Abstract
Imbalance of the gut microbiota plays an important role in the pathogenesis of various diseases. Although many clinical studies have analyzed the gut microbiota, the definition of normal gut microbiota remains unclear. In this study, we aim to establish the average gut microbiota in the healthy Japanese population. Using 16S ribosomal RNA gene sequencing, we analyzed gut microbial data from fecal samples obtained from 6,101 healthy Japanese individuals. Based on their ages, the individuals were divided into three groups: young, middle-age, and old. Individuals were further categorized according to body mass index (BMI) into lean, normal, and obese groups. The α and β diversities in the old group were significantly higher than those in the young and middle-age groups. The Firmicutes/Bacteroidetes ratio of subjects in the obese category was significantly lower compared with those of subjects in the lean and normal categories in the young and middle-age groups. Genus Bacteroides was the dominant gut microbiota across all the BMI categories in all the age groups. Among the top ten genera, the abundances of Bacteroides, Bifidobacterium, Anaerostipes, Blautia, Dorea, Fusicatenibacter, Lachnoclostridium, and Parabacteroides were significantly lower in the old group than in the young and middle-age groups. The correlation network at the genus level revealed different microbe-microbe interactions associated with age and BMI. We determined the average Japanese gut microbiota, and this information could be used as a reference. The gut microbiota greatly differs based on the life stage and metabolic status of the host, and this gives rise to a variety of host–gut microbe interactions that can lead to an increased susceptibility to disease.
Keywords: 16S rRNA, gut microbiota, large cohort, Japanese population
INTRODUCTION
Several clinical studies have clearly demonstrated that the gut microbiota plays a pivotal role not only in maintaining human health but also in the development of many diseases, such as inflammatory bowel disease, liver cirrhosis, rheumatoid arthritis, type 2 diabetes, and cardiovascular disease [1,2,3,4,5,6,7]. As the gut microbiota has a causal relationship with disease progression, modulation of the gut microbiota is gaining attention as a novel therapeutic approach [8]. For instance, clinical intervention studies have reported the potential of modulating the gut microbiota using oral antibiotics [9, 10]. Furthermore, fecal microbiota transplantation is performed as a standard treatment procedure for Clostridioides difficile infection [11]. These reports shed light on the innovative strategies that are available for treating diseases via gut microbial modulation. Moreover, analysis of the gut microbiota based on 16S rRNA sequencing is becoming cheaper and easier; hence, the gut microbiota may serve as a novel diagnostic marker in the near future. To date, a large number of clinical studies have analyzed gut microbiota in various diseases and compared them to detect the characteristics associated with differential abundances between patients with and without disease. As the gut microbial profile depends on several factors, such as age, body weight, local food, and lifestyle [12], it is critical for clinicians and researchers to know the “average” gut microbiota of individuals so that they can compare gut microbiota between heathy and diseased subjects to detect differences and elucidate links between disease progression and specific gut microbiota. Furthermore, elucidation of outlier gut microbiota may lead to the identification of hidden factors, including specific diseases. However, reports analyzing the gut microbiotas of healthy populations, especially the Japanese population, are limited, and the sample sizes have been too small [13,14,15,16,17,18].
In this study, we aimed to investigate the average gut flora in healthy Japanese individuals stratified by age and body mass index (BMI). We analyzed the Mykinso cohort data and determined the average gut microbiota to establish reference ranges. The Mykinso cohort is a part of the product services of Cykinso, Inc. (Tokyo, Japan) and is one of the largest cohorts in Japan used to study the life expectancy of healthy individuals using gut microbial data and related information [19]. Individuals interested to learn about their own gut microbiota can use the Mykinso commercial service (https://mykinso.com/). We believe that our results greatly contribute to the definition of normal gut microbiota, which will help in distinguishing the gut microbiota of diseased subjects from those of healthy individuals.
MATERIALS AND METHODS
Study population
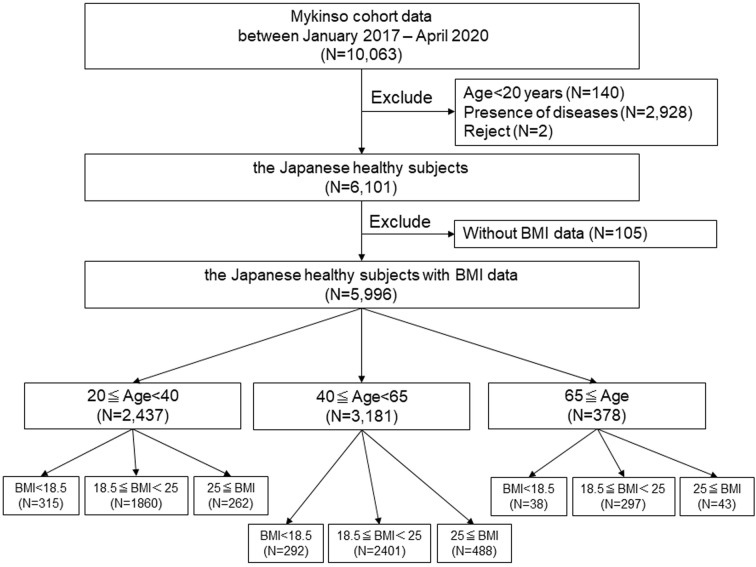
The study initially included 10,063 individuals registered in the Mykinso cohort from January 2017 to April 2020. Among them, 140 individuals were excluded due to their ages being less than 20, 2,928 individuals were excluded due to the presence of certain diseases as defined by a questionnaire, and two individuals were excluded due to their refusal to participate. Ultimately, 6,101 individuals were included in the study (Fig. 1). All participants provided written informed consent for enrollment, and the study was conducted according to the principles of the Declaration of Helsinki. The study was approved by the Cykinso Research Ethics Committee (No. LD-001-04 and LD-002-03) and registered with the UMIN Clinical Trials Registry (no. UMIN000028887 and UMIN000028888).
