Abstract
Successful reproduction in the Brassicaceae is mediated by a complex series of interactions between the pollen and the pistil, and some species have an additional layer of regulation with the self-incompatibility trait. While the initial activation of the self-incompatibility pathway by the pollen S-locus protein 11/S locus cysteine-rich protein and the stigma S Receptor Kinase is well characterized, the downstream mechanisms causing self-pollen rejection are still not fully understood. In previous studies, we detected the presence of autophagic bodies with self-incompatible (SI) pollinations in Arabidopsis lyrata and transgenic Arabidopsis thaliana lines, but whether autophagy was essential for self-pollen rejection was unknown. Here, we investigated the requirement of autophagy in this response by crossing mutations in the essential AUTOPHAGY7 (ATG7) and ATG5 genes into two different transgenic SI A. thaliana lines in the Col-0 and C24 accessions. By using these previously characterized transgenic lines that express A. lyrata and Arabidopsis halleri self-incompatibility genes, we demonstrated that disrupting autophagy weakened their SI responses in the stigma. When the atg7 or atg5 mutations were present, an increased number of SI pollen was found to hydrate and form pollen tubes that successfully fertilized the SI pistils. Additionally, we confirmed the presence of GFP-ATG8a-labeled autophagosomes in the stigmatic papillae following SI pollinations. Together, these findings support the requirement of autophagy in the self-incompatibility response and add to the growing understanding of the intracellular mechanisms employed in the transgenic A. thaliana stigmas to reject self-pollen.
In self-incompatible transgenic Arabidopsis thaliana lines, autophagy is an integral part of the cellular mechanisms in the stigma to efficiently block fertilization by self-incompatible pollen.
Introduction
Reproduction in the Brassicaceae is a tightly controlled process, with many components contributing to successful pollen–pistil interactions (reviewed in Mizuta and Higashiyama, 2018; Johnson et al., 2019; Adhikari et al., 2020; Abhinandan et al., 2021). The Brassicaceae dry stigma lacks surface secretions and instead regulates the delivery of resources to the pollen shortly after pollination. This leads to the selective hydration of compatible pollen grains, followed by the penetration of the pollen tube into the stigmatic barrier to grow down the reproductive tract (reviewed in Dickinson, 1995; Palanivelu and Tsukamoto, 2012; Adhikari et al., 2020; Abhinandan et al., 2021). It is also at this initial cell–cell communication stage that self-incompatible (SI) pollen is recognized and blocked by the stigma to prevent inbreeding (reviewed in Nasrallah, 2019; Durand et al., 2020; Abhinandan et al., 2021). A recent live-cell imaging study in Arabidopsis thaliana compared pollen hydration rates of compatible and SI pollen grains and found that while compatible pollen grains rapidly hydrated, SI pollen failed to fully hydrate which then led to a failure in pollen germination or a failure of pollen tubes to penetrate the stigmatic papilla cell wall (Rozier et al., 2020). Furthermore, actin foci were present in the stigmatic papillae at the site of emerging pollen tubes for compatible pollen only. The actin focalization is likely associated with the stigmatic papilla cellular responses to enable compatible pollen hydration and pollen tube penetration (Rozier et al., 2020).
The Brassicaceae SI response is initiated by the binding of the S-haplotype-specific pollen ligand, S-locus protein 11/S locus cysteine-rich protein (SP11/SCR) to the corresponding stigma-specific S Receptor Kinase (SRK). This leads to the phosphorylation of the SRK kinase domain and activation of the downstream signaling responses, ultimately leading to the rejection of the SI pollen grain (reviewed in Nasrallah, 2019; Abhinandan et al., 2021). Not all Brassicaceae species possess this trait due to mutations in the key self-incompatibility genes, SP11/SCR and SRK (reviewed in Durand et al., 2020). For example, the transition of A. thaliana to a fully selfing species is associated with mutations in the SCR and SRK genes (Kusaba et al., 2001; Shimizu et al., 2008; Boggs et al., 2009; Tsuchimatsu et al., 2010; Guo et al., 2011; Tsuchimatsu et al., 2017). In contrast, other family members like Arabidopsis lyrata and Arabidopsis halleri have intact SCR and SRK genes, which result in a robust self-incompatibility response upon self-pollination (Kusaba et al., 2001; Schierup et al., 2001; Goubet et al., 2012).
A number of studies have reintroduced the self-incompatibility trait back into A. thaliana by introducing A. lyrata or A. halleri SP11/SCR and SRK transgenes (Nasrallah et al., 2004; Boggs et al., 2009; Indriolo et al., 2014a, 2014b; Iwano et al., 2015; Zhang et al., 2019; Rozier et al., 2020). However, the success of this approach was accession-dependent; for example, transgenic experiments in A. thaliana Col-0 failed to produce SI plants, whereas experiments using A. thaliana C24 were very successful (Nasrallah et al., 2004; Boggs et al., 2009; Iwano et al., 2015). The cause of this was linked to inverted repeats within the A. thaliana SRKA pseudogene which triggers RNA silencing of the introduced SRK transgenes (Fujii et al., 2020). For the Col-0 accession, the inverted repeat was found in a conserved region of the predicted pseudoSRKA kinase domain resulting in the silencing of closely related SRK transgenes. For the C24 accession, the inverted repeat was found in a more polymorphic region of the predicted pseudoSRKA ectodomain and appeared to be less effective at inducing RNA silencing of related SRK transgenes (Fujii et al., 2020).
We have found that a strong SI response can also be recapitulated in A. thaliana Col-0 when the A. lyrata or A. halleri ARM Repeat Containing 1 (ARC1) transgene was added along with the A. lyrata or A. halleri SP11/SCR and SRK transgenes (Indriolo et al., 2014a, 2014b; Zhang et al., 2019). The ARC1 E3 ubiquitin ligase is proposed to act directly downstream of SRK in the self-incompatibility signaling pathway (Gu et al., 1998; Stone et al., 1999, 2003); reviewed in Abhinandan et al. (2021). A large part of the ARC1 gene has been deleted in the A. thaliana genome, and knockdowns of ARC1 in SI Brassica napus and A. lyrata resulted in a partial breakdown of self-incompatibility (Stone et al., 1999; Indriolo et al., 2012). ARC1 is proposed to target cellular components that are required in the stigma to accept compatible pollen, such as the EXO70A1 exocyst subunit (Samuel et al., 2009) and Brassica Glyoxalase I (GLO1) and Phospholipase D (Sankaranarayanan et al., 2015; Scandola and Samuel, 2019). SI pollinations in transgenic A. thaliana SI-C24 plants also induced a strong and rapid cytosolic calcium influx in the stigmatic papillae (Iwano et al., 2015). These combined self-incompatibility responses are proposed to disrupt the secretory activity in the stigmatic papillae which would be required for compatible pollen acceptance (Elleman and Dickinson, 1996; Samuel et al., 2009; Safavian and Goring, 2013; Safavian et al., 2015); reviewed in Dickinson (1995), Goring (2017), and Abhinandan et al. (2021).
In previous work, we uncovered signs of autophagy activation in SI A. lyrata and transgenic A. thaliana SI-Col-0 stigmatic papillae within 10 min of SI pollinations, but not with compatible pollinations (Safavian and Goring, 2013; Indriolo et al., 2014a, 2014b). Transmission electron microscopy (TEM) images revealed structures resembling autophagic bodies in the vacuoles of stigmatic papilla following SI pollinations (Safavian and Goring, 2013; Indriolo et al., 2014a, 2014b). In contrast, TEM images of compatible pollinations revealed vesicle-like structures in the stigmatic papillae under the compatible pollen contact site suggestive of secretory activity (Safavian and Goring, 2013; Indriolo et al., 2014a, 2014b). The presence of autophagosomes in A. lyrata SI pollinations was further confirmed using the GFP-ATG8a autophagosome marker (Safavian and Goring, 2013). Although these studies indicated that autophagy was activated following SI pollinations, it was unclear whether autophagy is essential for the rejection of SI pollen. Here, we have used two different transgenic A. thaliana SI lines (in the Col-0 and C24 accessions; Iwano et al., 2015; Zhang et al., 2019) to test the requirement of the autophagy system for self-pollen rejection. When loss-of-function mutants in the AUTOPHAGY7 (ATG7) and ATG5 genes (Chung et al., 2010; Ding et al., 2018) were combined with the SI-Col-0 and SI-C24 lines, we observed a partial breakdown of the self-incompatibility response in both accessions. This indicates that autophagy is an additional intracellular mechanism by which SI pollen is effectively rejected in both transgenic A. thaliana SI lines.
