Background:
In most eukaryotic cells, the mitochondrial DNA (mtDNA) is transmitted uniparentally and present in multiple copies derived from the clonal expansion of maternally inherited mtDNA. All copies are therefore near-identical, or homoplasmic. The presence of >1 mtDNA variant in the same cytoplasm can arise naturally or result from new medical technologies aimed at preventing mitochondrial genetic diseases and improving fertility. The latter is called divergent nonpathologic mtDNA heteroplasmy (DNPH). We hypothesized that DNPH is maladaptive and usually prevented by the cell.
Methods:
We engineered and characterized DNPH mice throughout their lifespan using transcriptomic, metabolomic, biochemical, physiologic, and phenotyping techniques. We focused on in vivo imaging techniques for noninvasive assessment of cardiac and pulmonary energy metabolism.
Results:
We show that DNPH impairs mitochondrial function, with profound consequences in critical tissues that cannot resolve heteroplasmy, particularly cardiac and skeletal muscle. Progressive metabolic stress in these tissues leads to severe pathology in adulthood, including pulmonary hypertension and heart failure, skeletal muscle wasting, frailty, and premature death. Symptom severity is strongly modulated by the nuclear context.
Conclusions:
Medical interventions that may generate DNPH should address potential incompatibilities between donor and recipient mtDNA.
Keywords: DNA, mitochondrial; haplotypes; heart diseases; heteroplasmy; hypertension, pulmonary; mice; oxidative phosphorylation
Clinical Perspective.
What Is New?
Divergent nonpathologic mitochondrial DNA (mtDNA) heteroplasmy causes cell metabolic stress that can lead to cell death.
Heart, lung, skeletal muscle, and eye tissues cannot resolve divergent nonpathologic mtDNA heteroplasmy, leading to functional impairment of mitochondria.
Divergent nonpathologic mtDNA heteroplasmy causes metabolic heart disease with pulmonary hypertension and frailty in mature adult mice.
What Are the Clinical Implications?
New medical technologies may cause the coexistence of mtDNA of different origins (divergent nonpathologic mtDNA heteroplasmy) in the same cell.
This study calls for careful selection of compatible donor mtDNA for mitochondrial therapy.
In mammals, mitochondrial DNA (mtDNA) encodes 13 proteins that must physically match with 70 subunits encoded in the nuclear genome to build a functional oxidative phosphorylation (OXPHOS) system. Under normal circumstances, mtDNA transmission in mammals is exclusively maternal, given that the paternal mitochondria are actively degraded after oocyte fertilization. Therefore, all mtDNA copies of an organism derive from the clonal expansion of the low number of copies present in the maternal progenitor germ cell from which the oocyte has originated. This fact led to the assumption that all cells in each organism are homoplasmic, meaning they express identical copies of mtDNA. However, high-resolution sequencing has revealed a background of low-frequency mutations that arise by replication errors, leading to the presence of >1 mtDNA species, or heteroplasmy.1
Heteroplasmy has received renewed attention with the advent of new medical technologies that may cause the coexistence of mtDNA of different origins in the same cell. Natural heteroplasmy2 is distinctly different from medically induced heteroplasmy, leading us to define the latter as divergent nonpathologic heteroplasmy (DNPH). Given the heterologous nature of mitochondrial donors, mitochondrial therapies can result in the coexistence of divergent nonpathologic mtDNA variants within recipient cells. DNPH may be a consequence of mitochondrial replacement therapy to prevent transmission of mtDNA-linked diseases3,4; fertility treatments involving the injection of oocyte cytoplasm from young donors into oocytes of subfertile women5; or the use of purified mitochondria or donor cells to deliver mitochondria to treat a number of diseases including myocardial and cerebral ischemia/reperfusion injury, pulmonary arterial hypertension, spinal cord injury, and models of Parkinson disease.6
Animals with identical nuclear genomes but different mtDNA haplotypes (conplastic) generate functionally different OXPHOS systems that shape organismal metabolism,7 supporting the conclusion that different wild-type mtDNA haplotypes produce substantially different phenotypes. However, debate persists about whether heteroplasmy poses an intrinsic risk to the health of the offspring owing to negative consequences arising from the conflict between alternative mtDNA variants. A spectrum of physiologic consequences has been reported, from none8–10 to mild physiologic effects such as transient hypertension11 and alterations in behavior and cognition.12
DNPH between 2 nonpathologic mtDNA haplotypes permits healthy development and the generation of stable and fertile heteroplasmic mouse colonies.10,12–15 mtDNA segregation or heteroplasmic changes in most tissues is an intriguing observation with no apparent functional significance. This led to the view that heteroplasmy among nonpathologic mtDNA haplotypes is benign and of negligible concern when induced medically. However, most studies focused on young animals, with no exhaustive and systematic longitudinal investigation into the physiology and lifespan of DNPH animals. We show that DNPH negatively affects mitochondrial function, with impairment of tissues that cannot resolve heteroplasmy. Progressive metabolic stress in these tissues leads to severe pathology in adulthood, including pulmonary hypertension and heart failure, skeletal muscle wasting, frailty, and premature death. We demonstrate that symptom severity and pathology phenotype are strongly modulated by the expression of mitochondrial nuclear-encoded genes.
Methods
Detailed methods are provided in the Supplemental Material. All animal studies conformed to European Union Directive 86/609/European Economic Community and Recommendation 2007/526/European Economic Community regarding the protection of animals used for experimental and other scientific purposes, enforced in Spanish law under Real Decreto 1201/2005. Approval of the different experimental protocols required estimation of minimum sample size and definition of the randomization and blinding criteria. There are no human participants in this study. The data, analytic methods, and study materials will be made available by the corresponding author to other researchers for purposes of reproducing the results or replicating the procedure. Datasets of the raw files, protein databases, search parameters, and results reported in this article are available in the PeptideAtlas repository (ftp://ftp.peptideatlas.org/; username: PASS01085; password: TC4334fh). Transcriptomic raw reads and TMM (trimmed mean of M values)–normalized counts have been deposited in Gene Expression Omnibus (accession number GSE93920).
Statistical Analysis
Unless specified otherwise, statistical analysis and graph production were performed in GraphPad Prism 9. Datasets were compared by analysis of variance or nonparametric analysis as appropriate. To perform multiple pairwise comparisons, we used analysis of variance followed by the Tukey test. For nonparametric Kruskal-Wallis tests, we corrected for multiple comparisons using the Dunn test. Differences were considered statistically significant at P<0.05 (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001). Data are presented as mean±SD or mean±SEM.
Results
DNPH and Homoplasmic Animals Are Phenotypically Different
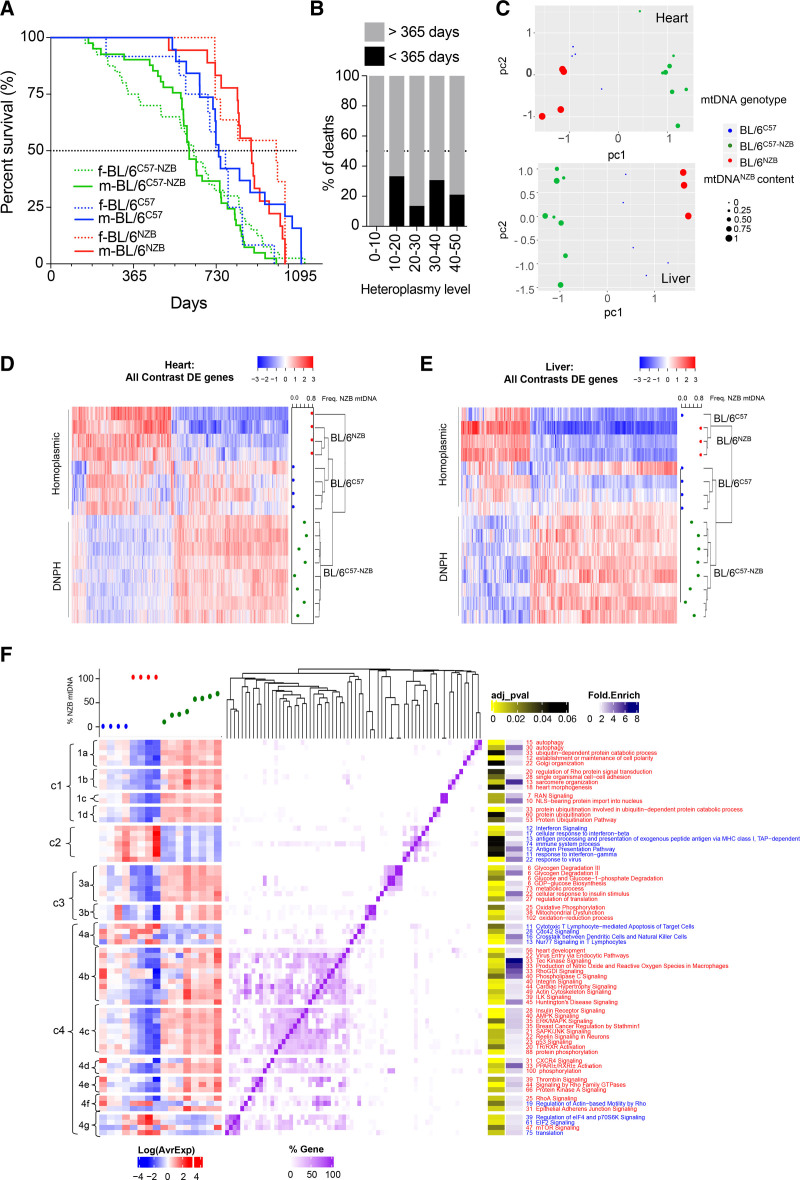
We conducted a longitudinal survival study to assess the physiologic consequences of heteroplasmy with 2 different wild-type mtDNAs, each of which would generate healthy mice in homoplasmy.7 mtDNA haplotypes C57 and NZB differ by 10 noncoding region single nucleotide polymorphisms (SNPs), 8 ribosomal RNA SNPs, 4 transfer RNA mutations, and 12 SNPs that result in missense mutations in the respiratory complex I (nicotinamide adenine dinucleotide–ubiquinone oxidoreductase [Nd] chain 1 = 3; Nd chain 2 = 2; Nd chain 4l = 1; Nd chain 4 = 1; Nd chain 5 = 2; and Nd chain 6 = 3 SNPs), complex III (cytochrome b = 1 SNP), and complex IV (Co1 = 1 SNP).7 DNPH mice had elevated mortality rates at a relatively young age, compromising health and leading to a markedly shorter lifespan compared with homoplasmic animals (Figure 1A). Heteroplasmy levels are calculated on the basis of the proportion of each mtDNA haplotype present (range, 0% to 50%; 0% = only 1 haplotype present; 50% = both haplotypes present at equal levels). Data showed elevated rates of premature death (<1 year) when heteroplasmy levels were >10% (Figure 1B).
Figure 1.
Mice with divergent nonpathologic mitochondrial DNA heteroplasmy show shortened life span and have a differential transcriptome profile. A, Longevity curves of heteroplasmic and homoplasmic animals separated by sex (female [f], dotted lines; male [m], continuous lines). Data from homoplasmic mice taken from Latorre-Pellicer et al.7 BL/6C57: 19 male, 12 female; BL/6NZB: 18 male, 11 female; BL/6C57-NZB: 41 male, 40 female. ****P<0.0001 for log-rank (Mantel-Cox; χ2=36.85; df=4), Gehan-Breslow-Wilcoxon test. B, Heteroplasmy level at weaning (being 50% of the maximum heteroplasmy level) and frequency of premature death (<365 days) of the animals monitored in A (0%–10%, n=4; 10%–20%, n=9; 20%–30%, n=22; 30%–40%, n=26; 40%–50%, n=19). C, Principal component analysis score plot showing the clustering gene expression of cardiac and liver samples from homoplasmic (n=4) and heteroplasmic (n=8) mice. Each dot corresponds to a different animal and dot size represents the % of NZB mitochondrial DNA (mtDNA) for heteroplasmic mice (green). BL/6C57 transcriptomes are marked in blue and BL/6NZB in red. D and E, Clustered heat maps of the profiles of differentially expressed (DE) heart and liver genes in all contrasts (adjusted P<0.05 for heart samples and adjusted P<0.2 for liver samples). Expression Z scores are shown for each sample on the blue–red scale (see also Tables S1 and S2). The frequency of NZB mtDNA is indicated at the right of each panel. F, Functional analysis of HDE (differentially expressed in the heart) genes between conditions. In the chart, significantly enriched (adjusted P<0.2) Gene Ontology biological processes obtained from the DAVID (Database for Annotation, Visualization and Integrated Discovery) platform and Canonical Pathways from the Ingenuity platform are clustered according to the number of shared genes (purple heat map). Numbers accompanying functional category names indicate the number of HDE genes in each category: blue for downregulated functions/pathways in heteroplasmic versus homoplasmic mice and red for upregulation. The left panel represents the transcriptional profile of the HDE genes present in each category across the different conditions, indicated as % NZB mtDNA content on top of the plot. DNPH indicates divergent nonpathologic heteroplasmy.
