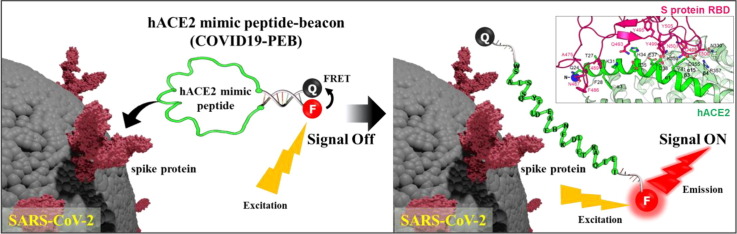
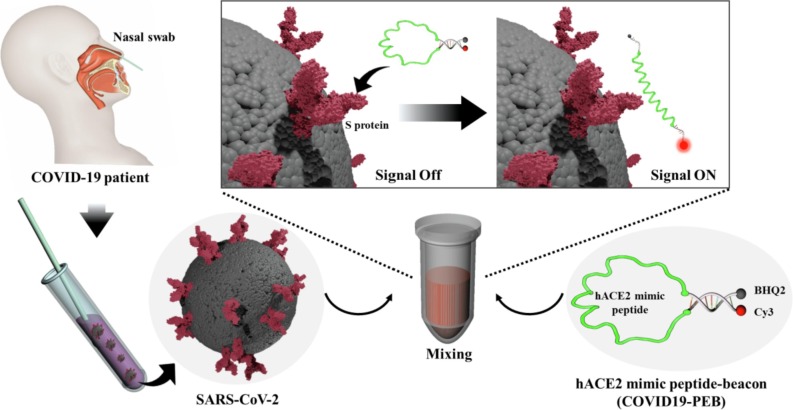
Graphical abstract
COVID 19 -PEB is developed to simply detect SARS-CoV-2 by introducing FRET-based molecular beacon to the hACE2 receptor mimic peptide, which has excellent binding ability with the spike protein. This allows direct detection of SARS-CoV-2 without complicated pretreatment, and in particular, it is also very simple to check whether or not SARS-CoV-2 is infected.
Abbreviations: SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; hACE2, Human angiotensin-converting enzyme 2; COVID19-PEB, hACE2 mimic peptide beacon; FRET, Fluorescence resonance energy transfer; Oligo-BHQ2, Black hole quencher 2 (BHQ2)-modified oligonucleotide; Oligo-Cy3, Organic fluorophore (Cy3)-modified oligonucleotide
Keywords: COVID-19, SARS-CoV-2, Spike protein RBD, hACE2 mimic peptide, FRET probe
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has led to a pandemic of acute respiratory disease, namely coronavirus disease (COVID-19). This disease threatens human health and public safety. Early diagnosis, isolation, and prevention are important to suppress the outbreak of COVID 19 given the lack of specific antiviral drugs to treat this disease and the emergence of various variants of the virus that cause breakthrough infections even after vaccine administration. Simple and prompt testing is paramount to preventing further spread of the virus. However, current testing methods, namely RT-PCR, is time-consuming. Binding of the SARS-CoV-2 spike (S) glycoprotein to human angiotensin-converting enzyme 2 (hACE2) receptor plays a pivotal role in host cell entry. In the present study, we developed a hACE2 mimic peptide beacon (COVID19-PEB) for simple detection of SARS-CoV-2 using a fluorescence resonance energy transfer system. COVID19-PEB exhibits minimal fluorescence in its ‘‘closed’’ hairpin structure; however, in the presence of SARS-CoV-2, the specific recognition of the S protein receptor-binding domain by COVID19-PEB causes the beacon to assume an ‘‘open’’ structure that emits strong fluorescence. COVID19-PEB can detect SARS-CoV-2 within 3 h or even 50 min and exhibits strong fluorescence even at low viral concentrations, with a detection limit of 4 × 103 plaque-forming unit/test. Furthermore, in SARS-CoV-2-infected patient samples confirmed using polymerase chain reaction, COVID19-PEB accurately detected the virus. COVID19-PEB could be developed as a rapid and accurate diagnostic tool for COVID-19.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a member of a large family of coronaviruses; it is a highly transmissible and pathogenic coronavirus that emerged in Wuhan City, Hubei Province, China, in late 2019 and has led to a pandemic of acute respiratory disease, commonly known as coronavirus disease (COVID-19). This virus has infected approximately 200 million people and caused 4 million deaths worldwide.[1], [2], [3], [4] The common symptoms of COVID-19 include cold, fever, dry cough, tiredness, and headache. COVID-19 may cause respiratory disorders and severe pneumonia in severe cases, particularly in older adults with underlying diseases.[5], [6], [7], [8], [9] No definitive cure is yet available for this novel highly infectious virus.[10], [11] Currently, it is being managed with active immunization across the countries using vaccines developed by AstraZeneca, Pfizer, and Moderna, among others. Hence, preventive measures such as wearing masks and maintaining social distance need to be strictly followed to curb the spread of SARS-CoV-2. Moreover, infected patients need to be identified, isolated, and treated separately. Selective and prompt testing is required to prevent the further spread of the virus.[12], [13], [14].
Therefore, various new technologies have been introduced for easier and quick identification of infected patients;[15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28] However, the current gold standard for virus diagnostic testing is still molecular diagnostic using quantitative real-time polymerase chain reaction (qRT-PCR), as recommended by the World Health Organization (WHO) and the American Center for Disease Control and Prevention (CDC). In the qRT-PCR method, a specific gene of SARS-CoV-2 is amplified from respiratory samples (e.g., nasal fluid, saliva, or sputum) of patients with suspected infection. This method exhibits high sensitivity (low detection limit) and specificity; however, it is time-consuming (several hours to days, depending on specimen collection, transportation to the laboratory, and process optimization) and requires sophisticated laboratory infrastructure and well-trained personnel. The complex nature of these tests makes them unsuitable for rapid, large-scale diagnosis, further limiting their availability and distribution.[29], [30], [31] To trace and curb the rapid spread of infection, simple diagnostic technology is required that performs mass field-testing of crude samples without additional extraction.[32] Antigen-antibody reaction-based lateral flow tests, also known as lateral flow immune-chromatographic assay or rapid tests, can detect the presence of target antigens (protein, antibody) in clinical samples without the need for specialized equipment; in particular, these tests are economical and provide quick results within 15–30 min.[33], [34], [35], [36], [37], [38], [39] SARS-CoV-2 antigen-detecting rapid diagnostic test kits were approved by the WHO for emergency use on March 17, 2021.[40] This assay can be divided into two methods: an antibody capable of directly binding the antigen derived from the infecting virus particles or detecting immunoglobulin (IgG, IgM, or a combination of both) generated upon immune response to infection in the body. In the latter method, rapid testing is impossible because the infection can be detected after forming the therapeutic antibody. Therefore, the former method of detecting viral antigen using a specific antibody, is being explored as an early detection method.[41], [42], [43], [44], [45] Currently, the rapid antigen diagnostic test using lateral flow has a great advantages as a quick and simple diagnostic method because it can visually confirm whether or not there is an infection. However, there is concern that false-negative or false-positive results have been reported, which may aggravate confusion in response to infectious diseases. Therefore, many developers are conducting research to improve it.
