Abstract
Peanut is an important oil and economic crop widely cultivated in the world. It has special characteristics such as blooming on the ground but bearing fruits underground. During the peg penetrating into the ground, it is subjected to mechanical stress from the soil at the same time. It has been proved that mechanical stress affects plant growth and development by regulating the ethylene signaling-related genes. In this study, we identified some genes related to ethylene signal of peanut, including 10 ethylene sensors, two constitutive triple responses (CTRs), four ethylene insensitive 2 (EIN2s), four ethylene insensitive 3 (EIN3s), six EIN3-binding F-box proteins (EBFs), and 188 Apetala2/ethylene-responsive factors (AP2/ERFs). One hundred and eighty-eight AP2/ERFs were further divided into four subfamilies, 123 ERFs, 56 AP2s, 6 Related to ABI3/VP1 (RAVs), and three Soloists, of them one hundred and seventy AP2/ERF gene pairs were clustered into segmental duplication events in genome of Arachis hypogaea. A total of 134, 138, 97, and 150 AhAP2/ERF genes formed 210, 195, 166, and 525 orthologous gene pairs with Arachis duranensis, Arachis ipaensis, Arabidopsis thaliana, and Glycine max, respectively. Our transcriptome results showed that two EIN3s (Arahy.J729H0 and Arahy.S7XF8N) and one EBFs (Arahy.G4JMEM) were highly expressed when mechanical stress increased. Among the 188 AhAP2/ERF genes, there were 31 genes with the fragments per kilobase of exon model per million mapped fragments (FPKM) ≥ 100 at least one of the 15 samples of Tifrunner. Among them, three AhAP2/ERFs (Arahy.15RATX, Arahy.FAI7YU, and Arahy.452FBF) were specifically expressed in seeds and five AhAP2/ERFs (Arahy.HGAZ7D, Arahy.ZW7540, Arahy.4XS3FZ, Arahy.QGFJ76, and Arahy.AS0C7C) were highly expressed in the tissues, which responded mechanical stress, suggesting that they might sense mechanical stress. Mechanical stress simulation experiment showed that three AhAP2/ERFs (Arahy.QGFJ76, Arahy.AS0C7C, and Arahy.HGAZ7D) were sensitive to mechanical stress changes and they all had the conservative repressor motif (DLNXXP) in the C-terminus, indicated that they might transmit mechanical stress signals through transcriptional inhibition. This study reveals the regulatory landscape of ethylene signal-related genes in peanut, providing valuable information for the mining of target genes for further study.
Keywords: peanut, ethylene signal, mechanical stress, EIN3, AP2/ERF, transcriptome
Introduction
Cultivated peanut [Arachis hypogaea (A. hypogaea) L.], belonging to family Leguminosae, originated in South America (Bertioli et al., 2019). It is an important oil and economic crop, supplying oil and protein for human beings, which widely planted in tropical and subtropical areas. As the harvesting organ of peanut, pod is the key factor affecting peanut yield and quality (Liu et al., 2020). However, the molecular mechanism of peanut pod growth and development is still not clear; an in-depth understanding of this process has great significance for increasing peanut yield and improving peanut quality.
The peanut pod has unique characteristics such as it blooms on the ground but bears fruits underground. After pollination, the embryo temporarily stops developing and the ovary stalk continuously elongates, pushing the ovary toward the ground, called “gynophore” (Kumar et al., 2019). When the ovary entering into the soil, the environmental conditions change, including dark conditions, mechanical stress, moisture and nutrition, etc. The embryo continues to develop and eventually forms a pod. Previous studies have shown that mechanical stress is one of the important factors affecting the development of peanut pods (Ziv and Zamski, 1975; Zamski and Ziv, 1976; Kumar et al., 2019).
It has been proved that ethylene plays a remarkable role in growth and developmental changes in a process triggered by mechanical stress (Goeschl et al., 1966; Okamoto and Takahashi, 2019; Wu et al., 2020). Mechanical stress results in a rapid increase in the amount of ethylene and the activity of 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, which is a key enzyme in ethylene synthesis (Eisinger, 1983; Biro and Jaffe, 1984; Emery et al., 1994). During the penetration, the amount of ethylene released from buried gynophore was twice as much as the aerial gynophore (Shlamovitz et al., 1995), indicating that ethylene is involved in the pod development.
Ethylene signal transduction begins with ethylene sensor family and ends with ethylene insensitive 3/ethylene insensitive 3-like (EIN3/EIL) and ethylene-responsive factor (ERF) family (Potuschak et al., 2003). Ethylene is perceived by five membrane-bound receptors: two ethylene receptors (ETR1 and ETR2), two ethylene response sensors (ERS1 and ERS2), and one ethylene insensitive 4 (EIN4). At low ethylene concentration, the receptors bind to constitutive triple response 1 (CTR1), which then phosphorylates ethylene insensitive 2 (EIN2). Due to the degradation of phosphorylated EIN2, the downward transmission of ethylene signal is inhibited. When the concentration of ethylene increases, ethylene molecules bind to receptors, inactivating CTR1 and unable to phosphorylate EIN2. At the same time, the N-terminal of the dephosphorylated EIN2 is cutoff and transported from the endoplasmic reticulum to the nucleus, stabilizing the transcription factor EIN3/EIL1 and resulting in an increased protein level of EIN3/EIL1 in nucleus. Furthermore, EIN3/EIL1 transmits ethylene signals by regulating the expression of ERFs and other ethylene-responsive genes (Guo and Ecker, 2004; Chen et al., 2005). EIN3-binding F-box protein (EBF) is located in the nucleus and interacts with EIN3/EIL; it regulated the ethylene signaling by modulating the stability of EIN3/EIL1 proteins. Overexpression of EBF1 results in plants insensitive to ethylene (Potuschak et al., 2003).
Apetala2 (AP2)/ERF superfamily is a large class of transcription factors in plants and shares a conserved AP2 domain (Perata, 2013). This superfamily is divided into three subfamilies: ERF, AP2, and RAV (Sakuma et al., 2002; Nakano et al., 2006). AP2/ERF transcription factors have been reported to be widely involved in the mechanical stimulation during the growth and development of plants (Nishiuchi et al., 2002; Feng et al., 2020).
What genes are involved in ethylene signaling pathway during the peanut pod development? According to the sequencing of tetraploid cultivated peanut (Bertioli et al., 2019; Chen et al., 2019; Zhuang et al., 2019), genes of ethylene signaling pathway, including the whole AP2/ERF superfamily in cultivated peanut, were identified in this study. Their gene structure, chromosome distribution, evolutionary relationships, and transcriptional expression during the pod development were analyzed. These results provide valuable information for clarifying regulatory mechanisms of mechanical stress in the process of peanut pod development.
Materials and Methods
Identification of Genes Involved in Ethylene Signaling Pathway in Arachis hypogaea
The genome data of A. hypogaea cv. Tifrunner, a runner-type peanut (registration number CV-93, PI 644011), were downloaded from https://www.peanutbase.org/home. The genomic data of Arabidopsis AP2/ERF sequences were downloaded from the arabidopsis information resource (TAIR).1 The ethylene pathway-related genes identified in Arabidopsis thaliana (A. thaliana) were used as clue to blast the peanut genome and these obtained genes were confirmed by the Conserved Domain Database (CDD).2 For AP2/ERF gene family, the Markov (HMM) file of AP2 domain (PF00847) downloaded from the Pfam database3 was used to retrieve AP2/ERF genes through hidden Markov marker and profiles (HMMER) version 3.0. The final family members of AP2/ERF were identified by a combination of basic local alignment search tool (BLAST) and HMM.
Analysis of Phylogenetic, Gene Structure, Promoter, and Chromosomal Mapping
The longest transcripts of protein sequences of genes were selected for bioinformatics analysis and alternative splicing was obtained through genome annotations. MEGA and PlantCare4 were used to construct evolutionary trees [using maximum likelihood (ML)] and identify the putative cis-regulatory elements. The visualization of these information and chromosomal mapping were realized through TBtools (Chen et al., 2020).
