Abstract
The emergence of variants and the reports of co-infection caused by Candida auris in COVID-19 patients adds a further complication to the global pandemic situation. To date, no effective therapy is available for C. auris infections. Thus, characterization of therapeutic targets and designing effective vaccine candidates using subtractive proteomics and immune-informatics approaches is useful tool in controlling the emerging infections associated with SARS-CoV-2. In the current study, subtractive proteomics-assisted annotation of the vaccine targets was performed, which revealed seven vaccine targets. An immunoinformatic-driven approach was then employed to map protein-specific and proteome-wide immunogenic peptides (CTL, B cell, and HTL) for the design of multi-epitope vaccine candidates (MEVCs). The results demonstrated that the vaccine candidates possess strong antigenic features (>0.4 threshold score) and are classified as non-allergenic. Validation of the designed MEVCs through molecular docking, in-silico cloning, and immune simulation further demonstrated the efficacy of the vaccines by producing immune factor titers (ranging from 2500 to 16000 au/mL) i.e., IgM, IgG, IL-6, and Interferon-α. In conclusion, the current study provides a strong impetus in designing anti-fungal strategies against Candida auris.
Keywords: Candida auris, COVID-19, Subtractive proteomics, Vaccines, Immune simulation
1. Introduction
Candida auris infections, and outbreaks caused by diverse fungal groups, pose a significant threat to human health, and have affected almost 1.7 billion people worldwide. These infections are associated with the immune invasion properties of these pathogens, causing skin and mucosa diseases [1,2]. Candida species are the major source of nosocomial infections, and 0.4 million people are infected by it annually, with more than 40% of patients succumbing to it [1,2]. Candida auris was first isolated in 2009 from a female patient's ear discharge in Japan. This species is nested within the Clavispora clade of the family Metschnikowiaceae (order Saccharomycetales), and four major lineages/clades have been reported: South Asian, South African, South American, and East Asian [2]. The members of these lineages are associated with the spread of C. auris infections and outbreaks [[3], [4], [5]]. With the discovery of C. auris, the rapid expansion of its lineages across the globe has been evident from the infection cases reported from 6 continents [6]. These nosocomial outbreaks can be linked to the prolific spread of C. auris across health facilities, with its persistence in the human host and on surfaces [3,7,8]. C. auris linked nosocomial outbreaks were first observed in South Korea, followed by reports from India, South Africa, Kuwait, Venezuela, the USA, and many European countries [[9], [10], [11], [12], [13], [14], [15]].
C. auris has been isolated from patients admitted to the hospitals, and it has been reported that this pathogen favors colonizing the skin [16]. According to one study, 5% of nosocomial infections reported in intensive care units (ICUs) in India are linked with C. auris [17]. This pathogen has caused multiple infection outbreaks in hospitals [16] and is considered one of the most prolific pathogens to colonize ICUs, and therefore plays a vital role in the transmission of nosocomial infections [18].
After December 2019, due to the fast expansion of SARS-CoV-2 (COVID 19), hospitals have made additional efforts to accommodate the huge influx of patients. Of these patients, 5–30% were critically ill and were provided mechanical ventilators in ICUs [19,20]. Due to the influx of patients, proper compliance of guidelines was not followed, leading to substantial contamination of ICUs and the development of co-infections. C. auris co-infection has been associated with 50% of deaths caused by COVID 19, making it a major contributor to the death rate of critically ill COVID patients [21,22]. Moreover, as C. auris is one of the most capable colonizers of the ICU environment, it has an increased chance of co-infection in critically ill COVID patients. Fungal co-infection was reported in SARS patients treated with heavy doses of corticosteroids resulting in immunosuppression and the establishment of mycosis. The ability of C. auris to develop multidrug resistance and to grow at high temperature (42 °C), with rapid expansion and high mortality rate, makes it an emerging and deadly pathogen.