To assess DNPH molecularly, we performed RNA sequencing from heart and liver tissues from 12-week-old homoplasmic and DNPH mice. In DNPH mice, the presence of ≥10% heteroplasmy levels induced defined transcriptomic signatures (Figure 1C –1E). Hierarchical clustering of cardiac transcriptional profiles on the basis of differentially expressed genes confirmed a significant difference between the strains (adjusted P<0.05; Figure 1D and Table S1). Mice with maximal heteroplasmy levels clustered separately from those with lower heteroplasmy levels. In addition, DNPH and homoplasmic liver samples did not show any significant difference at gene level; however, clustering of the samples on the basis of the profiles from genes with an adjusted P<0.2 indicates that even though there are no significant differences at individual gene level, globally the DNPH is producing a particular molecular profile that is different from that of homoplasmic mice (Figure 1C and 1E and Table S1).
We then performed functional enrichment analysis on differentially expressed genes on the basis of the categories provided by Gene Ontology and Ingenuity Pathway Analysis, comparing homoplasmic and heteroplasmic hearts (Figure 1F and Table S2). Four main clusters of differentially regulated pathways were identified. Cluster 1 is subdivided into 4 subclusters: autophagy (cluster 1a), intracellular membrane organization (cluster 1b), nuclear transport (cluster 1c), and ubiquitination (cluster 1d). Each of the cluster 1 pathways was upregulated and may reflect hyperactivation of intracellular repair mechanisms. Cluster 2 showed downregulation of immune response pathways. Cluster 3 reflected the increase in metabolic processes leading to the use of glucose (cluster 3a) and the overactivation of mitochondrial genes, especially OXPHOS and mitochondrial dysfunction genes (cluster 3b). Pathways associated with cluster 3 demonstrated increased glucose uptake and impaired OXPHOS function in heteroplasmic cardiac tissue. Cluster 4 was more heterogeneous, illustrating the activation of a variety of general signaling pathways. Several of these pathways are related to cell stress and nutrient sensing (Tec kinase, SAPK/JNK [stress-activated protein kinases/c-Jun N-terminal kinases], p53, and AMPK [5′-adenosine monophosphate–activated protein kinase] signaling). The mTOR (mammalian target of rapamycin), eIF4 (eukaryotic initiation factor 4)/P70S6K, and eIF2 (eukaryotic initiation factor 2) pathways were altered (Figure 1F), consistent with the fact that mTORC1 (mammalian target of rapamycin complex 1) stress responses are found to be involved in mtDNA diseases.16 Overall, there was no linear correlation between the transcriptional response to DNPH and the degree of heteroplasmy; rather, these programs appear to be triggered when heteroplasmy is present at levels >10%.
We noted that DNPH had a sex-specific effect on body appearance, with no weight difference observed between ~100-day-old male and female heteroplasmic animals (Figure S1A–S1C). Heteroplasmic mice had elongated tibias and femurs (Figure S1C). DNPH mice also demonstrated accelerated aging compared with homoplasmic animals, with early onset of kyphosis being evident from 25 weeks in female and 60 weeks in male mice (Figure S1D–S1E). Kyphosis angle did not correlate with the degree of heteroplasmy (Figure S1F).
DNPH Causes Diffuse Loss of Cardiomyocytes at Early Age
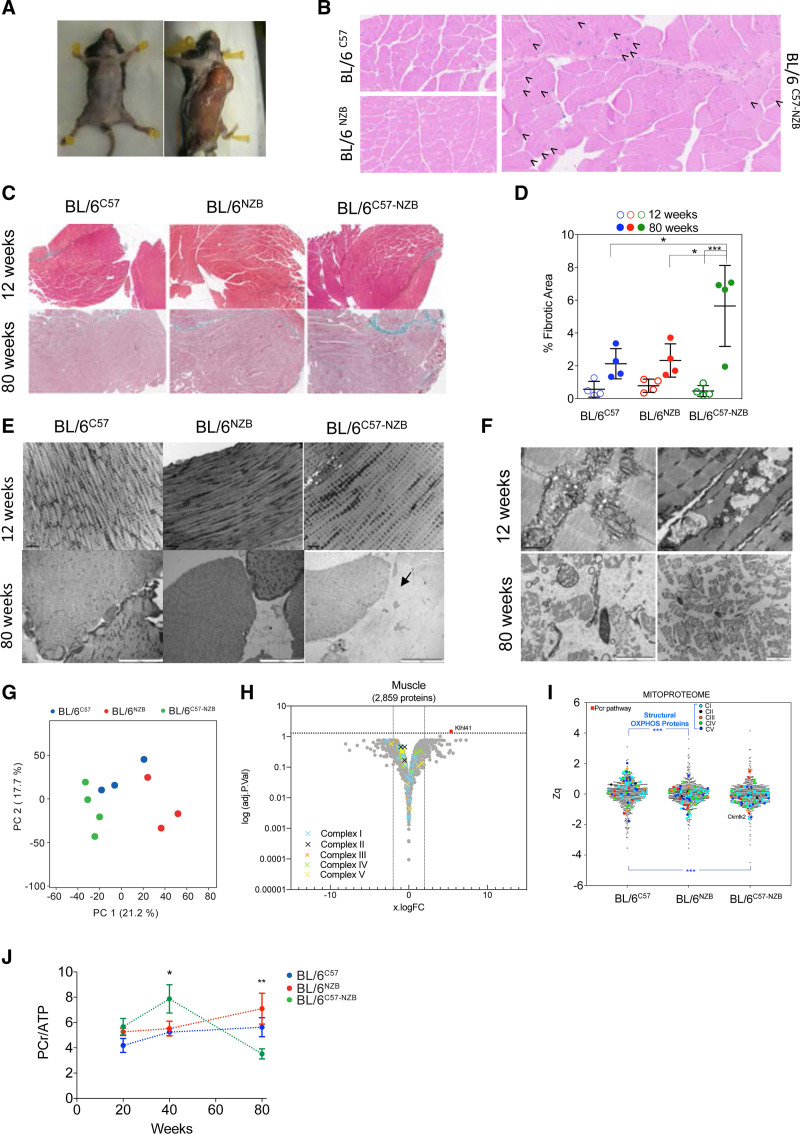
Plasma analysis revealed abnormal creatine kinase and myocardial band of creatine kinase in 12-week-old heteroplasmic mice (Figure 2A). These measures reverted to normal in older animals, suggesting that cardiomyocyte death is a juvenile event, presumably controlled by an adaptive response (Figure 2A). Evans blue staining showed cardiomyocyte death only in heteroplasmic animals (Figure S2A–S2C). Cardiomyocyte death was dispersed and symmetrically distributed between the right and left ventricles, consistent with a lack of fibrosis. In young mice, there were no differences in liver histopathology between strains (Figure S2D). In contrast, liver tissue from 80-week-old BL/6C57 mice demonstrated steatosis, whereas that of BL/6C57-NZB and BL/6NZB mice showed signs of inflammation (Figure S2D). Twelve-week-old heteroplasmic mice had impaired mitochondrial ATP (adenosine triphosphate) synthesis in hepatic and cardiac tissue (Figure S2E). This function was restored only in the liver of 20-week-old mice, presumably because hepatic cells can resolve heteroplasmy14 (Figure S2E).
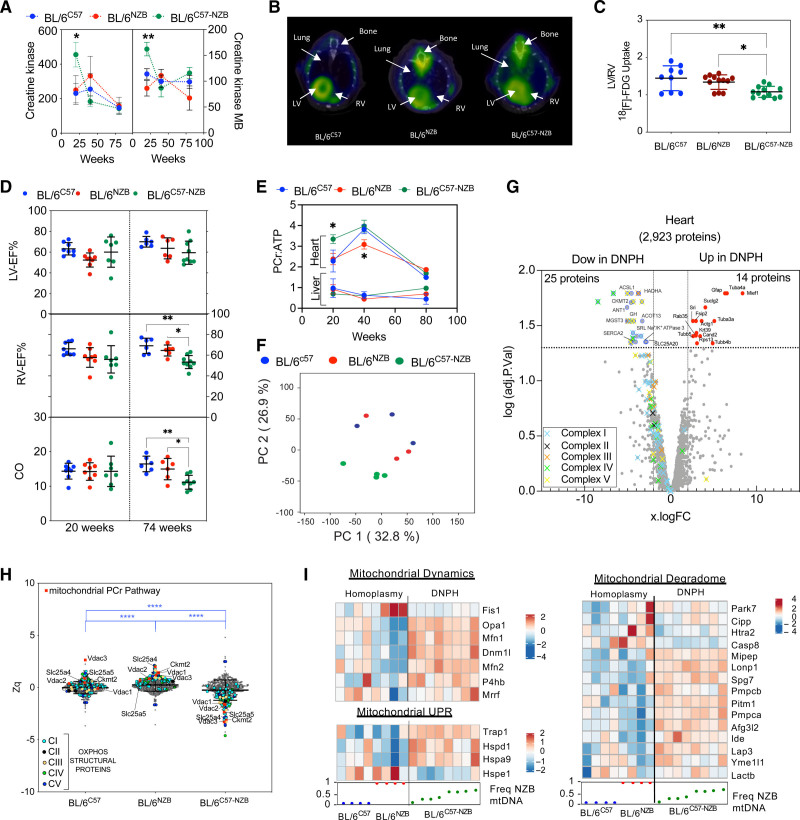
Figure 2.
