Abstract
Primary myelofibrosis (PMF) is a neoplasm prone to leukemic transformation, for which limited treatment is available. Among individuals diagnosed with PMF, the most prevalent mutation is the JAK2V617F somatic point mutation that activates the Janus kinase 2 (JAK2) enzyme. Our earlier reports on hyperactivity of β1 integrin and enhanced adhesion activity of the α2β1 complex in JAK2V617F megakaryocytes (MKs) led us to examine the new hypothesis that this mutation leads to posttranslational modification via changes in glycosylation. Samples were derived from immunoprecipitation of MKs obtained from Vav1-hJAK2V617F and WT mice. Immunoprecipitated fractions were separated by SDS-PAGE and analyzed using LC-MS/MS techniques in a bottom-up glycoproteomics workflow. In the immunoprecipitate, glycopeptiforms corresponding to 11 out of the 12 potential N-glycosylation sites of integrin β1 and to all nine potential glycosylation sites of integrin α2 were observed. Glycopeptiforms were compared across WT and JAK2V617F phenotypes for both integrins. The overall trend observed is that JAK2V617F mutation in PMF MKs leads to changes in β1 glycosylation; in most cases, it results in an increase in the integrated area of glycopeptiforms. We also observed that in mutated MKs, changes in integrin α2 glycosylation were more substantial than those observed for integrin β1 glycosylation, a finding that suggests that altered integrin α2 glycosylation may also affect activation. Additionally, the identification of proteins associated to the cytoskeleton that were co-immunoprecipitated with integrins α2 and β1 demonstrated the potential of the methodology employed in this study to provide some insight, at the peptide level, into the consequences of integrin activation in MKs. The extensive and detailed glycosylation patterns we uncovered provide a basis for future functional studies of each site in control cells as compared to JAK2V617F-mutated cells. Data are available via ProteomeXchange with identifier PXD030550.
Keywords: integrins, megakaryocytes, primary myelofibrosis, LC-MS/MS, glycopeptiforms
Abbreviations: BM, bone marrow; co-IP, co-immunoprecipitation; ECM, extracellular matrix; ER, Endoplasmic Reticulum; ETD, electron transfer dissociation; EThcD, electron-transfer/higher-energy collision-induced dissociation; FDR, false discovery rate; HCD, higher-energy collision–induced dissociation; JK2, Janus kinase 2; MPN, myeloproliferative neoplasm; PMF, primary myelofibrosis
Graphical Abstract
Highlights
-
•
β1, α2 Integrins are isolated from bone marrow of WT and JAK2V617F mouse megakaryocytes.
-
•
Glycopeptiforms are characterized at 11 of 12 potential N-linked sites on integrin β1.
-
•
Glycopeptiforms are characterized at all nine potential N-linked sites on integrin α2.
-
•
JAK2V617F megakaryocytes had glycosylation changes in both proteins at multiple sites.
In Brief
Changes in glycosylation were documented at multiple sites in integrins β1 and α2 isolated from bone marrow of WT and myelofibrotic (JAK2V617F) mouse megakaryocytes. Glycopeptiforms at 11 out of the 12 potential N-glycosylation sites of integrin β1 and at all nine potential glycosylation sites on integrin α2 were characterized.
Clonal proliferation of hematopoietic stem cells leads to disorders known as myeloproliferative neoplasms (MPNs), among which primary myelofibrosis (PMF) has the greatest potential for leukemic transformation (1) and a median survival of 4 to 7 years (2). About 60% of PMF patients carry the JAK2V617F somatic point mutation rendering the Janus kinase 2 (JAK2) enzyme constitutively active (1). The occurrence of permanently activated JAK2 on hematopoietic stem cells modifies the bone marrow (BM) niche components and consequently supports clonal expansion (1). Current treatments are few, and each carries one or more disadvantages. These treatments include medication for anemia, elective splenectomy (25% operative morbidity reported by Davies et al. (3) in an 18-years-long study), administration of a JAK2 inhibitor (decreases spleen enlargement and fibrosis, but treatment is effective only in some patients), and allogenic hematopoietic cell transplant (eligibility is limited (1)). A better understanding of the physiopathology of PMF is necessary to improve and ultimately supplement the limited number of treatment options currently available.
Recently, Matsuura et al. (4) examined integrin-mediated adhesion to fibronectin of megakaryocytes (MKs) carrying the JAK2V627F mutation, in order to expand the present understanding of megakaryocytosis in this pathology. Integrins are adhesion receptors that link cells to components of the extracellular matrix (ECM) and are, thereby, responsible for intracellular signal transduction (5, 6). Integrins function as transmembrane heterodimers composed of one α and one β subunit (5, 7). Twenty-four different integrin dimers have been reported; these are assembled from 18 α-subunits and eight β-subunits, among which 12 contain integrin β1 (7, 8). The N-terminal domain of the heterodimer constitutes the ligand-specific receptor site for elements of the ECM (7). For example, integrin α2β1 binds to collagen, and α5β2 binds to fibronectin (7). Upon activation, the short cytoplasmic C-terminal domains of integrin recruit diverse cytoplasmic proteins, resulting in the formation of large protein complexes (5, 8). These complexes are responsible for anchoring cells to the actin cytoskeleton and for hosting signal transduction events, thus promoting cell migration, proliferation, and differentiation (5, 8). Garcia et al. (6) demonstrated that, in muscle cells, the transition between proliferation and differentiation is controlled by the levels of integrin α5β1 bound to fibronectin. Moreover, mechanical forces play a role in integrin activities. The assembly of integrins on the cell surface can be regulated by forces imposed via bulky cell-surface components such as the glycoproteins with highly branched antennae in the cancer glycocalyx (9). Mamidi et al. (10) have demonstrated that integrin-induced mechanosignaling directs cell fate during development of the pancreas. For PMF initiated by the occurrence of JAK2V617F+, Matsuura et al. (4) demonstrated that cell adhesion to fibronectin via α5β1 integrin contributes to the proliferation of MKs in the BM. They found that although there was no significant change in the expression level of the β1 subunit in MKs derived from JAK2V617F+ mice when compared to WT MKs, the activated form of integrin β1 was more prevalent in the JAK2V617F+ phenotype. Further, the adhesion of JAK2V617F+ MKs to collagen though α2β1 was also found to be enhanced as compared to control cells (11). These findings suggest that posttranslational modifications are likely to underlie the increased activity of β1 in JAK2V617F PMF.
Integrins are glycoproteins that bear numerous N-linked glycans as covalent biosynthetic modifications at one or more sites. Significantly, 70% of proteins in eukaryotic cells undergo glycosylation, a cotranslational or posttranslational modification that modulates protein three-dimensional shape, stability, trafficking, and interactions (12, 13). In a cotranslational process that takes place in the endoplasmic reticulum (ER), nascent proteins can be N-glycosylated (at the side chain amide of asparagine residues within the consensus sequence Asn-X-Ser/Thr and sometime Cys, X being any amino acid, except proline) by transfer of an oligosaccharide, usually Glc3Man9GlcNAc2, from its phospholipid-linked precursor, through the action of oligosaccharyltransferase A. An immature already folded (glyco)protein may be posttranslationally N-glycosylated, at the same sequons given above, through the actions of oligosaccharyltransferase-B in the ER (14, 15). In both cases, the initial glycan is remodeled during transit through the ER and Golgi (12, 16), and the glycoprotein is subjected to quality control as it progresses. In addition, it can be O-glycosylated (at the hydroxyl group on serine or threonine residues) in the Golgi. These glycans can also undergo further processing before release from the Golgi. Once at the cell surface, soluble enzymes may initiate further modifications of glycan structures (17, 18). The final glycoform distributions depend on the availability and levels of relevant glycosidases, glycosyl transferases and nucleotide sugars, and the accessibility of the individual glycosylation sites on each protein (12).
Considering the hyperactivity of β1 integrin in JAK2V617F MK, we sought here to examine the new hypothesis that this mutation leads to changes in β1 glycosylation, using targeted glycoproteomics. Some previous studies have addressed overall N-glycosylation of integrin β1, and site occupancy has been linked to its expression level and function (19) and to its heterodimerization, for example, with integrin α5 (19, 20). Schultz et al. have related the degree of sialylation of integrin β1 glycans to metastasis of cancer cells (21). DiBuduo et al. have observed increased sialylation on integrin β1 in MKs derived from MPN patients (22). However, little site-specific glycan structural information has been published and, to our knowledge, disease-related changes in site-specific glycoform patterns have not been investigated. We report here the glycopeptiform profiles for 11 of the 12 potential sites in integrin β1 from control and JAK2V61F MK. Since the methodology we developed also provided information on integrin α2 glycosylation, we profiled that as well. We determined that all nine potential sites on integrin α2 are occupied and compared the glycoform profiles at each glycosylation site to look for differences between control and JAK2V617F MK.
Experimental Procedures
Experimental Design and Statistical Rationale
All samples analyzed were derived from a Vav1-hJAK2V617F mouse colony (JAK2V167F), and age-, sex-, strain-matching controls (WT mice). Three biological replicate experiments, in each case involving the analysis of 3.08 × 105 to 1.80 × 106 MKs obtained after culture of cells from the BM obtained from the hind limbs of three to five mice, age 12 to 16 weeks, were performed at approximately 4-month intervals; no technical replicate was acquired for the mass spectrometric data due to the limited amount of these samples, and the resulting very small pools of integrins isolated from the mouse MKs. For each phenotype, the sample was separated into three aliquots based on migration along an SDS-PAGE gel, and each of the bands was subjected to in-gel proteolytic digestion (reported as upper and lower 100–150 kDa, 150–250 kDa in supplemental Tables S1 and S2) prior to LC-MS/MS analysis, and thus any given sample was characterized by three raw data files. Glycopeptiform identification result files were combined manually (only upper and lower 100–150 kDa were used for integrin α2), Log2-fold change (FC) values were calculated using Excel spreadsheets, and the mean of relative integrated areas for glycopeptiforms was calculated using Prism GraphPad software.
JAK2V617F Transgenic Mice
Vav1-hJAK2V617F (JAK2V617F) mice were gift from Dr Zhizhuang Joe Zhao (University of Oklahoma). This mouse line has the hallmarks of PMF, including expansion of the MK lineage, a fibrotic BM, and splenomegaly (23, 24). Expansion and housing of the mouse colony and of matching controls were carried out at Boston University School of Medicine. Age-, sex-, and genotype-matched mice were used as controls. All studies involving mice were approved by the Boston University Institutional Animal Care and Use Committee. Animal housing conditions and treatment protocols were approved by the Institutional Animal Care and Use Committee of Boston University School of Medicine.
Mouse BM MK Culturing
BM was cultured as previously described (25) with pegylated MK growth and development factor (25 ng/ml, PEG-MGDF). MKs were purified using a bovine serum albumin gradient followed by cell counting, as described by Eliades et al. (25).
