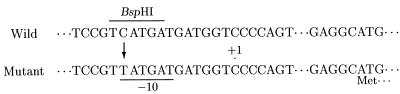Abstract
We determined the molecular basis for the enhanced expression of the aac(3)-Xa gene encoding an aminoglycoside 3-N-acetyltransferase in Streptomyces griseus. A C→T substitution was identified at the putative promoter of the mutant gene. RNA analyses demonstrated that the substitution caused a marked increase in the production of the gene-specific transcripts. Therefore, it seemed very likely that the aac(3)-Xa gene was activated by the substitution resulting in the emergence of a stronger promoter.
Aminoglycoside resistance genes are frequently found on mobile genetic elements known as plasmids and transposons, but in some cases they are found on chromosomes. The latter case includes genes for aminoglycoside acetyltransferase (AAC) enzymes, for example, the aac(2′)-Ia gene of Providencia stuartii (22), the aac(2′)-Ib gene of Mycobacterium fortuitum (1), the aac(2′)-Ic gene of Mycobacterium tuberculosis (2), the aac(2′)-Id gene of Mycobacterium smegmatis (2), the aac(6′)-Ic gene of Serratia marcescens (25), the aac(6′)-Ig gene of Acinetobacter haemolyticus (19), the aac(6′)-Ii gene of Enterococcus faecium (5), and the aac(6′)-Ij gene of Acinetobacter sp. strain 13 (18). Since these chromosomal aac genes are universally present in the respective species, it has been suggested that chromosomal AAC enzymes have other metabolic functions in addition to aminoglycoside acetylation. However, their primary role is still unknown, with an exception. It has been demonstrated that AAC(2′)-Ia contributes to the O-acetylation of peptidoglycan in P. stuartii (20).
Our studies have focused on the mechanism of aminoglycoside resistance in actinomycetes, especially on AAC enzymes (9, 10, 13, 14). Actinomycetes are a source of wide varieties of antibiotic resistance genes as well as a treasure box of antibiotic-synthesizing pathways. We have been investigating multiple resistances to aminoglycosides in aminoglycoside-producing actinomycetes. Consequently, we demonstrated that there are aminoglycoside resistances that do not contribute to the self resistance of aminoglycoside producers and thereby can be designated secondary aminoglycoside resistance (13). An aminoglycoside resistance gene in a streptomycin-producing soil isolate, Streptomyces griseus SS-1198, is a case in point. This gene was initially discovered by the emergence of a kanamycin (Km)-resistant mutant through the protoplast regeneration treatment of the wild-type strain SS-1198, which was unable to grow in the presence of 5 μg of Km/ml. In previous studies, we have characterized a high level (1,000 μg/ml) of Km resistance by the mutant SS-1198PR. Cloning experiments revealed that the Km resistance gene, aac(3)-Xa (formerly kan), encodes an aminoglycoside 3-N-acetyltransferase, AAC(3) (12). DNA hybridization experiments have demonstrated that the aac(3)-Xa gene was located in the chromosome and was present in all strains of S. griseus tested (11). On the other hand, a wild-type allele from strain SS-1198 has also been cloned (16). Restriction analysis has shown no substantial structural difference between wild-type and mutant gene fragments (16), suggesting that the Km resistance phenotype of the mutant gene was due to a point mutation of the wild-type gene. In this paper, we identified the location of the point mutation by determining the sequence differences between them and demonstrated the marked enhancement of the aac(3)-Xa gene expression by the mutant.
Bacterial strains and plasmids.
S. griseus SS-1198 is a soil isolate, and SS-1198PR strain is a Km-resistant mutant obtained through protoplast regeneration treatment of SS-1198 (12, 16). Streptomyces lividans TK21 and pIJ702 (17) were used for recombinant DNA experiments as a host strain and a vector plasmid, respectively.
Mapping the mutation.
