Abstract
Collagens are the major structural component in animal extracellular matrices and are critical signaling molecules in various cell-matrix interactions. Its unique triple helical structure is enabled by tripeptide Gly-X-Y repeats. Understanding of sequence requirements for animal-derived collagen led to the discovery of prokaryotic collagen-like protein in the early 2000s. These prokaryotic collagen-like proteins are structurally similar to mammalian collagens in many ways. However, unlike the challenges associated with recombinant expression of mammalian collagens, these prokaryotic collagen-like proteins can be readily expressed in E. coli and are amenable to genetic modification. In this review article, we will first discuss the properties of mammalian collagen and provide a comparative analysis of mammalian collagen and prokaryotic collagen-like proteins. We will then review the use of prokaryotic collagen-like proteins to both study the biology of conventional collagen and develop a new biomaterial platform. Finally, we will describe the application of Scl2 protein, a streptococcal collagen-like protein, in thromboresistant coating for cardiovascular devices, scaffolds for bone regeneration, chronic wound dressing and matrices for cartilage regeneration.
Keywords: collagen, prokaryotic collagen-like protein, streptococcal collagen-like protein, integrin, integrin-targeting biomaterials, biomaterials, hydrogel
Introduction
Collagen is the most abundant protein found in humans and other animals (Shoulders and Raines 2009; Pallela, Ehrlich, and Bhatnagar 2016). It is a major component of the extracellular matrix (ECM) that provides structural stability (Chang and Buehler 2014). Collagen also regulates cell and tissue biology by interacting with various cellular receptors and other extracellular components (Frantz, Stewart, and Weaver 2010). Its ubiquity within every animal signifies its importance in the formation and maturation of an organism; thus, it has been the target of many studies from the earlier 1900s through today (Siegfried 1902). These studies have improved the understanding of the structural features of collagens including the Gly-X-Y repeat necessary for its characteristic triple helical structure (Qiu et al., 2021; Chen, Chen, and Horng 2011). Understanding the sequence requirement for the triple helix formation led to the discovery that organisms other than animals such as prokaryotes can produce collagen-like proteins as well (Xu et al., 2002; Lukomski et al., 2000). Many different species of bacteria have been shown to produce collagen-like proteins, referred in this review article as prokaryotic collagen-like proteins, though the predominantly studied proteins come from the Streptococcus and the Bacillus families of bacteria (Lukomski et al., 2017; Qiu et al., 2021). The discovery of these prokaryotic collagen-like proteins has provided a new experimental platform. The streptococcal collagen-like protein, Scl2.28, provides a “blank slate” for protein engineering studies due to its inability to bind to any known ligand (Xu et al., 2002). The ability to recombinantly produce collagen-like molecule, based on Scl2.28 in E. coli has enabled the usage of collagen-like protein for multiple engineering applications from endothelialization of vascular grafts to bone regeneration (Cosgriff-Hernandez et al., 2010; Parmar et al., 2016a; Parmar et al., 2016b; Becerra-Bayona et al., 2018; Post et al., 2019).
In this review, we will first discuss the properties of mammalian collagen and provide a comparative analysis of mammalian collagen and prokaryotic collagen-like proteins. We will then review the use of prokaryotic collagen-like proteins to both study the biology of conventional collagen and develop a new biomaterial platform. The application of prokaryotic collagen-like proteins in integrin-targeting hydrogels in a range of biomedical applications will be highlighted. Overall, this review lays a foundation for utilizing recombinant bacterial collagen to perform fundamental studies on the structure and function of mammalian collagen and pursue advanced biomaterials that orchestrate specific cellular behaviors.
Human and Mammalian Collagen
Based on genetic sequence, 28 different types of collagens have been documented in humans (Söderhäll et al., 2007). In addition, several proteins, such as C1q, ficolins, and adiponectin contain minor collagenous domains but are not usually regarded as collagens since their functions and overall structures are not related to conventional collagens (Ricard-Blum, 2011).
Structure of Collagen
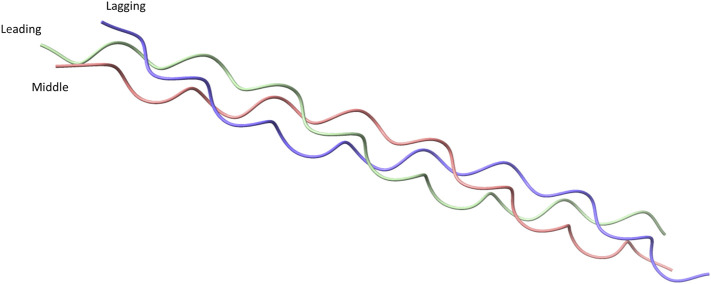
Collagens are defined by the presence of a unique Gly-X -Y tripeptide repeat where X is usually a proline and Y is usually hydroxyproline, which is required by eukaryotic collagen for stable triple-helix formation. Each collagen molecule is made up of three individual α-chains where each α-chain has the structure of a left-handed polyproline Type II-like helix. The three α-chains together form a tightly packed right-handed helix with a 1.5 nm diameter (Figure 1) (Lodish et al., 2000; Berisio et al., 2002; Adzhubei, Sternberg, and Makarov 2013; Qiu et al., 2021). Supercoiling of the individual α-chains in the trimeric structure leads to a rise of 2.9Å per residue and 3.33 residues per turn of the individual α-chains (Persikov et al., 2000). This packing requires each strand to rotate such that the glycine residues lie within the center (Lodish et al., 2000; Berisio et al., 2002). As anything larger than a glycine residue in the center would not fit in the trimeric helix due to steric hindrance (Chen, Chen, and Horng 2011).
FIGURE 1.
Crystal structure of a collagen peptide with the sequence (PPG)10, accessed from PDB ID: 1K6F. The crystal structure shows the structure of the triple helical collagen. Three individual peptides are shown with green representing the leading strand of collagen, pink representing the middle strand, and blue representing the lagging strand.
Collagen molecules can be formed by three identical α-chains or by a mixture of two to three different α-chains. The α-chain composition of different collagens is reviewed elsewhere (Birk and Peter, 2011; Ricard-Blum, 2011). Formation of homotrimer or heterotrimer is directed by a 20 to 250 amino acids long non-collagenous domain (Snellman et al., 2000; Boudko, Engel, and Bächinger 2012; Boudko and Bächinger 2016; Sharma et al., 2017). When collagen α-chains align to form a trimer, a staggered formation is produced where each α-chain has glycine residues in the center of the structure, with the leading strand being the first α-chain to have glycine on the inside of the helix. The middle and lagging strands are the second and third α-chain with the glycine on the inside of the helix, respectively (Figure 1). The stagger of α-chains must be specifically controlled because an incorrect stagger prevents appropriate interactions from occurring (Boudko and Bächinger 2016; 2012).
Collagen Superstructures
These triple helical collagen molecules then assemble into different supermolecular structures leading to the fibers and networks found in mammalian tissues. Based on the supermolecular structure and assemblies, collagen can be divided into different classes: fibrillar collagens, anchoring fibrils, beaded-filament forming collagens, network-forming collagen, fibrillar associated collagens with interrupted triple helices (FACIT), membrane associated collagen with interrupted triple helices (MACIT), multiplexins, and uncategorized collagens (Kucharz 1992; Ottani et al., 2002; Kielty and Grant 2003; Birk and Peter, 2011; Ricard-Blum, 2011). These different classes are reviewed below briefly.
Fibrillar collagens, which includes Type I, II, III, V, XI, XXIV, XXVII Collagens, is the most prominent class of collagen (Birk and Peter, 2011). Fibrillar collagens represent the largest percentage of collagen present in humans and are dominant structural components in the extracellular matrix in mammals (Gelse, Pöschl, and Aigner 2003). As the name suggests, fibrillar collagens form fibrils, which in turn form fascicles, and fascicles then assemble to form fibers (Fratzl 2003).
The non-fibrillar collagens form a variety of superstructures, encompassing all of the classes of collagen except the fibrillar collagen class. The beaded filament-forming collagen include Type VI, Type XXVI and Type XXVIII collagens, non-covalently assembling into thin and beaded filaments where globular domains appear as “beads” (Knupp and Squire 2003; Birk and Peter, 2011). The network-forming collagens, which includes Types IV, VIII, and X collagens, form non-covalent networks similar to the beaded filament-forming collagens but in a diamond or a hexagonal shape (Knupp and Squire 2003; Birk and Peter, 2011). Type IV Collagen; the archetype of network-forming collagens, is found in basement membrane, and is essential for basement membrane function (Khoshnoodi, Pedchenko, and Hudson 2008; Sand, Genovese, and Karsdal 2016). Anchoring fibril collagen i.e. Type VII Collagen, associates laterally via C-terminal of collagen monomers while the N-terminal attaches to the basement membrane (Chung and Uitto 2010; Birk and Peter, 2011). FACITs include Type IX, XII, XIV, XVI, XIX, XXI, and XXII Collagens, and interact with larger fibrillar collagens, such as Type I Collagen or Type II Collagen (Birk and Peter, 2011). In addition, Type IX Collagen can also incorporate into the fibrillar collagens of cartilage (Birk and Peter, 2011). MACITs, also known as membrane collagens, includes Type XIII, XVII, XXIII, and XXV collagens, and integrate into the cell membrane and anchor the cell to the respective basement membrane (Birk and Peter, 2011; Ricard-Blum, 2011). Multiplexin collagens, Type XV and XVIII, are integrated into the basement membrane (Birk and Peter, 2011).
Post-Translational Modifications of Collagen
All mammalian collagens undergo post-translational modifications that happen both before and after collagen is secreted from the cell. Individual α-chains are post-translationally modified before trimerization and secretion from the cell. These post-translation modifications include: 1) hydroxylation of some proline residues into 3-hydroxyproline or 4-hydroxyproline by prolyl-3-hydroxylase or prolyl-4-hydroxylase respectively, 2) hydroxylation of some lysine residues into hydroxylysine by lysyl hydroxylase, and 3) glycosylation of some of the hydroxylysine residues by hydroxylysyl galactosyltransferase and galactosylhydroxylysyl-glucosyltransferase (Gelse, Pöschl, and Aigner 2003). These post-translational modifications of individual α-chains cannot occur once the trimeric structure has been formed (Patino et al., 2002). Next, C-terminal non-collagenous regions are linked together by disulfide bonds catalyzed by protein disulfide isomerase to ensure correct strand alignment and trimer formation (Gelse, Pöschl, and Aigner 2003; Seibel, Robins, and Bilezikian 2006). Once the trimeric collagen molecule is assembled, it can be secreted out of the cell. Once outside the cell, the N-terminal and C-terminal pro-peptides of collagen molecule are cleaved by procollagen N-proteinase and procollagen C-proteinase, respectively (Patino et al., 2002). Finally, after the cleavage of N- and C-terminal propeptides, the collagen molecule is cross-linked to other collagen molecules through hydroxylysines, forming the fibril superstructure (Gelse, Pöschl, and Aigner 2003).
Prokaryotic Collagen-Like Proteins
For a long time, it was believed that non-eukaryotes cannot produce collagen. However, in the year 2000 Streptococcus pyogenes, a Gram-positive bacteria, was discovered to produce collagen-like protein named Streptococcal collagen-like protein A (SclA) (Lukomski et al., 2000). Later discoveries revealed that S. pyogenes encodes two genes, which produce two different proteins containing a collagen-like region: SclA and SclB (Whatmore 2001). These proteins have since been renamed, and are known today as Scl1 and Scl2 (Humtsoe et al., 2005). At the time, this discovery was surprising because it was believed that the characteristic triple helix cannot be formed without post-translational modification of proline into hydroxyproline, a post-translational mechanism specific to eukaryotes. Yet, Scl1 and Scl2 proteins were confirmed to have triple helical structure using circular dichroism (CD) where peak around 220 nm was observed (Xu et al., 2002). The peak at 220 nm is characteristic for the collagen triple-helical structure. In addition, rotary shadowing microscopy of Scl1 and Scl2 proteins showed a lollipop-like structure similar to human collagens (Xu et al., 2002; Adzhubei, Sternberg, and Makarov 2013). Since then, other bacterial species such as, Bacillus anthracis, Clostridium difficile, Clostridium perfringens, Legionella pneumophila, and Burkholderia have been shown to produce collagen-like proteins (Rasmussen, Jacobsson, and Björck 2003; Yu et al., 2014; Bachert et al., 2015; Schnicker and Dey 2016; Ellison et al., 2020). Both pathogenic and non-pathogenic soil bacteria, like Bacillus amyloliquefaciens, produces collagen-like proteins (Yu et al., 2014).
Admittedly in 1990, i.e., prior to the discovery of prokaryotic collagen-like proteins, viruses were discovered to produce collagen-like proteins, though these have been predominantly recognized in bacteriophages (Bamford and Bamford 1990; Rasmussen, Jacobsson, and Björck 2003). Some giant viruses even produce collagen and have the ability to hydroxylate and glycosylate their collagen, namely the mimivirus which infects amoebas (Luther et al., 2011; Yamada 2011). However, the bacteriophage collagens are only about 18 residues found within capsid proteins, and the collagens found within giant viruses rely on eukaryotic systems to be produced (Bamford and Bamford 1990). Consequently, the corresponding viral genes may not lend themselves to direct manipulation and expression in prokaryotic expression systems. These viral genes will not be further discussed in this review.
Domain Organization of Prokaryotic Collagen-Like Proteins
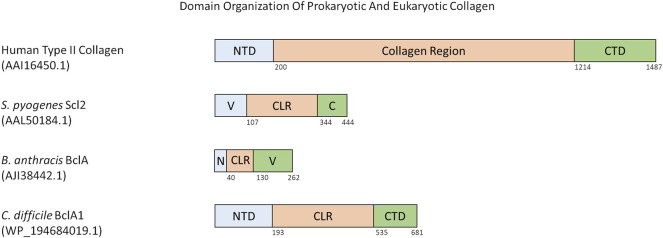
There are a few different domain organization formats of prokaryotic collagen-like proteins. Such as, the prototype Scl proteins from S. pyogenes, have an N-terminal variable domain that is able to nucleate trimerization of the collagen-like region with the characteristic Gly-X-Y repeats followed by a C-terminal non-collagenous repeat with motifs required for cell wall anchoring (Lukomski et al., 2017). The variable domain is significantly different between Scl1 and Scl2, and differs between M-types of S. pyogenes (Caswell et al., 2010). Both Scl1 and Scl2 proteins contain a variable number of Gly-X-Y repeats and sequence in their collagen-like region. For example, Scl1 protein from S. pyogenes M1 serotype contains 50 Gly-X-Y repeats whereas Scl2 from serotype M28 contains 79 Gly-X-Y repeats (Xu et al., 2002; Lukomski et al., 2017; Qiu et al., 2021) (Figure 2).
FIGURE 2.
Domain organization of various eukaryotic and prokaryotic collagens. All numbers in parenthesis below species name and protein name is the accession number for the protein as found in the NCBI Protein database. Numbers below the domains indicate the last residue of that domain. Abbreviations are as follows: NTD = N-terminal Domain, CTD = C-terminal Domain, V = Trimerization Domain, C = C-terminal domain and anchor to cell membrane, N = N-terminal Domain and anchor to cell membrane, CLR = Collagen-like Region.
Some prokaryotic collagen-like proteins have different domains flanking the collagen-like region. For example, the collagen-like proteins Bucl3 and Bucl8 from Burkholderia bacteria also contain the Talin-1 domain for binding to integrins, and outer membrane efflux protein domains to form an efflux channel in the outer membrane of the bacteria (Bachert et al., 2015; Grund et al., 2020). S. pneumoniae protein PclA has a collagen-like region interspersed with a non-collagen domain and other small interruptions, and a large C-terminal domain containing an LPXTG motif (Paterson et al., 2008). Within the Bacillus genus, a variety of collagen-like proteins have been reported (Qiu et al., 2021; Leski et al., 2009). These are named as Bcl proteins with a corresponding letter at the end e.g. BclA, BclB (Parenteau-Bareil, Gauvin, and Berthod 2010). Bcl proteins are composed of a N-terminal domain, a linker region present in some Bcl proteins, a collagen-like region of 27 to 1,158 residues, and finally a C-terminal domain that can have different predicted folds (Figure 2) (Leski et al., 2009). The similarly named BclA1, BclA2, and BclA3 found in C. difficile has a basic domain structure of a N-terminal domain between 5 and 193 residues, a collagen-like region with 354–422 residues, and a C-terminal domain with 131–148 residues (Figure 2) (Pizarro-Guajardo et al., 2014). Each N- and C-terminal domain of the BclA1, BclA2, and BclA3 proteins are currently not well studied and thus have unknown structures within C. difficile.
In summary, prokaryotic collagen-like proteins have diverse domain organization but generally contain a collagen-like region flanked by non-collagenous N-terminal and C-terminal domains. It is noticeable that a similar domain organization is also seen for fibrillar eukaryotic collagens (Figure 2).