SARS-CoV-2 has a single positive-strand RNA genome (29.8 kb) encoding four structural proteins: spike (S), envelope (E), matrix (M), and nucleocapsid (N) present in the virus particle, in addition to many nonstructural proteins.[46], [47], [48] In particular, the spike (S) protein receptor-binding domain (RBD) of SARS-CoV-2 allows the virus to enter the host cell by binding to the peptidase domain of human angiotensin-converting enzyme 2 (hACE2), one of the transmembrane proteins on the host cell surface.[49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61] In the present study, we designed and developed a hACE2 mimic peptide beacon (COVID19-PEB) for simple and rapid detection of SARS-CoV-2. First, the hACE2 mimic peptide sequence capable of specifically binding to the S protein RBD on the SARS-CoV-2 was identified.[62], [63], [64], [65], [66], [67], [68], [69] After that, COVID 19-PEB was synthesized by including this peptide in the central loop region; furthermore, two oligonucleotides with complementary sequences binding a phosphor and quencher, respectively, were added in the stem region. When SARS-CoV-2 is absent, COVID 19-PEB forms a hairpin structure, and the fluorescence signal is suppressed by fluorescence resonance energy transfer (FRET) effect (“OFF” state); however after SARS-CoV-2 was allowed to bind with the COVID 19-PEB peptide, the S protein RBD of SARS-CoV-2 bound firmly, leading to opening of the hairpin structure of the probe (“OPEN” state), thereby recovering and increasing the fluorescence signal within a short period. This system makes it easy to identify whether or not SARS-CoV-2 infection is present by simply mixing the sample with COVID 19-PEB and measuring the fluorescence intensity. It is also a system with excellent detection sensitivity and specificity.
2. Materials and methods
2.1. Materials
Oligomers were synthesized with a Cy3 donor-conjugated to its 9-terminus (Cy3-TTTTGGGGG-SH) and a black hole quencher 2 (BHQ2)-coupled oligomers to its 9-terminus as an acceptor (BHQ2-AAAACCCCC-Azide), which were obtained from Bioneer Inc. (Daejeon, Korea). Peptide substrate was conjugated from PEPTRON (Daejeon, Korea). Sodium ascorbate, copper (II) sulfate (CuSO4), and Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA) were obtained from Sigma-Aldrich Company (USA). Tris-HCl (pH 6.8) and Tris-EDTA (TE) buffer (pH 8.0) were purchased from Biosesang (Korea). All other chemicals and reagents were of analytical grade. SARS-CoV-2 (2019-nCoV) spike (S) protein RBD-His recombinant protein (Cat. no. 40592-V08B) was obtained from Sino Biological (China). Human nasal fluid and human saliva fluid were purchased from Innovative.
2.2. Preparation and characterization of COVID 19-PEB
Peptide beacon capable of detecting SARS-CoV-2 was synthesized with a SARS-CoV-2 -targeting hACE2 mimic peptide (peptide) and two oligomers: a donor-conjugated oligomer with Cy3 and an acceptor-conjugated oligomer with BHQ2. COVID 19-PEB was created using the following sequences: (4-pentynoyl)-I EEQA KTFL DKFN HEAE DLFY QSSL ASWK-(4-maleimidobutyryl), 5′-Azide-CCCCCAAAA-BHQ2-3′ (Oligo-BHQ2), and 5′-Cy3-TTTTGGGGG-SH-3′ (Oligo-Cy3) (Fig. S1). Peptide was dispersed in TE buffer (pH 8.0) at a concentration of 1 mM. Oligo-BHQ2 and Oligo-Cy3 were suspended in TE buffer (pH 8.0) and Tris-HCl (pH 6.8) at a concentration of 100 μM, respectively. After that, 100 μL of Oligo-BHQ2 was initially treated with 20 nmol CuSO4, 100 nmol THPTA, 1 μmol sodium ascorbate, and 10 μL peptide for 1 h at 4 °C to combine the pentynoyl group of the peptide and the azide group of Oligo-BHQ2 by click chemistry. The reacted peptide-BHQ2 was obtained by purifying using a NAP-5 column. Subsequently, 100 μL of Oligo-Cy3 was added to the purified peptide-BHQ2, the maleimide group of the peptide and the thiol group of Oligo-Cy3 were reacted, and the product was purified using a column. The synthesized COVID 19-PEB was annealed at 95 °C for 2 min and then slowly cooled to 25 °C to form the hairpin structure. The fluorescence intensity of COVID 19-PEB was measured using a Cytation™ 5 Cell Imaging Multi-Mode Reader (BioTek, USA) instrument at 530 nm (excitation) and 580 nm (emission) to confirm the efficacy of fluorescence quenching.
2.3. Measurement of the binding affinity of COVID 19-PEB toward SARS-CoV-2
We measured the binding affinity between COVID 19-PEB and SARS-CoV-2 using a biolayer interferometry-based biosensor BLItz system (FortéBio). First, the streptavidin biosensors (FortéBio) were immobilized with biotinylated hACE2 mimic peptide (Biotin-I EEQA KTFL DKFN HEAE DLFY QSSL ASWK; biotin-hACE2 mimic peptide) equilibrated in phosphate-buffered saline (PBS) for 2 min to establish a stable baseline; thereafter, they were immersed into 4 µL of 1 µM in S protein of SARS-CoV-2 and bovine serum albumin (BSA) as a control for 300 s to obtain association curves, respectively. Later, their binding affinities were calculated by fitting the curves using BLItz software. The biotinylated peptide was bound to the protein and then analyzed via western blotting to additionally confirm the binding of the peptide to the S protein of SARS-CoV-2. S protein and BSA was incubated with biotin-ACE2 mimic peptide for different durations (0.5, 1, 2, 4 h). After that, 10 μL of TE buffer and 10 μL of NuPAGE™ LDS Sample Buffer (4X) was added to 20 μL of the sample. A 20-μL aliquot of each sample was electrophoresed in 0.75 mm-thick sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels containing 10% acrylamide. Gels were run at 80–150 V for 120 min. After electrophoresis, these gels were blotted to the polyvinylidene fluoride membranes using a wet-type blotting system, Criterion Blotter with Plate Electrodes (Bio-Rad). The transfer buffer formulation used was as follows: 25 mM Tris-HCl at pH 8.3, 192 mM glycine, 0.05% SDS, 20% methanol. In general, the blotting step is followed by the blocking step in the western blotting procedure. The blotted membrane was briefly washed in TTBS (TBS [50 mM Tris-HCl at pH 7.4, 150 mM NaCl] containing 0.1% Tween 20) and soaked with blocking buffer (1% skim milk and 0.1% BSA in TTBS) for 5 min. After a 3-min washing step with TPBS (PBS [10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl at pH 7.4] containing 0.1% Tween 20), the membrane was incubated in 0.2% GA or 0.4% PFA in TPBS for 15 min. After that, the aldehyde-treated membrane was briefly washed thrice with TPBS and was then incubated in the blocking buffer for 30 min and probed with the anti-biotin horseradish peroxidase (HRP) for 90 min at 4 °C, followed by measurement of the chemiluminescence. The blotted membrane was briefly washed in TTBS (TBS [50 mM Tris-HCl at pH 7.4, 150 mM NaCl] containing 0.1% Tween 20) and soaked with blocking buffer (1% skim milk and 0.1% BSA in TTBS) for 5 min. After a 3-min washing step with TPBS (PBS [10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl at pH 7.4] containing 0.1% Tween 20), the membrane was incubated in 0.2% GA or 0.4% PFA in TPBS for 15 min. The aldehyde-treated membrane was briefly washed thrice with TPBS. After that, the membrane was incubated in the blocking buffer for 30 min and probed with the anti-biotin horseradish peroxidase (HRP) for 90 min at 4 °C, followed by measurement of the chemiluminescence.
