Abstract
Nogalamycin is an anthracycline antibiotic produced by Streptomyces nogalater. Its aglycone has a unique stereochemistry (7S, 9S, 10R) compared to that of most other anthracyclines (7S, 9R, 10R). The gene snoaL, encoding a nogalonic acid methyl ester cyclase for nogalamycin, was used to generate nogalamycinone, demonstrating that the single cyclase dictates the C-9 stereochemistry of anthracyclines.
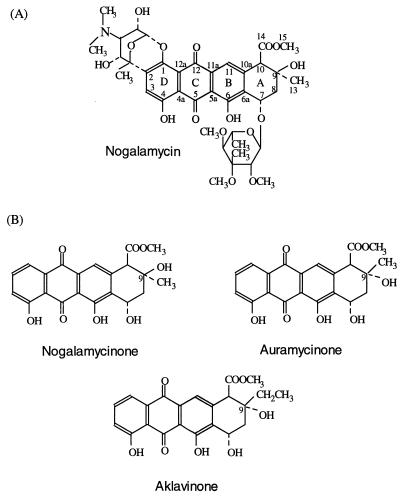
Most anthracyclines that have been investigated are biosynthesized via aklavinone as a key intermediate. Biosynthesis has been well demonstrated with daunomycins (e.g., see references 8 and 13), which, due to their clinical significance, are the best known anthracyclines. However, nogalamycin (Fig. 1A), produced by Streptomyces nogalater (ATCC 27451), is a distinctive anthracycline. Its aglycone (nogalamycinone) is synthesized via a polyketide biosynthetic pathway from 10 acetates (17). Nogalamycin has two sugar residues: a neutral sugar, nogalose, and a dimethyl amino sugar, nogalamine. The features that make nogalamycin different from most other anthracyclines are the attachment of nogalamine at both C-1 (by a typical glycosidic bond) and at C-2 (by an unusual C-C bond) (16) and the opposite stereochemistry at C-9 (1). In addition to the nogalamycin group, steffimycins are the only anthracyclines known to have the 9S configuration (2).
FIG. 1.
The structures of nogalamycin (A) and nogalamycinone, auramycinone, and aklavinone (B).
It has not been possible to produce nogalamycinone (Fig. 1B), either (i) chemically, because nogalamine could not be removed from the aglycone (only the bisanhydro form of the aglycone was obtained by treatment with strong base at elevated temperatures) (16), or (ii) by genetic engineering, due to the lack of the cyclase determining the unique stereochemistry at C-9. In this paper, we report the production of nogalamycinone by genetically engineered Streptomyces lividans TK24, clarifying an important intermediate of nogalamycin biosynthesis.
Strains, culture conditions, and DNA manipulation.
Streptomyces strains were grown at 30°C in tryptone soya broth containing thiostrepton (50 μg/ml) for preparation of plasmid DNA and in E1 medium containing glucose (20 g/liter), soluble starch (20 g/liter), Farmamedia (5 g/liter), yeast extract (2.5 g/liter), CaCO3 (3 g/liter), NaCl (1 g/liter), MgSO4 · 7H2O (1 g/liter), and K2HPO4 · 3H2O (1 g/liter) in tap water (pH 7.5) for production of anthracycline metabolites (19). A DNA fragment derived from a nogalamycin biosynthetic cluster containing a cyclase gene was cloned in a pIJ486-based plasmid and introduced into S. lividans TK24 (6). Streptomyces galilaeus mutant H039 producing aklavinone-(rhodinose)2–3 (19) was used as a host in attempts to produce C-9 stereoisomers of aklavinone. DNA isolation and manipulation were carried out by standard methods (6, 14).
Cloning and sequencing of the gene for nogalamycin cyclization.
