Abstract
A clinical strain of Acinetobacter baumannii (strain Ab RYC 52763/97) that was isolated during an outbreak in our hospital and that was resistant to all β-lactam antibiotics tested produced three β-lactamases: a TEM-1-type (pI, 5.4) plasmid-mediated β-lactamase, a chromosomally mediated OXA-derived (pI, 9.0) β-lactamase, and a presumptive chromosomal cephalosporinase (pI, 9.4). The nucleotide sequence of the chromosomal cephalosporinase gene shows for the first time the gene encoding an AmpC β-lactamase in A. baumannii. In addition, we report here the biochemical properties of this A. baumannii AmpC β-lactamase.
Acinetobacter spp. are recognized as important opportunist pathogens mainly in immunocompromised patients (9, 10). Their contribution to nosocomial infection has increased over the past three decades (6, 7, 27), and several outbreaks of hospital infection have been reported worldwide (5; G. Bou, G. Cerveró, D. Malpica, M. Pérez-Vázquez, L. De Rafael, and J. Martínez-Beltrán, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. K-120, 1998). Though prevalent in nature (4) and generally regarded as commensals of human skin and of the human respiratory tract (1, 15), they have been implicated as the cause of serious infectious diseases such as pneumonia, urinary tract infection, endocarditis, wound infection, meningitis, and septicemia, involving mostly patients with impaired host defenses (6, 9, 30). Antimicrobial treatment of serious infections due to Acinetobacter, particularly those caused by Acinetobacter baumannii, is complicated by the widespread multidrug resistance pattern of the organism (8, 18). With respect to β-lactam antibiotics, the most common mechanism of resistance is due to the synthesis of β-lactamases encoded either by the chromosome or by plasmids (9). So far, several class A and class D plasmid-encoded β-lactamases conferring different phenotypes of resistance to A. baumannii have been described (9, 24, 31, 32; G. Bou, A. Oliver, and J. Martínez-Beltrán, submitted for publication). Likewise, different chromosomally mediated cephalosporinases (pI, >8) have also been reported (9, 25), and sometimes these cephalosporinases have been particularly difficult to visualize by isoelectric focusing (24). In any case, nucleotide sequencing of the ampC gene has never been described. The main objective of this work was to clone and sequence the gene encoding the chromosomally mediated AmpC β-lactamase from a multiresistant A. baumannii clinical strain (Ab RYC 52763/97) isolated during an outbreak at our hospital in 1997 (Bou et al., 38th ICAAC). A detailed description of its biochemical characteristics is also provided.
The susceptibility testing of the Ab RYC 52763/97 strain was performed by an agar dilution method, as recommended by the National Committee for Clinical Laboratory Standards (23), using Mueller-Hinton agar and an inoculum size of 104 CFU per spot. Antibiotics were kindly provided as powders of a fixed potency by their corresponding manufacturers. The antibiotic susceptibility profiles of all strains included in this study are shown in Table 1. The strain Ab RYC 52763/97 was highly resistant to all β-lactam antibiotics tested, with minimum inhibitory concentrations (MICs) of imipenem and meropenem being 128 and 256 μg/ml, respectively.
TABLE 1.
