Summary
During embryo development, human primordial germ cells (hPGCs) express a naive gene expression program with similarities to pre-implantation naive epiblast (EPI) cells and naive human embryonic stem cells (hESCs). Previous studies have shown that TFAP2C is required for establishing naive gene expression in these cell types, however the role of additional naive transcription factors in hPGCs is not known. Here, we show that unlike TFAP2C, the naive transcription factors KLF4 and TFCP2L1 are not required for the induction of hPGC-like cells (hPGCLCs) from hESCs, and they have no role in establishing and maintaining the naive-like gene expression program with extended time in culture. Taken together, our results suggest a model whereby the molecular mechanisms that drive naive gene expression in hPGCs/hPGCLCs are distinct from those in the naive EPI/hESCs.
Keywords: Primordial Germ Cells, PGCs, hPGCs, Pluripotency, KLF4, TFCP2L1, stem cells
Introduction
The first three weeks of human embryo development are characterized by extensive developmental progression from a relatively simple blastocyst at the end of week one post-fertilization (pf), to a complex conceptus embedded in the wall of the uterus in which the nascent embryonic sac forms, the embryonic disc acquires symmetry and gastrulation occurs. In addition, during this time, primordial germ cells (PGCs) are specified and the extraembryonic compartment increases in cell number and complexity in order to facilitate the patterning of the developing embryo and sustain the pregnancy. As the embryo implants into the uterus, the pluripotent cells transition through key pluripotent states. Namely, the inner cell mass (ICM) cells of the blastocyst progress into pre-implantation and early post implantation naive and formative EPI cells, followed by a transition into post-implantation primed EPI cells (Smith, 2017). As the primed EPI undergoes gastrulation, the pluripotent gene expression program is repressed as somatic cell fates are established. In contrast to developing somatic cells, newly specified hPGCs express a naive-like pluripotent gene expression program similar to the ICM/naive-EPI, including expression of diagnostic transcription factors KLF4, TFCP2L1 and TFAP2C (Tang et al., 2015; Chen et al., 2018). This has led to the hypothesis that mechanisms controlling naive pluripotent gene expression in the ICM/EPI are similar to those responsible for naive-like gene expression in hPGCs.
The hPGC lineage is specified in the early post-implantation embryo during the pluripotent transition from naive to primed (for a comprehensive review of PGC specification see Hancock et al., 2021). Using the mouse as a model, PGCs are specified from embryonic (E) day 6.25 epiblast cells in vivo (Ohinata et al., 2005), and formative epiblast-like cells (EpiLCs) in vitro (Hayashi et al., 2011). Once specified, mouse PGCs exhibit a pluripotent-like gene expression program that can be reset to self-renewing pluripotency in vitro through the derivation of embryonic germ cells (EGCs) from in vitro cultured PGCs (Leitch et al., 2013). In hPGCs, the naive-like pluripotent gene expression program is maintained until around week 10 pf (Li et al., 2017), after which the hPGCs heterogeneously repress the naive-like program while differentiating into oogonia and pro-spermatogonia. Although hPGCs express diagnostic naive transcription factors that are required for naive pluripotent self-renewal, hPGCs are considered unipotent as their only fate is to differentiate into gametes. Failure to repress the naive gene expression program in hPGCs is speculated to be associated with development of germ cell tumors (Gell et al., 2018; Schmoll, 2002), and transformation of hPGCs into EGCs in vitro (Shamblott et al., 1998). However unlike EGCs in the mouse which have a similar gene expression program to ESCs (Sharova et al., 2007), hPGCs do not exhibit robust self-renewal in vitro unless transduced with lentiviruses expressing OCT4 and SOX2 which are speculated to supplement the effect of KLF4 and cMYC (Bazley et al., 2015). Taken together, understanding the acquisition and control of naive-like pluripotent gene expression in hPGCs may provide critical insights into the similarities and differences between functional pluripotency in the EPI, the unipotency of hPGCs in the embryo, and the transformation of hPGCs into germ cell tumors in vivo and EGCs in vitro.
In the last decade a number of groups have developed media formulations to stabilize the self-renewing state of naive, formative and primed human pluripotent cells (Chan et al., 2013; Gafni et al., 2013; Hanna et al., 2010; Kinoshita et al., 2021; Takashima et al., 2014; Theunissen et al., 2014; Ware et al., 2014). The naive state is modelled in vitro by culturing hESCs or human induced pluripotent stem cell (hiPSCs) in a media called t2i/L/Go (t2iLGo) or 5iLA/F (5iLAF) (Takashima et al., 2014; Theunissen et al., 2014). Naive cells are refractory to differentiation in vitro, and instead require Wnt inhibition to dismantle the naive gene expression program in order to establish the differentiation-competent formative state of pluripotency (Rostovskaya et al., 2019). Culture of human cells in the formative pluripotent state requires transitioning naive hESCs or ICM explants into low levels of Activn, the Tankyrase inhibitor XAV939 and a retinoic acid receptor inverse agonist (AloXR) (Kinoshita et al., 2021). Finally primed human pluripotency is stabilized under conventional culture conditions in KSR/FGF2 and these cells are poised to rapidly differentiate (Thomson et al., 1998).
Modelling hPGC specification in vitro involves differentiating hPGC-like cells (hPGCLCs) from hESCs/hiPSCs (Irie et al., 2015; Sasaki et al., 2015; Tang et al., 2015). In humans, the transcription factor TFAP2C is required to both establish the naive-like gene expression in hPGC-like cells (Chen et al., 2018) and to open naive enhancers to promote expression of the naive transcriptome in hESCs (Pastor et al., 2018). Similar to TFAP2C, KLF family members also regulate transcription in naive hESCs, with KLF4-KLF17 transcription factors responsible for binding young transposable elements (TE) at sites called TE Enhancers (TEENhancers) to drive expression of neighboring genes (Pontis et al., 2019). The transcription factor that functions synergistically with KLF4 to maintain naive pluripotency in t2iLGo is Transcription Factor CP2-like protein 1 (TFCP2L1) (Takashima et al., 2014). TFCP2L1 and KLF4 function downstream of Wnt/β-catenin to induce and maintain the naive pluripotent state in the mouse (Qiu et al., 2015). However, it is unclear whether KLF4 and TFCP2L1 regulate the specification and establishment of naive-like gene expression hPGCLCs.
In order to address this, we characterized KLF4 and TFCP2L1 expression with induction of hPGCLCs from hESCs, and used CRISPR/Cas9 gene editing to address the function of these transcription factors in hPGCLC induction and hESC reversion to the naive state in 5iLAF.
Results
KLF4 protein is expressed before TFCP2L1 during hPGCLC induction from hESCs.
To characterize KLF4 and TFCP2L1 we differentiated hPGCLCs from hESCs through an incipient mesoderm-like cell (iMeLC) intermediate (Sasaki et al., 2015). Fluorescence activated cell sorting (FACS) was used to isolate hPGCLCs at day 4 (D4) of differentiation using conjugated antibodies that recognize Integrin alpha-6 (ITGA6) and epithelial cell adhesion molecule (EPCAM) which are expressed on the cell surface of hPGCLCs and separates them from the differentiating somatic cells (Chen et al., 2017; Sasaki et al., 2015). Semi quantitative reverse transcriptase polymerase chain reaction (RT-PCR) was performed on the hESCs and hPGCLCs showing that hPGC markers PRDM1, SOX17 and TFAP2C are significantly up regulated in hPGCLCs relative to undifferentiated hESCs as previously reported (Irie, et al., 2015, Sasaki et al., 2015; Kojima et al., 2017) and this is also the case for KLF4 and TFCP2L1 (Fig. 1a).
Figure 1. KLF4 and TFCP2L1 are dynamically expressed during hPGCLC specification.
