Abstract
Smad proteins transduce signals downstream of transforming growth factor-β (TGF-β) and are one of the factors that regulate the expression of genes related to diseases affecting the skin. In the present study, we identified MAB21L4, also known as male abnormal 21 like 4 or C2orf54, as the most up-regulated targets of TGF-β and Smad3 in differentiated human progenitor epidermal keratinocytes using chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq). We found that TGF-β induced expression of the barrier protein involucrin (encoded by the IVL gene). Transcriptional activity of the IVL promoter induced by TGF-β was inhibited by MAB21L4 siRNAs. Further analysis revealed that MAB21L4 siRNAs also down-regulated the expression of several target genes of TGF-β. MAB21L4 protein was located mainly in the cytosol, where it was physically bound to Smad3 and a transcriptional corepressor c-Ski. siRNAs for MAB21L4 did not inhibit the binding of Smad3 to their target genomic regions but down-regulated the acetylation of histone H3 lys 27 (H3K27ac), an active histone mark, near the Smad3 binding regions. These findings suggest that TGF-β-induced MAB21L4 up-regulates the gene expression induced by TGF-β, possibly through the inhibition of c-Ski via physical interaction in the cytosol.
Keywords: TGF-β, epidermal keratinocytes, Smad3, c-Ski, ChIP-seq
Graphical Abstract
Graphical Abstract.
Abbreviations
- TGF-β
transforming growth factor-β
- ChIP
chromatin immunoprecipitation
- MAB21
male abnormal 21
The epidermis represents a barrier against hazardous environmental conditions and physical injury. In addition to the tight cellular interactions between epidermal cells, proteins derived from terminally differentiated keratinocytes are processed to form the outermost layer of the epidermis, called the cornified envelope, to provide the protective role of the skin (1). Involucrin and loricrin are examples of such proteins. Transforming growth factor-β (TGF-β) is a multi-functional cytokine with various physiological roles. The epidermis is regulated by TGF-β and its related cytokines, including activins and bone morphogenetic proteins (BMPs) (2). TGF-β forms a receptor complex at the cell membrane and the activated type I receptor, i.e. ALK5 phosphorylates Smad2 and Smad3 (Smad2/3). Phosphorylated Smad2/3 form a complex with Smad4, translocate to the nucleus and work as transcription factors (TFs) to regulate expression of the target genes of TGF-β. Extensive research has revealed that TGF-β inhibits proliferation of keratinocytes to maintain homeostasis of the skin. TGF-β family cytokines also play a role in the development and homeostasis of hair follicles. A differentiated epidermal keratinocyte cell line, denoted HaCaT, was established by Boukamp et al. (3) and has been widely used to analyse TGF-β signalling pathways in vitro. However, the precise mechanisms by which TGF-β regulates differentiated keratinocyte-derived proteins in the epidermis remain unclear.
In the present study, we performed an integrated analysis to identify the target genes of Smad3 using chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) in differentiated human epidermal keratinocytes. Based on these analyses, we identified MAB21L4, also known as male-abnormal 21 like 4 or C2orf54, as an important Smad3 target and verified that TGF-β and Smad3 regulate the expression of MAB21L4. We explored the function of MAB21L4 in HaCaT cells and found that MAB21L4 up-regulates the expression of involucrin and other target genes of TGF-β. Our findings revealed an epidermis-specific positive feedback loop of TGF-β signalling, illuminating the role of MAB21L4 and suggesting new possible therapeutic target for diseases affecting the skin.
Materials and Methods
Cell culture
HaCaT is a spontaneously immortalized human epidermal keratinocyte cell line that was established previously (3). HaCaT cells and 293T cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM #11965; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% foetal bovine serum, 100 U/ml penicillin G and 100 μg/ml streptomycin. Human progenitor epidermal keratinocyte (HPEK) cells were obtained from CELLnTEC (Bern, Switzerland) and cultured in Progenitor Cell Targeting medium (CnT-57 or CnT-PR; CELLnTEC) supplemented with 50 U/ml penicillin G and 50 μg/ml streptomycin to maintain their progenitor state. For induction of differentiation, cells were cultured in CnT-02 or CnT-PR-D supplemented with 1.2 mM CaCl2, according to the manufacturer’s instructions (CELLnTEC). Cells were grown in a humidified atmosphere with 5% CO2 at 37°C.
Reagents and antibodies
Recombinant human TGF-β (TGF-β3) was obtained from R&D systems (Minneapolis, MN, USA). The following antibodies were used: rabbit anti-phospho-Smad2 (p-Smad2; Cell Signaling Technology, Danvers, MA, USA), mouse anti-α-tubulin (DM1A; Sigma-Aldrich, St. Louis, MO, USA), rabbit anti-Smad3 (EP568Y, Abcam, Cambridge, UK), rabbit anti-Smad2 (EP567Y, Abcam), rabbit anti-involucrin (IVL) (ab53112, Abcam), mouse anti-Histone H3 acetyl Lys 27 (H3K27ac) (MABI0009, clone MABI0309, MAB Institute, Nagano, Japan), mouse anti-myc (9E10, Nacalai Tesque, Kyoto, Japan), mouse anti-FLAG (F3165, Sigma-Aldrich), mouse anti-HA (Invivogen, San Diego, CA, USA) and rabbit anti-HA (Y-11, Santa Cruz Biotechnology, Dallas, TX, USA).
An anti-MAB21L4 antibody (#11802) was raised by immunizing a rabbit with the amino-terminal domain of a recombinant human MAB21L4 fragment (amino acids 1–154, NP_001078906) expressed in DH5α cells transformed with the pGEX6P-1 vector. For preparation of the recombinant protein, cell lysate was purified using glutathione Sepharose 4B (GE healthcare, Chicago, IL, USA) and enzymatically processed using Turbo 3C protease (Fujifilm Wako Pure Chemical, Osaka, Japan) to elute the MAB21L4 fragment without the GST domain. After size separation by SDS-PAGE and CBB staining (CBB Stain One Super, Nacalai Tesque), the excised gel portion was directly injected subcutaneously into a rabbit by Eurofins Genomics Japan (Tokyo, Japan). Antisera were purified using Protein A by Eurofins Genomics Japan.
Plasmids
Plasmid expressing human MAB21L4 (NP_001078906) tagged with FLAG was cloned from the first strand cDNA of HaCaT cells using a standard PCR-based approach. Smad2, Smad3, Smad4, constitutively active ALK5 (ALK5-TD), c-Ski and SnoN expression vectors were prepared as described previously (4, 5). All insert sequences were verified by sequencing.
Immunoprecipitation and immunoblotting
Cells were lysed in cell lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1 mM phenylmethylsulfonyl fluoride, 1% aprotinin, 1% phosphatase inhibitor cocktail). Immunoprecipitation, sodium dodecyl sulphate gel electrophoresis and immunoblotting were performed as described previously (6), with a LAS-4000 lumino-image analyser used for image acquisition (Fujifilm, Tokyo, Japan).
Luciferase reporter assay
IVL-luc reporter was constructed using a standard PCR-based approach. The −2812 to +81 genomic region relative to the transcription start site (TSS) of the IVL gene was amplified by PCR using genomic DNA obtained from HaCaT cells as the template and primers containing restriction sites for NheI and BglII. The PCR product was cloned into the NheI–BglII site of the pGL4 luciferase reporter (Promega, Madison, WI, USA), which contains an adenoviral major late promoter (MLP) derived from (CAGA)9-luc (7). The sequence of the promoter was verified by comparing with the human genome (hg19).