Fig. 1.
Study population.
BMI: body mass index.
Age and BMI stratification
The subjects were divided according to their ages into young adults (20≤ age <40 years), middle-aged adults (40≤ age <65 years), or old adults (65 years ≤age). We further categorized the subjects according to the World Health Organization (WHO) BMI classification [20] as lean (BMI <18.5), normal (18.5≤ BMI <25), or obese (25≤ BMI).
Fecal sampling, DNA extraction, sequencing, and analyzing sequencing data
The detailed methods were described in elsewhere [19]. Briefly, we collected fecal samples using brush-type collection kits containing guanidine thiocyanate solution (TechnoSuruga Laboratory, Shizuoka, Japan), transported them at ambient temperature, and stored them at 4°C. DNA was extracted from the fecal samples using a DNeasy PowerSoil Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The amplicons of the V1V2 region were prepared using a forward primer (16S_27Fmod: TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG AGR GTT TGA TYM TGG CTC AG) and reverse primer (16S_338R: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GTG CTG CCT CCC GTA GGA GT) along with KAPA HiFi HotStart ReadyMix (Roche, Basel, Switzerland). Sequencing libraries were prepared according to the 16S library preparation protocol provided by Illumina (Illumina, San Diego, CA, USA). Dual index adapters for sequencing on the Illumina MiSeq platform were attached using a Nextera XT Index kit (Illumina, San Diego, CA, USA). Each sequencing library was diluted to 5 ng/μL. We mixed equal volumes of the libraries to obtain a final concentration of 4 nM. The DNA concentrations of the mixed libraries were measured by quantitative real-time polymerase chain reaction (qPCR) using KAPA SYBR FAST qPCR Master Mix (Roche, Basel, Switzerland), primer 1 (AAT GAT ACG GCG ACC ACC), and primer 2 (CAA GCA GAA GAC GGC ATA CGA). These libraries were sequenced in a 250-bp paired-end run using MiSeq Reagent Kit v2 (500 cycles).
Bioinformatics analysis
The data processing and assignment based on the QIIME2 pipeline (version 2020.8) [21] were performed using the following steps: (1) joining paired end reads, filtering, and denoising with DADA2 and (2) assigning taxonomic information to each amplicon sequence variant (ASV) using a naive Bayes classifier in the QIIME2 classifier. The classifier was trained with a robust taxonomy simplifier for SILVA (arts-SILVA), which was originally developed from the 16S rRNA taxonomy dataset based on SILVA 138 [22]. arts-SILVA was developed for the purpose of making Mykinso testing reports easier to understand for those who are not familiar with complex rules of taxonomic nomenclature. arts-SILVA simplifies resulting taxonomic assignments by removing some study-related labels, curating obvious mis-entries, and generalizing uncommon names in the SILVA database. To obtain arts-SILVA, the V1V2 regions of reference sequences of SILVA were extracted and then clustered according to the original manuscript for QIIME2 preparation of SILVA. Next, some unnecessary/seemingly miss-labeled entries were cleaned up (i.e., removed labels with little information such as “D_6__unclutured bacteria” and corrected duplicate entries such as “D_0__Bacteria;D_1__Bacteria Firmicutes” to “D_0__Bacteria;D_1__Firmicutes”). Then, useless taxa such as “D_6__human metagenome” were removed by manual inspection. After that, a consensus taxonomy was assigned to each cluster for which 100% of the assigned taxa were in 100% agreement, and finally, the label “Ambiguous taxa” was removed.
Statistics
R and RStudio (versions 3.5.1 and 1.1.456, respectively) were used for all data manipulation, analyses, and graphs. The R packages qiime2R and microbiome R were used for all analyses. The R packages tidyMicro (version 1.48) and ggplot2 were used for visualization.
Mykinso Research Cohort characteristics were summarized as frequencies (%) for categorical variables and means (standard deviations) or medians (interquartile range, IRQ) for continuous variables. Differences in these characteristics were assessed using Fisher’s exact test for categorical variables and t-tests for continuous variables.
Relative abundance (RA) was calculated as the number of sequenced reads for each taxon in a sample and was standardized by the total number of sequences generated for each sample. Only taxa that were present in at least 50% of the cohort and had an RA of at least 0.1% in at least one sample were included in the analyses. Sequence counts for taxa that did not meet these requirements were aggregated into an “Other” category. These filtering requirements were applied at the phylum, family, and genus levels. Sequence counts that could not be classified at the taxonomic level of interest were left as unclassified counts of the lowest level possible. The α diversity at 10,000 reads per sample was evaluated at the ASV level using the Shannon index, and significant differences were evaluated using ANOVA. The β diversity was used to evaluate differences in the community composition between samples using the Bray-Curtis distance method. Group differences in Bray-Curtis distances within each group were assessed using ANOVA. Differences in Bray-Curtis distances between groups were assessed using a non-parametric permutation-based multivariate analysis of variation (PERMANOVA) test using the vegan package with 999 permutations. We tested for significant impacts from the co-occurrences between age group and BMI class for this distance measure by including interaction terms between the two phenotypes in each model.
