Abstract
Over the past two decades, it is improved gut microbiota plays an important role in the health and disease pathogenesis. Metabolites, small molecules produced as intermediate or end products of microbial metabolism, is considered as one of the major interaction way for gut microbiota with the host. Bacterial metabolisms of dietary substrates, modification of host molecules or bacteria are the major source of metabolites. Signals from microbial metabolites affect immune maturation and homeostasis, host energy metabolism as well as mucosal integrity maintenance. Based on many researches, the composition and function of the microbiota can be changed, which is also seen in the metabolite profiles of patients with inflammatory bowel disease (IBD). Additionally, some specific classes of metabolites also can trigger IBD. In this paper, definition of the key classes of microbial-derived metabolites which are changed in IBD, description of the pathophysiological basis of association and identification of the precision therapeutic modulation in the future are the major contents.
Keywords: Inflammatory bowel disease, Microbial metabolites, Short chain fatty acids, Bile acids, Tryptophan
Core Tip: In the gastrointestinal tract of the human, a variety of microorganisms such as bacteria, fungi, viruses, archaea and protozoa are collected and considered as a community, containing a genome, suitable to the environment and the host. Microbiome, with wide functions, can ferment dietary fibres, defence pathogen, synthesize vitamin and promote immune maturation and metabolic homeostasis, which indicates microbiome is deeply integrated with human biology. However, because gut microbiota is associated with ancestral diet high in fibre, the dietary alteration resulted from western diet causes maladaptive change for this association, and products of microbial metabolism exist at a nexus between host and microbiome.
INTRODUCTION
In recent years, metabolomics based evaluation system has been used to search for biomarkers of inflammatory bowel disease (IBD) and explore its pathogenesis. It is becoming an important method of IBD research[1]. In the gastrointestinal tract of the human, a variety of microorganisms such as bacteria, fungi, viruses, archaea and protozoa are collected and considered as a community, containing a genome, suitable to the environment and the host[2]. Microbiome can ferment dietary fibres, defence pathogen, synthesize vitamin and promote immune maturation and metabolic homeostasis, which indicates microbiome is deeply integrated with human biology[3]. However, because gut microbiota is associated with ancestral diet high in fibre, the dietary alteration resulted from western diet causes maladaptive change for this association[4].
Researchers have begun to investigate the metabolites of IBD using metabonomics to explain the undetermined etiology and improve treatment outcomes[5]. In terms of diagnosis and evaluation, to date, there is no reliable laboratory test that can distinguish between the two subtypes of IBD, namely ulcerative colitis and Crohn's disease; Conventional inflammatory hematological indicators, such as platelet parameters, erythrocyte sedimentation rate (ESR), and C-reactive protein (CRP), can be used to judge the correlation with IBD inflammatory activity and predict treatment effect and prognosis[6]. However, such tests cannot screen IBD patients in a timely and effective manner. Therefore, new biomarkers for IBD are still being sought, and the emergence of metabolomics techniques can help better analyze and solve these key questions.
As an integral part of systems biology, metabolomics is a method of quantitative analysis of all metabolites in a living organism, imitating the research ideas of genomics and proteomics, and searching for the relative relationship between metabolites and physiological and pathological changes[7]. The research objects are mostly small molecules with a relative molecular weight of less than 1000.
Microbial metabolites are found in a variety of biological tissues such as faeces, urine, serum, and have diversized effects on these host physiology[8]. IBD patients can be classified based on the alterations in faecal, urinary and serum metabolomes, which shows a new mechanism and association is found[9]. Additionally, several metabolite classes become the key research, for finding the association with intestinal inflammation and IBD, such as bile acid erivatives[10]. It is noted that these may affect the host, and association is found among the host, dysbiotic microbiota and an altered metabolite milieu through the inflammation model involving diet, genetic risk and the microbiota[11].
A metabolome is a collection of metabolites in a cell, tissue, or organ, consisting of a series of molecules of different chemical types, such as peptides, carbohydrates, lipids, nucleic acids, and the catalytic products of foreign substances[12]. Metabolomics studies evaluate the effects of exogenous stimuli and explore their mechanisms by quantitatively analyzing the changes of endogenous metabolites in biological systems[13]. Complete metabolomics includes sample collection and preparation; Detection and identification of metabolites; Data analysis and modeling; To establish the relationship between temporal and spatial variation of metabolites and characteristics of organisms[14]. Metabonomics is the genomics, proteomics, transcriptome study after emerging "omics", metabonomics has the advantage of the medium and small molecular substances is the study of the biology to produce the final results and metabolism, while genome and proteome have a cumulative and compensation effect, they are effective tiny changes in metabolites is amplified, therefore, The identification of metabolites is easier and more accurate to reflect the state of biological system[15].
In this review, the characteristics description of gut metabolome, and the description of the integration of metabolomics with other data are involved[16]. Furthermore, untargeted researches of IBD, targeted metabolomics with the focus on short chain fatty acids (SCFAs), bile acids and tryptophan metabolism are included[17]. In final, small molecule discovery, diagnostic potential and therapeutic manipulation of the gut microbiome–metabolome axis as the future directions is dicussed[18].
CHARACTERIZATION OF THE GUT METABOLOME
Metabolites are comprehensive products of physiological processes, and metabolic phenotypes can reflect the organism's life state. Metabolomics technology reflects the body's response to physiological stimulation or gene modification through qualitative and quantitative analysis of real-time changes in endogenous substances[19]. Compared with other omics, it detects a relatively small number of substances, but amplifies small differences at the level of genes and proteins. At the same time, effectively reduce the interference of inactive genes and inactive proteins, higher accuracy. Therefore, metabolomics has developed rapidly in the last 20 years[20].
The human body is a super organism composed of host and microorganism. Intestinal flora participates in absorption, metabolism, immunity, defense and maintenance of intestinal stability[21]. However, for a long time, the important role of intestinal flora in maintaining human balance has been underestimated due to the large variety and number, complex interrelationship and lack of efficient and convenient analysis methods[22]. In addition to maintaining intestinal homeostasis through self-formation of mucosal barrier and multi-signaling pathway regulation, co-metabolites of host and intestinal flora are also a major regulation mode. Intestinal flora participates in host metabolism, producing a large number of small molecules and hormones, and metabolites enter the host body to participate in body circulation and affect host homeostasis[23]. For example, flagellin and lip-polysaccharide produced by intestinal flora regulate human fat metabolism through nuclear factor interleukin-3 and biological clock, so intestinal flora is both metabolic participant and regulator[24]. Changes in metabolites can also predict abnormalities in the gut microbiota, For example, the increase of organic acid content in urine suggests that Clostridium difficile, Faecalibacterium prausnitzii and Bifidobacterium SPP., Subdoligranulum SPP., and Lactobacillus SPP[25].
For metabolomics, there are several analytical techniques and platforms on the basis of mass spectrometry [26]. Metabolomics research strategies can be pursued in either targeted or untargeted ways. Targeted metabolomics accurately quantifies a group of known metabolites, whereas untargeted approaches cover as many metabolites as possible[27].
Metabolomics has the advantages of high throughput, high accuracy and high sensitivity, which can detect the changes of small molecule metabolites produced by the body in the dynamic process of metabolism qualitatively or quantitatively[28]. The analysis process of metabonomics includes three parts: sample preparation, data collection and data analysis and interpretation[29]. The samples for metabonomics analysis are mainly biological specimens, which are commonly used in urine, blood, saliva, feces extract, and local pathological tissues such as colon tissue. The metabolites in the samples are usually 1H-NMR and gas chromatography[30].
Gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry (LIQUID chromatography-mass spectrometry) Lc-ms) and other methods. Bioinformatics platform is used for data processing for data analysis and interpretation, and principal Component analysis (PCA) is the most commonly used method[31]. PCA and Partial least squares discriminant analysis (PLS-DA). Since the 1990s. Since the emergence of the late Era, metabolomics has been widely used in the study of drug toxicity and mechanism, microbial and plant metabolomics, nutrition science and disease diagnosis and other fields[32].
Notably, attention should be taken to the untargeted metabolomics, though this method is a promising method to discover program[33]. Firstly, no universal method exists in sample preprocessing and extraction, particular methods in allusion to particular families of metabolites[34]. As a result, several methods can be combined. Consequently, considering different sample matrix and molecules, extraction methods show different efficacy so that it is difficult to obtain precise quantification. Secondly, there are various pitfalls when spectral data from mass spectrometry experiments are converted into to chemically annotated compounds[35]. According to different levels of evidences, various compounds are still classified as unknowns[36]. To understand them, it requires manual expertise and computational algorithms. Thirdly, it is not enough to take databases such as Metlin as the references, because it only contains a small fraction of compounds[37]. Based on estimation, higher than 90% of spectral features in a microbiome study is not known. But, according to a recent study, a molecular class can be assigned from Human Metabolome Database based on 43% of metabolite features[38]. Additionally, it is a great challenge to annotate the unknown compound. Finally, various intermediate- or low-concentration compounds or proximally produced ones in the intestine, can not be well represented in the faecal metabolome[39]. Consequently, special requirements in the sample collection and standardization, annotation and data integration at a systematic biology level as well as careful design are needed when integrating discovery projects that incorporate untargeted metabolomics with other omics technologies[40].
However, targeted metabolomics own several strongpoints in extraction, known internal standards and more precise absolute quantification, even though it is limited to spectrum[41].
UNTARGRTED METABOLOMICS IN IBD
A large number of untargeted faecal metabolomic researches among the patients with IBD are published[42]. Though these researches involve different groups, including adults and children, and different subtypes and using different metabolomics approaches, some common themes are considered[43]. metabolomics can be used to classify the groups who are healthy and with IBD[44]. However, metabolomics is not an ideal approach to discriminate Crohn’s disease and ulcerative colitis.
From metabolite group, the description of dysregulation of bile acid metabolism, changes in the levels of amino acids, sphingolipids, polyaminesand reductions in levels of medium-chain fatty acids and SCFAs is consistent in faecal samples[45]. At the same time, variation in the metabolome is associated with the variation of microbiome between the healthy subjects and patients groups[46].
Bile acids can be divided into primary bile acids and secondary bile acids according to their sources, and free bile acids and conjugated bile acids according to their structural types[47]. Primary bile acids are produced in the liver and bind to taurine or glycine to form the corresponding conjugated bile acids, which are then released into the gallbladder by bile Salt export Pump for storage and further concentration[48]. After eating, bile acids are released into the duodenum, facilitating emulsification and absorption of lipids in the small intestine. At the terminal ileum, about 95% of the bile acids are reabsorbed and returned to the liver via the portal vein[49]. The cycle in which bile acids are secreted from the liver to the intestine, reabsorbed in the ileum, and then returned to the liver via the portal vein is called hepatoenteric circulation. 400-800 mg of unabsorbed bile acids enter the colon daily for further dissociation, hydroxylation, differential isomerization, and dehydroxylation, and the enzymes catalyzing these reactions are mostly produced by intestinal microorganisms[50]. The biliary saline hydrolysates involved in bile acid dissociation are mainly produced by Bacteroides, Clostridium, Lactobacillus, Bifidobacterium and Listeria. Dehydrogenase or heteroisomerase involved in Bile acids oxidation or heteroisomerase reaction are mainly Bacteroides, Eubacterium, Clostridium, Escherichia, Eggerthella., Peptostreptococcus and Ruminococcus[51]. A series of reactions of bile acids in colon play an important role in maintaining the diversity and dynamic balance of bile acid pools. It is worth noting that bile acids can also affect the composition of intestinal microorganisms through bacteriostasis while intestinal microorganisms produce enzymes catalyzing the metabolism of bile acids[52].