Structure of Collagen-Like Regions
Similar to vertebrate collagen, prokaryotic collagen-like proteins contain Gly-X-Y tripeptide repeats, where glycine is required every third residue. The X and Y positions of prokaryotic collagen-like proteins can be occupied by any amino acids, but some residues are preferred (see below). Despite the lack of hydroxyproline, prokaryotic collagen-like proteins form triple helical structures that show the characteristic 220 nm peak in CD (Xu et al., 2002). In addition, the ratio of intensity of the peak at 220 nm over intensity of the peak at 198 nm for the collagen-like region of Scl 2.28 from S. pyogenes is similar to that of mammalian collagen, indicating that the triple helix is fully formed. The collagen-like region of Scl 2.28 from S. pyogenes forms a triple-helix with a diameter of 4–5 nm (Yoshizumi et al., 2009). Similar to mammalian collagen, the collagen-like region of prokaryotic collagen-like proteins is resistant to trypsin digestion (Xu et al., 2010). Prokaryotic collagen-like proteins also appear as lollipop-like structures in rotary shadowing microscopy similar to human collagens (Xu et al., 2002; Adzhubei, Sternberg, and Makarov 2013).
Most bacteria cannot produce hydroxyproline. So how do bacteria produce stable collagen-like structures? Collagen-like region of S. pyogenes Scl proteins contain a large number of polar or charged residues in X and Y positions (Mohs et al., 2007). The most common residue in bacterial collagens present in the X position is proline, similar to vertebrate collagen, whereas threonine is the predominant residue in the Y (Rasmussen, Jacobsson, and Björck 2003; Ghosh et al., 2012). Polar residues in the Scl protein interact with the water molecules to create an extensive network of water-mediated hydrogen bonding, similar to the role that hydroxyproline has in eukaryotic collagen. Such networks are called hydration networks. It stabilizes the triple helix and provides Scl2 with a higher thermal melting temperature of 36°C (Mohs et al., 2007). NMR studies of rat-tail tendon and crystal structures of synthetic collagen peptides have shown that a hydration network occurs in human collagen as well (Migchelsen and Berendsen 1973; Mohs et al., 2007). Mohs et al. demonstrated the effect of hydration network on bacterial collagens by measuring the thermal melting temperature shift that occurs when bacterial collagen is placed in a neutral and an acidic buffers, respectively (Mohs et al., 2007). Scl2.28 protein, which has a low proline percentage but higher percentage of lysine and aspartic acid residues, has a melting temperature of 35.6°C at neutral pH and shifts to 24.2°C at 2.2 pH (Mohs et al., 2007). This large shift is believed to be caused by the loss of charge on aspartic acid and glutamic acid residues in Scl2.28, thus preventing the formation of an extensive hydration network (Mohs et al., 2007).
For those in the phylum Firmicutes, which includes Bacillus and Streptococcus, the collagen-like regions of the proteins are composed of approximately 16.42% threonine, 10.09% proline, 9.17% alanine, and 7.89% glutamine residues (Qiu et al., 2021). Firmicutes have the highest total ratio of threonine and glutamine residues, revealing the importance of hydration network formation for these collagen-like proteins to form stable triple helices (Qiu et al., 2021). Other phyla of bacteria have higher percentages of alanine or proline residues within the collagen-like regions (Qiu et al., 2021).
Post-Translation Modifications of Prokaryotic Collagen-Like Proteins
Prokaryotes are unable to generate all the post-translation modifications observed in vertebrates. While bacteria may not be able to employ all post-translational modifications required for collagen formation in mammals, they do employ some post-translational modifications that are thought to lead to stable triple-helical collagens (Sylvestre, Couture-Tosi, and Mock 2002; Dell et al., 2011). B. anthracis produces a prolyl-4-hydroxylase enzyme that converts proline to hydroxyproline in synthetic peptides, implying the ability for B. anthracis to improve the stability of their collagen proteins by introducing hydroxyproline. Although the bacillus enzyme does not work as consistently as eukaryotic prolyl-4-hydroxylase (Schnicker and Dey 2016). BclA is also known to be glycosylated in B. anthracis and B. cereus spores (Tan and Turnbough 2010; Thompson et al., 2012). It is believed this glycosylation of BclA protein improves hydration network formation and interaction because the sugar moiety is able to interact with multiple water molecules in solution (Sylvestre, Couture-Tosi, and Mock 2002; Boydston et al., 2005; Yu et al., 2014). Similarly, BclA1 protein from C. difficile has been shown to be glycosylated (Pizarro-Guajardo et al., 2014). Additionally, the C. difficile BclA1 protein is cleaved from a ∼72 kDa protein into a 33 kDa protein, though not much more is known about this process (Díaz-González et al., 2015).
Function of Prokaryotic Collagen-Like Proteins
Bacterial collagen-like proteins can have very different functions. Some of the functions are in the non-collagenous domains while others depend on the motifs in the collagen-like region. For example, Scl1 and Scl2 proteins from the M1 serotype of S. pyogenes can intertwine with wounded (denatured) human collagen found within wounded tissue, aiding in invasion and adherence to the wounded site during the infection process (Ellison et al., 2020). In addition, Scl1 protein binds to fibronectin and integrins (α2β1 and α11β1) and mediates bacterial adherence to the wound microenvironment, intracellular invasion, and evasion of the immune system (Caswell et al., 2008, 2010; Oliver-Kozup et al., 2011; Oliver-Kozup et al., 2013; Lukomski et al., 2017). The S. pneumoniae PclA protein has been shown to play a role in adherence and invasion of host cells (Paterson et al., 2008). BclA1 from C. difficile is required for colonization and infection in animals (Pizarro-Guajardo et al., 2016). B. anthracis BclA protein is known to play a role in stabilizing the exosporium of the spore and has reported melting temperatures of up to 95°C due to the very stable C-terminal domain (Boydston et al., 2005). Collagen-like protein expressed by B. amyloliquefaciens, a bacteria found in soil, enables it to adhere to plant roots where they promote plant growth (Zhao et al., 2015). Two of the four collagen-like proteins in B. amyloliquefaciens have also been reported to be present within the flagella of the cell and play a role in motility (Zhao et al., 2016).
Structural Similarities and Differences Between Eukaryotic and Prokaryotic Collagens
Both vertebrate and prokaryotic collagen-like proteins have Gly-X-Y repeats in their primary sequence. Despite the absence of hydroxyproline in prokaryotic collagen-like proteins, its’ individual chains form Type II helices as observed in mammalian collagen and form a triple-helical structure characteristic of mammalian collagen (Xu et al., 2002). Although mammalian collagen can form superstructures, such self-assemblies have not been observed for collagen-like proteins found in prokaryotes (Table 1). Proline occupies approximately one-third of the X position within the Gly-X-Y repeat of both human collagen and collagen-like proteins in Gram-positive bacteria (Ghosh et al., 2012). Human collagen often contains hydroxyproline in the Y position whereas Gram-positive bacteria often use threonine at the Y position (Ghosh et al., 2012) (Table 1). The differences between mammalian and bacterial collagens are summarized in Table 1.
TABLE 1.
Characteristics of eukaryotic collagens and prokaryotic collagen-like proteins.
| Eukaryotic collagen | Prokaryotic collagen-like protein | References | |
|---|---|---|---|
| Domain Organization | Has a N-terminal domain, a collagen region, and a C-terminal domain | Has a N-terminal domain, a collagen region, and a C-terminal domain | Qiu et al., 2021 |
| Might have some interruptions within the collagen region | Might have some interruptions within the collagen region | Bachert et al., 2015 | |
| Paterson et al., 2008 | |||
| Lukomski et al., 2017 | |||
| Leski et al., 2009 | |||
| Tan and Turnbough 2010 | |||
| Pizarro-Guajardo et al. (2014) | |||
| Residue Composition of collagen region (X/Y) | Gly-X-Y (Percentages based on human collagen) | Gly-X-Y | Rasmussen et al. (2003) |
| X: 31.1% Proline (fibrillar), 24.6% Proline (non-fibrillar) | X: 31.0% Proline (Gram-positive), 19% Proline (all bacteria) | Ghosh et al., 2012; Hynes 2012 | |
| Y: 33.5% Proline/hydroxyproline (fibrillar), 42.2% Proline/hydroxyproline (non-fibrillar) | Y: 48.3% Threonine (Gram-positive), 31.6% Threonine (all bacteria) | ||
| Post-translational modification | Hydroxylation of proline to hydroxyproline and lysine to hydroxylysine, glycosylation of hydroxylysine residues, removal of N-terminal and C-terminal pro-peptides, and cross-linking of lysine and hydroxylysine residues | In most cases, there is no hydroxylation of proline to hydroxyproline, no glycosylation of the collagen structure, and no cleavage of the protein | Schnicker and Dey 2016 |
| A limited number of bacteria have their own post-translational modifications, allowing for hydroxylation, glycosylation, or cleavage of proteins | Tan and Turnbough 2010 | ||
| Thompson et al., 2012 | |||
| Gelse et al. (2003) | |||
| Function | Provides mechanical properties (load bearing, tensile strength, and torsional stiffness), cell signaling either directly or through other ECM components, filtration, adhering materials together | Adhesion to specific environments, formation of biofilm, internalization into host, evasion of immune system, use for motility, and spore formation | Qiu et al., 2021 |
| Gelse et al. (2003) | |||
| Bachert et al., 2015 | |||
| Paterson et al., 2008 | |||
| Lukomski et al., 2017 | |||
| Leski et al., 2009 | |||
| Tan and Turnbough 2010 | |||
| Pizarro-Guajardo et al., 2014 | |||
| Zhao et al., 2015 | |||
| Zhao et al., 2016 | |||
| Hynes 2002 | |||
| Paten et al., 2019 | |||
| Shaw and Olsen 1991 | |||
| Izzi et al., 2020 | |||
| Brazel et al. (1988) | |||
| Location | Present within the matrix, with some cellular receptor binding (such as integrin) | N-terminal or C-terminally bound to bacteria cell wall, Bucl8 has two outer transmembrane domains | Qiu et al., 2021 |
| A few collagens can be found integrated within cell membranes | Bachert et al., 2015 | ||
| Lukomski et al., 2017 | |||
| Izzi et al. (2020) | |||
| Ability to form superstructures | Fibrillar collagen can come together to form larger fibrils, fascicles, and tendons | Unable to form larger superstructures, none are currently known | Qiu et al., 2021 |
| Gelse et al. (2003) | |||
| Boydston et al., 2005 | |||
| Canty and Kadler (2002) |
Using Prokaryotic Collagen-Like Protein to Investigate Eukaryotic Collagen Biology
Various tools have been used to investigate the specific interaction sites on eukaryotic collagen, e.g. imaging of complexes for locating binding sites, cyanogen bromide to cleave the collagen into smaller peptides, synthetic collagen peptides to determine residues involved in binding to a target, and there is production of recombinant and mammalian collagen using yeast and transgenic plants (Farndale et al., 2008; Brodsky and Kaplan 2013; Farndale 2019; Fertala 2020). These techniques each have their unique strengths and weaknesses. Cyanogen bromide cleavage might lead to cleavage of the binding site or generate too large of a collagen fragment to accurately locate the binding residues. The success of an imaging approach depends on the resolution and availability of reference structures (Perumal, Antipova, and Orgel 2008; Sweeney et al., 2008; Orgel et al., 2009). Synthetic collagen peptide works effectively for shorter sequences but has difficulty if the binding sequence is large. Although synthetic collagen peptides provide the opportunity to insert peptide binding sequences into a triple helical framework, binding regions larger than nine residues cannot be investigated (Farndale 2019). There have been limited advancement in producing recombinant collagen from yeast and transgenic plants; however, both of these routes are significantly more time-intensive and expensive than utilizing E. coli for production of bacterial recombinant collagen-like proteins (Brodsky and Kaplan 2013; Fertala 2020).
Prokaryotic collagen-like protein provides a novel strategy for investigating interactions of vertebrate collagen with other proteins. Prokaryotic collagen-like proteins can serve as a “host” to a “guest” eukaryotic collagen sequence. Collagen-like region from S. pyogenes protein Scl2.28 comprising 79 Gly-X-Y repeats does not interact with known collagen binding partners in vertebrates and has thus been named a “blank slate” (Qiu et al., 2021; Cereceres et al., 2015; Xu et al., 2002). This “blank slate” collagen can act as a “host” to “guest” sequence from vertebrae collagen sequences. This “host-guest” collagen, referred to as recombinant bacterial collagen in this review, can be used to insert sequences from human collagen allowing for the discovery of longer binding sequences or investigating multiple binding sequences within one protein. An et al. inserted 24 amino acids into blank slate Scl 2.28 protein to identify fibronectin binding site in Type II collagen (An et al., 2014). Previously, fibronectin binding site in Type II collagen was defined to be GLO GQR GER (Erat et al., 2009; 2010). Using recombinant collagen, An et al. determined that the binding site for fibronectin in Type II collagen is at least 18 amino acids long (Table 2). Similarly, recombinant bacterial collagen has been shown to be useful in studying the interactions between collagen and integrin, and describing the stability of a Type I Collagen homotrimer against MMP degradation (Humtsoe et al., 2005; An et al., 2014; Mekkat et al., 2018). As can be seen in Table 2, there are still many interactions within eukaryotic collagen that are not well-defined. Recombinant bacterial collagen could be used to identify the residues in vertebrate collagen that are involved in these various interactions.
TABLE 2.
Binding sites on human collagen to various ECM proteins and cellular receptors. Symbol Definition: * - computational modeling used to define residues, ∼ denotes protease cut site in the protein sequence, † protease cut site was determined using Mass Spectroscopy, “O/P” means that both hydroxyproline and proline have been used and were shown to interact, “x” represents any amino acid unless otherwise stated.
