Abstract
Most potato cultivars are susceptible to late blight disease caused by the oomycete pathogen Phytophthora infestans. Here we report that the genetic loss of host susceptibility is a new source of resistance to prevent or diminish pathogen infection. Previously, we showed that RNAi-mediated silencing of the potato susceptibility (S) genes StDND1, StDMR1, and StDMR6 leads to increased late blight resistance. The mechanisms underlying this S-gene-mediated resistance have thus far not been identified. In this study, we examined the infection process of P. infestans in StDND1-, StDMR1-, and StDMR6-silenced potato lines. Microscopic analysis showed that penetration of P. infestans spores was hampered in StDND1-silenced plants. In StDMR1- and StDMR6-silenced plants, P. infestans infection was arrested at a primary infection stage by enhanced cell death responses. Histochemical staining revealed that StDMR1- and StDMR6-silenced plants display elevated ROS levels in cells at the infection sites. Resistance in StDND1-silenced plants, however, seems not to rely on a cell death response as ROS accumulation was found to be absent at most inoculated sites. Quantitative analysis of marker gene expression suggests that the increased resistance observed in StDND1- and StDMR6-silenced plants relies on an early onset of salicylic acid- and ethylene-mediated signaling pathways. Resistance mediated by silencing StDMR1 was found to be correlated with the early induction of salicylic acid-mediated signaling. These data provide evidence that different defense mechanisms are involved in late blight resistance mediated by functional impairment of different potato S-genes.
Introduction
Late blight, caused by the oomycete Phytophthora infestans, is one of the most devastating diseases of potato worldwide. The pathogen produces sporangia, asexual spores that spread by wind and rain. Infection starts when a sporangium that has landed on a leaf germinates directly, or develops into a zoosporangium releasing zoospores that encyst and germinate. Thereafter, the emerging germ tube develops into an appressorium that penetrates the leaf cuticle and epidermis. Infection expands throughout the leaf tissue via hyphal growth, resulting in water-soaked lesions that turn black. After a few days, the infected leaf tissue becomes necrotic and starts to sporulate. Depending on environmental conditions, an unprotected potato field can be devastated within 10 days [1].
A strategy to control late blight is to grow resistant potato cultivars obtained by introgressing disease resistance (R) genes that are effective against diverse P. infestans isolates [1]. More than 35 potato R genes conferring late blight resistance have been identified to date, most of which encode nucleotide-binding domain, leucine-rich repeat-containing (NLR) proteins [2–6]. These intracellular proteins mount successive defense responses when they recognize corresponding avirulence (AVR) proteins secreted by the pathogen. These AVR proteins, known as RXLR effectors, display high evolutionary rates, and as a result P. infestans can rapidly escape NLR-mediated resistance, thereby limiting the durability of genetic resistance in cultivars possessing a single R gene [7, 8]. Stacking of R genes is a potential strategy to achieve broad-spectrum and durable late blight resistance [9], as shown in cultivar ‘Sarpo Mira’, which has at least five introgressed R genes (i.e. R3a, R3b, R4, Rpi-Smira1, and Rpi-Smira2) from different genetic sources [10]. There is still a risk that such R-gene-rich potato varieties are overcome by the pathogen if no regional resistance gene strategy is developed [10]. Next to R-gene-mediated resistance, infection by P. infestans can be halted by enhancing the overall host defense status, for example through transcriptional activation of defense-related genes, accumulation of defense metabolites and cell wall reinforcement [11–14].
Another approach to gaining disease resistance is impairment of plant susceptibility genes (S genes), a concept that has been exploited in the last decade to develop novel breeding strategies to control diverse crop diseases [15–17]. More than 150 S genes have been described in Arabidopsis, and there is increasing evidence that S-gene orthologs are present in diverse crop species [18–20]. In potato, for example, reduced expression via RNAi-mediated silencing of the S gene SYNTAXIN-RELATED1 (StSYR1), an ortholog of AtPEN1 in Arabidopsis, decreased susceptibility to P. infestans [21].
In our previous studies, we selected 11 Arabidopsis thalianaS genes and silenced their orthologs in the late blight-susceptible potato cultivar (cv.) ‘Désirée’. RNAi-mediated silencing of six of these genes, which included StDND1, StDMR1, and StDMR6, resulted in increased resistance to P. infestans [15, 18]. This showed that loss of function of a putative S gene can be exploited to hamper infection and leaf colonization by P. infestans, and can generate late blight resistance in potato.
StDND1, StDMR1, and StDMR6 are the orthologs of Arabidopsis DND1, DMR1, and DMR6, respectively. Arabidopsis DND1 (Defense, No Death 1) encodes a cyclic nucleotide-gated cation channel (CNGC) and the dnd1 mutant showed resistance to the bacterium Pseudomonas syringae pv. glycinea and the oomycete Hyaloperonospora arabidopsidis, as well as the fungal pathogens Botrytis cinerea and Alternaria brassicicola [22–27]. As indicated by the gene name “Defense, No Death”, resistance to P. syringae observed in the dnd1 mutant was not associated with a hypersensitive response [28], but did show a constitutively elevated expression level of the pathogenesis-related gene PR1 [23, 28]. Our previous results showed that silencing of DND1 orthologs in tomato and potato led to reduced susceptibility to B. cinerea, which was associated with impediment of conidial germination and attachment as well as hyphal growth [29]. Arabidopsis DMR1 (Downy Mildew Resistance 1) encodes a homoserine kinase that catalyzes the phosphorylation of homoserine to O-phospho-homoserine [30, 31]. Arabidopsis dmr1 mutants were found to be resistant to H. arabidopsidis, the powdery mildew fungus Oidium neolycopersici, and two Fusarium pathogens [32–34]. In dmr1 mutants, hyphal growth of H. arabidopsidis was arrested and underdeveloped haustoria were often surrounded by cell wall appositions containing callose [35]. The resistance observed in dmr1 mutants was shown to be associated with accumulation of homoserine [32–35]. Arabidopsis DMR6 (Downy Mildew Resistance 6) encodes a 2-oxoglutarate Fe(II)-dependent oxygenase [36]. Arabidopsis dmr6 mutants showed reduced susceptibility to H. arabidopsidis, P. syringae pv. tomato, and Phytophthora capsici [34–36]. In the dmr6 mutant, hyphal growth and haustoria development of H. arabidopsidis was observed, but the haustoria often had aberrant shapes and stayed immature [35]. Enhanced expression of defense-associated genes, including PR1, was suggested to contribute to the observed dmr6-mediated resistance [34, 36].