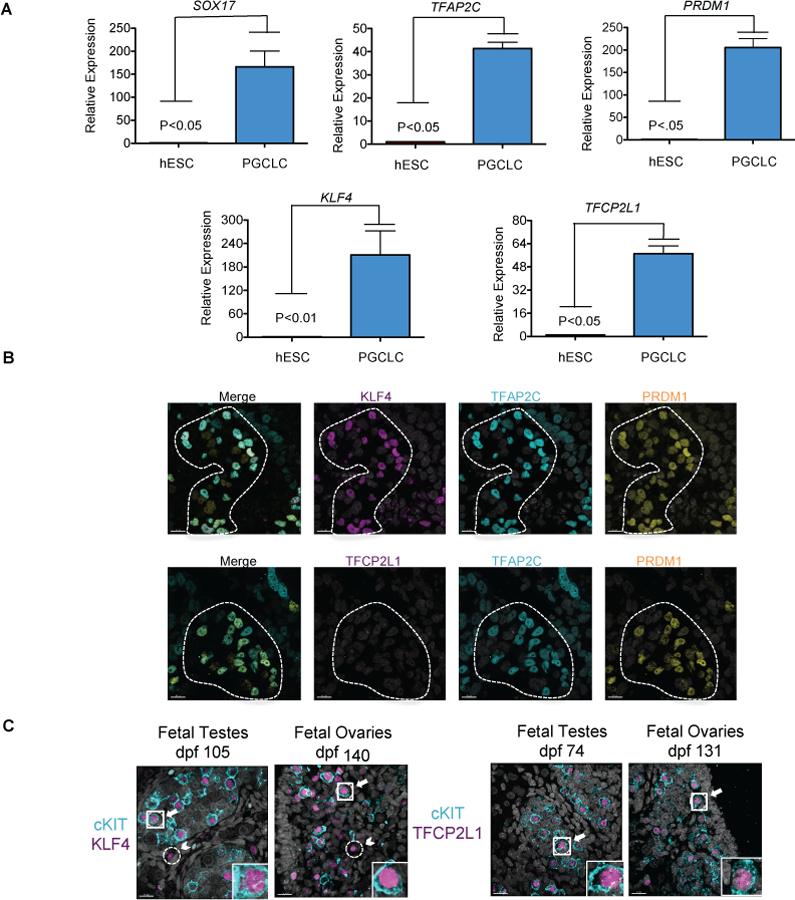
a. Quantitative real-time rt-PCR of KLF4 and TFCP2L1 in hESCs, and hPGCLCs from the UCLA1 hESC line at day 4 (D4) of differentiation. n=3 independent replicates of hESC and hPGCLCs. T-test was used to determine significance between these two groups. b. Representative immunofluorescence for KLF4 and TFCP2L1 in TFAP2C/PRDM1 double positive hPGCLCs at D4 (n= 8 aggregates of UCLA8). Scale bars show 15 microns. c. Representative immunofluorescence images of prenatal gonads (n=6) and testes (n=6) at indicated days post fertilization (dpf): Arrows highlight double positive hPGCs, Arrow head indicates single positive somatic cells
Next, we performed immunofluorescence of aggregates at D4 to evaluate KLF4 and TFCP2L1 protein expression in the newly induced hPGCLCs. We discovered that KLF4 is co-expressed with TFAP2C/PRDM1 in 75–90% of hPGCLCs, consistent with previous work showing more hPGCLCs to express KLF4 (Chen et al., 2018), while TFCP2L1 protein was not detected at this stage (Fig. 1b). To verify this result, we evaluated TFCP2L1 and KLF4 protein expression in hPGCLCs differentiated from UCLA2 (46, XY) hESCs (Supplementary Fig. 1a) and show again that KLF4 protein but not TFCP2L1 is detectable at D4 in hPGCLCs.
Given that both KLF4 and TFCP2L1 protein were previously identified in week 7 genital ridge BLIMP1+ hPGCs (Tang et al., 2015), we expanded upon this finding to better understand KLF4 and TFCP2L1 expression in cKIT+ hPGCs in vivo. To achieve this we examined either KLF4 or TFCP2L1 protein in cKIT+ hPGCs from day 74 (D74) to D140 pf in both male and female gonads (Fig. 1c). Although hPGC differentiation into oogonia and spermatogonia are initiated at around week 10 pf, cKIT+ hPGCs persist in the developing fetal gonad well into the second trimester (Gkountela et al., 2013). In 100% of human fetal testis (n=6) and fetal ovary (n=6) samples evaluated between D74-D140 we show that KLF4 and TFCP2L1 protein are expressed in the nucleus of cKIT+ hPGCs (Supplementary figure 1b). In addition, we discovered that KLF4 protein was expressed in rare cKIT negative cells, which were outside the seminiferous tubules of the fetal testes (Fig. 1c; arrowheads). Based on recent 10x Genomics data of prenatal ovaries and testes (Chitiashvili et al., 2020) the KLF4+/cKIT negative cells most likely correspond to rare subpopulations of granulosa progenitors, undefined interstitial cells, macrophages and endothelial cells. In contrast, TFCP2L1 protein was specific to cKIT+ hPGCs in both males and females (Fig. 1c). Taken together, KLF4 is upregulated with hPGCLC induction from hESCs and both KLF4 and TFCP2L1 are expressed throughout hPGC development in vivo with KLF4 also expressed in gonadal somatic cells.
KLF4 and TFCP2L1 are required to reset naive pluripotency in 5iLAF in vitro
To investigate the role of KLF4 and TFCP2L1 in hPGC development using the hPGCLC model, we created null mutant hESC sublines for each gene following picking and expanding CRISPR-Cas9 gene edited colonies. Specifically, we produced two KLF4 mutant hESC sublines in the UCLA1 (46, XX) hESC line (#35 and #43), and one in the H1 OCT4-GFP line (46, XY). For TFCP2L1, we produced two mutant sublines (#3 and #7) in the UCLA1 hESC line. Mutations in each of the sublines were confirmed by genotyping and Sanger sequencing of the isolated DNA (Supplementary Fig. 2a, b).
Previous studies showed that knockdown of KLF4 and TFCP2L1 in the naive media t2iLGo disrupts naive colony formation (Takashima et al., 2014). In order to evaluate the role of KLF4 and TFCP2L1 in under 5iLAF naive culture conditions, we reverted control and mutant hESC sublines to the naive state using 5iLAF and used FACS to quantify the percentage of CD75+ cells (naive) as well as CD24+ (primed) cells (Fig. 2a) (Collier et al., 2017). Using this approach, we found a significant reduction in the CD75+ / CD24-naive population relative to controls following naive reversion (Fig. 2b and supplementary figure 2c). The null mutant phenotype was confirmed by immunofluorescence for KLF4 and TFCP2L1 proteins, which were detectable in the dome shaped colonies of the control sublines but absence in the residual surviving mutant cells after naive reversion (Fig. 2c,d). To verify that the colonies in the control cultures were naive, we performed immunofluorescence for the naive marker KLF17 as well as the pan-pluripotent transcription factor OCT4 (Fig. 2c,d). These results show that KLF17 and OCT4 are expressed in the dome shaped colonies of the control cultures, whereas in the mutant sublines, KLF17 and OCT4 proteins were sporadically expressed in random cells not organized into colonies. Together these findings show that CRISPR/Cas9 gene editing of KLF4 and TFCP2L1 loci result in null mutations which cause a significant reduction in the capacity of primed hESCs to revert to the naive state in the media 5iLAF.
Figure 2. KLF4 and TFCP2L1 are required for naive pluripotency in 5iLAF.
