Abstract
The coronavirus disease 2019 (COVID-19) pandemic had grounded the world to a standstill. As the disease continues to rage two years on, it is apparent that effective therapeutics are critical for a successful endemic living with COVID-19. A dearth in suitable antivirals has prompted researchers and healthcare professionals to investigate existing and developmental drugs against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Although some of these drugs initially appeared to be promising for the treatment of COVID-19, they were ultimately found to be ineffective. In this review, we provide a retrospective analysis on the merits and limitations of some of these drugs that were tested against SARS-CoV-2 as well as those used for adjuvant therapy. While many of these drugs are no longer part of our arsenal for the treatment of COVID-19, important lessons can be learnt. The recent inclusion of molnupiravir and Paxlovid™ as treatment options for COVID-19 represent our best hope to date for endemic living with COVID-19. Our viewpoints on these two drugs and their prospects as current and future antiviral agents will also be provided.
Chemical compounds studied in this article: Chloroquine (PubChem CID: 2719), Hydroxychloroquine (PubChem CID: 3652), Ivermectin (PubChem CID: 6321424), Lopinavir (PubChem CID: 92727), Ritonavir (PubChem CID: 392622), Remdesivir (PubChem CID: 121304016), Dexamethasone (PubChem CID: 5743), Baricitinib (PubChem CID: 44205240), Molnupiravir (Pubchem CID: 145996610), Nirmatrelvir (PubChem CID: 155903259)
Keywords: Antivirals, COVID-19, Small molecule therapeutics, Drug discovery
Graphical Abstract
1. Introduction
The continuing battle against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been long drawn with no end in sight. Although significant advances in science have resulted from this pandemic – from a greater understanding of coronaviruses (especially SARS-CoV-2) to the development of rapid diagnostics and the game changing development and use of mRNA vaccines – no truly effective treatment for COVID-19 exists. In view of the emerging SARS-CoV-2 variants, it has become apparent that effective therapeutics are critical for successful endemic living with COVID-19. Although therapeutics can be in the form of biologics (e.g. Regeneron antibody treatment), effective orally active small molecule therapeutics are more likely to be useful in managing this disease. This is in view of the limited access to good healthcare for the poorer populations in the world. In addition, biologics are more likely to be sensitive to mutations emerging from variants of concern (VOCs) that can render them less effective [1]. Furthermore, the necessity of biologics for cold chain storage can hinder their distribution and handling. Small molecule therapeutics, on the other hand, can overcome the latter challenges due to their robustness, which aids their storage, distribution, and handling [2].
In retrospect, there is no shortage of reported potential small molecule therapeutics for the treatment of COVID-19 [3], [4], [5], [6]. The number of true contenders, however, are few and far between. In view of a typical long timeline (12–15 years) from initial drug discovery to drug approval, a popular approach in the search of small molecule therapeutics is to utilise drug repurposing. Should a known drug (or an advanced drug in development) be repurposed, the timeline for approval can be significantly shortened in view of the safety studies that would have already been carried out. From a purist’s view, drug repurposing is “a hypothesis-free drug repositioning” as succinctly phrased by Aled Edwards [7]. In this context, a sobering observation made by Edwards is that there has never been a discovery of an old drug for a new indication found in the past 20 years. This implies that while a hypothesis-free approach presents an unbiased standpoint for drug repositioning, the fact remains that a hypothesis-driven evaluation is evidently more fruitful and consequently adopted in the search for drugs to treat COVID-19.
Another sobering piece of information relates to the arsenal of FDA-approved small molecule anti-virals [8], [9]. As of 2020, there were 85 monotherapies, of which 72 target the virus while another 13 target the host. Of these, 69 were small molecule drugs [8]. Not surprisingly, a significant number of these drugs target chronic rather than acute viral infections such as human immunodeficiency virus-1 (HIV-1), hepatitis B virus (HBV) and hepatitis C virus (HCV) infections. The dearth of antiviral drugs may be surprising in view of the indisputable role of viruses in causing disease and death. However antiviral drug discovery is confounded by the following: (i) rapid mutation of the viruses making them a moving target; (ii) few targets that are unique to a virus as they hijack the host replication machinery for replication; (iii) the time window for the administration of an antiviral drug may be critical for efficacy thus giving rise to variable treatment outcomes; (iv) the lack of experimental animal models that can truly mimic a human host, from immune response to virulence and pathogenicity; (v) challenges associated with the lack of adequate diagnostics for many viruses. Early identification of promising lead compounds is typically carried out through in vitro studies, either through target-based approaches (e.g. inhibition of protein function) or phenotypic approaches (e.g. inhibition of cytopathic effect), but there are clearly significant gaps in translating in vitro outcomes to in vivo efficacies. At times, small patient pools also present additional challenges in drug development and clinical approvals as is evident in the case of brincidofovir for smallpox [10].
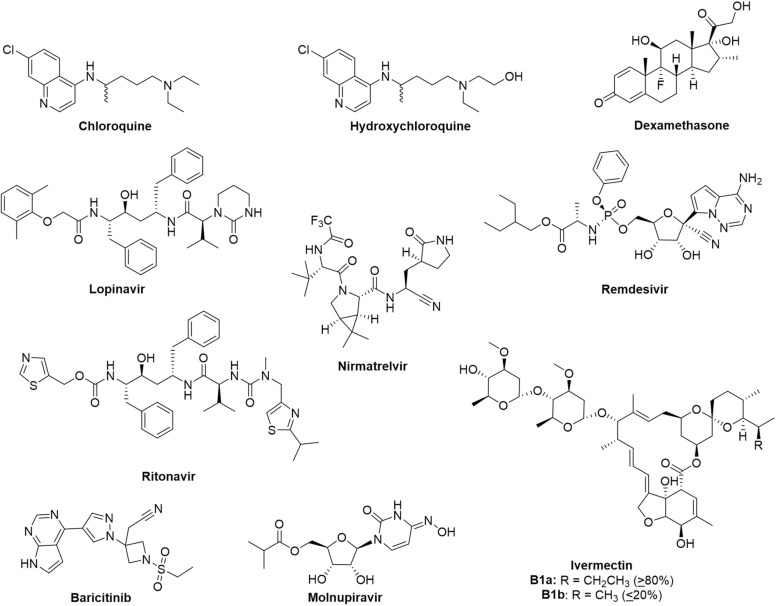
As COVID-19 rages two years on, we examine a selection of the most promising therapeutics that were reported in the early years of the pandemic that were explored as mono- or combination-therapy ( Fig. 1) for the treatment of COVID-19. These compounds were selected based on three criteria: (1) they are small molecules; (2) these treatment options were prominent and widely evaluated in 2020; and (3) their clinical efficacy has been evaluated and data is available as of 2021. Many of these drugs were reutilized from other indications, with postulation that they may retain similar roles in antiviral treatment or symptoms management in COVID-19 ( Table 1). Their merits and shortcomings in disease management will be discussed and evaluated. Some of these drugs were amongst the first that demonstrated in vitro efficacy against SARS-CoV-2, while others were highly publicised by the media and general scientific community. Advancement in the development of antiviral small molecule therapeutics has also progressed two antivirals, namely molnupiravir and nirmatrelvir (Fig. 1, Table 1), into clinical use. In this regard, we provide our viewpoint on these ten drugs from a retrospective analysis and assess the drug targets for their suitability as antiviral targets, especially towards future viral pathogens, as well as for management of the disease. In view of the rapid pace of developments on COVID-19 and treatments, our perspective is not meant to be comprehensive, but aims to highlight some valuable lessons that can be learnt for drug discovery and development of therapeutics.
Fig. 1.
Chemical structures of the most promising drug candidates for the treatment of COVID-19. Some of these compounds are being explored as combination drugs.
Table 1.
Summary of the drugs discussed, which include monotherapy and combination-therapy. Their previous indications are specified where appropriate, along with their postulated roles in SARS-CoV-2.
| Drug | Trade Name | Previous Indication | Postulated role/target in SARS-CoV-2 |
|---|---|---|---|
| Chloroquine | Aralen | Malaria, Rheumatoid arthritis | Inhibit viral entry |
| Hydroxychloroquine | Plaquenil | Malaria, Rheumatoid arthritis | Inhibit viral entry |
| Ivermectin | Stromectol | Antiparasitic | Inhibit nuclear import via IMPα/β |
| Lopinavir-Ritonavir | Kaletra | HIV-1 Mpro | SARS-CoV-2 Mpro |
| Remdesivir | Veklury | EBOV RdRp | SARS-CoV-2 RdRp |
| Dexamethasone | Decadron | Immunomodulatory | Management of pro-inflammatory cytokines |
| Baricitinib | Olumiant | Rheumatoid arthritis, Janus kinase inhibitor | Management of pro-inflammatory cytokines |
| Molnupiravir | Lagevrio | – | SARS-CoV-2 RdRp |
| Nirmatrelvir-Ritonavir | Paxlovid | – | SARS-CoV-2 Mpro |
2. SARS-CoV-2 replication pathway
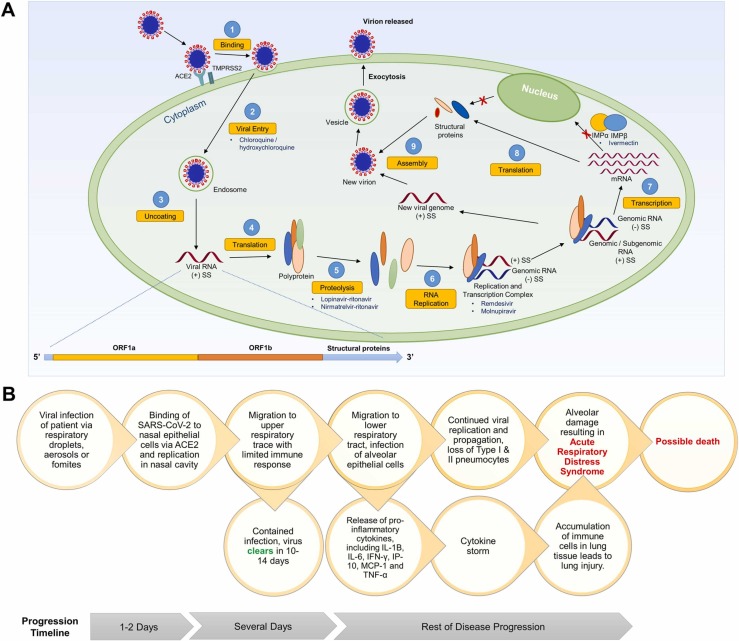
SARS-CoV-2 is an enveloped positive-sense single-stranded RNA virus, belonging to the beta-coronavirus genus of the Coronaviridae family [11]. The infection and replication cycle of SARS-CoV-2 is broadly similar to that of SARS-CoV and MERS-CoV [12], [13], [14]. Briefly, infection begins when the S1 subunit of the spike protein binds to the host receptor via the receptor-binding domain. For MERS-CoV, this receptor is the dipeptidyl peptidase-4, whereas for SARS-CoV and SARS-CoV-2, angiotensin-converting enzyme 2 (ACE2) receptor is the crucial host receptor [15]. The higher affinity of SARS-CoV-2 for ACE2 as compared to SARS-CoV may account for its greater transmissibility [16]. The S2 subunit of the spike proteins is responsible for viral entry and fusion of viral and host membranes [15]. Infections on host cellular surfaces are mediated by serine protease transmembrane serine protease 2 (TMPRSS2), while endosomal entry is mediated by endosomal cysteine protease cathepsin B/L [17]. Importantly, SARS-CoV-2 was found to use TMPRSS2-mediated entry as the primary viral entry route [15]. As priming by these proteases are critical for the fusion and subsequent entry of the virus into the host cell, they represent viable antiviral targets [12]. Uniquely, a polybasic site between the S1 and S2 subunit of SARS-CoV-2 can be cleaved by another protease known as furin, which may additionally account for the increased transmissibility of SARS-CoV-2 [18]. It has also been suggested that the spike proteins of coronaviruses are heavily glycosylated, which enables evasion of recognition by host immune antibodies [19]. Following the entry of the virus into the host cell, the genomic RNA is uncoated and released, and translation of two open reading frames ORF1a and ORF1b occurs. The resulting polyproteins are then processed by two cysteine proteases, papain-like protease (PLpro) and 3C-like protease (3CLpro; also known as main protease or Mpro), to smaller non-structural proteins (NSPs) needed for viral replication and transcription. RNA-dependent RNA polymerase (RdRp) in the replication and transcription complex (RTC) is a critical enzyme that is responsible for the formation of negative-sense genomic and subgenomic RNAs. Finally, new virions are assembled from the genomic material and structural proteins, which are then released from the cells through exocytosis ( Fig. 2A).
Fig. 2.
(A) Life cycle of SARS-CoV-2. The key steps are highlighted, and drugs that were proposed to inhibit those steps are indicated where appropriate. (B) Pathophysiology of COVID-19. In most cases, viral infection is limited to the URT, and the patient recovers within 14 days with no further complications. However, the viral infection can migrate to the LRT and lead to ARDS or a cytokine storm. This has been proposed as the main cause of death from COVID-19-related complications.
Antiviral approaches against SARS-CoV-2 can be categorised into the following: (1) inhibition of host receptor recognition; (2) inhibition of viral entry; (3) inhibition of viral protein maturation; (4) inhibition of viral genome replication; (5) inhibition of viral protein translation. These approaches will be further discussed in Section 4.
3. Pathophysiology of COVID-19
Besides targeting the virus, another approach in disease management aims to target and slow down disease progression. Typically, infection begins at the nasal epithelium followed by viral replication. Thereafter, the virus migrates down the upper respiratory tract (URT) [20]. In 80% of cases, the infection is contained in the URT, and the patient recovers uneventfully within 14 days. In the other 20%, the virus may migrate to the lower respiratory tract (LRT), which causes infection to pulmonary alveolar epithelial cells [20]. Potentially, this could lead to enhanced viral replication, as well as the upregulation of pro-inflammatory cytokines including interleukins IL-1, IL-6, IL-8 and IL-12, and tumour necrosis factor-α (TNF-α) [21]. The enhanced viral replication can lead to acute respiratory distress syndrome (ARDS), while the latter can progress to a cytokine storm with implications such as possible lung injury, alveolar damage, and development of ARDS. Together, this has been proposed as the main cause of death from COVID-19-related complications (Fig. 2B) [22].