Isolation of Integrins β1 and α2 From MKs
A nondenaturing lysis buffer composed of 10 mM Tris-HCl pH 7.4 (ThermoFisher Scientific), 150 mM NaCl, 0.1% to 0.25% TritonX (AmericanBio), 1 mM phenylmethylsulfonyl fluoride (ThermoFisher Scientific), 1 mM ethylenediaminetetraacetic acid (Millipore Sigma), and 200 μl of 10X MS-Safe protease and phosphatase inhibitor (ThermoFisher Scientific) was used to lyse between 3.08 × 105 and 1.80 × 106 MKs. Cell pellets were resuspended in the lysis buffer, passed through a 25G syringe (BD Precisionglide, ThermoFisher Scientific) 20 times, and incubated on ice for 20 min. Debris was pelleted down by centrifugation at 10,000 rpm for 10 min, and the protein concentrations in the lysates were measured using a BCA assay at 562 nm (ThermoFisher Scientific). Cell counts and total amount of protein measured for MK lysates in WT and JAK2V617F samples are reported in supplemental Table S3. Complexes containing integrins α2 and β1 were recovered from lysed cell pellets by co-immunoprecipitation (co-IP) using 1 μg of CD49b (integrin α2) monoclonal antibody (HMa2, eBioscience) for 50 μg of lysate. The co-IP reaction was carried out overnight, and immunoprecipitated protein complexes were purified using Dynabeads protein G (ThermoFisher Scientific). The purified co-IP fraction was recovered from Dynabeads by adding 4X SDS sample loading buffer (Millipore Sigma) and heating the mixture at 90 °C for 5 min, followed by centrifugation at 13,000g for 2 min. Integrin β1 was characterized in 1/10 of the co-IP eluate by Western blot using a 1:2000 solution of mouse anti-CD29 (integrin β1, BD Biosciences) and a 1:4000 solution of anti-mouse IgG HRP-linked (Cell Signaling Technology) as primary and secondary antibody, respectively. The proteins on the membrane were detected by chemiluminescence (Immobilon ECL Western HRP Substrate, Millipore Sigma) and analyzed on the ImageQuant LAS 4000 system (supplemental Fig. S1).
Sample Preparation for Mass Spectrometry
The components in lysates and co-IP fractions were resolved by SDS-PAGE (4–15% Mini-PROTEAN TGX, Bio-Rad). For each phenotype, the co-IP fraction was equally divided and loaded into three separate wells. The aliquots were processed individually, and bands from all three lanes that were judged to be equivalent based on their migration along the gel were combined into one sample prior to mass spectrometric analysis. Gel bands were cut according to the scheme shown in supplemental Fig. S2. The procedure for proteolytic digestion was adapted from the UCSF in-gel digestion protocol (26, 27). Briefly, the gel pieces were minced finely, and the proteins present in the gel pieces were reduced in 10 mM 1,4-dithiothreitol (Millipore Sigma) for 1 h at 56 °C, alkylated in 55 mM iodoacetamide (Millipore Sigma) for 45 min at room temperature in the dark, and digested at 37 °C overnight with 0.5 μg mass spectrometric grade trypsin/Lys-C (Promega). The resulting peptides were extracted from the gel pieces using a solution containing 5% formic acid and 50% acetonitrile (ThermoFisher Scientific) and subsequently dried under vacuum. Co-IP fractions were enriched for glycopeptides using a ZIC-HILIC resin (ProteoExtract Glycopeptide Enrichment Kit, Millipore Sigma) according to the protocol established by Alagesan et al. (28). Recombinant mouse integrin α2β1 protein expressed in CHO cells (R&D Systems) was used as a mass spectrometry standard.
LC-MS/MS Analysis
The first biological replicate was analyzed using a nanoAcquity UPLC system (Waters Corp.) coupled to a TriVersa NanoMate ion source (Advion) on a Q-Exactive HF mass spectrometer (ThermoFisher Scientific). The second and third replicates were analyzed using an Acquity M-Class UPLC (Waters) system coupled to a TriVersa NanoMate ion source mounted on a Fusion Lumos Tribrid Orbitrap mass spectrometer (ThermoFisher). Glycopeptide-enriched fractions were reconstituted in 5 μl buffer A (1% acetonitrile [ACN] and 0.1% formic acid [FA]). Unbound and lysate fractions were reconstituted in 10 μl of the same buffer. Peptide digest samples (2.5–3 μl) were loaded into a trapping column (Acquity M-Class UPLC 180 μm × 20 mm, 5 μm 100 Å BEH C18, Waters Corp.), washed with 100% buffer A for 4 min, and then peptides were separated using a capillary column (150 μm × 10 cm, 1.7 μm 130 Å BEH C18 [first biological replicate] or 75 μm × 10 cm, 1.8 μm 100 Å nanoEase M/Z HSS T3 C18 [second and third biological replicates] [both from Waters Corp.]) under a 120-min linear gradient of 2% to 40% buffer B (99% ACN and 0.1% FA) at a flow rate of 0.5 μl/min. The samples were subjected to nanoelectrospray ionization using a capillary voltage of 1.7 kV, and the ions were introduced in the mass analyzer through a capillary tube maintained at 320 °C. On the Q Exactive HF mass spectrometer, the S-lens RF level was set to 55%; mass spectra were recorded in profile mode over the range m/z 300 to 2000 at a resolution of 60,000 @ m/z 200. The 20 most abundant ions (1.0 × 105 intensity threshold, charge states 2–6) were selected for tandem mass spectrometry (MS/MS) over the scan range m/z 200 to 2000, using the isolation width 2.0 m/z, and were fragmented by higher-energy collision–induced dissociation (HCD) under nitrogen gas at normalized stepped collision energies of 27 and 32%. The second and third biological replicates were analyzed using the Orbitrap Fusion Lumos system, in order to take advantage of its higher sensitivity. The Fusion Lumos instrument was operated with the ion source RF level set to 30%; mass spectra were recorded in profile mode over the range m/z 350 to 2000 at a resolution of 120,000 @ m/z 200. A 3-s cycle time (2.0 × 104 intensity threshold, charge states 2–6) was selected for MS/MS analysis at a resolution of 60,000 @ m/z 200, using a 1.6 m/z isolation width window. Precursors were fragmented by HCD at normalized stepped collision energies of 20, 30, and 35%. Because of the extremely limited sample amounts, only a single LC-MS/MS run was performed for each biological replicate. In addition to HCD, electron-transfer/higher-energy collision–induced dissociation (EThcD) MS/MS spectra of integrins α2 and β1 obtained from recombinant mouse integrin α2β1 protein (R&D Systems) were collected. These confirmed the site-specific glycoform compositions assigned during analysis of the biological samples. Acquisition of EThcD data was triggered by the detection of at least two glycan oxonium ions in MS/MS spectra generated by HCD (35 % normalized collision energy). Calibrated charge-dependent electron transfer dissociation (ETD) parameters were used, with a supplemental collision energy of 20% and orbitrap resolution of 30,000 @ m/z 200.
Data Analysis
The protein content of unbound fractions from the ZIC-HILIC enrichment for glycopeptides and lysate fractions for MK samples was evaluated using Peaks Studio X+ software (Bioinformatics Solutions Inc). Data were searched against the UniProt protein database for Mus musculus (54,424 validated proteins, May 2019). Data were searched with 10-ppm error tolerance for precursor ions and 0.02-Da mass error tolerance for fragment ions. The protease was specified as Trypsin/Lys-C, allowing a maximum of three missed cleavages in the semi-specific digestion mode. Carbamidomethylation of cysteine was chosen as a fixed modification; oxidation of methionine, pyroglutamic acid from aminoterminal glutamine, and deamidation of asparagine were set as variable modifications, with a maximum of three variable modifications per peptide. The false discovery rate (FDR) was estimated using the decoy fusion method. Protein search results were filtered with a −10 logP minimum score of 20 and two unique peptides; peptide search results were filtered at 0.1% FDR. Data obtained from glycopeptide-enriched samples (from the co-IP fractions) were analyzed using Byonic, version 3.8-11 software (Protein Metrics). For each LC-MS/MS run, data were searched against the UniProt protein database for M. musculus (54,424 validated proteins, May 2019) and sequences for mouse integrins α2 and β1 in three separate searches. Once the presence of a glycoprotein is established, searching against a focused database greatly decreases the search space, and this maximizes the number of possible glycoproteoforms retrieved. Digestion and instrument search parameters used for Byonic searches were identical to the ones used in Peaks Studio X+ software. Carbamidomethylation of cysteine was set as a fixed modification, oxidation of methionine as the rare 1 modification, and deamidation of asparagine as the rare 2 modification, with a maximum of two total rare modifications. The Byonic glycan database of 309 N-glycans was used as common glycan modification; no sodium adduction was considered. The protein search results were filtered to 1% FDR. Glycopeptides identified by Byonic were examined manually, and the only matches accepted as valid were those for which both oxonium ions and at least two full-length peptide ions with one N-acetyl hexosamine or multiple saccharide units attached were present in the spectrum. Integrated areas, obtained from extracted ion chromatograms corresponding to the glycopeptiforms of interest, were reported manually. In order to compare glycopeptiforms in myelofibrotic mice to WT, the total signal corresponding to the sum of all glycopeptiforms that span each N-linked site was normalized to the total ion current for each of the biological replicates.
