Abstract

The large family of polypeptide GalNAc-transferases (GalNAc-Ts) controls with precision how GalNAc O-glycans are added in the tandem repeat regions of mucins (e.g., MUC1). However, the structural features behind the creation of well-defined and clustered patterns of O-glycans in mucins are poorly understood. In this context, herein, we disclose the full process of MUC1 O-glycosylation by GalNAc-T2/T3/T4 isoforms by NMR spectroscopy assisted by molecular modeling protocols. By using MUC1, with four tandem repeat domains as a substrate, we confirmed the glycosylation preferences of different GalNAc-Ts isoforms and highlighted the importance of the lectin domain in the glycosylation site selection after the addition of the first GalNAc residue. In a glycosylated substrate, with yet multiple acceptor sites, the lectin domain contributes to orientate acceptor sites to the catalytic domain. Our experiments suggest that during this process, neighboring tandem repeats are critical for further glycosylation of acceptor sites by GalNAc-T2/T4 in a lectin-assisted manner. Our studies also show local conformational changes in the peptide backbone during incorporation of GalNAc residues, which might explain GalNAc-T2/T3/T4 fine specificities toward the MUC1 substrate. Interestingly, we postulate that a specific salt-bridge and the inverse γ-turn conformation of the PDTRP sequence in MUC1 are the main structural motifs behind the GalNAc-T4 specificity toward this region. In addition, in-cell analysis shows that the GalNAc-T4 isoform is the only isoform glycosylating the Thr of the immunogenic epitope PDTRP in vivo, which highlights the relevance of GalNAc-T4 in the glycosylation of this epitope. Finally, the NMR methodology established herein can be extended to other glycosyltransferases, such as C1GalT1 and ST6GalNAc-I, to determine the specificity toward complex mucin acceptor substrates.
Keywords: O-GalNAc glycosylation, GalNAc-Ts, mucin-1, NMR, molecular dynamics, cell analysis
Mucin-type (GalNAc-type) O-glycosylation is one of the most abundant and diverse forms of posttranslational modifications (PTMs) and is differentially regulated by a large family of polypeptide GalNAc-transferases (GalNAc-Ts).1 GalNAc-Ts are type-II membrane proteins that catalyze the transfer of N-acetylgalactosamine (GalNAc) from UDP-GalNAc to Ser/Thr residues (and possibly Tyr).1 While 20 GalNAc-Ts isoforms in humans initiate the O-GalNAc-type glycosylation (hereafter, O-glycosylation) with distinct, although partly overlapping, kinetic properties and acceptor substrate specificities,2 other types of protein O-glycosylation pathways are initiated by a less number of enzymes (ranging between one to four enzymes).3−5 No clear consensus motifs for O-glycosylation have been reported, making this PTM the most complex and differentially regulated type of protein glycosylation. GalNAc-Ts are localized in the Golgi apparatus and distributed among all organs with different expression levels in each tissue.1,5,6 Thus, the available repertoire of GalNAc-Ts and their localization determine which proteins are O-glycosylated and where O-glycans are attached. In this context, some glycoproteins with fewer acceptor sites are glycosylated by specific GalNAc-Ts (nonredundant glycosylation),7−11 while complex glycoproteins with multiple acceptor sites (e.g., mucins) are glycosylated by multiple GalNAc-Ts (redundant glycosylation).12
GalNAc-Ts contains a region in the Golgi lumen displaying an N-terminal catalytic that adopts a GT-A fold13 and belongs to CAZy family 27 and connected to a C-terminal lectin domain by a short flexible linker,14−17 which dictates the relative orientation between the catalytic and lectin domains. The GalNAc-Ts are unique among metazoan GTs because of the C-terminal GalNAc-binding lectin domain, which adopts a β-trefoil fold built from three homologous repeat subdomains, named α, β, and γ.17 However, only one or two specific subdomains (α, β, or γ) actively bind to the GalNAc moiety. For example, in GalNAc-T2, -T3, and -T4, only the α-subdomain recognizes the sugar moiety.2,14,18−22
GalNAc-Ts can be classified into two classes based on their short-range (acceptor sites adjacent to prior glycosites) and long-range (acceptor sites far from prior glycosites) glycosylating capabilities. The short-range glycosylation preferences account for the glycosylation of glycopeptide substrates where a proximal glycosite to the acceptor site binds to the catalytic domain promoting glycosylation (one to three residues from the prior glycosite).23,24 In contrast, the long-range glycosylation preferences refer to the binding of a prior glycosite to the lectin domain, which subsequently facilitates the recognition of the distant acceptor site (6 to ∼17 residues away from the acceptor site) by the catalytic domain, promoting the glycosylation event.12 Both glycosylation preferences operate on N- or C-terminal directions, and specifically, for the long-range glycosylation preferences, this behavior is dictated by the GalNAc-T isoform-specific interdomain flexible linker motion.25 GalNAc-T3/T4/T6/T12 preferentially glycosylate remote C-terminal sites from the prior N-terminal GalNAc site,2,14,18−22 whereas GalNAc-T1/T2/T14 exhibit the opposite preference.15,26,27
Much of the knowledge about the GalNAc-Ts family is derived from in vitro enzyme studies with the characterization of products by mass spectrometry (MS) analysis.18,21,26,28−30 In addition, studies of isogenic cell lines, animal models, and individuals with impaired GalNAc-T2/T3 mutations have provided information on the nonredundant functions of several GalNAc-T isoforms.31−33 At the molecular level, insights into the catalytic mechanism of GalNAc-Ts have been revealed by X-ray crystallography, NMR, and molecular dynamics (MD) simulations23,25,34,35 by employing short (glyco)peptides. Specifically, our previous results suggest that GalNAc-Ts adopt a UDP-GalNAc-dependent induced-fit mechanism.34
However, our understanding of the molecular basis of GalNAc-Ts substrate specificity and especially how these enzymes act in a coordinated manner on very complex substrates such as mucins are barely known. The best-studied model of coordinated O-glycosylation has been performed on the mucin 1 (MUC1) tandem repeats (TRs), which consist of repeating 20-mer units of GVT3S4APDT8RPAPGS14T15APPAH. In vitro and in cell studies have demonstrated that several GalNAc-Ts (GalNAc-T1/T2/T3/T11) initiate O-glycosylation in MUC1 and add GalNAc moieties to T3, T15, and S14 with different preferences and order of glycosylation.11,36 On the contrary, GalNAc-T4 is the only isoform requiring prior O-glycosylation of at least one site by the above isoforms to glycosylate in vitro the remaining acceptor sites, S4 and T8.10,22
O-Glycosylation induces specific alterations in the chemical environments of protein residues, which can be readily monitored by high-resolution NMR spectroscopy,37 allowing the determination of the glycosylation specificity for different acceptor sites. In this context, Gariépy and co-authors applied NMR to ascertain the order and kinetics of GalNAc-introduction in a MUC1-5TR substrate by GalNAc-T1 isoform.38 NMR allows the determination of how glycosylation affects the protein structure conformation, and in cases of clustered modifications, how one glycosylation site affects the subsequent glycosylation reaction. Particularly, Ser and Thr O-GalNAc glycosylation was reported to elicit extended-like structures in short MUC1 glycopeptides.39−42
On this basis, herein, we exploit NMR spectroscopy to characterize the full process of MUC1 O-glycosylation by GalNAc-T2/T3/T4 and to investigate the role of the lectin domain to guide catalysis (Figure 1).
Figure 1.

MUC1 O-glycosylation by multiple GalNAc-Ts. (A) MUC14TR template used in this study. (B) 3D structure of the GalNAc-Ts isoforms used in this study and complexed with a glycopeptide (GalNAc-T2 PDB ID: 5AJP; GalNAc-T3 PDB ID: 6S24; GalNAc-T4 PDB ID: 6H0B). The lectin binding site is highlighted and displays GalNAc in yellow sticks and the essential Asp residue, conserved in all isoforms, in pink sticks. The Asp established H-bonds with OH-3 and OH-4 of GalNAc, which are identified as dashed blue lines.
The glycosylation process of a four TR domain of MUC1 (MUC14TR) (Figure 1A) was explored by using the wild-type GalNAc-T2, -T3, and -T4 isoforms and their corresponding impaired lectin mutants GalNAc-T2D458R, GalNAc-T3D517H, and GalNAc-T4D459H, which contain a critical mutation in the conserved Asp residue located at the α-subdomain of the lectin domain (Figure 1B). Previous studies demonstrated that the mutation of this Asp to a positive charged residue (Arg or His) precludes the binding of GalNAc by the lectin domain.18,21,43
Through monitoring of the chemical shift perturbation (CSP) of the amide resonances in 1H/15N-HSQC spectra of the 15N-isotopically labeled MUC14TR, our studies revealed a stepwise mechanism for each isoform, confirming the distinct glycosylation preferences employed by each isoform and highlighting the relevance of the lectin domain in these enzymes. The alterations in the conformation of MUC14TR upon GalNAc incorporation along the O-glycosylation process were also monitored, and their implication on the GalNAc-Ts activity was also inferred. Finally, cellular assays were performed to define the GalNAc-T isoforms involved in glycosylating S4 and T8 in the MUC1 structure, revealing GalNAc-T4 isoform as the only isoform able of glycosylating the T8 of the immunogenic epitope PDTRP.
