Abstract
Extracellular vesicles (EVs) derived from various stem cell sources induce cardioprotective effects during ischemia-reperfusion injury (IRI). These have been attributed mainly to the anti-apoptotic, pro-angiogenic, miRNA cargo within the stem cell-derived EVs. However, the mechanisms of EV-mediated endothelial signaling to cardiomyocytes, as well as their therapeutic potential toward ischemic myocardial injury, are not clear. EV content beyond miRNA that may contribute to cardioprotection have not been fully illuminated. This study characterized the protein cargo of human vascular endothelial EVs (EEVs) to identify lead cardioactive proteins and assessed the effect of EEVs on human laminar cardiac tissues (hlCTs) exposed to IRI. We mapped the protein content of human vascular EEVs and identified proteins that were previously associated with cellular metabolism, redox state, and calcium handling, among other processes. Analysis of the protein landscape of human cardiomyocytes revealed corresponding modifications induced by EEVs treatment. To assess their human-specific cardioprotection in vitro, we developed a human heart-on-a-chip (hHOC) IRI assay using human stem cell-derived, engineered cardiac tissues. We found that EEVs alleviated cardiac cell death as well as the loss in contractile capacity during and after simulated IRI in an uptake- and dose-dependent manner. Moreover, we found that EEVs increased the respiratory capacity of normoxic cardiomyocytes. These results suggest that vascular EEVs rescue hlCTs exposed to IRI possibly by supplementing injured myocytes with cargo that supports multiple metabolic and salvage pathways, and therefore may serve as a multi-targeted therapy for IRI.
One Sentence Summary:
Endothelial extracellular vesicles containing cardioprotective proteins rescue engineered human cardiac tissue in an ischemia-reperfusion injury model.
Introduction
Each year, over half a million Americans suffer from myocardial infarction (MI) (1). The most effective therapeutic intervention requires timely and complete restoration of arterial blood flow to the affected area (2). However, this process may induce further cardiomyocyte death and contribute to the final infarct size (3), the primary determinant of left ventricular recovery prospects and potential of heart failure (4). These conjugated events, termed ischemia–reperfusion injury (IRI), eventually escalate to the development of ischemic heart disease (2, 4, 5). Thus, additional cardioprotective strategies are needed (6). The cellular response to IRI involves multiple pathophysiological mechanisms, such as calcium and proton overload, oxidative stress, endoplasmic reticulum stress, mitochondrial dysfunction, activation of apoptotic pathways (5), and myocardial hibernation (7). This complex set of processes impedes the development of effective therapies. Additionally, patients with MI often suffer other comorbidities, which affect the efficacy of treatment. Therefore, due to the complexity of the disease, it is suggested that optimal cardioprotection may require synergistic, multi-targeted therapy (8). Further, the species-dependency of the response to IRI (5, 9) renders it difficult to translate preclinical findings to clinical protocols, imposing a pressing need for developing human-specific preclinical assays.
Several pharmacological, mechanical, and cell-based therapies have been investigated in recent years. Ischemic postconditioning, involving short cycles of ischemia and reperfusion immediately after reflow, has been shown to exert a protective effect (2), while tissue revascularization approaches (10, 11) salvage viable myocardium but fail to restore apoptotic tissue. Moreover, pharmacological interventions that target isolated molecules or signaling pathways have shown unsatisfactory results. For example, calcium channel blockers, antioxidants, and anti-inflammatory agents have proven ineffective in reducing infarct size and improving clinical metrics (12). Similarly, agents targeting mitochondria and modulation of nitric oxide signaling have failed in recent clinical trials (4). Cell therapy approaches are generally safe, yet their efficacy remains controversial (13). Several clinical trials have found little benefit in transplantation of bone marrow-derived stem cells for treating myocardial infarction in patients (14, 15). Nevertheless, recent data suggest that a broad spectrum of factors secreted by stem cells of various sources may be highly therapeutic in cardiovascular disease (16, 17). Specifically, stem cell-derived extracellular vesicles (EVs), particularly exosomes, are now believed to be the key source of beneficial effects related to stem cells. These EVs have been shown to simultaneously trigger angiogenic and anti-apoptotic processes mediated by specific miRNA cargo (18–23). Interestingly, it has recently been demonstrated that naïve, non-stem cell exosomes alleviated cardiomyocyte death after IRI. This has been suggested to occur via activation of the membrane receptor-dependent ERK1/2 pathway (24, 25). In view of this evidence, it is unclear whether these EVs exert their paracrine effect by binding to membrane receptors that initiate intracellular signaling cascades or by directly delivering RNA and protein cargo across tissues and organs.
Vascular endothelial cells, which are abundant in perfused tissues, act as ‘first responders’ to hypoxic stress (26): They directly interface the parenchymal cells, and therefore are highly involved in paracrine signaling to body tissues, including the highly vascularized heart (27). Similar to other cell types (28, 29), endothelial cells secrete EVs, such as exosomes, which convey messages in the form of protein and RNA cargo to recipient cells (e.g., cardiomyocytes). We hypothesized that endothelial cell-derived extracellular vesicles (EEVs) contain cargo that can impart direct protective effects on cardiac muscle. To assess whether EEVs’ protein content could play a role mediating such effects, we mapped the vascular EEVs’ proteome. Doing so, we found a diverse population of over 1800 proteins with a high abundance of likely candidates for cytoprotection in ischemia-reperfusion injury, including several proteins previously associated with ischemic preconditioning. We tested the preconditioning-like cardioprotective effect exerted by these EEVs on human tissue using a human tissue heart-on-chip (hHOC) IRI assay. In the hHOC model, human cardiac myocytes (hCMs) are assembled into a layer of ventricular-like muscle, and tissue contractile function is continuously monitored during and after ischemic conditions using embedded sensors (28, 29). We found that EEVs derived from normoxic and hypoxic endothelial cell cultures increased respiratory capacity in normoxic cardiomyocytes and limited myocardial cell death and loss of contractile function associated with IRI. Proteomics analysis demonstrated that EEV treatment resulted in a shift of the protein landscape of myocytes from an IR-injured to non-injured profile. Furthermore, we observed that the EEV-mediated cardioprotective effect was dose-dependent and required internalization. In light of the reported findings, we propose that EEVs protect human laminar cardiac tissue (hlCT) from IRI in part by supplementing injured myocytes with proteins that support multiple metabolic and salvage pathways—thus paving a way for new therapeutic approaches. Simultaneously, this study demonstrates the use of the hHOC system for modeling IRI in vitro, with real time data for assessing therapeutic strategies and their dynamic effects on IRI.
Results
Endothelium-derived extracellular vesicles isolation and characterization
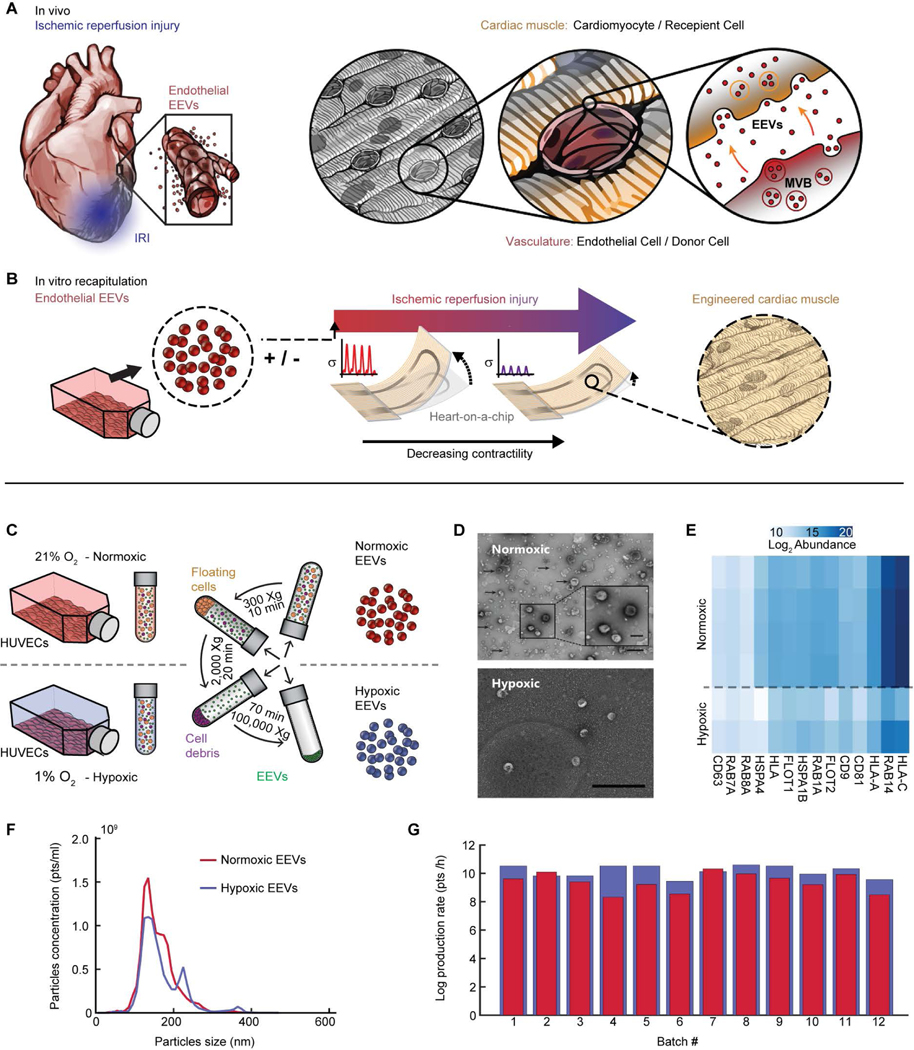
Endothelial-derived EVs may directly influence cardiac muscle viability and contractile function in IRI (Fig. 1A). To elucidate such effects, we produced EVs from human umbilical vein endothelial cell (HUVEC) culture and designed an in vitro system whereby contractile function of hCM-derived laminar cardiac tissues is measured continuously in a controlled environment (Fig. 1B). To test whether EV secretion from endothelial cells is modulated in response to hypoxia, we isolated EVs from normoxic (21% O2) and hypoxic (1% O2) HUVEC cultures (Fig. 1C). Transmission electron microscopy (TEM) micrographs support the presence of EVs in the acquired pellet (Fig. 1D). Proteomics analysis further confirmed that the isolated fraction contained subpopulations of exosomes by showing an abundance of known exosomal markers (30), such as tetraspanins (CD9, CD63, CD81), HSP70, RAB proteins, and others (Fig. 1E) (31). The average vesicle diameter was 163±10 nm and 151±7 nm for normoxic and hypoxic EEVs, respectively; both within the upper size range for exosomes (Fig. 1F). EEV yield per isolation (25–30 T175 culture flasks) was in the range of 109-1011, depending on HUVEC culturing conditions. The EEV production rate was 6.5±1.7-times higher for hypoxic compared to normoxic EEVs (Fig. 1G). Overall, our data suggest that both normoxic and hypoxic endothelial cultures produce heterogeneous populations of EVs (e.g., exosomes and microvesicles) that can be isolated using differential ultracentrifugation.
Fig. 1. Simulating ischemia-reperfusion injury to assess cardioprotection by endothelial EVs.
(A) Schematic of the vasculature-rich heart tissue lined with endothelial cells interfacing the cardiomyocytes. This allows for paracrine signaling affecting cardiomyocytes in health and in ischemic stress conditions. EEVs can be internalized by target cells, and transfer messages by delivering proteins and RNA. (B) Schematic of heart–on–a–chip, which allows for real time measurements of contractile function, with human laminar cardiac tissues (hlCTs) to simulate IRI in the presence of EEVs. (C) Schematic diagram showing EEV isolation by a series of centrifugation steps. EVs were isolated from either normoxic (21% O2) or hypoxic (1% O2) HUVEC cultures. (D) TEM images of normoxic (top) and hypoxic (bottom) EVs confirm expected cup shape; scale bar= 500 nm; close-up scale bar= 200 nm. (E) Mass spectrometry analysis confirmed the extracted pellet contained EVs including exosomes, by identifying known exosomal markers. (F) Size distribution for normoxic (red) and hypoxic (blue) vesicles. (G) EEVs production rate calculated from the EEV yield divided by hours in culture before harvest, for each isolation batch.