Gene Duplication Analysis
Genomic data of Arachis duranensis, Arachis ipaensis, and Glycine max can be downloaded from the following website https://www.peanutbase.org/home and https://v1.legumefederation.org/data/v2/Glycine/max. Interspecific and intraspecific syntenic analysis was performed by using Multiple Collinearity Scan toolkit (MCScanX) with the default parameters. The visualization is achieved through TBtools (Chen et al., 2020).
Plant Material, Transcriptome, and Expression Analysis of Mechanical Stress Simulation Experiment
Haihua 1, a peanut cultivar, was planted at the Experimental Station of Institute of Advanced Agricultural Sciences of Peking University. The pod shells were the tissue that felt mechanical stress; thus, the pericarp of pods with a transverse diameter of about 15 mm (about 10 days after penetration into soil) was selected as experimental materials (Supplementary Figure 1A). The pods just dug out of the soil were the control group (CK, day 1). The pods out of the soil bagged with black breathable paper bag were the treatment group, simulating mechanical stress loss (Supplementary Figure 1B). Samples harvested after 10 (day 1), 34 (day 2), and 58 (day 3) h of treatment, termed as D1, D2, and D3, respectively. The pods were buried back into the soil and harvested after 48 h for simulating mechanical stress recovery, termed as recovery (RE) (day 5) (Supplementary Figure 1C). Each group had 15 pods, three biological replicates of each group. The pod shells of pods from the control group and the treatment group were isolated and stored at −80°C.
Ribonucleic acid was isolated from the thoroughly ground samples of pod shell with Takara MiniBEST Plant RNA Extraction Kit (Takara, Dalian, China). The complementary DNA (cDNA) libraries were constructed and then sequenced on the Illumina NovaSeq 6000. The original image data obtained were transformed into sequence data by consensus assessment of sequence and variation (CASAVA) base recognition. High-quality sequence data were obtained by removing reads containing adapter and low-quality sequences. Reference genome and gene model annotation files were downloaded from https://www.peanutbase.org/home and paired-end clean reads were aligned to the reference genome using Hisat2 (version 2.0.5). The RNA sequencing (RNA-Seq) data of this experiment had been deposited in the National Center for Biotechnology Information (NCBI) [sequence read archive (SRA) accession: PRJNA770556]. Besides for our own transcriptome data, gene expression analysis also referenced the previous transcriptome publications, a developmental transcriptome of Tifrunner (Clevenger et al., 2016).
Quantitative PCR Validation
The cDNA was synthesized with the PrimeScript RT Reagent Kit (Takara, Dalian, China). The gene primers were designed by Primer35 and the elongation factor 1B (Arahy.E3HYWR) was used for normalization (Supplementary Table 1). The qPCR was performed on the ABI 7500 Fast using the TB Green® Premix Ex Taq™ (Takara, Dalian, China), with three replicates. The reaction conditions were: 95°C for 5 min, 40 thermal cycles of 95°C for 30 s, and then 60°C for 30 s. The 2–ΔΔCT method was used for relative quantification analysis (Livak and Schmittgen, 2001).
Prediction of Target Genes and Interacted Proteins
Prediction of target genes and interaction network were referenced to homologous genes in A. thaliana according to http://planttfdb.gao-lab.org/index.php and https://cn.string-db.org, respectively (Tian et al., 2019; Szklarczyk et al., 2021). Only interactions that had been demonstrated by experimental evidence were shown.
Results
Identification and Expression Analysis of Ethylene Sensors, Constitutive Triple Responses, Ethylene Insensitive 2, Ethylene Insensitive 3/Ethylene Insensitive 3-Like, and Binding F-Box Proteins
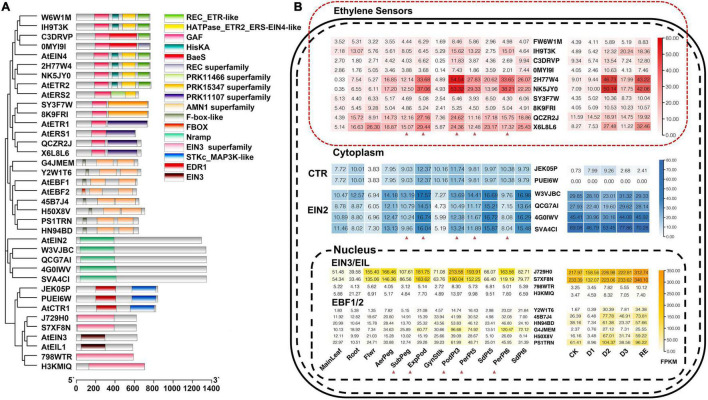
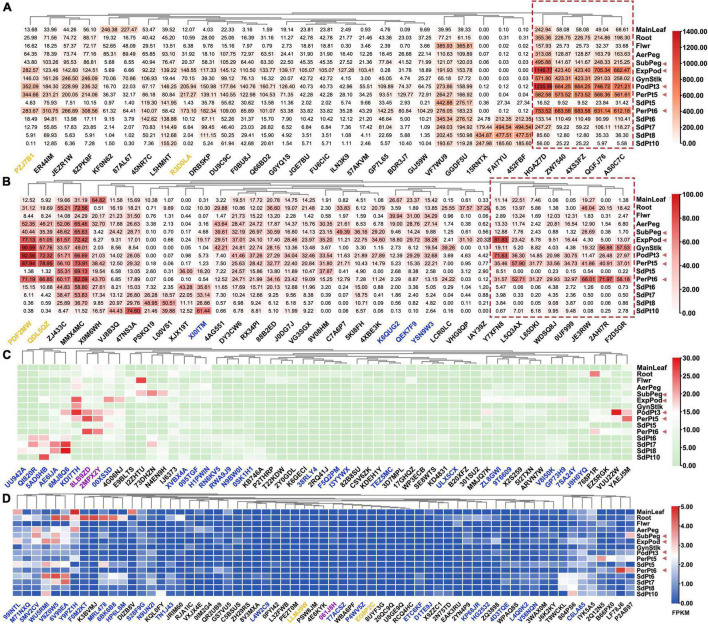
According to homologous genes in Arabidopsis, ten ethylene sensors, two CTRs, four EIN2s, four EIN3/EILs, and six EBFs were identified in A. hypogaea cv. Tifrunner (Figure 1A and Supplementary Table 2). They were located on the corresponding chromosomes of the A and B subgenomes, each A gene corresponded to a B gene (Supplementary Figure 2). Expression analysis showed that the expression of two ethylene sensors (Arahy.2H77W4 and Arahy.NK5JY0) in pod was significantly higher than that in other tissues and increased after penetration and expansion. Mechanical stress simulation experiment showed that they increased after 34 h of losing pressure and then decreased. When the pressure was restored, their expression rose again (Figure 1B). This result indicated that Arahy.2H77W4 and Arahy.NK5JY0 might be the main ethylene sensors functioning in the pod development. Besides, the expression of two AhEIN3/EILs (Arahy.J729H0 and Arahy.S7XF8N) in reproductive organ, especially in pericarps, was significantly higher than leaf and root. Because pericarp first sensed the changes from the peripheral environment, they suggesting the significance of Arahy.J729H0 and Arahy.S7XF8N in the pod development (Figure 1B). In addition, an AhEBFs (Arahy.G4JMEM) also showed similar expression pattern with AhEIN3/EILs.
FIGURE 1.
Conserved domain and expression of ethylene sensors, constitutive triple responses (CTRs), ethylene insensitive 2 (EIN2s), ethylene insensitive 3/ethylene insensitive 3-like (EIN3/EILs), and EIN3-binding F-box proteins (EBFs). (A) Conserved domain of ethylene sensors, CTRs, EIN2s, EIN3/EILs, and EBFs in Arachis hypogaea (A. hypogaea) genome; (B) Developmental expression analysis of ethylene sensors, CTRs, EIN2s, EIN3/EILs, and EBFs in A. hypogaea genome. The red triangle indicates the tissues that experience mechanical stress.