2.4. Viruses
SARS-CoV-2 was provided by the Korea National Institute of Health and cultured in a BL-3 laboratory of the Korea Research Institute of Bioscience and Biotechnology (KRIBB). Human coronavirus OC43 (HCoV-OC43) was obtained from Korea Centers for Disease Control and Prevention (KCDC); both influenza viruses, influenza A (pH1N1) virus (Human/Canada/2009) and influenza B viruses (Victoria Brisbane/20/2009), were provided by the BioNano Health Guard Research Center (H-GUARD), and these were used as controls. All virus titers were determined by 50% tissue culture infective dose (TCID50) in confluent cells in 96-well microplates. The theoretical relationship between TCID50 and PFU is approximately 0.7 PFU = 1 TCID50 based on the Poisson distribution, which describes the random events (virus particles) that occur at a known average rate (virus titer) in a fixed space (the amount of virus medium in a well). The virus measurement unit was changed from TCID50 to PFU and was expressed in the graph.
2.5. SARS-CoV-2 detection using COVID19-PEB
COVID19-PEB was tested against various viruses (SAR-CoV-2, HCoV-OC43, Influenza A, and Influenza B viruses) to confirm the specific detection for SARS-CoV-2. Each virus was added to every well in the 96-well black plate and treated with COVID 19-PEB (10 nmol in 100 µL). The fluorescence of the plate was then measured with a microplate reader at various time intervals at room temperature (25 °C), using wavelengths of 530 nm (excitation) and 580 nm (emission). Furthermore, whether COVID 19-PEB can detect target viruses present in body fluids (e.g., saliva and nasal fluid) without nonspecific reactions was confirmed using SARS-CoV-2 in 10% human nasal fluid and saliva. The detection method was performed as previously described.
2.6. Measurement of SARS-CoV-2 detection ability of COVID 19-PEB in patient samples
We used 60 clinical samples (nasal fluids) from SARS-CoV-2 infected patients (30 positive samples and 30 negative samples) provided by the Severance Hospital, Seoul, Korea (IRB approval number: 4–2020-0465) to verify the availability in the real field. After treating each clinical sample with COVID 19-PEB (10 nmol in 100 µL) in a 96-well plate, each fluorescence was measured after 1, 2, and 4 h at a room temperature (25 °C), respectively, and further compared and expressed as a heat map image to distinguish the positive and negative samples.
2.7. Statistical analysis
Data were analyzed statistically and p-value was obtained by performing two-way ANOVA followed by turkey test using GraphPad Prism 8 software. Statistical significance was set at p < 0.05; *<0.05, **<0.005 and ***<0.0005.
3. Result and discussion
We have developed a hACE2 mimic peptide beacon (COVID 19-PEB), a smart probe capable of simple and specific detection of SARS-CoV-2 (Fig. 1 ).
Fig. 1.
Schematic illustration of the simple detection method of SARS-CoV-2 using the hACE2 mimic peptide-based molecular beacon (COVID 19-PEB).
3.1. Design of a peptide mimicking hACE2 and confirmation of interaction between the designed peptide and SARS-CoV-2 S protein
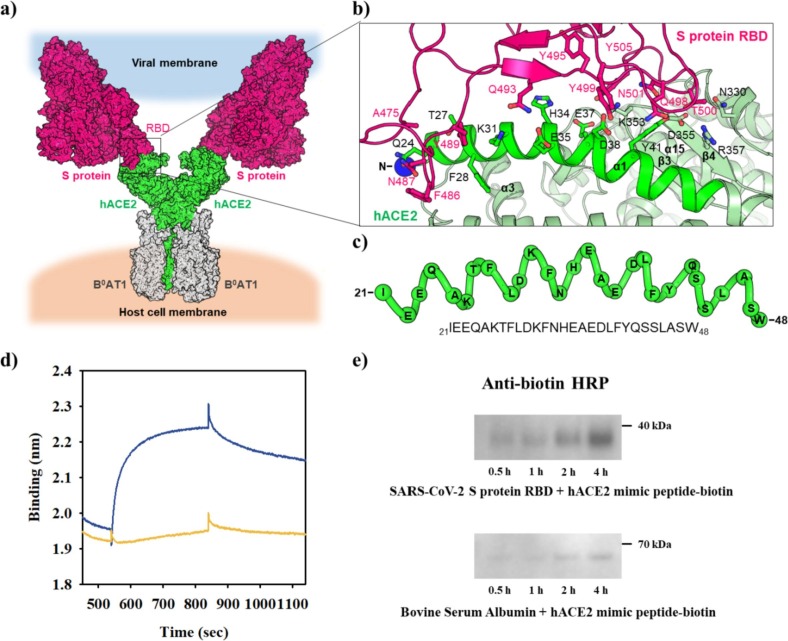
The S protein of SARS-CoV-2 is crucial in recognizing hACE2 for cell entry (Fig. 2 a).[47] To develop a diagnostic tool for SARS-CoV-2, we analyzed the binding between the S protein RBD and hACE2 ectodomain (Fig. 2a and b; Fig. S2). Seventeen residues of RBD tightly interacted with the 19 residues of hACE2 via one salt bridge, 11 hydrogen bonds, and 107 hydrophobic links (Fig. 2b; Fig. S2). The structural analysis clearly revealed that the hACE2 α1 helix is a key component to interact with RBD (Fig. 2b; Fig. S2).[64], [70] Thus, we selected the α1 helix (residues 21–48) to devise a SARS-CoV-2-specific sensing COVID 19-PEB (Fig. 2c). Moreover, a binding test was performed using BLItz to confirm whether the hACE2 mimic peptide (peptide) sequence obtained via the aforementioned structural analysis has a specific affinity for the S protein of SARS-CoV-2. The binding affinity of this peptide was significantly higher in the S protein RBD (with a dissociation constant (Kd) of 6.2 nM) than in the control (BSA) (Fig. 2d). The binding was further validated by immunoassay using anti-biotin-horseradish peroxidase (HRP) as the second antibody after combining biotinylated peptide with S protein RBD (37 kDa). BSA (67 kDa) was used as a control that did not reveal any binding (Fig. 2e and Fig. S3).
Fig. 2.
Overall structure of the hACE2 and SARS-CoV-2 S protein complex. a) Binding model of the hACE2 and SARS-CoV-2 S protein. The superimposition of protein structures generated the model structure, that is, SARS-CoV-2 S protein (PDB ID, 6vsd), SARS-CoV-2 S protein RBD bound with hACE2 (PDB ID, 6m0j), and hACE2 interacting with the amino acid transporter B0AT1 (PDB ID, 6 m18). hACE2 (green), B0AT1 (gray), and S protein (magenta) are shown in surface representation. b) Binding interface of SARS-CoV-2 S protein RBD and hACE2. hACE2 (green) and S protein RBD (magenta) are shown in the cartoon diagram. Residues involved in the interaction between the two proteins are highlighted as sticks (PDB ID, 6 m18). c) The hACE2 α1 helix (residues 21–48; green) is critical for the interaction with S protein RBD. d) Binding affinities of the hACE2 mimic peptide for SARS-CoV-2 S protein RBD (blue) and bovine serum albumin (yellow). e) Western blot showing the binding of biotin-hACE2 mimic peptide with SARS-CoV-2 S protein RBD and bovine serum albumin.
