Abstract
Florfenicol is an antibiotic approved for veterinary use in cattle in the United States in 1996. Although this drug is not used in poultry, we have detected resistance to florfenicol in clinical isolates of avian Escherichia coli. Molecular typing demonstrated that the florfenicol resistance gene, flo, was independently acquired and is plasmid encoded.
Florfenicol is a synthetic, fluorinated analogue of chloramphenicol which lacks chloramphenicol's associated human health risk (11). It has been used in Asia for aquaculture since the 1980's (12). In early 1996, an injectable formulation of florfenicol was approved for the treatment of bovine respiratory disease in the United States. It has not yet been approved for poultry, and, in fact, an animal feed formulation is not available.
Florfenicol is bacteriostatic, and its mechanism of action is similar to that of chloramphenicol (7, 22). The mechanism of resistance to florfenicol is unknown but is associated with the flo determinant, a highly conserved gene sequence detected in Salmonella enterica serovar Typhimurium DT104 (4, 6) and in the fish pathogen Pasteurella piscicida (Photobacterium damsela) (15). The flo gene confers resistance to both chloramphenicol and florfenicol (4, 14).
Resistance to chloramphenicol is most commonly mediated by mono- and diacetylation via chloramphenicol acetyltransferase (CAT) enzymes, which prevents the binding of chloramphenicol to the 50S ribosomal subunit (21). None of the genes encoding CAT has been shown to confer resistance to florfenicol, and there is no homology between the CATs and Flo (9). Another mediator of chloramphenicol resistance, the cmlA gene of Pseudomonas aeruginosa, is believed to be a nonenzymatic efflux pump (3). CmlA is approximately 50% similar in amino acid sequence to Flo (4), but it is not known whether cmlA confers resistance to florfenicol.
Our study examined the prevalence of florfenicol resistance in clinical avian Escherichia coli isolates. We hypothesized that there were preexisting genes in these bacterial isolates that conferred resistance to florfenicol and that this resistance might limit the future usefulness of the drug in other veterinary species. We report here the presence and incidence of the florfenicol resistance gene, flo, in avian E. coli.
The characteristics of the avian E. coli isolates are presented in Table 1. Of the 100 isolates cultured from litter and from clinical and postmortem material, 11 were found to be resistant to chloramphenicol by disc diffusion (30 μg). All 11 chloramphenicol-resistant isolates were multidrug resistant, and 4 of these isolates, avian E. coli isolates 5334, 5790, 5840, and 6468, were also resistant to florfenicol by disc diffusion (<21-mm-diameter zone of inhibition with a 30-μg disc). S. enterica serovar Typhimurium DT104, for which the MIC of florfenicol was 64 μg/ml, was used as the positive control (4). E. coli K-12 containing cmlA (3) exhibited a florfenicol MIC of 2 μg/ml, as did E. coli DH5α, the negative control. All of the flo-containing isolates exhibited florfenicol MICs of at least 32 μg/ml. The florfenicol MIC for two isolates which did not contain flo was 8 μg/ml. The breakpoints for florfenicol resistance recently adopted for bovine respiratory pathogens are 2 (sensitive), 4 (intermediate), and 8 (resistant) μg/ml (T. Shyrock, Chairholder, Veterinary Antimicrobial Susceptibility Testing Subcommittee, Nation Committee for Clinical Laboratory Standards, personal communication, 1999).
TABLE 1.
Characteristics of chloramphenicol-resistant avian E. colia isolates
| Isolate | Source | Site of isolation | Clinical signs | Antibiotic resistanceb | Gene(s)c detected by DNA hybridization
|
MIC (μg/ml) of:
|
||
|---|---|---|---|---|---|---|---|---|
| flo | int | Chloramphenicol | Florfenicol | |||||
| A35 | Va. | Litter sample | None | TE S SU NA SR EN | − | + | 200 | 4 |
| A410 | Va. | Litter sample | None | TE S SU NA SR EN | − | + | 400 | 4 |
| 3518 | Ga. | Yolk sac | Colibacillosis | AU TE S NA | − | + | 100 | 8 |
| 5334 | N.C. | Trachea | None | TE NE S SU | + | − | 100 | 64 |
| 5361 | Ga. | Peritoneum | Colibacillosis | AU TE S SU NA SR EN | − | + | 200 | 8 |
| 5790 | Ga. | Trachea | Respiratory disease | AU TE S SU | + | + | 100 | 32 |
| 5965 | Ga. | Trachea | Airsacculitis | AU TE S SU | − | + | 400 | 4 |
| 5967 | Ga. | Trachea | Airsacculitis | AU TE S SU | − | + | 200 | 4 |
| 6005 | Ga. | Trachea | Airsacculitis | AU TE GE S SU | − | + | 100 | 4 |
| 6468 | Ga. | Trachea | Respiratory disease | AU TE GE S SU | + | + | 100 | 32 |
| 5840 | Ga. | Trachea | Respiratory disease | TE S SU | + | − | 100 | 64 |
| Salmonella DT104 | —d | TE S SU | + | + | 100 | 64 | ||
| DH5α | NA | − | − | 0.78 | 2 | |||
None of the E. coli isolates were resistant to ceftiofur, an antibiotic that is routinely included in antibiotic susceptibility tests of poultry isolates.