Interestingly, in our mechanical stress simulation experiment using cultivar Haihua 1, all the six AhEBFs decreased after 10 h of mechanical stress loss, then significantly increased on the second day of mechanical stress loss, then slightly decreased on the third day, and increased again when mechanical stress was restored 2 days later (Figure 1B). They response to the changes of the external environment and there was no positive or negative correlation between their expression patterns and mechanical stress, so it was speculated that AhEBFs might be act as signal transducers, sending the external signals to downstream genes. If the external environment did not change, their expression would not change dramatically.
Evolution and Alternative Splicing Analysis of Apetala2/Ethylene- Responsive Factor Transcription Factors in Arachis hypogaea
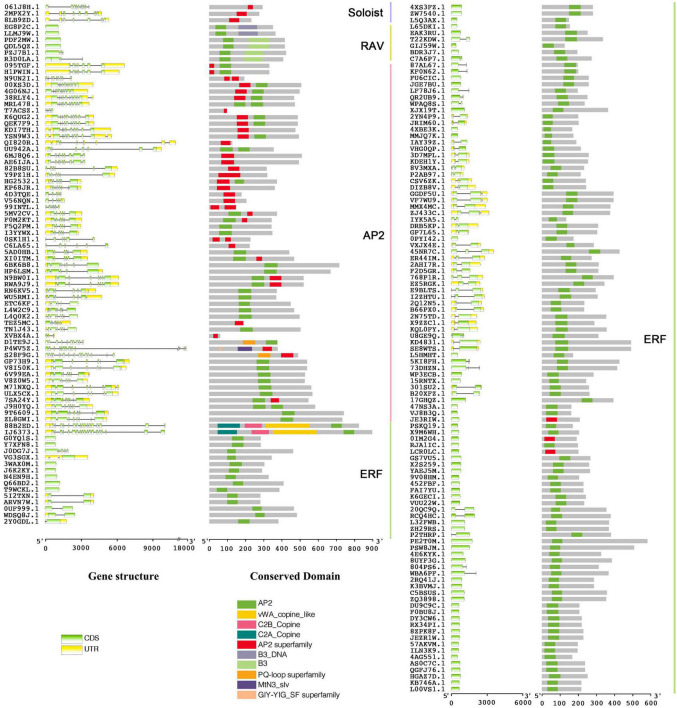
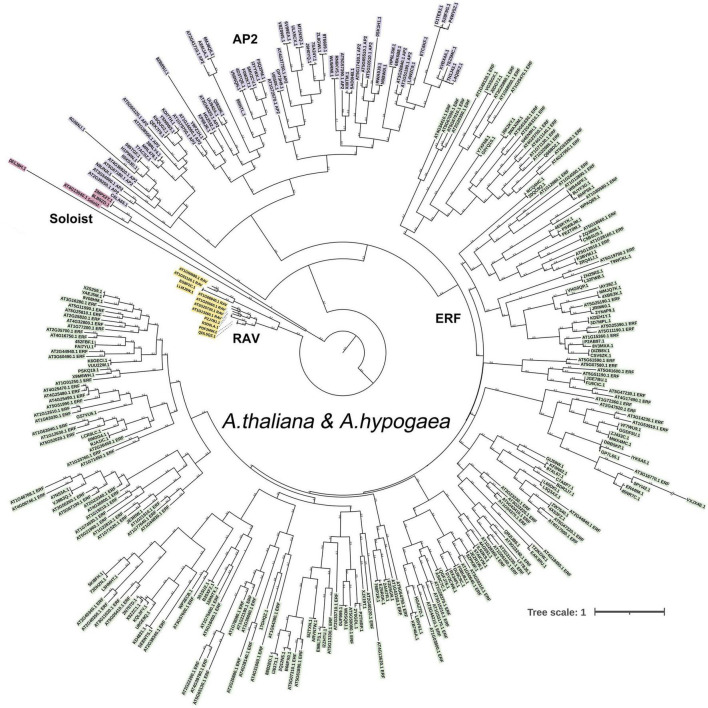
Combining BLAST and HMM results, there were 188 AP2/ERF genes identified in A. hypogaea cv. Tifrunner. The CDD and phylogenetic analysis showed that 123 genes belonged to ERF subfamily, 56 genes belonged to AP2 subfamily, six genes belonged to RAV subfamily, and three genes belonged to Soloist. Among AP2 subfamily, 21 genes contained two AP2 domains and 35 genes contained one AP2 domain (Figures 2, 3). Gene structure showed that most of AP2 subfamily members and three Soloists had numerous introns. However, ERF subfamily members contained only 2 or less introns, except Arahy.88B2ED and Arahy.IJ6373 (Figure 2).
FIGURE 2.
Gene structure and conserved domain of 188 Apetala2/ethylene-responsive factors (AhAP2/ERFs) in A. hypogaea genome.
FIGURE 3.
Phylogenetic tree of AhAP2/ERFs and AtAP2/ERFs.
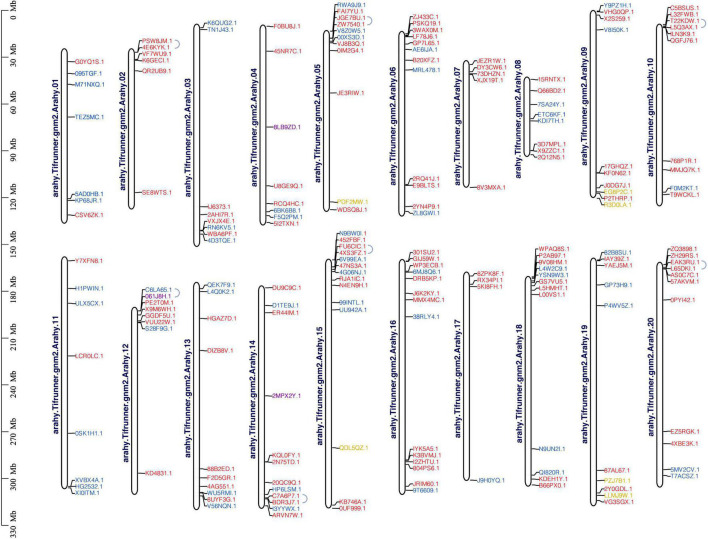
One hundred and eighty-eight AP2/ERF genes were unevenly distributed on 20 chromosomes, but they did not form a correspondence between A and B subgenome (Figure 4 and Supplementary Figure 3). The possible causes were the loss of gene annotation or the increase of members after the formation of tetraploid. The maximum number of AP2/ERFs were located on chromosomes 15 and 16, both with 14 AP2/ERFs. Chromosome 17 had the minimum number of AP2/ERF, only 4. Three Soloists were distributed on chromosomes 4, 12, and 14, respectively. The six RAV family members were distributed on chromosomes 5, 9, 15, and 19. Chromosomes 2 and 7 only had ERF family members (Figure 4 and Supplementary Figure 3). Fourteen AhAP2/ERF genes were clustered into seven pairs of tandem duplication event on chromosomes 2, 5, 10, 12, 14, 15, and 20. The correspondence between 2, 5, 10 and 12, 15, 20 suggests that tandem repetition preceded the formation of the tetraploid (Figure 4). One hundred and seventy AP2/ERF gene pairs were clustered into segmental duplication events in genome of A. hypogaea cv. Tifrunner, suggesting the high conservation of the AP2/ERF gene family, which played crucial roles in the expansion of AP2/ERF gene family (Supplementary Figure 3 and Supplementary Table 3).
FIGURE 4.
Chromosomal distribution and tandem duplication of AhAP2/ERFs in A. hypogaea genome. Red gene ID: ERFs; blue gene ID: AP2; yellow gene ID: RAV; purple gene ID: Soloist.