Differences in the RAs of phylum-, family-, and genus-level taxa were evaluated using generalized linear models (GLMs) assuming a negative binomial distribution and log link function; the total number of sequences was used as an offset. Benjamini and Hochberg’s false discovery rate (FDR) correction was then applied to adjust for multiple comparisons within each taxon level, with significance defined as FDR p-value ≤0.05. For each taxon, a GLM model was first fit, including an interaction term between BMI class and age group. If the interaction term was significant (FDR p<0.05), it was determined that the effect of BMI class on the taxa was modified by age, and results were reported for these models. If the interaction term was not significant, the interaction term was removed from the model, and the independent effect of BMI class, after adjusting for age, was modeled and reported. Stacked bar charts were constructed using estimated taxa RAs obtained from the GLMs to visually display the estimated microbiota compositions by phenotypic group. The top 10 most abundant taxa were plotted, while the remaining estimated taxa counts were aggregated into an “Other” category for readability.
Correlation network analysis was performed using SparCC [23] to calculate pairwise correlations between taxonomic features. In the resulting correlation network, nodes represent taxonomic features, and edges represent correlations greater than the correlation threshold (i.e., 0.3) between pairs of taxa; blue and red colors indicate positive and negative correlations, respectively. Chord diagrams were displayed using the R circle package [24].
RESULTS
Study population
A total of 6,101 healthy, disease-free Japanese subjects were enrolled in the study. After excluding subjects without BMI data, 5,996 subjects were included in the analysis and divided into three groups according to their ages: young 2,437 (40.6%), middle age 3,181 (53.1%), and old 378 (6.3%; Fig. 1). Baseline characteristics, including age, sex, BMI, alcohol consumption, and smoking status, are shown in Table 1. The percentage of females did not differ significantly among the three groups. The mean BMI was significantly different among the groups (young 21.5 ± 3.1, middle age 22.2 ± 3.2, old 21.9 ± 2.9; p<0.001). Alcohol consumption and smoking differed significantly among the groups (Table 1).
Table 1. Patient characteristics.
| 20≤Age<40 | 40≤Age<65 | 65≤Age | p value | |
|---|---|---|---|---|
| (N=2,437) | (N=3,181) | (N=378) | ||
| Age (years) | 31.3 (5.5) | 49.0 (6.5) | 72.6 (6.8) | <0.001 |
| Sex (female), n (%) | 1,531 (62.8%) | 2,008 (63.1%) | 222 (58.7%) | 0.2 |
| Body Mass Index (kg/m2) | 21.5 (3.1) | 22.2 (3.2) | 21.9 (2.9) | <0.001 |
| Alcohol, n (%) | ||||
| Never | 1,422 (58.3%) | 1,615 (50.8%) | 219 (57.9%) | <0.001 |
| Current | 929 (38.1%) | 1,424 (44.8%) | 128 (33.9%) | |
| Past | 82 (3.4%) | 138 (4.3%) | 28 (7.4%) | |
| Unknown | 4 (0.2%) | 4 (0.1%) | 3 (0.8%) | |
| Smoking, n (%) | ||||
| Never | 1,902 (78.0%) | 2,029 (63.8%) | 242 (64.0%) | <0.001 |
| Current | 201 (8.3%) | 288 (9.1%) | 21 (5.6%) | |
| Past | 330 (13.5%) | 857 (26.9%) | 108 (28.6%) | |
| Unknown | 4 (0.2%) | 7 (0.2%) | 7 (1.8%) |
( )=standard deviation: S.D.
We further divided the subjects in each age group according to their BMIs. The percentage of females was significantly higher in the lean subgroup than those in the normal and obese subgroups in all age groups (Table 2). Age differed significantly in the young group, but not in the middle-age or old group. The mean BMIs of the lean and obese subjects were 17.52 ± 0.77 kg/m2 and 27.9 ± 2.8 kg/m2 in the young group, 17.5 ± 0.9 kg/m2 and 27.7 ± 2.7 kg/m2 in the middle-age group, and 17.0 kg/m2 ± 1.3 and 27.1 ± 2.4 kg/m2 in the old group, respectively. In the young and middle-age groups, alcohol consumption and smoking were significantly different among the three BMI subgroups. In contrast, in the old group, no such significant differences were observed.
Table 2. Patient characteristics in each age category.