| Target protein | Collagen binding site | Investigated using | References |
|---|---|---|---|
| Decorin | “Gx1x2GDR GEx3GP site, where x1 = K or A, x2 = N, K, S, or P, x3 = P or T” * | Full-length Type I Collagen Fibril | Orgel et al. (2009) |
| Biglycan | Known to share a binding domain with decorin, though exact residues are unknown | Full-length Type I Collagen | Wiberg et al. (2001) |
| Discoidin Domain Receptor 1 | GVMGFO | Synthetic collagen peptide | Xu et al., 2011 |
| Discoidin Domain Receptor 2 | GVMGFO +2 other sites | Synthetic collagen peptide | Konitsiotis et al., 2008 |
| Orgel and Madhurapantula (2019) | |||
| Integrin α1β1 | GF″O/P″GEN, GF″O/P″GER, GLOGER, GASGER, GVOGEA, and GLOGEN | Synthetic collagen peptide & Prokaryotic collagen-like protein | Xu et al., 2000 |
| Hamaia et al., 2012 | |||
| Knight et al., 2000 | |||
| Cosgriff-Hernandez et al. (2010) | |||
| Integrin α2β1 | GF″O/P″GER | Synthetic collagen peptide & Prokaryotic collagen-like protein | Humtsoe et al., 2005 |
| Xu et al., 2000 | |||
| Knight et al., 2000 | |||
| Cosgriff-Hernandez et al., 2010 | |||
| Knight et al. (1998) | |||
| Integrin α10β1 | GFOGER | Synthetic collagen peptide | Hamaia et al. (2017) |
| Integrin α11β1 | GFOGER | Synthetic collagen peptide | Sweeney et al., 2008 |
| Zhang et al. (2003) | |||
| Fibronectin | GLAGQR GIVGLP GQRGER | Prokaryotic collagen-like protein & Synthetic collagen peptide | An et al., 2014 |
| Erat et al. (2009) | |||
| Secreted protein, acidic and rich in cysteine known as “SPARC” | GPOGPS GPRGQO GVMGFO GPKGND GAO | Cyanogen bromide peptides & Synthetic collagen peptide | Giudici et al. (2008) |
| Osteopontin | Binds Type I Collagen at an unknown binding site | Full-length Type I Collagen | Liu et al., 2007 |
| GpVI | GAOGLR GGAGPO GPEGGK GAAGPO GPO, (POG)10 | Synthetic collagen peptide | Jarvis et al., 2008 |
| Horii et al. (2006) | |||
| Collagen V | Cross-linked in LPQPPQ EKAHDG GRY region | Full-Length Type I Collagen | Sweeney et al. (2008) |
| Platelet derived growth factor-BB | Binds Type 1 Collagen at an unknown binding site | Full-length Type I Collagen | Lin et al., 2006 |
| Endo180/uPARAP | GPPGPP GPPGPP GPPSAG FDFSFL PQPPQE KAHDGG RYYRA | Full-length Collagen | Sweeney et al., 2008 |
| Thomas et al., 2005 | |||
| Jürgensen et al. (2011) | |||
| Thrombospondin1 | Known to bind to Type I, VI, XI Collagen, and to Endostatin | Full-length Collagen | Faye et al. (2009) |
| THBS1 | |||
| Procollagen C-proteinase enhancer-1 (PCPE-1) | Binds to the C-terminal propeptide region, the lysine residues C-terminal of the BMP-1 cut site on α1 Type III Collagen | Type I Procollagen, & recombinant Type III Collagen C-propeptide | Kessler and Adar 1989 |
| Bourhis et al. (2013) | |||
| Bone morphogenetic protein-1 (BMP-1) | YRA ∼ DDA NVVRDR D α1 (I) | Type I and III Procollagen | Kessler et al., 1996 |
| YRA ∼ DQP RSAPSL R α2 (I) | Lindahl et al. (2011) | ||
| YYG ∼ DEP MDFKIN T α1 (III) | |||
| SMADs | C-terminal propeptide of Type I, III, and V Collagen | Recombinant Type I, III, and V Collagen C-propeptide | Ellis et al. (2003) |
| Osteoclast-associated receptor (OSCAR) | GxOGPx GFx, GPOGPA GFO, GAOGAS GDR | Synthetic collagen peptide | Zhou et al., 2016 |
| Barrow et al., 2011 | |||
| An and Brodsky (2016) | |||
| Bone SialoProtein-2 | In the triple-helical region of Type I Collagen, and a binding site within Type XI Collagen KKKSNY TKKKRT LATNSK KKSKM, KKKSNY TKKKRT LATNSK KK | Type I Collagen (treated with pepsin), & peptides of non-collagen regions of Type XI Collagen | Baht et al. (2008) |
| Gorski et al. (2021) | |||
| Periostin | Binds Type I and V Collagen with an unknown binding site | Full-length Type I and V Collagen | Takayama et al., 2006 |
| Norris et al., 2007 | |||
| Kii and Ito 2017 | |||
| Kudo and Kii (2018) | |||
| Pigment epithelium-derived factor (PEDF) | KGHRGF SGL, KGHRGY SGL | Synthetic collagen peptide | Sekiya et al., 2011 |
| Broadhead et al. (2010) | |||
| Heparin | GBBGB, where “B” is a base residue | Type I Collagen, Prokaryotic collagen-like protein, & Synthetic collagen peptide | Capila and Linhardt 2002 |
| GRPGKR GKQGQK, RGTPGK PGPRGQ RGPTGP RGERGPR, GRKGR, GKRGK | Sweeney et al., 1998 | ||
| Peng et al., 2014 | |||
| Ricard-Blum et al., 2006 | |||
| Parmar et al. (2016a) | |||
| Heparan-Sulfate | KGHRGF, RGTPGK PGPRGQ RGPTGP RGERGPR | Type I Collagen & Synthetic collagen peptide | Sweeney et al., 2008 |
| Capila and Linhardt 2002 | |||
| Sweeney et al., 1998 | |||
| Ricard-Blum et al. (2006) | |||
| Leukocyte associated Ig-like receptor-1 (LAIR-1) | GAOGLR GGAGPO GPEGGK GAAGPO GPO | Synthetic collagen peptide | Lebbink et al., 2009 Brondijk et al. (2010) |
| Heat Shock Protein-47 (HSP47) | xGxRG | Synthetic collagen peptide | Koide et al. (2006) |
| Fibromodulin | KGHR | Synthetic collagen peptide | Di Lullo et al., 2002 |
| Kalamajski et al. (2016) | |||
| Trypsin-1 | Cleaves after lysine or arginine, except when followed by proline | Human collagen, 90% Type I and 10% Type III | Mirigian et al., 2013 |
| Only works on non-triple helical collagen | Simpson (2006) | ||
| Elastase 2 | KLK ∼ AR, FVR ∼ NK, And GDR ∼ GL as found in Type XVII Collagen, GPLGIA GITGAR GLA in Type III Collagen | Full-length Type XVII Collagen† & recombinant Type III Collagen | Lin et al., 2012 Williams and Olsen (2009) |
| Matrix Metalloproteinase-1 (MMP-1) | GPQG ∼ LA GQRGIV GLP | Prokaryotic collagen-like protein, Full-length Type I Collagen | (Birkedal-Hansen et al., 1993; Yu et al., 2012) |
| MMP-2 | GPQG ∼ LA GQRGVV GLP | Full-length Type I Collagen | Sweeney et al., 2008 |
| Birkedal-Hansen et al. (1993) | |||
| MMP-8 | GPQG ∼ LA GQRGVV GLP | Full-length Type I Collagen | Sweeney et al., 2008 |
| Birkedal-Hansen et al. (1993) | |||
| MMP-13 | G ∼ LAGQR GIVGLO GQRGER, GLOGER GRTGPA GAAGAR | Synthetic collagen peptide | Howes et al. (2014) |
| Prolyl-4-hydroxylase (P4H) | “X”PG, with “X” being preferred to be Proline | Synthetic collagen peptide | Rapaka et al., 1978 |
| Kivirikko et al., 1972 | |||
| Gorres and Raines (2010) | |||
| von Willebrand Factor (VWF) | RGQOGV MGF | Synthetic collagen peptide | Lisman et al. (2006) |
| Nidogen | Can bind 80 nm away from the C-terminus of Type IV Collagen, and with the ectodomain of Type XIII Collagen | Full-length Type IV Collagen and recombinant Type XIII Collagen ectodomain | Aumailley et al., 1989 |
| Tu et al. (2002) |
The stability of the triple helix structure in different regions of the recombinant bacterial collagen can be controlled by choosing triplets with different predicted melting temperature (Persikov, Ramshaw, and Brodsky 2005). This change in stability of recombinant bacterial collagen can be used to alter the affinity of various interactions (Persikov, Ramshaw, and Brodsky 2005). The nature, number and spacing of specific binding motifs can also be easily manipulated in the recombinant bacterial collagen sequence (Peng et al., 2014). When heparin and integrin binding sites were inserted into blank slate Scl 2.28 protein, recombinant bacterial collagen binding to both proteins could be observed (Peng et al., 2014). This strategy can be used to replicate a collagen sequence that has multiple binding partners and will allow determination of competition for binding to the collagen sequence between multiple ECM components.
While recombinant bacterial collagens as a tool provide a great opportunity to learn more about collagen interactions with other molecules, it has some drawbacks. Recombinant bacterial collagens cannot perfectly replicate eukaryotic collagen yet. The recombinant bacterial collagens produced in E. coli do not have the same post-translational modifications as eukaryotic collagen. The lack of collagen monomer cross-linking prevents the formation of larger superstructures found in eukaryotes (Canty and Kadler 2002; Gelse, Pöschl, and Aigner 2003). Additionally, prokaryotic collagen-like proteins are homotrimers, therefore, studying interactions of heterotrimeric eukaryotic collagens remains a challenge.
Some of these issues are currently being addressed, such as the lack of hydroxyproline within prokaryotic collagen-like proteins (Buechter et al., 2003; Pinkas et al., 2011; Schnicker and Dey 2016). Schnicker and Dey expressed B. anthracis prolyl-4-hydroxylase enzyme in E. coli (and purified) to hydroxylate (P-P-G)5 and (P-P-G)10 peptides (Schnicker and Dey 2016). While hydroxylation of the peptides using B. anthracis prolyl-4-hydroxylase was observed, both the X-position and Y-position prolines were hydroxylated with the Y-position prolines being favored, which might not replicate the exact eukaryote proline hydroxylation (Schnicker and Dey 2016). Using a different approach, Buechter et al. produced a fragment of Type III collagen with hydroxyproline in E. coli by supplementing growth media with hydroxyproline and sodium chloride. However, this method does not discriminate in X and Y positions in Gly-X-Y tripeptide (Buechter et al., 2003). Pinkas et al. created a strain of E. coli that expresses human prolyl-4-hydroxylase enzyme and allows for the conversion of α-ketoglutarate into l-ascorbate (Pinkas et al., 2011). Using this strain, the authors demonstrated that the proline residues within the P-P-G peptide could be converted into hydroxyproline but less hydroxylation was observed with longer (P-P-G) repeats (Pinkas et al., 2011). While great strides have been made to produce recombinant bacterial collagen with hydroxyproline in E. coli, future work is needed to produce longer recombinant bacterial collagen segments with incorporated hydroxyproline at Y position with consistent results.
Heterotrimeric collagens have not been as well researched as homotrimeric vertebrae collagens. It is difficult to achieve the correct stoichiometric ratio in solution with both synthetic peptides and recombinant collagen. Moreover, individual strands in the heterotrimer must be placed in the correct stagger (Figure 1). Drs. Boudko and Bachinger generated heterotrimeric recombinant collagen in E. coli using segments of α1 and α2 chains of Type 1 collagen by using the second non-collagenous domain (NC2) from Type IX Collagen to control trimerization (Boudko and Bächinger 2016; 2012). Stagger of the recombinant collagen could be controlled by using the 35 residue long NC2 domain of Type IX Collagen and led to the desired chains being in the leading, middle and lagging positions (Boudko and Bächinger 2016).
Novel Biomaterials: Bioactive Hydrogels Based on Prokaryotic Collagen-Like Proteins
Collagen, the most abundant protein in the body, has been used as scaffolds for regeneration and repair of a variety of tissues, such as skin (Carver et al., 1995; Brown et al., 2002; Helary, Zarka, and Giraud-Guille 2012; Shokrgozar et al., 2012), cartilage (Li et al., 2020), bone (Salasznyk et al., 2004), vasculature (Stamati et al., 2014; Copes et al., 2019), and neural tissues (Suri and Schmidt 2010). Despite its broad use, mammalian collagens have significant limitations as scaffolds. As an animal-derived product, there is significant batch-to-batch variability due to harvest techniques, post-processing procedures, and storage conditions. Mammalian collagen products also suffer from a short product shelf life, diminishing bioactivity after processing, and potential immunogenicity (Cao and Xu 2008; Schmidt et al., 2016; Dhavalikar et al., 2020). Despite the aforementioned limitations, collagens contain substantial regenerative potential due to its native cell interactions, cell-responsive degradation, and tunable properties (Antoine et al., 2014). As such, there is an urgent need to find a replacement biomaterial that can provide these cellular cues while circumventing the limitations of current collagen scaffolds.
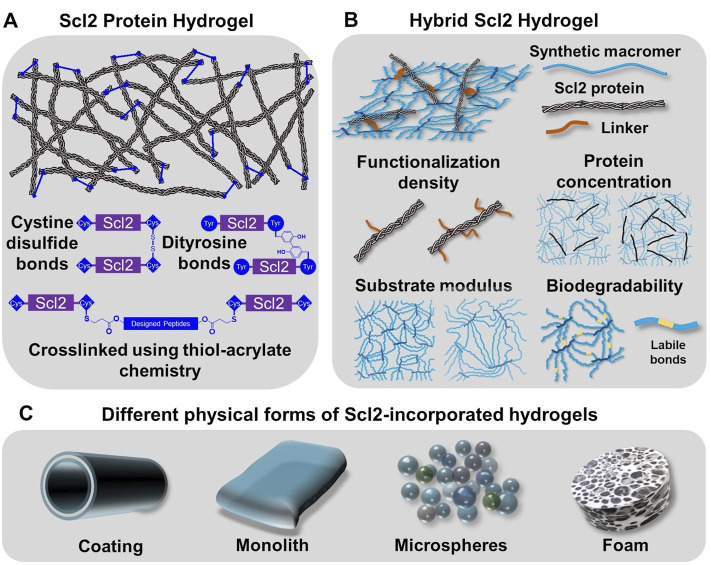
Previous sections have introduced the discovery of prokaryotic collagen-like proteins and the structural and functional resemblance to collagen that make it a strong candidate for bioengineering applications. Recombinant expression of prokaryotic collagen-like proteins provides a manufacturing process with minimal batch variability and scale-up potential. In addition, these prokaryotic collagen-like proteins generally have no inherent binding motifs for integrins and other extracellular matrix molecules which provides a unique opportunity for the engineering of specific cellular responses (Humtsoe et al., 2005; Kim et al., 2005; Xu et al., 2002). Selective insertion of multivalent binding sites for integrins and other molecules into the recombinant bacterial collagen has been used to generate targeted cell interactions to guide regeneration. In this section, we discuss strategies to design hydrogels based on recombinant bacterial collagen in biomedical applications, namely the Scl2.28 protein and Scl2 proteins derived from it (details included in the Supplementary Table S1). First, modification of Scl2 proteins to form hydrogels will be described. Hybrid Scl2 protein hydrogels will then be reviewed with the discussion of the effect of different fabrication parameters on the resultant cell-material interactions. Finally, the fabrication of the hybrid Scl2 hydrogels into different forms of hydrogel products will be described to demonstrate the versatility of this biomaterial platform (Figure 3).
FIGURE 3.
Incorporation of Scl2 proteins into hydrogels as alternative materials to animal-derived collagen matrices. (A) Scl2 protein hydrogel network with varied crosslinking modalities for enhanced gel stability. (B) Hybrid Scl2 hydrogels with the Scl2 protein anchored into a synthetic hydrogel network via a functionalization linker and the different parameters used to modulate hydrogel properties. (C) Different physcial forms of hybrid Scl2 hydrogels used in biomedical applications.
Scl2 Protein Hydrogels
Collagen matrices are typically crosslinked to form a functional gel and tune its physical properties and resorption rate. Given the similarities in structure and functional residues, prokaryotic collagen-like proteins like Scl2 proteins have the potential to form hydrogel network via the same chemical or enzyme-induced crosslinking methods. However, Peng et al. found that after freeze-drying and treatment with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) for chemical crosslinking, Scl2 protein-based hydrogel sponges were unstable in water and resulted in structural disintegration (Peng et al., 2010). It was hypothesized that the instability of the EDC-crosslinking hydrogel was attributed to the smaller molecular weight of Scl2 protein as compared to collagen and fewer functional residues. A glutaraldehyde vapor was then applied as an alternative crosslinking mechanism. Although this generated stable hydrogels, there was limited control over crosslink density and hydrogel properties (Peng, Glattauer, and Ramshaw 2017). To resolve this problem, researchers introduced peptide sequences with functional residues to create additional crosslinking sites and stabilize the hydrogel network (Stoichevska et al., 2016; Stoichevska et al., 2017). Stoichevska et al. proposed introducing chemically functional residues to Scl2 proteins and forming crosslinks via oxidation (Stoichevska et al., 2016; Stoichevska et al., 2017). As shown in Figure 3A, cysteine or tyrosine residues were introduced to either or both C-terminal and N-terminal of the collagen-like domain. The hydrogel crosslinking was designed to form when cysteine residues form disulfide bonds or adjacent tyrosine residues form dityrosine bonds via photo-initiated oxidation. In this way, the collagen-like domain was unmodified and the specific binding sites for cell attachment via integrins and interactions with other extracellular matrix molecules were preserved. The results demonstrated that the tyrosine crosslinking resulted in gel formation while cysteine-based disulfide bonding did not form a hydrogel, although it significantly increased the number of intramolecular crosslinks (Winterbourn et al., 2004). This work also suggested different roles with intramolecular crosslinks forming small soluble aggregates and intermolecular crosslinks leading to hydrogel network formation. Similarly, Parmar et al. modified both the C- and N-termini of Scl2.28 proteins with an additional sequence, GGPCPPC (Parmar et al., 2016a). The cysteine residue was then designed to react with two different types of peptides that contained substrates for either matrix metalloproteinase 7 (MMP7) or aggrecanase. The linkage between the Scl2.28 proteins to these peptides was formed via thiol-acrylate “click” chemistry that required only mild reaction conditions and preserved protein bioactivity. This gave rise to a stable Scl2 protein network with MMP7- or aggrecanase-cleavable bonds, allowing for cell-responsive degradation of the hydrogel product after implantation.
In summary, typical collagen crosslinking methods are insufficient to make Scl2 protein hydrogels without additional modification of the protein to increase crosslinking sites due to its reduced length as compared to collagen. The incorporation of enzymatically labile sequences in the crosslinkers provides a well-defined hydrogel platform of Scl2 protein with high adaptability and an adjustable degradation profile. Although this provides a promising biomaterial platform, many of these studies did not demonstrate the ability of tuning the protein hydrogel mechanical properties. Furthermore, the use of enzymatically labile peptides produced by solid-phase synthesis reduces the cost-effectiveness of these protein hydrogels and creates challenges in large-scale production.
Hybrid Scl2 Hydrogels
To expand the available physical properties to Scl2-based hydrogels, researchers created hybrid Scl2 hydrogels by incorporating Scl2 proteins into a synthetic hydrogel network (Cosgriff-Hernandez et al., 2010). Cosgriff-Hernandez et al. reported the first Scl2 protein functionalization method and incorporation into a hydrogel network to study the effect of different binding motifs for integrins α1β1 and α2β1 (Cosgriff-Hernandez et al., 2010). Cell culture studies with native and modified C2C12 cells demonstrated that an Scl2 protein modified with a collagen-derived GFPGER binding motif interacted with both α1β1 and α2β1 integrins; whereas, an Scl2 protein modified with a unique GFPGEN binding motif interacted only with α1β1 integrins. Endothelial cells adhered to and spread on both Scl2 proteins, but smooth muscle cells did not adhere to Scl2 proteins without the α2β1 integrin interaction (Cosgriff-Hernandez et al., 2010). In addition to selective cell interactions, these hybrid Scl2 hydrogels also provide a unique opportunity to decouple the Scl2 ligand presentation from other gel physical properties such as matrix stiffness and resorption rate. Scl2 proteins are first functionalized with a polyethylene glycol (PEG)-based linker that contains an acrylate or acrylamide group that allows it to be anchored into a PEG-based hydrogel (Figure 3B) (Cosgriff-Hernandez et al., 2010). PEG-based hydrogels have been widely used in numerous biomedical applications, such as cell-laden scaffolds, wound dressings, implant coatings, and interstitial tissue substitutes. The antifouling function of PEG hydrogels limits non-specific protein adsorption such that the protein presentation to cells is limited to the Scl2 protein (Chen et al., 2019). The insertion of Scl2 protein has minimal effect on the network formation of the polymeric hydrogel, allowing for the modulation of ligand presentation independent of substrate bulk properties. The following sections detail the effect of hydrogel properties on cell-material interactions.