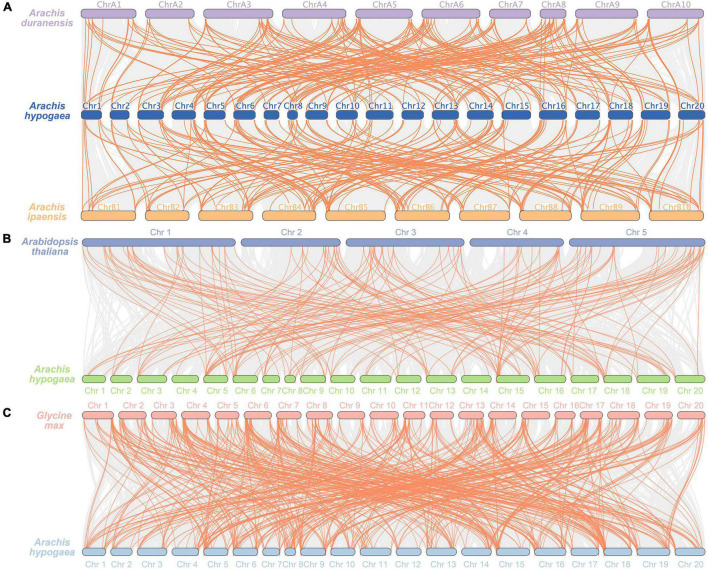
In order to further study the phylogenetic and evolutionary relationships of AP2/ERF gene family in A. hypogaea, three comparative syntenic maps associated with Arachis duranensis, Arachis ipaensis, A. thaliana, and Glycine max were performed (Figure 5). A total of 134 and 138 AhAP2/ERF genes formed 210 and 195 orthologous gene pairs with 71 and 77 AP2/ERF genes in Arachis duranensis and Arachis ipaensis, respectively. AhAP2/ERF genes that were not found orthologous gene pairs with Arachis duranensis or Arachis ipaensis may indicate that these genes appeared after the heterozygote was formed (Figure 5A and Supplementary Table 4). A total of 97 and 150 AhAP2/ERF genes formed 166 and 525 orthologous gene pairs with 75 and 206 AP2/ERF genes in A. thaliana and Glycine max, respectively (Figures 5B,C and Supplementary Table 4). Apparently, soybeans had a closer relatedness with peanuts. Some AhAP2/ERF genes formed two or more orthologous gene pairs with all the four species, such as Arahy.00XS3D and Arahy.095TGF, suggesting the conservation in evolution and the importance of gene function (Supplementary Table 4).
FIGURE 5.
Synteny analysis of AP2/ERF genes between A. hypogaea and four other plant species. Gray lines indicated the syntenic gene pairs within genome, while tangerine lines highlighted the syntenic AP2/ERF gene pairs. (A) Synteny analysis of AP2/ERF genes between A. hypogaea, Arachis duranensis, and Arachis ipaensis; (B) Synteny analysis of AP2/ERF genes in A. hypogaea vs. Arabidopsis thaliana (A. thaliana); (C) Synteny analysis of AP2/ERF genes in A. hypogaea vs. Glycine max.
According to the genome annotations, we found that a total of 12 AhAP2/ERF genes had alternative splices (ASs), including exon skip (ES), alternate promoter (AP), alternate terminator (AT), and retained intron (RI). AP or AT was found to exist in 6 AhAP2/ERFs, which was the main splicing method; RI was found to exist in 5 AhAP2/ERFs and ES was found to exist in 2 AhAP2/ERFs (Supplementary Figure 4).
Developmental Transcriptome Changes of Apetala2/Ethylene-Responsive Factor Genes
Referenced to developmental transcriptome map of Tifrunner from previous transcriptome publications (Clevenger et al., 2016), 188 AhAP2/ERF genes were divided into four categories according to their highest FPKM: (a) 31 AhAP2/ERFs, FPKM ≥ 100 in at least one of the 15 samples; (b) 35 AhAP2/ERFs, 30 ≤ FPKM ≤ 100 in at least one of the 15 samples; (c) 57 AhAP2/ERFs, 5 ≤ FPKM ≤ 30 in at least one of the 15 samples; and (d) 65 AhAP2/ERFs, 0 ≤ FPKM ≤ 5 in all the 15 samples (Figure 6 and Supplementary Table 5). The high expression genes (categories a and b) were mainly ERF members, accounting for 80% of the number of highly expressed genes. Fifty-two of 56 AP2 genes and all the Soloists were in the low expression categories (categories c and d). There were two RAVs and 29 ERFs in “a” category and three AhAP2/ERFs (Arahy.15RATX, Arahy.FAI7YU, and Arahy.452FBF) were specifically expressed in seeds and highly expressed in late seed maturation. Five AhAP2/ERFs (Arahy.HGAZ7D, Arahy.ZW7540, Arahy.4XS3FZ, Arahy.QGFJ76, and Arahy.AS0C7C) were highly expressed in Expod, PodPt3, PerPt5, and PerPt6, which were the tissues that sensed mechanical stress, suggesting that they might be related to mechanical stress sensing (Figure 6A). Similar expression patterns were found in 8 AhAP2/ERFs from “b” category. Interestingly, their expression in seeds were relatively lower than other tissues (Figure 6B), indicating that they might be related to response to external changes.
FIGURE 6.
Gene express patterns of 188 FcAP2/ERF genes in 15 samples in A. hypogaea cv. Tifrunner. Main leaf, main stem leaf; Root, roots of 10 days postemergence; Flwr, petals, keel, and hypanthium sepals; AerPeg, elongating aerial pegs; Subpeg, elongating subterranean pegs; GynStlk, Pattee 1 stalk of gynophore; PodPt3, Pattee 3 pod; PerPt5, Pattee 5 pericarp; SdPt5, Pattee 5 seed; PerPt6, Pattee 6 pericarp; SdPt6, Pattee 6 seed; SdPt7, Pattee 7 seed; SdPt8, Pattee 8 seed; SdPt10, Pattee 10 seed (Clevenger et al., 2016). Black gene ID: ERFs; blue gene ID: AP2; yellow gene ID: RAV; purple gene ID: Soloist. They were divided into the four groups, red dotted boxes indicated FcAP2/ERFs that might be involved in mechanical pressure signal transduction. The red triangle indicates the tissues that experience mechanical stress. (A) The maximum FPKM value was greater than 100; (B) The maximum FPKM value was between 30 and 100; (C) The maximum FPKM value was between 5 and 30; (D) The maximum FPKM value was less than 5.
Promoter Analysis of Highly Expressed Apetala2/Ethylene-Responsive Factor Genes
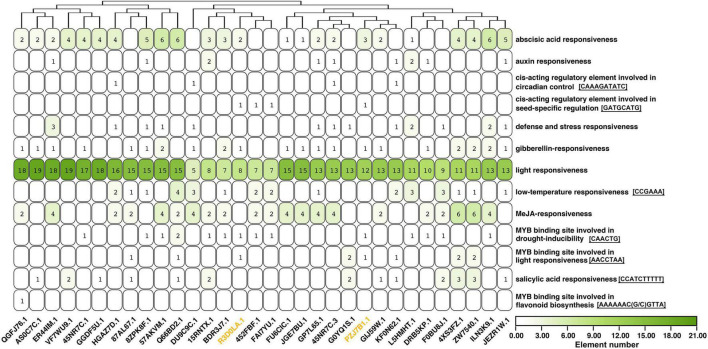
The 2,000 bp upstream sequences of 31 AhAP2/ERFs (FPKM ≥ 100 in at least one of the 15 samples) were extracted for promoter analysis. The most abundant elements were light responsiveness, followed by abscisic acid (ABA) responsiveness and methyl jasmonate (MeJA) responsiveness, suggesting that their expression was mainly regulated by light, ABA, and MeJA (Figure 7 and Supplementary Table 6). Four AhAP2/ERFs (Arahy.HGAZ7D, Arahy.DU9C9C, Arahy.45NR7C.3, and Arahy.KF0N62) contained a circadian control element (CAAAGATATC motif). Among them, Arahy.HGAZ7D and Arahy.DU9C9C were highly expressed in the tissues that sensed mechanical stress, especially Arahy.HGAZ7D (Figure 6A), indicating that circadian rhythms were involved in the pod development. There were 13 AhAP2/ERFs contained at least one salicylic acid-responsiveness element (CCATCTTTTT motif). Arahy.4XS3FZ and its homolog (Arahy.ZW7540) had the most number (3) and their protein sequence showed 43.4 and 41.9% similarity to NtERF1 and AtERF2, which were involved in disease resistance pathways, suggesting Arahy.4XS3FZ and Arahy.ZW7540 might be related to disease resistance. In addition, there were four AhAP2/ERF (Arahy.R3D0LA, Arahy.452FBF, Arahy.FAI7YU, and Arahy.PZJ7B1) that contained a cis-acting regulatory element involved in seed-specific regulation (GATGCATG motif). Significantly, Arahy.R3D0LA and Arahy.PZJ7B1 were slowly expressed in seeds, while Arahy.452FBF and Arahy.FAI7YU were specifically expressed in the seed (Figures 6A, 7).