MICS of β-lactams
| Antibiotic | MIC (μg/ml)a
|
|||
|---|---|---|---|---|
| A. baumannii RYC 52763/97 | E. coli TG1 | E. coli TG1 (pGER1) | E. coli TG1 (pGER2) | |
| Ampicillin | >1,024 | 4 | 128 | >1,024 |
| Ampicillin plus clavulanic acidb | >1,024 | 4 | 64 | 128 |
| Ampicillin plus sulbactamb | >1,024 | 2 | 16 | >1,024 |
| Ampicillin plus tazobactamb | >1,024 | 2 | 32 | 64 |
| Ticarcillin | >1,024 | 4 | 32 | >1,024 |
| Cefazolin | >256 | 8 | >256 | >256 |
| Cefuroxime | >256 | 4 | >256 | >256 |
| Cefoxitin | >256 | 4 | 4 | 4 |
| Cefotaxime | >256 | ≤0.125 | 4 | 8 |
| Cefotaxime plus clavulanic acid | >256 | ≤0.125 | 2 | 4 |
| Cefotaxime plus sulbactam | >256 | ≤0.125 | 0.25 | 2 |
| Cefotaxime plus tazobactam | >256 | ≤0.125 | 0.5 | 1 |
| Ceftazidime | >256 | ≤0.125 | 16 | 32 |
| Ceftazidime plus clavulanic acid | >256 | ≤0.125 | 8 | 16 |
| Ceftazidime plus sulbactam | >256 | ≤0.125 | 1 | 8 |
| Ceftazidime plus tazobactam | >256 | ≤0.125 | 2 | 4 |
| Cefepime | 256 | ≤0.125 | 0.25 | 0.5 |
| Aztreonam | >256 | ≤0.125 | 1 | 4 |
| Imipenem | 128 | 0.25 | 0.25 | 0.25 |
| Meropenem | 256 | 0.03 | 0.03 | 0.03 |
Enzymes are as follows: for A. baumannii RYC 52763/97, AmpC plus a β-lactamase with a pI of 9.0 plus TEM-1; for E. coli TG1(pGER1), AmpC; and for E. coli TG1(pGER2), AmpC plus TEM-1. Clone E. coli TG1(pGER2) harbored the ampC gene in the pUC18 multicopy plasmid (containing the blaTEM-1 resistance marker).
Concentration of β-lactamase inhibitor was 4 μg/ml.
β-Lactamases were analyzed by isoelectric focusing as described by Matthew et al. (22). The sonicated extract of strain Ab RYC 52763/97 contained three β-lactamases, as follows: one yielded a band with a pI of 5.4 and was cloned after amplification with bla-TEM primers C1 (5′-GGGAATTCTCGGGGAAATGTGCGCGGAAC) and C2 (5′-GGGATCCGAGTAAACTTGGTCTGACAG) (TEM-1 type); a second focused at a pI of 9.0, and after cloning (clone pBMB-1), the gene nucleotide sequence showed homology with those of OXA-derived β-lactamases (Bou et al., submitted); and the third focused at a pI of 9.4 and likely corresponded to a chromosomal cephalosporinase. With the A. baumannii clinical strain no transfer of resistance phenotype was observed in conjugation experiments. By alkaline lysis (28), a plasmid of 22 kb was isolated in this strain. By enzymatic restriction and ligation techniques (28) only a TEM-1 gene was found in this plasmid.
Chromosomal DNA of Ab RYC 52763/97 was prepared by the method of Grimont and Grimont (16), digested with HindIII (Boehringer-Mannheim, Mannheim, Germany), and ligated into the HindIII site of pBGS18− (29). Recombinant plasmids were introduced into Escherichia coli TG1 by transformation with CaCl2 (28), and transformants were selected on Luria-Bertani agar plates supplemented with ampicillin (25 μg/ml) and kanamycin (50 μg/ml). Three E. coli transformants were obtained on selective medium supplemented with the antibiotics mentioned above. They harbored a recombinant plasmid, designated pGER1, with an insert of about 2.2 kb.
The β-lactam susceptibility pattern of the three transformants was identical, displaying resistance to ampicillin, cefazolin, cefuroxime, and, to a lesser extent, ceftazidime, while the susceptibility to the remaining β-lactams tested was differently affected by the presence of the recombinant plasmid pGER1 (Table 1). The MICs of ticarcillin, cefotaxime, cefepime, and aztreonam for E. coli TG1(pGER1) were increased, whereas the MICs of cefoxitin, imipenem, and meropenem remained unchanged. Regarding the effect of the β-lactamase inhibitors sulbactam and tazobactam, the MICs of ampicillin, cefotaxime, and ceftazidime decreased by 4- to 16-fold, in contrast with the very slight effect obtained with clavulanic acid. By isoelectric focusing, the single band of β-lactamase activity in the E. coli transformant cofocused with the β-lactamase (pI, 9.4) of the wild-type strain (results not shown).