3.2. Confirmation of COVID19-PEB manufacturing and interaction with SARS-CoV-2 S protein
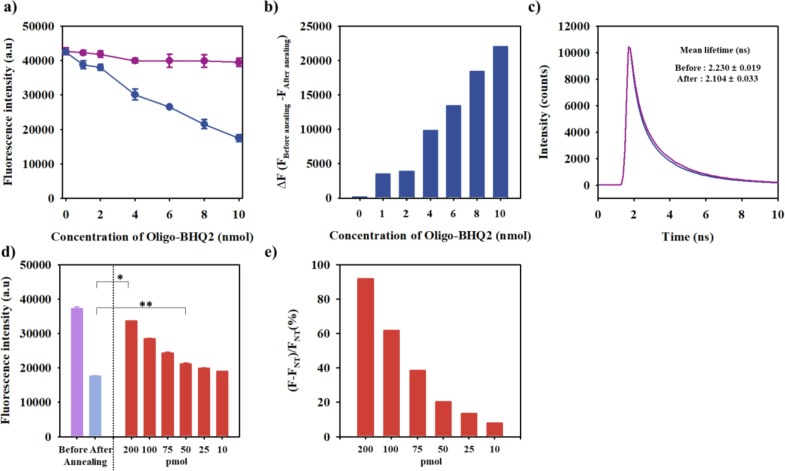
COVID 19-PEB that could specifically detect SARS-CoV-2 based on FRET was prepared using this hACE2 mimic peptide by verifying its specific recognition to the S protein of SARS-CoV-2; herein, hACE2 mimic peptide was chemically conjugated with two oligonucleotides, a black hole quencher 2 (BHQ2)-modified oligonucleotide (Oligo-BHQ2) and an organic fluorophore (Cy3)-modified oligonucleotide (Oligo-Cy3) (Fig. S1). The special advantage of the hairpin structured peptide beacon is that it applies to most peptides interacting with the target proteins, and it is a platform that can apply almost unlimited amounts of interesting phosphors. In particular, COVID 19-PEB comprises two oligonucleotides composed of complementary sequences, designed to form a stable hairpin structure after annealing, thereby resulting in a remarkably close distance between the fluorophore donor (Cy3, maximum emission at 530 nm) and the fluorophore acceptor (BHQ2, absorption at 560 nm). This result indicates that stronger quenching of the fluorescence occurred after annealing (close form) than before (open form) annealing due to the FRET effect in which the excitation energy of Oligo-Cy3 fluorescence was absorbed by the quencher (BHQ2) of Oligo-BHQ2. The quenching efficiency of COVID 19-PEB was determined by binding it to a fixed concentration (10 nmol) of Oligo-Cy3, increasing the concentration of Oligo-BHQ2, changing the fluorescence intensity before and after annealing. The fluorescence intensity before annealing did not differ significantly because the amount of Oligo-Cy3 was constant even though the amount of Oligo-BHQ2 increased; however, the fluorescence intensity decreased as the amount of Oligo-BHQ2 increased after annealing (Fig. 3 a). The quenching ability was the highest when the amount of Oligo-BHQ2 was 10 nmol (Fig. 3b), and this was selected as the optimal condition for the experiment. Additionally, by measuring the decay in fluorescence lifetime of COVID 19-PEB, it was confirmed that the decrease (quenching) in fluorescence intensity was due to the FRET effect. When the amounts of Oligo-Cy3 and Oligo-BHQ2 were fixed at 10 nmol, the fluorescence lifetime before and after annealing COVID 19-PEB were measured as 2.23 ± 0.02 ns and 2.10 ± 0.03 ns, respectively (Fig. 3c). This result indicated that COVID 19-PEB revealed both decreasing and increasing trends in fluorescence intensity due to the FRET effect. Thus, COVID19-PEB formed a stable hairpin structure without the target S proteins of SARS-CoV-2, thereby suppressing the fluorescence intensity (Fig. 3d). However, COVID 19-PEB treated with SARS-CoV-2 or S protein RBD significantly increased the fluorescence intensity time-dependently. After treatment of each sample (SARS-CoV-2, S protein, or BSA as a control) with COVID 19-PEB (10 nmol), the fluorescence intensity was measured every 10 min for 180 min (3 h) to evaluate the optimal sensing time of the SARS-CoV-2 S protein with COVID 19-PEB (Fig. S4). In the presence of the target S protein RBD or SARS-CoV-2, the fluorescence intensity gradually increased for 50 min, followed by a rapid increase afterward up to 3 h; moreover, the intensity change could be measured even within 1 h (Fig. S4). The fluorescence change was hardly observed in the BSA used as a control. Peptides without oligonucleotides can directly interact with the S protein RBD, which takes a relatively short time. (Fig. 3d) However, our probe is a FRET-based detection system that shows an increase in fluorescence intensity only after interacting with SARS-CoV-2. Therefore, the time taken for signal development depends on whether or not the oligonucleotide is bound to the peptide. Therefore, we tested various concentrations of the S protein using COVID 19-PEB. Their fluorescence intensities were measured after sensing 100 µL of S proteins (200, 100, 75, 50, 25, and 10 pmol) using COVID 19-PEB. After that, the relative fluorescence intensity was calculated based on the fluorescence intensity in the absence of target protein (NT, nontreatment). It was observed that the fluorescence intensity gradually increased with the increasing concentration of the S proteins (Fig. 3e). In particular, the detection limit (LOD) of COVID 19-PEB was calculated to be 52.5 pmol.[71] Collectively, these data suggest that the specific interaction of hACE2 residues within COVID 19-PEB with the SARS-CoV-2 S protein RBD may disrupt the hairpin structure. Subsequently, this event may trigger the formation of the hACE2 α1 helix structure binding to S protein RBD, leading to the quenched Cy3 opening for signal activation.
Fig. 3.
Fluorescence intensity of COVID19-PEB molecules prepared by conjugating a fixed concentration of Oligo-Cy3 with increasing concentrations of Oligo-BHQ2 (0, 1, 2, 4, 6, 8, and 10 nmol): a) COVID19-PEB fluorescence before annealing (violet) and after annealing (blue) and b) changes in fluorescence intensity (ΔF = Fbefore annealing − Fafter annealing). c) Fluorescence decay curves COVID19-PEB before annealing (violet) and after annealing (blue), Fluorescence detection of S protein RBD using COVID19-PEB: d) fluorescence intensity *<0.05, **<0.005 (n = 3), and e) relative fluorescence intensity (FNT, the fluorescence intensity in the absence of target protein; NT, nontreatment).