Mass spectrometry assessment of EEV protein content
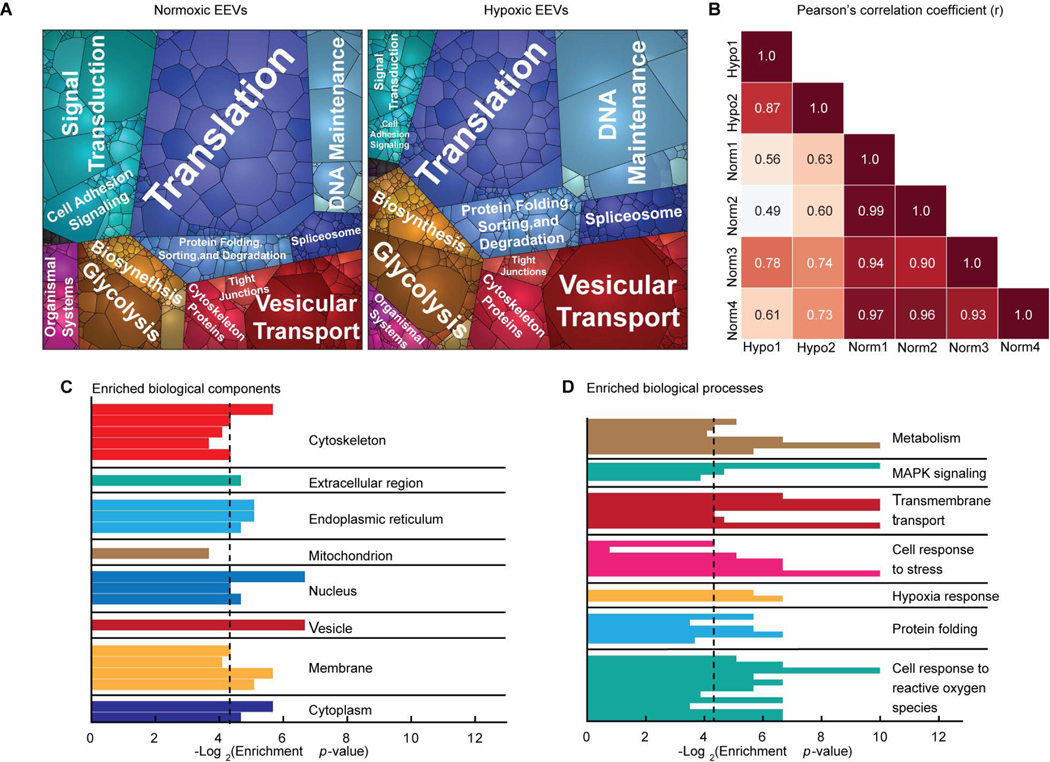
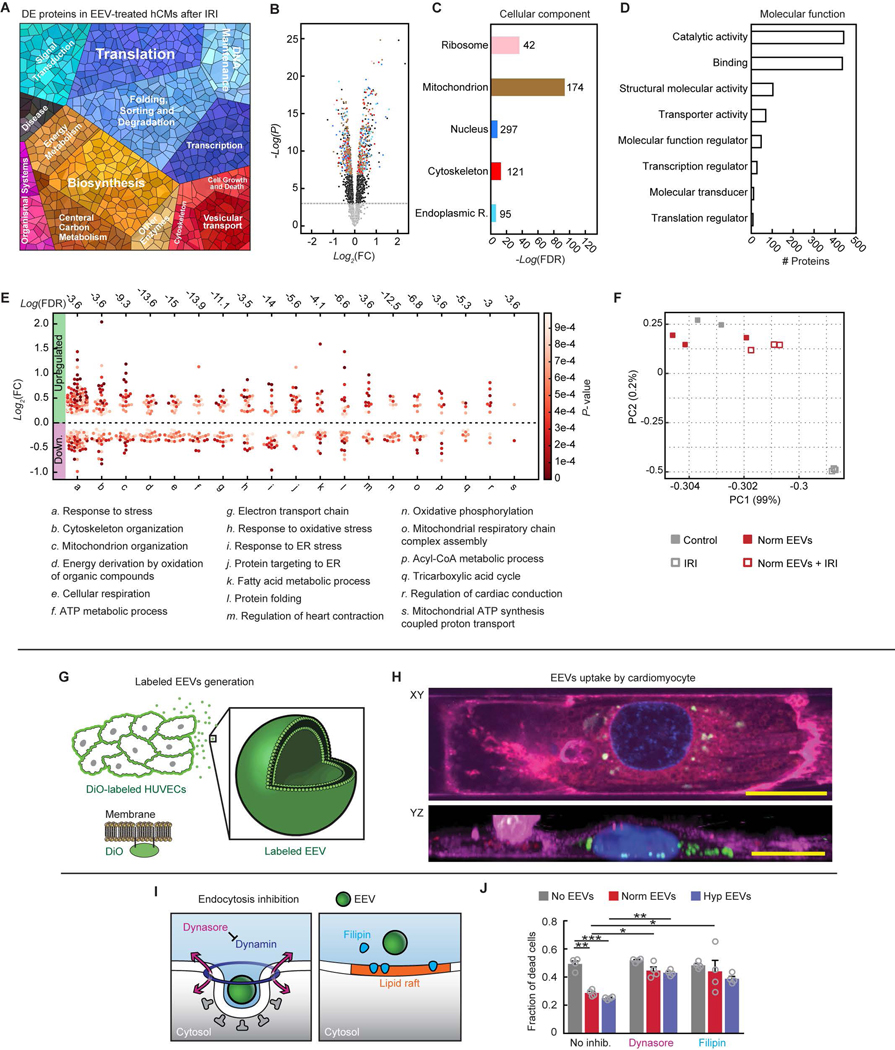
To characterize the putative EV-mediated signaling between endothelial cells and cardiomyocytes, we mapped and analyzed the endothelial cell-derived EV protein cargo. After removing common protein contaminants, including some of bovine origin (table S1), we identified >1,800 proteins in both the normoxic and hypoxic EEV samples (data file S1). The number of identified proteins is consistent with recent proteomic studies of other EVs’ proteomes (32). To assess the proteins’ biological functions and potential physiological relevance to cardioprotection, we used a battery of bioinformatics analyses. The proteomaps (Fig. 2A) exhibited a high degree of diversity in the biological functions of the EEVs proteomes, particularly with respect to signaling intermediates, metabolic proteins, and protein expression/protein folding factors. A strikingly high abundance of ribosomal and protein folding-related proteins was presented in the maps. This result suggests that EEVs may enhance protein synthesis and regulation of protein structure, as well as play a role in translation, transcription, DNA repair, regulation of cell growth, and apoptosis (33–35). Ribosomal proteins have previously been reported in EV fractions isolated using ultracentrifugation (36) and sucrose gradient techniques (37, 38). Yet, these were absent in fractions isolated using size exclusion chromatography (39) or high-resolution density gradients (31). Therefore, EV isolation methods have fundamental influence on detectable content.
Fig. 2. Endothelial EVs characterization.
(A) Proteomaps illustrating the KEGG orthology terms and relative differences in mass abundance for the 1,820 proteins identified in normoxic (left, N=4) and hypoxic (right, N=2) EVs. (B) Sample-wise Pearson’s correlation analysis revealed a high degree of similarity between the global expression profiles for the normoxic (Norm) and hypoxic (Hypo) EV samples. Correlation coefficients between the four normoxic samples were greater than 0.9, and 0.87 for the two hypoxic samples. Gene ontology enrichment analysis identified statistically overrepresented (C) cellular localization and (D) biological function in the protein expression data set. Black vertical lines indicate P-value of 0.05.
We then compared protein content between EEVs derived from normoxic and hypoxic conditions. Strong positive correlations were found between the technical replicates within these sample groups, and moderately positive correlations were found between the normoxic and hypoxic samples (Fig. 2B). These correlations suggest consistency between sample runs and similarity between the normoxic and hypoxic EEVs. Comparing the protein content of normoxic and hypoxic EEV samples, we identified fewer than 100 proteins that were significantly (p≤0.05) differentially expressed between normoxic and hypoxic conditions (fig. S1A). The majority of differentially expressed proteins were related to genetic information processing. Smaller subsets of the differentially expressed proteins where related to metabolism, cellular processes, and environmental information processing (fig. S1B).
Subsequently, to identify the cellular compartments (Fig. 2C) and biological processes (Fig. 2D) that were significantly (P ≤ 0.05) overrepresented in the data set, we used gene ontology enrichment analysis. Overrepresented clusters included cytoskeleton, endoplasmic reticulum, membrane and nuclear proteins in the compartment cluster, and metabolism, transport, protein folding, and cellular response to stress and ROS in the biological processes cluster (table S2, A and B). Metabolic proteins were related to glucose metabolism, such as glycolytic enzymes and lactate dehydrogenase (LDH), as well as fatty acid metabolism and cellular respiration.
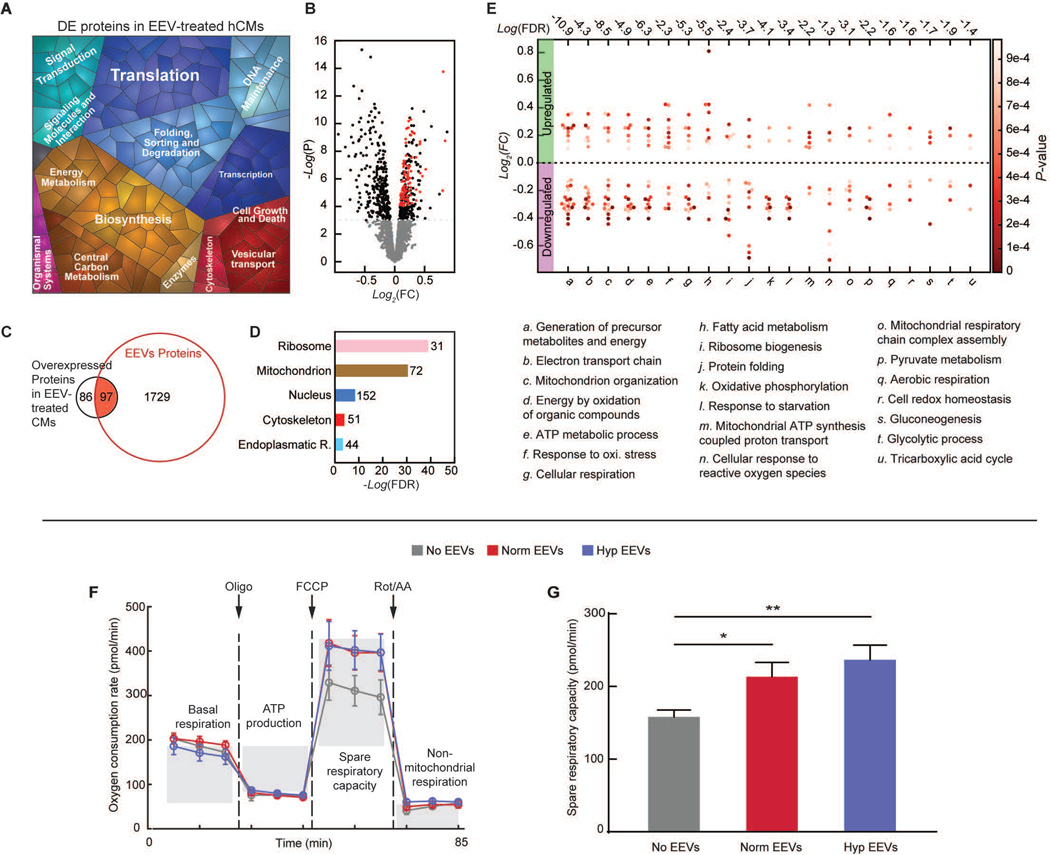
To investigate the direct effect of the EEVs on hCMs, we mapped and analyzed the protein content of normoxic EEV-loaded versus untreated hCMs. A proteomap representing the significantly (P ≤ 0.05) differentially expressed proteins (DEPs, data file S2) is presented in Fig. 3A. The signature of this proteomap is similar to the one representing the EEVs cargo (Fig. 2A). Overall, 394 significantly DEPs were detected, 183 of which were overexpressed in the EEV-treated myocytes. Of the 183 overexpressed proteins, 97 proteins were enriched in EEVs (Fig. 3, B and C, overlapping proteins labeled in red). Again, we used gene ontology enrichment analysis to identify the cellular compartments and biological processes. Overrepresented clusters of the DEPs included cytoskeleton, endoplasmic reticulum, nuclear and mitochondrial proteins (Fig. 3D), as did the EEVs. Further analysis of the DEPs revealed changes in various biological processes including cellular response to stress, metabolism, respiration, and energy derivation (Fig. 3E), as well as translation and signal transduction (Fig. 3A). Major molecular function and biological process classification of the DEPs are depicted in figs. S2 and S3.