Results
Immunoprecipitation of MK Integrin β1 Using Antibodies Against Either Anti-Integrin β1 or Anti-Integrin α2
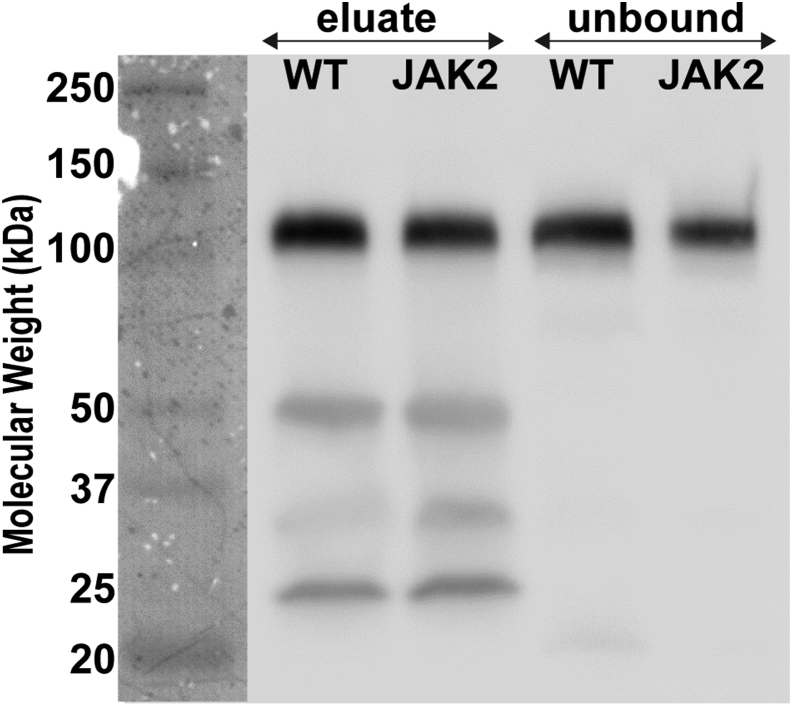
Toward our initial goal of defining the glycosylation profile of integrin β1 in MKs, we tested two approaches for immunoprecipitation of this protein from the cell lysates. We initially tested an anti-integrin β1 cytosolic antibody (AB1952, Millipore Sigma), but subsequent SDS-PAGE gel electrophoresis of the immunoprecipitate yielded only a very faint band, situated between 100 and 150 kDa (not shown). Next, we tested in parallel a mouse monoclonal IgG1 for integrin β1 conjugated to agarose beads and a CD49b (integrin α2) monoclonal antibody in solution (as detailed under Experimental Procedures). As shown in supplemental Fig. S1, the eluate recovered from the anti-integrin β1 IgG1-coated beads did not yield a visible gel band. Yet, a strong signal for integrin β1 was detected in the corresponding unbound and first wash fractions. In contrast, a better yield of immunoprecipitated integrin β1 was obtained when the anti-integrin α2 antibody was used, and a weaker Western blot signal was obtained from the corresponding unbound fraction. This result suggests that most of the integrin β1 is strongly coupled to integrin α2 in MKs and/or that the integrin β1 antibodies target an area that is naturally occupied by other integrins. Therefore, in the following experiments, we analyzed samples derived from co-IP with anti-CD479b (Fig. 1). For each phenotype—WT and JAK2V617F+—five 14-weeks-old male mice were used, yielding 3 to 4 × 105 MK cells, and 1/10 of the eluate was taken for the Western blot analysis shown in Figure 1. In both WT and JAK2V617F+ samples, the bands producing a Western blot signal for integrin β1 lie between 100 and 150 kDa. Mass spectrometry analyses of bands in the region corresponding to 100 to 250 kDa on an SDS-PAGE gel revealed integrin β1 (supplemental Fig. S2).
Fig. 1.
Western blot analysis (using anti-β1) followed by chemiluminescent detection of immunoprecipitated integrin β1 at approximately 130 kDa isolated from megakaryocytes (MKs) derived from 13- to 14-weeks-old WT and JAK2(V617F) male mice. Immunoprecipitation (IP) was carried out using anti-α2 integrin. Unbound fractions following co-IP are also represented. JAK2, Janus kinase 2.
Integrin β1 Glycopeptides in WT MKs
Integrin β1 glycopeptiforms were characterized in MKs of WT and myelofibrotic mice. Integrin β1 co-IP fractions resolved by SDS-PAGE, represented in supplemental Fig. S2, were enriched for glycopeptides and analyzed by LC-MS/MS. We have coined the term glycopeptiform to describe each individual peptide containing a given glycosylation site modified with a discrete glycoform, in order to refer to site-specific glycosylation microheterogeneity in a concise manner. Assignments for a total of 1009 glycopeptiforms were manually validated for integrin β1. The glycopeptides eluted between 8 and 44 min; the molecular weights of the precursors ranged from ∼2000 to 5700 Da. About two-thirds of the precursors that met the selection criteria for MS/MS analysis were triply charged, and the remainder were doubly or quadruply charged. We manually validated peptide spectral matches for glycopeptiforms rather than rely solely on Byonic scores and/or |logProb| values (absolute value of log base 10 of integrin β1 p-value) because at the time of our study, the scoring system seemed to be more geared toward peptides than toward glycopeptides. Overall, 80% of the validated spectral matches for glycopeptiforms had a Byonic score ranging from 57 to 193, and 75% of the spectral matches had a |logProb| value greater than 1. In the biological samples, 11 out of the 12 potential N-glycosylation sites in integrin β1 were determined to be occupied; among these, five were represented in at least two biological replicates across both phenotypes where each biological sample represented MKs pooled from five age-, sex-, and strain-matched mice. (Fig. 2). We compared the glycopeptiform distributions for these five N-glycosylation sites (N212, N406, N481, N520 and N669).
Fig. 2.
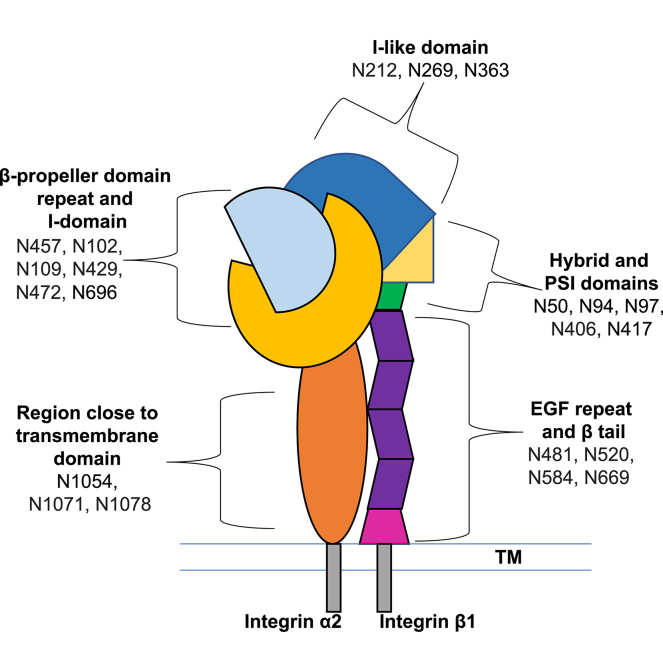
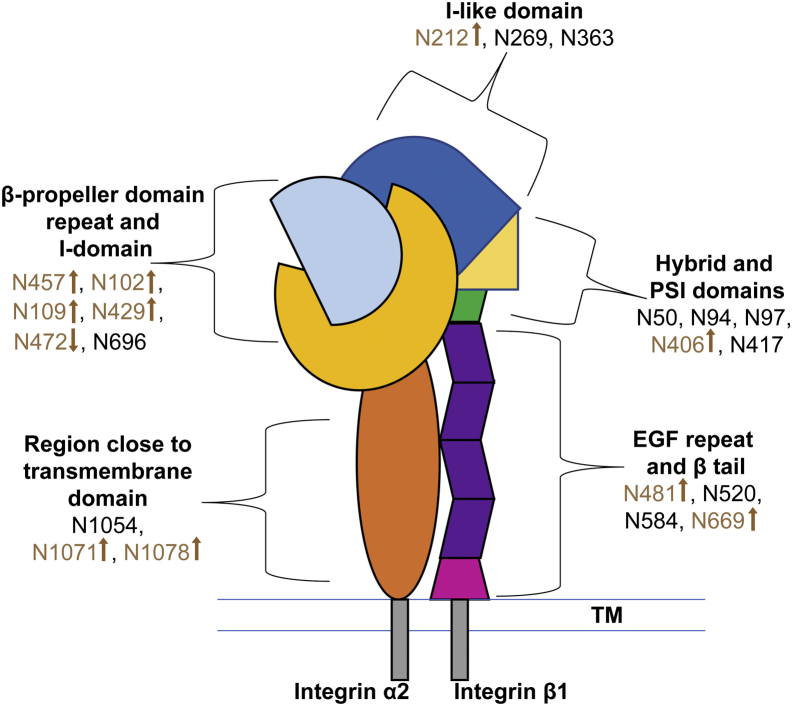
Schematic diagram of the integrin heterodimer α2β1. Twelve integrin β1 potential N-glycosylation sites spanning all domains are represented: I-like (“the head” in contact with the ECM, N212, N269, and N363), PSI and hybrid (N50, N94, N97, N406, and N417), membrane proximal (EGF repeat and β-tail, N481, N520, N584 and N669). Integrin α2 has nine potential N-glycosylation sites situated on or in close proximity to the β-propeller domain repeat and I-domain (in contact with the ECM, N102, N109, N429, N457, N472, and N696) and on a region close to the transmembrane (TM) (N1054, N1071 and N1078). Integrin α2 is significantly longer than integrin β1 (1178 versus 778 amino acids); the major difference is a proposed beta-barrel region lacking in potential N-linked sites, located close to the TM region, but prior to the last three N-linked sites (see supplemental Fig. S6). Arrows indicate direction of change in the relative abundance of glycopeptiforms.