Results and Discussion
NMR Analysis of 15N-MUC14TR Glycosylation by the GalNAc-T2 Isoform—Role of the Lectin Domain
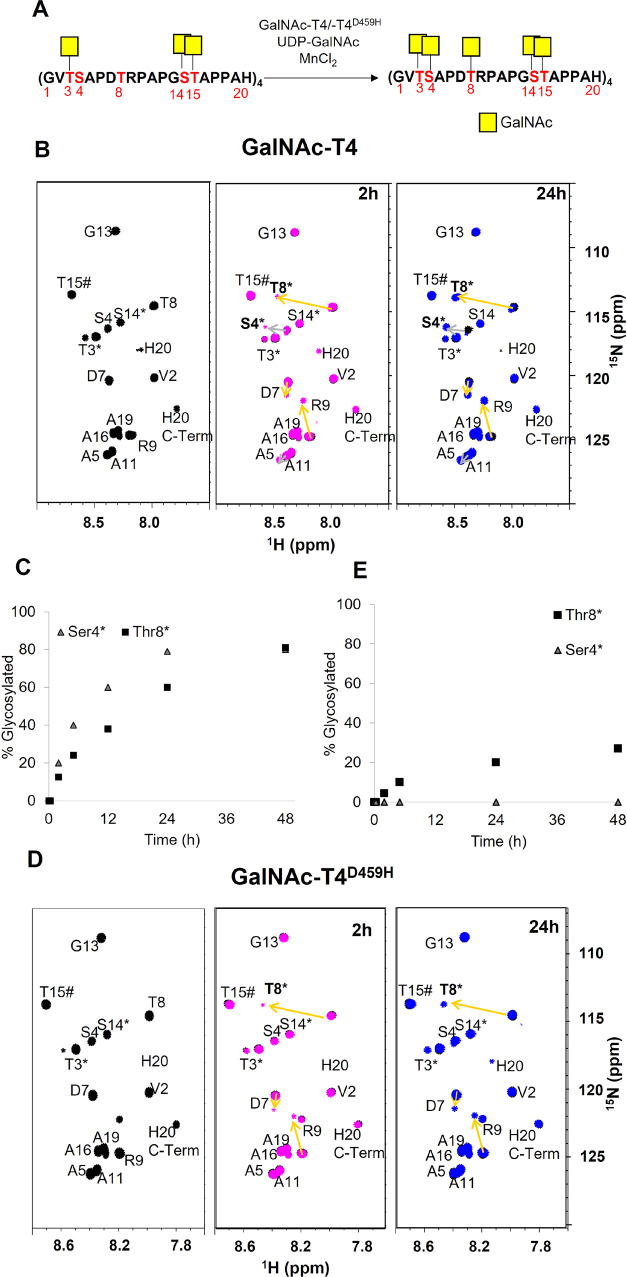
15N-MUC14TR was used as a probe to explore MUC1 O-glycosylation by NMR (see material and methods for details, Figures S1 and S2 and Table S1 of the Supporting Information). The 15N-MUC14TR molecular weight was verified by MS, and its NMR spectrum was assigned by standard procedures (Figure S2 and Table S1). The reaction with GalNAc-T2 and GalNAc-T2D458R impaired lectin mutant was carried under an excess of UDP-GalNAc and was followed over time through the acquisition of a series of 1H/15N-HSQC spectra. Figure 2 shows the selected 1H/15N-HSQC spectra of MUC14TR before and after addition of UDP-GalNAc at 20 min and 24 h. In the 1H/15N-HSQC spectra before the addition of UDP-GalNAc, apart from the N-terminals T3 and S4 and C-terminals A19 and H20 for which two peaks are visible, only one amide peak is visible for each of the residues in the same relative position on the MUC14TR (Figure 2B,C black spectra and Figure S2). The 1H,15N combined CSPs observed on the 1H/15N-HSQC spectra reflect the GalNAc transfer to MUC14TR (Figure S3A). In agreement with previously reported results,36 the first glycosylation events in the MUC14TR take place at the T15 moieties (T15*, where * denotes that the residue is covalently attached to GalNAc), as deduced by the strong CSPs observed at these residues (0.38 ppm) and their vicinal amino acids, with A16 experiencing a considerable CSP (0.28 ppm). Then, glycosylation at T3 is evidenced by its CSP (0.32 ppm) along with those observed for S4 (0.23 ppm), A5 (0.13 ppm), and V2 (0.08 ppm). Finally, glycosylation at S14 is verified by its corresponding CSP (0.15 ppm) along with those observed for the glycosylated T15* (0.23 ppm) (Figure 2B identified as T15#). Furthermore, CSPs are also detected for A16 (0.11 ppm) and G13 (0.10 ppm). The measurement of the cross-peak volumes in the 1H/15N-HSQC spectra allowed the quantification of the % glycosylation of each acceptor site in all four TRs. Indeed, after 20 min of reaction (fast glycosylation), complete glycosylation at T15 (blue arrows) and T3 (green arrows) was observed (Figure 2B). In contrast, only ∼70% of S14 residues (orange arrows) were glycosylated. Thus, one of the four TR domains is not glycosylated at S14 in MUC14TR and requires 24 h (exhaustive glycosylation) for completion (Figure 2B). MS analysis together with the full NMR assignment of the purified product confirmed that T15, T3, and S14 were fully glycosylated in all TRs, while S4 and T8 remained nonglycosylated (Figure S4 and Table S2).
Figure 2.
Glycosylation of MUC14TR catalyzed by the GalNAc-T2 and its GalNAc-T2D458R mutant. Table S5 details the experimental conditions. (A) Scheme of the MUC14TR glycosylation event in the presence of the GalNAc-T2 enzyme. The glycosylation sites are displayed in red in the amino acid sequence of MUC14TR; (B,C) 1H,15N-HSQC spectra recorded during the glycosylation process of MUC14TR by the GalNAc-T2 (B) and its GalNAc-T2D458R mutant (C) without UDP-GalNAc (black) and after 20 min (red) and 24 h (blue), using an excess of the donor UDP-GalNAc. Arrows indicate the observed CSP that occur as a result of the GalNAc additions at T15 (blue arrows), T3 (green arrows), and S14 (orange arrows). The * labeling in the amino acids on the spectra indicates that either Thr or Ser residues are glycosylated, and # labeling indicates shifts on glycosylated T15 after glycosylation of S14.
The combined 1H,15N-CSPs (Figures S3A) observed for the neighboring residues to the glycosylation site suggest local alterations of the chemical and structural environment of MUC14TR due to the incorporation of GalNAc units, which is consistent with previous observations with short MUC1 Tn-glycopeptides.39−42 The 1H and 15N NMR chemical shifts for the nonglycosylated MUC14TR are indicative of a major random coil-like conformational ensemble. However, besides the multiple conformations in solution, local conformations, typically found in the extended region of the Ramachandran plot for the three potential glycosylation regions of MUC1, VTSA (mixture of β-strand/inverse γ turn), DTR (inverse γ turn), and GSTA (polyproline II-like), were previously identified by Kinarsky et al.(39) Thus, according to the authors, nonglycosylated MUC1 has in these regions a natural tendency to adopt extended-like conformations. Herein, the chemical shift index (CSI)44,45 of the 13Cα,13Cβ of the naked MUC14TR and the glycosylated (MUC14TR-T3*S*14T15*) were calculated and compared (Figure S3B, gray and green, respectively), clearly indicating local deviations around VTS and GSTA from the random coil (small negative CSI values) toward an extended conformation (due to an increase of negative CSI values) after the introduction of GalNAc.
By using the GalNAc-T2D458R lectin mutant and after 20 min of reaction (Figure 2C), CSPs were only detected for T15 and the residues in its vicinity, and only after 90 min, glycosylation at T3 (∼20%) was detected (Figure S5). Even after 24 h, glycosylation at T3 was not completed (ca. 80%) (Figures 2C and S5), while only a very small fraction of S14 sites was glycosylated (5%). Thus, the glycosylation at T15 sites takes place prior to glycosylation at T3, and although the GalNAc-T2D458R mutant can glycosylate T3 and S14 residues, its catalytic efficiency is affected by the mutation at the lectin domain. These results suggest that GalNAc-T2 uses its lectin domain to drive glycosylation at S14 but also to a certain degree at T3 in MUC1. This agrees with previous studies, showing that GalNAc-T2 glycosylation at S14 requires the lectin domain,18 and further suggests that also the glycosylation of T3 is assisted by the lectin domain.
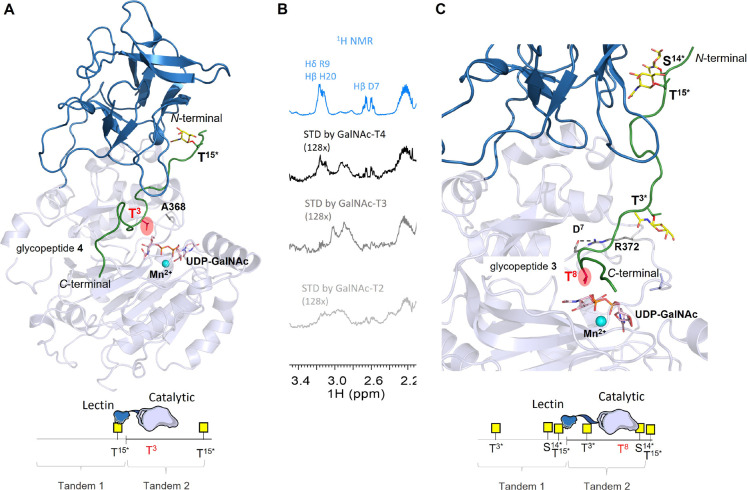
The preference of GalNAc-T2 toward T15 versus T3 in MUC14TR was also supported through NMR experiments limiting the UDP-GalNAc concentration with respect to the number of GalNAc-T2 potential acceptor sites in MUC14TR. First, we used the corresponding molar equivalents of UDP-GalNAc to glycosylate 4 out of the 20 putative glycosylation sites available at MUC14TR. The quantification of the cross-peak volumes in the 1H/15N-HSQC allowed assessing that, while ∼54% of the T15 residues were glycosylated (roughly two TR domains), only ∼7% of GalNAc-incorporation was observed at T3 (Figure S6). When the concentration of the UDP-GalNAc donor is sufficient to glycosylate 10 acceptor sites, all T15 residues were fully glycosylated, while T3 was glycosylated in only ∼56% and S14 did not show any trace of glycosylation (Figure S6). These results clearly demonstrate that GalNAc-T2 first glycosylates the T15 residues and initiates the addition of GalNAc at T3 before completing the glycosylation of all T15 residues. These observations are in agreement with the conclusions reported for MUC1 peptide fragments.18,36 Therefore, the reaction at the T3 site is assisted by the binding of the lectin domain of the enzyme to a prior glycosylated T15, evidencing that GalNAc-T2 displays a long-range N-terminal preference. To support this observation, MD simulations of 0.5 μs of GalNAc-T2 complexed with UDP-GalNAc/Mn2+ and MUC1 monoglycopeptide GVTSAPDTRPAPGST*APPA (1) were performed (underlined is the acceptor site and * indicates the glycosylated amino acid). Initially, we run extensive unrestrained MD simulations on the complexes. However, under these conditions and due to the flexibility of the protein and the glycopeptides, the residue to be glycosylated was not properly oriented in the catalytic domain, observing that the peptide fragment close to this domain explored additional areas of the protein. Therefore, a distance restriction between the oxygen of the hydroxyl group of the reactive Ser/Thr residue and C1 of GalNAc in UDP-GalNAc (distance O–C1 < 4.5 Å) was set in all MD simulations to maintain the correct orientation of the peptides in complex with the enzymes. The putative 3D structure of complex GalNAc-T2/1 was then generated using as starting coordinates the X-ray structure of a complex of GalNAc-T2 in an extended conformation (PDB entry 2FFU) and glycopeptide 1 (Figure 3A). The GalNAc at T15* of glycopeptide 1 was bound to the lectin domain. The complex GalNAc-T2/1 was stable throughout the MD trajectory (Figure S7A) suggesting that glycosylation at T3 can be lectin assisted, as inferred from the NMR experiments. According to the MD simulations, the methyl group of T3 is engaged in a CH−π interaction with the aromatic ring of F372 (distance 4.8 ± 0.4 Å, Figure 3A). Preference for glycosylation of Thr over Ser residues by GalNAc-T2 was previously observed.46,47 In this context, the stabilizing interaction between T3 and F372 is a structural feature that can explain why GalNAc-T2 prefers to glycosylate T3 rather than S4.