3.3. Confirmation of affinity between the synthesized COVID19-PEB and SARS-CoV-2 virus
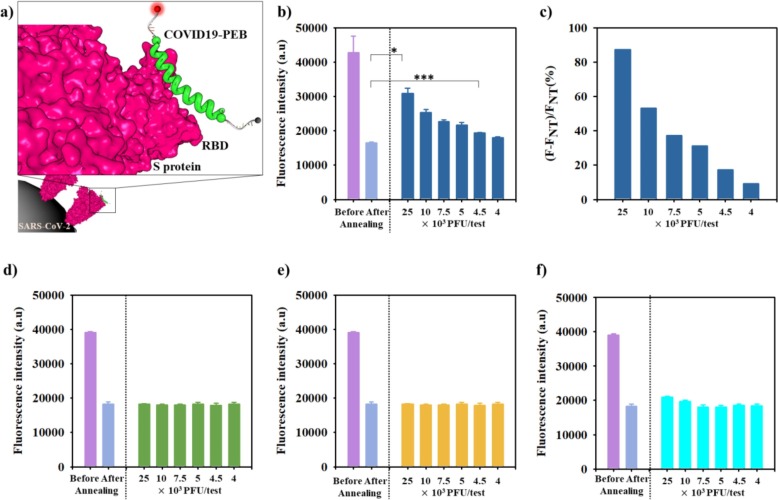
After confirming Cy3 opening following the specific interaction between the hACE2 residue in COVID 19-PEB and the SARS-CoV-2 S protein RBD, we performed experiment to detect SARS-CoV-2 virus (Fig. 4 a). Importantly, it is necessary to confirm that COVID 19-PEB can selectively detect only SARS-CoV-2 and no other viruses. We assessed the selective response of COVID 19-PEB relative to influenza A and B virus and a coronavirus that does not bind to ACE2. Since these viruses present similar symptoms when infected, their accurate diagnosis is necessary to treat appropriate antiviral agents. Fig. 4a illustrates the mechanism of action of COVID 19-PEB while detecting SARS-CoV-2. After culturing SARS-CoV-2, the change in fluorescence intensities was measured before and after adding COVID 19-PEB prepared by annealing (Fig. 4b and c). As previously described, after annealing, the fluorescence intensity of COVID 19-PEB was markedly suppressed (quenched) by the FRET effect compared with that before annealing. Furthermore, the fluorescence intensity of the annealed COVID 19-PEB without virus (nontreatment) was named FNT. After SARS-CoV-2 detection, the fluorescence intensity (F) increased proportionally after SARS-CoV-2 addition (Fig. 4b). The relative fluorescence intensities, calculated by ΔF/FNT (%) (ΔF = F − FNT), were significantly increased (250 × 104 PFU/mL: 87.3 %, 100 × 104 PFU/mL: 53.3 %, 7.5 × 104 PFU/mL: 37.2 %, 5 × 104 PFU/mL: 31.1%, 4.5 × 104 PFU/mL: 17.2 % and 4 × 104 PFU/mL: 9.1%) (Fig. 4c). However, no change in fluorescence signal was observed for control viruses (influenza A virus, influenza B virus, and HCoV-OC43) (Fig. 4 d–f).
Fig. 4.
a) Scheme showing fluorescence signal after COVID19-PEB interaction with the S protein of SARS-CoV-2. Fluorescence detection of decreasing concentrations of SARS-CoV-2 using a fixed concentration of COVID19-PEB: b) fluorescence intensity *<0.05, ***<0.0005 (n = 3) and c) relative fluorescence intensity. Fluorescence detection intensity of d) influenza A, e) influenza B and f) Coronavirus OC43 viruses based on concentration using COVID19-PEB (n = 3).
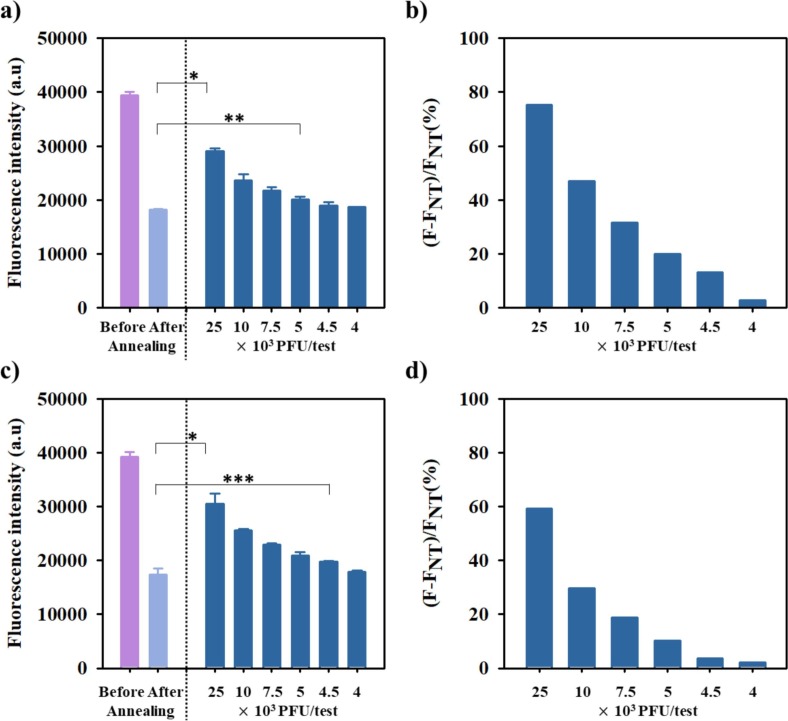
To test whether COVID 19-PEB is effective for testing real biofluids (saliva or nasal fluid), we performed an experiment using the virus in 10% nasal fluid and 10% saliva, respectively, in the manner mentioned above. FRET-based probes may cause a nonspecific reaction with a viscous fluid such as biofluid, thereby resulting in an unwanted increase in the fluorescence signal; however, COVID 19-PEB efficiently detected SARS-CoV-2 in the biofluid, with high sensitivity (Fig. 5 ).
Fig. 5.
Fluorescence detection of decreasing concentrations of SARS-CoV-2 in 10% saliva using fixed concentration of COVID19-PEB: a) total fluorescence intensity *<0.05, **<0.005 ***<0.0005 (n = 3) and b) specific fluorescence; same values are shown for SARS-CoV-2 in 10% nasal secretions: c) and d). (FNT: fluorescence intensity in the absence of SARS-CoV-2 (nontreatment, NT)).
3.4. Testing of clinical samples from SARS-CoV-2 patients with a detection system using COVID19-PEB
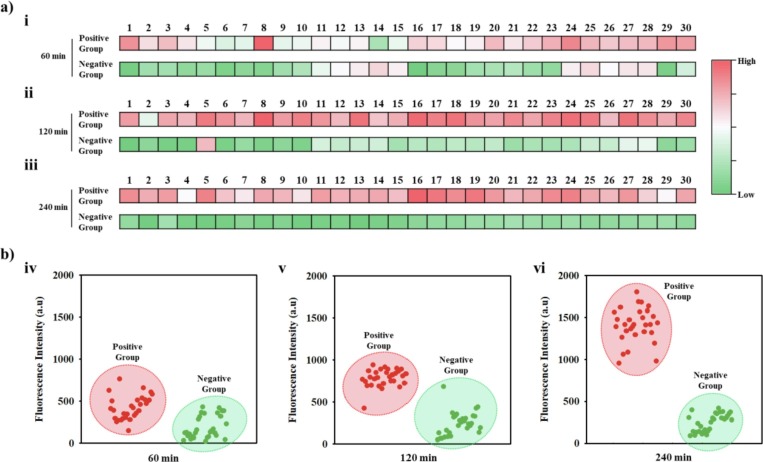
We tested the nasopharyngeal aspirate samples collected from 30 SARS-CoV-2 patients using COVID 19-PEB. Samples were collected from January 29 to February 16, 2021, at the Yonsei University Health Center, Severance Hospital, Korea, from patients diagnosed as positive for SARS-CoV-2 via qRT-PCR (Table S1). Similar to the SARS-CoV-2 test mentioned earlier, each patient sample (100 µL) without additional pretreatment was mixed with COVID 19-PEB, followed by measuring the fluorescence intensity every hour (60, 120, and 240 min). As a control group, 30 samples confirmed as negative in the qRT-PCR test result were also tested. This result is expressed as a heat map. The transition from green to red indicates the increase in fluorescence intensity; dark red indicates high fluorescence intensity, and dark green indicates low fluorescence intensity (Fig. 6 ). Furthermore, SARS-CoV-2 positive patients presented more red color on the heat map than the negative patients (Fig. 6a), and the fluorescence intensity expressed as a heat map was numerically graphed (Fig. 6b). With an increase in time, the contrast became clearer, and specifically at 240 min (4 h) after treatment, the positive and negative patients could be effectively distinguished. The results mentioned above indicate that our system could easily identify SARS-CoV-2 patients.
Fig. 6.