Divergent nonpathologic mitochondrial DNA heteroplasmy causes cardiac metabolic stress, leading to heart failure. A, Creatine kinase (CK) and CK-MB in plasma (10 homoplasmic animals per indicated age; 12-week-old heteroplasmic mice, n=20; 40- and 80-week-old heteroplasmic mice, n=10). Data from different individuals at different ages given as mean±SEM, 2-way analysis of variance test with Tukey multiple comparation test; *P<0.05, ** P<0.01. B and C, [18F]-fluorodeoxyglucose positron emission tomography–computed tomography cardiac analysis. (B) Representative axial fused positron emission tomography–computed tomography images and (C) quantitative analysis of the increase in relative right ventricle (RV) uptake of the glucose analogue [18F]-fluorodeoxyglucose in 20-week-old homoplasmic and heteroplasmic mice. *P<0.05, **P<0.01 (1-way analysis of variance; 4–5 mice per group). D, Magnetic resonance imaging analysis of heart function in 20- or 74-week-old male mice of the indicated strains. Each dot represents an individual mouse. *P<0.05, **P<0.01 (1-way analysis of variance). E, Longitudinal 31P magnetic resonance spectroscopy analysis of the phosphocreatine (PCr):ATP (adenosine triphosphate) ratio in heart and liver of the indicated mouse strain. *P<0.05 (2-way analysis of variance test with Tukey multiple comparation test; n=3–8). F through H, Heart proteomic analyses in 12-week-old mice. F, Score plot of the principal component (PC) analysis of the heart proteome. Each dot represents an individual mouse. G, Volcano plot illustrating proteins with significantly differential abundance between homoplasmic and heteroplasmic hearts (3 homoplasmic and 4 heteroplasmic heart samples). The log (adjusted P value) is plotted against the log fold change (logFC; fold difference between homoplasmic and heteroplasmic mice); 25 proteins are significantly underrepresented and 14 are significantly overrepresented. On the x axis, vertical lines denote ±2-fold change; on the y axis, the horizontal line denotes the significance threshold (P<0.05 before logarithmic transformation). H, Quantitative MitoProteome analysis highlighting structural oxidative phosphorylation (OXPHOS) components and mitochondrial elements of the PCr pathway. Zq indicates standardized log2 ratios of the indicated proteins: ACSL1 (acyl–coenzyme A synthetase long chain family member 1), SLC25A20 (carnitine/acylcarnitine translocase), HADHA (hydroxyacyl–coenzyme A dehydrogenase trifunctional multienzyme subunit alpha), and ACOT13 (hydroxyacyl–coenzyme A dehydrogenase trifunctional multienzyme subunit alpha). Of the 5 nonmitochondrial reduced proteins, 3 were highly involved in cellular ATP-consuming processes (SERCA2 [sarcoplasmic/endoplasmic reticulum calcium ATPase 2] and SRL [sarcalumenin] in Ca2+ transport and ATP1A3 [sodium–potassium ATPase catalytic subunit α3]) and 2 in signaling (GH [growth hormone] and MGST3 [microsomal glutathione S-transferase 3]). ****P<0.0001 (1-way analysis of variance; 3 homoplasmic and 4 heteroplasmic mice). I, logFC heat maps of mitochondrial dynamics, unfolded protein (UPR), and mitochondrial degradome in divergent nonpathologic heteroplasmy cardiac tissue. RNA sequencing data analysis of 12-week-old homoplasmic and heteroplasmic hearts. CO indicates cardiac output; DNPH, divergent nonpathologic heteroplasmy; LV, left ventricle; LV-EF, left ventricular ejection fraction; mtDNA, mitochondrial DNA; and RV-EF, right ventricular ejection fraction.
DNPH Causes Progressive Heart Mitochondrial Malfunction
We next performed in vivo analysis of glucose metabolism in 20-week-old mice by [18F]-fluorodeoxyglucose positron emission tomography–computed tomography. DNPH altered cardiac glucose uptake, predominantly in the right ventricle (Figure 2B and 2C). Magnetic resonance imaging of 20-week-old murine hearts revealed no differences between strains in the right or left ventricular ejection fraction or cardiac output (Figure 2D). However, the right ventricular ejection fraction and cardiac output were significantly impaired in 74-week-old DNPH mice (Figure 2D). Despite normal cardiac function in 20-week-old heteroplasmic mice, 31P magnetic resonance spectroscopy revealed altered energy metabolism, with an abnormal elevation in the phosphocreatine (PCr):ATP ratio (Figure 2E and Figure S2F). This alteration matches with a reduced mitochondrial ATP synthesis capacity in DNPH mice (Figure S2E). The metabolic shift toward glucose utilization may represent an adaptation in cardiomyocytes that prevents energetic deficits owing to insufficient mitochondrial performance. Conversely, the hepatic PCr:ATP ratio was unaffected at any age (Figure 2E). These results indicate that a sustained reduction in the contribution to the ATP pool is an early hallmark of DNPH mitochondria. This is resolved in the liver but not in the heart. Therefore, the heart compensates by shifting toward glycolysis.
Proteomic analyses of whole hearts and livers from mice with >30% heteroplasmy level confirmed significant differences between the proteomes of DNPH and homoplasmic hearts (Figure 2F and Tables S1 and S2) and livers (Figure S2G and Tables S1 and S2). Structural OXPHOS system components were significantly reduced in DNPH hearts (Figure 2G). In addition, we found a reduction in proteins involved in mitochondrial generation of creatine phosphate (CKMT2 [creatine kinase mitochondrial 2] and ANT1 [adenine nucleotide translocator 1]) and in β-oxidation and fatty acid metabolism (ACSL1 [acyl–coenzyme A synthetase long chain family member 1], SLC25A20 [carnitine/acylcarnitine translocase], HADHA [hydroxyacyl–coenzyme A dehydrogenase trifunctional multienzyme subunit alpha], and ACOT13 [acyl–coenzyme A thioesterase 13]). SERCA2 (sarcoplasmic/endoplasmic reticulum calcium ATPase 2) and SRL (sarcalumenin; both involved in Ca2+ transport) and ATP1A3 (sodium–potassium ATPase catalytic subunit α3; involved in ATP-consuming processes) were also reduced in DNPH animals (Figure 2G). The upregulated proteins included cytoskeletal reorganization, membrane vesicle trafficking, and phagosome formation, consistent with altered autophagy flux suggested by transcriptomics (Figure 2G and Figure 1F). Ingenuity Pathway Analysis in DNPH liver proteomics revealed downregulation of mTOR/EIF2 signaling, whereas PPAR (peroxisome proliferator-activated receptor)–alpha/RXR (retinoid X receptor)–alpha signaling was activated (for proteomic data, see Methods and Tables S1 and S2). DNPH liver showed no significant alteration of the OXPHOS proteome (Figure S2H).
Analysis of the MitoProteome confirmed simultaneous reduction in nuclear- and mtDNA-encoded OXPHOS protein subunits in DNPH hearts (Figure 2H) but not livers (Figure S2I), suggesting a reduced mitochondrial capacity to provide energy in DNPH hearts (Figure 2E–2H). Supporting this notion, the heteroplasmic cardiac MitoProteome showed reductions in several key components of mitochondrial phosphocreatine production. These reduced proteins included both ADP (adenosine diphosphate)/ATP translocase isoforms in the inner mitochondrial membrane (ANT1 [adenine nucleotide translocator 1] is specific to striated muscle, whereas ANT2 [adenine nucleotide translocator 2] is ubiquitously expressed in mice); mitochondrial creatine kinase (CKMT2 [creatine kinase mitochondrial 2]), which is essential for maintaining elevated myocardial phosphate metabolism17; and the mitochondrial outer membrane pore components (VDAC1 [voltage-dependent anion-selective channel 1], VDAC2 [channel 2], and VDAC3 [channel 3]), which are responsible for the release of phosphocreatine from the mitochondrial intermembrane space. OXPHOS structural proteins were not altered in heteroplasmic liver (Figure S2I). RNA sequencing analyses also revealed upregulation in the expression of genes involved in mitochondrial dynamics, quality control, and unfolded protein response in cardiac tissue of 12-week-old DNPH mice (Figure 2I). The observed differences suggest mitochondrial impairment and damage in the hearts, but not livers, of DNPH animals.
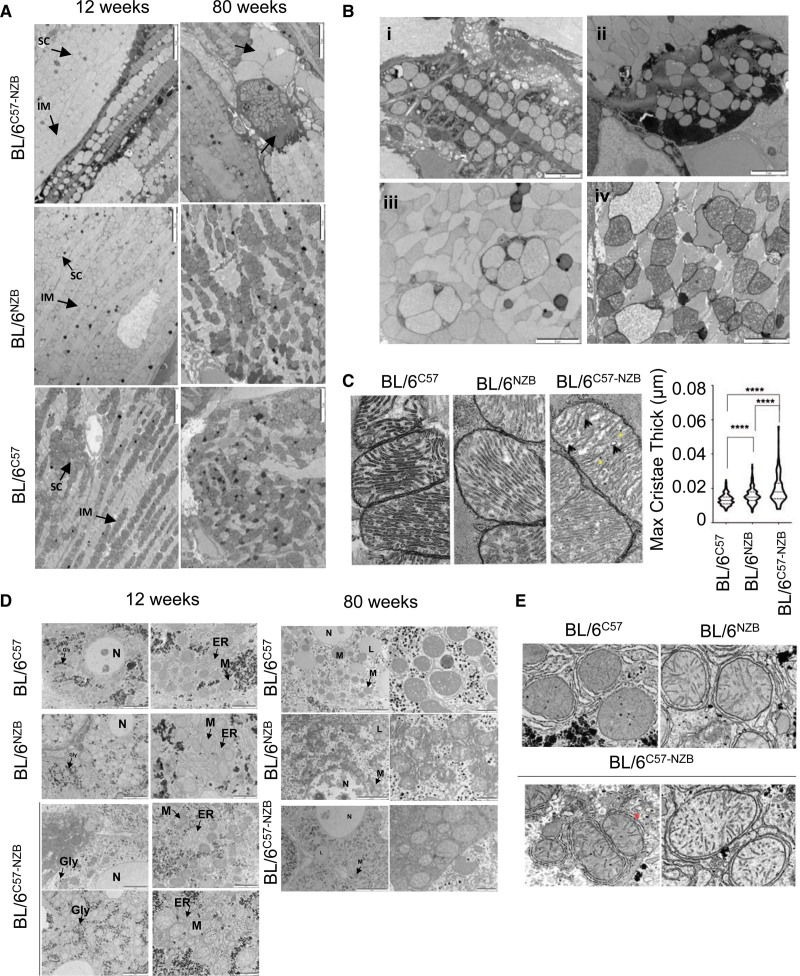
The metabolic alterations in DNPH hearts were accompanied by ultrastructural changes (Figure 3A and 3B). Transmission electron microscopy (TEM) of heteroplasmic hearts from 12-week-old mice revealed necrotic fibers, sarcomere disruption, intercalated disc and intercardiomyocyte connection abnormalities, and multiple signs of disrupted mitochondria (Figure 3B). Heteroplasmic hearts from 80-week-old mice showed a variety of ultrastructural alterations, fibrotic areas, and mitochondrial damage (Figure 3A and 3B). We also observed alterations in mitochondrial inner membrane ultrastructure, inducing increased cristae widening, in DNPH samples (Figure 3C). These changes indicate a severe energy insufficiency that likely triggers the cardiac metabolic shift.
Figure 3.
Divergent nonpathologic mitochondrial DNA heteroplasmy alters cardiac ultrastructure. A, Transmission electron microscopy analysis of the heart in 12- and 80-week-old homoplasmic and heteroplasmic mice. Arrows indicate abnormal cardiac fibers and scars. Images are representative of 5 mice per genotype. Scale bars: 5 µm. B, Representative transmission electron microscopy images showing specific features observed in the hearts of 12-week-old heteroplasmic mice: (i) necrotic fiber; (ii) damaged cardiomyocytes; (iii) mitochondria in phagosomes; (iv) vacuolated mitochondria and mitochondrial swelling. Images are representative of 5 heteroplasmic mice. Scale bars: 2 µm. C, Transmission electron microscopy images (left) and quantitative analysis (right) of cardiac mitochondrial cristae thickness. ****P<0.0001 (nonparametric Kruskal-Wallis test; BL/6C57, n=409; BL/6NZB, n=299; BL/6C57-NZB cristae, n=219). D and E, Electron microscopy analyses in liver tissue. Representative transmission electron microscopy images of hepatocytes (D) and liver mitochondrial ultrastructure (E) from male mice of the indicated age and strain (n=4 to 5 mice per genotype). Red asterisk represents an interaction at the interface of endoplasmic reticulum (ER) and mitochondria (M). Gly indicates glycogen; IM, intermyofibrillar mitochondria; L, lipid droplets; N, nucleus; and SC, subsarcolemmal mitochondria.
In the liver, TEM revealed differences in mitochondrial electron density between BL/6C57 and BL/6NZB mitochondria, and both patterns were apparent in DNPH animals, although there was no evidence of mitochondrial damage (Figure 3D). We also detected increased interactions between the endoplasmic reticulum and mitochondria in BL/6NZB and BL/6C57-NZB liver samples (Figure 3D and 3E).