In our attempts to clarify the molecular genetics of nogalamycin biosynthesis, we expanded the previously characterized nogalamycin gene cluster (15, 20, 21). A fragment from this previously cloned nogalamycin biosynthetic region was used as a probe to screen for nogalamycin biosynthetic genes in a library of S. nogalater genomic DNA. Subcloning of a 10-kb BglII fragment from one of the hybridizing clones, pFDSno5, into pIJ486 gave pSY42. Plasmid pSY42 was subcloned in pUC19 for sequencing. The automatic ABI DNA sequenator (Perkin-Elmer) was used according to the manufacturer's instructions to locate a cyclase for nogalamycin biosynthesis. The sequence was analyzed with the GCG sequence analysis software package (version 8; Genetics Computer Group, Madison, Wis.), and an open reading frame for a cyclase, designated snoaL, was revealed. The translation table was modified to also accept GTG as a start codon, and codon usage was analyzed with published data (18). The deduced protein SnoaL, consisting of 134 amino acids, showed a remarkable similarity to the analogous cyclases derived from the aklavinone pathway. Compared to the consensus sequence determined by the LINEUP program from the deduced amino acid sequences of dnrD (9), dauD (4), rdmA (10), and acmA (7), similarity and identity were 84 and 72%, respectively. The major difference between nogalonic acid methyl ester (NAME) and aklanonic acid methyl ester (AAME) cyclases is that 10 highly conserved amino acids found at the amino terminus of AAME cyclases are missing in the NAME cyclase. The alignment of the deduced amino acid sequences is shown in Fig. 2.
FIG. 2.
Alignment of amino acid sequences (PILEUP) of five cyclases: DnrD (L40425 [9]) and DauD (L35154 [4]) from S. peucetius, RdmA (not completely sequenced) from Streptomyces purpurascens (U10405 [10]), AcmA from S. galilaeus (AF043550 [7]), and SnoaL from S. nogalater.
Expression of snoaL.
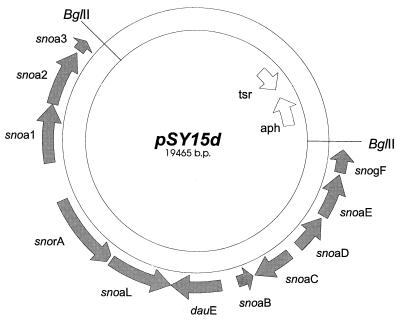
We have recently assembled an auramycinone biosynthetic gene cluster by stepwise cloning of genes from three different Streptomyces species (7). Auramycinone (Fig. 1B) was produced in S. lividans TK24 carrying the pIJ486-based plasmid pSY15b, which contains the following genes from S. nogalater: sno1 to -3 for minimal polyketide synthase (PKS), snoaB for mono-oxygenase, snoaC for methyl transferase, snoaD for polyketide reductase, snoaE for aromatase, snorA for an activator, and snogF for 3′,5′-epimerase involved in the deoxyhexose pathway, in addition to the genes for an AAME cyclase (acmA from Streptomyces galilaeus) and a C-7 ketoreductase (dauE from Streptomyces peucetius ATCC 27952, designated according to Dickens et al. [5]). In this work, the plasmid pSY15d (Fig. 3) was constructed by replacing the gene acmA encoding AAME cyclase in pSY15b with the NAME cyclase gene snoaL. Structural analysis (described below) confirmed that pSY15d caused the production of 9-epiauramycinone, called nogalamycinone (Fig. 1B), in S. lividans TK24. This indicates that only one gene product, the cyclase, is responsible for the C-9 stereochemistry of the anthracyclinone, since auramycinone differs from nogalamycinone only in its stereochemistry at C-9. Aklavinone, the anthracyclinone of aclacinomycins produced by S. galilaeus, and auramycinone have the same stereochemistry at C-9, differing only in the substituent, which is an ethyl or a methyl group, respectively (Fig. 1B). Thus, AAME cyclase catalyzes the cyclization, resulting in a typical R configuration at C-9. In contrast, NAME cyclase is responsible for the rare 9S configuration found in nogalamycinone. The aldol reaction closing the ring A (11) is highly specific, because a single Streptomyces strain does not produce 9-epi forms of an aglycone.