Figure 3.
Putative 3D models to explain the glycosylation of T3 and S14 of MUC14TR catalyzed by GalNAc-T2. (A) 3D view of complex GalNAc-T2/1 derived from a representative frame obtained from 0.5 μs of MD simulation. A key CH−π interaction between the β-methyl group of T3 and the aromatic ring of F372 is shown in dashed lines (distance 4.8 ± 0.4 Å). (B) 3D view of the binding site of GalNAc-T2/2 derived from a representative frame obtained from 0.5 μs of MD simulation. A relevant hydrogen bond, 38% populated through the whole trajectory, between the GalNAc at T6 of 2 (T15 in MUC14TR) and W265 is highlighted in dashed lines. In both structures, the catalytic domain is displayed in orange and the lectin domain is depicted in green. In the glycopeptides, the GalNAc units are shown in yellow sticks. UDP-GalNAc is displayed in gray sticks and Mn2+ is shown as a cyan sphere. Relevant residues of the glycopeptides and some residues of catalytic domain are also shown as sticks and in a different color. Schematic representation of the sequential glycosylation process at T3 and S14 of MUC14TR catalyzed by GalNAc-T2 is also displayed. These models highlight the N-terminal preference of GalNAc-T2.
Finally, with UDP-GalNAc donor concentration enough to glycosylate all putative 20 acceptor sites, it was possible to reach full T15 and T3 glycosylation; however, only 45% of S14 was glycosylated (Figure S6). These data indicate that glycosylation at S14 is only initiated when all T15 and T3 residues have already been glycosylated. Moreover, the severe impairment of S14 glycosylation by the GalNAcD458R mutation suggests that the S14 glycosylation might be promoted by a prior binding of the glycosylated T3 of the following TR to the lectin domain, a mechanism compatible with a long-range N-terminal preference. This lectin dependence to glycosylate S14 suggests that the glycosylation of the S14 located at TR4 of MUC14TR (TR4 S14) should have a considerably slower rate than the glycosylation of the other three S14 acceptor sites. The absence of a fifth tandem repeat in our construct precludes the glycosylation of TR4 S14 in a lectin-dependent manner. In fact, under an excess of UDP-GalNAc, GalNAc-T2 only takes 20 min (fast glycosylation) to glycosylate the three S14 residues (11 GalNAc units are attached to MUC14TR, providing a mass of 9831 m/z; Figure S3), when the last S14 residue requires 24 h (exhaustive glycosylation) to be glycosylated (note that at 16 h, the last S14 is still not glycosylated since only 85% of the S14 residues are glycosylated, data not shown). Due to the degeneracy of S14 cross peaks in MUC14TR, it was not possible to assure with absolute certainty that the last S14 to be glycosylated corresponds to TR4 S14. However, considering the lectin dependence of GalNAc-T2 to glycosylate S14, it is tempting to speculate that the last S14 residue to be glycosylated should be located at the TR4 of MUC14TR.
To understand the molecular basis of S14 glycosylation, we performed guided 0.5 μs MD simulations of GalNAc-T2 complexed with UDP-GalNAc/Mn2+ and a MUC1 diglycopeptide PAPGST*APPAHGVT*SAP (2) (Figure 3B). In this diglycopeptide, the relative position of S at GST*AP and glycosylated T* at GVT*SA are equivalent to S14 in one tandem repeat and T3* in the following tandem repeat of MUC14TR. Therefore T* in GVT*SA (T3* in a TR+1 of MUC14TR) is bound to the lectin domain to perform glycosylation at S in the GST*AP motif (S14 in a TR of MUC14TR). The computational study shows that the distance of the hydroxyl group of S at GST*AP (equivalent to S14) to C1 of UDP-GalNAc (distance <4.5 Å) is stable through the whole trajectory (Figures 3B and S7B). A crucial hydrogen bond (38% populated along the MD trajectory) between the carbonyl group of GalNAc of the adjacent glycosylated T* in GST*AP (T15* in a TR of MUC14TR) and the NH of the sidechain of W265 could be behind the stable and short distance between the S residue at GST*AP (S14 in a TR of MUC14TR) and UDP-GalNAc and should be an essential feature for the reaction to occur. In conclusion, our data pinpoints the N-terminal preference of GalNAc-T2 to glycosylate T3 and S14 in MUC14TR through a mechanism assisted by the lectin domain.
NMR Analysis of 15N-MUC14TR Glycosylation by the GalNAc-T3 Isoform and Its Comparison to that of GalNAc-T2
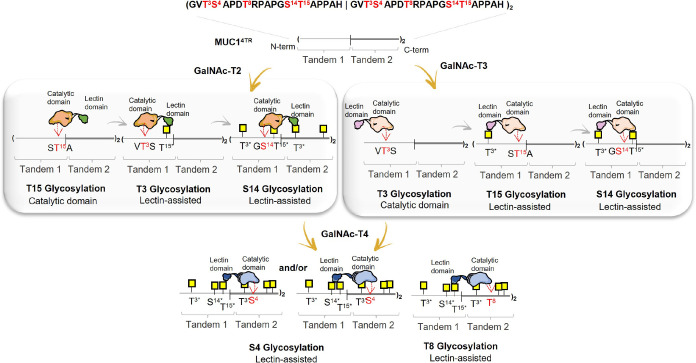
Although GalNAc-T3 glycosylates the same three MUC1 glycosylation sites found for GalNAc-T2, the order of glycosylation is different.36 To compare the glycosylation process of MUC14TR by GalNAc-T3 and its corresponding GalNAc-T3D517H mutant, we used the same methodology as described above for GalNAc-T2. GalNAc-T3 glycosylates first T3, then T15, and finally S14. Interestingly, after 20 min of reaction time, three out of four T3 and T15 residues were glycosylated, corresponding to the three TRs, while less than 5% of S14 was glycosylated (Figure S8). After 90 min, glycosylation at T3 and T15 residues was completed (100%). In contrast, for GalNAc-T3D517H, after 20 min (fast glycosylation), only glycosylation at T3 residues occurred (65%, corresponding to ∼3 TR domains), while after 90 min, glycosylation at T3 and T15 residues reached 100 and ∼25%, respectively. After 24 h (exhaustive glycosylation), the glycosylation of S14 residues is ∼70% in the presence of GalNAc-T3, while it is not detected in the presence of GalNAc-T3D517H (Figure S8). Moreover, even after 72 h, glycosylation at S14 is still not completed (ca. 88%) while remaining undetected with the mutant (Figure S8). Therefore, while GalNAc-T3-mediated glycosylation at T3 is only dependent on the natural affinity of the catalytic domain toward the GVTS region of MUC1, remote glycosylation at T15 and S14 is clearly assisted by the lectin domain. The observed long-range glycosylation by GalNAc-T3 agrees with the contribution of the lectin domain to the recognition of the GalNAc moiety located at T3, which promotes the GalNAc transfer to the T15 and S14 residues. This result is in agreement with the preference of GalNAc-T3 to glycosylate C-terminal remote acceptor sites prior to N-terminal glycosites.28,48
Interestingly, a comparison of the glycosylation rates on MUC14TR between both isoforms shows that GalNAc-T3 is slower to glycosylate S14 than GalNAc-T2. These differences in the ability to incorporate GalNAc at S14 agrees with a previous report using kinetics studies assisted by MS that demonstrate that GalNAc-T2 glycosylates S14 faster than GalNAc-T1/T3 isoforms.36 These differences are likely, not due to the different directionality of the lectin-mediated long-range glycosylation preferences since GalNAc-T1 and T2 isoforms display the same type of long-range glycosylation preferences.15,26,27 More likely, the divergent rates for the GalNAc-T3 and T2 isoforms are due to how the prior glycosylated sites affect the conformation and/or presentation of each TR domain, which determine the recognition by the catalytic domain of these isoforms. To evaluate this hypothesis, the conformation of MUC14TR-T3*T15* in solution was investigated. The preparation of MUC14TR-T3*T15* and its NMR and MS characterization are described in SI and displayed in Figure S9 and Table S3, respectively. The 1H,15N-CSP of MUC14TR-T3*T15* using naked MUC14TR as reference (Figure S10A) and the comparison of the 13C-CSI of MUC14TR-T3*T15* with the naked MUC14TR were scrutinized (Figure S10B), showing that glycosylation at T3 and T15 modulates the conformation of the surrounding residues. Specifically, the 13C-CSI difference for S14 before and after glycosylation at T15 is rather large (Figure S10B) and suggests a decrease in the flexibility of S14 and a shift toward an extended conformation. Previously, Kinarsky et al.(39) suggested that the GS14T15A region changes from polyproline II-like toward an inverse γ-turn conformation upon glycosylation of T15 in MUC1 short glycopeptides. In fact, similar behavior has been previously observed also by us.49 Based on this, it is tempting to propose that GalNAc-T2 has more affinity to the inverse γ-turn conformation of GS14T15*A and preferentially glycosylates the S14 acceptor site in this region than GalNAc-T3 and T1.