Fig. 3. Endothelial EVs’ effects on hCMs.
(A) Proteomap illustrating the KEGG orthology terms and relative differences in mass abundance for the significantly (P-value <0.05) differentially expressed proteins (DEPs) in EEV-treated (N=3 samples), compared with untreated (N=2 samples), hCMs. (B) Volcano plot presenting the fold change and p-value of the DEPs. The red dots represent proteins that were significantly overexpressed in the EEV-treated myocytes and also enriched in the EEVs. (C) A Venn diagram demonstrating the amount of the DEPs in the EEV-treated myocytes, relative to the normoxic EEVs’ proteome, and the overlap. (D) Cellular localization in the protein expression data set. The numbers next to the bars represent the number of significantly (P≤0.05) enriched proteins related to each of the depicted cellular compartments. (E) Dominant biological processes (BPs) in the DEPs. Each dot represents a single protein, whereas the clusters represent BPs. The red color scale represents the individual p-value for the enrichment of each protein in the data set. The dashed horizontal line represents 0-fold change, while all the dots above the line represent overexpressed proteins. The numbers above each cluster represent the false discovery rate, namely the confidence level in the representation of the specific BP. (F) Oxygen consumption rate (OCR, mean±s.e.m) of hCMs treated with vehicle (N=6), normoxic (Norm) EEVs (N=6), and hypoxic (Hyp) EEVs (N=6) was measured using Seahorse extracellular flux analyzer, while altering mitochondrial function to delineate the different components of respiration. (G) Spare respiratory capacity (mean±s.e.m) was calculated as the difference between the maximum and baseline OCR. Significance relative to untreated control was tested with one-way ANOVA. Statistical significance was presented by *P≤ 0.05, **P ≤ 0.01. P-values are detailed in data file S4.
Due to the observed enrichment of metabolic process and mitochondrial proteins in the EEVs and in the EEV-loaded hCMs, we sought to assess the effects of EEVs on mitochondrial function in hCMs. To probe this, we used an extracellular flux analyzer to measure oxygen consumption rates (OCRs) (40). Our results show that baseline OCR was similar in the EEV-treated and control groups (Fig. 3F). When hCMs were treated with oligomycin, an ATP synthase inhibitor, the measured OCR values were still similar in the different experimental groups. However, when the mitochondrial membrane potential was depleted using carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), a notably higher OCR was observed for EEV-treated cultures, indicative of a larger spare respiratory capacity (Fig. 3, F and G). Lastly, non-mitochondrial oxygen consumption was similar in the EEV-treated and control cultures (Fig. 3F), as seen after electron transport was blocked using antimycin A and rotenone. In summary, EEVs significantly (p=0.0195, for normoxic and p=0.0019 for hypoxic EEVs) increase the spare respiratory capacity in hCMs while baseline and ATP synthesis are unaltered.
Since the EEVs and EEV-treated hCMs were also enriched in proteins related to cellular response to mitochondrial stress, we sought to measure intracellular reactive oxygen species (ROS) of hCMs in response to an oxidative stress-inducing drug. Menadione is a compound that undergoes redox cycling with formation of ROS. Measuring intracellular ROS after menadione treatment demonstrated slower increase in ROS in hCMs pre-treated with EEVs (fig. S4).
Together, our results support that EEV treatment of myocytes lead to changes in protein profile associated with various cellular components and biological processes. Metabolic process and mitochondrial proteins were particularly enriched in EEVs, resulting in EEV-treated hCMs having larger bioenergetic reserve and being slightly more resistant to ROS.
Cardioprotective proteins in EEVs
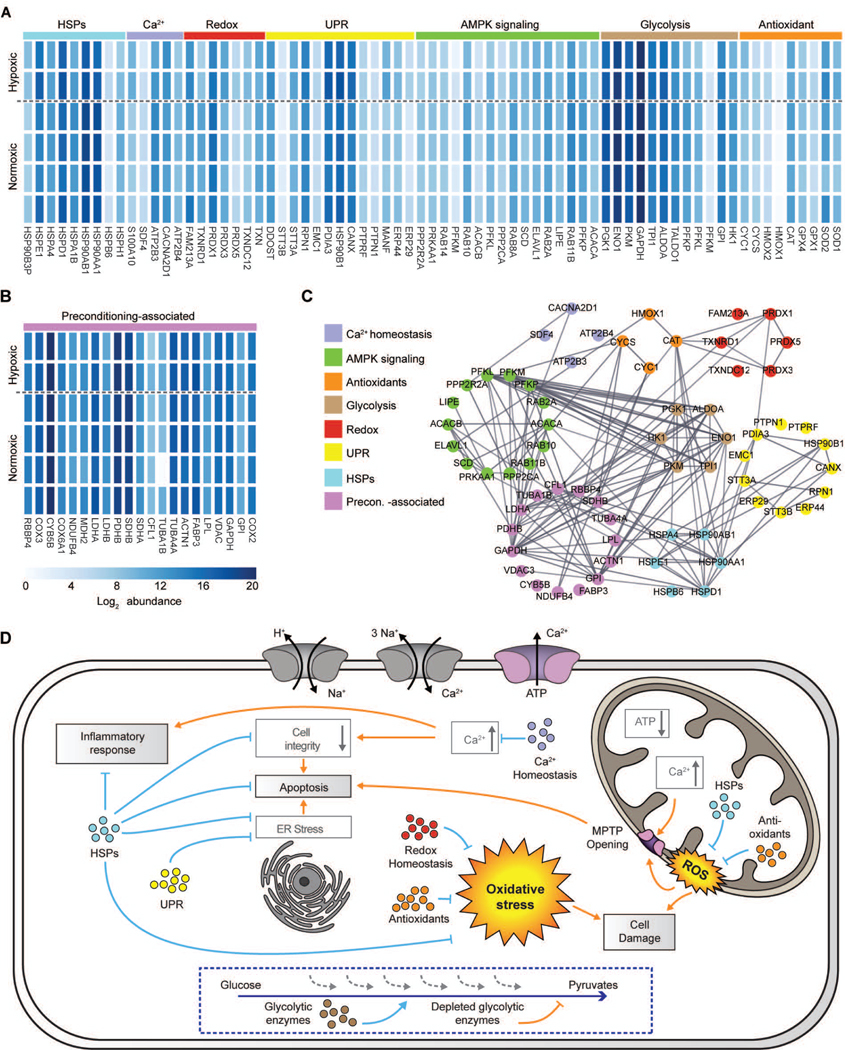
We then asked whether EEVs contained proteins that can target one or more of the pathophysiological mechanisms initiated by IRI. This process leads to cellular organelle and cytoskeletal damage, dysfunction, and ultimately cell death via multiple pathways (5). We identified seven groups of proteins related to these IRI-induced deleterious mechanisms in both normoxic and hypoxic EEV samples (Fig. 4A and table S3): (i) nine antioxidants; (ii) nine out of the ten enzymes participating in glycolysis; (iii) sixteen AMPK-related proteins; (iv) thirteen proteins involved in the protective unfolded protein response (UPR) pathway; (v) seven proteins involved in cellular redox homeostasis; (vi) nine heat shock proteins (HSPs) that are known to affect multiple pathological processes and ameliorate myocardial IRI; and (vii) five proteins related to calcium homeostasis.
Fig. 4. Identification of cardioprotection-related proteins in normoxic and hypoxic EEVs.
(A) Heat map the log2 mass abundance values for the groups of proteins enriched in the normoxic and hypoxic EEV expression profiles associated with IRI response, and possibly involved in cardioprotection by mitigating IRI-related pathological processes. (B) Heatmap illustrating the log2 mass abundance values for the group of proteins enriched in the normoxic and hypoxic EEV protein expression profiles associated with preconditioning. (C) Protein-protein interaction network illustrating known interactions between the proteins comprising the groups of IRI response and preconditioning-related proteins observed in the normoxic and hypoxic protein expression profiles. (D) Schematic of potential cellular mechanisms of cardioprotection mediated by the groups of proteins discovered in the EEV samples, and the related IRI-induced pathological processes that are potentially mitigated. The different groups of proteins are represented in color coding corresponding to their representation in panel A. Orange arrows stand for pathological pathways and blue arrows indicate the counteractions.
In addition to proteins that may target the pathophysiological mechanisms initiated by IRI, we identified a group of proteins in the EEVs that are known to be downregulated in cardiomyocytes during IRI (Fig. 4B). These proteins are simultaneously known to be positively affected by ischemic preconditioning treatment. Ischemic preconditioning was previously found to upregulate 30 out of 64 genes that are downregulated after myocardial IRI (41). Of these, 21 respective proteins were found in the EEVs. The majority of these are involved in energy production and conversion, as well as metabolism. In addition, proteins respective to 6 out of the 34 downregulated genes that were not affected by preconditioning were found in EEVs. These findings suggest that EEVs may induce a cardioprotective effect partly via pathways similar to those associated with ischemic preconditioning.
Protein-protein interaction network analysis of the protein clusters (Fig. 4C) identified a high degree of connectivity between the preconditioning-associated proteins and the seven identified protein groups that may target IRI pathophysiological mechanisms. Particularly, these are proteins heavily involved in cellular metabolism and repair (i.e., “glycolysis”, “AMPK”, and “HSP” protein groups). The observed high connectivity implies synergistic activity of the different clusters in modifying the cellular response to IRI, whereby multiple pathways can be targeted by EEVs to induce a cardioprotective effect. Together, these EV-residing proteins may assist in preservation of cellular integrity, structure, and function, as well as inhibit the onset of apoptotic signaling initiated by pathological IRI (Fig. 4D).
Effect of EEVs on cardiac tissue viability after simulated ischemia-reperfusion injury
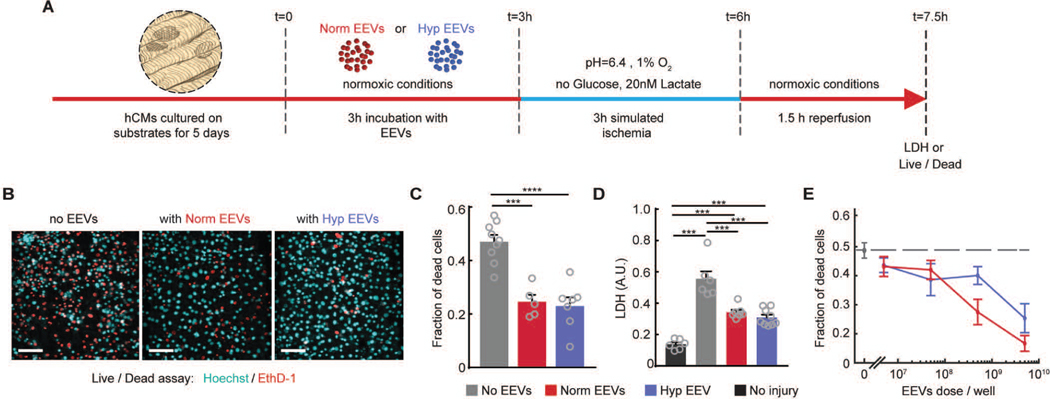
To test whether EEVs could be cardioprotective in hCMs during IRI, we investigated their cardioprotective effect on cardiomyocytes derived from human stem cells. To simulate ischemia on hCMs, we replaced the cardiac cell culture medium with glucose-deprived, lactate-containing, and acidic “ischemia buffer”, followed by exposure to 1% O2 environment. After 3 hours under ischemic conditions, reperfusion was simulated by returning to standard cell culture medium and restoring the O2 to 21% for 1.5 hours before conducting viability measurements. Live/dead cell assays showed nearly 50% cell death, indicating extensive damage to the hCMs.