Antibiotic resistance was determined by disc diffusion. Abbreviations for tetracyclines: AU, chlortetracycline (15 μg) and TE, tetracycline (30 μg). Abbreviations for quinolones/fluoroquinolones: NA, nalidixic acid (30 μg); SR, sarafloxacin (5 μg); and EN, enrofloxacin (5 μg). Abbreviation for sulfonamide: SU, sulfisoxazole (250 μg). Abbreviations for aminoglycosides: GE, gentamicin (10 μg); NE, neomycin (30 μg); and S, streptomycin (10 μg).
Isolates were not probed for the presence of cat genes.
—, data from reference 6.
The florfenicol-resistant E. coli isolates came from clinical samples sent from different poultry farms in Georgia and North Carolina. To determine whether they represent dissemination of a clonal strain, random amplification of polymorphic DNA (RAPD) by the method of Maurer et al. was employed (17). RAPD analysis showed four distinct patterns, or RAPD types (data not shown), suggesting that florfenicol resistance is not limited to a particular strain of avian E. coli.
DNA-DNA colony hybridizations with probes specific for cmlA, flo, and int were done to correlate the presence of these genes with florfenicol resistance. PCR was used to generate the DNA probes; Table 2 lists the primers employed and the expected sizes of the PCR products. The identities of the PCR products were confirmed by DNA sequencing. The PCR mixture consisted of 2 mM MgCl2, 0.2 mM deoxyribonucleoside triphosphates (digoxigenin labeled), 50 pmol of each oligonucleotide primer, and 0.5 U of Taq polymerase (Boehringer Mannheim, Indianapolis, Ind.). The program parameters for the hot-air thermocycler were 30 cycles of (i) 94°C for 1 s, (ii) 40°C for 1 s, and (iii) 72°C for 15 s. The PCR products were purified by using WIZARD DNA Clean-Up System kits (Promega) and combined with hybridization buffer, containing 0.75 M sodium chloride, 1% nonfat dry milk, 0.1% N-laurylsarcosine, and 0.2% sodium dodecyl sulfate; they were kept frozen at −20°C until use. Bacterial cells were patched onto nylon membranes with toothpicks, and DNA-DNA hybridizations were performed as described by Sambrook et al. (20), with hybridizations and washes being done at 68°C. Hybridizing DNA fragments were detected by using an anti-digoxigenin antibody-alkaline phosphatase conjugate with a color substrate solution of 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate (XP).
TABLE 2.
PCR-generated DNA probes for detecting presence of integrons, cmlA, and flo
| Probe | Oligonucleotide primersa | Sequence accession no. | Product size (bp) | Template source | Reference for primers |
|---|---|---|---|---|---|
| flo | F, 5′-TATCTCCCTGTCGTTCCAG-3′; R, 5′-AGAACTCGCCGATCAATG-3′ | D37826 | 399 | S. enterica serovar Typhimurium (DT104) | This study |
| cmlA | F, 5′-CCGCCACGGTGTTGTTGTTATC-3′; R, 5′-CACCTTGCCTGCCCATCATTAG-3′ | M64556 | 698 | Plasmid R1033 | This study |
| int | F, 5′-CCTCCCGCACGATGATC-3′; R, 5′-TCCACGCATCGTCAGGC-3′ | 280 | Plasmid pDU202 | 25 |
F, forward primer; R, reverse primer.