Results
Generation of SI A. thaliana transgenic lines carrying the atg7 and atg5 mutations to disrupt autophagy
To investigate the involvement of autophagy in self-incompatibility, two different transgenic A. thaliana SI lines were used to cross with atg7 and atg5 autophagy mutants. ATG7 and ATG5 are essential components in the formation of autophagosomes (reviewed in (Ding et al., 2018), and both the atg7 and atg5 mutations lead to a loss of autophagy (Thompson et al., 2005; Chung et al., 2010; Young et al., 2019). The first SI line is in the Col-0 accession (SI-Col-0) and carries the A. halleri SCR13, SRK13, and ARC1 transgenes in a single T-DNA (Ah-SCR13 Ah-SRK13 Ah-ARC1 #2 line; Zhang et al., 2019). This SI-Col-0 line was crossed to the previously characterized atg7-2 mutant in the Col-0 accession (Chung et al., 2010). The F1 plants from this cross were selfed, and all pollination assays were carried out in the F2 generation. To confirm that any observed effects were not specific to the loss of ATG7, the previously characterized atg5-1 mutation in the Col-0 accession (Thompson et al., 2005) was also crossed into the SI-Col-0 line, and taken to the F2 generation for pollination assays. The second SI line is in the C24 accession (SI-C24) and carries the A. lyrata SCRb and SRKb transgenes (SI-C24 #15-1 line; Iwano et al., 2015). As there were no atg7 mutants available in the C24 accession, CRISPR/Cas9 technology was used to generate additional atg7 mutants. Deletion of the ATG7 gene was confirmed through PCR and sequencing of genomic regions flanking gRNA target sites (Supplemental Figure S1). The atg7-9 C24 mutant displayed the same overall wild-type plant and flower morphology (Supplemental Figure S2) as seen for the atg7-2 Col-0 mutant. The C24 atg7-9 mutant was crossed with the SI-C24 line, and following selfing of the F1 plants, all pollination assays were carried out in F2 generation (T4 generation for the atg7-9 mutation). For all three sets of F2 progeny, genotyping was first carried out to identify the transgenic plants carrying the SI transgenes, followed by genotyping for the atg7 or atg5 mutations to identify plants that were wild-type or homozygous for the atg mutations.
Disrupting autophagy breaks down the initial SI response of blocking pollen hydration
To assess the early stages of pollen rejection, the pollen hydration checkpoint was first investigated. SI pollen from the parental SI-Col-0 or SI-C24 transgenic lines were used to pollinate pistils from the respective F2 progeny segregating for the atg7 or atg5 mutations, and pollen hydration assays were carried out by measuring the increase in pollen diameter at 10-min postpollination (Lee et al., 2020). Positive controls of the SI-Col-0 and SI-C24 pollen placed on compatible wild-type Col-0 and C24 pistils, respectively, resulted in a large increase in diameter as a result of pollen hydration (Figure 1, A and B). In contrast, self-pollen placed on the SI-Col-0 and SI-C24 pistils resulted in very little pollen hydration at 10-min postpollination (Figure 1, A and B). This lack of hydration was consistent between the SI-Col-0 and SI-C24 lines and indicative of the early recognition and rejection of SI pollen. Importantly, stigmas from the atg7-2, atg5-1, and atg7-9 mutants showed normal SI-Col-0 and SI-C24 pollen hydration, respectively, indicating that disruption in the autophagic process did not perturb the compatible postpollination responses (Figure 1, A and B).
Figure 1.
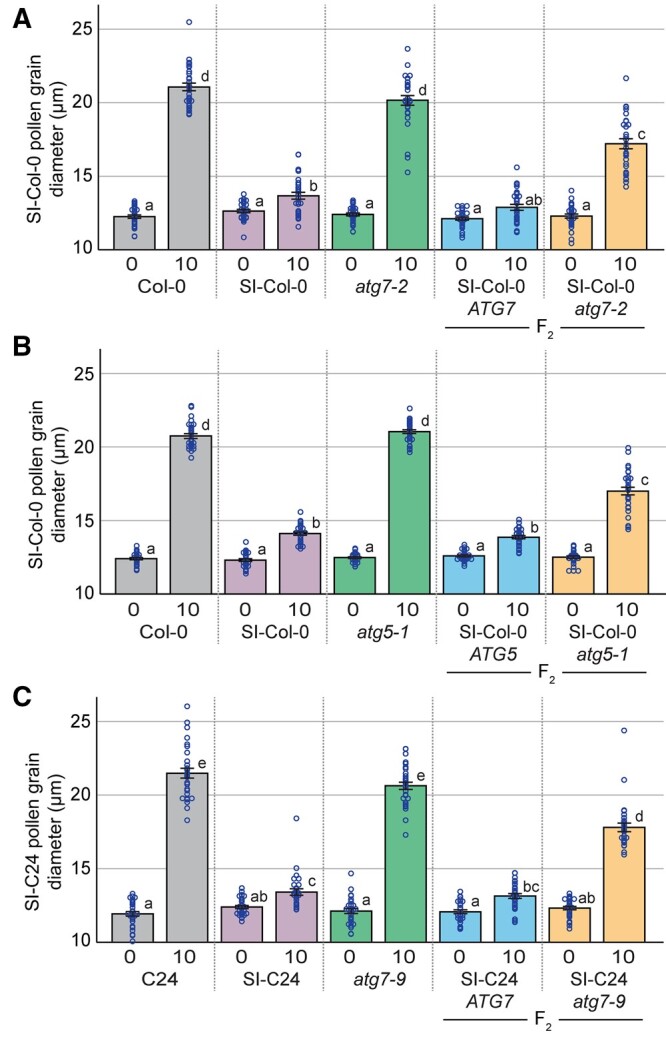
Pollen hydration assays on control and SI F2 progeny segregating for the atg7 and atg5 mutations. Pollen hydration assays for (A), SI-Col-0 pollen grains placed on stigmas from Col-0, SI-Col-0, atg7-2, and F2 progeny from the SI-Col-0 x atg7-2 cross; (B) SI-Col-0 pollen grains placed on stigmas from Col-0, SI-Col-0, atg5-1, and F2 progeny from the SI-Col-0 x atg5-1 cross; and (C) SI-C24 pollen grains placed on stigmas from C24, SI-C24, atg7-9, and F2 progeny stigmas from the SI-C24 × atg7-9 cross. Pollen hydration was measured by taking pollen grain diameters at 0- and 10-min postpollination. Data are shown as mean ± se with all data points displayed. n = 30 pollen grains per line, P < 0.05 (one-way ANOVA with Tukey’s HSD post-hoc test).
For the F2 progeny from the SI-Col-0 x atg7-2 cross, the SI-Col-0 plants homozygous for wild-type ATG7 showed the same small increase in pollen hydration as the control SI pollinations at 10-min postpollination. In contrast, when SI-Col-0 pollen was applied to the SI-Col-0 stigmas that were homozygous for the atg7-2 mutation, an increase in pollen hydration was observed, indicating an incomplete rejection of the SI pollen (Figure 1A). Similarly, pollen hydration assays on the F2 progeny from the SI-Col-0 × atg5-1 cross showed that hydration of SI-Col-0 pollen on SI-Col-0 stigmas wild-type for ATG5 displayed a small increase while SI-Col-0 pollen hydration on SI-Col-0 stigmas homozygous for the atg5-1 mutation was significantly increased (Figure 1B). A comparable set of pollen hydration experiments on the F2 progeny from the SI-C24 × atg7-9 cross also showed a similar trend (Figure 1C). SI-C24 stigmas that were wild-type for ATG7 supported similar small increases in SI-C24 pollen hydration as the control SI pollinations, and the introduction of the atg7-9 mutation into this background resulted in increased hydration of the SI pollen. Together, these results indicate that autophagy is required to fully prevent the hydration of SI pollen.