For promoter-reporter assays, cells were transfected with the reporter plasmid vectors and siRNAs using Lipofectamine 2000, according to the manufacturer’s instructions (Thermo Fisher Scientific). At 24 h after transfection, cells were treated with 1 ng/ml TGF-β for 24 h. Luciferase activity was measured using an LB940FP luminometer (Berthold Technologies, Bad Wildbad, Germany).
Immunofluorescence staining
HaCaT cells were seeded on a two-well chamber glass slide (Thermo Fisher Scientific) after coating with collagen I (Nitta Gelatin, Osaka, Japan) and transfected with FLAG-tagged MAB21L4. After 24 h, cells were fixed with ice-cold 50% acetone/50% methanol for 5 min. Cells were then washed with phosphate-buffered saline and blocked with BlockingOne (Nacarai Tesque). Anti-FLAG antibody was used as the first antibody at 5 μg/ml and Alexa Fluor 488-conjugated anti-mouse IgG (Thermo Fisher Scientific) was used as the secondary antibody (1:500). After mounting with Antifade mounting medium with DAPI (Vector laboratories, Burlingame, CA, USA), images were captured with a fluorescence microscope BZ-X710 (Keyence, Osaka, Japan). For each condition, randomly selected two images were obtained.
RNA interference
siRNAs against human MAB21L4 (#1, 5′-UCCUGCUGGACAAGUUCCAGGUCUU-3′; #2, 5′-ACGGCCUGACCUUUGGGCCACCUGAA-3′) and control siRNA (Cat. 12,935–112) were synthesized by Thermo Fisher Scientific. siRNAs were introduced into HaCaT cells using Lipofectamine 2000 or Lipofectamine RNAiMAX reagents (Thermo Fisher Scientific) according to the manufacturer’s protocol.
RNA isolation and quantitative RT-PCR
Total RNA was extracted using an RNeasy Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. First-strand cDNA was synthesized using PrimeScript2 reverse transcriptase (TakaraBio, Shiga, Japan). Quantitative RT-PCR (qRT-PCR) analysis was performed using FastStart Universal SYBR Green Master Mix with ROX (Roche Diagnostics, Basel, Switzerland) and the ABI PRISM 7500Fast Sequence Detection System or StepONE Plus real time PCR system (Thermo Fisher Scientific). Primer sequences for qRT-PCR are shown in Supplementary Table S1.
RNA-seq
RNA-seq was performed as described previously (8). Reference files of the human reference sequence assembly (NCBI Build 37/hg19) and the GTF annotation file were obtained from iGenomes (http://support.illumina.com/sequencing/sequencing_software/igenome.html). Gene expression levels were estimated as fragments per kilobase of exon per million fragments mapped (FPKM) values using Tophat/Cufflinks (versions 2.0.8 and 2.0.2, respectively), using the default parameter settings.
ChIP assay, ChIP-sequencing and data analysis
Chromatin isolation, sonication, immunoprecipitation using anti-Smad3, anti-Smad2 and anti-H3K27ac antibodies and quantification by real-time PCR were conducted as described previously (9). Primer sequences are shown in Supplementary Table S2. The amount of immunoprecipitated DNA relative to the input was calculated for all samples, and enrichment values were calculated relative to the %input values at SOBP locus, which serves as a negative control region. For high-throughput sequencing analysis (ChIP-seq), libraries were constructed, amplified with 15 cycles of PCR, size-selected and sequenced using an Illumina Genome Analyzer (Illumina, San Diego, CA, USA) or Ion Proton/Ion PGM sequencers (Thermo Fisher Scientific), according to the manufacturer’s protocols.
The human reference sequence assembly (NCBI Build 37/hg19) and GTF annotation files were obtained from iGenomes. ChIP-seq data sets were aligned using Bowtie (version 0.12.7) with the command ‘-S -a --best --strata -v 1 -m 1’ (for Smad3) or Bowtie2 (for Smad2 and H3K27ac) with the default parameters. Antibody-enriched genomic regions were identified using MACS2 software (model-based analysis of ChIP-seq) with a q-value threshold of 0.0001. Motif enrichment analysis was performed using Centrimo with the default parameters (CentriMo version 5.3.0; http://meme-suite.org/tools/centrimo) (10).
Accession numbers
Raw sequencing data can be accessed via the GEO repository (GSE180252).
Statistical analysis
Dunnett tests were used to compare multiple samples to the control, using GraphPad Prism (Usaco, Tokyo, Japan).
Results
Identification of Smad3 binding regions in human epidermal keratinocytes
HPEK cells undergo differentiation when cultured with differentiation medium. Upon induction of differentiation following 6 days of culture, the cells expressed genes representing the differentiated state of the epidermis: KRT5 (encoding keratin 5) as a marker of the basal layer, KRT1 and KRT10 as markers of the spinous layer and IVL (encoding involucrin) as a marker of the granular layer (Fig. 1A).
Fig. 1.
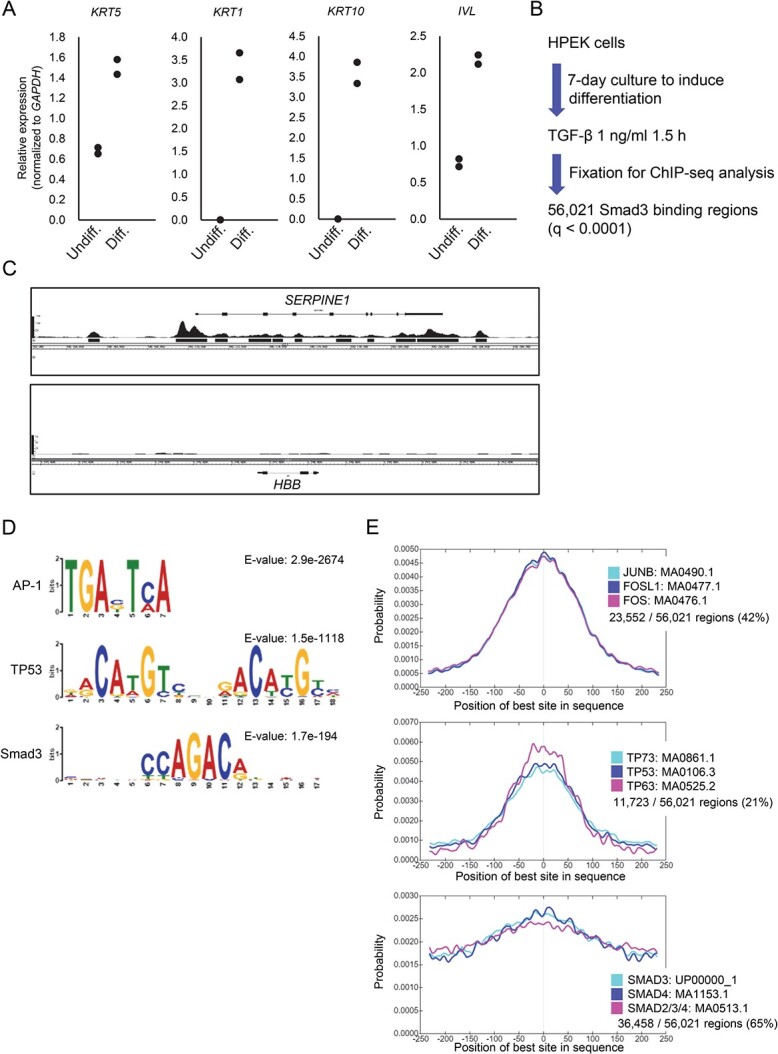
Smad3 binding regions in epidermal keratinocytes. (A) Expression of epidermal differentiation markers in HPEK cells. Total RNA was isolated after the induction of differentiation of HPEK cells for 6 days, and the expression of keratinocyte differentiation markers was compared to that of undifferentiated progenitor cells using qRT-PCR. Data were obtained from two biological replicates. (B) Experimental design of Smad3 ChIP-seq analysis of differentiated HPEK cells. Cells were cultured in differentiation medium for 7 days and then stimulated with TGF-β for 1.5 h before fixation. Smad3 binding regions were determined by ChIP and ChIP-seq. (C) Smad3 binding signals at the known target of TGF-β. Peak signals at the SERPINE1 locus show enrichment of Smad3 binding signals, and black bars show significant binding regions at q < 0.0001. The HBB locus is shown as a negative control. (D) The top enriched motif groups, AP-1 and TP53, according to CentriMo. Smad binding motifs were also identified. (E) Relative position of the enriched motifs alongside the peak summit of the Smad3 binding regions. The top 3 motifs included in each motif group are shown. Percentages show the frequency of motifs in the Smad3 binding regions. Undiff., undifferentiated cells; Diff., differentiated cells.