FIGURE 7.
Promoter analysis of 31 highly expressed AhAP2/ERF genes. Black gene ID: ERFs; yellow gene ID: RAV. Only elements with the same sequence were displayed. The number represents the number of conserved domains and those with zero number is not marked.
Expression Changes of Apetala2/Ethylene-Responsive Factor Genes in Response to Mechanical Stress
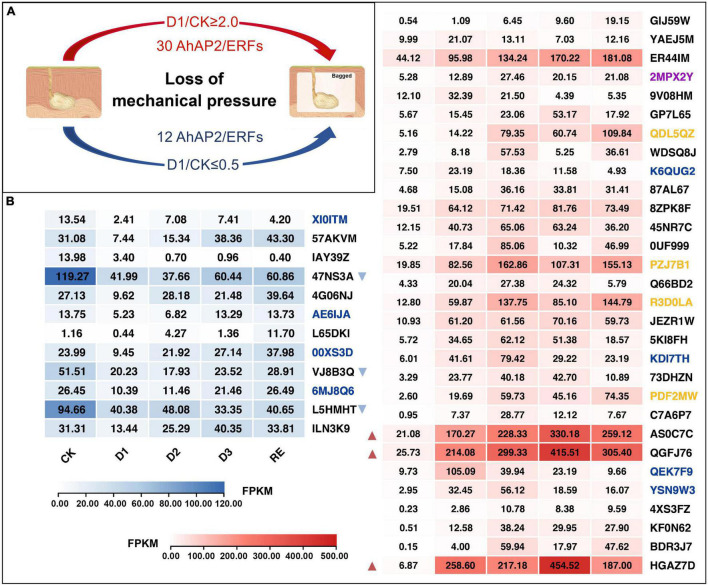
Mechanical stress simulation experiment showed that 30 AhAP2/ERFs were upregulated 2-fold change and 12 AhAP2/ERFs were downregulated 2-fold change after out of the soil (losing the mechanical stress) (Figure 8A). In the downregulated group, there were three AhAP2/ERFs (Arahy.47NS3A, Arahy.VJ8B3Q, and Arahy.L5HMHT) with higher expression levels and downregulated by 0.35, 0.39, and 0.43 times in D1 vs. CK, respectively. In the upregulated group, there were also three AhAP2/ERFs (Arahy.QGFJ76, Arahy.AS0C7C, and Arahy.HGAZ7D) with higher expression levels and upregulated by 8.08, 8.32, and 37.63 times in D1 vs. CK, respectively (Figure 8B). Coincidentally, these three unregulated AhAP2/ERFs were included in the five AhAP2/ERFs that were highly expressed in Expod, PodPt3, PerPt5, and PerPt6. Four RAV members with higher expression levels were upregulated by 2.76–7.57 times in D1 vs. CK (Figures 6A, 8B).
FIGURE 8.
Expression changes of AhAP2/ERF genes in mechanical stress simulation experiment. Black gene ID: ERFs; blue gene ID: AP2; yellow gene ID: RAV; purple gene ID: Soloist. CK: control group, pods just out of the soil; D1: treatment group, 10 h bagging; D2: 34 h bagging; D3: 58 h bagging; RE: pods buried back into the soil for 48 h (Supplementary Figure 1C: Flowchart). (A) Number of differentially expressed AhAP2/ERF genes after 10 h loss of mechanical stress; (B) The FPKM value of AhAP2/ERF genes. The red and blue triangles indicated genes with high upregulation or downregulation, respectively.
Validate the Transcriptome of Mechanical Stress Simulation by Real-Time Quantitative PCR
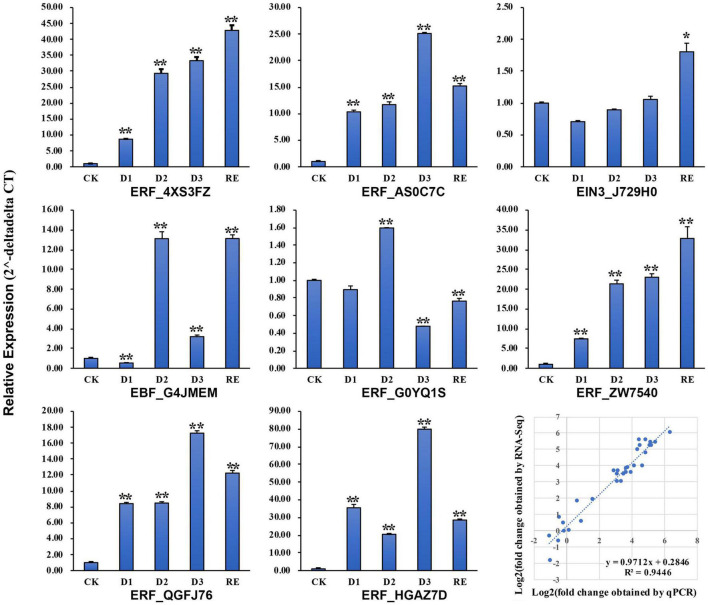
Eight genes including 1 EIN3, 1 EBF, and 6 AP2/ERFs were selected to verify the transcriptome results by qRT-PCR. The expression patterns of those genes were consistent with the transcriptome results (Figure 9) and linear regression analysis revealed a high correlation coefficient (R2 = 0.94; Figure 9).
FIGURE 9.
Validation of the RNA sequencing (RNA-Seq) results by real-time quantitative PCR (qRT-PCR). *, **Significantly different at p ≤ 0.05 and 0.01, respectively.
Protein Sequence Alignment, Target Genes, and Protein Interaction Networks
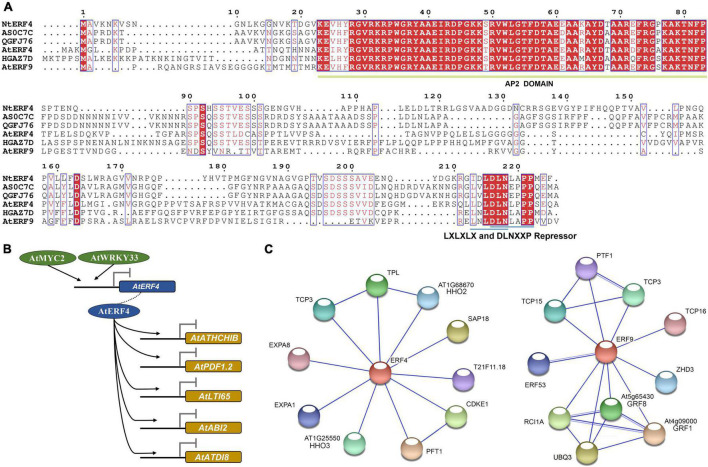
To explore the possible genetic functions of key three AhAP2/ERFs, their protein sequences were further studied. The protein sequences of Arahy.QGFJ76 and Arahy.AS0C7C showed only one amino acid difference and they were homologous genes located on chromosomes 10 (subgenome A) and 20 (subgenome B), respectively. The protein sequence similarity between Arahy.HGAZ7D and Arahy.QGFJ76 or Arahy.AS0C7C was 75% and they contained one AP2 domain, belonging to ERF subfamily (Figure 10A). Furthermore, the protein sequences of Arahy.QGFJ76 and Arahy.AS0C7C showed 46.6 and 46.2% similarity to NtERF4 and the protein sequences of Arahy.HGAZ7D showed 44.7, 46.0, and 47.3% similarity to NtERF4, AtERF4, and AtERF9, respectively. NtERF4, AtERF4, and AtERF9 were all the transcriptional repressors. Coincidentally, repressor motifs (LXLXLX and DLNXXP) were also found in the C-terminus of these three AhAP2/ERFs (Figure 10A).