Very recently, studies have shown that CRISPR-generated mutants in these three individual genes led to resistance to various pathogens in different crops [37–40]. In previous studies, we showed that individual silencing of StDND1, StDMR1, and StDMR6 increases potato resistance against P. infestans. To analyze how these S-gene-silenced plants prime defense responses to arrest colonization we monitored the infection process of P. infestans by microscopic and histological examination, and determined expression profiles of defense marker genes at early infection stages. Our results show that P. infestans infection in diverse S-gene-silenced potato plants is hindered at different stages.
Results
Increased resistance to multiple P. infestans isolates in StDND1-, StDMR1-, and StDMR6-silenced plants
Previously, we demonstrated that silencing of StDND1, StDMR1, or StDMR6 in potato results in enhanced resistance to P. infestans isolate Pic99189 [15], and additionally to three genetically diverse isolates, Pic99177, USA618, and EC1, upon silencing of StDND1 [18]. In this study, we tested all three S-gene-silenced potato lines with multiple P. infestans isolates (Supplementary Table S1). The reduction in S-gene expression was quantified by qRT–PCR (Fig. 1a) and resistance was assessed in detached leaf assays by measuring lesion size at 3–7 days post-inoculation (dpi) in two independent experiments. All three tested isolates were able to establish infection on the susceptible control cv. ‘Désirée’ efficiently, resulting in sporulating lesions at 7 dpi (Fig. 1b). In contrast, lesion development on the S-gene silenced plants was largely hampered. StDND1- and StDMR6-silenced plants showed no lesion growth upon inoculation with Pic99177 and, when inoculated with the aggressive isolates USA618 and EC1, the lesions were significantly smaller as compared with those on cv. ‘Désirée’. No lesion growth was observed on leaves of StDMR1-silenced plants inoculated with all three isolates (Fig. 1b and c). Dark necrotic spots surrounding the inoculation site were visible on StDMR1- and StDMR6-silenced plants at 6 dpi, whereas StDND1-silenced plants did not show such response (Fig. 2a). These results confirm that silencing StDND1 leads to resistance to multiple P. infestans isolates [18] and show that the same holds true for the other two S genes, StDMR1 and StDMR6. Further, as in our previous studies [15, 18] dwarfness was observed in StDMR1-silenced plants and autonecrosis in some of the StDND1-silenced plants.
Figure 1.
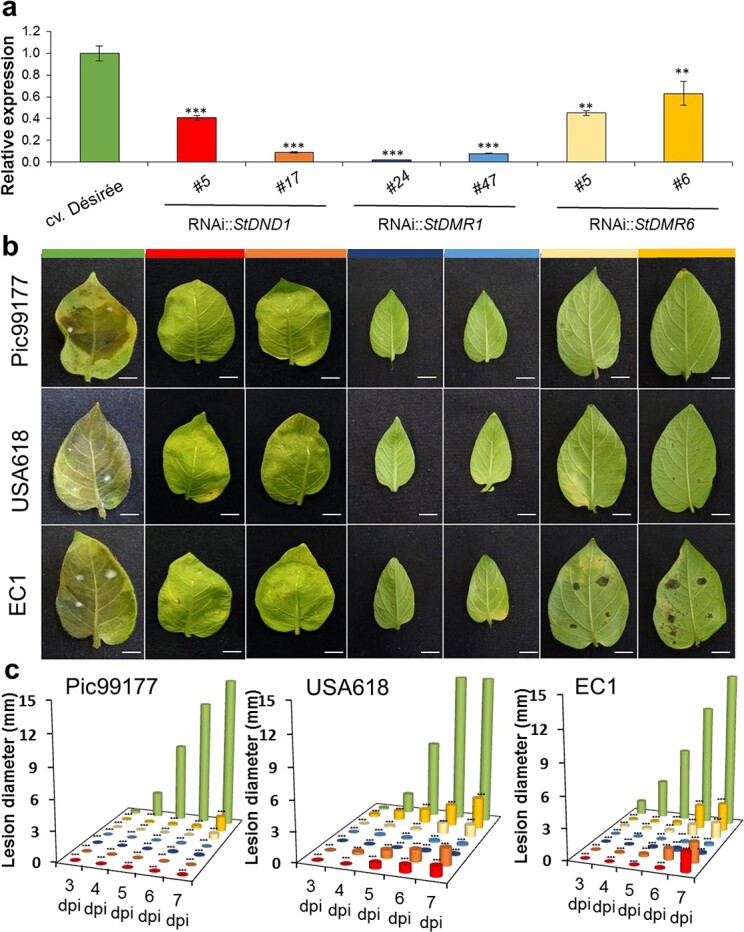
StDND1-, StDMR1-, and StDMR6-silenced potato lines show enhanced resistance to multiple P. infestans isolates. a Relative S-gene expression in leaves of potato cv. ‘Désirée’ and two independent potato transformants per RNAi genotype. b Disease symptoms on leaves of cv. ‘Désirée’ and S-gene-silenced lines after inoculation with the P. infestans isolates Pic99177, USA618, and EC1 at 7 dpi. Scale bars represent 10 mm. c Lesion development on leaves inoculated with P. infestans isolates Pic99177, USA618, and EC1. Three plants were used for each potato genotype, and each was spot-inoculated with 12 droplets of P. infestans inoculum. Asterisks indicate significant differences from the recipient cv. ‘Désirée’ (**P < .01; ***P < .001). Two independent experiments were performed with similar results.
Figure 2.