To locate the mutation site of the mutant aac(3)-Xa gene of strain SS-1198PR, hybrid genes were constructed by swapping restriction fragments of the wild-type gene with those of the mutant gene. The 1.8-kb BglII-BamHI fragment, containing the wild-type or mutant gene, was divided into 0.5-kb BglII-BamHI and 1.3-kb BamHI fragments which contained mainly upstream regions and structural genes, respectively. After the insertion of these fragments into pIJ702 (17) with all possible combinations, the hybrid genes were tested for their ability to confer Km resistance to S. lividans TK21. As shown in Table 1, a high level (1,000 μg/ml) of Km resistance was obtained only with genes containing the 0.5-kb BglII-BamHI fragment derived from the mutant gene. By contrast, genes containing the 0.5-kb fragment from the wild-type gene conferred resistance to Km at concentrations as low as 50 μg/ml. Thus, the high level of Km resistance conferred by hybrid genes depended upon the origin of the 0.5-kb BglII-BamHI fragment, while no effect was observed with the 1.3-kb BamHI fragment. This result indicates that the mutation responsible for the Km resistance must be present within the 0.5-kb BglII-BamHI fragment, in which the putative promoter of the mutant aac(3)-Xa gene is contained (15).
TABLE 1.
Kanamycin resistance conferred to S. lividans TK21 by hybrid genes
| Plasmid | Classification of gene fragment
|
Resistance level (μg/ml)a | |
|---|---|---|---|
| 0.5-kb BglII-BamHI | 1.3-kb BamHI | ||
| pANT3-1 | Mutant | Mutant | 1,000 |
| pANT201 | Mutant | Wild type | 1,000 |
| pANT4-5 | Wild type | Wild type | 50 |
| pANT205 | Wild type | Mutant | 50 |
| pIJ702b | <5 | ||
Maximum growth allowance concentrations on yeast extract-malt extract agar (Difco) plate were determined after incubation at 27°C for 3 days.
Vector plasmid.
Comparing nucleotide sequences between the wild-type and mutant genes.
To identify the precise position of the mutation, we determined the nucleotide sequence of the 0.5-kb BglII-BamHI fragments of the wild-type aac(3)-Xa gene and compared it with that of the mutant gene (15). It turned out that a C:G pair at the position of −12 of the wild-type gene was replaced by a T:A pair in the mutant gene (Fig. 1). It was noted that the substitution occurred at the putative −10 sequence of the mutant aac(3)-Xa gene (Fig. 1) (15).
FIG. 1.
Comparison between upstream sequences of the wild-type and mutant aac(3)-Xa genes. Single base substitution is represented by an arrow. The BspHI site of the wild-type sequence is overlined and the putative −10 sequence of the mutant gene is underlined. The transcriptional starting point of the mutant gene was treated as the +1 position (15).
We also determined the nucleotide sequence of the 1.3-kb BamHI fragment of the wild-type gene and compared the sequence with that of the mutant gene. No difference between them was observed (data not shown).
Further evidence for the single base substitution could be obtained from restriction fragment length polymorphism, because the substitution of T for C in the mutant aac(3)-Xa gene resulted in the lack of a BspHI site in the wild-type sequence (Fig. 1). Total DNAs (5 μg each) from the wild-type and mutant strains were digested with BspHI and SphI, electrophoresed on 0.8% agarose gel, blotted onto a nylon membrane (Amersham Hybond-N+), and hybridized with the 32P-labeled 0.5-kb BglII-BamHI fragment of the mutant aac(3)-Xa gene under the conditions described by Church and Gilbert (4). For the wild-type strain, SS-1198, two bands (2.4 and 7.9 kb) were detected, but only one band (10.3 kb) was observed for the mutant strain SS-1198PR (data not shown). This result clearly shows the lack of the BspHI site in the mutant aac(3)-Xa gene. Consequently, the substitution was confirmed at the genomic DNA level.
Detection of mRNA by slot blotting and reverse transcriptase (RT)-PCR.