| 20≤Age<40 | p value | 40≤Age<65 | p value | 65≤Age | p value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (N=2,437) | (N=3,181) | (N=378) | ||||||||||
| Lean | Normal | Obese | Lean | Normal | Obese | Lean | Normal | Obese | ||||
| N=315 | N=1,860 | N=262 | N=292 | N=2,401 | N=488 | N=38 | N=297 | N=43 | ||||
| Age (years) | 30.8 (5.5) | 31.3 (5.5) | 32.0 (5.1) | 0.047 | 49 (6) | 49 (7) | 49 (6) | >0.9 | 75.1 (9.7) | 72.5 (6.4) | 71.3 (6.1) | 0.073 |
| Sex (female), n (%) | 249 (79.0%) | 1,175 (63.2%) | 107 (40.8%) | <0.001 | 253 (86.6%) | 1,534 (63.9%) | 221 (45.3%) | <0.001 | 30 (78.9%) | 172 (57.9%) | 20 (46.5%) | 0.01 |
| Body Mass Index (kg/m2) | 17.5 (0.8) | 21.3 (1.7) | 27.9 (2.8) | <0.001 | 17.5 (0.9) | 21.6 (1.8) | 27.7 (2.7) | <0.001 | 17.0 (1.3) | 21.8 (1.7) | 27.1 (2.4) | <0.001 |
| Shannon-index | 6.18 (0.60) | 6.23 (0.58) | 6.09 (0.63) | 0.005 | 6.30 (0.61) | 6.29 (0.60) | 6.14 (0.61) | <0.001 | 6.20 (0.62) | 6.44 (0.57) | 6.33 (0.59) | 0.082 |
| Alcohol, n (%) | ||||||||||||
| Never | 215 (68.2%) | 1,077 (57.9%) | 130 (49.6%) | <0.001 | 173 (59.2%) | 1,201 (50.0%) | 241 (49.4%) | 0.026 | 27 (71.1%) | 164 (55.2%) | 28 (65.1%) | 0.3 |
| Current | 88 (27.9%) | 723 (38.9%) | 118 (45.0%) | 104 (35.6%) | 1,097 (45.7%) | 223 (45.7%) | 9(23.7%) | 108 (36.4%) | 11 (25.6%) | |||
| Past | 12 (3.8%) | 56 (3.0%) | 14 (5.4%) | 14(4.8%) | 101 (4.2%) | 23(4.7%) | 2 (5.3%) | 22 (7.4%) | 4 (9.3%) | |||
| Unknown | 0 (0.0%) | 4 (0.2%) | 0 (0.0%) | 1 (0.3%) | 2 (0.1%) | 1 (0.2%) | 0 (0.0%) | 3 (1.0%) | 0 (0.0%) | |||
| Smoking, n (%) | ||||||||||||
| Never | 255 (81.0%) | 1,477 (79.4%) | 170 (64.9%) | <0.001 | 225 (77.1%) | 1,536 (64.0%) | 268 (54.9%) | <0.001 | 29 (76.3%) | 190 (64.0%) | 23 (53.5%) | 0.1 |
| Current | 25(7.9%) | 136 (7.3%) | 40 (15.3%) | 15(5.1%) | 201 (8.4%) | 72 (14.8%) | 2 (5.3%) | 15 (5.1%) | 4 (9.3%) | |||
| Past | 35 (11.1%) | 243 (13.1%) | 52 (19.8%) | 52 (17.8%) | 658 (27.4%) | 147 (30.1%) | 5(13.1%) | 88 (29.6%) | 15 (34.9%) | |||
| Unknown | 0 (0.0%) | 4 (0.2) | 0 (0.0%) | 0 (0.0%) | 6 (0.2%) | 1 (0.2%) | 2 (5.3%) | 4 (1.3%) | 1 (2.3%) | |||
( )=standard deviation: S.D.
Gut microbiota diversity
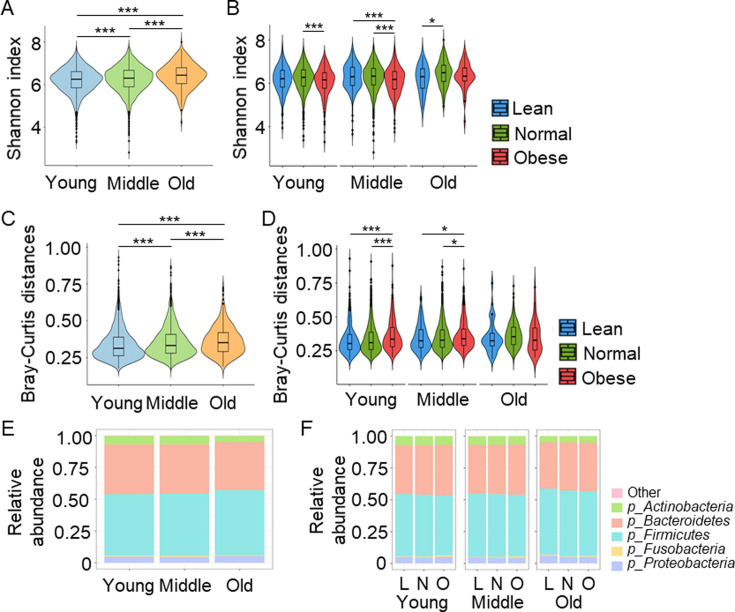
The α diversity, as assessed by Shannon-Wiener index, was significantly increased in the old group relative to the young and middle-age groups (young 6.21 ± 0.59 vs. middle age 6.27 ± 0.60 vs. old 6.41 ± 0.59; p<0.001; Fig. 2A). The α diversity in the middle-age group was significantly increased compared with the young group (p<0.001; Fig. 2A). In the BMI subcluster analysis, significantly increased α diversity was observed in the subjects categorized as normal relative to those categorized as obese in the young (normal 6.23 ± 0.58, obese 6.09 ± 0.63; p<0.01) and middle-age groups (normal 6.29 ± 0.60, obese 6.14 ± 0.61; p<0.001; Fig. 2B). In the old age group, the α diversity in the normal category was significantly higher compared with that in the lean category (lean 6.20 ± 0.62 vs. normal 6.33 ± 0.59); p<0.05); however, no significant difference was observed between the lean and obese categories or between the normal and obese categories (Fig. 2B).
Fig. 2.
Gut microbiota diversity at the phylum level.