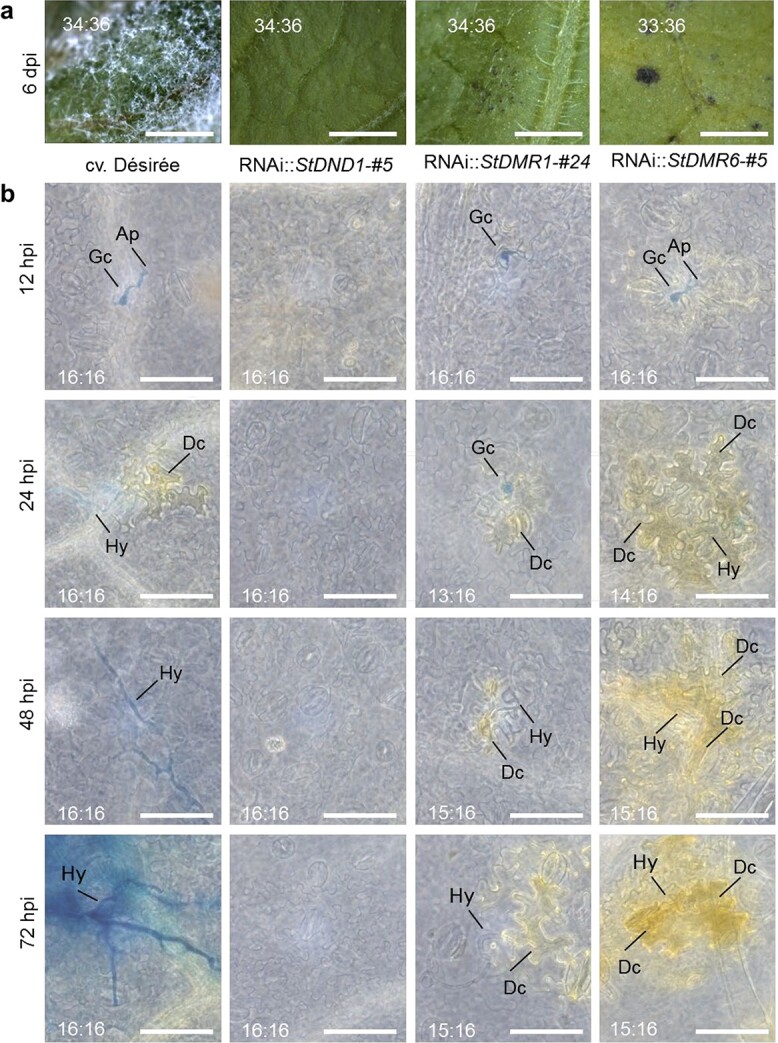
S-gene silencing in potato hampers P. infestans at different infection stages. a Disease symptoms on leaves of cv. ‘Désirée’ and S-gene silenced plants after inoculation with the P. infestans GUS-transformant EY6. Each image represents a single inoculation site at 6 dpi. The numbers in each image represent the ratio of inoculated sites with similar symptoms to the total number of inoculated sites in two independent experiments. Scale bars represent 5 mm. b Microscopic images of GUS-stained infection sites after inoculation with P. infestans EY6. The numbers in each image represent the ratio of inoculated sites with similar responses to the total number of inoculated sites in two independent experiments. Scale bars represent 100 μm. Ap, appressoria; Gc, germinated cyst; Hy, hypha; Dc, dead cell. Similar results were obtained with another set of RNAi lines (RNAi::StDND1-#17, RNAi::StDMR1-#47, RNAi::StDMR6-#6).
Hindered P. infestans infection in StDND1-, StDMR1-, and StDMR6-silenced potato plants
To follow the infection process in more detail, leaves were inoculated with zoospores of the transgenic P. infestans reporter strains 14-3-GFP and EY6 (GUS labeling) and microscopically monitored at different time points of infection [0–96 hours post-inoculation (hpi)]. To monitor strain EY6, a histological study was performed at seven time points (Supplementary Fig. S1). At the two earliest time points, i.e. 0 and 6 hpi, no zoospores were found on any of the genotypes, indicating that they were not attached to the leaf surface and thus were likely washed off during slide preparation. At 12 hpi, germinating cysts with primary appressoria were observed (Fig. 2; Supplementary Fig. S2). Leaf tissue below infection sites on StDMR6-silenced plants started turning yellow at 12 hpi (Fig. 2). From 24 hpi onwards, intracellular hyphae were observed in leaves of cv. ‘Désirée’, whereas on StDMR1- and StDMR6-silenced plants cysts with a short germination tube and intercellular hyphae were found (Fig. 2; Supplementary Fig. S2). Exceptions were StDND1-silenced plants, on which no P. infestans was observed at any time point (Fig. 2).
Growth and proliferation of P. infestans were analyzed in detail by monitoring isolate 14-3-GFP at eight time points (Supplementary Fig. S1). Three hours after inoculation with P. infestans cysts, germination was observed on all plants, except on StDND1-silenced lines (Supplementary Fig. S3a). On cv. ‘Désirée’ plants, hyphal elongation started at 16 hpi. Extensively branched hyphae with collapsed cells underneath became evident at 48 hpi (Supplementary Fig. S3b). Mycelium developed sporangiophores from 72 hpi onwards, releasing numerous sporangia at 96 hpi. On StDMR1- and StDMR6-silenced plants, the development of P. infestans was similar to that observed on cv. ‘Désirée’ at 6 and 16 hpi (Supplementary Fig. S3a). However, at later time points hyphal elongation was arrested (Supplementary Fig. S3b). This was associated with local cell death at the infection sites that became apparent at 16 hpi (Supplementary Fig. S3a). On StDND1-silenced plants, only germinated cysts with short germ tubes were found (Supplementary Fig. S3).
StDMR1- and StDMR6-silenced plants display early ROS accumulation upon P. infestans inoculation
To further determine the different interaction stages, we investigated ROS accumulation in leaves inoculated with P. infestans EY6 (Supplementary Fig. S1), using 3,3-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining to monitor generation of hydrogen peroxide (H2O2) and superoxide anions (O2−), respectively. On the StDMR1- and StDMR6-silenced plants, H2O2 and O2− accumulation was observed at the inoculation sites starting from 6 hpi (Fig. 3). On cv. ‘Désirée’ plants, O2− and H2O2 production became evident starting from 12 and 24 hpi, respectively (Fig. 3; Supplementary Fig. S4). Compared with cv. ‘Désirée’ plants, where ROS accumulation expanded beyond the inoculation sites at 48 hpi, it was limited to cells at the original inoculation site until 72 hpi on StDMR1-silenced plants and until 48 hpi on StDMR6-silenced plants (Fig. S4). In inoculated StDND1-silenced plants there was no accumulation of H2O2 or O2− (Fig. 3; Supplementary Fig. S4).
Figure 3.