NMR Analysis of 15N-MUC14TR Glycosylation by the GalNAc-T4 Isoform—Differences in S4 and T8 Glycosylation
GalNAc-T4 isoform is the only isoform identified that is able to glycosylate in vitro S4 and T8 in the MUC14TR structure.10,22,50,51 To further explore this event by NMR, we used the GalNAc-glycopeptide MUC14TR-T3*S14*T15* as the acceptor substrate (Figure 4). The CSP analysis is now focused on the effects of glycosylation at S4 and T8 (Figures 4B,C and S11). Glycosylation at T8 induces a strong CSP (0.51 ppm, yellow arrow) at this residue along with others at R9 (0.29 ppm, yellow arrow) and D7 (0.10 ppm, yellow arrow) (Figures 4B and S11). In contrast, glycosylation at S4 only provides moderate CSP at the corresponding S4 (0.18 ppm, gray arrow) and A5 (0.07 ppm, gray arrow) (Figures 4B and S11).
Figure 4.
Glycosylation of MUC14TR-T3*S14*T15* by GalNAc-T4 and the GalNAc-T4D459H mutant. Table S5 details the experimental conditions. (A) Scheme of the MUC14TR-T3*S14*T15* glycosylation event catalyzed by GalNAc-T4. The glycosylation sites are displayed in red. (B,D) 1H/15N-HSQC of the MUC14TR-T3*S14*T15* product in the presence of GalNAc-T4 (B) and GalNAc-T4D459H (D). In black, only with MnCl2, and in magenta and blue, after addition of UDP-GalNAc (in excess) at 2 and 24 h, respectively. The arrows indicate CSP that occurs as result of GalNAc additions at S4 (gray arrows) and at T8 (yellow arrows). The * labeling in the amino acid on the spectra indicates glycosylation. (C,E) Time course of relative % of glycosylation at S4 and T8 residues by GalNAc-T4 (C) and GalNAc-T4D459H (E) (see the Experimental Section for details).
After 5 h of reaction time, S4 and T8 residues were glycosylated 40 and 24%, respectively, while after 48 h, ∼75–80% of the S4 and T8 residues were glycosylated (Figure 4C) indicating that one glycosylation is lacking for both S4 and T8. Nevertheless, the mass analysis of the product purified by HPLC displayed two masses, ∼11,255.6 m/z and ∼11,458.6 m/z, which corresponded to two different glycosylated MUC14TR that account for 18 and 19 GalNAc units, respectively (Figure S12).
The NMR assignment of the major product with 19 glycosylated sites out of 20 allowed us to unequivocally identify that the TR1 S4 is not glycosylated (Figures S12 and Table S4), suggesting that glycosylation of S4 by GalNAc-T4 is lectin domain-assisted and requires the binding of the lectin domain to a GalNAc moiety present at the preceding MUC1 TR domain. This is compatible with a long-range C-terminal preference. In this scenario, the GalNAc-T4 lectin domain should bind the GalNAc moiety at either S14 or T15 at a given TR of MUC14TR-T3*S14*T15* product to drive the glycosylation process onto S4 of the following TR domain. The analysis with the GalNAc-T4D459H lectin mutant was also performed (Figures 4D,E and S13). While S4 glycosylation was completely abolished in the presence of the mutant, a small fraction of T8 glycosylation occurred very slowly and progressively with the time of incubation (after 24 h, 20% of the T8 residues were glycosylated, reaching a maximum of 35% after 72 h). These results confirm the importance of the lectin domain in the glycosylation of S4 and demonstrate that the T8 glycosylation process is also severely affected in the presence of GalNAc-T4D459H (Figure 4), strongly suggesting that the glycosylation of T8 also takes place in a lectin domain-assisted manner. The results described above for MUC14TR-T3*S14*T15* indicate that S4 and T8 are glycosylated by GalNAc-T4 in a lectin-dependent manner. However, it was not inferred whether the lectin domain binds at the previous glycosylated S14 or T15 to drive the glycosylation events at S4 or T8. Therefore, to uncover how the glycosylation takes place at those acceptor sites, 0.5 μs MD simulations were performed using 3D models of GalNAc-T4 complexed with UDP-GalNAc/Mn2+ and MUC1 triglycopeptide PGS*T*APPAHGVT*SAPDTRPAPG (3). In these models, the lectin domain was bound to either GalNAc at S or T of the GSTA region (equivalent to S14* or T15* of a preceding TR of MUC14TR) to drive glycosylation at S in GVT*SA (equivalent to S4 in a TR of MUC14TR) or T in PDTRP (equivalent to T8 in a TR of MUC14TR) (Figures 5 and 6).
Figure 5.
Putative 3D models obtained by MD simulations to explain the glycosylation of S4 and T8 in MUC14TR catalyzed by GalNAc-T4. (A,B) Glycosylation of S4. A 3D view of two complexes of GalNAc-T4/3 derived from a representative frame obtained from 0.5 μs of MD simulation in which GalNAc-T15 (A) or GalNAc-S14 (B) is accommodated in the lectin domain, respectively. A hydrogen bond between the carbonyl group of GalNAc at T3 and the main chain of F284, populated in 30% during MD trajectory, is established in (A). (C) Glycosylation of T8. 3D view of GalNAc-T4/3 derived from a representative frame obtained from 0.5 μs of MD simulation in which GalNAc-T15 (C) is accommodated in the lectin domain. The methyl group of T8 is involved in CH−π interactions with the aromatic ring of F364 (6.3 ± 0.5 Å). In all structures, the catalytic domain is shown in light blue and the lectin domain in dark blue. In the glycopeptides, the GalNAc units are shown in yellow sticks and the glycopeptide backbone in green ribbon. UDP-GalNAc is displayed in gray sticks and Mn2+ is shown as a cyan sphere. Relevant residues of the glycopeptides and some residues of catalytic domain are also shown as sticks and in a different color. Schematic representation of the glycosylation process at S4 and T8 of MUC14TR catalyzed by GalNAc-T4 is also displayed. These models highlight the C-terminal preference of GalNAc-T4.
Figure 6.
Putative 3D models obtained by MD simulations to explain the glycosylation of MUC14TR catalyzed by GalNAc-T4. (A) 3D view of two complexes of GalNAc-T4/4 derived from a representative frame obtained from 0.5 μs of MD simulations. Hydrophobic interaction between the methyl groups of T3 and A368 (4.7 ± 0.6 Å) is observed, which may favor the proper orientation of this threonine residue for efficient glycosylation. (B) Close view of STD–NMR experiments for short diglycopeptide (MUC1-T3*T15*). STD–NMR experiments were performed at 298 K in the presence of UDP (75 μM) and MnCl2 (150 μM) with a molar ratio of 75:1 diglycopeptide:GalNAc-Ts (GalNAc-T2, -T3, and -T4). The reference spectrum (1H NMR) is displayed in blue, while the STD spectrum (STD) is displayed in black for GalNAc-T4, in dark gray for GalNAc-T3, and in gray for GalNAc-T2. (C) Close view of GalNAc-T4/3 derived from a representative frame obtained from 0.5 μs of MD simulation in which GalNAc-T15 is accommodated in the lectin domain. Interestingly, D7 is engaged in a high populated salt-bridge interaction with R372 (71 and 88%) (dashed line). In the structures, the catalytic domain is shown in light blue and the lectin domain in dark blue. In the glycopeptides, the GalNAc units are shown in yellow sticks and the glycopeptide backbone in green ribbon. UDP-GalNAc is displayed in gray sticks and Mn2+ is shown as a cyan sphere. Relevant residues of the glycopeptides and some residues of catalytic domain are also shown as sticks and in a different color. Relevant residues of the glycopeptide and the protein are also shown as sticks. Schematic representation of the glycosylation process at T3 and T8 of MUC14TR catalyzed by GalNAc-T4 is also displayed. These models highlight the C-terminal preference of GalNAc-T4.
According to 0.5 μs guided MD simulations (with a distance between the hydroxyl group of S4 and C1 of UDP-GalNAc <4.5 Å), glycosylation of S4 is feasible when the lectin domain accommodates either GalNAc at T15* (Figures 5A and S14A) or S14* (Figures 5B and S14B). In both possibilities, the GalNAc moiety at T3* interacts through transient hydrogen bonds with several residues of the catalytic domain. Nevertheless, when T15* is bound to the lectin domain, a hydrogen bond populated around 30% along the MD trajectory is established between the carbonyl group of GalNAc at T3* and the NH of F284 of the enzyme (Figure 5A), which could allow for the proper orientation of S4 for the glycosylation. The MD simulations (with a distance between the hydroxyl group of T8 and C1 of UDP-GalNAc < 4.5 Å) indicate that T8 is not properly located in the active site when S14* is bound to the lectin domain, suggesting that the glycosylation in T8 likely occurs with the assistance of the lectin domain bound preferentially at T15* of the preceding TR domain of MUC14TR (Figures 5C and S14C). In this latest binding mode, GalNAc at T3* forms transient hydrogen bonds with the enzyme, and the methyl group of T8 is involved in a CH−π interaction with the aromatic ring of F364 (6.3 ± 0.5 Å), which may favor the correct orientation of this residue for optimal glycosylation (Figure 5C). In the long-range glycosylation, the T8 residue in MUC14TR is located 13 residues apart in the sequence from the glycosylated T15* of the preceding TR, while S4 is much closer (9 residues), being the shorter distance within the optimal range for GalNAc-T4 catalysis. Previous studies demonstrate that GalNAc-T4 prefers to glycosylate acceptor residues located 7–11 residues away from a prior glycosite,28,48 which might explain the slight preference of GalNAc-T4 to glycosylate S4 versus T8 residues in MUC14TR.
