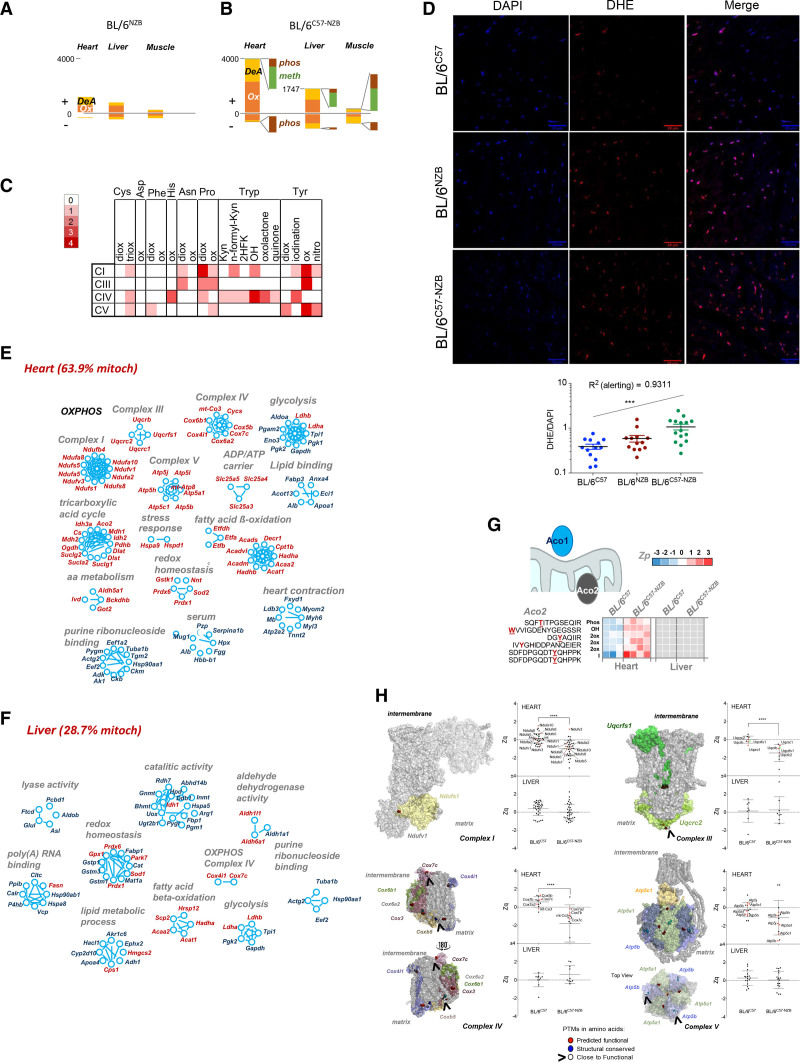
Figure 5.
Divergent nonpathologic mitochondrial DNA heteroplasmy causes generalized oxidative damage to cardiac mitochondrial proteins. A and B, Quantification of peptides containing posttranslational modifications (PTMs) that were significantly more abundant (+) or less abundant (–) in the indicated tissues of (A) BL/6NZB mice (n=3) or (B) BL/6C57-NZB mice (n=4) relative to BL/6C57 mice (n=3). C, Frequency of modified amino acid and type of oxidative alteration in proteins of the indicated respiratory complexes. Red intensity is proportional to PTM abundance; white represents undetected PTM. D, Representative dihydroethidium staining (top) and derived reactive oxygen species quantification (bottom) in heart slices of 20-week-old mice of the indicated genotype. Each dot in the chart represents a different tissue slide. ***P<0.001 (1-way analysis of variance test for linear trend; n=4 per strain). E and F, Inventory of proteins harboring PTMs grouped by function in (E) heart and (F) liver (12-week-old homoplasmic mice, n=3; 12-week-old heteroplasmic mice, n=4). Red text indicates mitochondrial localization. G, Aco2 (aconitase 2) peptides with the indicated PTMs and quantitative changes observed between BL/6C57 homoplasmic and BL/6C57-NZB heteroplasmic heart and liver tissues. Color is graded according to the log2 transformation of the ratio between the sample and the internal standard peptide level intensities (Zp). Gray indicates that the PTM was not detected. H, Protein models showing subunits of the indicated respiratory complexes. PTMs on amino acids with predicted functional relevance (exposed residues with a conservation score = 8 or 9) are highlighted in red. PTMs on structural residues (buried residues with a conservation score = 9) are highlighted in blue. Modified residues close to a functional residue are shown in white. The graphs show the standardized log2 ratios of the indicated complex proteins (Zq) or the standardized, protein-corrected log2 ratios of peptides (Zpq) in heart and liver between the BL/6C57 homoplastic and BL/6C57-NZB heteroplasmic mice. Colored dots indicate proteins exposed to the matrix side of the inner mitochondrial membrane (red) or to the intermembrane space (green) harboring PTMs. 2HFK indicates Trp to 2-hydroxy formyl kynurenine; 2ox, dioxidation; 3ox, trioxidation; DeA, deamidation; Kyn, Trp to kynurenine; meth, methylation; OH, Trp to hydroxytryptophan; Ox, oxidation; OXPHOS, oxidative phosphorylation; and phos, phosphorylation.