C. auris genome size ranges from 12.1 Mb to 12.7 Mb and consists of over 5500 genes [23,24]. Subtractive proteomics approaches aided by computational algorithms are used to select the most suitable proteins, among this large pool of data, to design suitable vaccines and to combat nosocomial infections caused by C. auris. Computational vaccine design is an effective approach to check all required parameters, such as antigenicity, allergenicity, physicochemical properties, and thermostability, making this approach cost-effective, thermodynamically stable, and safe [25]. In this study, outer membrane proteins which are considered highly virulent, antigenic, and essential for host-pathogen interactions were investigated [26]. The selected proteins were then used to predict cytotoxic T lymphocyte (CTL), helper T lymphocyte (HTL), and B-cell epitopes, and we have designed protein-specific and whole proteome-wide multi-epitope subunit vaccines against C. auris. This study may help to combat nosocomial infections and reduce the chances of co-infection with COVID. The designed candidate vaccines were also evaluated using molecular docking, and immune simulation strategies. However, further experimental validations are required to confirm the efficacy and safety of the proposed vaccine designs.
2. Materials and methods
2.1. Retrieval of Candida auris proteome and subtractive proteomics
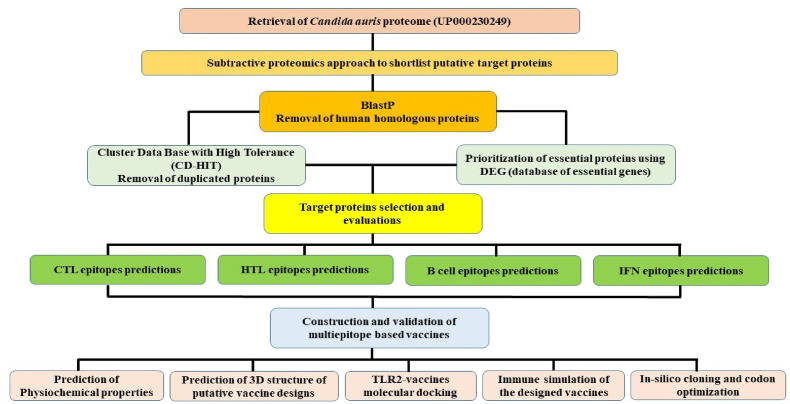
Candida auris standard reference proteome (UniProt ID: UP000230249) containing 5409 proteins was retrieved from UniProtKB (https://www.uniprot.org/) [27] and analyzed by using the subtractive proteomics to identify proteins that might be used in vaccine development against Candida auris infection. Firstly, the BLASTp tool using non-redundant protein sequences (nr) database with default parameters (e-value: 10−5) was used to identify the human host (Homo sapiens, ID: 9606) and pathogen homologous proteins. To avoid the autoimmune response of the host, the non-homologous proteins were utilized while the homologous proteins were eliminated [28]. To prevent overlapping's in the epitope selection owing to sequence similarity, the Cluster Data Base with High Tolerance (CD-HIT) suite (http://weizhongli-lab.org/cd-hit/) was used to remove the duplicated proteins with an 80% percent identity (cut-off value: 0.8) [29]. This cut-off number has already been utilized by many researchers and has been submitted to the database to report the extremely high-quality matches [30]. The methodological workflow adopted in this study is shown in Fig. 1 .
Fig. 1.
The methodological workflow showing different steps adopted in the design of the study.
2.2. Prioritization of the essential proteins
The DEG (database of essential genes) (http://tubic.tju.edu.cn/deg/) contains information on key proteins and genes that pathogens need to survive. These essential proteins are promising candidates for vaccine development. Therefore, the important protein of Candida auris was found by using the DEG online tools [31]. However, the CELLO online server (http://cello.life.nctu.edu.tw/cello.html) having 88% accuracy and based on SVM was used for the identification of subcellular localization of important proteins [32]. According to scientific logic, the extracellular proteins are measured as highly appropriate for vaccine development because of their critical role in the attachments of the pathogen to the host cell surface and induction of pathogenicity [33]. Selected pathogenic proteins were also tested for antigenicity using the webserver VaxiJen (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) by selecting fungi as the target organism at a threshold of 0.4 and used for further investigations [34]. The ProtParam weight calculator program was used to compute the molecular weight of chosen proteins [35].