Disruption of autophagy increases SI pollen tube penetration
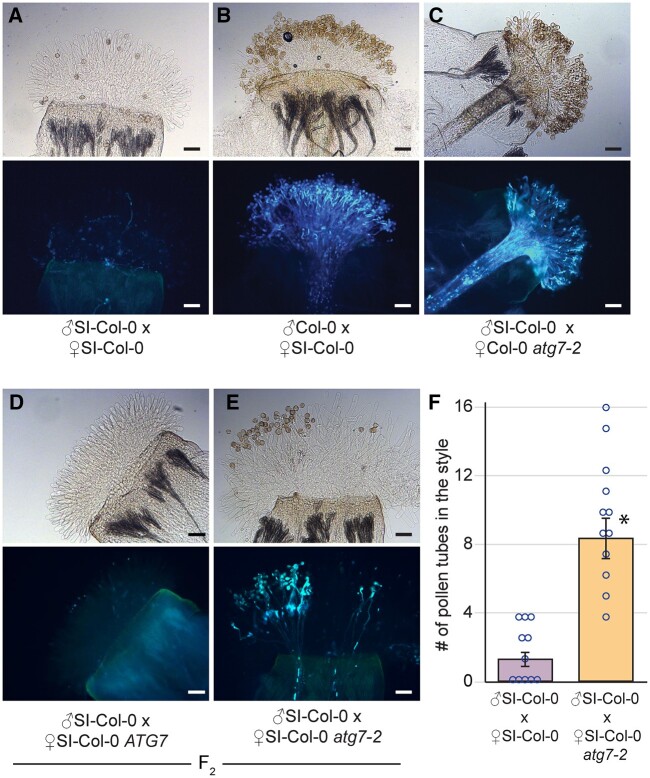
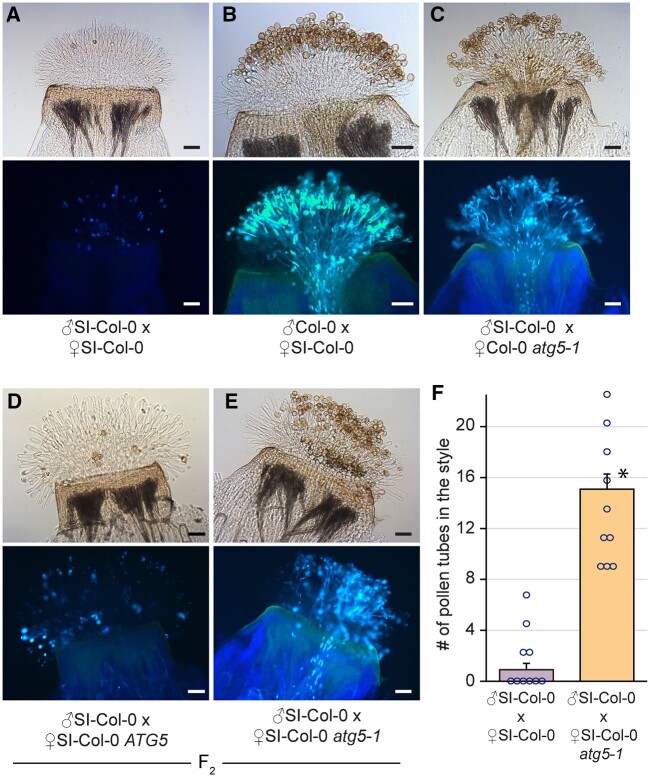
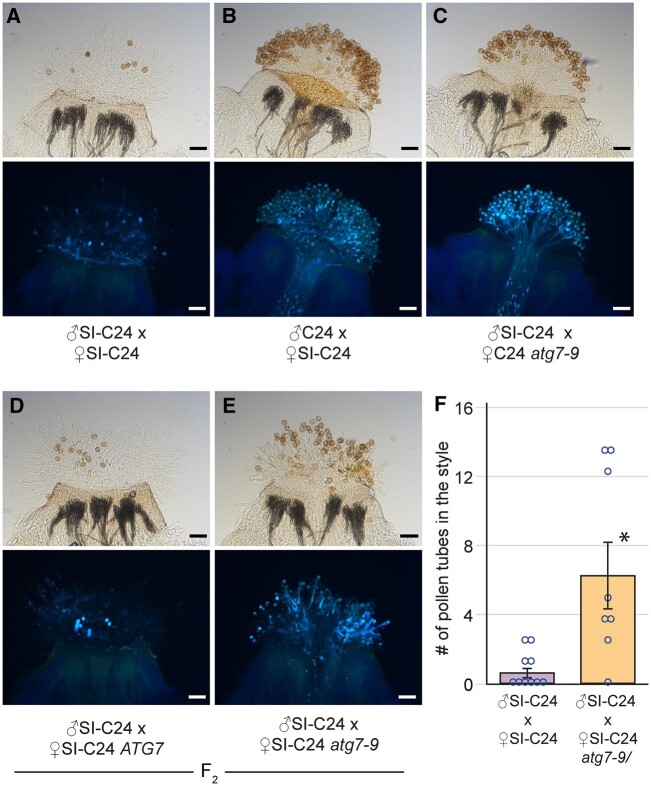
With the observed increase in SI pollen hydration, the next stage of preventing SI pollen germination and pollen tube penetration was investigated. To determine if this block was affected by the atg7 and atg5 mutations, pollinated pistils were collected at 2- and 24-h postpollination and stained with aniline blue to visualize pollen tubes. At 2-h postpollination, SI-Col-0 and SI-C24 pollen was completely rejected by the respective SI pistils, and no pollen tubes were visible (Supplemental Figures S3 and S4). Although we did not see any obvious difference at 2-h postpollination, we did observe an occasional pollen tube that penetrated the stigma barrier of the SI-Col-0 and SI-C24 atg homozygous mutants. To address whether we could observe a greater number of SI-Col-0 and SI-C24 pollen tubes after a longer period, pollinated pistil samples were then harvested at 24-h postpollination. A later time point following pollination is more reflective of the natural pollination process and provides more time for the SI pollen to germinate and grow into the pistil. Self-pollination of the SI-Col-0 and SI-C24 parental lines resulted in an absence of pollen tubes (Figures 2, A, 3, A, and 4, A), while pollinations with compatible wild-type pollen led to an abundance of pollen tubes growing in the pistils (Figures 2, B, 3, B, and 4, B). The results indicated that the SI-Col-0 and SI-C24 pistils were able to fully reject SI pollen and accept wild-type compatible pollen at 24-h postpollination. The SI-Col-0 and SI-C24 pollen also produced numerous pollen tubes when placed on the atg mutant pistils, indicating that the absence of autophagy did not affect these compatible pollinations (Figures 2, C and 3, C). The impact of the atg mutations on self-incompatibility at 24-h postpollination was examined by applying the SI-Col-0 and SI-C24 pollen on their respective F2 progeny homozygous for the atg mutations. When SI-Col-0 atg7-2 stigmas or SI-Col-0 atg5-1 stigmas were pollinated with SI-Col-0 pollen, there was a general increase in the number of pollen grains adhering to the stigma and forming pollen tubes when compared to the SI-Col-0 pistils (Figures 2, D–F and 3, D–F). The pollinations were repeated for the SI-C24 F2 plants, and again the SI-C24 atg7-9 pistils did show some SI-C24 pollen adhering to the stigmatic surface and successfully growing pollen tubes through the pistil while the SI-C24 ATG7 pistils rejected the SI-C24 pollen (Figure 4, D–F). The number of pollen tubes was counted in the SI-Col-0 and SI-C24 pistils that were wild-type or homozygous for the atg mutations, and a significant increase in the number of pollen tubes were counted in the homozygous atg7 and atg5 mutants (Figures 2, F, 3, F, and 4, F). Thus, the 24-h postpollination samples demonstrated that autophagy is required for fully blocking SI pollen germination and pollen tube growth through the SI pistil.