Since the point mutation at the corresponding region of the −10 sequence of the mutant aac(3)-Xa gene was confirmed, it was expected that the mutation affected transcription level. To analyze this possibility, RNA was extracted from log phase cells of the wild-type and mutant strains (8), and slot blot hybridization experiments were carried out with the 0.5-kb BglII-BamHI fragment of the aac(3)-Xa gene. No transcripts in the wild-type cells were detectable under the conditions used (Fig. 2, lane 1), while the mutant clearly produced aac(3)-Xa RNA (Fig. 2, lane 2).
FIG. 2.
Slot blot hybridization. Lanes 1 and 2, RNA from the wild-type (sensitive) S. griseus strain SS-1198 and chromosomal mutant (resistant) of this strain, S. griseus SS-1198PR, respectively; lane 3, S. lividans TK21 carrying the cloned mutant gene on the plasmid pANT3-1 (12); lane 4, the plasmidless S. lividans TK21. The slot blot was probed with the 32P-labeled 0.5-kb BglII-BamHI fragment of the aac(3)-Xa gene.
The wild-type sequence, CATGAT (Fig. 1), corresponding to the putative −10 sequence of the mutant gene, falls into the CANNAT class that has been found in other Streptomyces −10 regions (3). This led us to suspect that the wild-type gene must produce a small amount of transcripts. In order to demonstrate the presence of aac(3)-Xa RNA in the wild-type strain, we carried out RT-PCR, which is a more sensitive method than filter hybridization. Total RNA was extracted from log phase cells with Sepasol-RNA I (Nacalai Tesque) according to the manufacturer's instructions and treated with DNase I (Takara Shuzo). Heat-denatured RNA samples (≃2 μg) were reverse transcribed with 100 U of ReverTra Ace (TOYOBO) and 2.5 μM KANP6 or STRP2 primer in a final volume of 20 μl at 42°C for 30 min, 50°C for 30 min, and 55°C for 30 min. PCR was carried out with a 2-μl aliquot of the mixture, 1 μM concentrations of each primer, 2.5 U of AmpliTaq (GeneAmp PCR reagent kit; Perkin-Elmer), 1.5 mM MgCl2, and 10% (vol/vol) dimethyl sulfoxide. PCR conditions were 98°C for 3 min, 98°C for 1 min, 60°C for 1 min, 72°C for 2 min for 35 cycles, followed by 72°C for 5 min. Reaction products were separated on a 1.5% agarose gel.
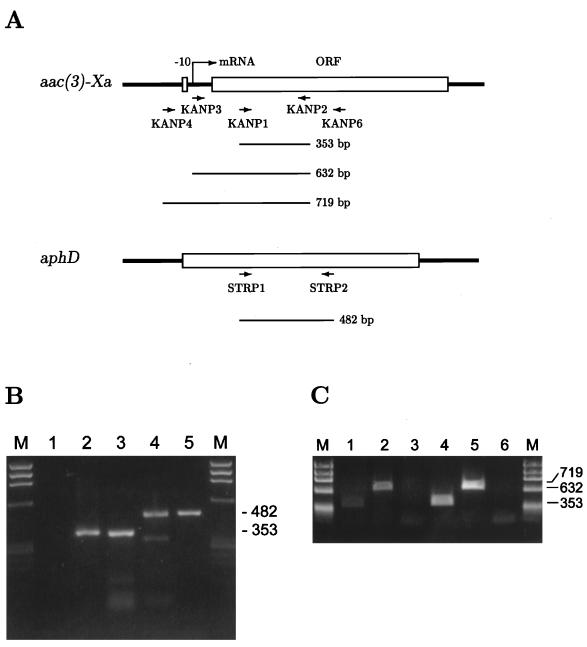
RT-PCR experiments were performed with the KANP1 and KANP2 primers, which were located at a mid-region of the aac(3)-Xa gene (Fig. 3A). RNase A-treated samples and chromosomal DNA were also used as negative and positive controls, respectively, in addition to the amplification of aphD RNA (6) for monitoring the reaction. As expected, a PCR product of 353 bp was amplified. This result indicates that the aac(3)-Xa gene was expressed in the wild-type strain. However, it was possible that read-through products of a gene upstream of the aac(3)-Xa gene would be amplified. We therefore carried out RT-PCR experiments with a KANP3 or KANP4 primer (Fig. 3A), whose 5′ ends were located just at or 87 bp upstream from the transcriptional starting point of the mutant aac(3)-Xa gene (15), respectively, instead of with the KANP1 primer. In both strains, KANP3-directed product (632 bp) was amplified but KANP4-directed product (719 bp) was not. Thus, it turned out that only aac(3)-Xa gene-specific transcripts were detected in these experiments. The results suggest that the transcription starting point of the wild-type gene is the same as or in close proximity to that of the mutant gene. This would be supported by missing promoter-like sequences in the region-spanning primers KANP3 and KANP4.