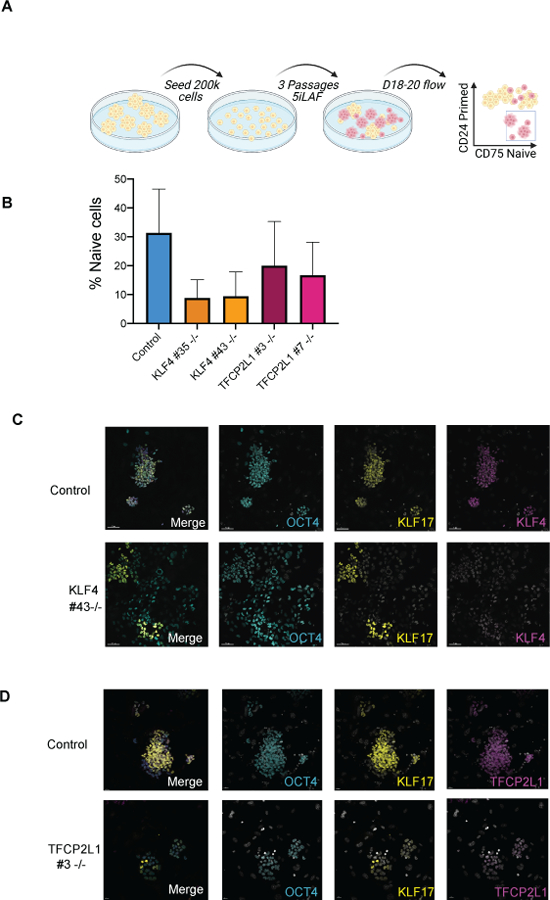
a. Experimental schematic for reversion of hESCs from the primed to naive state. b. Quantification of CD75 positive, CD24 negative (naive) populations in the KLF4 and TFCP2L1 mutants and controls as evaluated by flow cytometry between D17–20 (n=3 each mutant cell line) c. Representative immunofluorescence image at passage 3 (D17-D19) reversions with KLF4 control and KO subline Scale bars show 50 microns. d. Representative immunofluorescence image at passage 3 (D17-D19) reversions with TFCP2L1 control and KO subline. Scale bars show 20 microns.
KLF4 and TFCP2L1 are not required for naive pluripotent gene expression in hPGCLCs
Next, we evaluated the role of KLF4 in hPGC development by differentiating hPGCLCs from control and mutant hESC sublines. To quantify hPGCLC induction, we performed FACS for ITGA6/EPCAM at D4 (Fig. 3a–c) and show that all three KLF4 mutant sublines (UCLA1 #35, #43; H1 #39) produce ITGA6/EPCAM double positive hPGCLCs at percentages equivalent to their respective controls. Similarly, hPGCLC induction in the TFCP2L1 mutant sublines (#3, #7) were also unaffected relative to control (Fig. 3d and e). This suggests that hPGCLC induction from primed hESCs does not require KLF4 or TFCP2L1.
Figure 3. KLF4 and TFCP2L1 are not involved in hPGCLC induction from hESCs.
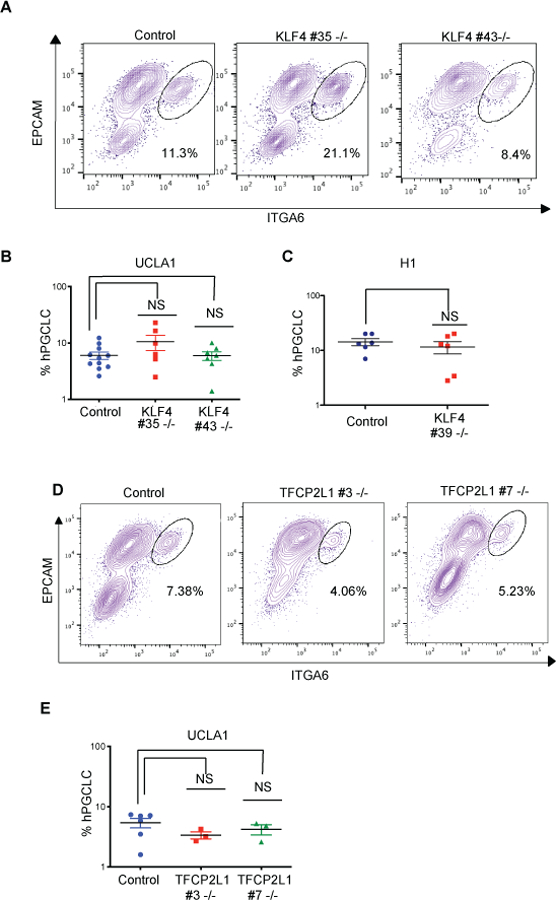
a. Representative FACS plots showing EPCAM/ITGA6 double-positive hPGCLCs in control and KLF4 mutant aggregates at D4. b. Quantification of the percentage ITGA6/EPCAM hPGCLCs in control (n= 12 biological replicates) and KLF4 UCLA1 mutant sublines (n=6 and n=7 biological replicates respectively). c. Quantification of the percentage ITGA6/EPCAM hPGCLCs in control (n= 6 biological replicates) and KLF4 H1 mutant subline (n=6 biological replicates). d. Representative FACS plots showing EPCAM/ITGA6 double-positive hPGCLCs in TFCP2L1 control and mutant aggregates at D4. e. Quantification of the percentage ITGA6/EPCAM hPGCLCs in control (n= 8 biological replicates) and TFCP2L1 UCLA1 mutant sublines (n=3 and n=3 biological replicates respectively).
Even though hPGCLC specification had occurred in the absence of KLF4, it is conceivable that naive gene expression in the resulting hPGCLCs is de-regulated. To address this, we performed RNA-Seq of ITGA6/EPCAM FACS-isolated KLF4 null mutant (H1 #39) and H1 control hPGCLCs at D4 of differentiation and show that the KLF4 null mutant hPGCLC transcriptome is overall very similar to that of control cells (Supplementary figure 3b). The rare exceptions include KLF4, (Supplementary Fig. 3c), which we also confirmed was not expressed at the protein level in hPGCLCs (Supplementary Fig. 3a). The remaining differentially expressed genes (DEGs) were unrelated to naive pluripotent gene expression (Supplementary figure 3c). To highlight the similarity in gene expression between KLF4 mutant and control hPGCLCs, we displayed diagnostic genes of hPGC development, naive and primed pluripotency, shared pluripotency and somatic cell differentiation on a heatmap (Supplementary Figure 3d). This display highlights the similarity in naive, primed and shared pluripotent gene expression across the data sets, as well as a robust early hPGC gene expression program similar to hPGCs in vivo. Taken together, these results indicate that while KLF4 is highly upregulated upon hPGCLC specification, it is not functionally required for induction of hPGCLCs from hESCs and is not required for establishing the transcriptional state of naive-like pluripotency in hPGCLCs.
TFCP2L1 and KLF4 are not required for hPGCLC maintenance or proliferation
Next, we used the extended culture system (Gell et al., 2020) to evaluate whether KLF4 and TFCP2L1 protein regulate hPGCLC survival and proliferation after induction of the initial hPGCLC population in the aggregates. To achieve this, we used FACS to isolate D4 hPGCLCs with ITGA6/EPCAM, and cultured the hPGCLCs for an additional 10–21 days in extended culture (D4CX) (Fig. 4a). During the course of extended culture hPGCLCs do not revert to EGCs (Gell et al., 2020), and maintain germ cell identity as shown by co-expression of SOX17, PRDM1, and TFAP2C (Fig. 4b). Furthermore, by D10 of extended culture, TFCP2L1 protein is detectable in PRDM1/TFAP2C double positive hPGCLCs (Fig. 4c), and continues to be positive through D4C21 (Supplementary Fig. 4b). This result indicates that unlike KLF4 which is expressed in D4 hPGCLCs (Fig. 1b, Supplementary Fig. 1a) and through D4C21 hPGCLCs in extended culture (Supplementary Fig. 4a), TFCP2L1 turns on later, by D4C10 (Fig. 4c) and continues to be expressed at D4C21 (Supplementary Fig. 4b).
Figure 4. hPGCLCs survive in extended culture and express TFCP2L1.