To elucidate the transcriptional role of TGF-β in both undifferentiated and differentiated epidermal keratinocytes, we performed RNA-seq analysis of HPEK cells stimulated with TGF-β (Supplementary Fig. S1). Gene set enrichment analysis revealed that the genes related to epithelial-to-mesenchymal transition, apical junction and TGF-β signalling were up-regulated by TGF-β stimulation in both undifferentiated and differentiated cells (Supplementary Fig. S1A–C). The genes related to K-Ras signalling, allograft rejection and androgen and oestrogen responses were enriched in differentiated cells, regardless of TGF-β stimulation (Supplementary Fig. S1D, E).
We next obtained ChIP-seq data of Smad3 binding sites in the differentiated HPEK cells stimulated with TGF-β for 1.5 h (Fig. 1B). Significant Smad3 binding peaks were identified at the well-known target gene locus of TGF-β, SERPINE1, but not in the HBB locus, which served as the negative control region (Fig. 1C). We identified 56,021 Smad3 binding regions with a q-value of <0.0001. Motif analysis of the Smad3 binding sites revealed the enrichment of binding sequences of several TFs that are known co-regulators of Smad signalling (Fig. 1D, E). Motif centrality analysis suggested that AP-1 binding sites and TP53 family binding sites were concentrated towards the peaks of Smad3 binding positions (Fig. 1E). Smad binding sites were also concentrated towards the peaks to a lesser extent, which likely reflects the existence of two or more Smad binding motifs within a binding region (9, 11), or the low complexity of the Smad binding motif compared to AP-1 and TP53 binding motifs.
Identification of MAB21L4 as a target gene of TGF-β/Smad3
We then used a keratinocyte cell line, HaCaT, and obtained Smad3 ChIP-seq data to compare with the data obtained from the differentiated HPEK cells. Because Smad2 is also the downstream signalling molecule of TGF-β signalling that forms complex with Smad3 and Smad4, we also obtained ChIP-seq data of Smad2 in HaCaT cells to compare with the Smad3 data in the same experimental condition. As a result, 22,194 and 18,691 binding regions were determined from Smad3 and Smad2 ChIP-seq, respectively (Fig. 2A). We found that 72.8% and 72.2% of the Smad3 and Smad2 binding regions, respectively, in HaCaT cells were shared with the Smad3 binding regions in HPEK cells. We also found that 69.2% of the Smad3 binding regions were shared with the Smad2 binding regions in HaCaT cells.
Fig. 2.
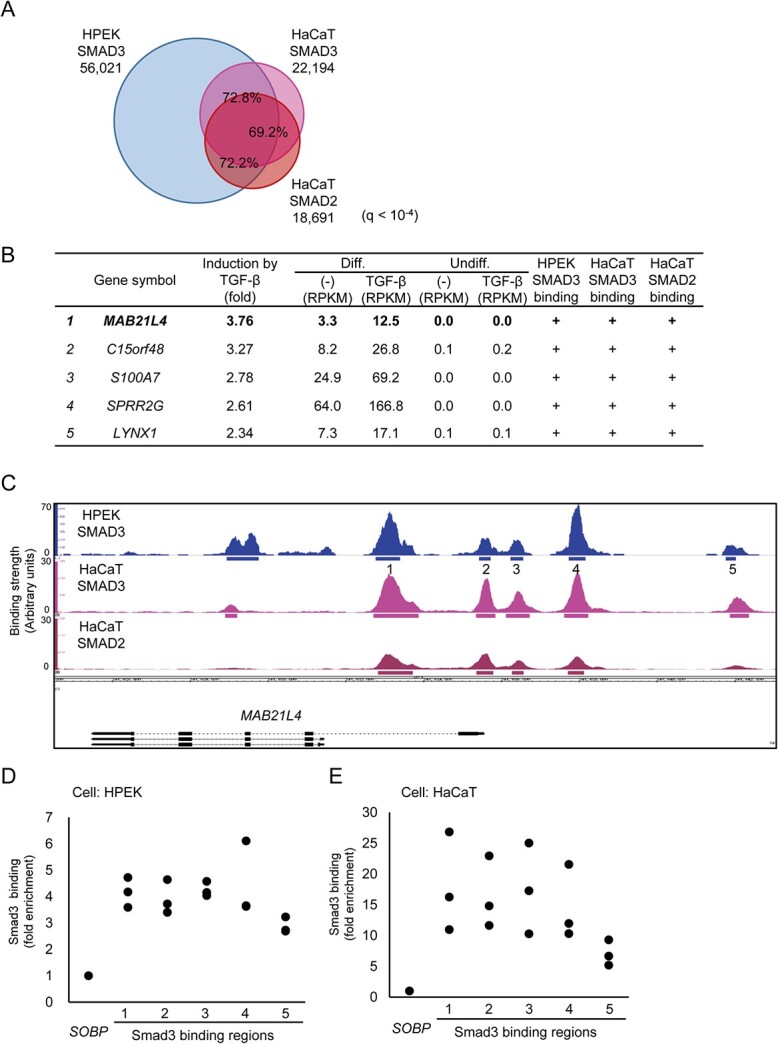
Identification of MAB21L4 as a target of Smad3. (A) Comparison of the Smad3 and Smad2 binding regions in HPEK and HaCaT cells. ChIP-seq data of HaCaT cells stimulated with 1 ng/ml of TGF-β3 for 1.5 h were obtained for comparison to the data obtained in Fig. 1. Significant binding regions were determined at q < 10−4 and overlap analysis was performed by using bedtools (https://bedtools.readthedocs.io/en/latest/). (B) MAB21L4 was identified as the most up-regulated target gene of TGF-β and Smad3 in differentiated HPEK cells. Genes with expression levels of at least 10 FPKM in any of the differentiated HPEK samples were included in the analysis. The data were then sorted by the order of induction by TGF-β. Genes exclusively expressed in the differentiated HPEK cells (FPKM < 1 in undifferentiated HPEK cells) are shown. ‘Smad3 binding’ is defined as significant Smad3 binding within 100 kb from the gene. (C) Smad3 and Smad2 binding regions at the MAB21L4 locus are shown. There are three variants (a longer form and lower two shorter forms) of MAB21L4. Numbers show the regions analysed by ChIP-qPCR in (D) and (E). (D) ChIP-qPCR analysis of Smad3 binding at the MAB21L4 locus. Differentiated HPEK cells were treated with TGF-β for 1.5 h and fixed. The SOBP locus was used as the negative control region (9). Data were obtained from three biological replicates. (E) ChIP-qPCR analysis of Smad3 binding in an epidermal keratinocyte cell line HaCaT. ChIP was performed as in (D). Data were obtained from three biological replicates.