FIG. 3.
RT-PCR analyses. (A) Schematic representation of the locations of the primers. PCR products are indicated by bars with their expected size. Nucleotide sequences of the primers are follows: KANP1, 5′-ACTCTGTTGGTCACCTGCGG-3′; KANP2, 5′-TGCAGCAGCGTCATCGTGTC-3′; KANP3, 5′-CCCCAGTCCGTGTTCCGG-3′; KANP4, 5′-CTGATAGTCGAGAAAGGCCC-3′; KANP6, 5′-AGCATGGAACCGACGATCAC-3′; STRP1, 5′-ACCACGACGAGGAGAGCAG-3′; STRP2, 5′-TTCCGTCAGCAGGTCGAAG-3′. (B) Amplification of aac(3)-Xa RNA with KANP1 and KANP2 (lanes 1 to 3) and of aphD RNA with STRP1 and STRP2 (lanes 4 and 5) in the wild-type strain. Two micrograms of RNA was reverse transcribed with KANP6 (lanes 1 and 2) or STRP2 primer (lane 4), and a 2-μl aliquot was used for amplification. RNase A-treated RNA (lane 1) and 250 ng of total DNA (lanes 3 and 5) were used as negative and positive controls, respectively. HaeIII digestion of φX174 DNA served as the size standard (lanes M). (C) Amplification of aac(3)-Xa RNA with KANP1 (lanes 1 and 4), KANP3 (lanes 2 and 5) and KANP4 (lanes 3 and 6) as 5′ primers in the wild-type (lanes 1 to 3) and mutant (lanes 4 to 6) strains HaeIII digestion of φX174 DNA served as the size standard (lanes M).
Concluding remarks.
We identified the single base substitution responsible for the activation of the aac(3)-Xa gene. Slot blot analysis demonstrated that this mutation resulted in a marked increase in the transcription level of the aac(3)-Xa gene in the mutant strain, compared with that in the wild-type strain. The substitution of T for C in the mutant gene led to the formation of a sequence, TATGAT, that is more similar to the prokaryote [TATAAT] and Streptomyces [TA(G/C)(G/C/A)(G/T)T] −10 consensus sequences (3). Moreover, searching the transcription factor database (7) showed that there was no recognition site for a prokaryotic transcription regulator in the proximity of the mutation site (data not shown). It is therefore suggested that the substitution of T for C at the first position (−12) of the putative −10 sequence resulted in the emergence of a stronger promoter. However, we cannot rule out the possibility of the mutation creating a new promoter rather than creating a stronger promoter because the 5′ ends of aac(3)-Xa gene-specific transcripts were not identified in the wild-type strain.
aac(3)-Xa RNA could be detected in the wild-type cells by using RT-PCR, whereas the wild-type strain SS-1198 is sensitive to Km. It seems likely that the expression of the aac(3)-Xa gene in the wild-type cells is too weak to contribute to its Km resistance. In contrast, the cloning of the wild-type gene with a high-copy-number vector demonstrated that the wild-type gene could confer a low level (50 μg/ml) of Km resistance to S. lividans, suggesting that the difference between resistance levels conferred by the wild-type gene and the mutant genes would be due not to derepression in the heterologous host but to a gene dosage effect. However, we cannot rule out the presence of negative regulators, such as aar factors which regulate the expression of the aac(2′)-Ia gene in P. stuartii (21–24). We have currently found that protoplast regeneration treatment is not necessary for obtaining Km-resistant mutants, because we isolated such mutants without protoplasting (unpublished data). In preliminary experiments, the majority of the mutants have DNA amplification as a possible mechanism for Km resistance (16). One such mutant was found to have the same base substitution described in this paper. However, several mutants have neither DNA amplification nor promoter mutations. These observations may suggest the existence of a novel regulatory factor, such as a repressor, for the expression of the aac(3)-Xa gene in S. griseus. Further studies to analyze this possibility are in progress.