The pathogenesis of IBD is not completely clear, genetic factors and environmental factors are important factors leading to mucosal immune response disorders. With the development of metabolomics, the researchers observed significant changes in a variety of metabolites in IBD patients compared with healthy subjects, such as fecal amino acids, bile acids, sphinomyelic acid, medium chain fatty acids, short chain fatty acids, and polyamines[53]. The change of intestinal bile acid profile is closely related to the development of IBD. A study involving 155 patients with IBD and 65 healthy controls showed that more than 2700 metabolites differed between patients with IBD and healthy people[54]. The metabolites of IBD patients were mainly manifested as sphenolipids and bile acids, while the bile acids enriched in feces of IBD patients were mainly conjugated bile acids and sulphated bile acids, while the content of secondary bile acids decreased. The analysis of serum bile acid content in patients with IBD showed that the serum bile acid metabolism was disorder, especially the secondary bile acid content in patients with active stage was significantly reduced[55].
The diversity of intestinal bile acids is closely related to intestinal microbes. Although the causal relationship between intestinal flora and IBD is still unclear, the disorder of bile acid metabolism in IBD patients is often accompanied by changes in intestinal microbial composition[56]. The diversity of intestinal flora in IBD patients decreased, and the composition ratio of intestinal flora also changed, which was mainly manifested as the decreased abundance of Firmicutes[57]. The proportion of Clostridia changed significantly, the abundance of Roseburia and Faecalibacterium decreased, while that of Ruminococcus gnavus increased. The ratio of Faecalibacterium prausntizii and Escherichia coli decreased. In turn, increased bile acid content in the gut promotes the growth of biliary tolerant bacteria such as Bilophila Wadsworthi and promotes TH1-mediated mucosal immune response[58].
Untargeted metabolomics researches are also carried out among the human biological samples, involving urine, serum and/or plasma and intestinal biopsy samples[59]. Compared with control group, the level of urinary mammalian–microbial co-metabolite hippurate is reduced, and tricarboxylic acid cycle intermediates and amino acid metabolism in IBD group are altered[60]. Generally, it is possible to distinguish the healthy subjects and the controlled ones, even though it is not completely clear to know the differences between metabolomes of patients with Crohn’s disease and those with ulcerative colitis[61].
The new research results involve loss of secondary bile acids, vitamins B3 and B5 and SCFAs, and expansion of acylcarnitines and polyunsaturated fatty acids[62]. But, it is found that the result is associated with dysbiotic subset of samples. This dysbiotic subset was defined as being outside the ninetieth percentile of the healthy cohort, and ‘excursions’ into this state were weakly correlated with inflammatory activity[63]. This indicates that multimodal functional assessment is important in explaining the longitudinal relationship, with a higher understanding than a single classification[64]. The study on the mechanism in the future can answer the question of clinical relevance involving the determination of biomarkers of treatment response and disease course[65].
TARGETED METABOLOMICS IN IBD
Biles acids
Bile acids is synthesized from cholesterol by the liver in a multi-enzyme process, involving two products[66]. These primary bile acids exhibit amphipathic properties, which are beneficial for the lipid digestion and absorption in the small intestine[67]. When reaching the distal ileum, 95% of them are reabsorbed, and the rest is recycled and replaced, which are controlled by fibroblast growth factor[68]. Bile acids not only can regulate own synthesis, but also can exert many metabolic and immune effects through binding a series of receptors such as farnesoid X receptor (FXR), TGR5 as well as constitutive androstane receptor[69]. TGR5 is beneficial for improving insulin sensitivity, reduce energy expenditure in muscle and brown adipose tissue, and relax the gallbladder[70]. TGR5 also reduces Kupffer cell response to lipopolysaccharide by nuclear factor-κB inhibition and release of IL-1, IL-6 and tumor necrosis factor (TNF) from peripheral blood monocytes in humans. Activation of FXR has diverse effects on host metabolism[71].
Hepatic bile acid synthesis is regulated by the FXR-FGF15/19 signaling pathway. Activation of this signaling pathway reduces the expression of enzymes related to hepatic bile acid synthesis and reduces bile acid synthesis[72]. It has been found that reduced FGF19 levels in Crohn's disease (CD) patients lead to reduced activation of FXR, which inhibits the FXR-FGF15/19 pathway and leads to increased bile acid synthesis in the liver, leading to the development of intestinal inflammation[73]. Therefore, compared with the normal population, the enterohepatic circulation is blocked in IBD patients, and the activation of signal pathways that negatively regulate the synthesis of intrahepatic bile acids is reduced, leading to an increase in the total amount of bile acids in the intestinal lumen, which is also one of the causes of intestinal inflammation[73].
Restoration of bile acid pools was found to increase colon RORγ Treg cell numbers and improve host susceptibility to inflammatory colitis through bile acid nuclear receptors[74]. Recent studies have shown that ursodeoxycholic acid, as a candidate drug for nonalcoholic fatty liver disease[75], can improve intestinal barrier function, reduce intestinal inflammation, and regulate intestinal microbiota composition. Ursodeoxycholic acid and its metabolite, shicholic acid, can reduce the severity of intestinal inflammation and inhibit the expression of mucosal cytokines in a DSS-induced mouse model of colitis[76]. It has also been found that ursodeoxycholic acid and shicholic acid can inhibit the cleavage of caspase-3, a protease related to colon epithelial apoptosis[77]. Therefore, ursodeoxycholic acid may improve intestinal inflammation by inhibiting the apoptosis of epithelial cells. However, long-term high dose (28-30 mg/kg/d) ursodeoxycholic acid exposure is not protective for Ulcerative colitis (UC) patients and may increase the risk of colorectal cancer[78]. The role of ursodeoxycholic acid in the treatment of IBD is still controversial and has not been confirmed by strong clinical evidence, so further studies are needed.
Although some reports suggest that secondary bile acids such as deoxycholic acid and shicholic acid may be cytotoxic molecules that cause oxidative stress, membrane damage, interference with DNA repair, mucosal inflammation and colon cancer[79], some studies have found that Clostridium hiranonis promotes the production of secondary bile acids[80]. Relieves DSS-induced colitis because secondary bile acids at physiological concentrations inhibit the growth of E. coli and B. perfringens in vitro[81]. Clostridium scindens may also be involved in the induction of CD relief in children by producing secondary bile acids. Therefore, secondary bile acids may be a promising therapeutic agent for IBD and deserve further study[82].
MICROBIALLY TRANSFORMED BILE ACIDS IN IBD
The pathogenesis of IBD is related to genetics, environment, intestinal microecology and immunity, but the specific biological mechanism is still unclear. As an important part of intestinal microecology, intestinal flora can directly affect intestinal environmental homeostasis and participate in bile acid metabolism, while the abnormal bile acid metabolism also affects the quality and quantity of intestinal flora, and both of them are involved in the occurrence and development of intestinal inflammation[83].
For IBD patients, due to bile acid malabsorption and the inherently bidirectional nature of such interaction, it is not easy to observe the effect of dysbiosis on bile acid dysmetabolism[84]. Some researchers including Devkota carry out a ground-breaking study find that the propotion of taurine-conjugated bile acids in the mice with the diet high in milk-derived fat is higher than that in the ones with low-fat diet, which leads to the bloom of B. wadsworthia[85]. Due to the development of colitis, the change happens on IL10−/− mice, which indicates that western diet, altered host metabolites, dysbiosis are associated with inflammation in the genetically susceptible host[86].
Recent studies have found that bile acids are a powerful driver of intestinal microbiota maturation in newborns[87]. The effects of bile acids on intestinal flora are bidirectional, promoting the growth of bacteria dependent on bile acid metabolism on the one hand, and inhibiting the growth of bacteria sensitive to bile on the other hand[88]. Bile acids have a direct lactating effect on bacterial cell membranes and can also play an antibacterial role by activating the synthesis of antimicrobial substances mediated by FXR[89]. FXR is expressed in liver, intestine, fat, vascular wall, pancreas, kidney and other tissues[90]. Activation of intestinal FXR can induce the encoding of antimicrobial target genes including angiogenin, carbonic anhydrase 12 and inducible nitric oxide synthase, thus exerting antibacterial effect[91]. In the process of deoxycholic acid induced intestinal inflammation, the diversity of intestinal flora decreased significantly[92]. At the level of bacteroidetes, the proportion of Firmicutes increased while the proportion of Bacteroidetes decreased. At the genus level, the proportion of Bacteroidetes increased, while the proportion of Clostridium XIVA decreased, indicating that high concentration of deoxycholic acid could aggravate the imbalance of intestinal flora[93].
Additionally, using synthetic agonist to directly stimulating FXR shows anti-inflammatory effect, and shows protection function in chemically induced colitis, but susceptibility to chemical injury is increased as the result of FXR−/− mice exhibit[94]. It should be noted that if microbial bile acid metabolizing genes is lost, the cell subtype in vivo may be reduced[95].
Repeated exposure of intestinal epithelial cells to high concentrations of bile acids is an important risk factor for IBD, and IBD patients have high levels of deoxycholic acid in the intestine[96]. In rats with colitis induced by Trinitrobenzenesulfonic acid (TNBS), apical sodium-dependent bile acid transporter (ASBT) expression decreased[97]. In IBD patients, ileal inflammation blocks hepatoenteric circulation of bile acids, leading to reduced ileal reabsorption, which may be due to inhibition of ASBT promoter expression by inflammatory cytokines, thus increasing fecal bile acids[98].
In recent years, intestinal flora has been regarded as an "endocrine organ" that regulates host physiological functions by producing metabolites such as bile acids and short-chain fatty acids. Bile acid metabolism mainly occurs in hepatocytes and intestinal flora[99].
Animal studies have shown that deoxycholic acid reduces intestinal abundance of Clostridium, a species that produces bile acid hydrolases that convert Tauro-Muricholic acid into cholic acid[100]. The decrease of this genus resulted in the accumulation of taurocholic acid, which is an FXR antagonist and inhibits the FXR-mediated signaling pathway[101]. FXR interacts with downstream fibroblast growth factor15/19 (FGF15/19) to regulate bile acid metabolism through the enterohepatic signaling pathway[102]. Once FXR and FGF15/19 signal pathways are interfered, the intestinal liver circulation of bile acids in the body is disturbed, resulting in increased bile acid levels in the intestinal lumen. High concentration of bile acids can promote the occurrence of gastrointestinal inflammation by damaging the DNA of intestinal cells[103].
In many studies, the altered faecal bile acid in IBD is described repeatedly. Considering potential confounders, Ganesan et al[104] perform a study and focus on the patients with isolated colonic IBD. Recent studies have shown that deoxycholic acid mediates intestinal ecological imbalance, damages the intestinal mucosal barrier, and ultimately leads to intestinal inflammation. The disturbance of intestinal microbiome - bile acid dialogue can damage intestinal barrier function and activate inflammatory signaling pathway, leading to the occurrence and development of intestinal inflammation[105].
TRYPTOPHAN
Through eating poultry, fish and dairy foods, the humans obtain tryptophan. This is an essential, aromatic amino acid and the precursor for the synthesis of several important bioactive molecules. Tryptophan metabolism is mainly produced in the gastrointestinal tract[106]. Amino acids play an important role in keeping your gut healthy. Tryptophan is an essential amino acid associated with autoimmune, and its metabolic pathway is related to the pathogenesis of inflammatory bowel disease[107].