By evaluating the RNA-seq data of HPEK cells, we found that both undifferentiated and differentiated HPEK cells respond to TGF-β at the transcriptional level, with no remarkable differences observed in the enriched gene sets (Supplementary Fig S1A, C). We therefore next focused on individual target genes of Smad3 that were specific to the differentiated HPEK cells. MAB21L4 was the most up-regulated target of Smad3 of unknown function and was found only in the differentiated HPEK and HaCaT cells but not in the undifferentiated HPEK cells (Fig. 2B). We identified several Smad3 and Smad2 binding regions both upstream and downstream of the TSS of MAB21L4 (Fig. 2C). We selected five nearby regions and verified Smad3 binding using ChIP-qPCR (Fig. 2D). We also used HaCaT cells to confirm that Smad3 binds to the same regions (Fig. 2E). Based on the data obtained from a public gene expression database (12), MAB21L4 is expressed in the skin and oesophagus, suggesting that it is expressed mainly in the stratified squamous epithelium (Supplementary Fig. S2A).
Regulation of MAB21L4 by TGF-β and related signalling pathways
Next, we evaluated the effect of TGF-β and other signal transduction pathways related to TGF-β and/or epidermal keratinocytes on the expression of MAB21L4. The expression of MAB21L4 was strongly induced in HPEK cells after 6 days of culture with differentiation medium (Fig. 3A). MAB21L4 mRNA and protein were detectable in differentiated HaCaT cells, even at steady state (Fig. 3B and C), and stimulation with TGF-β resulted in the rapid and transient induction of MAB21L4. RNA-seq data showed that, of the MAB21 family genes, only MAB21L4 was expressed in HPEK cells (Fig. 3D). TP63, a TP53 family gene, is one of the most important TFs of the epidermis. Both alleles of the TP53 gene were spontaneously mutated in HaCaT cells, and we previously reported the relationship between TP63, mutant TP53, RAS and TGF-β signalling using this cell line (13). Using previously published microarray data, we found that the expression of MAB21L4 was down-regulated by TP53 siRNA (Supplementary Fig. S2B), as well as by RAS signalling (Supplementary Fig. S2C). Notably, some of these effects were observed even without TGF-β stimulation. These results suggest that the expression of MAB21L4 was regulated by several signalling pathways in epidermal keratinocytes.
Fig. 3.
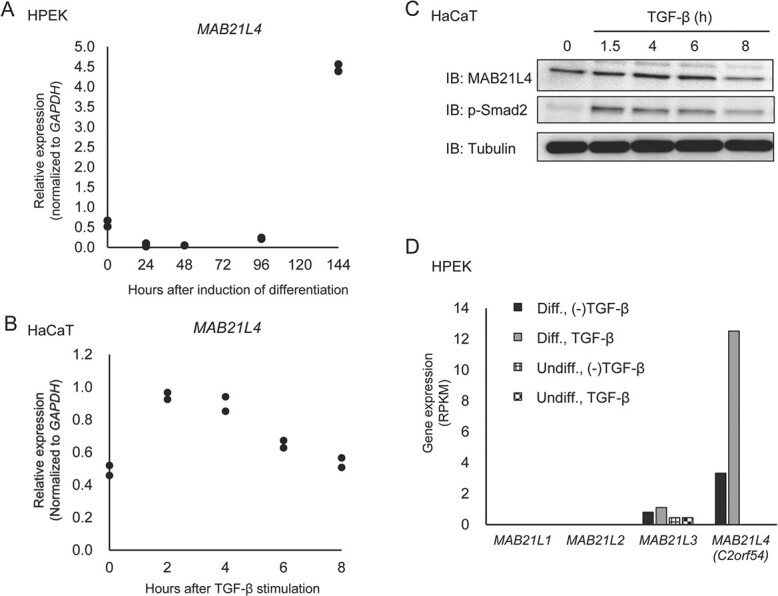
Regulation of MAB21L4 expression. (A) MAB21L4 is expressed after a delay following the induction of differentiation in HPEK cells. Total RNA was serially isolated after the induction of differentiation of HPEK cells as indicated, and the expression of MAB21L4 was determined by qRT-PCR. Data were obtained from two biological replicates. (B) Up-regulation of MAB21L4 mRNA by TGF-β in HaCaT cells. Cells were stimulated with TGF-β as indicated. Data were obtained from two biological replicates. (C) The change in the amount of MAB21L4 protein after TGF-β stimulation in HaCaT cells. The experiment was repeated with the similar results, and the representative data are shown. (D) Expression of MAB21-family mRNAs in HaCaT cells. RPKM values of each mRNA were obtained from the RNA-seq data used in Supplementary Fig. S1. IB, immunoblotting.
MAB21L4 up-regulates TGF-β-induced involucrin expression
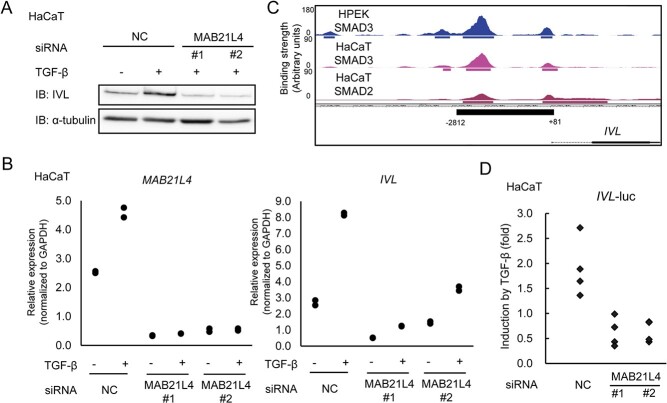
Based on the finding that the expression of MAB21L4 was strongly up-regulated in the late phase after the induction of differentiation, we knocked down the expression of MAB21L4 in HaCaT cells and performed qRT-PCR to elucidate the function of MAB21L4. We found that the amount of involucrin protein and mRNA induced by TGF-β stimulation decreased in MAB21L4-depleted cells (Fig. 4A, B). We cloned the promoter region of the IVL gene, which contains Smad3 and Smad2 binding regions (Fig. 4C), into the upstream of an adenoviral MLP in a luciferase-reporter plasmid to evaluate the effect of MAB21L4 siRNA on TGF-β-induced transcription of IVL. We found that the up-regulation of transcriptional activity induced by TGF-β was inhibited in MAB21L4-depleted cells (Fig. 4D), suggesting that TGF-β-induced MAB21L4 in turn regulates the IVL transcription induced by TGF-β.
Fig. 4.