4. Approved drugs and investigational drugs evaluated for the treatment of COVID-19
Treatment for COVID-19 can be categorised as (i) antiviral treatment, and/or (ii) adjuvant therapy. These two approaches will be discussed separately in the following sections.
4.1. Antiviral treatment
4.1.1. Chloroquine / hydroxychloroquine
Amongst the many drugs explored for the treatment of COVID-19, chloroquine (CQ) and hydroxychloroquine (HCQ) (Fig. 1) were possibly the first to be publicly hailed as a potential treatment. Originally discovered in the 1966, CQ was used as an antiparasitic agent against malaria [23]. CQ increases the endosomal and/or lysosomal pH, which in turn inhibits key enzymatic processes that eventually leads to the death of the parasite, most commonly Plasmodium falciparum [24]. The antiviral properties of CQ against HIV-1 were proposed in 1990, primarily through the same mechanism of increasing the endosomal pH [25]. Specifically, CQ/HCQ may inhibit the pH-dependent conformational changes in the structural proteins of viruses that are essential for binding and fusion to host receptors and endocytosis. They may also inhibit key enzymatic activities involved in the post-translational modifications of the glycoproteins on the viral envelope, leading to attenuated viral virulence [26]. Beyond HIV-1, CQ has also demonstrated antiviral effects against other RNA viruses, including coronaviruses such as SARS-CoV. CQ was found to inhibit SARS-CoV in vitro with an EC50 value of 8.8 µM, which corresponds to the effective plasma concentrations of the drug used for malaria treatment [27]. Mechanistically, CQ has been shown to inhibit the terminal glycosylation of the cellular receptor, ACE2, in addition to elevating the endosomal pH. This disrupts the binding of SARS-CoV to the host cell receptor and interferes with viral entry and virulence [28]. In addition, changes in endosomal pH can inhibit cathepsin L-mediated priming, thereby preventing viral entry through this pathway [29]. Due to the similarities between SARS-CoV and SARS-CoV-2, CQ was investigated as a potential antiviral against the novel coronavirus [30], [31], [32]. HCQ, a derivative of CQ, is thought to behave similarly as CQ but with much reduced toxicities in animals [33]. The additional anti-inflammatory properties of HCQ were also touted to be useful for mitigating cytokine storms that may occur with COVID-19 infection (Fig. 2B), thus making this proposed dual action drug highly desirable for treating both the symptoms and the infection itself [34].
The in vitro studies with CQ and HCQ on SARS-CoV-2 reported EC50 values of 2.71–7.36 µM and 4.51–12.96 µM respectively [34], [35]. These positive outcomes, however, failed to be translated into clinical efficacy. Based on a recent systematic review with meta-analysis on 12 randomised clinical trials (RCTs) with 8569 adult participants (with search up to September, 2020), HCQ has indicated little to no effect on virological clearance and clinical worsening (compared to mechanical ventilation) and mortality in COVID-19 patients [36]. The same review indicated an approximate three-fold increase in the risk of drug-related adverse events like gastrointestinal disorders and prolongation of the corrected QT (QTc) intervals indicating cardiovascular irregularities, which further discouraged the continuation of trials involving CQ/HCQ for COVID-19 treatment [36]. Earlier systematic reviews with meta-analysis on RCTs alone (7 trials, 4984 patients) [37] and with observational studies (12 cohort studies + 3 RCTs, 10,659 patients) [38] also concluded that there was no benefit in using CQ/HCQ for improving clinical outcomes of COVID-19. Large-scale RCTs with results published later such as the Randomized Evaluation of COVID-19 Therapy (RECOVERY) collaborative trial (in the UK, with 4716 hospitalized patients) [39] and the WHO Solidarity trial (multinational, with 954 hospitalized patients received HCQ) [40] likewise demonstrated no mortality benefit of CQ/HCQ, which was consistently observed across various age groups and disease severity.
This apparent discrepancy between in vitro and clinical outcomes is suggested to stem from cell line-dependent effects of CQ observed against SARS-CoV-2 [41]. While SARS-CoV-2 may mediate viral entry through both pH-dependent cathepsin L and pH-independent TMPRSS2 pathways, viral entry and infection in airway epithelial cells occurred predominantly through the latter. In contrast, viral entry in cell lines expressing low levels of TMPRSS2 (e.g. Vero E6 African green monkey kidney-derived cells) proceed via cathepsin L, for which CQ/HCQ has superior suppression effects [29]. Consequently, this may have overpredicted the effects of CQ/HCQ and limited their clinical relevance [41].
While these 4-aminoquinoline drugs are no longer part of the arsenal for the treatment of COVID-19, there are important lessons to be learnt. Retrospective analyses of CQ and HCQ revealed that these drugs lacked in vitro to in vivo translatability in SARS-CoV. Yet, this was not addressed or evaluated following the end of the SARS epidemic [42]. The urgency for therapeutics against the SARS-CoV-2 arose when infection numbers surged worldwide and this prompted clinical trials on CQ/HCQ despite their lack of efficacy as antiviral and prophylactic agents [43], [44], [45]. RCTs and systematic reviews merely reaffirmed the ineffectiveness of CQ/HCQ for the treatment of COVID-19 [36], [37], [38], [39], [40]. The evaluation of CQ/HCQ against SARS-CoV-2 herein clearly exemplifies the need for careful evaluation of drugs according to its safety and efficacy profile before proceeding to clinical trials. Furthermore, these studies have also highlighted the importance of understanding the pathology of viral infections to aid in the selection of appropriate cell lines for antiviral studies [41].
4.1.2. Lopinavir-ritonavir
Another potential strategy to develop an effective antiviral is to disrupt the maturation of viral proteins via the inhibition of a key protease. Lopinavir (Fig. 1) is a peptidomimetic drug that is commonly used against HIV-1. As an aspartate protease inhibitor, it targets the main protease of HIV-1 and prevents the polyprotein cleavage, thereby disrupting virion maturation and ending the infection and replication cycle [46]. Although lopinavir can be used as a monotherapy, the drug is often used in combination with low doses of a CYP450 inhibitor ritonavir (Fig. 1) in clinical settings to slow down its hepatic metabolism [47]. Lopinavir, with or without ritonavir, has demonstrated antiviral activities against SARS-CoV, MERS-CoV and HIV-1 [48], [49], [50]. Lopinavir reduced SARS-CoV viral titer in fetal rhesus kidney-4 (FRhK-4) cells with an IC50 value of 6.36 µM [49] and was also found to inhibit the purified SARS-CoV Mpro with an IC50 value of 50 µM [50]. Similarly, lopinavir treatment inhibited MERS-CoV replication in human hepatocellular carcinoma Huh-7 cells with an EC50 value of 8.0 µM [48]. The broad-spectrum antiviral activity of lopinavir makes it an attractive drug candidate for the treatment of COVID-19.
Lopinavir reduced SARS-CoV-2 viral titer in Vero E6 cells with an EC50 value of 26.63 µM [51]. The drug was presumed to target the Mpro of SARS-CoV-2, similar to its antiviral action in SARS-CoV. To study this, lopinavir and ritonavir were evaluated for SARS-CoV-2 Mpro inhibitory activity using a cell-based assay in HEK293T cells [52]. Interestingly, treatment with lopinavir alone showed no inhibitory activity at non-toxic concentrations, while treatment with ritonavir alone gave an IC50 value of 13.7 µM. A combination of lopinavir-ritonavir resulted in an IC50 value of 10.9 µM. However, neither lopinavir, ritonavir nor lopinavir-ritonavir combination displayed any inhibitory activity when tested against the purified Mpro [52]. In view of the latter, it was not surprising that lopinavir-ritonavir treatment in SARS-CoV-2-infected ferrets also showed no significant reduction in in vivo viral titers [45]. Despite this, clinical trials commenced in view of the lack of effective COVID-19 treatment. An initial study involving 199 patients revealed that lopinavir-ritonavir combination therapy (400 mg and 100 mg, respectively; twice daily) was not effective, and viral loads were identical between treatment group and standard care group at different time points taken for up to 28 days [53]. Larger clinical trials that followed reported similar results. The study led by the RECOVERY collaborative group involving 1616 patients reported no significant improvement in the mortality rate amongst severe COVID-19 cases after oral treatment of lopinavir-ritonavir (400 mg and 100 mg, respectively; twice daily) for 10 days. In addition, one serious adverse event of elevated alanine aminotransferase stemming from lopinavir treatment was recorded from the treatment group [54]. A systematic clinical review on lopinavir-ritonavir (7 trials, 8432 participants) similarly concluded that there was no clinical benefit in terms of mortality, virological clearance and radiological improvements as compared with standard supportive care in COVID-19 patients [55]. The interim findings of the WHO Solidarity trial also found minimal clinical benefit in hospitalized patients treated with lopinavir-ritonavir (200 mg and 50 mg, respectively; twice daily) [40]. Together, these findings concluded the ineffectiveness of the lopinavir-ritonavir combination for the treatment of COVID-19.
Retrospective analyses of the use of lopinavir in COVID-19 treatment suggest several factors that may contribute to the failure of the drug as a therapeutic. Firstly, the lack of inhibitory activity by lopinavir against the purified Mpro of SARS-CoV-2 suggest that this compound is not a direct inhibitor of Mpro [52]. However, the exact molecular target has not been elucidated.
Secondly, lopinavir is reported to have high plasma binding of > 95%, leaving the remaining 5% as unbound drug to act on viral targets [56]. This limits the clinical translation of the drug for COVID-19 treatment. To compensate for this, Cattaneo et al. proposed an adjustment to the clinical dosing in order to achieve the in vitro IC50 value. This corresponds to a dosing of approximately 20-fold higher than the in vitro reported data. However, as noted by the authors, the adjusted dosing would not be feasible due to the potential toxicity risks to patients [56].
Thirdly, the clinical evaluation for lopinavir in COVID-19 treatment commenced in spite of the lack of inhibitory activity by lopinavir against the purified SARS-CoV-2 Mpro and the unsatisfactory in vivo results for lopinavir treatment [45], [52]. Pre-clinical studies were designed to guide clinical studies in predicting the efficacy and dosing of drugs. In view of the unsatisfactory results from the pre-clinical studies however, this was not possible. This may have resulted in the ineffectiveness of lopinavir in COVID-19 treatment.
It is noteworthy that the clinical effectiveness of protease inhibitors are, in general, limited to early-stage disease when the viral load is low, and inhibition of viral replication is crucial. Most clinical trials for lopinavir were evaluated on severe COVID-19 cases, where the viral load is high, and the drug is less effective. To overcome this limitation, protease inhibitors may find synergism with other therapies that act on different stages of disease progression [57]. While viral proteases are good targets and their inhibition is an excellent antiviral strategy, this episode demonstrated the importance of target validation. In the case of lopinavir, while the drug successfully inhibited the main protease of HIV-1 and Mpro of SARS-CoV, this did not extend to the Mpro of SARS-CoV-2. Furthermore, the high plasma binding of lopinavir [56], coupled with the high concentration required to inhibit viral replication in vitro [51], also suggest the importance of considering the pharmacokinetic properties of drugs in their clinical translatability and relevance.
4.1.3. Remdesivir
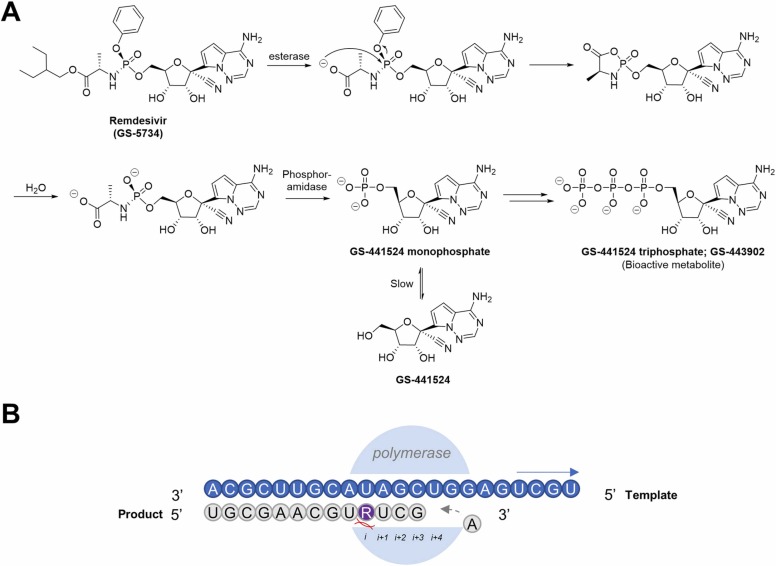
GS-5734 (Remdesivir) is a prodrug that rapidly metabolizes to GS-441524 monophosphate, followed by phosphorylation by the host cell to a bioactive analog of adenosine triphosphate ( Fig. 3A) [58]. As part of a library initially developed by Gilead Sciences against HCV, the drug acts by targeting the RdRp [59]. Kinetic studies performed on recombinant SARS-CoV-2 RdRp revealed an enhanced efficiency in incorporating GS-441524 triphosphate (GS-443902) as opposed to ATP [60]. Following its incorporation into the elongation strand (position i) during viral replication, the machinery stalls while incorporating a nucleotide into the i + 4 position due to steric clashes between the 1’-CN group of the incorporated GS-441524 and the protein (Fig. 3B) [61], [62]. As a result, the incorporated GS-441524 effectively inhibits viral RNA synthesis. Being a relatively late player into the game, remdesivir was first used against the Ebola virus (EBOV), where it demonstrated excellent in vitro potency and retained in vivo activity in animal infection models [58]. Owing to the importance of the RdRp in RNA synthesis and viral replication, remdesivir was en route to becoming a promising broad-spectrum antiviral against RNA viruses [63].
Fig. 3.