Ligand Concentration and Protein Functionalization Density
To understand the effect of ligand concentration and protein functionalization density on the cell-material interactions, Browning et al. compared cell behavior at different Scl2-2 protein (Supplementary Table S1) concentrations (Browning et al., 2014) and varying PEG linker densities on the Scl2 protein backbone (Browning et al., 2013). As the protein loading concentration increased, cell adhesion, proliferation, migration rate, and spreading increased at differing rates. By controlling the molar ratio of the linker to lysine residues to 0.1:1, 0.5:1, and 1:1, Scl2 proteins modified with integrin-binding motif GFPGER with varied functionalization densities were prepared and then incorporated into PEG diacrylate hydrogels. There was no significant difference in concentration of functionalized Scl2 proteins on swelling and mechanical properties of hydrogels. The initial incorporation and retention of the protein in the hydrogel was monitored for 6 weeks. The results demonstrated a decrease in protein content for all the functionalization densities with the proteins at the lowest functionalization density displaying the greatest protein loss (Browning et al., 2013). This was attributed to the hydrolytic degradation of the acrylate ester bonds and fewer linkages to the hydrogel network. Notably, reduced functionalization significantly improved cell adhesion and spreading to the bioactive hydrogels despite the reduced ligand concentration (Browning et al., 2013). It was hypothesized that the linker created steric hindrance around the ligand and reduced integrin binding. To overcome the steric hindrance caused by protein functionalization, Cereceres et al. redesigned the Scl2 protein to reduce steric hindrance to the integrin-binding site, GFPGER, by moving the reactive residues into the globular region and away from the integrin-binding sites (Cereceres et al., 2015). This newly engineered Scl2 protein hydrogel displayed a higher cell adhesion as compared to the original bioactive hydrogel.
Substrate Modulus and Degradation Kinetics
Substrate stiffness has been identified as a key factor in directing cell adhesion, migration, and differentiation (Engler et al., 2006; Hynes 2009; O’Brien 2011; Lu, Weaver, and Werb 2012). Given this established mechanoresponsive behavior, cell behavior was characterized on Scl2 hydrogels of two different substrate moduli as controlled by PEG macromer molecular weight (10 vs 3.4 kDa) (Browning et al., 2014). Endothelial cell adhesion, migration, and spreading increased significantly on gels with a higher modulus. Unlike collagen hydrogels that often couple substrate stiffness and ligand presentation (e.g., increasing collagen concentration), the substrate modulus of these hydrogels is tunable independent of variations in protein functionalization and loading concentration (Browning et al., 2014). This uniquely permits the study of individual roles of substrate modulus and ligand presentation on integrin-mediated focal adhesion complex formation and the corollary effect on cell behavior.
In addition to substrate stiffness, these hybrid gels also offer a broader range of degradation kinetics to meet the needs of different applications. For example, a wound hydrogel scaffold can be designed to degrade to complement the rate of regeneration; whereas a hydrogel coating of cardiovascular devices should preserve the structure and functions over the life of the device (10–20 years) to provide long-term efficacy. Browning et al. developed a non-degradable hydrogel network of PEG-diacrylamide that replaced the hydrolytically labile ester bonds of PEG diacrylate (Browning et al., 2013). To increase the Scl2 protein retention in the hydrogel, a new biostable linker was designed to eliminate hydrolytically labile groups. This biostable linker markedly increased the retention of the protein in the hydrogel over 6 weeks while maintaining equivalent bioactivity. Researchers have also developed a degradable PEG-based hydrogel network for regenerative applications (Cereceres et al., 2015). In this work, more labile thio-β ester bonds were incorporated into the hydrogel network. Compositional control was used to generate a range of different degradation rates from days to months without changing the hydrogel bulk properties such as swelling or modulus. The degradable linkage can also be utilized to release the incorporated Scl2 protein locally.
Hydrogel Fabrication
In addition to the hydrogel chemistry variables described above, Figure 3C illustrates some commonly utilized fabrication methods to generate hydrogel slabs, coatings, microspheres, and foams. Most commonly, PEG-Scl2 hydrogels (PEG-based hydrogels containing Scl2 proteins) are formed in molds via photopolymerization to generate bulk hydrogel slabs, tubes, or sheets (Cosgriff-Hernandez et al., 2010; Becerra-Bayona et al., 2018). Browning et al. developed a multilayer vascular graft by coating an electrospun polyurethane mesh with PEG-Scl2 hydrogels via UV-initiated polymerization with a tubular mold (Browning et al., 2012). However, difficulty in fabricating thin coatings and uneven coverage of the hydrogel on the mesh surface constrained the clinical application of the graft. Wancura et al. developed a new hydrogel coating fabrication method using redox-initiated polymerization and diffusion-mediated crosslinking (Wancura et al., 2020). The authors achieved the formation of uniform hydrogel coating layers with controlled thickness. In addition, this fabrication method allowed for the formation of multilayer hydrogel with distinct features and coating of the hydrogel on devices of complex structures and materials. PEG-Scl2 hydrogel microspheres were fabricated via a co-fluidics method to generate injectable wound dressings (Cereceres et al., 2015). Finally, Scl2 protein hydrogel sponges were fabricated and used to deliver hyaluronic acid and chondroitin sulfate and temporally control the sponge degradation to induce chondrogenesis for the articular cartilage repair (Parmar et al., 2016b).
In summary, Scl2 protein and hybrid PEG-Scl2 hydrogels have shown great potential to replace animal-derived collagen matrices. As a new biomaterial platform, Scl2 proteins offer significant advantages in selective cell interactions and tunable physical properties and forms. The next section will elaborate on the specific applications of these Scl2-based hydrogels in the regeneration and repair of different tissues.
Application of SCL2 Proteins as Integrin-Targeting Materials
A key goal in biomaterial design for regenerative applications is directing cellular behavior. Integrin-targeting biomaterials provide a mechanism to mimic native cell-extracellular matrix interactions (Dhavalikar et al., 2020). Various extracellular matrix-derived proteins or peptide sequences have been utilized to target and bind specific integrins to direct cell proliferation, migration, and differentiation. The directed cell responses can facilitate numerous tissue repair processes, including ECM reorganization, angiogenesis, and tissue remodeling, to achieve improved and accelerated tissue repair or regeneration. Insertion of different binding motifs into the Scl2 protein backbone has been used to achieve selective integrin interactions (Stoichevska et al., 2016; Peng et al., 2014; An et al., 2016; 2013; 2014). As compared to collagen and other protein-based biomaterials that have multiple cellular interactions, these Scl2 proteins can be utilized to target specific cell behavior. In addition to providing improved medical devices, these new biomaterials also provide new tools to elucidate complex regenerative processes.
In this section, we will review recent studies applying these integrin-targeting materials based on Scl2 proteins to clinical applications. This includes thromboresistant coatings of cardiovascular devices, scaffolds for bone regeneration, chronic wound dressings, and matrices for cartilage repair.
Thromboresistant Coatings for Cardiovascular Devices
Sustained thromboresistance has been a challenge in the field of cardiovascular devices that continues to limit clinical outcomes. The integrin-targeting nature of Scl2 proteins provides an opportunity to target endothelial cell interactions while limiting platelet attachment and activation. The hybrid Scl2 hydrogel combines the antifouling nature of PEG hydrogels for acute thromboresistance with the cell selectivity of the Scl2 proteins to promote endothelialization for sustained thromboresistance. Browning et al. investigated the ability of these hybrid Scl2 hydrogels to direct endothelial cell interactions (Browning et al., 2012; Browning et al., 2013; Browning et al., 2014). In these studies, researchers modulated protein-incorporated hydrogels by tuning the ligand concentration, adjusting protein functionalization density, changing the substrate stiffness, and constructing a biostable hydrogel network and functionalized linkage. Endothelial cell adhesion, spreading, and migration were then characterized, showing that the cell-material interaction was significantly affected by the accessibility and number of binding motifs on the Scl2 proteins and the bioactivity retention of proteins. Furthermore, PEG-Scl2 hydrogels showed minimal levels of platelet adhesion and activation indicating its thromboresistance (Browning et al., 2011).
To further understand the hemostatic regulation by different integrin-binding interactions, Post et al. fabricated bioactive hydrogels with different sets of integrin binding sites by incorporating different proteins, including Scl2GFPGER, collagen, and gelatin (Post et al., 2019). Combining the results of the flow cytometry and the human umbilical vein endothelial cell (HUVEC) attachment quantification after blocking integrins with antibodies, the researchers were able to demonstrate the specific integrin-binding sites of different proteins. The platelet activation study showed that the PEG-Scl2 hydrogel was the least pro-thrombotic and most anti-thrombotic among varied types of hydrogels, suggesting that GFPGER-binding integrins α1β1 and α2β1 supported a more anti-thrombotic HUVEC phenotype (Post et al., 2019). Platelet adhesion and activation results then verified that fewer platelets adhered to PEG-Scl2 hydrogel than the other types of protein-incorporated hydrogels. Additionally, HUVECs on PEG-Col hydrogel (PEGDA hydrogel incorporating natural collagen)and PEG-Scl2 hydrogel demonstrated similar PAC-1 expression levels to the PEGDA blank hydrogel that stimulated the minimal platelet activation (Post et al., 2019). These results suggest that integrins α1β1 and α2β1-mediated HUVEC responses can reduce the platelet adhesion and activation, and biomaterials targeting these integrins can potentially avoid deleterious coagulation cascade, thrombus formation, and vascular occlusion as previous studied (Radke et al., 2018). This highlighted the important roles of integrin-binding interaction and provided guidelines for biomaterial design, including the spatial and temporal exposure of specific integrin-binding sites to orchestrate target cellular processes at different tissue regenerative stages.
The integrin-binding ligand not only affects the adhesion, proliferation, and migration of human aortic endothelial cells, but also makes a difference in endothelial cell differentiational markers of VE-cadherin, PECAM-1, NOS3, TM, and E-selectin (Munoz-Pinto et al., 2015). These markers are correlated to the endothelization processes, such as the formation of the tight junction and the establishment of a confluent monolayer of cells. Both PECAM-1 and VE-cadherin expression and protein production levels of the endothelial cells cultured on the PEG-Scl2 hydrogels were not significantly different from that of PEG-Collagen hydrogels. The protein production of TM and E-selectin demonstrated no variance between hydrogels with the Scl2 proteins and collagens. However, PEG-Scl2 hydrogel did not induce the same expression level of NOS3 as the PEG-collagen hydrogel, which is significant for the vascular homeostasis. This was due to the other various binding sites on the natural collagen molecule that can promote the NOS3 expression and production. Still, the versatility of Scl2 proteins allowed for further modification that can implement the same effect on this specific marker expression, such as insertion of heparin-binding site. The studies were then extended to more endothelial cell phenotypes, that were endothelial progenitor cells (EOCs) and human umbilical vein endothelial cells (HUVECs) (Munoz-Pinto et al., 2015). Both gene expression and protein production results demonstrated a similar or improved endothelial maturation level of EOCs cultured on PEG-Scl2 hydrogel compared to that cultured on PEG-Collagen hydrogel. The PEG-Scl2 hydrogel had a similar or improved EOC homeostatic marker expression to the PEG-Collagen hydrogel. These results supported further exploration of PEG-Scl2 hydrogel as an advanced thromboresistant vascular graft coating. An increase in integrin binding sites of the protein significantly improved the cell adhesion and retention on the hydrogel substrate while maintaining a relatively high cell migration rate (Quiroz et al., 2018). EOCs cultured on PEG-Scl2 hydrogel with three integrin-binding sites displayed improved gene expression and protein production levels of intermediate differentiation markers compared to that of PEG-Collagen hydrogels. This work indicated that PEG-Scl2 hydrogels have the potential to improve endothelial cell differentiation with amplified integrin-mediated interactions when the cell adhesion, retention, and migration rate were suitable for the formation of a cellular monolayer.
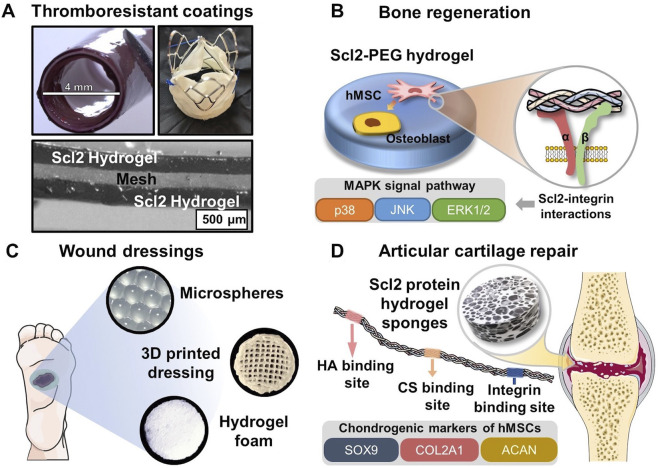
Overall, these studies demonstrate that the incorporated Scl2 proteins can induce targeted integrin-mediated interactions with different types of endothelial cells to promote cell adhesion and migration, as well as hemostatic phenotypes. As shown in Figure 4A, the well-designed thromboresistant Scl2 protein hydrogel can be coated on the surface of cardiovascular grafts and heart valves with customized bioactivity and potential for sustained thromboresistance.
FIGURE 4.
Biomedical applications of Scl2-containing biomaterials with targeted integrin interactions. (A) PEG-Scl2 hydrogel coatings of cardiovascular devices to enhance thromboresistance; (B) PEG-Scl2 hydrogel that revealed integrin-mediated hMSC osteogenic differentiation via the MAPK signal pathway (C) Different types of dressings containing Scl2 proteins to enhance chronic wound healing; (D) PEG-Scl2 protein hydrogel with binding sites for chondroitin sulfate and hyaluronic acid to improve cartilage repair.
Scaffolds for Bone Regeneration
Collagen I has been widely shown to support mesenchymal stem cell (MSC) osteogenic differentiation in both two-dimensional (2D) and three-dimensional (3D) environments (Donzelli et al., 2007; Chamieh et al., 2016; Chan et al., 2010; Lund et al., 2008; Schneider et al., 2010; Chiu et al., 2014; Somaiah et al., 2015). As such, the application of collagen-mimetic proteins to bone scaffold design has arisen. In Becerra-Bayona et al., Scl2 proteins were used to elucidate the impact of individual collagen motifs on associated cellular responses (Becerra-Bayona et al., 2018). As shown in Figure 4B, human MSCs (hMSCs) were cultured within PEG hydrogels containing either Scl2GFPGER or Scl2GFPGEN in the absence of osteogenic supplements. PEG-Scl2GFPGER gels were associated with increased hMSC osterix expression, osteopontin production, and calcium deposition relative to the PEG-Scl2GFPGEN gels (Becerra-Bayona et al., 2018). Results indicated that integrin α2 signaling may have an increased osteogenic effect relative to integrin α1. Since integrin α1β1 and α2β1 both trigger the MAPK pathway, ERK, JNK, and p38 activation were also assessed to evaluate potential mechanisms underlying observed responses. Inhibition results suggested that p38 activity triggered by integrin α2β1 but not α2β1 plays a key role in collagen-supported hMSC osteogenesis (Becerra-Bayona et al., 2018). Similarly, Reyes et al. found that surfaces coated with triple helical GFOGER (analogous to Scl2GFPGER but without the flexibility afforded by recombinant expression) induced similar levels of expression of osteogenic markers runx2, osteocalcin, and bone sialoprotein by MC3T3-E1 as native collagen I (Reyes and García 2004). These triple helical GFOGER peptide coatings have further been passively adsorbed onto polymeric scaffolds and have also been found to significantly accelerate and increase bone formation in non-healing femoral defects compared to uncoated scaffolds and empty defects (Wojtowicz et al., 2010). Further studies with the more versatile Scl2 proteins will allow for improved design of collagen-based scaffolds.
Chronic Wound Dressings
Natural collagen has been applied to different skin substitute products or wound dressings due to its compatibility and bioactive cues (Singer and Clark 1999). As an advantageous replacement for natural collagen, Scl2 proteins with binding sites of integrins α1β1 and α2β1 can direct keratinocyte migration and proliferation, ECM deposition, and wound contraction in the later stage of wound healing. As discussed in the previous section, Cereceres et al. redesigned Scl2 protein to have the integrin-binding motif, GFPGER, away from functionalization sites (Cereceres et al., 2015). By characterizing the relative α1 integrin-domain binding, the authors were able to prove that the relocation of integrin binding sites significantly reduced the steric blocking by the protein functionalization. As a result, the redesigned eColGFPGER hydrogel demonstrated a critical increase in fibroblast adhesion compared to the original Scl2GFPGER hydrogel, suggesting increased integrin-mediated interactions. The well-defined hydrogel platform with the tunable presentation of integrin binding sites allows for a comprehensive investigation of the roles of different integrin-mediated signal pathways in wound healing. This contains a group of integrins that have been identified to influence different stages of wound healing, including formerly mentioned α1β1 and α2β1, as well as α3β1 and α11β1 (Koivisto et al., 2014; Larjava et al., 1996; Liu et al., 2010; Longmate and DiPersio 2014; Choma et al., 2007; Thannickal et al., 2003; Dhavalikar et al., 2020). Also, wound dressings of different forms (Figure 4C), including hydrogel microspheres, 3D-printed hydrogel dressing with hierarchical, and highly porous hydrogel foam, can be employed to deliver the Scl2 protein to the wound bed while providing moisture balance for improved healing potential.