Figure 2.
Representative images of aniline blue-stained SI-Col-0 or SI-Col-0 atg7-2 pistils at 24 h following pollination. A–E, All stigmas were hand-pollinated with SI-Col-0 pollen or wild-type Col-0 pollen as indicated and collected at 24-h postpollination for aniline blue staining. The genotypes of the pistils are indicated for each panel. Pollen tubes penetrating through the stigma and style indicate a breakdown of self-incompatibility as seen in (E). For each panel, top image = brightfield, bottom image = aniline blue. Scale bar = 100 µm. F, Bar graph showing the mean number of pollen tubes at 24-h postpollination. Data are shown as mean ± se with all data points displayed. n = 11–12 pistils per line. At 24-h, pistils from SI-Col-0 atg7-2 mutants allowed a significantly higher number of SI-Col-0 pollen tubes compared to the SI-Col-0 pistils. (P < 0.05, Student’s t test).
Figure 3.
Representative images of aniline blue-stained SI-Col-0 or SI-Col-0 atg5-1 pistils at 24 h following pollination. A–E, All stigmas were hand-pollinated with SI-Col-0 pollen or wild-type Col-0 pollen as indicated and collected at 24-h postpollination for aniline blue staining. The genotypes of the pistils are indicated for each panel. Pollen tubes penetrating through the stigma and style indicate a breakdown of self-incompatibility as seen in (E). For each panel, top image = brightfield, bottom image = aniline blue. Scale bar = 100 µm F, Bar graph showing the mean number of pollen tubes at 24 h postpollination. Data are shown as mean ± se with all data points displayed. n = 10 pistils per line. At 24 h, pistils from SI-Col-0 atg5-1 mutants allowed a significantly higher number of SI-Col-0 pollen tubes compared to the SI-Col-0 pistils (P < 0.05, Student’s t test).
Figure 4.
Representative images of aniline blue-stained SI-C24 or SI-C24 atg7-9 pistils at 24 h following pollination. A–E, All stigmas were hand-pollinated with SI-C24 pollen or wild-type C24 pollen as indicated and collected at 24-h postpollination for aniline blue staining. The genotypes of the pistils are indicated for each panel. Pollen tubes penetrating through the stigma and style indicate a breakdown of self-incompatibility as seen in (E). For each part, top image = brightfield, bottom image = aniline blue. Scale bar = 100 µm. F, Bar graph showing the mean number of pollen tubes at 24-h postpollination. Data are shown as mean ± se with all data points displayed. n = 8–10 pistils per line. At 24-h, pistils from SI-C24 atg7-9 mutants allowed a significantly higher number of SI-C24 pollen tubes compared to the SI-C24 pistils (P < 0.05, Student’s t test).
Loss of autophagy increases seed set in the SI lines
With the observation that self-incompatibility is partially lost in the SI-Col-0 atg7-2, SI-Col-0 atg5-1, and SI-C24 atg7-9 mutant pistils, resulting in more pollen tubes growing through the pistils, the impact of this on seed set was assessed. Manual pollinations were carried out using SI-Col-0 and SI-C24 pollen on pistils from the respective parental lines and F2 progeny homozygous for the atg7 or atg5 mutations, and 2-week-old siliques were harvested for seed counting (Figure 5; Supplemental Figure S5). As expected for compatible pollinations in Col-0, C24, and the atg mutants, abundant seeds/silique were produced (Figure 5, A–C). As well, self-pollinations of SI-Col-0 and SI-C24 pistils resulted in a very limited seed yield consistent with the rejection of SI pollen (Figure 5, A–C). When the SI-Col-0 atg7-2 pistils were pollinated with SI-Col-0 pollen, there was a significant increase in yield to 27 seeds/silique compared to 12.8 seeds/silique for SI-Col-0 indicating an incomplete rejection of the SI-Col-0 pollen by the SI-Col-0 atg7-2 pistils (Figure 5A). Similarly, when SI-Col-0 atg5-1 pistils were pollinated with SI-Col-0 pollen, there was an increased average yield of 31.9 seeds/silique compared to 10.8 seeds/silique for SI-Col-0 (Figure 4B). Finally, similar trends in seed yields were observed in the C24 accession. SI pollinations with SI-C24 pistils resulted in a low average of 6.8 seeds/silique as expected yet when the SI-C24 atg7-9 pistils were pollinated with SI-C24 pollen, there was a significant increase in seed yields to 17.2 seeds/silique (Figure 5C). Altogether, these results indicate that loss of autophagy was sufficient to disrupt the self-incompatibility pathway allowing for the growth of more SI pollen tubes through the SI-Col-0 and SI-C24 pistils carrying atg7 or atg5 mutations to initiate fertilization and seed production, and thus, confirming that autophagy is required for full rejection of self-pollen.
Figure 5.
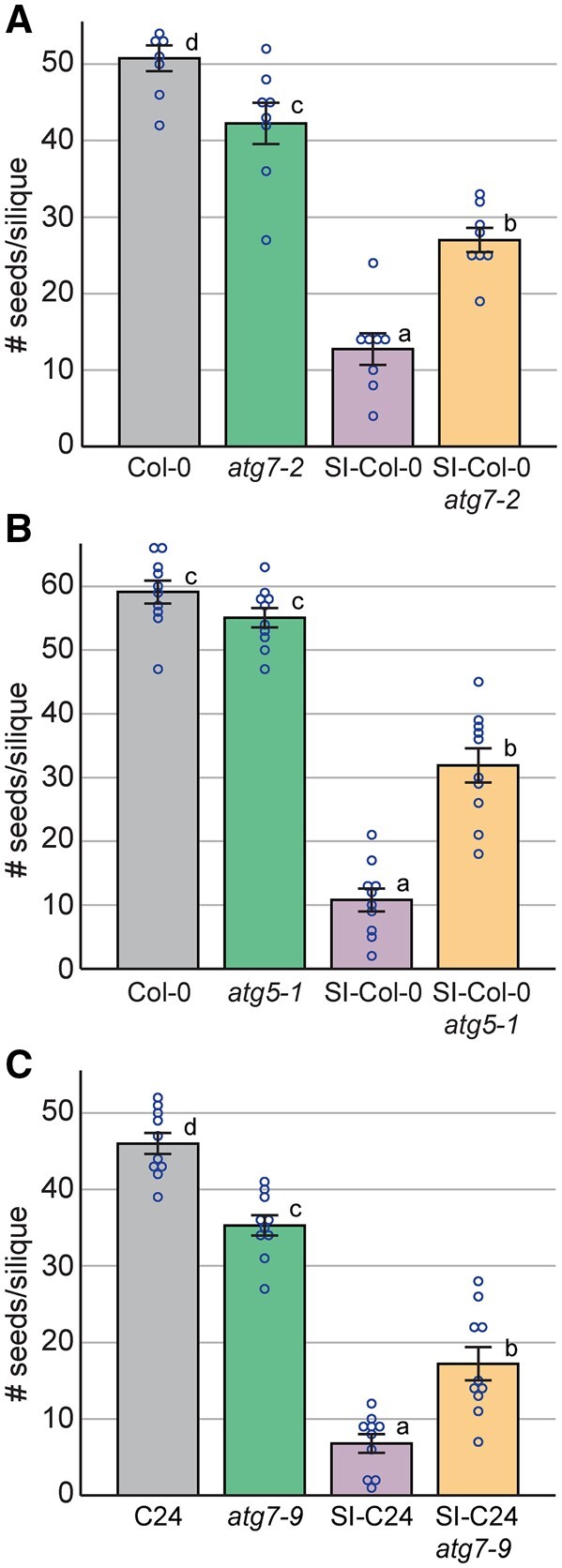
Seeds per silique counts for control and SI F2 progeny with the atg7 or atg5 mutations. Seeds per silique counts following manual pollination with (A), (B), SI-Col-0 pollen or (C), SI-C24 pollen on pistils with the indicated genotypes. Siliques were harvested at 2-week postpollination and cleared for seeds counts. Data are shown as mean ± se with all data points displayed. n = 8–10 siliques per line. Compared to the control SI pollinations, significant increases in the number of seeds per silique were observed for the SI plants that were homozygous for the atg7 or atg5 mutations, P < 0.05 (one-way ANOVA with Tukey’s HSD post-hoc test).