Role of the GalNAc-T4 Catalytic Domain in Glycosylating MUC14TR
Glycosylation of S4 and T8 of MUC1 by GalNAc-T4 is a process dominated by its lectin long-range preference. However, other features may modulate the GalNAc-T4 activity: (1) conformational changes in MUC1 structure induced by the presence of GalNAc in adjacent sites of the glycosylation site and (2) the preference of the catalytic domain toward specific peptide regions and/or glycopeptides (catalytic-domain-dependent glycosylation). To evaluate how the prior glycosites may affect the subsequent GalNAc-T4 glycosylation reactions, additional experiments were performed using MUC14TR-T3*T15* (Figure S15) and MUC14TR-T15* (Figure S16) as acceptor substrates. In the presence of MUC14TR-T3*T15*, GalNAc-T4 prefers to glycosylate S4 (GVT*SA) over S14 (GST*AP) or T8 (PDTRP) (Figure S15). After 24 h, 60% of S4, ∼15% in S14, and 8% of T8 residues were glycosylated. This result supports that GalNAc-T4 lectin domain likely binds to the GalNAc moiety at T15* of a preceding TR to drive glycosylation at S4 of the following TR. By comparing the GalNAc incorporation in MUC14TR-T3*T15* versus MUC14TR-T3*S14*T15* by GalNAc-T4 (Figures 4B and S15), differences in glycosylation of the two acceptor substrates were observed. The slower glycosylation of MUC14TR-T3*T15* versus MUC14TR-T3*S14*T15* indicates that S14 glycosylation must be priorly glycosylated by GalNAc-T2/-T3 to maximize the full glycosylation of MUC1 and exemplify the importance of the complementary and hierarchical functions of GalNAc-Ts during the O-glycosylation process of MUC1. Interestingly, when using the MUC14TR-T15* as the acceptor substrate, GalNAc-T4 first glycosylates T3 over S4, both located at the GVTSA motif (Figure S16). MD simulations performed on the complex between the enzyme and a glycopeptide as an acceptor substrate, PGST*APPAHGVTSAPDTRPAPG (4), show that a hydrophobic contact between the methyl group of T3 and A368 may help to align this residue for glycosylation, which could be the reason for the preference of T3 related to S4 (Figure 6A). These two residues are in the same region of MUC1 peptide (sharing a similar chemical environment), and in both cases, glycosylation is assisted by the lectin domain (T3 and S4 are eight and nine residues away from T15*, respectively). Thus, the less efficient capacity of GalNAc-T4 to glycosylate Ser than Thr residues is likely due to the poorer binding to the catalytic domain, as shown by our calculations. In fact, as verified by other GalNAc-Ts (e.g., T2 and T3), also for GalNAc-T4, a preference was shown to glycosylate Thr over Ser in random peptides.46 On the other hand, from the conformational perspective, the 13C-CSI analysis of MUC14TR-T3*S14*T15* versus the naked MUC14TR (Figure S4B) clearly indicates that glycosylation at T3 affects the conformation of the S4 neighboring residue. Our 13C-CSI analysis shows that the presence of GalNAc at T3 increases the population of β-strand-like conformation at the GVTS motif and agrees with the conformational analysis of MUC1 short glycopeptides previously reported.39,52 This alteration in the local conformation of S4 might also play a role in the glycosylation of S4 by GalNAc-T4. Additionally, as previously mentioned, the presence of the neighboring GalNAc at T3* contributes to additional interactions with the catalytic domain favoring the glycosylation of the contiguous S4 (Figure 6A, h-bond between the carbonyl of GalNAc and NH of F284). The presence of GalNAc at T3* as a prerequisite for glycosylation of S4 at the GVTSA region of MUC1 agrees with the absence of glycosylation at S4 observed for the MUC1 glycopeptide mutant in which T3 was mutated by a valine residue.22 The existence of this short-range glycosylation preference by GalNAc-T4 was previously reported by us using short model glycopeptides.23
In the case of T8 glycosylation (located at the PDTRP sequence), GalNAc-T4 glycosylates this residue in the same proportion as S4 at GVT*SA when the acceptor substrate is MUC14TR-T3*S14*T15*(Figure 4B). In this acceptor substrate, the glycosylation of T8 and S4 seems to occur independently. In addition, a small fraction of T8 glycosylation is detected in the case of the mutant GalNAc-T4D458H (Figure 4D,E). Surprisingly, in the presence of MUC14TR-T3*T15*, T8 glycosylation occurs in the same proportion as that of S14 at GST*AP. CSP and CSI analyses between MUC14TR-T3*T15 and MUC14TR-T3*S14*T15* (Figure S17) illustrate that S14 displays negligible effects on the chemical environment and conformation of PDTRP MUC14TR, suggesting that GalNAc-S14 does not modulate the glycosylation of T8 at the PDTRP motif. Previous studies suggest that the DTR fragment at MUC1 falls in the inverse γ-turn geometry’s area of the extended conformations in the Ramachandran plot.39 This conformation is likely important for the recognition of the GalNAc-T4 catalytic domain according to our results, explaining why the GalNAc-T4D459H still glycosylates T8. Overall, these data suggest that GalNAc-T4 has some natural preference to glycosylate the PDTRP region in the absence of a functional lectin domain. To further confirm this hypothesis, we performed saturation transfer difference (STD)–NMR experiments with GalNAc-T2, -T3, and -T4 using a short diglycopeptide containing one TR (MUC1-T3*T15*) (Figures 6B and S18).
STD signals corresponding to side chain protons of D7 and R9 at the PDTRP region were only detected in the case of GalNAc-T4. No STD signals were observed within the PDTRP sequence for either GalNAc-T2 or T3 isoform, demonstrating the specific recognition of the catalytic domain of GalNAc-T4 toward the inverse γ-turn PDTRP region around T8. Interestingly, the T8 at the PDTRP region is the only acceptor site at MUC1 flanked by two charged residues, which implies that these charge residues are likely specifically recognized by the GalNAc-T4 catalytic domain. Indeed, the analysis of the MD simulations on glycopeptide 3 in complex with GalNAc-T4 (Figure 6C) points out a significant salt-bridge between D7 at the PDTRP region of 3 and the R372 residue of GalNAc-T4 (high populated salt-bridge around 70–90% along the MD trajectory). Remarkably, this residue of GalNAc-T4 is not conserved in GalNAc-T2 and T3, which could be a mechanism of this enzyme to recognize the most immunogenic domain of MUC1. In short, GalNAc-T4 needs its lectin domain to efficiently glycosylate both S4 and T8 residues. However, other structural features such as the conformation and composition of the peptide sequence together with GalNAc-T4 short-range glycosylation preference finely tune MUC1 O-glycosylation by GalNAc-T4.
Elucidation of the GalNAc-T Isoforms Glycosylating S4 and T8 Residues in Cells
It is well accepted that GalNAc-T1, -T2, and -T3 are the major isoforms glycosylating in vitro and in vivo T3, S14, and T15 in MUC1.36,51 Although it is well accepted that GalNAc-T4 is capable of glycosylation S4 and T8in vitro,10,22 it is not clear whether GalNAc-T4 glycosylates these acceptor sites in vivo. One study showed that overexpression of GalNAc-T4 in CHO cells appears to enhance T8 glycosylation at the PDTRP sequence,50 and more recently, knock out (KO) of the GALNT4 gene in HEK293 cells revealed selective loss of the O-glycan at the Thr in the PDTR motif (T8) of the MUC1 tandem repeats when a reporter construct with seven TRs (MUC17TRs) was expressed.53 Since loss of GalNAc-T4 in HEK293 cells did not appear to affect O-glycosylation at the Ser in VTSA (S4), we wanted to further confirm this finding using an alternative bottom-up MS approach. The traditional bottom-up MS strategy for MUC1 TRs is based on use of endo-Asp digestion with cleavage in the PDTR motif, and an O-glycan at the Thr site may bias the cleavage and result of the analysis. We therefore took advantage of the glycomucinase BT4244 from Bacteroides thetaiotaomicron previously shown to cleave N-terminal to Ser or Thr residues with an α-GalNAc O-glycan attached.54 We recently demonstrated that this enzyme efficiently cleaves the Tn glycoform of the MUC17TRs reporter with predominant cleavage in between the O-glycan diads at the VT3S4A and GS14T15A motifs.55 The BT4244 glycomucinase efficiently cleaves the MUC1 reporter expressed in HEK293KOCOSMC/GALNT4 with O-glycosylation limited to α-GalNAc O-glycans,56 and we could confirm that the T8 in PDT8R was not glycosylated in HEK293 cells with the KO of GALNT4, while the S4 in VTS4A was efficiently O-glycosylated (Figure 7). This suggests that S4 may be O-glycosylated by other GalNAc-T isoforms in HEK293 cells despite those past studies with in vitro enzyme assays that have failed to identify such activities. Nevertheless, our results clearly confirm that GalNAc-T4 is the only isoform capable of glycosylating T8 in PDTRP in vivo and completing the O-glycosylation of the five glycosites in MUC1.
Figure 7.
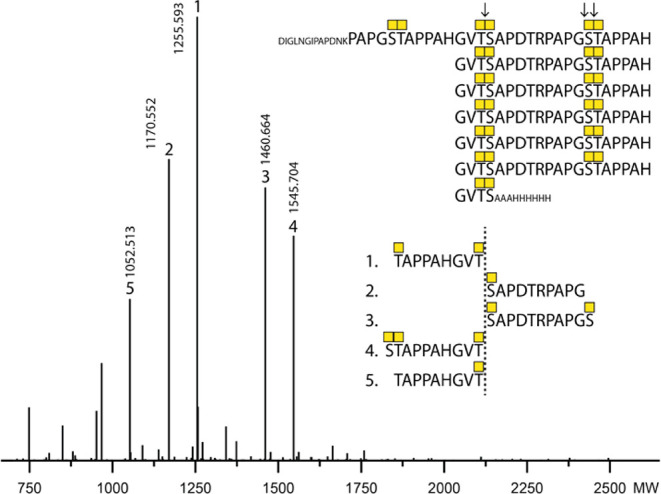
Exclusive contribution of GALNT4 in cells to MUC1 O-glycosylation at T8 in the PDTR site. Deconvoluted bottom-up analysis of the isolated MUC17TR reporter produced in HEK293KOCOSMC/GALNT4 using BT4244 glycomucinase digestion. The BT4244 glycomucinase digestion sites are indicated by arrows. All identified glycosites by MS/MS ETD analysis are illustrated with numbers assigned to each peak based on decreasing abundance. Only peaks with abundance above 10% were assigned.