A. Shannon-Wiener index in the indicated groups. B. Shannon-Wiener index in the indicated groups. C. Bray-Curtis distances in the indicated groups. D. Bray-Curtis distances in the indicated groups. E. Distribution of gut microbiota at the phylum level in the indicated groups. F. Distribution of gut microbiota at the phylum level in the indicated groups. *p<0.05. ***p<0.001. L: lean; N: normal; O: obese.
The β diversity was estimated using the Bray-Curtis distances, and that in the old group was significantly higher than that in the middle-age and young groups (young 0.31, range 0.26–0.39, middle age 0.33, 0.28–0.41, old 0.35, 0.29–0.42; p<0.001; Fig. 2C). In the young and middle-age groups, the β diversity in the obese individuals was significantly higher than that in the normal or lean individuals (Fig. 2D).
Gut microbiota at the phylum level
The RAs of the top five phyla detected in the gut microbiota of the three groups are shown in Fig. 2E and Supplementary Table 1. The abundance of Bacteroidetes was significantly different among the groups and decreased according to age (young 39.23%, range 34.67–43.61%, middle age 38.81%, 34.46–43.21%, old 37.78%, 32.75–42.94%; p=0.003). The abundance of Firmicutes was also significantly different among the groups and increased according to age (young 48.63%, range 43.41–53.75%, middle age 49.28%, 44.04–54.42%, old 51.66%, 46.02–56.51%; p<0.001), thus leading to an increased Firmicutes/Bacteroidetes ratio in the old group (Supplementary Table 1). The abundances of Actinobacteria, Fusobacteria, and Proteobacteria were also significantly different among the groups (Supplementary Table 1).
In the BMI subgroup analysis shown in Fig. 2F and Supplementary Table 2, the abundance of Bacteroidetes in the middle-age group was significantly different (lean 38.76%, range 34.12–42.39%, normal 38.70%, 34.29–43.13%, obese 39.43%, 35.52–43.81%; p<0.003). The abundances of Firmicutes in the young and middle age groups were significantly different, but no significant difference was observed in the old group (Supplementary Table 2). The Firmicutes/Bacteroidetes ratio was significantly lower in the obese group than in the lean and normal groups (lean 1.26, range 1.03–1.52, normal 1.24, 1.02–1.50, obese 1.20, 0.93–1.47; p=0.029). This trend was also observed in the middle-age group (lean 1.26, 1.07–1.58, normal 1.27, 1.04–1.56, obese 1.21, 1.01–1.49; p=0.003).
Gut microbiota at the genus level
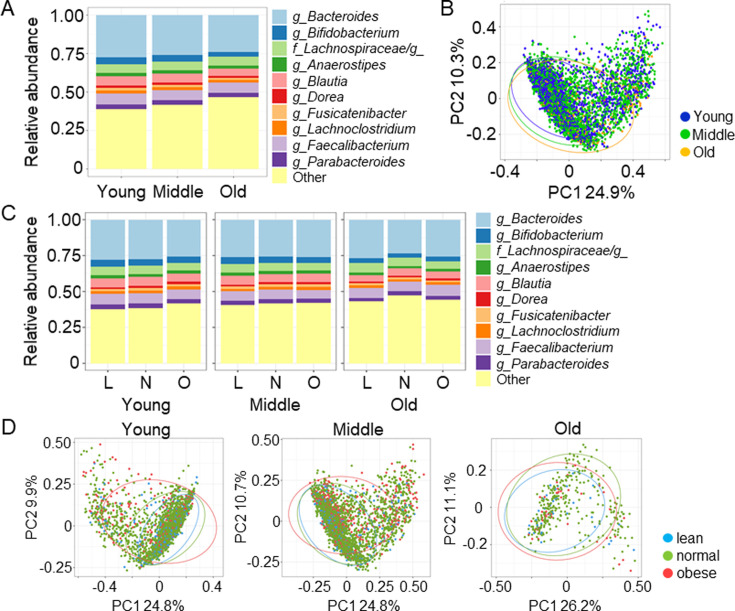
The RAs of the top 10 genera detected in the gut microbiota of the three groups are shown in Fig. 3A and Supplementary Table 3. The abundance of Bacteroides in the young group was significantly higher than those in the middle-age and old groups (young 29.13%, range 21.16–34.84%, middle age 27.51%, 18.39–33.42%, old 25.30%, 15.55–32.56%; p<0.001). The abundance of Bifidobacterium in the young group was also significantly higher than those in the middle age and old (young 3.72, range 1.26–6.87, middle age 3.27, 1.02–6.78, old 1.97, 0.60–4.28; p<0.001). Other genera such as Anaerostipes, Blautia, Dorea, Fusicatenibacter, Lachnoclostridium, Faecalibacterium, and Parabacteroides showed significant differences among the three groups (Supplementary Table 3). The results of principal coordinate analysis at the genus level showed that the three groups did not differ drastically in the abundance of major gut bacteria and were consistent with higher β diversity in the old group (Fig. 3B).
Fig. 3.
Gut microbiota at the genus level.
A. Distribution of gut microbiota at the genus level in the indicated groups. B. Principal coordinate analysis at the genus level was performed to compare the distribution of the gut microbiota. C. Distribution of gut microbiota at the genus level in the indicated groups. D. Principal coordinate analysis at the genus level in the indicated groups. L: lean; N: normal; O: obese.