Confirmation of SARS-CoV-2 presence in clinical samples using COVID19-PEB a) Heat map and b) fluorescence distribution graph of fluorescence intensity after detecting SARS-CoV-2 in clinical samples using COVID19-PEB. Clinical samples were collected from human body fluids infected with SARS-CoV-2. (Positive samples: n = 30 and negative samples: n = 30). Detection time: 60 min (i and iv), 120 min (ii and v), and 240 min (iii and vi).
4. Conclusion
In summary, we developed a highly sensitive detection method for SARS-CoV-2 using COVID 19-PEB that directly detects SARS-CoV-2 by producing intense fluorescence; thus, this approach provides an alternative to genetic techniques, such as qRT-PCR assays. Moreover, this method detects low concentrations of SARS-CoV-2 within 3 h and is comparatively faster than the qRT-PCR assay time. Thus, COVID 19-PEB has great potential for use in multiple assays of SARS-CoV-2, where the multiple fluorophores and hACE2 mimic peptide sequences allow detection of interactions with specific S protein RBD. Recently, due to genetic mutation, the SARS-CoV-2 variants with increased infectivity (alpha, beta, gamma, and delta) have posed serious concerns. Most SARS-CoV-2 mutant viruses have mutated S protein RBD, allowing them to bind more aggressively to hACE2 in the host cells.[72], [73], [74], [75], [76], [77] Thus, whether diagnostic methods using bioreceptors (antibodies, peptides, and aptamers) targeting the SARS-CoV-2 S protein RBD can detect these variants remains unclear. Furthermore, this diagnostic method, based on a hACE2–mimic peptide, exhibiting affinity for SARS-CoV-2, could detect the binding affinity despite the presence of viral mutations. Additionally, this peptide exhibits a strong binding affinity to the S proteins of SARS-CoV-2 variants (alpha, beta, gamma, and delta) (Fig. S5 and S6). Although COVID 19-PEB does not effectively discriminate among the genetic mutations in SARS-CoV-2, it is useful for sensitive and rapid detection of SARS-CoV-2 and its variants.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by National R&D Programs through National Research Foundation (NRF) of Korea funded by Ministry of Science and ICT (MSIT) of Korea (NRF-2021M3E5E3080379, NRF-2021M3H4A1A02051048, NRF-2018M3A9E2022821, NRF-2021M3E5E3080844, NRF-2022R1C1C1008815, and NRF-2020R1A2C1010453), Global Frontier Program through Center for BioNano Health-Guard funded by MSIT of Korea (H-GUARD_2014M3A6B2060507 and H-GUARD_2013M3A6B2078950), Technology Development Program for Biological Hazards Management in Indoor Air through Korea Environment Industry & Technology Institute (KEITI) funded by Ministry of Environment (ME) of Korea (2021003370003), Industrial Technology Alchemist Program of the Ministry of Trade, Industry, and Energy (MOTIE) of Korea (20012435), Nanomedical Devices Development Program of National Nano Fab Center (CSM2105M101), National Research Council of Science & Technology (NST) grant by MSIT (No. CAP20012-000), and the KRIBB Research Initiative Program (1711134081).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cej.2022.136143.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. The New England Journal of Medicine. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Telenti A., Arvin A., Corey L., Corti D., Diamond M.S., Garcia-Sastre A., Garry R.F., Holmes E.C., Pang P.S., Virgin H.W. After the pandemic: perspectives on the future trajectory of COVID-19. Nature. 2021;596:495–504. doi: 10.1038/s41586-021-03792-w. [DOI] [PubMed] [Google Scholar]
- 3.Zhu N.a., Zhang D., Wang W., Li X., Yang B.o., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pekar J., Worobey M., Moshiri N., Scheffler K., Wertheim J.O. Timing the SARS-CoV-2 index case in Hubei province. Science. 2021;372(6540):412–417. doi: 10.1126/science.abf8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oxley T.J., Mocco J., Majidi S., Kellner C.P., Shoirah H., Singh I.P., De Leacy R.A., Shigematsu T., Ladner T.R., Yaeger K.A., Skliut M., Weinberger J., Dangayach N.S., Bederson J.B., Tuhrim S., Fifi J.T. Large-Vessel Stroke as a Presenting Feature of Covid-19 in the Young. The New England Journal of Medicine 382. 2020;382(20):e60. doi: 10.1056/NEJMc2009787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan A.T., Brownstein J.S. Putting the Public Back in Public Health — Surveying Symptoms of Covid-19. The New England Journal of Medicine 383. 2020;383(7):e45. doi: 10.1056/NEJMp2016259. [DOI] [PubMed] [Google Scholar]
- 9.Solomon C.G., Berlin D.A., Gulick R.M., Martinez F.J. Severe Covid-19. N Engl J Med. 2020;383(25):2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 10.Tummino T.A., Rezelj V.V., Fischer B., Fischer A., O’Meara M.J., Monel B., Vallet T., White K.M., Zhang Z., Alon A., Schadt H., O’Donnell H.R., Lyu J., Rosales R., McGovern B.L., Rathnasinghe R., Jangra S., Schotsaert M., Galarneau J.-R., Krogan N.J., Urban L., Shokat K.M., Kruse A.C., García-Sastre A., Schwartz O., Moretti F., Vignuzzi M., Pognan F., Shoichet B.K. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021;373(6554):541–547. doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drayman N., DeMarco J.K., Jones K.A., Azizi S.-A., Froggatt H.M., Tan K., Maltseva N.I., Chen S., Nicolaescu V., Dvorkin S., Furlong K., Kathayat R.S., Firpo M.R., Mastrodomenico V., Bruce E.A., Schmidt M.M., Jedrzejczak R., Muñoz-Alía M.Á., Schuster B., Nair V., Han K.-Y., O’Brien A., Tomatsidou A., Meyer B., Vignuzzi M., Missiakas D., Botten J.W., Brooke C.B., Lee H., Baker S.C., Mounce B.C., Heaton N.S., Severson W.E., Palmer K.E., Dickinson B.C., Joachimiak A., Randall G., Tay S. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science. 2021;373(6557):931–936. doi: 10.1126/science.abg5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahmani A.M., Mirmahaleh S.Y.H. Coronavirus disease (COVID-19) prevention and treatment methods and effective parameters: A systematic literature review. Sustain Cities Soc. 2021;64:102568–102585. doi: 10.1016/j.scs.2020.102568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girum T., Lentiro K., Geremew M., Migora B., Shewamare S. Global strategies and effectiveness for COVID-19 prevention through contact tracing, screening, quarantine, and isolation: a systematic review, Trop. Med Health. 2020;48:91–105. doi: 10.1186/s41182-020-00285-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiss C., Carriere M., Fusco L., Capua I., Regla-Nava J.A., Pasquali M., Scott J.A., Vitale F., Unal M.A., Mattevi C., Bedognetti D., Merkoçi A., Tasciotti E., Yilmazer A., Gogotsi Y., Stellacci F., Delogu L.G. Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. ACS Nano. 2020;14(6):6383–6406. doi: 10.1021/acsnano.0c03697. [DOI] [PubMed] [Google Scholar]