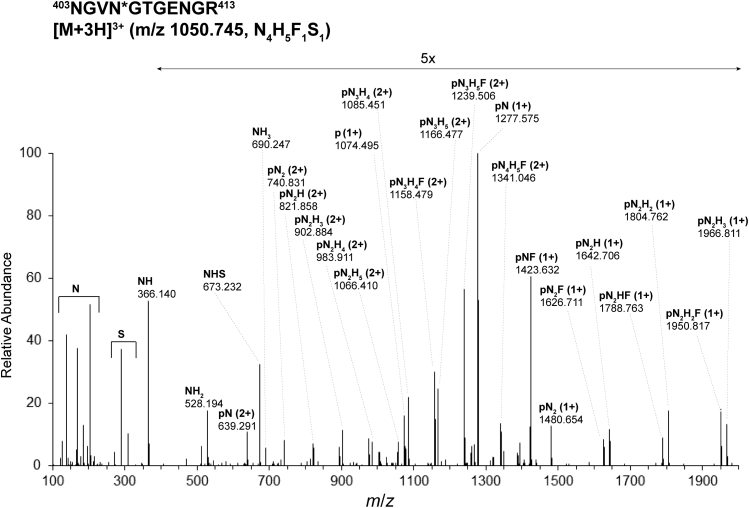
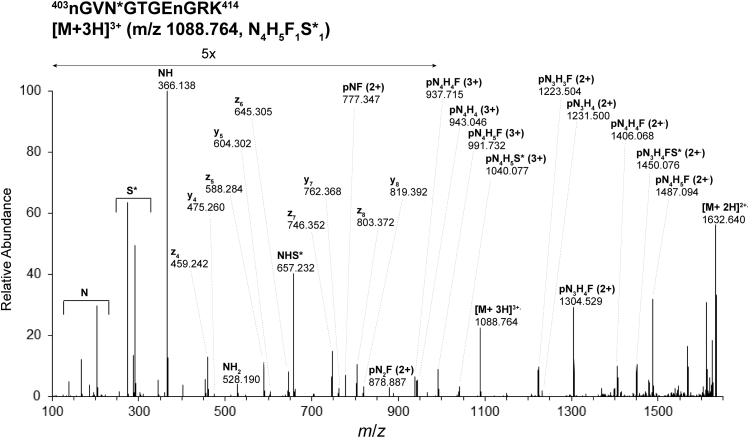
Assignments of integrin β1 glycopeptiforms in MKs of WT and myelofibrotic mice made by the proteomics software packages were validated by manual examination of HCD MS/MS and EThcD MS/MS spectra. The HCD MS/MS spectrum obtained for the precursor [M + 3H]3+, m/z 1050.745, obtained for glycopeptide 403NGVN∗GTGENGR413 bearing the glycoform with the composition of four N-acetyl glucosamine (N), five hexose (H), one fucose (F), and one N-glycolyl neuraminic acid (S) residues (N4H5F1S1) at N406 of integrin β1 isolated from JAK2(V617F+) 13- to 14- weeks-old male mice MKs is shown in Figure 3 and is characteristic of this dataset. The presence of glycan oxonium ions in the range m/z 100 to 700 is consistent with the proposed glycan composition: N or HexNAc, m/z 204.086, S or NeuGc, m/z 307.090, N1H1S1, m/z 673.232. The MS/MS spectrum includes a series of doubly charged intact peptide ions with one or multiple saccharide units attached; for example, from pN2+ (p for the peptide backbone) to pN3H42+ for integrin β1 m/z 1085.451 glycopeptide 403NGVN∗GTGENGR413 at N406 (Fig. 3). Overall, comprehensive fragmentation, for both control and JAK2V617F+ samples provided information on the glycan composition and some structural information on the glycans and allowed site-specific characterization of glycopeptides at 11 of the 12 potential N-linked sites on MK integrin β1. Glycan sites and compositions assigned via HCD MS/MS analysis of the biological samples were confirmed using EThcD MS/MS. For example, the spectrum of the glycopeptide 403nGVN∗GTGEnGRK414 bearing the glycoform N4H5F1S∗1 ([M+3H]3+, m/z 1088.764) (S∗ = N-acetyl neuraminic acid, NeuAc; n = deamidation of Asn to Asp) at N406 of integrin β1 (Fig. 4) was obtained during analysis of recombinant mouse integrin α2β1 protein expressed in CHO cells (R&D Systems). The sequence of the peptide backbone is fully defined in the EThcD MS/MS spectrum (Fig. 4) by the series of y and z ions in the range m/z 450 to 820, and the glycan composition is characterized by series of triply and doubly charged ions between m/z 770 and m/z 1640. Here, as in the spectra shown in Figures 3 and 5, the presence of the pNF fragment indicates the presence of a glycopeptiform with core fucosylation.
Fig. 3.
Higher-energy collision–induced dissociation (HCD) MS/MS spectrum of glycopeptide403NGVN∗GTGENGR413bearing the glycoform N4H5F1S1([M + 3H]3+, m/z 1050.745) at N406 of integrin β1 isolated from MKs of JAK2(V617F) 13- to 14-weeks-old male mice. The precursor ion [M + 3H]3+ was fragmented by HCD at normalized stepped collision energies of 20, 30, and 35%. F, fucose (deoxyHex); Hex, H, hexose; HexNAc, N, N-acetylhexosamine; JAK2, Janus kinase 2; NeuGc, S, N-glycolyl neuraminic acid; p, the intact peptide backbone.
Fig. 4.
Electron-transfer/higher-energy collision-induced dissociation (EThcD) MS/MS spectrum of glycopeptide403nGVN∗GTGEnGRK414bearing the glycoform N4H5F1S∗1([M + 3H]3+, m/z 1088.764) at N406 of integrin β1 obtained from recombinant mouse integrin α2β1 protein (R&D Systems) confirming the glycosylation site and glycoform composition assigned during analysis of the biological sample. Acquisition of EThcD spectra was triggered by the detection of at least two glycan oxonium ions in the MS/MS spectra generated by HCD (35% normalized collision energy). Calibrated charge-dependent ETD parameters were used with a supplemental collision energy of 20% and orbitrap resolution of 30,000 @ m/z 200. Lowercase n in a peptide sequence indicates that an asparagine that has been modified by deamidation (to an aspartic acid). F, fucose (deoxyHex); Hex, H, hexose; HexNAc, N, N-acetylhexosamine; NeuAc, S∗, N-acetyl neuraminic acid; p, the intact peptide backbone.
Fig. 5.
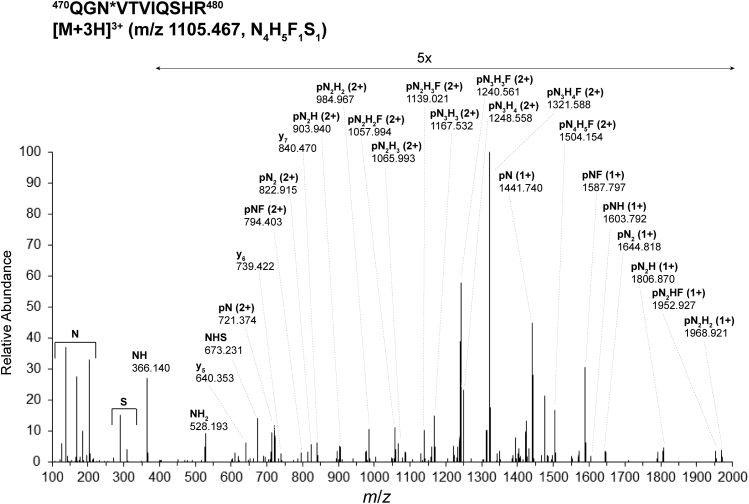
Higher-energy collision–induced dissociation (HCD) MS/MS spectrum of glycopeptide470QGN∗VTVIQSHR480([M + 3H]3+m/z 1105.467) bearing the glycoform N4H5F1S1at N472 of integrin α2 isolated from MKs of JAK2(V617F) 13- to 14-weeks-old male mice. This spectrum derives from the same raw data file as the one used for Figure 3. F, fucose (deoxyHex); H, hexose (Hex); JAK2, Janus kinase 2; N, N-acetylhexosamine (HexNAc); p, the intact peptide backbone; S, N-glycolyl neuraminic acid (NeuGc).
Integrin β1 Glycopeptiforms in JAK2V617F+ MKs
In order to examine the hypothesis that the JAK2V617F mutation in PMF MKs leads to changes in β1 glycosylation, we focused on four sites for which side-by-side comparison of glycopeptiforms (reported in Table 1) was possible: N212 located in the I-like domain, N406 located in the hybrid domain, N481 located in the EGF repeat domain, and N669 located in the β-tail domain. These glycosylation sites were probed since they have been related to cell migration and/or cell spreading on fibronectin (N212, N481 and N669) (29, 30, 31, 32) and to integrin β1 activation (N406) (29, 30, 31). The integrin β1 N-glycan microheterogeneity we determined at sites N212, N406, N481, and N669 is illustrated in Table 1. Glycans at the latter two sites have undergone extensive processing, with the result that the compositions range from N3H3 to N4H5F1S2 at N481 and N4H5F1 to N5H6F1S3 at N669. All sialylated compositions contain NeuGc (S), the dominant form in mice, and only one glycopeptide at N669 carries both NeuGc and NeuAc (S∗). Based on our dataset, we estimate that ∼95 to 98% of sialic acids detected in the mouse MKs analyzed in this study are NeuGc. In order to compare glycopeptiforms in myelofibrotic mice to WT, the total signal corresponding to the sum of all glycopeptiforms that span each N-linked site was normalized to the total ion current for each of the biological replicates; the results were tabulated and are depicted in supplemental Fig. S3. Each biological replicate is depicted as a single point, and the mean is indicated by a dash. Most of the biological replicates form a tight cluster around the same values; even when there is more distance between the replicates, trends in WT versus JAK2V617F hold. Log2-FC of glycopeptiforms for JAK2V617F with respect to WT were calculated in order to evaluate whether or not the mutation in PMF MKs leads to changes in β1 glycosylation (Table 2). A total of 20 glycopeptiforms across N212, N406, N481, and N669 of integrin β1 are compared across JAK2V617F and WT phenotypes. Only seven glycopeptiforms yielded a change (FC > 1), remarkably at least one at each of the four N-glycosylation sites for which the data are summarized in Table 2. At N212, located on the I-like domain in contact with the ECM, 2/3 of the reported glycopeptiforms, all fucosylated and sialylated, increase in JAK2 PMF (glycan compositions N4H5F1S1 and N4H5F1S2 on 203(LRNPc)TSEQN∗cTSPFSYK220). The most significant change, increase in JAK2 PMF, was observed at N406 (2.48 FC, hybrid and PSI domains) for the glycopeptiform N3H6F1S1, 403NGVN∗GTGENGR(K)414. On the membrane proximal domain of integrin β1, three glycopeptiforms at N481 and one at N669 increase in the case of the JAK2V617F+ mutation. Note here that N669 is the only site where an apparent decrease in glycosylation is observed. The overall trend observed is that JAK2V617F mutation in PMF MKs leads to changes in β1 glycosylation; in most cases, it results in an overall increase in the integrated area of glycopeptiforms, whereas the distribution of glycoforms remains similar; the changes that are observed represent increase in the degree of sialylation.
Table 1.
