DR, Paige KN (2015) Plasticity in ploidy: a generalized response to stress. Trends Plant Sci 20: 165–175 [DOI] [PubMed] [Google Scholar]
- Schouten HJ, Tikunov Y, Verkerke W, Finkers R, Bovy A, Bai Y, Visser RGF (2019) Breeding has increased the diversity of cultivated tomato in the Netherlands. Front Plant Sci 10: 1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K, et al. (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35: 441–443 [DOI] [PubMed] [Google Scholar]
- Sikorskaite S, Rajamaki ML, Baniulis D, Stanys V, Valkonen JP (2013) Protocol: optimised methodology for isolation of nuclei from leaves of species in the Solanaceae and Rosaceae families. Plant Methods 9: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulders MJM, Rus-Kortekaas W, Gilissen LJW (1994) Development of polysomaty during differentiation in diploid and tetraploid tomato (Lycopersicon esculentum) plants. Plant Science 97: 53–60 [Google Scholar]
- Sugimoto K, Temman H, Kadokura S, Matsunaga S (2019) To regenerate or not to regenerate: factors that drive plant regeneration. Curr Opin Plant Biol 47: 138–150 [DOI] [PubMed] [Google Scholar]
- Svitashev S, Schwartz C, Lenderts B, Young JK, Mark Cigan A (2016) Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat Commun 7: 13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe I, Labib G, Melchers G (1971) Regeneration of whole plants from isolated mesophyll protoplasts of tobacco. Die Naturwissenschaften 58: 318–320 [Google Scholar]
- Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Minambres M, Walther D, Schulze WX, Paz-Ares J, et al. (2015) Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 1: 15025. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Ashman TL, Soltis PS, Soltis DE (2021) Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell 33: 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan MG, Hutton SF, Ibrahem RM, Kormelink R, Visser RG, Scott JW, Edwards JD, Bai Y (2013) The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet 9: e1003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Yifhar T, Pekker I, Peled D, Friedlander G, Pistunov A, Sabban M, Wachsman G, Alvarez JP, Amsellem Z, Eshed Y (2012) Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24: 3575–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lin T, Meng X, Du H, Zhang J, Liu G, Chen M, Jing Y, Kou L, Li X, et al. (2021) A route to de novo domestication of wild allotetraploid rice. Cell 184: 1156–1170 e1114 [DOI] [PubMed] [Google Scholar]
- Yun T, Li H, Chang P-C, Lin MF, Carroll A, McLean CY (2021) Accurate, scalable cohort variant calls using DeepVariant and GLnexus. Bioinformatics 36: 5582–5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Li T, Dittman JD, Su J, Li R, Gassmann W, Peng D, Whitham SA, Liu S, Yang B (2020) CRISPR/Cas9-based gene editing using egg cell-specific promoters in Arabidopsis and soybean. Front Plant Sci 11: 800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsogon A, Cermak T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol 36: 1211–1216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Illumina sequencing reads generated for this study have been deposited at NCBI under BioProject PRJNA768623.