To test the hypothesis that EEVs can improve the survivability of human myocardium after IRI, we treated hCMs with two equal doses of normoxic and hypoxic EEVs. One dose was given 3 hours prior to the onset of ischemia and the second right after 3 hours ischemia (before reperfusion) (Fig. 5A). End-point quantification of the fraction of dead cells in IRI-challenged engineered myocardial tissue (Fig. 5B) demonstrated a statistically significant (p=0.0001 for normoxic and P<0.0001 for hypoxic EEVs) ~50% increase in tissue viability in the presence of either normoxic and hypoxic EEVs (Fig. 5C). Notably, preconditioning hCMs with 10 minutes ischemia and 10 minutes reperfusion increased tissue viability by ~23% relative to untreated controls (fig. S5). In addition, secreted LDH, an enzyme indicating cellular damage, was lower in EEV-treated tissues (Fig. 5D). The effect of EEVs on cardiac tissue viability proved to be dose-dependent, as demonstrated after exposure of hCMs to increasing concentrations of normoxic or hypoxic EEVs (5×106, 5×107, 5×108 and 5×109 EEVs/well) prior to IRI (Fig. 5E). The fraction of dead cells decreased with increasing EEV concentration relative to untreated cells, for both normoxic and hypoxic EEV populations. Notably, the cardioprotective effect did not reach a plateau in the tested range, implying that stronger cardioprotection might be possible. However, we did not increase EEV concentrations due to technical limitations of upscaling EEV production.
Fig. 5. EEVs are protective in myocardial ischemia-reperfusion injury.
(A) The experimental timeline illustrating timing of EEV treatment, ischemia, and reperfusion periods. (B) Viability was assessed at end point using an Ethidium-based live/dead assay. Representative images shown for tissues untreated (left), treated with normoxic (Norm, middle), and hypoxic (Hyp, right) EEVs (Nuclei of live cells: Cyan; dead cells: red; 100μm scale bar). (C) Fraction of dead cells after reperfusion was quantified from the live/dead assay images. >5 field of views for each sample were imaged with N=9 for control; N=5 for normoxic; N=7 for hypoxic EEV treatment. Tested with one-way ANOVA. (D) Injury after reperfusion was assessed by LDH biochemical assay. LDH readout was compared between an uninjured group to the control and EEV-treated groups following IRI, with N=6 for uninjured, N=6 for control; N=7 for normoxic; N=9 for hypoxic EEV treatment. Data is represented as mean±s.e.m and tested with one-way ANOVA. Statistical significance was presented by **(P≤0.01), ***(P≤0.001). P-values are detailed in data file S4. (E) Fraction of dead cells following IRI as a function of EEV concentration (N=3 for each group)
To elucidate the cellular processes involved in the EEV-mediated rescue of viability, we compared the protein profiles of IRI-challenged hCMs with and without EEV treatment. We found ~1350 proteins that were significantly (P ≤ 0.05) differentially expressed in the EEV-treated population (data file S3) and analyzed their cellular component and biological process classifications. The proteomap was similar to that derived from the EEV samples, with DEPs including metabolic proteins, and protein expression/protein folding factors (Fig. 6A). The DEPs’ cellular component (Fig. 6, B and C) and molecular function (Fig. 6D, fig. S6) classifications showed dominant mitochondrial and catalytic activity-related proteins. This implies differences in the metabolic landscape of the EEV-treated injured cells. The biological processes related to the DEPs include cellular response to stress, respiration, energy derivation, glucose, and fatty acid metabolism, as well as protein targeting and folding (Fig. 6E, fig. S7). Principal component analysis of injured / uninjured and EEV-treated / untreated hCMs demonstrated that EEVs only slightly affect the protein profile of uninjured cells, whereas the larger modifications induced by IRI were partially rectified by the EEV treatment (Fig. 6F).
Fig. 6. Endothelial EVs’ effects on IR-injured hCMs.
(A) Proteomap illustrating the KEGG orthology terms and relative differences in mass abundance for the significantly (P-value <0.05) differentially expressed proteins (DEPs) in EEV treated (N=3 samples), compared with untreated (N=3 samples) IR-injured hCMs. (B) Volcano plot presenting the fold change and P-value of the DEPs. The colored dots represent proteins of the cellular components represented in panel C. (C) Cellular localization in the protein expression data set. The numbers next to the bars represent the number of significantly enriched proteins related to each of the depicted cellular compartments. (D) Molecular function in the protein expression data set shows modification of catalytic activity and binding. (E) Dominant biological processes (BPs) in the DEPs. Each dot represents a single protein, whereas the clusters represent BPs. The red color scale represents the individual P-value for each protein in the data set. The dashed horizontal line represents 0-fold change, while all the dots above the line represent overexpressed proteins. The numbers above each cluster represent the false discovery rate, namely the confidence level in the representation of the specific BP. (F) Two-dimensional principle component analysis of four treatment groups: Control – only hCMs (N=2), normoxic EEV-treated hCMs (Norm EEVs, N=3), IR-injured hCMs (IRI, N=3), and normoxic EEV-treated + IR-injured hCMs (Norm EEVs + IRI, N=3). (G) Schematic of production of DiO-labeled EEVs by prior labeling of cultured HUVECs’ plasma membranes. DiO-labeled EEVs could be detected using confocal microscopy. (H) XY and reconstructed YZ planes are presented 45 minutes after adding EEVs, showing internalization of the labeled EEVs (20 μm scale bars). Nuclei: blue; plasma membrane of cardiomyocytes: magenta; EEVs: green. (I) Schematic of two strategies to inhibit endocytosis in cardiomyocytes. Dynasore (left) inhibits dynamin, and Filipin (right) interrupts the lipid rafts. (J) Fraction of dead cells following IRI in the different treatment groups (9 experiment groups, N=4 for each group). Data is represented as mean±s.e.m, and tested with two-way ANOVA. Statistical significance was presented by *(P≤0.05), **(P≤0.01), ***(P≤0.001), ****(P≤0.0001). P-values are detailed in data file S4.
The EEV-dependent effect on hCMs’ viability and protein expression after IRI may be mediated by cargo delivered into the cell via endocytosis or activation of a ligand-receptor response on the cell membrane without EEV internalization (24). To determine whether endothelial cell-derived EVs are internalized by cardiomyocytes, we produced fluorescent EEVs by labeling HUVECs with DiO, a green lipophilic tracer that is transferred to the cell membrane (Fig. 6, G and H). Using time-lapse microscopy, we observed internalization of these labeled EEVs by hCMs within 45 min (Fig. 6H, fig. S8, movies S1–S3). Therefore, we hypothesized that the cardioprotective effect of the EEVs was mediated by post-internalization cargo release. To test this hypothesis, we assessed whether inhibition of endocytosis in cardiomyocytes reduces the EEV-induced protective effect observed during IRI. Endocytosis can occur via multiple pathways, some of which include inwards budding of the plasma membrane and formation of early endosomes – the first compartment of the endocytic pathway. The inward budding of the plasma membrane can occur in specific domains and by different mechanisms. We targeted two endocytosis routes: dynamin- and lipid raft-mediated endocytosis (Fig. 6I). Dynamin-mediated endocytosis was inhibited by using dynasore (42), which prevents dynamin from tightening around the inward budding vesicle neck (e.g., clathrin-coated pit) and the formation of an endosome (43). Lipid raft-mediated endocytosis was inhibited by using filipin (44). Lipid rafts are cholesterol-rich plasma membrane subdomains that form an inward budding vesicle. Filipin prevents shuttling into the cell by cholesterol sequestration which interrupts the rafts (45). Although neither dynasore nor filipin had any direct effect on cell viability among untreated control samples, they both significantly (P-values detailed in data file S4) reduced the EEV-mediated cardioprotection as measured by an increased fraction of dead cells (Fig. 6J). Together, our results support the notion that EEVs interfere with the fatal IRI-associated pathophysiological pathways and mediate their cardioprotective effect by releasing their cargo within cardiomyocytes.
Effect of EEVs on cardiac tissue contractile function during ischemia-reperfusion injury
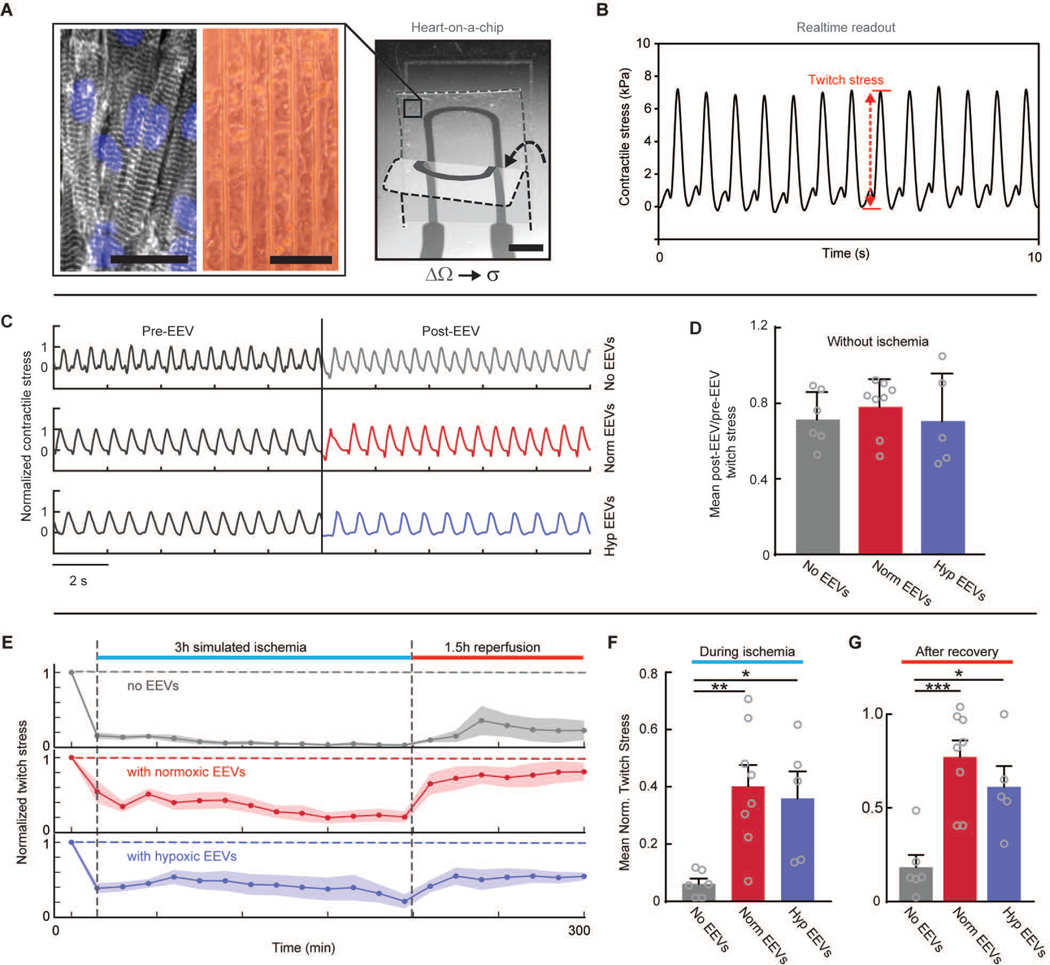
Recovery of viable tissue contractile function after IRI is a key factor in determining the outcomes of myocardial infarction and the possible development of heart failure. To determine whether EEVs aid in preservation and recovery of contractile function after IRI, we used an instrumented hHOC (28, 29) platform. The hHOC platform features engineered hlCTs on soft cantilever substrates. The tissues are guided into an anisotropic and laminar architecture that recapitulates the native myocardial muscle, through micropatterned cues on the cantilever surface (28, 29, 46, 46). In the instrumented hHOC, each cantilever further contains an embedded strain gauge (Fig. 7A and fig. S9). Upon myocardial tissue contraction, the cantilever deflects, leading to a resistance change in the embedded strain sensor that is directly proportional to tissue contractile stress (Fig. 7B). Continuous recording of the gauge resistance thus provides readout of the contractile stress of an engineered cardiac tissue, before, during, and following simulated IRI. Conditioning of hlCTs with normoxic or hypoxic EEVs had no direct effect on baseline contractile activity (Fig. 7C–D). However, upon exposing the engineered cardiac tissues to ischemia, we observed an almost immediate cessation of tissue contraction and a minor recovery of twitch stress following reperfusion (Fig. 7E, top panel). However, EEV-treated tissues continued contracting during simulated ischemia and displayed significant (p=0.0005 and p=0.0172 for normoxic and hypoxic EEVs) recovery of twitch stress following reperfusion (Fig. 7E). While the contractile twitch stress in untreated tissues decreased to below 10% of baseline during simulated ischemia, tissue contractile stress decreased to merely 40±8% and 36±10% of baseline for cardiac tissues conditioned with normoxic and hypoxic EEVs, respectively (Fig. 7F). Furthermore, twitch stress recovery after reperfusion was 77±9% and 61±11% for normoxic and hypoxic EV-conditioned tissues respectively, whereas in untreated tissues, twitch stress recovery was only 18±7% of baseline (Fig. 7G).
