Abstract
Vaccines for SARS-CoV-2 have been hugely successful in alleviating hospitalization and deaths caused by the newly emerged coronavirus that is the cause of COVID. However, although the parentally administered vaccines are very effective at reducing severe disease, they do not induce sterilizing immunity. As the virus continues to circulate around the globe, it is still not clear how long protection will last, nor whether variants will emerge that escape vaccine immunity. Animal models can be useful to complement studies of antigenicity of novel variants and inform decision making about the need for vaccine updates. The Syrian golden hamster is the preferred small animal model for SARS-CoV-2 infection. Since virus is efficiently transmitted between hamsters, we developed a transmission challenge model that presents a more natural dose and route of infection than the intranasal challenge usually employed. Our studies demonstrate that an saRNA vaccine based on the earliest Wuhan-like virus spike sequence induced neutralizing antibodies in sera of immunized hamsters at similar titres to those in human convalescent sera or vaccine recipients. The saRNA vaccine was equally effective at abrogating clinical signs in animals who acquired through exposure to cagemates infected either with a virus isolated in summer 2020 or with a representative Alpha (B.1.1.7) variant isolated in December 2020. The vaccine also reduced shedding of infectious virus from the nose, further reinforcing its likely effectiveness at reducing onwards transmission. This model can be extended to test the effectiveness of vaccination in blocking infections with and transmission of novel variants as they emerge.
Keywords: SARS-CoV-2, Variant, saRNA, Hamster, Transmission
1. Introduction
SARS-CoV-2 is the causative agent of COVID-19, a disease that has swept the world causing more than 5 million deaths since its emergence into humans from an animal source in late 2019. Numerous vaccines have been developed in an unprecedented effort to bring the pandemic under control [1]. Amongst the most efficacious and widely used are RNA vaccines that encode the spike protein(S) the major viral antigen of SARS-CoV-2. Antibodies to S induced by the vaccines can neutralize the infectivity of the virus by blocking its access to the virus’ entry receptor, ACE2, on target cells [2]. Other antibodies that bind but do not directly neutralize virus infectivity, as well as T cell responses to S, likely also contribute to the protection from disease, infection and reduction in onwards transmission that COVID vaccines can confer [3], [4].
During the first 9 months of circulation in humans, SARS-CoV-2 was relatively evolutionarily static. A point mutation in spike, D614G, emerged early in the pandemic, and the B lineage (B.1) bearing this change became predominant across the world [5], [6]. Since late 2020, numerous variants have evolved, mostly from the B.1 lineage. These include the Alpha Variant of Concern (VOC) that originated in the UK in September 2020 and led to a second wave of infections in the UK in early 2021 [6]. Alpha rapidly spread across the world becoming the predominant variant in many countries. Further VOCs have emerged including Beta, Gamma, Omicron and Delta. Laboratory data indicate some degree of antigenic distance between the original Wuhan S protein and the S proteins of the VOCs such that sera from human vaccinees neutralize VOCs less efficiently. Since there is a correlation between neutralizing antibody and vaccine efficacy, this poses the question of whether vaccine updates will be necessary [4]. It is critical to ensure that the vaccines in use in the field, all of which currently encode spike protein from the original Wuhan-like virus, can cross-protect against variants [7]. As further variants continue to emerge, it will be important to establish laboratory models that can predict whether antigenic distance between the vaccine immunogen and the variant spike protein compromises vaccine efficacy.
The favoured small animal model for SARS-CoV-2 infection is the Syrian Golden hamster. The murine ACE2 receptor contains a number of amino acid differences between human ACE2 in the receptor:spike interface meaning that mice are refractory to SARS-CoV-2 infection. In contrast, the hamster ACE2 sequence is much closer to that of humans [8]. As a result, hamsters are susceptible to inoculation with very low doses of unadapted human SARS-CoV-2 isolates, and readily transmit virus to other hamsters either by direct contact between cohoused animals or indirect contact to animals in adjacent cages [9].
Hamsters have been used extensively in preclinical studies of COVID vaccines including adenovirus vectored vaccines and mRNA vaccines [10], [11], [12], [13].
Here we show that the Syrian Golden hamster is a suitable model for assessing the efficacy of self-amplifying RNA (saRNA) vaccines comprised of a Venezuelan Equine Encephalitis Virus (VEEV) amplicon encoding the SARS-CoV-2 spike protein, a construct we previously demonstrated capable of inducing antibodies in mice that could efficiently neutralize SARS-CoV-2 [14]. Immunization of hamsters with saRNA induced serum neutralising antibodies that cross neutralized both an historic D614G virus isolate from summer 2020 (B.1.238), or an isolate of Alpha VOC (B.1.1.7) virus. We employed a transmission challenge model, exposing vaccinated hamsters to infected donors cohoused in the same cage. Thus, virus exposure is via a natural route and the challenge dose reflects that capable of transmission. Immunization with a Wuhan-like spike encoded saRNA vaccine efficiently protected against weight loss induced by either the D614G or the Alpha VOC virus. Exposed animals still became infected but shedding of infectious virus was reduced. This model can now be utilized to assess vaccine protection against further variants as they emerge.
