1. Introduction
Effective immune responses develop after a well-orchestrated series of events that include recognition, immune cell interactions and activation/inhibition of signaling pathways. The signaling lymphocyte activation molecule family (SLAMF) of cell surface receptors, which consists of nine transmembrane proteins (SLAMF1-9) expressed at different levels, are involved in viral and bacterial recognition, serve as co-stimulatory molecules at immune synapses, and modulate myeloid and lymphocyte development. SLAMF receptors are homophilic receptors, with the exception of SLAMF2 and SLAMF4, and are only expressed on hematopoietic cells. Their adaptors, SLAM associated protein (SAP) and Ewing’s sarcoma-associated transcript 2 (EAT-2), bind to the cytoplasmic tails and control the functions and magnitude of SLAMF receptor signaling. In this review, we summarize the current knowledge on the role of SLAMF receptors in regulating immune functions and recent findings describing how SLAMF receptors can be exploited as drug targets in human malignancies.
2. A Background of the SLAM Family Members and adaptors SAP and EAT-2
2.1. Structure
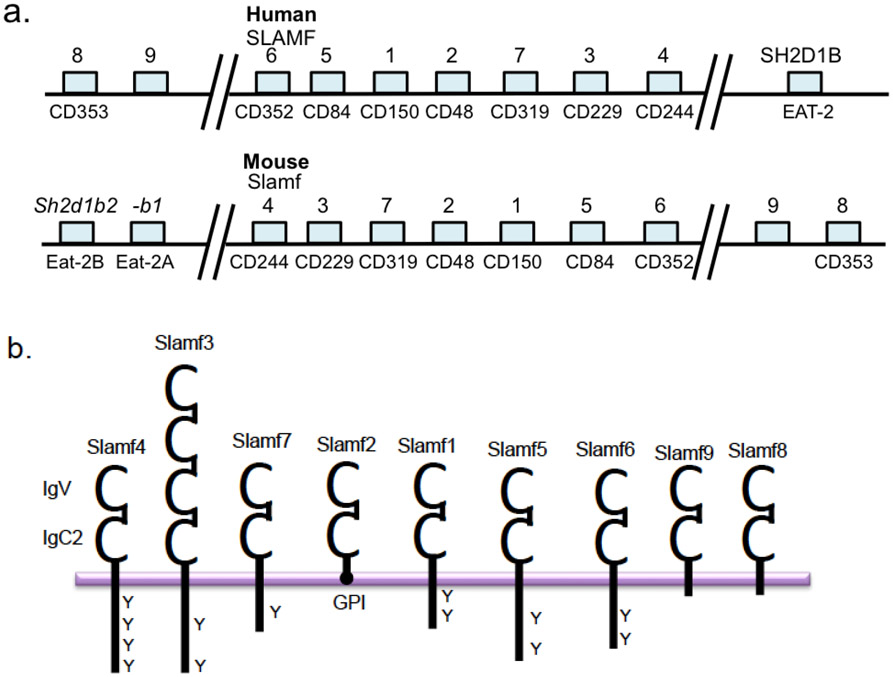
The SLAM family of immune cell surface receptors is a member of the CD2 subfamily of the immunoglobulin (Ig) superfamily consisting of nine members, SLAMF1-9 [1-4]. SLAMF receptors are type I transmembrane glycoproteins comprised of an extracellular membrane containing an N-terminal V-Ig domain followed by a C2-Ig domain in the extracellular region (this set is duplicated in SLAMF3), a transmembrane region, and an intracellular cytoplasmic tail containing tyrosine based switch motifs (ITSM). Notable exceptions to this structure include SLAMF2, which has a glycosyl-phosphatidyl-inositol (GPI) membrane anchor and like SLAMF8 and SLAMF9 lack ITSM motifs [5-8]. Binding of SLAM associated adaptors; SAP and EAT-2, to cytoplasmic tails of various SLAMFs regulate their function on different immune cells. Expression of SLAMFs and their adaptors is restricted to hematopoietic cells. In addition, the gene loci are located on chromosome 1 in both mice and humans, except SAP, which is located on the X chromosome [9](Figure 1).
Figure 1.
Signaling lymphocyte activation molecule family (SLAMF) of genes and protein. a. Organizational overview of SLAMF gene cluster on chromosome 1 in human and mouse. b. SLAMF members consist of an IgV/IgC2 ectodomain, which is duplicated in SLAMF3. While SLAMF2 and SLAMF4 bind each other, other SLAMF receptors are homophilic. Six members of the family contain varying lengths of cytoplasmic tail with ITSM motifs (Y) that can recruit and bind the adaptors SAP and/or EAT-2.
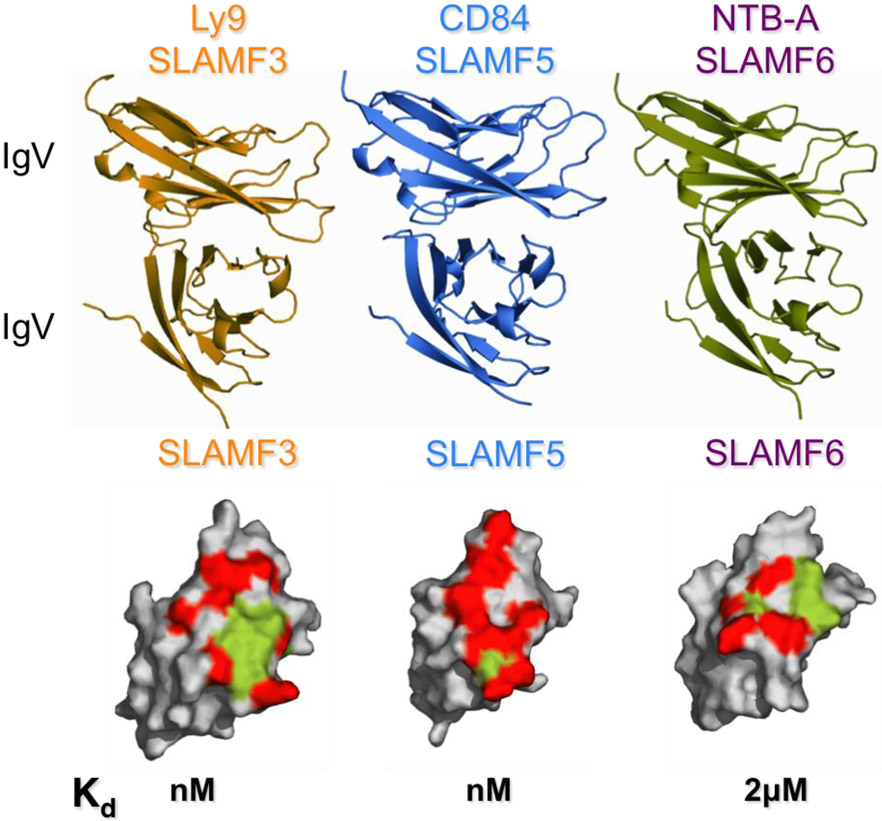
All SLAMFs are homophilic receptors aside from SLAMF2 and SLAMF4, which bind each other [10-12]. The determination of the SLAMF3, SLAMF5 and SLAMF6 crystal structures revealed in-trans interactions through their IgV domains (SLAMF3 unpublished data, generously donated by Profs. Steve Almo and Stanley Nathenson, Albert Einstein College of Medicine (Figure 2) [13, 14]. Engagement of SLAMF receptors on immune cells (e.g. APC - T cell) trigger inhibitory or activating signals that modulate cellular responses. Within these homophilic and heterophilic interactions, the binding affinities for each SLAMF varies (SLAMF3 nM, SLAMF5 sub-μM, SLAMF6 ~ 2 μM, SLAMF2/4 ~4 μM, SLAMF1 ~200 μM) which likely contributes to functional differences within the family of receptors [12-15]. In addition to being self-ligands, SLAMF1 also serves as an entry receptor for Measles virus [16, 17] while SLAMF1, SLAMF2 and SLAMF6 have been demonstrated to interact with bacterial components [18-21] (reviewed in detail ref [22]).
Figure 2.
Homophilic engagement of SLAMF3, SLAMF5 and SLAMF6 occurs via interactions of the IgV domains. Specificity of homophilic binding is determined by different surface characteristics. All three SLAMF receptors show different binding affinities. Green = hydrophobic, red= hydrophilic amino acids
2.2. SAP and X-linked Lymphoproliferative Disease (XLP)
First identified in 1975, X-linked lymphoproliferative disease (XLP) (also known as Duncan’s disease) is an extremely rare primary immunodeficiency that mainly manifests in males and is primarily characterized by extreme susceptibility to infection with Epstein-Barr virus (EBV) [23]. However, most XLP patients infected with EBV develop fatal or fulminant infectious mononucleosis due to dysregulated immune responses, which leads to clonal proliferation and expansion of T and B cells. NK and CD8+ T cell functional defects have been identified in XLP patients likely contributing to the inability to control EBV infections [24-27]. Patients usually exhibit progressive loss of serum IgG and develop B cell lymphomas and dys-gammaglobulinemia [28-31].
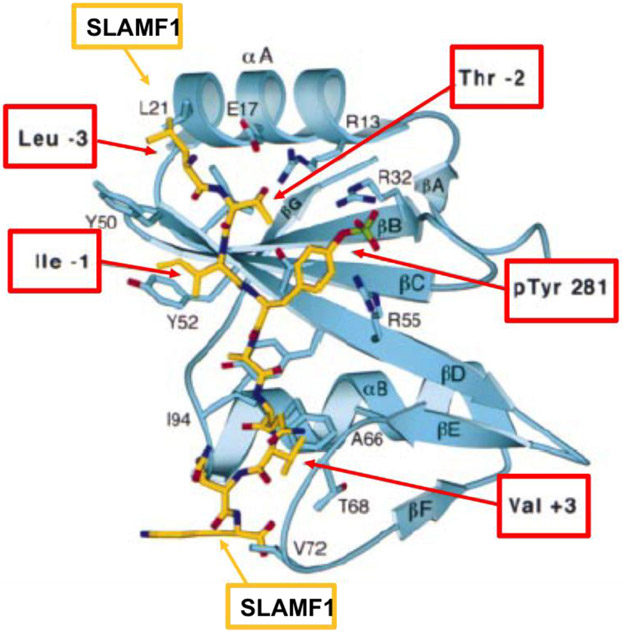
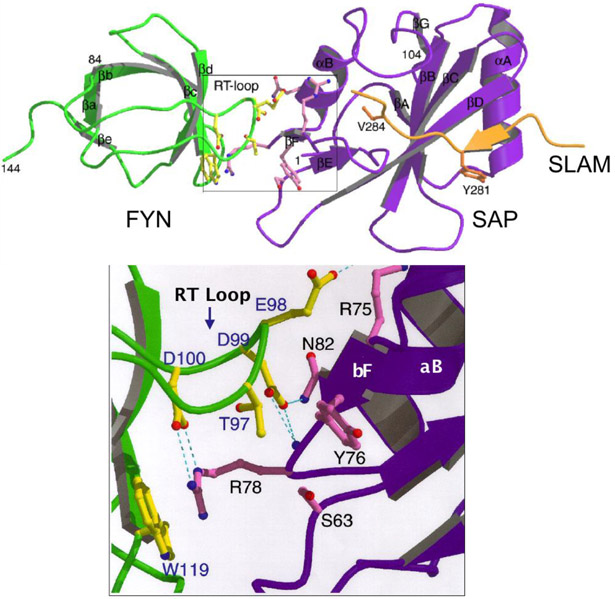
Not until twenty years after the description of XLP, the genetic cause was determined by the Terhorst lab: mutations in or deletion of the SH2D1A gene, which encodes a 15 kD cytoplasmic protein SAP consisting of a single Src homology 2 (SH2) domain and a 28 amino acid tail [32-35]. In the same publication, SAP was shown to bind to SLAMF1 and subsequent studies showed binding of SAP to ITSM motifs in the cytoplasmic tail of six of the SLAMF receptors (Figure 3) [1, 30, 34, 35]. SAP was then identified to be required for recruitment and activation of Src-family kinase FynT upon SLAM ligation [36]. Subsequently, the crystal structure of the SLAM-SAP-Fyn-SH3 ternary complex revealed that SAP binds the FynT SH3 domain through a non-canonical surface interaction and couples Fyn to SLAM receptors (Figure 4) [37].
Figure 3. Ribbon diagram showing SAP/SLAMF1 pY281 complex.
The bound SLAMF1 phosphopeptide is shown in a stick representation (yellow). Selected SAP residues that form the binding site are shown in blue. SLAMF1 residues N-terminal to pY281 make additional interactions with SAP at pY −3 and pY −1 (positions relative to pTyr281)[34, 35].
Figure 4. SAP couples Fyn to SLAMF receptors.
SAP binds the Fyn SH3 domain through a non-canonical surface interaction (zoomed area). SLAM peptide binds SAP in a 3-pronged mode by via the b-strand of N-terminal, Tyr 281 and Val 284.
SAP is mainly expressed in T cells, NK cells, NKT cells and eosinophils while expression in B cells is found only in some cases [33, 34, 38-40]. Of note, later research also identified mutations in XIAP, X-linked inhibitor of apoptosis, which are associated with XLP like disease manifestations in a small number of families [41, 42].
Studies from SAP deficient mice shed light on understanding the basis of XLP. Naïve CD4+ T cells from SAP deficient mice exhibited reduced production of peptide-MHC or T cell receptor (TCR) driven TH2 cytokines [43-46]. In addition, germinal center formation is significantly impaired and antibody secreting cells (ASCs) and memory B cells are lost leading to severely reduced levels of serum IgG and IgE [43, 47-49]. Later, these defects were also identified in XLP patients [50, 51].
In SAP deficient mice there appeared to a B cell defect, which was dependent on the genetic background [52] as evidenced by adoptive transfer studies of SAP deficient B cells and WT CD4+ T cells [47]. Although the B cell defect seems secondary to that of CD4+ T cells [48, 49, 51], in vitro experiments suggested an intrinsic defect in class switching [40].
CD8+ T cells and NK cell functions are also altered in SAP deficient mice and XLP patients. While human in vitro studies suggested SAP is required for induction of cytotoxicity by CD8+ T cells [53], SAP deficient mice presented more virus specific CD8+ T cells in lymphocytic choriomeningitis virus (LCMV) and γ-herpesvirus 68 infections, suggesting SAP inhibits CD8+ T cell responses [54, 55]. Finally, SAP was found to be indispensable for NKT cell development as SAP deficient mice and XLP patients lack NKT cells [56-58].
In summary, SAP signaling is critical for mounting proper immune responses. Moreover, SAP, as well as EAT-2, regulates signaling through SLAMFs by setting thresholds for activation/inhibition and modulating cell-cell interactions and responses. Here, we will highlight the roles of SLAMF receptors in regulating normal immune responses and how they are involved in hematological malignancies.
3. Immune cell Functions of SLAMF Receptors
SLAMF receptors are adhesion molecules that are involved in development of lymphocytes and in orchestrating innate and adaptive immune responses. While contribution of each SLAMF member is unique, they can have compensating or opposing roles in function, which will be discussed in this section.
3.1. SLAMF1 (CD150, SLAM)
Current understanding of the biology of SLAMF1 (SLAM, CD150) demonstrates the importance of this receptor at multiple levels during an immune response. The cytoplasmic tail of SLAMF1 can recruit SAP and EAT-2 upon immune cell activation in both mice and humans [35, 59]. SLAMF1 is widely expressed on B and T cells, macrophages, dendritic cells, platelets and hematopoietic stem cells [60-64]. Expression on B cells starts from pre-B cell stage and is upregulated in plasma cells [65]. In B cell malignancies, SLAMF1 demonstrated heterogeneous expression, with differentially expressed isoforms that may contribute to disease pathogenesis [66].
SLAMF1 ligation during T cell activation is critical for the fate of the T cell. Upon T cell stimulation, SLAMF1 co-localizes with the T cell receptor and the tyrosine residues on the cytoplasmic tail are phosphorylated [67]. However, binding of SAP to the cytoplasmic tail can be in both a phosphotyrosine-independent and dependent manner [67]. Initial studies using monoclonal antibodies against SLAMF1 in conjunction with TCR stimulation induced IFNγ secretion and promoted TH1 responses and even induced switching of TH2 cells to TH0 phenotype [60, 68, 69]. SLAMF1−/− mice exhibited no difference in IFNγ production compared to WT controls, but interestingly IL-4 and IL-13 cytokine levels were reduced indicating compromised TH2 responses [44, 62]. During germinal center (GC) reactions, T follicular helper cells (TFH) residing within the GCs use SAP signaling, while SLAMF1 is required for IL-4 production by TFH cells to provide optimal help to B cells [70]. This discrepancy in vitro and in vivo suggests that there may be other SLAMF receptors that can compensate in vivo for TH1 responses in the absence of SLAMF1. The role of SLAMF1 in NKT cells will be discussed in the SLAMF6 chapter.
The findings that upon encountering inflammatory stimuli with lipopolysaccharide (LPS), IL-1β or IFNγ, SLAMF1 is upregulated on antigen presenting cells (APCs), i.e. macrophages and dendritic cells (DCs), led to the notion that SLAMF1 may have a role in regulating APC functions [62, 63, 71]. Interestingly, SLAMF1 was found to interact with the OmpC and OmpF bacterial membrane proteins of Gram-negative bacteria to facilitate phagocytosis in macrophages [18]. It further regulated the activity of NADPH oxidase (NOX2) complex in phagolysosomal compartments and facilitated the recruitment of autophagy complex containing Beclin-1, Vps34, Vps15 and UVRAG [20]. As autophagy is a universal process by which cells remove misfolded proteins, clear damaged organelles and also eliminate intracellular pathogens [72], expression and signaling through SLAMF1 may play a role in proper regulation of this process. Additionally, a recent study demonstrated that SLAMF1 enhances type I interferon production upon encountering Gram-negative bacteria by modulating MyD88 in a TLR4 independent signaling manner [73]. Taken together, these findings suggest SLAMF1 is a target for controlling inflammatory responses to Gram-negative bacteria through multiple mechanisms.
3.1.1. SLAMF1 is implicated in various Autoimmune Diseases
Autoimmunity is in general defined by the loss of immune tolerance to self-antigens and causes destruction of healthy cells. SLAMF1 has been associated with human inflammatory bowel syndromes including Crohn’s disease as well as experimental murine models of colitis [74-76]. Inflamed colon sections from Crohn’s disease patients showed enhanced SLAMF1 expression on monocytes and macrophages driven by Toll like receptor (TLR) responses to bacterial ligands as compared to healthy gut tissue [74]. Involvement of SLAMF1 was also demonstrated in an experimental model of colitis where antibodies blocking SLAMF1 homophilic interactions reduced colitis in mice [75].