MAB21L4 up-regulates involucrin expression. (A) Up-regulation of involucrin protein induced by TGF-β was abrogated by MAB21L4 siRNA. HaCaT cells were transfected with two different MAB21L4 siRNAs and stimulated with TGF-β for 24 h. The experiment was repeated with the similar results, and the representative data are shown. (B) The effect of MAB21L4 siRNA on the expression of MAB21L4 mRNA in HaCaT cells. HaCaT cells were transfected with MAB21L4 siRNAs and stimulated with TGF-β for 24 h. Data were obtained from two biological replicates. (C) Smad3 and Smad2 binding regions at the promoter of the IVL locus. Significant binding regions are represented as in Fig. 2C. The region (−2812 to +81 from the TSS) used in the promoter-reporter analysis shown in (D) is also indicated in the bottom panel. (D) MAB21L4 siRNAs down-regulated TGF-β-induced transcriptional activity of the IVL promoter. HaCaT cells were transfected with luciferase reporter plasmids and siRNA as indicated. Cells were then stimulated with TGF-β or left untreated. Data were obtained from four biological replicates.
MAB21L4 regulates target gene expression of TGF-β
We further evaluated the effect of MAB21L4 siRNA on gene expression using qRT-PCR to determine whether MAB21L4 also regulates other targets of TGF-β signalling. The expression of SERPINE1 and CDKN1A were up-regulated by TGF-β, but were down-regulated by MAB21L4 siRNAs (Fig. 5A). Co-immunoprecipitation analysis suggested that MAB21L4 physically interacts with Smad3, but not with Smad2 (Fig. 5B). However, the binding strength was much weaker when compared with the interaction between Smad3 and Smad2. MAB21L4 is located primarily in the cytosol (Fig. 5C and D), suggesting its regulatory function on TGF-β signalling outside the nucleus. We next evaluated the binding of Smad3 to the target genomic regions using ChIP-qPCR. Unexpectedly, recruitment of Smad3 to the known binding regions at SERPINE1 and CDKN1A loci was not significantly affected by MAB21L4 siRNA (Fig. 5E).
Fig. 5.
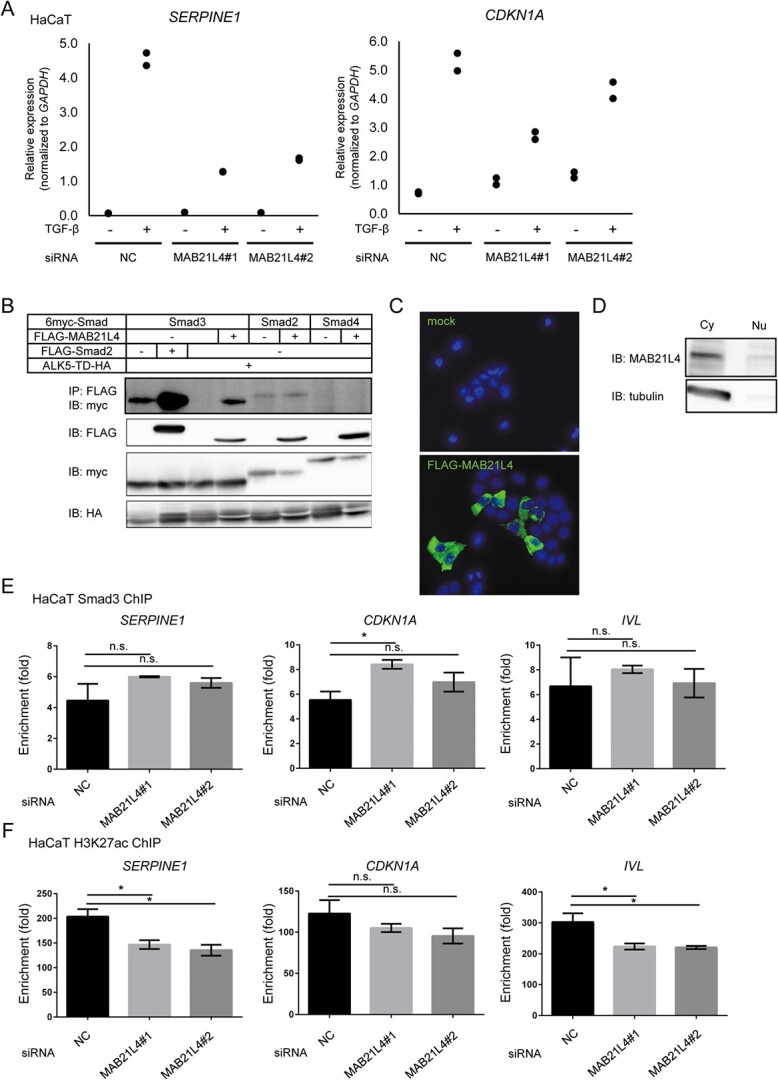
MAB21L4 up-regulates TGF-β target genes. (A) The effect of MAB21L4 siRNA on the TGF-β-induced expression of SERPINE1 and CDKN1A in HaCaT cells. The same samples shown in Fig. 4B were used for this evaluation. (B) MAB21L4 protein physically binds to Smad proteins; 293T cells were transfected with the expression plasmids as indicated and lysed for co-immunoprecipitation analysis. The experiment was repeated with the similar results and the representative data are shown. (C) Cytoplasmic localization of FLAG-MAB21L4 protein in transfected HaCaT cells. Cells were transfected with the plasmids encoding FLAG-tagged MAB21L4 or empty vector. Data were obtained from the two microscopic fields, and the representative images are shown. (D) Cell fractionation assay showing the localization of endogenous MAB21L4 protein. HaCaT cells were lysed with NE-PER (Thermo Fisher Scientific) to obtain the cytoplasmic and nuclear fractions. The experiment was repeated with similar results, and the representative data are shown. (E) Effect of knockdown of MAB21L4 on the Smad3 binding to the target genomic regions. HaCaT cells transfected with siRNA as indicated were stimulated with TGF-β for 1.5 h and harvested for ChIP-qPCR analysis. Data represents the mean ± standard deviation of two biological replicates. (F) Down-regulation of H3K27ac by MAB21L4 siRNAs. HaCaT cells were transfected with the siRNAs as indicated and were stimulated with TGF-β for 1.5 h. Data represents the mean ± standard deviation of two biological replicates. Note that the most enriched genomic positions of Smad3 and H3K27ac appeared different at the IVL locus and the different primers were used between Fig. 5E and F, as shown in Supplementary Table S2. NC, control siRNA; IP, immunoprecipitation; IB, immunoblotting; ALK5-TD, constitutively active ALK5; n.s.: not significant. *P < 0.05 by Dunnett test.
Based on the down-regulation of target gene expression (Fig. 5A), we then evaluated a histone tail modification, H3K27ac, around the Smad3 binding regions. H3K27ac is the established marker associated with active transcription. Indeed, H3K27ac was significantly suppressed by MAB21L4 siRNAs at SERPINE1 and IVL loci, suggesting transcriptional repression (Fig. 5F). These results suggested that MAB21L4 expression leads to elevated transcriptional activity without up-regulation of the Smad3 binding. These observations prompted us to evaluate the possibility that MAB21L4 functions via functional interaction with histone acetylation/deacetylation machinery.
c-Ski and SnoN are the representative proteins that physically bind to Smad family proteins and suppress transcription via recruitment of HDAC complex (14, 15). Previous reports have shown as alternative regulatory mechanisms that c-Ski and SnoN bind to the Smad family proteins in the cytosol to inhibit the formation of active Smad complex (16) or their nuclear localization (17, 18). It is also reported that stimulation with TGF-β induces rapid degradation of c-Ski and SnoN to induce transcription (5, 19, 20). We thus evaluated whether the MAB21L4 protein physically binds to c-Ski and SnoN. We found that MAB21L4 bound to c-Ski regardless of the TGF-β signal activation (Fig. 6A). SnoN also bound to MAB21L4, but not as strong as c-Ski. We also found the decrease in the amounts of c-Ski and SnoN proteins in the MAB21L4 co-transfected cells (Fig. 6A, second panel). The cycloheximide (CHX) chase assay suggested that turnover of the c-Ski protein was shortened by MAB21L4 expression (Fig. 6B).