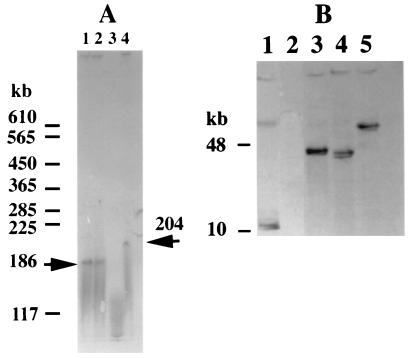
Results of the DNA colony hybridizations are shown in Table 1. Only one isolate, E. coli 6468, contained cmlA, whereas all four florfenicol-resistant isolates contained flo. The int gene probe revealed that 9 of 11 isolates were positive for the int sequence, indicating that integron-related genes were commonly present in multidrug-resistant clinical isolates. DNA-DNA hybridization was also used to assess whether the flo gene was plasmid associated. The flo gene was used to probe plasmids isolated from the flo-positive avian E. coli isolates (Fig. 1). Plasmid DNA was isolated by the S1 nuclease method of Barton et al. (2) and separated by pulsed-field gel electrophoresis (pulse time, 2 to 40 s; voltage, 6 V/cm; 25 h). The DNA was transferred from the agarose gel to a nylon membrane with a vacuum blotter (Bio-Rad, Hercules, Calif.) according to the manufacturer's recommendations. The procedure for DNA-DNA hybridizations was performed as described above. Three of the four isolates contained flo on high-molecular-weight plasmids of 186 and 204 kb. The florfenicol resistance determinant appears to be present in a variety of large-molecular-weight XbaI DNA fragments in avian E. coli, in contrast to the mapping of the flo resistance gene to a 10-kb XbaI fragment of S. enterica serovar Typhimurium DT104 (Fig. 1). Therefore, its location in avian E. coli may be similar to its placement in large-molecular-weight R plasmids in Pasteurella piscicida (14); however, the differences in sizes of the plasmids and fragments suggest that the gene was independently acquired.
FIG. 1.
Localization of the florfenicol resistance gene flo to large-molecular-weight plasmids in avian E. coli (A) Large-sized plasmids (>100 kb) were separated by pulsed-field gel electrophoresis and probed with labeled flo. Lanes: 1, avian E. coli isolate 5334; 2, isolate 5790; 3, isolate 5840; 4, isolate 6468. Arrows identify plasmid bands recognized by the probe. (B) The location of flo in the avian E. coli chromosome was mapped by pulsed-field gel electrophoresis. E. coli genomic DNA was cut with XbaI, separated by pulsed-field gel electrophoresis, and probed with labeled flo. Lanes: 1, S. enterica serovar Typhimurium DT104; 2, E. coli K12 LE392; 3 to 5, avian E. coli isolates 5334 (lane 3), 5790 (lane 4), and 6468 (lane 5).
Two of the four florfenicol-resistant isolates also contained int, the DNA integrase gene that is characteristic of integrons, which are transmissible elements deemed important in the horizontal transfer of antibiotic resistance genes. In Salmonella strain DT104, flo is chromosomally located between two integrons (6). Many of the antibiotic resistance genes found in gram-negative bacteria are located within integrons, which are mobile genetic elements (18). The integrase acts as a site-specific DNA recombinase in the insertion of antibiotic resistance genes into these elements (8, 18). For example, cmlA is present within the integron of Tn1696, which makes up part of the Pseudomonas aeruginosa IncP plasmid R1033 (3). The cmlA drug resistance gene does not appear to be responsible for high-level florfenicol resistance, since we found that only one florfenicol-resistant E. coli isolate possessed the gene and since E. coli containing cmlA was sensitive to florfenicol.
Our study demonstrates the persistence of chloramphenicol resistance in avian E. coli, although this drug has not been used therapeutically in food animals since its use was officially banned in 1988 (13). We also demonstrated that a low percentage (4%) of clinical avian E. coli isolates already display resistance to florfenicol, although the drug is not used therapeutically in chickens. In fact, a feed formulation is not currently available in the United States. Poultry production is rather unique in the United States since the processing company owns the birds and the feed; farmers are contracted to house the animals. The processing company employs veterinarians who are responsible for vaccination and medication of the birds in the face of illness. Therefore, the attending veterinarian prescribes the antibiotics used for treatment of disease, and it is improbable that the birds from which we isolated florfenicol-resistant E. coli had ever been exposed to florfenicol.
Antimicrobials are useful therapeutic agents only if the drug concentrations achieved in the serum and tissue are greater than the MIC of the drug. Florfenicol attains a maximum concentration of 3 μg/ml in the serum of feeder calves (16). The manufacturer of florfenicol reports an MIC of 1 μg/ml or less against 90% of the bacterial isolates from natural infections in cattle (5, 11, 23). Pharmacokinetic studies have shown that the peak plasma florfenicol concentration in ducks and chickens is approximately 3 μg/ml, with similar levels in the liver, kidney, and lung tissues (1, 10, 19). Our study showed that all 11 chloramphenicol-resistant avian E. coli isolates had florfenicol MICs of greater than 3 μg/ml, suggesting that this antimicrobial agent may not be therapeutically successful in some cases.