Pathology of rheumatoid arthritis (RA), characterized by chronic inflammation of the joints, has also been associated with increased expression levels of SLAMF1 on T cells in the synovial fluid and tissue of patients compared to that of healthy individuals [77]. This study suggested SLAMF1 homophilic interactions between T cells and synovial mononuclear cells may contribute to cytokine profile changes in these patients. Additionally, another study found EBV-associated RNA and DNA in the synovial fluid of RA patients and speculated that SAP and SLAM interactions and/or signaling may play an important role due to the link between SAP signaling and EBV infections [78].
Another autoimmune disease where SLAMF1 has been implicated is multiple sclerosis (MS), a condition where immune cells attack myelin sheath [79]. The percentage of T cells expressing SLAMF1 was increased in the blood of MS patients, but the contribution of this to the disease is not yet known [80].
3.2. SLAMF3 (Ly9, CD229)
3.2.1. Structure and Expression of SLAMF3
SLAMF3 is a unique member of the family with its two sets of extracellular IgV/IgC2 domains [81, 82]. While several isoforms of SLAMF3 exist, their contribution to immune responses is not known. Homophilic interaction occurs via the N-terminal IgV domains and induces localization at the immune synapses indicating that SLAMF3 may be important in T-B cell interactions [83]. Indeed, SLAMF3 is expressed on all subsets of B cells and T cells [84, 85]. High expression on NK cells and monocytes are restricted to murine cells [81, 86, 87]. A SLAMF3 homolog, A33, has been identified in genomes of squirrel monkey CMV (SMCMV) and owl monkey CMV (OMCMV), that infect New world monkeys[88]. This was acquired by retrotranscription of virus-host coevolution by the new world monkey CMV.
Similar to other family members, the cytoplasmic tail of SLAMF3 contains ITSM motifs for binding of SAP and EAT-2. Interestingly, SAP has been demonstrated to bind only to human SLAMF3 cytoplasmic tail but not mouse [7, 59, 89]. In addition, Grb2 (growth factor receptor-bound protein 2), a ubiquitous adaptor protein, as well as the μ2 chain of AP-2 adaptor complex, was demonstrated to bind the phosphorylated cytoplasmic tail of SLAMF3 independent of SAP binding site [90, 91]. Regulated by TCR stimulation, recruitment and binding of Grb2 to the SLAMF3 cytoplasmic tail facilitates internalization of the receptor [91]. This interaction appears to be unique among the SLAMF members.
3.2.2. Insights into immune cell functions
Involvement and importance of SLAMF3 in regulating immune cell interactions and functions came from studies using SLAMF3-deficient (Ly9−/−) mice. T cells from Ly9−/− mice presented a mild TH2 defect in vitro along with reduced proliferation upon suboptimal anti-CD3 stimulations [92]. While Ly9−/− mice mounted normal T and B cell responses during in vivo viral infection with LCMV [92], the proportion of innate memory CD8+ T cells significantly increased upon post-infection with murine cytomegalovirus (MCMV) [93]. Furthermore, invariant NKT (iNKT) cells are increased in Ly9−/− mice suggesting SLAMF3 is an inhibitory receptor for expansion of innate CD8+ T cells and iNKT cell development [93].
While B cell development in the bone marrow is normal in Ly9−/− mice, splenic transitional 1, marginal zone and B1a B cells are expanded [94]. In accordance with this increase, T cell independent antibody responses after immunization with 2,4,6-trinitrophenyl-Ficoll were increased. Injecting monoclonal antibodies against SLAMF3 (αSLAMF3) in WT mice resulted in loss of marginal zone B cells and innate like B cells along with down-regulation of CD19/CD21/CD81 complex in an Fc independent manner. Furthermore, aged Ly9−/− mice presented with spontaneous autoantibody production, indicating loss of self tolerance, regardless of background [95]. These findings demonstrate SLAMF3 is a negative regulator of immune responses and may be relevant in targeting B cell related diseases.
3.3. SLAMF6 (CD352, Ly108, NTB-A)
3.3.1. Structure, expression and ligands
SLAMF6 (human: NTB-A, mouse: Ly108) structural details are reviewed elsewhere [3]. In mice, different alternatively spliced forms exist: Ly108.1 and Ly108.2 were first identified, containing one and two additional unique tyrosine motifs, respectively [96, 97]. Later a novel isoform Ly108-H1 was discovered [98]. Tyrosine phosphorylation of the cytoplasmic tail of SLAMF6 leads to recruitment of SAP with high affinity binding, but in the absence of phosphorylation, SAP cannot be recruited [3, 99, 100]. Binding of SAP activates downstream signaling by recruiting the Src family kinase Fyn [37, 101, 102]. This interaction prevents the tyrosine phosphatases SHP1 and/or SHP2 from binding to the cytoplasmic tail and their subsequent negative regulation [34, 35].
SLAMF6 is expressed on a wide variety of immune cells including T cells (also TFH), B cells, NK cells (expressed in human only), double positive thymocytes, eosinophils and neutrophils (mouse only) [6, 38, 96, 103-106]. Furthermore, high expression of SLAMF6 has been determined in various B cell lymphomas, i.e. mantle cell and follicular lymphomas [107]. Expression on normal as well as malignant cells suggests that SLAMF6 may be involved in distinct cell-cell interactions in different microenvironments and may be a useful therapeutic target.
Similar to SLAMF3, a SLAMF6 homolog, S1, with a 97% amino acid sequence identity in its ligand binding N-terminal Ig domain have been identified in SMCMV and OMCMV [88]. This was acquired by retrotranscription of virus-host coevolution by the new world monkey CMV. This suggests an immune evasion mechanism of viruses by acquiring the host SLAMF receptors that retain the ligand binding capacity. This allows the interference with host SLAMF functions and induces immunomodulatory actions.
In addition to being a self-ligand, SLAMF6 detects viral and bacterial components as well. For instance, human and mouse SLAMF6 bind the outer membrane proteins OmpC and OmpF, of E.coli, S. typhimurium, and in part to Citrobacter rodentium [18, 21]. Human SLAMF6 can recognize the influenza haemagglutinin (HA) and Vpu protein of HIV-1, both of which affect NK cell cytotoxicity mediated by SLAMF6 [108, 109].
3.3.2. SLAMF6 signaling in T and B cells
Co-stimulatory molecules play an important role in activation and initiation of proper T cell responses upon TCR engagement. CD28 is the best-known secondary signal necessary for T cell activation. However, studies from CD28 deficient mice suggested additional co-stimulatory molecules play a role as antigen dependent T cell responses were still intact in these mice [110, 111]. In the absence of CD28, stimulation and crosslinking of SLAMF6 with monoclonal antibodies in vitro leads to recruitment of SAP to the phosphorylated cytoplasmic tail, and subsequent events lead to T cell proliferation and cytokine production [112]. When SLAMF6 was blocked in vivo using a soluble SLAMF6-Fc fusion protein, B cell isotype switching to IgG2a and IgG3 was blocked [112]. In fact, injections of anti-SLAMF6 to WT mice immunized with NP-OVA or in a chronic graft versus host disease (cGVHD) model inhibited T and B cell responses in these distinct in vivo models [113, 114].
Involvement of SLAMF6 in regulating immune responses was demonstrated using mice deficient in SLAMF6 [115]. Expression of the SLAMF6 extracellular domain was disturbed by removal of exons 2 and 3 (Ly108 ΔE2+3) [115]. In vivo infection of Ly108 ΔE2+3 mice with L. mexicana led to delayed formation of lesions as well as significantly smaller lesions compared to WT controls, indicating a role for Ly108 in innate and adaptive immune responses.
Under normal circumstances, T cell expansion upon infections is constrained by a mechanism called restimulation-induced cell death (RICD), which induces apoptosis of effector T cells during the peak of an immune response [116]. Upon restimulation of these T cells, pro-apoptotic molecules are upregulated, such as FAS ligand and BIM, to induce apoptosis. This mechanism was found to be defective in patients with XLP, mainly due to absence of SAP and defective signaling through SLAMF6 [117]. In RICD, SAP/SLAMF6 signaling was shown to augment TCR signaling to achieve the threshold necessary for RICD. This was facilitated by SAP’s recruitment of Lck, but not Fyn, to the cytoplasmic tail [118].
SLAMF6 is also implicated in trogocytosis, the transfer of membrane patches from target to effector T cells [119, 120]. The trogocytosis capacity of CD8+ T cell clones from melanoma patients correlated with their cytotoxic capacity. Furthermore, higher cytotoxic capacity was correlated with increased phosphorylation of SLAMF6 [119]. Enhanced cytotoxicity of these CD8+ T cells could be blocked by an antagonist αSLAMF6, indicating that SLAMF6 co-stimulation plays an active role in T cell functional diversity. As CD8+ T cells from XLP patients are defective in cytolytic activity against EBV-infected B cell targets, SLAMF6 as well as SAP may be involved [53, 121-123]. Mice deficient in SAP presented altered T cell responses including short-lived T-B cell interactions, defective germinal center formation, and humoral immunity [2, 100, 124]. As CD8+ T cells are critical in controlling infections, the role of SAP as well as contributions of SLAMFs were investigated. Studies using SAP deficient mouse T cells showed inefficient killing of B cell target cells, but not others, suggesting SAP is especially critical in T-B cell interactions [125]. These defects were at the level of immune synapse organization and inefficient actin clearance. Conjugation of OVA-pulsed B cells with WT or SAP−/− cytotoxic CD8+ T lymphocytes (CTL) showed significant SHP-1 localization at the immune synapse with SAP−/− CTL compared to WT CTL as quantified from immunofluorescence microscopy images [125]. This suggested that in the absence of SAP, SHP1 is recruited to the cytoplasmic tail of SLAMF6 [126]. When B cell targets were SLAMF6 deficient, killing ability of SAP−/− T cells was rescued demonstrating that SLAMF6 is an important regulator of SAP dependent responses from CTLs. Our current understanding of how SLAMF6 may play a role in T-B cell interactions is summarized in Figure 5.
Figure 5.
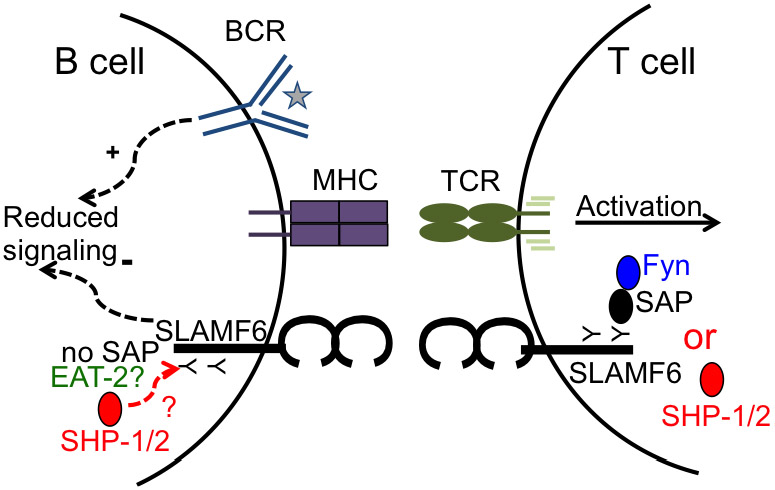
SLAMF6 localizes at the immune synapse. Ligation of SLAMF6 on a T cell recruits binding of SAP to the ITSM on the cytoplasmic tail. SAP recruits Fyn and induces activation. SAP has the highest affinity for cytoplasmic tail of SLAMs and blocks binding of SHP-1/2. In the absence of SAP, SHP-1/2 binds to cytoplasmic tail of SLAMF6 and induces negative signaling on T cells [126]. B cells do not express SAP. Whether EAT-2 is expressed or it blocks recruitment of SHP-1/2 is not known. However, ligation of SLAMF6 with an antibody in B cells appear to induce negative signals, which may be due to binding of SHP-1/2.
3.3.3. SLAMF6 controls neutrophil functions
Disruption of Ly108 expression in mice (Ly108 ΔE2+3) also revealed a role for Ly108 in neutrophil functions [115]. In response to infections with Salmonella typhimurium, neutrophils exhibited significantly reduced reactive oxygen species (ROS) production and bacterial killing with increased IL-6, IL-12 and TNF-α production compared to WT controls. Mechanisms of Ly108 signaling involved in ROS production is still not known. While neutrophils are not implicated in XLP and human neutrophils do not appear to express SLAMF6, it may be of interest to investigate whether the expression can be induced upon activation.
3.3.4. SLAMF6 is an activating receptor on human NK cells
Activating signals induced upon ligation of NTB-A on an NK cell induces phosphorylation of its cytoplasmic tail, which is Src kinase dependent, and leads to recruitment of SAP, EAT-2 and SHP1/2 [127, 128]. Full SLAMF6 dependent activation of NK cell cytotoxicity depends on simultaneous binding of SAP and EAT-2 to the phosphorylated tyrosine residues on the cytoplasmic tail [128]. Crosslinking of human SLAMF6 with antibodies stimulate NK cells for target cell killing in cytotoxicity assays, and homophilic interaction of ligands induces NK cell cytotoxicity against target cells with subsequent IFNγ and TNFα secretion [103, 127]. However, if the SLAMF6 homophilic interaction between an NK cell and a target cell is blocked using an anti-SLAMF6 antibody, NK cell cytotoxicity is inhibited [106]. Interestingly, blocking the homophilic interaction between neighboring NK cells has no effect on proliferation or cytotoxicity [129]. Instead, homophilic interaction with a MHC-I negative target cell induces potent cytotoxicity of NK cells. This selective mechanism is probably in place to ensure NK cells do not end up killing each other.
While expression of SLAMF receptors is restricted to hematopoietic cells, a recent study demonstrated that SLAMF6 enhanced activation of NK cells against a non-hematopoietic target cell [130]. This was regulated by SAP, which uncoupled SHP-1 binding from the cytoplasmic tail of SLAMF6, diminishing the effect of the receptor on NK cell responsiveness to non-hematopoietic cells. This proved that SLAM-SAP pathways could also influence NK cell education [130].
3.3.5. SLAMF6 regulates NKT cell development
Natural killer T (NKT) cells represent a subset of T lymphocytes that develop from double positive (DP) CD4+CD8+ precursor cells in the thymus [131]. While positive selection of conventional T cells is mediated by interactions with thymic epithelial cells, NKT cells are selected by lipid antigens presented by CD1d on other DP thymocytes [132, 133]. These cells express an invariant TCR and can rapidly secrete cytokines following infections [134].
With the severe lack of NKT cells in XLP patients, SAP was hypothesized to be involved in NKT cell development [1]. Indeed, mouse studies showed that SAP expression in NKT cells is necessary for cognate help to B cells[135]. As binding of SAP and recruitment of Fyn is induced by signaling through SLAMF receptors, SLAMF1 and SLAMF6, which are expressed on DP thymocytes, their possible role in NKT cell development was investigated. Mouse studies from single knockouts (SLAMF1−/−, SLAMF6−/−) revealed only a modest effect on NKT cell development, probably due to overlapping functions of these two receptors [62, 115]. Supporting this hypothesis, mice reconstituted with double mutant (SLAMF1−/−SLAMF6−/−) bone marrow chimeras had severely impaired NKT cell development [105]. Furthermore, expression of promyelocytic zinc finger (PLZF), a transcription factor required for development of invariant NKT (iNKT) and other innate like T lymphocytes, was demonstrated to be modulated by co-stimulation through SLAMF6 [136-138]. These studies together underline the importance of SLAMF6 in NKT cell development. Other SLAMF members were also found to regulate NKT cell development in a SAP dependent and independent manner [139].
It has also been determined that SLAMF6 acts as a SAP-dependent on and off switch for stable T-B cell interactions [126]. In the absence of SAP, SLAMF6 recruited the negative regulator SHP-1 at the T-B cell synapse limiting adhesion. Interestingly, when both SAP and SLAMF6 were removed (SAP−/−SAMF6−/−), both CD4+ T cell function as well as NKT cell differentiation was restored. Therefore, SLAMF6 also serves an important role in sending positive and negative signals depending on the competition between SAP and SHP-1 binding, which in turn regulates T cell help and NKT cell development.
3.3.6. SLAMF6 as a susceptibility gene for Systemic Lupus Erythematosus
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that mainly affects females and is characterized by the production of autoantibodies against self antigens [140]. Generation of these antibodies results in clinical manifestations including arthritis, kidney damage, skin disease and blood cell abnormalities. Both genetic and environmental factors are known to contribute to disease manifestation [140, 141].
Analysis of the SLAM/CD2 gene cluster revealed extensive polymorphisms, and among 35 inbred mouse strains two stable haplotypes were identified [97, 142]. Haplotype 1 is found in C57BL/6J (B6) and related strains while haplotype 2 is found in autoimmune-prone mouse strains, i.e. NZW[97]. It appeared that autoimmunity was induced only when the haplotype 2 SLAM locus was expressed in B6 background [97]. Sle1 was identified on murine chromosome 1 as a cause of loss of tolerance and autoantibody production. Fine mapping of the Sle1 locus identified 4 loci contributing to the disease manifestation, one of which was the most potent: Sle1b [97]. The congenic B6.Sle1b mouse strain presented with spontaneous autoimmunity and production of antinuclear antibodies (ANAs), indicating a failure to maintain central and peripheral tolerance [97, 143, 144]. Among the SLAMF genes, SLAMF6 was the strongest candidate for lupus due to the expression of alternatively spliced variants: Ly108-1 and Ly108-2. Of the two, Ly108-1 had a greater tendency to be phosphorylated at the cytoplasmic tail for SAP, Fyn recruitment and signaling than Ly108-2 isoform [145]. Kumar et al. and colleagues demonstrated that the Ly108-1 isoform was more abundant in B6.Sle1b mice compared to B6 mice, which was found to sensitize immature B cells to deletion and RAG re-expression [146]. This study provided the idea that SLAMF6 serves as a regulatory checkpoint for self-reactive B cells to protect from autoimmunity.