Fig. 6.
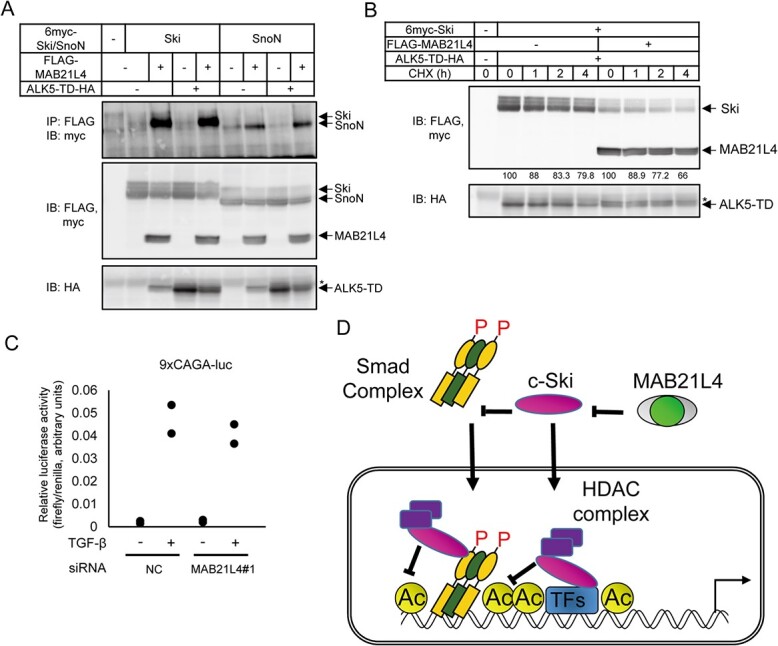
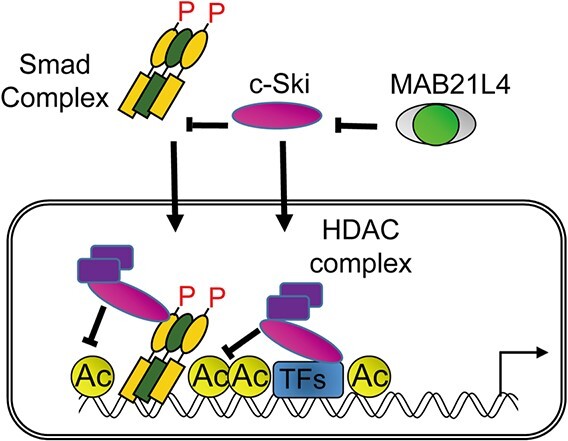
MAB21L4 physically interacts with c-Ski and SnoN proteins. (A) Co-immunoprecipitation analysis showing the physical interaction of c-Ski and SnoN with MAB21L4 protein; 293T cells were transfected as indicated. 6myc-tagged c-Ski and SnoN proteins co-immunoprecipitated with anti-FLAG antibody was detected in the top panel. The blotted membrane was serially used for detection of different antigens and thus anti-HA image shows residual anti-FLAG and anti-myc bands shown as *. (B) Effect of co-expression of MAB21L4 protein on the turnover of c-Ski protein; 293T cells were transfected with the expression plasmids as indicated. Cells were then treated with CHX at 100 μg/ml for the indicated times before cell lysis. Amounts of the c-Ski protein relative to the samples without CHX treatment (%) are shown below the top panel. (C) Effect of MAB21L4 siRNA on Smad-specific transcriptional activity. HaCaT cells were transfected with a luciferase reporter plasmid and siRNA. Cells were then treated with TGF-β. Data were obtained from two biological replicates. (D) A model of mechanism of enhanced target gene expression by MAB21L4 (also shown in Graphical Abstract). MAB21L4 protein physically binds to c-Ski in the cytosol to inhibit the known repressor function of c-Ski, either inhibition of the nuclear translocation of Smad complex or recruitment of HDAC complex on the target genomic regions. We cannot exclude the possibility that c-Ski inhibits the transcription via other TFs, independent of Smad complex. P, phosphorylation of Smad proteins; Ac, acetylation of histone H3, specifically lys 27, shown in this study. *, residual anti-FLAG bands as in Fig. 6A.
TGF-β-specific transcriptional reporter assay also suggested that, unlike the natural IVL promoter-reporter (Fig. 4D), MAB21L4 siRNA did not inhibit the transcriptional activity of the Smad-binding reporter 9xCAGA (Fig. 6C), suggesting the requirement of natural regulatory DNA sequence for MAB21L4 to enhance transcription.
Taken together, these results suggest that MAB21L4 up-regulates the expression of the target genes of TGF-β through in part the inhibition of c-Ski and SnoN.
Discussion
The present study identified involucrin as an up-regulated target of MAB21L4. Importantly, the previous publications showed that although involucrin is one of the major components of the cornified envelope, targeted deletion of involucrin did not affect the development of the epidermis and hair follicles (21). In contrast, Koch et al. reported that the loss of loricrin, another barrier protein of the skin, resulted in several skin abnormalities and a delay in the formation of the skin barrier at birth. Nevertheless, in case of the adult mice, loricrin depletion did not show any defects in barrier function, possibly due to the compensatory expression of involucrin and other barrier proteins (22). Considering the molecular function of MAB21L4 on TGF-β signalling, the effect of MAB21L4 on the expression of epidermal genes may depend on TGF-β signalling, because the expression of involucrin, but not loricrin, is up-regulated by TGF-β, based on our RNA-seq data. Further studies are required to elucidate the roles of MAB21L4 on diseases of the skin, especially in the context of TGF-β-dependent processes, as reported previously (23).
MAB21L4 belongs to the MAB21 family of proteins, which are required for the development of dorsal structures, including eye morphogenesis. Previous studies have been undertaken on MAB21L1, MAB21L2 and their orthologues (24–29). Mab21-l3 is expressed in multiciliate cells and ionocytes and is required for the expression of lineage-specific master genes during xenopus development (30). Mechanistically, MAB21L2 may physically interact with Smad1 and Smad4 to inhibit BMP signalling (31). Amino acid sequence analysis suggests that MAB21 proteins have a common domain with nucleotidyltransferase (NTase) activity, which suggests that they may exert important biological functions by processing RNA, DNA and proteins (32, 33). However, the biological role of MAB21L4 has not been previously elucidated.
The present study found that MAB21L4 enhances TGF-β signalling, possibly by binding to c-Ski and SnoN (Fig. 6D). The predicted NTase activity might contribute to the process by modulating the transcriptional corepressors c-Ski and SnoN. As a result, it is possible that the function of MAB21L4 is inhibition of the formation of an inactive Smad complex that contains c-Ski and the HDAC complex (34). Of note, unlike MAB21L4, c-Ski inhibits the transcriptional activity of the 9xCAGA reporter (17), suggesting that MAB21L4 might not suppress the other mechanisms of Smad signal inhibition by c-Ski, i.e. active Smad complex formation in the cytosol and its nuclear translocation.