Autophagosomes are produced following self-pollination in the SI-C24 line
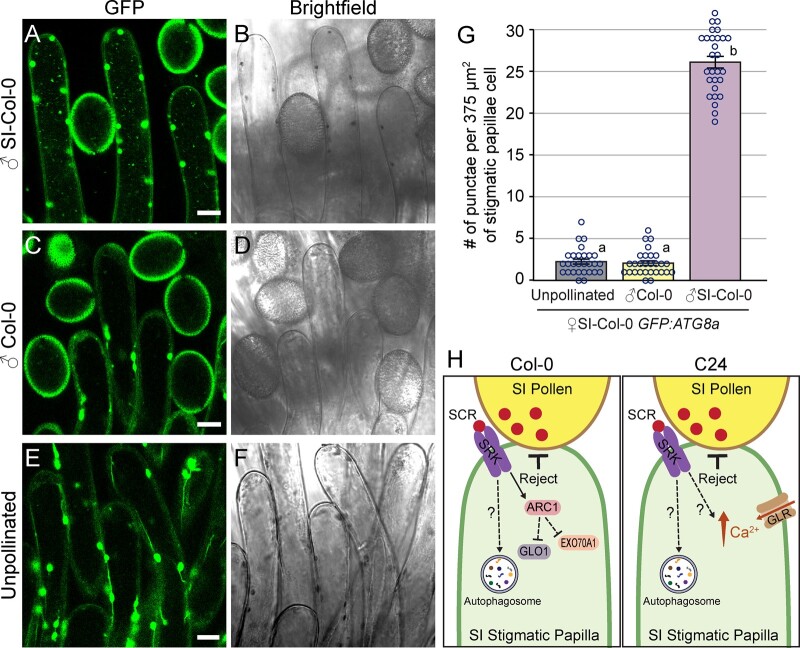
Previously, autophagosomes were detected by TEM following SI pollinations in A. lyrata and in the transgenic A. thaliana SI-Col-0 transformed with A. lyrata S-locus genes (Safavian and Goring, 2013; Indriolo et al., 2014a, 2014b). To confirm that autophagosomes are also forming in the SI-Col-0 Ah-SCR13 Ah-SRK13 Ah-ARC1 #2 line used in this study, GFP-ATG8a was used as an autophagosome marker (Thompson et al., 2005). Col-0 plants carrying the ProUBQ10:GFP-ATG8a construct (Kim et al., 2013) were crossed with the SI-Col-0 line, and the subsequent F1 generation was used for imaging. The SI-Col-0 GFP-ATG8a stigmas were imaged as unpollinated or manually pollinated with either Col-0 or SI-Col-0 pollen. At 15-min postpollination, many GFP punctae were observed in the SI-Col-0 GFP-ATG8a stigmatic papillae contacting SI-Col-0 pollen (Figure 6A). Conversely, few puncta were observed in either unpollinated SI-Col-0 GFP-ATG8a stigmatic papillae or those pollinated with Col-0 pollen, consistent with a low background level of autophagy (Figure 6, C and E). The number of puncta were counted for both SI-Col-0 pollinated, Col-0 pollinated and unpollinated SI-Col-0 GFP-ATG8a stigmatic papillae (Figure 6G), and there was a significant increase in the number of GFP-ATG8a-labeled punctae in the SI-pollinated SI-Col-0 GFP-ATG8a stigmatic papillae indicating a strong induction of autophagy. These observations confirm our previous findings that autophagy is activated in the stigmatic papillae with SI pollinations when compared to a compatible pollinated or unpollinated stigma (Safavian and Goring, 2013; Indriolo and Goring, 2014).
Figure 6.
Autophagosomes are more abundant in SI-Col-0 GFP-ATG8a papillae following pollination with SI-Col-0 pollen. A–F, Representative images showing that more GFP-ATG8a-labeled autophagosomes marked by GFP-ATG8a are visible at 15-min postpollination in the SI-Col-0 GFP-ATG8a papillae pollinated with SI-Col-0 pollen (A and B) compared to Col-0 pollinated (C and D) or unpollinated (E and F) SI-Col-0 GFP-ATG8a papillae. Scale bar = 10 μm. G, Bar graph showing the mean number of autophagosomes present in each sample. Data are shown as mean ± se with all data points displayed. n = 28 papillae from six stigmas for each set of data. At 15-min postpollination with SI-Col-0 pollen, there is a significant increase in the number of GFP-labeled punctae in the SI-Col-0 GFP-ATG8a papillae (P < 0.05) (One-way ANOVA with Tukey’s HSD post-hoc test). The large punctae are chloroplasts with autofluorescence and were not counted. H, Models for SI pollen rejection in the transgenic A. thaliana accessions. Following the self-incompatibility determinants, SP11/SCR and SRK, the specific intracellular cellular mechanisms by which SI pollen is rejected in transgenic A. thaliana SI-Col-0 and SI-C24 lines are still not well-understood. These models summarize the known components for each transgenic accession (Samuel et al., 2009; Indriolo et al., 2014a, 2014b; Iwano et al., 2015; Kenney et al., 2020). Here, we show that autophagy is a shared requirement in the stigmatic papillae for both transgenic SI accessions to fully enable SI pollen rejection.
Discussion
Self-incompatibility plays an important regulatory role in many outcrossing Brassicaceae species where producing inbred progeny would be disadvantageous (Shimizu and Tsuchimatsu, 2015; Durand et al., 2020). Previous studies have uncovered many important steps in the self-incompatibility pathway, but there are still outstanding questions on the cellular mechanisms by which self-pollen is rejected (reviewed in Goring, 2017; Jany et al., 2019; Abhinandan et al., 2021). In previous studies, we observed the presence of autophagosomes in the cytoplasm and autophagic bodies in the vacuole following SI pollinations in SI A. lyrata and transgenic A. thaliana SI-Col-0 lines (Safavian and Goring, 2013; Indriolo and Goring, 2014). What was not known, however, was whether autophagy was necessary for SI pollen rejection. In this study, we addressed this question by crossing two different autophagy-deficient mutants (atg7 and atg5) into two different transgenic A. thaliana SI-lines and demonstrated that a functioning autophagy system is required in the stigma for the full rejection of SI pollen. It is important to note that disrupting autophagy did not cause a full loss of self-incompatibility indicating that other cellular responses are required in conjunction with autophagy for SI pollen rejection. Importantly, this partial breakdown in the stigma was seen for all stages from increased pollen hydration to more pollen tubes growing through the pistil to increase seed yields. Finally, the requirement of autophagy in the rejection of SI pollen was consistent between the three different autophagy-deficient mutants (atg7-2, atg7-9, and atg5) and the two different A. thaliana accessions (Col-0 and C24) used in the SI lines indicating that this is a shared mechanism for SI pollen rejection.