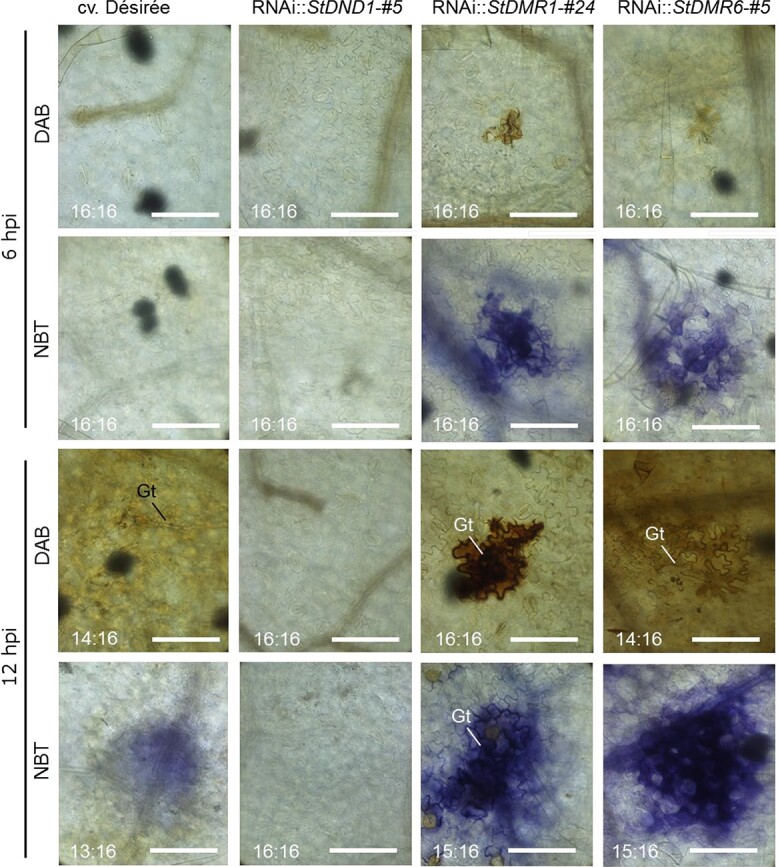
S-gene silencing enhances ROS accumulation at early time points of infection. Microscopic images of potato leaves stained with DAB (H2O2) and NBT (O2−) at 6 and 12 hpi with P. infestans EY6. Each image represents a single inoculation site. The numbers in each image represent the ratio of inoculated sites with similar responses to the total number of inoculated sites in two independent experiments. Scale bars represent 100 μm. Gt, germ tube. Similar results were obtained with another set of RNAi lines (RNAi::StDND1-#17; RNAi::StDMR1-#47; RNAi::StDMR6-#6).
Changes in defense-related gene expression in S-gene-silenced plants
To confirm the microscopic observations and validate our sampling procedure, we quantified pathogen biomass by qRT–PCR at various times after inoculation with strain EY6 (Fig. 4; Supplementary Fig. and Fig S5). In cv. ‘Désirée’ plants there was a steady increase in P. infestans biomass starting from 3 hpi, reaching a plateau at 24 hpi. In contrast, P. infestans biomass was low in leaves of RNAi plants, confirming increased resistance to P. infestans acquired by silencing of StDND1, StDMR1, and StDMR6 (Fig. 4; Supplementary Fig. S5).
Figure 4.
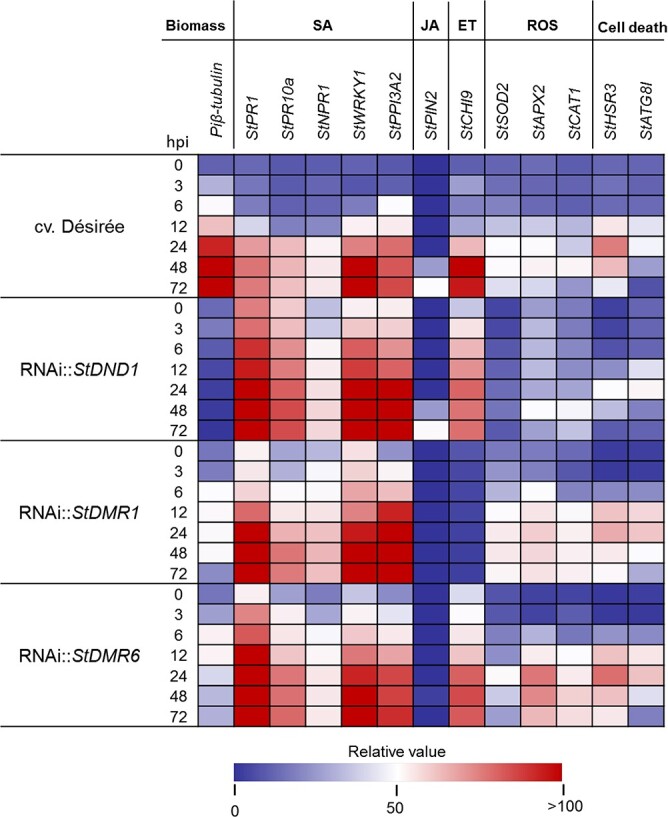
Expression profiles of potato defense marker genes in S-gene-silenced potato lines during P. infestans infection. Relative expression of defense genes in StDND1-, StDMR1-, and StDMR6-silenced potato lines and its recipient cv. ‘Désirée’ upon inoculation with P. infestans EY6. Values are average gene expression levels measured in three biological replicates, each consisting of three technical repeats (Supplementary Figs S5–S10). Expression of potato EF1a (StEF1a) was used as endogenous control, and values were calculated relative to expression levels at 0 hpi in cv. ‘Désirée’. Pathogen biomass in leaves was quantified relative to Piβ-tubulin expression.
To determine which signaling pathways play a role in the acquired resistance, we examined the expression of several defense-related marker genes by qRT–PCR (Fig. 4 and; Supplementary Figs S6 and S8). These included StPR1, StPR10a, StNPR1, StWRKY1, and StPP13A2 for the salicylic acid (SA) pathway, StPIN2 for the jasmonate (JA) pathway, and StCHI9 for the ethylene (ET) pathway [41–43]. Induction of expression of SA marker genes was observed in cv. ‘Désirée’ starting from 12 hpi, in contrast to earlier induction in the S-gene-silenced lines. The JA-responsive marker StPIN2 was slightly upregulated in cv. ‘Désirée’ and StDND1-silenced plants at 48 and 72 hpi while no or hardly any expression was observed in StDMR1- and StDMR6-silenced plants. The ET marker gene StCHI9 showed a clear increase in expression in cv. ‘Désirée’ from 24 hpi onwards. An earlier, but weaker, induction of StCHI9 expression was detected in StDND1- and StDMR6-silenced plants. These data suggest that in StDMR1-silenced plants only the SA-mediated signaling pathway contributes to increased resistance to P. infestans, while in StDND1- and StDMR6-silenced plants ET-mediated signaling may contribute as well.