Dietary tryptophan can be metabolized by intestinal flora into a series of indole-metabolites, such as indole-acetic acid, indole-3-acetaldehyde, indole-3-aldehydes, indole-acrylics, and indole-3-propionic acid, which act as ligands for aromatic hydrocarbon receptors, which have been implicated in the pathogenesis of IBD[108]. Indoles, indoles propionic acid and indoles acrylic acid may reduce intestinal permeability and affect mucosal homeostasis by binding progesterone X receptor. Indoleformaldehyde can activate aromatic hydrocarbon receptors on intestinal immune cells and increase IL-22 production. Indole derivatives bind to aromatic hydrocarbon receptors to produce IL-10, so oral indole-3-propionic acid protects mice from DSS-induced colitis[108]. Therefore, the disorder of intestinal flora can destroy immune regulation through its metabolites and promote the progress of intestinal inflammation.
Dietary tryptophan follows one of three main routes, and the majority is metabolized in the kynurenine pathway. and its rate-limiting enzymes include indoleamine 2,3-dioxygenase 1 in mucosal and immune cells and tryptophan 2,3-dioxygenase in the liver[109]. Serotonin pathway is considered as the second major host route, of which, the rate-limiting enzyme tryptophan hydroxylase 1 in enterochromaffin cell plays a controlled role. In final, tryptophan can be metabolized by the gut microbiota into a range of indole metabolites[110]. It is important that microbiota and microbial metabolite also play a regulation role for both host tryptophan pathways[111].
In allusion to metabolites from gut microbiome,GLP1 is released due to the stimulation of indole, but indole derivatives can act as agonists for AhR.Diet-derived AhR agonists has a great effect on maintaining the microbial load and composition and immune tolerance in the proximal small intestine[112]. On contrary, in the distal small intestine and colon, microbiotaderived AhR agonists has a great effect. Existing researches show that limited mount of bacteria can produce AhR agonists, whereas when C. sporogenes is produced, which triggers IPA. Its production is associated with fldAIBC phenyllactate gene cluster, beneficial for maintaining barrier function and inhibiting mucosal TNF production[113].
TRYPTOPHAN METABOLISM
A study involving 535 patients with IBD shows that tryptophan metabolism is increased, and disease activity is inversely correlated with tryptophan levels[114]. Based on this analysis, it is suggested metabolism is increased through kynurenine pathway[115]. The result is also observed in the previous studies. Previous studies have reported that the expression of AhR in inflamed mucosal samples from patients with Crohn’s disease is decreased, and lack of dietary tryptophan has the association with worsening colitis in mouse models[116].
Tryptophan is one of the eight essential amino acids in the human body. When tryptophan is ingested, it enters different metabolic pathways, including protein anabolism and tryptophan catabolism[117]. The tryptophan catabolism pathway includes: 5-ht pathway (about 95%), canisurine pathway (about 1%-2%), and bacterial decomposition pathway[118]. These pathways work together to maintain the balance of the body's environment and keep the body healthy. At the same time, as a drug, tryptophan is widely used in the treatment of many diseases[119]. For example, tryptophan supplementation can reduce mood swings and irritability in women with premenstrual syndrome, improve sleep quality in patients with sleep disorders, treat depression, and may be used as an adjunct therapy for smokers to quit smoking. Recent studies have confirmed that the metabolic pathway of tryptophan regulates gastrointestinal function[120], and changes in tryptophan metabolism can lead to gastrointestinal dysfunction and disease, such as inflammatory bowel disease, irritable bowel syndrome, celiac disease and diverticulitis. In previous studies, tryptophan levels in UC patients were significantly lower than those in healthy volunteers. The same results were seen in UC animal models. However, the researchers did not make a strict distinction between the active stage and the remission stage[121]. By comparing the differences of tryptophan in different periods of UC patients (active stage, remission stage) and healthy volunteers, this study further demonstrated that tryptophan plays a key role in the occurrence and development of UC[122]. In the experiment, we also found that serum tryptophan level was negatively correlated with inflammatory markers ESR and CRP in active UC patients (P < 0.05). Based on the changes of serum tryptophan in active and remission stages of UC, as well as the correlation between serum tryptophan level in active stage and inflammatory markers, serum tryptophan level in patients with UC may have important significance for monitoring the activity of UC. This also fully demonstrates the value of tryptophan in UC research[123].
Additionally, researchers carry out a research to examine the effect of mucinutilizing bacteria, and confirm that commensal P. russellii plays a role in reducing the susceptibility to colitis by metabolizing tryptophan to IA, through which, goblet cell differentiation is improved and inflammatory signal is reduced[124]. The same pathway occurs for IA in P. russellii. A novel finding that fldAIBC phenyllactate gene cluster was also identified in P. russellii is described. This is a new finding, making a link between a bacterium with mucin- and tryptophan-metabolizing abilities to improved epithelial integrity, so that the researchers can know the decrease in phenyllactate gene cluster in ulcerative colitis in the condition of metagenomics study involving IBD and non-IBD control individuals[125].
Intestinal flora involved in tryptophan metabolism is also an important signal pathway to regulate the immune system. It has been confirmed that tryptophan metabolism is weakened in the absence of intestinal bacteria, which is manifested as increased tryptophan content and decreased active metabolites. Intestinal tryptophan metabolites can regulate the activity of Aryl hydrocarbon receptor (AhR) in immune cells, which is a key factor to maintain immune balance[126]. Ah R can effectively balance T regulatory cells and Th17. Indole is a product of tryptophan expressed by enterobacteriaceae tryptophan enzyme, which can protect the integrity of intestinal bacterial membrane, reduce the adhesion of pathogenic bacteria, promote the production of anti-inflammatory factor interleukin-10, and reduce the production of inflammatory factors. Indole-3-propionic acid (INdole-3-propionic acid) is a product of indoles under the action of intestinal flora, and its production requires the participation of Clostridium sporogenes. Indole-3-propionic acid can inhibit inflammatory signals through progesterone X receptors and improve the integrity of the intestinal barrier[127].
SCFAs
SCFAs is a kind of beneficial metabolite and derived from microbiota-accessible carbohydrates[128]. After microbial fermentation, acetate, propionate and butyrate are produced, also other gases such as gases methane and hydrogen sulfide are included[129]. Different diets affect the proportions 50%-70% for acetate, 10%-20% for propionate and the biggest for butyrate. In allusion to dietary intervention, if the humans take more animal-based diets, the protein fermentation is rapidly changed and dysbiosis is caused[130].
SCFAs are organic fatty acids composed of one to six carbon atoms. The main production site of SCFAs is the colon. SCFAs are mostly produced by the enzymatic hydrolysis of carbohydrates in undigested and absorbed food residues by anaerobic bacteria in the colon, mainly including acetic acid, propionic acid and butyric acid, with a concentration of about 20-140 in the colon The type and quantity of mmol/L. sccfAs were mainly determined by the composition of intestinal flora, digestion time, host-microbial metabolic flux and fiber content in host food[131]. SCFAs produced by fermentation participate in the metabolism of different organs in human body and play different functions. Acetic acid produced by bacterial fermentation can be absorbed and utilized by the host and is an important source of host energy, providing about 10% of the total daily energy of the human body. After absorbed by blood, propionic acid catabolizes in the liver, participating in the process of pyruvate reversal into glucose, and may inhibit the synthesis of fat; Butyric acid is mainly used by epithelial cells and is the main energy source of epithelial cells[132].
Notably, it is observed typical dysbiosis in IBD is correlated with the lost bacterial species, which is consistent with the general result that faecal SCFA level is reduced in the metabolomics studies of human IBD[133]. If the section of colon is surgically failed, diversion colitis is caused and can be treated with SCFA enemas. Butyrate by enema is effective to treat ulcerative colitis to a certain degree, and butyrate-producing dietary fibre additives may be beneficial for the maintenance of remission[134].
SCFAs and IBD
If the butyrate-producing species in IBD patients is lost, it is observed that butyrate oxidation is impaired. On the basis off some researches, among the patients with IBD, the level of faecal SCFAs is reduced at different degrees[135]. This result is consistent with the research on the quantitative PCR targeting the butyryl-CoA: Acetate-CoA-transferase gene[136]. While the result for active ulcerative colitis and Crohn’s disease indicates that the intermediate molecule lactate is increased[137]. But, due to the fact that only approximately 5% of SCFAs remain in faeces, there is no significant difference in the alterations in transit, absorption and utilization between the controlled subjects and healthy ones[138]. As a result, to know that anti-inflammatory associations with butyrateproducing bacteria may be caused by other mechanisms is also beneficial for the further study[139].
SCFAs are endogenous substances that have been widely studied for the correlation between IBD metabolomics and intestinal flora. SCFAs are products of indigestible carbohydrates under the action of Clostridium plusseri, Rothberry, Eubacterium and Bifidobacteria, mainly including acetic acid, propionic acid, butyric acid and isobutyric acid[140]. The content of SCFAs in feces can reflect the status of intestinal flora. It has been reported that the fecal butyric acid, isobutyric acid and acetic acid in patients with active IBD decreased significantly, and the contents of Clostridium coccoides, Clostridium leptum, Clostridium pralei and R.i. Ntestinalis decreased. Correlation analysis showed that butyric-producing bacteria Clostridium plerii and enterorothburiae were positively correlated with butyric acid, isobutyric acid and acetic acid, while abnormal enterobacteria tended to be normal during remission[141]. Therefore, this study suggested that the decrease of SCFAs content caused by intestinal flora disturbance was related to the incidence of IBD. The abnormal metabolism of butyric acid, which is the energy source of colon epithelial cells, and acetic acid, which is involved in adipogenesis and gluconeogenesis, suggests that inflammation leads to disturbance of intestinal energy metabolism. Acetic acid and butyric acid exert anti-inflammatory effects by activating G protein receptor 41 (GPR41) and GPR43 and inhibiting histone deacetylases. Regulating intestinal production of short-chain fatty acids has become a new target for the treatment of IBD[142].
According to a mouse study, the association between the low level and dietary fibre and susceptibility to dextran sulfate sodium (DSS)-induced colitis is constructed, in which, diet-derived SCFAs plays an important role[143]. Based on the result of DSS model, if the fatty acid receptor GPR43 in mice is lost, refractory colitis is triggered[144]. The same finding is seen in the constituents of the gut microbiota[145]. It is indicated that overactivation of the NLRP1A receptor in mice leads to loss of butyrate-producing Clostridiales, with increased IL-18 and IFNγ production[146]. In final, it is observed the gene expression of NLRP1 in inflamed regions of the distal colon in patients with ulcerative colitis is increased, and a majority of bacterial operational taxonomic units were of the order Clostridiales, which is consist with the mechanistic observation in a human cohort[147] (Figure 1).
Figure 1.
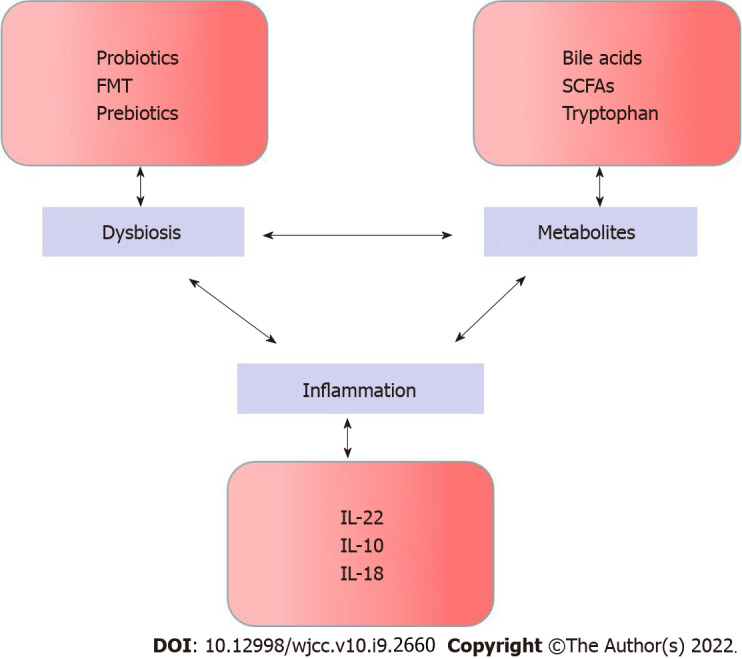
The cycle of circular causality in inflammatory bowel disease. FMT: Fecal microbiota transplantation; SCFAs: Short chain fatty acids.