Average relative areas of described glycopeptiforms characterized in at least two out of the three biological replicates that span N-linked glycosylation sites N212, N406, N481, and N669 of integrin β1 isolated from WT and JAK2(V617F) male mice MKs derived from 13- to 14-weeks-old male WT and JAK2(V617F) mice
| Integrin β1 | Rel. areaa | JAK2(V16F) | Integrin β1 | Rel. areaa | JAK2(V16F) |
|---|---|---|---|---|---|
| N212 | N481 | ||||
| HexNAc(4)Hex(5)Fuc(1)NeuGc(1) [N4H5F1S1] | HexNAc(3)Hex(3) [N3H3] | ||||
| C.TSEQN∗cTSPFSYK.N | 5.63E-05 | 1.11E-04 | H.EGN∗GTFEcGAcR.c | 9.35E-04 | 2.56E-03 |
| R.NPcTSEQN∗cTSPFSYK.N | 1.95E-04 | 3.36E-04 | HexNAc(4)Hex(3) [N4H3] | ||
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | H.EGN∗GTFEcGAcR.c | 7.55E-03 | 1.36E-02 | ||
| C.TSEQN∗cTSPFSYK.N | 2.04E-04 | 1.87E-04 | HexNAc(4)Hex(4) [N4H4] | ||
| K.LRNPcTSEQN∗cTSPFSYK.N | 2.95E-05 | - | H.EGN∗GTFEcGAcR.c | 3.07E03 | 4.05E-03 |
| N.PcTSEQN∗cTSPFSYK.N | - | 9.89E-04 | HexNAc(4)Hex(4)NeuGc(1) [N4H4S1] | ||
| R.NPcTSEQN∗cTSPFSYK.N | 5.16E-04 | 5.96E-04 | H.EGN∗GTFEcGAcR.c | 3.29E-04 | 1.69E03 |
| N406 | HexNAc(4)Hex(5) [N4H5] | ||||
| HexNAc(3)Hex(6)Fuc(1)NeuGc(1) [N3H6F1S1] | H.EGN∗GTFEcGAcR.c | 6.51E-04 | 1.95E-03 | ||
| K.NGVN∗GTGENGR.K | 2.09E-05 | 1.11E-04 | HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | ||
| K.NGVN∗GTGENGRK.C | 1.11E-04 | 8.96E-04 | K.cHEGN∗GTFEcGAcR.c | 1.47E-04 | 1.09E-04 |
| HexNAc(4)Hex(5)Fuc(1) [N4H5F1] | HexNAc(4)Hex(5)NeuGc(1) [N4H5S1] | ||||
| K.NGVN∗GTGENGR.K | 7.19E-05 | 8.91E-05 | H.EGN∗GTFEcGAcR.c | 5.38E-04 | 9.67E-04 |
| K.NGVN∗GTGENGRK.c | 6.35E-05 | 7.74E-05 | N669 | ||
| HexNAc(4)Hex(5)Fuc(1)NeuGc(1) [N4H5F1S1] | HexNAc(4)Hex(5)Fuc(1) [N4H5F1] | ||||
| K.NGVN∗GTGENGR.K | 1.10E-03 | 1.69E-03 | K.DTcAQEcSHFN∗LTK.V | 1.14E-04 | 5.04E-05 |
| K.NGVN∗GTGENGRK.c | 1.24E-03 | 1.52E-03 | HexNAc(4)Hex(5)Fuc(1)NeuGc(1) [N4H5F1S1] | ||
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | K.DTcAQEcSHFN∗LTK.V | 7.14E-04 | 5.27E-04 | ||
| K.NGVN∗GTGENGR.K | 3.17E-04 | 1.69E-04 | HexNAc(4)Hex(5)Fuc(1)NeuAc(1)NeuGc(1) [N4H5F1S∗1S1] | ||
| K.NGVN∗GTGENGRK.c | 2.72E-04 | 4.62E-04 | K.DTcAQEcSHFN∗LTK.V | 2.83E-04 | 7.34E-04 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | |||||
| K.DTcAQEcSHFN∗LTK.V | 4.28E-03 | 3.52E-03 | |||
| HexNAc(5)Hex(6)Fuc(1)NeuGc(2) [N5H6F1S2] | |||||
| K.DTcAQEcSHFN∗LTK.V | 1.84E-04 | 7.88E-05 | |||
| HexNAc(5)Hex(6)Fuc(1)NeuGc(3) [N5H6F1S3] | |||||
| K.DTcAQEcSHFN∗LTK.V | 2.92E-04 | 3.00E-04 | |||
Peptide sequences, including sequences resulting from alternative proteolytic cleavage, covering N-glycosylation sites N212, N406, N481, and N669 are indicated. Lowercase c in a peptide sequence indicates a cysteine that has been modified by carboxymethylation prior to the proteolytic digestion.
Abbreviations: F, fucose (deoxyHex); Hex, H, hexose; HexNAc, N, N-acetylhexosamine; NeuAc, S∗, N-acetyl neuraminic acid; NeuGc, S, N-glycolyl neuraminic acid.
Average Log2-fold change of the relative area of glycopeptiforms characterized in at least two out of the three biological replicates and reported in Table 1.
Table 2.
Average Log2-fold change of the relative area of integrin β1 glycopeptiforms characterized in at least two out of the three biological replicates and reported in Table 1
| Integrin β1 | JAK2(V617F) Log2-fold change/WTa |
|---|---|
| N212 203(LNRPc)TSEQN∗cTSPFSYK220 | |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(1) [N4H5F1S1] | 1.01 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | 1.07 |
| HexNAc(4)Hex(6)Fuc(1)NeuGc(2) [N4H6F1S2] | 0.17 |
| N406 403NGVN∗GTGENGR(K)414 | |
| HexNAc(3)Hex(6)Fuc(1)NeuGc(1) [N3H6F1S1] | 2.48 |
| HexNAc(4)Hex(5)Fuc(1) [N4H5F1] | −0.36 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(1) [N4H5F1S1] | 0.57 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | 0.32 |
| N481 477A(cH)EGN∗GTFEcGAcR490 | |
| HexNAc(3)Hex(3) [N3H3] | 0.73 |
| HexNAc(4)Hex(3) [N4H3] | 1.39 |
| HexNAc(4)Hex(4) [N4H4] | 0.65 |
| HexNAc(4)Hex(4)Fuc(1) [N4H4F1] | 1.92 |
| HexNAc(4)Hex(5) [N4H5] | 1.36 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | 0.81 |
| HexNAc(4)Hex(5)NeuGc(1) [N4H5S1] | 0.94 |
| N669 659DTcAQEcSHFN∗LTK672 | |
| HexNAc(4)Hex(5)Fuc(1) [N4H5F1] | −0.86 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(1) [N4H5F1S1] | 0.20 |
| HexNAc(4)Hex(5)Fuc(1)NeuAc(1)NeuGc(1) [N4H5F1S∗1S1] | 1.42 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | −0.29 |
| HexNAc(5)Hex(6)Fuc(1)NeuGc(2) [N5H6F1S2] | −0.60 |
| HexNAc(5)Hex(6)Fuc(1)NeuGc(3) [N5H6F1S3] | 0.11 |
Peptide sequences, including sequences resulting from alternative proteolytic cleavage, covering N-glycosylation sites N212, N406, N481 and N669 are indicated. Lowercase c in a peptide sequence indicates a cysteine that has been modified by carboxymethylation prior to the proteolytic digestion.
Abbreviations: F, fucose (deoxyHex); Hex, H, hexose; HexNAc, N, N-acetylhexosamine; NeuAc, S∗, N-acetyl neuraminic acid; NeuGc, S, N-glycolyl neuraminic acid.
Average Log2-fold change of the relative area of glycopeptiforms characterized in at least two out of the three biological replicates and reported in Table 1.
Integrin α2 Glycopeptides in WT Versus JAK2V617F+ MKs
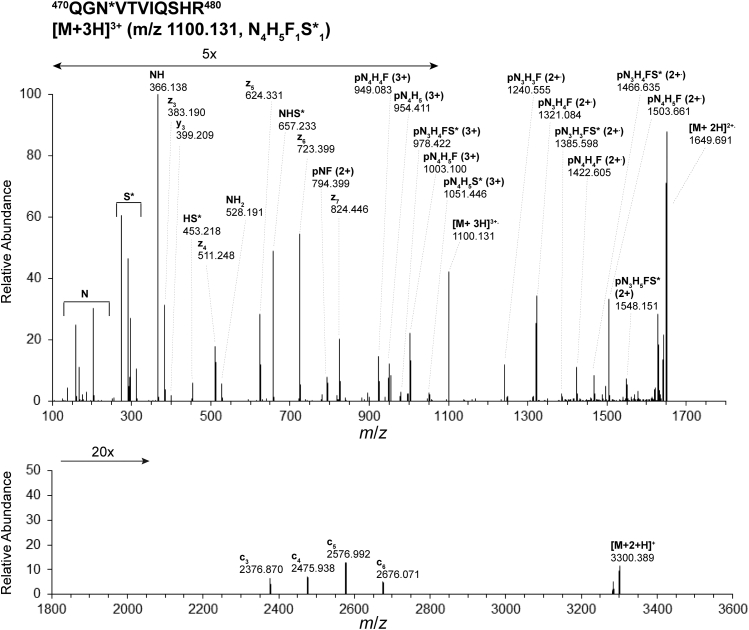
Our initial efforts to isolate integrin β1 from MKs revealed that most of integrin β1 is tightly associated with integrin α2 and/or that the integrin β1 antibodies target an epitope that is common to multiple integrins. The analysis of samples derived from co-IP with anti-CD479b (anti-integrin α2) (Fig. 1) had the advantage that it yielded information on α2 glycosylation in addition to information on our primary target, integrin β1. For integrin α2, only gel bands situated between 100 kDa and 150 kDa (supplemental Fig. S2) are included in this analysis, and assignments of 476 MS scans to α2-integrin glycopeptiforms were validated. Similar to the results we obtained for integrin β1, the glycopeptiforms assigned to integrin α2 eluted between 13 and 46 min; their molecular weights ranged from 1789 to 3433 Da. Integrin α2 has nine potential N-glycosylation sites (Fig. 2) situated on or in close proximity to the β-propeller domain repeat and I-domain (in contact with the ECM, N457, N102, N109, N429, N472, and N696) and proximal to the transmembrane region (N1054, N1071, and N1078). All nine N-glycosylation sites were characterized in at least one biological replicate; seven were represented in at least two biological replicates across both phenotypes (N109, N429, N1071, N1078, N102, N457, and N472) and subsequently used for comparison between JAK2 PMF and WT. Integrin α2 N-glycan microheterogeneity is illustrated in supplemental Table S4. Unlike the integrin β1 glycopeptiforms, many integrin α2 glycopeptides carry glycans which have undergone minimal processing (reported as H5-10, supplemental Table S4). Noticeably, 4/7 reported N-sites carry exclusively glycopeptiforms with only hexose (H5-10) residues beyond the chitobiose core (N109, N429, N1071 and N1078, supplemental Fig. S4), N102 is the only site that lacks glycoforms with H5-10 compositions. Reported sequences for glycopeptides and glycan compositions were confirmed by the examination of HCD MS/MS and EThcD MS/MS spectra (Figs. 5 and 6). Additionally, the EThcD MS/MS spectrum of glycopeptide 470QGN∗VTVIQSHR480 ([M + 3H]3+ m/z 1100.136) bearing the glycoform N4H5F1S∗1 at N472 of integrin α2 (Fig. 6, bottom panel) included a series of c fragments (c3 to c6) indicative of the glycosylation site at N472 on the peptide backbone. A total of 16 glycopeptiforms across N109, N102, N429, N457, N472, N1071, and N1078 of integrin α2 are compared across JAK2V617F and WT phenotypes. Changes in glycosylation in the occurrence of JAK2 PMF was observed for all glycopeptiforms (Table 3). The most significant changes were observed for glycan compositions N2H5-10. At N109, N429, N1071, N1078, N102, and N457, an increase in glycosylation site occupancy was observed (1.20–2.11 FC). At N472 (470QGN∗VTVIQSH(R)480), the abundances of all glycoforms decreased, with the exception of N3H5F1, which increased in JAK2 PMF. Interestingly, the greatest decrease in abundance was observed for N3H6S∗1 (−3.52 FC), the only composition observed with NeuAc instead of NeuGc, the predominant sialic acid in mice. N472 is situated in the region of the β-propeller domain repeat and I-domain of integrin α2 that is, most likely, interacting with integrin β1 upon formation of the heterodimer. Overall, in myelofibrotic MKs, the changes observed for integrin α2 glycosylation were more substantial than those observed for integrin β1 glycosylation.