To further validate the oxidative burst and cell death responses observed in inoculated StDMR1- and StDMR6-silenced plants, we also evaluated the expression of marker genes associated with these events (Fig. 4; Supplementary Figs S9 and S10). We included three genes encoding ROS-scavenging or ROS-generating enzymes, i.e. superoxide dismutase (StSOD2), ascorbate peroxidase (StAPX2), and catalase (StCAT1), and two cell death-related genes, StHSR3 and StATG8I. Compared with cv. ‘Désirée’, slightly higher induction of expression of these genes was observed in StDMR1- and StDMR6-silenced plants from 6 hpi onwards. In contrast, expression of these marker genes was hardly detectable in StDND1-silenced plants (Fig. 4), and thus these expression patterns match the ROS accumulation observed in the respective RNAi plants (Figs 2 and 3; Supplementary Fig. S3).
Discussion
This study shows that broad-spectrum resistance to P. infestans resistance can be efficiently achieved by silencing of StDND1, StDMR1, or StDMR6 in potato [5, 18]. This is in line with several recent studies that realized resistance to a number of pathogens by CRISPR editing these three individual genes in different crops [37–40], demonstrating the importance of S-genes in breeding crops with improved disease resistance.
Compared with our previous studies and those reported in the literature, this study gathered insight into the infection process of P. infestans on S-gene-silenced potato plants by making use of GUS(β-glucuronidase)- and GFP(green fluorescent protein)-labeled P. infestans strains. On StDND1-silenced plants, penetration of P. infestans was hampered and growth was arrested after cyst germination. On StDMR1- and StDMR6-silenced plants, growth of P. infestans was hindered after germ tube emergence, which was associated with cell death and ROS accumulation at the inoculation sites (Fig. 5).
Figure 5.
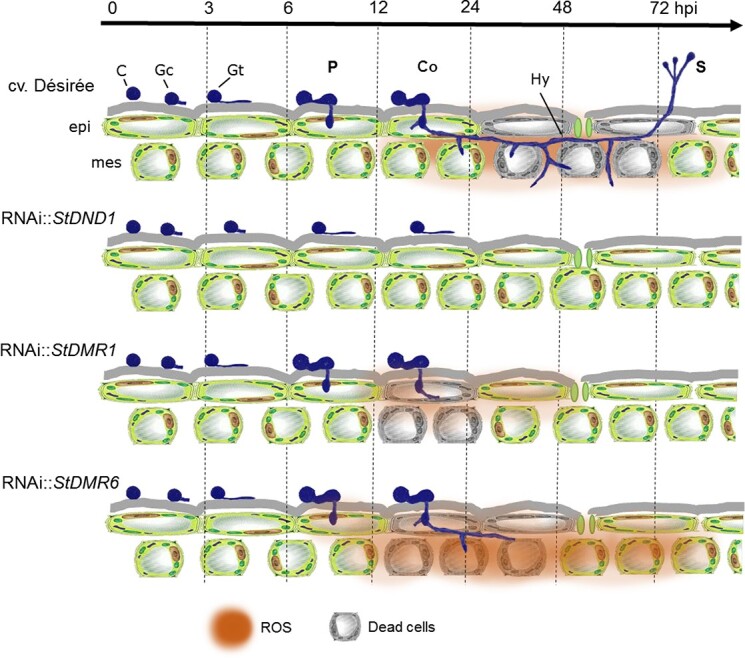
S-gene silencing in potato arrests colonization by P. infestans at different stages of early infection. Model depicting differences in P. infestans colonization on leaves of cv. ‘Désirée’ (susceptible; top panel) and S-gene-silenced potato lines (resistant; lower panels). On StDND1-silenced lines, P. infestans is arrested prior to host penetration of epidermal cells (P). StDMR1-silenced plants display strong epidermal cell death at sites of penetration, whereas StDMR6-silenced plants arrest P. infestans colonization (Co) by enhanced cell death in the mesophyll layer. P. infestans is able sporulate (S) to complete its lifecycle on cv. ‘Désirée’ within 72 hpi. C, cyst; Gc, germinated cyst; Gt, germ tube formation.
For monitoring the activity of defense-related signaling pathways we made use of marker genes. Resistance mediated by silencing StDMR1 was found to be correlated with the early induction of SA-mediated signaling. Although delayed germination and no penetration were observed on StDND1-silenced plants, it is intriguing to notice that, in both StDND1- and StDMR6-silenced plants, the increased resistance was associated with an early onset of SA- and ET-mediated signaling pathways. It thus seems that P. infestans germ tubes release signals that potentially induce defense signaling. The association between the microscopic observations and defense-related gene expression in the S-gene-silenced plants may offer leads for further exploration of the mechanisms underlying enhanced resistance to P. infestans.
The dnd1 mutant of Arabidopsis exhibits enhanced resistance to avirulent isolates of P. syringae, which is not associated with a hypersensitive response [27, 28]. In agreement with this, our study demonstrated that a hypersensitive response does not contribute to the acquired resistance to P. infestans by silencing the potato DND1 ortholog. Instead, all (germinated) cysts of the GUS-expressing line EY6 were washed off during slide preparation, suggesting that cyst attachment to the plant surface of StDND1-silenced plants is blocked. This lack of attachment was also observed when the StDND1-silenced plants were inoculated with B. cinerea [29]. It might be that the leaf surface of StDND1-silenced plants is chemically and/or physically altered. However, we have no direct evidence to prove the lack of cyst attachment. When no washing steps were included in preparing samples with the GFP-labeled isolate, cysts germinated and produced short germ tubes (Supplementary Fig. S3). Further, similar expression profiles were found for both the StDND1- and StDMR6-silenced plants. These observations may indicate that cysts do attach to the leaf surface of StDND1-silenced plants but are unsuccessful in penetration. A recent study showed that Phytophthora species, including P. infestans, use a slicing mechanism to invade their hosts. The tip of the germ tube transforms into a knife-like structure that cuts through the plant surface at an oblique angle [45]. Using this strategy, Phytophthora is able to penetrate without brute force and with minimal consumption of energy. This study further implied that according to the laws of mechanics Phytophthora is unable to penetrate the plant without first attaching itself tightly to the leaf surface. Taking this into consideration, we speculate that the leaf surface of StDND1-silenced plants is physically altered, and as a result the pathogen cannot attach and penetrate. The DND1 gene encodes a protein that is a member of the CNGC family but as yet there is no indication of direct involvement of CNGC family members in cell wall biogenesis or cuticle formation. CNGCs play roles in conducting Ca2+ into plant cells and are involved in various physiological processes [27]. The Arabidopsis dnd1 mutant displays elevated SA levels, which are likely required for its resistance to a broad range of pathogens [23]. In the StDND1-silenced potato plants, we observed a constitutively elevated expression of SA marker genes indicative of elevated SA levels (Fig. 5; Supplementary Fig. S6, [18]). The elevated levels of PR1 expression in StDND1-silenced plants may lead to increased content of PR1 protein, which has been shown to have inhibitory activity against P. infestans [44]. Further, the ET marker gene StCHI9 was induced upon P. infestans infection in the StDND1-silenced potato plants (Fig. 5: Supplementary Fig. S8). This suggests that the increased resistance to P. infestans found in the StDND1-silenced potato plants relies on both SA- and ET-mediated signaling pathways.