There are several limitations to the present study. First, we need more comprehensive analyses, e.g. ChIP-seq and RNA-seq, after inactivation of MAB21L4 gene, to elucidate the central mechanisms of MAB21L4 function in epidermal keratinocytes. We cannot exclude the possibility that MAB21L4 may regulate non-Smad pathways or other signalling pathways that modulate TGF-β signalling. Second, biological significance of the expression of MAB21L4 in epidermal keratinocytes and in the other stratified epithelia need to be evaluated in animal models. Third, the effect of MAB21L4 siRNAs on the amount c-Ski protein is not fully consistent with the previous studies. c-Ski is rapidly degraded after TGF-β stimulation; however, the effect of MAB21L4 on c-Ski/SnoN turnover is minimal and found even in the absence of TGF-β signal activation. In addition, only a part of c-Ski function is inhibited by MAB21L4. We thus cannot exclude the possibility that MAB21L4 regulates c-Ski through a different mechanism from degradation. Finally, Smad2- or Smad3-specific functions have been revealed based on their different binding affinities to the cofactors (35, 36). Although many of the Smad2 and Smad3 binding regions are shared in the present study, the effect of MAB21L4 on Smad2 binding may differ from that on Smad3.
Taken together, MAB21L4 contributes to epidermis-specific TGF-β signalling to regulate IVL expression. Comprehensive analyses of the effect of MAB21L4 on the cell type-specific Smad3 binding (37, 38), epigenomic state and gene expression are required in the future to elucidate the mechanism of MAB21L4 action.
Supplementary Material
Acknowledgements
We thank the members of the Department of Molecular Pathology for technical assistance and discussion.
Funding
This work was supported by KAKENHI Grants-in-Aid for Scientific Research (S) [15H05774] and (A) [20H00513] (K. Miyazono) from the Japan Society for the Promotion of Science (JSPS) and Takeda Science Foundation (D.K.). Y.T. is supported by a Research Fellowship for Young Scientists (DC) and the Graduate Program for Leaders in Life Innovation from the JSPS.
Contributor Information
Tomohiro Ogami, Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan.
Yusuke Tamura, Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan.
Kim Toss, Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan.
Keiko Yuki, Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan.
Masato Morikawa, Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan.
Shuichi Tsutsumi, Genome Science Division, Research Center for Advanced Science and Technology, The University of Tokyo, Komaba 4-6-1, Meguro-ku, Tokyo 153-8904, Japan.
Hiroyuki Aburatani, Genome Science Division, Research Center for Advanced Science and Technology, The University of Tokyo, Komaba 4-6-1, Meguro-ku, Tokyo 153-8904, Japan.
Keiji Miyazawa, Department of Biochemistry, Graduate School of Medicine, University of Yamanashi, 1110 Shimokato, Chuo, Yamanashi 409-3898, Japan.
Kohei Miyazono, Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan.
Daizo Koinuma, Department of Molecular Pathology, Graduate School of Medicine, The University of Tokyo, Hongo 7-3-1, Bunkyo-ku, Tokyo 113-0033, Japan.
Author Contributions
T.O., K. Miyazawa, D.K. and K. Miyazono designed the study; T.O., Y.T., K.Y., K.T. and D.K. performed the experiments and analysed the data; D.K., K.Y., M.M., S.T. and H.A. performed next-generation sequencing analysis; T.O., D.K. and K. Miyazono wrote the manuscript; and D.K. and K. Miyazono coordinated all aspects of this work.
Supplementary Data
Supplementary Data are available at JB Online.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
References
- 1. Candi, E., Schmidt, R., and Melino, G. (2005) The cornified envelope: a model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6, 328–340 [DOI] [PubMed] [Google Scholar]
- 2. Kahata, K., Dadras, M.S., and Moustakas, A. (2018) TGF-beta family signaling in epithelial differentiation and epithelial-mesenchymal transition. Cold Spring Harb. Perspect. Biol. 10, a022194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boukamp, P., Petrussevska, R.T., Breitkreutz, D., Hornung, J., Markham, A., and Fusenig, N.E. (1988) Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106, 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koinuma, D., Shinozaki, M., Komuro, A., Goto, K., Saitoh, M., Hanyu, A., Ebina, M., Nukiwa, T., Miyazawa, K., Imamura, T., and Miyazono, K. (2003) Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 22, 6458–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagano, Y., Mavrakis, K.J., Lee, K.L., Fujii, T., Koinuma, D., Sase, H., Yuki, K., Isogaya, K., Saitoh, M., Imamura, T., Episkopou, V., Miyazono, K., and Miyazawa, K. (2007) Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J. Biol. Chem. 282, 20492–20501 [DOI] [PubMed] [Google Scholar]
- 6. Koinuma, D., Shinozaki, M., Nagano, Y., Ikushima, H., Horiguchi, K., Goto, K., Chano, T., Saitoh, M., Imamura, T., Miyazono, K., and Miyazawa, K. (2011) RB1CC1 protein positively regulates transforming growth factor-beta signaling through the modulation of Arkadia E3 ubiquitin ligase activity. J. Biol. Chem. 286, 32502–32512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dennler, S., Itoh, S., Vivien, D., ten Dijke, P., Huet, S., and Gauthier, J.M. (1998) Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Morikawa, M., Koinuma, D., Mizutani, A., Kawasaki, N., Holmborn, K., Sundqvist, A., Tsutsumi, S., Watabe, T., Aburatani, H., Heldin, C.H., and Miyazono, K. (2016) BMP sustains embryonic stem cell self-renewal through distinct functions of different Kruppel-like factors. Stem Cell Rep. 6, 64–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koinuma, D., Tsutsumi, S., Kamimura, N., Taniguchi, H., Miyazawa, K., Sunamura, M., Imamura, T., Miyazono, K., and Aburatani, H. (2009) Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor beta signaling. Mol. Cell. Biol. 29, 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bailey, T.L., and Machanick, P. (2012) Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 40, e128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Itoh, Y., Koinuma, D., Omata, C., Ogami, T., Motizuki, M., Yaguchi, S.I., Itoh, T., Miyake, K., Tsutsumi, S., Aburatani, H., Saitoh, M., Miyazono, K., and Miyazawa, K. (2019) A comparative analysis of Smad-responsive motifs identifies multiple regulatory inputs for TGF-beta transcriptional activation. J. Biol. Chem. 294, 15466–15479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fagerberg, L., Hallstrom, B.M., Oksvold, P., Kampf, C., Djureinovic, D., Odeberg, J., Habuka, M., Tahmasebpoor, S., Danielsson, A., Edlund, K., Asplund, A., Sjostedt, E., Lundberg, E., Szigyarto, C.A., Skogs, M., Takanen, J.O., Berling, H., Tegel, H., Mulder, J., Nilsson, P., Schwenk, J.M., Lindskog, C., Danielsson, F., Mardinoglu, A., Sivertsson, A., von Feilitzen, K., Forsberg, M., Zwahlen, M., Olsson, I., Navani, S., Huss, M., Nielsen, J., Ponten, F., and Uhlen, M. (2014) Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics 13, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vasilaki, E., Morikawa, M., Koinuma, D., Mizutani, A., Hirano, Y., Ehata, S., Sundqvist, A., Kawasaki, N., Cedervall, J., Olsson, A.K., Aburatani, H., Moustakas, A., Miyazono, K., and Heldin, C.H. (2016) Ras and TGF-beta signaling enhance cancer progression by promoting the DeltaNp63 transcriptional program. Sci. Signal. 9, ra84. [DOI] [PubMed] [Google Scholar]