A later study identified a new isoform of Ly108 present in haplotype 1 but not haplotype 2: Ly108-H1 [98]. This isoform was found to regulate SLE in a CD4+ T cell dependent manner and expression of this isoform in lupus-prone mice significantly suppressed autoimmunity. Deletion of SLAMF6 in Chr1b also disrupted the autoimmune phenotype. In addition, autoimmunity in B6.Sle1b mice correlated with expansion of an osteopontin-expressing TFH cell subset, which was suppressed when the Ly108-H1 isoform was expressed in these mice [147]. Overall, these studies pointed to the importance of alternative splicing in pathogenesis of SLE and that both B and T cell tolerance is compromised.
3.4. SLAMF2, 4, 5, 7 and 8
SLAMF2 (CD48) has multiple binding partners besides SLAMF4, including CD2, CD58 and bacterial lectin FimH [11, 148, 149]. Roles of SLAMF2 in regulating immunity and tolerance are reviewed in detail excellently elsewhere [148]. Briefly, SLAMF2 is constitutively expressed nearly on all hematopoietic cells and is upregulated upon activation on some subsets [148, 150, 151]. SLAMF2 interacts with protein tyrosine kinase Lck in T cells and its crosslinking induces TCR activation and IL-2 production [152, 153]. SLAMF2 interaction in cis with CD2 also facilitates TCR signaling by bringing Lck and linker for activation of T cells (LAT) to the TCR/CD3 complex. Furthermore, SLAMF2/CD2 interactions contribute to APC-T cell adhesion and synapse organization as well as regulate effector functions of CD8+ T cells, highlighting that SLAMF2 is important in immune cell responses at different levels [154, 155]. However, SLAMF4 (2B4) also can contribute to these various responses by being the ligand for SLAMF2. Originally identified as an activating NK cell receptor, SLAMF4 is also expressed on CD8+ T cells, γδ+T cells and monocytes [156, 157]. Crosslinking with monoclonal antibodies against SLAMF4 and interactions of SLAMF4 on NK cells and SLAMF2 on target cells induces NK cell mediated cytotoxicity [156]. This perforin-mediated killing by NK cells is governed by phosphorylation of SLAMF4 by associating with LAT in the lipid rafts and subsequent recruitment of SAP to the immune synapse [158, 159]. However, in the absence of SAP, phosphatases SHP-1/2 bind to the phosphorylated tyrosine residues inducing inhibitory signals on NK cell cytotoxicity [160]. This helps explain why NK cell cytotoxicity is impaired in XLP patients. This duality of SLAMF4 signaling in NK cell functions is reviewed in detail elsewhere [161].
SLAMF5 is expressed on T cells, B cells, monocytes, DCs and platelets [61, 63, 86, 162-164]. High and low expression of SLAMF5 on B cells dictates their subset. Human memory B cells express high SLAMF5 (CD84hi) along with the memory marker CD27, somatically mutated Ig variable genes, and exhibit increased proliferation compared to other CD84lo subsets[165]. High expression of SLAMF5 in memory B cells may be the consequence of signals induced upon binding of SAP and EAT-2 to the cytoplasmic tail of SLAMF5.
Activation of T cells is also dependent on SLAMF5, similar to that observed in other SLAMF receptors. Crosslinking of the TCR and ligation of SLAMF5 induces T cell activation and IFNγ secretion [163]. Activation of SLAMF5 leads to phosphorylation of its cytoplasmic tail by Lck, which recruits SAP [166]. Mouse studies involving GC – TFH interactions revealed that SLAMF5 interaction is important for prolonged cell-cell contact and optimal TFH and GC formation [104], stressing the relevance and importance of this receptor at the immune synapses and regulating immune cell functions.
In addition to its role in regulating T and B cells, SLAMF5 is shown to modulate downstream signaling of TLR4 upon LPS stimulation in macrophages and regulate effector function and cell fate decisions [167].
SLAMF7 is expressed on the surface of NK cells, NKT cells, B cells, DCs, macrophages and activated T cells in mice, while in humans its expression is induced after maturation of DCs [168, 169]. SLAMF7 binds EAT-2, but not SAP [169-171], and two isoforms of SLAMF7 have been identified: CS-1 long (CS1-L) and CS-1 short (CS1-S) [172]. While CS1-S does not bind EAT-2, CS1-L binds EAT-2 and induces NK cell mediated cytotoxicity. Presence or absence of EAT-2 dictates whether SLAMF7 induces positive or negative signals on NK cell activation [169, 171]. As EAT-2 is not expressed in T cells, signaling through SLAMF7 induces inhibitory signals in T cell responses [169]. SLAMF7 also acts as a negative regulator of proinflammatory responses in activated human monocytes [173]. Additionally, expression and stimulation of SLAMF7 isoform CS1-L on B cells induces B cell proliferation and autocrine signaling [174]. SLAMF7 expression on multiple myeloma cells has emerged as a therapeutic target, which will be discussed later.
SLAMF8 (CD353, BLAME) is expressed on myeloid cells, including monocytes, macrophages, DCs and neutrophils upon activation with bacteria or IFNγ [175, 176]. Studies using SLAMF8 deficient mice revealed that SLAMF8 may regulate protein kinase C (PKC) activity in macrophages, which phosphorylates p40phox leading to enhanced NADPH Oxidase (NOX2) responses upon exposure to bacteria [176]. In addition, migration of macrophages and neutrophils are accelerated in SLAMF8−/− mice and inhibition of NOX2 responses by diphenyleneiodonium (DPI) inhibited this migration [177]. These studies suggest SLAMF8 regulates inflammatory responses of myeloid cells in a NOX2 dependent manner.
While SLAMF8 signaling is SAP independent, de Calisto et al. demonstrated the lack of SLAMF8 led to the expansion of innate CD8+ T cells that expressed the transcription factor Eomes and produced IFNγ upon stimulation [139]. These mice also had increased proportion of PLZFhi NKT cells in the thymus.
In conclusion, different SLAMF receptors play distinct roles in regulating immune responses and lymphocyte development.
4. SLAMF receptors in Hematological Malignancies
Hematologic malignancies are cancers that affect the blood, lymphatic system and the bone marrow. Malignant cells arise from blood cells of common lymphoid and myeloid progenitor origin and are categorized under three main subsets: leukemias (acute and chronic), lymphomas and myelomas [178]. Chromosomal translocations are a common cause of these diseases unlike solid tumors, and thus are commonly used as diagnostic factors.
As SLAMF members are expressed on a variety of normal immune cells that form the tumor microenvironment as well as cells that become malignant, it is plausible to hypothesize that SLAMFs may play a role in all these complex interactions. In fact, in recent years many investigators have demonstrated relevant roles for SLAMFs, specifically SLAMF6 in chronic lymphocytic leukemia and SLAMF7 in multiple myeloma (MM), both of which will be discussed in further detail.
4.1. The Pathogenesis of Chronic Lymphocytic Leukemia
Chronic lymphocytic leukemia (CLL) is the most common leukemia in western countries accounting for 40% of all leukemias. It is a malignancy of mature CD5+ B cells that accumulate in the blood, bone marrow and secondary lymphoid organs [179]. Phenotypically they are similar to antigen-experienced B cells expressing CD19, CD5, CD23, CD25, CD69 and CD71, and the memory B cell marker CD27 [180-182]. CLL is broadly divided into 2 subsets depending on the immunoglobulin (Ig) heavy chain mutation status (IGHV). CLL B cells with unmutated IGHV (U-CLL) derive from mature CD5+ B cells, whereas CLL cells with mutated IGHV (M-CLL) derive from CD5+CD27+ post-germinal center B cell subsets [183].
CLL patients can have varying clinical outcomes depending on the aggressiveness of the disease. While some patients are stable and are only observed over time without any need for treatment, some experience more aggressive disease and require immediate treatment. Multiple factors play role in determining the course of the disease. Mutational status of CLL B cells is an important prognostic factor and patients with U-CLL show a more aggressive disease and shorter survival time compared to M-CLL patients [184, 185]. Chromosomal alterations are another parameter for disease outcome. Patients with 13q deletions that include the miRNAs miR15a and miR16-1 are usually associated with favorable disease outcome [186]. Mouse models with deletion of the miR15a and miR16-1 locus mimic many features of human CLL [187]. Chromosomal deletion of 17p and 11q harbor the p53 and ataxia telangiectasia mutated (ATM) genes and are associated with poor disease outcome [186, 188]. Trisomy 12 is found in ~ 15% of CLL patients and signifies an intermediate prognosis. In addition to chromosomal aberrations, high expression of CD38 and ZAP70 in CLL B cells is associated with a poor outcome and shorter time to treatment [184, 189, 190]. Besides these clinical markers that are associated with disease outcome of CLL patient, studies using the application of whole exome sequencing have identified recurrent somatic mutations that are involved in DNA damage, mRNA processing, WNT and Notch signaling and chromatin modifications that can affect B cell signal transductions [191-194].
4.1.1. CLL Microenvironment
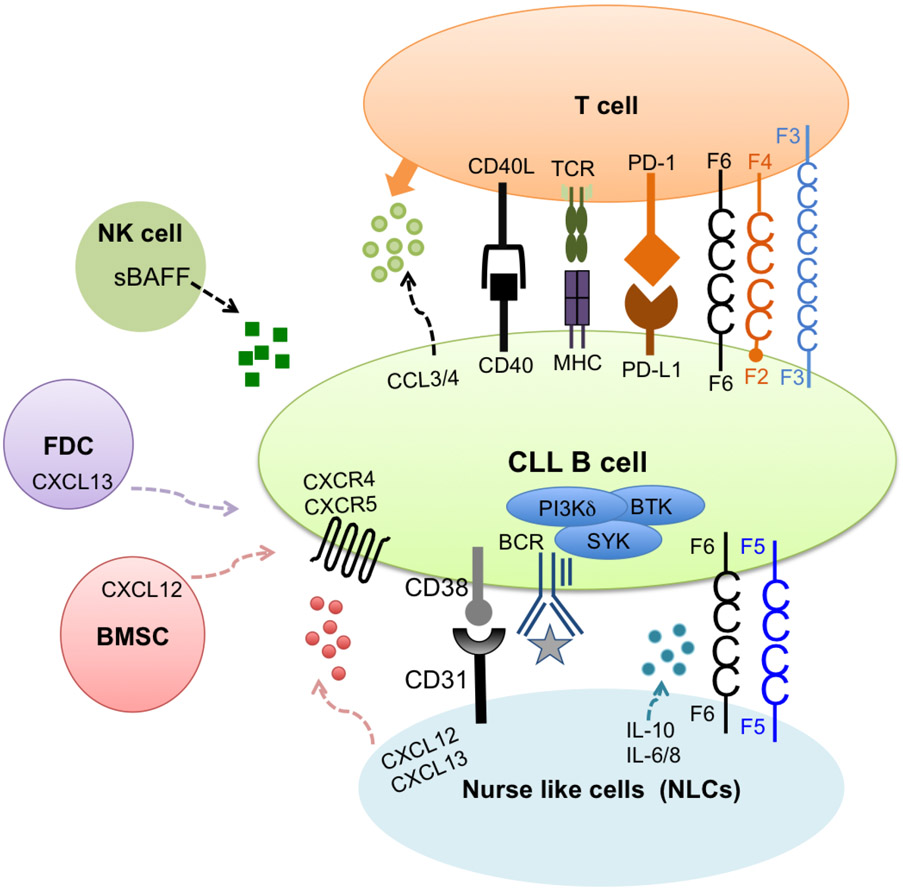
Cells of the immune system coevolve with the tumor and provide the tumor a friendly microenvironment for survival of CLL cells. Culturing CLL B cells in vitro after isolation from peripheral blood mononuclear cells (PBMCs) leads to the induction of spontaneous apoptosis indicating that CLL B cells are highly dependent on the microenvironment for survival signals [195, 196]. Only when cultured with bone marrow stromal cells, CLL B cells survived in vitro. This suggests that the tumor microenvironment in vivo has a profound effect on survival and expansion of CLL leukemic cells [179]. Some of the key interactions between CLL B cells and the tumor microenvironment are highlighted in Figure 6.
Figure 6. The CLL Microenvironment.
Contact between CLL cells and nurse like cells (NLCs) is established by chemokine receptors and adhesion molecules expressed on CLL cells and ligands on NLCs. The CD38-CD31 axis promotes CLL survival. CXCR4/CXCL12 chemokine gradient allows shuffling of CLL cells between circulation and secondary lymphoid organs to receive survival and proliferation signals. SLAMF5 and SLAMF6 are expressed on the surface of NLCs (Yigit, unpublished data). The relevant contribution of these receptors to CLL survival requires further investigation. T cells are another major contributor to CLL survival. Secretion of chemokines CCL3/4 by CLL cells attracts T cells nearby. CD40/CD40L interaction promotes survival and PD-1/PD-L1 pathway favors immune evasion of CLL cells. NK cells, bone marrow stromal cells (BMSCs) and follicular dendritic cells (FDCs) also contribute to CLL survival.
One of the CLL supporting cells is the ‘nurse like cells’ (NLCs), which are of monocyte origin [197]. These cells were found to differentiate in vitro from PBMCs of CLL patients and secrete chemokines such as CXCL12 and CXCL13 for their survival in vitro [197, 198]. These chemokines induce chemotaxis for migration of CLL cells in and out of secondary lymphoid organs in vivo. CXCL12 mediated signaling is increased by CD38 expressed on CLL cells further promoting survival [199, 200]. NLCs also activate the B cell receptor signaling and NF-κB pathway for survival [201].
The B cell receptor (BCR) signaling pathway is a key survival factor for CLL [202]. The BCR is composed of an antigen-specific surface membrane Ig (smIg) along with Igα/Igβ heterodimers. Engagement of the BCR triggers phosphorylation of Igα/Igβ and recruitment of Lyn, which in turn activates kinases SYK, BTK and PI3K. This leads to the activation of downstream signaling cascades including phospholipase C gamma 2 (PLCγ2), calcium signaling, NF-κB and mitogen-activated protein kinase (MAPK) pathways. These signaling events promote survival and proliferation of B cells [203]. IGHV mutation status of CLL cells determines the responsiveness of BCR engagement. While CLL B cells from U-CLL patients are more responsive to BCR stimulation and mostly recognize autoantigens [204, 205], M-CLL cells show constitutive phosphorylation of ERK kinase along with reduced surface BCR leading to an “anergic” phenotype [206, 207]. In addition to activated BCR signaling, ligation of CD40/CD40L on malignant B cells and T cells also promote survival of CLL cells [208].
Another hallmark of CLL is dysfunctional T cells in the tumor microenvironment. Although there is an overall expansion in the T cell compartment, the normal CD4/CD8 ratio appears to be inverted due to differential sensitivity of CD4+ and CD8+ T cells to Fas/Fas Ligand induced cell death [209-213]. Within CD4 T cells, frequency of regulatory T cells (Treg) is increased further supporting leukemic expansion [214-216]. CD4+ and CD8+ T cells show many defects in CLL including T cell exhaustion, inability to form immune synapses and impaired cytotoxic function [217-219]. Exhausted CD8 T cell state correlates with increased expression of exhaustion markers such as programmed death-1 (PD-1), CD160, SLAMF4 and KLRG1[217, 220-222]. Several studies hypothesized one factor for exhaustion may be cytomegalovirus (CMV) infection that influences and expands the CMV-specific CD4+ and CD8+ T cell subsets in healthy individuals and is also expanded in CMV-seropositive CLL patients [223-225]. However, within the exhausted T cell pool, CMV-specific T cells showed reduced expression of exhaustion markers as well as retained cytotoxic capacity and cytokine production compared to other exhausted T cells [226, 227]. These data indicated that there might be other factors influencing changes in the T cell compartment in CLL.
PD-1/PD-L1 is a major pathway contributing to the known T cell defects in CLL [228]. PD-L1 is overexpressed on CLL cells and myeloid derived suppressor cells (MDSCs), which further up-regulates PD-1 on T cells [229, 230]. Within CD4 and CD8 T cell populations, naïve T cells are reduced while effector memory CD4+ T cells and terminally differentiated CD8+ T cells are increased, which clinically corresponds with disease aggressiveness [228]. Up-regulated PD-1 inhibits IFNγ secretion skewing the immune responses to a dysregulated TH2 response [228]. These T cell defects are recapitulated in the TCL1 transgenic (Eμ-TCL1) mouse model of CLL [231-233], allowing a useful platform to study and understand the contribution of PD1/PD-L1 pathway in CLL pathogenesis and how this pathway can be targeted therapeutically.
4.1.2. Therapeutic options in CLL
Allogeneic hematopoietic stem cell transplantation (HSCT) represents one of the oldest treatments for hematological malignancies which set the foundation for the development of cancer immunotherapy [234]. Performed for the first time in 1968, high doses of radiation and chemotherapy were given to the patient that wiped out the entire immune system followed by donor HSC transplantation for repopulation of the hematopoietic system. Extensive studies over the years provided great insight into the efficiency of the ability of donor immune cells to eliminate recipient tumor cells. This is known as ‘graft versus leukemia’ (GVL) effect. GVL potency was further appreciated by the finding that post- HSCT, donor lymphocyte infusions (DLI) induced remarkable responses and remissions without radiation or chemotherapy, in leukemias [235, 236].
Targeting BCR signaling using small inhibitor molecules has dramatically improved treatment options for CLL patients [202]. The BTK inhibitor, ibrutinib (PCI-32765) binds irreversibly to a cysteine residue (Cys-481) in the BTK kinase domain and inhibits its phosphorylation and enzymatic activity [237]. Ibrutinib inhibited proliferation and stromal cell contact of CLL cells and reduced their viability in vitro [238]. Ibrutinib also prevented tissue homing in response to chemokines CXCL12 and CXCL13 in vitro and in a mouse model of CLL [239]. This inhibition of tissue homing chemokines directly correlated with a transient lymphocytosis in CLL patients undergoing ibrutinib treatment, which allowed CLL cells to move from secondary lymphoid organs into the circulation and induced cell death [240]. Because ibrutinib not only binds to BTK in CLL B cells but also to ITK in T cells, its immunomodulatory role was also investigated in CLL patients [241]. In patients treated with ibrutinib, the increased T cell numbers normalized and production of inflammatory cytokines were reduced, and the T cell repertoire diversity increased [242, 243]. Ibrutinib also reduced Treg numbers [244]. Expression of PD-1 and PD-L1 upon ibrutinib treatment markedly decreased, improving activated effector T cell functions [245, 246].
While ibrutinib targeted CLL B and T cells, a second-generation highly selective BTK inhibitor, acalabrutinib, was produced [247]. Pharmacodynamics and proteomic analysis appeared to be similar on leukemic cells compared to ibrutinib, while off target effects on T cells was more pronounced using ibrutinib than acalabrutinib [248, 249].