OTHER ASSOCIATED METABOLITES
Fatty acids showed significant changes in faeces of UC patients, including a significant decrease in short chain fatty acids (SCFA, such as acetic acid, propionic acid and butyric acid) and medium chain fatty acids (valeric acid, hexanoic acid, heptanic acid, captanic acid and nonanoic acid)[148]. Fatty acids participate in energy supply through β -oxidation, and butyric acid oxidation can provide 60% energy for colon cells, again suggesting abnormal energy metabolism in UC state. SCFA plays an anti-inflammatory role through tumor necrosis factor α and NF-κB pathways and the activation of GPR41 and GPR43. The decrease of SCFA induced by inflammation can make inflammation persist. Medium chain fatty acids activate peroxisome proliferators to activate receptors for anti-inflammatory effects[149].
Methylamine, trimethylamine, and trimethylamine oxide are products of choline action by gastrointestinal flora, and their reduction indicates that choline metabolism is disturbed[150]. Choline is an important component of cell membrane and the key to maintaining cell membrane integrity. Choline increases in feces of UC patients, while glycerophosphoryl choline decreases as a choline metabolite, indicating that the integrity of intestinal cell membrane changes in UC patients and inflammation may lead to intestinal mucosal damage[151]. Low density lipoprotein (LDL) and very low densit lipoprotein (VLDL) are products of fat metabolism in the liver. The decrease in serum levels of both is caused by inflammation that increases the activity of phospholipase A2, promotes the hydrolysis of phospholipids in LDL and VLDL, and inhibits the expression of lipase that hydrolyzes triglycerides[152].
Succinate metabolism in IBD is a promising direction. Succinate is the intermediate of host cells and the microbiota. In the host, succinate acts as an important pro-inflammatory signal and has been shown to be a key mediator, via IL-1β, of macrophage response to lipopolysaccharide[153]. For the patients with Crohn’s disease, the level of succinate in the serum is increased, and increased result is obtained for the expression of the SUCNR1, Sucnr1–/– mice are protected against fibrosis and 2,4,6-trinitrobenzene sulfonic acid-induced colitis[154]. On contrary, the level of urinary succinate among the IBD patients is decreased, lower than that of the controlled subjects[155]. The level of faecal succinate in the patients with ulcerative colitis and Crohn’s disease is higher than that in the subjects with no IBD and in DSS-induced colitis at the peak period of weight loss[156]. It is analyzed from the perspective of microbiota that succinate-utilizing Phascolarctobacterium is less abundant in both ulcerative colitis and Crohn’s disease than in healthy subjects. Some researchers make efforts to find the effect of important metabolite and the role of gut microbiota[157].
FUTURE DIRECTIONS
Small-molecule discovery
The family involving G protein-coupled receptor (GPCR) ligands, N-acyl amides, is identified[158]. These are encoded by the microbiota and that are agonists to receptors that have important implications for gastrointestinal and metabolic physiology, such as the endocannabinoid receptor GPR119[159]. A study carried out by Settanni et al[160] uses bioinformatics mining to identify biologically conserved gene clusters in metagenomes by the computational and synthetic biology pipeline.
Using another different ‘chemistry forward’ method and relying on a high-throughput GPCR reporter platform, human gut microbiota is derived from 11 patients with IBD, aiming to identify relevant metabolites. This study not only identify the activation of GPCRs in a wide range based on the constituents analysis of the human gut microbiota, but also identify Morganella morganii converted the aromatic amino acid l-phenylalanine into phenethylamine, and the clear in vivo association between M. morganiiproduced and Lactobacillus reuteri-produced histamine and colonic transit time[161]. By relying on Human Microbiome Project dataset, these research results are obtained based on the deep mining of metagenomic assemblies, so as to determine the increase level of histidine decarboxylase genes in the patients with Crohn’s disease[162]. Using these methods, information as well as techniques from different sources to examine the microbiota for novel compounds, it is of great significance to identify bioactive molecules relevant to IBD[162].
THERAPEUTIC OPTIONS
Next-generation probiotics. Studies have shown that dysbiosis has the association with disease, which motivate the researchers to perform combined therapy involving the host process and microbiome[163]. Intestinal probiotics Akkermansia Muciniphila can induce homeostasis IgG production and antigen-specific T cell response in mice, regulate immune homeostasis, and improve the symptoms of DSS induced colitis in mice.
A recent study found that a hydrolyzed protein diet alleviates DSS-induced colitis by regulating the imbalance of intestinal flora and increasing the production of secondary bile acids by Clostridium hiranonis[164]. Other studies found that exogenous fucose can significantly reduce the inflammatory response of DSS colitis mice, and it was found that after the administration of fucose, the imbalance of intestinal flora was improved, so as to restore the normal synthesis of bile acid pools in the body and liver, and then reduce the colitis of mice.
However, potential next-generation probiotics (NGPs) invove commensals in the gut of adults, so researchers pay more attention to its metabolite production. Candidate NGPs, according to the identification of its role in specific condition based on the analysis of gut microbiome, is featured with fastidiousness, or extreme sensitiveness of oxygen[165]. As a result, how to isolate, cultivate characterize and formulate them is of a great challenge. It is possible for NGPs and other similar products performed with evaluation under the regulation of LBPs, so that investigation for the new drug procedures based on FDA152 is required[165].
In the previous studies, the potential of targeted microbiome–metabolite therapeutics in the form of the colonization resistance is found[162]. Some studies show C. difficile spore germination receptor binds to the primary bile acid CA derivatives and is inhibited by the secondary bile acid deoxycholic acid (DCA). When C. scindens to mice at risk of Clostridium difficile infection (CDI) is administrated, the resistance to CDI is improved[162].
Faecal microbiota transplantation
It is interesting that bile acid profiles show marked alteration in CDI and restored after successful fecal microbiota transplantation (FMT) which is effective in treating recurred CDI[158]. some studies demonstrate that FMT is effective in reducing the remission in ulcerative colitis. Fecal microbiota transplantation is the transplantation of faecal flora from a healthy donor into the gastrointestinal tract of a patient and can correct IBD-related intestinal flora disorders. Some scholars have found that fecal bacteria transplantation may be a safe and effective method for the treatment of refractory CD. Six weeks after receiving fecal bacteria transplantation, the severity index of Crohn's disease under endoscopy decreased significantly with CD, and the level of C-reactive protein also decreased significantly compared with the control group[162]. Patients who were in remission after fecal bacteria transplantation had more Eubacterium Hallii and Roseburia Inulivorans in their faeces and colon, which increased short-chain fatty acid biosynthesis and secondary bile acid levels. Specific bacterial and metabolic pathways in fecal bacteria transplantation are involved in inducing remission, and these findings may be of great reference value for the design of microbial therapies for IBD. The sample size may also affect the experimental results. Therefore, whether fecal transplantation is effective in alleviating symptoms of IBD patients needs to be verified by clinical studies with larger samples.
Metabolites
LBPs is a partially attractive due to its ability in getting high concentration of metabolites in the intestine, so oral metabolites or their precursors may be a great challenge in pharmacology[154]. At present, a placebo-controlled, crossover trial in allusion to tryptophan metabolites is undergoing, aiming to determine the efficiency of oral 5-hydroxytryptophan administration on fatigue among the patients with IBD[165]. For diabetes, niacin is delivered to the gut microbiota by microcapsules, which indicates that the level of gut Bacteroidetes is increased, and the marker of insulin resistance in healthy control volunteers is improved. This is an interesting finding, which demonstrates some preclinical and clinical promise is effective in treating ulcerative colitis.
CONCLUSION
Based on analysis involving technology, observational data and experimental insight, it is illuminated that microbial metabolites is beneficial for the pathogenesis of IBD. With the change of lifestyle and dietary pattern, the incidence of IBD is increasing year by year. High-fat diet changes the intestinal microecological homeostasis and also affects the metabolism of bile acids in the body, so that the role of the dialogue mechanism between intestinal flora and bile acids in gastrointestinal diseases has attracted more and more attention. In conclusion, existing studies have shown that intestinal flora imbalance or bile acid metabolism disorder caused by various external factors can make intestinal flora, bile acid metabolism and IBD form a vicious circle of mutual influence. For the prevention and treatment of IBD, it is necessary to further explore the mechanism of intestinal flora, bile acids and their receptors in IBD. The scientific and reasonable application of probiotics, antibiotics, bile acids and their derivatives to prevent IBD will be the key problem to be solved in the future.
In this review, untargeted metabolomics and microbiome analyses are summarized, suggesting that the pattern of alteration for metabolite profiles is consisteds. Among of them, bile acids, SCFAs and tryptophan metabolites are analyzed in details. This study emphasizes the unknowns in the faecal metabolome, so creating large multinational cohorts of patients with IBD and controls, and of pipelines for microorganism–metabolite discovery and evaluation is encouraged. From the perspective of future direction, some promising fields are identified to achieve the objective. It is noted that how to integrate dietary and targeted microbiota manipulation or even microbiota reconstitution to realize the goals is a great challenge. Gut microbiota-derived metabolites is possible to become a promising direction.
Footnotes
Conflict-of-interest statement: The authors declare that there are no conflicts of interest.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: October 12, 2021
First decision: December 3, 2021
Article in press: February 27, 2022
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Rimondi A, Sassaki LY S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
Contributor Information
Lie Zheng, Department of Gastroenterology, Shaanxi Hospital of Traditional Chinese Medicine, Xi’an 710003, Shaanxi Province, China.
Xin-Li Wen, Department of Gastroenterology, Shaanxi Hospital of Traditional Chinese Medicine, Xi’an 710003, Shaanxi Province, China.
Sheng-Lei Duan, Department of Gastroenterology, Shaanxi Hospital of Traditional Chinese Medicine, Xi’an 710003, Shaanxi Province, China. 281930369@qq.com.