In order to perform the sequencing reactions the 2.2-kb insert from the plasmid pGER1 was cloned in the pUC18 multicopy plasmid (29), resulting in the plasmid pGER2. Sequencing was carried out with the Taq DyeDeoxiTerminator Cycle Sequencing Kit and with primers specific to the coding sequence, and the sequence was analyzed in an automatic DNA sequencer (377 Abi-Prims; Perkin-Elmer). The entire sequence of the fragment was determined twice for accuracy.
The sequenced fragment was 2,195 bp long and contained only one open reading frame (ORF) (Fig. 1). An ATG codon initiated an 1,152-bp ORF which ended with a TAA codon (383 amino acids long). The initiation codon was preceded by a Shine-Dalgarno ribosome-binding sequence, AGGAG. GenBank database searches with this ORF revealed similarities with several class C chromosomal and plasmid-mediated β-lactamases (Table 2). The highest identity (40.5 to 42.3%) was observed with the following β-lactamases: AmpC of Aeromonas sobria, CMY-1, AmpC of Serratia marcescens, FOX-2, FOX-3, and AmpC of Pseudomonas aeruginosa; 35 to 40% identity was seen for AmpC β-lactamases from E. coli, Proteus stuartii, Yersinia enterocolitica, Morganella morganii, Enterobacter cloacae, and Citrobacter freundii. The deduced peptide sequence contained conserved motifs found in serine β-lactamases (14), as follows: the SXSK motif of the active site of AmpC, the class C typical motif YXN, and the KTG domain. The nucleotide sequence of the flanking regions of the ampC gene (about 400 bp on each side) did not show inverted repeated sequences suggestive of the presence of a transposable element. This ampC gene was probably not inserted in an integron, since the 59-base element (specific to gene cassettes inserted in integron structures) was not observed on the flanking regions.
FIG. 1.
Nucleotide sequence of the ampC gene. The deduced amino acid sequence of AmpC β-lactamase is shown below the nucleotide triplets. The boldface ATG and TAA represent the initiation and termination codons, respectively. A putative Shine-Dalgarno (S.D.) ribosomal recognition site (underlined) is indicated. The positions of the primers used to sequence the gene are indicated by underlining with arrowheads. The β-lactamase active site S-V-S-K, the conserved triad K-T-G, and the class C typical motif Y-X-N are presented in boldface. A putative terminator sequence is underlined.
TABLE 2.
Percent identity of amino acid sequences of A. baumannii AmpC and other class C β-lactamases
| β-Lactamase (reference) | % Identity with:
|
||||||
|---|---|---|---|---|---|---|---|
| A. baumannii AmpC | A. sobria AmpC | CMY-1 | S. marcescens AmpC | FOX-2 | FOX-3 | P. aeruginosa AmpC | |
| A. baumannii AmpC | 100 | 42.3 | 41.8 | 41.7 | 41.3 | 41.0 | 40.5 |
| A. sobria AmpC (26) | 100 | 74.5 | 46.9 | 77.2 | 75.3 | 55.5 | |
| CMY-1 (2) | 100 | 48.7 | 74.3 | 72.4 | 57.9 | ||
| S. marcescens AmpCa | 100 | 46.6 | 46.1 | 48.8 | |||
| FOX-2 (3) | 100 | 96.3 | 54.7 | ||||
| FOX-3 (20) | 100 | 54.4 | |||||
| P. aeruginosa AmpC (19) | 100 | ||||||
EMBL accession no. AB008454.