Rituximab, a monoclonal antibody against the B cell surface antigen CD20, is widely used in the treatment of B cell malignancies including CLL [250-252]. The primary mode of action of rituximab includes ADCC and complement dependent cytotoxicity (CDC) as well as direct anti-proliferative and pro-apoptotic effects [253, 254]. While major advances were brought by rituximab, relapse and resistance to treatment are eventually seen in patients. One potential reason is the removal of bound CD20 complexes from the surface of CLL cells by trogocytosis [255, 256]. Administering rituximab together with ibrutinib led to better responses, but follow up studies identified that ibrutinib interferes with the effect of rituximab by downregulating CD20 on the cell surface of CLL cells [257, 258]. More efficient combinations with rituximab may be required for better and durable effects of this immunotherapy agent.
Lenalidomide is an immunomodulatory agent that affects the tumor microenvironment and the immune system. In particular, it corrects CLL B cell - T cell immunological synapse formation and down-regulates PD-1 on T cells [218, 229, 259]. Lenalidomide normalizes total T cell and Treg numbers, similar to ibrutinib, in vivo [260]. Combining lenalidomide with αCD20 improved ADCC activity of NK cells in vitro, and this combination also demonstrated efficacy in clinical trials [261, 262]. Other small inhibitor molecules targeting BCR signaling, monoclonal antibodies and immunomodulatory drugs used in treatment of CLL patients are discussed in detail elsewhere [202, 263].
4.2. SLAMF Receptors in CLL
The importance of SLAMF receptors in regulating innate and adaptive immune responses makes them relevant candidates in context of various diseases including chronic lymphocytic leukemia. SLAMF receptors, particularly SLAMF1, SLAMF5 and SLAMF6, are expressed on both human and mouse CLL cells [264]. Two separate studies have identified surface expression of SLAMF1 with longer time to treatment and overall increased survival in CLL patient cohorts using either cluster analysis or multivariate prognostic models [265, 266]. Kaplan Meyer curves predicted longer treatment free survival rates in SLAMF1+ CLL patients (6% cut-off applied) compared to SLAMF1− patients [267]. SLAMF1 surface expression appeared to be important for an intact autophagy pathway. As demonstrated in previous studies evaluating the role of SLAMF1 in recruiting NOX2/Vps34/Beclin/UVRAG complex in phagosomes [20], ligation of SLAMF1 using an agonistic antibody triggered ROS accumulation and induced formation of this macrocomplex in a CLL cell line, MEC-1. When SLAMF1 was silenced in primary CLL cells, they became resistant to autophagy inducing agents demonstrating that this may be the underlying reason for unfavorable outcome in SLAMF1 low CLL patients as they may be more resistant to various drugs targeting this pathway.
An additional study revealed a link between SLAMF1 and CD180 in modulating transcriptional program in CLL cells by regulating expression levels of some transcription factors [268]. Furthermore, an isoform of SLAMF1 that lacks the transmembrane domain was identified and found to co-localize with CD180 on the cell surface [269]. Co-ligation of SLAMF1 and CD180 on CLL cells led to inhibition of the Akt and MAPK pathways that disrupts survival signals in CLL B cells. These studies suggest that SLAMF1 may be important as a diagnostic marker as well as an interesting therapeutic target.
The observation that SLAMF5 was overexpressed on CLL B cells compared to healthy B cells suggested that SLAMF5 might be involved in survival of CLL cells in the tumor microenvironment [270, 271]. Blocking SLAMF5 in vitro and in vivo disrupted the tumor – tumor microenvironment interactions inducing cell death of CLL cells [272]. It would be relevant to study the SLAMF5 interaction between CLL B and T cells in immune synapse formation and cytotoxicity as studies using WT mice indicated SLAMF5 is required for optimal T-B cell interaction in germinal centers [104].
SLAMF6 expression is high on normal T and B cells and studies have already indicated its importance in B-T cell signaling. Therefore it was plausible to hypothesize that monoclonal antibodies targeting SLAMF6 may be of therapeutic interest in CLL [264]. An adoptive transfer model of an aggressive TCL1 clone, TCL1-192 [273], into SCID (severe combined immune deficiency) mice, which lack T and B cells, was injected with anti-SLAMF6 upon leukemic. This led to significantly reduced leukemic burden by inducing antibody dependent cellular cytotoxicity (ADCC) and reduced proximal B cell receptor signaling [264]. Interestingly, the antibody was unable induce ADCC in peritoneal cavity (PerC) of mice, due to possibly different microenvironments and signaling compared to blood or spleen [264, 274]. One such finding was the elevated reactive oxygen species (ROS) production in PerC CLL B and normal B1a B cells compared to the cells residing in spleen [274]. While inhibition of ROS limited leukemic expansion in this niche, the finding that ROS is reduced upon BTK inhibitor, ibrutinib and leukemic infiltration to the blood, prompted us to do a combination therapy. By combining anti-SLAMF6 with ibrutinib, leukemic cells were pushed out of this niche into the circulation making them targetable by the antibody. This led to an overall greater reduction in leukemic burden than either regimen alone. Exploration of SLAMF6 as a therapeutic target would be of great interest in CLL and other B cell malignancies.
4.3. Targeting SLAMF7 in Multiple Myeloma
Multiple myeloma (MM) is a malignancy of plasma cells characterized by monoclonal expansion in bone marrow and accumulation of monoclonal antibodies produced by malignant plasma cells that result in organ damage, including bone lesions, renal diseases and anemia [275, 276]. In the past decade, the front line therapy for MM included proteasome inhibitors (i.e. bortezomib) and immunomodulatory agents that resulted in overall survival of 9 months emphasizing a need for additional treatment options [277, 278]. Use of monoclonal antibodies in treatment of MM opened a new era in treatment for this disease [279-281]. One of these antibodies (elotuzumab) targets SLAMF7, which is highly expressed on MM cells [168, 282]. In plasma cells and MM cells, EAT-2 is normally not expressed. However, in approximately ~30-50% of MM patients, amplification of the long arm of chromosome 1q leads to expression of EAT-2, which is actually associated with poor prognosis and unresponsiveness to therapy [283, 284]. In addition, SLAMF7 is implicated to have a tumor-supporting effect in MM, which is further induced upon EAT-2 expression. When SLAMF7 expression was inhibited on MM cells, adhesion to bone marrow stromal cells was reduced [282]. A similar reduction was observed when elotuzumab was used making it an ideal candidate for use in clinic with MM patients.
Several mechanisms of action have been identified for elotuzumab. Primarily the antibody induces ADCC [168], activates NK cell cytotoxicity by binding to SLAMF7 on NK cells [285], and inhibits tumor cell stromal cell interaction [282]. While elotuzumab alone had no clinical effect in advanced MM patients, combination with proteasome inhibitors or immunomodulatory drugs improved progression free survival rates [286, 287]. Blocking PD-1 further increased efficacy of elotuzumab and enhanced NK and CD8 T cell activity in mouse models of MM [288]. In addition, a SLAMF7 specific peptide inducing antigen-specific cytotoxic T cells was identified, which may be used as a novel immunotherapy for MM [289]. A recent study demonstrated the effectiveness of SLAMF7-CAR-T cells to eliminate relapsed/refractory MM cells [290] . Overall, all these approaches underline the importance of targeting SLAMF7 in MM.
5. Conclusions
SLAMF receptors and their adaptors play vital roles in maintaining a balanced immune response and their interrupted functions are associated with various diseases. Ability of SLAMF receptors to play activating and inhibitory roles depending on the signals they receive or the cell-cell interactions they encounter makes them an important target when thinking about designing therapeutics. Monoclonal antibodies that can block or engage SLAMF-SLAMF interactions are being targeted in diseases such as CLL and MM. Further understanding the fundamentals on how SLAMF members play a role in the bigger picture will make them once step closer to being targets in many more diseases.
References
- [1].Engel P, Eck MJ, Terhorst C, The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease, Nature reviews. Immunology, 3 (2003) 813–821. [DOI] [PubMed] [Google Scholar]
- [2].Detre C, Keszei M, Romero X, Tsokos GC, Terhorst C, SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions, Seminars in immunopathology, 32 (2010) 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Calpe S, Wang N, Romero X, Berger SB, Lanyi A, Engel P, Terhorst C, The SLAM and SAP gene families control innate and adaptive immune responses, Advances in immunology, 97 (2008) 177–250. [DOI] [PubMed] [Google Scholar]
- [4].Ma CS, Deenick EK, The role of SAP and SLAM family molecules in the humoral immune response, Annals of the New York Academy of Sciences, 1217 (2011) 32–44. [DOI] [PubMed] [Google Scholar]
- [5].Wang N, Morra M, Wu C, Gullo C, Howie D, Coyle T, Engel P, Terhorst C, CD150 is a member of a family of genes that encode glycoproteins on the surface of hematopoietic cells, Immunogenetics, 53 (2001) 382–394. [DOI] [PubMed] [Google Scholar]
- [6].Fraser CC, Howie D, Morra M, Qiu Y, Murphy C, Shen Q, Gutierrez-Ramos JC, Coyle A, Kingsbury GA, Terhorst C, Identification and characterization of SF2000 and SF2001, two new members of the immune receptor SLAM/CD2 family, Immunogenetics, 53 (2002) 843–850. [DOI] [PubMed] [Google Scholar]
- [7].Tovar V, del Valle J, Zapater N, Martin M, Romero X, Pizcueta P, Bosch J, Terhorst C, Engel P, Mouse novel Ly9: a new member of the expanding CD150 (SLAM) family of leukocyte cell-surface receptors, Immunogenetics, 54 (2002) 394–402. [DOI] [PubMed] [Google Scholar]
- [8].Yokoyama S, Staunton D, Fisher R, Amiot M, Fortin JJ, Thorley-Lawson DA, Expression of the Blast-1 activation/adhesion molecule and its identification as CD48, Journal of immunology, 146 (1991) 2192–2200. [PubMed] [Google Scholar]
- [9].Wu C, Sayos J, Wang N, Howie D, Coyle A, Terhorst C, Genomic organization and characterization of mouse SAP, the gene that is altered in X-linked lymphoproliferative disease, Immunogenetics, 51 (2000) 805–815. [DOI] [PubMed] [Google Scholar]
- [10].Latchman Y, McKay PF, Reiser H, Identification of the 2B4 molecule as a counter-receptor for CD48, Journal of immunology, 161 (1998) 5809–5812. [PubMed] [Google Scholar]
- [11].Brown MH, Boles K, van der Merwe PA, Kumar V, Mathew PA, Barclay AN, 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48, The Journal of experimental medicine, 188 (1998) 2083–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Velikovsky CA, Deng L, Chlewicki LK, Fernandez MM, Kumar V, Mariuzza RA, Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family, Immunity, 27 (2007) 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yan Q, Malashkevich VN, Fedorov A, Fedorov E, Cao E, Lary JW, Cole JL, Nathenson SG, Almo SC, Structure of CD84 provides insight into SLAM family function, Proceedings of the National Academy of Sciences of the United States of America, 104 (2007) 10583–10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cao E, Ramagopal UA, Fedorov A, Fedorov E, Yan Q, Lary JW, Cole JL, Nathenson SG, Almo SC, NTB-A receptor crystal structure: insights into homophilic interactions in the signaling lymphocytic activation molecule receptor family, Immunity, 25 (2006) 559–570. [DOI] [PubMed] [Google Scholar]
- [15].Mavaddat N, Mason DW, Atkinson PD, Evans EJ, Gilbert RJ, Stuart DI, Fennelly JA, Barclay AN, Davis SJ, Brown MH, Signaling lymphocytic activation molecule (CDw150) is homophilic but self-associates with very low affinity, The Journal of biological chemistry, 275 (2000) 28100–28109. [DOI] [PubMed] [Google Scholar]
- [16].Tatsuo H, Ono N, Tanaka K, Yanagi Y, SLAM (CDw150) is a cellular receptor for measles virus, Nature, 406 (2000) 893–897. [DOI] [PubMed] [Google Scholar]
- [17].Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y, Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM, Nature structural & molecular biology, 18 (2011) 135–141. [DOI] [PubMed] [Google Scholar]
- [18].Berger SB, Romero X, Ma C, Wang G, Faubion WA, Liao G, Compeer E, Keszei M, Rameh L, Wang N, Boes M, Regueiro JR, Reinecker HC, Terhorst C, SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages, Nature immunology, 11 (2010) 920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Baorto DM, Gao Z, Malaviya R, Dustin ML, van der Merwe A, Lublin DM, Abraham SN, Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic, Nature, 389 (1997) 636–639. [DOI] [PubMed] [Google Scholar]
- [20].Ma C, Wang N, Detre C, Wang G, O'Keeffe M, Terhorst C, Receptor signaling lymphocyte-activation molecule family 1 (Slamf1) regulates membrane fusion and NADPH oxidase 2 (NOX2) activity by recruiting a Beclin-1/Vps34/ultraviolet radiation resistance-associated gene (UVRAG) complex, The Journal of biological chemistry, 287 (2012) 18359–18365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].van Driel B, Wang G, Liao G, Halibozek PJ, Keszei M, O'Keeffe MS, Bhan AK, Wang N, Terhorst C, The cell surface receptor Slamf6 modulates innate immune responses during Citrobacter rodentium-induced colitis, International immunology, 27 (2015) 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].van Driel BJ, Liao G, Engel P, Terhorst C, Responses to Microbial Challenges by SLAMF Receptors, Front Immunol, 7 (2016) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Purtilo DT, Cassel CK, Yang JP, Harper R, X-linked recessive progressive combined variable immunodeficiency (Duncan's disease), Lancet, 1 (1975) 935–940. [DOI] [PubMed] [Google Scholar]
- [24].Sullivan JL, Byron KS, Brewster FE, Purtilo DT, Deficient natural killer cell activity in x-linked lymphoproliferative syndrome, Science, 210 (1980) 543–545. [DOI] [PubMed] [Google Scholar]
- [25].Harada S, Bechtold T, Seeley JK, Purtilo DT, Cell-mediated immunity to Epstein-Barr virus (EBV) and natural killer (NK)-cell activity in the X-linked lymphoproliferative syndrome, International journal of cancer, 30 (1982) 739–744. [DOI] [PubMed] [Google Scholar]
- [26].Argov S, Johnson DR, Collins M, Koren HS, Lipscomb H, Purtilo DT, Defective natural killing activity but retention of lymphocyte-mediated antibody-dependent cellular cytotoxicity in patients with the X-linked lymphoproliferative syndrome, Cellular immunology, 100 (1986) 1–9. [DOI] [PubMed] [Google Scholar]
- [27].Rousset F, Souillet G, Roncarolo MG, Lamelin JP, Studies of EBV-lymphoid cell interactions in two patients with the X-linked lymphoproliferative syndrome: normal EBV-specific HLA-restricted cytotoxicity, Clinical and experimental immunology, 63 (1986) 280–289. [PMC free article] [PubMed] [Google Scholar]
- [28].Harrington DS, Weisenburger DD, Purtilo DT, Malignant lymphoma in the X-linked lymphoproliferative syndrome, Cancer, 59 (1987) 1419–1429. [DOI] [PubMed] [Google Scholar]
- [29].Egeler RM, de Kraker J, Slater R, Purtilo DT, Documentation of Burkitt lymphoma with t(8;14) (q24;q32) in X-linked lymphoproliferative disease, Cancer, 70 (1992) 683–687. [DOI] [PubMed] [Google Scholar]
- [30].Morra M, Howie D, Grande MS, Sayos J, Wang N, Wu C, Engel P, Terhorst C, X-linked lymphoproliferative disease: a progressive immunodeficiency, Annual review of immunology, 19 (2001) 657–682. [DOI] [PubMed] [Google Scholar]
- [31].Sumegi J, Huang D, Lanyi A, Davis JD, Seemayer TA, Maeda A, Klein G, Seri M, Wakiguchi H, Purtilo DT, Gross TG, Correlation of mutations of the SH2D1A gene and epstein-barr virus infection with clinical phenotype and outcome in X-linked lymphoproliferative disease, Blood, 96 (2000) 3118–3125. [PubMed] [Google Scholar]
- [32].Coffey AJ, Brooksbank RA, Brandau O, Oohashi T, Howell GR, Bye JM, Cahn AP, Durham J, Heath P, Wray P, Pavitt R, Wilkinson J, Leversha M, Huckle E, Shaw-Smith CJ, Dunham A, Rhodes S, Schuster V, Porta G, Yin L, Serafini P, Sylla B, Zollo M, Franco B, Bolino A, Seri M, Lanyi A, Davis JR, Webster D, Harris A, Lenoir G, de St Basile G, Jones A, Behloradsky BH, Achatz H, Murken J, Fassler R, Sumegi J, Romeo G, Vaudin M, Ross MT, Meindl A, Bentley DR, Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene, Nature genetics, 20 (1998) 129–135. [DOI] [PubMed] [Google Scholar]
- [33].Nichols KE, Harkin DP, Levitz S, Krainer M, Kolquist KA, Genovese C, Bernard A, Ferguson M, Zuo L, Snyder E, Buckler AJ, Wise C, Ashley J, Lovett M, Valentine MB, Look AT, Gerald W, Housman DE, Haber DA, Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome, Proceedings of the National Academy of Sciences of the United States of America, 95 (1998) 13765–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C, The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM, Nature, 395 (1998) 462–469. [DOI] [PubMed] [Google Scholar]
- [35].Poy F, Yaffe MB, Sayos J, Saxena K, Morra M, Sumegi J, Cantley LC, Terhorst C, Eck MJ, Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition, Molecular cell, 4 (1999) 555–561. [DOI] [PubMed] [Google Scholar]