The aac(3)-Xa gene exists in all strains of S. griseus tested (11), suggesting that the gene has a primary function other than aminoglycoside modification. This would agree with the result that the gene was expressed in the wild-type strain. Although only a small amount of the gene-specific RNA was detected in the wild-type strain, the expression of the gene may be controlled by specific timing. Identification of such timing may yield interesting information on the regulation of the acc(3)-Xa gene.
Acknowledgments
We thank N. Summers and K. Summers for correcting the manuscript and the late K. Kawaguchi for technical assistance.
REFERENCES
- 1.Ainsa J A, Martin C, Gicquel B, Gomez-Lus R. Characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase gene from Mycobacterium fortuitum. Antimicrob Agents Chemother. 1996;40:2350–2355. doi: 10.1128/aac.40.10.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ainsa J A, Perez E, Pelicic V, Berthet F X, Gicquel B, Martin C. Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: characterization of the aac(2′)-Ic gene from Mycobacterium tuberculosis and the aac(2′)-Id gene from Mycobacterium smegmatis. Mol Microbiol. 1997;24:431–441. doi: 10.1046/j.1365-2958.1997.3471717.x. [DOI] [PubMed] [Google Scholar]
- 3.Bourn W R, Babb B. Computer assisted identification and classification of streptomycete promoters. Nucleic Acids Res. 1995;23:3696–3703. doi: 10.1093/nar/23.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa Y, Galimand M, Leclercq R, Duval J, Courvalin P. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob Agents Chemother. 1993;37:1896–1903. doi: 10.1128/aac.37.9.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Distler J, Ebert A, Mansouri K, Pissowotzki K, Stockmann M, Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1992;15:8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh D. New developments of a transcription factors database. Trends Biochem Sci. 1991;16:445–447. doi: 10.1016/0968-0004(91)90173-s. [DOI] [PubMed] [Google Scholar]
- 8.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: The John Innes Foundation; 1985. pp. 214–220. [Google Scholar]
- 9.Hotta K, Sunada A, Ishikawa J, Mizuno S, Ikeda Y, Kondo S. The novel enzymatic 3"-N-acetylation of arbekacin by an aminoglycoside 3-N-acetyltransferase of Streptomyces origin and the resulting activity. J Antibiot. 1998;51:735–742. doi: 10.7164/antibiotics.51.735. [DOI] [PubMed] [Google Scholar]
- 10.Hotta K, Zhu C-B, Ogata T, Sunada A, Ishikawa J, Mizuno S, Ikeda Y, Kondo S. Enzymatic 2′-N-acetylation of arbekacin and antibiotic activity of its product. J Antibiot. 1996;49:458–464. doi: 10.7164/antibiotics.49.458. [DOI] [PubMed] [Google Scholar]
- 11.Hotta K, Ishikawa J. Strain- and species-specific distribution of the streptomycin gene cluster and kan-related sequences in Streptomyces griseus. J Antibiot. 1988;41:1116–1123. doi: 10.7164/antibiotics.41.1116. [DOI] [PubMed] [Google Scholar]
- 12.Hotta K, Ishikawa J, Ichihara M, Naganawa H, Mizuno S. Mechanism of increased kanamycin-resistance generated by protoplast regeneration of Streptomyces griseus. I. Cloning of a gene segment directing a high level of an aminoglycoside 3-N-acetyltransferase activity. J Antibiot. 1988;41:94–103. doi: 10.7164/antibiotics.41.94. [DOI] [PubMed] [Google Scholar]
- 13.Hotta K, Ishikawa J, Ogata T, Mizuno S. Secondary aminoglycoside resistance in aminoglycoside-producing strains of Streptomyces. Gene. 1992;115:113–117. doi: 10.1016/0378-1119(92)90548-4. [DOI] [PubMed] [Google Scholar]