The synthesis of chromosomal AmpC β-lactamases in some gram-negative rods such as E. cloacae, C. freundii, and P. aeruginosa may be inducible and involves genes such as the repressor ampR and ampD. The first is located immediately upstream from the ampC gene and is divergently transcribed (19). Sequencing of the flanking regions of the gene encoding the AmpC β-lactamase produced by the Ab RYC 52763/97 strain did not show homology with the ampR regulatory gene. Induction experiments with cefoxitin (one-half of the MIC) performed with the E. coli transformant carrying the ampC gene and with the original Ab RYC 52763/97 strain did not show an increase in the synthesis of the AmpC β-lactamase, determined as specific enzymatic activity, when the inducer was added (data not shown). These experiments indicated that the A. baumannii AmpC β-lactamase in this strain is noninducible.
The substrate profile of the AmpC β-lactamase was determined with the enzyme partially purified by G-75 Sephadex (Pharmacia Fine Chemicals AB, Uppsala, Sweden). Initial hydrolysis rates were monitored spectrophotometrically (UVIKON-930) at 25°C in 0.05 M phosphate buffer (pH 7.4), as described previously (12). The Vmax values indicate that cephaloridine is hydrolyzed more quickly than ampicillin and that cefoxitin and imipenem are not hydrolyzed at detectable levels (Table 3). Inhibition studies showed that the remaining β-lactamase activities in the presence of 100 μM each inhibitor were as follows: clavulanic acid, 91%; sulbactam, 40%; and tazobactam, 45%. The 50% inhibitory concentrations (IC50s) were obtained from semilogarithmic plots of enzymatic activity against inhibitor concentration. The IC50s were 270 μM for clavulanic acid, 7 μM for sulbactam, and 8 μM for tazobactam. These results correlated with the decrease in the MICs of ampicillin, cefotaxime, and ceftazidime in combination with these inhibitors against E. coli TG1 harboring AmpC β-lactamase (pGER1).
TABLE 3.
Kinetic and inhibition parameters of the A. baumannii AmpC β-lactamasea
| Antibiotic | Km (μM) | Vmax (μmol/ min/μl) | Relative Vmaxb | IC50 (μM) |
|---|---|---|---|---|
| Cephaloridine | 960 | 5,000 | 100 | |
| Ampicillin | 38 | 380 | 7.6 | |
| Ceftazidime | 14 | 52 | 1.0 | |
| Cefotaxime | 7.0 | 25 | 0.5 | |
| Cefoxitin | <1 | <0.01 | ||
| Imipenem | <1 | <0.01 | ||
| Clavulanic acid | 270c | |||
| Sulbactam | 7c | |||
| Tazobactam | 8c |
Kinetic experiments were performed using a 7.45-mg/ml proteic extract with a specific activity of 0.064 μmol of nitrocefin hydrolyzed/min/μg of protein.
Obtained by normalizing the value for each antibiotic to that for cephaloridine (set to 100).
Nitrocefin (25 μg/ml) was used as the substrate following 10 min preincubation of enzyme and inhibitor.
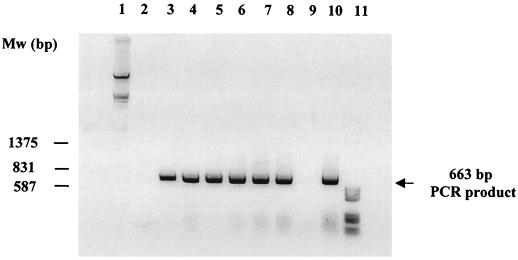
In order to detect the ampC gene in different A. baumannii isolates, a PCR assay was performed. Six genotypically different A. baumannii strains (REP-PCR tested) with different antibiotic susceptibility patterns that had been collected in our hospital during the previous five years were used. The reactions were carried out with a 50-μl volume of a reaction mixture containing 20 mM Tris-HCl (pH 8.8), 100 mM potassium chloride, 2.0 mM magnesium chloride, 200 μM deoxynucleotide triphosphate, 50 pmol of each primer, 500 ng of the chromosomal DNA, and 2.5 U of Taq polymerase (Roche). The primers of the ampC coding region, P1 (5′-TAAACACCACATATGTTCCG) and P2 (5′-ACTTACTTCAACTCGCGACG), were used. Amplification reactions were submitted to the following program: initial denaturation (4 min at 94°C) followed by 30 cycles of denaturation (1 min at 94°C), annealing (1 min at 50°C), and extension (2 min at 72°C), with a single extension of 10 min at 72°C. The amplified 663-bp product was resolved by electrophoresis in a 1.5% (wt/vol) agarose gel containing ethidium bromide (50 μg/ml). As shown in Fig. 2, the ampC gene was extensively spread between the six A. baumannii isolates but not in Acinetobacter junii. This result strongly suggests that the AmpC β-lactamase may play a role in the β-lactam resistance of A. baumannii.