Fig. 7. HUVEC-derived EVs rescue contractile function during IRI.
(A) A representative image of hCM-derived tissue in the HOC system. Left image shows immunostain for nuclei blue) and sarcomeric alpha-actinin (gray). Middle image shows an optical microscopy tissue culture image (100 μm scale bars). On the right, a single PDMS cantilever with an embedded strain gauge is depicted (2 mm scale bar). As the hlCT contracts the cantilever bends, stretching the strain gauge and leading to increased resistance. Two different bending positions are overlaid to demonstrate the cantilever’s motion. The relative resistance changes (ΔΩ) are proportional to the generated twitch stress (σ). (B) An example trace showing measured twitch stress. (C) Example traces of contractile stress before and after 3-hours of EEV incubation in untreated, normoxic, and hypoxic EEV-treated tissues. The contractile stress was normalized to average amplitude before treatment. (D) Mean ratio between average stress before treatment to average stress after a 3-hour EEV incubation period, in control (N=6), normoxic (N=8), and hypoxic (N=5) EEV-treated tissues. No significant differences were found (one-way ANOVA). (E) Twitch stress amplitude normalized to baseline values throughout the simulated IRI. The bold traces present the average values over all the tested samples and shaded areas represent the SEM. (F) Average twitch stress during ischemia relative to baseline. (G) Average twitch stress recovery relative to baseline for the three experimental groups. Error bars represent s.e.m. N=6 for control; N=8 for normoxic; and N=5 for hypoxic EEV-treated groups. Tested with one-way ANOVA. There was no significant difference between the two groups treated with EEVs. Statistical significance was presented by *(P≤0.05), **(P≤0.01), ***(P≤0.001). P-values are detailed in data file S4.
In addition to testing the response of human cardiac tissues to the EEV treatment, we constructed neonatal rat ventricular myocytes (NRVMs) tissues in the same HOC system (fig. S10A) and exposed them to the same simulated IRI protocol and EEV treatment. Interestingly, EEVs were internalized in the NRVMs (fig. S10B), yet their protective effect on NRVM-derived tissues viability and contractility was notably smaller (fig. S10C–E).
Discussion
The key finding of this study is that vascular endothelial cell-derived extracellular vesicles exert a protective effect against myocardial IRI in human tissue. Importantly, this cardioprotective effect is manifested as a significant reduction in cell death, partial conservation of the cellular proteome, as well as preservation of tissue contractility as demonstrated using our IRI hHOC assay.
In control tissues without EEV treatment, the loss of contractile function was greater than the loss in viability. This mirrors in vivo IRI, where tissue contractile function is impaired not only due to cardiomyocyte death, but also due to ATP depletion, abnormal calcium concentrations, and damage to contractile filaments of the surviving myocytes (5). In EEV-treated tissues, both viability and contractile function were improved, and the rescue effect on contractile function was greater than the effect on viability. Proteomics analysis revealed that EEVs shifted the protein profiles of IR-injured hCMs closer to their uninjured signature, indicating that EEVs likely exert their cardioprotective effects by both increasing survivability of cardiomyocytes and preserving cellular processes, function, and integrity.
Previous studies have reported cardioprotective effects of exosomes derived from various cell sources (18–21, 24, 25), but the mechanisms of action and the origin of protective exosomes remain elusive. Vicencio et al. (24) reported that plasma-derived exosomes were cardioprotective both in vivo and in vitro, using adult rats and adult rat myocytes. In that study, the authors did not observe exosome uptake, and proposed that exosomal HSP70 may bind to Toll-like receptor 4 to activate the protective ERK1/2-HSP27 signaling pathway. Other studies reported uptake of stem cell-derived (21) or cardiac progenitor cell (CPC)-derived (20) protective exosomes in cardiomyocytes. Barile et al. (20) suggested that CPC-derived exosomes induced cardioprotective effects in vitro and in a mouse MI model, by delivery of anti-apoptotic and pro-angiogenic miRNAs. Similarly, Khan et al. (21) reported that mouse embryonic stem cell-derived exosomes improved ejection function and reduced infarct size and apoptosis in a mouse model of myocardial infarction. Notably, they reported that the effect involved neovascularization and expansion of cardiac progenitor cells mediated by the embryonic stem cell-specific miRNA-294. In both studies, the protective effect could not be recapitulated using fibroblast-derived exosomes. Here, we used endothelial cells as a source for EVs. Primary human endothelial cells are relatively easy to obtain, expand, and culture, providing a convenient source for this therapeutic approach. Our study shows that: (i) the protective effect does not require stem cells or stem cells derivatives (EVs), at the site of action; (ii) the effect is caused by an intracellular mechanism, as it requires uptake; and (iii) the protective effect is associated with both increased survivability and improved contractile function of cardiomyocytes by a complex mechanism. Notably, neither filipin nor dynasore fully suppressed the EEV-mediated rescue of viability. This result implies that EEV uptake by hCMs is mediated by multiple routes of endocytosis. Since the EEVs contain diverse protein and nucleic acids cargo, it is impossible to delineate, at this point, the differential effects of the various proteins or RNA cargo. However, a recent study by Perez-Boza et al. (48) mapped the RNA landscape of HUVEC-derived EVs isolated by ultracentrifugation and compared it with the RNA content of the parent cells. We examined this dataset for specific miRNA previously associated with cardioprotection (table S4). Notably, miR-146a, miR-210 and miR-294, which have been linked to cell survival (19–21), were not detected in the EEVs. On the other hand, pro-vasculogenic miR-132 (20) and the pro-contractile miR-21–5p (49) were not in higher abundancy with respect to the parent cells. Therefore, the beneficial effects of the EEVs could be mediated in part by miR-132 and miR-21–5p, or other mRNA content. However, as our in-vitro system consisted of only hCMs, pro-vasculogenic and anti-inflammatory effects would not contribute to the cardioprotection observed here. Moreover, we observed a range of effects beyond increased survival, including improved function during and following ischemia, increased cellular respiratory reserve, and slightly decreased ROS production in menadione-challenged cells. This implies that multiple factors likely contribute to the net effect.
Analysis of EEV proteome supports that protein content could play a key role in such a multifaceted mechanism (table S3). A multitude of the identified EEV proteins are involved in cellular homeostasis and preservation. Antioxidants, a high abundance of heat shock proteins (50), UPR intermediates, and ER-resident chaperones (51, 52), as well as ribosomal proteins such as Rps3a that regulate apoptosis (34, 35, 53), were harbored in the EEVs, all of which are known to increase cell survivability. Furthermore, we found several proteins that support recovery of contractile structures and metabolic function, such as glycolytic enzymes and AMPK signaling intermediates which can collectively increase glycolytic activity (54, 55) and ATP production in the post-ischemic myocardium (56–59). Interestingly, we found that EEV treatment resulted in modulation of the expression of proteins related to various metabolic and energy derivation pathways, as well as increased reserve respiratory capacity, which has been shown to be regulated by AMPK (60). This increased reserve respiratory capacity can be utilized by the cells in cases of metabolic stress and has been shown to increase their survival by adapting to metabolic stress (60, 61). Moreover, heat shock proteins can improve the repair of structural proteins after ischemia-induced cytoskeletal damage (62) and assist in suppressing oxidative stress (63–65). Additionally, antioxidants may mitigate oxidative stress and prevent the associated damage to structural proteins and organelles. Consistent with these findings, the protein expression profile of injured hCMs treated with EEVs differ from the untreated injured cells and is more similar to uninjured cardiomyocytes.
In addition to significant post-IRI recovery of contractile function, we also observed that the electrically paced, EEV-treated tissues maintained contractile function during the prolonged ischemic phase of IRI, whereas control tissues ceased contracting almost immediately. This is in accordance with the observed increase in maximum respiratory capacity for EEV-treated hCMs. Interestingly, EEVs contained high abundance of LDH and other proteins involved in lactate metabolism. These proteins can promote the conversion of lactate to pyruvate and provide an anaerobic metabolic substrate. Consistently, upregulation of proteins involved in pyruvate metabolism, gluconeogenesis, and galactose metabolism pathways were observed in ischemia-reperfusion injured hCMs that were pre-treated with EEVs, as compared to injured, untreated cells. To note, in the hibernating myocardial tissue that can sustain ischemia, lactate can be used as a metabolic substrate and enable contraction in response to inotropic stimulus (7, 66).
We found that normoxic and hypoxic EEVs had similar protein content. Major DEPs comprised only 5% of the entire cargo and were related to genetic information processing and metabolism. However, the functional difference in the two EEV populations is still not clear since they had similar effects on cardioprotection, basal respiration, ATP production, spare respiratory capacity, and non-mitochondrial respiration. The extrapolated EEV production rate was higher under hypoxic conditions, in accordance with prior findings (67, 68). It is unknown whether EEVs play a role as cardioprotective agents against IRI in vivo, however this observation, together with the observed dose dependency, suggests that an innate signal modulation by quantity could be possible.
In this study, we used a recently published human ‘Heart-on-a-Chip’ in-vitro IRI assay (28, 29) to examine the effects of endothelial cell-derived EVs on the outcome of myocardial IRI. Embedded sensors in the hHOC allowed for continuous monitoring of tissue contractile function from within a closed environment. This enabled us to observe the isolated, dynamic functional response of hlCTs to IRI and concomitant EEV treatment in vitro. Although the engineered hlCTs in the hHOC system generated contractile activity and responded to simulated IRI as expected, there are also inherent limitations. These include: (i) the partial immaturity of stem cell derived hCMs; (ii) the homogeneity of engineered myocardial tissues (i.e., only hCMs); and (iii) the configuration of hCMs as two-dimensional monolayers, which do not fully recapitulate the three-dimensional, multi-cellular adult heart. Nevertheless, we previously showed that recapitulating the extracellular matrix (ECM) and anisotropy of the native myocardium promotes tissue maturation as measured by structural organization, contractile performance and sarcomeric gene expression profiles (46). We have also recently shown that such tissue models can be used for rapid screening of pharmacological agents and confirmed that they respond appropriately to a wide range of known cardiac and cardiotoxic compounds (29). Moreover, in this study, we showcase the advantage of using such in vitro systems by demonstrating that the cardioprotective effect exerted by EEVs on laminar hCMs could not be recapitulated with NRVM-derived tissues. This approach may enable us to choose an appropriate in vivo model for further testing of the suggested therapeutic approach.