In Arabidopsis dmr1 mutants, pathogen resistance without visible hypersensitive response [35] to H. arabidopsidis, O. neolycopersici, Fusarium culmorum, and Fusarium graminearum was found to be associated with homoserine accumulation [32, 33, 35]. Our results show clear cell death of single epidermal cells of StDMR1-silenced potato plants at the site of P. infestans inoculation. This ensures pathogen arrest at a very early infection stage, when cysts start to germinate. P. infestans is a hemibiotroph that requires an initial biotrophic phase in which nutrients from living host cells are acquired via intracellular haustoria [46]. A rapid host cell death observed in StDMR1-silenced plants thus strongly hinders this crucial biotrophic growth phase (Fig. 3). In contrast to Arabidopsis dmr1 mutants [31], induction of SA marker gene expression and ROS accumulation were detected in the StDMR1-silenced potato plants.
Arabidopsis dmr6 mutants were shown to have a reduced susceptibility to H. arabidopsidis, P. syringae (only at adult stage), and P. capsici [34–36]. In the dmr6 mutant, haustorium formation of H. arabidopsidis was found to be severely affected; haustoria had aberrant shapes and stayed immature [35]. Here we found that death of multiple cells occurred on StDMR6-silenced plants around 16 hpi after P. infestans inoculation, which was associated with induced expression of SA and ET marker genes. DMR6 encodes a 2-oxoglutarate Fe(II)-dependent oxygenase, suggesting that the observed resistance is caused by the accumulation of a toxic DMR6 substrate or the absence of a DMR6 metabolic product required for pathogen growth [34]. Later, it was shown this was accompanied by elevated SA levels, and that DMR6 functions in a feedback mechanism to tightly control the SA level [36].
Potato breeding for late blight resistance has so far relied on the introgression of dominant resistance (R) genes from crossable relatives [1]. However, R-gene-mediated resistance is not always durable; P. infestans can easily avoid recognition by mutating or deleting RXLR effector genes so that NLRs encoded by dominant R-genes are not activated to mount effective defense [2]. Thus, we previously proposed an alternative/additive strategy to enhance late blight resistance by impairing plant S-genes that are required by P. infestans for successful host colonization [47]. In potato, several of these S-genes have been identified, including genes encoding a KRBP1 protein [K-homology (KH) RNA-binding protein], three isoforms of the PP1c protein (phosphatase PP1 catalytic subunits) and an NRL1 protein [non-phototrophic hypocotyl 3/root phototropism 2 (NPH3/RPT2)-like protein]. These three host proteins are targeted by the P. infestans RXLR effectors Pi04314, Pi04089, and Pi02860 to promote infection [48–50]. Some S-genes function as negative regulators of plant defense, which are often upregulated by pathogens to suppress plant defense [19]. Therefore, silencing of S-gene expression by RNAi can avoid the suppression of plant defense by pathogens. RNAi is especially powerful in polyploid crops like potato since it results in a dominant inherited resistance [15]. Recent advances in genome editing, including CRISPR/Cas9 technology, make S-gene editing a promising strategy for resistance breeding, even in tetraploid potato and other polyploid crops [51].
The three S-genes studied here were not identified by screening potato for susceptibility factors towards late blight disease. Instead, they were pinpointed as potential candidates because they are orthologs of S-genes identified in Arabidopsis. Our results showed that these orthologs can also be functionally conserved across plant species. For example, downregulation of DND1 expression in potato and tomato led to resistance to powdery mildew, late blight, and grey mold disease [18, 29]. Downregulation of DMR1 expression in tomato and pepper was shown to enhance resistance to powdery mildew and P. capsici, respectively [33, 52]. Similarly, tomato dmr6 mutants generated by CRISPR/Cas9 were found to be resistant to P. capsici, as well as P. syringae pv. tomato [53]. These results, including those presented here, show that impairment of orthologous S-genes in diverse crop species potentially leads to broad-spectrum resistance to multiple/diverse pathogens. However, further studies are needed to fully unravel the molecular basis of resistance mediated by loss-of-function mutation of S-genes in different plant species.
Materials and methods
Pathogen growth and inoculum preparation
The P. infestans isolates used in this study (Supplementary Table S1) are genetically diverse isolates, including Pic99177, USA618, EC1, EY6 (88069), and 14–3-GFP (H30P02). They were cultured on rye sucrose agar medium in the dark at 15°C for 10–14 days. Agar plates fully covered with sporulating mycelium were flooded with cold water (4°C). Sporangiospore suspensions were harvested and incubated at 4°C for 1–2 hours to induce zoospore release. Zoospores were isolated by filtration through 15-μm nylon mesh. Experimental procedures are visualized in Supplementary Fig. S1.
Plant growth conditions and infection assays
Three-week-old in vitro-propagated potato plantlets were transferred to soil and grown in a greenhouse at 25 ± 2°C with 75% relative humidity and a 16:8 hour (light/dark) photoperiod. S-gene-silenced lines RNAi::StDND1-#5, −#17; RNAi::StDMR6-#5, −#6; and RNAi::StDMR1-#24, −#47 were previously described by Sun et al. [15], and are derivatives of potato cv. ‘Désirée’ (R0), which was used as non-transgenic control throughout the experiments. Mature potato plants with fully developed composite leaves were used for detached leaf assays as described by Vleeshouwers et al. [54]. Harvested leaves were arranged in floral foam and placed in plastic boxes with a transparent lid. Abaxial leaf surfaces were inoculated with 10-μl droplets of inoculum (2.5 × 104 zoospores/ml), and subsequently incubated at 18°C with a 16/8 hour photoperiod. Lesion diameters were measured 3–7 dpi using a caliper with a digital display (DIGI-MET®, Helios Preisser, Germany). Disease assays were repeated twice, each consisting of >30 inoculation spots per potato line (Supplementary Fig. S1).