(A) Metabolism of prodrug remdesivir (GS-5734) to GS-441524 monophosphate, which is phosphorylated in host cells to the bioactive triphosphate of GS-441524 (GS-443902). (B) When GS-441524 (represented in purple) is incorporated into the replication strand (represented in grey), the machinery stalls while trying to incorporate the 4th nucleotide due to steric clashes between the 1’-CN group of GS-441524 and the polymerase, resulting in its delayed chain termination mechanism of action.
Remdesivir has outstanding in vitro and in vivo antiviral effects against SARS-CoV and MERS-CoV [64]. In human airway epithelial (HAE) cells, remdesivir was reported to have IC50 values of 0.069 µM and 0.074 µM against SARS-CoV and MERS-CoV respectively; in primate models, lower viral loads were detected in the lungs with milder occurrences of lung lesions [64], [65]. Against SARS-CoV-2, remdesivir has also shown promising results in inhibiting the virus in both Vero E6 (EC50 = 0.77 µM) and Huh-7 cells (EC99 = 1.85 µM) [35]. Lower lung viral loads and reduced lung damage were also observed in rhesus macaques treated with remdesivir as compared to those treated with vehicle controls. Notably, reduced viral titers in the bronchoalveolar lavages at 12 h post-treatment from the initial dose indicated clinical benefit for commencing remdesivir treatment in early-stage disease to prevent progression to pneumonia [66]. Impressively, the antiviral effects against SARS-CoV-2 are predominant despite the presence of an exonuclease (ExoN) at the N-terminal domain of nsp14, which functions to correct mismatched nucleosides. Based on the structure of the RdRp in the GS-441524-stalled state, GS-441524 is still concealed within the nsp12 core polymerase during the incorporation of the nucleotide at the i + 4 position (Fig. 3B). This makes the mismatched position inaccessible for the ExoN to initiate excision [67]. Furthermore, it is also postulated that the incorporation of the next three base pairs following GS-441524 occurs rapidly and this could circumvent the proofreading mechanism [68].
To understand the implications of remdesivir resistance in coronaviruses, two mutants, F476L and V553L, were generated by passaging of parent nucleoside GS-441524 in murine hepatitis virus (MHV). Both mutations were identified to be in a conserved region across all coronaviruses in the nsp12 core polymerase-coding region within the RdRp [69]. This involved structural changes to the nsp12 polymerase domain, which is likely to alter the stringency of nucleotide incorporation [70]. When these sequence mutations were introduced into the SARS-CoV genome (yielding mutants F480L and V557L, respectively) a similarly conferred resistance for the virus against remdesivir was obtained. This suggests the involvement of a similar mechanism of resistance following remdesivir exposure. Observations for the impaired ability of the mutants to infect new cells also confirm the high barrier of resistance [69].
However, mixed results were reported from clinical studies involving the use of remdesivir for the treatment of COVID-19. While case reports and small-cohort observational studies for hospitalized patients were encouraging for clinical improvement when remdesivir was used [71], these results were not replicated in larger placebo-controlled RCTs. Similar doubt remains when reported clinical improvement for hospitalized patients was not statistically significant ([72]; 237 hospitalized patients from multi-centers in China) or are of uncertain clinical importance ([73]; 596 hospitalized patients, multinational). The later clinical trial [73] was also criticized for reporting bias in clinical status in terms of measuring outcomes and missing outcome data [74], adding to the uncertainty for the use of remdesivir. A more elaborate clinical trial showed positive effects of remdesivir in shortening time to recovery, but not in severe COVID-19 patients who required mechanical ventilation or extracorporeal membrane oxygenation (ECMO) at the start of enrollment ([75]; with 1062 hospitalized patients, double-blinded placebo-controlled RCT). The demonstrated effectiveness was however not reproduced in the larger randomized WHO Solidarity Trial ([40]; with 2750 hospitalized patients receiving remdesivir intervention). While there is no evidence suggesting benefits of remdesivir in the reduction of mortality in hospitalized patients ([40], [72], [73], [75]; 4 RCTs with a total of 7142 patients), early evidence has suggested the effectiveness of the drug in slowing down disease progression towards hospitalization or mortality in the outpatient setting [76].
As the effectiveness of remdesivir treatment is observed to be limited to cases with mild COVID-19, it may be possible that the metabolism of the drug is hindered in individuals with severe COVID-19, which may contribute to its ineffectiveness in many of the studies conducted. Another possibility is that remdesivir alone is not sufficient for disease management in later stages of the disease where the immune system of the host has been compromised, and managing inflammation and cytokine storms become as important as inhibiting viral replication (see Section 4.1). A workaround is to administer a combination of remdesivir with an immunomodulator such as corticosteroids, and this had proven to be effective in a hamster model of SARS-CoV-2 [77]. A plausible candidate to fill this role is dexamethasone, which will be discussed in a later section. A combination of remdesivir and Janus kinase (JAK) inhibitor baricitinib was also observed to aid patient recovery in clinical settings [78], prompting FDA authorization for use in COVID-19 treatment for hospitalized adults and pediatric patients above the age of two who require external respiratory support [79].
In terms of drug cocktails being considered, a combination of remdesivir and a nsp14-ExoN inhibitor may also be a viable treatment solution [80]. Although previous studies demonstrated that remdesivir is able to evade the proofreading machinery, high concentrations of UTP could potentially negate the inhibitory effects of remdesivir and resume RNA elongation [62]. Consequently, this may allow the nsp14-ExoN proofreader to correct the mismatches. Furthermore, the nsp14-ExoN of SARS-CoV-2 has recently been reported to be involved in its virulence. Specifically, the nsp14-ExoN of SARS-CoV-2 can inhibit the synthesis of host immunogenic proteins, thereby evading host immune response [81]. The development of nsp14-ExoN inhibitors have recently gained popularity, although to date, none has advanced into clinical trials [82], [83], [84]. Preliminary in vitro compound screenings have identified PF-03882845, Inauhzin and Trifluperidol as potential nsp14-ExoN inhibitors that display synergism with remdesivir [85].
Despite the many setbacks, remdesivir has received FDA approval for use in adult patients hospitalized with COVID-19 [86] and in outpatient treatment for mild-to-moderate disease [87]. However, remdesivir has a major shortcoming in the form of its administration, i.e. via intravenous (IV) injections, which poses restrictions in its handling to only clinical professionals. The inconsistent efficacy of remdesivir in clinical trials [40], persistence of parent nucleoside GS-441524 as the metabolite in circulation [66], [88], and effectiveness of GS-441524 in veterinary settings against feline coronavirus (FCoV) [89], [90], [91] have led to studies where GS-441524 was evaluated for its suitability in COVID-19 treatment [92]. While the drug can be administered orally [93], GS-441524 suffers from a slow first-phosphorylation step that is rectified in part by the McGuigan prodrug on remdesivir [59], [94]. Several research groups have also investigated various derivatives and strategies to make oral versions of remdesivir. Jubilant Pharma has recently reported the completion of the pharmacokinetic and safety studies of their remdesivir oral formulation [95]. GS-621763, an oral prodrug of GS-441524, has also been shown to be effective against SARS-CoV-2 in vitro (EC50 = 0.11–0.73 µM in Vero E6 cells), and led to a near-complete reduction of viral titers in ferrets following a 10 mg/kg treatment twice daily for 3 days [96]. Lipidation is also a promising strategy for prodrug development and for altering pharmacokinetic properties. Lipid prodrugs of GS-441524 possessed good oral bioavailability and can be easily activated by cells in the body. The most active compound ODBG-P-RVn (1-O-octadecyl-2-O-benzyl-glycero-3-phosphate remdesivir nucleoside) also showed in vivo efficacy against SARS-CoV-2, hence possessing the potential to be developed into oral antivirals [97]. Besides lipidation, deuteration can also augment the pharmacokinetic properties of drugs such as in the case of VV116, a 7-deuterated derivative of GS-441524. Importantly, pre-clinical results have concluded that the compound may be suitable as an oral drug candidate [98]. As of December 2021, VV116 has entered clinical trials and has been approved for emergency use authorization (EUA) in Uzbekistan.
Altogether, the elaborate account of remdesivir has elicited two learning points in drug development. Firstly, a combined understanding of the disease pathology and the pharmacology of a drug is essential to ensure its timely administration. The responses of the host post-infection, especially cytokine storms, may lead to severe and critically ill clinical progression of COVID-19 patients and as such, late administration of virus-targeting drugs or direct-acting antivirals would render the treatment ineffective as preventing viral replication would no longer be clinically relevant. Secondly, the availability of the drug to be self-administered by the general population is critical to ensure timely treatment and to manage disease progression. In the case of remdesivir, this is hindered by the need for IV administration. The development of an oral formulation of remdesivir is likely to overcome this challenge. Perhaps, a revamped familiarity may reemerge in the near future, and hopefully, it promises better prospects.
4.1.4. Ivermectin
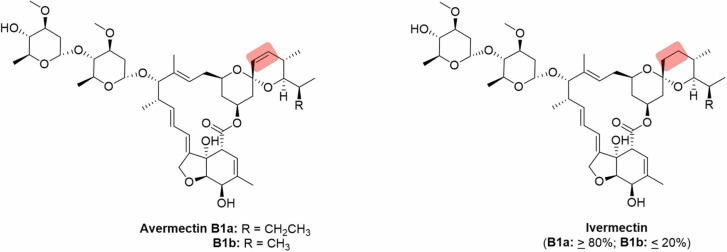
Ivermectin belongs to a class of compounds collectively known as avermectins, a group of macrolides produced by Streptomyces avermitilis [99]. As a semi-synthetic derivative of avermectin ( Fig. 4), ivermectin possesses enhanced safety and antiparasitic properties [100], and was most notably used in the treatment of onchocerciasis or river blindness, and lymphatic filariasis [101], [102]. The discovery of ivermectin as an inhibitor of importin α/β heterodimer (IMP α/β)-mediated nuclear import opened possibilities to explore the drug as an antiviral. This process is believed to be critical for infection by many different RNA viruses [103], [104], [105]. Excellent in vitro antiviral activity against the HIV-1 and dengue virus (DENV) were demonstrated, both of which rely heavily on the IMP α/β nuclear import in their respective viral replication pathways [106]. In addition, ivermectin also show antiviral effects against zika virus (ZIKV) and chikungunya virus (CHIKV), although the mechanisms of inhibition in these viruses are unknown [107], [108].
Fig. 4.
Avermectin and its semi-synthetic derivative ivermectin. Highlighted parts denote the structural difference between the two compounds.
In the context of SARS-CoV-2, a decrease in the release of virions of up to 93% was observed in vitro after a 24 h treatment with 5 µM of ivermectin [109]. No toxicity was observed at that concentration. Although the mechanism of antiviral action of ivermectin in coronaviruses was not confirmed, inhibition of viral replication was similarly proposed to occur through the inhibition of importin (Fig. 2A). A molecular docking study of ivermectin against common antiviral targets such as IMPα, helicase ADP and NDB sites, proteases, and proteins of the RTC identified the RdRp (−10.4 kcal/mol), helicase (NCB site) (−9.6 kcal/mol) and IMPα (−9.0 kcal/mol) as the top three targets as ranked by binding energies [110]. While docking studies are not confirmative of actual drug-protein interactions, the results suggested that binding between ivermectin and IMPα may be possible. IMPα in the IMP α/β heterodimer is reported to house the cargo and recognize the nuclear localization signal [111]. Its inhibition therefore translates to the overall inhibition of the IMP α/β nuclear import pathway. This suggests that inhibition of IMPα by ivermectin could inhibit nuclear import of SARS-CoV-2 viral proteins should it rely on IMP α/β nuclear import for viral replication [109].
While the drug seemed promising, a major flaw was detected in the initial in vitro studies against SARS-CoV-2. Specifically, the concentration of ivermectin used was 50 times above the acceptable level used in clinical practice (700 µg/kg), which limited clinical translatability to humans [112]. Plagiarism concerns from early clinical data further raised questions on the effectiveness of the drug, eventually resulting in the retraction of the preprint article [113]. Furthermore, completed clinical trials of ivermectin (14 trials, 1678 participants) were plagued with small sample sizes and hence lack of statistical power, inconsistent trial designs (varied treatment dose/duration and non-randomized trials) and high risk of bias in reporting study outcomes [114]. The present very-low to low-certainty evidence makes the effect and safety of ivermectin in preventing and treating COVID-19 inconclusive [114]. The serious errors underlying the published studies supporting the use of ivermectin in COVID-19 treatment were also revealed in a recent correspondence by Sheldrick and co-workers, namely non-randomization of interventions, falsification of patient numbers and data, duplication of positive results, inaccurate calculations and lack of scrutinization from local authorities [115], [116]. Other clinical data revealed no significant benefits from ivermectin therapy in alleviating symptoms of mild COVID-19 [117]. The lack of credible scientific data from these studies was likely driven by a publication bias fueled by the hype around ivermectin as a “miracle” anti-COVID drug. Adverse effects from ivermectin without clear benefits and more seriously, an opportunity cost that prevents ivermectin believers from getting vaccination or standard care has implications in the current management of COVID-19 pandemic [115], [116].
At present, ivermectin continues to rank amongst the most controversial drugs in the wake of COVID-19 drug-repurposing efforts. The story of ivermectin and COVID-19 highlights an important lesson in drug discovery and development. Clinical studies are often influenced by the outcomes of animal studies, which in turn draws reference from in vitro results. In this instance, the in vivo exposure of ivermectin that can be safely achieved in humans was well below that required based on the in vitro assay data. This highlights the importance of animal models for predicting and defining the efficacious and toxic doses. In other words, any flaws in experimental design between in vitro validation to clinical translation could snowball into larger and more severe problems, ultimately affecting efficacy and/or toxicity in humans. The choice of a suitable pharmacotherapy for a disease should therefore carefully weigh its potential efficacy and toxicity in the early pre-clinical phase to avoid wastage of resources channeled into large clinical studies. It is thus imperative to ensure that sufficient efficacy and toxicity data has been obtained in pre-clinical studies before considering its further clinical application.
The downfall of ivermectin was a clear mismatch between in vitro and clinical feasibility, further fueled by impartiality and eventually data manipulation. Furthermore, the role of the IMPα/β heterodimer, the postulated target of ivermectin in coronavirus replication, had not been validated. Further investigation of the viral replication cycle of coronaviruses revealed that replication is independent of the importin nuclear transporters [12].