Matrices for Cartilage Regeneration
A series of studies were conducted by Parmar et al. on developing Scl2 protein sponges with inserted binding sites for hyaluronic acid (HA), chondroitin sulfate (CS), heparin, or integrin (Figure 4D) (Parmar et al., 2016a; Parmar et al., 2016b; Parmar et al. 2017). Specifically, the researchers introduced different cleavage linkages to the protein hydrogel sponge network to achieve temporal control over the hydrogel degradation, modulating the chondrogenesis of hMSCs (Parmar et al., 2016a). The results indicated that groups with a ratio of MMP7-cleavable and aggrecanase-cleavage linkages at 25:75 and 50:50 significantly upregulated chondrogenic expression levels, rendering the highest production levels of sulfated glycosaminoglycan (sGAG) and collagen. Additionally, the hMSC-laden protein hydrogel sponges of these two groups maintained their mechanical integrity and strength after 6 weeks, which was never seen in other degradable cartilage scaffold work. Another study from the group investigated the effect of HA and chondroitin sulfate binding site ratio on cell responses and chondrogenesis (Parmar et al., 2016b). In vitro characterization identified that the hydrogel foam groups with HA-Scl2 and HA:CS (75:25)-Scl2 proteins (Supplementary Table S1) had drastically upregulated expression levels of chondrogenesis markers COL2A1, ACAN, and SOX9 with greatest sGAG and collagen production. Importantly, these works have provided a versatile Scl2 protein hydrogel platform with tunable binding sites and adjustable degradation profiles, demonstrating potential applications in a broader tissue engineering context. However, although the integrin-binding sites were inserted in the employed Scl2 protein, the authors had not identified the integrins of interest in chondrogenic differentiation of hMSCs. Other research indicated that β1 integrins, including α1β1, α2β1, and α3β1 of collagen receptors, were activated during chondrogenic differentiation (Takahashi et al., 2003; Goessler et al., 2008). Thus, a future direction of Scl2 protein hydrogel application in articular cartilage repair can be the investigation of different integrin-mediated cell behaviors by Scl2 proteins with specific binding sites.
This section reviewed several applications of Scl2 protein hydrogels in repairing and regenerating damaged tissues of the vasculature, bone, skin, and articular cartilage. With highly tunable mechanical properties and customized bioactivity, these Scl2-based hydrogels demonstrated the versatility of this new biomaterial platform. Furthermore, this novel class of biomaterials is capable of extending our understanding of the biological processes in cellular behaviors and tissue recovery and guiding the future biomaterial design for more advanced solutions in regenerative medicine.
Current Challenges and Future Perspectives
The selective insertions of specific integrin-binding motifs into Scl2 proteins provided numerous opportunities for advancing the understanding of integrin-ligand interactions, integrin-mediated signal pathways, and resultant cellular behaviors. Native collagens have a collection of integrin binding sites that can be countered, compounded, or have synergetic effects after interacting with specific integrins. With no inherent binding motif, Scl2 proteins can be inserted with one or more types of ligands as designed. Previous studies have utilized engineered Scl2 proteins to facilitate the investigation of multiple critical integrin-mediated cell behaviors in bone regeneration, endothelialization, and hemostatic regulation with vascular devices (Browning et al., 2012; Becerra-Bayona et al., 2018; Post et al., 2019; Wancura et al., 2020). Moreover, isolated fundamental integrin-ligand interactions in re-epithelialization, ECM redeposition, and wound contraction demand much attention and further probing for advanced wound healing modalities. It is also necessary to identify and isolate fundamental integrin interactions that play central roles in the chondrogenic differentiation of hMSCs and the essential glycosaminoglycan network reconstruction. Researchers have also discovered that varied types of cells express distinct sets of integrins, and these cells demonstrate different attachment affinities to other binding motifs (Cosgriff-Hernandez et al., 2010; Post et al., 2019). Hence, Scl2 proteins with inserted binding motifs allow for possible mapping of the relative affinities between different combinations of integrins and binding motifs in the context of multiple cells active in a particular circumstance or different cell phenotypes during differentiation. Substrates coated with recombinant Scl2 proteins can select cells from a co-culture system or target cells at defined differentiation stages. Regardless, recombinant bacterial collagens with inserted integrin-binding sites offer the opportunities to compare different integrin-ligand binding affinities, revealing the roles of isolated integrin-ligand interactions in the given tissue engineering field guide the biomaterial design for enhanced clinical outcomes.
The previous sections show examples of Scl2 proteins to develop biomaterials that guide integrin-mediated cellular activities for advanced tissue engineering. Therefore, the ligand presentation to the target cell population is essential for biomaterial design. It has been demonstrated that a change in the integrin ligand number of Scl2 proteins could result in different cell adhesion, migration rate, and marker expression levels (Quiroz et al., 2018). Also, the designed incorporation of recombinant bacterial collagens into highly tunable synthetic polymer hydrogels enabled the improvement and modulation of ligand exposure to the integrins (Cosgriff-Hernandez et al., 2010; Browning et al., 2011; Browning et al., 2013; Browning et al., 2014). However, cells interact with all the components of their surrounding environment dynamically, so spatial and temporal controls of specific integrin interaction play pivotal roles in understanding cell behaviors and directing tissue engineering. At the subcellular level, grouping multiple ligands together in islands can effectively promote integrin clustering and amplify targeted cell responses. The substrate mechanical properties, surface topography, distribution, and density of ligands can also be modulated to render different integrin-ligand binding modalities, leading to cell adhesion, migration, differentiation, and ECM production changes. In addition, there is a gap between 2D and 3D cell culture systems as 3D culturing has increased surface area for integrin binding and mimics the cell native niche with a dynamic interface. Integrin clusters are more prominent with longer lifetimes in 3D environments (Lepzelter et al., 2012). Cutting-edge nanopatterning techniques allow for a refined design of the biomaterial surface with a much higher resolution and versatile geometries and demonstrate a great promise in controlling the ligand accessibility to targeted cells. Substrates with temporally controlled degradation profiles induced by hydrolysis or enzyme cleavage potentiate the sequential exposure of different integrin-binding sites on the embedded Scl2 proteins to modulate specific cell behaviors in different regenerative stages. Biomaterial design and development have been advanced by the synthetic polymer science of growing versatility and continuously progressing fabrication techniques. However, there are remaining critical challenges in translating in vitro findings to in vivo results and applying the materials or devices to damaged tissues. For evaluating the effect of a chosen integrin interaction with its binding site, transgenic mice with ablation or deletion of targeted integrin genes have been studied in vivo. However, they are limited by unknown outcomes caused by mutagenesis. Also, fundamental biological factors must be considered in the context of a dynamic in vivo environment. Key players, including multiple other cells, plenty of growth factors, and remodeling ECM, can engender ambiguous or unreliable conclusions by compensating or competing with the ligands presented by the biomaterial. Researchers must make scrupulous assumptions and analyze the outcomes critically to avoid false conclusions in elucidating the roles of integrins in the cell response to designed biomaterials.
Animal-derived collagens have been widely used in biomedical applications due to their biocompatibility, biodegradability, and easy accessibility. However, its use has been hindered by batch-to-batch variability and limited potential for production scaling-up. The discovery of prokaryotic collagen-like proteins has provided opportunities to meet natural collagens’ structural and functional needs in various applications while improving production rates and reducing product variability. With selectively inserted binding sites, prokaryotic collagen-like protein has been shown in multiple publications to interact with extracellular matrix molecules and bind to targeted integrins, leading to appropriate cellular responses. This technology provides a versatile platform to investigate collagen binding sites, characterize their interaction with extracellular matrix proteins, elucidate specific cellular responses to the prokaryotic collagen-like protein-incorporated material, and guide the design of advanced biomaterials for tissue engineering.
Acknowledgments
The authors would like to thank Dr. Lakshmi Prasad Dasi and Sanchita Bhat from the Georgia Institute of Technology for their help in providing the image of thromboresistant coatings for cardiovascular devices.
Author Contributions
JP, ZL and MHa wrote the manuscript. JP and ZL contributed to the manuscript equally. SA, MG, EC-H and MHo edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funds from the Hamill Foundation and Texas A&M University. Funding was provided by National Institutes of Health (Grant Nos. R21 EB020978 and R01 EB013297) and National Science Foundation (Grant Nos. 1822196 and 1346807).
Conflict of Interest
MHo and EC-H report a stakeholder interest in ECM Biosurgery which seeks to commercialize Designer Collagens based on the Scl2 protein.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.840939/full#supplementary-material
References
- Adzhubei A. A., Sternberg M. J. E., Makarov A. A. (2013). Polyproline-II Helix in Proteins: Structure and Function. J. Mol. Biol. 425 (12), 2100–2132. 10.1016/j.jmb.2013.03.018 [DOI] [PubMed] [Google Scholar]
- An B., Abbonante V., Xu H., Gavriilidou D., Yoshizumi A., Bihan D., et al. (2016). Recombinant Collagen Engineered to Bind to Discoidin Domain Receptor Functions as a Receptor Inhibitor. J. Biol. Chem. 291 (9), 4343–4355. 10.1074/jbc.M115.674507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B., Abbonante V., Yigit S., Balduini A., Kaplan D. L., Brodsky B. (2014). Definition of the Native and Denatured Type II Collagen Binding Site for Fibronectin Using a Recombinant Collagen System. J. Biol. Chem. 289 (8), 4941–4951. 10.1074/jbc.M113.530808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B., Brodsky B. (2016). Collagen Binding to OSCAR: The Odd Couple. Blood 127 (5), 521–522. 10.1182/blood-2015-12-682476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B., DesRochers T. M., Qin G., Xia X., Thiagarajan G., Brodsky B., et al. (2013). The Influence of Specific Binding of Collagen-Silk Chimeras to Silk Biomaterials on hMSC Behavior. Biomaterials 34 (2), 402–412. 10.1016/j.biomaterials.2012.09.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine E. E., Vlachos P. P., Rylander. M. N. (2014). Review of Collagen I Hydrogels for Bioengineered Tissue Microenvironments: Characterization of Mechanics, Structure, and Transport. Tissue Eng. B: Rev. 20 (6), 683–696. 10.1089/ten.teb.2014.0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumailley M., Wiedemann H., Mann K., Timpl R. (1989). Binding of Nidogen and the Laminin-Nidogen Complex to Basement Membrane Collagen Type IV. Eur. J. Biochem. 184 (1), 241–248. 10.1111/j.1432-1033.1989.tb15013.x [DOI] [PubMed] [Google Scholar]
- Bachert B. A., Choi S. J., Snyder A. K., Rio R. V. M., Durney B. C., Holland L. A., et al. (2015). A Unique Set of the Burkholderia Collagen-like Proteins Provides Insight into Pathogenesis, Genome Evolution and Niche Adaptation, and Infection Detection. PLOS ONE 10 (9), e0137578. 10.1371/journal.pone.0137578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baht G., Hunter G., Goldberg H. (2008). Bone Sialoprotein-Collagen Interaction Promotes Hydroxyapatite Nucleation. Matrix Biol. 27 (7), 600–608. 10.1016/j.matbio.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Bamford J. K. H., Bamford D. H. (1990). Capsomer Proteins of Bacteriophage PRD1, a Bacterial Virus with a Membrane. Virology 177 (2), 445–451. 10.1016/0042-6822(90)90508-o [DOI] [PubMed] [Google Scholar]
- Barrow A. D., Raynal N., Andersen T. L., Slatter D. A., Bihan D., Pugh N., et al. (2011). OSCAR Is a Collagen Receptor that Costimulates Osteoclastogenesis in DAP12-Deficient Humans and Mice. J. Clin. Invest. 121 (9), 3505–3516. 10.1172/JCI45913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra‐Bayona S. M., Guiza‐Arguello V. R., Russell B., Höök M., Hahn M. S., Hahn M. S. (2018). Influence of Collagen‐based Integrin α 1 and α 2 Mediated Signaling on Human Mesenchymal Stem Cell Osteogenesis in Three Dimensional Contexts. J. Biomed. Mater. Res. 106 (10), 2594–2604. 10.1002/jbm.a.36451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisio R., Vitagliano L., Mazzarella L., Zagari A. (2002). Crystal Structure of the Collagen Triple helix Model [(Pro-Pro-Gly)(10)](3). Protein Sci. 11 (2), 262–270. 10.1110/ps.32602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birk D. E., Brückner P. (2011). “Collagens, Suprastructures, and Collagen Fibril Assembly,” in The Extracellular Matrix: An Overview. Editor Mecham R. P. (Berlin, Heidelberg: Springer; ), 7777–115115. Biology of Extracellular Matrix. 10.1007/978-3-642-16555-9_3 [DOI] [Google Scholar]
- Birkedal-Hansen H., Moore W. G. I., Bodden M. K., Windsor L. J., Birkedal-Hansen B., DeCarlo A., et al. (1993). Matrix Metalloproteinases: A Review. Crit. Rev. Oral Biol. Med. 4 (2), 197–250. 10.1177/10454411930040020401 [DOI] [PubMed] [Google Scholar]
- Boudko S. P., Bächinger. H. P. (2016). Structural Insight for Chain Selection and Stagger Control in Collagen. Sci. Rep. 6 (1), 37831. 10.1038/srep37831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko S. P., Bächinger. H. P. (2012). The NC2 Domain of Type IX Collagen Determines the Chain Register of the Triple Helix. J. Biol. Chem. 287 (53), 44536–44545. 10.1074/jbc.M112.417543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudko S. P., Engel J., Bächinger H. P. (2012). The Crucial Role of Trimerization Domains in Collagen Folding. Int. J. Biochem. Cel Biol. 44 (1), 21–32. 10.1016/j.biocel.2011.09.009 [DOI] [PubMed] [Google Scholar]
- Bourhis J.-M., Vadon-Le Goff S., Afrache H., Mariano N., Kronenberg D., Thielens N., et al. (2013). Procollagen C-Proteinase Enhancer Grasps the Stalk of the C-Propeptide Trimer to Boost Collagen Precursor Maturation. Proc. Natl. Acad. Sci. 110 (16), 6394–6399. 10.1073/pnas.1300480110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boydston J. A., Chen P., Steichen C. T., Turnbough C. L. (2005). Orientation within the Exosporium and Structural Stability of the Collagen-like Glycoprotein BclA of Bacillus Anthracis. J. Bacteriol. 187 (15), 5310–5317. 10.1128/JB.187.15.5310-5317.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazel D., Pollner R., Oberbäumer I., Kohn K. (1988). Human Basement Membrane Collagen (Type IV). The Amino Acid Sequence of the alpha2(IV) Chain and its Comparison with the alpha1(IV) Chain Reveals Deletions in the alpha1(IV) Chain. Eur. J. Biochem. 172 (1), 35–42. 10.1111/j.1432-1033.1988.tb13852.x [DOI] [PubMed] [Google Scholar]