- 15.Steiner D.J., Cognetti J.S., Luta E.P., Klose A.M., Bucukovski J., Bryan M.R., Schmuke J.J., Nguyen-Contant P., Sangster M.Y., Topham D.J., Miller B.L. Array-based analysis of SARS-CoV-2, other coronaviruses, and influenza antibodies in convalescent COVID-19 patients. Biosens Bioelectron. 2020;169:112643–112650. doi: 10.1016/j.bios.2020.112643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahapatra S., Chandra P. Clinically practiced and commercially viable nanobio engineered analytical methods for COVID-19 diagnosis. Biosens Bioelectron. 2020;165:112361–112373. doi: 10.1016/j.bios.2020.112361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon J., Kwon H.-J., Yong D., Lee I.-C., Kim H., Kang H., Lim E.-K., Lee K.-S., Jung J., Park H.G., Kang T. Colorimetric Detection of SARS-CoV-2 and Drug-Resistant pH1N1 Using CRISPR/dCas9. ACS Sens. 2020;5(12):4017–4026. doi: 10.1021/acssensors.0c01929. [DOI] [PubMed] [Google Scholar]
- 18.Sheikhzadeh E., Eissa S., Ismail A., Zourob M. Diagnostic techniques for COVID-19 and new developments. Talanta. 2020;220:121392–121408. doi: 10.1016/j.talanta.2020.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Häffner S.M., Parra-Ortiz E., Browning K.L., Jørgensen E., Skoda M.W.A., Montis C., Li X., Berti D., Zhao D., Malmsten M. Membrane Interactions of Virus-like Mesoporous Silica Nanoparticles. ACS Nano. 2021;15(4):6787–6800. doi: 10.1021/acsnano.0c10378. [DOI] [PubMed] [Google Scholar]
- 20.Wang C., Wang S., Chen Y., Zhao J., Han S., Zhao G., Kang J., Liu Y., Wang L., Wang X., Xu Y., Wang S., Huang Y.i., Wang J., Zhao J. Membrane Nanoparticles Derived from ACE2-Rich Cells Block SARS-CoV-2 Infection. ACS Nano. 2021;15(4):6340–6351. doi: 10.1021/acsnano.0c06836. [DOI] [PubMed] [Google Scholar]
- 21.Li J., Lillehoj P.B. Microfluidic Magneto Immunosensor for Rapid, High Sensitivity Measurements of SARS-CoV-2 Nucleocapsid Protein in Serum. ACS Sens. 2021;6(3):1270–1278. doi: 10.1021/acssensors.0c02561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson D., Lei Y. Mini review: Recent progress in RT-LAMP enabled COVID-19 detection. Sensors and Actuators Reports. 2020;2:100017–100026. doi: 10.1016/j.snr.2020.100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F., Yang J., He R., Yu X., Chen S., Liu Y., Wang L., Li A., Liu L., Zhai C., Ma L. PfAgo-based detection of SARS-CoV-2. Biosens Bioelectron. 2021;177:112932–112937. doi: 10.1016/j.bios.2020.112932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voss W.N., Hou Y.J., Johnson N.V., Delidakis G., Kim J.E., Javanmardi K., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., Chou C.-W., Abbasi S.A., Pickens W., George K., Boutz D.R., Towers D.M., McDaniel J.R., Billick D., Goike J., Rowe L., Batra D., Pohl J., Lee J., Gangappa S., Sambhara S., Gadush M., Wang N., Person M.D., Iverson B.L., Gollihar J.D., Dye J.M., Herbert A.S., Finkelstein I.J., Baric R.S., McLellan J.S., Georgiou G., Lavinder J.J., Ippolito G.C. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science. 2021;372(6546):1108–1112. doi: 10.1126/science.abg5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F.u., Ramirez S.A.S.d., Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(37):22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo C.H., Jang S., Shin G., Jung G.Y., Lee J.W. Sensitive fluorescence detection of SARS-CoV-2 RNA in clinical samples via one-pot isothermal ligation and transcription. Nat Biomed Eng. 2020;4(12):1168–1179. doi: 10.1038/s41551-020-00617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang B.-H., Lee Y., Yu E.-S., Na H., Kang M., Huh H.J., Jeong K.-H. Ultrafast and Real-Time Nanoplasmonic On-Chip Polymerase Chain Reaction for Rapid and Quantitative Molecular Diagnostics. ACS Nano. 2021;15(6):10194–10202. doi: 10.1021/acsnano.1c02154. [DOI] [PubMed] [Google Scholar]
- 28.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., Fusco D., Drouin A., Yin X., Hu T., Ning B. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens Bioelectron. 2020;164:112316–112322. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dziabowska K., Czaczyk E., Nidzworski D. Detection Methods of Human and Animal Influenza Virus-Current Trends. Biosensors (Basel) 2018;8:1–24. doi: 10.3390/bios8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roda A., Cavalera S., Di Nardo F., Calabria D., Rosati S., Simoni P., Colitti B., Baggiani C., Roda M., Anfossi L. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens Bioelectron. 2021;172:112765–112770. doi: 10.1016/j.bios.2020.112765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheong J., Yu H., Lee C.Y., Lee J.-u., Choi H.-J., Lee J.-H., Lee H., Cheon J. Fast detection of SARS-CoV-2 RNA via the integration of plasmonic thermocycling and fluorescence detection in a portable device. Nat Biomed Eng. 2020;4(12):1159–1167. doi: 10.1038/s41551-020-00654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim E., Lim E.-K., Park G., Park C., Lim J.-W., Lee H., Na W., Yeom M., Kim J., Song D., Haam S. Advanced Nanomaterials for Preparedness Against (Re-)Emerging Viral Diseases. Adv Mater. 2021;33(47):2005927. doi: 10.1002/adma.202005927. [DOI] [PubMed] [Google Scholar]
- 33.Mahshid S.S., Flynn S.E., Mahshid S. The potential application of electrochemical biosensors in the COVID-19 pandemic: A perspective on the rapid diagnostics of SARS-CoV-2. Biosens Bioelectron. 2021;176:112905–112914. doi: 10.1016/j.bios.2020.112905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 35.Kang Y.-F., Sun C., Zhuang Z., Yuan R.-Y., Zheng Q., Li J.-P., Zhou P.-P., Chen X.-C., Liu Z., Zhang X., Yu X.-H., Kong X.-W., Zhu Q.-Y., Zhong Q., Xu M., Zhong N.-S., Zeng Y.-X., Feng G.-K., Ke C., Zhao J.-C., Zeng M.-S. Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano. 2021;15(2):2738–2752. doi: 10.1021/acsnano.0c08379. [DOI] [PubMed] [Google Scholar]
- 36.Pinals R.L., Ledesma F., Yang D., Navarro N., Jeong S., Pak J.E., Kuo L., Chuang Y.-C., Cheng Y.-W., Sun H.-Y., Landry M.P. Rapid SARS-CoV-2 Spike Protein Detection by Carbon Nanotube-Based Near-Infrared Nanosensors. Nano Lett. 2021;21(5):2272–2280. doi: 10.1021/acs.nanolett.1c00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X., Wang X., Li S., Luo W., Zhang X., Wang C., Chen Q., Yu S., Tai J., Wang Y.i. Rapid, Ultrasensitive, and Highly Specific Diagnosis of COVID-19 by CRISPR-Based Detection. ACS Sens. 2021;6(3):881–888. doi: 10.1021/acssensors.0c01984. [DOI] [PubMed] [Google Scholar]
- 38.Zhao B., Che C., Wang W., Li N., Cunningham B.T. Single-step, wash-free digital immunoassay for rapid quantitative analysis of serological antibody against SARS-CoV-2 by photonic resonator absorption microscopy. Talanta. 2021;225:122004–122011. doi: 10.1016/j.talanta.2020.122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bobrin V.A., Chen S.-P., Grandes Reyes C.F., Sun B., Ng C.K., Kim Y., Purcell D., Jia Z., Gu W., Armstrong J.W., McAuley J., Monteiro M.J. Water-Borne Nanocoating for Rapid Inactivation of SARS-CoV-2 and Other Viruses. ACS Nano. 2021;15(9):14915–14927. doi: 10.1021/acsnano.1c05075. [DOI] [PubMed] [Google Scholar]
- 40.Who SARS-CoV-2 antigen-detecting rapid diagnostic tests, Disease Control in Humanitarian Emergencies, Emergencies Preparedness. WHO Headquarters (HQ) 2020 [Google Scholar]