FIG. 3.
Diagram of the plasmid pSY15d. The ORFs are shown by arrows. sno or dau indicates that the ORF is cloned from S. nogalater or S. peucetius, respectively. The genes encode the following proteins: snoa3, acyl carrier protein; snoa2, ketosynthase II; snoa1, ketosynthase I; snorA, activator; snoaL, nogalonic acid methyl ester cyclase; dauE, aklaviketone reductase; snoaB, mono-oxygenase; snoaC, methyl transferase; snoaD, polyketide reductase; snoaE, aromatase; snoaF, 3′,5′-epimerase involved in the deoxyhexose pathway. The gene for SnorA is incomplete but functional.
Plasmid pSYEL56, carrying snoaL cloned in a pIJ486 vector downstream of the ermE promoter (3), was constructed in order to produce C-9 stereoisomers of anthracycline molecules that normally have the typical 9R stereochemistry. Plasmid pSYEL56 was first introduced into TK24, and after isolation from TK24, it was subsequently introduced into the S. galilaeus mutant H039 to see if it would promote formation of a 9S derivative of aklavinone-(rhodinose)2–3. However, the products of H039/pSYEL56 did not differ from the products of H039 when analyzed by high-performance liquid chromatography (HPLC). Because aklavinone has an ethyl group at C-9 instead of a methyl group like auramycinone and nogalamycinone (Fig. 1B), it is possible that aklanonic acid methyl ester is not an acceptable substrate for NAME cyclase.
Structural elucidation of nogalamycinone.
TK24/pSY15d was fermented on a 10-liter scale by using E1 medium (28°C, 330 rpm, 450 liters/min). After 160 h, the culture was extracted with dichloromethane-methanol (3:1) at pH 6. The organic layer was evaporated to dryness. A viscous residue was flashed through a SiO2 column and eluted with dichloromethane-methanol (10:0 to 9:1). Fractions containing nogalamycinone were further purified on a preparative reversed-phase column (LichroCART RP-18, 5 μm; mobile phase; acetonitrile–1% AcOH in water [1:1]) with a Merck-Hitachi HPLC.
Structural determination was accomplished by nuclear magnetic resonance (NMR) and spectral comparison with nogalamycinone and auramycinone, whose stereochemistries have been established. NMR spectra were recorded on a JEOL JNM-L400 spectrometer at 25°C. 1H and 13C samples were internally referenced to tetramethylsilane. Proton and carbon assignments were based on a conventional nuclear Overhauser enhancement (NOE) difference and heteronuclear shift quantum correlation (HSQC) and heteronuclear multiple-bond connectivity (HMBC) measurements. Spectra of nogalamycinone indicated a structure similar to that of auramycinone, except with respect to ring A. Ring A in anthracyclines has been shown to have two possible low-energy conformers, an α and β half-chair. However, the hydrogen bond between O7 and H(O9) stabilizes the α half-chair conformation of auramycinone (12). In nogalamycinone, the ring A adopted the other minimum energy conformation, a β half-chair, since the hydrogen bonding was inaccessible. Diagnostic couplings (Table 1) for β A-ring puckering were measured together with the absence of typical 4J between H8eq. and H10eq. Inversion of the stereochemistry at C-9 was deduced from a series of 1d NOE difference measurements. Strong NOE (20%) was observed between H7/H8eq., and moderate (5%) NOE was observed between 9-CH3/H10eq. The spectra indicated that auramycinone is similar to nogalamycinone, except that it has the opposite stereochemistry at C-9.
TABLE 1.