DNPH Causes Pulmonary Hypertension
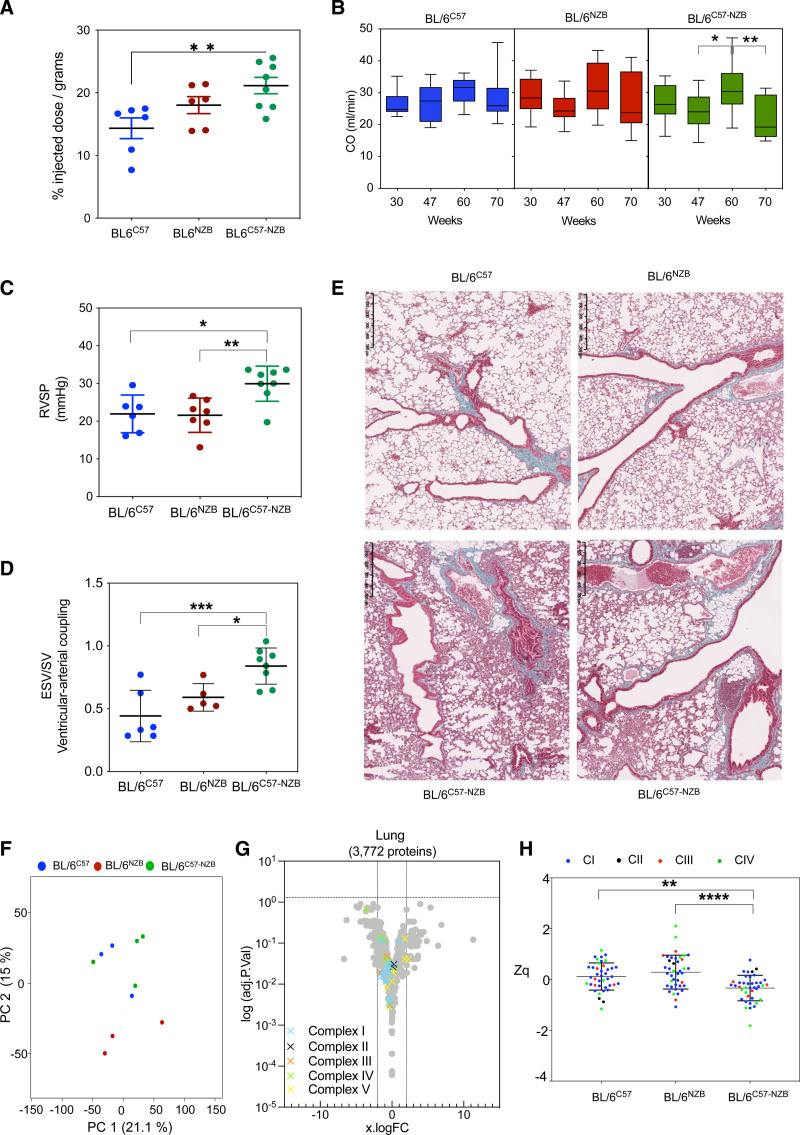
Aged heteroplasmic mice display right ventricular impairment, prompting us to conduct a detailed study of lung function. Glucose uptake was increased in the lungs of 20-week-old heteroplasmic mice, detected using [18F]-fluorodeoxyglucose positron emission tomography–computed tomography (Figure 2B) and confirmed ex vivo by total tissue uptake quantification (Figure 4A). The glycolytic switch in the right ventricle reduces contractility and promotes the inactivation of mitochondria-mediated apoptosis in pulmonary artery smooth muscle cells.18 Right ventricular ejection fraction is an excellent prognostic indicator for pulmonary arterial hypertension.19 Magnetic resonance imaging analysis of cardiac function revealed a decrease in right ventricular ejection fraction in heteroplasmic mice >70 weeks of age (Figure 2D). Echocardiography showed that aortic cardiac output remained constant in homoplasmic BL/6C57 and BL/6NZB mice, whereas aortic cardiac output increased in 60-week-old heteroplasmic mice (Figure 4B). This may reflect a compensatory increase in blood supply in heteroplasmic mice to meet oxygen demands when cardiac and pulmonary function are stunted. Cardiac output was decreased in DNPH animals >70 weeks of age (Figure 2D and Figure 4B).
Figure 4.
Divergent nonpathologic mitochondrial DNA heteroplasmy causes pulmonary hypertension. A, Ex vivo quantitative analysis of [18F]-fluorodeoxyglucose uptake in lung tissue of 20-week-old mice (6 homoplasmic mice and 8 heteroplasmic mice; *P<0.05, 1-way analysis of variance test). B, In vivo longitudinal echocardiography study of cardiac output (CO) assessed in the aorta. Data represented as mean±SD. *P<0.05, **P<0.01, 2-way analysis of variance test with Tukey multiple comparison test (12–14 mice per genotype and age). C, In vivo measurement for the right ventricular systolic pressure volume (RVSP) after right ventricle catheterization in 74-week-old mice. Each dot represents an individual mouse. D, Magnetic resonance imaging assessment of the right ventricular–-arterial coupling (end systolic volume/stroke volume [ESV/SV]) in 74-week-old mice. Each dot represents an individual mouse. *P<0.05, **P<0.01, 1-way analysis of variance test. E, Representative lung histologic sections from 80-week-old homoplasmic and heteroplasmic mice stained with Masson trichrome staining (4–5 mice per genotype). F through H, Lung proteomic analysis in 12-week-old mice. F, Principal component (PC) analysis. Each dot corresponds to an individual mouse. G, Volcano plot illustrating proteins with significantly differential abundance between homoplasmic and heteroplasmic lung tissues (4 homoplasmic mice and 5 heteroplasmic mice). The log (adjusted P value) is plotted against the log fold change (logFC; fold difference between homoplasmic and heteroplasmic mice). On the x axis, vertical lines denote ±2-fold change; on the y axis, the horizontal line denotes the significance threshold (P<0.05). H, Expression analysis of structural oxidative phosphorylation components. Data are presented as mean±SD. **P<0.01, ****P<0.0001 (1-way analysis of variance test).
We next assessed right ventricular systolic pressure by heart catheterization in young and older mice. In young animals, these measurements showed no significant differences between strains. In contrast, right ventricular systolic pressure was increased in DNPH mice >70 weeks of age, the stage at which animals with the most severe symptoms begin to die (Figure 4C). This increase correlates with the decrease in right ventricular ejection fraction at the same age. As expected, end systolic volume/stroke volume, a measure related to ventricular–arterial coupling,20 also increased (Figure 4D). Histopathologic analysis of the lungs of older DNPH mice contained many inflammatory foci and showed evidence of vascular and epithelial remodeling and hyaline membrane formation, associated with secondary pulmonary hypertension (Figure 4E).
Principal component analysis for proteomics on lung tissue from 12-week-old homoplasmic and DNPH mice did not reveal major differences when compared with homoplasmic young animals (Figure 4F). No differential expression of individual proteins between strains was detected (Figure 4G). However, we found a significant decrease in mitochondrial respiratory chain proteins as a group (Figure 4H). Pulmonary hypertension has been linked to mutations in COX5A (cytochrome c oxidase subunit 5A) of mitochondrial complex IV.21 Our analyses detected a significant reduction in the complex IV subunit in the lungs of DNPH mice (Figure 4H).
TEM analysis of the lungs of 12-week-old heteroplasmic mice revealed abnormal mitochondria ultrastructure (damaged, vacuolated, and swollen) in the alveolar system, specifically in type II alveolar epithelial cells (Figure S3A). A characteristic feature of heteroplasmic mice was collagen accumulation in the alveolar walls in young animals. mtDNA damage can increase alveolar epithelial cell apoptosis and is associated with pulmonary fibrosis.22 TEM analysis of type II alveolar epithelial cells in heteroplasmic mice also revealed an excess of lamellar bodies (Figure S3B and S3C) and abnormal convoluted nuclei (Figure S3A and S3B). Immunohistochemical analyses of lung tissue revealed extensive F4/80+ areas in heteroplasmic samples, indicating abnormal abundance of alveolar macrophages attributable to immune activation and infiltration (Figure S3D and S3E).
DNPH thus has different effects on mitochondrial physiology in the liver, heart, and lung. Whereas the liver maintains healthy mitochondria, cardiac and pulmonary mitochondria progressively deteriorate, inducing sustained cell stress.
DNPH Causes Oxidative Damage in Heart and Triggers a Stress Response
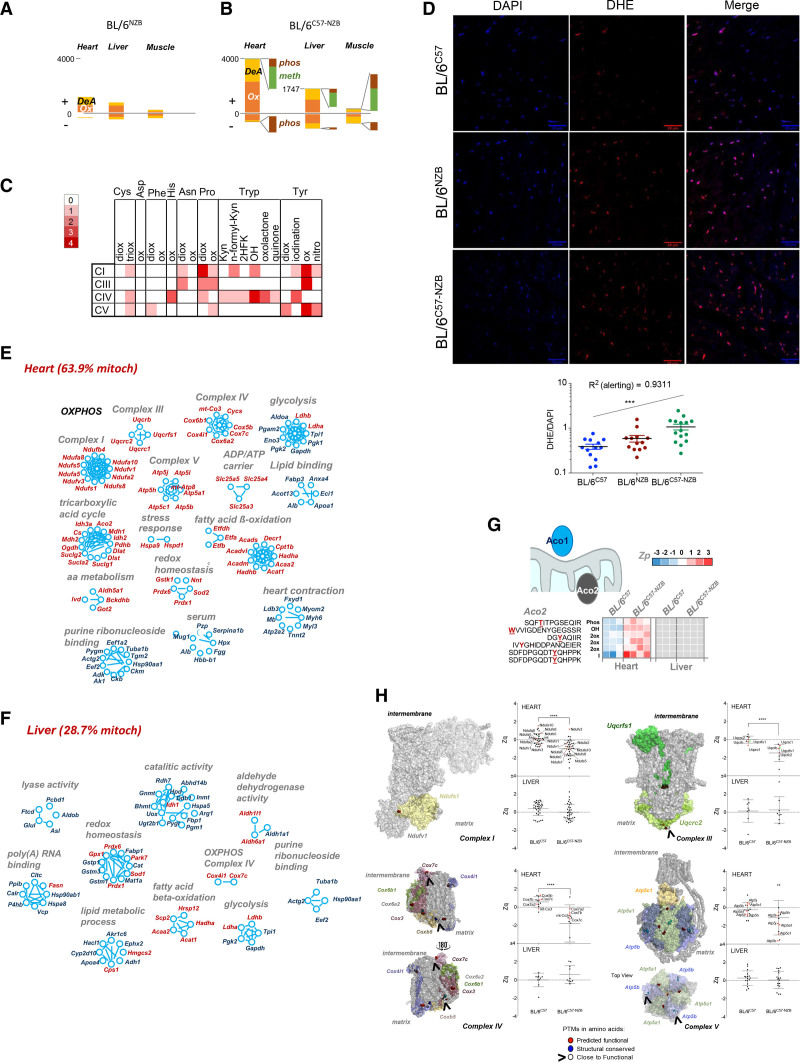
We next characterized the DNPH-induced posttranslational modification (PTM) landscape.23 In 12-week-old animals, homoplasmic BL/6C57 and BL/6NZB mice showed no major differences in PTMs in liver, skeletal muscle, and heart (Figure 5A). In contrast, the DNPH condition induced PTM changes significantly more frequently in the heart (Figure 5B). Most DNPH-induced PTMs in the heart were oxidative, mainly affecting Tyr, Trp, and Pro amino acids (Figure 5C), but also Asn, His, Phe, Asp, and Cys residues (Figure 5C). The abundant oxidative modifications in DNPH hearts may be a consequence of the increased oxidative stress detected in heart sections stained with dihydroethidium (Figure 5D). Higher levels of reactive oxygen species were previously documented in DNPH-derived mouse embryonic fibroblasts and mouse adult fibroblasts (MEF).14,15
Figure 5.