Fig. 6.
Electron-transfer/higher-energy collision-induced dissociation (EThcD) MS/MS spectrum of glycopeptide470QGN∗VTVIQSHR480([M + 3H]3+m/z 1100.136) bearing the glycoform N4H5F1S∗1at N472 of integrin α2 obtained from recombinant mouse integrin α2β1 protein (R&D Systems) confirming the glycosylation site and glycoform composition assigned during analysis of the biological sample. Acquisition of EThcD spectra was triggered by the detection of at least two glycan oxonium ions in the MS/MS spectra generated by HCD (35 % normalized collision energy). Calibrated charge-dependent ETD parameters were used with a supplemental collision energy of 20%, and orbitrap resolution of 30,000 @ m/z 200. The top panel comprises ions in the range m/z 100 to 1700; the bottom panel displays the range m/z 1800 to 3600, highlighting c3 to c6 fragments indicative of the glycan localization on the peptide backbone. F, fucose (deoxyHex); Hex, H, hexose; HexNAc, N, N-acetylhexosamine; JAK2, Janus kinase 2; NeuAc, S∗, N-acetyl neuraminic acid; p, the intact peptide backbone.
Table 3.
Average Log2 fold change of the relative area of integrin α2 glycopeptiforms characterized in at least two out of the three biological replicates and reported in supplemental Table S2
| Integrin α2 | JAK2(V617F+) Log2-fold change/WTa |
|---|---|
| N109 106TN∗MSLGL(TLTR)118 | |
| HexNAc(2)Hex(5–10)[H(5–10)] | 1.98 |
| N429 429N∗HSSF(LGYSVAA)440 | |
| HexNAc(2)Hex(5–10)[H(5–10)] | 1.99 |
| N1071 1066AEYFIN∗VTTR1075 | |
| HexNAc(2)Hex(5–10)[H(5–10)] | 2.11 |
| N1078 1076VWN∗R1079 | |
| HexNAc(2)Hex(5–10)[H(5–10)] | 1.48 |
| N102 92(LNLQN)SASISN∗VTIEK107 | |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | 1.20 |
| N457 456AN∗YTGQIVL(YSVNK)469 | |
| HexNAc(2)Hex(5–10)[H(5–10)] | 1.38 |
| HexNAc(3)Hex(6)[N3H6] | 1.47 |
| N472 470QGN∗VTVIQSH(R)480 | |
| HexNAc(2)Hex(5–10)[H(5–10)] | −2.56 |
| HexNAc(3)Hex(6) [N3H6] | −2.14 |
| HexNAc(3)Hex(5)Fuc(1) [N3H5F1] | 1.66 |
| HexNAc(3)Hex(6)Fuc(1) [N3H6F1] | −1.63 |
| HexNAc(3)Hex(6)NeuAc(1) [N3H6S∗1] | −3.52 |
| HexNAc(3)Hex(6)Fuc(1)NeuGc(1) [N3H6F1S1] | −1.26 |
| HexNAc(4)Hex(5)Fuc(1) [N4H5F1] | −1.56 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(1) [N4H5F1S1] | −1.05 |
| HexNAc(4)Hex(5)Fuc(1)NeuGc(2) [N4H5F1S2] | −1.49 |
Peptide sequences, including sequences resulting from alternative proteolytic cleavage, covering N-glycosylation sites N109, N429, N1071, N1078, N102, N457, and N472 are indicated.
Abbreviations: F, fucose (deoxyHex); Hex, H, hexose; HexNAc, N, N-acetylhexosamine; NeuAc, S∗, N-acetyl neuraminic acid; NeuGc, S, N-glycolyl neuraminic acid.
Average Log2-fold change of the relative area of glycopeptiforms characterized in at least two out of the three biological replicates and reported in supplemental Table S2.
Discussion
Integrin β1 plays a key role in MKs through its partnering with other integrins such as α2 or α5, whose complexes with integrin β1 mediate MK binding to collagen or fibronectin, respectively. In an earlier study, we showed that integrin α5β1-mediated binding to fibronectin contributes to MK lineage expansion under JAK2V617F+ mutation, and β1 presents in an active form in the mutated cells compared to controls (4). We also found increased adhesion of JAK2V617F+ MKs to collagen via α2β1 integrin, as compared to control samples (11). In studying potential protein modifications that might contribute to β1 activation in JAK2V617F+MKs, we focused on glycosylation, in consideration of the published reports on effects of this type of β1 integrin modification on its function that are discussed herein. Integrin β1 has 12 potential N-glycosylation sites (29) spanning all domains: I-like (“the head” in contact with the ECM, N212, N269, and N363), PSI and hybrid (N50, N94, N97, N406, and N417), membrane proximal (EGF repeat and β-tail, N481, N520, N584, and N669). Hou et al., Gu et al., Pan et al., and Seales et al. (29, 30, 31, 32) among others have extensively studied the impact of integrin β1 N-glycosylation on diverse biological functions such as activation, cell membrane complex formation, or cellular signaling. They conducted global and/or domain-specific studies. Alteration of the activity of glycosyltransferases (30) or use of N-glycosylation mutants (29) resulted in increase or inhibition of cell migration and adhesion, respectively. Inhibition of α2,6-sialylation decreased integrin β1 activation (29), presumably by altering its binding to fibronectin (31, 32). Lastly, Hou et al. (29) showed that formation of the αnβ1 heterodimer depends on the overall N-glycosylation profile of integrin β1.
Challenges in the detailed analysis of glycoproteins are mostly due to the inherent lower abundance of the individual proteoforms of cotransitionally and posttransitionally modified proteins (33). Additionally, because of the structural heterogeneity represented among the glycoproteoforms for a given protein and consequent heterogeneity of the proteolytic glycopeptiforms derived from it, a single modification site requires the consideration of multiple m/z values and requires lower detection limits in order to observe the full range of species (33, 34). Global analysis of protein glycosylation as a means of understanding disease physiopathology has been undertaken by many groups over the last three decades (33, 34, 35). As technologies in mass spectrometry advance, the glycoproteomics field has been moving from global glycosylation profiles (glycosylation site occupancy mapping, glycoprotein abundance and overall glycan compositions) (35) to trying to establish cell line-, protein-, and site-specific glycosylation profiles (34, 36, 37, 38). The last approach is necessary to understand how the dynamic phenomena detected as site-specific changes in N-linked glycosylation patterns correlate with functional changes observed at the cellular level (34). We aimed here to explore the site-specific N-glycosylation of integrins α2 and β1, also in order to gain an understanding of how changes in the populations of glycoforms at individual glycosylation sites correlate with the physiological changes observed in myelofibrotic mice. The mouse glycoproteome has been studied by several groups (33, 34, 35, 36, 37, 38, 39). Some have established profiles of proteins modified by a specific type of glycan structure (epitope) such as Lewis x (Galβ1-4(Fucα1-3)GlcNAc-R) (39) or sialic acid (35) at the organism level (both structural features are commonly associated with diseases, especially cancer). The examination of epitopes can contribute to understanding of the dynamics of glycosylation during investigation of a biological process and is of great interest for relating glycoproteomics data to genetic data. Danzer et al. (38) have published a detailed study of the N-glycoproteome of mouse pancreatic β-cells, establishing a list of 317 glycosylated proteins, but their study did not include definition of the glycosylation sites for these proteins. However, the results from their work were published a decade ago, at a time when the tools for capturing site-specific heterogeneity at a large scale were not yet available. In a study that combined CID and ETD MS/MS, Medzihradszky et al. (40) showed that the site-specific glycosylation patterns of mouse proteins vary at both the tissue and the subcellular level. Others such as Lee et al. (36) and Riley et al. (37) have established profiles at the organ level, looking at the glycoproteomes of mouse liver membrane and mouse brain tissue, respectively. Riley et al. (37) used activated ion ETD techniques to characterize 1545 N-glycosylation sites spanning 771 glycoproteins from mouse brain tissue. They detected a total of eight glycopeptiforms for integrin β1, with the following site-specific glycan compositions: HexNAc2Hex5 at N50; HexNAc2Hex9 at N481; and HexNAc2Hex5–9, as well as HexNAc4Hex4dHex2 and HexNAc4Hex5dHex2NeuAc1, at N669. For integrin α2, they reported a single site-specific glycan composition, HexNAc2Hex5 at N472. With respect to primary cells, Lewandrowski et al. (41) determined occupancy (but not glycan structures) on human platelet integrin β1 sites N417 and N669, as well as the integrin α2 sites N112, N343, N619, N1057, and N1074, using electrostatic repulsion hydrophilic interaction chromatography in combination with mass spectrometry. With MALDI-qTOF tandem mass spectrometry, Ethier et al. (42), characterized the entire released glycan pool and determined the site-specific glycopeptiforms for site N868 of integrin α5 extracted from human placenta as an α5β1 heterodimer. Thus, overall, site-specific investigations of N-glycosylation for integrins have been very limited and, to our knowledge, determination of the N-glycosylation of integrins expressed on MKs has not been reported.
Although a crystal structure of α5β1 integrin headpiece in complex with RGD peptide has been determined (43), there is no crystal structure currently available for integrin α2 nor for the integrin heterodimer α2β1, but the recent release of AlphaFold by Jumper et al. (44) allowed us to localize N-glycosylation sites on the structures calculated by the program. Supplemental Fig. S6 represents integrin α2 (top) and integrin β1 (bottom) with similar projections. Looking at integrin α2, it is interesting to note that the two most processed N-linked sites (N102 and N472) are each in close proximity with one of the two least processed sites (N109 and N429, respectively). The difference in processing is most likely due to variances in accessibility. In the future, additional information about the alignment between the two integrins upon formation of the heterodimer and its response to site-specific glycoform modification or elimination of specific potential glycosylation site(s) would further the understanding of the interrelationships between the conformations and activities of integrin α2 and β1 and their correlation to physiological changes observed in myelofibrotic mice.