2.3. Mapping immunogenic peptides in the target proteins
An online web server NetCTL 1.2 (http://www.cbs.dtu.dk/services/NetCTL/) [36] was used to predict Cytotoxic T-lymphocytes. The key values used by the server for the prediction of CTL are transportation efficiency, TAP (Transport Associated with Antigen Processing), MHC I binding peptides prediction, and proteasomal C terminal cleavage. The threshold value was set to 0.75 for the prediction of CTL epitopes. The IEDB online server (http://www.iedb.org/) was used to predict HTL epitopes using the seven reference alleles set of human HLAs [37,38]. The IC50 value determines the affinity score of each peptide. Binding affinity is strong for peptides with an IC50 value less than 50 nM. Peptides with an IC50 of less than 500 nM are intermediates, whereas peptides with an IC50 of fewer than 5000 nM indicate poor epitope binding affinity. The peptides' affinity scores and percentile rankings are inversely linked, which indicates that the higher the binding affinity, the lower the percentile rank, and vice versa. The Algpred server (http://www.imtech.res.in/raghava/algpred2/) [39,40] was used to evaluate the Allergenicity of each MHC-II predicted epitopes. Similarly, the VexiJen server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used to assess the antigenicity of each epitope [34]. As a result, non-allergenic epitopes with high antigenicity were chosen for putative vaccine development. Next, we used the ABCPred (https://webs.iiitd.edu.in/raghava/abcpred/) online server to predict the linear B-cell epitopes. The mentioned server has >65% accuracy, 0.49 sensitivity and 0.75 specificity [41]. For linear B-cell epitope prediction, the user employs the Kernel Method by analyzing SVM (Support Vector Machine) and uses AAP (amino acid pair) antigenicity. The Ellipro service (http://tools.iedb.org/ellipro/) was also used to predict discontinuous B-cell epitopes, with findings based on Thornton's method and a residue clustering algorithm [42]. Additionally, IFN-epitope (http://crdd.osdd.net/raghava/ifnepitope/) server was used to investigate interferon-gamma triggering MHC-II epitopes. This online tool uses the SVM score and motif for each input epitope to predict interferon-inducing properties [43].
2.4. Construction of multi epitopes subunit vaccine
The final putative vaccine constructs were designed using B cell, CTL, and HTL epitopes with high binding affinity, non-allergenicity, and antigenicity. Based on published data [37,39], the selected epitopes were linked together using different linkers. This included the use of epitopes specific AAY, GPGPG, and KK linkers to join the putative CTL, HTL, and B-cell epitopes, respectively. The strategy was used in the target proteins specific vaccine design. Additionally, a proteome-wide vaccine was also constructed in this study. These linkers are very important in epitope presentation as well as prevention of neoepitopes and enhance immunogenicity by stabilizing the protein structure. The EAAK linker was used to link the adjuvant to the N-terminal of the final candidate vaccine constructs [44].
2.5. Prediction of physicochemical properties
The ProtParam online tool (http://web.expasy.org/protparam/) was used to estimate the physicochemical features of constructed vaccines such as instability index, theoretical PI, in vivo and invitro half-life, amino acid composition, and GRAVY [35].
2.6. Prediction of 3D structure of the designed vaccines
The Robetta online server (http://robetta.bakerlab.org) was used to predict the tertiary structure of the putative vaccine [45]. Robetta server uses a comparative modeling approach based on BLAST, FFAS03, or 3d-jury to identify templates for the particular amino acid sequence. The Robetta server has been rated as one of the most correct and stable servers since 2014. It makes use of Continuous Automated Model Evaluation (CAMEO) to compute up to 20 pre-released PDB targets per week. This server's average LDDT (Local Distance Difference Test) score is around 69, indicating a highly independent score [45].