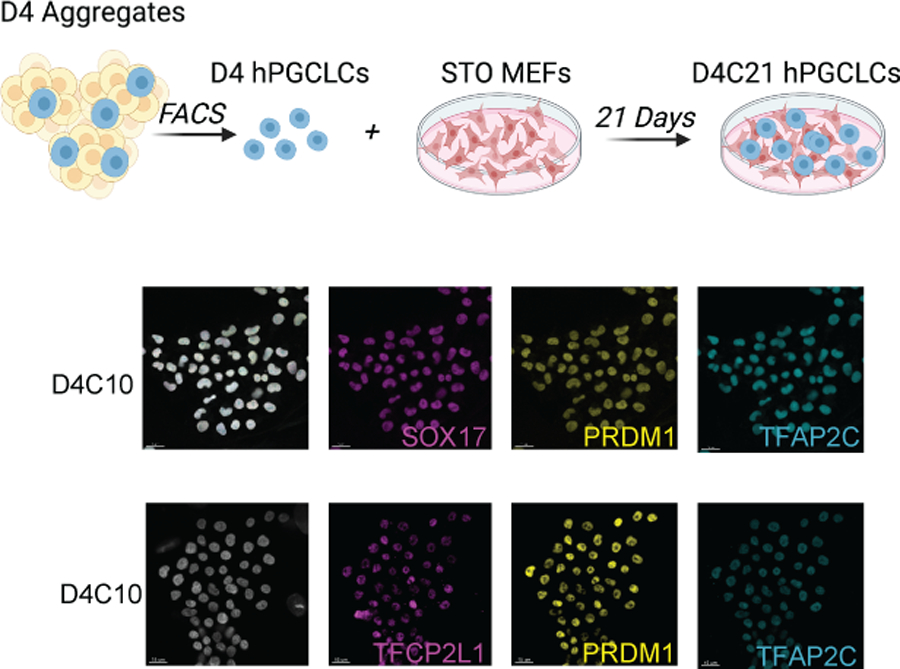
a. Schematic of D4 hPGCLCs grown in extended culture (C). Analysis of extended culture is referred to as D4CX, with CX representing days in extended culture and D4 = 4 days of PGCLC differentiation in the aggregates. b. Representative immunofluorescence of UCLA2 hPGCLCs at D4C10. c. Representative immunofluorescence image of wildtype UCLA2 PGCLCs at D4C10 (n= 10 colonies analyzed) scale bars = 15 microns.
Given that both TFCP2L1 and KLF4 protein are expressed in hPGCLCs during extended culture, we sought to evaluate the role of KLF4 and TFCP2L1 in hPGCLC proliferation by exposing cells to 5-ethynyl-2’-deoxyuridine (EdU) at D4C21, and calculating the percentage of EdU+ cells in the PRDM1/TFAP2C double positive hPGCLC colonies. Under control conditions, we show that approximately 30% of PRDM1/TFAP2C positive hPGCLCs incorporate EdU, and are therefore in S-phase of the cell cycle during the 4-hour window of EdU exposure (Fig. 5a–d). Analysis of EdU incorporation in hPGCLCs derived from the TFCP2L1 mutant sublines (#3 and #7), and the KLF4 mutant sublines (#35 and #43) show that the percentage of EdU+ cells is equivalent to the controls. Additionally, we counted the total number of hPGCLC colonies as defined by a cluster of at least 4 PRDM1/TFCP2L1 double positive cells within ~10 microns of each other. Although the normalized colony numbers of control hPGCLC extended cultures was variable, all data obtained from the knockout hPGCLC extended cultures were within the range of controls. We therefore concluded that knockout of KLF4 or TFCP2L1 did not strongly impact survival of hPGCLCs after 21-day maintenance in extended culture (Supplementary figure 4 a–c). Together these results show that KLF4 and TFCP2L1 protein are expressed in hPGCs and hPGCLCs however each are dispensable for hPGCLC induction, survival, proliferation and establishment of naive-like pluripotency (Fig. 5e).
Figure 5. KLF4 and TFCP2L1 are not required for proliferation in extended culture.
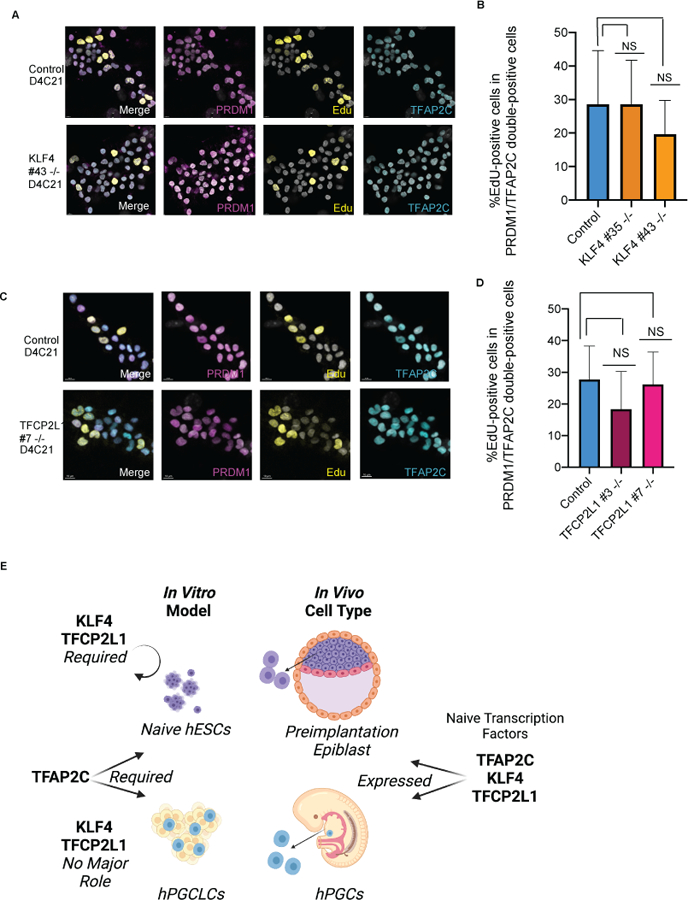
a. Representative immunofluorescence image of KLF4 control and KO hPGCLCs with EdU staining. Scale bars = 7 microns. b. Quantification of percentage of EdU+ cells within a hPGCLC colony. Error bars indicate n=4–10 colonies analyzed. c. Representative immunofluorescence image of TFCP2L1 control and KO hPGCLCs with EdU staining. Scale bars = 10 microns. d. Quantification of percentage of EdU+ cells within a hPGCLC colony. Error bars indicate n=4–10 colonies analyzed. e. Model showing expression patterns in vivo and divergent functional roles of the naïve transcription factors TFAP2C, KLF4 and TFCP2L1 in vitro models.
Discussion
Our results highlight notable differences between the mechanism responsible for establishing naive pluripotent gene expression in naive EPI/hESCs and hPGCs/hPGCLCs. In previous studies, it was shown that TFAP2C binds to and opens naive enhancers to regulate the establishment and maintenance of naive ground state pluripotency in 5iLAF and t2iLGo, as well as the specification and reacquisition of naive-like ground state pluripotency in hPGCLCs (Chen et al., 2018; Chen et al., 2019; Pastor et al., 2018). In the current study, we evaluated two additional regulators of naive hESC pluripotency, KLF4 and TFCP2L1, which similar to TFAP2C regulate self-renewal of naive cells in t2iLGo (Takashima et al., 2014). Here we show that KLF4 and TFCP2L1 also regulate naive pluripotency when reverting primed cells to the naive state in the media 5iLAF. However, unlike TFAP2C, our data showed that KLF4 and TFCP2L1 are not required for hPGCLC induction or maintenance in culture. This suggests that even though a common naive gene expression program can be identified in naive hESCs and the hPGCs, only TFAP2C has a role in establishing the naive-like state of pluripotency in both hPGCLCs and hESCs, whereas KLF4 and TFCP2L1 function only in t2iLGo and 5iLAF naive hESCs.
