Abstract
Obtaining a parts list of the sensory components of the retina is vital to understand the effects of light in behavior, health, and disease. Rods, cones, and intrinsically photosensitive retinal ganglion cells (ipRGCs) are the best described photoreceptors in the mammalian retina, but recent functional roles have been proposed for retinal neuropsin (Opn5)—an atypical opsin. However, little is known about the pattern of Opn5 expression in the retina. Using cre (Opn5cre ) and cre‐dependent reporters, we uncover patterns of Opn5 expression and find that Opn5 is restricted to retinal ganglion cells (RGCs). Opn5‐RGCs are nonhomogenously distributed through the retina, with greater densities of cells located in the dorsotemporal quadrant. In addition to the local topology of these cells, using cre‐dependent AAV viral tracing, we surveyed their central targets and found that they are biased towards image‐forming and image‐stabilizing regions. Finally, molecular and electrophysiological profiling reveal that Opn5‐RGCs comprise previously defined RGC types that respond optimally to edges and object‐motion (F‐mini‐ONs, HD2, HD1, LEDs, ooDSRGCs, etc.). Together, these data describe the second collection of RGCs that express atypical opsins in the mouse, and expand the roles of image‐forming cells in retinal physiology and function.
Using a combination of genetic reporters, viral tracing, molecular analyses, and electrophysiological profiling, this study elucidates the Neuropsin‐expressing Retinal Ganglion Cell (Opn5‐RGC) types of the mouse retina. These neurons are spatially nonhomogenous and stratify their dendrites in the outer sublamina of the IPL. Additionally, Opn5‐RGCs are high‐density, motion and edge‐encoding RGC types that project to image‐forming and image‐stabilizing central targets in the brain. This study diversifies the potential photoreceptive compartments of the mouse eye.
1. INTRODUCTION
Retinal ganglion cells (RGCs) comprise a diverse collection of 45 or more projection neuron types that link retinal events, such as the detection of photons, to various brain targets involved in processing information from the visual scene (Baden et al., 2020; Tran et al., 2019). The majority of RGC types encode specific “features” (object or global motion, contrast, etc.) through unique circuitry that begins with photoreception via rod and cone photoreceptors (Dhande et al., 2015). However, a fraction of these RGCs can also detect photons autonomously through the expression of an atypical photopigment, melanopsin, or Opn4. These cells, termed intrinsically photosensitive retinal ganglion cells (ipRGCs), comprise six RGC types (M1–M6) that vary in cellular physiology, morphology, circuitry, sensitivity to light, and thus, their influence on visual behaviors (Aranda & Schmidt, 2021). Through the integration of rod/cone input and melanopsin phototransduction, ipRGCs regulate behaviors ranging from circadian photoentrainment, sleep, mood, spatial learning, pattern recognition, and visual perception (Aranda & Schmidt, 2021; Do, 2019; Lucas et al., 2012).
In addition to melanopsin, another atypical photopigment, neuropsin (Opn5), is expressed in the retina of vertebrates ranging from zebrafish to humans (Davies et al., 2015; Tarttelin et al., 2003). Functionally characterized as a UVA (380 nm)‐sensitive opsin in humans and mice (Kojima et al., 2011), retinal neuropsin signaling has been implicated in autonomous entrainment of the retinal clock (Buhr et al., 2015), behavioral entrainment to scotopic violet light (Ota et al., 2018), regression of the hyaloid vessels in the eye (Nguyen et al., 2019), and is required for short‐wavelength‐mediated suppression of myopia development in mice (Jiang et al., 2021). Though neuropsin signaling and function appear important in diverse aspects of ocular physiology and health, little is known beyond its expression in a subset of RGCs (Buhr et al., 2015; Jiang et al., 2021; Nguyen et al., 2019). Our understanding of light's influence on physiology and health has stemmed in large part from the evaluation of the unique projections and characteristics of M1–M6 ipRGCs. Given the emerging importance of neuropsin, determining expression patterns, the repertoire of neuropsin‐expressing cell types, and their projection targets will be equally important.
In this study, we assess the retinal patterns of Opn5 expression in mice. Consistent with previous reports (Buhr et al., 2015; Nguyen et al., 2019), we find that Opn5 is largely restricted to RGCs, but these cells exhibit a unique spatial distribution pattern with enrichment in the dorsotemporal (DT) retina. Using viral‐tracing, we establish that the Opn5‐RGC population projects to different brain regions than their ipRGC counterparts, targeting image‐forming and image‐stabilizing centers in the brain. Using combinations of molecular markers and functional recordings, we identify the types of Opn5‐RGCs. Most are ON‐OFF RGC types involved in motion, edge, and spot detection. Together these findings expand the cellular complement and characteristics of opsin‐expressing RGCs, with properties largely differing from their Opn4‐expressing ipRGC counterparts.
SIGNIFICANCE STATEMENT
Photopigment‐expressing retinal ganglion cells (RGCs), such as ipRGCs, are the primary conduit for the effects of light on several subcortical visual behaviors such as circadian entrainment, sleep, learning, depression, etc. Recent work has implicated Opn5, another RGC‐expressed photopigment in entrainment of the retinal clock, myopia development, and vascular regression. In this present study, we analyze the cellular composition, characteristics, and typology of RGCs that express Opn5. We identify them as small receptive field RGC types that detect spots and motion and project primarily to image‐forming visual targets in the brain. These findings expand the photoreceptive compartment in the retina and suggest putative cell types that regulate short‐wavelength light's effects on retinal circadian clocks, myopia, and vascular development in the retina.
2. RESULTS
2.1. Opn5cre specifically marks RGCs
To assess the patterns of neuropsin expression in the retina, we activated cre‐dependent reporters (Ai6, Ai9, and Ai14) using an Opn5cre knock‐in allele (Nguyen et al., 2019). The retina is composed of three nuclear layers and two neuropils (Figure 1a, schematic) which contain the cell bodies, dendrites, and axons of collections of cell classes. Retinal cross‐sections from Opn5cre/+ ; Ai14 (R26‐LSL‐tdTomato) mice (Figure 1a, middle) revealed that labeling was largely restricted to the ganglion cell layer (GCL) which consists of RGCs and displaced amacrine cells (ACs) (Masland, 2011). When viewed in flat mount preparations (Figure 1a, right), tdTomato‐labeled axonal fasciculations could be observed projecting toward the optic disc. To better determine whether recombined cells were indeed restricted to the GCL, we quantified cellular density across the three nuclear layers in whole‐mount retinal preparations (Figure 1a). This showed that the majority of labeled cells were restricted to the GCL (520.8 ± 41.4 cells/mm2, n = 7) with a small fraction in the inner nuclear layer (INL, 5.9 ± 1.1 cells/mm2, n = 6) and no detectable cells in the outer nuclear layer (ONL, n = 7).
FIGURE 1.
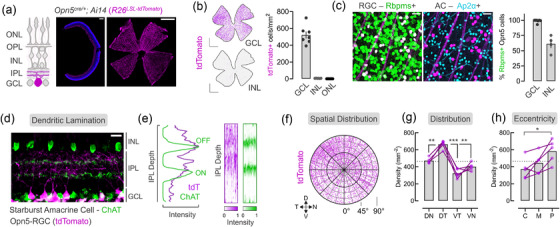
Opn5cre specifically labels retinal ganglion cells (RGCs) with distinct spatial characteristics. (a) Schematic representation of retinal laminar structure. Representative reporter expression (tdTomato, magenta) from Opn5cre; Ai14 retina in cross‐section (middle) or flat mount (right). (b) Distribution and density of reporter+ cells within the GCL, INL, and ONL (n = 6 animals). (c) Immunofluorescent labeling of RGC marker (Rbpms, green) and amacrine (AC) maker (Ap2a, cyan) in the GCL with quantification (n = 5 GCL, n = 4 INL). (d) Lamination patterns of Opn5‐RGCs (magenta) and ChAT+ Starburst Amacrine Cells (green). (e) Quantification of normalized dendrite fluorescent intensity across the IPL from an individual ROI (left) with multi‐ROI heatmap representations (right; 6–11 ROIs/animal, n = 3 animals). (f‐h) Spatial cellular distribution of Opn5‐RGCs, represented as a polar plot of the GCL (f), quantified at each spatial location— dotted line represents global average density (g), and retinal eccentricity dotted line represents global average density (h)(C–central, M–middle, P–peripheral). GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; D, dorsal; N, nasal; V, ventral; T, temporal; DN, dorsonasal; DT, dorsotemporal; VN, ventronasal; VT, ventrotemporal (*p < 0.05, **p < 0.01, ***p > 0.001, G–RM one‐way ANOVA with Tukey's post‐hoc analysis, H–one‐way ANOVA with Tukey's post‐hoc analysis). Scale bar = 15 μm (d), 25 μm (c), 200 μm (A, middle), 1000 μm (A‐right, B)
This pattern of Opn5 expression is similar to that observed from two independent Opn5lacZ lines (Figure 2a) which show restricted GCL labeling (Buhr et al., 2015). In addition to alternative reporter lines, we confirmed that the Opn5cre ; Ai14 line reports cells expressing Opn5 transcript by using RNAScope on freshly harvested hypothalamic unfixed preoptic area tissue and retina (Figure 2b‐e). In both tissues, tdTomato+ cells had markedly higher Opn5 transcript than non‐tdTomato (control) cells (Figure 2c, e), suggesting that this genetic line reports Opn5 patterns accurately. The detection of Opn5 transcript in tdTomato‐labeled cells indicates active expression patterns of Opn5 in adult cells, and that patterns of Opn5 expression are not due to developmental activity at the Opn5 locus (lineage‐mark) or an artifact of knocking in cre‐recombinase.
FIGURE 2.