References
- 1.Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 2017;179:204–222. doi: 10.1016/j.trsl.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkinson EM, Ilhan ZE, Herbst-Kralovetz MM. Microbiota-drug interactions: Impact on metabolism and efficacy of therapeutics. Maturitas. 2018;112:53–63. doi: 10.1016/j.maturitas.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Khalsa J, Duffy LC, Riscuta G, Starke-Reed P, Hubbard VS. Omics for Understanding the Gut-Liver-Microbiome Axis and Precision Medicine. Clin Pharmacol Drug Dev. 2017;6:176–185. doi: 10.1002/cpdd.310. [DOI] [PubMed] [Google Scholar]
- 4.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, Gewirtz AT. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 7.Etienne-Mesmin L, Chassaing B, Gewirtz AT. Tryptophan: A gut microbiota-derived metabolites regulating inflammation. World J Gastrointest Pharmacol Ther. 2017;8:7–9. doi: 10.4292/wjgpt.v8.i1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan S, Ding Y, Saedi N, Choi M, Sridharan GV, Sherr DH, Yarmush ML, Alaniz RC, Jayaraman A, Lee K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018;23:1099–1111. doi: 10.1016/j.celrep.2018.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bringer MA, Gabrielle PH, Bron AM, Creuzot-Garcher C, Acar N. The gut microbiota in retinal diseases. Exp Eye Res. 2022;214:108867. doi: 10.1016/j.exer.2021.108867. [DOI] [PubMed] [Google Scholar]
- 10.Ding Y, Yanagi K, Cheng C, Alaniz RC, Lee K, Jayaraman A. Interactions between gut microbiota and non-alcoholic liver disease: The role of microbiota-derived metabolites. Pharmacol Res. 2019;141:521–529. doi: 10.1016/j.phrs.2019.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Rio D, Zimetti F, Caffarra P, Tassotti M, Bernini F, Brighenti F, Zini A, Zanotti I. The Gut Microbial Metabolite Trimethylamine-N-Oxide Is Present in Human Cerebrospinal Fluid. Nutrients. 2017;9 doi: 10.3390/nu9101053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, Rezzi S, Ross A, Kochhar S, Holmes E, Nicholson JK. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648–652. doi: 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen LN, Roager HM, Casas ME, Frandsen HL, Gosewinkel U, Bester K, Licht TR, Hendriksen NB, Bahl MI. Glyphosate has limited short-term effects on commensal bacterial community composition in the gut environment due to sufficient aromatic amino acid levels. Environ Pollut. 2018;233:364–376. doi: 10.1016/j.envpol.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Hou Y, Wang G, Zheng X, Hao H. Gut Microbial Metabolites of Aromatic Amino Acids as Signals in Host-Microbe Interplay. Trends Endocrinol Metab. 2020;31:818–834. doi: 10.1016/j.tem.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Krishnan S, Alden N, Lee K. Pathways and functions of gut microbiota metabolism impacting host physiology. Curr Opin Biotechnol. 2015;36:137–145. doi: 10.1016/j.copbio.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maes M, Vojdani A, Geffard M, Moreira EG, Barbosa DS, Michelin AP, Semeão LO, Sirivichayakul S, Kanchanatawan B. Schizophrenia phenomenology comprises a bifactorial general severity and a single-group factor, which are differently associated with neurotoxic immune and immune-regulatory pathways. Biomol Concepts. 2019;10:209–225. doi: 10.1515/bmc-2019-0023. [DOI] [PubMed] [Google Scholar]
- 18.Zierer J, Jackson MA, Kastenmüller G, Mangino M, Long T, Telenti A, Mohney RP, Small KS, Bell JT, Steves CJ, Valdes AM, Spector TD, Menni C. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. 2018;50:790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain A, Li XH, Chen WN. An untargeted fecal and urine metabolomics analysis of the interplay between the gut microbiome, diet and human metabolism in Indian and Chinese adults. Sci Rep. 2019;9:9191. doi: 10.1038/s41598-019-45640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 21.Lawson CE, Harcombe WR, Hatzenpichler R, Lindemann SR, Löffler FE, O'Malley MA, García Martín H, Pfleger BF, Raskin L, Venturelli OS, Weissbrodt DG, Noguera DR, McMahon KD. Common principles and best practices for engineering microbiomes. Nat Rev Microbiol. 2019;17:725–741. doi: 10.1038/s41579-019-0255-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parra-Llorca A, Gormaz M, Alcántara C, Cernada M, Nuñez-Ramiro A, Vento M, Collado MC. Preterm Gut Microbiome Depending on Feeding Type: Significance of Donor Human Milk. Front Microbiol. 2018;9:1376. doi: 10.3389/fmicb.2018.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakshmanan V, Ray P, Craven KD. Rhizosphere Sampling Protocols for Microbiome (16S/18S/ITS rRNA) Library Preparation and Enrichment for the Isolation of Drought Tolerance-Promoting Microbes. Methods Mol Biol. 2017;1631:349–362. doi: 10.1007/978-1-4939-7136-7_23. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi J, Matsuura M, Hisamatsu T. Safety evaluation of ustekinumab for moderate-to-severe ulcerative colitis. Expert Opin Drug Saf. 2022;21:1–8. doi: 10.1080/14740338.2021.1980536. [DOI] [PubMed] [Google Scholar]
- 25.Shade A, Dunivin TK, Choi J, Teal TK, Howe AC. Strategies for Building Computing Skills To Support Microbiome Analysis: a Five-Year Perspective from the EDAMAME Workshop. mSystems. 2019;4 doi: 10.1128/mSystems.00297-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dheilly NM, Bolnick D, Bordenstein S, Brindley PJ, Figuères C, Holmes EC, Martínez Martínez J, Phillips AJ, Poulin R, Rosario K. Parasite Microbiome Project: Systematic Investigation of Microbiome Dynamics within and across Parasite-Host Interactions. mSystems. 2017;2 doi: 10.1128/mSystems.00050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heimann TM, Swaminathan S, Slater GI, Kurtz RJ. Perianal Fistula After Ileoanal Pouch in Patients With Ulcerative Colitis: A Review of 475 Patients Operated on at a Major IBD Center. Dis Colon Rectum. 2022;65:76–82. doi: 10.1097/DCR.0000000000002114. [DOI] [PubMed] [Google Scholar]
- 28.Sandborn WJ, Danese S. Ozanimod for Ulcerative Colitis. Reply. N Engl J Med. 2022;386:194–195. doi: 10.1056/NEJMc2117224. [DOI] [PubMed] [Google Scholar]
- 29.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;6:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kho ZY, Lal SK. The Human Gut Microbiome - A Potential Controller of Wellness and Disease. Front Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsugawa H. Advances in computational metabolomics and databases deepen the understanding of metabolisms. Curr Opin Biotechnol. 2018;54:10–17. doi: 10.1016/j.copbio.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Barko PC, McMichael MA, Swanson KS, Williams DA. The Gastrointestinal Microbiome: A Review. J Vet Intern Med. 2018;1:9–25. doi: 10.1111/jvim.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li DS, Huang QF, Guan LH, Zhang HZ, Li X, Fu KL, Chen YX, Wan JB, Huang M, Bi HC. Targeted bile acids and gut microbiome profiles reveal the hepato-protective effect of WZ tablet (Schisandra sphenanthera extract) against LCA-induced cholestasis. Chin J Nat Med. 2020;18:211–218. doi: 10.1016/S1875-5364(20)30023-6. [DOI] [PubMed] [Google Scholar]
- 34.Yin S, Guo P, Hai D, Xu L, Shu J, Zhang W, Khan MI, Kurland IJ, Qiu Y, Liu Y. Optimization of GC/TOF MS analysis conditions for assessing host-gut microbiota metabolic interactions: Chinese rhubarb alters fecal aromatic amino acids and phenol metabolism. Anal Chim Acta. 2017;995:21–33. doi: 10.1016/j.aca.2017.09.042. [DOI] [PubMed] [Google Scholar]
- 35.Schönfeld P, Wojtczak L. Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J Lipid Res. 2016;57:943–954. doi: 10.1194/jlr.R067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyagawa Y, Mori T, Goto K, Kawahara I, Fujiwara-Tani R, Kishi S, Sasaki T, Fujii K, Ohmori H, Kuniyasu H. Intake of medium-chain fatty acids induces myocardial oxidative stress and atrophy. Lipids Health Dis. 2018;17:258. doi: 10.1186/s12944-018-0908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyudinina AY, Ivankova GE, Bojko ER. Priority use of medium-chain fatty acids during high-intensity exercise in cross-country skiers. J Int Soc Sports Nutr. 2018;15:57. doi: 10.1186/s12970-018-0265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yunus IS, Jones PR. Photosynthesis-dependent biosynthesis of medium chain-length fatty acids and alcohols. Metab Eng. 2018;49:59–68. doi: 10.1016/j.ymben.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 39.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA 3rd IBDMDB Investigators, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aden K, Rehman A, Waschina S, Pan WH, Walker A, Lucio M, Nunez AM, Bharti R, Zimmerman J, Bethge J, Schulte B, Schulte D, Franke A, Nikolaus S, Schroeder JO, Vandeputte D, Raes J, Szymczak S, Waetzig GH, Zeuner R, Schmitt-Kopplin P, Kaleta C, Schreiber S, Rosenstiel P. Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology. 2019;157:1279–1292.e11. doi: 10.1053/j.gastro.2019.07.025. [DOI] [PubMed] [Google Scholar]
- 41.Weng YJ, Gan HY, Li X, Huang Y, Li ZC, Deng HM, Chen SZ, Zhou Y, Wang LS, Han YP, Tan YF, Song YJ, Du ZM, Liu YY, Wang Y, Qin N, Bai Y, Yang RF, Bi YJ, Zhi FC. Correlation of diet, microbiota and metabolite networks in inflammatory bowel disease. J Dig Dis. 2019;20:447–459. doi: 10.1111/1751-2980.12795. [DOI] [PubMed] [Google Scholar]
- 42.Bjerrum JT, Wang Y, Hao F, Coskun M, Ludwig C, Günther U, Nielsen OH. Metabonomics of human fecal extracts characterize ulcerative colitis, Crohn's disease and healthy individuals. Metabolomics. 2015;11:122–133. doi: 10.1007/s11306-014-0677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 44.Clerc F, Novokmet M, Dotz V, Reiding KR, de Haan N, Kammeijer GSM, Dalebout H, Bladergroen MR, Vukovic F, Rapp E IBD-BIOM Consortium, Targan SR, Barron G, Manetti N, Latiano A, McGovern DPB, Annese V, Lauc G, Wuhrer M. Plasma N-Glycan Signatures Are Associated With Features of Inflammatory Bowel Diseases. Gastroenterology. 2018;155:829–843. doi: 10.1053/j.gastro.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 45.Keshteli AH, van den Brand FF, Madsen KL, Mandal R, Valcheva R, Kroeker KI, Han B, Bell RC, Cole J, Hoevers T, Wishart DS, Fedorak RN, Dieleman LA. Dietary and metabolomic determinants of relapse in ulcerative colitis patients: A pilot prospective cohort study. World J Gastroenterol. 2017;23:3890–3899. doi: 10.3748/wjg.v23.i21.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dawiskiba T, Deja S, Mulak A, Ząbek A, Jawień E, Pawełka D, Banasik M, Mastalerz-Migas A, Balcerzak W, Kaliszewski K, Skóra J, Barć P, Korta K, Pormańczuk K, Szyber P, Litarski A, Młynarz P. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J Gastroenterol. 2014;20:163–174. doi: 10.3748/wjg.v20.i1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daniluk U, Daniluk J, Kucharski R, Kowalczyk T, Pietrowska K, Samczuk P, Filimoniuk A, Kretowski A, Lebensztejn D, Ciborowski M. Untargeted Metabolomics and Inflammatory Markers Profiling in Children With Crohn's Disease and Ulcerative Colitis-A Preliminary Study. Inflamm Bowel Dis. 2019;25:1120–1128. doi: 10.1093/ibd/izy402. [DOI] [PubMed] [Google Scholar]