1H and 13C NMR spectroscopic data for auramycinone and nogalamycinone in CDCl3
| Site | NMR result for:
|
|||
|---|---|---|---|---|
| Auramycinone
|
Nogalamycinone
|
|||
| 1H (at 400 MHz) | 13C (at 100 MHz) | 1H (at 400 MHz) | 13C (at 100 MHz) | |
| 1 | 7.69, 1H, dd, 7.5, 1.3 | 120.2 | 7.76, 1H, dd, 7.5, 1.2 | 119.8 |
| 2 | 7.62, 1H, dd, 8.2, 7.5 | 137.4 | 7.67, 1H, dd, 8.3, 7, 5 | 137.4 |
| 3 | 7.23, 1H, dd, 8.2, 1.3 | 124.8 | 7.28, 1H, dd, 8.3, 1.2 | 124.8 |
| 4 | 162.4 | 162.5 | ||
| 4-OH | 11.77, 1H, s | 11.86, 1H, s | ||
| 4a | 115.5 | 115.6 | ||
| 5 | 192.3 | 192.7 | ||
| 5a | 114.4 | 114.6 | ||
| 6 | 161.1 | 160.9 | ||
| 6-OH | 12.55, 1H, s | 12.76, 1H, s | ||
| 6a | 132.4 | 134.1 | ||
| 7 | 5.33, 1H, dd, 5.1, 3.2 | 62.4 | 5.40, 1H, t, 7.0 | 64.0 |
| 7-OH | Exchange, broadened | 3.66, 1H, brs | ||
| 8A | 2.58, 1H, dd, 14.9, 5.1 | 37.0 | 2.66, 1H, dd, 13.9, 7.0 | 40.9 |
| 8B | 2.22, 1H, ddd, 14.9, 3.2, 1.5 | 1.89, 1H, dd, 13.9, 7.1 | ||
| 9 | 69.9 | 70.5 | ||
| 9-OH | 4.28, 1H, s | 3.49, 1H, brs | ||
| 10 | 4.04, 1H, dd, 1.5, 0.3 | 57.8 | 3.93, 1H, d, 0.8 | 56.0 |
| 10a | 142.3 | 142.1 | ||
| 11 | 7.55, 1H, d, 0.3 | 121.2 | 7.51, 1H, d, 0.8 | 120.1 |
| 11a | 133.2 | 133.3 | ||
| 12 | 180.9 | 180.9 | ||
| 12a | 132.6 | 132.1 | ||
| 13 | 1.41, 3H, s | 27.5 | 1.44, 3H, s | 28.7 |
| 14 | 171.3 | 173.0 | ||
| 15 | 3.70, 3H, s | 52.5 | 3.90, 3H, s | 52.6 |
Nucleotide sequence accession number.
The nucleotide sequence determined in this paper has been submitted to the GenBank database under accession no. AF187532.
Acknowledgments
We thank Paula Mykkänen for revising the English usage in this paper.
This research was supported by the Emil Aaltonen Foundation, by the Academy of Finland, by the Technology Development Center of Finland (TEKES), and by the EU foundation.
REFERENCES
- 1.Arora S K. Molecular structure, absolute stereochemistry, and interactions of nogalamycin, a DNA-binding anthracycline antitumour antibiotic. J Am Chem Soc. 1983;105:1328–1332. [Google Scholar]
- 2.Arora S K. Molecular structure, stereochemistry and interactions of steffimycin B, a DNA-binding anthracycline antibiotic. J Biomol Struct Dyn. 1985;3:377–385. doi: 10.1080/07391102.1985.10508424. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38:215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 4.Dickens M L, Ye J, Strohl W R. Analysis of clustered genes encoding both early and late steps in daunomycin biosynthesis by Streptomyces sp. strain C5. J Bacteriol. 1995;177:536–543. doi: 10.1128/jb.177.3.536-543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickens M L, Ye J, Strohl W R. Cloning, sequencing, and analysis of aklaviketone reductase from Streptomyces sp. strain C5. J Bacteriol. 1996;178:3384–3388. doi: 10.1128/jb.178.11.3384-3388.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopwood D A, Bibb M J, Chater K F, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: John Innes Foundation; 1985. [Google Scholar]
- 7.Kantola, J., T. Kunnari, A. Hautala, J. Hakala, K. Ylihonko, and P. Mäntsälä. Elucidation of anthracyclinone biosynthesis by stepwise cloning of genes for anthracyclines from three different Streptomyces spp. Microbiology, in press. [DOI] [PubMed]