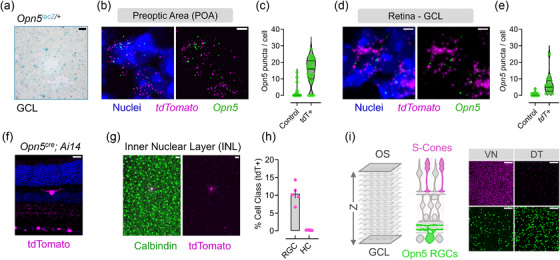
Expression of Opn5‐transcript and spatial distribution via genetic reporters. (a) GCL X‐gal labeling of whole‐mount retina in the Opn5lacZ (Buhr et al., 2015; Zhang et al., 2020) reporter line. (b‐c) Transcript detection of Opn5 (green) in Opn5cre; Ai14 (magenta) hypothalamic POA neurons (b), quantified in (c) compared to tdTomato‐ cells (control, n = 3 animals). (d) Same as (b‐c) but detection of Opn5 transcript in tdTomato+ cells of the GCL in the retina (n = 3 animals). (f) Retinal cross‐section highlighting labeled (magenta) cells in the GCL but sparsely in the INL/OPL regions. (g) Calbindin (green) colocalization with INL tdTomato+ cells in the Opn5cre; Ai14 line. (h) Percent of each cell class (RGC or HC–horizontal cell) labeled in the Opn5cre line (RGC n = 5, HC n = 6). (I, left) Schematic representation of optical sections via confocal imaging for S‐cones (magenta) and Opn5‐RGCs (Green), with representative optical z‐slices in the ventronasal (VN) and dorsotemporal (DT) retina. Scale bar = 5 μm (b, d), 20 μm (F, G), 50 μm (A), 100 μm (I)
Subsequently, we labeled Opn5cre/+ ; Ai14 retina with markers of RGCs (Rbpms) and ACs (Ap2α) and assessed tdTomato+ GCL and INL cells (Figure 1c). While the majority of tdTomato‐labeled cells in the GCL (98.8 ± 1.0%, n = 5) and INL (61.1± 6.8%, n = 4) expressed Rbpms, a fraction of the INL cells expressed neither RGC nor AC markers (data not shown). Further assessment of retina in cross‐section and whole mount suggests the remaining labeled cells were horizontal cells (HCs, Figure 2f–g) which are inhibitory interneurons located in the INL with neuronal processes in the outer plexiform layer (OPL). In the mouse retina, HCs represent a single type and given limited expression of tdTomato in the Opn5cre line, these cells likely represent a stochastic expression of the Opn5 locus. Given the large bias of labeled cells in the GCL over the INL (∼99:1), and the percentage of each cell class labeled in our analysis (Figure 2h), we suggest that approximately 10% of RGCs express the neuropsin photopigment with limited expression in other inner retinal neurons. Thus, we refer to these cells as Opn5‐RGCs.
2.2. Opn5‐RGCs display ON & OFF dendritic lamination and spatial biases in the distribution
RGCs receive presynaptic input from bipolar cell (BC) axons and AC dendrites in the inner plexiform layer (IPL, Figure 1a). Here, parallel synaptic channels convey information about light increments (ON pathway) or decrements (OFF pathway), which are spatially segregated into five sublaminae (S1–S5) (Sanes & Zipursky, 2020). ChAT+ starburst amacrine cell (SAC) laminate their dendrites within S2 and S4 and can be used to demarcate sublaminae. To determine Opn5‐RGC dendritic lamination patterns, we used ChAT labeling in retinal cross‐sections from Opn5cre ; Ai14 mice (Figure 1d). Assessment of tdTomato+ processes revealed prominent lamination between the SAC bands (ON‐OFF region), and dendritic distribution into OFF sublaminae (S1–S2). Quantitative assessment of fluorescent intensity along the IPL (Figure 1e) highlighted the strongest tdTomato signal located between S1–S3, beyond the ON‐SAC band. Repeating this analysis across multiple IPL regions (n = 3 animals), we visualized multiple intensity‐depth curves as heatmaps for Opn5‐RGCs (magenta) and ON/OFF‐SACs (green) (Figure 1e). While diffuse dendritic signal was observed from Opn5‐RGCs beyond the ON‐SAC band (S4), we find two possible (yet diffuse) bands that laminate in the ON‐OFF region between the SACs (S3) and with OFF‐SAC dendrites (S1‐2). Collectively, these data suggest Opn5‐RGCs receive both ON and OFF synaptic inputs.
2.3. Opn5‐RGCs display spatial biases in topographical distribution
Photoreceptors and opsins have nonhomogeneous distribution patterns across the retina. These unique topographies are thought to arise due to evolutionary pressures posed by the visuoecological niche of the organism (Baden et al., 2020). In mice, S‐cone opsin is expressed in a gradient with enrichment in the ventronasal (VN) retina, and is positioned to survey the upper visual field including the sky (Nadal‐Nicolás et al., 2020; Qiu et al., 2021). By contrast, M1 and M2 ipRGCs are enriched in the dorsal retina (Steven Hughes et al., 2013). To determine if Opn5‐RGCs also display nonuniform patterns of spatial distribution, we assessed labeled cell densities across the polar axis (dorsal–D, nasal–N, ventral–V, temporal–T) and eccentricity (central, middle, periphery) within the GCL (Figure 1f). Analysis of each quadrant of the retina revealed a significant enrichment of Opn5‐RGCs in the DT quadrant (Figure 1g). In addition to using landmarks and regional labeling strategies to quantify each quadrant, we used immunolabeling of S‐cone opsin to confirm DT Opn5‐RGC enrichment (Figure 2i). When analyzed across retinal eccentricities (central, middle, and peripheral retina from the optic disc), Opn5‐RGCs subtly increased in density along this centrifugal axis (Figure 1h). This DT enrichment pattern has been reported before for Alpha‐ON sustained and Alpha‐OFF sustained RGCs, where this spatial bias may enhance sampling of frontal visual space (Bleckert et al., 2014), and could be involved in binocular vision (Johnson et al., 2021). Thus, similar to some other RGC types, including some ipRGCs, Opn5‐RGCs display nonhomogeneous distribution in the retina.
2.4. Opn5‐RGCs project to image‐forming and image‐stabilizing centers
RGCs encode different features of the visual environment as spikes and transmit this information to more than 40 regions in the mouse brain (Dhande et al., 2015; Martersteck et al., 2017; Morin & Studholme, 2014). To determine the projection patterns of Opn5‐RGCs, and thus, their potential input to visual behaviors, we assessed known retinorecipient brain targets for the presence of labeled tdTomato processes in Opn5cre ; Ai14 mice (Figure 3a–c). In the hypothalamus (Figure 3a), a major target of retinal input is the suprachiasmatic nucleus (SCN) which receives almost exclusive input from ipRGCs (Baver et al., 2008; Beier et al., 2021). While we observed dense innervation in the optic tract below the SCN, there was little evidence of projections into the SCN from Opn5‐RGCs (Figure 3a). Furthermore, we identified a set of tdTomato‐labeled fibers overlying optic tract RGC axons, and these appear to project to the supraoptic nucleus (SON, Figure 3a). While RGCs have been shown to innervate the peri‐SON (Beier et al., 2021; Hattar et al., 2006), Opn5 is also expressed in hypothalamic QPLOT neurons that are presynaptic to the SON, and these processes may reflect dendritic or axonal innervation by central QPLOT hypothalamic neurons (Upton et al., 2021; K. X. Zhang et al., 2020).
FIGURE 3.
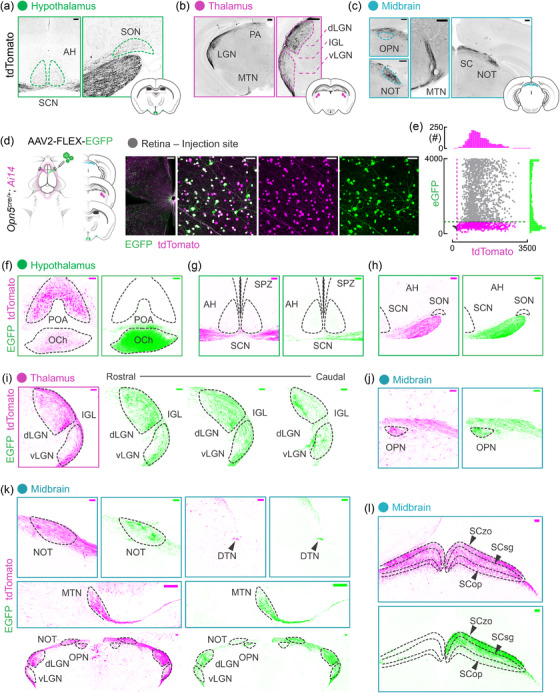
Central projection patterns of Opn5‐RGCs. (a‐c) tdTomato (greyscale‐inverted) neuronal process assessment in the (a) hypothalamus (SCN, suprachiasmatic nucleus; AH, anterior hypothalamus; SON, supraoptic nucleus), thalamus (dLGN, dorsal lateral geniculate nucleus; IGL, intergeniculate leaflet; vLGN, ventral geniculate nucleus), and midbrain (OPN, olivary pretectal nucleus; NOT, nucleus of the optic tract; MTN, medial terminal nucleus; SC, superior colliculus). (d‐e) Viral‐mediated Opn5‐RGC tracing schematic depicting delivery method/material (AAV2‐FLEX‐eGFP; n = 5 animals), specific inflection of retinal tdTomato+ (magenta) cells and associated signal intensity quantification (n = 2 animals). (f‐h) Traced eGFP+ (green) projections and native reporter (tdTomato, magenta) in the hypothalamus reporting additional Opn5+ domain (F, POA, preoptic area) with Opn5‐RGC axons in the optic chiasm (F, OCh). (g) Projections of Opn5‐RGCs along the ventral hypothalamus (SCN, AH, SPZ–subparaventricular zone) and (h) lateral hypothalamus (SON). (I) Native and labeled Opn5‐RGC axonal innervation of the LGN complex along the rostral‐caudal axis. (j, k) Labeling of projections to midbrain structures such as the OPN, NOT, MTN, and DTN (dorsal terminal nucleus). (l) Opn5‐RGC projections in the SC with outlined layers (zo, zonal; sg, superficial grey; op, optic). Scale bar = 50 μm (D), 100 μm (A, F‐L), 200 μm (B, C, D–wholemount)
A major target of RGCs is the lateral geniculate complex of the thalamus, comprised of the dorsal and ventral lateral geniculate nucleus (dLGN, vLGN) and the intergeniculate leaflet (IGL). We found diffusely labeled fibers in the dLGN, distributed between lateral shell and medial core (Figure 3b). Additionally, we found sparse but detectible fibers in the lateral vLGN and terminals absent from the IGL (Figure 3b). Within the midbrain, RGC projections innervate a diverse array of targets depending on their type and encoded features (Dhande et al., 2015). We observed sparsely labeled fibers from Opn5‐RGCs in the olivary pretectal nucleus (OPN), which has established roles in the pupillary light reflex, gating the amount of light entering the eye (Figure 3c). Denser fibers were observed innervating the nucleus of the optic tract (NOT, Figure 3c), medial terminal nucleus (MTN, Figure 3c), and the dorsal terminal nucleus (DTN, Figure 3k). These nuclei are collectively termed the accessory optic system (AOS) and are involved in gaze/image‐stabilization by producing compensatory eye movements during slow visual‐field motion (Dhande et al., 2015; Sun et al., 2015). Approximately 85% of RGCs innervate the superior colliculus (SC) of the midbrain, which integrates all major sensory modalities to regulate innate behaviors (Ellis et al., 2016; Hofbauer & Dräger, 1985; Reinhard et al., 2019). Consistent with the notion that most RGCs project to this region, we found dense Opn5‐RGC innervation throughout the structure, with the strongest signal in the superficial layers (Figure 3c) (Martersteck et al., 2017).