FIGURE 10.
Protein sequence alignment, target genes, and interaction network analysis of key AP2/ERFs according to A. thaliana homologs. (A) Protein sequence alignment of AtERF4, AtERF9, NtERF4, Arahy.AS0C7C, Arahy.QGFJ76, and Arahy.HGAZ7D; (B) Target genes of Arabidopsis homologs; (C) Interaction network relationships of Arabidopsis homologs.
In Arabidopsis, the homologous gene AtERF4 of Arahy.HGAZ7D could bind to the promoters of AtATHCHIB, AtPDF1.2, AtLTI65, AtABI2, and AtATDI8 to response to hormonal signals such as ethylene, jasmonic acid, and ABA and inhibit their expressions. Furthermore, the expression of AtERF4 was inhibited by Arabidopsis thaliana myelocytomatosis 2 (AtMYC2) and Arabidopsis thaliana WRKY (WRKY amino acid sequence at the N-terminus) 33 (AtWRKY33) (Figure 10B). Besides, interaction network analysis showed that AtERF4 could interacted with 10 proteins and might form two protein complexes, one with AtTCP3, Arabidopsis thaliana TOPLESS (AtTPL), and Arabidopsis thaliana hypersensitivity to low phosphate-elicited primary root shortening 1 homolog2 (AtHHO2) and the other with Arabidopsis thaliana cyclin-dependent kinase E-1 (AtCDKE1) and Arabidopsis thaliana photochrome and flowering time 1 (AtPFT1) (Figure 10B). Although, the target gene of AtERF9 had not been elucidated, AtERF9 could also interacted with 10 proteins and might form two protein complexes, one with AtTCP3, AtTCP15, and Arabidopsis thaliana plastid transcription factor 1 (AtPTF1)/TCP13 and the other with Arabidopsis thaliana growth-regulating factor 1 (AtGRF1), AtGRF8, Arabidopsis thaliana rare cold inducible 1A (AtRCI1A), and Polyubiquitin 3 (UBQ3) (Figure 10C). Both the AtERF4 and AtERF9 interacted with T-complex protein (TCPs). TCP transcription factors play an important role in plant organ morphogenesis by regulating the expression of boundary-specific genes (Koyama et al., 2007). Therefore, it was speculated that AhERFs might regulate the morphological change from gynophore to pod through TCPs.
Discussion
The ethylene signal-related genes in peanut were systematically analyzed in this study. Ethylene signal-related genes including AP2/ERFs in cultivated peanut were identified. Combined developmental- and mechanical stress-related transcriptome provides a comprehensive understanding of these gene changes related to peg penetration and pod development. Two AhEIN3s, one AhEBFs, and three AhAP2/ERFs may be the key genes for their high expression and expression differences.
This study revealed transcriptional changes of ethylene signal genes in peanut. However, the final executor of life activities is protein and there is a large amount of posttranscriptional regulation between transcription and protein (Floris et al., 2009). Therefore, transcription levels and protein levels are often inconsistent (Cui et al., 2019). Besides, posttranslational protein modifications including phosphorylation, ubiquitination, sumoylation, etc., are the core of many cellular signaling events (Dai Vu et al., 2018), they are greatly increasing the complexity of biological events. Currently, there are few studies on posttranscriptional protein modification of EIN3 and AP2/ERFs. However, advances in mass spectrometry have made it possible to discover more posttranscriptional modifications. Therefore, the posttranslational protein modifications will be necessary for further study of key genes.
Ethylene signal-related genes including AP2/ERFs in cultivated peanut were all located at the two ends of chromosomes (Supplementary Figure 2 and Figure 4), which are consistent with the previous description in the cultivated peanut genome; nonautonomous long terminal repeat (LTR) retrotransposons and Ty3-gypsy elements are mainly distributed in the middle of chromosomes, while genes are mainly distributed in the end (Bertioli et al., 2019; Chen et al., 2019). Comparative genomic analysis of wheat and rice showed that gene evolution occurred preferentially at the end of chromosomes and was associated with replication and differentiation associated with high recombination rates (See et al., 2006). Other comparative studies also showed that the region around telomeres was unstable and a hot spot in chromosome evolution, containing extensive rearrangement and segmental gene duplication (Eichler and Sankoff, 2003; Kellis et al., 2003). Therefore, the genes of cultivated peanut, such as those of other crops, are mainly distributed at both the ends of the chromosome, which are conducive to their own better evolution and expansion.
Our results showed that there were differences in the gene structure of different subfamilies of AhAP2/ERFs (Figure 2). It is particularly interesting that AP2 subfamily members contained more introns than other subfamilies. This also occurs in other plant such as pear (Li et al., 2018), tartary buckwheat (Liu et al., 2019), longan (Xu et al., 2020), sugarcane (Li et al., 2020), poppy (Yamada et al., 2020), creat (Yao et al., 2020), and pineapple (Zhang et al., 2021). There are two hypotheses about the origin of introns. One is that the intron is already present when the gene appears and the other is that the intron is inserted through transposition during evolution (Rodríguez-Trelles et al., 2006). Although the origin is still unknown, most early eukaryotic genes are rich in intron. Therefore, from an evolutionary perspective, intron poverty is later than intron abundance (Liu et al., 2021).
A single gene can expand the potential informational content of genomes, which plays a major role in the generation of proteomic and functional diversity in organisms (Blencowe, 2006). Introns play an important role in AS, which is ubiquitous under stress conditions (Chaudhary et al., 2019), as previously reported. Intron retention is a major phenomenon in AS in Arabidopsis and transcripts that retain introns are mainly related to stress and external/internal stimuli (Ner-Gaon et al., 2004). Nevertheless, the contribution of AS to proteome complexity is still elusive in plants. Why introns preferred AP2 subfamily in AP2/ERF family? It is speculated that AP2 subfamily is a reserved family of AP2/ERF and evolutionarily earlier than ERF subfamily. AP2 subfamily maintains variability in response to external changes, while ERF has evolved to maintain basal growth and development. The highly expressed genes were mainly members of ERF family, which seemed to support this hypothesis (Figure 6A). More details need further research to reveal.
Most of published data have been demonstrated that ethylene is involved in the plant growth and development (Bleecker and Kende, 2000; Dubois et al., 2018). Moreover, there is ample evidence that different stimulus, such as wounding, mechanical stress, waterlogging, and submergence, induce the burst of ethylene (Khan et al., 2020; Zhao et al., 2020). Mechanical stress has proved to play an important role in shaping architecture of plants (Hamant and Traas, 2010; Landrein and Ingram, 2019; Okamoto et al., 2021). Ethylene is the first plant growth regulator to be implicated in the response to mechanical stress (Goeschl et al., 1966). More recently, it has been reported that before the seedlings are unearthed, the pressure from the soil stimulates ethylene production and represses polygalacturonase involved in expansion 3 (PGX3) to facilitate seedling emergence from the soil in a dose-dependent manner through the transcription factor EIN3 (Wu et al., 2020). When peg penetrates into the soil, the mechanical stress increases and the light decreases. Our results showed that the expression of EIN3 had been at a high level (Figure 1B). EIN3 primarily binds to ethylene-related elements present in the promoter of AP2/ERFs to activate their transcription (Solano et al., 1998). In peanut, whether it regulates some AP2/ERFs or other genes in a dose-dependent manner? How this process is cross-regulated with light and other hormones? It requires further research.
Piezo is the most well-characterized mechanosensors in mammals. Upon activation, the Piezo channel facilitates the entry of calcium ion (Ca2+) inside the cell, translating the cellular signals (Coste et al., 2012). The sequences of Piezo family proteins are highly conserved in plants, in both the moss (Physcomitrium patens) and the small flowering plant Arabidopsis; the mutations of plant Piezo sensors change vacuolar morphology and growth patterns of apical growth cells (Radin et al., 2021). Moreover, AtPiezo responds to mechanical stimuli at the transcriptional level, playing a role in the perception of mechanical force in plant root cap and the flow of Ca2+ is involved in this process (Fang et al., 2021a,b; Mousavi et al., 2021). In peanut, four AhPiezo were identified, but they did not show any significant changes in transcription levels during mechanical stress changes (Supplementary Figure 5). Whether Piezo was involved in the formation of peanut pods and whether Piezo was associated with other hormonal signals are an interesting question in plants, waiting for our further exploration.