2. Results
2.1. saRNA is immunogenic in hamsters
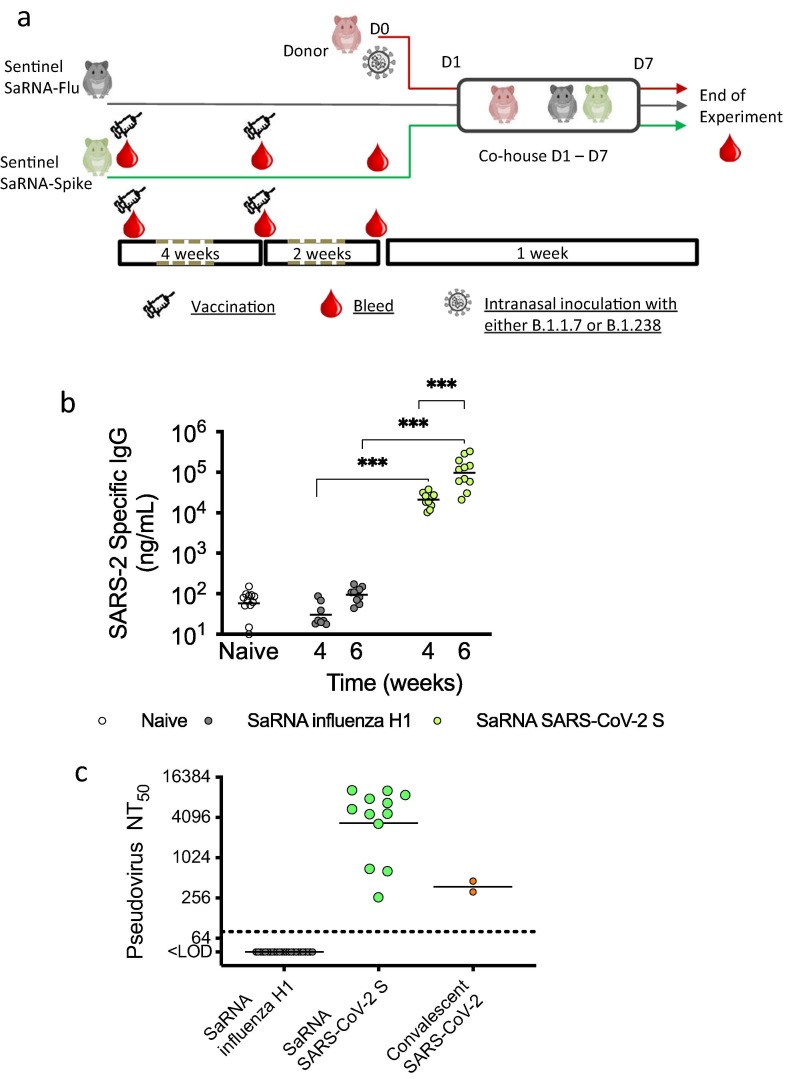
To investigate the efficacy of a saRNA SARS-CoV-2 vaccine, we used Syrian golden hamsters as an in vivo model (Fig. 1 a). We immunised 12 hamsters with 5 μg saRNA vaccine encoding the Wuhan-like SARS-CoV-2 spike protein and 12 hamsters with the same dose of an irrelevant saRNA vaccine against the haemagglutinin (HA) protein from a pH1N1 influenza virus, A/California/04/2009. Both saRNAs were delivered in a lipid nanoparticle as previously described for immunization of mice [14]. After 4 weeks, they received a booster dose of the same amount and same specificity as the priming dose. All animals were bled prior to immunization, at 4 weeks after the first dose and at 2 weeks after the boost, prior to exposure to infected cage mates. S-specific IgG was detected in sera of hamsters SARS-CoV-2 S immunized animals at 4 weeks after the first dose and titres were significantly increased 2 weeks after the boost (Fig. 1b). All SARS-CoV-2 S saRNA-vaccinated hamsters showed serum neutralizing antibody levels against D614G virus similar to or higher than those in two human convalescent sera collected more than 3 weeks after recovery from first wave infection whereas there was no S specific IgG nor neutralizing activity detected in pre immunization sera (not shown) or in sera from influenza HA vaccinated hamsters (Fig. 1b and c). For a subset of hamsters (7/12), there was sufficient serum volume to re-test the neutralizing activity against the Alpha VOC, and titres against this variant were not significantly different than against the D614G virus (Fig S1a). This is in line with our data using sera from human vaccinees following a single dose of Pfizer vaccine that did not find any difference in titres against D614G vs Alpha variant (Fig S1b).
Fig. 1.