- 48.Sairenji T, Collins KL, Evans DV. An Update on Inflammatory Bowel Disease. Prim Care. 2017;44:673–692. doi: 10.1016/j.pop.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 49.Abegunde AT, Muhammad BH, Bhatti O, Ali T. Environmental risk factors for inflammatory bowel diseases: Evidence based literature review. World J Gastroenterol. 2016;22:6296–6317. doi: 10.3748/wjg.v22.i27.6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J Gastroenterol. 2014;20:1192–1210. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scoville EA, Allaman MM, Brown CT, Motley AK, Horst SN, Williams CS, Koyama T, Zhao Z, Adams DW, Beaulieu DB, Schwartz DA, Wilson KT, Coburn LA. Alterations in Lipid, Amino Acid, and Energy Metabolism Distinguish Crohn's Disease from Ulcerative Colitis and Control Subjects by Serum Metabolomic Profiling. Metabolomics. 2018;14:17. doi: 10.1007/s11306-017-1311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishiumi S, Izumi Y, Yoshida M. Alterations in Docosahexaenoic Acid-Related Lipid Cascades in Inflammatory Bowel Disease Model Mice. Dig Dis Sci. 2018;63:1485–1496. doi: 10.1007/s10620-018-5025-4. [DOI] [PubMed] [Google Scholar]
- 53.Suh JH, Degagné É, Gleghorn EE, Setty M, Rodriguez A, Park KT, Verstraete SG, Heyman MB, Patel AS, Irek M, Gildengorin GL, Hubbard NE, Borowsky AD, Saba JD. Sphingosine-1-Phosphate Signaling and Metabolism Gene Signature in Pediatric Inflammatory Bowel Disease: A Matched-case Control Pilot Study. Inflamm Bowel Dis. 2018;24:1321–1334. doi: 10.1093/ibd/izy007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wlodarska M, Luo C, Kolde R, d'Hennezel E, Annand JW, Heim CE, Krastel P, Schmitt EK, Omar AS, Creasey EA, Garner AL, Mohammadi S, O'Connell DJ, Abubucker S, Arthur TD, Franzosa EA, Huttenhower C, Murphy LO, Haiser HJ, Vlamakis H, Porter JA, Xavier RJ. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe. 2017;22:25–37.e6. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, Bertha M, Cohen M, Garber J, Khalili H, Gevers D, Ananthakrishnan AN, Kugathasan S, Lander ES, Blainey P, Vlamakis H, Xavier RJ, Huttenhower C. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017;9:103. doi: 10.1186/s13073-017-0490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu LC. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J Biomed Sci. 2018;25:79. doi: 10.1186/s12929-018-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu YJ, Tang B, Wang FC, Tang L, Lei YY, Luo Y, Huang SJ, Yang M, Wu LY, Wang W, Liu S, Yang SM, Zhao XY. Parthenolide ameliorates colon inflammation through regulating Treg/Th17 balance in a gut microbiota-dependent manner. Theranostics. 2020;10:5225–5241. doi: 10.7150/thno.43716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandana UK, Barlaskar NH, Gulzar ABM, Laskar IH, Kumar D, Paul P, Pandey P, Mazumder PB. Linking gut microbiota with the human diseases. Bioinformation. 2020;16:196–208. doi: 10.6026/97320630016196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E, McCullagh J, Sansom SN, Arancibia-Cárcamo CV, Uhlig HH, Powrie F. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity. 2019;50:432–445.e7. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larraufie P, Martin-Gallausiaux C, Lapaque N, Dore J, Gribble FM, Reimann F, Blottiere HM. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci Rep. 2018;8:74. doi: 10.1038/s41598-017-18259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Primec M, Klemenak M, Di Gioia D, Aloisio I, Bozzi Cionci N, Quagliariello A, Gorenjak M, Mičetić-Turk D, Langerholc T. Clinical intervention using Bifidobacterium strains in celiac disease children reveals novel microbial modulators of TNF-α and short-chain fatty acids. Clin Nutr. 2019;38:1373–1381. doi: 10.1016/j.clnu.2018.06.931. [DOI] [PubMed] [Google Scholar]
- 62.Tuncil YE, Thakkar RD, Marcia ADR, Hamaker BR, Lindemann SR. Divergent short-chain fatty acid production and succession of colonic microbiota arise in fermentation of variously-sized wheat bran fractions. Sci Rep. 2018;8:16655. doi: 10.1038/s41598-018-34912-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun N, Wang L, Tong L, Zhou X, Liu L, Sun Y, Zhou S. Comparison of Structural and Functional Characterizations of Arabinoxylans from Different Wheat Processing Varieties. Plant Foods Hum Nutr. 2019;74:376–382. doi: 10.1007/s11130-019-00734-w. [DOI] [PubMed] [Google Scholar]
- 64.Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, Stappenbeck TS. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou B, Yuan Y, Zhang S, Guo C, Li X, Li G, Xiong W, Zeng Z. Intestinal Flora and Disease Mutually Shape the Regional Immune System in the Intestinal Tract. Front Immunol. 2020;11:575. doi: 10.3389/fimmu.2020.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim M, Qie Y, Park J, Kim CH. Gut Microbial Metabolites Fuel Host Antibody Responses. Cell Host Microbe. 2016;20:202–214. doi: 10.1016/j.chom.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, Sparwasser T, Bérard M, Cerf-Bensussan N, Eberl G. A Weaning Reaction to Microbiota Is Required for Resistance to Immunopathologies in the Adult. Immunity. 2019;50:1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 68.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 69.Laserna-Mendieta EJ, Clooney AG, Carretero-Gomez JF, Moran C, Sheehan D, Nolan JA, Hill C, Gahan CGM, Joyce SA, Shanahan F, Claesson MJ. Determinants of Reduced Genetic Capacity for Butyrate Synthesis by the Gut Microbiome in Crohn's Disease and Ulcerative Colitis. J Crohns Colitis. 2018;12:204–216. doi: 10.1093/ecco-jcc/jjx137. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Leong LEX, Keating RL, Kanno T, Abell GCJ, Mobegi FM, Choo JM, Wesselingh SL, Mason AJ, Burr LD, Rogers GB. Opportunistic bacteria confer the ability to ferment prebiotic starch in the adult cystic fibrosis gut. Gut Microbes. 2019;10:367–381. doi: 10.1080/19490976.2018.1534512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hove H, Mortensen PB. Influence of intestinal inflammation (IBD) and small and large bowel length on fecal short-chain fatty acids and lactate. Dig Dis Sci. 1995;40:1372–1380. doi: 10.1007/BF02065554. [DOI] [PubMed] [Google Scholar]
- 72.Pekmez CT, Dragsted LO, Brahe LK. Gut microbiota alterations and dietary modulation in childhood malnutrition - The role of short chain fatty acids. Clin Nutr. 2019;38:615–630. doi: 10.1016/j.clnu.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 73.Wang S, Dong W, Liu L, Xu M, Wang Y, Liu T, Zhang Y, Wang B, Cao H. Interplay between bile acids and the gut microbiota promotes intestinal carcinogenesis. Mol Carcinog. 2019;58:1155–1167. doi: 10.1002/mc.22999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao H, Kang S. The Role of the Gut Microbiome in Energy Balance With a Focus on the Gut-Adipose Tissue Axis. Front Genet. 2020;11:297. doi: 10.3389/fgene.2020.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cani PD, Van Hul M, Lefort C, Depommier C, Rastelli M, Everard A. Microbial regulation of organismal energy homeostasis. Nat Metab. 2019;1:34–46. doi: 10.1038/s42255-018-0017-4. [DOI] [PubMed] [Google Scholar]
- 77.Schmalle V, Lorentz A. Role of the microbiota in circadian rhythms of the host. Chronobiol Int. 2020;37:301–310. doi: 10.1080/07420528.2020.1726374. [DOI] [PubMed] [Google Scholar]
- 78.Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B, Lequin O, Kharrat P, Thomas G, Rainteau D, Aubry C, Breyner N, Afonso C, Lavielle S, Grill JP, Chassaing G, Chatel JM, Trugnan G, Xavier R, Langella P, Sokol H, Seksik P. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut. 2016;65:415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barra M, Danino T, Garrido D. Engineered Probiotics for Detection and Treatment of Inflammatory Intestinal Diseases. Front Bioeng Biotechnol. 2020;8:265. doi: 10.3389/fbioe.2020.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tully DC, Rucker PV, Chianelli D, Williams J, Vidal A, Alper PB, Mutnick D, Bursulaya B, Schmeits J, Wu X, Bao D, Zoll J, Kim Y, Groessl T, McNamara P, Seidel HM, Molteni V, Liu B, Phimister A, Joseph SB, Laffitte B. Discovery of Tropifexor (LJN452), a Highly Potent Non-bile Acid FXR Agonist for the Treatment of Cholestatic Liver Diseases and Nonalcoholic Steatohepatitis (NASH) J Med Chem. 2017;60:9960–9973. doi: 10.1021/acs.jmedchem.7b00907. [DOI] [PubMed] [Google Scholar]
- 81.Deutschmann K, Reich M, Klindt C, Dröge C, Spomer L, Häussinger D, Keitel V. Bile acid receptors in the biliary tree: TGR5 in physiology and disease. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1319–1325. doi: 10.1016/j.bbadis.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 82.Joyce SA, Gahan CG. Disease-Associated Changes in Bile Acid Profiles and Links to Altered Gut Microbiota. Dig Dis. 2017;35:169–177. doi: 10.1159/000450907. [DOI] [PubMed] [Google Scholar]
- 83.Vaughn BP, Kaiser T, Staley C, Hamilton MJ, Reich J, Graiziger C, Singroy S, Kabage AJ, Sadowsky MJ, Khoruts A. A pilot study of fecal bile acid and microbiota profiles in inflammatory bowel disease and primary sclerosing cholangitis. Clin Exp Gastroenterol. 2019;12:9–19. doi: 10.2147/CEG.S186097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darnaud M, Dos Santos A, Gonzalez P, Augui S, Lacoste C, Desterke C, De Hertogh G, Valentino E, Braun E, Zheng J, Boisgard R, Neut C, Dubuquoy L, Chiappini F, Samuel D, Lepage P, Guerrieri F, Doré J, Bréchot C, Moniaux N, Faivre J. Enteric Delivery of Regenerating Family Member 3 alpha Alters the Intestinal Microbiota and Controls Inflammation in Mice With Colitis. Gastroenterology. 2018;154:1009–1023.e14. doi: 10.1053/j.gastro.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 85.Rau M, Stieger B, Monte MJ, Schmitt J, Jahn D, Frey-Wagner I, Raselli T, Marin JJ, Müllhaupt B, Rogler G, Geier A. Alterations in Enterohepatic Fgf15 Signaling and Changes in Bile Acid Composition Depend on Localization of Murine Intestinal Inflammation. Inflamm Bowel Dis. 2016;22:2382–2389. doi: 10.1097/MIB.0000000000000879. [DOI] [PubMed] [Google Scholar]
- 86.Xu M, Cen M, Shen Y, Zhu Y, Cheng F, Tang L, Hu W, Dai N. Deoxycholic Acid-Induced Gut Dysbiosis Disrupts Bile Acid Enterohepatic Circulation and Promotes Intestinal Inflammation. Dig Dis Sci. 2021;66:568–576. doi: 10.1007/s10620-020-06208-3. [DOI] [PubMed] [Google Scholar]