Divergent nonpathologic mitochondrial DNA heteroplasmy causes generalized oxidative damage to cardiac mitochondrial proteins. A and B, Quantification of peptides containing posttranslational modifications (PTMs) that were significantly more abundant (+) or less abundant (–) in the indicated tissues of (A) BL/6NZB mice (n=3) or (B) BL/6C57-NZB mice (n=4) relative to BL/6C57 mice (n=3). C, Frequency of modified amino acid and type of oxidative alteration in proteins of the indicated respiratory complexes. Red intensity is proportional to PTM abundance; white represents undetected PTM. D, Representative dihydroethidium staining (top) and derived reactive oxygen species quantification (bottom) in heart slices of 20-week-old mice of the indicated genotype. Each dot in the chart represents a different tissue slide. ***P<0.001 (1-way analysis of variance test for linear trend; n=4 per strain). E and F, Inventory of proteins harboring PTMs grouped by function in (E) heart and (F) liver (12-week-old homoplasmic mice, n=3; 12-week-old heteroplasmic mice, n=4). Red text indicates mitochondrial localization. G, Aco2 (aconitase 2) peptides with the indicated PTMs and quantitative changes observed between BL/6C57 homoplasmic and BL/6C57-NZB heteroplasmic heart and liver tissues. Color is graded according to the log2 transformation of the ratio between the sample and the internal standard peptide level intensities (Zp). Gray indicates that the PTM was not detected. H, Protein models showing subunits of the indicated respiratory complexes. PTMs on amino acids with predicted functional relevance (exposed residues with a conservation score = 8 or 9) are highlighted in red. PTMs on structural residues (buried residues with a conservation score = 9) are highlighted in blue. Modified residues close to a functional residue are shown in white. The graphs show the standardized log2 ratios of the indicated complex proteins (Zq) or the standardized, protein-corrected log2 ratios of peptides (Zpq) in heart and liver between the BL/6C57 homoplastic and BL/6C57-NZB heteroplasmic mice. Colored dots indicate proteins exposed to the matrix side of the inner mitochondrial membrane (red) or to the intermembrane space (green) harboring PTMs. 2HFK indicates Trp to 2-hydroxy formyl kynurenine; 2ox, dioxidation; 3ox, trioxidation; DeA, deamidation; Kyn, Trp to kynurenine; meth, methylation; OH, Trp to hydroxytryptophan; Ox, oxidation; OXPHOS, oxidative phosphorylation; and phos, phosphorylation.
Approximately 65% of the proteins affected by PTM alterations in the heart were mitochondrial proteins, compared with <30% in the liver (Figure 5E and 5F). In addition to OXPHOS proteins, oxidation of mitochondrial proteins also included tricarboxylic acid cycle components—β-oxidation, amino acid metabolism, ADP/ATP carriers, and mitochondrial chaperones—in DNPH hearts. Several nonmitochondrial proteins were also differentially modified, notably including serum components and proteins involved in glycolysis, lipid binding, purine ribonucleotide binding, and heart contraction (Figure 5E). DNPH animals demonstrated increased oxidative PTMs in both stress response proteins (Hspa9 [heat shock protein family A member 9], Hspd1 [heat shock protein family D member 1]) and reactive oxygen species–regulating proteins (Gstk1 [glutathione S-transferase kappa 1], Nnt [nicotinamide nucleotide transhydrogenase], Sod2 [superoxide dismutase 2], Prdx1 [peroxiredoxin-1], Prdx6 [peroxiredoxin-6]). The enzyme aconitase is particularly reactive oxygen species–sensitive, existing as a mitochondrial matrix isoform (Aco2 [aconitase 2]) and a cytoplasmic isoform (Aco1 [aconitase 1]). Aco2 was severely modified in heteroplasmic hearts but was unaffected in heteroplasmic liver (Figure 5G), whereas Aco1 remained unmodified in both tissues. Heteroplasmic hearts also showed an increase in oxidative PTMs in proteins of the respiratory complexes I, III, IV, and V (Figure 5H), correlating with reductions in these proteins. No differentially modified respiratory subunits were found in DNPH livers (Figure 5H). Among modified residues, 70% were conserved or interacted with conserved amino acids. PTM landscape analysis predicted that most of these residues would have a functional effect.23
Generalized Mitochondrial Function Impairment in Tissues That Fail to Resolve DNPH
The striking divergence in the effects of DNPH between different organs suggests that the impaired mitochondrial function in the heart and lung of heteroplasmic mice might be attributable to the lack of mitochondrial segregation. In addition to liver, 2 other tissues that segregate mtDNA haplotypes—brown adipose tissue and kidney—showed no signs of increased damage in 20- and 80-week-old heteroplasmic mice compared with homoplasmic controls (Figure S4A and S4B).
Unlike liver, brown adipose tissue, and kidney, skeletal muscle cannot resolve heteroplasmy.14 Our DNPH mice showed a massive loss of skeletal muscle with age (Figure 6A). Histologic analysis of skeletal muscle in 80-week-old heteroplasmic mice showed a high proportion of central nuclei (Figure 6B), abnormally high levels of fibrosis (Figure 6C and 6D), and evident abnormalities in muscle fiber ultrastructure, including disruption of the myofibrillar banding patterns and loss of mitochondria and their inner and outer membrane ultrastructure (Figure 6E and 6F). Proteomic analyses of muscle samples from 12-week-old mice confirmed the specific profile of heteroplasmic mice (Figure 6G). Volcano plot analysis of the complete proteome showed no overall differences in OXPHOS proteins between homoplasmic and heteroplasmic skeletal muscle in young animals (Figure 6H). Detailed MitoProteome analyses revealed a greater abundance of complex I structural subunits in BL/6C57 muscle than in BL/6NZB or BL/6C57-NZB mice (Figure 6I). The lack of major OXPHOS component alterations matches the absence of skeletal muscle impairment at this age, suggesting mitochondrial performance is maintained in skeletal muscle. Supporting this conclusion,31P magnetic resonance spectroscopy revealed a normal PCr:ATP ratio in skeletal muscle of 12-week-old heteroplasmic animals (Figure 6J) and the muscle content of ANT1 and ANT2, CKMT2, and VDAC proteins was unaffected (Figure 6H). However, progressive loss of muscle mass was accompanied by an abnormally high PCr:ATP ratio in heteroplasmic skeletal muscle of 40-week-old mice, suggesting a drop in mitochondrial bioenergetic efficiency (Figure 6J). Old heteroplasmic mice showed a sharp drop in the PCr:ATP ratio (Figure 6J), matching the massive loss of muscle mass in these animals (Figure 6A). We also documented rear leg paralysis in 4 out of 81 older heteroplasmic mice (Videos S1 and S2).
Figure 6.
Analysis of skeletal muscle. A, Representative images showing the loss of muscle mass in heteroplasmic animals. B, Representative hematoxylin and eosin staining on skeletal muscle sections from 80-week-old homoplasmic and heteroplasmic mice. Arrows indicate central nuclei. C and D, Analysis of skeletal muscle collagen and fibrosis in mice of the indicated age and mouse strain. C, Representative images of skeletal muscle histologic sections stained with Masson trichrome staining at the indicated ages and (D) quantification of the fibrotic area. Age (P<0.001) and genotype (P<0.05) as well as their interaction (P<0.01) are significantly different assessed by 2-way analysis of variance. Multiple comparisons corrected by Tukey (n=4 mice per genotype and age). E and F, Electron microscopy analyses of skeletal muscle samples from 12- and 80-week-old mice (3–4 mice per genotype and age). Black arrow indicates a fibrotic area. G through I, Proteomic analysis of skeletal muscle in 12-week-old mice. G, Score plot of the principal component (PC) analysis. H, Volcano plot illustrating proteins with significantly differential abundance between homoplasmic and heteroplasmic gastrocnemius (3 homoplasmic and 4 heteroplasmic samples). The log (adjusted P value) is plotted against the log fold change (logFC; fold change homoplasmic/heteroplasmic animals). On the x axis, vertical lines denote ±2-fold change; on the y axis, the horizontal line denotes the significance threshold (P<0.05 prior to logarithmic transformation). I, MitoProteome analysis highlighting structural oxidative phosphorylation components and mitochondrial elements of the phosphocreatine (PCr) pathway (3–4 mice per genotype). J, 31P magnetic resonance spectroscopy analysis of the PCr/ATP (adenosine triphosphate) ratio in the skeletal muscle (soleus) of different individuals at the indicated age. *P<0.05, **P<0.01, 2-way analysis of variance test. Multiple comparisons corrected by Tukey test (n=5–9). In D and G, dots represent individual mice. In D and J, data are presented as mean±SD. OXPHOS indicates oxidative phosphorylation.
The eye was also unable to resolve heteroplasmy. Corneal opacity was evident in adult heteroplasmic animals (Figure S5A and S5B). The retinas of 12-week-older heteroplasmic mice had structural abnormalities and were abnormally thin (Figure S5C and S5D), but no differences were found in the histologic analysis of the cornea. However, TEM analysis revealed that corneal ultrastructure in DNPH mice was also altered, including the structure and thickness of the Descemet membrane (Figure S5E). Descemet membrane is a specialized basement membrane of the endothelium and its thickness increases with age and after endothelial damage.24 We observed accumulation of engulfed mitochondria in vesicles that may be autolysosomes, suggesting stalled mitophagy in the cornea of heteroplasmic animals (Figure S5E).
DNPH-Induced Pathology Is Modulated by the Nuclear Context
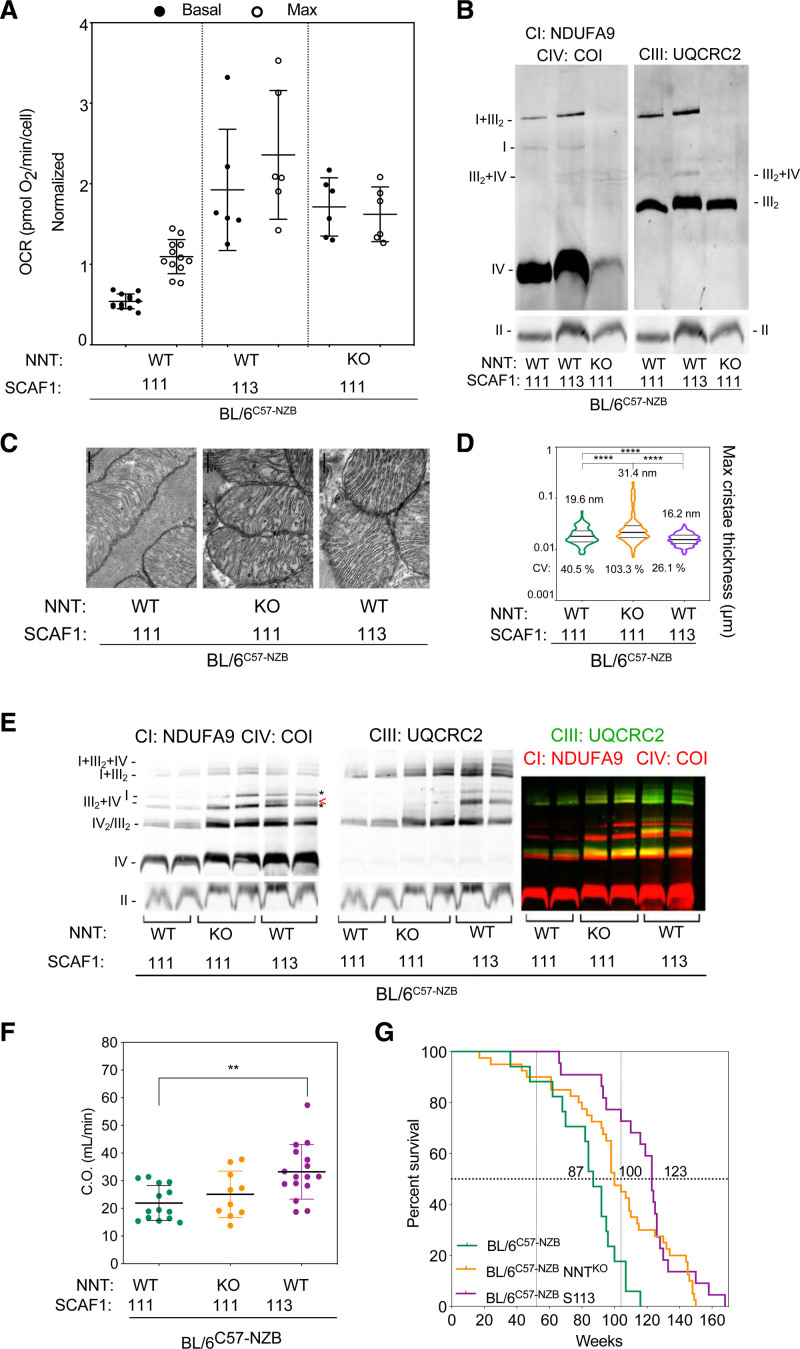
We have observed that several nuclear features strongly affect mtDNA segregation in DNPH animals.14 Examples affecting mtDNA germline and postnatal segregation include mild alterations in mitochondrial reactive oxygen species signaling/detoxification owing to the absence of NNT and impairment of the superassembly of respiratory complexes owing to the lack of functional SCAF1 (supercomplex assembly factor 1). We analyzed whether the DNPH phenotype is determined by the expression of NNT or of functional SCAF1 (SCAF1[113]). The reference nuclear context of our DNPH mice is C57BL/6 with SCAF1(111) and wild-type NNT. Control MAF had lower baseline and maximum oxygen consumption rates than DNPH MAF that were genetically modified to express SCAF1(113)—BL/6C57-NZB:SCAF1(113), with wild-type NNT (Figure 7A). Blue native electrophoresis analysis showed that BL/6C57-NZB DNPH control MAF had less complex I and a higher ratio of free complex I to complex I+complex III2 than homoplasmic MAF derived from BL/6C57 animals.14 Most complex I in DNPH BL/6C57-NZB MAF expressing the functional form of SCAF1(113) was associated with complex III2 (Figure 7B). Lack of NNT in DNPH MAF induced a sharp reduction in the expression and assembly of complex I and complex IV (Figure 7B). The total amount of free complex III2 in DNPH MAF lacking NNT was like the other DNPH and homoplasmic cell lines.14 As expected, supercomplexes III2+IV were only present in cells expressing SCAF1(113), as shown in Figure 7B. The use of galactose could only be compared for DNPH MAF expressing SCAF1(113) and homoplasmic MAF (Figure S6A). With galactose as substrate, DNPH-SCAF1(113) MAF showed a substantial increase in supercomplex III2+IV (the Q respirasome) and the appearance of I+III2+IV (the N respirasome), but this was not observed in homoplasmic cells (Figure S6A). Seahorse assays conducted with glucose, galactose, or fatty acid–rich medium revealed that the respiratory capacity of mitochondria in DNPH-SCAF1(113) was higher than that of mitochondria in DNPH-SCAF1(111) MAF for all carbon sources (Figure S6B).