While ivermectin was ineffective as a small molecule therapeutic for COVID-19, this may not necessarily be the end for the drug. Recent studies have shown that ivermectin can modulate IL-6:IL-10 ratio in lung tissues [118]. This ratio had been proposed to be a predictive marker for disease progression in hospitalised patients, where low IL-6:IL-10 ratio is indicative of less severe disease progression [119]. As such, the study suggests that ivermectin treatment may present clinical benefit for managing SARS-CoV-2 infections in patients [118]. More importantly, ivermectin may still be a viable treatment option for infections caused by viruses that rely heavily on the IMP α/β nuclear import pathway [103], [106], which includes members of the flavivirus, retrovirus and alphavirus families.
4.2. Adjuvant treatment
4.2.1. Dexamethasone
While most of the drugs aforementioned were explored as antivirals, managing the severity of disease is just as critical in preventing deaths. Since severe immune responses may lead to cytokine storms in SARS-CoV-2 infection, the importance of using corticosteroids for their potent immunomodulatory properties is highlighted. Amongst many options, one popular choice is dexamethasone (Fig. 1).
Briefly, corticosteroids work by binding to glucocorticoid receptors in the cytosol. Once activated, the glucocorticoid-receptor complex can translocate to the cell nucleus to regulate the expression of pro-inflammatory proteins [120]. For dexamethasone in particular, it could inhibit the production of pro-inflammatory cytokines such as IL-1, IL-2, IL-6, IL-8, IFN-γ and TNF-α [121].
Clinical evaluation of dexamethasone treatment has yielded favourable outcomes for disease recovery with conclusive evidence. A systematic review on the use of corticosteroids for the treatment of COVID-19 analyzed 11 RCTs with 8075 participants; out of the 3072 participants randomized to receive corticosteroids, more than 75% of them received dexamethasone [122]. It was reported with a moderate level of certainty that systemic corticosteroids added on to standard care has a marginal benefit in reducing all-cause mortality as compared to standard care alone in hospitalized COVID-19 patients [122]. In addition, another recent RCT led by the RECOVERY collaborative group similarly reported an improvement in 28-day mortality (482 deaths in treatment group vs 1110 deaths in control group) for hospitalized patients with COVID-19 who received 6 mg dexamethasone once daily compared to those who only received standard of care [123].
Today, dexamethasone is listed as an essential medicine by WHO due to its suitability for multiple indications [124]. Yet, its use for the management of COVID-19 is not without its problems. Due to its immunosuppressant properties, inappropriate prescribing of the drug may result in delayed viral clearance and compromise host immunity, resulting in false impressions of disease recovery [125], [126], [127]. Notably, this has been demonstrated in the history of using dexamethasone against various types of coronavirus-induced pneumonia, where conflicting results were observed from SARS and MERS. Specifically, while dexamethasone could significantly reduce mortality and duration of hospitalization in severe cases of SARS-CoV infection [128], no improvement in mortality was noted for MERS-CoV infections [129], [130].
Although the use of dexamethasone could undoubtedly reduce mortality in patients with severe COVID-19 [123], the fact remains that the drug does not cure the disease but merely alleviate its symptoms. The usage of dexamethasone in the management of different human coronavirus infections, from SARS to MERS and COVID-19, has emphasized that its prescription must not be indiscriminate. Instead, a case-by-case evaluation guided by several considerations must be made prior to its prescription, such as the need to evaluate the pros and cons for the administration of the drug, pre-existing medical conditions of the patients, severity of the disease, and the dose and duration of the treatment [131]. Recent developments have also suggested that dexamethasone can in fact bind to ACE2 of the host, which can allow the corticosteroid to act as an inhibitor for viral entry [132]. However, this observation has not been validated in in vivo models.
4.2.2. Baricitinib
Janus kinase (JAK) inhibitors are a class of compounds that possess inhibitory properties against cytokines to attenuate and modify immune response. Amongst JAK inhibitors, baricitinib, an FDA-approved drug for the treatment of rheumatoid arthritis, has received much interest lately as a potential anti-inflammatory agent for suppressing elevated cytokine levels in severe SARS-CoV-2 infection [133]. Due to its high binding affinity for AP2-associated protein kinase 1 (AAK1), a regulator of endocytosis [134], baricitinib was proposed to inhibit viral entry into host cells. Collectively, all these have identified baricitinib as a potential dual-action drug [135].
Baricitinib treatment in primary human hepatocytes and lung organoids was observed to reverse the effects of interferon-α2 (IFN-α2)-induced gene upregulation, including the increased expression of pro-inflammatory cytokines in a cytokine storm and ACE2 induction [136]. The sensitivity of ACE2 expression in liver cells to IFN-α2 signalling hence allowed baricitinib to moderate SARS-CoV-2 entry in the liver. In the same in vitro study, baricitinib was also shown to efficiently block viral entry at nanomolar concentrations through the inhibition of specific numb-associated kinases (NAKs) including AAK1 and cyclin G-associated kinase (GAK) [136]. Clinical trials with baricitinib have shown promising outcomes as outlined below. In a systematic review and meta-analysis (13 studies, 3977 participants), baricitinib treatment was associated with significant reduction in mortality by day 28 in hospitalized patients with COVID-19, without increasing the occurrence of adverse events [137]. Similarly in another systematic review with meta-analysis, (12 studies, 3564 patients, including 2 double-blinded multicentre RCTs with 2558 patients, 6 observational cohort studies and 4 non-randomised trials), baricitinib demonstrated consistent reduction in mortality rate across subgroups and improvement in clinical status as indicated by lower rate of intensive care unit (ICU) admissions and less worsening to mechanical ventilation [138]. For the two RCTs included in this systematic review, they are, by far, the only two large-scale double-blinded trials on baricitinib with results available – the Adaptive COVID-19 Treatment Trial 2 (ACTT-2) [78] with 1033 hospitalised patients on the combination therapy of remdesivir (200 mg loading on day 1 + 100 mg for days 2–10) and baricitinib (4 mg daily dose for 14 days) and the COV-BARRIER trial [139] (multinational, 1525 hospitalized patients) on baricitinib (4 mg daily dose for 14 days), as compared to standard of care. While both trials concluded that there was a reduction in all-cause mortality with baricitinib use in severe patients who require baseline oxygen support, the COV-BARRIER trial also reported synergism when baricitinib is used with systemic corticosteroids (i.e. dexamethasone), in reducing death amongst hospitalised adults [139]. The ACTT-2 trial additionally showed a reduction in severe adverse events (e.g. multiple organ dysfunction syndrome, shock and acute kidney injury) with the combination therapy [78].
In view of these promising clinical data, WHO has recently recommended baricitinib as an add-on therapy with corticosteroids in severe patients with COVID-19 pneumonia [140]. Furthermore, as mentioned in Section 4.3, the combination therapy of baricitinib and remdesivir have also been granted approval for the treatment of COVID-19 in hospitalized adults or paediatric patients above the age of two who require mechanical ventilation or ECMO [79]. The impressive safety profile of baricitinib also warrants further investigation of its use with other virus-targeting agents [141], [142].
Of note, the ability of baricitinib to moderate SARS-CoV-2 entry into lung cells was a prediction made by artificial intelligence [135]. Studies have since showed that the modulatory effect of baricitinib for viral entry work in liver cells as well [136]. The dual-action role of baricitinib against SARS-CoV-2 infections is an impressive feat that demonstrates the practicality of a single drug acting via multiple mechanisms of action, which may be impossible to predict without the aid of computational studies. This highlights the growing role of computer-aided drug discovery in streamlining processes of target identification and drug reutilization.
5. New small molecule antiviral treatment – a glimpse of hope?
The urgency of the pandemic had prompted many drugs to be reutilized as possible treatment options for COVID-19, albeit many were found to be ineffective. Compounds developed by traditional medicinal chemistry approaches began to gain traction, including molnupiravir and nirmatrelvir, which are briefly described below.
5.1. Molnupiravir
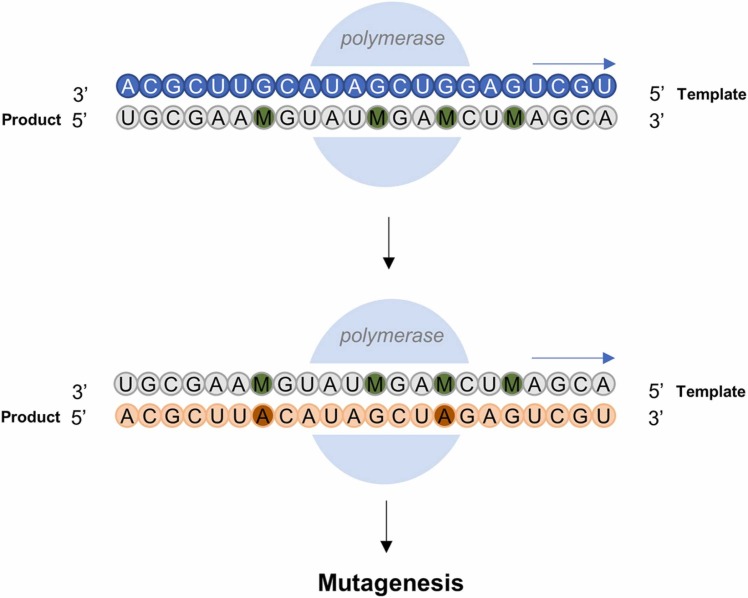
Amongst the antivirals that have been tested against SARS-CoV-2 thus far, the prodrug remdesivir undoubtedly holds the most promise. Recently, another nucleoside prodrug molnupiravir (MK-4482; EIDD-2801; Fig. 1) was observed to exhibit significant improvement to non-hospitalized COVID-19 patients [143]. This isopropyl ester prodrug is metabolized to β-D-N4-hydroxycytidine (NHC; EIDD-1931) [144], [145]. NHC is primarily a mimetic of cytidine (C), although its tautomerism allows it to also mimic uridine (U) [146]. This allows the RdRp to effectively assimilate it into the replication strand, pairing it with guanidine (G). When the NHC-containing strand is used as a template for further replication, A (adenosine) or G (but not C or U) may be paired to NHC, thereby inducing mutagenesis which is lethal to the virus if left unchecked ( Fig. 5) [147]. Initial clinical evaluation of the drug saw a 50% improvement in hospitalization risk and mortality in non-hospitalized patients with mild to severe disease, urging for a premature termination of recruitment into the Phase 3 MOVe-Out trials in view of application for emergency usage [148]. However, updates on the MOVe-Out trials had since reported a mere 30% improvement in hospitalization risk – a significant drop from the initial 50% reported [149]. The drug was also not associated with any clinical improvements in hospitalized patients with severe COVID-19, which prompted the termination of the MOVe-In trial [143].
Fig. 5.
The metabolite of Molnupiravir, NHC (represented in green) is a primary mimetic for cytidine, pairing with guanidine during viral replication. Tautomerism of NHC can allow it to occasionally mimic uridine, thus pairing with adenosine. When the NHC-containing strand is used as a template for further replication, adenosine or guanidine may be paired with NHC, thereby inducing mutagenesis.
Prior to Merck’s revelation of molnupiravir, research groups in collaboration with Emory Institute of Drug Development (EIDD) had demonstrated the in vitro potency of the drug against SARS-CoV, MERS-CoV and SARS-CoV-2 inoculated in human epithelial cells [150], [151]. The safety and pharmacokinetic profiles of molnupiravir were also carefully examined prior to patient recruitment for clinical trials [152]. It is postulated that molnupiravir is likely to escape interference by nsp14-ExoN due to the apparent stability of NHC-G and NHC-A base pairs, which may not induce correction by the proofreading machinery [147], [153]. The drug has also demonstrated effectiveness against infection models of SARS-CoV-2 in mice, both as an antiviral treatment and a prophylaxis [154]. Evidence also suggests that the drug may prevent the spread of the virus in ferret infection models [145]. Taken together, this suggests that the drug may be able to prevent and treat mild COVID-19 cases, and limit transmissibility in the community.
Besides these successes, molnupiravir also holds an important advantage over remdesivir. While remdesivir must be administered intravenously by clinical professionals, molnupiravir can be formulated as an oral pill. In theory, this should greatly improve logistical accessibility and distribution and hence treatment compliance. Merck and Ridgeback have signed deals with regional drug manufacturers in hopes to lower the price of the drug [155], but it is unknown if this move is sufficient to improve accessibility.
Another lingering concern involves the toxicity of molnupiravir – can the drug induce mutagenesis in the host as well? Initial studies have disputed this [150], but recent studies warn of possible host mutagenesis in view of the common DNA and RNA precursors utilized by both the host and virus. This mutagenesis, though, may be confined to dividing cells where DNA is constantly synthesized, owing to the mechanism of action of the drug [156]. These mutagenic effects have been observed in mammalian cells [156], but its effects to the entire organism have not been verified. In addition, experts have also cautioned for potential drug resistance. Due to the high viral loads over the duration of an infection, almost every possible mutation is highly likely to occur. While direct mutations to the RdRp remain unlikely, survival pressure that relies on fitness advantage may result in stronger variants or tolerance to molnupiravir [157], [158].
Molnupiravir received EUA by the FDA on 23 December 2021, following its first authorization for use as a COVID-19 therapeutic in the U.K. 7 weeks prior. It should be noted that both authorizations are exclusively for adult patients with mild-to-moderate disease and within the first 5 days of disease onset [155], [159]. While the outcomes for the clinical evaluation of molnupiravir were not spectacular [149], the drug still undoubtedly led to improvement in patient recovery and can potentially be a useful prophylaxis against SARS-CoV-2 infections.