The rationale for using both the SI-Col-0 and SI-C24 lines was to determine if there were any accession- or transgene-specific associations with autophagy. The SI-Col-0 carries the A. halleri SCR13, SRK13, and ARC1 transgenes (Zhang et al., 2019), while the SI-C24 line carries the A. lyrata SCRb and SRKb transgenes (Iwano et al., 2015). In addition to two different S-haplotypes from two different Arabidopsis species, the SI-Col-0 line also carries the A. halleri ARC1 transgene. Overall, our results indicate that autophagy is a shared intracellular response in these two transgenic accessions for SI pollen rejection (see models in Figure 6H). The importance of this discovery is related to how little is really known about the intracellular mechanisms employed by these transgenic A. thaliana SI-lines. Much of our current understanding of the self-incompatibility pathway relies on knowledge gained from the compatible pollen response pathway in combination with studies on the Brassica self-incompatibility system (reviewed in Abhinandan et al., 2021). Many studies have verified the requirements of SP11/SCR and SRK transgenes for conferring the self-incompatibility trait in A. thaliana (Nasrallah et al., 2004; Boggs et al., 2009; Indriolo et al., 2014a, 2014b; Iwano et al., 2015; Zhang et al., 2019; Rozier et al., 2020), but the intracellular mechanisms by which self-pollen rejection occurs in these transgenic A. thaliana SI-lines are still poorly understood (Figure 6H). A role for a rapid cytosolic calcium influx and GLUTAMATE RECEPTOR-LIKE (GLR) genes in the stigmatic papillae was identified using the transgenic A. thaliana SI-C24 plants, but how they are activated in response to SI-pollen is not known (Iwano et al., 2015). As well, EXO70A1 and GLO1 have been linked to the compatible pollen responses in A. thaliana Col-0 and SI-Col-0 plants, respectively (Samuel et al., 2009; Kenney et al., 2020), and could be potentially targeted by ARC1 in the A. thaliana SI-Col-0 plants (Figure 6H). The fact that both accessions require an intact autophagy system in the stigma for full SI pollen rejection indicates that autophagy activation does not require ARC1, though the potential involvement of another related PUB E3 ligase cannot be ruled out (Indriolo et al., 2012; Indriolo and Goring, 2016).
The question then arises on how autophagy is activated and the purpose of autophagy activation in self-incompatibility. SI pollen carries the signals to be recognized as both SI pollen and compatible pollen, and it has been hypothesized that self-incompatibility blocks compatible pollen responses in the stigma (Dickinson, 1995). With the working model that secretory activity in the stigma is required for compatible pollen acceptance (reviewed in Dickinson, 1995; Goring, 2017; Abhinandan et al., 2021), several of the self-incompatibility cellular responses such as EX070A1 inhibition (Samuel et al., 2009; Sankaranarayanan et al., 2015), a strong and rapid calcium influx (Iwano et al., 2015), and the absence of actin focalization (Rozier et al., 2020) are consistent with blocking secretion (reviewed in Goring, 2017). Autophagy could also be an additional step to clear any secretory activity that was initially activated. With the already established connection of the exocyst for compatible pollen responses (Samuel et al., 2009; Safavian et al., 2014, 2015; Jamshed et al., 2020) and the targeting of EXO70A1 in Brassica self-incompatibility (Samuel et al., 2009), it may be that some of the exocyst subunits are redirected toward autophagy during the self-incompatibility response. For example, EXO70B2 is another exocyst subunit involved in secretion, and phosphorylation during immune responses was found to redirect EXO70B2 to autophagosomes for degradation, and this was proposed to reduce secretion (Brillada et al., 2021). Finally, the exocyst subunit, SEC5, has been linked to Rho of Plants GTPase signaling and autophagosome formation under stress conditions (Lin et al., 2021).
In conclusion, this study now firmly places autophagy as part of the cellular mechanisms leading to SI pollen rejection. Future studies will need to focus on how autophagy is activated in the self-incompatibility pathway and whether there are specific cytoplasmic components targeted for autophagic destruction. Understanding the mechanisms underlying self-incompatibility is important as seed yield of many important crop species is directly tied to their reproductive success, and the ability to modulate selfing would be a powerful tool in increasing yield.
Materials and methods
Plant materials and growth conditions
All A. thaliana seeds (Col-0, C24, transgenic lines, T-DNA insertion mutants and CRISPR/Cas9 deletion mutants) were sterilized and cold stratified at 4°C for 3 days. Seedlings were germinated on half Murashige and Skoog (1/2 MS) medium plates supplemented with 0.4% (w/v) phytoagar (pH 5.8) and 1% (w/v) sucrose. Seedlings were grown at 22°C under 16-h light for 7 days, and then transferred to Sunshine #1 soil supplemented with Plant Prod All Purpose Fertilizer 20-20-20 (1 g/L water). All plants were grown in chambers under a 16-h light/8-h dark growth cycle at 22°C, with humidity kept under 50%.
For the Col-0 accession experiments, the Ah-SCR13 Ah-SRK13 Ah-ARC1 #2 Col-0 line (Zhang et al., 2019) was crossed to the atg7-2 mutant (Col-0 accession; Chung et al., 2010) and atg5-1 mutant (Thompson et al., 2005). For the C24 accession experiments, the SI-C24 #15-1 line (with Al-SCRb AlSRKb; Iwano et al., 2015) was crossed to the atg7-9 mutant (C24 accession; see below). All transgenic lines were genotyped using primers found in Supplemental Table S1. For the C24 accession, plants were genotyped for AlSCRb, AlSRKb, and atg7-9. Progeny was analyzed in the F2 generation for all three crosses. For SI-Col-0 GFP-ATG8a, the Ah-SCR13 Ah-SRK13 Ah-ARC1 #2 Col-0 line (Zhang et al., 2019), was crossed into the ProUBQ10:GFP-ATG8a line (Col-0 accession; Kim et al., 2013), and the resulting F1 progeny was used for all imaging.
Plasmid construction and plant transformation
The atg7-9 CRISPR/Cas9 deletion mutation was generated in C24 using previously described methods in the Col-0 accession (Doucet et al., 2019a, 2019b). The final pBEE401E vector contained two guide RNAs targeting exons 3 and 8 of ATG7 (Supplemental Figure S1). A. thaliana C24 plants were transformed using floral dip (Clough and Bent, 1998), and plants were dipped twice, 3 days apart, to maximize transformation efficiency. T1 seeds were collected and germinated on soil, and 1-week-old seedlings were sprayed with BASTA herbicide to select for transformants. PCR was used to confirm transformants (BASTA marker) and the presence of atg7 mutations (deletions of the exons 3–8 region; Supplemental Figure S1). These atg7 deletion mutants were then verified through sequencing of regions flanking the deletions. The atg7 mutants showed a wild-type plant and flower morphology, along with the accelerated senescence typically seen in atg mutants (Thompson et al., 2005; Avila-Ospina et al., 2014). One homozygous mutant, atg7-9, from the T2 generation was crossed to the SI-C24 #15-1 line, and all data were collected in the F2 generation of this cross (F4 generation for atg7-9).
Confocal microscopy
A. thaliana SI-Col-0 plants carrying the ProUBQ10:GFP-ATG8a construct were manually pollinated using pollen from the SI-Col-0 line or wild-type Col-0. Pistils were removed at 10-min postpollination and images of autophagosomes were taken at 15-min postpollination. All images were captured using a Leica TCS SP8 confocal microscope using a 40× oil-immersion objective. Imaging was completed using a 488-nm laser at 35% intensity, captured using either a HyD detector for GFP (495–545-nm emission, 100% gain, 2.0 AU pinhole diameter) or a photomultiplier tube detector for bright field (700–850 V). Leica LAS AF lite in combination with ImageJ was used for image processing and puncta counting. GFP punctae were counted in stigmatic papillae cell areas directly adjacent to SI-Col-0 pollen or Col-0 pollen, and an equivalent cell area in unpollinated samples.