Figure 7.
Effect of the nuclear context on the divergent nonpathologic mitochondrial DNA heteroplasmy phenotype. A, Respiratory performance at high glucose concentration (25 mM) by mouse adult fibroblasts from unmodified heteroplasmic mice (BL/6C57-NZB, containing SCAF1[111] and wild-type NNT [nicotinamide nucleotide transhydrogenase]) and 2 genetically modified strains containing SCAF1(111) and lacking NNT (BL/6C57-NZB:NNTKO) or containing SCAF1(113) and wild-type NNT (BL/6C57-NZB: S113). The chart plot shows basal and maximum oxygen consumption rates (OCRs) measured using Seahorse (5 to 6 independent assays per cell clone). Data are presented as mean±SEM. B, Blue native electrophoresis of digitonin-solubilized mitochondria from the indicated cell clones. Left: complex I (NDUFA9), complex IV (COI). Right: complex III (UQCRC2). Bottom: complex II (FP70). C and D, Transmission electron microscopy analysis of cardiac mitochondrial ultrastructure and cristae thickness. Images in (C) are representative of the indicated clones and the chart (D) shows quantification of 409 BL/6C57-NZB cristae, 268 BL/6C57-NZB:NNTKO cristae, and 288 BL/6C57-NZB:S113 cristae. Violin plot shows the frequency distribution of the data; lines represent median and quartiles. ****P<0.0001 (nonparametric Kruskal-Wallis test). E, Blue native electrophoresis of digitonin-solubilized cardiac mitochondria from 12-week-old mice of the indicated strains. Left: complex I (NDUFA9), complex IV (COI). Asterisks indicate the position where CIV migrates without CIII and the arrowhead the position of SC III2+IV. Middle: complex III (UQCRC2). Right: merged immunodetection of the indicated proteins (green, CIII; red, CI and CIV). Bottom: complex II (FP70). F, Cardiac output echocardiography analysis in 74-week-old male mice of the indicated genotype. Each dot represents an individual mouse. **P<0.01 (t test, BL/6C57-NZB:S113 or BL/6C57-NZB:NNTKO versus control heteroplasmic mice: BL/6C57-NZB, n=14; BL/6C57-NZB:S113, n=16; and BL/6C57-NZB:NNTKO, n=10). G, Survival curves of the heteroplasmic strains. For the comparison of BL/6C57-NZB and BL/6C57-NZB:S113 mice, ****P<0.0001 by log-rank test (Mantel-Cox; χ2=22.29; df=1) and ****P<0.0001 by Gehan-Breslow-Wilcoxon test. For the comparison of BL/6C57-NZB and BL/6C57-NZB:NNTKO, **P<0.01 by log-rank test (Mantel-Cox; χ2=9.322; df=1) and *P<0.05 by Gehan-Breslow-Wilcoxon test. BL/6C57-NZB mice (results taken from Figure 1A): BL/6C57-NZB:NNTKO mice, 22 male and 18 female; and BL/6C57-NZB:S113 mice, 13 male and 9 female. SCAF indicates supercomplex assembly factor 1.
To investigate the mitochondrial network status in the heteroplasmic MAF models, we immunodetected TOM20 on the mitochondrial outer membrane. BL/6C57-NZB MAF contained fewer mitochondria than other DNPH cells, which were evenly distributed in the cytoplasm (Figure S6C). Similar to its effect on baseline respiration, expression of SCAF1(113) in DNPH MAF restored other homoplasmic features, such as overall mitochondrial content and the concentration of perinuclear mitochondria (Figure S6C). NNT ablation in DNPH MAF (BL/6C57-NZB:NNTKO, S111) exacerbated mitochondrial fragmentation and these cells accumulated mitochondria in the cell periphery and in abundant cytoskeleton-positive extracellular vesicles (Figure S6C). This high peripheral and vesicular concentration of mitochondria is suggestive of removal of mitochondria by exocytosis and may be related to the surprising loss of almost all the NZB mtDNA from DNPH-NNTKO MAF during the first passages after immortalization (Figure S6C). The high instability of heteroplasmy in the NNTKO nuclear background makes it difficult to analyze the recovery of respiration in these cells. Note that the rapid elimination of NZB mtDNA did not allow DNPH-NNTKO MAF to escape glucose dependence because NNT is required to maintain glutaminolysis to replenish tricarboxylic acid intermediates through anaplerotic reactions.25 In the absence of NNT, glycolysis-derived pyruvate is used, making NNT-deficient cells highly sensitive to glucose deprivation.25 Homoplasmic NNTKO MAF cannot survive on galactose.25 Therefore, mild modifications of the nuclear response to heteroplasmy can substantially affect the phenotype of adult mouse fibroblasts in culture.
To examine the impact of the nuclear effect at the organismal level, we analyzed cardiac tissue ultrastructure in the heteroplasmic strains by TEM (Figure S6D). Cardiomyocytes in DNPH-NNTKO hearts had short sarcomeres and contained relatively few lipid droplets. In contrast, reexpression of SCAF1(113) in DNPH mice improved cardiomyocyte lipid mobilization, revealed by increased abundance of lipid droplets and their distribution close to intermyofibrillar mitochondria (Figure S6D). Mitochondria in DNPH-NNTKO hearts had irregular cristae with a significant increase in mean maximum thickness (Figure 7C and 7D). In contrast, mitochondria in DNPH-SCAF1(113) hearts showed improved cristae organization, shape, and density (Figure 7C and 7D), which is intimately linked to the structure and function of the respiratory chain.26 Blue native electrophoresis analysis of the DNPH models suggested that the OXPHOS impairment in heteroplasmic cardiac mitochondria was partially compensated by the loss of NNT or the reexpression of SCAF1(113), as shown in Figure 7E. Furthermore, only DNPH-SCAF1(113) hearts contained III2+IV, accompanied by elevations in all complexes and subsarcolemmal mitochondria (Figure 7E). Echocardiography showed that DNPH-SCAF1(113) mice, but not DNPH-NNTKO mice, were protected against the decline in the descending aortic blood flow and cardiac output observed in aged DNPH mice (Figure 7F and Figure 2D).
NNT ablation or SCAF1(113) expression improved the general appearance of aged DNPH mice. Both strains (at >350 days) showed a healthier appearance and were more active than BL/6C57-NZB mice (Figure S6E). Improved cardiac function in the modified heteroplasmic strains was accompanied by an increase in median lifespan (DNPH: 87 weeks; DNPH-NNTKO: 100 weeks; DNPH-SCAF1[113]: 127 weeks) and maximum lifespan (DNPH: 116 weeks; DNPH-NNTKO: 150 weeks; DNPH-SCAF1[113]: 158 weeks), as shown in Figure 7G. Heteroplasmy triggers a stress response in the cells but the severity of the DNPH phenotype is dependent on the nuclear genetic background.
Discussion
mtDNA heteroplasmy is prevented in nature. Uniparental transmission is the norm in eukaryotes and is achieved by active elimination of male mtDNA27,28; by positive selection during germline maturation and embryogenesis15; and by active mtDNA selection in most tissues.9,10,12,29,30 DNPH negatively affects behavior and cognition.12 However, behavioral changes are restricted to animals, whereas DNPH prevention is a near-universal feature of eukaryotes.31 We propose that DNPH is negative for the cell and leads to the progressive and nonrandom selection of 1 mtDNA variant. On tissues where DNPH remains largely stable, especially the heart, lung, and skeletal muscle, it compromises cellular energy budgets and translates into reduced organismal fitness.
Different combinations of mtDNA haplotypes induce different phenotypes in a spectrum ranging from absence of pathology to varying degrees of disease. In most reports, NZB mtDNA was mixed with Balb/c mtDNA,9,11 129 mtDNA,12 or C57 mtDNA. Balb/c, 129, and C57 mtDNA variants are practically identical and phenotypes are unlikely to vary widely between these models.32,33 Previous studies of mitochondrial DNPH used 3 different nuclei derived from inbred strains: Balb/cByJ, with 6.25% to 25% NZB,9,11 and 129/SvEv backcrossed (10 generations) to C57BL/6J from Jackson Laboratory.12 The C57BL/6J nuclear background lacks 2 OXPHOS system components: NNT and functional SCAF1(113).34 The C57BL/6JOlaHsd background only lacks functional SCAF1 (expresses the mutant SCAF1[111]). We demonstrate that expression of functional SCAF1 or NNT has a major effect on the DNPH phenotype, delaying or reducing disease severity and explaining discrepancies in the literature. Nevertheless, chronic DNPH compromises mitochondrial function and produces a pathologic phenotype independent of the nuclear context.
The compromised cell fitness in heteroplasmic tissues provides an evolutionary rationale for the existence of such diverse and redundant mechanisms to prevent heteroplasmy. In young DNPH animals, an abnormal loss of cardiomyocytes leads to weakened but functionally subclinical cardiac performance. Compounding this effect, parallel damage of pulmonary tissue attributable to unresolved heteroplasmy forces the right ventricle to overwork, leading to symptomatic manifestations of right ventricular deficiency. The left ventricle also has tissue damage, but this is comparatively minor. The liver exhibits successful adaptation to heteroplasmy, which leads to the selective elimination of one of the mtDNA variants likely by selective mitophagy.14,35 It remains unclear why some tissues fail to eliminate heteroplasmy. The heart, skeletal muscle, bladder, and eye are composed of postmitotic cells rich in mitochondria and with a highly structured cytoplasm, features that can hinder mtDNA segregation. Nevertheless, cells recognize DNPH and adapt their transcriptome and proteome profiles regardless of their capacity for mtDNA selection. However, the pathways involved in this adaptation differ between tissues, being centered on downregulation of mitochondrial-dependent metabolism in the heart, contrasting with a more overall homeostatic adaptation in the liver. The accumulation in cardiac tissue damage, together with an overall shift in cardiomyocyte metabolism toward increased glycolysis, severely compromises the right ventricular/pulmonary axis. This effect manifests itself early in life and is compounded by the natural decline in function during aging. In several other tissues that are unable to resolve heteroplasmy, including ocular and skeletal muscle tissue, this metabolically dependent effect of aging accelerating organ failure was observed.