5.2. PaxlovidTM (Nirmatrelvir-ritonavir)
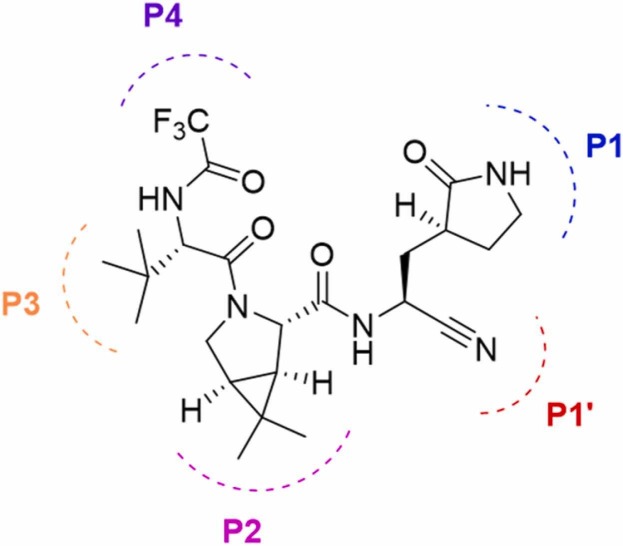
As drug discovery research progresses with the pandemic, Pfizer announced a novel protease inhibitor designed to target the Mpro of SARS-CoV-2 – a different target from the nucleos(t)ide drugs remdesivir and molnupiravir. Sold under the brand name PaxlovidTM, the treatment comprises of nirmatrelvir (Fig. 1) and low-dose ritonavir to slow down hepatic metabolism. Nirmatrelvir is a peptidomimetic with a glutamine-mimicking pyrrolidone moiety in the P1 domain and a cyano group in the P1’ domain ( Fig. 6), which fits in the S1 and S1’ pockets of the Mpro of SARS-CoV-2 respectively. While the cyano groups of both remdesivir and nirmatrelvir are key to their respective mechanisms of action, these cyano groups have different functions in their inhibitory mechanisms. In remdesivir, the cyano group causes steric hindrance that stalls the RdRp machinery while in nirmatrelvir, the cyano group is an electrophilic warhead that forms a thioimidate adduct with the catalytic cysteine of Mpro, effectively making the compound a reversible covalent inhibitor. The in vitro and in vivo efficacies of nirmatrelvir against SARS-CoV-2, as well as its pharmacokinetic profiles, have been reported [160].
Fig. 6.
Structure of peptidomimetic Nirmatrelvir. P4 to P1 denotes the positions of the sequence from the N-terminus to the C-terminus.
Like molnupiravir, PaxlovidTM is also formulated for oral administration [161]. Interim results for phase II-III clinical evaluation of PaxlovidTM (1219 patients) reported that the drug reduced hospitalization risk and mortality by 89% in patients with mild COVID-19 when taken within three days of onset of symptoms [162]. In view of that, PaxlovidTM has also been granted EUA by the FDA on 22 December 2021 for mild-to-moderate disease in patients with minimum age of 12 [163].
In view of all the data presented, PaxlovidTM may be the most promising treatment option against COVID-19. Unlike nucleoside analogues, protease inhibitors have more specific interactions with their targets, which limits their otherwise broad-spectrum antiviral activity. However, the risk of off-target interactions with other nucleophilic residues in peptides and proteins in the host body are also additional challenges for a covalent inhibitor [164].
6. Concluding remarks and future perspective
On hindsight, mankind should have been more prepared for a pandemic. The outbreaks of SARS (2003, (+)-sense RNA virus, Coronaviridae family), swine flu (2009, (-)-sense RNA virus, Orthomyxoviridae family), MERS (2012, (+)-sense RNA virus, Coronaviridae family), Ebola (2014, ss-(-)-sense RNA virus, Filoviridae family) and Zika (2016, (+)-sense RNA virus, Flaviviridae family) show how easily infectious diseases that are originated from viruses can rapidly spread in this highly connected world. When we finally emerge from this pandemic, we must be prepared for the next one which will again be an unseen enemy. This preparation should be in the continued study of viruses and therapeutics despite the potential lack of financial gains in the short term. In view of the rapid mutation rates seen in viruses, multi-targeted broad-spectrum antivirals would be highly coveted.
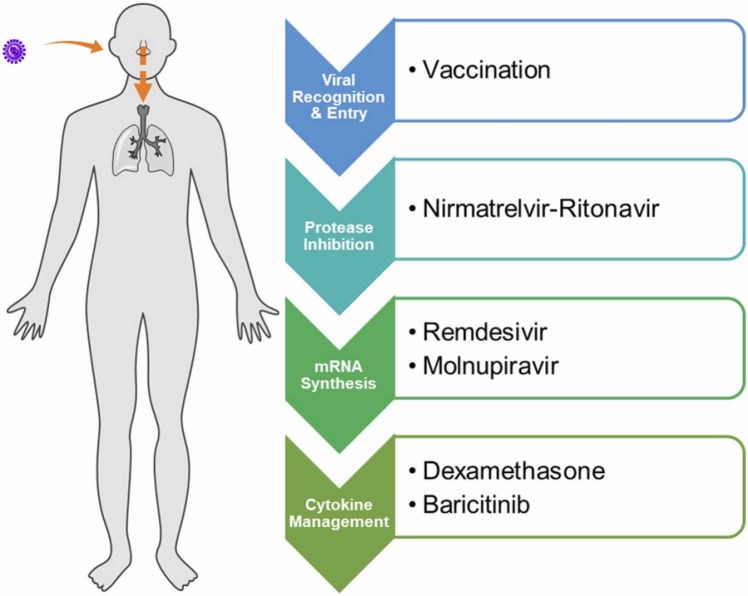
The development of antiviral agents poses intrinsic challenges that are easily overlooked. One major reason may be an initial lack of understanding in the virus pathology. Moving forward, one hopes that the lessons uncovered herein could aid in drug development against viral pathogens. These include (i) the selection of appropriate cell lines for antiviral studies, (ii) validation of viral targets, (iii) a proper workflow to demonstrate translatability from in vitro to in vivo efficacy and ultimately clinical relevance, (iv) identification of the best window for therapeutic intervention to achieve optimal outcome, and lastly, (v) the timely administration of antiviral treatment that could otherwise render the drugs ineffective. While vaccination remains our best preventive measure against SARS-CoV-2 infections, the availability of therapeutics is equally crucial in managing treatment for people who are vulnerable, especially the elderly as well as the immunocompromised. Promising treatment options at present include protease inhibitor PaxlovidTM, nucleos(t)ide analogues remdesivir and molnupiravir, and immunomodulators dexamethasone and baricitinib ( Fig. 7).
Fig. 7.
Effective treatment options against COVID-19 thus far and the phases of SARS-CoV-2 pathophysiology for which they act on.
While many celebrate the prospects of an end to the disease that brought the world to a standstill, history has taught us to be wary. Potential toxicity of molnupiravir by mutagenesis to organs and germline in the host should be examined. If ultimately proven to be effective and safe, both molnupiravir and nirmatrelvir could be a huge step forward for mankind in the treatment of COVID-19. Despite their promise, the interactions of molnupiravir and PaxlovidTM with other drugs are not yet well understood. This is especially true for PaxlovidTM, which contains the CYP450 inhibitor ritonavir, hence reducing tolerability for patients on multiple medications [165]. Another lingering concern involves the development of resistance to these drugs, especially in individuals with weakened immunity which could result in prolonged SARS-CoV-2 infection and subsequent treatment duration, or those who are unable to withstand the full treatment regimen [165]. Concerns on the effectiveness of these drugs against emerging VOCs have also gained traction. Thankfully, remdesivir, molnupiravir and nirmatrelvir have all shown promising results against the Omicron variant (B.1.1.529) and past VOCs [166], which further highlights the relevance of small molecule therapeutics in the ongoing fight against viral infections.
Lest we forget the lives that have been lost in this current war against COVID-19, we must continue to build our arsenal of small molecule therapeutics, especially orally available ones, against viral diseases. For that to come to fruition, government authorities and funding agencies must be willing to support drug discovery and development efforts against infectious pathogens. International collaboration in drug development research should also be encouraged to share scientific developments and improve interconnectedness in the scientific community. Lastly, a strong emphasis should also be placed towards sufficiently powered clinical trials in times of a pandemic to ensure confidence and validity in the results obtained. The ball should be in our court the next time round.
CRediT authorship contribution statement
Wei Shen Ho: Conceptualization, Writing – original and all subsequent drafts, Visualization. Ruirui Zhang: Writing – sections in original draft, Reviewing selected sections. Yeong Lan Tan: Writing and reviewing selected sections in revised drafts. Christina L. L. Chai: Conceptualization, Writing – original draft preparation and reviewing of all drafts, Supervision.
Declaration of competing interest
The authors declare no competing financial interest.
Acknowledgements
This work was supported by a MOE Tier 1 Grant (A-0004633-00-00) from the Ministry of Education, Singapore and an NUS Research Scholarship. Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not reflect the views of the Ministry of Education, Singapore and National University of Singapore, Singapore.
References
- 1.Khandia R., Singhal S., Alqahtani T., Kamal M.A., El-Shall N.A., Nainu F., Desingu P.A., Dhama K. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res. 2022;209 doi: 10.1016/j.envres.2022.112816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu Y.B., Briggs K.T., Taraban M.B., Brinson R.G., Marino J.P. Grand challenges in pharmaceutical research series: ridding the cold chain for biologics. Pharm. Res. 2021;38(1):3–7. doi: 10.1007/s11095-021-03008-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naveja J.J., Madariaga-Mazon A., Flores-Murrieta F., Granados-Montiel J., Maradiaga-Cecena M., Alaniz V.D., Maldonado-Rodriguez M., Garcia-Morales J., Senosiain-Pelaez J.P., Martinez-Mayorga K. Union is strength: antiviral and anti-inflammatory drugs for COVID-19. Drug Discov. Today. 2021;26(1):229–239. doi: 10.1016/j.drudis.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarighi P., Eftekhari S., Chizari M., Sabernavaei M., Jafari D., Mirzabeigi P. A review of potential suggested drugs for coronavirus disease (COVID-19) treatment. Eur. J. Pharmacol. 2021;895 doi: 10.1016/j.ejphar.2021.173890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D., Hu J., Li D., Yang W., Yin S.F., Qiu R. Reviews on biological activity, clinical trial and synthesis progress of small molecules for the treatment of COVID-19. Top. Curr. Chem. 2021;379(1):52. doi: 10.1007/s41061-020-00318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiwari V., Beer J.C., Sankaranarayanan N.V., Swanson-Mungerson M., Desai U.R. Discovering small-molecule therapeutics against SARS-CoV-2. Drug Discov. Today. 2020;25(8):1535–1544. doi: 10.1016/j.drudis.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards A. What are the odds of finding a COVID-19 drug from a lab repurposing screen? J. Chem. Inf. Model. 2020;60(12):5727–5729. doi: 10.1021/acs.jcim.0c00861. [DOI] [PubMed] [Google Scholar]
- 8.Tompa D.R., Immanuel A., Srikanth S., Kadhirvel S. Trends and strategies to combat viral infections: a review on FDA approved antiviral drugs. Int. J. Biol. Macromol. 2021;172:524–541. doi: 10.1016/j.ijbiomac.2021.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhuri S., Symons J.A., Deval J. Innovation and trends in the development and approval of antiviral medicines: 1987-2017 and beyond. Antivir. Res. 2018;155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan-Tack K., Harrington P., Bensman T., Choi S.-Y., Donaldson E., O’Rear J., McMillan D., Myers L., Seaton M., Ghantous H., Cao Y., Valappil T., Birnkrant D., Struble K. Benefit-risk assessment for brincidofovir for the treatment of smallpox: U.S. Food and drug administration’s evaluation. Antivir. Res. 2021;195 doi: 10.1016/j.antiviral.2021.105182. [DOI] [PubMed] [Google Scholar]
- 11.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelrahman Z., Li M., Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza a respiratory viruses. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020;21(1):224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomes C.P., Fernandes D.E., Casimiro F., da Mata G.F., Passos M.T., Varela P., Mastroianni-Kirsztajn G., Pesquero J.B. Cathepsin L in COVID-19: from pharmacological evidences to genetics. Front. Cell. Infect. Microbiol. 2020;10(777) doi: 10.3389/fcimb.2020.589505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020;10(1):14991. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang L., Xie X., Tu Z., Fu J., Xu D., Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target. Ther. 2021;6(1):255. doi: 10.1038/s41392-021-00679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hojyo S., Uchida M., Tanaka K., Hasebe R., Tanaka Y., Murakami M., Hirano T. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 2020;40(1):37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mallucci L. Effect of chloroquine on lysosomes and on growth of mouse hepatitis virus (MHV-3) Virology. 1966;28(3):355–362. doi: 10.1016/0042-6822(66)90046-8. [DOI] [PubMed] [Google Scholar]
- 24.Slater A.F.G. Chloroquine: mechanism of drug action and resistance in plasmodium falciparum. Pharmacol. Ther. 1993;57(2):203–235. doi: 10.1016/0163-7258(93)90056-j. [DOI] [PubMed] [Google Scholar]
- 25.Tsai W.-P., Nara P.L., Kung H.-F., Oroszlan S. Inhibition of human immunodeficiency virus infectivity by chloroquine. AIDS Res. Hum. Retrovir. 1990;6(4):481–489. doi: 10.1089/aid.1990.6.481. [DOI] [PubMed] [Google Scholar]