- [36].Latour S, Gish G, Helgason CD, Humphries RK, Pawson T, Veillette A, Regulation of SLAM-mediated signal transduction by SAP, the X-linked lymphoproliferative gene product, Nature immunology, 2 (2001) 681–690. [DOI] [PubMed] [Google Scholar]
- [37].Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ, SAP couples Fyn to SLAM immune receptors, Nature cell biology, 5 (2003) 155–160. [DOI] [PubMed] [Google Scholar]
- [38].Munitz A, Bachelet I, Fraenkel S, Katz G, Mandelboim O, Simon HU, Moretta L, Colonna M, Levi-Schaffer F, 2B4 (CD244) is expressed and functional on human eosinophils, Journal of immunology, 174 (2005) 110–118. [DOI] [PubMed] [Google Scholar]
- [39].Kis LL, Nagy N, Klein G, Klein E, Expression of SH2D1A in five classical Hodgkin's disease-derived cell lines, International journal of cancer, 104 (2003) 658–661. [DOI] [PubMed] [Google Scholar]
- [40].Al-Alem U, Li C, Forey N, Relouzat F, Fondaneche MC, Tavtigian SV, Wang ZQ, Latour S, Yin L, Impaired Ig class switch in mice deficient for the X-linked lymphoproliferative disease gene Sap, Blood, 106 (2005) 2069–2075. [DOI] [PubMed] [Google Scholar]
- [41].Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, Fischer A, de Saint Basile G, Latour S, XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome, Nature, 444 (2006) 110–114. [DOI] [PubMed] [Google Scholar]
- [42].Marsh RA, Villanueva J, Zhang K, Snow AL, Su HC, Madden L, Mody R, Kitchen B, Marmer D, Jordan MB, Risma KA, Filipovich AH, Bleesing JJ, A rapid flow cytometric screening test for X-linked lymphoproliferative disease due to XIAP deficiency, Cytometry. Part B, Clinical cytometry, 76 (2009) 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wu C, Nguyen KB, Pien GC, Wang N, Gullo C, Howie D, Sosa MR, Edwards MJ, Borrow P, Satoskar AR, Sharpe AH, Biron CA, Terhorst C, SAP controls T cell responses to virus and terminal differentiation of TH2 cells, Nature immunology, 2 (2001) 410–414. [DOI] [PubMed] [Google Scholar]
- [44].Davidson D, Shi X, Zhang S, Wang H, Nemer M, Ono N, Ohno S, Yanagi Y, Veillette A, Genetic evidence linking SAP, the X-linked lymphoproliferative gene product, to Src-related kinase FynT in T(H)2 cytokine regulation, Immunity, 21 (2004) 707–717. [DOI] [PubMed] [Google Scholar]
- [45].Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL, Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP, Proceedings of the National Academy of Sciences of the United States of America, 98 (2001) 7449–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, Antonellis A, Koretzky GA, Gardner K, Schwartzberg PL, SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1, Immunity, 21 (2004) 693–706. [DOI] [PubMed] [Google Scholar]
- [47].Morra M, Barrington RA, Abadia-Molina AC, Okamoto S, Julien A, Gullo C, Kalsy A, Edwards MJ, Chen G, Spolski R, Leonard WJ, Huber BT, Borrow P, Biron CA, Satoskar AR, Carroll MC, Terhorst C, Defective B cell responses in the absence of SH2D1A, Proceedings of the National Academy of Sciences of the United States of America, 102 (2005) 4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R, SAP is required for generating long-term humoral immunity, Nature, 421 (2003) 282–287. [DOI] [PubMed] [Google Scholar]
- [49].Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL, SH2D1A regulates T-dependent humoral autoimmunity, The Journal of experimental medicine, 200 (2004) 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Malbran A, Belmonte L, Ruibal-Ares B, Bare P, Massud I, Parodi C, Felippo M, Hodinka R, Haines K, Nichols KE, de Bracco MM, Loss of circulating CD27+ memory B cells and CCR4+ T cells occurring in association with elevated EBV loads in XLP patients surviving primary EBV infection, Blood, 103 (2004) 1625–1631. [DOI] [PubMed] [Google Scholar]
- [51].Ma CS, Hare NJ, Nichols KE, Dupre L, Andolfi G, Roncarolo MG, Adelstein S, Hodgkin PD, Tangye SG, Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells, The Journal of clinical investigation, 115 (2005) 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Detre C, Yigit B, Keszei M, Castro W, Magelky EM, Terhorst C, SAP modulates B cell functions in a genetic background-dependent manner, Immunology letters, 153 (2013) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dupre L, Andolfi G, Tangye SG, Clementi R, Locatelli F, Arico M, Aiuti A, Roncarolo MG, SAP controls the cytolytic activity of CD8+ T cells against EBV-infected cells, Blood, 105 (2005) 4383–4389. [DOI] [PubMed] [Google Scholar]
- [54].Chen G, Tai AK, Lin M, Chang F, Terhorst C, Huber BT, Signaling lymphocyte activation molecule-associated protein is a negative regulator of the CD8 T cell response in mice, Journal of immunology, 175 (2005) 2212–2218. [DOI] [PubMed] [Google Scholar]
- [55].Kim IJ, Burkum CE, Cookenham T, Schwartzberg PL, Woodland DL, Blackman MA, Perturbation of B cell activation in SLAM-associated protein-deficient mice is associated with changes in gammaherpesvirus latency reservoirs, Journal of immunology, 178 (2007) 1692–1701. [DOI] [PubMed] [Google Scholar]
- [56].Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL, Regulation of NKT cell development by SAP, the protein defective in XLP, Nature medicine, 11 (2005) 340–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R, Signaling lymphocytic activation molecule-associated protein controls NKT cell functions, Journal of immunology, 174 (2005) 3153–3157. [DOI] [PubMed] [Google Scholar]
- [58].Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, de Saint-Basile G, Latour S, Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product, The Journal of experimental medicine, 201 (2005) 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, Coyle AJ, Denny C, Yaffe MB, Engel P, Eck MJ, Terhorst C, Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells, The EMBO journal, 20 (2001) 5840–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cocks BG, Chang CC, Carballido JM, Yssel H, de Vries JE, Aversa G, A novel receptor involved in T-cell activation, Nature, 376 (1995) 260–263. [DOI] [PubMed] [Google Scholar]
- [61].Nanda N, Andre P, Bao M, Clauser K, Deguzman F, Howie D, Conley PB, Terhorst C, Phillips DR, Platelet aggregation induces platelet aggregate stability via SLAM family receptor signaling, Blood, 106 (2005) 3028–3034. [DOI] [PubMed] [Google Scholar]
- [62].Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C, The cell surface receptor SLAM controls T cell and macrophage functions, The Journal of experimental medicine, 199 (2004) 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kruse M, Meinl E, Henning G, Kuhnt C, Berchtold S, Berger T, Schuler G, Steinkasserer A, Signaling lymphocytic activation molecule is expressed on mature CD83+ dendritic cells and is up-regulated by IL-1 beta, Journal of immunology, 167 (2001) 1989–1995. [DOI] [PubMed] [Google Scholar]
- [64].Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ, SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells, Cell, 121 (2005) 1109–1121. [DOI] [PubMed] [Google Scholar]
- [65].De Salort J, Sintes J, Llinas L, Matesanz-Isabel J, Engel P, Expression of SLAM (CD150) cell-surface receptors on human B-cell subsets: from pro-B to plasma cells, Immunology letters, 134 (2011) 129–136. [DOI] [PubMed] [Google Scholar]
- [66].Gordiienko IM, Shlapatska LM, Kovalevska LM, Sidorenko SP, Differential expression of CD150/SLAMF1 in normal and malignant B cells on the different stages of maturation, Experimental oncology, 38 (2016) 101–107. [PubMed] [Google Scholar]
- [67].Howie D, Simarro M, Sayos J, Guirado M, Sancho J, Terhorst C, Molecular dissection of the signaling and costimulatory functions of CD150 (SLAM): CD150/SAP binding and CD150-mediated costimulation, Blood, 99 (2002) 957–965. [DOI] [PubMed] [Google Scholar]
- [68].Castro AG, Hauser TM, Cocks BG, Abrams J, Zurawski S, Churakova T, Zonin F, Robinson D, Tangye SG, Aversa G, Nichols KE, de Vries JE, Lanier LL, O'Garra A, Molecular and functional characterization of mouse signaling lymphocytic activation molecule (SLAM): differential expression and responsiveness in Th1 and Th2 cells, Journal of immunology, 163 (1999) 5860–5870. [PubMed] [Google Scholar]
- [69].Aversa G, Chang CC, Carballido JM, Cocks BG, de Vries JE, Engagement of the signaling lymphocytic activation molecule (SLAM) on activated T cells results in IL-2-independent, cyclosporin A-sensitive T cell proliferation and IFN-gamma production, Journal of immunology, 158 (1997) 4036–4044. [PubMed] [Google Scholar]
- [70].Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S, Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150), Journal of immunology, 185 (2010) 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bleharski JR, Niazi KR, Sieling PA, Cheng G, Modlin RL, Signaling lymphocytic activation molecule is expressed on CD40 ligand-activated dendritic cells and directly augments production of inflammatory cytokines, Journal of immunology, 167 (2001) 3174–3181. [DOI] [PubMed] [Google Scholar]
- [72].Kelekar A, Autophagy, Annals of the New York Academy of Sciences, 1066 (2005) 259–271. [DOI] [PubMed] [Google Scholar]
- [73].Yurchenko M, Skjesol A, Ryan L, Richard GM, Kandasamy RK, Wang N, Terhorst C, Husebye H, Espevik T, SLAMF1 is required for TLR4-mediated TRAM-TRIF-dependent signaling in human macrophages, The Journal of cell biology, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Theil D, Farina C, Meinl E, Differential expression of CD150 (SLAM) on monocytes and macrophages in chronic inflammatory contexts: abundant in Crohn's disease, but not in multiple sclerosis, Journal of clinical pathology, 58 (2005) 110–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].van Driel B, Liao G, Romero X, O'Keeffe MS, Wang G, Faubion WA, Berger SB, Magelky EM, Manocha M, Azcutia V, Grisham M, Luscinskas FW, Mizoguchi E, de Waal Malefyt R, Reinecker HC, Bhan AK, Wang N, Terhorst C, Signaling lymphocyte activation molecule regulates development of colitis in mice, Gastroenterology, 143 (2012) 1544–1554 e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Abadia-Molina AC, Ji H, Faubion WA, Julien A, Latchman Y, Yagita H, Sharpe A, Bhan AK, Terhorst C, CD48 controls T-cell and antigen-presenting cell functions in experimental colitis, Gastroenterology, 130 (2006) 424–434. [DOI] [PubMed] [Google Scholar]
- [77].Isomaki P, Aversa G, Cocks BG, Luukkainen R, Saario R, Toivanen P, de Vries JE, Punnonen J, Increased expression of signaling lymphocytic activation molecule in patients with rheumatoid arthritis and its role in the regulation of cytokine production in rheumatoid synovium, Journal of immunology, 159 (1997) 2986–2993. [PubMed] [Google Scholar]
- [78].Sawada S, Takei M, Epstein-Barr virus etiology in rheumatoid synovitis, Autoimmunity reviews, 4 (2005) 106–110. [DOI] [PubMed] [Google Scholar]
- [79].Hafler DA, Multiple sclerosis, The Journal of clinical investigation, 113 (2004) 788–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ferrante P, Fusi ML, Saresella M, Caputo D, Biasin M, Trabattoni D, Salvaggio A, Clerici E, de Vries JE, Aversa G, Cazzullo CL, Clerici M, Cytokine production and surface marker expression in acute and stable multiple sclerosis: altered IL-12 production and augmented signaling lymphocytic activation molecule (SLAM)-expressing lymphocytes in acute multiple sclerosis, Journal of immunology, 160 (1998) 1514–1521. [PubMed] [Google Scholar]
- [81].de la Fuente MA, Tovar V, Villamor N, Zapater N, Pizcueta P, Campo E, Bosch J, Engel P, Molecular characterization and expression of a novel human leukocyte cell-surface marker homologous to mouse Ly-9, Blood, 97 (2001) 3513–3520. [DOI] [PubMed] [Google Scholar]
- [82].Sandrin MS, Gumley TP, Henning MM, Vaughan HA, Gonez LJ, Trapani JA, McKenzie IF, Isolation and characterization of cDNA clones for mouse Ly-9, Journal of immunology, 149 (1992) 1636–1641. [PubMed] [Google Scholar]
- [83].Romero X, Zapater N, Calvo M, Kalko SG, de la Fuente MA, Tovar V, Ockeloen C, Pizcueta P, Engel P, CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse, Journal of immunology, 174 (2005) 7033–7042. [DOI] [PubMed] [Google Scholar]
- [84].Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR, T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells, Journal of immunology, 173 (2004) 68–78. [DOI] [PubMed] [Google Scholar]
- [85].Vinuesa CG, Tangye SG, Moser B, Mackay CR, Follicular B helper T cells in antibody responses and autoimmunity, Nature reviews. Immunology, 5 (2005) 853–865. [DOI] [PubMed] [Google Scholar]
- [86].Romero X, Benitez D, March S, Vilella R, Miralpeix M, Engel P, Differential expression of SAP and EAT-2-binding leukocyte cell-surface molecules CD84, CD150 (SLAM), CD229 (Ly9) and CD244 (2B4), Tissue antigens, 64 (2004) 132–144. [DOI] [PubMed] [Google Scholar]
- [87].Sintes J, Vidal-Laliena M, Romero X, Tovar V, Engel P, Characterization of mouse CD229 (Ly9), a leukocyte cell surface molecule of the CD150 (SLAM) family, Tissue antigens, 70 (2007) 355–362. [DOI] [PubMed] [Google Scholar]
- [88].Perez-Carmona N, Farre D, Martinez-Vicente P, Terhorst C, Engel P, Angulo A, Signaling Lymphocytic Activation Molecule Family Receptor Homologs in New World Monkey Cytomegaloviruses, Journal of virology, 89 (2015) 11323–11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Sayos J, Martin M, Chen A, Simarro M, Howie D, Morra M, Engel P, Terhorst C, Cell surface receptors Ly-9 and CD84 recruit the X-linked lymphoproliferative disease gene product SAP, Blood, 97 (2001) 3867–3874. [DOI] [PubMed] [Google Scholar]
- [90].Martin M, Del Valle JM, Saborit I, Engel P, Identification of Grb2 as a novel binding partner of the signaling lymphocytic activation molecule-associated protein binding receptor CD229, Journal of immunology, 174 (2005) 5977–5986. [DOI] [PubMed] [Google Scholar]
- [91].Del Valle JM, Engel P, Martin M, The cell surface expression of SAP-binding receptor CD229 is regulated via its interaction with clathrin-associated adaptor complex 2 (AP-2), The Journal of biological chemistry, 278 (2003) 17430–17437. [DOI] [PubMed] [Google Scholar]
- [92].Graham DB, Bell MP, McCausland MM, Huntoon CJ, van Deursen J, Faubion WA, Crotty S, McKean DJ, Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice, Journal of immunology, 176 (2006) 291–300. [DOI] [PubMed] [Google Scholar]
- [93].Sintes J, Cuenca M, Romero X, Bastos R, Terhorst C, Angulo A, Engel P, Cutting edge: Ly9 (CD229), a SLAM family receptor, negatively regulates the development of thymic innate memory-like CD8+ T and invariant NKT cells, Journal of immunology, 190 (2013) 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Cuenca M, Romero X, Sintes J, Terhorst C, Engel P, Targeting of Ly9 (CD229) Disrupts Marginal Zone and B1 B Cell Homeostasis and Antibody Responses, Journal of immunology, 196 (2016) 726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].de Salort J, Cuenca M, Terhorst C, Engel P, Romero X, Ly9 (CD229) Cell-Surface Receptor is Crucial for the Development of Spontaneous Autoantibody Production to Nuclear Antigens, Front Immunol, 4 (2013) 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Peck SR, Ruley HE, Ly108: a new member of the mouse CD2 family of cell surface proteins, Immunogenetics, 52 (2000) 63–72. [DOI] [PubMed] [Google Scholar]
- [97].Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR Jr., Morel L, Wakeland EK, Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus, Immunity, 21 (2004) 769–780. [DOI] [PubMed] [Google Scholar]
- [98].Keszei M, Detre C, Rietdijk ST, Munoz P, Romero X, Berger SB, Calpe S, Liao G, Castro W, Julien A, Wu YY, Shin DM, Sancho J, Zubiaur M, Morse HC 3rd, Morel L, Engel P, Wang N, Terhorst C, A novel isoform of the Ly108 gene ameliorates murine lupus, The Journal of experimental medicine, 208 (2011) 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Veillette A, SLAM-family receptors: immune regulators with or without SAP-family adaptors, Cold Spring Harbor perspectives in biology, 2 (2010) a002469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Cannons JL, Tangye SG, Schwartzberg PL, SLAM family receptors and SAP adaptors in immunity, Annual review of immunology, 29 (2011) 665–705. [DOI] [PubMed] [Google Scholar]
- [101].Li C, Iosef C, Jia CY, Han VK, Li SS, Dual functional roles for the X-linked lymphoproliferative syndrome gene product SAP/SH2D1A in signaling through the signaling lymphocyte activation molecule (SLAM) family of immune receptors, The Journal of biological chemistry, 278 (2003) 3852–3859. [DOI] [PubMed] [Google Scholar]
- [102].Latour S, Roncagalli R, Chen R, Bakinowski M, Shi X, Schwartzberg PL, Davidson D, Veillette A, Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation, Nature cell biology, 5 (2003) 149–154. [DOI] [PubMed] [Google Scholar]
- [103].Flaig RM, Stark S, Watzl C, Cutting edge: NTB-A activates NK cells via homophilic interaction, Journal of immunology, 172 (2004) 6524–6527. [DOI] [PubMed] [Google Scholar]
- [104].Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodriguez J, Cheng J, Wakeland EK, Germain RN, Schwartzberg PL, Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84, Immunity, 32 (2010) 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A, Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development, Immunity, 27 (2007) 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Falco M, Marcenaro E, Romeo E, Bellora F, Marras D, Vely F, Ferracci G, Moretta L, Moretta A, Bottino C, Homophilic interaction of NTBA, a member of the CD2 molecular family: induction of cytotoxicity and cytokine release in human NK cells, European journal of immunology, 34 (2004) 1663–1672. [DOI] [PubMed] [Google Scholar]