Nonpathologic mouse mtDNA haplotypes can differentially modulate organismal metabolism, which may provide a molecular explanation for the maladaptive effect of DNPH.7 This observation has been extended to humans in a large-cohort study performed using tissue samples from the UK Biobank.36 Results indicated that many mtDNA single nucleotide variants lead to unexpected effects on human physiology.36 Nucleus–mitochondrial genetic variants also have been described in human cardiovascular and pulmonary diseases.37–39
The detrimental effects of DNPH observed in this study call for careful selection of donor mtDNA for mitochondrial replacement therapies. Even the most promising method for replacement of oocyte mitochondria carrying known pathologic mtDNA mutations may fail to achieve 100% replacement. Recipient cells have a strong capacity to select and amplify preexisting mtDNA (which is often initially undetectable), resulting in DNPH.40,41 Rejuvenation of oocytes by injection of donor cytoplasm involves the same difficulties.42 A similar risk may be present in therapies using purified mitochondria or transfer mitochondria from a donor targeting damaged cells to treat cardiopulmonary or neurologic diseases.43–46 Any therapeutic strategy that might result in DNPH should ensure a match between donor and recipient mtDNA haplotypes.47
Article Information
Acknowledgments
The authors thank Dr R. Martínez-de-Mena, M.M. Muñoz-Hernandez, A. Gonzalez-Guerra, I. Martinez-Carrascoso, E.R. Martínez-Jimenez, and Dr C. Jimenez for technical assistance; Drs J.M. Fernández-Toro and L.M. Criado for generation of heteroplasmic mice; Dr A. Dopazo and the Genomics Unit of the Centro Nacional de Investigaciónes Cardiovasculares for RNA sequencing analyses; M. Cueva and R. Alvarez for mouse work; A. Molina-Iracheta and R. Doohan for histology; and S. Bartlett (Centro Nacional de Investigaciónes Cardiovasculares) and Dr D.T. Cain (University of Oxford) for English editing.
Sources of Funding
Dr Lechuga-Vieco was supported by a Predoctoral Fellowship from Ministerio de Ciencia e Innovación (SVP-2013-068089) and by a Postdoctoral Fellowship from the European Molecular Biology Organization (ALTF115-2019). This study was supported by grants to Dr Enríquez from Ministerio de Ciencia e Innovación (grants SAF2015-65633-R and RTI2018-099357-B-I00) and Humand Frontier Science Program (grant RGP0016/2018). Dr Ruíz-Cabello is supported by grants from the Ministerio de Economía, Industria y Competitivida (grant SAF2017-84494-C2-R), Programa Red Guipuzcoana de Ciencia, Tecnología e Información (grant 2018-CIEN-000058-01), and the Gobierno Vasco, Dpto Industria, Innovación, Comercio y Turismo under the ELKARTEK Program (grant KK-2019/bmG19). Dr Ruíz-Cabello received funding from the BBVA Foundation (Ayudas a Equipos de Investigación Científica Biomedicina 2018). CIC biomaGUNE is supported by the Maria de Maeztu Units of Excellence Program from the Spanish State Research Agency (grant MDM-2017-0720). Dr Santos has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement 796721. Dr Vázquez received funding from Ministerio de Ciencia e Innovación (grants PGC2018-097019-B-I00 and PRB3-IPT17/0019/0003-ISCIII-SGEFI/ERDF, ProteoRed), the Fundació MaratóTV3 (grant 122/C/2015), and “la Caixa” Banking Foundation (project code HR17-00247). The CNIC is supported by the Instituto de Salud Carlos III (ISCIII), the Ministerio de Ciencia e Innovación (MCIN), and the Pro Centro CNIC Foundation and is a Severo Ochoa Center of Excellence (grant CEX2020-001041-S funded by MICIN/AEI/10.13039/501100011033).
Disclosures
Drs Lechuga-Vieco and Latorre-Pellicer performed most of the mouse and cellular experimental work. Dr Acín-Pérez and R. Justo-Méndez performed Seahorse and adenosine triphosphate synthesis analyses. Dr Calvo, R. Magni, and Dr Vázquez performed the proteomic analysis and the posttranslational modification analysis together with Drs Bagwan and Bonzon-Kulichenko and performed the structural modeling of the respiratory complexes and the conservation of the modified residues. Drs Torroja and Sánchez-Cabo analyzed the genomic and transcriptomic data. Drs García-Gil and Lechuga-Vieco performed and analyzed electron microscopy imaging. Drs Pellico, Lechuga-Vieco, Benito, and Ruíz-Cabello performed and analyzed the in vivo imaging data. Dr Santos performed the in vivo assessment of ventricular pressure. Drs Enríquez and Lechuga-Vieco participated in the design of the experimental work, the integrated analysis of the results, and the writing of the manuscript. Dr Simon edited the manuscript. Dr Enríquez directed and designed the research.
Supplemental Material
Expanded Methods
Figures S1–S6
Tables S1 and S2
Videos S1 and S2
References 48–59
Supplementary Material
Nonstandard Abbreviations and Acronyms
- Aco1
- aconitase 1
- Aco2
- aconitase 2
- ACOT13
- acyl–coenzyme A thioesterase 13
- ACSL1
- acyl–coenzyme A synthetase long chain family member 1
- ADP
- adenosine diphosphate
- AMPK
- adenosine monophosphate–activated protein kinase
- ANT1
- adenine nucleotide translocator 1
- ANT2
- adenine nucleotide translocator 2
- ATP
- adenosine triphosphate
- ATP1A3
- sodium–potassium ATPase catalytic subunit α3
- CKMT2
- creatine kinase mitochondrial 2
- DNPH
- divergent nonpathologic heteroplasmy
- eIF2
- eukaryotic initiation factor 2
- eIF4
- eukaryotic initiation factor 4
- Gstk1
- glutathione S-transferase kappa 1
- HADHA
- hydroxyacyl–coenzyme A dehydrogenase trifunctional multienzyme subunit alpha
- Hspa9
- heat shock protein family A member 9
- Hspd1
- heat shock protein family D member 1
- JNK
- c-Jun N-terminal kinase
- MAF
- mouse adult fibroblasts
- mtDNA
- mitochondrial DNA
- mTOR
- mammalian target of rapamycin
- mTORC1
- mammalian target of rapamycin complex 1
- Nd
- nicotinamide adenine dinucleotide–ubiquinone oxidoreductase
- NNT
- nicotinamide nucleotide transhydrogenase
- OXPHOS
- oxidative phosphorylation
- PCr
- phosphocreatine
- PPAR
- peroxisome proliferator-activated receptor
- Prdx1
- peroxiredoxin-1
- Prdx6
- peroxiredoxin-6
- PTM
- posttranslational modification
- RXR
- retinoid X receptor
- SAPK
- stress-activated protein kinases
- SCAF1
- supercomplex assembly factor 1
- SERCA2
- sarcoplasmic/endoplasmic reticulum calcium ATPase 2
- SLC25A20
- carnitine/acylcarnitine translocase
- SNP
- single nucleotide polymorphism
- Sod2
- superoxide dismutase 2
- SRL
- sarcalumenin
- TEM
- transmission electron microscopy
- VDAC
- voltage-dependent anion-selective channel
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.121.056286.
For Sources of Funding and Disclosures, see page 1100.
Circulation is available at www.ahajournals.org/journal/circ
Contributor Information
Ana Victoria Lechuga-Vieco, Email: avlecvie@gmail.com.
Ana Latorre-Pellicer, Email: ana.latorre.pellicer@gmail.com.
Enrique Calvo, Email: ecalvo@cnic.es.
Carlos Torroja, Email: ctorroja@cnic.es.
Juan Pellico, Email: juan.pellico@kcl.ac.uk.
Rebeca Acín-Pérez, Email: rbkacin@gmail.com.
Arnoldo Santos, Email: asantosoviedo@gmail.com.
Navratan Bagwan, Email: navratan.bagwan@gmail.com.
Elena Bonzon-Kulichenko, Email: ebonzon@cnic.es.
Ricardo Magni, Email: ricardo.magni@externo.cnic.es.
Marina Benito, Email: marina.benito.vicente@gmail.com.
Raquel Justo-Méndez, Email: raquel.justo@cnic.es.
Anna Katharina Simon, Email: katja.simon@imm.ox.ac.uk.
Jesús Vázquez, Email: jesus.vazquez@cnic.es.
Jesús Ruíz-Cabello, Email: jesus.vazquez@cnic.es.