- 8.Lomovskaya N, Doi-Katayama Y, Filippini S, Nastro C, Fonstein L, Gallo M, Colombo A L, Hutchinson C R. The Streptomyces peucetius dpsY and dnrX genes govern early and late steps of daunorubicin and doxorubicin biosynthesis. J Bacteriol. 1998;180:2379–2386. doi: 10.1128/jb.180.9.2379-2386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madduri K, Hutchinson C R. Functional characterization and transcriptional analysis of a gene cluster governing early and late steps in daunorubicin biosynthesis in Streptomyces peucetius. J Bacteriol. 1995;177:3879–3884. doi: 10.1128/jb.177.13.3879-3884.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemi J, Mäntsälä P. Nucleotide sequences and expression of genes from Streptomyces purpurascens that cause the production of new anthracyclines in Streptomyces galilaeus. J Bacteriol. 1995;177:2942–2945. doi: 10.1128/jb.177.10.2942-2945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Hagan D. The polyketide metabolites. Chichester, England: Ellis Horwood Limited; 1991. [Google Scholar]
- 12.Penco S, Vigevani A, Tosi C, Fusco R, Borghi D, Arcamone F. Conformational flexibility of ring A in a series of substituted anthracyclines: 1H-n.m.r. and quantum mechanical studies. Anticancer Drug Des. 1986;1:161–165. [PubMed] [Google Scholar]
- 13.Rajgarhia V B, Strohl W R. Minimal Streptomyces sp. strain C5 daunorubicin polyketide biosynthesis genes required for aklanonic acid biosynthesis. J Bacteriol. 1997;179:2690–2696. doi: 10.1128/jb.179.8.2690-2696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Torkkell S, Ylihonko K, Hakala J, Skurnik M, Mäntsälä P. Characterization of Streptomyces nogalater genes encoding enzymes involved in glycosylation steps in nogalamycin biosynthesis. Mol Gen Genet. 1997;256:203–209. doi: 10.1007/s004380050562. [DOI] [PubMed] [Google Scholar]
- 16.Wiley P F, Kelly R B, Caron E L, Wiley V H, Johnson J H, MacKellar F A, Mizsak S A. Structure of nogalamycin. J Am Chem Soc. 1977;99:542–549. doi: 10.1021/ja00444a038. [DOI] [PubMed] [Google Scholar]
- 17.Wiley P F, Elrod D W, Marshall V P. Biosynthesis of the anthracycline antibiotics nogalamycin and steffimycin B. J Org Chem. 1978;43:3457–3461. [Google Scholar]
- 18.Wright F, Bibb M J. Codon usage in the G+C-rich Streptomyces genome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 19.Ylihonko K, Hakala J, Niemi J, Lundell J, Mäntsälä P. Isolation and characterization of aclacinomycin A-non-producing Streptomyces galilaeus (ATCC 31615) mutants. Microbiology. 1994;140:1359–1365. doi: 10.1099/00221287-140-6-1359. [DOI] [PubMed] [Google Scholar]
- 20.Ylihonko K, Hakala J, Kunnari T, Mäntsälä P. Production of hybrid anthracycline antibiotics by heterologous expression of Streptomyces nogalater nogalamycin biosynthesis genes. Microbiology. 1996;142:1965–1972. doi: 10.1099/13500872-142-8-1965. [DOI] [PubMed] [Google Scholar]
- 21.Ylihonko K, Tuikkanen J, Jussila S, Cong L, Mäntsälä P. A gene cluster involved in nogalamycin biosynthesis from Streptomyces nogalater: sequence analysis and complementation of early-blocked mutations in the anthracycline pathway. Mol Gen Genet. 1996;251:113–120. doi: 10.1007/BF02172908. [DOI] [PubMed] [Google Scholar]