Among current interventions to reduce ischemic damage, ischemic preconditioning is the most efficient intervention in reducing infarct size and is known to improve cell survival and recovery of mechanical function by activating multiple pathways (69, 70). Preconditioning has been shown to decrease the fraction of dead cells by up to 50% and improve recovery of contractile force two-fold, following only 90 minutes of ischemia and reperfusion in human in-vitro/ex-vivo models (69, 70). In comparison, we found that the fraction of dead cells was similarly reduced by 50% and that recovery of contraction was approximately four times higher in EEV-treated tissues than in control tissues following 3 hours of simulated ischemia and 90 minutes reperfusion. Additionally, preconditioning hlCTs with 10 minutes ischemia and 10 minutes reperfusion, followed by prolonged (3 hours) ischemia and reperfusion (90 minutes) resulted in lesser viability compared to EEV treated, injured hlCTs. This comparison suggests that the EEV-induced cardioprotective effect could be at least as effective as the protective effect induced by preconditioning.
Our findings indicate that EEVs could constitute a multi-target therapy for IRI, but several challenges are apparent for translating EVs into a clinically relevant therapy. For instance, the cell source used here for EEV derivation was venous endothelial cells. Although they produce an apparent cardioprotective effect, other cell types, for example, arterial endothelial cells, may be more relevant as a cell source for EVs. An adaptation of our current experimental design, where an assessment of the therapeutic effect of EEVs when administered before, or immediately, after reperfusion, is an important experimental milestone enroute to future clinical deployment of EEVs. Similarly, studying the most effective time window for a cardioprotective effect will be necessary for translation of the findings presented herein. A question remains regarding the duration of EEV cargo-induced effects, and how they vary with time after treatment. As a further obstacle for clinical translation, traditional cell culture may not be sufficiently scalable for high throughput production of EVs. Finally, issues such as dosing efficiency, clearance, targeted delivery and EVs cargo optimization to increase therapeutic efficiency, must still be evaluated. To optimize the treatment an appropriate in vivo model should be designed, which could be facilitated by the HOC system as demonstrated here with both human- and NRVM-derived tissues. In that sense, the present results may represent a starting point for production of therapeutic EV-mimetics.
Materials and Methods
Study design
The aims of this study were to characterize the protein cargo of human vascular EEVs, to identify lead cardioactive proteins, and assess the effect of EEVs on hlCTs exposed to IRI. Based on analysis of EEV protein contents, we hypothesized that these EEVs are cardioprotective against IRI. We designed a hHOC system to test the effects of EEVs on hlCT contractile function and used hCMs cultures to test the effects of EEVs on myocyte respiration, viability after IRI, and EEV uptake. Cardiac cultures or tissues in hHOC systems were randomly assigned for the various treatment groups in all the assays. The investigators were not blinded on any of the reported experiments. No sample size calculations were performed. Standard statistical methods were used to determine significance, as detailed in data file S4. We generally assumed Normal distribution with the exception of OCR data. No data were excluded unless apparent failure was observed in the following cases: (i) breakage of HOC strain sensor; (ii) cardiac tissue delamination from HOC substrate; or (iii) bacterial infections. No outliers were removed.
Experiments with hCMs in cultures or in HOC systems had N≥3 biological replicates for each group. Technical replicates were not considered as independent samples, thus multiple technical replicates of the same one biological sample were counted as N=1. Experiments were repeated multiple times in multiple occasions. For proteomics data, EEVs proteomics were derived from 4 samples of normoxic EEVs and 2 samples of hypoxic EEVs. Proteomic analyses of cardiomyocytes were derived from 3 samples for each treatment group, except for the untreated, uninjured control group (2 samples).
Extracellular vesicles isolation
EVs were isolated by ultracentrifugation, as previously published (71). Briefly, 12 h after passage 5 or 6, HUVECs were washed once with warm PBS and the standard endothelial growth medium was replaced with EGM containing 2% exosome-depleted fetal bovine serum (Invitrogen, Carlsbad, CA, USA). After 48 h, culture medium was collected for “normoxic EVs” isolation. Fresh exosome-free medium was then added to the flasks, and cells were then subjected to 3 h of hypoxia (1% O2). Then, culture medium was collected for “hypoxic EVs” isolation. The culture medium was divided into 50 mL conical tubes and centrifuged at 300 g, 4°C, for 10 min. The supernatant was collected and centrifuged at 2000 g, 4°C for 20 min. Supernatant was collected and centrifuged at 10,000 g, 4°C, for 30 min. Finally, the supernatant was placed in thin wall, Ultra-Clear, 38.5 mL tubes and centrifuged at 100,000 g, 4°C, for 70 min, in an XPN-80 Beckman Coulter ultracentrifuge, equipped with an SW 32 Ti rotor (Beckman Coulter). The supernatant was discarded, and the pellet in each of the tubes was re-suspended in sterile PBS. All the “normoxic EVs” suspensions were pooled in a single tube, and the “hypoxic EVs” suspensions in another. Then, a second ultracentrifugation cycle was carried out under the same conditions. The EVs pellet was then re-suspended in 500 μL PBS, or in 150 μL lysis buffer (Covaris, Protein extraction buffer TP), if the samples were designated for proteins analysis. Particle Analysis and concentration assessment using NanoSight (NS300, Malvern) were performed with 2 μL of each sample. Samples were stored in −80°C until use or kept on ice if brought to protein analysis or used on the same day.
Extracellular vesicles labeling
EV-producing endothelial cells were initially labeled using a lipophilic tracer prior to the last passage, before being placed in the exosome-free, serum-enriched medium. Staining medium was prepared by adding 5 μL DiO or DiD stock solution (Vybrant Cell-Labeling Solutions, Life Technologies) per 1 mL of growth medium. Culture medium was drained from T175 flasks containing confluent, adherent HUVECs at P4 or P5. The cells in each flask were then incubated with 3 mL of the staining solution, for 15 min, at 37°C. After incubation, the staining solution was drained, followed by washing cells twice with growth medium for 10 min each. After the second wash, cells were dissociated and passaged 1:3. The culture medium was then switched to growth medium supplemented with 2% exosome-depleted fetal bovine serum after 24 h. Culture media was collected for EEV isolation after 48 h, as described above. Following EEV isolation, 20 μL of labeled and non-labeled EEV suspensions were added to a black 384-well plate and loaded into a plate reader for fluorescence intensity measurement.
Simulated ischemia reperfusion injury (IRI)
To simulate IRI, cardiomyocytes were exposed to 3 h ischemic conditions and 1.5 h under reperfusion treatment. Ischemia was simulated by switching the complete culture medium to a Tyrode’s-based “ischemia buffer” with (mM): KCl 4, NaCl 115, CaCl 1.8, HEPES 5, MgCl2 1, NaH2PO4 0.33, sodium lactate 20, with no glucose, and pH 6.4. O2 was set to 1%. For reperfusion, O2 was increased back to 21% and medium was changed back to complete culture medium, pH 7.4. To test EEVss for a possible cardioprotective effect, cardiomyocytes were incubated with 109 EEVs/well (“normoxic” or “hypoxic”), 3 h prior to onset of ischemia, with an additional, identical dose of EEVs at the onset of simulated ischemia, with the “ischemia buffer”. Reperfusion was simulated by switching the “ischemia buffer” back to complete culture medium at 21% O2 for 1.5 h. Finally, after reperfusion, media were collected for LDH measurement (Cytotox 96, Promega), and viability assays were performed. Functional contractile measurements were conducted throughout the simulated IRI.
Ischemic preconditioning
To assess and compare the rescue mediated by preconditioning compared to EEV treatment, we conducted the IRI protocol, as described above, with hCMs that were exposed to 10 minutes ischemia and 10 minutes reperfusion prior to the prolonged ischemia. hCMs were pre-plated, seeded in well plates and cultured for 6 days, as described above. On day 6, the cells were washed 3 times with warm, sterile PBS and ‘ischemia buffer’ (described above) was added to the wells. The well plates were then transferred to a 1% oxygen incubator for 10 minutes. After 10 minutes of simulated ischemia, the ‘ischemia buffer’ was removed from the wells and replaced by the normal culture media. The well-plates were then transferred to an incubator with 21% oxygen for 10 minutes, after which the prolonged ischemia and reperfusion protocol was implemented. A live/dead assay was performed right after the 1.5 h reperfusion period. In the same experiment we used a second well plate with hCMs that were not preconditioned as a control.
Heart-on-chip real-time contractility assays
The HOC systems, multi-well devices with instrumented muscular thin film cantilever substrates, were fabricated as previously described (29). Briefly, multilayered Polydimethylsiloxane (PDMS) films (21–23 μm) were cast onto glass substrates using a spin-coating technique. An embedded flexible titanium-gold thin film (24 nm) strain gauge was deposited using an E-beam evaporator (Denton) and a custom-made shadow mask. Ridges (5 μm-tall and 4 μm-wide, spaced by 30 μm grooves) guide the formation of anisotropic cardiac thin film tissues. Cantilevers sized 3.2 mm x 4.2 mm were defined using a CO2 laser (Epilog). A Poly(N-isopropylacrylamide) film ensured that cantilevers could detach from glass substrates when cooled briefly to room temperature. After UVO treatment and FN incubation, hCMs were seeded on hHOC devices as described above, and cultured for 6 days prior to IRI experiments. Real-time contractility data was recorded using a multichannel DMM system (Keithley 3706a) and analyzed using a custom Matlab (Mathworks inc.) code as previously described (29).
Extracellular vesicles uptake assays
EV internalization by cardiomyocytes was visualized by time-lapse confocal microscopy. hCMs were cultured as described above and seeded onto circular 18 mm coverslips, which are coated with micromolded PDMS. After 5 days in culture, the plasma membrane and nuclei were labeled using CellMask Deep Red Plasma Membrane Stain (Life Technologies) and Hoescht (Life Technologies), respectively, according to the products’ recommended manuals. Following staining, the coverslip was transferred to an imaging chamber filled with 1 mL Live Cell Imaging Buffer (Life Technologies). The microscope (Olympus IX83) was set to perform confocal imaging at several positions along the coverslip, over a period of 3 h, using a 60× objective (Olympus). Immediately after the first time point at which images were acquired, 109 DiO-labeled EEVs were added to the well, and the imaging continued. The acquired images were processed using ImageJ 2.0.0 and Imaris.
Uptake inhibition assay
Clathrin-mediated endocytosis was inhibited by administration of 80 μM dynasore (Sigma Aldrich) 30 min before adding EEVs to the cardiac tissue culture wells (72). The lipid raft/caveolae-related endocytic pathway was disrupted by administration of 1 μg/mL filipin (Sigma Aldrich), 30 min prior to addition of EEVs (73). To test the effects of EEVs uptake inhibition on the EEV-mediated cardioprotection, IRI was simulated after a 3 h incubation with “normoxic” and “hypoxic” EEVs under the inhibited endocytosis and control conditions. Following IRI, tissue viability was quantified by performing the viability assay described above.
Preparation of hCMs for proteomics analysis
Human stem cell-derived cardiomyocytes were thawed, pre-plated, cultured, and dissociated as described above. 5M hCMs were re-seeded in 8 wells in a density of 105 cells/cm2, and cultured for 6 days. On day 6, the media was changed for all the wells. Half of the wells were treated with 3×1010 EEVs (5×109 EEVs per 105 cells, as used for the rest of the experiments). The other half were treated with vehicle. After 3 h, 2 of the EEV- treated and 2 vehicle- treated wells were exposed to IRI, as described above. This procedure resulted in four experimental groups: (1) hCMs, (2) hCMs+EEVs, (3) hCMs+IRI, (4) hCMs+EEVs+IRI, with two wells per group. Using 0.25% trypsin-EDTA, cells were harvested from the wells for further mass spectroscopy analysis. After centrifugation, the supernatant was removed, pellets were washed with PBS, and then centrifuged again. This was repeated for 3 times overall. After the final centrifugation step, the supernatant was removed and the pellets were transferred to −80°C.