Assays for pollen hydration, aniline blue staining, and seed set
All postpollination assays used manual pollinations and were conducted as described in Lee et al. (2020). For the SI-C24 plants, late stage 12 pistils were first emasculated and wrapped in plastic wrap overnight prior to pollination. For the SI-Col-0 plants, the pistils did not require emasculation due to the approach herkogamy phenotype (Zhang et al. 2019), and freshly opened flowers with unpollinated pistils were selected for these assays. All pollinations were completed at humidity levels under 50% for consistency. Pollen hydration assays were conducted by removing and mounting pistils on 1/2 MS, and lightly pollinating the stigmatic surface using a single anther. Images were taken at 0- and 10-min postpollination using a Nikon sMz800 microscope, and pollen grains on the stigmatic surface were randomly selected for measurements using the NIS-elements imaging software. For the aniline blue staining, one anther was used to lightly pollinate each pistil, and the pistils were then collected at 2- and 24-h postpollination for staining as described in Lee et al. (2020). The aniline blue-stained pistils were mounted on a slide with sterile water, flattened with the coverslip, and imaged at 10× magnification on a Zeiss Axioscope2 Plus fluorescent microscope. Aniline blue images were captured using the UV laser (150 ms exposure, 2 gain), and bright-field images were captured using the built-in halogen lamp (20 ms exposure, 2 gain). For seed counts, pistils were manually pollinated with a single anther and were left for 10–14 days. The maturing siliques were then removed and cleared in 70% (v/v) ethanol for 3–5 days, or until seeds were visible (Beuder et al., 2020). Siliques were then mounted on a dry slide with the septum facing upwards and imaged using a Nikon sMz800 microscope, and seeds were counted using the NIS-elements imaging software.
Statistical analyses
One-way analysis of variance (ANOVA) with Tukey’s honest significant difference (HSD) post-hoc tests were performed using SPSS (IBM, Armonk, NY, USAs), and Student’s t tests were performed using Excel (Microsoft Redmond, WA, USA).
Accession numbers
Gene identifiers for the autophagy genes are as follows: ATG5, At5g17290, and ATG7 At5g45900.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Generation of the ATG7 CRISPR/Cas9 deletion mutation in the C24 accession.
Supplemental Figure S2 . Representative images of flowering plants for the SI-C24 and atg7-9 mutant plants in the C24 accession.
Supplemental Figure S3 . Representative images of aniline blue-stained pistils at 2 h following pollination with SI-Col-0 or Col-0 pollen.
Supplemental Figure S4 . Representative images of aniline blue-stained pistils at 2 h following pollination with SI-C24 or C24 pollen.
Supplemental Figure S5. Representative images of cleared siliques used in seed counting for control and SI plants with the atg7 mutation.
Supplemental Table S1. Primer sequences.
Supplementary Material
Acknowledgments
We thank Laura Canales and Arjun Sharma for technical assistance, and members of the Goring lab for critically reading this article. We are very grateful to Seiji Takayama (Nara Institute of Science and Technology; University of Tokyo) for providing the SI-C24 #15-1 seeds (A. thaliana C24), Richard Vierstra (Washington University in St Louis), and Peter Bozhkov (Swedish University of Agricultural Sciences) for providing atg7-2 and atg5-1 mutant seeds (A. thaliana Col-0), and Marisa Otegui (University of Wisconsin-Madison) for providing the ProUBQ10:GFP-ATG8a seeds (A. thaliana Col-0).
Funding
This research was supported by a grant from Natural Sciences and Engineering Research Council of Canada to D.R.G., S.R.M. was supported by the Rustom H. Dastur Graduate Scholarship, and H.K.L. was supported by an Ontario Graduate Scholarship.
Conflict of interest statement. None declared.
Contributor Information
Stuart R Macgregor, Department of Cell & Systems Biology, University of Toronto, Toronto, Canada M5S 3B2.
Hyun Kyung Lee, Department of Cell & Systems Biology, University of Toronto, Toronto, Canada M5S 3B2.
Hayley Nelles, Department of Cell & Systems Biology, University of Toronto, Toronto, Canada M5S 3B2.
Daniel C Johnson, Department of Cell & Systems Biology, University of Toronto, Toronto, Canada M5S 3B2.
Tong Zhang, National Key Laboratory of Crop Genetic Improvement, National Center of Rapeseed Improvement in Wuhan, Huazhong Agricultural University, Wuhan 430070, China.
Chaozhi Ma, National Key Laboratory of Crop Genetic Improvement, National Center of Rapeseed Improvement in Wuhan, Huazhong Agricultural University, Wuhan 430070, China.
Daphne R Goring, Department of Cell & Systems Biology, University of Toronto, Toronto, Canada M5S 3B2; Centre for the Analysis of Genome Evolution & Function, University of Toronto, Toronto, Canada M5S 3B2.
D.R.G. conceived the project. D.R.G., S.R.M., H.K.L., H.N., D.C.J., and T.Z. conceptualized and designed the experiments; S.R.M. performed crosses, generated and analyzed data for the SI-C24 atg7-9 plants, SI-Col-0 atg5-1 plants, and the SI-Col-0 atg7-2 GFP-ATG8a imaging. H.K.L. and S.R.M. performed crosses, generated and analyzed data for the SI-Col-0 atg7-2 plants. H.N. and D.C.J. performed preliminary work on the SI lines and imaging; T.Z. generated the SI-Col-0 line. D.R.G. and C.M. supervised the experiments. S.R.M., H.K.L., and D.R.G. wrote the manuscript. All authors commented on the manuscript before submission. D.R.G. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Daphne Goring (d.goring@utoronto.ca).
References
- Abhinandan K, Sankaranarayanan S, Macgregor S, Goring DR, Samuel MA (2021) Cell-cell signaling during the Brassicaceae self-incompatibility response. Trends Plant Sci S1360-1385(21)00303-4, https://doi.org/10.1016/j.tplants.2021.10.011 [DOI] [PubMed] [Google Scholar]
- Adhikari PB, Liu X, Wu X, Zhu S, Kasahara RD (2020) Fertilization in flowering plants: an odyssey of sperm cell delivery. Plant Mol Biol 103: 9–32 [DOI] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C (2014) Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65: 3799–3811 [DOI] [PubMed] [Google Scholar]
- Beuder S, Dorchak A, Bhide A, Moeller SR, Petersen BL, MacAlister CA (2020) Exocyst mutants suppress pollen tube growth and cell wall structural defects of hydroxyproline O-arabinosyltransferase mutants. Plant J 103: 1399–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs NA, Nasrallah JB, Nasrallah ME (2009) Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. Plos Genet 5: e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillada C, Teh OK, Ditengou FA, Lee CW, Klecker T, Saeed B, Furlan G, Zietz M, Hause G, Eschen-Lippold L, et al. (2021) Exocyst subunit Exo70B2 is linked to immune signaling and autophagy. Plant Cell 33: 404–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dickinson H (1995) Dry stigmas, water and self-incompatibility in Brassica. Sex Plant Reprod 8: 1–10 [Google Scholar]
- Ding X, Zhang X, Otegui MS (2018) Plant autophagy: new flavors on the menu. Curr Opin Plant Biol 46: 113–121 [DOI] [PubMed] [Google Scholar]
- Doucet J, Lee HK, Udugama N, Xu J, Qi B, Goring DR (2019a) Investigations into a putative role for the novel BRASSIKIN pseudokinases in compatible pollen-stigma interactions in Arabidopsis thaliana. BMC Plant Biol 19: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet J, Truong C, Frank-Webb E, Lee HK, Daneva A, Gao Z, Nowack MK, Goring DR (2019b) Identification of a role for an E6-like 1 gene in early pollen-stigma interactions in Arabidopsis thaliana. Plant Reprod 32: 307–322 [DOI] [PubMed] [Google Scholar]