Mechanistically, TFCP2L1 has been shown to target the KLF4 promoter, and to also bind KLF4 protein (Wang et al., 2019), highlighting that these transcription factors function in combinatorial roles in naive hESCs. In the current study we observed KLF4 protein at the time of hPGCLC specification whereas TFCP2L1 protein was expressed later, suggesting that TFCP2L1 protein most likely does not act upstream of KLF4 in hPGCLC induction. This uncoupling of TFCP2L1 protein expression from KLF4 at the time of hPGCLC specification may be necessary to avoid re-establishment of functional naive pluripotency, and therefore a protective mechanism to reduce the risk of germ cell tumors. Indeed the TFCP2L1 locus is in a haplotype block associated with testicular germ cell tumors (Wang et al., 2017).
Our results show that neither TFCP2L1 nor KLF4 regulate proliferation and survival of the hPGCLC population in extended culture. Success of the hPGCLC extended culture system depends on addition of cAMP agonists including Forskolin and Rolipram (Gell et al., 2020), which increase the intracellular content of cAMP. This is important because in placental cell lines, the expression of TFCP2L1 protein is associated with cAMP signaling (Henderson et al., 2008). Collectively this could suggest a relationship between TFCP2L1 protein expression and intracellular cAMP which could be investigated in future studies.
Our studies indicate that KLF4 is dispensable for early hPGCLC development, either at the time of hPGCLC specification or during extended culture. In the mouse, KLF4 is reported to function redundantly with other KLF family members including KLF2 and KLF5 to regulate naive pluripotency by co-binding critical pluripotent transcription factors including POU5f1, SOX2, NANOG, ESRRB (Jiang et al., 2008). Our RNA-Seq of hPGCLCs, together with previously published data sets of FACS isolated hPGCs (Chen et al., 2019; Tang et al., 2015) indicates that KLF5 KLF11, KLF13 and KLF16 are all highly expressed in hPGCLCs/hPGCs, whereas KLF2 expression is below the limit of detection. Therefore, it could be hypothesized that similar to the mouse, other KLF family members substitute for KLF4 in regulating hPGC development. However, none of the KLF family members (including KLF17) were upregulated as a consequence of KLF4 knockout in hPGCLCs.
In summary, the requirement for KLF4 and TFCP2L1 in establishing naive pluripotency in human pluripotent stem cells but not in hPGCLCs likely highlights a critical mechanistic difference in these closely related embryonic cell types, and illustrates that establishment and maintenance of naive-like pluripotency in hPGCLCs is critically dependent on TFAP2C. Improved late-stage modeling of hPGC development including expression of gonadal hPGC markers, together with a more detailed understanding of naive-like transcription factors in germ cell development and pluripotency, collectively will provide a better understanding of causes of infertility and germ cell tumors in the context of healthy embryonic development.
Experimental Procedures
Human fetal samples
All prenatal gonads were obtained from the University of Washington Birth Defects Research Laboratory (BDRL), under the regulatory oversight of the University of Washington IRB approved Human Subjects protocol combined with a Certificate of Confidentiality from the Federal Government. All consented material was donated anonymously and carried no personal identifiers, therefore the use of the de-identified fetal tissue at UCLA was deemed exempt by the UCLA IRB under 45 CRF 46.102(f). Developmental age was documented by BDRL as days post fertilization using prenatal intakes, foot length, Streeter’s Stages and crown-rump length. All prenatal gonads documented with birth defect or chromosomal abnormality were excluded from this study.
Human ESC culture
The hESC lines in this study are as follows: UCLA1 (46, XX), UCLA2 (46, XY)(Diaz Perez et al., 2012), UCLA8 (46, XX)(Chen et al., 2017), and H1 OCT4-GFP (H1) (46, XY)(Gkountela et al., 2015). All hESCs were cultured on mitomycin C-inactivated mouse embryonic fibroblasts (MEFs) and split every 7 days using Collagenase type IV (GIBCO, 17104–019). hESC media was comprised of 20% knockout serum replacement (KSR) (GIBCO, 10828–028), 100mM L-Glutamine (GIBCO,25030–081), 1x MEM Non-Essential Amino Acids (NEAA) (GIBCO, 11140–050), 55mM 2-Mercaptoethanol (GIBCO, 21985–023), 10ng/mL recombinant human FGF basic (Proteintech HZ1285), 1x Penicillin-Streptomycin (GIBCO, 15140–122), and 50ng/mL primocin (InvivoGen, ant-pm-2) in DMEM/F12 media (GIBCO, 11330–032). All hESC lines used in this study are registered with the National Institute of Health Human Embryonic Stem Cell Registry and are available for research use with NIH funds. hESCs used in this study were routinely tested for mycoplasma (Lonza, LT07–418). All experiments were approved by the UCLA Embryonic Stem Cell Research Oversight Committee.
Realtime PCR
Undifferentiated hESCs, iMeLCs, D4 hPGCLCs (EPCAM/ITGA6), and D4 somatic cells (the EPCAM/ITGA6 negative cells) were re-suspended in 350 uL RLT buffer (QIAGEN) and RNA was extracted using RNeasy micro kit (QIAGEN). RNA was converted to cDNA using SuperScript® II Reverse Transcriptase (Invitrogen). Real-time quantitative PCR was performed using TaqMan® Universal PCR Master Mix (Applied Biosystems), and with Taqman probes detecting expression of GAPDH, TFAP2C, SOX17, PRDM1, KLF4, and TFCP2L1. Expression levels for genes of interest were normalized to housekeeping gene GAPDH in each cell type. To quantify relative expression in each cell type, expression levels was normalized to the expression level of hESCs. hPGCLCs were compared to hESCs in biological triplicate, and to iMeLCs and somatic cells in duplicate. P-values were calculated using two-tailed student T test.
Immunofluorescence
Aggregates collected at D4, and human fetal tissue samples were fixed in 4% PFA for 1 hour, washed twice for 15 minutes in PBS, stained with hemotoxylin, and mounted in histogel (Thermo Scientific). Samples were embedded into paraffin blocks and cut onto slides in 5 um-thick sections. Slides were deparaffinized and rehydrated through a series of xlene and ethanol series. For antigen retrieval, slides were heated to 95C in Tris-EDTA solution (10 mM Tris Base, 1 mM EDTA solution, .05% Tween-20, pH9.0). Sections were permeabilized (.05% Triton-100 in PBS) for 20 minutes and blocked in PBS containing 10% normal donkey serum for 1 hour. Primary antibodies incubated overnight at 4C. Antibodies included anti-TFAP2C (sc12762; 1:100), anti-SOX17 (GT15094; 1:100), anti-Blimp1 (9115S; 1:100), anti-KLF4 (AF3640; 1:100), anti-TFCP2L1 (AF5726; 1:100), and (cKIT A405 1:100). The next day, slides were washed, blocked for an additional 30 minutes, and stained with secondary antibody for 1 hour in their corresponding species-specific secondary antibody. Secondary antibodies included donkey anti-mouse 488 IgG (715–546-150; 1:200), donkey anti-mouse 594 IgG (A21447; 1:200), donkey anti-rabbit 488 IgG (711–545-152; 1:200), donkey anti-rabbit 594 IgG (711–585-152; 1:200), donkey anti-rabbit 647 IgG (711–605-152; 1:200), donkey anti-goat 488 IgG (705–546-147; 1:200), donkey anti-goat 594 IgG KLF4, TFCP2L1 (705–586-147; 1:200), donkey anti-goat 647 IgG KLF4, TFCP2L1 (A21447; 1:200). Dapi (xxx; 1:1000) was added during secondary antibody incubation and samples were mounted in ProLong Gold antifade reagent (Invitrogen).
hPGCLCs at D4C10 or D4C21 on chamber slides, and 5iLAF cells split onto coverslips in culture at P3 were washed and fixed in 4% PFA for 10 minutes. Cells were washed, permeabilized, blocked, and stained as described above. Primary antibodies were anti-KLF17 (042649 1:200), and anti-OCT4 (sc-5279 1:100). Secondary antibodies were incubated for 30 minutes.