Sample-wise Pearson correlation analysis
Pearson Product Moment Correlation (76) was used to assess the degree of the linear correlation between pairwise comparisons of the global protein expression profiles of the normoxic and hypoxic EEV samples. Pairwise comparisons were performed using the corr() function provided in the PANDAS version 0.19.0–2 Python package, and visualized using the Seaborn statistical data visualization Python package, version 0.7.1–1.
Statistical analysis
Data are expressed as the mean±s.e.m for all biological replicates. Technical replicates of the same biological replicate were averaged and treated as N=1 (i.e., N indicates the number of biological replicates). Statistical analyses were performed using one-way or two-way ANOVA followed by the post hoc Tukey’s test with GraphPad Prism 7 software (GraphPad Software). P-Values less than 0.05 (*), 0.01 (**), 0.001 (***) or 0.0001 (****) were considered statistically significant. Statistical analyses and exact P-values for all the presented data are detailed in data file S4.
Supplementary Material
Table S1. Common contaminant proteins detected in EEV samples.
Table S2. Gene Ontology terms and enrichment P-values for over-represented protein groups presented in figure 2C–D with corresponding color coding.
Table S3. Cardioprotective function of protein groups represented in EEVs.
Table S4. Possible cardioprotective miRNA cargo in previously published studies.
Fig. S1. Differentially-expressed EEVs protein content in normoxic and hypoxic conditions.
Fig. S2. Molecular function GO classification of differentially expressed proteins in hCMs treated with EEVs compared with control, untreated hCMs.
Fig. S3. Biological process GO classification of differentially expressed proteins in hCMs treated with EEVs compared with control, untreated hCMs.
Fig. S4. ROS measurement in Menadione-challenged hCMs un/treated with EEVs.
Fig. S5. Preconditioning vs. EEV treatment effect on viability following IRI.
Fig. S6. Molecular function GO classification of differentially expressed proteins in injured hCMs pre-treated with EEVs compared with injured, untreated hCMs.
Fig. S7. Biological process GO classification of differentially expressed proteins in injured hCMs pre-treated with EEVs compared with injured, untreated hCMs.
Fig. S8. EEV uptake by human cardiomyocytes
Fig. S9. Heart-on-a-chip device for contractility measurements.
Fig. S10. Non-significant cardioprotective effect of human EEVs on neonate rat ventricular myocytes-based heart-on-a-chip assay.
Data file S4. Summary of statistical analyses and P-values.
Data file S2. Differentially expressed proteins in EEV-treated vs. untreated hCMs.
Data file S3. Differentially expressed proteins in IR-injured, EEV-treated vs. injured, untreated hCMs.
Data file S1. Total EEVs proteome raw data.
Mov. S2. EEV uptake by human cardiomyocytes (cell 2).
Mov. S3. EEV uptake by human cardiomyocytes (cell 3).
Mov. S1. EEV uptake by human cardiomyocytes (cell 1).
Acknowledgments:
We thank Dr. F. Pasqualini for valuable discussions, shared insights, and detailed comments on the manuscript. We thank T. C. Ferrante for advising on microscopy and helping with image processing using IMARIS, and M. Rosnach for his help with the illustrations. We thank Prof. M. A. Moses and Dr. G. Morad for allowing us to use NanoSight NS300. We thank Dr. Y. Blinder for assissting with hCMs proteomics data visualization. We thank Mr. D. Drennan and Ms. A. Greer for providing technical assistance. We thank The Harvard Center for Nano Scale Systems for training and use of cleanroom facilities.
Funding: This work was sponsored by the John A. Paulson School of Engineering and Applied Sciences at Harvard University, the Wyss Institute for Biologically Inspired Engineering at Harvard University, Harvard Materials Research Science and Engineering Center grant DMR-1420570, and the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Numbers UH3TR000522 and 1-UG3-HL-141798-01. H.A.M.A. would like to thank the American Chemical Society for generous support through the Irving S. Sigal Postdoctoral Fellowship. M.M.C.B. was supported by the Human Frontier Science Foundation as a cross-disciplinary postdoctoral fellow (LT000900/2013-C).
Footnotes
Competing interests: B.B. is a scientific consultant for Merck. The authors declare no competing financial interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. All proteomics raw data are deposited at: https://massive.ucsd.edu/ProteoSAFe/private-dataset.jsp?task=3421c03b031d4315a46dc8c1875ef63d. Scripts for proteomics data analysis are available at https://github.com/spsheehy/Proteomics and https://github.com/harvardinformatics/YadidMetal_May_2020. Scripts for contractility data analysis were previously published (26).
References and notes
- 1.Mozaffarian D, Benjamin E, Go A, Arnett D, Blaha M, Cushman M, Das S, de Ferranti S, Després J, Fullerton H, Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation 133, e38 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Hausenloy DJ, Yellon DM, Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J Clin Invest 123, 92–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yellon DM, Hausenloy DJ, Myocardial reperfusion injury. N Engl J Med 357, 1121–1135 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Hausenloy DJ, Botker HE, Engstrom T, Erlinge D, Heusch G, Ibanez B, Kloner RA, Ovize M, Yellon DM, Garcia-Dorado D, Targeting reperfusion injury in patients with ST-segment elevation myocardial infarction: trials and tribulations. Eur Heart J 38, 935–941 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalogeris T, Baines CP, Krenz M, Korthuis RJ, Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol 298, 229–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heusch G, Rassaf T, Time to Give Up on Cardioprotection? A Critical Appraisal of Clinical Studies on Ischemic Pre-, Post-, and Remote Conditioning. Circ Res 119, 676–695 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Heusch G, Schulz R, Rahimtoola SH, Myocardial hibernation: a delicate balance. Am J Physiol Heart Circ Physiol 288, H984–999 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Davidson SM, Ferdinandy P, Andreadou I, Bøtker HE, Heusch G, Ibáñez B, Ovize M, Schulz R, Yellon DM, Hausenloy DJ, Multitarget strategies to reduce myocardial ischemia/reperfusion injury: JACC review topic of the week. J Am Coll Cardiol 73, 89–99 (2019). [DOI] [PubMed] [Google Scholar]
- 9.McGonigle P, Ruggeri B, Animal models of human disease: challenges in enabling translation. Biochem Pharmacol 87, 162–171 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Post MJ, Laham R, Sellke FW, Simons M, Therapeutic angiogenesis in cardiology using protein formulations. Cardiovasc Res 49, 522–531 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Anavekar NS, Chareonthaitawee P, Narula J, Gersh BJ, Revascularization in Patients With Severe Left Ventricular Dysfunction: Is the Assessment of Viability Still Viable? J Am Coll Cardiol 67, 2874–2887 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Hausenloy DJ, Bøtker HE, Condorelli G, Ferdinandy P, Garcia-Dorado D, Heusch G, Lecour S, Van Laake LW, Madonna R, Ruiz-Meana M, Translating cardioprotection for patient benefit: position paper from the Working Group of Cellular Biology of the Heart of the European Society of Cardiology. Cardiovas Res 98, 7–27 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Banerjee MN, Bolli R, Hare JM, Clinical studies of cell therapy in cardiovascular medicine: recent developments and future directions. Circ Res 123, 266–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quyyumi AA, Vasquez A, Kereiakes DJ, Klapholz M, Schaer GL, Abdel-Latif A, Frohwein S, Henry TD, Schatz RA, Dib N, PreSERVE-AMI: a randomized, double-blind, placebo-controlled clinical trial of intracoronary administration of autologous CD34+ cells in patients with left ventricular dysfunction post STEMI. Circ Res 120, 324–331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher SA, Doree C, Mathur A, Taggart DP, Martin-Rendon E, Stem cell therapy for chronic ischaemic heart disease and congestive heart failure. Cochrane Database Syst Rev 12, CD007888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duran JM, Makarewich CA, Sharp TE, Starosta T, Zhu F, Hoffman NE, Chiba Y, Madesh M, Berretta RM, Kubo H, Bone-derived stem cells repair the heart after myocardial infarction through transdifferentiation and paracrine signaling mechanisms. Circ Res 113, 539–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai M, Shen R, Song L, Lu M, Wang J, Zhao S, Tang Y, Meng X, Li Z, He Z-X, Bone marrow mesenchymal stem cells (BM-MSCs) improve heart function in swine myocardial infarction model through paracrine effects. Scientific reports 6, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, Salto-Tellez M, Timmers L, Lee CN, El Oakley RM, Pasterkamp G, de Kleijn DP, Lim SK, Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res 4, 214–222 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Ibrahim AG, Cheng K, Marban E, Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports 2, 606–619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, Popescu LM, Torre T, Siclari F, Moccetti T, Vassalli G, Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res 103, 530–541 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VN, Benedict C, Ramirez V, Lambers E, Ito A, Gao E, Misener S, Luongo T, Elrod J, Qin G, Houser SR, Koch WJ, Kishore R, Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res 117, 52–64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng X, Chen L, Chen W, Yang J, Yang Z, Shen Z, Mesenchymal Stem Cell-Derived Exosomes Improve the Microenvironment of Infarcted Myocardium Contributing to Angiogenesis and Anti-Inflammation. Cell Physiol Biochem 37, 2415–2424 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Li X, Chen C, Wei L, Li Q, Niu X, Xu Y, Wang Y, Zhao J, Exosomes derived from endothelial progenitor cells attenuate vascular repair and accelerate reendothelialization by enhancing endothelial function. Cytotherapy 18, 253–262 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Vicencio JM, Yellon DM, Sivaraman V, Das D, Boi-Doku C, Arjun S, Zheng Y, Riquelme JA, Kearney J, Sharma V, Plasma exosomes protect the myocardium from ischemia-reperfusion injury. Journal of the American College of Cardiology 65, 1525–1536 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Davidson SM, Riquelme JA, Zheng Y, Vicencio JM, Lavandero S, Yellon DM, Endothelial cells release cardioprotective exosomes that may contribute to ischaemic preconditioning. Scientific reports 8, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michiels C, Arnould T, Remacle J, Endothelial cell responses to hypoxia: initiation of a cascade of cellular interactions. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research 1497, 1–10 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Hsieh PC, Davis ME, Lisowski LK, Lee RT, Endothelial-cardiomyocyte interactions in cardiac development and repair. Annu. Rev. Physiol 68, 51–66 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park S-J, Kotikian A, Nesmith AP, Campbell PH, Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater 16, 303–308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lind JU, Yadid M, Perkins I, O’Connor BB, Eweje F, Chantre CO, Hemphill MA, Yuan H, Campbell PH, Vlassak JJ, Cardiac microphysiological devices with flexible thin-film sensors for higher-throughput drug screening. Lab on a Chip 17, 3692–3703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ExoCarta: [cited 2020 6 Aug]. Available from: http://exocarta.org/exosome_markers_new.