As discussed in the introduction, the biosynthesis of N-glycoproteins involves a stepwise assembly of the Glc3Man9GlcNAc2-dolichol phosphate-linked precursor, prior to its cotranslational or posttranslational transfer to the nascent protein and sequential trimming by glucosidases and mannosidases in the ER, and further modification by glycosyl hydrolases and transferases in the Golgi to produce the mature hybrid and complex carbohydrate structures, and quality control is imposed (45). The presence of N-glycans contributes toward proper protein folding. The makeup of the pools of glycan-modifying enzymes is species- and tissue-specific. Their activities can vary in the same cell under different physiological or mutation conditions, and between different cell types, yielding variable N-glycan structures and/or different levels of glycosylation at different sites within the same protein (37, 40, 46). Several hematopoietic malignancies, such as leukemia, have been associated with dysregulated expression of glycosyltransferase and/or glycosidase genes (47), leading to altered glycan properties, such as polysialylation (48, 49) and hyperfucosylation (50) or failure of receptors, e.g., CD79a, to reach the cell surface, combined with effects on IgM (51). In MKs, MPL, the thrombopoietin receptor, bears N-glycosylation essential for its function (52), but this process is incomplete in MPNs (53). Interference with sialylation affects cell adhesion and other properties of tumor cells (49). Calreticulin, a calcium-dependent lectin that facilitates proper folding of newly synthesized N-glycoproteins, is mutated in some forms of MPN. Calreticulin-mutant cells display functional changes in MPN N-glycan pattern (54). Considering this precedent, it is possible that JAK2 mutation and a hyperactive JAK signaling in PMF lead to changes in some of the enzymes responsible for the glycosylation pattern we uncovered in the current study. Future investigations should probe for possible modifications in levels of expression and/or activity of various glycosyltransferases and/or glycosidases in the state of JAK2V617F-induced PMF.
Overall, the coverage we achieved for integrin β1 was greater than coverage for integrin α2 across phenotypes and gel bands. Further examination of the MK co-IP fractions (SDS-PAGE gel bands situated between 100 kDa and 250 kDa) using Peaks Studio X+ software revealed the presence of 47 proteins in the WT fractions and 59 proteins in the JAK2 fractions, in addition to integrins β1 and α2, with associated sequence coverage ranging from 5 to 21% (supplemental Table S5). Other integrins found in these gel bands from the MK immunoprecipitates included integrins β3, αIIb, and α6. Integrins have short cytoplasmic C-terminal domains, where, upon activation of the heterodimer, cytoplasmic proteins are recruited, resulting in a large protein complex anchoring the cell to the actin cytoskeleton and hosting signal transduction events (5, 8). In the analyzed co-IP fractions, along with integrins β1 and α2, actin (cytoplasmic 1 and 2) was significantly present (∼22% coverage). Additionally, proteins that link integrins to the actin cytoskeleton such as talin (∼5% coverage), vinculin (∼6% coverage), and alpha-actinin (∼10% coverage) were also characterized these fractions. We note here that no significant difference was observed between WT and JAK2 phenotypes with respect to coverage for these cytoskeleton-associated proteins. Calderwood et al. (55) have demonstrated how a fragment of talin head domain (Glu-186 to Gln-435) binds to the tail of integrin β3, resulting in the activation of the heterodimer αIIbβ3. Three peptides belonging to talin head domain were identified in MK co-IP fractions: 197FFYSDQNVDSR207, 221DDILNGSHPVSFDK234, and 429STVLQQQYNR438. A peptide from integrin β1 tail was identified (677EKLPQPVQVDPVTHCK692) in the same fraction where peptides from talin starting at Asp-221 and Ser-429 were identified. Although, talin F3 subdomain (Gly-309 to Ser-406), which had been reported to have the highest affinity for integrin β3 tail (55), was not identified in MK co-IP fractions, the identification of structural features for talin and the integrin β1 tail has the potential, in follow-up studies, to provide some insight into the mechanisms of integrin activation in MKs.
Taken together, the glycosylation patterns we uncovered lead us to propose future investigations involving functional studies via mutagenesis of individual N-linked sites on both α2 and β1 integrins, as well as probing for possible modifications in the levels of expression and/or activity of various glycosyltransferases, glycosidases, and chaperones that could reveal pathway disruptions underlying JAK2V617F-induced PMF.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (56) via the PRIDE (57) partner repository with the dataset identifier PXD030550 and are reported in compliance with MIRAGE standards (58).
Supplemental data
This article contains supplemental data (46).
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We thank Dr Shinobu Matsuura for early discussions on her published study concerning ß1 increased activation in MPN megakaryocytes and antibodies used.
Funding and additional information
This research is supported by the National Institutes of Health (NIH) under NIH grants R01 HL136363 and R21 AR072748 to K. R. and NIH grants R24 GM134210, S10 OD021651, and S10 OD021728 to C. E. C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions
M. M. G., C. M. W., A. K., C. E. C., and K. R. methodology; M. M. G. software; M. M. G. and V. L. S. data acquisition, M. M. G. data curation, M. M. G. validation; M. M. G., C. M. W., A. J. P., V. L. S., and A. K. investigation; M. M. G., C. M. W., C. E. C., and K. R. writing-reviewing and editing; V. L. S., C. E. C., and K. R. formal analysis; C. E. C. and K. R. conceptualization; C. E. C. and K. R. supervision; C. E. C. and K. R. funding acquisition; C. E. C. and K. R. resources.
Footnotes
Present address for Maissa M. Gaye: Waters Corporation, R&D, Chemistry Technology Center, Milford, Massachusetts, USA.
Present address for Christina M. Ward: Analytical Development, Sarepta Therapeutics, Andover, Massachusetts, USA.
Present address for Andrew J. Piasecki: Department of Biology, Northeastern University, Boston, Massachusetts, USA.
Present address for Vanessa L. Stahl: Department of Biology, University of Massachusetts Amherst, Amherst, Massachusetts, USA.
Contributor Information
Catherine E. Costello, Email: cecmsms@bu.edu.
Katya Ravid, Email: kravid@bu.edu.
Supplemental Data
References
- 1.Leiva O., Ng S.K., Chitalia S., Balduini A., Matsuura S., Ravid K. The role of the extracellular matrix in primary myelofibrosis. Blood Cancer J. 2017;7 doi: 10.1038/bcj.2017.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cervantes F., Dupriez B., Pereira A., Passamonti F., Reilly J.T., Morra E., Vannucchi A.M., Mesa R.A., Demory J.L., Barosi G., Rumi E., Tefferi A. New prognostic scoring system for primary myelofibrosis based on a study of the International Working Group for Myelofibrosis Research and Treatment. Blood. 2009;113:2895–2901. doi: 10.1182/blood-2008-07-170449. [DOI] [PubMed] [Google Scholar]
- 3.Davies I.L.I., Cho J., Lewis M.H. Splenectomy results from an 18-year single centre experience. Ann. R. Coll. Surg. Engl. 2014;96:147–150. doi: 10.1308/003588414X13814021677593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuura S., Thompson C.R., Ng S.K., Ward C.M., Karagianni A., Mazzeo C., Malara A., Balduini A., Ravid K. Adhesion to fibronectin via α5β1 integrin supports expansion of the megakaryocyte lineage in primary myelofibrosis. Blood. 2020;135:2286–2291. doi: 10.1182/blood.2019004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark E.A., Brugge J.S. Integrins and signal-transduction pathways - the road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 6.Garcia A.J., Vega M.D., Boettiger D. Modulation of cell proliferation and differentiation through substrate-dependent changes in fibronectin conformation. Mol. Biol. Cell. 1999;10:785–798. doi: 10.1091/mbc.10.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu J.G., Taniguchi N. Regulation of integrin functions by N-glycans. Glycoconj. J. 2004;21:9–15. doi: 10.1023/B:GLYC.0000043741.47559.30. [DOI] [PubMed] [Google Scholar]
- 8.van der Flier A., Sonnenberg A. Function and interactions of integrins. Cell Tissue Res. 2001;305:285–298. doi: 10.1007/s004410100417. [DOI] [PubMed] [Google Scholar]
- 9.Paszek M.J., DuFort C.C., Rossier O., Bainer R., Mouw J.K., Godula K., Hudak J.E., Lakins J.N., Wijekoon A.C., Cassereau L., Rubashkin M.G., Magbanua M.J., Thorn K.S., Davidson M.W., Rugo H.S., et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mamidi A., Prawiro C., Seymour P.A., de Lichtenberg K.H., Jackson A., Serup P., Semb H. Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature. 2018;564:114–118. doi: 10.1038/s41586-018-0762-2. [DOI] [PubMed] [Google Scholar]
- 11.Abbonante V., Chitalia V., Rosti V., Leiva O., Matsuura S., Balduini A., Ravid K. Upregulation of lysyl oxidase and adhesion to collagen of human megakaryocytes and platelets in primary myelofibrosis. Blood. 2017;130:829–831. doi: 10.1182/blood-2017-04-777417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., Schnaar R.L., Seeberger P.H. 3rd Ed. Cold Spring Harbor Press; New York, NY: 2017. Essentials of Glycobiology. [PubMed] [Google Scholar]
- 13.Apweiler R., Hermjakob H., Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 14.Shrimal S., Cherepanova N.A., Gilmore R. Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin. Cell Dev. Biol. 2015;41:71–78. doi: 10.1016/j.semcdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramírez A.S., Kowal J., Locher K.P. Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B. Science. 2019;366:1372–1375. doi: 10.1126/science.aaz3505. [DOI] [PubMed] [Google Scholar]