Immunization of hamsters with saRNA and challenge by direct contact exposure to infected animals. (a) Schematic of immunization and challenge schedule. Groups of n = 6 animals were immunized by I.M. administration with two doses, given 4 weeks apart, of saRNA encoding influenza HA (control group) of SARS-CoV-2 S. Two weeks after the second dose, immunized hamsters were cohoused with infected donor hamsters one day after they had been intranasally inoculated with 103 pfu SARS-CoV-2 either WT (D614G) variant (B.1.238) or Alpha VOC (B.1.1.7). (b)SARS-CoV-2 S specific IgG antibody levels in sera of naïve or immunized hamsters after first and second dose of saRNA vaccine measured by ELISA. p value established using a paired student’s t test. (c)Neutralizing antibody levels in sera of immunized hamsters collected 2 weeks after second dose of saRNA vaccine measured using a lentivirus pseudotype neutralization assay.
2.2. saRNA SARS-CoV-2 spike vaccine protects against weight loss after exposure to SARS-CoV-2 variant infected cage mates
Twelve naïve animals were infected with SARS-CoV-2 virus by direct intranasal inoculation. Six hamsters received 103 PFU of a D614G isolate from UK collected in summer 2020, B.1.238, and six received 103 PFU of an isolate of the Alpha variant (GISAID ID: EPI_ISL_693401) propagated from a swab collected in London, UK in December 2020 [15]. At 24 h post infection, previously vaccinated animals (2 weeks after booster dose) were introduced in donors’ cages to assess the direct contact transmission from infected donors. Each cage contained three hamsters; either a B.1.238 or Alpha VOC infected donor animal, co-housed with one SARS-CoV-2 spike vaccinated and one influenza HA vaccinated animal (Fig. 1a). All animals were monitored for clinical signs and weighed daily. Nasal washes were collected daily for 7 days to quantify viral loads in the upper respiratory tract. Virus shedding in the nose was titrated by plaque assays and E gene RT-qPCR.
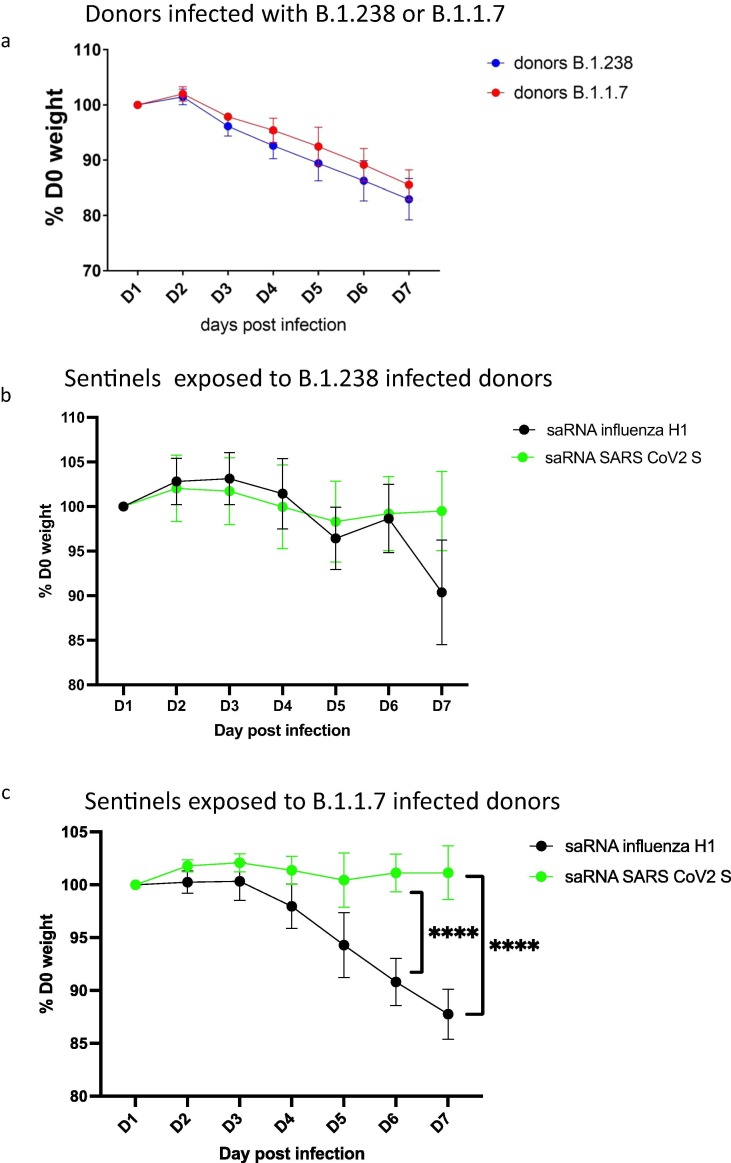
All donor animals that were directly infected with either virus lost weight from day 3 (Fig. 2 a). There was no significant difference in mean weight loss between the donors inoculated with B.1.238 or Alpha VOC (p = 0.0710, Two-way ANOVA). Weight loss continued until the experiment was terminated at day 7 for both viruses, when the mean weight of B.1.238 infected donors was 82.96% starting weight (S.D. = 3.718) and for Alpha variant infected donors the mean was 85.53% (S.D. = 2.70) starting weight.