For EdU analysis, cells in culture were incubated with EdU for 4 hours fixed, and detected using Click-iT™ EdU Cell Proliferation Kit for Imaging, Alexa Flour™ 488 dye before permeabilization.
Image quantification
hPGCLCs in aggregates were quantified in IMARIS 8.1 (Bitplane). For KLF4 and quantification, we counted the percentage of cells that were KLF4+ in the TFCP2C, PRDM1 double-positive hPGCLC population. This was repeated in 3 cell lines. In human fetal samples, we counted the percentage of cKIT positive hPGCs positive for KLF4 or TFCP2L1. This was repeated in 6 biological samples each of prenatal testes and ovaries.
In extended culture, total hPGCLC colonies in each well were counted and normalized to the number of cells plated for each sample. Each was performed with 3 technical replicates. For EdU quantification, the number of EdU positive cells in each hPGCLC colony was counted and EdU percentage of each hPGCLC colony was compared between the mutant and control-dervied hPGCLCs. Error bars on graphs indicated standard error.
hESC mutants made by CRISPR/Cas9
To make null-mutations for KLF4 and TFCP2L1, pairs of gRNAs were designed to target the functionally important, most N-terminus coding region of each gene. Guides were designed using https://zlab.bio/guide-design-resources, and cloned into PX459 vector (Ran et al., 2013). Pairs of guides were designed approximately at a 3 kB distance from each other in the genome. 4–6 days before nucleofection, UCLA1 or H1-OCT4-GFP hESCs were purified from MEFs and transferred to matrigel (BD) in mTeSR media (stemcell tech). At 70% confluence, cells were electroporated with 4 ug of each gRNA pair was using P3 Primary Cell 4D-Nucleofector® X Kit according to the manufacturer’s instructions (Lonza, V4XP-3024). 1 day following recovery, cells were dissociated with Accutase and replated on DR4 MEFs in a 6-well plate. Cells were treated with puromycin at a concentration of .35 ug/mL for 1 day. Once colonies emerge after 6–8 days, they are dissociated and plated at 10k and 50k densities in 10 cm plates. 48 colonies were picked when at the desired density afte 10 days. Colonies were split in half after 4 days. To determine homozygous mutants, we genotyped genomic DNA and chose colonies with the expected shorter band. Genotyping primers and gRNA sequences are listed in Supplementary table 1. To confirm bi-allelic mutantions, mutant bands were cloned into Blunt-PCR-Cloning vector using Zero Blunt PCR Cloning Kit (TheroFisher, K270020). 5–10 colonies from each band were picked and sequenced.
hPGCLC induction
hPGCLCs were induced as described previously (Chen et al., 2017). 1 hour before plating, 12-well plates are treated with human plasma fibronection (Invitrogen). hESCs were washed and dissociated in 0.05% trypsin for 5 minutes, and quenched with MEF media. The MEFs were removed by plating in 10-cm cell culture dishes, twice for 5 minutes each. Purified hESCs were spun, filtered with 100um filter, and seeded at a density of 200k per 12-well in iMeLC media including 15% KSR (GIBCO, 10828–028), 1x NEAA (GIBCO, 11140–050), 0.1mM 2-Mercaptoethanol (GIBCO, 21985–023), 1x Penicillin-Streptomycin-Glutamine (GIBCO, 10378–016), 1mM sodium pyruvate (GIBCO, 11360–070), 50ng/mL Activin A (Peprotech, AF-120–14E), 3mM CHIR99021 (Stemgent, 04–0004), 10mM of ROCKi (Y27632, Stemgent, 04–0012-10), and 50ng/mL primocin in Glasgow’s MEM (GMEM) (GIBCO, 11710–035). After 24 hours, cells were dissociated with trypsin, inactivated with trypsin inhibitor (Sigma), resuspended in PGLCC media, and plated in ultra-low cell attachment U-bottom 96-well plates (Corning) at a density of 3k cells/well. hPGCLC media is comprised of 15% KSR (GIBCO, 10828–028), 1x NEAA (GIBCO, 11140–050), 0.1mM 2-Mercaptoethanol (GIBCO, 21985–023), 1x Penicillin-Streptomycin-Glutamine (GIBCO, 10378–016), 1mM sodium pyruvate (GIBCO, 11360–070), 10ng/mL human LIF (Millipore, LIF1005), 200ng/mL human BMP4 (R&D systems, 314-BP), 50ng/mL human EGF (R&D systems, 236-EG), 10mM of ROCKi (Y27632, Stemgent, 04–0012-10), and 50ng/mL primocin in Glasgow’s MEM (GMEM) (GIBCO, 11710–035).
Flow cytometry and fluorescence activated cell sorting
D4 aggregates were collected and dissociated with .05% trypsin for 10 minutes. The dissociated cells were stained with conjugated cell-surface antibodies for at least 15 minutes. Antibodies included anti-ITA6-BV421 (BioLegend, 313624 1:60), anti-EPCAM-488 (BioLegend 324210; 1:60) for UCLA-derived lines and anti-ITA6–488 (BioLegend, 313608 1:60), anti-EPCAM-APC (BioLegend 324208; 1:60) for H1-derived lines. After at least 15 minutes, cells were washed with FACS buffer (1% BSA in PBS), resuspended in FACS buffer with 7AAD (BD PharMingen 559925; 1:40). Single-cell suspensions of hESCs were used as single-color compensation controls and evaluated each with 7AAD, anti-ITA6-BC421, and anti-EPCAM-488 for UCLA1-derived lines and anti-ITA6–488, anti-EPCAM-APC, and unstained GFP-OCT4-expressing cells for the H1-OCT4-GFP derived lines. For gating controls, fluorescence-minus-one controls were made against each fluorophore, staining the residual cells from the aggregate supernatant with each antibody, minus its respective control. Gating was then established based on the absence of signal in the cell population of interest. Double-positive ITGA6, EPCAM cells were analyzed using an ARIA-H Fluorescence Activated Cell Sorter and sorted into either 350 uL of FR10 media or RLT buffer. Analysis was performed using FlowJo version 10.
For flow cytometry, as described previously (Collier et al., 2017), cells in 5iLAF at passage 3 were dissociated using accutase, passed through a 40um strainer (BD) and resuspended in FACS buffer at equal cell numbers. Conjugated antibodies including anti-CD75-APC (ThermoFisher 50–0759-41; 1:20), anti-CD24-BV421 (BD 562789 1:40), anti-CD90.2-APC-Cy7 (BioLegend 105327 1:20), and fixable live-dead-APC-Cy7 (Fisher 50–169-66) were diluted in staining buffer (BD 563794) and used to resuspend cell pellet, staining in the dark for at least 15 minutes. Live, non-mouse cells, through exclusion of CD90.2 positive mouse cells, were gated and analyzed for their percentage of CD75 positive, CD24 negative populations. Analyses was performed on an LSR Fortessa cytometer in duplicate.
Primed to Naive Reversion
Cells were reverted to the naive ground state in 5iLAF as described previously (Theunissen et al., 2014). At day 7, primed hESCs were dissociated into single cells with accutase and re-plated on MEFs in hESC media with Y27632 (Stemgent, 04–0012-10) at a density of 200k cells/well per 6-well plate. After one day, media was changed to 5iLAF media including a 50/50 mixture of DMEM/F12 (Gibco 11320–033) and Neurobasal (Gibco 21103–049), 1X N2 (Gibco 17502–048), 1X B27 (17504–044), 20 ng/mL rhLIF (Millipore LIF1005), 1 mM GlutaMAX (Gibco 35050–061), 1% NEAA (Gibco 11140–050), .1 mM 2-Mercaptoethanol (GIBCO, 21985–023), 1x Penicillin-Streptomycin (Gibco 15140–122), 50 ug/mL BSA (Gibco A10008–01), 1 mM PD0325901 (Stemgent 04–006-02), 1 mM IM-12 (BML-WN102–0005), .05 mM SB590885 (R&D 2650/10), 1 uM WH-4–023 (A Chemtek H620061), 10 uM Y-27632, 20 ng/mL Activin A (Peprotech AF-120–14E), 8 ng/mL FGF2 (Proteintech HZ1285), .50% KSR (Gibco 10828–028), and 1X primocin (Invitrogen (ant-pm-2). Media is changed daily and cells are passaged every 5 days at a ratio between 1:1 and 1:3 until robust colonies emerge.