- 14. Stroschein, S.L., Wang, W., Zhou, S., Zhou, Q., and Luo, K. (1999) Negative feedback regulation of TGF-beta signaling by the SnoN oncoprotein. Science 286, 771–774 [DOI] [PubMed] [Google Scholar]
- 15. Nomura, T., Khan, M.M., Kaul, S.C., Dong, H.D., Wadhwa, R., Colmenares, C., Kohno, I., and Ishii, S. (1999) Ski is a component of the histone deacetylase complex required for transcriptional repression by mad and thyroid hormone receptor. Genes Dev. 13, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu, J.W., Krawitz, A.R., Chai, J., Li, W., Zhang, F., Luo, K., and Shi, Y. (2002) Structural mechanism of Smad4 recognition by the nuclear oncoprotein Ski: insights on Ski-mediated repression of TGF-beta signaling. Cell 111, 357–367 [DOI] [PubMed] [Google Scholar]
- 17. Nagata, M., Goto, K., Ehata, S., Kobayashi, N., Saitoh, M., Miyoshi, H., Imamura, T., Miyazawa, K., and Miyazono, K. (2006) Nuclear and cytoplasmic c-Ski differently modulate cellular functions. Genes Cells 11, 1267–1280 [DOI] [PubMed] [Google Scholar]
- 18. Krakowski, A.R., Laboureau, J., Mauviel, A., Bissell, M.J., and Luo, K. (2005) Cytoplasmic SnoN in normal tissues and nonmalignant cells antagonizes TGF-beta signaling by sequestration of the Smad proteins. Proc. Natl. Acad. Sci. U. S. A. 102, 12437–12442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le Scolan, E., Zhu, Q., Wang, L., Bandyopadhyay, A., Javelaud, D., Mauviel, A., Sun, L., and Luo, K. (2008) Transforming growth factor-beta suppresses the ability of Ski to inhibit tumour metastasis by inducing its degradation. Cancer Res. 68, 3277–3285 [DOI] [PubMed] [Google Scholar]
- 20. Levy, L., Howell, M., Das, D., Harkin, S., Episkopou, V., and Hill, C.S. (2007) Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 27, 6068–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Djian, P., Easley, K., and Green, H. (2000) Targeted ablation of the murine involucrin gene. J. Cell Biol. 151, 381–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koch, P.J., de Viragh, P.A., Scharer, E., Bundman, D., Longley, M.A., Bickenbach, J., Kawachi, Y., Suga, Y., Zhou, Z., Huber, M., Hohl, D., Kartasova, T., Jarnik, M., Steven, A.C., and Roop, D.R. (2000) Lessons from loricrin-deficient mice: compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J. Cell Biol. 151, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ashcroft, G.S., Yang, X., Glick, A.B., Weinstein, M., Letterio, J.L., Mizel, D.E., Anzano, M., Greenwell-Wild, T., Wahl, S.M., Deng, C., and Roberts, A.B. (1999) Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat. Cell Biol. 1, 260–266 [DOI] [PubMed] [Google Scholar]
- 24. Mariani, M., Corradi, A., Baldessari, D., Malgaretti, N., Pozzoli, O., Fesce, R., Martinez, S., Boncinelli, E., and Consalez, G.G. (1998) Mab21, the mouse homolog of a C. elegans cell-fate specification gene, participates in cerebellar, midbrain and eye development. Mech. Dev. 79, 131–135 [DOI] [PubMed] [Google Scholar]
- 25. Lau, G.T., Wong, O.G., Chan, P.M., Kok, K.H., Wong, R.L., Chin, K.T., Lin, M.C., Kung, H.F., and Chow, K.L. (2001) Embryonic XMab21l2 expression is required for gastrulation and subsequent neural development. Biochem. Biophys. Res. Commun. 280, 1378–1384 [DOI] [PubMed] [Google Scholar]
- 26. Kennedy, B.N., Stearns, G.W., Smyth, V.A., Ramamurthy, V., van Eeden, F., Ankoudinova, I., Raible, D., Hurley, J.B., and Brockerhoff, S.E. (2004) Zebrafish rx3 and mab21l2 are required during eye morphogenesis. Dev. Biol. 270, 336–349 [DOI] [PubMed] [Google Scholar]
- 27. Merello, E., De Marco, P., Moroni, A., Raso, A., Calevo, M.G., Consalez, G.G., Cama, A., and Capra, V. (2004) Molecular genetic analysis of human homologs of Caenorhabditis elegans mab-21-like 1 gene in patients with neural tube defects. Birth Defects Res. A Clin. Mol. Teratol. 70, 885–888 [DOI] [PubMed] [Google Scholar]
- 28. Yamada, R., Mizutani-Koseki, Y., Koseki, H., and Takahashi, N. (2004) Requirement for Mab21l2 during development of murine retina and ventral body wall. Dev. Biol. 274, 295–307 [DOI] [PubMed] [Google Scholar]
- 29. Saito, Y., Kojima, T., and Takahashi, N. (2012) Mab21l2 is essential for embryonic heart and liver development. PLoS One 7, e32991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takahashi, C., Kusakabe, M., Suzuki, T., Miyatake, K., and Nishida, E. (2015) mab21-l3 regulates cell fate specification of multiciliate cells and ionocytes. Nat. Commun. 6, 6017. [DOI] [PubMed] [Google Scholar]
- 31. Baldessari, D., Badaloni, A., Longhi, R., Zappavigna, V., and Consalez, G.G. (2004) MAB21L2, a vertebrate member of the male-abnormal 21 family, modulates BMP signaling and interacts with SMAD1. BMC Cell Biol. 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuchta, K., Knizewski, L., Wyrwicz, L.S., Rychlewski, L., and Ginalski, K. (2009) Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 37, 7701–7714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kranzusch, P.J. (2019) cGAS and CD-NTase enzymes: structure, mechanism, and evolution. Curr. Opin. Struct. Biol. 59, 178–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suzuki, H., Yagi, K., Kondo, M., Kato, M., Miyazono, K., and Miyazawa, K. (2004) c-ski inhibits the TGF-beta signaling pathway through stabilization of inactive Smad complexes on Smad-binding elements. Oncogene 23, 5068–5076 [DOI] [PubMed] [Google Scholar]
- 35. Miyazono, K.I., Moriwaki, S., Ito, T., Kurisaki, A., Asashima, M., and Tanokura, M. (2018) Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors. Sci. Signal 11, eaao7227 [DOI] [PubMed] [Google Scholar]
- 36. Aragon, E., Wang, Q., Zou, Y., Morgani, S.M., Ruiz, L., Kaczmarska, Z., Su, J., Torner, C., Tian, L., Hu, J., Shu, W., Agrawal, S., Gomes, T., Marquez, J.A., Hadjantonakis, A.K., Macias, M.J., and Massague, J. (2019) Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-beta signaling. Genes Dev. 33, 1506–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Isogaya, K., Koinuma, D., Tsutsumi, S., Saito, R.A., Miyazawa, K., Aburatani, H., and Miyazono, K. (2014) A Smad3 and TTF-1/NKX2-1 complex regulates Smad4-independent gene expression. Cell Res. 24, 994–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mizutani, A., Koinuma, D., Tsutsumi, S., Kamimura, N., Morikawa, M., Suzuki, H.I., Imamura, T., Miyazono, K., and Aburatani, H. (2011) Cell type-specific target selection by combinatorial binding of Smad2/3 proteins and hepatocyte nuclear factor 4alpha in HepG2 cells. J. Biol. Chem. 286, 29848–29860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.