Fig. 2.
Weight loss in directly inoculated and exposed and vaccinated hamsters. (a)Weight loss in donor hamsters (n = 6 per group) directly inoculated on day 0 with 103 pfu WT D614G (B.1.238) (blue) or with Alpha VOC (B.1.1.7) (red). (b) Weight loss in hamsters vaccinated with saRNA encoding influenza H1 HA (black) or SARS-CoV-2 S protein (green) and exposed to donors infected with WT D614G (B.1.238) SARS-CoV-2. (c) Weight loss in hamsters vaccinated with saRNA encoding influenza H1 HA (black) or SARS-CoV-2 S protein (green) and exposed to donors infected with Alpha VOC (B.1.1.7) SARS-CoV-2. p value established using Šídák's multiple comparisons test after 2-way ANOVA. **** p < 0.0001.
In the vaccinated sentinel groups, influenza HA-vaccinated animals lost weight after exposure to the infected donor animals whereas the SARS-CoV-2 spike-vaccinated hamsters’ weights remained stable (Fig. 2 b and c). For the B.1.238 group, the HA vaccinated hamsters had a mean weight at 6 days after exposure of 90.37% starting weight (S.D. = 5.87%) whereas the SARS-CoV-2 spike vaccinated hamsters retained 99.50% their starting weight (S.D. = 4.44%) but this difference was not statistically significant) (p = 0.5702, Two-way ANOVA) (Fig. 2b). Weight loss occurred faster and was higher in the Alpha VOC exposed HA-vaccinated group (Fig. 2c) (p < 0.0001, Two-way ANOVA), compared to the spike-vaccinated group. The weight loss was significantly greater on Day 5 (p = T 0.0272, Šídák's multiple comparisons test), Day 6 (p < 0.0001) and Day 7 (p < 0.0001) in the HA vaccinated group than in the spike vaccinated group.
2.3. Transmission was less efficient to SARS-CoV-2 saRNA vaccinated hamsters.
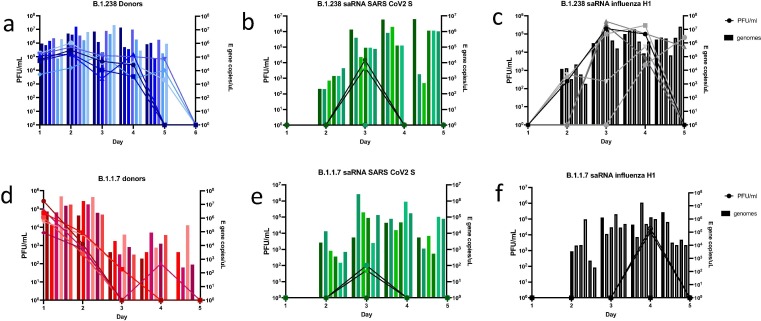
All donors were robustly infected and infectious virus was detected in nasal washes at high titres by day 1 post infection (Fig. 3 a, d). Donors infected with B.1.238 shed infectious virus in the nasal wash to higher peak titre (p = 0.0121, t test with log transformation) and higher total amount (P < 0.0001, area under the curve (AUC), t test), compared to those infected with Alpha VOC.
Fig. 3.
Viral loads detected in hamster nasal wash by plaque assay and E gene qPCR. Infectious titres measured by plaque assay in Vero cells are indicated by lines. Viral RNA loads measured by qRT-PCR for E gene are indicated by histogram bars for each individual hamster n = 6 per group. (a) B.1.238 infected donors. (b) saRNA SARS-CoV-2 S vaccinated hamsters exposed to B.1.238 donors. (c) saRNA influenza H1 HA vaccinated hamsters exposed to B.1.238 donors. (d) B.1.1.7 infected donors. (e) saRNA SARS-CoV-2 S vaccinated hamsters exposed to B.1.1.7 donors. (f) saRNA influenza H1 vaccinated hamsters exposed to B.1.1.7 donors.
Transmission occurred to all co-housed animals as evidenced by detection of SARS-CoV-2 E gene RNA in nasal wash following exposure to infected donors (Fig. 3b, c, e and f). However, the total viral RNA load in the SARS-CoV-2 spike-immunized groups was lower than in the influenza HA immunized groups (AUC, p = 0.0021 for B.1.238 group and p = 0.0033 for Alpha VOC group, t test), indicating that virus replication was curtailed by vaccination. In addition, transient infectious virus was only detected in the nasal wash of 2/6 SARS-CoV-2 spike immunized hamsters exposed to B.1.238 donors on day 3 (Fig. 3b) vs more prolonged shedding in all 6 influenza HA immunized animals (Fig. 3c). In the Alpha VOC challenged groups, only 2/6 animals shed infectious virus in the nose in either SARS-CoV-2 spike (Fig. 3e) or the control influenza HA immunized groups (Fig. 3f) but titres were 2 log10 lower after SARS-CoV-2 S immunization.