RNA sequencing library preparation and data analysis
Total RNA was extracted from H1-OCT4-GFP sorted D4 hPGCLCs using RNeasy micro kit (Qiagen 74004). Total RNA was reverse transcribed and cDNA was amplified using Nugen RNA-Seq System V2 (Nugen, 7102–32). DNA was extracted using MinElute PCR purification kit (Qiagen) and quantified using the Qubit dsDNA High-Sensitivity Kit (Life Technologies). Amplified cDNA was fragmented using Covaris S220 Focused-ultrasonicator. RNA-sequencing libraries were generated using Nugen Rapid Library Systems (Nugen 0320–32). Libraries were subjected to single-end 125 bp sequening on HiSeq4000 with 6 indexed libraries per lane.
RNA sequencing analysis
Raw reads in qseq format obtained from the sequencer were first converted to fastq files with a customized perl script. Read quality was evaluated with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). High-quality reads were aligned to the hg19 human reference genome using Tophat (v 2.0.13) by using “-no-coverage-search” option, allowing up to two mismatches, and only keeping unique reads. The number of unique mappable reads was quantified by HTseq (v 0.5.4) under default parameters (Supplementary Table 2). Expression levels were determined by RPKM (reads per kilobase of exons per million aligned reads) in R using customized scripts. For RNAseq of published datasets, GSE76970 (Chen et al., 2017) and GSE93126 (Pastor et al., 2016), processed data of the raw read counts of each gene was utilized, with the same downstream analysis.
Extended culture
D4 hPGCLCs were cultured (C) for an additional 10 or 21 days as described previously(Gell et al., 2020). Sorted hPGCLCs were plated at densities ranging 200–3000 cells in either a chamber well (D4C10) or 24-well (D4C21) in FR10 media. FR10 medium(Ohta et al., 2017) contains 10% KSR, 2.5% FBS (Thermo Fisher Scientific, SH3007003), 1× NEAA (Gibco, 11140–050), 1 mM sodium pyruvate (Gibco, 11360070), 2 mM L-glutamine (Gibco, 25030081), 0.1 mM 2-mercaptoethanol (Gibco, 21985–023), 1× penicillin streptomycin (Gibco, 15140–122), 100 ng/mL SCF (PeproTech, 250–03), 10 μM forskolin (Sigma, F6886), 10 μM rolipram (Sigma, R6520), and 50 ng/mL primocin in Glasgow’s MEM (Gibco, 11710–035). For D4C21, hPGCLCs were dissociated at D4C10 using .05% trypsin for 3 minutes. Cells are spun down at 1.6 rpm for 5 minutes, carefully resuspended, and plated at a 1:2 ratio in chamber well. Media was replaced daily until readout.
Supplementary Material
Acknowledgements
The authors would like to thank Felicia Codrea, Jessica Scholes, and Jeffery Calimlim for FACS and Jinghua Tang for banking and culturing of the UCLA hESC lines. This work is supported by funds from an anonymous donor as well as NIH/NICHD R01 HD079546 (ATC), NIH/NIGMS P01 GM099134 (KP), and a Faculty Scholar grant from the Howard Hughes Medical Institute (KP). GH acknowledges the support of the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA Training Program for supporting this work. Human fetal tissue research is supported by a grant to Ian Glass at the University of Washington Birth Defects laboratory 5R24HD000836-53. Human conceptus tissue requests can be made to bdrl@u.washington.edu.
Footnotes
Declaration of Interests
The authors declare no competing interests
References
- Bazley FA, Liu CF, Yuan X, Hao H, All AH, De Los Angeles A, Zambidis ET, Gearhart JD, Kerr CL, 2015. Direct Reprogramming of Human Primordial Germ Cells into Induced Pluripotent Stem Cells: Efficient Generation of Genetically Engineered Germ Cells. Stem Cells Dev 24, 2634–2648. 10.1089/scd.2015.0100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y-S, Göke J, Ng J-H, Lu X, Gonzales KAU, Tan C-P, Tng W-Q, Hong Z-Z, Lim Y-S, Ng H-H, 2013. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13, 663–675. 10.1016/j.stem.2013.11.015 [DOI] [PubMed] [Google Scholar]
- Chen D, Liu W, Lukianchikov A, Hancock GV, Zimmerman J, Lowe MG, Kim R, Galic Z, Irie N, Surani MA, Jacobsen SE, Clark AT, 2017. Germline competency of human embryonic stem cells depends on eomesodermin. Biol. Reprod 97, 850–861. 10.1093/biolre/iox138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Liu W, Zimmerman J, Pastor WA, Kim R, Hosohama L, Ho J, Aslanyan M, Gell JJ, Jacobsen SE, Clark AT, 2018. The TFAP2C-Regulated OCT4 Naive Enhancer Is Involved in Human Germline Formation. Cell Reports 25, 3591–3602.e5. 10.1016/j.celrep.2018.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Sun N, Hou L, Kim R, Faith J, Aslanyan M, Tao Y, Zheng Y, Fu J, Liu W, Kellis M, Clark A, 2019. Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell Reports 29, 4568–4582.e5. 10.1016/j.celrep.2019.11.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitiashvili T, Dror I, Kim R, Hsu F-M, Chaudhari R, Pandolfi E, Chen D, Liebscher S, Schenke-Layland K, Plath K, Clark A, 2020. Female human primordial germ cells display X-chromosome dosage compensation despite the absence of X-inactivation. Nat Cell Biol 22, 1436–1446. 10.1038/s41556-020-00607-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier AJ, Panula SP, Schell JP, Chovanec P, Plaza Reyes A, Petropoulos S, Corcoran AE, Walker R, Douagi I, Lanner F, Rugg-Gunn PJ, 2017. Comprehensive Cell Surface Protein Profiling Identifies Specific Markers of Human Naive and Primed Pluripotent States. Cell Stem Cell 20, 874–890.e7. 10.1016/j.stem.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Perez SV, Kim R, Li Z, Marquez VE, Patel S, Plath K, Clark AT, 2012. Derivation of new human embryonic stem cell lines reveals rapid epigenetic progression in vitro that can be prevented by chemical modification of chromatin. Hum Mol Genet 21, 751–764. 10.1093/hmg/ddr506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z, Mukamel Z, Krupalnik V, Zerbib M, Geula S, Caspi I, Schneir D, Shwartz T, Gilad S, Amann-Zalcenstein D, Benjamin S, Amit I, Tanay A, Massarwa R, Novershtern N, Hanna JH, 2013. Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286. 10.1038/nature12745 [DOI] [PubMed] [Google Scholar]
- Gell JJ, Liu W, Sosa E, Chialastri A, Hancock G, Tao Y, Wamaitha SE, Bower G, Dey SS, Clark AT, 2020. An Extended Culture System that Supports Human Primordial Germ Cell-like Cell Survival and Initiation of DNA Methylation Erasure. Stem Cell Reports 10.1016/j.stemcr.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell JJ, Zhao J, Chen D, Hunt TJ, Clark AT, 2018. PRDM14 is expressed in germ cell tumors with constitutive overexpression altering human germline differentiation and proliferation. Stem Cell Res 27, 46–56. 10.1016/j.scr.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Li Z, Vincent JJ, Zhang KX, Chen A, Pellegrini M, Clark AT, 2013. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol 15, 113–122. 10.1038/ncb2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao W-W, Hargan-Calvopiña J, Chen P-Y, Clark AT, 2015. DNA Demethylation Dynamics in the Human Prenatal Germline. Cell 161, 1425–1436. 10.1016/j.cell.2015.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, Wang W, Li R, Yan J, Zhi X, Zhang Y, Jin H, Zhang W, Hou Y, Zhu P, Li J, Zhang L, Liu S, Ren Y, Zhu X, Wen L, Gao YQ, Tang F, Qiao J, 2015. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell 161, 1437–1452. 10.1016/j.cell.2015.05.015 [DOI] [PubMed] [Google Scholar]