In the BMI subgroup analysis, the abundance of Bacteroides did not differ among the three BMI groups in any age group (Fig. 3C). The abundance of Bifidobacterium in obese subjects was significantly lower in the middle-age group than in the lean or normal group (lean 3.56, 1.17–7.10, normal 3.39, 1.06–6.8, obese 2.79, 0.76–6.37; p=0.027; Fig. 3C). The results of principal coordinate analysis identified and described gut microbiota profiles at the genus level in three BMI categories in each age group (Fig. 3D).
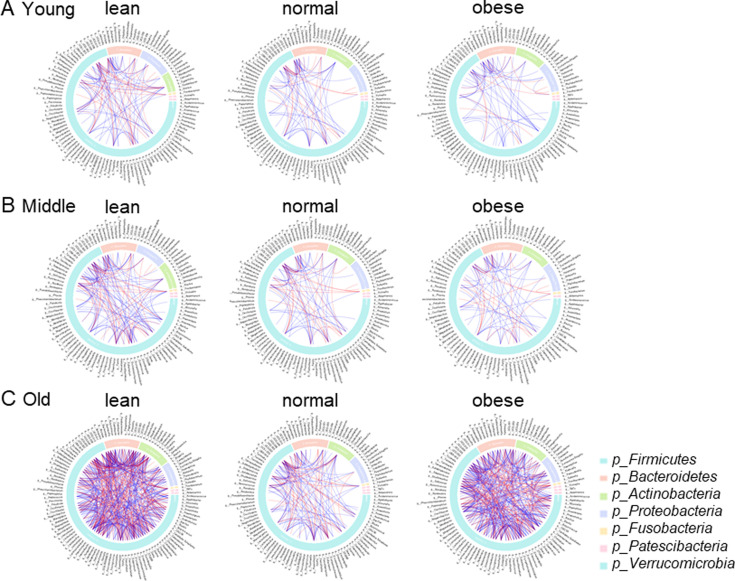
Correlation of microbes
Microbiota-microbiota correlation at the genus level in the three BMI categories in each age group was analyzed to assess the stability of the gut microbiota (Fig. 4). We observed gut microbial interactions in the case of both the same phylum and different phyla, and many lines were seen in Firmicutes, which was the most dominant phylum. Interestingly, the correlation was unique to each age and BMI group. In the young and middle-age groups, we observed relatively many lines in lean subjects compared with normal and obese subjects, which may have reflected lower β diversity in the lean group. In the old age group, many lines were observed, especially in lean and obese subjects, indicating an imbalanced or unstable microbiota configuration in that age group.
Fig. 4.
Gut microbial network correlation at the genus level.
Microbe-microbe interactions were analyzed at the genus level. The blue and red lines indicate positive and negative correlations, respectively. Gut microbiota were placed according to their phylum-level classifications. A. Data of subjects in the young group. B. Data of subjects in the middle-age group. C. Data of subjects in the old group.
DISCUSSION
Growing evidence suggests that the gut microbiota plays a key role in the development of many diseases. However, the normal gut microbiota in the Japanese population has yet to be defined. The goal of using the Mykinso cohort data was to develop a clinical screening method that enables the accurate and rapid detection of a large number of microorganisms in stool samples. It was also to develop quantitative techniques that can be used to improve the current understanding of the relationship between lifestyle and bacterial flora, as well as to develop a novel method of providing lifestyle advice based on gut health. Our first report of average Japanese gut microbiota, which used 2,675 samples from the Mykinso cohort data, included only individuals with ages from 20 to 60 [19]. Taking into consideration the long average life expectancy of the Japanese, gut microbial data of subjects over 60 years of age are essential. In this study, we identified the average gut microbiota in each age group using the largest set of Japanese gut microbial date (5,996 samples). We further classified subjects into three BMI subgroups to investigate the impact of BMI on gut microbiota. The main findings of the present study are as follows: (1) Both α and β diversity increased with age. (2) The Firmicutes/Bacteroidetes ratio was lower in obese subjects compared with lean and normal subjects in the young and middle-age groups. (3) Although the average gut microbiota was different at the genus level among subjects categorized in each age and BMI group, the genus Bacteroides was the most dominant bacterium in all groups. (4) A variety of gut microbiome networks were observed in the age and BMI groups. This indicates that the gut microbiota changes over time based on life stage and metabolic status; thus, matching ages and BMIs would be a good strategy when comparing gut microbiota in clinical settings. Furthermore, considering the host-gut microbe interactions, our results may help to understand how gut microbiota shape the aging and metabolic health of the host.
To date, several studies have aimed to elucidate gut microbiota in the healthy Japanese population and provided intriguing evidence regarding the average gut microbiota [14, 15, 17, 18], although the numbers of subjects were relatively small compared with our study (367 [14], 516 [15], 277 [17] and 1,946 [18]). When comparing our results with those of previous studies, we found both consistency and inconsistency in our findings. For instance, we reported increased Firmicutes abundance in old age (Fig. 2E). This is similar to previous reports indicating that the abundance of Firmicutes continued to increase sequentially with age over a span of 20 years [14, 17]. In addition, the Firmicutes/Bacteroidetes ratio, which has been reported to increase in obese subjects compared with lean subjects [25,26,27], did not increase in obese subjects in our study (Fig. 2F and Supplementary Table 2). Takagi et al. reported no correlation between the Firmicutes/Bacteroidetes ratio and BMI in the Japanese population [17], thus confirming that the Firmicutes/Bacteroidetes ratio did not increase in Japanese obese subjects. On the contrary, although Oki et al. reported that the abundances of Christensenellaceae, Mogibacteriaceae, and Rikenellaceae were higher in lean subjects (BMI <25) than in obese subjects (BMI > 30) [15], we did not observe this phenomenon. This may be due to the classification of the subjects; we divided our subjects into three groups (BMI <18.5, 18.5 ≤BMI ≤25, and BMI >25). Considering that overweight/obesity falls within BMI ≥25, according to WHO criteria for Asians [20], our BMI categorization was reasonable, and our results would be useful in clinical practice. With regards to gender, we did not investigate the impact of gender on gut microbiota, as a previous report showed that gender was not strongly correlated with the general composition of the gut microbiota in the Japanese population [15].