In total 4 saRNA SARS-CoV-2 S immunized animals who became infected on exposure shed infectious virus, 2/6 in the B.1.238 group (cage 1 and 3 FigS2) and 2/6 in the B.1.1.7 group (cage 9 and 12 supplementary Fig. 3). We investigated further any explanation for this apparent ‘breakthrough’. These four animals did not make lower neutralizing antibody responses than the rest (Fig S 4). The donors in the cages where these ‘breakthroughs’ occurred did not shed higher infectious virus (Fig S2 and S3). However, it was notable that there were only two influenza HA immunized sentinels who shed infectious virus in the Alpha VOC group and these were cohoused in the same cages (cages 9 and 12 Fig S3) as the SARS-CoV2-S immunized animals who shed infectious virus. The breakthrough was not likely to be explained by infection acquired from the influenza HA immunized animals since they did not shed infectious virus until day 4, whereas the S immunized animals shed on day 3. However, it might be that the donors in those cages were more likely to transmit their virus for other reasons.
2.4. Vaccination with saRNA SARS-CoV-2 S limits virus detected in lower respiratory tract
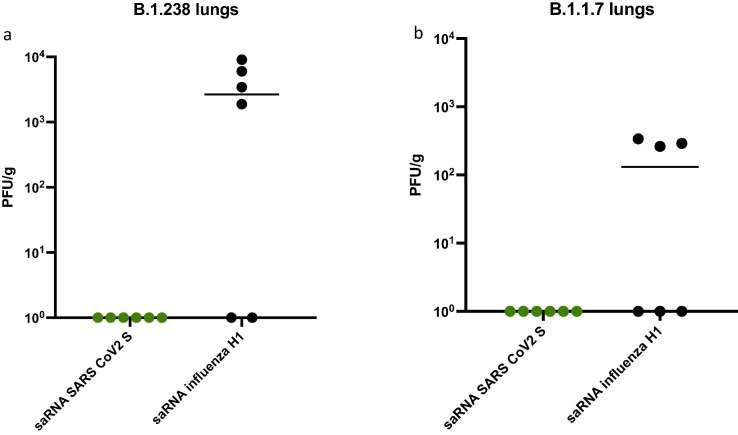
All remaining hamsters were culled on day 7 post infection of donors and lung tissue was harvested. Viral RNA was detected in 4/6 of saRNA influenza H1 vaccinated animals exposed to B.1.238 (Fig. 4 a) and 3/6 saRNA influenza H1 vaccinated animals exposed to Alpha VOC (Fig. 4b). None of the hamsters immunized with saRNA SARS-CoV-2 S vaccine had any detectable virus in their lungs at day 7 (Fig. 4a and b).
Fig. 4.
Infectious virus in lungs at 6 days post exposure for sentinel animals. Infectious virus measured by plaque assay in Vero cells from lung homogenates of immunized hamsters exposed to donors infected with (a) WT D614G (B.1.238) or (b) Alpha VOC (B.1.1.7) SARS-CoV-2.
3. Discussion
As new SARS-CoV-2 variants emerge worldwide it is imperative that we establish systems to monitor whether vaccines will remain effective. Here we investigate whether an saRNA vaccine that encodes the original Wuhan-like SARS-CoV-2 spike, typical of those in current use, confers cross-protection against the earliest SARS-CoV-2 variant, the Alpha VOC, also known as B.1.1.7 following challenge of vaccinated animals by a natural exposure route. SaRNA offers potential advantages over mRNA as the amplification kinetics can promote antigen expression for up to 21 days and offer potential for low dose RNA vaccination [14].
Using an in vivo contact transmission model in Syrian Golden hamsters, we demonstrated that a prime-boost vaccination with a Wuhan-like spike saRNA vaccine protected against severe disease, measured by weight loss, following infection by exposure to a D614G ‘wild type’ virus or the Alpha VOC. There was no virus detected in the lungs of any of the animals in the spike vaccine groups suggesting replication in the lung had been limited. All hamsters receiving the saRNA SARS-CoV-2 spike vaccine showed neutralising antibody responses against SARS-CoV-2 at least equivalent to that seen in sera from convalescent or vaccinated humans. Neutralizing titres in a subset of vaccinated hamsters’ sera was equally efficient against the Alpha VOC (sFig S1a) as against first wave virus. Indeed we and others have previously shown that human post-vaccination sera also efficiently cross neutralizes the Alpha VOC (Fig S1b) [15]. Since the level of protection against severe disease conferred by vaccination has been correlated with the neutralizing antibody response [4], the protection against weight loss conferred by the saRNA vaccine against exposure to Alpha VOC was not unexpected. Indeed, real world vaccine effectiveness of RNA vaccines based on the Wuhan-like spike against severe disease caused by Alpha VOC has been measured at 93% in the UK [16].