- 87.Ticho AL, Malhotra P, Dudeja PK, Gill RK, Alrefai WA. Bile Acid Receptors and Gastrointestinal Functions. Liver Res. 2019;3:31–39. doi: 10.1016/j.livres.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guard BC, Suchodolski JS. HORSE SPECIES SYMPOSIUM: Canine intestinal microbiology and metagenomics: From phylogeny to function. J Anim Sci. 2016;94:2247–2261. doi: 10.2527/jas.2015-0029. [DOI] [PubMed] [Google Scholar]
- 89.Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C, Kasper DL. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature. 2020;577:410–415. doi: 10.1038/s41586-019-1865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo J, Shao J, Yang Y, Niu X, Liao J, Zhao Q, Wang D, Li S, Hu J. Gut Microbiota in Patients with Polycystic Ovary Syndrome: a Systematic Review. Reprod Sci. 2022;29:69–83. doi: 10.1007/s43032-020-00430-0. [DOI] [PubMed] [Google Scholar]
- 91.Van den Bossche L, Borsboom D, Devriese S, Van Welden S, Holvoet T, Devisscher L, Hindryckx P, De Vos M, Laukens D. Tauroursodeoxycholic acid protects bile acid homeostasis under inflammatory conditions and dampens Crohn's disease-like ileitis. Lab Invest. 2017;97:519–529. doi: 10.1038/labinvest.2017.6. [DOI] [PubMed] [Google Scholar]
- 92.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, Sauk JS, Wilson RG, Stevens BW, Scott JM, Pierce K, Deik AA, Bullock K, Imhann F, Porter JA, Zhernakova A, Fu J, Weersma RK, Wijmenga C, Clish CB, Vlamakis H, Huttenhower C, Xavier RJ. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Torres J, Palmela C, Brito H, Bao X, Ruiqi H, Moura-Santos P, Pereira da Silva J, Oliveira A, Vieira C, Perez K, Itzkowitz SH, Colombel JF, Humbert L, Rainteau D, Cravo M, Rodrigues CM, Hu J. The gut microbiota, bile acids and their correlation in primary sclerosing cholangitis associated with inflammatory bowel disease. United European Gastroenterol J. 2018;6:112–122. doi: 10.1177/2050640617708953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lucas López R, Grande Burgos MJ, Gálvez A, Pérez Pulido R. The human gastrointestinal tract and oral microbiota in inflammatory bowel disease: a state of the science review. APMIS. 2017;125:3–10. doi: 10.1111/apm.12609. [DOI] [PubMed] [Google Scholar]
- 95.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lorenzo-Zúñiga V, Bartolí R, Planas R, Hofmann AF, Viñado B, Hagey LR, Hernández JM, Mañé J, Alvarez MA, Ausina V, Gassull MA. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 2003;37:551–557. doi: 10.1053/jhep.2003.50116. [DOI] [PubMed] [Google Scholar]
- 97.Hajiagha MN, Taghizadeh S, Asgharzadeh M, Dao S, Ganbarov K, Köse Ş, Kafil HS. Gut Microbiota and Human Body Interactions; Its Impact on Health: A Review. Curr Pharm Biotechnol. 2022;23:4–14. doi: 10.2174/1389201022666210104115836. [DOI] [PubMed] [Google Scholar]
- 98.Van den Bossche L, Hindryckx P, Devisscher L, Devriese S, Van Welden S, Holvoet T, Vilchez-Vargas R, Vital M, Pieper DH, Vanden Bussche J, Vanhaecke L, Van de Wiele T, De Vos M, Laukens D. Ursodeoxycholic Acid and Its Taurine- or Glycine-Conjugated Species Reduce Colitogenic Dysbiosis and Equally Suppress Experimental Colitis in Mice. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.02766-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baars A, Oosting A, Knol J, Garssen J, van Bergenhenegouwen J. The Gut Microbiota as a Therapeutic Target in IBD and Metabolic Disease: A Role for the Bile Acid Receptors FXR and TGR5. Microorganisms. 2015;3:641–666. doi: 10.3390/microorganisms3040641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Milshteyn A, Colosimo DA, Brady SF. Accessing Bioactive Natural Products from the Human Microbiome. Cell Host Microbe. 2018;23:725–736. doi: 10.1016/j.chom.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martín R, Miquel S, Benevides L, Bridonneau C, Robert V, Hudault S, Chain F, Berteau O, Azevedo V, Chatel JM, Sokol H, Bermúdez-Humarán LG, Thomas M, Langella P. Functional Characterization of Novel Faecalibacterium prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. prausnitzii as a Next-Generation Probiotic. Front Microbiol. 2017;8:1226. doi: 10.3389/fmicb.2017.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Beurden YH, de Groot PF, van Nood E, Nieuwdorp M, Keller JJ, Goorhuis A. Complications, effectiveness, and long term follow-up of fecal microbiota transfer by nasoduodenal tube for treatment of recurrent Clostridium difficile infection. United European Gastroenterol J. 2017;5:868–879. doi: 10.1177/2050640616678099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chang CJ, Lin TL, Tsai YL, Wu TR, Lai WF, Lu CC, Lai HC. Next generation probiotics in disease amelioration. J Food Drug Anal. 2019;27:615–622. doi: 10.1016/j.jfda.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ganesan K, Chung SK, Vanamala J, Xu B. Causal Relationship between Diet-Induced Gut Microbiota Changes and Diabetes: A Novel Strategy to Transplant Faecalibacterium prausnitzii in Preventing Diabetes. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19123720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rossi O, van Berkel LA, Chain F, Tanweer Khan M, Taverne N, Sokol H, Duncan SH, Flint HJ, Harmsen HJ, Langella P, Samsom JN, Wells JM. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep. 2016;6:18507. doi: 10.1038/srep18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Michonneau D, Latis E, Curis E, Dubouchet L, Ramamoorthy S, Ingram B, de Latour RP, Robin M, de Fontbrune FS, Chevret S, Rogge L, Socié G. Metabolomics analysis of human acute graft-versus-host disease reveals changes in host and microbiota-derived metabolites. Nat Commun. 2019;10:5695. doi: 10.1038/s41467-019-13498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Amedei A, Morbidelli L. Circulating Metabolites Originating from Gut Microbiota Control Endothelial Cell Function. Molecules. 2019;24 doi: 10.3390/molecules24213992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ji Y, Gao Y, Chen H, Yin Y, Zhang W. Indole-3-Acetic Acid Alleviates Nonalcoholic Fatty Liver Disease in Mice via Attenuation of Hepatic Lipogenesis, and Oxidative and Inflammatory Stress. Nutrients. 2019;11 doi: 10.3390/nu11092062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Armand L, Andriamihaja M, Gellenoncourt S, Bitane V, Lan A, Blachier F. In vitro impact of amino acid-derived bacterial metabolites on colonocyte mitochondrial activity, oxidative stress response and DNA integrity. Biochim Biophys Acta Gen Subj. 2019;1863:1292–1301. doi: 10.1016/j.bbagen.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 110.Zhang S, Zhao Y, Ohland C, Jobin C, Sang S. Microbiota facilitates the formation of the aminated metabolite of green tea polyphenol (-)-epigallocatechin-3-gallate which trap deleterious reactive endogenous metabolites. Free Radic Biol Med. 2019;131:332–344. doi: 10.1016/j.freeradbiomed.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Álvarez-Cilleros D, Ramos S, Goya L, Martín MÁ. Colonic metabolites from flavanols stimulate nitric oxide production in human endothelial cells and protect against oxidative stress-induced toxicity and endothelial dysfunction. Food Chem Toxicol. 2018;115:88–97. doi: 10.1016/j.fct.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 112.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327–339.e4. doi: 10.1053/j.gastro.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li KY, Wang JL, Wei JP, Gao SY, Zhang YY, Wang LT, Liu G. Fecal microbiota in pouchitis and ulcerative colitis. World J Gastroenterol. 2016;22:8929–8939. doi: 10.3748/wjg.v22.i40.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.De Preter V, Machiels K, Joossens M, Arijs I, Matthys C, Vermeire S, Rutgeerts P, Verbeke K. Faecal metabolite profiling identifies medium-chain fatty acids as discriminating compounds in IBD. Gut. 2015;64:447–458. doi: 10.1136/gutjnl-2013-306423. [DOI] [PubMed] [Google Scholar]
- 115.Pierre JF, Hinterleitner R, Bouziat R, Hubert NA, Leone V, Miyoshi J, Jabri B, Chang EB. Dietary antioxidant micronutrients alter mucosal inflammatory risk in a murine model of genetic and microbial susceptibility. J Nutr Biochem. 2018;54:95–104. doi: 10.1016/j.jnutbio.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Landy J, Walker AW, Li JV, Al-Hassi HO, Ronde E, English NR, Mann ER, Bernardo D, McLaughlin SD, Parkhill J, Ciclitira PJ, Clark SK, Knight SC, Hart AL. Variable alterations of the microbiota, without metabolic or immunological change, following faecal microbiota transplantation in patients with chronic pouchitis. Sci Rep. 2015;5:12955. doi: 10.1038/srep12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 118.Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017;18:2. doi: 10.1186/s12865-016-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Little R, Wine E, Kamath BM, Griffiths AM, Ricciuto A. Gut microbiome in primary sclerosing cholangitis: A review. World J Gastroenterol. 2020;26:2768–2780. doi: 10.3748/wjg.v26.i21.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gasaly N, de Vos P, Hermoso MA. Impact of Bacterial Metabolites on Gut Barrier Function and Host Immunity: A Focus on Bacterial Metabolism and Its Relevance for Intestinal Inflammation. Front Immunol. 2021;12:658354. doi: 10.3389/fimmu.2021.658354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang C, Zhao Y, Im S, Nakatsu C, Jones-Hall Y, Jiang Q. Vitamin E delta-tocotrienol and metabolite 13'-carboxychromanol inhibit colitis-associated colon tumorigenesis and modulate gut microbiota in mice. J Nutr Biochem. 2021;89:108567. doi: 10.1016/j.jnutbio.2020.108567. [DOI] [PubMed] [Google Scholar]
- 122.Bischoff SC, Kaden-Volynets V, Filipe Rosa L, Guseva D, Seethaler B. Regulation of the gut barrier by carbohydrates from diet - Underlying mechanisms and possible clinical implications. Int J Med Microbiol. 2021;311:151499. doi: 10.1016/j.ijmm.2021.151499. [DOI] [PubMed] [Google Scholar]
- 123.Shi J, Du P, Xie Q, Wang N, Li H, Smith EE, Li C, Liu F, Huo G, Li B. Protective effects of tryptophan-catabolizing Lactobacillus plantarum KLDS 1.0386 against dextran sodium sulfate-induced colitis in mice. Food Funct. 2020;11:10736–10747. doi: 10.1039/d0fo02622k. [DOI] [PubMed] [Google Scholar]