References
- 1.Stewart JB, Chinnery PF. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat Rev Genet. 2021;22:106–118. doi: 10.1038/s41576-020-00284-x [DOI] [PubMed] [Google Scholar]
- 2.van den Ameele J, Li AYZ, Ma H, Chinnery PF. Mitochondrial heteroplasmy beyond the oocyte bottleneck. Semin Cell Dev Biol. 2019;97:156–166. doi: 10.1016/j.semcdb.2019.10.001 [DOI] [PubMed] [Google Scholar]
- 3.Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, Woodward J, Li Y, Ramsey C, Kolotushkina O, Mitalipov S. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craven L, Tuppen HA, Greggains GD, Harbottle SJ, Murphy JL, Cree LM, Murdoch AP, Chinnery PF, Taylor RW, Lightowlers RN, et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woods DC, Tilly JL. Autologous germline mitochondrial energy transfer (AUGMENT) in human assisted reproduction. Semin Reprod Med. 2015;33:410–421. doi: 10.1055/s-0035-1567826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roushandeh AM, Kuwahara Y, Roudkenar MH. Mitochondrial transplantation as a potential and novel master key for treatment of various incurable diseases. Cytotechnology. 2019;71:647–663. doi: 10.1007/s10616-019-00302-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, Sánchez-Cabo F, Torroja C, Acín-Pérez R, Calvo E, Aix E, González-Guerra A, Logan A, et al. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature. 2016;535:561–565. doi: 10.1038/nature18618 [DOI] [PubMed] [Google Scholar]
- 8.Battersby BJ, Shoubridge EA. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum Mol Genet. 2001;10:2469–2479. doi: 10.1093/hmg/10.22.2469 [DOI] [PubMed] [Google Scholar]
- 9.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet. 1997;16:93–95. doi: 10.1038/ng0597-93 [DOI] [PubMed] [Google Scholar]
- 10.Burgstaller JP, Johnston IG, Jones NS, Albrechtová J, Kolbe T, Vogl C, Futschik A, Mayrhofer C, Klein D, Sabitzer S, et al. MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Rep. 2014;7:2031–2041. doi: 10.1016/j.celrep.2014.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Acton BM, Lai I, Shang X, Jurisicova A, Casper RF. Neutral mitochondrial heteroplasmy alters physiological function in mice. Biol Reprod. 2007;77:569–576. doi: 10.1095/biolreprod.107.060806 [DOI] [PubMed] [Google Scholar]
- 12.Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M, Crimi M, Waymire K, Lin CS, Masubuchi S, Friend N, Koike M, et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151:333–343. doi: 10.1016/j.cell.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146 [DOI] [PubMed] [Google Scholar]
- 14.Lechuga-Vieco AV, Latorre-Pellicer A, Johnston IG, Prota G, Gileadi U, Justo-Méndez R, Acín-Pérez R, Martínez-de-Mena R, Fernández-Toro JM, Jimenez-Blasco D, et al. Cell identity and nucleo-mitochondrial genetic context modulate OXPHOS performance and determine somatic heteroplasmy dynamics. Sci Adv. 2020;6:eaba5345. doi: 10.1126/sciadv.aba5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latorre-Pellicer A, Lechuga-Vieco AV, Johnston IG, Hämäläinen RH, Pellico J, Justo-Méndez R, Fernández-Toro JM, Clavería C, Guaras A, Sierra R, et al. Regulation of mother-to-offspring transmission of mtDNA heteroplasmy. Cell Metab. 2019;30:1120–1130.e5. doi: 10.1016/j.cmet.2019.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suomalainen A, Battersby BJ. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cell Biol. 2018;19:77–92. doi: 10.1038/nrm.2017.66 [DOI] [PubMed] [Google Scholar]
- 17.Spindler M, Niebler R, Remkes H, Horn M, Lanz T, Neubauer S. Mitochondrial creatine kinase is critically necessary for normal myocardial high-energy phosphate metabolism. Am J Physiol Heart Circ Physiol. 2002;283:H680–H687. doi: 10.1152/ajpheart.00800.2001 [DOI] [PubMed] [Google Scholar]
- 18.Ryan JJ, Archer SL. Emerging concepts in the molecular basis of pulmonary arterial hypertension: part I: metabolic plasticity and mitochondrial dynamics in the pulmonary circulation and right ventricle in pulmonary arterial hypertension. Circulation. 2015;131:1691–1702. doi: 10.1161/CIRCULATIONAHA.114.006979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talati M, Hemnes A. Fatty acid metabolism in pulmonary arterial hypertension: role in right ventricular dysfunction and hypertrophy. Pulm Circ. 2015;5:269–278. doi: 10.1086/681227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Álvarez A, Fernández-Friera L, García-Ruiz JM, Nuño-Ayala M, Pereda D, Fernández-Jiménez R, Guzmán G, Sanchez-Quintana D, Alberich-Bayarri A, Pastor-Escuredo D, et al. Noninvasive monitoring of serial changes in pulmonary vascular resistance and acute vasodilator testing using cardiac magnetic resonance. J Am Coll Cardiol. 2013;62:1621–1631. doi: 10.1016/j.jacc.2013.07.037 [DOI] [PubMed] [Google Scholar]
- 21.Baertling F, Al-Murshedi F, Sánchez-Caballero L, Al-Senaidi K, Joshi NP, Venselaar H, van den Brand MA, Nijtmans LG, Rodenburg RJ. Mutation in mitochondrial complex IV subunit COX5A causes pulmonary arterial hypertension, lactic acidemia, and failure to thrive. Hum Mutat. 2017;38:692–703. doi: 10.1002/humu.23210 [DOI] [PubMed] [Google Scholar]
- 22.Kim SJ, Cheresh P, Jablonski RP, Williams DB, Kamp DW. The role of mitochondrial DNA in mediating alveolar epithelial cell apoptosis and pulmonary fibrosis. Int J Mol Sci. 2015;16:21486–21519. doi: 10.3390/ijms160921486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bagwan N, Bonzon-Kulichenko E, Calvo E, Lechuga-Vieco AV, Michalakopoulos S, Trevisan-Herraz M, Ezkurdia I, Rodríguez JM, Magni R, Latorre-Pellicer A, et al. Comprehensive quantification of the modified proteome reveals oxidative heart damage in mitochondrial heteroplasmy. Cell Rep. 2018;23:3685–3697.e4. doi: 10.1016/j.celrep.2018.05.080 [DOI] [PubMed] [Google Scholar]
- 24.Tuft SJ, Williams KA, Coster DJ. Endothelial repair in the rat cornea. Invest Ophthalmol Vis Sci. 1986;27:1199–1204. [PubMed] [Google Scholar]
- 25.Gameiro PA, Laviolette LA, Kelleher JK, Iliopoulos O, Stephanopoulos G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J Biol Chem. 2013;288:12967–12977. doi: 10.1074/jbc.M112.396796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, et al. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013;155:160–171. doi: 10.1016/j.cell.2013.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878 [DOI] [PubMed] [Google Scholar]
- 28.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333 [DOI] [PubMed] [Google Scholar]
- 29.Meirelles FV, Smith LC. Mitochondrial genotype segregation in a mouse heteroplasmic lineage produced by embryonic karyoplast transplantation. Genetics. 1997;145:445–451. doi: 10.1093/genetics/145.2.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda K, Takahashi S, Onishi A, Hanada H, Imai H. Replicative advantage and tissue-specific segregation of RR mitochondrial DNA between C57BL/6 and RR heteroplasmic mice. Genetics. 2000;155:777–783. doi: 10.1093/genetics/155.2.777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lane N. The problem with mixing mitochondria. Cell. 2012;151:246–248. doi: 10.1016/j.cell.2012.09.028 [DOI] [PubMed] [Google Scholar]
- 32.Bayona-Bafaluy MP, Acín-Pérez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu P, Pérez-Martos A, Fernández-Silva P, Bai Y, Enríquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003; 31:5349–5355. doi: 10.1093/nar/gkg739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goios A, Pereira L, Bogue M, Macaulay V, Amorim A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007;17:293–298. doi: 10.1101/gr.5941007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enríquez JA. Mind your mouse strain. Nat Metab. 2019;1:5–7. doi: 10.1038/s42255-018-0018-3 [DOI] [PubMed] [Google Scholar]
- 35.Lechuga-Vieco AV, Justo-Méndez R, Enríquez JA. Not all mitochondrial DNAs are made equal and the nucleus knows it. IUBMB Life. 2021; 3:511–529. doi: 10.1002/iub.2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yonova-Doing E, Calabrese C, Gomez-Duran A, Schon K, Wei W, Karthikeyan S, Chinnery PF, Howson JMM. An atlas of mitochondrial DNA genotype-phenotype associations in the UK Biobank. Nat Genet. 2021;53:982–993. doi: 10.1038/s41588-021-00868-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bray AW, Ballinger SW. Mitochondrial DNA mutations and cardiovascular disease. Curr Opin Cardiol. 2017;32:267–274. doi: 10.1097/HCO.0000000000000383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraja AT, Liu C, Fetterman JL, Graff M, Have CT, Gu C, Yanek LR, Feitosa MF, Arking DE, Chasman DI, et al. Associations of mitochondrial and nuclear mitochondrial variants and genes with seven metabolic traits. Am J Hum Genet. 2019;104:112–138. doi: 10.1016/j.ajhg.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaz Fragoso CA, Manini TM, Kairalla JA, Buford TW, Hsu FC, Gill TM, Kritchevsky SB, McDermott MM, Sanders JL, Cummings SR, et al. Mitochondrial DNA variants and pulmonary function in older persons. Exp Gerontol. 2019;115:96–103. doi: 10.1016/j.exger.2018.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hyslop LA, Blakeley P, Craven L, Richardson J, Fogarty NM, Fragouli E, Lamb M, Wamaitha SE, Prathalingam N, Zhang Q, et al. Towards clinical application of pronuclear transfer to prevent mitochondrial DNA disease. Nature. 2016;534:383–386. doi: 10.1038/nature18303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang E, Wu J, Gutierrez NM, Koski A, Tippner-Hedges R, Agaronyan K, Platero-Luengo A, Martinez-Redondo P, Ma H, Lee Y, et al. Mitochondrial replacement in human oocytes carrying pathogenic mitochondrial DNA mutations. Nature. 2016;540:270–275. doi: 10.1038/nature20592 [DOI] [PubMed] [Google Scholar]
- 42.Barritt J, Willadsen S, Brenner C, Cohen J. Cytoplasmic transfer in assisted reproduction. Hum Reprod Update. 2001;7:428–435. doi: 10.1093/humupd/7.4.428 [DOI] [PubMed] [Google Scholar]
- 43.Plotnikov EY, Khryapenkova TG, Galkina SI, Sukhikh GT, Zorov DB. Cytoplasm and organelle transfer between mesenchymal multipotent stromal cells and renal tubular cells in co-culture. Exp Cell Res. 2010;316:2447–2455. doi: 10.1016/j.yexcr.2010.06.009 [DOI] [PubMed] [Google Scholar]
- 44.Islam MN, Das SR, Emin MT, Wei M, Sun L, Westphalen K, Rowlands DJ, Quadri SK, Bhattacharya S, Bhattacharya J. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat Med. 2012;18:759–765. doi: 10.1038/nm.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hayakawa K, Esposito E, Wang X, Terasaki Y, Liu Y, Xing C, Ji X, Lo EH. Transfer of mitochondria from astrocytes to neurons after stroke. Nature. 2016;535:551–555. doi: 10.1038/nature18928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCully JD, Cowan DB, Pacak CA, Toumpoulis IK, Dayalan H, Levitsky S. Injection of isolated mitochondria during early reperfusion for cardioprotection. Am J Physiol Heart Circ Physiol. 2009;296:H94–H105. doi: 10.1152/ajpheart.00567.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Røyrvik EC, Burgstaller JP, Johnston IG. mtDNA diversity in human populations highlights the merit of haplotype matching in gene therapies. Mol Hum Reprod. 2016;22:809–817. doi: 10.1093/molehr/gaw062 [DOI] [PubMed] [Google Scholar]
- 48.Theophilus BDM, Rapley R, Wartiovaara A, Syvänen A-C. PCR mutation detection protocols. Methods Mol Biol. 2002;187:57–63. doi: 10.1007/978-1-60761-947-512013752 [Google Scholar]
- 49.Johnston IG, Jones NS. Evolution of cell-to-cell variability in stochastic, controlled, heteroplasmic mtDNA populations. Am J Hum Genet. 2016;99:1150–1162. doi: 10.1016/j.ajhg.2016.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: an open platform for biomedical image analysis. Mol Reprod Dev. 2015;82:518–529. doi: 10.1002/mrd.22489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fernández-Vizarra E, Ferrín G, Pérez-Martos A, Fernández-Silva P, Zeviani M, Enríquez JA. Isolation of mitochondria for biogenetical studies: an update. Mitochondrion. 2010;10:253–262. doi: 10.1016/j.mito.2009.12.148 [DOI] [PubMed] [Google Scholar]
- 53.Vives-Bauza C, Yang L, Manfredi G. Methods in cell biology. Methods Cell Biol. 2007; 80:155–171. doi: 10.1016/S0091-679X(06)80007-5 [DOI] [PubMed] [Google Scholar]
- 54.Martínez-Bartolomé S, Navarro P, Martín-Maroto F, López-Ferrer D, Ramos-Fernández A, Villar M, García-Ruiz JP, Vázquez J. Properties of average score distributions of SEQUEST: the probability ratio method. Mol Cell Proteomics. 2008;7:1135–1145. doi: 10.1074/mcp.M700239-MCP200 [DOI] [PubMed] [Google Scholar]
- 55.Navarro P, Vázquez J. A refined method to calculate false discovery rates for peptide identification using decoy databases. J Proteome Res. 2009;8:1792–1796. doi: 10.1021/pr800362h [DOI] [PubMed] [Google Scholar]
- 56.García-Marqués F, Trevisan-Herraz M, Martínez-Martínez S, Camafeita E, Jorge I, Lopez JA, Méndez-Barbero N, Méndez-Ferrer S, Del Pozo MA, Ibáñez B, et al. A novel systems-biology algorithm for the analysis of coordinated protein responses using quantitative proteomics. Mol Cell Proteomics. 2016;15:1740–1760. doi: 10.1074/mcp.M115.055905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trevisan-Herraz M, Bagwan N, García-Marqués F, Rodriguez JM, Jorge I, Ezkurdia I, Bonzon-Kulichenko E, Vázquez J. SanXoT: a modular and versatile package for the quantitative analysis of high-throughput proteomics experiments. Bioinformatics. 2019;35:1594–1596. doi: 10.1093/bioinformatics/bty815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Navarro P, Trevisan-Herraz M, Bonzon-Kulichenko E, Núñez E, Martínez-Acedo P, Pérez-Hernández D, Jorge I, Mesa R, Calvo E, Carrascal M, et al. General statistical framework for quantitative proteomics by stable isotope labeling. J Proteome Res. 2014;13:1234–1247. doi: 10.1021/pr4006958 [DOI] [PubMed] [Google Scholar]
- 59.Ben Chorin A, Masrati G, Kessel A, Narunsky A, Sprinzak J, Lahav S, Ashkenazy H, Ben-Tal N. ConSurf-DB: An accessible repository for the evolutionary conservation patterns of the majority of PDB proteins. Protein Sci. 2020;29:258–267. doi: 10.1002/pro.3779 [DOI] [PMC free article] [PubMed] [Google Scholar]