- 14.Hotta K, Ogata T, Ishikawa J, Okanishi M, Mizuno S, Morioka M, Naganawa H, Okami Y. Mechanism of multiple aminoglycoside resistance of kasugamycin-producing Streptomyces kasugaensis MB273: involvement of two types of acetyltransferases in resistance to astromicin group antibiotics. J Antibiot. 1996;49:682–688. doi: 10.7164/antibiotics.49.682. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa J, Hotta K. Nucleotide sequence and transcriptional start point of the kan gene encoding an aminoglycoside 3-N-acetyltransferase from Streptomyces griseus SS-1198PR. Gene. 1991;108:127–132. doi: 10.1016/0378-1119(91)90497-y. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa J, Koyama Y, Mizuno S, Hotta K. Mechanism of increased kanamycin-resistance generated by protoplast regeneration of Streptomyces griseus. II. Mutational alteration and gene amplification. J Antibiot. 1988;41:104–112. doi: 10.7164/antibiotics.41.104. [DOI] [PubMed] [Google Scholar]
- 17.Katz E, Thompson C J, Hopwood D A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983;129:2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- 18.Lambert T, Gerbaud G, Courvalin P. Characterization of the chromosomal aac(6′)-Ij gene of Acinetobacter sp. 13 and the aac(6′)-Ih plasmid gene of Acinetobacter baumannii. Antimicrob Agents Chemother. 1994;38:1883–1889. doi: 10.1128/aac.38.9.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambert T, Gerbaud G, Galimand M, Courvalin P. Characterization of Acinetobacter haemolyticus aac(6′)-Ig gene encoding an aminoglycoside 6′-N-acetyltransferase which modifies amikacin. Antimicrob Agents Chemother. 1993;37:2093–2100. doi: 10.1128/aac.37.10.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payei K G, Rather P N, Clarke A J. Contribution of gentamicin 2′-N-acetyltransferase to the O acetylation of peptide glycan in Providencia stuartii. J Bacteriol. 1995;177:4303–4310. doi: 10.1128/jb.177.15.4303-4310.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rather P N, Orosz E. Characterization of aarA, a pleiotropic negative regulator of the 2′-N-acetyltransferase in Providencia stuartii. J Bacteriol. 1994;176:5140–5144. doi: 10.1128/jb.176.16.5140-5144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rather P N, Orosz E, Shaw K J, Hare R, Miller G. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J Bacteriol. 1993;175:6492–6498. doi: 10.1128/jb.175.20.6492-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rather P N, Paradise M R, Parojcic M M, Patel S. A regulatory cascade involving AarG, a putative sensor kinase, controls the expression of the 2′-N-acetyltransferase and an intrinsic multiple antibiotic resistance (Mar) response in Providencia stuartii. Mol Microbiol. 1998;28:1345–1353. doi: 10.1046/j.1365-2958.1998.00900.x. [DOI] [PubMed] [Google Scholar]
- 24.Rather P N, Solinsky K A, Paradise M R, Parojcic M M. aarC, an essential gene involved in density-dependent regulation of the 2′-N-acetyltransferase in Providencia stuartii. J Bacteriol. 1997;179:2267–2273. doi: 10.1128/jb.179.7.2267-2273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw K J, Rather P N, Sabatelli F J, Mann P, Munayyer H, Mierzwa R, Petrikkos G L, Hare R S, Miller G H, Bennett P, Downey P. Characterization of the chromosomal aac(6′)-Ic gene from Serratia marcescens. Antimicrob Agents Chemother. 1992;36:1447–1455. doi: 10.1128/aac.36.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]