- [107].Korver W, Singh S, Liu S, Zhao X, Yonkovich S, Sweeney A, Anton K, Lomas WE 3rd, Greenwood R, Smith A, Tran DH, Shinkawa P, Jimenez M, Yeung P, Aguilar G, Palencia S, Vatta P, Mueller M, Zhan X, Newton EM, Liu Y, Zhao J, Emtage P, Levy MD, Hsi ED, Funk WD, Abo A, The lymphoid cell surface receptor NTB-A: a novel monoclonal antibody target for leukaemia and lymphoma therapeutics, British journal of haematology, 137 (2007) 307–318. [DOI] [PubMed] [Google Scholar]
- [108].Shah AH, Sowrirajan B, Davis ZB, Ward JP, Campbell EM, Planelles V, Barker E, Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu, Cell host & microbe, 8 (2010) 397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Duev-Cohen A, Bar-On Y, Glasner A, Berhani O, Ophir Y, Levi-Schaffer F, Mandelboim M, Mandelboim O, The human 2B4 and NTB-A receptors bind the influenza viral hemagglutinin and co-stimulate NK cell cytotoxicity, Oncotarget, 7 (2016) 13093–13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Lucas PJ, Negishi I, Nakayama K, Fields LE, Loh DY, Naive CD28-deficient T cells can initiate but not sustain an in vitro antigen-specific immune response, Journal of immunology, 154 (1995) 5757–5768. [PubMed] [Google Scholar]
- [111].Shahinian A, Pfeffer K, Lee KP, Kundig TM, Kishihara K, Wakeham A, Kawai K, Ohashi PS, Thompson CB, Mak TW, Differential T cell costimulatory requirements in CD28-deficient mice, Science, 261 (1993) 609–612. [DOI] [PubMed] [Google Scholar]
- [112].Valdez PA, Wang H, Seshasayee D, van Lookeren Campagne M, Gurney A, Lee WP, Grewal IS, NTB-A, a new activating receptor in T cells that regulates autoimmune disease, The Journal of biological chemistry, 279 (2004) 18662–18669. [DOI] [PubMed] [Google Scholar]
- [113].H.P. wang N, Yigit B, Zhao H, Keeffe MSO, Sage P, Sharpe A, Terhorst C, Negative Regulation of humoral immunity due to interplay between the SLAMF1, SLAMF5, and SLAMF6 receptors, Front. Immunol, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wang N, Keszei M, Halibozek P, Yigit B, Engel P, Terhorst C, Slamf6 negatively regulates autoimmunity, Clinical immunology, 173 (2016) 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Howie D, Laroux FS, Morra M, Satoskar AR, Rosas LE, Faubion WA, Julien A, Rietdijk S, Coyle AJ, Fraser C, Terhorst C, Cutting edge: the SLAM family receptor Ly108 controls T cell and neutrophil functions, Journal of immunology, 174 (2005) 5931–5935. [DOI] [PubMed] [Google Scholar]
- [116].Snow AL, Pandiyan P, Zheng L, Krummey SM, Lenardo MJ, The power and the promise of restimulation-induced cell death in human immune diseases, Immunological reviews, 236 (2010) 68–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Snow AL, Marsh RA, Krummey SM, Roehrs P, Young LR, Zhang K, van Hoff J, Dhar D, Nichols KE, Filipovich AH, Su HC, Bleesing JJ, Lenardo MJ, Restimulation-induced apoptosis of T cells is impaired in patients with X-linked lymphoproliferative disease caused by SAP deficiency, The Journal of clinical investigation, 119 (2009) 2976–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Katz G, Krummey SM, Larsen SE, Stinson JR, Snow AL, SAP facilitates recruitment and activation of LCK at NTB-A receptors during restimulation-induced cell death, Journal of immunology, 192 (2014) 4202–4209. [DOI] [PubMed] [Google Scholar]
- [119].Uzana R, Eisenberg G, Sagi Y, Frankenburg S, Merims S, Amariglio N, Yefenof E, Peretz T, Machlenkin A, Lotem M, Trogocytosis is a gateway to characterize functional diversity in melanoma-specific CD8+ T cell clones, Journal of immunology, 188 (2012) 632–640. [DOI] [PubMed] [Google Scholar]
- [120].Joly E, Hudrisier D, What is trogocytosis and what is its purpose?, Nature immunology, 4 (2003) 815. [DOI] [PubMed] [Google Scholar]
- [121].Sharifi R, Sinclair JC, Gilmour KC, Arkwright PD, Kinnon C, Thrasher AJ, Gaspar HB, SAP mediates specific cytotoxic T-cell functions in X-linked lymphoproliferative disease, Blood, 103 (2004) 3821–3827. [DOI] [PubMed] [Google Scholar]
- [122].Hislop AD, Palendira U, Leese AM, Arkwright PD, Rohrlich PS, Tangye SG, Gaspar HB, Lankester AC, Moretta A, Rickinson AB, Impaired Epstein-Barr virus-specific CD8+ T-cell function in X-linked lymphoproliferative disease is restricted to SLAM family-positive B-cell targets, Blood, 116 (2010) 3249–3257. [DOI] [PubMed] [Google Scholar]
- [123].Palendira U, Low C, Chan A, Hislop AD, Ho E, Phan TG, Deenick E, Cook MC, Riminton DS, Choo S, Loh R, Alvaro F, Booth C, Gaspar HB, Moretta A, Khanna R, Rickinson AB, Tangye SG, Molecular pathogenesis of EBV susceptibility in XLP as revealed by analysis of female carriers with heterozygous expression of SAP, PLoS biology, 9 (2011) e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN, SAP-controlled T-B cell interactions underlie germinal centre formation, Nature, 455 (2008) 764–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Zhao F, Cannons JL, Dutta M, Griffiths GM, Schwartzberg PL, Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis, Immunity, 36 (2012) 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S, The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development, Immunity, 36 (2012) 986–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Bottino C, Falco M, Parolini S, Marcenaro E, Augugliaro R, Sivori S, Landi E, Biassoni R, Notarangelo LD, Moretta L, Moretta A, NTB-A [correction of GNTB-A], a novel SH2D1A-associated surface molecule contributing to the inability of natural killer cells to kill Epstein-Barr virus-infected B cells in X-linked lymphoproliferative disease, The Journal of experimental medicine, 194 (2001) 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Eissmann P, Watzl C, Molecular analysis of NTB-A signaling: a role for EAT-2 in NTB-A-mediated activation of human NK cells, Journal of immunology, 177 (2006) 3170–3177. [DOI] [PubMed] [Google Scholar]
- [129].Stark S, Watzl C, 2B4 (CD244), NTB-A and CRACC (CS1) stimulate cytotoxicity but no proliferation in human NK cells, International immunology, 18 (2006) 241–247. [DOI] [PubMed] [Google Scholar]
- [130].Wu N, Zhong MC, Roncagalli R, Perez-Quintero LA, Guo H, Zhang Z, Lenoir C, Dong Z, Latour S, Veillette A, A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education, Nature immunology, 17 (2016) 387–396. [DOI] [PubMed] [Google Scholar]
- [131].Bendelac A, Savage PB, Teyton L, The biology of NKT cells, Annual review of immunology, 25 (2007) 297–336. [DOI] [PubMed] [Google Scholar]
- [132].Starr TK, Jameson SC, Hogquist KA, Positive and negative selection of T cells, Annual review of immunology, 21 (2003) 139–176. [DOI] [PubMed] [Google Scholar]
- [133].Berg LJ, Signaling pathways that regulate T cell development and differentiation, Journal of immunology, 189 (2012) 5487–5488. [DOI] [PubMed] [Google Scholar]
- [134].Berg LJ, Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development, Nature reviews. Immunology, 7 (2007) 479–485. [DOI] [PubMed] [Google Scholar]
- [135].Detre C, Keszei M, Garrido-Mesa N, Kis-Toth K, Castro W, Agyemang AF, Veerapen N, Besra GS, Carroll MC, Tsokos GC, Wang N, Leadbetter EA, Terhorst C, SAP expression in invariant NKT cells is required for cognate help to support B-cell responses, Blood, 120 (2012) 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Dutta M, Kraus ZJ, Gomez-Rodriguez J, Hwang SH, Cannons JL, Cheng J, Lee SY, Wiest DL, Wakeland EK, Schwartzberg PL, A role for Ly108 in the induction of promyelocytic zinc finger transcription factor in developing thymocytes, Journal of immunology, 190 (2013) 2121–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant'Angelo DB, The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions, Nature immunology, 9 (2008) 1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A, The transcription factor PLZF directs the effector program of the NKT cell lineage, Immunity, 29 (2008) 391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].De Calisto J, Wang N, Wang G, Yigit B, Engel P, Terhorst C, SAP-Dependent and -Independent Regulation of Innate T Cell Development Involving SLAMF Receptors, Front Immunol, 5 (2014) 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Tsokos GC, Systemic lupus erythematosus, The New England journal of medicine, 365 (2011) 2110–2121. [DOI] [PubMed] [Google Scholar]
- [141].Moulton VR, Suarez-Fueyo A, Meidan E, Li H, Mizui M, Tsokos GC, Pathogenesis of Human Systemic Lupus Erythematosus: A Cellular Perspective, Trends in molecular medicine, 23 (2017) 615–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Limaye N, Belobrajdic KA, Wandstrat AE, Bonhomme F, Edwards SV, Wakeland EK, Prevalence and evolutionary origins of autoimmune susceptibility alleles in natural mouse populations, Genes and immunity, 9 (2008) 61–68. [DOI] [PubMed] [Google Scholar]
- [143].Morel L, Blenman KR, Croker BP, Wakeland EK, The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes, Proceedings of the National Academy of Sciences of the United States of America, 98 (2001) 1787–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Shmerling RH, Autoantibodies in systemic lupus erythematosus--there before you know it, The New England journal of medicine, 349 (2003) 1499–1500. [DOI] [PubMed] [Google Scholar]
- [145].Zhong MC, Veillette A, Control of T lymphocyte signaling by Ly108, a signaling lymphocytic activation molecule family receptor implicated in autoimmunity, The Journal of biological chemistry, 283 (2008) 19255–19264. [DOI] [PubMed] [Google Scholar]
- [146].Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C, Regulation of B cell tolerance by the lupus susceptibility gene Ly108, Science, 312 (2006) 1665–1669. [DOI] [PubMed] [Google Scholar]
- [147].Keszei M, Detre C, Castro W, Magelky E, O'Keeffe M, Kis-Toth K, Tsokos GC, Wang N, Terhorst C, Expansion of an osteopontin-expressing T follicular helper cell subset correlates with autoimmunity in B6.Sle1b mice and is suppressed by the H1-isoform of the Slamf6 receptor, FASEB journal : official publication of the Federation of American Societies for Experimental Biology, 27 (2013) 3123–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].McArdel SL, Terhorst C, Sharpe AH, Roles of CD48 in regulating immunity and tolerance, Clinical immunology, 164 (2016) 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Arulanandam AR, Moingeon P, Concino MF, Recny MA, Kato K, Yagita H, Koyasu S, Reinherz EL, A soluble multimeric recombinant CD2 protein identifies CD48 as a low affinity ligand for human CD2: divergence of CD2 ligands during the evolution of humans and mice, The Journal of experimental medicine, 177 (1993) 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Tissot C, Rebouissou C, Klein B, Mechti N, Both human alpha/beta and gamma interferons upregulate the expression of CD48 cell surface molecules, Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research, 17 (1997) 17–26. [DOI] [PubMed] [Google Scholar]
- [151].Kis-Toth K, Tsokos GC, Engagement of SLAMF2/CD48 prolongs the time frame of effective T cell activation by supporting mature dendritic cell survival, Journal of immunology, 192 (2014) 4436–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Moran M, Miceli MC, Engagement of GPI-linked CD48 contributes to TCR signals and cytoskeletal reorganization: a role for lipid rafts in T cell activation, Immunity, 9 (1998) 787–796. [DOI] [PubMed] [Google Scholar]
- [153].Patel VP, Moran M, Low TA, Miceli MC, A molecular framework for two-step T cell signaling: Lck Src homology 3 mutations discriminate distinctly regulated lipid raft reorganization events, Journal of immunology, 166 (2001) 754–764. [DOI] [PubMed] [Google Scholar]
- [154].Milstein O, Tseng SY, Starr T, Llodra J, Nans A, Liu M, Wild MK, van der Merwe PA, Stokes DL, Reisner Y, Dustin ML, Nanoscale increases in CD2-CD48-mediated intermembrane spacing decrease adhesion and reorganize the immunological synapse, The Journal of biological chemistry, 283 (2008) 34414–34422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Chavin KD, Qin L, Lin J, Woodward J, Baliga P, Kato K, Yagita H, Bromberg JS, Anti-CD48 (murine CD2 ligand) mAbs suppress cell mediated immunity in vivo, International immunology, 6 (1994) 701–709. [DOI] [PubMed] [Google Scholar]
- [156].Garni-Wagner BA, Purohit A, Mathew PA, Bennett M, Kumar V, A novel function-associated molecule related to non-MHC-restricted cytotoxicity mediated by activated natural killer cells and T cells, Journal of immunology, 151 (1993) 60–70. [PubMed] [Google Scholar]
- [157].Nakajima H, Cella M, Langen H, Friedlein A, Colonna M, Activating interactions in human NK cell recognition: the role of 2B4-CD48, European journal of immunology, 29 (1999) 1676–1683. [DOI] [PubMed] [Google Scholar]
- [158].Tangye SG, Cherwinski H, Lanier LL, Phillips JH, 2B4-mediated activation of human natural killer cells, Molecular immunology, 37 (2000) 493–501. [DOI] [PubMed] [Google Scholar]
- [159].Tangye SG, Phillips JH, Lanier LL, Nichols KE, Functional requirement for SAP in 2B4-mediated activation of human natural killer cells as revealed by the X-linked lymphoproliferative syndrome, Journal of immunology, 165 (2000) 2932–2936. [DOI] [PubMed] [Google Scholar]
- [160].Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C, Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244), Blood, 105 (2005) 4722–4729. [DOI] [PubMed] [Google Scholar]
- [161].Waggoner SN, Kumar V, Evolving role of 2B4/CD244 in T and NK cell responses during virus infection, Frontiers in immunology, 3 (2012) 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].de la Fuente MA, Pizcueta P, Nadal M, Bosch J, Engel P, CD84 leukocyte antigen is a new member of the Ig superfamily, Blood, 90 (1997) 2398–2405. [PubMed] [Google Scholar]
- [163].Martin M, Romero X, de la Fuente MA, Tovar V, Zapater N, Esplugues E, Pizcueta P, Bosch J, Engel P, CD84 functions as a homophilic adhesion molecule and enhances IFN-gamma secretion: adhesion is mediated by Ig-like domain 1, Journal of immunology, 167 (2001) 3668–3676. [DOI] [PubMed] [Google Scholar]
- [164].Zaiss M, Hirtreiter C, Rehli M, Rehm A, Kunz-Schughart LA, Andreesen R, Hennemann B, CD84 expression on human hematopoietic progenitor cells, Experimental hematology, 31 (2003) 798–805. [DOI] [PubMed] [Google Scholar]
- [165].Tangye SG, van de Weerdt BC, Avery DT, Hodgkin PD, CD84 is up-regulated on a major population of human memory B cells and recruits the SH2 domain containing proteins SAP and EAT-2, European journal of immunology, 32 (2002) 1640–1649. [DOI] [PubMed] [Google Scholar]
- [166].Tangye SG, Nichols KE, Hare NJ, van de Weerdt BC, Functional requirements for interactions between CD84 and Src homology 2 domain-containing proteins and their contribution to human T cell activation, Journal of immunology, 171 (2003) 2485–2495. [DOI] [PubMed] [Google Scholar]
- [167].Sintes J, Romero X, de Salort J, Terhorst C, Engel P, Mouse CD84 is a pan-leukocyte cell-surface molecule that modulates LPS-induced cytokine secretion by macrophages, Journal of leukocyte biology, 88 (2010) 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Hsi ED, Steinle R, Balasa B, Szmania S, Draksharapu A, Shum BP, Huseni M, Powers D, Nanisetti A, Zhang Y, Rice AG, van Abbema A, Wong M, Liu G, Zhan F, Dillon M, Chen S, Rhodes S, Fuh F, Tsurushita N, Kumar S, Vexler V, Shaughnessy JD Jr., Barlogie B, van Rhee F, Hussein M, Afar DE, Williams MB, CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma, Clinical cancer research : an official journal of the American Association for Cancer Research, 14 (2008) 2775–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Cruz-Munoz ME, Dong Z, Shi X, Zhang S, Veillette A, Influence of CRACC, a SLAM family receptor coupled to the adaptor EAT-2, on natural killer cell function, Nature immunology, 10 (2009) 297–305. [DOI] [PubMed] [Google Scholar]
- [170].Bouchon A, Cella M, Grierson HL, Cohen JI, Colonna M, Activation of NK cell-mediated cytotoxicity by a SAP-independent receptor of the CD2 family, Journal of immunology, 167 (2001) 5517–5521. [DOI] [PubMed] [Google Scholar]
- [171].Tassi I, Colonna M, The cytotoxicity receptor CRACC (CS-1) recruits EAT-2 and activates the PI3K and phospholipase Cgamma signaling pathways in human NK cells, Journal of immunology, 175 (2005) 7996–8002. [DOI] [PubMed] [Google Scholar]
- [172].Lee JK, Boles KS, Mathew PA, Molecular and functional characterization of a CS1 (CRACC) splice variant expressed in human NK cells that does not contain immunoreceptor tyrosine-based switch motifs, European journal of immunology, 34 (2004) 2791–2799. [DOI] [PubMed] [Google Scholar]
- [173].Kim JR, Horton NC, Mathew SO, Mathew PA, CS1 (SLAMF7) inhibits production of proinflammatory cytokines by activated monocytes, Inflammation research : official journal of the European Histamine Research Society … [et al.], 62 (2013) 765–772. [DOI] [PubMed] [Google Scholar]
- [174].Lee JK, Mathew SO, Vaidya SV, Kumaresan PR, Mathew PA, CS1 (CRACC, CD319) induces proliferation and autocrine cytokine expression on human B lymphocytes, Journal of immunology, 179 (2007) 4672–4678. [DOI] [PubMed] [Google Scholar]