- 26.Rolain J.-M., Colson P., Raoult D. Recycling of chloroquine and its hydroxyl analogue to face bacterial, fungal and viral infections in the 21st century. Int. J. Antimicrob. Agents. 2007;30(4):297–308. doi: 10.1016/j.ijantimicag.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keyaerts E., Vijgen L., Maes P., Neyts J., Ranst M.V. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323(1):264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2(1):69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17(1) doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Touret F., de Lamballerie X. Of chloroquine and COVID-19. Antivir. Res. 2020;177 doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colson P., Rolain J.-M., Lagier J.-C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55(4) doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devaux C.A., Rolain J.-M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McChesney E.W. Animal toxicity and pharmacokinetics of hydroxychloroquine sulfate. Am. J. Med. 1983;75(1):11–18. doi: 10.1016/0002-9343(83)91265-2. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh B., Ryan H., Kredo T., Chaplin M., Fletcher T. Chloroquine or hydroxychloroquine for prevention and treatment of COVID‐19. Cochrane Database Syst. Rev. 2021;2 doi: 10.1002/14651858.CD013587.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elavarasi A., Prasad M., Seth T., Sahoo R.K., Madan K., Nischal N., Soneja M., Sharma A., Maulik S.K., Shalimar P., Garg Chloroquine and hydroxychloroquine for the treatment of COVID-19: a systematic review and meta-analysis. J. Gen. Intern. Med. 2020;35(11):3308–3314. doi: 10.1007/s11606-020-06146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pathak D.S.K., Salunke D.A.A., Thivari D.P., Pandey A., Nandy D.K., Harish D., Ratna V.K., Pandey D.S., Chawla D.J., Mujawar D.J., Dhanwate D.A., Menon D.V. No benefit of hydroxychloroquine in COVID-19: results of systematic review and meta-analysis of randomized controlled trials”. Diabetes Metab. Syndr. 2020;14(6):1673–1680. doi: 10.1016/j.dsx.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horby P., Mafham M., Linsell L., et al. Effect of hydroxychloroquine in hospitalized patients with Covid-19. N. Engl. J. Med. 2020;383(21):2030–2040. doi: 10.1056/NEJMoa2022926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan H., Peto R., Henao-Restrepo A.-M., Preziosi M.-P., Sathiyamoorthy V., Abdool Q., Karim, e. al, Repurposed antiviral drugs for covid-19 — Interim WHO solidarity trial results. N. Engl. J. Med. 2021;384(6):497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., Gassen N.C., Müller M.A., Drosten C., Pöhlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585(7826):588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- 42.Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K.S., Sidwell R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-CoV inhibitors for inhibition of SARS-Cov replication in BALB/c mice. Antivir. Chem. Chemother. 2006;17(5):275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 43.Maisonnasse P., Guedj J., Contreras V., Behillil S., Solas C., Marlin R., Naninck T., Pizzorno A., Lemaitre J., Gonçalves A., Kahlaoui N., Terrier O., Fang R.H.T., Enouf V., Dereuddre-Bosquet N., Brisebarre A., Touret F., Chapon C., Hoen B., Lina B., Calatrava M.R., van der Werf S., de Lamballerie X., Le R. Grand, Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature. 2020;585(7826):584–587. doi: 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]
- 44.Rosenke K., Jarvis M.A., Feldmann F., Schwarz B., Okumura A., Lovaglio J., Saturday G., Hanley P.W., Meade-White K., Williamson B.N., Hansen F., Perez-Perez L., Leventhal S., Tang-Huau T.-L., Callison J., Haddock E., Stromberg K.A., Scott D., Sewell G., Bosio C.M., Hawman D., de Wit E., Feldmann H. Hydroxychloroquine prophylaxis and treatment is ineffective in macaque and hamster SARS-CoV-2 disease models. JCI Insight. 2020;5(23) doi: 10.1172/jci.insight.143174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park S.-J., Yu K.-M., Kim Y.-I., Kim S.-M., Kim E.-H., Kim S.-G., Kim Eun J., Casel Mark Anthony B., Rollon R., Jang S.-G., Lee M.-H., Chang J.-H., Song M.-S., Jeong Hye W., Choi Y., Chen W., Shin W.-J., Jung Jae U., Choi Young K., Palese P. Antiviral efficacies of FDA-approved drugs against SARS-CoV-2 infection in ferrets. mBio. 2020;11(3):e01114–e01120. doi: 10.1128/mBio.01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su B., Wang Y., Zhou R., Jiang T., Zhang H., Li Z., Liu A., Shao Y., Hua W., Zhang T., Wu H., He S., Dai L., Sun L. Efficacy and tolerability of lopinavir/ritonavir- and efavirenz-based initial antiretroviral therapy in HIV-1-infected patients in a tertiary care hospital in Beijing, China. Front. Pharmacol. 2019;10:1472. doi: 10.3389/fphar.2019.01472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walmsley S., Bernstein B., King M., Arribas J., Beall G., Ruane P., Johnson M., Johnson D., Lalonde R., Japour A., Brun S., Sun E. Lopinavir–ritonavir versus nelfinavir for the initial treatment of HIV infection. N. Engl. J. Med. 2002;346(26):2039–2046. doi: 10.1056/NEJMoa012354. [DOI] [PubMed] [Google Scholar]
- 48.Wilde A.Hd, Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., Nieuwkoop Sv, Bestebroer T.M., v.d. Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of middle east respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58(8):4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu C.M., Cheng V.C.C., Hung I.F.N., Wong M.M.L., Chan K.H., Chan K.S., Kao R.Y.T., Poon L.L.M., Wong C.L.P., Guan Y., Peiris J.S.M., Yuen K.Y. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu C.-Y., Jan J.-T., Ma S.-H., Kuo C.-J., Juan H.-F., Cheng Y.-S.E., Hsu H.-H., Huang H.-C., Wu D., Brik A., Liang F.-S., Liu R.-S., Fang J.-M., Chen S.-T., Liang P.-H., Wong C.-H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA. 2004;101(27):10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choy K.-T., Wong A.Y.-L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P.-H., Huang X., Peiris M., Yen H.-L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahdi M., Mótyán J.A., Szojka Z.I., Golda M., Miczi M., Tőzsér J. Analysis of the efficacy of HIV protease inhibitors against SARS-CoV-2′s main protease. Virol. J. 2020;17(1):190. doi: 10.1186/s12985-020-01457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C., Trial A. of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horby P.W., Mafham M., Bell J.L., Linsell L., Staplin N., Emberson J., Palfreeman A., Raw J., Elmahi E., Prudon B., Green C., Carley S., Chadwick D., Davies M., Wise M.P., Baillie J.K., Chappell L.C., Faust S.N., Jaki T., Jefferey K., Lim W.S., Montgomery A., Rowan K., Juszczak E., Haynes R., Landray M.J. Lopinavir–ritonavir in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2020;396(10259):1345–1352. doi: 10.1016/S0140-6736(20)32013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patel T.K., Patel P.B., Barvaliya M., Saurabh M.K., Bhalla H.L., Khosla P.P. Efficacy and safety of lopinavir-ritonavir in COVID-19: a systematic review of randomized controlled trials. J. infect. Public Health. 2021;14(6):740–748. doi: 10.1016/j.jiph.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cattaneo D., Cattaneo D., Gervasoni C., Corbellino M., Galli M., Riva A., Gervasoni C., Clementi E., Clementi E. Does lopinavir really inhibit SARS-CoV-2? Pharmacol. Res. 2020;158 doi: 10.1016/j.phrs.2020.104898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krumm Z.A., Lloyd G.M., Francis C.P., Nasif L.H., Mitchell D.A., Golde T.E., Giasson B.I., Xia Y. Precision therapeutic targets for COVID-19. Virol. J. 2021;18(1):66. doi: 10.1186/s12985-021-01526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.-S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531(7594):381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho A., Saunders O.L., Butler T., Zhang L., Xu J., Vela J.E., Feng J.Y., Ray A.S., Kim C.U. Synthesis and antiviral activity of a series of 1′-substituted 4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg. Med. Chem. Lett. 2012;22(8):2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dangerfield T.L., Huang N.Z., Johnson K.A. Remdesivir is effective in combating COVID-19 because It Is a better substrate than ATP for the viral RNA-dependent RNA polymerase. iScience. 2020;23(12) doi: 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. Mechanism of inhibition of ebola virus RNA-Dependent RNA polymerase by remdesivir. Viruses. 2019;11(4):326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bravo J.P.K., Dangerfield T.L., Taylor D.W., Johnson K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell. 2021;81(7) doi: 10.1016/j.molcel.2021.01.035. 1548-1552.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lo M.K., Jordan R., Arvey A., Sudhamsu J., Shrivastava-Ranjan P., Hotard A.L., Flint M., McMullan L.K., Siegel D., Clarke M.O., Mackman R.L., Hui H.C., Perron M., Ray A.S., Cihlar T., Nichol S.T., Spiropoulou C.F. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci. Rep. 2017;7(1):43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sheahan T.P., Sims Amy C., Graham Rachel L., Menachery Vineet D., Gralinski Lisa E., Case James B., Leist Sarah R., Pyrc K., Feng Joy Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke Michael O., Mackman Richard L., Spahn Jamie E., Palmiotti Christopher A., Siegel D., Ray Adrian S., Cihlar T., Jordan R., Denison Mark R., Baric Ralph S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9(396):eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA. 2020;117(12):6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williamson B.N., Feldmann F., Schwarz B., Meade-White K., Porter D.P., Schulz J., van Doremalen N., Leighton I., Yinda C.K., Pérez-Pérez L., Okumura A., Lovaglio J., Hanley P.W., Saturday G., Bosio C.M., Anzick S., Barbian K., Cihlar T., Martens C., Scott D.P., Munster V.J., de Wit E. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature. 2020;585(7824):273–276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Höbartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12(1):279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson K.A., Dangerfield T. Chapter Two - Mechanisms of inhibition of viral RNA replication by nucleotide analogs. Enzymes. 2021;49:39–62. doi: 10.1016/bs.enz.2021.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R., Subbarao K., Gallagher T., Enjuanes L. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9(2) doi: 10.1128/mBio.00221-18. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shannon A., Le N.T.-T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X., Nicastri E., Oda R., Yo K., Quiros-Roldan E., Studemeister A., Redinski J., Ahmed S., Bernett J., Chelliah D., Chen D., Chihara S., Cohen S.H., Cunningham J., D’Arminio Monforte A., Ismail S., Kato H., Lapadula G., L’Her E., Maeno T., Majumder S., Massari M., Mora-Rillo M., Mutoh Y., Nguyen D., Verweij E., Zoufaly A., Osinusi A.O., DeZure A., Zhao Y., Zhong L., Chokkalingam A., Elboudwarej E., Telep L., Timbs L., Henne I., Sellers S., Cao H., Tan S.K., Winterbourne L., Desai P., Mera R., Gaggar A., Myers R.P., Brainard D.M., Childs R., Flanigan T. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382(24):2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Y. Wang, D. Zhang, G. Du, R. Du, J. Zhao, Y. Jin, S. Fu, L. Gao, Z. Cheng, Q. Lu, Y. Hu, G. Luo, K. Wang, Y. Lu, H. Li, S. Wang, S. Ruan, C. Yang, C. Mei, Y. Wang, D. Ding, F. Wu, X. Tang, X. Ye, Y. Ye, B. Liu, J. Yang, W. Yin, A. Wang, G. Fan, F. Zhou, Z. Liu, X. Gu, J. Xu, L. Shang, Y. Zhang, L. Cao, T. Guo, Y. Wan, H. Qin, Y. Jiang, T. Jaki, F.G. Hayden, P.W. Horby, B. Cao, C. Wang, Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial, Lancet 395(10236) (2020) 1569–1578. [DOI] [PMC free article] [PubMed]
- 73.C.D. Spinner, R.L. Gottlieb, G.J. Criner, J.R. Arribas López, A.M. Cattelan, A. Soriano Viladomiu, O. Ogbuagu, P. Malhotra, K.M. Mullane, A. Castagna, L.Y.A. Chai, M. Roestenberg, O.T.Y. Tsang, E. Bernasconi, P. Le Turnier, S.-C. Chang, D. SenGupta, R.H. Hyland, A.O. Osinusi, H. Cao, C. Blair, H. Wang, A. Gaggar, D.M. Brainard, M.J. McPhail, S. Bhagani, M.Y. Ahn, A.J. Sanyal, G. Huhn, F.M. Marty, f.t.G.-U.–.-. Investigators, Effect of Remdesivir vs Standard Care on Clinical Status at 11 Days in Patients With Moderate COVID-19: A Randomized Clinical Trial, JAMA 324(11) (2020) 1048–1057. [DOI] [PMC free article] [PubMed]
- 74.Ansems K., Grundeis F., Dahms K., Mikolajewska A., Thieme V., Piechotta V., Metzendorf M.I., Stegemann M., Benstoem C., Fichtner F. Remdesivir for the treatment of COVID‐19. Cochrane Database Syst. Rev. 2021;8 doi: 10.1002/14651858.CD014962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., M.-d. Oh G.M., Ruiz-Palacios T., Benfield G., Fätkenheuer M.G., Kortepeter R.L., Atmar C.B., Creech J., Lundgren A.G., Babiker S., Pett J.D., Neaton T.H., Burgess T., Bonnett M., Green M., Makowski A., Osinusi S., Nayak H.C., Lane Remdesivir for the treatment of covid-19 — final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gottlieb R.L., Vaca C.E., Paredes R., Mera J., Webb B.J., Perez G., Oguchi G., Ryan P., Nielsen B.U., Brown M., Hidalgo A., Sachdeva Y., Mittal S., Osiyemi O., Skarbinski J., Juneja K., Hyland R.H., Osinusi A., Chen S., Camus G., Abdelghany M., Davies S., Behenna-Renton N., Duff F., Marty F.M., Katz M.J., Ginde A.A., Brown S.M., Schiffer J.T., Hill J.A. Early remdesivir to prevent progression to severe Covid-19 in outpatients. N. Engl. J. Med. 2021;386(4):305–315. doi: 10.1056/NEJMoa2116846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ye Z.-W., Yuan S., Chan J.F.-W., Zhang A.J., Yu C.-Y., Ong C.P., Yang D., Chan C.C.-Y., Tang K., Cao J., Poon V.K.-M., Chan C.C.-S., Cai J.-P., Chu H., Yuen K.-Y., Jin D.-Y. Beneficial effect of combinational methylprednisolone and remdesivir in hamster model of SARS-CoV-2 infection. Emerg. Microbes Infect. 2021;10(1):291–304. doi: 10.1080/22221751.2021.1885998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.A.C. Kalil, T.F. Patterson, A.K. Mehta, K.M. Tomashek, C.R. Wolfe, V. Ghazaryan, V.C. Marconi, G.M. Ruiz-Palacios, L. Hsieh, S. Kline, V. Tapson, N.M. Iovine, M.K. Jain, D.A. Sweeney, H.M. El Sahly, A.R. Branche, J. Regalado Pineda, D.C. Lye, U. Sandkovsky, A.F. Luetkemeyer, S.H. Cohen, R.W. Finberg, P.E.H. Jackson, B. Taiwo, C.I. Paules, H. Arguinchona, N. Erdmann, N. Ahuja, M. Frank, M.-d. Oh, E.-S. Kim, S.Y. Tan, R.A. Mularski, H. Nielsen, P.O. Ponce, B.S. Taylor, L. Larson, N.G. Rouphael, Y. Saklawi, V.D. Cantos, E.R. Ko, J.J. Engemann, A.N. Amin, M. Watanabe, J. Billings, M.-C. Elie, R.T. Davey, T.H. Burgess, J. Ferreira, M. Green, M. Makowski, A. Cardoso, S. de Bono, T. Bonnett, M. Proschan, G.A. Deye, W. Dempsey, S.U. Nayak, L.E. Dodd, J.H. Beigel, Baricitinib plus Remdesivir for Hospitalized Adults with Covid-19, N. Engl. J. Med. 384(9) (2020) 795–807.