While qualitative mapping of lineage‐marked projections provides us with putative Opn5‐RGC targets, due to expression of Opn5 in nuclei within the brain (preoptic area, inferior olive), we opted to confirm RGC targets using viral labeling (Figure 3d) (K. X. Zhang et al., 2020). To trace Opn5‐RGC projections, we injected an AAV carrying a cre‐dependent eGFP reporter construct (AAV2‐FLEX‐eGFP) into the vitreous cavity of Opn5cre ; Ai14 mice. Assessment of the injected retina revealed GFP expression restricted to tdTomato‐labeled Opn5 cells, with up to 80% of cells coexpressing GFP (Figure 3d–e). Consistent with our analysis of labeling in the Opn5cre ; Ai14 line, we observed strong reporter expression in the optic chiasm and optic tract (Figure 3f–h), with no apparent GFP+ fibers in the SCN (Figure 3g). We found limited GFP+ fibers in the tract overlying the optic tract (Figure 3h), suggesting these may emerge from a different Opn5‐expressing population (K. X. Zhang et al., 2020). Replicating our findings from lineage‐marked axons in Figure 3a–c, we found major overlap between GFP+ fibers and tdTomato+ fibers in the dLGN and vLGN (Figure 3i), OPN (Figure 3j), NOT/MTN/DTN (Figure 3k), and SC (Figure 3l). In the SC, while all layers (SCzo, SCsg, and SCop) appeared to contain tdTomato+ fibers, GFP+ tracing was strongest in the superficial layers (SCzo & SCsg) while faint in the SCop (Figure 3l). These SC labeling results may reflect a lack of viral labeling of the deeper projecting RGC types (Martersteck et al., 2017; Morin & Studholme, 2014). We could not find any indication of Opn5‐RGC innervation of targets such as the ventral medial hypothalamus, zona incerta, perilateral habenula (pLHb), and posterior pretectal nucleus (data not shown).
Notably, these targets are largely different from those of ipRGCs (Beier et al., 2021; Hattar et al., 2006). When subject to similar viral labeling (Figure 4a–f) and consistent with prior studies, ipRGCs densely innervate the SCN (Figure 4b), the IGL, medial vLGN (Figure 4d), and the OPN (Figure 4e). Sparse ipRGC innervation was observed in the pLHb and dLGN (Figure 4c, d). These differences in innervation of opsin‐expressing RGCs are obvious when entire thalamic and midbrain structures are compared (Figure 4f). Thus, while ipRGCs mainly project to nuclei involved in circadian entrainment (SCN, IGL) and nonimage forming visual behaviors (pLHb, vLGN), the majority of Opn5‐RGCs target image‐forming (dLGN), sensory‐motor integration (SC) and image‐stabilizing (NOT, MTN, DTN) regions.
FIGURE 4.
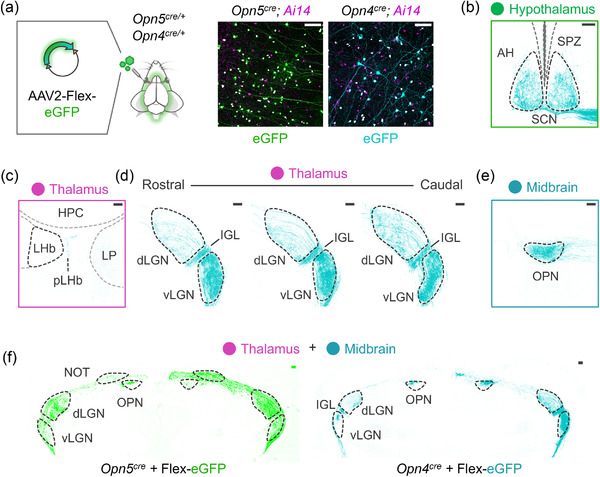
ipRGC projection patterns reflect a unique collection of targets compared to Opn5‐RGCs. (a) Viral‐mediated Opn5‐RGC/Opn4‐RGC tracing schematic depicting delivery method/material (AAV2‐FLEX‐eGFP; n = 4 animals), with specific infection of retinal tdTomato+ (magenta). (b) Traced projections (cyan) to a major ipRGC targe–tSCN. (c‐d) Projections of labeled ipRGCs innervating thalamic nuclei such as the pLHb (sparsely), dLGN (sparsely), IGL (dense), vLGN (dense). (e) Traced ipRGC projections to the midbrain OPN. (f) Thalamic and midbrain structures innervated by traced Opn5‐RGCs (green, same as Figure 2k) and ipRGCs (cyan). LH, bilateral habenula; pLHb, perilateral habenula; HPC, hippocampus; LP, lateral posterior nucleus, additional abbreviations are described in Figure 2). Scale bar = 100 μm (A‐F)
2.5. Molecular and functional fingerprinting of Opn5‐RGCs
Given the projection patterns of Opn5‐RGCs, they appear to be distinct from M1–M6 ipRGCs. However, it is unclear if these cells comprise a single type or reflect a collection of described types. Retinal neurons of a functional type distribute themselves in a regular mosaic pattern (Rodieck, 1991). This spatial arrangement allows for optimal sampling of visual space by limiting overlap between circuit units. To determine whether Opn5‐RGCs comprise single or multiple types, we computed parameters that distinguish between mosaic arrangements and random distribution of cells and compared this to ChAT+ cells in the INL (a known AC type; OFF‐SACs) and Opn4cre ; Ai14 ipRGCs in the GCL (6 RGC types) (Figure 5a). For each category of cell, we computed the density recovery profile (Figure 5b–d, top row) and the nearest neighbor distance (nnd) distribution (Figure 5b–d, bottom row). OFF‐SACs displayed a steadily increasing cell density as the radial distance increases from a reference cell in the density recovery profile, and a distribution of neighbors that was Gaussian (Figure 5b) (Keeley et al., 2020). Both are suggestive of nonrandom (and thus more regular) distribution patterns. However, both opsin‐expressing RGC cells displayed density recovery profiles with steep increases to average density and Poisson nnd distributions similar to random point patterns (Figure 5c, d) (Keeley et al., 2020). An additional metric, the packing factor (p), describes the efficiency of points packed into an array where 0 < p < 1 (0 = random array; 1 = hexagonal array of points). Comparing the packing factor for OFF‐SACs, Opn5‐RGCs, and ipRGCs revealed a significant difference between the known regular mosaic (OFF‐SAC) and both opsin‐expressing RGC distributions (Figure 5e). Together these data strongly suggest that like ipRGCs, Opn5‐RGCs comprise more than one RGC type.
FIGURE 5.
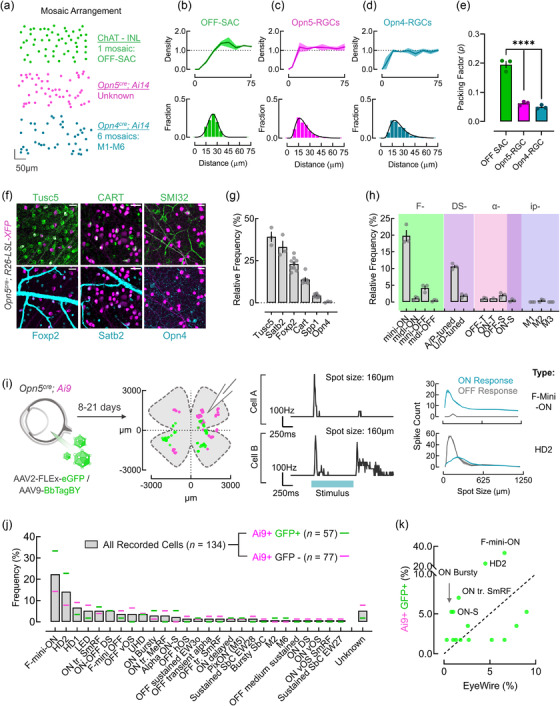
Molecular and functional classification of Opn5 RGCs. (a) Spatial distribution of labeled OFF‐starbursts (ChAT+; green), Opn5‐RGCs (magenta), and ipRGCs (cyan). (b‐d) Quantified spatial characteristics: normalized density recovery profile (DRP, top) and nearest neighbor distances (nnd, bottom) for OFF‐Starbursts (B), Opn5‐RGCs (c), and ipRGCs/Opn4‐RGC (d). Packing factor (p) estimation for each cell type from array coordinates (n = 3 animals). (f) Immunofluorescent images of Opn5cre; R26‐LSL‐XFP (magenta) retina labeled against markers of major RGC types, Tusc5, Satb2, Foxp2, Cart, SMI32, Opn4. (g) Quantification of report cell overlap with RGC type marker in F (Tusc5; n = 3, Satb2; n = 3, Foxp2; n = 9, Cart; n = 6, SMI32 or Spp1; n = 9, Opn4; n = 3). (h) Relative frequency of RGC types represented in the Opn5cre; Ai14 line using a combination of markers (see Supporting information Figure S3) (n = 3 animals per cell type). (i) Typological assessment of Opn5‐RGCs using natively labeled (magenta) or virally labeled (green) cells following intravitreal injection of AAV2‐FLEX‐eGFP or AAV9‐BbTagBY, with spatial locations of each recorded cell. (I, Right) Examples firing rates from two cells in whole‐cell mode when exposed to a 160 μm spot of light over their receptive fields, and spatial tuning curves to spots of varying sizes. (j) Typological classification of RGC types encountered in the Opn5cre that were virally labeled (Ai9+ GFP+ or green, n = 57 cells) or natively fluorescent (Ai9+ GFP‐ or magenta, n = 77). RGC types were normalized as a proportion of each fluorescent category (virally labeled or native). (k) Correlation of RGC type frequency (%) of virally labeled cells compared to expected frequencies based on the Eyewire dataset (Bae et al., 2018) (****p < 0.0001; One‐way ANOVA with Tukey's post‐hoc analysis). Scale bar = 25 μm (F)
To determine the typological repertoire of Opn5‐RGCs, we first used available molecular markers that broadly mark major RGC subclasses with partial overlap (Tusc5: T5‐RGCs, Foxp2: F‐RGCs, Cart: ooDSRGCs, Spp1/SMI32: Alpha‐RGCs, and Opn4: M1‐M3 ipRGCs) or have been shown to mark unidentified RGC types, for example, Satb2 (Figure 5f, g) (Kay et al., 2011; Krieger et al., 2017; Rousso et al., 2016; Sweeney et al., 2019; Tran et al., 2019). Assessment of labeled cells in the Opn5cre line crossed to R26‐LSL‐XFP reporter strains revealed their composition as 39.2 ± 3.0% Tusc5+ (n = 3), 33.1 ± 3.7% Satb2+ (n = 3), 23.0 ± 1.3% Foxp2+ (n = 9), 13.8 ± 1.3% Cart+ (n = 6), 4.2 ± 0.3% Spp1+ (n = 9), and 0.5 ± 0.2% Opn4+ (n = 3) (Figure 5g). However, because of the overlapping nature of some markers, we further divided the representation of marker combinations of Opn5‐RGCs capturing 78.5% of Opn5‐RGCs (Figure 6b).