- [175].Kingsbury GA, Feeney LA, Nong Y, Calandra SA, Murphy CJ, Corcoran JM, Wang Y, Prabhu Das MR, Busfield SJ, Fraser CC, Villeval JL, Cloning, expression, and function of BLAME, a novel member of the CD2 family, Journal of immunology, 166 (2001) 5675–5680. [DOI] [PubMed] [Google Scholar]
- [176].Wang G, Abadia-Molina AC, Berger SB, Romero X, O'Keeffe MS, Rojas-Barros DI, Aleman M, Liao G, Maganto-Garcia E, Fresno M, Wang N, Detre C, Terhorst C, Cutting edge: Slamf8 is a negative regulator of Nox2 activity in macrophages, Journal of immunology, 188 (2012) 5829–5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Wang G, van Driel BJ, Liao G, O'Keeffe MS, Halibozek PJ, Flipse J, Yigit B, Azcutia V, Luscinskas FW, Wang N, Terhorst C, Migration of myeloid cells during inflammation is differentially regulated by the cell surface receptors Slamf1 and Slamf8, PloS one, 10 (2015) e0121968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Bachireddy P, Burkhardt UE, Rajasagi M, Wu CJ, Haematological malignancies: at the forefront of immunotherapeutic innovation, Nature reviews. Cancer, 15 (2015) 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Ten Hacken E, Burger JA, Microenvironment interactions and B-cell receptor signaling in Chronic Lymphocytic Leukemia: Implications for disease pathogenesis and treatment, Biochimica et biophysica acta, 1863 (2016) 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Chiorazzi N, Rai KR, Ferrarini M, Chronic lymphocytic leukemia, The New England journal of medicine, 352 (2005) 804–815. [DOI] [PubMed] [Google Scholar]
- [181].Damle RN, Ghiotto F, Valetto A, Albesiano E, Fais F, Yan XJ, Sison CP, Allen SL, Kolitz J, Schulman P, Vinciguerra VP, Budde P, Frey J, Rai KR, Ferrarini M, Chiorazzi N, B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes, Blood, 99 (2002) 4087–4093. [DOI] [PubMed] [Google Scholar]
- [182].Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H, Freedman A, Inghirami G, Cro L, Baldini L, Neri A, Califano A, Dalla-Favera R, Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells, The Journal of experimental medicine, 194 (2001) 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Seifert M, Sellmann L, Bloehdorn J, Wein F, Stilgenbauer S, Durig J, Kuppers R, Cellular origin and pathophysiology of chronic lymphocytic leukemia, The Journal of experimental medicine, 209 (2012) 2183–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, Buchbinder A, Budman D, Dittmar K, Kolitz J, Lichtman SM, Schulman P, Vinciguerra VP, Rai KR, Ferrarini M, Chiorazzi N, Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia, Blood, 94 (1999) 1840–1847. [PubMed] [Google Scholar]
- [185].Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK, Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia, Blood, 94 (1999) 1848–1854. [PubMed] [Google Scholar]
- [186].Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, Dohner K, Bentz M, Lichter P, Genomic aberrations and survival in chronic lymphocytic leukemia, The New England journal of medicine, 343 (2000) 1910–1916. [DOI] [PubMed] [Google Scholar]
- [187].Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R, The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia, Cancer cell, 17 (2010) 28–40. [DOI] [PubMed] [Google Scholar]
- [188].Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S, From pathogenesis to treatment of chronic lymphocytic leukaemia, Nature reviews. Cancer, 10 (2010) 37–50. [DOI] [PubMed] [Google Scholar]
- [189].Crespo M, Bosch F, Villamor N, Bellosillo B, Colomer D, Rozman M, Marce S, Lopez-Guillermo A, Campo E, Montserrat E, ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia, The New England journal of medicine, 348 (2003) 1764–1775. [DOI] [PubMed] [Google Scholar]
- [190].Wiestner A, Rosenwald A, Barry TS, Wright G, Davis RE, Henrickson SE, Zhao H, Ibbotson RE, Orchard JA, Davis Z, Stetler-Stevenson M, Raffeld M, Arthur DC, Marti GE, Wilson WH, Hamblin TJ, Oscier DG, Staudt LM, ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile, Blood, 101 (2003) 4944–4951. [DOI] [PubMed] [Google Scholar]
- [191].Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, Cresta S, Gargiulo E, Forconi F, Guarini A, Arcaini L, Paulli M, Laurenti L, Larocca LM, Marasca R, Gattei V, Oscier D, Bertoni F, Mullighan CG, Foa R, Pasqualucci L, Rabadan R, Dalla-Favera R, Gaidano G, Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation, The Journal of experimental medicine, 208 (2011) 1389–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, Bassaganyas L, Baumann T, Juan M, Lopez-Guerra M, Colomer D, Tubio JM, Lopez C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernandez JM, Puente DA, Freije JM, Velasco G, Gutierrez-Fernandez A, Costa D, Carrio A, Guijarro S, Enjuanes A, Hernandez L, Yague J, Nicolas P, Romeo-Casabona CM, Himmelbauer H, Castillo E, Dohm JC, de Sanjose S, Piris MA, de Alava E, San Miguel J, Royo R, Gelpi JL, Torrents D, Orozco M, Pisano DG, Valencia A, Guigo R, Bayes M, Heath S, Gut M, Klatt P, Marshall J, Raine K, Stebbings LA, Futreal PA, Stratton MR, Campbell PJ, Gut I, Lopez-Guillermo A, Estivill X, Montserrat E, Lopez-Otin C, Campo E, Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia, Nature, 475 (2011) 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E, Diop M, Scourzic L, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Kikushige Y, Davi F, Lambert J, Gautheret D, Merle-Beral H, Sutton L, Dessen P, Solary E, Akashi K, Vainchenker W, Mercher T, Droin N, Ogawa S, Nguyen-Khac F, Bernard OA, Acquired initiating mutations in early hematopoietic cells of CLL patients, Cancer discovery, 4 (2014) 1088–1101. [DOI] [PubMed] [Google Scholar]
- [194].Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, Zhang W, Vartanov AR, Fernandes SM, Goldstein NR, Folco EG, Cibulskis K, Tesar B, Sievers QL, Shefler E, Gabriel S, Hacohen N, Reed R, Meyerson M, Golub TR, Lander ES, Neuberg D, Brown JR, Getz G, Wu CJ, SF3B1 and other novel cancer genes in chronic lymphocytic leukemia, The New England journal of medicine, 365 (2011) 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Panayiotidis P, Jones D, Ganeshaguru K, Foroni L, Hoffbrand AV, Human bone marrow stromal cells prevent apoptosis and support the survival of chronic lymphocytic leukaemia cells in vitro, British journal of haematology, 92 (1996) 97–103. [DOI] [PubMed] [Google Scholar]
- [196].Lagneaux L, Delforge A, Bron D, De Bruyn C, Stryckmans P, Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells, Blood, 91 (1998) 2387–2396. [PubMed] [Google Scholar]
- [197].Burger JA, Tsukada N, Burger M, Zvaifler NJ, Dell'Aquila M, Kipps TJ, Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1, Blood, 96 (2000) 2655–2663. [PubMed] [Google Scholar]
- [198].Burkle A, Niedermeier M, Schmitt-Graff A, Wierda WG, Keating MJ, Burger JA, Overexpression of the CXCR5 chemokine receptor, and its ligand, CXCL13 in B-cell chronic lymphocytic leukemia, Blood, 110 (2007) 3316–3325. [DOI] [PubMed] [Google Scholar]
- [199].Vaisitti T, Aydin S, Rossi D, Cottino F, Bergui L, D'Arena G, Bonello L, Horenstein AL, Brennan P, Pepper C, Gaidano G, Malavasi F, Deaglio S, CD38 increases CXCL12-mediated signals and homing of chronic lymphocytic leukemia cells, Leukemia, 24 (2010) 958–969. [DOI] [PubMed] [Google Scholar]
- [200].Deaglio S, Aydin S, Grand MM, Vaisitti T, Bergui L, D'Arena G, Chiorino G, Malavasi F, CD38/CD31 interactions activate genetic pathways leading to proliferation and migration in chronic lymphocytic leukemia cells, Molecular medicine, 16 (2010) 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Burger JA, Quiroga MP, Hartmann E, Burkle A, Wierda WG, Keating MJ, Rosenwald A, High-level expression of the T-cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation, Blood, 113 (2009) 3050–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Burger JA, Chiorazzi N, B cell receptor signaling in chronic lymphocytic leukemia, Trends in immunology, 34 (2013) 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Shaffer AL 3rd, Young RM, Staudt LM, Pathogenesis of human B cell lymphomas, Annual review of immunology, 30 (2012) 565–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Allsup DJ, Kamiguti AS, Lin K, Sherrington PD, Matrai Z, Slupsky JR, Cawley JC, Zuzel M, B-cell receptor translocation to lipid rafts and associated signaling differ between prognostically important subgroups of chronic lymphocytic leukemia, Cancer research, 65 (2005) 7328–7337. [DOI] [PubMed] [Google Scholar]
- [205].Binder M, Muller F, Jackst A, Lechenne B, Pantic M, Bacher U, Zu Eulenburg C, Veelken H, Mertelsmann R, Pasqualini R, Arap W, Trepel M, B-cell receptor epitope recognition correlates with the clinical course of chronic lymphocytic leukemia, Cancer, 117 (2011) 1891–1900. [DOI] [PubMed] [Google Scholar]
- [206].Muzio M, Apollonio B, Scielzo C, Frenquelli M, Vandoni I, Boussiotis V, Caligaris-Cappio F, Ghia P, Constitutive activation of distinct BCR-signaling pathways in a subset of CLL patients: a molecular signature of anergy, Blood, 112 (2008) 188–195. [DOI] [PubMed] [Google Scholar]
- [207].Apollonio B, Scielzo C, Bertilaccio MT, Ten Hacken E, Scarfo L, Ranghetti P, Stevenson F, Packham G, Ghia P, Muzio M, Caligaris-Cappio F, Targeting B-cell anergy in chronic lymphocytic leukemia, Blood, 121 (2013) 3879–3888, S3871-3878. [DOI] [PubMed] [Google Scholar]
- [208].Kitada S, Zapata JM, Andreeff M, Reed JC, Bryostatin and CD40-ligand enhance apoptosis resistance and induce expression of cell survival genes in B-cell chronic lymphocytic leukaemia, British journal of haematology, 106 (1999) 995–1004. [DOI] [PubMed] [Google Scholar]
- [209].Tinhofer I, Marschitz I, Kos M, Henn T, Egle A, Villunger A, Greil R, Differential sensitivity of CD4+ and CD8+ T lymphocytes to the killing efficacy of Fas (Apo-1/CD95) ligand+ tumor cells in B chronic lymphocytic leukemia, Blood, 91 (1998) 4273–4281. [PubMed] [Google Scholar]
- [210].Goolsby CL, Kuchnio M, Finn WG, Peterson L, Expansions of clonal and oligoclonal T cells in B-cell chronic lymphocytic leukemia are primarily restricted to the CD3(+)CD8(+) T-cell population, Cytometry, 42 (2000) 188–195. [PubMed] [Google Scholar]
- [211].Serrano D, Monteiro J, Allen SL, Kolitz J, Schulman P, Lichtman SM, Buchbinder A, Vinciguerra VP, Chiorazzi N, Gregersen PK, Clonal expansion within the CD4+CD57+ and CD8+CD57+ T cell subsets in chronic lymphocytic leukemia, Journal of immunology, 158 (1997) 1482–1489. [PubMed] [Google Scholar]
- [212].Mellstedt H, Choudhury A, T and B cells in B-chronic lymphocytic leukaemia: Faust, Mephistopheles and the pact with the Devil, Cancer immunology, immunotherapy : CII, 55 (2006) 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Gorgun G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG, Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells, The Journal of clinical investigation, 115 (2005) 1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].D'Arena G, Laurenti L, Minervini MM, Deaglio S, Bonello L, De Martino L, De Padua L, Savino L, Tarnani M, De Feo V, Cascavilla N, Regulatory T-cell number is increased in chronic lymphocytic leukemia patients and correlates with progressive disease, Leukemia research, 35 (2011) 363–368. [DOI] [PubMed] [Google Scholar]
- [215].Giannopoulos K, Schmitt M, Kowal M, Wlasiuk P, Bojarska-Junak A, Chen J, Rolinski J, Dmoszynska A, Characterization of regulatory T cells in patients with B-cell chronic lymphocytic leukemia, Oncology reports, 20 (2008) 677–682. [PubMed] [Google Scholar]
- [216].Dasgupta A, Mahapatra M, Saxena R, A study for proposal of use of regulatory T cells as a prognostic marker and establishing an optimal threshold level for their expression in chronic lymphocytic leukemia, Leukemia & lymphoma, 56 (2015) 1831–1838. [DOI] [PubMed] [Google Scholar]
- [217].Wherry EJ, T cell exhaustion, Nature immunology, 12 (2011) 492–499. [DOI] [PubMed] [Google Scholar]
- [218].Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W, Byrd JC, Gribben JG, Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug, The Journal of clinical investigation, 118 (2008) 2427–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Palma M, Gentilcore G, Heimersson K, Mozaffari F, Nasman-Glaser B, Young E, Rosenquist R, Hansson L, Osterborg A, Mellstedt H, T cells in chronic lymphocytic leukemia display dysregulated expression of immune checkpoints and activation markers, Haematologica, 102 (2017) 562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Nunes C, Wong R, Mason M, Fegan C, Man S, Pepper C, Expansion of a CD8(+)PD-1(+) replicative senescence phenotype in early stage CLL patients is associated with inverted CD4:CD8 ratios and disease progression, Clinical cancer research : an official journal of the American Association for Cancer Research, 18 (2012) 678–687. [DOI] [PubMed] [Google Scholar]
- [221].Tonino SH, van de Berg PJ, Yong SL, ten Berge IJ, Kersten MJ, van Lier RA, van Oers MH, Kater AP, Expansion of effector T cells associated with decreased PD-1 expression in patients with indolent B cell lymphomas and chronic lymphocytic leukemia, Leukemia & lymphoma, 53 (2012) 1785–1794. [DOI] [PubMed] [Google Scholar]
- [222].Gothert JR, Eisele L, Klein-Hitpass L, Weber S, Zesewitz ML, Sellmann L, Roth A, Pircher H, Duhrsen U, Durig J, Expanded CD8+ T cells of murine and human CLL are driven into a senescent KLRG1+ effector memory phenotype, Cancer immunology, immunotherapy : CII, 62 (2013) 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, Nayak L, Moss PA, Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals, Journal of immunology, 169 (2002) 1984–1992. [DOI] [PubMed] [Google Scholar]
- [224].Mackus WJ, Frakking FN, Grummels A, Gamadia LE, De Bree GJ, Hamann D, Van Lier RA, Van Oers MH, Expansion of CMV-specific CD8+CD45RA+CD27− T cells in B-cell chronic lymphocytic leukemia, Blood, 102 (2003) 1057–1063. [DOI] [PubMed] [Google Scholar]
- [225].Pourgheysari B, Bruton R, Parry H, Billingham L, Fegan C, Murray J, Moss P, The number of cytomegalovirus-specific CD4+ T cells is markedly expanded in patients with B-cell chronic lymphocytic leukemia and determines the total CD4+ T-cell repertoire, Blood, 116 (2010) 2968–2974. [DOI] [PubMed] [Google Scholar]
- [226].Riches JC, Davies JK, McClanahan F, Fatah R, Iqbal S, Agrawal S, Ramsay AG, Gribben JG, T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production, Blood, 121 (2013) 1612–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].te Raa GD, Pascutti MF, Garcia-Vallejo JJ, Reinen E, Remmerswaal EB, ten Berge IJ, van Lier RA, Eldering E, van Oers MH, Tonino SH, Kater AP, CMV-specific CD8+ T-cell function is not impaired in chronic lymphocytic leukemia, Blood, 123 (2014) 717–724. [DOI] [PubMed] [Google Scholar]
- [228].Brusa D, Serra S, Coscia M, Rossi D, D'Arena G, Laurenti L, Jaksic O, Fedele G, Inghirami G, Gaidano G, Malavasi F, Deaglio S, The PD-1/PD-L1 axis contributes to T-cell dysfunction in chronic lymphocytic leukemia, Haematologica, 98 (2013) 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Ramsay AG, Clear AJ, Fatah R, Gribben JG, Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: establishing a reversible immune evasion mechanism in human cancer, Blood, 120 (2012) 1412–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Jitschin R, Braun M, Buttner M, Dettmer-Wilde K, Bricks J, Berger J, Eckart MJ, Krause SW, Oefner PJ, Le Blanc K, Mackensen A, Mougiakakos D, CLL-cells induce IDOhi CD14+HLA-DRlo myeloid-derived suppressor cells that inhibit T-cell responses and promote TRegs, Blood, 124 (2014) 750–760. [DOI] [PubMed] [Google Scholar]
- [231].Gassner FJ, Zaborsky N, Catakovic K, Rebhandl S, Huemer M, Egle A, Hartmann TN, Greil R, Geisberger R, Chronic lymphocytic leukaemia induces an exhausted T cell phenotype in the TCL1 transgenic mouse model, British journal of haematology, 170 (2015) 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Bresin A, D'Abundo L, Narducci MG, Fiorenza MT, Croce CM, Negrini M, Russo G, TCL1 transgenic mouse model as a tool for the study of therapeutic targets and microenvironment in human B-cell chronic lymphocytic leukemia, Cell death & disease, 7 (2016) e2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].McClanahan F, Riches JC, Miller S, Day WP, Kotsiou E, Neuberg D, Croce CM, Capasso M, Gribben JG, Mechanisms of PD-L1/PD-1-mediated CD8 T-cell dysfunction in the context of aging-related immune defects in the Emicro-TCL1 CLL mouse model, Blood, 126 (2015) 212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Jenq RR, van den Brink MR, Allogeneic haematopoietic stem cell transplantation: individualized stem cell and immune therapy of cancer, Nature reviews. Cancer, 10 (2010) 213–221. [DOI] [PubMed] [Google Scholar]
- [235].Khouri IF, Bassett R, Poindexter N, O'Brien S, Bueso-Ramos CE, Hsu Y, Ferrajoli A, Keating MJ, Champlin R, Fernandez-Vina M, Nonmyeloablative allogeneic stem cell transplantation in relapsed/refractory chronic lymphocytic leukemia: long-term follow-up, prognostic factors, and effect of human leukocyte histocompatibility antigen subtype on outcome, Cancer, 117 (2011) 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Wu CJ, Ritz J, Induction of tumor immunity following allogeneic stem cell transplantation, Advances in immunology, 90 (2006) 133–173. [DOI] [PubMed] [Google Scholar]