Conclusion
In this study, the ethylene signal-related genes in peanut (A. hypogaea L.) were analyzed, including conserved domains, gene structure, evolutions, AS, cis-elements, and expression patterns. Ten ethylene sensors, two CTRs, four EIN2s, four EIN3s, six EBFs, and 188 AP2/ERFs were identified and their expression pattern was revealed according to published transcriptome data of Tifrunner and our own mechanical stress simulation transcriptome. The change of mechanical stress affected the expression of genes related to ethylene signaling pathway, in particular two AhEIN3s, one AhEBFs, and three AhAP2/ERFs were mostly affected, suggesting that they might play an important role in the pod development of peanut.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA770556.
Author Contributions
XL and YC conceived this study. XL and XD supervised the research. YC performed the experiments. YC, XL, JB, and XH analyzed the data. XL, YC, XD, JB, YG, and FX prepared the manuscript. All the authors have read and approved the manuscript for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This study was supported by the grants from Taishan Scholar Foundation of Shandong Province (tsqn202103161) and Shandong Science and Technology Innovation Funds for XL. YC was supported in part by the Postdoctoral Fellowship of Peking-Tsinghua Center for Life Sciences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.828482/full#supplementary-material
Experimental materials and illustrations of mechanical pressure simulation experiment.
Chromosomal distribution of ethylene sensors, constitutive triple responses (CTRs), ethylene insensitive 2 (EIN2s), ethylene insensitive 3/ethylene insensitive 3-like (EIN3/EILs), and EIN3-binding F-box proteins (EBFs) in Arachis hypogaea (A. hypogaea) genome.
Gene location and intragenomic collinearity of AhAP2/ERFs. Gray lines indicated all the collinearity in A. hypogaea genome, while colored lines highlighted the collinearity of AhAP2/ERFs.
Alternative splice (AS) analysis of Apetala2/ethylene-responsive factor (AP2/ERF) genes in A. hypogaea. ES, exon skip; AP, alternate promoter; AT, alternate terminator; RI, retained intron. The red inverted triangle indicated the difference of AS.
Protein sequence alignment and express pattern of AhPiezo.
The primers of quantitative PCR (qPCR).
The FPKM values of ethylene signal genes during developmental stage and in response to mechanical stress.
Segmental duplication Apetala2/ethylene-responsive factor (AhAP2/ERF) gene pairs in Arachis hypogaea (A. hypogaea) genome.
Segmental duplication AhAP2/ERF gene pairs in A. hypogaea and four other plant species.
The FPKM values of AhAP2/ERFs during developmental stage.
Promoter elements information.
The FPKM values of AhAP2/ERFs in response to mechanical stress.
References
- Bertioli D. J., Jenkins J., Clevenger J., Dudchenko O., Schmutz J., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat Genet. 51 877–884. 10.1038/s41588-019-0405-z [DOI] [PubMed] [Google Scholar]
- Biro R., Jaffe M. (1984). Thigmomorphogenesis: ethylene evolution and its role in the changes observed in mechanically perturbed bean plants. Physiol. Plant 62 289–296. 10.1111/j.1399-3054.1984.tb04575.x [DOI] [PubMed] [Google Scholar]
- Bleecker A. B., Kende H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16 1–18. 10.1146/annurev.cellbio.16.1.1 [DOI] [PubMed] [Google Scholar]
- Blencowe B. J. (2006). Alternative splicing: new insights from global analyses. Cell 126 37–47. 10.1016/j.cell.2006.06.023 [DOI] [PubMed] [Google Scholar]
- Chaudhary S., Jabre I., Reddy A. S., Staiger D., Syed N. H. (2019). Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 24 496–506. 10.1016/j.tplants.2019.02.006 [DOI] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen X., Lu Q., Liu H., Zhang J., Hong Y., et al. (2019). Sequencing of cultivated peanut. Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant. 12 920–934. 10.1016/j.molp.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Chen Y. F., Etheridge N., Schaller G. E. (2005). Ethylene signal transduction. Ann. Bot. 95 901–915. 10.1093/aob/mci100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevenger J., Chu Y., Scheffler B., Ozias-Akins P. (2016). A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 7:1446. 10.3389/fpls.2016.01446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B., Xiao B., Santos J. S., Syeda R., Grandl J., Spencer K. S., et al. (2012). Piezo proteins are pore-forming subunits of mechanically activated channels. Nature 483 176–181. 10.1038/nature10812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Wang Z., Chen S., Vainstein A., Ma H. (2019). Proteome and transcriptome analyses reveal key molecular differences between quality parameters of commercial-ripe and tree-ripe fig (Ficus carica L.). BMC Plant Biol. 19:146. 10.1186/s12870-019-1742-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Vu L., Gevaert K., De Smet I. (2018). Protein language: post-translational modifications talking to each other. Trends Plant Sci. 23 1068–1080. 10.1016/j.tplants.2018.09.004 [DOI] [PubMed] [Google Scholar]
- Dubois M., Van den Broeck L., Inzé D. (2018). The pivotal role of ethylene in plant growth. Trends Plant Sci. 23 311–323. 10.1016/j.tplants.2018.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E. E., Sankoff D. (2003). Structural dynamics of eukaryotic chromosome evolution. Science 301 793–797. 10.1126/science.1086132 [DOI] [PubMed] [Google Scholar]
- Eisinger W. (1983). Regulation of pea internode expansion by ethylene. Annu. Rev. Plant Physiol. 34 225–240. 10.1146/annurev.pp.34.060183.001301 [DOI] [Google Scholar]
- Emery R., Reid D., Chinnappa C. (1994). Phenotypic plasticity of stem elongation in two ecotypes of Stellaria longipes: the role of ethylene and responses to wind. Plant Cell Environ. 17 691–700. 10.1111/j.1365-3040.1994.tb00161.x [DOI] [Google Scholar]
- Fang X., Liu B., Shao Q., Huang X., Li J., Luan S., et al. (2021a). AtPiezo plays an important role in root cap mechanotransduction. Int. J. Mol. Sci. 22:467. 10.3390/ijms22010467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Zhang Y., Cheng B., Luan S., He K. (2021b). Evidence for the involvement of AtPiezo in mechanical responses. Plant Signal. Behav. 16:1889252. 10.1080/15592324.2021.1889252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng K., Hou X. L., Xing G. M., Liu J. X., Duan A. Q., Xu Z. S., et al. (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit Rev. Biotechnol. 40 750–776. 10.1080/07388551.2020.1768509 [DOI] [PubMed] [Google Scholar]
- Floris M., Mahgoub H., Lanet E., Robaglia C., Menand B. (2009). Post-transcriptional regulation of gene expression in plants during abiotic stress. Int. J. Mol. Sci. 10 3168–3185. 10.3390/ijms10073168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeschl J. D., Rappaport L., Pratt H. K. (1966). Ethylene as a factor regulating the growth of pea epicotyls subjected to physical stress. Plant Physiol. 41 877–884. 10.1104/pp.41.5.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ecker J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7 40–49. 10.1016/j.pbi.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Hamant O., Traas J. (2010). The mechanics behind plant development. New Phytol. 185 369–385. 10.1111/j.1469-8137.2009.03100.x [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. (2003). Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423 241–254. 10.1038/nature01644 [DOI] [PubMed] [Google Scholar]
- Khan M. I. R., Trivellini A., Chhillar H., Chopra P., Ferrante A., Khan N. A., et al. (2020). The significance and functions of ethylene in flooding stress tolerance in plants. Environ. Exp. Bot. 179:104188. 10.1016/j.envexpbot.2020.104188 [DOI] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19 473–484. 10.1105/tpc.106.044792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Pandey M. K., Roychoudhry S., Nayyar H., Kepinski S., Varshney R. K. (2019). Peg biology: deciphering the molecular regulations involved during peanut peg development. Front Plant Sci. 10:1289. 10.3389/fpls.2019.01289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrein B., Ingram G. (2019). Connected through the force: mechanical signals in plant development. J. Exp. Bot. 70 3507–3519. 10.1093/jxb/erz103 [DOI] [PubMed] [Google Scholar]