However, the parentally administered saRNA vaccine was much less effective at blocking infection. Indeed, all exposed animals became infected regardless of vaccination status. It is important to recognise that animals were challenged at 2 weeks following second vaccination to replicate efficacy assessment in clinical trials, however responses may have continued to improve over subsequent weeks. Lack of efficacy against infection may reflect the poor utility of parenteral vaccination to induce mucosal IgA. Although not measured in this study due to lack of an available assay it would be important to determine mucosal response in future studies. The likelihood that vaccinated but infected animals would transmit onwards was reduced since viral shedding was reduced in the spike vaccinated groups. This was seen at the level of reduced viral RNA load in nasal wash but, more importantly, the number of animals in the spike vaccine groups that shed infectious virus were lower as were the levels of infectious virus shed. The outcome in the hamster model is thus is reminiscent of the situation reported following RNA vaccination in humans, were the real-world vaccine effectiveness (VE) against infection is lower than against disease, but transmission is still reduced when the antigenic match between vaccine and infecting virus is good. For example, in contrast to the 93% VE against severe disease [16] the VE against infection is 80% for the Alpha VOC [17] whereas protection against infection and onwards transmission with Delta VOC, which is more antigenically distinct from the Wuhan-like vaccine, is lower still [18], [19].
The Syrian Golden hamster model has been used effectively by others to show vaccine efficacy against different variants including Alpha and Beta VOCs [10], [11], [13], [20], [21]. In other studies, vaccinated animals were challenged directly with an artificially high dose of virus. All vaccines protected against severe disease but reduction in viral loads was not significant when intramuscular vaccines were given and animals were challenged with the more antigenically distant Beta VOC [20]. Here we challenged vaccinated hamsters via a contact transmission model. This allows infection of the sentinel vaccinated animals by different routes including airborne exposure, fomite exposure and direct contact, as well as potentially through saliva since the animals share food and water. All sentinel animals, whether vaccinated with S or control vaccine, acquired infection, however weight loss and virus shedding were reduced by vaccination. Although this route of challenge might result in some variation in the dose received, and therefore a wider spread of data in some groups, the route and dose of virus might be considered more natural than a direct intranasal inoculation. In future studies it will be important to understand the level of protection conferred by the variety of COVID vaccines against both severe disease but also infection and transmission of more antigenically distinct variants such as Beta, Delta and Omicron VOCs. We propose that the hamster transmission model described here might be suitable to monitor variants for vaccine escape and contribute to decision making around the need for vaccine updates.
4. Methods
4.1. Biosafety and ethics statement
All work performed was approved by the local genetic manipulation (GM) safety committee of Imperial College London, St. Mary’s Campus (centre number GM77), and the Health and Safety Executive of the United Kingdom, under reference CBA1.77.20.1. Animal research was carried out under a United Kingdom Home Office License, P48DAD9B4.
4.2. Cells and viruses
African green monkey kidney (Vero) cells (Nuvonis Technologies) were maintained in OptiPRO SFM (Life Technologies) containing 2X GlutaMAX (Gibco). Maintained at 37 °C, 5% CO2. Human embryonic kidney cells (293T; ATCC; ATCC CRL-11268) were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco), 10% fetal calf serum (FCS), 1× non-essential amino acids (NEAA; Gibco), 1× penicillin–streptomycin (P/S; Gibco). Stably transduced ACE2-expressing 293T cells were produced as previously described [22], [23].
Viruses were isolated by inoculating 100ul of neat swab material onto 24-well plates of Vero cells, incubating at 37 °C, 5% CO2 for 1 h before adding 1 ml OptiPRO SFM supplemented with 2X Glutamax, 1% P/S and 1% amphotericin and incubating again for 5–7 days until cytopathic effect was observed. Isolates were passaged twice in Vero cells and used for subsequent experiments.
| Name used in text | Virus name | GISAID Accession ID | PANGO lineage |
|---|---|---|---|
| B.1.1.7 /Alpha VOC | hCoV-19/England/204690005/2020 | EPI_ISL_693401 | B.1.1.7 |
| WT D614G | hCoV-19/England/20M325595N/2020 | n/a | B.1.238 |
4.3. Plaque assays
Nasal wash samples were serially diluted in OptiPRO SFM, 2X GlutaMAX (1:10) and added to Vero cell monolayers for 1 h at 37 °C. Inoculum was then removed and cells were overlayed with DEMEM containing 0.2% w/v bovine serum albumin, 0.16% w/v NaHCO3, 10 mM HEPES, 2 mM L-Gutamine, 1X P/S and 0.6% w/v agarose. Plates were incubated at 37 °C, 5% CO2 for 3 days. The overlay was then removed, and monolayers were stained with crystal violet solution for 1 h at room temperature. Plates were washed with tap water then dried and virus plaques were counted.