FIGURE 6.
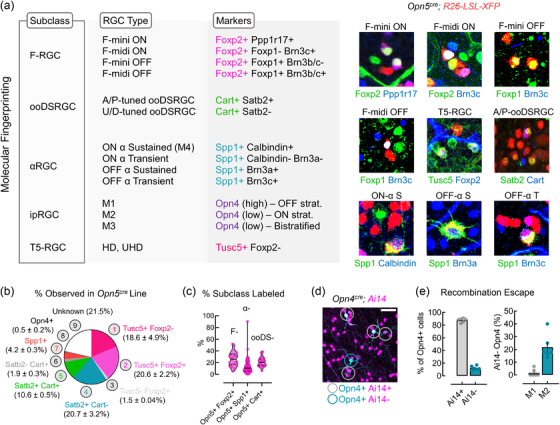
Molecular classification strategies for opsin‐expressing RGCs. (a) Molecular classification scheme used in this study to assign RGC type based on combinations of 2+ markers. (b) Percent observation of Opn5‐RGC cells corresponding to particular molecular type using molecular markers in (a) within the Opn5cre line (data is reported as mean ± SEM). (c) Percent of each RGC subclass (F–F‐RGC, α–Alphas, ooDS–ON‐OFF DSRGC) positive for Opn5 (F–n = 3, alpha–n = 9, ooDS–n = 3). (d) Immunofluorescent labeling of melanopsin (Opn4, cyan) and tdTomato (magenta) in flat mount Opn4cre; Ai14 retina. (e) Percent of immunolabeled ipRGCs (M1–M3) positive or negative for reporter expression (Ai14±). (f) Percent of reporter negative (Ai14‐) ipRGC types (M1, M2) from Opn4cre; Ai14 whole‐mount retina (n = 6 animals). Scale bar = 50 μm (D)
While single markers define broader RGC subclasses (Tusc5–T5‐RGCs, Foxp2–F‐RGCs, Cart–ON‐OFF‐DSRGCs, etc.), combinations of markers have been used to further define RGC types (Figure 6a) (Krieger et al., 2017; Rousso et al., 2016). Using this combinatorial approach with nonoverlapping RGC subclass‐ and type‐specific markers, we determined the RGC typology of Opn5‐RGCs (Figure 5h). Within the Foxp2+ subset of Opn5‐RGCs, the majority appeared to be F‐mini‐ON (Foxp2+ Ppp1r17+), with a minimal representation of F‐mini‐OFF, F‐midi‐ON, and F‐midi‐OFF types (Figure 5h) (Rousso et al., 2016). Within the Cart+ subset, cells largely coexpressed Satb2, suggesting they are anterior‐/posterior‐tuned ON‐OFF DSRGCs (Figure 5h) (Dhande et al., 2019). Alpha RGCs and ipRGC types did not show any obvious patterns of overrepresentation in the Opn5cre line, but a slight overrepresentation of Alpha OFF‐Transient RGCs was observed. Given these results, Opn5 expression appears to be specific to RGC types within diverse subclasses and does not represent an entire subclass by itself (Figure 6c). Furthermore, defining whether the entire RGC type (F‐mini‐ONs, A‐/P‐tuned ooDSRGCs) expresses Opn5 is difficult owing to the escape rate of recombination. For example, when assessing Opn4cre ; Ai14 retina stained against melanopsin (Figure 6d), recombination escape is apparent with ∼12.3 ± 1.3% of Opn4+ cells (n = 6 animals) remaining unlabeled by the reporter. Additionally, when each ipRGC type is assessed, this phenomenon is overrepresented in M2s (21.8 ± 3.9% escape, n = 6) which express lower levels of melanopsin than M1s (1.7 ± 1.2% escape, n = 6) (Aranda & Schmidt, 2021). Given that Opn5 is barely detected in pan‐RGC single‐cell RNA sequencing datasets (Figure 7a, b) (Tran et al., 2019), it is currently unknown if Opn5 expression is ubiquitous among F‐mini‐ON and A/P‐tuned ooDSRGCs. Detectable Opn5 transcript expression (Figure 7a, b) is confined to cluster C7 and C10. While these represent unknown RGC types, both C7 and C10 are transcriptomically similar to Alpha‐RGCs, DS‐RGCs, and T5‐RGC subclasses (Tran et al., 2019).
FIGURE 7.
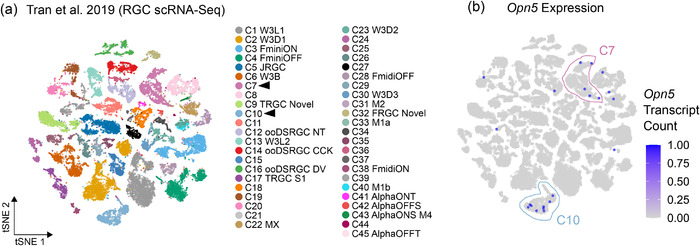
Pan‐RGC single‐cell RNA‐Seq (Tran et al., 2019). (a) 2D t‐stochastic neighbor embedding (tSNE) of RGC single‐cell RNA sequencing data (Tran et al., 2019) with cluster labels pulled from Tran et al. (2019) and Ding & Regev (2021). Arrowheads correspond to clusters with 2 < cells with detectable Opn5 expression. (b) Expression of un‐normalized Opn5 transcript counts projected onto RGC clusters
These marker combinations only allow us to identify about 13 out of more than 45 types of RGCs in the mouse eye, and less than half of the Opn5‐RGC population. Many RGC types are known either through the use of other cre‐mouse lines (e.g., JamB‐CreERT2 [Kim et al., 2008]), transgenic reporters (e.g., W3 RGCs in the TYW3 line [Y. Zhang et al., 2012]), scRNA‐Seq (Rheaume et al., 2018; Tran et al., 2019), and through electrophysiological responses to visual stimuli (Jacoby & Schwartz, 2017; Mani & Schwartz, 2017). We sought to more comprehensively assess Opn5‐RGC types and their visual response properties through electrophysiological profiling and mapping responses to known functional types (Goetz et al., 2021; Parmhans et al., 2021). To this end, we virally labeled Opn5‐RGCs in Opn5cre ; Ai9 mice (using either AAV2‐Flex‐eGFP or AAV9‐BbTagBY) and recorded from Ai9+ eGFP+ (active cre‐expressing) or Ai9+ eGFP‐ (lineage) cells (Figure 5i). In cell‐attached recording mode, we recorded spike trains in response to spots of varying sizes, moving bars, etc. (see Materials &Methods) and based on cellular responses, we assigned these cells to known functional RGC types (Figure 5j). Recordings and assignments from both virally traced and lineage cells were largely consistent, with F‐mini‐ON (Cooler & Schwartz, 2021; Rousso et al., 2016) and HD2 (Jacoby & Schwartz, 2017) being overrepresented in each group. Additionally, of the RGC types that represent >5% frequency (average of Ai9+ GFP+ and Ai9+ GFP‐ groups), we find that four of five types are ON‐OFF RGCs that respond optimally to edges, small‐spots, and motion (F‐mini‐ON, HD2, HD1, Local Edge Detector/LED, and ON‐OFF DSRGC). As edge or spot detector RGC types (F‐mini‐ON, HD2) are found at high densities in the retina, an apparent enrichment of these cells could be an artificial inflation (Kim et al., 2008; Rousso et al., 2016). To rule out this possibility, we compared the frequency of each RGC type in our recording dataset to their frequencies in the Eyewire museum (Figure 5k) (Bae et al., 2018). Consistent with the notion that F‐mini‐ON and HD2 RGCs were enriched in our electrophysiological survey of Opn5‐RGCs, we found these RGC types at rates well above their expected frequencies (Figure 5K).
3. DISCUSSION
In this study, we provide the first classification of neuropsin‐expressing RGCs (Opn5‐RGCs) from their characteristics to typology using genetic (Opn5cre ), molecular, viral, and electrophysiological approaches. In the absence of knockout‐validated antibodies to Opn5, we have used genetic reporters to identify Opn5‐expressing RGCs. While this analysis was restricted to a single cre knock‐in mouse line, many lines of evidence suggest that our method of assessment captures the expression patterns of native Opn5. In addition to the Opn5cre line, mice with LacZ knocked into the Opn5 locus display GCL‐restricted labeling and colocalization with Rbpms, an RGC‐specific marker (Buhr et al., 2015). Cre‐recombinase lines crossed to cre‐activated reporters report the history of gene expression as opposed to active expression patterns. Using fluorescence in situ hybridization (RNAScope) against Opn5 revealed specific expression of transcript in reporter+ cells within the preoptic area of the hypothalamus and retina (Figure 2) highlighting active expression within cre‐activated cells. Consistently, when injected with viruses that require active cre‐expression to label cells, Opn5‐RGCs were infected at high densities (Figure 3) and the functional cell‐type distributions were similar between lineage‐marked and actively expressing cells (Figure 5i–k). Taken together, these cells appear to be high‐density RGCs with unique retinal spatial characteristics and output projections when compared with the other opsin‐expressing RGC subclass, ipRGCs.
3.1. Spatial and dendritic characteristics of Opn5‐RGCs
Within the GCL, Opn5‐RGCs appear at high densities ranging from 370 cells/mm2 in the ventral retina to almost 670 cells/mm2 at their highest in the DT quadrant. This spatial pattern is also displayed by Alpha ON‐S, Alpha OFF‐S, M1, M2, and SPIG1+ DSRGC types in the mouse retina (Bleckert et al., 2014; Steven Hughes et al., 2013; Yonehara et al., 2009). Patterning of Opn5‐expressing cells across retinal space is not unique to mice, as Opn5 expression in ray‐finned fish retina (zebrafish, medaka, and spotted gar) is highest within the ventral zone and limited in the dorsal region (Sato et al., 2016). These differences in distribution may have arisen through selective evolutionary pressures associated with terrestrial and aquatic visual needs (Baden et al., 2020).