- [237].Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, Li S, Pan Z, Thamm DH, Miller RA, Buggy JJ, The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy, Proceedings of the National Academy of Sciences of the United States of America, 107 (2010) 13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Herman SE, Gordon AL, Hertlein E, Ramanunni A, Zhang X, Jaglowski S, Flynn J, Jones J, Blum KA, Buggy JJ, Hamdy A, Johnson AJ, Byrd JC, Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765, Blood, 117 (2011) 6287–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Ponader S, Chen SS, Buggy JJ, Balakrishnan K, Gandhi V, Wierda WG, Keating MJ, O'Brien S, Chiorazzi N, Burger JA, The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo, Blood, 119 (2012) 1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Herman SE, Niemann CU, Farooqui M, Jones J, Mustafa RZ, Lipsky A, Saba N, Martyr S, Soto S, Valdez J, Gyamfi JA, Maric I, Calvo KR, Pedersen LB, Geisler CH, Liu D, Marti GE, Aue G, Wiestner A, Ibrutinib-induced lymphocytosis in patients with chronic lymphocytic leukemia: correlative analyses from a phase II study, Leukemia, 28 (2014) 2188–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Dubovsky JA, Beckwith KA, Natarajan G, Woyach JA, Jaglowski S, Zhong Y, Hessler JD, Liu TM, Chang BY, Larkin KM, Stefanovski MR, Chappell DL, Frissora FW, Smith LL, Smucker KA, Flynn JM, Jones JA, Andritsos LA, Maddocks K, Lehman AM, Furman R, Sharman J, Mishra A, Caligiuri MA, Satoskar AR, Buggy JJ, Muthusamy N, Johnson AJ, Byrd JC, Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes, Blood, 122 (2013) 2539–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Yin Q, Sivina M, Robins H, Yusko E, Vignali M, O'Brien S, Keating MJ, Ferrajoli A, Estrov Z, Jain N, Wierda WG, Burger JA, Ibrutinib Therapy Increases T Cell Repertoire Diversity in Patients with Chronic Lymphocytic Leukemia, Journal of immunology, 198 (2017) 1740–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Niemann CU, Herman SE, Maric I, Gomez-Rodriguez J, Biancotto A, Chang BY, Martyr S, Stetler-Stevenson M, Yuan CM, Calvo KR, Braylan RC, Valdez J, Lee YS, Wong DH, Jones J, Sun C, Marti GE, Farooqui MZ, Wiestner A, Disruption of in vivo Chronic Lymphocytic Leukemia Tumor-Microenvironment Interactions by Ibrutinib--Findings from an Investigator-Initiated Phase II Study, Clinical cancer research : an official journal of the American Association for Cancer Research 22 (2016) 1572–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Podhorecka M, Goracy A, Szymczyk A, Kowal M, Ibanez B, Jankowska-Lecka O, Macheta A, Nowaczynska A, Drab-Urbanek E, Chocholska S, Jawniak D, Hus M, Changes in T-cell subpopulations and cytokine network during early period of ibrutinib therapy in chronic lymphocytic leukemia patients: the significant decrease in T regulatory cells number, Oncotarget, 8 (2017) 34661–34669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Long M, Beckwith K, Do P, Mundy BL, Gordon A, Lehman AM, Maddocks KJ, Cheney C, Jones JA, Flynn JM, Andritsos LA, Awan F, Fraietta JA, June CH, Maus MV, Woyach JA, Caligiuri MA, Johnson AJ, Muthusamy N, Byrd JC, Ibrutinib treatment improves T cell number and function in CLL patients, The Journal of clinical investigation, 127 (2017) 3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Kondo K, Shaim H, Thompson PA, Burger JA, Keating M, Estrov Z, Harris D, Kim E, Ferrajoli A, Daher M, Basar R, Muftuoglu M, Imahashi N, Alsuliman A, Sobieski C, Gokdemir E, Wierda W, Jain N, Liu E, Shpall EJ, Rezvani K, Ibrutinib modulates the immunosuppressive CLL microenvironment through STAT3-mediated suppression of regulatory B-cell function and inhibition of the PD-1/PD-L1 pathway, Leukemia, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Herman SEM, Montraveta A, Niemann CU, Mora-Jensen H, Gulrajani M, Krantz F, Mantel R, Smith LL, McClanahan F, Harrington BK, Colomer D, Covey T, Byrd JC, Izumi R, Kaptein A, Ulrich R, Johnson AJ, Lannutti BJ, Wiestner A, Woyach JA, The Bruton Tyrosine Kinase (BTK) Inhibitor Acalabrutinib Demonstrates Potent On-Target Effects and Efficacy in Two Mouse Models of Chronic Lymphocytic Leukemia, Clinical cancer research : an official journal of the American Association for Cancer Research, 23 (2017) 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Patel VK, Lamothe B, Ayres ML, Gay J, Cheung JP, Balakrishnan K, Ivan C, Morse J, Nelson M, Keating MJ, Wierda WG, Marszalek JR, Gandhi V, Pharmacodynamics and proteomic analysis of acalabrutinib therapy: similarity of on-target effects to ibrutinib and rationale for combination therapy, Leukemia, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Patel V, Balakrishnan K, Bibikova E, Ayres M, Keating MJ, Wierda WG, Gandhi V, Comparison of Acalabrutinib A Selective Bruton Tyrosine Kinase Inhibitor, with Ibrutinib in Chronic Lymphocytic Leukemia Cells, Clinical cancer research : an official journal of the American Association for Cancer Research, 23 (2017) 3734–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Hallek M, Fischer K, Fingerle-Rowson G, Fink AM, Busch R, Mayer J, Hensel M, Hopfinger G, Hess G, von Grunhagen U, Bergmann M, Catalano J, Zinzani PL, Caligaris-Cappio F, Seymour JF, Berrebi A, Jager U, Cazin B, Trneny M, Westermann A, Wendtner CM, Eichhorst BF, Staib P, Buhler A, Winkler D, Zenz T, Bottcher S, Ritgen M, Mendila M, Kneba M, Dohner H, Stilgenbauer S, International Group of I, German G Chronic Lymphocytic Leukaemia Study, Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial, Lancet, 376 (2010) 1164–1174. [DOI] [PubMed] [Google Scholar]
- [251].Hainsworth JD, Monoclonal antibody therapy in lymphoid malignancies, The oncologist, 5 (2000) 376–384. [DOI] [PubMed] [Google Scholar]
- [252].Eichhorst B, Robak T, Montserrat E, Ghia P, Hillmen P, Hallek M, Buske C, Committee EG, Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up, Annals of oncology : official journal of the European Society for Medical Oncology, 26 Suppl 5 (2015) v78–84. [DOI] [PubMed] [Google Scholar]
- [253].Chao MP, Treatment challenges in the management of relapsed or refractory non-Hodgkin's lymphoma - novel and emerging therapies, Cancer management and research, 5 (2013) 251–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Bachy E, Salles G, Are we nearing an era of chemotherapy-free management of indolent lymphoma?, Clinical cancer research : an official journal of the American Association for Cancer Research, 20 (2014) 5226–5239. [DOI] [PubMed] [Google Scholar]
- [255].Beum PV, Lindorfer MA, Taylor RP, Within peripheral blood mononuclear cells, antibody-dependent cellular cytotoxicity of rituximab-opsonized Daudi cells is promoted by NK cells and inhibited by monocytes due to shaving, Journal of immunology, 181 (2008) 2916–2924. [DOI] [PubMed] [Google Scholar]
- [256].Beum PV, Peek EM, Lindorfer MA, Beurskens FJ, Engelberts PJ, Parren PW, van de Winkel JG, Taylor RP, Loss of CD20 and bound CD20 antibody from opsonized B cells occurs more rapidly because of trogocytosis mediated by Fc receptor-expressing effector cells than direct internalization by the B cells, Journal of immunology, 187 (2011) 3438–3447. [DOI] [PubMed] [Google Scholar]
- [257].Da Roit F, Engelberts PJ, Taylor RP, Breij EC, Gritti G, Rambaldi A, Introna M, Parren PW, Beurskens FJ, Golay J, Ibrutinib interferes with the cell-mediated anti-tumor activities of therapeutic CD20 antibodies: implications for combination therapy, Haematologica, 100 (2015) 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Pavlasova G, Borsky M, Seda V, Cerna K, Osickova J, Doubek M, Mayer J, Calogero R, Trbusek M, Pospisilova S, Davids MS, Kipps TJ, Brown JR, Mraz M, Ibrutinib inhibits CD20 upregulation on CLL B cells mediated by the CXCR4/SDF-1 axis, Blood, 128 (2016) 1609–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Kater AP, Tonino SH, Egle A, Ramsay AG, How does lenalidomide target the chronic lymphocytic leukemia microenvironment?, Blood, 124 (2014) 2184–2189. [DOI] [PubMed] [Google Scholar]
- [260].Lee BN, Gao H, Cohen EN, Badoux X, Wierda WG, Estrov Z, Faderl SH, Keating MJ, Ferrajoli A, Reuben JM, Treatment with lenalidomide modulates T-cell immunophenotype and cytokine production in patients with chronic lymphocytic leukemia, Cancer, 117 (2011) 3999–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Wu L, Adams M, Carter T, Chen R, Muller G, Stirling D, Schafer P, Bartlett JB, lenalidomide enhances natural killer cell and monocyte-mediated antibody-dependent cellular cytotoxicity of rituximab-treated CD20+ tumor cells, Clinical cancer research : an official journal of the American Association for Cancer Research, 14 (2008) 4650–4657. [DOI] [PubMed] [Google Scholar]
- [262].Badoux XC, Keating MJ, Wen S, Wierda WG, O'Brien SM, Faderl S, Sargent R, Burger JA, Ferrajoli A, Phase II study of lenalidomide and rituximab as salvage therapy for patients with relapsed or refractory chronic lymphocytic leukemia, Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 31 (2013) 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Jain N, O'Brien S, Targeted therapies for CLL: Practical issues with the changing treatment paradigm, Blood reviews, 30 (2016) 233–244. [DOI] [PubMed] [Google Scholar]
- [264].Yigit B, Halibozek PJ, Chen SS, O'Keeffe MS, Arnason J, Avigan D, Gattei V, Bhan A, Cen O, Longnecker R, Chiorazzi N, Wang N, Engel P, Terhorst C, A combination of an anti-SLAMF6 antibody and ibrutinib efficiently abrogates expansion of chronic lymphocytic leukemia cells, Oncotarget, 7 (2016) 26346–26360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Zucchetto A, Cattarossi I, Nanni P, Zaina E, Prato G, Gilestro M, Marconi D, Bulian P, Rossi FM, Del Vecchio L, Omede P, Geuna M, Del Poeta G, Gattei V, Cluster analysis of immunophenotypic data: the example of chronic lymphocytic leukemia, Immunology letters, 134 (2011) 137–144. [DOI] [PubMed] [Google Scholar]
- [266].Schweighofer CD, Coombes KR, Barron LL, Diao L, Newman RJ, Ferrajoli A, O'Brien S, Wierda WG, Luthra R, Medeiros LJ, Keating MJ, Abruzzo LV, A two-gene signature SKI and SLAMF1, predicts time-to-treatment in previously untreated patients with chronic lymphocytic leukemia, PloS one, 6 (2011) e28277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Bologna C, Buonincontri R, Serra S, Vaisitti T, Audrito V, Brusa D, Pagnani A, Coscia M, D'Arena G, Mereu E, Piva R, Furman RR, Rossi D, Gaidano G, Terhorst C, Deaglio S, SLAMF1 regulation of chemotaxis and autophagy determines CLL patient response, The Journal of clinical investigation, 126 (2016) 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Gordiienko I, Shlapatska L, Kholodniuk VM, Kovalevska L, Ivanivskaya TS, Sidorenko SP, CD150 and CD180 are involved in regulation of transcription factors expression in chronic lymphocytic leukemia cells, Experimental oncology, 39 (2017) 291–298. [PubMed] [Google Scholar]
- [269].Gordiienko I, Shlapatska L, Kholodniuk V, Sklyarenko L, Gluzman DF, Clark EA, Sidorenko SP, The interplay of CD150 and CD180 receptor pathways contribute to the pathobiology of chronic lymphocytic leukemia B cells by selective inhibition of Akt and MAPK signaling, PloS one, 12 (2017) e0185940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Binsky-Ehrenreich I, Marom A, Sobotta MC, Shvidel L, Berrebi A, Hazan-Halevy I, Kay S, Aloshin A, Sagi I, Goldenberg DM, Leng L, Bucala R, Herishanu Y, Haran M, Shachar I, CD84 is a survival receptor for CLL cells, Oncogene, 33 (2014) 1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Huang PY, Best OG, Almazi JG, Belov L, Davis ZA, Majid A, Dyer MJ, Pascovici D, Mulligan SP, Christopherson RI, Cell surface phenotype profiles distinguish stable and progressive chronic lymphocytic leukemia, Leukemia & lymphoma, 55 (2014) 2085–2092. [DOI] [PubMed] [Google Scholar]
- [272].Marom A, Barak AF, Kramer MP, Lewinsky H, Binsky-Ehrenreich I, Cohen S, Tsitsou-Kampeli A, Kalchenko V, Kuznetsov Y, Mirkin V, Dezorella N, Shapiro M, Schwartzberg PL, Cohen Y, Shvidel L, Haran M, Becker-Herman S, Herishanu Y, Shachar I, CD84 mediates CLL-microenvironment interactions, Oncogene, 36 (2017) 628–638. [DOI] [PubMed] [Google Scholar]
- [273].Chen SS, Batliwalla F, Holodick NE, Yan XJ, Yancopoulos S, Croce CM, Rothstein TL, Chiorazzi N, Autoantigen can promote progression to a more aggressive TCL1 leukemia by selecting variants with enhanced B-cell receptor signaling, Proceedings of the National Academy of Sciences of the United States of America, 110 (2013) E1500–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Yigit B, Wang N, Chen SS, Chiorazzi N, Terhorst C, Inhibition of reactive oxygen species limits expansion of chronic lymphocytic leukemia cells, Leukemia, 31 (2017) 2273–2276. [DOI] [PubMed] [Google Scholar]
- [275].Palumbo A, Anderson K, Multiple myeloma, The New England journal of medicine, 364 (2011) 1046–1060. [DOI] [PubMed] [Google Scholar]
- [276].Mahindra A, Laubach J, Raje N, Munshi N, Richardson PG, Anderson K, Latest advances and current challenges in the treatment of multiple myeloma, Nature reviews. Clinical oncology, 9 (2012) 135–143. [DOI] [PubMed] [Google Scholar]
- [277].Kumar SK, Lee JH, Lahuerta JJ, Morgan G, Richardson PG, Crowley J, Haessler J, Feather J, Hoering A, Moreau P, LeLeu X, Hulin C, Klein SK, Sonneveld P, Siegel D, Blade J, Goldschmidt H, Jagannath S, Miguel JS, Orlowski R, Palumbo A, Sezer O, Rajkumar SV, Durie BG, International Myeloma Working G, Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: a multicenter international myeloma working group study, Leukemia, 26 (2012) 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Dimopoulos MA, Richardson PG, Moreau P, Anderson KC, Current treatment landscape for relapsed and/or refractory multiple myeloma, Nature reviews. Clinical oncology, 12 (2015) 42–54. [DOI] [PubMed] [Google Scholar]
- [279].van de Donk NW, Kamps S, Mutis T, Lokhorst HM, Monoclonal antibody-based therapy as a new treatment strategy in multiple myeloma, Leukemia, 26 (2012) 199–213. [DOI] [PubMed] [Google Scholar]
- [280].Raje N, Longo DL, Monoclonal Antibodies in Multiple Myeloma Come of Age, The New England journal of medicine, 373 (2015) 1264–1266. [DOI] [PubMed] [Google Scholar]
- [281].Lonial S, Durie B, Palumbo A, San-Miguel J, Monoclonal antibodies in the treatment of multiple myeloma: current status and future perspectives, Leukemia, 30 (2016) 526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Tai YT, Dillon M, Song W, Leiba M, Li XF, Burger P, Lee AI, Podar K, Hideshima T, Rice AG, van Abbema A, Jesaitis L, Caras I, Law D, Weller E, Xie W, Richardson P, Munshi NC, Mathiot C, Avet-Loiseau H, Afar DE, Anderson KC, Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu, Blood, 112 (2008) 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Shaughnessy JD Jr., Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, Stewart JP, Kordsmeier B, Randolph C, Williams DR, Xiao Y, Xu H, Epstein J, Anaissie E, Krishna SG, Cottler-Fox M, Hollmig K, Mohiuddin A, Pineda-Roman M, Tricot G, van Rhee F, Sawyer J, Alsayed Y, Walker R, Zangari M, Crowley J, Barlogie B, A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1, Blood, 109 (2007) 2276–2284. [DOI] [PubMed] [Google Scholar]
- [284].Klein U, Jauch A, Hielscher T, Hillengass J, Raab MS, Seckinger A, Hose D, Ho AD, Goldschmidt H, Neben K, Chromosomal aberrations +1q21 and del(17p13) predict survival in patients with recurrent multiple myeloma treated with lenalidomide and dexamethasone, Cancer, 117 (2011) 2136–2144. [DOI] [PubMed] [Google Scholar]
- [285].Collins SM, Bakan CE, Swartzel GD, Hofmeister CC, Efebera YA, Kwon H, Starling GC, Ciarlariello D, Bhaskar S, Briercheck EL, Hughes T, Yu J, Rice A, Benson DM Jr., Elotuzumab directly enhances NK cell cytotoxicity against myeloma via CS1 ligation: evidence for augmented NK cell function complementing ADCC, Cancer immunology, immunotherapy : CII, 62 (2013) 1841–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Jakubowiak AJ, Benson DM, Bensinger W, Siegel DS, Zimmerman TM, Mohrbacher A, Richardson PG, Afar DE, Singhal AK, Anderson KC, Phase I trial of anti-CS1 monoclonal antibody elotuzumab in combination with bortezomib in the treatment of relapsed/refractory multiple myeloma, Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 30 (2012) 1960–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Lonial S, Vij R, Harousseau JL, Facon T, Moreau P, Mazumder A, Kaufman JL, Leleu X, Tsao LC, Westland C, Singhal AK, Jagannath S, Elotuzumab in combination with lenalidomide and low-dose dexamethasone in relapsed or refractory multiple myeloma, Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 30 (2012) 1953–1959. [DOI] [PubMed] [Google Scholar]
- [288].Bezman NA, Jhatakia A, Kearney AY, Brender T, Maurer M, Henning K, Jenkins MR, Rogers AJ, Neeson PJ, Korman AJ, Robbins MD, Graziano RF, PD-1 blockade enhances elotuzumab efficacy in mouse tumor models, Blood advances, 1 (2017) 753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Bae J, Song W, Smith R, Daley J, Tai YT, Anderson KC, Munshi NC, A novel immunogenic CS1-specific peptide inducing antigen-specific cytotoxic T lymphocytes targeting multiple myeloma, British journal of haematology, 157 (2012) 687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Gogishvili T, Danhof S, Prommersberger S, Rydzek J, Schreder M, Brede C, Einsele H, Hudecek M, SLAMF7-CAR T cells eliminate myeloma and confer selective fratricide of SLAMF7(+) normal lymphocytes, Blood, 130 (2017) 2838–2847. [DOI] [PubMed] [Google Scholar]