- 124.Bunt DV, Minnaard AJ, El Aidy S. Potential Modulatory Microbiome Therapies for Prevention or Treatment of Inflammatory Bowel Diseases. Pharmaceuticals (Basel) 2021;14 doi: 10.3390/ph14060506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pascoal LB, Rodrigues PB, Genaro LM, Gomes ABDSP, Toledo-Teixeira DA, Parise PL, Bispo-Dos-Santos K, Simeoni CL, Guimarães PV, Buscaratti LI, Elston JGA, Marques-Souza H, Martins-de-Souza D, Ayrizono MLS, Velloso LA, Proenca-Modena JL, Moraes-Vieira PMM, Mori MAS, Farias AS, Vinolo MAR, Leal RF. Microbiota-derived short-chain fatty acids do not interfere with SARS-CoV-2 infection of human colonic samples. Gut Microbes. 2021;13:1–9. doi: 10.1080/19490976.2021.1874740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Singhal S, Rani V. Study to Explore Plant-Derived Trimethylamine Lyase Enzyme Inhibitors to Address Gut Dysbiosis. Appl Biochem Biotechnol. 2022;194:99–123. doi: 10.1007/s12010-021-03747-x. [DOI] [PubMed] [Google Scholar]
- 127.Li B, Zhang H, Shi L, Li R, Luo Y, Deng Y, Li S, Liu Z. Saccharomyces boulardii alleviates DSS-induced intestinal barrier dysfunction and inflammation in humanized mice. Food Funct. 2022;13:102–112. doi: 10.1039/d1fo02752b. [DOI] [PubMed] [Google Scholar]
- 128.Komatsu Y, Shimizu Y, Yamano M, Kikuchi M, Nakamura K, Ayabe T, Aizawa T. Disease progression-associated alterations in fecal metabolites in SAMP1/YitFc mice, a Crohn's disease model. Metabolomics. 2020;16:48. doi: 10.1007/s11306-020-01671-5. [DOI] [PubMed] [Google Scholar]
- 129.Melhem H, Kaya B, Ayata CK, Hruz P, Niess JH. Metabolite-Sensing G Protein-Coupled Receptors Connect the Diet-Microbiota-Metabolites Axis to Inflammatory Bowel Disease. Cells. 2019;8 doi: 10.3390/cells8050450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ong HS, Yim HCH. Microbial Factors in Inflammatory Diseases and Cancers. Adv Exp Med Biol. 2017;1024:153–174. doi: 10.1007/978-981-10-5987-2_7. [DOI] [PubMed] [Google Scholar]
- 131.Yang W, Cong Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell Mol Immunol. 2021;18:866–877. doi: 10.1038/s41423-021-00661-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nishida A, Nishino K, Sakai K, Owaki Y, Noda Y, Imaeda H. Can control of gut microbiota be a future therapeutic option for inflammatory bowel disease? World J Gastroenterol. 2021;27:3317–3326. doi: 10.3748/wjg.v27.i23.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xiu W, Chen Q, Wang Z, Wang J, Zhou Z. Microbiota-derived short chain fatty acid promotion of Amphiregulin expression by dendritic cells is regulated by GPR43 and Blimp-1. Biochem Biophys Res Commun. 2020;533:282–288. doi: 10.1016/j.bbrc.2020.09.027. [DOI] [PubMed] [Google Scholar]
- 134.Qiu X, Macchietto MG, Liu X, Lu Y, Ma Y, Guo H, Saqui-Salces M, Bernlohr DA, Chen C, Shen S, Chen X. Identification of gut microbiota and microbial metabolites regulated by an antimicrobial peptide lipocalin 2 in high fat diet-induced obesity. Int J Obes (Lond) 2021;45:143–154. doi: 10.1038/s41366-020-00712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Markandey M, Bajaj A, Ilott NE, Kedia S, Travis S, Powrie F, Ahuja V. Gut microbiota: sculptors of the intestinal stem cell niche in health and inflammatory bowel disease. Gut Microbes. 2021;13:1990827. doi: 10.1080/19490976.2021.1990827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.DeLuca JA, Allred KF, Menon R, Riordan R, Weeks BR, Jayaraman A, Allred CD. Bisphenol-A alters microbiota metabolites derived from aromatic amino acids and worsens disease activity during colitis. Exp Biol Med (Maywood) 2018;243:864–875. doi: 10.1177/1535370218782139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ward JBJ, Lajczak NK, Kelly OB, O'Dwyer AM, Giddam AK, Ní Gabhann J, Franco P, Tambuwala MM, Jefferies CA, Keely S, Roda A, Keely SJ. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am J Physiol Gastrointest Liver Physiol. 2017;312:G550–G558. doi: 10.1152/ajpgi.00256.2016. [DOI] [PubMed] [Google Scholar]
- 138.Verdugo-Meza A, Ye J, Dadlani H, Ghosh S, Gibson DL. Connecting the Dots Between Inflammatory Bowel Disease and Metabolic Syndrome: A Focus on Gut-Derived Metabolites. Nutrients. 2020;12 doi: 10.3390/nu12051434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Song D, Lai L, Ran Z. Metabolic Regulation of Group 3 Innate Lymphoid Cells and Their Role in Inflammatory Bowel Disease. Front Immunol. 2020;11:580467. doi: 10.3389/fimmu.2020.580467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wojciech L, Tan KSW, Gascoigne NRJ. Taming the Sentinels: Microbiome-Derived Metabolites and Polarization of T Cells. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21207740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mogilnicka I, Ufnal M. Gut Mycobiota and Fungal Metabolites in Human Homeostasis. Curr Drug Targets. 2019;20:232–240. doi: 10.2174/1389450119666180724125020. [DOI] [PubMed] [Google Scholar]
- 142.Loo YT, Howell K, Chan M, Zhang P, Ng K. Modulation of the human gut microbiota by phenolics and phenolic fiber-rich foods. Compr Rev Food Sci Food Saf. 2020;19:1268–1298. doi: 10.1111/1541-4337.12563. [DOI] [PubMed] [Google Scholar]
- 143.Banfi D, Moro E, Bosi A, Bistoletti M, Cerantola S, Crema F, Maggi F, Giron MC, Giaroni C, Baj A. Impact of Microbial Metabolites on Microbiota-Gut-Brain Axis in Inflammatory Bowel Disease. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22041623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Coman V, Vodnar DC. Gut microbiota and old age: Modulating factors and interventions for healthy longevity. Exp Gerontol. 2020;141:111095. doi: 10.1016/j.exger.2020.111095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zeng SL, Li SZ, Xiao PT, Cai YY, Chu C, Chen BZ, Li P, Li J, Liu EH. Citrus polymethoxyflavones attenuate metabolic syndrome by regulating gut microbiome and amino acid metabolism. Sci Adv. 2020;6:eaax6208. doi: 10.1126/sciadv.aax6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kc D, Sumner R, Lippmann S. Gut microbiota and health. Postgrad Med. 2020;132:274. doi: 10.1080/00325481.2019.1662711. [DOI] [PubMed] [Google Scholar]
- 147.Yu F, Jiang R, Han W, Zhan G, Xu X, Jiang X, Wang L, Xiang S, Zhou Q, Liu C, Zhu B, Hua F, Yang C. Gut microbiota transplantation from db/db mice induces diabetes-like phenotypes and alterations in Hippo signaling in pseudo germ-free mice. Aging (Albany NY) 2020;12:24156–24167. doi: 10.18632/aging.104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Woldeamlak B, Yirdaw K, Biadgo B. Role of Gut Microbiota in Type 2 Diabetes Mellitus and Its Complications: Novel Insights and Potential Intervention Strategies. Korean J Gastroenterol. 2019;74:314–320. doi: 10.4166/kjg.2019.74.6.314. [DOI] [PubMed] [Google Scholar]
- 149.Napolitano M, Covasa M. Microbiota Transplant in the Treatment of Obesity and Diabetes: Current and Future Perspectives. Front Microbiol. 2020;11:590370. doi: 10.3389/fmicb.2020.590370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rasmussen TS, Koefoed AK, Jakobsen RR, Deng L, Castro-Mejía JL, Brunse A, Neve H, Vogensen FK, Nielsen DS. Bacteriophage-mediated manipulation of the gut microbiome - promises and presents limitations. FEMS Microbiol Rev. 2020;44:507–521. doi: 10.1093/femsre/fuaa020. [DOI] [PubMed] [Google Scholar]
- 151.Barba C, Soulage CO, Caggiano G, Glorieux G, Fouque D, Koppe L. Effects of Fecal Microbiota Transplantation on Composition in Mice with CKD. Toxins (Basel) 2020;12 doi: 10.3390/toxins12120741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gilbert B, Schrenzel J. Fecal microbiota transplantation : current status and prospects. Rev Med Suisse. 2019;15:976–983. [PubMed] [Google Scholar]
- 153.Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Kheradman R, Heuman D, Wang J, Gurry T, Williams R, Sikaroodi M, Fuchs M, Alm E, John B, Thacker LR, Riva A, Smith M, Taylor-Robinson SD, Gillevet PM. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology. 2017;66:1727–1738. doi: 10.1002/hep.29306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Simpson CA, Diaz-Arteche C, Eliby D, Schwartz OS, Simmons JG, Cowan CSM. The gut microbiota in anxiety and depression - A systematic review. Clin Psychol Rev. 2021;83:101943. doi: 10.1016/j.cpr.2020.101943. [DOI] [PubMed] [Google Scholar]
- 155.Merra G, Noce A, Marrone G, Cintoni M, Tarsitano MG, Capacci A, De Lorenzo A. Influence of Mediterranean Diet on Human Gut Microbiota. Nutrients. 2020;13 doi: 10.3390/nu13010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Konjevod M, Nikolac Perkovic M, Sáiz J, Svob Strac D, Barbas C, Rojo D. Metabolomics analysis of microbiota-gut-brain axis in neurodegenerative and psychiatric diseases. J Pharm Biomed Anal. 2021;194:113681. doi: 10.1016/j.jpba.2020.113681. [DOI] [PubMed] [Google Scholar]
- 157.Duttaroy AK. Role of Gut Microbiota and Their Metabolites on Atherosclerosis, Hypertension and Human Blood Platelet Function: A Review. Nutrients. 2021;13 doi: 10.3390/nu13010144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fu Y, Wang Y, Gao H, Li D, Jiang R, Ge L, Tong C, Xu K. Associations among Dietary Omega-3 Polyunsaturated Fatty Acids, the Gut Microbiota, and Intestinal Immunity. Mediators Inflamm. 2021;2021:8879227. doi: 10.1155/2021/8879227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wan MLY, Co VA, El-Nezami H. Dietary polyphenol impact on gut health and microbiota. Crit Rev Food Sci Nutr. 2021;61:690–711. doi: 10.1080/10408398.2020.1744512. [DOI] [PubMed] [Google Scholar]
- 160.Settanni CR, Bibbò S, Ianiro G, Rinninella E, Cintoni M, Mele MC, Cammarota G, Gasbarrini A. Gastrointestinal involvement of autism spectrum disorder: focus on gut microbiota. Expert Rev Gastroenterol Hepatol. 2021;15:599–622. doi: 10.1080/17474124.2021.1869938. [DOI] [PubMed] [Google Scholar]
- 161.Badgeley A, Anwar H, Modi K, Murphy P, Lakshmikuttyamma A. Effect of probiotics and gut microbiota on anti-cancer drugs: Mechanistic perspectives. Biochim Biophys Acta Rev Cancer. 2021;1875:188494. doi: 10.1016/j.bbcan.2020.188494. [DOI] [PubMed] [Google Scholar]
- 162.Zhou A, Lei Y, Tang L, Hu S, Yang M, Wu L, Yang S, Tang B. Gut Microbiota: the Emerging Link to Lung Homeostasis and Disease. J Bacteriol. 2021;203 doi: 10.1128/JB.00454-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Rawat K, Singh N, Kumari P, Saha L. A review on preventive role of ketogenic diet (KD) in CNS disorders from the gut microbiota perspective. Rev Neurosci. 2021;32:143–157. doi: 10.1515/revneuro-2020-0078. [DOI] [PubMed] [Google Scholar]
- 164.Fan X, Jin Y, Chen G, Ma X, Zhang L. Gut Microbiota Dysbiosis Drives the Development of Colorectal Cancer. Digestion. 2021;102:508–515. doi: 10.1159/000508328. [DOI] [PubMed] [Google Scholar]
- 165.Gasmi A, Mujawdiya PK, Pivina L, Doşa A, Semenova Y, Benahmed AG, Bjørklund G. Relationship between Gut Microbiota, Gut Hyperpermeability and Obesity. Curr Med Chem. 2021;28:827–839. doi: 10.2174/0929867327666200721160313. [DOI] [PubMed] [Google Scholar]