In the mouse, Opn5‐RGCs functionally appear to be F‐mini‐ON, HD2, HD1, and ooDSRGC types, of which the F‐mini‐ON RGCs are collectively enriched in the central and ventral retina. Additionally, studies using a genetic line that marks members of the HD subclass (also known as W3 cells) have reported a ventral enrichment for these RGC types (Y. Zhang et al., 2012). While our results represent a departure from assessed distribution patterns, reports of F‐mini‐ON density along the dorsal‐ventral axis have been represented as a percent of maximum density (Rousso et al., 2016) and it is unclear whether the W3 line marks entire RGC types or it reflects differential transgenic reporter across retinal space, limiting labeling of these cells in the dorsal retina. Thus, it is difficult to make accurate comparisons between RGC type distribution patterns in our line versus reported RGC types. It is also possible that the spatial patterns of Opn5‐RGCs are due to variable genetic activity of the cre across retinal space. However, our analysis of Opn5 transcript and cre‐specificity (Figure 2d, e) was done across the dorsoventral axis, and all cre‐recombined cells contained higher Opn5 transcript numbers than surrounding tdTomato‐negative cells.
Beyond their spatial distribution, Opn5‐RGCs laminate their dendrites in mixed ON/OFF (S3) and OFF sublaminae of the IPL (S1‐2) (Figure 1d, e). Consistent with their lamination patterns, Opn5‐RGCs comprise RGC types with similar dendritic lamination patterns and stereotypical ON‐OFF responses to visual stimuli projected onto their receptive fields (Figure 5i–k). In contrast, all characterized ipRGCs receive ON inputs even though three types possess OFF stratifying dendrites (Do, 2019; Dumitrescu et al., 2009; Schmidt & Kofuji, 2011).
3.2. Retinorecipient targets of Opn5‐RGCs
Based on our viral labeling approach, Opn5‐RGCs collectively target well‐established image‐forming and image‐stabilizing nuclei such as the dLGN (patterned/conscious vision), the SC (sensorimotor integration and innate behaviors), and the AOS (gaze/image‐stabilization, head/eye movements). While comprehensive, it is possible that certain central targets were not sampled with this approach, due to limited cre‐expression in Opn5‐RGCs. On the other hand, sampling of certain targets may be due to effects of cre‐expression in non‐Opn5 RGCs. This is unlikely, as almost all cre‐recombined cells had detectable Opn5 transcript when assessed via RNA‐Scope (Figure 2d, e).
Opn5 has been shown to regulate behavioral entrainment of the circadian clock to scotopic violet light and SCN cfos induction following violet light induction (Ota et al., 2018). Based on these behaviors, one would expect that like M1 and M2 ipRGCs, Opn5‐RGCs might innervate the SCN of the hypothalamus (Beier et al., 2021). However, analysis of projections in the Opn5cre ; Ai14 brain and viral tracing (Figure 3g) revealed an absence of SCN innervation. Thus, any influence of Opn5 on behavioral entrainment likely originates from regulation of the autonomous retinal clock (Buhr et al., 2015), or polysynaptic pathways involving Opn5‐RGC input.
Tracing to the MTN is particularly unexpected, as our molecular and electrophysiological analyses failed to uncover an ON‐DSRGC population that is a major input to the MTN (Gauvain & Murphy, 2015; Yonehara et al., 2009). However, it has been previously reported that ooDSRGCs labeled in the BD‐Cre line (Kay et al., 2011) project to the dLGN, NOT, and MTN. While a role for Opn5 in image‐forming vision has yet to be established, previous studies have reported a lack of function for Opn5 in the optokinetic reflex (Buhr et al., 2015). However, because the projectors used to generate drifting gratings in these experiments are likely limited in the UVA region of the spectrum, it is difficult to exclude a role for Opn5 in this behavioral pathway.
In addition to image‐forming and image‐stabilizing regions, we noted innervation of nonimage forming centers such as the OPN (pupillary light reflex) and vLGN (mood, spatial learning). While rod, cone, and various modes of ipRGC signaling account for both the transient and sustained components of the pupillary light reflex (Keenan et al., 2016), recent work suggests that the OPN core receives substantial input from small receptive field M6 ipRGCs (Levine & Schwartz, 2020; Quattrochi et al., 2019) and, thus, could participate in alternative behaviors such as predation. Moving forward, functional and behavioral analyses that consider the spectral composition of ambient and triggered illumination will help further define the potential roles of Opn5‐RGCs in image‐forming and nonimage‐forming visual behaviors.
3.3. Opn5‐RGCs encode edges, spots, and motion
Our molecular and functional analysis of Opn5‐RGC types reveals a correspondence between these cells and established RGCs that encode features such as edges (F‐mini‐ON), spots (HD1, HD2, LED), and directional motion (ooDSRGCs) (Figure 5). From our electrophysiological survey, six RGC types reflect 60% of the Opn5‐RGC population (F‐mini‐ON, HD2, HD1, LED, ON tr. SmRF, and ooDSRGCs). The remaining cells are scattered among 21 additional RGC types which are not especially overrepresented in the Opn5cre line.
Molecular fingerprinting also captured similar proportions of F‐mini‐ON (molecular = 19.8%; functional = 22.3%), HD1 + HD2 + ON tr. SmRF + UHD (molecular = 18.5% Tusc5+ Foxp2‐; functional = 30.3%), and ooDSRGCs (molecular = 13.8%; functional = 5.2%) in the Opn5cre line, increasing our confidence in the proposed functional types. Beyond these listed types, however, it is difficult to make accurate comparisons between our two forms of fingerprinting as comprehensive studies that link functional RGC properties to gene expression patterns are ongoing (Goetz et al., 2021). Our molecular analysis of Opn5‐RGCs revealed roughly 40% were Tusc5+. Multiple lines of evidence suggest that Tusc5 RGCs (T5‐RGCs) are analogous with high‐definition, strongly surround‐suppressed RGC types (F‐mini‐ON, HD1, HD2, UHD, etc.) which correspond to the most enriched functional RGC types observed in the Opn5cre line. W3‐RGCs labeled in the TYW3 reporter line specifically express Tusc5 and have shared functional responses to visual stimuli and dendritic stratification when compared to the functional HD subclass (Bae et al., 2018; Jacoby & Schwartz, 2017; Tran et al., 2019; Y. Zhang et al., 2012). Second, using identical stimuli to this article, Goetz et al., 2021 identified HD1, HD2, UHD, F‐mini‐ON, F‐mini‐OFF, ON tr. SmRF, and OFF tr. SmRF RGC types to be Tusc5+ through electrophysiological profiling followed by scRNA‐Seq (Func‐Seq). Given these correspondences, we strongly believe that Opn5‐RGCs comprise six or more RGC types that encode edges, motion, and have strong surround‐suppression.
3.4. Functional roles of Opn5‐RGCs
The functional roles of Opn5 in retinal physiology are akin to those of Opn4 in ipRGCs. Opn5 is required for entrainment of the autonomous retinal clock (Buhr et al., 2015), behavioral rhythms (Ota et al., 2018), vascular development (Nguyen et al., 2019), and suppression of experimental myopia (Jiang et al., 2021). By extension, the discovery that Opn5 is expressed in cells that encode edges and motion seems to suggest broader roles for these RGCs beyond feature detection. Diverse functions beyond feature‐encoding are not limited to opsin‐expressing RGCs, but appear to be a fundamental aspect of RGCs, at least during development (D'Souza & Lang, 2020). Given the overall results of our study, some important questions remain to be addressed, namely (1) what are the unique roles of each Opn5‐RGC type in Opn5‐regulated retinal physiology and development, and (2) what mechanism does Opn5 employ to regulate these diverse behaviors? In multielectrode array recordings of explanted retina, no electrical activity can be detected in the absence of rod, cone, and melanopsin signaling (S. Hughes et al., 2016; Polosukhina et al., 2012). Consistent with these findings, our own patch‐clamp recordings failed to identify an intrinsic photocurrent (data not shown) when assessing Opn5‐RGCs, suggesting a unique mechanism of signaling that may segregate electrical activity (visual channels) from Opn5‐influenced functions. To this end, molecular and biochemical analyses of Opn5‐mediated signaling will be vital to understand how this unique subset of opsin‐expressing RGCs regulates retinal physiology, behavior, and vision.
4. MATERIALS AND METHODS
4.1. Mice
Animals were housed in a pathogen‐free vivarium in accordance with the guidelines established by Cincinnati Children's Hospital Medical Center (CCHMC), Baylor College of Medicine IACUC, and Northwestern University IACUC. Genetically modified mice used in this study include Opn5Cre (Nguyen et al., 2019), Opn5lacZ (Buhr et al., 2015), Opn4Cre (Ecker et al. 2010, a gift from S. Hattar), Ai6 [JAX #007906], Ai9 [JAX #007909], Ai14 [JAX #007914] (Madisen et al., 2010), and cAMPer [JAX #032205]. Mice between 1 and 6 months old from both sexes were used in all experiments.
4.2. Tissue processing (whole mount)
Retinal tissue was processed as previously described (Nguyen et al, 2019). Briefly, mice were euthanized via isoflurane inhalation followed by cervical dislocation. Eyes were enucleated and fixed for 30 min in 4% paraformaldehyde in PBS, rinsed with PBS, and stored at 4°C until use. For whole‐mount analysis, retinae were dissected out of the eye, cleaned of debris, and cut into petals. For experiments where retinal topography was studied, a small mark was made on the limbus of the eye (at the ventral pole) prior to enucleation. During retinal dissection, a deep cut was made from the periphery to the optic disc to facilitate identification of the dorsal, nasal, temporal, and ventral poles. For immunofluorescence, retinae were incubated in 0.5% Triton‐X in PBS (0.5% PBST) for 15 min, followed by antigen retrieval in 50% acetone in dH2O, washed twice in PBS, and blocked in 0.5% donkey serum in 0.5% PBST for 30 min. Retinae were then incubated in primary antibody for 3 to 5 days at 4°C on an orbital rocker. Primary antibodies used for whole‐mount retina analysis include those against Chk anti‐GFP (1:1000, abcam ab290, RRID:AB_303395), Rb anti‐dsRed (1:1000, TakaraBio 632496, RRID:AB_10013483), Gt anti‐ChAT (1:300, EMD Millipore AB144P, RRID:AB_2079751), Rb anti‐Rbpms (1:500, Abcam ab152101), Ms anti‐Ap2a (1:300, Development Studies Hybridoma Bank 3B5, RRID:AB_528084), Rb anti‐Opn1sw (1:500, EMD Millipore ABN1660), Ms anti‐Calbindin (1:300, Abcam ab82812, RRID:AB_1658451), Chk anti‐Calbindin (1:500, Novus Biologicals NBP2‐50028), Rb anti‐Tusc5 (1:500, Santa Cruz BioTechnology sc‐377025), Ms anti‐Satb2 (1:300, Santa Cruz BioTechnology sc‐81376, RRID:AB_1129287), Rb anti‐Foxp1 (1:1000, CST 2005S), Shp anti‐Foxp2 (1:1000, R&D Systems AF5647‐SP), Ms anti‐Foxp2 (1:300, EMD Millipore MABE415, RRID:AB_2721039), Rb anti‐Foxp2 (1:1000, Abcam ab16046, RRID:AB_2107107), Rb anti‐Cart (1:2000, Sigma Aldrich HPA046278, RRID:AB_10961570), Rb anti‐Cart (1:250, Phoenix Pharmaceuticals H‐003‐62, RRID:AB_2313614), Gt anti‐Osteopontin (1:1000, R&D Systems AF808, RRID:AB_2194992), Ms anti‐SMI32 (1:500, Enzo Lifescience ENZ‐ABS219‐0100), Rb anti‐Melanopsin (N15, a gift from Ignacio Provencio), Rb anti‐Ppp1r17 (1:500, Thermofisher PA5‐61599, RRID:AB_2645859), Ms anti‐Brn3a (1:500, EMD Millipore MAB1585, RRID:AB_94166), Gt anti‐Brn3b/c (1:500, Santa Cruz BioTechnology sc‐6026, RRID:AB_673441), Ms anti‐Brn3c (1:200, Santa Cruz Biotechnology sc‐81980, RRID:AB_2167543). Retinae were washed with PBS (30 min × 3–5) and incubated in secondary antibodies (Jackson Immuno Research) and Hoechst for 2 days at 4°C. Finally, retinae were washed with PBS (30 min × 3–5) and mounted in Fluorogel on a glass slide and imaged on a Nikon A1 inverted microscope.