Conclusions
Nature has deployed a vast number of GalNAc-Ts isoforms to deal with very complex protein substrates, like mucins, ensuring their function in living organisms. Deciphering the molecular determinants behind the glycosylation of mucins by GalNAc-Ts is crucial to understand the mode of action and the fine specificity of GalNAc-Ts toward mucins and is essential to explain how the complex mucinome is created. Herein, we provide NMR-derived structural insights into the full process of MUC1 O-glycosylation involving consecutive steps catalyzed by GalNAc-T2/T3 and T4. Apart from confirming the order by which GalNAc-T2/T3 and T4 glycosylate the different acceptor sites in the MUC1 TRs, we also demonstrate that the lectin-mediated functions of these GalNAc-Ts are strongly involved in all subsequent glycosylation reactions after the first GalNAc-addition. The lectin domain strongly contributes to define the site, the order, and the orientation of glycosylation in a complex mucin template such as MUC14TR (Figure 8). Since GalNAc-T1 and T2 are rather ubiquitously expressed and are the workhorses to initiate the O-glycoproteome in cells,33 we predict that these together with GalNAc-T3 (and its close paralog GalNAc-T6) in cells will drive the initial O-glycosylation of MUC1, while GalNAc-T4 serves to complete glycosylation. This hierarchy in GalNAc-Ts seems to be fundamental to obtain a fully glycosylated MUC1 product. Furthermore, our NMR data show local conformational changes in TRs during the incorporation of GalNAc residues, which may correlate with the preferences of some of GalNAc-Ts toward specific (glyco)peptides conformations (e.g., GalNAc-T2 glycosylates Ser residues at the glycosylated GST*AP better than GalNAc-T3/T1).
Figure 8.
Overview of the proposed mechanism for MUC1 GalNAc O-glycosylation by GalNAc-T2, -T3, and -T4.
We have also demonstrated that GalNAc-T4 is the unique isoform responsible to glycosylate the PDTRP region of MUC1 in cells, a process that depends significantly on its lectin domain and to a lesser extent on the exquisite affinity of its catalytic domain to the PDTRP motif. The inverse γ-turn conformation adopted by the DTR sequence in solution39 concomitantly with a specific salt-bridge that can be established between the Asp of PDTRP and the Arg of the enzyme might be the main structural features behind GalNAc-T4 preference toward this region. The PDTRP motif of the MUC1 structure is highly immunogenic, and most of the anti-MUC1 antibodies specifically target this peptide region at MUC1.57,58 Thus, the evidence that only GalNAc-T4 isoform can glycosylate this region in vivo points to the importance of the regulation of GalNAc-T4 expression in the glycosylation of the PDTRP region of MUC1. In summary, this work highlights the relevance of the lectin domain in the MUC1 glycosylation process, which clearly pinpoints the lectin domain of GalNAc-Ts as an attractive target for the rational design of inhibitors toward GalNAc-Ts. Finally, the NMR methodology and protocols applied herein are robust and can be applied to determine the specificity of other glycosyltransferases, such as C1GalT1 and ST6GalNAc-I, toward MUC1 or other mucin acceptor substrates.
Acknowledgments
The authors acknowledge Fundação para a Ciência e a Tecnologia (FCT-Portugal) for funding the projects, IF/00780/2015, PTDC/BIA-MIB/31028/2017, and UCIBIO project (UIDP/04378/2020 and UIDB/04378/2020), and Associate Laboratory Institute for Health and Bioeconomy—i4HB project (LA/P/0140/2020). The authors also thank FCT-Portugal for the PhD grant attributed to ASG (SFRH/BD/140394/2018) and A.D. (PD/BD/142847/2018) under the PTNMRPhD Program (PD00065/2013) and for the Norma transitória DL 57/2016 Program Contract to J.S.D. The NMR spectrometers are part of the National NMR Facility supported by FCT-Portugal (ROTEIRO/0031/2013-PINFRA/22161/2016, cofinanced by FEDER through COMPETE 2020, POCI and PORL and FCT through PIDDAC). RHG thanks ARAID, the Spanish Ministry of Science, Innovation and Universities (BFU2016-75633-P and PID2019-105451GB-I00), and Gobierno de Aragón (E34_R17 and LMP58_18 to R.H-G.) with FEDER (2014–2020) funds for “Building Europe from Aragón” for the financial support. F.C. thanks Agencia Estatal Investigación (AEI, Spain) (grant RTI2018-099592-B-C21). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Grant Agreement no. 956544. J.J.-B. and H.C. thank EU for funding (Tollerant ITN, GA-642157). F.M. and J.J.-B. acknowledge the COST Action GLYCONanoProbes (CA18132). J.J.-B also thanks the European Research Council for the financial support (ERC-2017-AdG, project number 788143-RECGLYCANMR), AEI (Spain, grant RTI218-094751-B-C21), and CIBER, an initiative of Instituto de Salud Carlos III (ISCIII), Madrid, Spain. H.C. acknowledges the Lundbeck Foundation and the Danish National Research Foundation (DNRF107).
Glossary
Abbreviations
- GalNAc-Ts
polypeptide GalNAc-transferases
- MUC14TR
mucin1 with four tandem repeats
- MUC17TR
mucin1 with seven tandem repeats
- NMR
nuclear magnetic resonance
- CSP
combined chemical shift perturbations
- STD
saturation transfer difference
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.1c00529.
(PDF)
Author Contributions
H.C. expressed and purified the MUC14TR and performed and analyzed the NMR experiments. A.D., A.S.G., C.O.S., R.F., and J.S.D. helped on the expression and purification of MUC14TR and the assignment of glycosylated MUC14TR products. M.R. expressed and purified the enzymes and mutants. I.C. synthesized the glycopeptide. L.S., Y.N., and S.Y.V. performed the cell assays and MS analysis. F.C. performed the MDs simulations. F.M. designed the research. F.M. and R.H-G. wrote the article together with the main contributions of F.C., E.J.C., J.J-B., Y.N., H.C., and H.C. All authors read and approved the final manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Bennett E. P.; Mandel U.; Clausen H.; Gerken T. A.; Fritz T. A.; Tabak L. A. Control of Mucin-Type O-Glycosylation: A Classification of the Polypeptide GalNAc-Transferase Gene Family. Glycobiology 2012, 22, 736–756. 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill D. J.; Clausen H.; Bard F. Location, Location, Location: New Insights into O-GalNAc Protein Glycosylation. Trends Cell Biol. 2011, 21, 149–158. 10.1016/j.tcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Hurtado-Guerrero R.; Davies G. J. Recent Structural and Mechanistic Insights into Post-Translational Enzymatic Glycosylation. Curr. Opin. Chem. Biol. 2012, 16, 479–487. 10.1016/j.cbpa.2012.10.013. [DOI] [PubMed] [Google Scholar]
- Narimatsu Y.; Joshi H. J.; Nason R.; Van Coillie J.; Karlsson R.; Sun L.; Ye Z.; Chen Y.-H.; Schjoldager K. T.; Steentoft C.; Furukawa S.; Bensing B. A.; Sullam P. M.; Thompson A. J.; Paulson J. C.; Büll C.; Adema G. J.; Mandel U.; Hansen L.; Bennett E. P.; Varki A.; Vakhrushev S. Y.; Yang Z.; Clausen H. An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol. Cell 2019, 75, 394–407. 10.1016/j.molcel.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager K. T.; Narimatsu Y.; Joshi H. J.; Clausen H. Global View of Human Protein Glycosylation Pathways and Functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- White T.; Bennett E. P.; Takio K.; Sørensen T.; Bonding N.; Clausen H. Purification and CDNA Cloning of a Human UDP-N-Acetyl-Alpha-D-Galactosamine:Polypeptide N-Acetylgalactosaminyltransferase. J. Biol. Chem. 1995, 270, 24156–24165. 10.1074/JBC.270.41.24156. [DOI] [PubMed] [Google Scholar]
- Chefetz I.; Kohno K.; Izumi H.; Uitto J.; Richard G.; Sprecher E. GALNT3, a Gene Associated with Hyperphosphatemic Familial Tumoral Calcinosis, Is Transcriptionally Regulated by Extracellular Phosphate and Modulates Matrix Metalloproteinase Activity. Biochim. Biophys. Acta, Mol. Basis Dis. 2009, 1792, 61–67. 10.1016/j.bbadis.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K.; Jeanneau C.; Tarp M. A.; Benet-Pagès A.; Lorenz-Depiereux B.; Bennett E. P.; Mandel U.; Strom T. M.; Clausen H. Polypeptide GalNAc-Transferase T3 and Familial Tumoral Calcinosis: Secretion of Fibroblast Growth Factor 23 Requires O-Glycosylation. J. Biol. Chem. 2006, 281, 18370–18377. 10.1074/jbc.M602469200. [DOI] [PubMed] [Google Scholar]
- Khetarpal S. A.; Schjoldager K. T.; Christoffersen C.; Raghavan A.; Edmondson A. C. Loss of Function of GALNT2 Lowers High-Density Lipoproteins in Humans, Nonhuman Primates, and Rodents. Cell Metab. 2016, 24, 234–245. 10.1016/j.cmet.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett E. P.; Hassan H.; Mandel U.; Mirgorodskaya E.; Roepstorff P.; Burchell J.; Taylor-Papadimitriou J.; Hollingsworth M. a.; Merkx G.; van Kessel A. G.; Eiberg H.; Steffensen R.; Clausen H. Cloning of a Human UDP- N -Acetyl-α-d-Galactosamine:Polypeptide N -Acetylgalactosaminyltransferase That Complements Other GalNAc-Transferases in Complete O -Glycosylation of the MUC1 Tandem Repeat. J. Biol. Chem. 1998, 273, 30472–30481. 10.1074/jbc.273.46.30472. [DOI] [PubMed] [Google Scholar]