- Li P., Chai Z., Lin P., Huang C., Huang G., Xu L., et al. (2020). Genome-wide identification and expression analysis of AP2/ERF transcription factors in sugarcane (Saccharum spontaneum L.). BMC Genomics 21:685. 10.1186/s12864-020-07076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Tao S., Wei S., Ming M., Huang X., Zhang S., et al. (2018). The mining and evolutionary investigation of AP2/ERF genes in pear (Pyrus). BMC Plant Biol. 18:46. 10.1186/s12870-018-1265-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Lyu H. M., Zhu K., Van de Peer Y., Cheng Z. M. (2021). The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 105 1072–1082. 10.1111/tpj.15088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Sun W., Ma Z., Zheng T., Huang L., Wu Q., et al. (2019). Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat (Fagopyum Tataricum). BMC Plant Biol. 19:84. 10.1186/s12870-019-1681-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Wang J., Wan S., Peng Z., Li X. (2020). Research progress of peanut pod development and its regulation mechanisms. Chinese J. Oil Crop Sci. 06 940–950. 10.19802/j.issn.1007-9084.2020234 [DOI] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mousavi S. A., Dubin A. E., Zeng W. Z., Coombs A. M., Do K., Ghadiri D. A., et al. (2021). PIEZO ion channel is required for root mechanotransduction in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 118:e2102188118. 10.1073/pnas.2102188118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 411–432. Epub. 10.1104/pp.105.073783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H., Halachmi R., Savaldi-Goldstein S., Rubin E., Ophir R., Fluhr R. (2004). Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 39 877–885. 10.1111/j.1365-313X.2004.02172.x [DOI] [PubMed] [Google Scholar]
- Nishiuchi T., Suzuki K., Kitajima S., Sato F., Shinshi H. (2002). Wounding activates immediate early transcription of genes for ERFs in tobacco plants. Plant Mol. Biol. 49 473–482. 10.1023/a:1015553232309 [DOI] [PubMed] [Google Scholar]
- Okamoto T., Takahashi T. (2019). Ethylene signaling plays a pivotal role in mechanical-stress-induced root-growth cessation in Arabidopsis thaliana. Plant Signal. Behav. 14:1669417. 10.1080/15592324.2019.1669417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T., Takatani S., Motose H., Iida H., Takahashi T. (2021). The root growth reduction in response to mechanical stress involves ethylene-mediated microtubule reorganization and transmembrane receptor-mediated signal transduction in Arabidopsis. Plant Cell Rep. 40 575–582. 10.1007/s00299-020-02653-6 [DOI] [PubMed] [Google Scholar]
- Perata P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199 639–649. 10.1111/nph.12291 [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., et al. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115 679–689. 10.1016/s0092-8674(03)00968-1 [DOI] [PubMed] [Google Scholar]
- Radin I., Richardson R. A., Coomey J. H., Weiner E. R., Bascom C. S., Li T., et al. (2021). Plant PIEZO homologs modulate vacuole morphology during tip growth. Science 373 586–590. 10.1126/science.abe6310 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Trelles F., Tarrío R., Ayala F. J. (2006). Origins and evolution of spliceosomal introns. Annu. Rev. Genet. 40 47–76. 10.1146/annurev.genet.40.110405.090625 [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J. G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. (2002). DNA- binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290 998–1009. 10.1006/bbrc.2001.6299 [DOI] [PubMed] [Google Scholar]
- See D. R., Brooks S., Nelson J. C., Brown-Guedira G., Friebe B., Gill B. S. (2006). Gene evolution at the ends of wheat chromosomes. Proc. Natl. Acad. Sci. U.S.A. 103 4162–4167. 10.1073/pnas.0508942102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlamovitz N., Ziv M., Zamski E. (1995). Light, dark and growth regulator involvement in groundnut (Arachis hypogaea L.) pod development. Plant Growth Regul. 16 37–42. 10.1007/BF00040505 [DOI] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12 3703–3714. 10.1101/gad.12.23.3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A. L., Nastou K. C., Lyon D., Kirsch R., Pyysalo S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49 D605–D612. 10.1093/nar/gkaa1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Yang D. C., Meng Y. Q., Jin J. P., Gao G. (2019). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48 D1104–D1113. 10.1093/nar/gkz1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Li Y., Lyu M., Luo Y., Shi H., Zhong S. (2020). Touch-induced seedling morphological changes are determined by ethylene-regulated pectin degradation. Sci. Adv. 6:eabc9294. 10.1126/sciadv.abc9294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen X., Chen Y., Zhang Q., Su L., Chen X., et al. (2020). Genome-wide identification of miRNAs and their targets during early somatic embryogenesis in Dimocarpus longan Lour. Sci. Rep. 10:4626. 10.1038/s41598-020-60946-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Nishida S., Shitan N., Sato F. (2020). Genome-wide identification of AP2/ERF transcription factor-encoding genes in California poppy (Eschscholzia californica) and their expression profiles in response to methyl jasmonate. Sci. Rep. 10:18066. 10.1038/s41598-020-75069-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W., An T., Xu Z., Zhang L., Gao H., Sun W., et al. (2020). Genomic-wide identification and expression analysis of AP2/ERF transcription factors related to andrographolide biosynthesis in Andrographis paniculata. Ind. Crops Prod. 157:112878. 10.1016/j.indcrop.2020.112878 [DOI] [Google Scholar]
- Zamski E., Ziv M. (1976). Pod formation and its geotropic orientation in the peanut, Arachis hypogaea L., in relation to light and mechanical stimulus. Ann. Bot. 40 631–636. 10.1093/oxfordjournals.aob.a085173 [DOI] [Google Scholar]
- Zhang H., Pan X., Liu S., Lin W., Li Y., Zhang X. (2021). Genome-wide analysis of AP2/ERF transcription factors in pineapple reveals functional divergence during flowering induction mediated by ethylene and floral organ development. Genomics 113 474–489. 10.1016/j.ygeno.2020.10.040 [DOI] [PubMed] [Google Scholar]
- Zhao C., Zhang H., Song C., Zhu J. K., Shabala S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation. 1:100017. 10.1016/j.xinn.2020.100017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang W., Chen H., Yang M., Wang J., Pandey M. K., et al. (2019). The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nat. Genet. 51 865–876. 10.1038/s41588-019-0402-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv M., Zamski E. (1975). Geotropic responses and pod development in gynophore explants of peanut (Arachis hypogaea L.) cultured in vitro. Ann. Bot. 39 579–583. 10.1093/oxfordjournals.aob.a084968 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental materials and illustrations of mechanical pressure simulation experiment.
Chromosomal distribution of ethylene sensors, constitutive triple responses (CTRs), ethylene insensitive 2 (EIN2s), ethylene insensitive 3/ethylene insensitive 3-like (EIN3/EILs), and EIN3-binding F-box proteins (EBFs) in Arachis hypogaea (A. hypogaea) genome.
Gene location and intragenomic collinearity of AhAP2/ERFs. Gray lines indicated all the collinearity in A. hypogaea genome, while colored lines highlighted the collinearity of AhAP2/ERFs.
Alternative splice (AS) analysis of Apetala2/ethylene-responsive factor (AP2/ERF) genes in A. hypogaea. ES, exon skip; AP, alternate promoter; AT, alternate terminator; RI, retained intron. The red inverted triangle indicated the difference of AS.
Protein sequence alignment and express pattern of AhPiezo.
The primers of quantitative PCR (qPCR).
The FPKM values of ethylene signal genes during developmental stage and in response to mechanical stress.
Segmental duplication Apetala2/ethylene-responsive factor (AhAP2/ERF) gene pairs in Arachis hypogaea (A. hypogaea) genome.
Segmental duplication AhAP2/ERF gene pairs in A. hypogaea and four other plant species.
The FPKM values of AhAP2/ERFs during developmental stage.
Promoter elements information.
The FPKM values of AhAP2/ERFs in response to mechanical stress.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA770556.