- 31.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Reassessment of exosome composition. Cell 177, 428–445. e418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nasiri Kenari A, Kastaniegaard K, Greening DW, Shambrook M, Stensballe A, Cheng L, Hill AF, Proteomic and Post-Translational Modification Profiling of Exosome-Mimetic Nanovesicles Compared to Exosomes. Proteomics 19, 1800161 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Lu H, Zhu YF, Xiong J, Wang R, Jia Z, Potential extra-ribosomal functions of ribosomal proteins in Saccharomyces cerevisiae. Microbiol Res 177, 28–33 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Naora H, Involvement of ribosomal proteins in regulating cell growth and apoptosis: translational modulation or recruitment for extraribosomal activity? Immunol Cell Biol 77, 197–205 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Naora H, Takai I, Adachi M, Naora H, Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol 141, 741–753 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morhayim J, van de Peppel J, Demmers JA, Kocer G, Nigg AL, van Driel M, Chiba H, van Leeuwen JP, Proteomic signatures of extracellular vesicles secreted by nonmineralizing and mineralizing human osteoblasts and stimulation of tumor cell growth. The FASEB Journal 29, 274–285 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Bec N, Bonhoure A, Henry L, Berry L, Larroque C, Coux O, Stoebner PE, Vidal M, Proteasome 19S RP and translation preinitiation complexes are secreted within exosomes upon serum starvation. Traffic, (2019). [DOI] [PubMed] [Google Scholar]
- 38.Zhang W, He P, Wang S, Adili A, Chen Z, Zhang C-Y, Jiang X, Li J, Zhang Y, Characterization of Protein Profiling and mRNA Expression of LLC Exosomes. The Protein Journal, 1–12 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Turchinovich A, Drapkina O, Tonevitsky A, Transcriptome of extracellular vesicles: State-of-the-art. Frontiers in immunology 10,(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dranka BP, Benavides GA, Diers AR, Giordano S, Zelickson BR, Reily C, Zou L, Chatham JC, Hill BG, Zhang J, Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radical Biology and Medicine 51, 1621–1635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HK, Kang SW, Jeong SH, Kim N, Ko JH, Bang H, Park WS, Choi TH, Ha YR, Lee YS, Youm JB, Ko KS, Rhee BD, Han J, Identification of potential target genes of cardioprotection against ischemia-reperfusion injury by express sequence tags analysis in rat hearts. J Cardiol 60, 98–110 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Hazan-Halevy I, Rosenblum D, Weinstein S, Bairey O, Raanani P, Peer D, Cell-specific uptake of mantle cell lymphoma-derived exosomes by malignant and non-malignant B-lymphocytes. Cancer letters 364, 59–69 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T, Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell 10, 839–850 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Montecalvo A, Larregina AT, Shufesky WJ, Beer Stolz D, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood, The Journal of the American Society of Hematology 119, 756–766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Auriac A, Willemetz A, Canonne-Hergaux F, Lipid raft-dependent endocytosis: a new route for hepcidin-mediated regulation of ferroportin in macrophages. Haematologica 95, 1269–1277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheehy SP, Grosberg A, Qin P, Behm DJ, Ferrier JP, Eagleson MA, Nesmith AP, Krull D, Falls JG, Campbell PH, Toward improved myocardial maturity in an organ-on-chip platform with immature cardiac myocytes. Experimental Biology and Medicine 242, 1643–1656 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20, 616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pérez-Boza J, Lion M, Struman I, Exploring the RNA landscape of endothelial exosomes. RNA 24, 423–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayourian J, Ceholski DK, Gorski PA, Mathiyalagan P, Murphy JF, Salazar SI, Stillitano F, Hare JM, Sahoo S, Hajjar RJ, Exosomal microRNA-21–5p mediates mesenchymal stem cell paracrine effects on human cardiac tissue contractility. Circ Res 122, 933–944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamin IJ, McMillan DR, Stress (heat shock) proteins: molecular chaperones in cardiovascular biology and disease. Circ Res 83, 117–132 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, Glembotski CC, Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res 98, 1186–1193 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Belmont PJ, Chen WJ, San Pedro MN, Thuerauf DJ, Gellings Lowe N, Gude N, Hilton B, Wolkowicz R, Sussman MA, Glembotski CC, Roles for endoplasmic reticulum-associated degradation and the novel endoplasmic reticulum stress response gene Derlin-3 in the ischemic heart. Circ Res 106, 307–316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hwang IK, Yoo KY, Kim DW, Kim SY, Park JH, Ryoo ZY, Kim J, Choi SY, Won MH, Ischemia-induced ribosomal protein S3 expressional changes and the neuroprotective effect against experimental cerebral ischemic damage. J Neurosci Res 86, 1823–1835 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Nishino Y, Miura T, Miki T, Sakamoto J, Nakamura Y, Ikeda Y, Kobayashi H, Shimamoto K, Ischemic preconditioning activates AMPK in a PKC-dependent manner and induces GLUT4 up-regulation in the late phase of cardioprotection. Cardiovasc Res 61, 610–619 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Li J, Hu X, Selvakumar P, Russell III RR, Cushman SW, Holman GD, Young LH, Role of the nitric oxide pathway in AMPK-mediated glucose uptake and GLUT4 translocation in heart muscle. American Journal of Physiology-Endocrinology and Metabolism 287, E834–E841 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Jeremy RW, Ambrosio G, Pike MM, Jacobus WE, Becker LC, The functional recovery of post-ischemic myocardium requires glycolysis during early reperfusion. J Mol Cell Cardiol 25, 261–276 (1993). [DOI] [PubMed] [Google Scholar]
- 57.Jeremy RW, Koretsune Y, Marban E, Becker LC, Relation between glycolysis and calcium homeostasis in postischemic myocardium. Circ Res 70, 1180–1190 (1992). [DOI] [PubMed] [Google Scholar]
- 58.Van Emous JG, Vleggeert-Lankamp CL, Nederhoff MG, Ruigrok TJ, Van Echteld CJ, Postischemic Na(+)-K(+)-ATPase reactivation is delayed in the absence of glycolytic ATP in isolated rat hearts. Am J Physiol Heart Circ Physiol 280, H2189–2195 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Vanoverschelde JL, Janier MF, Bakke JE, Marshall DR, Bergmann SR, Rate of glycolysis during ischemia determines extent of ischemic injury and functional recovery after reperfusion. Am J Physiol 267, H1785–1794 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Pfleger J, He M, Abdellatif M, Mitochondrial complex II is a source of the reserve respiratory capacity that is regulated by metabolic sensors and promotes cell survival. Cell death & disease 6, e1835-e1835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sansbury BE, Jones SP, Riggs DW, Darley-Usmar VM, Hill BG, Bioenergetic function in cardiovascular cells: the importance of the reserve capacity and its biological regulation. Chemico-biological interactions 191, 288–295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jennings R, Murry CE, Steenbergen JC, Reimer K, Development of cell injury in sustained acute ischemia. Circulation 82, II2–12 (1990). [PubMed] [Google Scholar]
- 63.Polla B, Kantengwa S, Francois D, Salvioli S, Franceschi C, Marsac C, Cossarizza A, Mitochondria are selective targets for the protective effects of heat shock against oxidative injury. Proceedings of the national academy of sciences 93, 6458–6463 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Currie RW, Karmazyn M, Kloc M, Mailer K, Heat-shock response is associated with enhanced postischemic ventricular recovery. Circ Res 63, 543–549 (1988). [DOI] [PubMed] [Google Scholar]
- 65.Mocanu MM, Steare SE, Evans MC, Nugent JH, Yellon DM, Heat stress attenuates free radical release in the isolated perfused rat heart. Free Radic Biol Med 15, 459–463 (1993). [DOI] [PubMed] [Google Scholar]
- 66.Heusch G, Hibernating myocardium. Physiol Rev 78, 1055–1085 (1998). [DOI] [PubMed] [Google Scholar]
- 67.King HW, Michael MZ, Gleadle JM, Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 12, 421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salomon C, Kobayashi M, Ashman K, Sobrevia L, Mitchell MD, Rice GE, Hypoxia-induced changes in the bioactivity of cytotrophoblast-derived exosomes. PLoS One 8, e79636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ikonomidis JS, Tumiati LC, Weisel RD, Mickle DA, Li R-K, Preconditioning human ventricular cardiomyocytes with brief periods of simulated ischaemia. Cardiovascular research 28, 1285–1291 (1994). [DOI] [PubMed] [Google Scholar]
- 70.Speechly-Dick M, Grover G, Yellon D, Does ischemic preconditioning in the human involve protein kinase C and the ATP-dependent K+ channel? Studies of contractile function after simulated ischemia in an atrial in vitro model. Circ Res 77, 1030–1035 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Thery C, Amigorena S, Raposo G, Clayton A, Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3 22 (2006). [DOI] [PubMed] [Google Scholar]
- 72.Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, Simons M, Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci 124, 447–458 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Morgelin M, Belting M, Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem 288, 17713–17724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu C, Nebert DW, Update on genome completion and annotations: Protein Information Resource. Human genomics 1, 229 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mi H, Muruganujan A, Casagrande JT, Thomas PD, Large-scale gene function analysis with the PANTHER classification system. Nature protocols 8, 1551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, Weitz DA, Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. The Journal of cell biology 173, 733–741 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic acids research 43, D447–D452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wiśniewski JR, Zougman A, Nagaraj N, Mann M, Universal sample preparation method for proteome analysis. Nature methods 6, 359–362 (2009). [DOI] [PubMed] [Google Scholar]
- 80.Kall L, Storey JD, Noble WS, Non-parametric estimation of posterior error probabilities associated with peptides identified by tandem mass spectrometry. Bioinformatics 24, i42–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liebermeister W, Noor E, Flamholz A, Davidi D, Bernhardt J, Milo R, Visual account of protein investment in cellular functions. Proceedings of the National Academy of Sciences 111, 8488–8493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Criddle DN, Gillies S, Baumgartner-Wilson HK, Jaffar M, Chinje EC, Passmore S, Chvanov M, Barrow S, Gerasimenko OV, Tepikin AV, Menadione-induced reactive oxygen species generation via redox cycling promotes apoptosis of murine pancreatic acinar cells. Journal of Biological Chemistry 281, 40485–40492 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Javadov S, Karmazyn M, Escobales N, Mitochondrial permeability transition pore opening as a promising therapeutic target in cardiac diseases. J Pharmacol Exp Ther 330, 670–678 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Chen Z, Siu B, Ho YS, Vincent R, Chua CC, Hamdy RC, Chua BH, Overexpression of MnSOD protects against myocardial ischemia/reperfusion injury in transgenic mice. J Mol Cell Cardiol 30, 2281–2289 (1998). [DOI] [PubMed] [Google Scholar]
- 85.Shibata R, Sato K, Pimentel DR, Takemura Y, Kihara S, Ohashi K, Funahashi T, Ouchi N, Walsh K, Adiponectin protects against myocardial ischemia-reperfusion injury through AMPK- and COX-2-dependent mechanisms. Nat Med 11, 1096–1103 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Benhar M, Forrester MT, Stamler JS, Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol 10, 721–732 (2009). [DOI] [PubMed] [Google Scholar]
- 87.Dhalla NS, Elmoselhi AB, Hata T, Makino N, Status of myocardial antioxidants in ischemia-reperfusion injury. Cardiovasc Res 47, 446–456 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Common contaminant proteins detected in EEV samples.
Table S2. Gene Ontology terms and enrichment P-values for over-represented protein groups presented in figure 2C–D with corresponding color coding.
Table S3. Cardioprotective function of protein groups represented in EEVs.
Table S4. Possible cardioprotective miRNA cargo in previously published studies.
Fig. S1. Differentially-expressed EEVs protein content in normoxic and hypoxic conditions.
Fig. S2. Molecular function GO classification of differentially expressed proteins in hCMs treated with EEVs compared with control, untreated hCMs.
Fig. S3. Biological process GO classification of differentially expressed proteins in hCMs treated with EEVs compared with control, untreated hCMs.
Fig. S4. ROS measurement in Menadione-challenged hCMs un/treated with EEVs.
Fig. S5. Preconditioning vs. EEV treatment effect on viability following IRI.
Fig. S6. Molecular function GO classification of differentially expressed proteins in injured hCMs pre-treated with EEVs compared with injured, untreated hCMs.
Fig. S7. Biological process GO classification of differentially expressed proteins in injured hCMs pre-treated with EEVs compared with injured, untreated hCMs.
Fig. S8. EEV uptake by human cardiomyocytes
Fig. S9. Heart-on-a-chip device for contractility measurements.
Fig. S10. Non-significant cardioprotective effect of human EEVs on neonate rat ventricular myocytes-based heart-on-a-chip assay.
Data file S4. Summary of statistical analyses and P-values.
Data file S2. Differentially expressed proteins in EEV-treated vs. untreated hCMs.
Data file S3. Differentially expressed proteins in IR-injured, EEV-treated vs. injured, untreated hCMs.
Data file S1. Total EEVs proteome raw data.
Mov. S2. EEV uptake by human cardiomyocytes (cell 2).
Mov. S3. EEV uptake by human cardiomyocytes (cell 3).
Mov. S1. EEV uptake by human cardiomyocytes (cell 1).