2.7. TLR2-vaccine docking
Using the HawkDock server (http://cadd.zju.edu.cn/hawkdock/), the designed vaccine was docked with the TLR2 (Human Toll-like receptor-2). The server employs a hybrid docking method and requires an amino acid sequence as input for analysis, setting it apart from other similar servers. The HawkDock server provides experimental information on the protein-protein binding site and small-angle x-ray drip quickly and authentically [46]. The putative vaccine design was modeled using Robetta before protein-protein docking and the TLR-2 structure was acquired from the ALPHAFOLD-2 using ID: AF-O60603-F1 [47]. The structure was prepared before the docking by removing water molecules, heteroatom, and other atoms.
2.8. Immune simulation
To create an immunogenic profile of the designed vaccine, we performed the immune simulation by using the online server called C-ImmSim (http://150.146.2.1/C-IMMSIM/index.php) [48]. C-ImmSim is a dynamic agent-based simulator for assessing the immunological responses of the body to antigens. C-ImmSim uses an agent-based approach and the specific scoring matrix PSSM to predict immune interactions and epitopes. Following the submission of the designed vaccine to the online server with the default simulation settings, the production of antibodies, interferon, and cytokines were measured; however, the same server was also used to check the reactions of both Th1 and Th2 [49].
2.9. In-silico cloning and codon optimization
We used the Java codon adaptation tool (JCat tool) for codon optimization and reversed translation to produce the multi-epitope subunit vaccine in an appropriate vector [50]. The Jcat program is also used to guarantee that the vaccination sequence is expressed at a high level in an appropriate vector. Three choices, such as bacterial ribosome binding sites, Rho-independent transcription termination, and restriction enzyme cleavage sites, were selected in this tool. Jcat determines the GC content and CAI score of the constructed vaccine to optimized the reversely transcribed vaccine construct to a bacterial expression system [51]. The XhoI and EcoRI, restriction enzymes were manually inserted into the putative vaccine sequence, and the sequence was subsequently cloned onto the pET-28a (+) plasmid using Snapgene software [26,52].
3. Results and discussion
3.1. Proteome subtraction
Annotation of pathogen proteomes substantially assist mining for therapeutic targets and the development of therapeutic strategies. Identification and characterization of target proteins are important to aid vaccine target prioritization and for designing strategies for the treatment of different diseases. As such, proteome- and genome-wide, and functional genomics-based identification and validation of therapeutic biomarkers are widely used for different diseases [38,53]. In this study, we used a subtractive proteomics pipeline to identify the putative vaccine targets for the multiple drug-resistant fungi, Candida auris, which has been reported in SARS-CoV-2 hospitalized patients in India, USA, and other parts of the world. For this purpose, the whole proteome, containing 5409 proteins, of Candida auris species was screened through CELLO (version 2.5), a cellular localization prediction tool, to screen out extracellular proteins. As previously reported, extracellular proteins are prioritized as targets for vaccine design as they perform a key role in viral pathogenesis [26]. A total of 135 proteins were found to be extracellular. Furthermore, identifying non-homologous sequences and removing homologous proteins using BLASTp determined that 38 proteins were homologous to human proteins while 97 proteins were identified as non-homologous. The non-homologous sequences were further screened for paralogs using CD-HIT and a single sequence (OA2HIA3C7) was removed as it shared 91.6% identity with its paralog (A0A2H0ZN80). The remaining 96 proteins were subjected to functional characterization using the DEG database to find genes essential to the pathogenesis and survival of C. auris. This determined that only 13 of these proteins are essential and have a critical role in pathogenesis. Next, we evaluated the antigenic and allergenic properties of these shortlisted targets, which showed that only 7 proteins were determined as highly antigenic and non-allergenic, shown in Table 1 . These target proteins include a mitochondrial import receptor subunit, two putative beta-glucanase/Beta-glucan synthetases, two 1,3-beta-glucanosyltransferases, a uricase, and a putative SUN family protein.