4.4. E gene qPCR
Viral RNA was extracted from Hamster nasal wash supernatants using the Qiagen Viral RNA mini kit, according to manufacturer’s instructions.
Quantitative real-time RT-PCR (qRT-PCR) was then performed using AgPath RT-PCR (Life Technologies) kit on a QuantStudio(TM) 7 Flex System with the primers for E gene used in (Corman et al., 2020). A standard curve was also generated using dilutions viral RNA of known copy number to allow quantification of E gene copies in the samples from Ct values. E gene copies per ml of original virus supernatant were then calculated.
4.5. Hamster transmission studies
Hamster transmission studies were performed in a containment level 3 laboratory, using ISO Rat900 Individually Ventilated Cages (IVC) (Techniplast, U.K). Outbred Syrian Hamsters (4–6 weeks old), weighing 70–140 g were used. Prior to the study hamsters were confirmed to be seronegative against SARS-CoV-2. Twelve hamsters were immunized twice, four weeks apart with 5ug saRNA SARS-CoV-2 S, intramuscularly in 100 μl. An additional twelve hamsters were immunised twice, four weeks apart, with 5ug saRNA influenza H1, intramuscularly in 100 μl. Twelve donor hamsters were intranasally inoculated with 50 μl of 103 PFU of either B.1.1.7 or B.1.238 virus while lightly anaesthetised with isoflurane. To assess direct contact transmission one previously immunized hamster with saRNA SARS-CoV-2 S and one previously immunised with saRNA influenza H1 were introduced into each cage 1-day post initial inoculation and cohoused continuously thereafter. All animals were nasal washed daily, while lightly anaesthetised under isoflurane, by instilling 400 μl of PBS into the nostrils, the expectorate was collected into disposable 50 ml falcon tubes. Hamsters were weighed daily post-infection.
4.6. Pseudovirus production and neutralisation assays
SARS-CoV-2 spike-bearing lentiviral pseudotypes (PV) were generated as described previously [22], [15]. Briefly, 10 cm dishes of HEK 293Ts were transfected using lipofectamine 3000 (Thermo) with a mixture of pCSFLW, pCAGGS-GAGPOL and either D614G or B.1.1.7 spike proteins expressed in pcDNA3.1 [14]. 24 h post transfection, supernatant was discarded and replaced with fresh media. Pseudovirus-containing supernatant were subsequently collected at 48 and 72 h post-transfection and pooled, filtered through a 0.45 μm filter, aliquoted and frozen at −80 °C. Pseudovirus neutralisation assays were performed by incubating serial dilutions of heat-inactivated hamster convalescent antisera with a constant amount of pseudovirus. Antisera/pseudovirus mix was then incubated at 37 °C for 1 h and overlayed into 96 well plates of HEK 293T-ACE2 cells. 48 h later assays were then lysed and read on a FLUOstar Omega plate reader (BMF Labtech) using the Luciferase Assay System (Promega).
4.7. Live virus neutralisation assay
The ability of sera to neutralise SARS-CoV-2 virus was assessed by neutralisation assay on Vero cells. Sera were serially diluted in OptiPRO SFM (Life Technologies) and incubated for 1 h at RT with 100 TCID50/well of SARS-CoV-2 variants and transferred to 96-well plates pre-seeded with Vero-E6 cells. Serum dilutions were performed in duplicate. Plates were incubated at 37 °C, 5% CO2 for 42 h before fixing cells in 4% PFA. Cells were treated with methanol 0.6% H2O2 and stained for 1 h with a 1:3000 dilution of 40143-R019 rabbit mAb to SARS-CoV-2 nucleocapsid protein (Sino Biological). A 1:3000 dilution of sheep anti-rabbit HRP conjugate (Sigma) was then added for 1 h. TMB substrate (Europa Bioproducts) was added and developed for 20 mins before stopping the reaction with 1 M HCl. Plates were read at 450 nm and 620 nm and the concentration of serum needed to reduce virus signal by 50% was calculated to give NT50 values.
4.8. Statistical analysis
Statistical analysis was performed using Graphpad Prism. Neutralising antibody titres were log10 transformed and tested using a paired Student’s test. Weight loss was compared using 2-Way ANOVA, followed by Šídák's multiple comparisons test. For all tests, a value of p < 0.05 was considered significant.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Wendy Barclay reports financial support was provided by Medical Research Council. Robin Shattock reports financial support was provided by Bill and Melinda Gates Foundation. Ruthiran Kugasathan reports financial support was provided by Wellcome Trust. Robin Shattock reports a relationship with VXT that includes: board membership, consulting or advisory, and equity or stocks. Robin Shattock has patent pending to VXT.