- Broadhead M. L., Becerra S. P., Choong P. F. M., Dass. C. R. (2010). The Applied Biochemistry of PEDF and Implications for Tissue Homeostasis. Growth Factors 28 (4), 280–285. 10.3109/08977191003604513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky B., Kaplan D. L. (2013). Shining Light on Collagen: Expressing Collagen in Plants. Tissue Eng. A 19 (13–14), 1499–1501. 10.1089/ten.tea.2013.0137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondijk T. H., de Ruiter T., Ballering J., Wienk H., Lebbink R. J., van Ingen H., et al. (2010). Crystal Structure and Collagen-Binding Site of Immune Inhibitory Receptor LAIR-1: Unexpected Implications for Collagen Binding by Platelet Receptor GPVI. Blood 115 (7), 1364–1373. 10.1182/blood-2009-10-246322 [DOI] [PubMed] [Google Scholar]
- Brown R., Sethi K. K., Gwanmesia I., Raemdonck D., Eastwood M., Mudera V. (2002). Enhanced Fibroblast Contraction of 3D Collagen Lattices and Integrin Expression by TGF-Β1 and -β3: Mechanoregulatory Growth Factors? Exp. Cel Res. 274 (2), 310–322. 10.1006/excr.2002.5471 [DOI] [PubMed] [Google Scholar]
- Browning M. B., Dempsey D., Guiza V., Becerra S., Rivera J., Russell B., et al. (2012). Multilayer Vascular Grafts Based on Collagen-Mimetic Proteins. Acta Biomater. 8 (3), 1010–1021. 10.1016/j.actbio.2011.11.015 [DOI] [PubMed] [Google Scholar]
- Browning M. B., Guiza V., Russell B., Rivera J., Cereceres S., Höök M., et al. (2014). Endothelial Cell Response to Chemical, Biological, and Physical Cues in Bioactive Hydrogels. Tissue Eng. Part A 20 (23–24), 3130–3141. 10.1089/ten.tea.2013.0602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning M. B., Russell B., Rivera J., Höök M., Cosgriff-Hernandez E. M. (2013). Bioactive Hydrogels with Enhanced Initial and Sustained Cell Interactions. Biomacromolecules 14 (7), 2225–2233. 10.1021/bm400634j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning M. B., Wilems T., Hahn M., Cosgriff-Hernandez E. M. (2011). Compositional Control of Poly(ethylene Glycol) Hydrogel Modulus Independent of Mesh Size. J. Biomed. Mater. Res. 98A (2), 268–273. 10.1002/jbm.a.33109 [DOI] [PubMed] [Google Scholar]
- Buechter D. D., Paolella D. N., Leslie B. S., Brown M. S., Mehos K. A., Gruskin E. A. (2003). Co-translational Incorporation ofTrans-4-Hydroxyproline into Recombinant Proteins in Bacteria. J. Biol. Chem. 278 (1), 645–650. 10.1074/jbc.M209364200 [DOI] [PubMed] [Google Scholar]
- Canty E. G., Kadler K. E. (2002). Collagen Fibril Biosynthesis in Tendon: A Review and Recent Insights. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 133 (4), 979–985. 10.1016/S1095-6433(02)00212-X [DOI] [PubMed] [Google Scholar]
- Cao H., Xu S.-Y. (2008). Purification and Characterization of Type II Collagen from Chick Sternal Cartilage. Food Chem. 108 (2), 439–445. 10.1016/j.foodchem.2007.09.022 [DOI] [PubMed] [Google Scholar]
- Capila I., Linhardt R. J. (2002). Heparin-Protein Interactions. Angew. Chem. Int. Edition 41 (3), 390–412. [DOI] [PubMed] [Google Scholar]
- Carver W., Molano I., Reaves T. A., Borg T. K., Terracio L. (1995). Role of the ?1?1 Integrin Complex in Collagen Gel Contraction In Vitro by Fibroblasts. J. Cel. Physiol. 165 (2), 425–437. 10.1002/jcp.1041650224 [DOI] [PubMed] [Google Scholar]
- Caswell C. C., Barczyk M., Keene D. R., Lukomska E., Gullberg D. E. (2008). Identification of the First Prokaryotic Collagen Sequence Motif that Mediates Binding to Human Collagen Receptors, Integrins α2β1 and α11β1. J. Biol. Chem. 283 (52), 36168–36175. 10.1074/jbc.M806865200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell C. C., Oliver-Kozup H., Han R., Lukomska E., Lukomski S. (2010). Scl1, the Multifunctional Adhesin of Group AStreptococcus, Selectively Binds Cellular Fibronectin and Laminin, and Mediates Pathogen Internalization by Human Cells. FEMS Microbiol. Lett. 303 (1), 61–68. 10.1111/j.1574-6968.2009.01864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereceres S., Touchet T., Browning M. B., Smith C., Rivera J., Höök M., et al. (2015). Chronic Wound Dressings Based on Collagen-Mimetic Proteins. Adv. Wound Care 4 (8), 444–456. 10.1089/wound.2014.0614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamieh F., Collignon A.-M., Coyac B. R., Lesieur J., Ribes S., Sadoine J., et al. et al (2016). Accelerated Craniofacial Bone Regeneration through Dense Collagen Gel Scaffolds Seeded with Dental Pulp Stem Cells. Sci. Rep. 6 (1), 38814. 10.1038/srep38814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B. P., Hui T. Y., Wong M. Y., Yip K. H. K., Chan G. C. F. (2010). Mesenchymal Stem Cell-Encapsulated Collagen Microspheres for Bone Tissue Engineering. Tissue Eng. C: Methods 16 (2), 225–235. 10.1089/ten.tec.2008.0709 [DOI] [PubMed] [Google Scholar]
- Chang S.-W., Buehler M. J. (2014). Molecular Biomechanics of Collagen Molecules. Mater. Today 17 (2), 70–76. 10.1016/j.mattod.2014.01.019 [DOI] [Google Scholar]
- Chen Q., Yu S., Zhang D., Zhang W., Zhang H., Zou J., et al. (2019). Impact of Antifouling PEG Layer on the Performance of Functional Peptides in Regulating Cell Behaviors. J. Am. Chem. Soc. 141 (42), 16772–16780. 10.1021/jacs.9b07105 [DOI] [PubMed] [Google Scholar]
- Chen Y.-S., Chen C.-C., Horng J.-C. (2011). Thermodynamic and Kinetic Consequences of Substituting Glycine at Different Positions in a Pro-hyp-gly Repeat Collagen Model Peptide. Biopolymers 96 (1), 60–68. 10.1002/bip.21470 [DOI] [PubMed] [Google Scholar]
- Chiu L.-H., Lai W.-F. T., Chang S.-F., Wong C.-C., Fan C.-Y., Fang C.-L., et al. (2014). The Effect of Type II Collagen on MSC Osteogenic Differentiation and Bone Defect Repair. Biomaterials 35 (9), 2680–2691. 10.1016/j.biomaterials.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Choma D. P., Milano V., Pumiglia K. M., Michael DiPersio C. (2007). Integrin α3β1-Dependent Activation of FAK/Src Regulates Rac1-Mediated Keratinocyte Polarization on Laminin-5. J. Invest. Dermatol. 127 (1), 31–40. 10.1038/sj.jid.5700505 [DOI] [PubMed] [Google Scholar]
- Chung H. J., Uitto J. (2010). Type VII Collagen: The Anchoring Fibril Protein at Fault in Dystrophic Epidermolysis Bullosa. Dermatol. Clin. 28 (1), 93–105. 10.1016/j.det.2009.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copes F., Pien N., Van Vlierberghe S., Boccafoschi F., Mantovani D. (2019). Collagen-Based Tissue Engineering Strategies for Vascular Medicine. Front. Bioeng. Biotechnol. 7, 166. 10.3389/fbioe.2019.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgriff-Hernandez E., Hahn M. S., Russell B., Wilems T., Munoz-Pinto D., Browning M. B., et al. (2010). Bioactive Hydrogels Based on Designer Collagens. Acta Biomater. 6 (10), 3969–3977. 10.1016/j.actbio.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Dell A., Galadari A., Sastre F., Hitchen P. (2010). Similarities and Differences in the Glycosylation Mechanisms in Prokaryotes and Eukaryotes. Int. J. Microbiol. 2010 (January). 1–14. 10.1155/2010/148178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhavalikar P., Robinson A., Lan Z., Jenkins D., Chwatko M., Salhadar K., et al. (2020). Review of Integrin‐Targeting Biomaterials in Tissue Engineering. Adv. Healthc. Mater. 9 (23), 2000795. 10.1002/adhm.202000795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lullo G. A., Sweeney S. M., Körkkö J., Ala-Kokko L., San Antonio J. D. (2002). Mapping the Ligand-Binding Sites and Disease-Associated Mutations on the Most Abundant Protein in the Human, Type I Collagen. J. Biol. Chem. 277 (6), 4223–4231. 10.1074/jbc.M110709200 [DOI] [PubMed] [Google Scholar]
- Díaz-González F., Milano M., Olguin-Araneda V., Pizarro-Cerda J., Tzeng S.-C., Maier C. S., et al. (2015). Protein Composition of the Outermost Exosporium-like Layer of Clostridium Difficile 630 Spores. J. Proteomics 123 (June), 1–13. 10.1016/j.jprot.2015.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzelli E., Salvadè A., Mimo P., Viganò M., Morrone M., Papagna R., et al. (2007). Mesenchymal Stem Cells Cultured on a Collagen Scaffold: In Vitro Osteogenic Differentiation. Arch. Oral Biol. 52 (1), 64–73. 10.1016/j.archoralbio.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Ellis L. R., Warner D. R., Greene R. M., Pisano M. M. (2003). Interaction of Smads with Collagen Types I, III, and V. Biochem. Biophysical Res. Commun. 310 (4), 1117–1123. 10.1016/j.bbrc.2003.09.130 [DOI] [PubMed] [Google Scholar]
- Ellison A. J., Dempwolff F., Kearns D. B., Raines R. T. (2020). Role for Cell-Surface Collagen of Streptococcus Pyogenes in Infections. ACS Infect. Dis. 6, 1843–1836. 10.1021/acsinfecdis.0c00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J., Sen S., Sweeney H. L., Discher D. E. (2006). Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 126 (4), 677–689. 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Erat M. C., Schwarz-Linek U., Pickford A. R., Farndale R. W., Campbell I. D., Vakonakis I. (2010). Implications for Collagen Binding from the Crystallographic Structure of Fibronectin 6FnI1-2FnII7FnI. JOURNAL Biol. Chem. 285 (44), 33764–33770. 10.1074/jbc.m110.139394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erat M. C., Slatter D. A., Lowe E. D., Millard C. J., Farndale R. W., Campbell I. D., et al. (2009). Identification and Structural Analysis of Type I Collagen Sites in Complex with Fibronectin Fragments. Proc. Natl. Acad. Sci. 106 (11), 4195–4200. 10.1073/pnas.0812516106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farndale R. W. (2019). Collagen-Binding Proteins: Insights from the Collagen Toolkits. Essays Biochem. 63 (3), 337–348. 10.1042/EBC20180070 [DOI] [PubMed] [Google Scholar]
- Farndale R. W., Lisman T., Bihan D., Hamaia S., Smerling C. S., Pugh N., et al. (2008). Cell-collagen Interactions: the Use of Peptide Toolkits to Investigate Collagen-Receptor Interactions. Biochem. Soc. Trans. 36 (2), 241–250. 10.1042/bst0360241 [DOI] [PubMed] [Google Scholar]
- Faye C., Chautard E., Olsen B. R., Ricard-Blum S. (2009). The First Draft of the Endostatin Interaction Network. J. Biol. Chem. 284 (33), 22041–22047. 10.1074/jbc.M109.002964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertala A. (2020). Three Decades of Research on Recombinant Collagens: Reinventing the Wheel or Developing New Biomedical Products? Bioengineering 7 (4), 155. 10.3390/bioengineering7040155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C., Stewart K. M., Weaver V. M. (2010). The Extracellular Matrix at a Glance. J. Cel Sci. 123 (24), 4195–4200. 10.1242/jcs.023820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratzl P. (2003). Cellulose and Collagen: From Fibres to Tissues. Curr. Opin. Colloid Interf. Sci. 8 (January), 32–39. 10.1016/S1359-0294(03)00011-6 [DOI] [Google Scholar]
- Gelse K., Pöschl E., Aigner T. (2003). Collagens-structure, Function, and Biosynthesis. Adv. Drug Deliv. Rev. 55 (January), 1531–1546. 10.1016/j.addr.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Ghosh N., McKillop T. J., Jowitt T. A., Howard M., Davies H., Holmes D. F., et al. (2012). Collagen-Like Proteins in Pathogenic E. Coli Strains. Plos One 7 (6), e37872. 10.1371/journal.pone.0037872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudici C., Raynal N., Wiedemann H., Cabral W. A., Marini J. C., Timpl R., et al. (2008). Mapping of SPARC/BM-40/Osteonectin-Binding Sites on Fibrillar Collagens. J. Biol. Chem. 283 (28), 19551–19560. 10.1074/jbc.M710001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessler U., Bugert P., Bieback K., Stern-Straeter J., Bran G., Hörmann K., et al. (2008). Integrin Expression in Stem Cells from Bone Marrow and Adipose Tissue during Chondrogenic Differentiation. Int. J. Mol. Med. 21 (3), 271–279. 10.3892/ijmm.21.3.271 [DOI] [PubMed] [Google Scholar]
- Gorres K. L., Raines R. T. (2010). Prolyl 4-Hydroxylase. Crit. Rev. Biochem. Mol. Biol. 45 (2), 106–124. 10.3109/10409231003627991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski J. P., Franz N. T., Pernoud D., Keightley A., Eyre D. R., Oxford J. T. (2021). A Repeated Triple Lysine Motif Anchors Complexes Containing Bone Sialoprotein and the Type XI Collagen A1 Chain Involved in Bone Mineralization. J. Biol. Chem. 296 (January), 100436. 10.1016/j.jbc.2021.100436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund M. E., Choi S. J., McNitt D. H., Barbier M., Hu G., LaSala P. R., et al. (2020). Burkholderia Collagen-like Protein 8, Bucl8, Is a Unique Outer Membrane Component of a Putative Tetrapartite Efflux Pump in Burkholderia Pseudomallei and Burkholderia Mallei. PLOS ONE 15 (11), e0242593. 10.1371/journal.pone.0242593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaia S. W., Luff D., Hunter E. J., Malcor J.-D., Bihan D., Gullberg D., et al. (2017). Unique Charge-dependent Constraint on Collagen Recognition by Integrin α10β1. Matrix Biol. 59 (May), 80–94. 10.1016/j.matbio.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaia S. W., Pugh N., Raynal N., Némoz B., Stone R., Gullberg D., et al. (2012). Mapping of Potent and Specific Binding Motifs, GLOGEN and GVOGEA, for Integrin α1β1 Using Collagen Toolkits II and III. J. Biol. Chem. 287 (31), 26019–26028. 10.1074/jbc.M112.353144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helary C., Zarka M., Giraud-Guille M. M. (2012). Fibroblasts within Concentrated Collagen Hydrogels Favour Chronic Skin Wound Healing. J. Tissue Eng. Regen. Med. 6 (3), 225–237. 10.1002/term.420 [DOI] [PubMed] [Google Scholar]
- Horii K., Kahn M. L., Herr A. B. (2006). Structural Basis for Platelet Collagen Responses by the Immune-type Receptor Glycoprotein VI. Blood 108 (3), 936–942. 10.1182/blood-2006-01-010215 [DOI] [PubMed] [Google Scholar]
- Howes J.-M., Bihan D., Slatter D. A., Hamaia S. W., Packman L. C., Knauper V., et al. (2014). The Recognition of Collagen and Triple-Helical Toolkit Peptides by MMP-13. J. Biol. Chem. 289 (35), 24091–24101. 10.1074/jbc.M114.583443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humtsoe J. O., Kim J. K., Xu Y., Keene D. R., Höök M., Lukomski S., et al. (2005). A Streptococcal Collagen-like Protein Interacts with the α2β1 Integrin and Induces Intracellular Signaling. J. Biol. Chem. 280 (14), 13848–13857. 10.1074/jbc.M410605200 [DOI] [PubMed] [Google Scholar]
- Hynes R. O. (2002). Integrins. Cell 110 (6), 673–687. 10.1016/s0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Hynes R. O. (2012). The Evolution of Metazoan Extracellular Matrix. J. Cel Biol. 196 (6), 671–679. 10.1083/jcb.201109041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. (2009). The Extracellular Matrix: Not Just Pretty Fibrils. Science 326 (5957), 1216–1219. 10.1126/science.1176009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzi V., Heljasvaara R., Heikkinen A., Karppinen S.-M., Koivunen J., Pihlajaniemi T. (2020). Exploring the Roles of MACIT and Multiplexin Collagens in Stem Cells and Cancer. Semin. Cancer Biol. 62 (May), 134–148. 10.1016/j.semcancer.2019.08.033 [DOI] [PubMed] [Google Scholar]
- Jarvis G. E., Raynal N., Langford J. P., Onley D. J., Andrews A., Smethurst P. A., et al. (2008). Identification of a Major GpVI-Binding Locus in Human Type III Collagen. Blood 111 (10), 4986–4996. 10.1182/blood-2007-08-108472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgensen H. J., Madsen D. H., Ingvarsen S., Melander M. C., Gårdsvoll H., Patthy L., et al. (2011). A Novel Functional Role of Collagen Glycosylation. J. Biol. Chem. 286 (37), 32736–32748. 10.1074/jbc.M111.266692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamajski S., Bihan D., Bonna A., Rubin K., Farndale R. W. (2016). Fibromodulin Interacts with Collagen Cross-Linking Sites and Activates Lysyl Oxidase. J. Biol. Chem. 291 (15), 7951–7960. 10.1074/jbc.M115.693408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler E., Adar R. (1989). Type I Procollagen C-Proteinase from Mouse Fibroblasts. Purification and Demonstration of a 55-kDa Enhancer Glycoprotein. Eur. J. Biochem. 186 (1–2), 115–121. 10.1111/j.1432-1033.1989.tb15184.x [DOI] [PubMed] [Google Scholar]