- Hintze J.; Ye Z.; Narimatsu Y.; Madsen T. D.; Joshi H. J.; Goth C. K.; Linstedt A.; Bachert C.; Mandel U.; Bennett E. P.; Vakhrushev S. Y.; Schjoldager K. T. Probing the Contribution of Individual Polypeptide GalNAc-Transferase Isoforms to the O-Glycoproteome by Inducible Expression in Isogenic Cell Lines. J. Biol. Chem. 2018, 293, 19064–19077. 10.1074/jbc.RA118.004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de las Rivas M.; Lira-Navarrete E.; Gerken T. A.; Hurtado-Guerrero R. Polypeptide GalNAc-Ts: From Redundancy to Specificity. Curr. Opin. Struct. Biol. 2019, 56, 87–96. 10.1016/j.sbi.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F. K.; Hazes B.; Raffo R.; DeSa D.; Tabak L. a. Structure-Function Analysis of the UDP-N-Acetyl-D-Galactosamine : Polypeptide N-Acetylgalactosaminyltransferase. J. Biol. Chem. 1999, 274, 6797–6803. 10.1074/jbc.274.10.6797. [DOI] [PubMed] [Google Scholar]
- Fritz T. A.; Raman J.; Tabak L. A. Dynamic Association between the Catalytic and Lectin Domains of Human UDP-GalNAc:Polypeptide α-N-Acetylgalactosaminyltransferase-2. J. Biol. Chem. 2006, 281, 8613–8619. 10.1074/jbc.M513590200. [DOI] [PubMed] [Google Scholar]
- Lira-Navarrete E.; Iglesias-Fernández J.; Zandberg W. F.; Compañón I.; Kong Y.; Corzana F.; Pinto B. M.; Clausen H.; Peregrina J. M.; Vocadlo D. J.; Rovira C.; Hurtado-Guerrero R. Substrate-Guided Front-Face Reaction Revealed by Combined Structural Snapshots and Metadynamics for the Polypeptide N-Acetylgalactosaminyltransferase 2. Angew. Chem., Int. Ed. 2014, 53, 8206–8210. 10.1002/anie.201402781. [DOI] [PubMed] [Google Scholar]
- Kubota T.; Shiba T.; Sugioka S.; Furukawa S.; Sawaki H.; Kato R.; Wakatsuki S.; Narimatsu H. Structural Basis of Carbohydrate Transfer Activity by Human UDP-GalNAc: Polypeptide α-N-Acetylgalactosaminyltransferase (Pp-GalNAc-T10). J. Mol. Biol. 2006, 359, 708–727. 10.1016/j.jmb.2006.03.061. [DOI] [PubMed] [Google Scholar]
- Hazes B. The (QxW)3 Domain: A Flexible Lectin Scaffold. Protein Sci. 1996, 5, 1490–1501. 10.1002/pro.5560050805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandall H. H.; Irazoqui F.; Tarp M. A.; Bennett E. P.; Mandel U.; Takeuchi H.; Kato K.; Irimura T.; Suryanarayanan G.; Hollingsworth M. A.; Clausen H. The Lectin Domains of Polypeptide GalNAc-Transferases Exhibit Carbohydrate-Binding Specificity for GalNAc: Lectin Binding to GalNAc-Glycopeptide Substrates Is Required for High Density GalNAc-O-Glycosylation. Glycobiology 2007, 17, 374–387. 10.1093/glycob/cwl082. [DOI] [PubMed] [Google Scholar]
- Tenno M.; Kézdy F. J.; Elhammer Å. P.; Kurosaka A. Function of the Lectin Domain of Polypeptide N-Acetylgalactosaminyltransferase 1. Biochem. Biophys. Res. Commun. 2002, 298, 755–759. 10.1016/S0006-291X(02)02549-4. [DOI] [PubMed] [Google Scholar]
- Fritz T. A.; Hurley J. H.; Trinh L.-B.; Shiloach J.; Tabak L. A. The Beginnings of Mucin Biosynthesis: The Crystal Structure of UDP-GalNAc:Polypeptide -N-Acetylgalactosaminyltransferase-T1. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 15307–15312. 10.1073/pnas.0405657101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J. W.; Bennett E. P.; Schjoldager K. T. B. G.; Meldal M.; Holmér A. P.; Blixt O.; Cló E.; Levery S. B.; Clausen H.; Wandall H. H. Lectin Domains of Polypeptide GalNAc Transferases Exhibit Glycopeptide Binding Specificity. J. Biol. Chem. 2011, 286, 32684–32696. 10.1074/jbc.M111.273722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H.; Reis C. A.; Bennett E. P.; Mirgorodskaya E.; Roepstorff P.; Hollingsworth M. A.; Burchell J.; Taylor-Papadimitriou J.; Clausen H. The Lectin Domain of UDP-N-Acetyl-D-Galactosamine: Polypeptide N-Acetylgalactosaminyltransferase-T4 Directs Its Glycopeptide Specificities. J. Biol. Chem. 2000, 275, 38197–38205. 10.1074/jbc.M005783200. [DOI] [PubMed] [Google Scholar]
- De Las Rivas M.; Daniel E. J. P.; Coelho H.; Lira-navarrete E.; Raich L.; Compañón I.; Diniz A.; Lagartera L.; Jiménez-Barbero J.; Clausen H.; Rovira C.; Marcelo F.; Corzana F.; Gerken T. A.; Hurtado-guerrero R. Structural and Mechanistic Insights into the Catalytic-Domain- Mediated Short-Range Glycosylation Preferences of GalNAc-T4. ACS Cent. Sci. 2018, 4, 1274–1290. 10.1021/acscentsci.8b00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A. J.; Daniel E. J. P.; Mahajan S. P.; Gray J. J.; Gerken T. A.; Tabak L. A.; Samara N. L. The Structure of the Colorectal Cancer-Associated Enzyme GalNAc-T12 Reveals How Nonconserved Residues Dictate Its Function. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 20404–20410. 10.1073/pnas.1902211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Rivas M.; Lira-Navarrete E.; Daniel E. J. P.; Companõn I.; Coelho H.; Diniz A.; Jiménez-Barbero J.; Peregrina J. M.; Clausen H.; Corzana F.; Marcelo F.; Jiménez-Osés G.; Gerken T. A.; Hurtado-Guerrero R. The Interdomain Flexible Linker of the Polypeptide GalNAc Transferases Dictates Their Long-Range Glycosylation Preferences. Nat. Commun. 2017, 8, 1959. 10.1038/s41467-017-02006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman J.; Fritz T. A.; Gerken T. A.; Jamison O.; Live D.; Liu M.; Tabak L. A. The Catalytic and Lectin Domains of UDP-GalNAc:Polypeptide α-N-Acetylgalactosaminyltransferase Function in Concert to Direct Glycosylation Site Selection. J. Biol. Chem. 2008, 283, 22942–22951. 10.1074/jbc.M803387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Navarrete E.; De Las Rivas M.; Compañón I.; Pallarés M. C.; Kong Y.; Iglesias-Fernández J.; Bernardes G. J. L.; Peregrina J. M.; Rovira C.; Bernadó P.; Bruscolini P.; Clausen H.; Lostao A.; Corzana F.; Hurtado-Guerrero R. Dynamic Interplay between Catalytic and Lectin Domains of GalNAc-Transferases Modulates Protein O-Glycosylation. Nat. Commun. 2015, 6, 6937. 10.1038/ncomms7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revoredo L.; Wang S.; Bennett E. P.; Clausen H.; Moremen K. W.; Jarvis D. L.; Ten Hagen K. G.; Tabak L. A.; Gerken T. A. Mucin-Type o-Glycosylation Is Controlled by Short- And Long-Range Glycopeptide Substrate Recognition That Varies among Members of the Polypeptide GalNAc Transferase Family. Glycobiology 2016, 26, 360–376. 10.1093/glycob/cwv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetaert D.; Richet C.; Gagnon J.; Boersma A.; Degand P. Studies of Acceptor Site Specificities for Three Members of UDP-GalNAc:N-Acetylgalactosaminyltransferases by Using a Synthetic Peptide Mimicking the Tandem Repeat of MUC5AC. Carbohydr. Res. 2001, 333, 165–171. 10.1016/S0008-6215(01)00135-5. [DOI] [PubMed] [Google Scholar]
- Kong Y.; Joshi H. J.; Schjoldager K. T. B. G.; Madsen T. D.; Gerken T. A.; Vester-Christensen M. B.; Wandall H. H.; Bennett E. P.; Levery S. B.; Vakhrushev S. Y.; Clausen H. Probing Polypeptide GalNAc-Transferase Isoform Substrate Specificities by in Vitro Analysis. Glycobiology 2015, 25, 55–65. 10.1093/glycob/cwu089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager K. T.; Joshi H. J.; Kong Y.; Goth C. K.; King S. L.; Wandall H. H.; Bennett E. P.; Vakhrushev S. Y.; Clausen H. Deconstruction of O-glycosylation—Gal NA C-T Isoforms Direct Distinct Subsets of the O-glycoproteome. EMBO Rep. 2015, 16, 1713–1722. 10.15252/embr.201540796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I.; Pallesen E. M.; Ye Z.; Vakhrushev S. Y.; Marinova I. N.; Nielsen M. I.; Kramer S. H.; Pedersen S. F.; Joshi H. J.; Bennett E. P.; Dabelsteen S.; Wandall H. H. O-glycan Initiation Directs Distinct Biological Pathways and Controls Epithelial Differentiation. EMBO Rep. 2020, 21, 1–17. 10.15252/embr.201948885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu Y.; Joshi H. J.; Schjoldager K. T.; Hintze J.; Halim A.; Steentoft C.; Nason R.; Mandel U.; Bennett E. P.; Clausen H.; Vakhrushev S. Y. Exploring Regulation of Protein O-Glycosylation in Isogenic Human HEK293 Cells by Differential O-Glycoproteomics. Mol. Cell. Proteomics 2019, 18, 1396–1409. 10.1074/mcp.RA118.001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Rivas M.; Coelho H.; Diniz A.; Lira-Navarrete E.; Compañón I.; Jiménez-Barbero J.; Schjoldager K. T.; Bennett E. P.; Vakhrushev S. Y.; Clausen H.; Corzana F.; Marcelo F.; Hurtado-Guerrero R. Structural Analysis of a GalNAc-T2 Mutant Reveals an Induced-Fit Catalytic Mechanism for GalNAc-Ts. Chem.—Eur. J. 2018, 24, 8382–8392. 10.1002/chem.201800701. [DOI] [PubMed] [Google Scholar]