FIG. 2.
Detection of the ampC gene in different A. baumannii isolates. The amplified products were resolved by 1.5% agarose gel electrophoresis and stained with ethidium bromide (50 μg/ml). Lanes 1 and 11, DNA molecular weight markers III and V, respectively (Boehringer-Mannheim); lane 2, negative control (genomic DNA of Listeria monocytogenes amplified with primers P1 and P2); lanes 3 to 8, A. baumannii strains (Ab 1 to 6); lane 9, A. junii strain; lane 10, positive control (strain Ab RYC 52763/97).
The presence of chromosomal β-lactamases in the genus Acinetobacter was first suggested by Matthew and Harris (21). By examining β-lactamases from 30 strains of Acinetobacter, Joly-Guillou et al. (17) revealed a cephalosporinase with a pI of >8.0 in all these strains. Afterwards, Blechschmidt et al. (11) reported the purification and biochemical characterization of an extracellular β-lactamase with a pI of 9.3 produced by Acinetobacter calcoaceticus. Other attempts have been carried out in order to characterize the A. baumannii cephalosporinase biochemically (25), but nothing has been reported about the gene encoding the enzyme. The present study describes the genetic and biochemical characteristics of the A. baumannii AmpC β-lactamase. The following important features are noteworthy. (i) The amino acid sequence is closely related to the class C cephalosporinases, and the highest similarity was obtained with the A. sobria AmpC β-lactamase. (ii) The enzyme shows a typical cephalosporinase substrate profile (13). (iii) Enzymatic inhibition assays and antibiotic susceptibility studies showed a moderate inhibitory effect of sulbactam and tazobactam against A. baumannii AmpC β-lactamase. As for clavulanic acid, only a very slight inhibitory effect was observed. (iv) No AmpC β-lactamase induction was observed when cefoxitin was added, and no ampR sequences were found on the flanking regions of the ampC gene. Nevertheless, whether a regulatory AmpR-like protein gene exists in A. baumannii, although not adjacent to the ampC gene, remains to be elucidated.
Certainly, resistance to β-lactam antibiotics in A. baumannii is a multifactorial problem involving resistance mechanisms in addition to β-lactamases (9); thus, in our study, the β-lactam MICs conferred by the AmpC β-lactamase, even by means of a multicopy plasmid carried by E. coli TG1, did not reach the high MICs observed for the Ab RYC 52763/97 clinical strain (MICs conferred by the OXA-derived β-lactamase [Bou et al., submitted] are not relevant when compared with the A. baumannii clinical strain). Note also that the MICs of the carbapenems were not affected by the presence of the AmpC β-lactamase even in a multicopy plasmid. Nevertheless, the presence of the ampC gene in all six different A. baumannii strains tested suggests that the AmpC β-lactamase, in combination with other β-lactam resistance mechanisms, may play an important role in β-lactam resistance in A. baumannii.
In summary, we report here for the first time the cloning, sequencing, and analysis of the ampC gene and the biochemical characterization of the AmpC β-lactamase from an A. baumannii clinical strain.