- Kessler E., Takahara K., Biniaminov L., Brusel M., Greenspan D. S. (1996). Bone Morphogenetic Protein-1: The Type I Procollagen C-Proteinase. Science 271 (5247), 360–362. 10.1126/science.271.5247.360 [DOI] [PubMed] [Google Scholar]
- Khoshnoodi J., Pedchenko V., Hudson B. G. (2008). Mammalian Collagen IV. Microsc. Res. Tech. 71 (5), 357–370. 10.1002/jemt.20564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielty C. M., Grant M. E. (2003). “The Collagen Family: Structure, Assembly, and Organization in the Extracellular Matrix,” in Connective Tissue and its Heritable Disorders (John Wiley & Sons; ), 159–221. 10.1002/0471221929.ch2 [DOI] [Google Scholar]
- Kii I., Ito H. (2017). Periostin and its Interacting Proteins in the Construction of Extracellular Architectures. Cell. Mol. Life Sci. 74 (23), 4269–4277. 10.1007/s00018-017-2644-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. K., Xu Y., Xu X., Keene D. R., Gurusiddappa S., Liang X., et al. (2005). A Novel Binding Site in Collagen Type III for Integrins α1β1 and α2β1. J. Biol. Chem. 280 (37), 32512–32520. 10.1074/jbc.M502431200 [DOI] [PubMed] [Google Scholar]
- Kivirikko K. I., Kishida Y., Sakakibara S., Prockop D. J. (1972). Hydroxylation of (X-Pro-Gly)n by Protocollagen Proline Hydroxylase Effect of Chain Length, Helical Conformation and Amino Acid Sequence in the Substrate. Biochim. Biophys. Acta (Bba) - Protein Struct. 271 (2), 347–356. 10.1016/0005-2795(72)90209-7 [DOI] [PubMed] [Google Scholar]
- Knight C. G., Morton L. F., Peachey A. R., Tuckwell D. S., Farndale R. W., Barnes M. J. (2000). The Collagen-Binding A-Domains of Integrins α1β1 and α2β1Recognize the Same Specific Amino Acid Sequence, GFOGER, in Native (Triple-Helical) Collagens. J. Biol. Chem. 275 (1), 35–40. 10.1074/jbc.275.1.35 [DOI] [PubMed] [Google Scholar]
- Knight C. G., Onley D. J., Peachey A. R., Messent A. J., Smethurst P. A., Tuckwell D. S., et al. (1998). Identification in Collagen Type I of an Integrin α2β1-binding Site Containing an Essential GER Sequence. J. Biol. Chem. 273 (50), 33287–33294. 10.1074/jbc.273.50.33287 [DOI] [PubMed] [Google Scholar]
- Knupp C., Squire J. M. (2003). Molecular Packing in Network-Forming Collagens. Scientific World JOURNALThe Scientific World J. 3, 558–577. 10.1100/tsw.2003.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide T., Nishikawa Y., Asada S., Yamazaki C. M., Takahara Y., Homma D. L., et al. (2006). Specific Recognition of the Collagen Triple Helix by Chaperone HSP47. J. Biol. Chem. 281 (16), 11177–11185. 10.1074/jbc.M601369200 [DOI] [PubMed] [Google Scholar]
- Koivisto L., Heino J., Häkkinen L., Larjava H. (2014). Integrins in Wound Healing. Adv. Wound Care 3 (12), 762–783. 10.1089/wound.2013.0436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konitsiotis A. D., Raynal N., Bihan D., Hohenester E., Farndale R. W., Leitinger B. (2008). Characterization of High Affinity Binding Motifs for the Discoidin Domain Receptor DDR2 in Collagen. J. Biol. Chem. 283 (11), 6861–6868. 10.1074/jbc.M709290200 [DOI] [PubMed] [Google Scholar]
- Kucharz E. J. (1992). “Structure, Heterogeneity, and Distribution,” in The Collagens: Biochemistry and Pathophysiology. Editor Kucharz E. J. (Berlin, Heidelberg: Springer; ), 5–29. 10.1007/978-3-642-76197-3_2 [DOI] [Google Scholar]
- Kudo A., Kii I. (2018). Periostin Function in Communication with Extracellular Matrices. J. Cel Commun. Signal. 12 (1), 301–308. 10.1007/s12079-017-0422-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larjava H., Haapasalmi K., Salo T., Wiebe C., Uitto V.-J. (1996). Keratinocyte Integrins in Wound Healing and Chronic Inflammation of the Human Periodontiurn. Oral Dis. 2 (1), 77–86. 10.1111/j.1601-0825.1996.tb00207.x [DOI] [PubMed] [Google Scholar]
- Lebbink R. J., Raynal N., de Ruiter T., Bihan D. G., Farndale R. W., Meyaard L. (2009). Identification of Multiple Potent Binding Sites for Human Leukocyte Associated Ig-like Receptor LAIR on Collagens II and III. Matrix Biol. 28 (4), 202–210. 10.1016/j.matbio.2009.03.005 [DOI] [PubMed] [Google Scholar]
- Lepzelter D., Bates O., Zaman M. (2012). Integrin Clustering in Two and Three Dimensions. Langmuir 28 (12), 5379–5386. 10.1021/la203725a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leski T. A., Caswell C. C., Klinke D. J., Bujnicki J. M., Hart S. J., Lukomski S. (2009). Identification and Classification of Bcl Genes and Proteins of Bacillus Cereus Group Organisms and Their Application in Bacillus Anthracis Detection and Fingerprinting. Appl. Environ. Microbiol. 75 (22), 7163–7172. 10.1128/AEM.01069-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chen R., Jia Z., Wang C., Xu Y., Li C., et al. (2020). Porous Fish Collagen for Cartilage Tissue Engineering. Am. J. Transl Res. 12 (10), 6107–6121. [PMC free article] [PubMed] [Google Scholar]
- Lin H., Chen B., Sun W., Zhao W., Zhao Y., Dai J. (2006). The Effect of Collagen-Targeting Platelet-Derived Growth Factor on Cellularization and Vascularization of Collagen Scaffolds. Biomaterials 27 (33), 5708–5714. 10.1016/j.biomaterials.2006.07.023 [DOI] [PubMed] [Google Scholar]
- Lin L., Betsuyaku T., Heimbach L., Li N., Rubenstein D., Shapiro S. D., et al. et al (2012). Neutrophil Elastase Cleaves the Murine Hemidesmosomal Protein BP180/Type XVII Collagen and Generates Degradation Products that Modulate Experimental Bullous Pemphigoid. Matrix Biol. 31 (1), 38–44. 10.1016/j.matbio.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl K., Barnes A. M., Fratzl-Zelman N., Whyte M. P., Hefferan T. E., Makareeva E., et al. (2011). COL1 C-Propeptide Cleavage Site Mutations Cause High Bone Mass Osteogenesis Imperfecta. Hum. Mutat. 32 (6), 598–609. 10.1002/humu.21475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman T., Raynal N., Groeneveld D., Maddox B., Peachey A. R., Huizinga E. G., et al. (2006). A Single High-Affinity Binding Site for von Willebrand Factor in Collagen III, Identified Using Synthetic Triple-Helical Peptides. Blood 108 (12), 3753–3756. 10.1182/blood-2006-03-011965 [DOI] [PubMed] [Google Scholar]
- Liu L., Qin C., Butler W. T., Ratner B. D., Jiang S. (2007). Controlling the Orientation of Bone Osteopontin via its Specific Binding with Collagen I to Modulate Osteoblast Adhesion. J. Biomed. Mater. Res. 80A (1), 102–110. 10.1002/jbm.a.30858 [DOI] [PubMed] [Google Scholar]
- Liu S., Shi-wen X., Blumbach K., Eastwood M., Denton C. P., Eckes B., et al. (2010). Expression of Integrin β1 by Fibroblasts Is Required for Tissue Repair In Vivo . J. Cel Sci. 123 (21), 3674–3682. 10.1242/jcs.070672 [DOI] [PubMed] [Google Scholar]
- Lodish H., Arnold B. S., Lawrence Z., Paul M., David B., James D. (2000). Collagen: The Fibrous Proteins of the Matrix. Molecular Cell Biology. 4th Edition. Available at: https://www.ncbi.nlm.nih.gov/books/NBK21582/ . [Google Scholar]
- Longmate W. M., DiPersio C. M. (2014). Integrin Regulation of Epidermal Functions in Wounds. Adv. Wound Care 3 (3), 229–246. 10.1089/wound.2013.0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Weaver V. M., Werb Z. (2012). The Extracellular Matrix: A Dynamic Niche in Cancer Progression. J. Cel Biol. 196 (4), 395–406. 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S., Bachert B. A., Squeglia F., Berisio R. (2017). Collagen-like Proteins of Pathogenic Streptococci. Mol. Microbiol. 103 (6), 919–930. 10.1111/mmi.13604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S., Nakashima K., Abdi I., Cipriano V. J., Ireland R. M., Reid S. D., et al. (2000). Identification and Characterization of the Scl Gene Encoding a Group A Streptococcus Extracellular Protein Virulence Factor with Similarity to Human Collagen. Infect. Immun. 68 (12), 6542–6553. 10.1128/IAI.68.12.6542-6553.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund A. W., Bush J. A., Plopper G. E., Stegemann J. P. (2008). Osteogenic Differentiation of Mesenchymal Stem Cells in Defined Protein Beads. J. Biomed. Mater. Res. 87B (1), 213–221. 10.1002/jbm.b.31098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther K. B., Hülsmeier A. J., Schegg B., Deuber S. A., Raoult D., Hennet T. (2011). Mimivirus Collagen Is Modified by Bifunctional Lysyl Hydroxylase and Glycosyltransferase Enzyme. J. Biol. Chem. 286 (51), 43701–43709. 10.1074/jbc.M111.309096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekkat A., Poppleton E., An B., Visse R., Nagase H., Kaplan D. L., et al. (2018). Effects of Flexibility of the α2 Chain of Type I Collagen on Collagenase Cleavage. J. Struct. Biol. 203 (3), 247–254. 10.1016/j.jsb.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migchelsen C., Berendsen H. J. C. (1973). Proton Exchange and Molecular Orientation of Water in Hydrated Collagen Fibers. An NMR Study of H2O and D2O. J. Chem. Phys. 59 (1), 296–305. 10.1063/1.1679805 [DOI] [Google Scholar]
- Mirigian L. S., Makareeva E., Koistinen H., Itkonen O., Sorsa T., Stenman U.-H., et al. (2013). Collagen Degradation by Tumor-Associated Trypsins. Arch. Biochem. Biophys. 535 (2), 111–114. 10.1016/j.abb.2013.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohs A., Silva T., Yoshida T., Amin R., Lukomski S., Inouye M., et al. (2007). Mechanism of Stabilization of a Bacterial Collagen Triple Helix in the Absence of Hydroxyproline. J. Biol. Chem. 282 (41), 29757–29765. 10.1074/jbc.M703991200 [DOI] [PubMed] [Google Scholar]
- Munoz-Pinto D. J., Guiza-Arguello V. R., Becerra-Bayona S. M., Erndt-MarinoBecerra-Bayona J., Samavedi S., Malmut S., et al. (2015). Collagen-Mimetic Hydrogels Promote Human Endothelial Cell Adhesion, Migration and Phenotypic Maturation. J. Mater. Chem. B 3 (40), 7912–7919. 10.1039/C5TB00990A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris R. A., Damon B., Mironov V., Kasyanov V., Ramamurthi A., Moreno-Rodriguez R., et al. (2007). Brook Damon, Vladimir Mironov, Vladimir Kasyanov, Anand Ramamurthi, Ricardo Moreno-Rodriguez, Thomas Trusk, et alPeriostin Regulates Collagen Fibrillogenesis and the Biomechanical Properties of Connective Tissues. J. Cel. Biochem. 101 (3), 695–711. 10.1002/jcb.21224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien F. J. (2011). Biomaterials & Scaffolds for Tissue Engineering. Mater. Today 14 (3), 88–95. 10.1016/S1369-7021(11)70058-X [DOI] [Google Scholar]
- Oliver-Kozup H., Martin K. H., Martin K. H., Schwegler-Berry D., Green B. J., Betts C., et al. (2013). The Group A Streptococcal Collagen-like Protein-1, Scl1, Mediates Biofilm Formation by Targeting the Extra Domain A-Containing Variant of Cellular Fibronectin Expressed in Wounded Tissue. Mol. Microbiol. 87 (3), 672–689. 10.1111/mmi.12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver-Kozup H. A., Elliott M., Bachert B. A., Martin K. H., Reid S. D., Schwegler-Berry D. E., et al. (2011). The Streptococcal Collagen-like Protein-1 (Scl1) Is a Significant Determinant for Biofilm Formation by Group a Streptococcus. BMC Microbiol. 11 (1), 262. 10.1186/1471-2180-11-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel J. P. R. O., Eid A., Antipova O., Bella J., Scott J. E. (2009). Decorin Core Protein (Decoron) Shape Complements Collagen Fibril Surface Structure and Mediates its Binding. PLOS ONE 4 (9), e7028. 10.1371/journal.pone.0007028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel J. P. R. O., Madhurapantula R. S. (2019). A Structural Prospective for Collagen Receptors Such as DDR and Their Binding of the Collagen Fibril. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1866 (11), 118478. 10.1016/j.bbamcr.2019.04.008 [DOI] [PubMed] [Google Scholar]
- Ottani V., Martini D., Franchi M., Ruggeri A., Raspanti M. (2002). Hierarchical Structures in Fibrillar Collagens. Micron 33 (January), 587–596. 10.1016/S0968-4328(02)00033-1 [DOI] [PubMed] [Google Scholar]
- Pallela R., Ehrlich H., Bhatnagar I. (2016). “Biomedical Applications of Marine Sponge Collagens,” in Marine Sponges: Chemicobiological and Biomedical Applications. Editors Pallela R., Ehrlich H. (New Delhi: Springer India; ), 373–381. 10.1007/978-81-322-2794-6_20 [DOI] [Google Scholar]
- Parenteau-Bareil R., Gauvin R., Berthod F. (2010). Collagen-Based Biomaterials for Tissue Engineering Applications. Materials 3 (3), 1863–1887. 10.3390/ma3031863 [DOI] [Google Scholar]
- Parmar P. A., Skaalure S. C., Chow L. W., St-Pierre J.-P., Stoichevska V., Peng Y. Y., et al. (2016a). Temporally Degradable Collagen-Mimetic Hydrogels Tuned to Chondrogenesis of Human Mesenchymal Stem Cells. Biomaterials 99 (August), 56–71. 10.1016/j.biomaterials.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar P. A., St‐Pierre J. P., Chow L. W., Puetzer J. L., Stoichevska V., Peng Y. Y., et al. (2016b). Harnessing the Versatility of Bacterial Collagen to Improve the Chondrogenic Potential of Porous Collagen Scaffolds. Adv. Healthc. Mater. 5 (13), 1656–1666. 10.1002/adhm.201600136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar P. A., St-Pierre J.-P., Chow L. W., Spicer C. D., Stoichevska V., Peng Y. Y., et al. (2017). Enhanced Articular Cartilage by Human Mesenchymal Stem Cells in Enzymatically Mediated Transiently RGDS-Functionalized Collagen-Mimetic Hydrogels. Acta Biomater. 51 (March), 75–88. 10.1016/j.actbio.2017.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paten J. A., Martin C. L., Wanis J. T., Siadat S. M., Figueroa-Navedo A. M., Ruberti J. W., et al. (2019). Molecular Interactions between Collagen and Fibronectin: A Reciprocal Relationship that Regulates De Novo Fibrillogenesis. Chem 5 (8), 2126–2145. 10.1016/j.chempr.2019.05.011 [DOI] [Google Scholar]
- Paterson G. K., Nieminen L., Jefferies J. M. C., Mitchell T. J. (2008). PclA, a Pneumococcal Collagen-like Protein with Selected Strain Distribution, Contributes to Adherence and Invasion of Host Cells. FEMS Microbiol. Lett. 285 (2), 170–176. 10.1111/j.1574-6968.2008.01217.x [DOI] [PubMed] [Google Scholar]
- Patino M. G., Neiders M. E., Andreana S., Noble B., Cohen R. E. (2002). Collagen: An Overview. Implant Dentistry 11 (3), 280–285. 10.1097/00008505-200207000-00014 [DOI] [PubMed] [Google Scholar]
- Peng Y. Y., Glattauer V., Ramshaw J. A. M. (20172017). Stabilisation of Collagen Sponges by Glutaraldehyde Vapour Crosslinking. Int. J. Biomater. 2017, 1–6. 10.1155/2017/8947823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. Y., Stoichevska V., Schacht K., Werkmeister J. A., Ramshaw J. A. M. (2014). Engineering Multiple Biological Functional Motifs into a Blank Collagen-like Protein Template fromStreptococcus Pyogenes. J. Biomed. Mater. Res. 102 (7), 2189–2196. 10.1002/jbm.a.34898 [DOI] [PubMed] [Google Scholar]
- Peng Y. Y., Yoshizumi A., Danon S. J., Glattauer V., Prokopenko O., Mirochnitchenko O., et al. (2010). A Streptococcus Pyogenes Derived Collagen-like Protein as a Non-cytotoxic and Non-immunogenic Cross-Linkable Biomaterial. Biomaterials 31 (10), 2755–2761. 10.1016/j.biomaterials.2009.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persikov A. V., Ramshaw J. A. M., Brodsky B. (2005). Prediction of Collagen Stability from Amino Acid Sequence. J. Biol. Chem. 280 (19), 19343–19349. 10.1074/jbc.M501657200 [DOI] [PubMed] [Google Scholar]
- Persikov A. V., Ramshaw J. A. M., Kirkpatrick A., Brodsky B. (2000). Amino Acid Propensities for the Collagen Triple-Helix. Biochemistry 39 (48), 14960–14967. 10.1021/bi001560d [DOI] [PubMed] [Google Scholar]