- 16.Robbins P.W., Hubbard S.C., Turco S.J., Wirth D.F. Proposal for a common oligosaccharide intermediate in synthesis of membrane glycoproteins. Cell. 1977;12:893–900. doi: 10.1016/0092-8674(77)90153-2. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs P.P., Sackstein R. CD44 and HCELL: Preventing hematogenous metastasis at step 1. FEBS Lett. 2011;585:3148–3158. doi: 10.1016/j.febslet.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen D.N., Xu B.K., Stanfield R.L., Bailey J.K., Horiya S., Temme J.S., Leon D.R., LaBranche C.C., Montefiori D.C., Costello C.E., Wilson I.A., Krauss I.J. Oligomannose glycopeptide conjugates elicit antibodies targeting the glycan core rather than its extremities. ACS Cent. Sci. 2019;5:237–249. doi: 10.1021/acscentsci.8b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isaji T., Sato Y., Fukuda T., Gu J. N-glycosylation of the I-like domain of beta1 integrin is essential for beta1 integrin expression and biological function: Identification of the minimal N-glycosylation requirement for alpha5beta1. J. Biol. Chem. 2009;284:12207–12216. doi: 10.1074/jbc.M807920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaji T., Sato Y., Zhao Y., Miyoshi E., Wada Y., Taniguchi N., Gu J. N-glycosylation of the beta-propeller domain of the integrin alpha5 subunit is essential for alpha5beta1 heterodimerization, expression on the cell surface, and its biological function. J. Biol. Chem. 2006;281:33258–33267. doi: 10.1074/jbc.M607771200. [DOI] [PubMed] [Google Scholar]
- 21.Schultz M.J., Swindall A.F., Bellis S.L. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012;31:501–518. doi: 10.1007/s10555-012-9359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Buduo C.A., Giannini S., Abbonante V., Rosti V., Hoffmeister K.M., Balduini A. Increased B4GALT1 expression is associated with platelet surface galactosylation and thrombopoietin plasma levels in MPNs. Blood. 2021;137:2085–2089. doi: 10.1182/blood.2020007265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xing S., Ho W.T.T., Zhao W.M., Ma J.M., Wang S.F., Xu X.S., Li Q.S., Fu X.Q., Xu M.J., Zhao Z.Z.J. Transgenic expression of JAK2(V617F) causes myeloproliferative disorders in mice. Blood. 2008;111:5109–5117. doi: 10.1182/blood-2007-05-091579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leiva O., Ng S.K., Matsuura S., Chitalia V., Lucero H., Findlay A., Turner C., Jarolimek W., Ravid K. Novel lysyl oxidase inhibitors attenuate hallmarks of primary myelofibrosis in mice. Int. J. Hematol. 2019;110:699–708. doi: 10.1007/s12185-019-02751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eliades A., Papadantonakis N., Bhupatiraju A., Burridge K.A., Johnston-Cox H.A., Migliaccio A.R., Crispino J.D., Lucero H.A., Trackman P.C., Ravid K. Control of megakaryocyte expansion and bone marrow fibrosis by lysyl oxidase. J. Biol. Chem. 2011;286:27630–27638. doi: 10.1074/jbc.M111.243113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenfeld J., Capdevielle J., Guillemot J.C., Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- 27.Hellman U., Wernstedt C., Góñez J., Heldin C.H. Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 28.Alagesan K., Khilji S.K., Kolarich D. It is all about the solvent: On the importance of the mobile phase for ZIC-HILIC glycopeptide enrichment. Anyl. Bioanyl. Chem. 2017;409:529–538. doi: 10.1007/s00216-016-0051-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou S., Hang Q., Isaji T., Lu J., Fukuda T., Gu J. Importance of membrane-proximal N-glycosylation on integrin β1 in its activation and complex formation. FASEB J. 2016;30:4120–4131. doi: 10.1096/fj.201600665R. [DOI] [PubMed] [Google Scholar]
- 30.Gu J., Isaji T., Sato Y., Kariya Y., Fukuda T. Importance of N-glycosylation on α5β1 integrin for its biological functions. Biol. Pharm. Bull. 2009;32:780–785. doi: 10.1248/bpb.32.780. [DOI] [PubMed] [Google Scholar]
- 31.Pan D., Song Y. Role of altered sialylation of the I-like domain of beta1 integrin in the binding of fibronectin to beta1 integrin: Thermodynamics and conformational analyses. Biophys. J. 2010;99:208–217. doi: 10.1016/j.bpj.2010.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seales E.C., Shaikh F.M., Woodard-Grice A.V., Aggarwal P., McBrayer A.C., Hennessy K.M., Bellis S.L. A protein kinase C/Ras/ERK signaling pathway activates myeloid fibronectin receptors by altering beta1 integrin sialylation. J. Biol. Chem. 2007;280:37610–37615. doi: 10.1074/jbc.M508476200. [DOI] [PubMed] [Google Scholar]
- 33.Xiao H.P., Sun F.X., Suttapitugsakul S., Wu R.H. Global and site-specific analysis of protein glycosylation in complex biological systems with mass spectrometry. Mass Spectrom. Rev. 2019;38:356–379. doi: 10.1002/mas.21586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Y., Zhang H. Glycoproteomics and clinical applications. Proteom. Clin. Appl. 2010;4:124–132. doi: 10.1002/prca.200900161. [DOI] [PubMed] [Google Scholar]
- 35.Kurogochi M., Matsushista T., Amano M., Furukawa J., Shinohara Y., Aoshima M., Nishimura S.I. Sialic acid-focused quantitative mouse serum glycoproteomics by multiple reaction monitoring assay. Mol. Cell. Proteomics. 2010;9:2354–2368. doi: 10.1074/mcp.M110.000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee A., Chick J.M., Kolarich D., Haynes P.A., Robertson G.R., Tsoli M., Jankova L., Clarke S.J., Packer N.H., Baker M.S. Liver membrane proteome glycosylation changes in mice bearing an extra-hepatic tumor. Mol. Cell. Proteomics. 2011;10:18. doi: 10.1074/mcp.M900538-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riley N.M., Hebert A.S., Westphall M.S., Coon J.J. Capturing site-specific heterogeneity with large-scale N-glycoproteome analysis. Nat. Commun. 2019;10:13. doi: 10.1038/s41467-019-09222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danzer C., Eckhardt K., Schmidt A., Fankhauser N., Ribrioux S., Wollscheid B., Muller L., Schiess R., Zullig R., Lehmann R., Spinas G., Aebersold R., Krek W. Comprehensive description of the N-glycoproteome of mouse pancreatic beta-cells and human islets. J. Proteome Res. 2012;11:1598–1608. doi: 10.1021/pr2007895. [DOI] [PubMed] [Google Scholar]
- 39.Noro E., Togayachi A., Sato T., Tomioka A., Fujita M., Sukegawa M., Suzuki N., Kaji H., Narimatsu H. Large-scale identification of N-glycan glycoproteins carrying Lewis x and site-specific N-glycan alterations in Fut9 knockout mice. J. Proteome Res. 2015;14:3823–3834. doi: 10.1021/acs.jproteome.5b00178. [DOI] [PubMed] [Google Scholar]
- 40.Medzihradszky K.F., Kaasik K., Chalkley R.J. Tissue-specific glycosylation at the glycopeptide level. Mol. Cell. Proteomics. 2015;14:2103–2110. doi: 10.1074/mcp.M115.050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewandrowski U., Lohrig K., Zahedi R.P., Wolters D., Sickmann A. Glycosylation site analysis of human platelets by electrostatic repulsion hydrophilic interaction chromatography. Clin. Proteomics. 2008;4:25–36. [Google Scholar]
- 42.Ethier M., Krokhin O., Ens W., Standing K.G., Wilkins J.A., Perreault H. Global and site-specific detection of human integrin alpha 5 beta 1 glycosylation using tandem mass spectrometry and the StrOligo algorithm. Rapid Commun. Mass Spectrom. 2005;19:721–727. doi: 10.1002/rcm.1844. [DOI] [PubMed] [Google Scholar]
- 43.Nagae M., Re S., Mihara E., Nogi T., Sugita Y., Takagi J. Crystal structure of α5β1 integrin ectodomain: Atomic details of the fibronectin receptor. J. Cell Biol. 2012;197:131–140. doi: 10.1083/jcb.201111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moremen K.W., Tiemeyer M., Nairn A.V. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.North S.J., Jang-Lee J., Harrison R., Canis K., Ismail M.N., Trollope A., Antonopoulos A., Pang P.C., Grassi P., Al-Chalabi S., Etienne A.T., Dell A., Haslam S.M. Mass spectrometric analysis of mutant mice. Methods Enzymol. 2010;478:27–77. doi: 10.1016/S0076-6879(10)78002-2. [DOI] [PubMed] [Google Scholar]
- 47.Hakomori S., Kannagi R. Glycosphingolipids as tumor-associated and differentiation markers. J. Natl. Cancer Inst. 1983;71:231–251. [PubMed] [Google Scholar]
- 48.Bhide G.P., Prehna G., Ramirez B.E., Colley K.J. The polybasic region of the polysialyltransferase ST8Sia-IV binds directly to the neural cell adhesion molecule, NCAM. Biochemistry. 2017;56:1504–1517. doi: 10.1021/acs.biochem.6b01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bull C., Boltje T.J., Wassink M., de Graaf A.M.A., van Delft F.L., den Brok M.H., Adema G.J. Targeting aberrant sialylation in cancer cells using a fluorinated sialic acid analog impairs adhesion, migration, and in vivo tumor growth. Mol. Cancer Ther. 2013;12:1935–1946. doi: 10.1158/1535-7163.MCT-13-0279. [DOI] [PubMed] [Google Scholar]
- 50.Blanas A., Sahasrabudhe N.M., Rodriguez E., van Kooyk Y., van Vliet S.J. Fucosylated antigens in cancer: An alliance toward tumor progression, metastasis, and resistance to chemotherapy. Front. Oncol. 2018;8:14. doi: 10.3389/fonc.2018.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vuillier F., Dumas G., Magnac C., Prevost M.C., Lalanne A.L., Oppezzo P., Melanitou E., Dighiero G., Payelle-Brogard A. Lower levels of surface B-cell-receptor expression in chronic lymphocytic leukemia are associated with glycosylation and folding defects of the mu and CD79a chains. Blood. 2005;105:2933–2940. doi: 10.1182/blood-2004-09-3643. [DOI] [PubMed] [Google Scholar]
- 52.Albu R.I., Constantinescu S.N. Extracellular domain N-glycosylation controls human thrombopoietin receptor cell surface levels. Front. Endocrinol. (Lausanne) 2011;2:71. doi: 10.3389/fendo.2011.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moliterno A.R., Spivak J.L. Posttranslational processing of the thrombopoietin receptor is impaired in polycythemia vera. Blood. 1999;94:2555–2561. [PubMed] [Google Scholar]
- 54.Cleyrat C., Darehshouri A., Steinkamp M.P., Vilaine M., Boassa D., Ellisman M.H., Hermouet S., Wilson B.S. Mpl traffics to the cell surface through conventional and unconventional routes. Traffic. 2014;15:961–982. doi: 10.1111/tra.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calderwood D.A., Yan B., de Pereda J.M., Alvarez B.G., Fujioka Y., Liddington R.C., Ginsberg M.H. The phosphotyrosine binding-like domain of talin activates integrins. J. Biol. Chem. 2002;277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 56.Deutsch E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S., Moritz R.L., Carver J.J., Wang M.X., Ishihama Y., Bandeira N., et al. The ProteomeXchange Consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017;45:D1100–D1106. doi: 10.1093/nar/gkw936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.W., Wang R., Hermjakob H. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Struwe W.B., Agravat S., Aoki-Kinoshita K.F., Campbell M.P., Costello C.E., Dell A., Ten F., Haslam S.M., Karlsson N.G., Khoo K.H., Kolarich D., Liu Y., McBride R., Novotny M.V., Packer N.H., et al. The minimum information required for a glycomics experiment (MIRAGE) project: Sample preparation guidelines for reliable reporting of glycomics datasets. Glycobiology. 2016;26:907–910. doi: 10.1093/glycob/cww082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (56) via the PRIDE (57) partner repository with the dataset identifier PXD030550 and are reported in compliance with MIRAGE standards (58).