4.3. Tissue processing (cryosections)
Retinae were harvested and fixed as described above. Following dissection out of the eye cup, retinae were incubated in 15% sucrose for 15 min followed by 30% sucrose incubation overnight (4°C). A total of 3–6 retinae from different animals were placed into OCT (optimal cutting temperature) solution, snap frozen, and sectioned at 14 μm. For RGC axon tracing to central targets, mice were transcardially perfused with PBS followed by 4% PFA. Brains were harvested and postfixed in 4% PFA overnight at 4°C. The following day, samples were washed in PBS, then incubated in 15% sucrose, and then 30% sucrose (each overnight at 4°C). Samples were snap frozen in OCT and sectioned at 30 μm. For immunofluorescence, sections were incubated similarly to whole‐mount retina except primary and secondary antibody incubation times. Primary antibodies were applied to slides and incubated overnight at room temperature. Secondary antibodies were applied and incubated for 2 h at room temperature.
4.4. X‐gal retinal labeling and RNAScope
Retinal X‐gal labeling of Opn5lacZ line has been described in detail in previous publications (Buhr et al., 2015; Zhang et al., 2020). To detect Opn5 transcript, brains and whole eyes were rapidly dissected from Opn5cre ; Ai14 mice, immersed in OCT, and snap frozen in liquid nitrogen. Fresh frozen sections of tissue were mounted on slides and placed at −80°C to adhere overnight. Tissue was then processed for Opn5 transcript detection using the RNAScope® Fluorescent Multiplex Assay (ACDBio) as per the manufacturer's protocol with minor modifications. To limit reagent quantities, sterile parafilm coverslips were placed over tissue and a third of the recommended volumes were used for all nonwashing reagents except for Protease IV. Probes used in this study include tdT‐C2 (#317041‐C2) and Mm‐Opn5 modified to C3 (#438071). Images were acquired using a 60× objective, tdT+ and tdT‐ cells were manually identified and the Opn5‐C3 signal was binarized and puncta were counted using a watershed/centroid, approach (Section 4.5).
4.5. Automated cell counting
Images from Opn5cre retina with cre‐dependent reporters (Ai6 or Ai14) and immunostaining were processed in ImageJ as follows. Single channels were separated and binarized using the threshold function. To delineate cells that were adjacent to each other but binarized to form a single cell, the watershed function from the BioVoxxel Toolbox was applied. Finally, cells were counted using the centroid measurement command in ImageJ. To determine dual‐channel signal intensity, as in Figure 2E, we used CellPose (Stringer et al., 2021) to generate masks of tdTomato+ and eGFP+ cells using default parameters. These masks were imported from CellPose to FIJI using an import macro into an ROI manager and each cell's signal intensity was recorded.
4.6. Dendritic lamination
Opn5cre/+; Ai14 retina were stained against Choline Acetyltransferase (ChAT) and dsRed (targeting tdTomato) as described above, imaged using a 40× objective to capture the entire IPL depth. For analysis in FIJI, maximum intensity projections were generated for each image, a 30 × 55 μm ROI was used to determine the intensity of each pixel from the inner to outer IPL for each channel and normalized to the maximum intensity value (Figure 1e). A total of 6–11 IPL ROIs were quantified per animal and displayed as a heatmap (Figure 1e).
4.7. Opn5‐RGC polar plot and eccentricity
Opn5‐RGC cell coordinates were imported into RStudio and represented in polar space using the R package Retistruct (http://davidcsterratt.github.io/retistruct/, Sterratt et al., 2013). For eccentricity analysis, Opn5cre/+; Ai6 retina were fixed and processed as described above. Whole retinal images were acquired and the average diameter (d) of the flat mounted retina was calculated in ImageJ using the line tool. Then, circles corresponding to the d, 2/3d, and 1/3d were drawn and centered on the optic disc to describe the boundaries used for eccentricity analysis. The areas between circles of size d and 2/3d demarcate the peripheral retina (P), between 2/3d and 1/3d represents the middle retina (M), and within the boundaries of circle of diameter 1/3d correspond to the central retina (C). Then, nonoverlapping ROIs (400 × 400 μm) were acquired from regions within the boundaries described and labeled cells were quantified. For experiments with retinal topology (dorsal, ventral, nasal, temporal locations), a deep ventral cut was made as a landmark when harvesting the retina as described above.
4.8. RGC‐cone retinal distribution
Opn5cre/+ ; Ai14 retina were stained against Opn1sw (S‐cone opsin) as described above. At least six fields were imaged in the dorsal and ventral hemiretina for each sample. A total of 200 μm2 ROIs were generated that scanned (in the z‐dimension) the inner retina (GCL) to the outer retina (OS/IS of photoreceptors) to generate regions for density quantification. To calculate normalized density for each cell type (S‐cones & Opn5 RGCs), each ROI's density was calculated (cells/10,000 μm2) and averaged across both dorsal and ventral domains. Each ROI density was divided by the average for that cell type and those values (normalized density) were visualized as a heatmap (Figure 2).
4.9. AAV‐mediated RGC projection tracing
Animals were anaesthetized with ventilated isoflurane (4%—induction) and maintained at 1–2% for the remainder of the procedure using a rebreather. Mice were subsequently placed under a dissecting microscope with proparacaine (company) drops applied to the eye, and a pilot incision was made at the limbus of the eye with a sterile gauge 27½ needle. A Hamilton syringe was then inserted into the pilot incision with the needle positioned in the vitreous cavity and 1 μL of AAV2‐pCAG‐FLEX‐EGFP‐WPRE (titer—6.5 × 1012 viral genomes [vg]/mL) was injected. An additional 30 s elapsed before the needle was removed. Depending on the experiment, mice were either injected in one or both eyes for RGC projection tracing. Following the injection, mice were observed until recovery from the anesthetic and returned to their home cage. Approximately 3–4 weeks following intravitreal injections, mice were sacrificed, and relevant tissue was harvested. AAV‐pCAG‐FLEX‐EGFP‐WPRE was a gift from Hongkui Zeng (Addgene viral prep # 51502‐AAV2; RRID:Addgene_51502).
4.10. Nearest neighbors distance and density recovery profile analysis
To determine the nearest neighbors distance profile for OFF‐Starbursts (n = 3), Opn5 RGCs (n = 3), and ipRGCs (n = 3), images from Opn5cre/+; Ai14 or wildtype C57B6/J retina were binarized and using the centroid function in ImageJ, coordinates from each cell were calculated. Then using an ImageJ macro nnd (https://icme.hpc.msstate.edu/mediawiki/index.php/Nearest_Neighbor_Distances_Calculation_with_ImageJ.html), centroid‐based coordinates were analyzed to derive the nnds between individual cells. For density recovery profile analysis, csv files were generated from the binarized coordinate data described above and analyzed as per Rodieck, 1991 using the R package sjeDRP (https://rdrr.io/github/sje30/sjedrp/).
4.11. RGC typology
Opn5cre mice were crossed with Ai9 mice. We then injected them with either an AAV9 BbTagBY under an eF1a promoter or an AAV2‐pCAG‐FLEX‐EGFP. Cells that expressed Opn5 at any time point during development were labeled with tdTomato from the Ai9 reporter and cells that express Opn5 in the adult animal were labeled with the virus.
The retinas were dissected in the dark, mounted flat, and perfused with AMES for recording (Jacoby et al., 2015). RGCs were targeted under 2‐Photon imaging for typology. Flash responses were recorded from darkness to a 200 μm diameter, 200 R*/rod/s spot. Spots of multiple sizes from 30 to 1200 μm in diameter at 200 R*/rod/s were used to determine receptive field size. Flash responses and spots of multiple sizes responses were compared to population data on rgctypes.org to confirm typology. Contrast responses to 200 and 2000 μm spots at a mean level of 1000 R*/rod/s were used to identify suppressed by contrast RGCs. Moving bars of 200 × 600 μm were presented from darkness at 200 R*/rod/s in 12 different directions of movement were used to identify direction‐selective RGCs. Flashed bars were 50 × 800 μm were shown from darkness at 200 R*/rod/s in 12 different orientations to identify ON Orientation Selective RGCs (A. Nath & Schwartz, 2016). Drifting gratings with a mean luminance of 1000 R*/rod/s were shown at 12 different directions to identify OFF Orientation Selective RGCs (Amurta Nath & Schwartz, 2017). For a detailed description of visual stimuli used to identify each type, refer to Goetz et al., 2021 (bioRxiv).
4.12. Pan‐RGC single‐cell transcriptomic analysis
Metadata and the count matrix from Tran et al., 2019 was retrieved from the Broad Institute's Single Cell Portal (https://singlecell.broadinstitute.org/single_cell/study/SCP509/mouse‐retinal‐ganglion‐cell‐adult‐atlas‐and‐optic‐nerve‐crush‐time‐series). Analysis was only performed on cells used to generate the RGC transcriptomic atlas and not those used in the optic nerve crush timecourse. Initially, analyses of these data were performed in RStudio using the Seurat version3 pipeline (Stuart et al., 2019). However, due to apparent batch effects of the data (not shown), we opted to separate each animal into a unique seurat object only to integrate them into a single dataset using the FindIntegrationAnchors() function (set to default parameters). To be consistent with the cell types described by Tran et al., 2019, we opted to use their cell cluster assignment, with additional labels from a more recent analysis (Ding & Regev, 2021).