- 79.Food and Drug Administration (FDA), FDA Authorizes Drug Combination for Treatment of COVID-19, 2020. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-drug-combination-treatment-covid-19. (Accessed 09 Mar 2022).
- 80.Khater S., Kumar P., Dasgupta N., Das G., Ray S., Prakash A. Combining SARS-CoV-2 proofreading exonuclease and RNA-dependent RNA polymerase inhibitors as a strategy to combat COVID-19: a high-throughput in silico screening. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.647693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hsu J.C.-C., Laurent-Rolle M., Pawlak J.B., Wilen C.B., Cresswell P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. USA. 2021;118(24) doi: 10.1073/pnas.2101161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Devkota K., Schapira M., Perveen S., Khalili Yazdi A., Li F., Chau I., Ghiabi P., Hajian T., Loppnau P., Bolotokova A., Satchell K.J.F., Wang K., Li D., Liu J., Smil D., Luo M., Jin J., Fish P.V., Brown P.J., Vedadi M. Probing the SAM binding site of SARS-CoV-2 Nsp14 in vitro using SAM competitive inhibitors guides developing selective bisubstrate inhibitors. SLAS Discov. 2021;26(9):1200–1211. doi: 10.1177/24725552211026261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Narayanan N., Nair D.T. Ritonavir may inhibit exoribonuclease activity of nsp14 from the SARS-CoV-2 virus and potentiate the activity of chain terminating drugs. Int. J. Biol. Macromol. 2021;168:272–278. doi: 10.1016/j.ijbiomac.2020.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahmed-Belkacem R., Sutto-Ortiz P., Guiraud M., Canard B., Vasseur J.-J., Decroly E., Debart F. Synthesis of adenine dinucleosides SAM analogs as specific inhibitors of SARS-CoV nsp14 RNA cap guanine-N7-methyltransferase, Eur. J. Med. Chem. 2020;201 doi: 10.1016/j.ejmech.2020.112557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Basu S., Mak T., Ulferts R., Wu M., Deegan T., Fujisawa R., Tan K.W., Lim C.T., Basier C., Canal B., Curran J.F., Drury L.S., McClure A.W., Roberts E.L., Weissmann F., Zeisner T.U., Beale R., Cowling V.H., Howell M., Labib K., Diffley J.F.X. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp14 RNA cap methyltransferase. Biochem. J. 2021;478(13):2481–2497. doi: 10.1042/BCJ20210219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Food and Drug Administration (FDA), Know Your Treatment Options for COVID-19, 2021. 〈https://www.fda.gov/consumers/consumer-updates/know-your-treatment-options-covid-19〉. (Accessed 31 Oct 2021).
- 87.Food and Drug Administration (FDA), FDA Takes Actions to Expand Use of Treatment for Outpatients with Mild-to-Moderate COVID-19, 2022. 〈https://www.fda.gov/news-events/press-announcements/fda-takes-actions-expand-use-treatment-outpatients-mild-moderate-covid-19〉. (Accessed 22 Mar 2022).
- 88.Yan V.C., Muller F.L. Advantages of the parent nucleoside GS-441524 over remdesivir for covid-19 Treatment. ACS Med. Chem. Lett. 2020;11(7):1361–1366. doi: 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dickinson P.J., Bannasch M., Thomasy S.M., Murthy V.D., Vernau K.M., Liepnieks M., Montgomery E., Knickelbein K.E., Murphy B., Pedersen N.C. Antiviral treatment using the adenosine nucleoside analogue GS-441524 in cats with clinically diagnosed neurological feline infectious peritonitis. J. Vet. Intern. Med. 2020;34(4):1587–1593. doi: 10.1111/jvim.15780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pedersen N.C., Perron M., Bannasch M., Montgomery E., Murakami E., Liepnieks M., Liu H. Efficacy and safety of the nucleoside analog GS-441524 for treatment of cats with naturally occurring feline infectious peritonitis. J. Feline Med. Surg. 2019;21(4):271–281. doi: 10.1177/1098612X19825701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murphy B.G., Perron M., Murakami E., Bauer K., Park Y., Eckstrand C., Liepnieks M., Pedersen N.C. The nucleoside analog GS-441524 strongly inhibits feline infectious peritonitis (FIP) virus in tissue culture and experimental cat infection studies. Vet. Microbiol. 2018;219:226–233. doi: 10.1016/j.vetmic.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Y., Cao L., Li G., Cong F., Li Y., Sun J., Luo Y., Chen G., Li G., Wang P., Xing F., Ji Y., Zhao J., Zhang Y., Guo D., Zhang X. Remdesivir metabolite GS-441524 effectively inhibits SARS-CoV-2 infection in mouse models. J. Med. Chem. 2021;65(4):2785–2793. doi: 10.1021/acs.jmedchem.0c01929. [DOI] [PubMed] [Google Scholar]
- 93.Xie J., Wang Z. Can remdesivir and its parent nucleoside GS-441524 be potential oral drugs? An in vitro and in vivo DMPK assessment. Acta Pharm. Sin. B. 11(6) ( 2021;(1607–1616) doi: 10.1016/j.apsb.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Murakami E., Tolstykh T., Bao H., Niu C., Steuer H.M.M., Bao D., Chang W., Espiritu C., Bansal S., Lam A.M., Otto M.J., Sofia M.J., Furman P.A. Mechanism of activation of PSI-7851 and its diastereoisomer PSI-7977. J. Biol. Chem. 2010;285(45):34337–34347. doi: 10.1074/jbc.M110.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jubilant Pharma, Jubilant Pharma announces the development of a novel oral formulation of Remdesivir with successful completion of safety and pharmacokinetic/absorption studies in animals and healthy human volunteers, 2021. (Accessed 29 Sep 2021).
- 96.Cox R.M., Wolf J.D., Lieber C.M., Sourimant J., Lin M.J., Babusis D., DuPont V., Chan J., Barrett K.T., Lye D., Kalla R., Chun K., Mackman R.L., Ye C., Cihlar T., Martinez-Sobrido L., Greninger A.L., Bilello J.P., Plemper R.K. Oral prodrug of remdesivir parent GS-441524 is efficacious against SARS-CoV-2 in ferrets. Nat. Commun. 2021;12(1):6415. doi: 10.1038/s41467-021-26760-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schooley Robert T., Carlin Aaron F., Beadle James R., Valiaeva N., Zhang X.-Q., Clark Alex E., McMillan E., Rachel L., Leibel Sandra N., McVicar Rachael, J., Xie F., Garretson Aaron T., Smith Victoria J., Murphy Y., Hostetler K. Rethinking remdesivir: synthesis, antiviral activity, and pharmacokinetics of oral lipid prodrugs. Antimicrob. Agents Chemother. 2021;65(10):e01155–21. doi: 10.1128/AAC.01155-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xie Y., Yin W., Zhang Y., Shang W., Wang Z., Luan X., Tian G., Aisa H.A., Xu Y., Xiao G., Li J., Jiang H., Zhang S., Zhang L., Xu H.E., Shen J. Design and development of an oral remdesivir derivative VV116 against SARS-CoV-2. Cell Res. 2021;31:1212–1214. doi: 10.1038/s41422-021-00570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Burg Richard W., Miller Brinton M., Baker Edward E., Birnbaum J., Currie Sara A., Hartman R., Kong Y.-L., Monaghan Richard L., Olson G., Putter I., Tunac Josefino B., Wallick H., Stapley Edward O., Oiwa R., Ōmura, S. Avermectins, New Family of Potent Anthelmintic Agents: Producing Organism and Fermentation. Antimicrob. Agents Chemother. 1979;15(3):361–367. doi: 10.1128/aac.15.3.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Campbell W.C. Ivermectinas an antiparasitic agent for use in humans. Annu. Rev. Microbiol. 1991;45(1):445–474. doi: 10.1146/annurev.mi.45.100191.002305. [DOI] [PubMed] [Google Scholar]
- 101.Crump A., Omura S. Ivermectin, ‘Wonder drug’ from Japan: the human use perspective. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2011;87(2):13–28. doi: 10.2183/pjab.87.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ribeiro F., Pereira C., Alves A., Marcon M. Treatment of human cavitary myiasis with oral ivermectin. Rev. Bras. Otorrinolaringol. 2001;67:755–761. [Google Scholar]
- 103.Pryor M.J., Rawlinson S.M., Butcher R.E., Barton C.L., Waterhouse T.A., Vasudevan S.G., Bardin P.G., Wright P.J., Jans D.A., Davidson A.D. Nuclear Localization of Dengue Virus Nonstructural Protein 5 Through Its Importin α/β–Recognized Nuclear Localization Sequences is Integral to Viral Infection. Traffic. 2007;8(7):795–807. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 104.Caly L., Wagstaff K.M., Jans D.A. Nuclear trafficking of proteins from RNA viruses: potential target for antivirals? Antivir. Res. 2012;95(3):202–206. doi: 10.1016/j.antiviral.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 105.Jans D.A., Martin A.J., Wagstaff K.M. Inhibitors of nuclear transport. Curr. Opin. Cell Biol. 2019;58:50–60. doi: 10.1016/j.ceb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 106.Wagstaff Kylie M., Sivakumaran H., Heaton Steven M., Harrich D., Jans David A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443(3):851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Barrows Nicholas J., Campos Rafael K., Powell S.T., Prasanth K.R., Schott-Lerner G., Soto-Acosta R., Galarza-Muñoz G., McGrath Erica L., Urrabaz-Garza R., Gao J., Wu P., Menon R., Saade G., Fernandez-Salas I., Rossi Shannan L., Vasilakis N., Routh A., Bradrick Shelton S., Garcia-Blanco Mariano A., Screen A. of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe. 2016;20(2):259–270. doi: 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Varghese F.S., Kaukinen P., Gläsker S., Bespalov M., Hanski L., Wennerberg K., Kümmerer B.M., Ahola T. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antivir. Res. 2016;126:117–124. doi: 10.1016/j.antiviral.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 109.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sen Gupta P.S., Biswal S., Panda S.K., Ray A.K., Rana M.K. Binding mechanism and structural insights into the identified protein target of COVID-19 and importin-α with in-vitro effective drug ivermectin. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1839564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Poon I.K.H., Jans D.A. Regulation of nuclear transport: central role in development and transformation? Traffic. 2005;6(3):173–186. doi: 10.1111/j.1600-0854.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- 112.Momekov G., Momekova D. Ivermectin as a potential COVID-19 treatment from the pharmacokinetic point of view: antiviral levels are not likely attainable with known dosing regimens. Biotechnol. Biotechnol. Equip. 2020;34(1):469–474. [Google Scholar]
- 113.Reardon S. Flawed ivermectin preprint highlights challenges of COVID drug studies. Nature. 2021;596(7871):173–174. doi: 10.1038/d41586-021-02081-w. [DOI] [PubMed] [Google Scholar]
- 114.Popp M., Stegemann M., Metzendorf M.I., Gould S., Kranke P., Meybohm P., Skoetz N., Weibel S. Ivermectin for preventing and treating COVID‐19. Cochrane Database Syst. Rev. 2021;7 doi: 10.1002/14651858.CD015017.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lawrence J.M., Meyerowitz-Katz G., Heathers J.A.J., Brown N.J.L., Sheldrick K.A. The lesson of ivermectin: meta-analyses based on summary data alone are inherently unreliable. Nat. Med. 2021;27:1853–1854. doi: 10.1038/s41591-021-01535-y. [DOI] [PubMed] [Google Scholar]
- 116.R. Schraer, J. Goodman, Ivermectin: How false science created a Covid 'miracle' drug, 2021. 〈https://www.bbc.com/news/health-58170809〉. (Accessed 26 Oct 2021).
- 117.López-Medina E., López P., Hurtado I.C., Dávalos D.M., Ramirez O., Martínez E., Díazgranados J.A., Oñate J.M., Chavarriaga H., Herrera S., Parra B., Libreros G., Jaramillo R., Avendaño A.C., Toro D.F., Torres M., Lesmes M.C., Rios C.A., Caicedo I. Effect of ivermectin on time to resolution of symptoms among adults with mild COVID-19: a randomized clinical trial. JAMA. 2021;325(14):1426–1435. doi: 10.1001/jama.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.de Melo G.D., Lazarini F., Larrous F., Feige L., Kornobis E., Levallois S., Marchio A., Kergoat L., Hardy D., Cokelaer T., Pineau P., Lecuit M., Lledo P.-M., Changeux J.-P., Bourhy H. Attenuation of clinical and immunological outcomes during SARS-CoV-2 infection by ivermectin. EMBO Mol. Med. 2021;13(8) doi: 10.15252/emmm.202114122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McElvaney O.J., Hobbs B.D., Qiao D., McElvaney O.F., Moll M., McEvoy N.L., Clarke J., O'Connor E., Walsh S., Cho M.H., Curley G.F., McElvaney N.G. A linear prognostic score based on the ratio of interleukin-6 to interleukin-10 predicts outcomes in COVID-19. eBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pandit S., Geissler W., Harris G., Sitlani A. Allosteric effects of dexamethasone and RU486 on glucocorticoid receptor-DNA interactions*. J. Biol. Chem. 2002;277(2):1538–1543. doi: 10.1074/jbc.M105438200. [DOI] [PubMed] [Google Scholar]
- 121.Ahmed M.H., Hassan A. Dexamethasone for the treatment of coronavirus disease (COVID-19): a review. SN Compr. Clin. Med. 2020;2(12):2637–2646. doi: 10.1007/s42399-020-00610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wagner C., Griesel M., Mikolajewska A., Mueller A., Nothacker M., Kley K., Metzendorf M.I., Fischer A.L., Kopp M., Stegemann M., et al. Systemic corticosteroids for the treatment of COVID‐19. Cochrane Database Syst. Rev. 2021;8 doi: 10.1002/14651858.CD014963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Horby P., Lim W.S., Emberson J.R. e. al, Dexamethasone in Hospitalized Patients with Covid-19. N. Engl. J. Med. 2020;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.World Health Organization, WHO model list of essential medicines - 22nd list, 2021, 2021.