Microscopy and histochemical staining
Host penetration and infection by P. infestans was assessed by imaging leaf disks (diameter 10 mm) punched out from spot-inoculated leaf areas. Bright-field images were taken with a Zeiss Stemi 508 stereomicroscope equipped with a digital camera. Accumulation of H2O2 and O2− was visualized by DAB and NBT staining, respectively [55]. To optimize staining, leaf disks were kept overnight in staining solutions. Tissue clearing was performed by boiling the leaf disks in 96% ethanol. The leaf disks were subsequently kept in 70% ethanol until examination. Histochemical GUS staining was performed as described by Sun et al. [29]. Fluorescence microscopy was performed with a Nikon 90i epifluorescence microscope equipped with a GFP-LP filter and a digital Nikon DS-5MC camera. For each time point, at least eight leaf disks per potato genotype were observed. Each bioassay was performed at least twice, with the exception of the histochemical GUS assay. Pictures show single infection sites.
Nucleic acid extraction and gene expression analyses
Leaf samples were flash-frozen, ground in liquid nitrogen, and stored at −80°C. Total RNA was extracted using a MagMAX-96 Total RNA Isolation Kit (Ambion) and treated with RNAse-free DNAse (Qiagen). RNA concentrations were measured with an Isogen Nanodrop Spectrophotometer ND-1000. Synthesis of cDNA was performed on 1 μg of total RNA using an iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT–PCR was performed on a C1000™ Thermal Cycler PCR system (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). Gene-specific primers were designed with the online tool Primer3 (Supplementary Table S2). Expression levels of marker genes were quantified with the 2-ΔΔCt method using StEF1α and Piβ-tubulin expression for normalization [15]. Quantitative RT–PCR assays were performed on three independent biological samples, each consisting of three technical replicates.
Statistical analysis
Data presented are means of at least three biological replicates with error bars indicating standard deviation. Statistical analyses for detached leaf assays and gene expression were conducted using one-tailed t-tests and Duncan’s multiple range test performed in SPSS, respectively.
Acknowledgements
We thank Dirk Jan Huigen and Gerda van Engelenhoven for maintaining the potato plant material, Ageeth van Tuinen and Marjon Arens for their assistance in disease assays. This work was financially supported by the University Fund Wageningen, TKI Uitgangsmaterialen (EZ-2012-07), the National Science Foundation of China (31801420), and the Scientific and Technological Project of Henan Province (202102110187).
Author contributions
K.S., K.B., E.J., F.G., and Y.B. designed this research. K.S., D.S., and K.B. collected and analyzed the data. K.S., K.B., and Y.B. wrote the manuscript. F.G., E.J., and R.G.F.V. edited the manuscript.
Data availability
All datasets supporting the conclusions of this article are included within the article (and its Supplementary material files).
Conflict of interest
The authors declare that they have no competing interests.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
References
- 1. Fry W. Phytophthora infestans: the plant (and R gene) destroyer. Mol Plant Pathol. 2008;9:385–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Elnahal ASM, Li J, Wang Xet al. Identification of natural resistance mediated by recognition of Phytophthora infestans effector gene Avr3aEM in potato. Front Plant Sci. 2020;11:919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Armstrong MR, Vosson J, Lim TYet al. Tracking disease resistance deployment in potato breeding by enrichment sequencing. Plant Biotechnol J. 2019;17:540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vossen JH, Arkel G, Bergervoet Met al. The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor Appl Genet. 2016;129:1785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodewald J, Trognitz B. Solanum resistance genes against Phytophthora infestans and their corresponding avirulence genes. Mol Plant Pathol. 2013;14:740–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vleeshouwers VG, Raffaele S, Vossen JHet al. Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol. 2011;49:507–31. [DOI] [PubMed] [Google Scholar]
- 7. Du Y, Weide R, Zhao Zet al. RXLR effector diversity in Phytophthora infestans isolates determines recognition by potato resistance proteins; the case study AVR1 and R1. Stud Mycol. 2018;89:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Champouret N, Bouwmeester K, Rietman Het al. Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol Plant Microbe Interact. 2009;22:1535–45. [DOI] [PubMed] [Google Scholar]
- 9. Zhu S, Li Y, Vossen JHet al. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 2012;21:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rietman H, Bijsterbosch G, Cano LMet al. Qualitative and quantitative late blight resistance in the potato cultivar Sarpo Mira is determined by the perception of five distinct RXLR effectors. Mol Plant Microbe Interact. 2012;25:910–9. [DOI] [PubMed] [Google Scholar]
- 11. Kuźnicki D, Meller B, Arasimowicz-Jelonek Met al. BABA-induced DNA methylome adjustment to intergenerational defense priming in potato to Phytophthora infestans. Front Plant Sci. 2019;10:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yogendra KN, Dhokane D, Kushalappa ACet al. StWRKY8 transcription factor regulates benzylisoquinoline alkaloid pathway in potato conferring resistance to late blight. Plant Sci. 2017;256:208–16. [DOI] [PubMed] [Google Scholar]
- 13. Du J, Verzaux E, Chaparro-Garcia Aet al. Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat Plants. 2015;1:15034. [DOI] [PubMed] [Google Scholar]
- 14. Llorente B, Lopez MG, Carrari Fet al. Downregulation of polyphenol oxidase in potato tubers redirects phenylpropanoid metabolism enhancing chlorogenate content and late blight resistance. Mol Breed. 2014;34:2049–63. [Google Scholar]
- 15. Sun K, Wolters AMA, Vossen JHet al. Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 2016;25:731–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pessina S, Pavan S, Catalano Det al. Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics. 2014;15:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pavan S, Schiavulli A, Appiano Met al. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor Appl Genet. 2011;123:1425–31. [DOI] [PubMed] [Google Scholar]
- 18. Sun K, Wolters AMA, Loonen AEHet al. Down-regulation of Arabidopsis DND1 orthologs in potato and tomato leads to broad-spectrum resistance to late blight and powdery mildew. Transgenic Res. 2016;25:123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van Schie CC, Takken FL. Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol. 2014;52:551–81. [DOI] [PubMed] [Google Scholar]