- Hancock GV, Wamaitha SE, Peretz L, Clark AT, 2021. Mammalian primordial germ cell specification. Development 148. 10.1242/dev.189217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R, 2010. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U.S.A 107, 9222–9227. 10.1073/pnas.1004584107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M, 2011. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532. 10.1016/j.cell.2011.06.052 [DOI] [PubMed] [Google Scholar]
- Henderson YC, Frederick MJ, Wang MT, Hollier LM, Clayman GL, 2008. LBP-1b, LBP-9, and LBP-32/MGR Detected in Syncytiotrophoblasts from First-Trimester Human Placental Tissue and Their Transcriptional Regulation. DNA and Cell Biology 27, 71–79. 10.1089/dna.2007.0640 [DOI] [PubMed] [Google Scholar]
- Irie N, Weinberger L, Tang WWC, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA, 2015. SOX17 is a critical specifier of human primordial germ cell fate. Cell 160, 253–268. 10.1016/j.cell.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan Y-S, Loh Y-H, Cai J, Tong G-Q, Lim C-A, Robson P, Zhong S, Ng H-H, 2008. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol 10, 353–360. 10.1038/ncb1698 [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Barber M, Mansfield W, Cui Y, Spindlow D, Stirparo GG, Dietmann S, Nichols J, Smith A, 2021. Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency. Cell Stem Cell 28, 453–471.e8. 10.1016/j.stem.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch HG, Nichols J, Humphreys P, Mulas C, Martello G, Lee C, Jones K, Surani MA, Smith A, 2013. Rebuilding pluripotency from primordial germ cells. Stem Cell Reports 1, 66–78. 10.1016/j.stemcr.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, Fan X, Wu X, Guo H, Wang X, Zhu X, Li R, Yan J, Wei Y, Zhao Y, Wang W, Ren Y, Yuan P, Yan Z, Hu B, Guo F, Wen L, Tang F, Qiao J, 2017. Single-Cell RNA-Seq Analysis Maps Development of Human Germline Cells and Gonadal Niche Interactions. Cell Stem Cell 20, 858–873.e4. 10.1016/j.stem.2017.03.007 [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, Saitou M, Surani MA, 2005. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207–213. 10.1038/nature03813 [DOI] [PubMed] [Google Scholar]
- Ohta H, Kurimoto K, Okamoto I, Nakamura T, Yabuta Y, Miyauchi H, Yamamoto T, Okuno Y, Hagiwara M, Shirane K, Sasaki H, Saitou M, 2017. In vitro expansion of mouse primordial germ cell‐like cells recapitulates an epigenetic blank slate. EMBO J 36, 1888–1907. 10.15252/embj.201695862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Chen D, Liu W, Kim R, Sahakyan A, Lukianchikov A, Plath K, Jacobsen SE, Clark AT, 2016. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell 18, 323–329. 10.1016/j.stem.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Liu W, Chen D, Ho J, Kim R, Hunt TJ, Lukianchikov A, Liu X, Polo JM, Jacobsen SE, Clark AT, 2018. TFAP2C regulates transcription in human naive pluripotency by opening enhancers. Nat Cell Biol 20, 553–564. 10.1038/s41556-018-0089-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D, 2019. Hominoid-Specific Transposable Elements and KZFPs Facilitate Human Embryonic Genome Activation and Control Transcription in Naive Human ESCs. Cell Stem Cell 24, 724–735.e5. 10.1016/j.stem.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Ye S, Ruiz B, Zhou X, Liu D, Zhang Q, Ying Q-L, 2015. Klf2 and Tfcp2l1, Two Wnt/β-Catenin Targets, Act Synergistically to Induce and Maintain Naive Pluripotency. Stem Cell Reports 5, 314–322. 10.1016/j.stemcr.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostovskaya M, Stirparo GG, Smith A, 2019. Capacitation of human naïve pluripotent stem cells for multi-lineage differentiation. Development 146. 10.1242/dev.172916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A, Kim R, Chronis C, Sabri S, Bonora G, Theunissen TW, Kuoy E, Langerman J, Clark AT, Jaenisch R, Plath K, 2017. Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell 20, 87–101. 10.1016/j.stem.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, Nakamura S, Sekiguchi K, Sakuma T, Yamamoto Takashi, Mori T, Woltjen K, Nakagawa M, Yamamoto Takuya, Takahashi K, Yamanaka S, Saitou M, 2015. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell 17, 178–194. 10.1016/j.stem.2015.06.014 [DOI] [PubMed] [Google Scholar]
- Schmoll H-J, 2002. Extragonadal germ cell tumors. Annals of Oncology, Educational Book of the 27th ESMO Congress, 18–22 October, 2002: Nice, France 13, 265–272. 10.1093/annonc/mdf669 [DOI] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Piao Y, Shaik N, Sullivan T, Stewart CL, Hogan BLM, Ko MSH, 2007. Global gene expression profiling reveals similarities and differences among mouse pluripotent stem cells of different origins and strains. Dev Biol 307, 446–459. 10.1016/j.ydbio.2007.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A, 2017. Formative pluripotency: the executive phase in a developmental continuum. Development 144, 365–373. 10.1242/dev.142679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, Reik W, Bertone P, Smith A, 2014. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254–1269. 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WWC, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA, 2015. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell 161, 1453–1467. 10.1016/j.cell.2015.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, Lungjangwa T, Imsoonthornruksa S, Stelzer Y, Rangarajan S, D’Alessio A, Zhang J, Gao Q, Dawlaty MM, Young RA, Gray NS, Jaenisch R, 2014. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 471–487. 10.1016/j.stem.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM, 1998. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- Vértesy Á, Arindrarto W, Roost MS, Reinius B, Torrens-Juaneda V, Bialecka M, Moustakas I, Ariyurek Y, Kuijk E, Mei H, Sandberg R, van Oudenaarden A, Chuva de Sousa Lopes SM, 2018. Parental haplotype-specific single-cell transcriptomics reveal incomplete epigenetic reprogramming in human female germ cells. Nature Communications 9, 1873. 10.1038/s41467-018-04215-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Xiaohu, Wang Xiaoxiao, Zhang S, Sun H, Li S, Ding H, You Y, Zhang X, Ye S-D, 2019. The transcription factor TFCP2L1 induces expression of distinct target genes and promotes self-renewal of mouse and human embryonic stem cells. J. Biol. Chem 294, 6007–6016. 10.1074/jbc.RA118.006341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, McGlynn KA, Rajpert-De Meyts E, Bishop DT, Chung CC, Dalgaard MD, Greene MH, Gupta R, Grotmol T, Haugen TB, Karlsson R, Litchfield K, Mitra N, Nielsen K, Pyle LC, Schwartz SM, Thorsson V, Vardhanabhuti S, Wiklund F, Turnbull C, Chanock SJ, Kanetsky PA, Nathanson KL, Testicular Cancer Consortium, 2017. Meta-analysis of five genome-wide association studies identifies multiple new loci associated with testicular germ cell tumor. Nat. Genet 49, 1141–1147. 10.1038/ng.3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, Tesar PJ, Okada J, Margaretha L, Sperber H, Choi M, Blau CA, Treuting PM, Hawkins RD, Cirulli V, Ruohola-Baker H, 2014. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A 111, 4484–4489. 10.1073/pnas.1319738111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