- Durand E, Chantreau M, Le Veve A, Stetsenko R, Dubin M, Genete M, Llaurens V, Poux C, Roux C, Billiard S, et al. (2020) Evolution of self-incompatibility in the Brassicaceae: lessons from a textbook example of natural selection. Evol Appl 13: 1279–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleman C, Dickinson H (1996) Identification of pollen of components regulating the responses in stigmatic Brassica oleracea. New Phytol 133: 197–205 [DOI] [PubMed] [Google Scholar]
- Fujii S, Shimosato-Asano H, Kakita M, Kitanishi T, Iwano M, Takayama S (2020) Parallel evolution of dominant pistil-side self-incompatibility suppressors in Arabidopsis. Nat Commun 11: 1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring DR (2017) Exocyst, exosomes, and autophagy in the regulation of Brassicaceae pollen-stigma interactions. J Exp Bot 69: 69–78 [DOI] [PubMed] [Google Scholar]
- Goubet PM, Berges H, Bellec A, Prat E, Helmstetter N, Mangenot S, Gallina S, Holl AC, Fobis-Loisy I, Vekemans X, et al. (2012) Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet 8: e1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Mazzurco M, Sulaman W, Matias DD, Goring DR (1998) Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YL, Zhao X, Lanz C, Weigel D (2011) Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol 157: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E, Goring DR (2014) A conserved role for the ARC1 E3 ligase in Brassicaceae self-incompatibility. Front Plant Sci 5: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E, Goring DR (2016) Yeast two-hybrid interactions between Arabidopsis lyrata S Receptor Kinase and the ARC1 E3 ligase. Plant Signal Behav 11: e1188233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E, Safavian D, Goring DR (2014a) The ARC1 E3 ligase promotes two different self-pollen avoidance traits in Arabidopsis. Plant Cell 26: 1525–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indriolo E, Safavian D, Goring DR (2014b) Signaling events in pollen acceptance or rejection in the Arabidopsis species. Sexual Reproduction in Animals and Plants, Springer, Berlin/Heidelberg, Germany, pp 255–271 [Google Scholar]
- Indriolo E, Tharmapalan P, Wright SI, Goring DR (2012) The ARC1 E3 ligase gene is frequently deleted in self-compatible Brassicaceae species and has a conserved role in Arabidopsis lyrata self-pollen rejection. Plant Cell 24: 4607–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano M, Ito K, Fujii S, Kakita M, Asano-Shimosato H, Igarashi M, Kaothien-Nakayama P, Entani T, Kanatani A, Takehisa M, et al. (2015) Calcium signalling mediates self-incompatibility response in the Brassicaceae. Nat Plants 1: 15128. [DOI] [PubMed] [Google Scholar]
- Jamshed M, Sankaranarayanan S, Abhinandan K, Samuel MA (2020) Stigma receptivity is controlled by functionally redundant MAPK pathway components in Arabidopsis. Mol Plant 13: 1582–1593 [DOI] [PubMed] [Google Scholar]
- Jany E, Nelles H, Goring DR (2019) The molecular and cellular regulation of Brassicaceae self-incompatibility and self-pollen rejection. Int Rev Cell Mol Biol 343: 1–35 [DOI] [PubMed] [Google Scholar]
- Johnson MA, Harper JF, Palanivelu R (2019) A fruitful journey: pollen tube navigation from germination to fertilization. Annu Rev Plant Biol 70: 809–837 [DOI] [PubMed] [Google Scholar]
- Kenney P, Sankaranarayanan S, Balogh M, Indriolo E (2020) Expression of Brassica napus GLO1 is sufficient to breakdown artificial self-incompatibility in Arabidopsis thaliana. Plant Reprod 33: 159–171 [DOI] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee HN, Kim SH, Shin KD, Chung T (2013) Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M, Dwyer K, Hendershot J, Vrebalov J, Nasrallah JB, Nasrallah ME (2001) Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. Lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–644 [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Macgregor S, Goring DR (2020) A toolkit for teasing apart the early stages of pollen-stigma interactions in Arabidopsis thaliana. Methods Mol Biol 2160: 13–28 [DOI] [PubMed] [Google Scholar]
- Lin Y, Zeng Y, Zhu Y, Shen J, Ye H, Jiang L (2021) Plant Rho GTPase signaling promotes autophagy. Mol Plant 14: 905–920 [DOI] [PubMed] [Google Scholar]
- Mizuta Y, Higashiyama T (2018) Chemical signaling for pollen tube guidance at a glance. J Cell Sci 131: jcs208447. [DOI] [PubMed] [Google Scholar]
- Nasrallah JB (2019) Self-incompatibility in the Brassicaceae: regulation and mechanism of self-recognition. Curr Top Dev Biol 131: 435–452 [DOI] [PubMed] [Google Scholar]
- Nasrallah ME, Liu P, Sherman-Broyles S, Boggs NA, Nasrallah JB (2004) Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc Natl Acad Sci USA 101: 16070–16074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivelu R, Tsukamoto T (2012) Pathfinding in angiosperm reproduction: pollen tube guidance by pistils ensures successful double fertilization. Wiley Interdisciplin Rev Dev Biol 1: 96–113 [DOI] [PubMed] [Google Scholar]
- Rozier F, Riglet L, Kodera C, Bayle V, Durand E, Schnabel J, Gaude T, Fobis-Loisy I (2020) Live-cell imaging of early events following pollen perception in self-incompatible Arabidopsis thaliana. J Exp Bot 71: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavian D, Goring DR (2013) Secretory activity is rapidly induced in stigmatic papillae by compatible pollen, but inhibited for self-incompatible pollen in the Brassicaceae. PLoS One 8: e84286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safavian D, Jamshed M, Sankaranarayanan S, Indriolo E, Samuel MA, Goring DR (2014) High humidity partially rescues the Arabidopsis thaliana exo70A1 stigmatic defect for accepting compatible pollen. Plant Reprod 27: 121–127 [DOI] [PubMed] [Google Scholar]
- Safavian D, Zayed Y, Indriolo E, Chapman L, Ahmed A, Goring DR (2015) RNA silencing of exocyst genes in the stigma impairs the acceptance of compatible pollen in Arabidopsis. Plant Physiol 169: 2526–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR (2009) Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankaranarayanan S, Jamshed M, Samuel MA (2015) Degradation of glyoxalase I in Brassica napus stigma leads to self-incompatibility response. Nat Plants 1: 15185. [DOI] [PubMed] [Google Scholar]
- Scandola S, Samuel MA (2019) A Flower-Specific Phospholipase D Is a Stigmatic Compatibility Factor Targeted by the Self-Incompatibility Response in Brassica napus. Curr Biol 29: 506–512 e504 [DOI] [PubMed] [Google Scholar]
- Schierup MH, Mable BK, Awadalla P, Charlesworth D (2001) Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158: 387–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu KK, Shimizu-Inatsugi R, Tsuchimatsu T, Purugganan MD (2008) Independent origins of self-compatibility in Arabidopsis thaliana. Mol Ecol 17: 704–714 [DOI] [PubMed] [Google Scholar]
- Shimizu KK, Tsuchimatsu T (2015) Evolution of selfing: recurrent patterns in molecular adaptation. Ann Rev Ecol Evol Syst 46: 593–622 [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Arnoldo M, Goring DR (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T, Goubet PM, Gallina S, Holl AC, Fobis-Loisy I, Berges H, Marande W, Prat E, Meng D, Long Q, et al. (2017) Patterns of polymorphism at the self-incompatibility locus in 1,083 Arabidopsis thaliana genomes. Mol Biol Evol 34: 1878–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimatsu T, Suwabe K, Shimizu-Inatsugi R, Isokawa S, Pavlidis P, Stadler T, Suzuki G, Takayama S, Watanabe M, Shimizu KK (2010) Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature 464: 1342–1346 [DOI] [PubMed] [Google Scholar]
- Young PG, Passalacqua MJ, Chappell K, Llinas RJ, Bartel B (2019) A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes. Autophagy 15: 941–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhou G, Goring DR, Liang X, Macgregor S, Dai C, Wen J, Yi B, Shen J, Tu J, Fu T, Ma C (2019) Generation of transgenic self-incompatible Arabidopsis thaliana shows a genus-specific preference for self-incompatibility genes. Plants 8: 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.