- 20. Bai Y, Pavan S, Zheng Zet al. Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of mlo function. Mol Plant Microbe Interact. 2008;21:30–9. [DOI] [PubMed] [Google Scholar]
- 21. Eschen-Lippold L, Landgraf R, Smolka Uet al. Activation of defense against Phytophthora infestans in potato by down-regulation of syntaxin gene expression. New Phytol. 2012;193:985–96. [DOI] [PubMed] [Google Scholar]
- 22. Su’udi M, Kim MG, Park SRet al. Arabidopsis cell death in compatible and incompatible interactions with Alternaria brassicicola. Mol Cells. 2011;31:593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Genger RK, Jurkowski GI, McDowell JMet al. Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol Plant Microbe Interact. 2008;21:1285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahn IP. Disturbance of the Ca2+/calmodulin-dependent signalling pathway is responsible for the resistance of Arabidopsis dnd1 against Pectobacterium carotovorum infection. Mol Plant Pathol. 2007;8:747–59. [DOI] [PubMed] [Google Scholar]
- 25. Jurkowski GI, Smith RK Jr, Yu ICet al. Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe Interact. 2004;17:511–20. [DOI] [PubMed] [Google Scholar]
- 26. Govrin EM, Levine A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr Biol. 2000;10:751–7. [DOI] [PubMed] [Google Scholar]
- 27. Clough SJ, Fengler KA, Yu ICet al. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci USA2000;97:9323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu IC, Parker J, Bent AF. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA1998;95:7819–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun K, Tuinen A, Kan JALet al. Silencing of DND1 in potato and tomato impedes conidial germination, attachment and hyphal growth of Botrytis cinerea. BMC Plant Biol. 2017;17:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stuttmann J, Hubbertan HM, Rietz Set al. Perturbation of Arabidopsis amino acid metabolism causes incompatibility with the adapted biotrophic pathogen Hyaloperonospora arabidopsidis. Plant Cell. 2011;23:2788–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Damme M, Zeilmaker T, Elberse Jet al. Downy mildew resistance in Arabidopsis by mutation of HOMOSERINE KINASE. Plant Cell. 2009;21:2179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brewer HC, Hawkins ND, Hammond-Kosack KE. Mutations in the Arabidopsis homoserine kinase gene DMR1 confer enhanced resistance to F. culmorum and F. graminearum. BMC Plant Biol. 2014;14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huibers RP, Loonen A, Gao Det al. Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS One. 2013;8:1–8, e67467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van Damme M, Huibers RP, Elberse Jet al. Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 2008;54:785–93. [DOI] [PubMed] [Google Scholar]
- 35. Van Damme M, Andel A, Huibers RPet al. Identification of Arabidopsis loci required for susceptibility to the downy mildew pathogen Hyaloperonospora parasitica. Mol Plant Microbe Interact. 2005;18:583–92. [DOI] [PubMed] [Google Scholar]
- 36. Zeilmaker TL, Ludwig NR, Elberse Jet al. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015;81:210–22. [DOI] [PubMed] [Google Scholar]
- 37. Hasley JAR, Navet N, Tian M. CRISPR/Cas9-mediated mutagenesis of sweet basil candidate susceptibility gene ObDMR6 enhances downy mildew resistance. PLoS One. 2021;16:e0253245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kieu NP, Lenman M, Wang ESet al. Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci Rep. 2021;11. 10.1038/s41598-021-83972-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Low YC, Lawton MA, Rong D. Validation of barley 2OGO gene as a functional orthologue of Arabidopsis DMR6 gene in Fusarium head blight susceptibility. Sci Rep. 2020;10. 10.1038/s41598-020-67006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang X, Low YC, Lawton MAet al. CRISPR-editing of sweet basil (Ocimum basilicum L.) homoserine kinase gene for improved downy mildew disease resistance. Front Genome Ed. 2021;3:629769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bouwmeester K, Han M, Blanco-Portales Ret al. The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in solanaceous plants. Plant Biotechnol J. 2014;12:10–6. [DOI] [PubMed] [Google Scholar]
- 42. Dammann C, Rojo E, Sanchez-Serrano JJet al. Abscisic acid and jasmonic acid activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J. 1997;11:773–82. [DOI] [PubMed] [Google Scholar]
- 43. Barry CS, Fox EA, Yen Het al. Analysis of the ethylene response in the epinastic mutant of tomato. Plant Physiol. 2001;127:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gamir J, Darwiche R, Vant Hof Pet al. The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J. 2017;89:502–9. [DOI] [PubMed] [Google Scholar]
- 45. Bronkhorst J, Kasteel M, Veen Set al. A slicing mechanism facilitates host entry by plant-pathogenic Phytophthora. Nat Microbiology. 2021;6:1000–6. [DOI] [PubMed] [Google Scholar]
- 46. Avrova AO, Boevink PC, Young Vet al. A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol. 2008;10:2271–84. [DOI] [PubMed] [Google Scholar]
- 47. Pavan S, Jacobsen E, Visser RGFet al. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol Breed. 2010;25:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boevink PC, Wang X, McLellan Het al. A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat Commun. 2016;7:10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X, Boevink P, McLellan Het al. A host KH RNA-binding protein is a susceptibility factor targeted by an RXLR effector to promote late blight disease. Mol Plant. 2015;8:1385–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang L, McLellan H, Naqvi Set al. Potato NPH3/RPT2-like protein StNRL1, targeted by a Phytophthora infestans RXLR effector, is a susceptibility factor. Plant Physiol. 2016;171:645–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peng A, Chen S, Lei Tet al. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J. 2017;15:1509–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rehrig WZ, Ashrafi H, Hill Tet al. CaDMR1 cosegregates with QTL Pc5.1 for resistance to Phytophthora capsici in pepper (Capsicum annuum). Plant Genome. 2014;7. 10.3835/plantgenome2014.03.0011. [DOI] [Google Scholar]
- 53. Toledo Thomazella DP, Brail Q, Dahlbeck Det al. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv. 2016;064824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vleeshouwers VG, Dooijeweert W, Keizer LCPet al. A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. Eur J Plant Pathol. 1999;105:241–50. [Google Scholar]
- 55. Wang Y, Bouwmeester K, Van de Mortel JEet al. A novel Arabidopsis-oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defense. Plant Cell Environ. 2013;36:224–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets supporting the conclusions of this article are included within the article (and its Supplementary material files).