Acknowledgments
This study was supported by funding from the Veterinary Medicine Experiment Station.
We thank Karen Jacobsen and Doug Kemp for direction regarding florfenicol usage information and Cynthia Liebert and Barry Harmon for suggestions regarding the manuscript.
REFERENCES
- 1.Afifi N A, Abo El-Sooud K. Tissue concentrations and pharmacokinetics of florfenicol in broiler chickens. Br Poult Sci. 1997;38:425–428. doi: 10.1080/00071669708418013. [DOI] [PubMed] [Google Scholar]
- 2.Barton B M, Harding G P, Zuccarelli A J. A general method for detecting and sizing large plasmids. Anal Biochem. 1995;226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 3.Bissonnette L, Champetier S, Buisson J-P, Roy P H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolton L F, Kelley L C, Lee M D, Fedorka-Cray P J, Maurer J J. Detection of multidrug-resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J Clin Microbiol. 1999;37:1348–1351. doi: 10.1128/jcm.37.5.1348-1351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booker C W. Evaluation of florfenicol for the treatment of undifferentiated fever in feedlot calves in western Canada. Can Vet J. 1997;38:555–560. [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannon M. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol and some fluorinated derivatives. J Antimicrob Chemother. 1990;26:307–317. doi: 10.1093/jac/26.3.307. [DOI] [PubMed] [Google Scholar]
- 8.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman C J, Foster T J. Nonenzymatic chloramphenicol resistance determinants specified by plasmids R26 and R55-1 in Escherichia coli K-12 do not confer high-level resistance to fluorinated analogs. Antimicrob Agents Chemother. 1982;22:912–914. doi: 10.1128/aac.22.5.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El-Banna H A. Pharmacokinetics of florfenicol in normal and Pasteurella-infected Muscovy ducks. Br Poult Sci. 1998;39:492–496. doi: 10.1080/00071669888656. [DOI] [PubMed] [Google Scholar]
- 11.Food and Drug Administration. 31 May 1996. NUFLOR® (florfenicol). FOI summary, NADA 141-063 (original). Food and Drug Administration, Laurel, Md.
- 12.Fukui H, Fujihara Y, Kano T. In vitro and in vivo antibacterial activities of florfenicol, a new fluorinated analog of thiamphenicol, against fish pathogens. Fish Pathol. 1987;22:201–207. [Google Scholar]
- 13.Gilmore A. Chloramphenicol and the politics of health. Can Med Assoc J. 1986;134:423–435. [PMC free article] [PubMed] [Google Scholar]
- 14.Kim E H, Yoshida T, Aoki T. Detection of R-plasmid-encoded with resistance to florfenicol in Pasteurella piscicida. Fish Pathol. 1993;28:165–170. [Google Scholar]
- 15.Kim E-H, Aoki T. Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscisida. Microbiol Immunol. 1996;40:665–669. doi: 10.1111/j.1348-0421.1996.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 16.Lobell R D. Pharmacokinetics of florfenicol following intravenous and intramuscular doses to cattle. J Vet Pharmacol Ther. 1994;17:253–258. doi: 10.1111/j.1365-2885.1994.tb00241.x. [DOI] [PubMed] [Google Scholar]
- 17.Maurer J J, Lee M D, Lobsinger C, Brown T, Maier M, Thayer S G. Molecular typing of avian Escherichia coli isolates by random amplification of polymorphic DNA. Avian Dis. 1998;42:431–451. [PubMed] [Google Scholar]
- 18.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 19.Rios A, Martinez-Larranaga M R, Anadon A. Plasma disposition of florfenicol in broiler chickens following intravenous administration. J Vet Pharmacol. 1997;20:182. [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Shaw W V. Bacterial resistance to chloramphenicol. Br Med Bull. 1984;40:36–41. doi: 10.1093/oxfordjournals.bmb.a071945. [DOI] [PubMed] [Google Scholar]
- 22.Syriopoulou V P, Harding A L, Goldmann D A, Smith A L. In vitro antibacterial activity of fluorinated analogs of chloramphenicol and thiamphenicol. Antimicrob Agents Chemother. 1981;19:294–297. doi: 10.1128/aac.19.2.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson D J. Efficacy of florfenicol for treatment of clinical and subclinical bovine mastitis. Am J Vet Res. 1996;57:526–528. [PubMed] [Google Scholar]