Nucleotide sequence accession number.
The nucleotide sequence of the ampC gene has been given the EMBL database accession no. AJ009979.
Acknowledgments
We thank L. de Rafael for his critical reading.
REFERENCES
- 1.Al-Khoja M S, Darrell J H. The skin as the source of Acinetobacter and Moraxella species occurring in blood cultures. J Clin Pathol. 1979;32:497–499. doi: 10.1136/jcp.32.5.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Stemplinger I, Jungwirth R, Wilhem R, Chong Y. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other β-lactamase genes. Antimicrob Agents Chemother. 1996;40:1926–1930. doi: 10.1128/aac.40.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind A, Wagner S, Jungwirth R, Schneider J, Meyer D. A novel class C β-lactamase (FOX-2) in Escherichia coli conferring resistance to cephamycins. Antimicrob Agents Chemother. 1997;41:2041–2046. doi: 10.1128/aac.41.9.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann P. Isolation of Acinetobacter from soil and water. J Bacteriol. 1968;96:39–42. doi: 10.1128/jb.96.1.39-42.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergogne-Bérézin E. The increasing significance of outbreaks of Acinetobacter spp.: the need for control and new agents. J Hosp Infect. 1995;30:441–452. doi: 10.1016/0195-6701(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 6.Bergogne-Bérézin E, Joly-Guillou M L, Vieu J F. Epidemiology of nosocomial infections due to Acinetobacter calcoaceticus. J Hosp Infect. 1987;10:105–113. doi: 10.1016/0195-6701(87)90135-6. [DOI] [PubMed] [Google Scholar]
- 7.Bergogne-Bérézin E, Joly-Guillou M L. An underestimated nosocomial pathogen, Acinetobacter calcoaceticus. J Antimicrob Chemother. 1985;16:535–538. doi: 10.1093/jac/16.5.535. [DOI] [PubMed] [Google Scholar]
- 8.Bergogne-Bérézin E, Joly-Guillou M L. Comparative activity of imipenem, ceftazidime and cefotaxime against Acinetobacter calcoaceticus. J Antimicrob Chemother. 1986;18(Suppl. E):35–39. doi: 10.1093/jac/18.supplement_e.35. [DOI] [PubMed] [Google Scholar]
- 9.Bergogne-Bérézin E, Towner K J. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergogne-Bérézin E, Joly-Guillou M L, Towner K J. History and importance of Acinetobacter spp.: role in infections, treatment and cost implications. In: Bergogne-Bérézin E, et al., editors. Acinetobacter: microbiology, epidemiology, infections, management. Boca Raton, Fla: CRC Press; 1996. pp. 1–12. [Google Scholar]
- 11.Blechschmidt B, Borneleit P, Kleber H P. Purification and characterization of an extracellular β-lactamase produced by Acinetobacter calcoaceticus. J Gen Microbiol. 1992;138:1197–1202. doi: 10.1099/00221287-138-6-1197. [DOI] [PubMed] [Google Scholar]
- 12.Bou G, Martínez-Beltrán J, Cerveró G, Pérez-Díaz J C. Biochemical and genetic characteristics of TEM-29B, a novel extended spectrum β-lactamase. FEMS Microbiol Lett. 1999;174:185–190. doi: 10.1111/j.1574-6968.1999.tb13567.x. [DOI] [PubMed] [Google Scholar]
- 13.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghuysen J M. Serine β-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 15.Glew R H, Moellering R C, Kunz L J. Infections with Acinetobacter calcoaceticus (Herellea vaginicola): clinical and laboratory studies. Medicine. 1977;56:79–97. doi: 10.1097/00005792-197703000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Grimont F, Grimont P A D. Ribosomal nucleic acid gene restriction patterns as potential taxonomic tools. Ann Inst Pasteur Microbiol. 1986;137B:165–175. doi: 10.1016/s0769-2609(86)80105-3. [DOI] [PubMed] [Google Scholar]