Interestingly, different microbe-microbe interactions were detected according to age and BMI. We believe this would have crucially important meaning from the perspective of future therapeutic modulation of gut microbiota. Specifically, prebiotics, probiotics, or fecal transplantation therapy may make vast alterations in subjects having many microbe-microbe interactions because each microbiota influences the composition of other microbes. Although our results show that many interactions exist in old age, suggesting fewer “core” gut microbiota, this may be due to the small number of subjects in the old age group. Further research is needed to confirm these results.
In clinical settings, our study results may help to detect “outlier” gut microbiota in the Japanese population and elucidate the previously unknown link between gut microbiota and metabolic health. Further, we could apply the data to future therapeutic strategies and health advice from the perspective of health and nutrition science. In the future, it may be necessary to elucidate gut microbial functional differences and the clinical validity of these reference values by comparing them with those of a clinical disease cohort to promote a better understanding of the association between gut microbiota and diseases.
The present study has several limitations that should be considered when interpreting the results. First, we analyzed the gut microbiota from only the Mykinso cohort data and did not include randomly selected healthy Japanese subjects. This may have caused a selection bias because the study population may not have been a true representation of the Japanese population in terms of food habits, residential area, and lifestyle. Second, although the total number of study participants was the highest reported to date, the number of participants aged ≥65 years was rather small compared with the other age groups. As the Mykinso cohort is a growing cohort, we could collect and analyze more gut microbiota in participants aged ≥65 years in the future. Third, we identified the healthy population only by using a simple questionnaire. Therefore, there was a possibility that hidden diseases existed in the study participants. Furthermore, the healthy population might have taken medications that affect gut microbiota. Fourth, we collected feces in tubes containing guanidine thiocyanate solution, which could have an impact on gut microbiota [28, 29]. Fifth, since the average gut microbiota do not necessarily reflect healthy gut microbiota, we did not refer to our analyzed gut microbiota as “healthy” or “normal” gut microbiota. Finally, a network analysis to depict the landscape of the Japanese gut microbiota or multivariable analysis to evaluate the importance of each variable with respect to the gut microbiota was not performed.
In conclusion, we described the composition of the average gut microbiota in three age groups and BMI groups of the healthy Japanese population. Our study aimed to understand the average gut microbiota in the Japanese population, and it may promote a better understanding of the gut microbiota of diseased individuals and how gut microbiota shape the aging and metabolic health of the host.
AUTHOR CONTRIBUTIONS
Conceptualization, N.Y., S.W., A.T., and T. Y.; methodology, N.Y., S.W., H.Y., A.T., and T.Y.; formal analysis, N.Y., S.W., H.Y.; writing – original draft, N.Y.; writing – review and editing, N.Y., S.W., H.Y., H.S., A.T., T.Y., and K.H.; resources, S.W. and A.T.; supervision, A.T. and K.H.; funding acquisition, A.T. and T.Y. All authors have approved the final version of this paper.
DATA AVAILABILITY STATEMENT
Requests for analyzed data should be addressed to the corresponding author.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
The data that support the findings of this study are available from the corresponding author upon reasonable request. As the present study does not involve research using public funds and the data set used in this study was not acquired exclusively for this manuscript and has been used in many other ongoing studies, we are not obligated to make the data open. We would like to finish our research projects before sharing the data with the scientific community. We would like to express our deepest gratitude to all participants of the Mykinso cohort who provided valuable data for our research. We thank Editage (www.editage.com) for English language editing.