- Perumal S., Antipova O., Orgel J. P. R. O. (2008). Collagen Fibril Architecture, Domain Organization, and Triple-Helical Conformation Govern its Proteolysis. Proc. Natl. Acad. Sci. 105 (8), 2824–2829. 10.1073/pnas.0710588105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkas D. M., Ding S., Raines R. T., Barron A. E. (2011). Tunable, Post-Translational Hydroxylation of Collagen Domains in Escherichia Coli. ACS Chem. Biol. 6 (4), 320–324. 10.1021/cb100298r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Guajardo M., Calderón-Romero P., Castro-Córdova P., Mora-Uribe P., Paredes-Sabja D. (2016). Ultrastructural Variability of the Exosporium Layer of Clostridium Difficile Spores. Appl. Environ. Microbiol. 82 (7), 2202–2209. 10.1128/AEM.03410-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizarro-Guajardo M., Olguín-Araneda V., Barra-Carrasco J., Brito-Silva C., Sarker M. R., Paredes-Sabja D. (2014). Characterization of the Collagen-like Exosporium Protein, BclA1, of Clostridium Difficile Spores. Anaerobe 25 (February), 18–30. 10.1016/j.anaerobe.2013.11.003 [DOI] [PubMed] [Google Scholar]
- Post A., Isgandarova S., Martinez-Moczygemba M., Hahn M., Russell B., Hook M., et al. (2019). Elucidation of Endothelial Cell Hemostatic Regulation with Integrin-Targeting Hydrogels. Ann. Biomed. Eng. 47 (3), 866–877. 10.1007/s10439-018-02194-w [DOI] [PubMed] [Google Scholar]
- Qiu Y., Zhai C., Chen L., Liu X., Yeo J. (2021). Current Insights on the Diverse Structures and Functions in Bacterial Collagen-like Proteins. ACS Biomater. Sci. Eng. 1, 1. 10.1021/acsbiomaterials.1c00018 [DOI] [PubMed] [Google Scholar]
- Quiroz J. F. D., Rodriguez P. D., Erndt-Marino J. D., Guiza V., Balouch B., Graf T., et al. (2018). Collagen-Mimetic Proteins with Tunable Integrin Binding Sites for Vascular Graft Coatings. ACS Biomater. Sci. Eng. 4 (8), 2934–2942. 10.1021/acsbiomaterials.8b00070 [DOI] [PubMed] [Google Scholar]
- Radke D., Jia W., Sharma D., Fena K., Wang G., Goldman J., et al. (2018). Tissue Engineering at the Blood‐Contacting Surface: A Review of Challenges and Strategies in Vascular Graft Development. Adv. Healthc. Mater. 7 (15), 1701461. 10.1002/adhm.201701461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaka R. S., Renugopalakrishnan V., Urry D. W., Bhatnagar R. S. (1978). Hydroxylation of Proline in Polytripeptide Models of Collagen: Stereochemistry of Polytripeptide-Prolyl Hydroxylase Interaction. Biochemistry 17 (14), 2892–2898. 10.1021/bi00607a030 [DOI] [PubMed] [Google Scholar]
- Rasmussen M., Jacobsson M., Björck L. (2003). Genome-Based Identification and Analysis of Collagen-Related Structural Motifs in Bacterial and Viral Proteins. J. Biol. Chem. 278 (34), 32313–32316. 10.1074/jbc.M304709200 [DOI] [PubMed] [Google Scholar]
- Reyes C. D., García A. J. (2004). α2β1integrin-specific Collagen-Mimetic Surfaces Supporting Osteoblastic Differentiation. J. Biomed. Mater. Res. 69A (4), 591–600. 10.1002/jbm.a.30034 [DOI] [PubMed] [Google Scholar]
- Ricard-Blum S., Beraud M., Raynal N., Farndale R. W., Ruggiero F. (2006). Structural Requirements for Heparin/Heparan Sulfate Binding to Type V Collagen. J. Biol. Chem. 281 (35), 25195–25204. 10.1074/jbc.M603096200 [DOI] [PubMed] [Google Scholar]
- Ricard-Blum S. (2011). The Collagen Family. Cold Spring Harbor Perspect. Biol. 3 (1), a004978. 10.1101/cshperspect.a004978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salasznyk R. M., Williams W. A., Boskey A., Batorsky A., Plopper G. E. (2004). Adhesion to Vitronectin and Collagen I Promotes Osteogenic Differentiation of Human Mesenchymal Stem Cells. J. Biomed. Biotechnol. 2004 (1), 24–34. 10.1155/s1110724304306017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand J. M. B., Genovese F., Karsdal M. A. (2016). “Type IV Collagen,” in Biochemistry of Collagens, Laminins and Elastin. Editor Karsdal M. A. (Academic Press; ), 31–41. 10.1016/B978-0-12-809847-9.00004-0 [DOI] [Google Scholar]
- Schmidt M. M., Dornelles R. C. P., Mello R. O., Kubota E. H., Mazutti M. A., Kempka A. P., et al. (2016). Collagen Extraction Process. Int. Food Res. J. 23 (3), 913–922. [Google Scholar]
- Schneider R. K., Puellen A., Kramann R., Raupach K., Bornemann J., Knuechel R., et al. (2010). The Osteogenic Differentiation of Adult Bone Marrow and Perinatal Umbilical Mesenchymal Stem Cells and Matrix Remodelling in Three-Dimensional Collagen Scaffolds. Biomaterials 31 (3), 467–480. 10.1016/j.biomaterials.2009.09.059 [DOI] [PubMed] [Google Scholar]
- Schnicker N. J., Dey M. (2016). Bacillus Anthracis Prolyl 4-Hydroxylase Modifies Collagen-like Substrates in Asymmetric Patterns. J. Biol. Chem. 291 (25), 13360–13374. 10.1074/jbc.M116.725432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel M. J., Robins S. P., Bilezikian J. P. (2006). Dynamics of Bone and Cartilage Metabolism: Principles and Clinical Applications. Elsevier. [Google Scholar]
- Sekiya A., Okano-Kosugi H., Yamazaki C. M., Koide T. (2011). Pigment Epithelium-Derived Factor (PEDF) Shares Binding Sites in Collagen with Heparin/Heparan Sulfate Proteoglycans. J. Biol. Chem. 286 (30), 26364–26374. 10.1074/jbc.M111.252684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U., Carrique L., Vadon-Le Goff S., Mariano N., Georges R.-N., Delolme F., et al. (2017). Structural Basis of Homo- and Heterotrimerization of Collagen I. Nat. Commun. 8 (1), 14671. 10.1038/ncomms14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L. M., Olsen B. R. (1991). FACIT Collagens: Diverse Molecular Bridges in Extracellular Matrices. Trends Biochem. Sci. 16 (January), 191–194. 10.1016/0968-0004(91)90074-6 [DOI] [PubMed] [Google Scholar]
- Shokrgozar M. A., Fattahi M., Bonakdar S., Ragerdi Kashani I., Majidi M., Haghighipour N., et al. (2012). Healing Potential of Mesenchymal Stem Cells Cultured on a Collagen-Based Scaffold for Skin Regeneration. Iran Biomed. J. 16 (2), 68–76. 10.6091/ibj.1053.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders M. D., Raines R. T. (2009). Collagen Structure and Stability. Annu. Rev. Biochem. 78 (1), 929–958. 10.1146/annurev.biochem.77.032207.120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried M. (1902). Reticulin and Collagen. J. Physiol. 28 (4), 319–324. 10.1113/jphysiol.1902.sp000919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. J. (2006). Fragmentation of Protein Using Trypsin. Cold Spring Harb Protoc. 2006 (5), pdb.prot4550. 10.1101/pdb.prot4550 [DOI] [Google Scholar]
- Singer A. J., Clark A. F. (1999). Cutaneous Wound Healing. New Engl. J. Med. 341 (10), 738–746. 10.1056/NEJM199909023411006 [DOI] [PubMed] [Google Scholar]
- Snellman A., Tu H., Vaisanen T., Kvist A.-P., Huhtala P., Pihlajaniemi T. (2000). A Short Sequence in the N-Terminal Region Is Required for the Trimerization of Type XIII Collagen and Is Conserved in Other Collagenous Transmembrane Proteins. EMBO J. 19 (19), 5051–5059. 10.1093/emboj/19.19.5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderhäll C., Marenholz I., Kerscher T., Rüschendorf F., Esparza-Gordillo J., Worm M., et al. (2007). Variants in a Novel Epidermal Collagen Gene (COL29A1) Are Associated with Atopic Dermatitis.” Edited by Gonçalo Abecasis. PLoS Biol. 5 (9), e242. 10.1371/journal.pbio.0050242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somaiah C., Kumar A., Mawrie D., Sharma A., Bhattacharyya J., Swaminathan R., et al. (2015). Collagen Promotes Higher Adhesion, Survival and Proliferation of Mesenchymal Stem Cells. Plos One 10 (12), e0145068. 10.1371/journal.pone.0145068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamati K., Priestley J. V., Mudera V., Cheema U. (2014). Laminin Promotes Vascular Network Formation in 3D In Vitro Collagen Scaffolds by Regulating VEGF Uptake. Exp. Cel Res. 327 (1), 68–77. 10.1016/j.yexcr.2014.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoichevska V., An B., Peng Y. Y., Yigit S., Vashi A. V., Kaplan D. L., et al. (2016). Formation of Multimers of Bacterial Collagens through Introduction of Specific Sites for Oxidative Crosslinking. J. Biomed. Mater. Res. A. 104 (9), 2369–2376. 10.1002/jbm.a.35772 [DOI] [PubMed] [Google Scholar]
- Stoichevska V., Peng Y. Y., Vashi A. V., Werkmeister J. A., Dumsday G. J., Ramshaw J. A. M. (2017). Engineering Specific Chemical Modification Sites into a Collagen-like Protein from Streptococcus Pyogenes. J. Biomed. Mater. Res. A. 105 (3), 806–813. 10.1002/jbm.a.35957 [DOI] [PubMed] [Google Scholar]
- Suri S., Schmidt C. E. (2010). Cell-Laden Hydrogel Constructs of Hyaluronic Acid, Collagen, and Laminin for Neural Tissue Engineering. Tissue Eng. Part A 16 (5), 1703–1716. 10.1089/ten.tea.2009.0381 [DOI] [PubMed] [Google Scholar]
- Sweeney S. M., Cynthia A. G., Gregg B., Antonio J. D. S. (1998). Defining the Domains of Type I Collagen Involved in Heparin- Binding and Endothelial Tube Formation. Proc. Natl. Acad. Sci. 95 (13), 7275–7280. 10.1073/pnas.95.13.7275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney S. M., Orgel J. P., McAuliffe J. D., Turner K. R., Di Lullo G. A., Chen Steven., et al. (2008). Candidate Cell and Matrix Interaction Domains on the Collagen Fibril, the Predominant Protein of Vertebrates. J. Biol. Chem. 283 (30), 21187–21197. 10.1074/jbc.M709319200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre P., Couture-Tosi E., Mock M. (2002). A Collagen-like Surface Glycoprotein Is a Structural Component of the Bacillus Anthracis Exosporium. Mol. Microbiol. 45 (1), 169–178. 10.1046/j.1365-2958.2000.03000.x [DOI] [PubMed] [Google Scholar]
- Takahashi I., Onodera K., Sasano Y., Mizoguchi I., Bae J.-W., Mitani H., et al. (2003). Effect of Stretching on Gene Expression of Β1 Integrin and Focal Adhesion Kinase and on Chondrogenesis through Cell-Extracellular Matrix Interactions. Eur. J. Cel Biol. 82 (4), 182–192. 10.1078/0171-9335-00307 [DOI] [PubMed] [Google Scholar]
- Takayama G., Arima K., Kanaji T., Toda S., Tanaka H., Shoji S., et al. (2006). Periostin: A Novel Component of Subepithelial Fibrosis of Bronchial Asthma Downstream of IL-4 and IL-13 Signals. J. Allergy Clin. Immunol. 118 (1), 98–104. 10.1016/j.jaci.2006.02.046 [DOI] [PubMed] [Google Scholar]
- Tan L., Turnbough C. L. (2010). Sequence Motifs and Proteolytic Cleavage of the Collagen-like Glycoprotein BclA Required for its Attachment to the Exosporium of Bacillus Anthracis. J. Bacteriol. 192 (5), 1259–1268. 10.1128/JB.01003-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal V. J., Lee D. Y., White E. S., Larios J. M., Chacon R., Horowitz J. C., et al. (2003). Myofibroblast Differentiation by Transforming Growth Factor-ॆ1 Is Dependent on Cell Adhesion and Integrin Signaling via Focal Adhesion Kinase. J. Biol. Chem. 278 (14), 12384–12389. 10.1074/jbc.M208544200 [DOI] [PubMed] [Google Scholar]
- Thomas E. K., Nakamura M., Wienke D., Isacke C. M., Pozzi A., Liang P. (2005). Endo180 Binds to the C-Terminal Region of Type I Collagen. J. Biol. Chem. 280 (24), 22596–22605. 10.1074/jbc.M501155200 [DOI] [PubMed] [Google Scholar]
- Thompson B. M., Hoelscher B. C., Adam D., Stewart G. C. (2012). Assembly of the BclB Glycoprotein into the Exosporium and Evidence for its Role in the Formation of the Exosporium ‘Cap’ Structure in Bacillus Anthracis. Mol. Microbiol. 86 (5), 1073–1084. 10.1111/mmi.12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H., Sasaki T., Snellman A., Walter G., Pirilä P., Timpl R., et al. (2002). The Type XIII Collagen Ectodomain Is a 150-Nm Rod and Capable of Binding to Fibronectin, Nidogen-2, Perlecan, and Heparin. J. Biol. Chem. 277 (25), 23092–23099. 10.1074/jbc.M107583200 [DOI] [PubMed] [Google Scholar]
- Wancura M., Talanker M., Toubbeh S., Bryan A., Cosgriff-Hernandez E. (2020). Bioactive Hydrogel Coatings of Complex Substrates Using Diffusion-Mediated Redox Initiation. J. Mater. Chem. B 8 (19), 4289–4298. 10.1039/D0TB00055H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatmore A. M. (2001). Streptococcus Pyogenes SclB Encodes a Putative Hypervariable Surface Protein with a Collagen-like Repetitive Structure. Microbiology (Reading, England) 147 (Pt 2), 419–429. 10.1099/00221287-147-2-419 [DOI] [PubMed] [Google Scholar]
- Wiberg C., Hedbom E., Khairullina A., Lamandé S. R., Oldberg A., Timpl R., et al. (2001). Biglycan and Decorin Bind Close to the N-Terminal Region of the Collagen VI Triple Helix. J. Biol. Chem. 276 (22), 18947–18952. 10.1074/jbc.M100625200 [DOI] [PubMed] [Google Scholar]
- Williams K. E., Olsen D. R. (2009). Matrix Metalloproteinase-1 Cleavage Site Recognition and Binding in Full-Length Human Type III Collagen. Matrix Biol. 28 (6), 373–379. 10.1016/j.matbio.2009.04.009 [DOI] [PubMed] [Google Scholar]
- Winterbourn C. C., Parsons-Mair H. N., Gebicki S., Gebicki J. M., Davies M. J. (2004). Requirements for Superoxide-dependent Tyrosine Hydroperoxide Formation in Peptides. Biochem. J. 381 (1), 241–248. 10.1042/bj20040259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtowicz A. M., Shekaran A., Oest M. E., Dupont K. M., Templeman K. L., Hutmacher D. W., et al. (2010). Coating of Biomaterial Scaffolds with the Collagen-Mimetic Peptide GFOGER for Bone Defect Repair. Biomaterials 31 (9), 2574–2582. 10.1016/j.biomaterials.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Yu Z., Inouye M., Brodsky B., Mirochnitchenko O. (2010). Expanding the Family of Collagen Proteins: Recombinant Bacterial Collagens of Varying Composition Form Triple-Helices of Similar Stability. Biomacromolecules 11 (2), 348–356. 10.1021/bm900894b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Raynal N., Stathopoulos S., Myllyharju J., Farndale R. W., Leitinger B. (2011). Collagen Binding Specificity of the Discoidin Domain Receptors: Binding Sites on Collagens II and III and Molecular Determinants for Collagen IV Recognition by DDR1. Matrix Biol. 30 (1), 16–26. 10.1016/j.matbio.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Gurusiddappa S., Rich R. L., Owens R. T., Keene D. R., Mayne R., et al. (2000). Multiple Binding Sites in Collagen Type I for the Integrins Α1β1 and Α2β1. J. Biol. Chem. 275 (50), 38981–38989. 10.1074/jbc.M007668200 [DOI] [PubMed] [Google Scholar]
- Xu Y., Keene D. R., Bujnicki J. M., Höök M., Lukomski S. (2002). Streptococcal Scl1 and Scl2 Proteins Form Collagen-like Triple Helices. J. Biol. Chem. 277 (30), 27312–27318. 10.1074/jbc.M201163200 [DOI] [PubMed] [Google Scholar]
- Yamada T. (2011). Giant Viruses in the Environment: Their Origins and Evolution. Curr. Opin. Virol. Virus. entry/Environmental Virol. 1 (1), 58–62. 10.1016/j.coviro.2011.05.008 [DOI] [PubMed] [Google Scholar]
- Yoshizumi A., Yu Z., Thiagarajan G., Ramshaw M., Inouye M., Brodsky B. (2009). Self-Association of Streptococcus Pyogenes Collagen-like Constructs into Higher Order Structures. Protein Sci. 18 (6), 1241–1251. 10.1002/pro.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., An B., John A., Ramshaw M., Brodsky B. (2014). Bacterial Collagen-like Proteins that Form Triple-Helical Structures. J. Struct. Biol. 186 (3), 451–461. 10.1016/j.jsb.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Visse R., Inouye M., Nagase H., Brodsky B. (2012). Defining Requirements for Collagenase Cleavage in Collagen Type III Using a Bacterial Collagen System. J. Biol. Chem. 287 (27), 22988–22997. 10.1074/jbc.M112.348979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.-M., Käpylä J., Santeri Puranen J., Graham Knight C., Tiger C.-F., Pentikäinen O. T., et al. (2003). Α11β1 Integrin Recognizes the GFOGER Sequence in Interstitial Collagens. J. Biol. Chem. 278 (9), 7270–7277. 10.1074/jbc.M210313200 [DOI] [PubMed] [Google Scholar]
- Zhao X., Wang R., Shang Q., Hao H., Li Y., Zhang Y., et al. (2016). The New Flagella-Associated Collagen-like Proteins ClpB and ClpC of Bacillus Amyloliquefaciens FZB42 Are Involved in Bacterial Motility. Microbiol. Res. 184 (March), 25–31. 10.1016/j.micres.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Zhao X., Wang Y., Shang Q., Li Y., Hao H., Zhang Y., et al. (2015). Collagen-Like Proteins (ClpA, ClpB, ClpC, and ClpD) Are Required for Biofilm Formation and Adhesion to Plant Roots by Bacillus Amyloliquefaciens FZB42. PLOS ONE 10 (2), e0117414. 10.1371/journal.pone.0117414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Jeanette L., Miller C., Conrady D. G., Barrow A. D., Chirgadze D. Y., et al. (2016). Structural Basis for Collagen Recognition by the Immune Receptor OSCAR. Blood 127 (5), 529–537. 10.1182/blood-2015-08-667055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.