- 17.Joly-Guillou M L, Bergogne-Bérézin E, Moreau N. Enzymatic resistance to β-lactams and aminoglycosides in Acinetobacter calcoaceticus. J Antimicrob Chemother. 1987;20:773–775. doi: 10.1093/jac/20.6.773. [DOI] [PubMed] [Google Scholar]
- 18.Joly-Guillou M L, Bergogne-Bérézin E, Vieu J F. Epidemiologie et résistance aux antibiotiques des Acinetobacter en milieu hospitalier. Presse Med. 1990;19:357–361. [PubMed] [Google Scholar]
- 19.Lodge J M, Minchin S D, Piddock L, Busby J W. Cloning, sequencing and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC β-lactamase. Biochem J. 1990;272:627–631. doi: 10.1042/bj2720627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchese A, Arlet G, Schito G C, Lagrange P H, Philippon A. Characterization of FOX-3, an AmpC-type plasmid-mediated β-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob Agents Chemother. 1998;42:464–467. doi: 10.1128/aac.42.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matthew M, Harris A M. Identification of β-lactamase by analytical isoelectric focusing: correlation with bacterial taxonomy. J Gen Microbiol. 1976;94:55–67. doi: 10.1099/00221287-94-1-55. [DOI] [PubMed] [Google Scholar]
- 22.Matthew M, Harris A M, Marshall M J, Ross G W. The use of analytical isoelectric focusing for detection and identification of β-lactamases. J Gen Microbiol. 1975;88:169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 24.Paton R, Miles R S, Hood J, Amyes S G B. ARI 1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents. 1993;2:81–88. doi: 10.1016/0924-8579(93)90045-7. [DOI] [PubMed] [Google Scholar]
- 25.Perilli M, Felici A, Oratore A, Cornaglia G, Bofiglio G, Rossolini G M, Amicosante G. Characterization of the chromosomal cephalosporinase produced by Acinetobacter lwoffii and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother. 1996;40:715–719. doi: 10.1128/aac.40.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasmussen B A, Keeney D, Yang Y, Bush K. Cloning and expression of a cloxacillin-hydrolyzing enzyme and a cephalosporinase from Aeromonas sobria AER 14M in Escherichia coli: requirement for an E. coli chromosomal mutation for efficient expression of the class D enzyme. Antimicrob Agents Chemother. 1994;38:2078–2085. doi: 10.1128/aac.38.9.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Retailliau H F, Hightower A W, Dixon R E, Allen J R. Acinetobacter calcoaceticus: a nosocomial pathogen with an unusual seasonal pattern. J Infect Dis. 1979;139:371–375. doi: 10.1093/infdis/139.3.371. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Spratt B G, Hedge P J, Heesen T S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 30.Tilley P A G, Roberts F J. Bacteremia with Acinetobacter species: risk factors and prognosis in different clinical settings. Clin Infect Dis. 1994;18:896–900. doi: 10.1093/clinids/18.6.896. [DOI] [PubMed] [Google Scholar]
- 31.Vahaboglu H, Öztürk R, Aygün G, Coşkunkan F, Yaman A, Kaygusuz A, Leblebicioglu H, Balik I, Aydin K, Otkun M. Widespread detection of PER-1-type extended-spectrum β-lactamases among nosocomial Acinetobacter and Pseudomonas aeruginosa isolates in Turkey: a nationwide multicenter study. Antimicrob Agents Chemother. 1997;41:2265–2269. doi: 10.1128/aac.41.10.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vila J, Navia M, Ruiz J, Casals C. Cloning and nucleotide sequence analysis of a gene encoding an OXA-derived β-lactamase in Acinetobacter baumannii. Antimicrob Agents Chemother. 1997;41:2757–2759. doi: 10.1128/aac.41.12.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]