Acknowledgments
Acknowledgements
The authors thank staff at Imperial College Central Biological Services for their expert help.
The swab from which Alpha VC B.1.1.7 was isolated was initially provided by Public Health England and we thank M. Zambon, R. Gopal and M. Patel for their help.
The swab from which WT D614G B.1.238 virus was isolated was initially provided through the ISARIC consortium and we acknowledge the ISARIC4C Investigators for their help (https://isaric4c.net/about/authors/).
We acknowledge the G2P-UK National Virology consortium funded by MRC/UKRI (grant ref: MR/W005611/1.) (United Kingdom) and The Bill and Melinda Gates Foundation [INV-016635] (United States) for funding. RK was supported by Wellcome fellowship no. 216353/Z/19/Z (United Kingdom).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2022.03.064.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 2.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M., Esswein S.R., Gristick H.B., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng S., et al., Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. medRxiv, 2021;p. 2021.06.21.21258528. [DOI] [PMC free article] [PubMed]
- 4.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27(7):1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 5.Volz E., et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184(1):64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peacock T.P., Penrice-Randal R., Hiscox J.A., Barclay W.S. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J Gen Virol. 2021;102(4) doi: 10.1099/jgv.0.001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cevik M., Grubaugh N.D., Iwasaki A., Openshaw P. COVID-19 vaccines: Keeping pace with SARS-CoV-2 variants. Cell. 2021;184(20):5077–5081. doi: 10.1016/j.cell.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conceicao C., et al., The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol, 2020;18(12): p. e3001016. [DOI] [PMC free article] [PubMed]
- 9.Rosenke K., Meade-White K., Letko M., Clancy C., Hansen F., Liu Y., et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerging Microbes Infect. 2020;9(1):2673–2684. doi: 10.1080/22221751.2020.1858177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Doremalen N., Purushotham J.N., Schulz J.E., Holbrook M.G., Bushmaker T., Carmody A., et al. Intranasal ChAdOx1 nCoV-19/AZD1222 vaccination reduces viral shedding after SARS-CoV-2 D614G challenge in preclinical models. Sci Transl Med. 2021;13(607) doi: 10.1126/scitranslmed.abh0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tostanoski L.H., Wegmann F., Martinot A.J., Loos C., McMahan K., Mercado N.B., et al. Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters. Nat Med. 2020;26(11):1694–1700. doi: 10.1038/s41591-020-1070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer M., Wang Y., Edwards D., Smith G.R., Rubenstein A.B., Ramanathan P., et al. Attenuated activation of pulmonary immune cells in mRNA-1273-vaccinated hamsters after SARS-CoV-2 infection. J Clin Invest. 2021;131(20) doi: 10.1172/JCI148036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bricker T.L., et al., A single intranasal or intramuscular immunization with chimpanzee adenovirus vectored SARS-CoV-2 vaccine protects against pneumonia in hamsters. bioRxiv; 2020. [DOI] [PMC free article] [PubMed]
- 14.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat Commun. 2020;11(1) doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown J.C., et al., Increased transmission of SARS-CoV-2 lineage B.1.1.7 (VOC 2020212/01) is not accounted for by a replicative advantage in primary airway cells or antibody escape. bioRxiv, 2021;p. 2021.02.24.432576.
- 16.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., et al. Effectiveness of Covid-19 Vaccines against the B.1.617.2 (Delta) Variant. N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pritchard E., Matthews P.C., Stoesser N., Eyre D.W., Gethings O., Vihta K.-D., et al. Impact of vaccination on new SARS-CoV-2 infections in the United Kingdom. Nat Med. 2021;27(8):1370–1378. doi: 10.1038/s41591-021-01410-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eyre DW. et al., The impact of SARS-CoV-2 vaccination on Alpha & Delta variant transmission. medRxiv 2021; p. 2021.09.28.21264260.
- 19.Singanayagam A., et al., Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. The Lancet Infectious Diseases. [DOI] [PMC free article] [PubMed]
- 20.Fischer R.J., van Doremalen N., Adney D.R., Yinda C.K., Port J.R., Holbrook M.G., et al. ChAdOx1 nCoV-19 (AZD1222) protects Syrian hamsters against SARS-CoV-2 B.1.351 and B.1.1.7. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-26178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer M., Wang Y., Edwards D., Smith G.R., Rubenstein A.B., Ramanathan P., et al. Attenuated activation of pulmonary immune cells in mRNA-1273–vaccinated hamsters after SARS-CoV-2 infection. J Clin Investig. 2021;131(20) doi: 10.1172/JCI148036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6(7):899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 23.Rebendenne A., Chaves Valadão A.L., Tauziet M., Maarifi G., Bonaventure B., McKellar J., et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J Virol. 2021;95(8) doi: 10.1128/JVI.02415-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.