- 41.Antiochia R. Developments in biosensors for CoV detection and future trends. Biosens Bioelectron. 2020;173:112777–112786. doi: 10.1016/j.bios.2020.112777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J.H., Choi M., Jung Y., Lee S.K., Lee C.S., Kim J., Kim J., Kim N.H., Kim B.T., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2. ACE2Biosens Bioelectron. 2021;171:112715–112724. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhalla N., Pan Y., Yang Z., Payam A.F. Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yakoh A., Pimpitak U., Rengpipat S., Hirankarn N., Chailapakul O., Chaiyo S. Paper-based electrochemical biosensor for diagnosing COVID-19: Detection of SARS-CoV-2 antibodies and antigen. Biosens Bioelectron. 2021;176:112912–112919. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorshkov K., Susumu K., Chen J., Xu M., Pradhan M., Zhu W., Hu X., Breger J.C., Wolak M., Oh E. Quantum Dot-Conjugated SARS-CoV-2 Spike Pseudo-Virions Enable Tracking of Angiotensin Converting Enzyme 2 Binding and Endocytosis. ACS Nano. 2020;14(9):12234–12247. doi: 10.1021/acsnano.0c05975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen E.C., Gilchuk P., Zost S.J., Suryadevara N., Winkler E.S., Cabel C.R., Binshtein E., Chen R.E., Sutton R.E., Rodriguez J., Day S., Myers L., Trivette A., Williams J.K., Davidson E., Li S., Doranz B.J., Campos S.K., Carnahan R.H., Thorne C.A., Diamond M.S., Crowe J.E., Jr. Convergent antibody responses to the SARS-CoV-2 spike protein in convalescent and vaccinated individuals. Cell Rep. 2021;36:109604–109628. doi: 10.1016/j.celrep.2021.109604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shang J., Ye G., Shi K.e., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369(6508):1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sztain T., Ahn S.-H., Bogetti A.T., Casalino L., Goldsmith J.A., Seitz E., McCool R.S., Kearns F.L., Acosta-Reyes F., Maji S., Mashayekhi G., McCammon J.A., Ourmazd A., Frank J., McLellan J.S., Chong L.T., Amaro R.E. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat Chem. 2021;13(10):963–968. doi: 10.1038/s41557-021-00758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11:2251–2256. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q., Xiang R., Huo S., Zhou Y., Jiang S., Wang Q., Yu F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct Target Ther. 2021;6:233–251. doi: 10.1038/s41392-021-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol. 2020;27(9):846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 56.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., Edwards C., Weimer E., Scherer E., Rouphael N., Edupuganti S., Weiskopf D., Tse L., Hou Y., Margolis D., Sette A., Collins M., Schmitz J., Baric R., Silva A. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:8413–8426. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.-C., Tiu C., Hu Z., Chen V.-W., Young B.E., Sia W.R., Tan Y.-J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.-F. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38(9):1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 59.Yan R., Zhang Y., Li Y., Xia L.u., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q.i., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 61.Youk J., Kim T., Evans K.V., Jeong Y.-I., Hur Y., Hong S.P., Kim J.H., Yi K., Kim S.Y., Na K.J., Bleazard T., Kim H.M., Fellows M., Mahbubani K.T., Saeb-Parsy K., Kim S.Y., Kim Y.T., Koh G.Y., Choi B.-S., Ju Y.S., Lee J.-H. Three-Dimensional Human Alveolar Stem Cell Culture Models Reveal Infection Response to SARS-CoV-2. Cell Stem Cell. 2020;27(6):905–919.e10. doi: 10.1016/j.stem.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han Y., Král P. Computational Design of ACE2-Based Peptide Inhibitors of SARS-CoV-2. ACS Nano. 2020;14(4):5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thurley S., Roglin L., Seitz O. Hairpin Peptide Beacon Dual-Labeled PNA-Peptide-Hybrids for Protein Detection. J. AM. CHEM. SOC. 2007;129:12693–12695. doi: 10.1021/ja075487r. [DOI] [PubMed] [Google Scholar]
- 64.Karoyan P., Vieillard V., Gomez-Morales L., Odile E., Guihot A., Luyt C.E., Denis A., Grondin P., Lequin O. Human ACE2 peptide-mimics block SARS-CoV-2 pulmonary cells infection. Commun Biol. 2021;4:197–205. doi: 10.1038/s42003-021-01736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han D.P., Penn-Nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350(1):15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okochi M., Sugita T., Tanaka M., Honda H. A molecular peptide beacon for IgG detection. RSC Adv. 2015;5(112):91988–91992. [Google Scholar]
- 67.Huang L., Sexton D.J., Skogerson K., Devlin M., Smith R., Sanyal I., Parry T., Kent R., Enright J., Wu Q.-L., Conley G., DeOliveira D., Morganelli L., Ducar M., Wescott C.R., Ladner R.C. Novel peptide inhibitors of angiotensin-converting enzyme 2. J Biol Chem. 2003;278(18):15532–15540. doi: 10.1074/jbc.M212934200. [DOI] [PubMed] [Google Scholar]
- 68.Liu Q., Wang J., Boyd B.J. Peptide-based biosensors. Talanta. 2015;136:114–127. doi: 10.1016/j.talanta.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 69.Lim E.-K., Guk K., Kim H., Chung B.-H., Jung J. Simple, rapid detection of influenza A (H1N1) viruses using a highly sensitive peptide-based molecular beacon. Chem Commun (Camb) 2016;52(1):175–178. doi: 10.1039/c5cc05684e. [DOI] [PubMed] [Google Scholar]
- 70.Cao L., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., Chen R.E., Carter L., Walls A.C., Park Y.-J., Strauch E.-M., Stewart L., Diamond M.S., Veesler D., Baker D. De novo design of picomolar sars-cov-2 miniprotein inhibitors. Science. 2020;370(6515):426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Armbruster D., Pry T. Limit of Blank, Limit of Detection and Limit of Quantitation. Clin Biochem Rev. 2008;29:1–4. [PMC free article] [PubMed] [Google Scholar]
- 72.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 73.Jakhmola S., Indari O., Kashyap D., Varshney N., Das A., Manivannan E., Jha H.C. Mutational analysis of structural proteins of SARS-CoV-2. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e06572. e06572-06585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yuan M., Huang D., Lee C.-C., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., Burton D.R., Reincke S.M., Prüss H., Kreye J., Nemazee D., Ward A.B., Wilson I.A. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373(6556):818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cai Y., Zhang J., Xiao T., Lavine C.L., Rawson S., Peng H., Zhu H., Anand K., Tong P., Gautam A., Lu S., Sterling S.M., Walsh R.M., Rits-Volloch S., Lu J., Wesemann D.R., Yang W., Seaman M.S., Chen B. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science. 2021;373(6555):642–648. doi: 10.1126/science.abi9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh J., Samal J., Kumar V., Sharma J., Agrawal U., Ehtesham N.Z., Sundar D., Rahman S.A., Hira S., Hasnain S.E. Structure-Function Analyses of New SARS-CoV-2 Variants B.1.1.7, B.1.351 and B.1.1.28.1: Clinical, Diagnostic, Therapeutic and Public Health Implications. Viruses. 2021;13:439–454. doi: 10.3390/v13030439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.