4.13. Statistical analysis and reproducibility
All statistical and image analyses were performed in GraphPad Prism Version 9.1.1 and ImageJ (FIJI), respectively. Experiments pertaining to electrophysiology were processed and analyzed in Matlab. Sample sizes (n) for each experiment are reported in the respective figure legends. All in‐text descriptive statistics are reported as mean ± SEM. Statistical tests performed in this manuscript include repeated measures (RM) one‐way ANOVA (Figure 1h), RM one‐way ANOVA with Tukey's post‐hoc analysis (Figure 1g), or ordinary one‐way ANOVA with Tukey's post‐hoc analysis (Figure 3e).
AUTHOR CONTRIBUTIONS
S.D. and G.W.S. designed the experiments. S.D., D.I.S., S.R.W., B.A.U, K.X.Z., R.D.M., and A.K.C. performed experiments and analysis. S.D. and R.A.L. wrote the paper. M.A.S., G.W.S, and R.A.L. reviewed the manuscript and provided coordinating project leadership.
CONFLICT OF INTEREST
The Lang lab has a sponsored research project with Bios Lighting Inc.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/cne.25272
ACKNOWLEDGMENTS
We thank Paul Speeg for excellent mouse colony management, Dr. Tudor Badea for gifting us anti‐Tusc5 antibody (SCB) aliquots. We thank Allie Pendery for reviewing this manuscript. This work was supported by NIH grants NEI R01 EY027711 and EY027077 to R.A.L., NIH DP2 EY026770 to G.W.S., F31 EY030737 to S.R.W., F31 EY030344 to D.I.S., and the Albert J. Ryan Predoctoral Fellowship to S.D. This work was also supported by funds from the Goldman Chair of the Abrahamson Pediatric Eye Institute at CCHMC.
D'Souza, S. P. , Swygart, D. I. , Wienbar, S. R. , Upton, B. A. , Zhang, K. X. , Mackin, R. D. , Casasent, A. K. , Samuel, M. A. , Schwartz, G. W. , & Lang, R. A. (2022). Retinal patterns and the cellular repertoire of neuropsin (Opn5) retinal ganglion cells. J Comp Neurol, 530(8), 1247–1262. 10.1002/cne.25272
DATA AVAILABILITY STATEMENT
Data will be made available upon reasonable request to the corresponding author.
REFERENCES
- Aranda, M. L. , & Schmidt, T. M. (2021). Diversity of intrinsically photosensitive retinal ganglion cells: Circuits and functions. Cellular and Molecular Life Sciences, 78(3), 889–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden, T. , Euler, T. , & Berens, P. (2020). Understanding the retinal basis of vision across species. Nature Reviews Neuroscience, 21(1), 5–20. [DOI] [PubMed] [Google Scholar]
- Bae, J. A. , Mu, S. , Kim, J. S. , Turner, N. L. , Tartavull, I. , Kemnitz, N. , Jordan, C. S. , Norton, A. D. , Silversmith, W. M. , Prentki, R. , Sorek, M. , David, C. , Jones, D. L. , Bland, D. , Sterling, A. L. R. , Park, J. , Briggman, K. L. , & Seung, H. S. (2018). Digital museum of retinal ganglion cells with dense anatomy and physiology. Cell, 173(5), 1293–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baver, S. B. , Pickard, G. E. , Sollars, P. J. , & Pickard, G. E. (2008). Two types of melanopsin retinal ganglion cell differentially innervate the hypothalamic suprachiasmatic nucleus and the olivary pretectal nucleus. European Journal of Neuroscience, 27(7), 1763–1770. [DOI] [PubMed] [Google Scholar]
- Beier, C. , Zhang, Z. , Yurgel, M. , & Hattar, S. (2021). Projections of ipRGCs and conventional RGCs to retinorecipient brain nuclei. Journal of Comparative Neurology, 529(8), 1863–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckert, A. , Schwartz, G. W. , Turner, M. H. , Rieke, F. , & Wong, R. O. (2014). Visual space is represented by nonmatching topographies of distinct mouse retinal ganglion cell types. Current Biology, 24(3), 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr, E. D. , Yue, W. W. S. , Ren, X. , Jiang, Z. , Liao, H.‐W. R. , Mei, X. , Vemaraju, S. , Nguyen, M. T. , Reed, R. R. , Lang, R. A. , Yau, K. W. , & Van Gelder, R. N. (2015). Neuropsin (OPN5)‐mediated photoentrainment of local circadian oscillators in mammalian retina and cornea. Proceedings of the National Academy of Sciences, 112(42), 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooler, S. , & Schwartz, G. W. (2021). An offset ON–OFF receptive field is created by gap junctions between distinct types of retinal ganglion cells. Nature Neuroscience, 24(1), 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, S. , & Lang, R. A. (2020). Retinal ganglion cell interactions shape the developing mammalian visual system. Development (Cambridge, England), 147(23), dev196535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, W. I. , Tamai, T. K. , Zheng, L. , Fu, J. K. , Rihel, J. , Foster, R. G. , Whitmore, D. , & Hankins, M. W. (2015). An extended family of novel vertebrate photopigments is widely expressed and displays a diversity of function. Genome Research, 25(11), 1666–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande, O. S. , Stafford, B. K. , Franke, K. , El‐Danaf, R. , Percival, K. A. , Phan, A. H. , Li, P. , Hansen, B. J. , Nguyen, P. L. , Berens, P. , Taylor, W. R. , Callaway, E. , Euler, T. , & Huberman, A. D. (2019). Molecular fingerprinting of On–Off direction‐selective retinal ganglion cells across species and relevance to primate visual circuits. Journal of Neuroscience, 39(1), 78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhande, O. S. , Stafford, B. K. , Lim, J.‐H. A. , & Huberman, A. D. (2015). Contributions of retinal ganglion cells to subcortical visual processing and behaviors. Annu Rev Vis Sci, 1(1), 291–328. [DOI] [PubMed] [Google Scholar]
- Ding, J. , & Regev, A. (2021). Deep generative model embedding of single‐cell RNA‐Seq profiles on hyperspheres and hyperbolic spaces. Nature Communications, 12(1), 2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do, M. T. H. (2019). Melanopsin and the intrinsically photosensitive retinal ganglion cells: Biophysics to behavior. Neuron, 104(2), 205–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitrescu, O. N. , Pucci, F. G. , Wong, K. Y. , & Berson, D. M. (2009). Ectopic retinal ON bipolar cell synapses in the OFF inner plexiform layer: Contacts with dopaminergic amacrine cells and melanopsin ganglion cells. Journal of Comparative Neurology, 517(2), 226–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker J. L., Dumitrescu O. N., Wong K. Y., Alam N. M., Chen S.‐K., LeGates T., Renna J. M., Prusky G. T., Berson D. M., Hattar S. (2010). Melanopsin‐Expressing retinal ganglion‐cell photoreceptors: cellular diversity and role in pattern vision. Neuron, 67(1), 49–60. 10.1016/j.neuron.2010.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, E. M. , Gauvain, G. , Sivyer, B. , & Murphy, G. J. (2016). Shared and distinct retinal input to the mouse superior colliculus and dorsal lateral geniculate nucleus. Journal of Neurophysiology, 116(2), 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauvain, G. , & Murphy, G. J. (2015). Projection‐specific characteristics of retinal input to the brain. Journal of Neuroscience, 35(16), 6575–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz, J. , Jessen, Z. F. , Jacobi, A. , Mani, A. , Cooler, S. , Greer, D. , Kadri, S. , Segal, J. , Shekhar, K. , Sanes, J. , & Schwartz, G. (2021). Unified classification of mouse retinal ganglion cells using function, morphology, and gene expression. bioRxiv , 2021.2006.2010.447922. 10.1101/2021.06.10.447922 bioRxiv [DOI] [PMC free article] [PubMed]
- Hattar, S. , Kumar, M. , Park, A. , Tong, P. , Tung, J. , Yau, K. W. , & Berson, D. M. (2006). Central projections of melanopsin‐expressing retinal ganglion cells in the mouse. Journal of Comparative Neurology, 497(3), 326–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofbauer, A. , & Dräger, U. C. (1985). Depth segregation of retinal ganglion cells projecting to mouse superior colliculus. Journal of Comparative Neurology, 234(4), 465–474. [DOI] [PubMed] [Google Scholar]
- Hughes, S. , Rodgers, J. , Hickey, D. , Foster, R. G. , Peirson, S. N. , & Hankins, M. W. (2016). Characterisation of light responses in the retina of mice lacking principle components of rod, cone and melanopsin phototransduction signalling pathways. Scientific Reports, 6, 28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, S. , Watson, T. S. , Foster, R. G. , Peirson, S. N. , & Hankins, M. W. (2013). Nonuniform distribution and spectral tuning of photosensitive retinal ganglion cells of the mouse retina. Current Biology, 23(17), 1696–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby J., Zhu Y., DeVries S. H., Schwartz G. W (2015). An Amacrine cell circuit for signaling steady illumination in the retina. Cell Reports, 13(12), 2663–2670. 10.1016/j.celrep.2015.11.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, J. , & Schwartz, G. W. (2017). Three small‐receptive‐field ganglion cells in the mouse retina are distinctly tuned to size, speed, and object motion. Journal of Neuroscience, 37(3), 610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X. , Pardue, M. T. , Mori, K. , Ikeda, S.‐i. , Torii, H. , D'Souza, S. , Lang, R. A. , Kurihara, T. , & Tsubota, K. (2021). Violet light suppresses lens‐induced myopia via neuropsin (OPN5) in mice. Proceedings of the National Academy of Sciences, 118(22), e2018840118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K. P. , Fitzpatrick, M. J. , Zhao, L. , Wang, B. , McCracken, S. , Williams, P. R. , & Kerschensteiner, D. (2021). Cell‐type‐specific binocular vision guides predation in mice. Neuron, 109(9), 1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, J. N. , De la Huerta, I. , Kim, I.‐J. , Zhang, Y. , Yamagata, M. , Chu, M. W. , Meister, M. , & Sanes, J. R. (2011). Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. Journal of Neuroscience, 31(21), 7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley, P. W. , Eglen, S. J. , & Reese, B. E. (2020). From random to regular: Variation in the patterning of retinal mosaics*. Journal of Comparative Neurology, 528(13), 2135–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, W. T. , Rupp, A. C. , Ross, R. A. , Somasundaram, P. , Hiriyanna, S. , Wu, Z. , Badea, T. C. , Robinson, P. R. , Lowell, B. B. , & Hattar, S. S. (2016). A visual circuit uses complementary mechanisms to support transient and sustained pupil constriction. Elife, 5, e15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I. J. , Zhang, Y. , Yamagata, M. , Meister, M. , & Sanes, J. R. (2008). Molecular identification of a retinal cell type that responds to upward motion. Nature, 452(7186), 478–482. [DOI] [PubMed] [Google Scholar]