REFERENCES
- 1.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, Terasawa K, Kashihara D, Hirano K, Tani T, Takahashi T, Miyauchi S, Shioi G, Inoue H, Tsujimoto G. 2013. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun 4: 1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, Forslund K, Hildebrand F, Prifti E, Falony G, Le Chatelier E, Levenez F, Doré J, Mattila I, Plichta DR, Pöhö P, Hellgren LI, Arumugam M, Sunagawa S, Vieira-Silva S, Jørgensen T, Holm JB, Trošt K, Kristiansen K, Brix S, Raes J, Wang J, Hansen T, Bork P, Brunak S, Oresic M, Ehrlich SD, Pedersen O, MetaHIT Consortium2016. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535: 376–381. [DOI] [PubMed] [Google Scholar]
- 4.Maeda Y, Takeda K. 2017. Role of gut microbiota in rheumatoid arthritis. J Clin Med 6: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. 2019. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med 25: 968–976. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, Hoshi N, Hatano N, Ozawa G, Sasaki N, Mizoguchi T, Amin HZ, Hirota Y, Ogawa W, Yamada T, Hirata KI. 2018. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation 138: 2486–2498. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida N, Yamashita T, Osone T, Hosooka T, Shinohara M, Kitahama S, Sasaki K, Sasaki D, Yoneshiro T, Suzuki T, Emoto T, Saito Y, Ozawa G, Hirota Y, Kitaura Y, Shimomura Y, Okamatsu-Ogura Y, Saito M, Kondo A, Kajimura S, Inagaki T, Ogawa W, Yamada T, Hirata KI. 2021. Bacteroides spp. promotes branched-chain amino acid catabolism in brown fat and inhibits obesity. iScience 24: 103342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan Y, Pedersen O. 2021. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol 19: 55–71. [DOI] [PubMed] [Google Scholar]
- 9.Reijnders D, Goossens GH, Hermes GD, Neis EP, van der Beek CM, Most J, Holst JJ, Lenaerts K, Kootte RS, Nieuwdorp M, Groen AK, Olde Damink SW, Boekschoten MV, Smidt H, Zoetendal EG, Dejong CHC, Blaak EE. 2016. Effects of gut microbiota manipulation by antibiotics on host metabolism in obese humans: a randomized double-blind placebo-controlled trial. Cell Metab 24: 63–74. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida N, Saito Y, Tsujimoto Y, Taito S, Banno M, Kataoka Y, Yamashita T, Hirata KI. 2020. The impact of antibiotics on the metabolic status of obese adults without bacterial infection: a systematic review and meta-analysis. Ann Transl Med 8: 1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, Speelman P, Dijkgraaf MGW, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368: 407–415. [DOI] [PubMed] [Google Scholar]
- 12.Sommer F, Bäckhed F. 2013. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11: 227–238. [DOI] [PubMed] [Google Scholar]
- 13.Nishijima S, Suda W, Oshima K, Kim SW, Hirose Y, Morita H, Hattori M. 2016. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res 23: 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. 2016. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oki K, Toyama M, Banno T, Chonan O, Benno Y, Watanabe K. 2016. Comprehensive analysis of the fecal microbiota of healthy Japanese adults reveals a new bacterial lineage associated with a phenotype characterized by a high frequency of bowel movements and a lean body type. BMC Microbiol 16: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pareek S, Kurakawa T, Das B, Motooka D, Nakaya S, Rongsen-Chandola T, Goyal N, Kayama H, Dodd D, Okumura R, Maeda Y, Fujimoto K, Nii T, Ogawa T, Iida T, Bhandari N, Kida T, Nakamura S, Nair GB, Takeda K. 2019. Comparison of Japanese and Indian intestinal microbiota shows diet-dependent interaction between bacteria and fungi. NPJ Biofilms Microbiomes 5: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takagi T, Naito Y, Inoue R, Kashiwagi S, Uchiyama K, Mizushima K, Tsuchiya S, Dohi O, Yoshida N, Kamada K, Ishikawa T, Handa O, Konishi H, Okuda K, Tsujimoto Y, Ohnogi H, Itoh Y. 2019. Differences in gut microbiota associated with age, sex, and stool consistency in healthy Japanese subjects. J Gastroenterol 54: 53–63. [DOI] [PubMed] [Google Scholar]
- 18.Park J, Kato K, Murakami H, Hosomi K, Tanisawa K, Nakagata T, Ohno H, Konishi K, Kawashima H, Chen YA, Mohsen A, Xiao JZ, Odamaki T, Kunisawa J, Mizuguchi K, Miyachi M. 2021. Comprehensive analysis of gut microbiota of a healthy population and covariates affecting microbial variation in two large Japanese cohorts. BMC Microbiol 21: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe S, Kameoka S, Shinozaki NO, Kubo R, Nishida A, Kuriyama M, Takeda AK. 2021. A cross-sectional analysis from the Mykinso Cohort Study: establishing reference ranges for Japanese gut microbial indices. Biosci Microbiota Food Health 40: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. Regional Office for the Western Pacific. 2000. The Asia-Pacific perspective: redefining obesity and its treatment. Health Communications Australia, Sydney. [Google Scholar]
- 21.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu YX, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vázquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, Wan Y, Wang M, Warren J, Weber KC, Williamson CHD, Willis AD, Xu ZZ, Zaneveld JR, Zhang Y, Zhu Q, Knight R, Caporaso JG. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman J, Alm EJ. 2012. Inferring correlation networks from genomic survey data. PLOS Comput Biol 8: e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. circlize Implements and enhances circular visualization in R. Bioinformatics 30: 2811–2812. [DOI] [PubMed] [Google Scholar]
- 25.Koliada A, Syzenko G, Moseiko V, Budovska L, Puchkov K, Perederiy V, Gavalko Y, Dorofeyev A, Romanenko M, Tkach S, Sineok L, Lushchak O, Vaiserman A. 2017. Association between body mass index and firmicutes/bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol 17: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathur R, Barlow GM. 2015. Obesity and the microbiome. Expert Rev Gastroenterol Hepatol 9: 1087–1099. [DOI] [PubMed] [Google Scholar]
- 28.Nishimoto Y, Mizutani S, Nakajima T, Hosoda F, Watanabe H, Saito Y, Shibata T, Yachida S, Yamada T. 2016. High stability of faecal microbiome composition in guanidine thiocyanate solution at room temperature and robustness during colonoscopy. Gut 65: 1574–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosomi K, Ohno H, Murakami H, Natsume-Kitatani Y, Tanisawa K, Hirata S, Suzuki H, Nagatake T, Nishino T, Mizuguchi K, Miyachi M, Kunisawa J. 2017. Method for preparing DNA from feces in guanidine thiocyanate solution affects 16S rRNA-based profiling of human microbiota diversity. Sci Rep 7: 4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Requests for analyzed data should be addressed to the corresponding author.