- 125.Theoharides T.C., Conti P. Dexamethasone for COVID-19? Not so fast. J. Biol. Regul. Homeost. Agents. 2020;34(3):1241–1243. doi: 10.23812/20-EDITORIAL_1-5. [DOI] [PubMed] [Google Scholar]
- 126.Wong C.K., Lam C.W.K., Wu A.K.L., Ip W.K., Lee N.L.S., Chan I.H.S., Lit L.C.W., Hui D.S.C., Chan M.H.M., Chung S.S.C., Sung J.J.Y. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 2004;136(1):95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mahallawi W.H., Khabour O.F., Zhang Q., Makhdoum H.M., Suliman B.A. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chen R.-c, Tang X.-p, Tan S.-y, Liang B.-l, Wan Z.-y, Fang J.-q, Zhong N. Treatment of severe acute respiratory syndrome with glucosteroids: the guangzhou experience. Chest. 2006;129(6):1441–1452. doi: 10.1378/chest.129.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Arabi Y.M., Mandourah Y., Al-Hameed F., Sindi A.A., Almekhlafi G.A., Hussein M.A., Jose J., Pinto R., Al-Omari A., Kharaba A., Almotairi A., Khatib K.A., Alraddadi B., Shalhoub S., Abdulmomen A., Qushmaq I., Mady A., Solaiman O., Al-Aithan A.M., Al-Raddadi R., Ragab A., Balkhy H.H., Harthy A.A., Deeb A.M., Mutairi H.A., Al-Dawood A., Merson L., Hayden F.G., Fowler R.A. Corticosteroid therapy for critically ill patients with middle east respiratory syndrome. Am. J. Respir. Crit. Care Med. 2018;197(6):757–767. doi: 10.1164/rccm.201706-1172OC. [DOI] [PubMed] [Google Scholar]
- 130.Lee K.H., Yoon S., Jeong G.H., Kim J.Y., Han Y.J., Hong S.H., Ryu S., Kim J.S., Lee J.Y., Yang J.W., Lee J., Solmi M., Koyanagi A., Dragioti E., Jacob L., Radua J., Smith L., Oh H., Tizaoui K., Cargnin S., Terrazzino S., Ghayda R.A., Kronbichler A., Shin J.I. Efficacy of corticosteroids in patients with SARS, MERS and COVID-19: a systematic review and meta-analysis. J. Clin. Med. 2020;9(8):2392. doi: 10.3390/jcm9082392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shang L., Zhao J., Hu Y., Du R., Cao B. On the use of corticosteroids for 2019-nCoV pneumonia. Lancet. 2020;395(10225):683–684. doi: 10.1016/S0140-6736(20)30361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang Y., Hu S., Wang J., Xue Z., Wang C., Wang N. Dexamethasone inhibits SARS-CoV-2 spike pseudotyped virus viropexis by binding to ACE2. Virology. 2021;554:83–88. doi: 10.1016/j.virol.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sorrell Fiona J., Szklarz M., Abdul Azeez Kamal R., Elkins Jon M., Knapp S. Family-wide structural analysis of human numb-associated protein kinases. Structure. 2016;24(3):401–411. doi: 10.1016/j.str.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Rawling M., Savory E., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395(10223):e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stebbing J., Sánchez Nievas G., Falcone M., Youhanna S., Richardson P., Ottaviani S., Shen Joanne X., Sommerauer C., Tiseo G., Ghiadoni L., Virdis A., Monzani F., Rizos Luis R., Forfori F., Avendaño Céspedes A., De Marco S., Carrozzi L., Lena F., Sánchez-Jurado Pedro M., Lacerenza Leonardo G., Cesira N., Caldevilla Bernardo D., Perrella A., Niccoli L., Méndez Lourdes S., Matarrese D., Goletti D., Tan Y.-J., Monteil V., Dranitsaris G., Cantini F., Farcomeni A., Dutta S., Burley Stephen K., Zhang H., Pistello M., Li W., Romero Marta M., Andrés Pretel F., Simón-Talero Rafaela S., García-Molina R., Kutter C., Felce James H., Nizami Zehra F., Miklosi Andras G., Penninger Josef M., Menichetti F., Mirazimi A., Abizanda P., Lauschke Volker M. JAK inhibition reduces SARS-CoV-2 liver infectivity and modulates inflammatory responses to reduce morbidity and mortality. Sci. Adv. 2021;7(1):eabe4724. doi: 10.1126/sciadv.abe4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang X., Shang L., Fan G., Gu X., Xu J., Wang Y., Huang L., Cao B. The efficacy and safety of janus kinase inhibitors for patients With COVID-19: a living systematic review and meta-analysis. Front. Med. 2022;8 doi: 10.3389/fmed.2021.800492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lin Z., Niu J., Xu Y., Qin L., Ding J., Zhou L. Clinical efficacy and adverse events of baricitinib treatment for coronavirus disease-2019 (COVID-19): a systematic review and meta-analysis. J. Med. Virol. 2022;94(4):1523–1534. doi: 10.1002/jmv.27482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marconi V.C., Ramanan A.V., de Bono S., Kartman C.E., Krishnan V., Liao R., Piruzeli M.L.B., Goldman J.D., Alatorre-Alexander J., de Cassia Pellegrini R., Estrada V., Som M., Cardoso A., Chakladar S., Crowe B., Reis P., Zhang X., Adams D.H., Ely E.W., Ahn M.-Y., Akasbi M., Alatorre-Alexander J., Altclas J.D., Ariel F., Ariza H.A., Atkar C., Bertetti A., Bhattacharya M., Briones M.L., Budhraja A., Burza A., Camacho Ortiz A., Caricchio R., Casas M., Cevoli Recio V., Choi W.S., Cohen E., Comulada-Rivera A., Cook P., Cornejo Juarez D.P., Daniel C., Degrecci Relvas L.F., Dominguez Cherit J.G., Ellerin T., Enikeev D., Erico Tanni Minamoto S., Estrada V., Fiss E., Furuichi M., Giovanni Luz K., Goldman J.D., Gonzalez O., Gordeev I., Gruenewald T., Hamamoto Sato V.A., Heo E.Y., Heo J.Y., Hermida M., Hirai Y., Hutchinson D., Iastrebner C., Ioachimescu O., Jain M., Juliani Souza Lima M.P., Khan A., Kremer A.E., Lawrie T., MacElwee M., Madhani-Lovely F., Malhotra V., Martínez Resendez M.F., McKinnell J., Milligan P., Minelli C., Moran Rodriguez M.A., Parody M.L., Paulin P., Pellegrini Rd.C., Pemu P., Procopio Carvalho A.C., Puoti M., Purow J., Ramesh M., Rea Neto A., Rea Neto A., Robinson P., Rodrigues C., Rojas Velasco G., Saraiva J.F.K., Scheinberg M., Schreiber S., Scublinsky D., Sevciovic Grumach A., Shawa I., Simon Campos J., Sofat N., Som M., Spinner C.D., Sprinz E., Stienecker R., Suarez J., Tachikawa N., Tahir H., Tiffany B., Vishnevsky A., Westheimer Cavalcante A., Zirpe K. Efficacy and safety of baricitinib for the treatment of hospitalised adults with COVID-19 (COV-BARRIER): a randomised, double-blind, parallel-group, placebo-controlled phase 3 trial. Lancet Respir. Med. 2021;9(12):1407–1418. doi: 10.1016/S2213-2600(21)00331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kmietowicz Z. Covid-19: WHO recommends baricitinib and sotrovimab to treat patients. BMJ. 2022;376:o97. doi: 10.1136/bmj.o97. [DOI] [PubMed] [Google Scholar]
- 141.Cantini F., Niccoli L., Matarrese D., Nicastri E., Stobbione P., Goletti D. Baricitinib therapy in COVID-19: A pilot study on safety and clinical impact. J. Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bechman K., Subesinghe S., Norton S., Atzeni F., Galli M., Cope A.P., Winthrop K.L., Galloway J.B. A systematic review and meta-analysis of infection risk with small molecule JAK inhibitors in rheumatoid arthritis. Rheumatology. 2019;58(10):1755–1766. doi: 10.1093/rheumatology/kez087. [DOI] [PubMed] [Google Scholar]
- 143.Co. Merck, Merck and Ridgeback Biotherapeutics Provide Update on Progress of Clinical Development Program for Molnupiravir, an Investigational Oral Therapeutic for the Treatment of, Mild-to-Moderate COVID-19, 2021.
- 144.Stuyver L.J., Whitaker T., McBrayer T.R., Hernandez-Santiago B.I., Lostia S., Tharnish P.M., Ramesh M., Chu C.K., Jordan R., Shi J., Rachakonda S., Watanabe K.A., Otto M.J., Schinazi R.F. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis c viruses in culture. Antimicrob. Agents Chemother. 2003;47(1):244–254. doi: 10.1128/AAC.47.1.244-254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021;6(1):11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gordon C.J., Tchesnokov E.P., Schinazi R.F., Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021;297(1) doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kabinger F., Stiller C., Schmitzová J., Dienemann C., Kokic G., Hillen H.S., Höbartner C., Cramer P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28(9):740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Co. Merck, Merck and Ridgeback’s Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalization or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3, Study (2021).
- 149.Merck & Co., Merck and Ridgeback Biotherapeutics Provide Update on Results from MOVe-OUT Study of Molnupiravir, an Investigational Oral Antiviral Medicine, in At Risk Adults With Mild-to-Moderate COVID-19 (2021).
- 150.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12(541):eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Toots M., Yoon J.-J., Cox Robert M., Hart M., Sticher Zachary M., Makhsous N., Plesker R., Barrena Alec H., Reddy Prabhakar G., Mitchell Deborah G., Shean Ryan C., Bluemling Gregory R., Kolykhalov Alexander A., Greninger Alexander L., Natchus Michael G., Painter George R., Plemper Richard K. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019;11(515):eaax5866. doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Painter W.P., Holman W., Bush J.A., Almazadi F., Malik H., Eraut N.C.J.E., Morin E.J., Szewczyk L.J., Painter G.R. Human safety, tolerability, and pharmacokinetics of molnupiravir, a novel broad-spectrum oral antiviral agent with activity against SARS-CoV-2. Antimicrob. Agents Chemother. 2021;65(5):e02428–20. doi: 10.1128/AAC.02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182(6):1560–1573. doi: 10.1016/j.cell.2020.07.033. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., Liu H., Madden V.J., Krzystek H.M., De C., White K.K., Gully K., Schäfer A., Zaman T., Leist S.R., Grant P.O., Bluemling G.R., Kolykhalov A.A., Natchus M.G., Askin F.B., Painter G., Browne E.P., Jones C.D., Pickles R.J., Baric R.S., Garcia J.V. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591(7850):451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Co. Merck, Merck and Ridgeback’s Molnupiravir, an Oral COVID-19 Antiviral Medicine, Receives First Authorization in the World (2021).
- 156.Zhou S., Hill C.S., Sarkar S., Tse L.V., Woodburn B.M.D., Schinazi R.F., Sheahan T.P., Baric R.S., Heise M.T., Swanstrom R. β-d-N4-hydroxycytidine Inhibits SARS-CoV-2 Through Lethal Mutagenesis But Is Also Mutagenic To Mammalian Cells. J. Infect. Dis. 2021;224(3):415–419. doi: 10.1093/infdis/jiab247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Padhi A.K., Shukla R., Saudagar P., Tripathi T. High-throughput rational design of the remdesivir binding site in the RdRp of SARS-CoV-2: implications for potential resistance. iScience. 2021;24(1) doi: 10.1016/j.isci.2020.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bloom J.D., Labthavikul S.T., Otey C.R., Arnold F.H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA. 2006;103(15):5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.I. Merck & Co., Merck and Ridgeback’s Molnupiravir Receives U.S. FDA Emergency Use Authorization for the Treatment of High-Risk Adults With Mild to Moderate COVID-19, 2021.
- 160.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 161.Pfizer Inc., Pfizer’s Novel COVID-19 Oral Antiviral Treatment Candidate Reduced Risk of Hospitalization or Death by 89% in Interim Analysis of Phase 2/3 EPIC-HR Study, 2021. 〈https://www.pfizer.com/news/press-release/press-release-detail/pfizers-novel-covid-19-oral-antiviral-treatment-candidate〉. (Accessed 10 Nov 2021).
- 162.Mahase E. Covid-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ. 2021;375:n2713. doi: 10.1136/bmj.n2713. [DOI] [PubMed] [Google Scholar]
- 163.Food and Drug Administration (FDA), FDA Authorizes First Oral Antiviral for Treatment of COVID-19, 2021. 〈https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-oral-antiviral-treatment-covid-19〉. (Accessed 09 Mar 2022).
- 164.Vandyck K., Deval J. Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr. Opin. Virol. 2021;49:36–40. doi: 10.1016/j.coviro.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ledford H. COVID antiviral pills: what scientists still want to know. Nature. 2021;599(7885):358–359. doi: 10.1038/d41586-021-03074-5. [DOI] [PubMed] [Google Scholar]
- 166.Vangeel L., Chiu W., De Jonghe S., Maes P., Slechten B., Raymenants J., André E., Leyssen P., Neyts J., Jochmans D. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antivir. Res. 2022;198 doi: 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]