- Kojima, D. , Mori, S. , Torii, M. , Wada, A. , Morishita, R. , & Fukada, Y. (2011). UV‐sensitive photoreceptor protein OPN5 in humans and mice. Plos One, 6(10), e26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, B. , Qiao, M. , Rousso, D. L. , Sanes, J. R. , & Meister, M. (2017). Four alpha ganglion cell types in mouse retina: Function, structure, and molecular signatures. Plos One, 12(7), e0180091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, J. N. , & Schwartz, G. W. (2020). The olivary pretectal nucleus receives visual input of high spatial resolution. bioRxiv , 2020.2006.2023.168054. 10.1101/2020.06.23.168054 bioRxiv [DOI]
- Lucas, R. J. , Lall, G. S. , Allen, A. E. , & Brown, T. M. (2012). Chapter 1 ‐ How rod, cone, and melanopsin photoreceptors come together to enlighten the mammalian circadian clock. In Kalsbeek A., Merrow M., Roenneberg T., & Foster R. G. (Eds.), Progress in brain research (Vol., 199, pp. 1–18). Elsevier. [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein Ed S., Zeng H. (2010). A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience, 13(1), 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani, A. , & Schwartz, G. W. (2017). Circuit mechanisms of a retinal ganglion cell with stimulus‐dependent response latency and activation beyond its dendrites. Current Biology, 27(4), 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martersteck, E. M. , Hirokawa, K. E. , Evarts, M. , Bernard, A. , Duan, X. , Li, Y. , Ng, L. , Oh, S. W. , Ouellette, B. , Royall, J. J. , Stoecklin, M. , Wang, Q. , Zeng, H. , Sanes, J. R. , & Harris, J. A. (2017). Diverse central projection patterns of retinal ganglion cells. Cell Reports, 18(8), 2058–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland, R. H. (2011). Cell populations of the retina: The Proctor lecture. Investigative Ophthalmology & Visual Science, 52(7), 4581–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, L. P. , & Studholme, K. M. (2014). Retinofugal projections in the mouse. Journal of Comparative Neurology, 522(16), 3733–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal‐Nicolás, F. M. , Kunze, V. P. , Ball, J. M. , Peng, B. T. , Krishnan, A. , Zhou, G. , Dong, L. , & Li, W. (2020). True S‐cones are concentrated in the ventral mouse retina and wired for color detection in the upper visual field. Elife, 9, e56840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, A. , & Schwartz, G. W. (2016). Cardinal orientation selectivity is represented by two distinct ganglion cell types in mouse retina. Journal of Neuroscience, 36(11), 3208–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, A. , & Schwartz, G. W. (2017). Electrical synapses convey orientation selectivity in the mouse retina. Nature Communications, 8(1), 2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, M. T. , Vemaraju, S. , Nayak, G. , Odaka, Y. , Buhr, E. D. , Alonzo, N. , Tran, U. , Batie, M. , Upton, B. A. , Darvas, M. , Kozmik, Z. , Rao, S. , Hegde, R. S. , Iuvone, P. M. , Van Gelder, R. N. , & Lang, R. A. (2019). An opsin 5‐dopamine pathway mediates light‐dependent vascular development in the eye. Nature Cell Biology, 21(4), 420–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, W. , Nakane, Y. , Hattar, S. , & Yoshimura, T. (2018). Impaired circadian photoentrainment in Opn5‐null mice. iScience, 6, 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmhans, N. , Fuller, A. D. , Nguyen, E. , Chuang, K. , Swygart, D. , Wienbar, S. R. , Lin, Y. , Kozmik, Z. , Dong, L. , Schwartz, G. W. , & Badea, T. C. (2021). Identification of retinal ganglion cell types and brain nuclei expressing the transcription factor Brn3c/Pou4f3 using a Cre recombinase knock‐in allele. Journal of Comparative Neurology, 529(8), 1926–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polosukhina, A. , Litt, J. , Tochitsky, I. , Nemargut, J. , Sychev, Y. , De Kouchkovsky, I. , Huang, T. , Borges, K. , Trauner, D. , Van Gelder, R. N. , & Kramer, R. H. (2012). Photochemical restoration of visual responses in blind mice. Neuron, 75(2), 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Y. , Zhao, Z. , Klindt, D. , Kautzky, M. , Szatko, K. P. , Schaeffel, F. , Rifai, K. , Franke, K. , Busse, L. , & Euler, T. (2021). Natural environment statistics in the upper and lower visual field are reflected in mouse retinal specializations. Current Biology, 31(15), 3233–3247. [DOI] [PubMed] [Google Scholar]
- Quattrochi, L. E. , Stabio, M. E. , Kim, I. , Ilardi, M. C. , Michelle Fogerson, P. , Leyrer, M. L. , & Berson, D. M. (2019). The M6 cell: A small‐field bistratified photosensitive retinal ganglion cell. Journal of Comparative Neurology, 527(1), 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard, K. , Li, C. , Do, Q. , Burke, E. G. , Heynderickx, S. , & Farrow, K. (2019). A projection specific logic to sampling visual inputs in mouse superior colliculus. Elife, 8, e50697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume, B. A. , Jereen, A. , Bolisetty, M. , Sajid, M. S. , Yang, Y. , Renna, K. , Sun, L. , Robson, P. , & Trakhtenberg, E. F. (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nature Communications, 9(1), 2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck, R. W. (1991). The density recovery profile: A method for the analysis of points in the plane applicable to retinal studies. Visual Neuroscience, 6(2), 95–111. [DOI] [PubMed] [Google Scholar]
- Rousso, D. L. , Qiao, M. , Kagan, R. D. , Yamagata, M. , Palmiter, R. D. , & Sanes, J. R. (2016). Two pairs of ON and OFF retinal ganglion cells are defined by intersectional patterns of transcription factor expression. Cell reports, 15(9), 1930–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes, J. R. , & Zipursky, S. L. (2020). Synaptic specificity, recognition molecules, and assembly of neural circuits. Cell, 181(3), 536–556. [DOI] [PubMed] [Google Scholar]
- Sato, K. , Yamashita, T. , Haruki, Y. , Ohuchi, H. , Kinoshita, M. , & Shichida, Y. (2016). Two UV‐sensitive photoreceptor proteins, Opn5m and Opn5m2 in ray‐finned fish with distinct molecular properties and broad distribution in the retina and brain. Plos One, 11(5), e0155339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T. M. , & Kofuji, P. (2011). Structure and function of bistratified intrinsically photosensitive retinal ganglion cells in the mouse. Journal of Comparative Neurology, 519(8), 1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterratt, D. C. , Lyngholm, D. , Willshaw, D. J. , & Thompson, I. D. (2013). Standard anatomical and visual space for the mouse retina: Computational reconstruction and transformation of flattened retinae with the Retistruct package. Plos Computational Biology, 9(2), e1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer, C. , Wang, T. , Michaelos, M. , & Pachitariu, M. (2021). Cellpose: A generalist algorithm for cellular segmentation. Nature Methods, 18(1), 100–106. [DOI] [PubMed] [Google Scholar]
- Stuart, T. , Butler, A. , Hoffman, P. , Hafemeister, C. , Papalexi, E. , Mauck, W. M. 3rd , Hao, Y. , Stoeckius, M. , Smibert, P. , & Satija, R. (2019). Comprehensive integration of single‐cell data. Cell, 177(7), 1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L.u O. , Brady, C. M. , Cahill, H. , Al‐Khindi, T. , Sakuta, H. , Dhande, O. S. , Noda, M. , Huberman, A. D. , Nathans, J. , & Kolodkin, A. L. (2015). Functional assembly of accessory optic system circuitry critical for compensatory eye movements. Neuron, 86(4), 971–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, N. T. , James, K. N. , Nistorica, A. , Lorig‐Roach, R. M. , & Feldheim, D. A. (2019). Expression of transcription factors divides retinal ganglion cells into distinct classes. Journal of Comparative Neurology, 527(1), 225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarttelin, E. E. , Bellingham, J. , Hankins, M. W. , Foster, R. G. , & Lucas, R. J. (2003). Neuropsin (Opn5): A novel opsin identified in mammalian neural tissue 1. FEBS Letters, 554(3), 410–416. [DOI] [PubMed] [Google Scholar]
- Tran, N. M. , Shekhar, K. , Whitney, I. E. , Jacobi, A. , Benhar, I. , Hong, G. , Yan, W. , Adiconis, X. , Arnold, M. E. , Lee, J. M. , Levin, J. Z. , Lin, D. , Wang, C. , Lieber, C. M. , Regev, A. , He, Z. , & Sanes, J. R. (2019). Single‐cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron, 104(6), 1039–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton, B. A. , D'Souza, S. P. , & Lang, R. A. (2021). QPLOT Neurons‐converging on a thermoregulatory preoptic neuronal population. Frontiers in Neuroscience, 15, 665762–665762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara, K. , Ishikane, H. , Sakuta, H. , Shintani, T. , Nakamura‐Yonehara, K. , Kamiji, N. L. , Usui, S. , & Noda, M. (2009). Identification of retinal ganglion cells and their projections involved in central transmission of information about upward and downward image motion. Plos One, 4(1), e4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. X. , D'Souza, S. , Upton, B. A. , Kernodle, S. , Vemaraju, S. , Nayak, G. , Gaitonde, K. D. , Holt, A. L. , Linne, C. D. , Smith, A. N. , Petts, N. T. , Batie, M. , Mukherjee, R. , Tiwari, D. , Buhr, E. D. , Van Gelder, R. N. , Gross, C. , Sweeney, A. , Sanchez‐Gurmaches, J. , … Lang, R. A. (2020). Violet‐light suppression of thermogenesis by opsin 5 hypothalamic neurons. Nature, 585(7825), 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Kim, I.‐J. , Sanes, J. R. , & Meister, M. (2012). The most numerous ganglion cell type of the mouse retina is a selective feature detector. Proceedings of the National Academy of Sciences of the United States of America, 109(36), E2391–E2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available upon reasonable request to the corresponding author.