- de las Rivas M.; Paul Daniel E. J.; Narimatsu Y.; Compañón I.; Kato K.; Hermosilla P.; Thureau A.; Ceballos-Laita L.; Coelho H.; Bernadó P.; Marcelo F.; Hansen L.; Maeda R.; Lostao A.; Corzana F.; Clausen H.; Gerken T. A.; Hurtado-Guerrero R. Molecular Basis for Fibroblast Growth Factor 23 O-Glycosylation by GalNAc-T3. Nat. Chem. Biol. 2020, 16, 351–360. 10.1038/s41589-019-0444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandall H. H.; Hassan H.; Mirgorodskaya E.; Kristensen A. K.; Roepstorff P.; Bennett E. P.; Nielsen P. A.; Hollingsworth M. A.; Burchell J.; Taylor-Papadimitriou J.; Clausen H. Substrate Specificities of Three Members of the Human UDP- N -Acetyl-α-d-Galactosamine:Polypeptide N -Acetylgalactosaminyltransferase Family, GalNAc-T1, -T2, and -T3. J. Biol. Chem. 1997, 272, 23503–23514. 10.1074/jbc.272.38.23503. [DOI] [PubMed] [Google Scholar]
- Smet-Nocca C.; Broncel M.; Wieruszeski J. M.; Tokarski C.; Hanouelle X.; Leroy A.; Landrieu I.; Rolando C.; Lippens G.; Hackenberger C. P. Identification of O-GlcNAc sites within peptides of the Tau protein and their impact on phosphorylation. Mol BioSyst. 2011, 7 (5), 1420–1429. 10.1039/c0mb00337a. [DOI] [PubMed] [Google Scholar]
- Brokx R. D.; Revers L.; Zhang Q.; Yang S.; Mal T. K.; Ikura M.; Gariépy J. Nuclear Magnetic Resonance-Based Dissection of a Glycosyltransferase Specificity for the Mucin MUC1 Tandem Repeat. Biochemistry 2003, 42, 13817–13825. 10.1021/bi0353070. [DOI] [PubMed] [Google Scholar]
- Kinarsky L.; Suryanarayanan G.; Prakash O.; Paulsen H.; Clausen H.; Hanisch F. G.; Hollingsworth M. A.; Sherman S. Conformational Studies on the MUC1 Tandem Repeat Glycopeptides: Implication for the Enzymatic O-Glycosylation of the Mucin Protein Core. Glycobiology 2003, 13, 929–939. 10.1093/glycob/cwg109. [DOI] [PubMed] [Google Scholar]
- Corzana F.; Busto J. H.; Jiménez-Osés G.; De Luis M. G.; Asensio J. L.; Jiménez-Barbero J.; Peregrina J. M.; Avenoza A. Serine versus Threonine Glycosylation: The Methyl Group Causes a Drastic Alteration on the Carbohydrate Orientation and on the Surrounding Water Shell. J. Am. Chem. Soc. 2007, 129, 9458–9467. 10.1021/ja072181b. [DOI] [PubMed] [Google Scholar]
- Madariaga D.; Martínez-Sáez N.; Somovilla V. J.; García-García L.; Berbis M. Á.; Valero-Gónzalez J.; Martín-Santamaría S.; Hurtado-Guerrero R.; Asensio J. L.; Jiménez-Barbero J.; Avenoza A.; Busto J. H.; Corzana F.; Peregrina J. M. Serine versus Threonine Glycosylation with α-o-GalNac: Unexpected Selectivity in Their Molecular Recognition with Lectins. Chem.—Eur. J. 2014, 20, 12616–12627. 10.1002/chem.201403700. [DOI] [PubMed] [Google Scholar]
- Bermejo I. A.; Usabiaga I.; Compañón I.; Castro-López J.; Insausti A.; Fernández J. A.; Avenoza A.; Busto J. H.; Jiménez-Barbero J.; Asensio J. L.; Peregrina J. M.; Jiménez-Osés G.; Hurtado-Guerrero R.; Cocinero E. J.; Corzana F. Water Sculpts the Distinctive Shapes and Dynamics of the Tumor-Associated Carbohydrate Tn Antigens: Implications for Their Molecular Recognition. J. Am. Chem. Soc. 2018, 140, 9952–9960. 10.1021/jacs.8b04801. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y.; Nudelman A. S.; Levery S. B.; Wandall H. H.; Bennett E. P.; Hindsgaul O.; Clausen H.; Nishimura S. I. Elucidation of the Sugar Recognition Ability of the Lectin Domain of UDP-GalNAc:Polypeptide N-Acetylgalactosaminyltransferase 3 by Using Unnatural Glycopeptide Substrates. Glycobiology 2012, 22, 429–438. 10.1093/glycob/cwr159. [DOI] [PubMed] [Google Scholar]
- Abu-Abed M.; Mal T. K.; Kainosho M.; MacLennan D. H.; Ikura M. Characterization of the ATP-Binding Domain of the Sarco(Endo)Plasmic Reticulum Ca2+-ATPase: Probing Nucleotide Binding by Multidimensional NMR. Biochemistry 2002, 41, 1156–1164. 10.1021/bi015703n. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Sykes B. D. The 13C Chemical-Shift Index: A Simple Method for the Identification of Protein Secondary Structure Using 13C Chemical-Shift Data. J. Biomol. NMR 1994, 4, 171–180. 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- Daniel E. J. P.; Las Rivas M.; Lira-Navarrete E.; García-García A.; Hurtado-Guerrero R.; Clausen H.; Gerken T. A. Ser and Thr Acceptor Preferences of the GalNAc-Ts Vary among Isoenzymes to Modulate Mucin-Type O-Glycosylation. Glycobiology 2020, 30, 910–922. 10.1093/glycob/cwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos D.; Freitas D.; Gomes J.; Magalhães A.; Steentoft C.; Gomes C.; Vester-Christensen M. B.; Ferreira J. A.; Afonso L. P.; Santos L. L.; Pinto de Sousa J.; Mandel U.; Clausen H.; Vakhrushev S. Y.; Reis C. A. Probing the O-Glycoproteome of Gastric Cancer Cell Lines for Biomarker Discovery. Mol. Cell. Proteomics 2015, 14, 1616–1629. 10.1074/mcp.M114.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T. A.; Revoredo L.; Thome J. J. C.; Tabak L. A.; Vester-Christensen M. B.; Clausen H.; Gahlay G. K.; Jarvis D. L.; Johnson R. W.; Moniz H. A.; Moremen K. The Lectin Domain of the Polypeptide GalNAc Transferase Family of Glycosyltransferases (PpGalNAc Ts) Acts as a Switch Directing Glycopeptide Substrate Glycosylation in an N- or C-Terminal Direction, Further Controlling Mucin Type O-Glycosylation. J. Biol. Chem. 2013, 288, 19900–19914. 10.1074/jbc.M113.477877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacÍas-León J.; Bermejo I. A.; Asín A.; García-García A.; Compañón I.; Jiménez-Moreno E.; Coelho H.; Mangini V.; Albuquerque I. S.; Marcelo F.; Asensio J. L.; Bernardes G. J. L.; Joshi H. J.; Fiammengo R.; Blixt O.; Hurtado-Guerrero R.; Corzana F. Structural Characterization of an Unprecedented Lectin-like Antitumoral Anti-MUC1 Antibody. Chem. Commun. 2020, 56, 15137–15140. 10.1039/d0cc06349e. [DOI] [PubMed] [Google Scholar]
- Olson F. J.; Bäckström M.; Karlsson H.; Burchell J.; Hansson G. C. A MUC1 Tandem Repeat Reporter Protein Produced in CHO-K1 Cells Has Sialylated Core 1 O-Glycans and Becomes More Densely Glycosylated If Coexpressed with Polypeptide-GalNAc-T4 Transferase. Glycobiology 2005, 15, 177–191. 10.1093/glycob/cwh158. [DOI] [PubMed] [Google Scholar]
- Hanisch F. G.; Reis C. A.; Clausen H.; Paulsen H. Evidence for Glycosylation-Dependent Activities of Polypeptide N-Acetylgalactosaminyltransferases RGalNAc-T2 and -T4 on Mucin Glycopeptides. Glycobiology 2001, 11, 731–740. 10.1093/glycob/11.9.731. [DOI] [PubMed] [Google Scholar]
- Martínez-Sáez N.; Peregrina J. M.; Corzana F. Principles of Mucin Structure: Implications for the Rational Design of Cancer Vaccines Derived from MUC1-Glycopeptides. Chem. Soc. Rev. 2017, 46, 7154–7175. 10.1039/c6cs00858e. [DOI] [PubMed] [Google Scholar]
- Nason R.; Büll C.; Konstantinidi A.; Sun L.; Ye Z.; Halim A.; Du W.; Sørensen D. M.; Durbesson F.; Furukawa S.; Mandel U.; Joshi H. J.; Dworkin L. A.; Hansen L.; David L.; Iverson T. M.; Bensing B. A.; Sullam P. M.; Varki A.; de Vries E.; de Haan C. A. M.; Vincentelli R.; Henrissat B.; Vakhrushev S. Y.; Clausen H.; Narimatsu Y. Display of the Human Mucinome with Defined O-Glycans by Gene Engineered Cells. Nat. Commun. 2021, 12, 4070. 10.1038/s41467-021-24366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noach I.; Ficko-Blean E.; Pluvinage B.; Stuart C.; Jenkins M. L.; Brochu D.; Buenbrazo N.; Wakarchuk W.; Burke J. E.; Gilbert M.; Boraston A. B. Recognition of Protein-Linked Glycans as a Determinant of Peptidase Activity. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E679–E688. 10.1073/pnas.1615141114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidi A.; Nason R.; Čaval T.; Sun L.; Sørensen D. M.; Furukawa S.; Ye Z.; Vincentelli R.; Narimatsu Y.; Vakhrushev S. Y.; Clausen H.. Exploring Glycosylation of Mucins with O-Glycodomain Reporters Expressed in Glycoengineered HEK293 Cells. J. Biol. Chem. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steentoft C.; Vakhrushev S. Y.; Vester-Christensen M. B.; Schjoldager K. T.-B. G.; Kong Y.; Bennett E. P.; Mandel U.; Wandall H.; Levery S. B.; Clausen H. Mining the O-Glycoproteome Using Zinc-Finger Nuclease-Glycoengineered SimpleCell Lines. Nat. Methods 2011, 8, 977–982. 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- Coelho H.; Matsushita T.; Artigas G.; Hinou H.; Cañada F. J.; Lo-Man R.; Leclerc C.; Cabrita E. J.; Jiménez-Barbero J.; Nishimura S. I.; Garcia-Martín F.; Marcelo F. The Quest for Anticancer Vaccines: Deciphering the Fine-Epitope Specificity of Cancer-Related Monoclonal Antibodies by Combining Microarray Screening and Saturation Transfer Difference NMR. J. Am. Chem. Soc. 2015, 137, 12438–12441. 10.1021/jacs.5b06787. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y.; Denda-Nagai K.; Takahashi Y.; Nagashima I.; Shimizu H.; Kishimoto T.; Noji M.; Shichino S.; Chiba Y.; Irimura T. Products of Chemoenzymatic Synthesis Representing MUC1 Tandem Repeat Unit with T-, ST- or STn-Antigen Revealed Distinct Specificities of Anti-MUC1 Antibodies. Sci. Rep. 2019, 9, 16641. 10.1038/s41598-019-53052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.