Table 1.
Putative vaccine target proteins shortlisted through subtractive proteomics pipeline.
| UniProt ID | Protein Name | Activity/Function |
|---|---|---|
| A0A2H0ZE14 | Putative mitochondrial import receptor subunit | Ion transport, Protein transport |
| A0A2H0ZXY4 | Putative beta-glucanase/Beta-glucan synthetase | carbohydrate metabolic process |
| A0A2H1A5Q4 | 1,3-beta-glucanosyltransferase | transferase activity |
| A0A2H1A279 | 1,3-beta-glucanosyltransferase | transferase activity |
| A0A2H1A7U2 | Putative beta-glucanase/Beta-glucan synthetase | cell wall biogenesis |
| A0A2H0ZDW4 | Uricase | urate oxidase activity |
| A0A2H1A5R4 | Putative SUN family protein | mitochondrial and cell wall functions |
These identified proteins are known to be critical for C. auris pathogenesis. The putative mitochondrial import receptor subunit protein, with an antigenicity score 0.665 and classified as non-allergenic, is a key mediator for regulating ion and protein transport. Similarly, the two beta-glucanase/Beta-glucan synthetase proteins, with antigenicity scores of 0.4504 and 0.9645 and also classified as non-allergenic, are involved in carbohydrate metabolism and cell wall synthesis. The two 1,3-beta-glucanosyltransferase proteins, with high antigenicity scores of 0.7396 and 0.6666, play a significant role in transferase activity and the uricase protein, with an antigenicity score of 0.5435, is important in the regulation of urate oxidase activity. Finally, the SUN family protein, with an antigenicity score of 0.7738, is also essential for regulating mitochondrial and cell wall functions. All seven target proteins sequences retrieved from Uniprot [54] were then subjected to epitope screening to find CTL, HTL, and B Cell epitopes.
3.2. Immune-based epitopes screening
T-lymphocytes and B-cells play an important role in the recognition of the antigenic components of pathogens. T-cells are equipped with cell surface T-cell receptors (TCRs), which help to recognise antigens. The T-cell epitope presentation is performed by MHC molecules and is vital to their recognition of antigens. Contrastingly, B-cell differentiation results in the soluble secretion of immunoglobulins which work as antibodies to mediate adaptive immunity. These B-cell-produced antibodies perform several functions when triggered by antigen-specific binding. Other vital functions, including neutralization and targeted destruction of pathogens, are also performed by B-Cells. T- and B-cell epitope prediction aims for the specific inclusion of highly antigenic protein components for enhanced antibody production and vaccine design. Identification of antigenic immune epitopes is a prerequisite to designing peptide-based ensemble vaccines.
Recent advancements in the field of computational modeling, and the evolution of structural vaccinology-specific algorithms for epitope predictions, are a great alternative to rational vaccine design [37,38]. These methods are cost-effective and less time-consuming as compared to experimental methods. Specifically, CTLs and B-cells are key to the regulation of cell-mediated and humoral immunity. Similarly, HTL epitopes are essential for inducing and maintaining effective antibody response. Here, we screened antigenicity of the seven shortlisted proteins in C. auris for potential CTL, B-Cell, and HTL epitopes. Among the screen of CTL epitopes, a total of 6, 32, 29, 17, 9, 10, and 9 MHC binders were identified for each protein, respectively. Similarly, the number of B-Cell epitopes identified for each protein were 36, 65, 59, 46, 29, 33, and 30, and there were 2583, 4634, 3752, 3094,1806, 2002, and 1883 HTL epitopes. These epitopes were further analyzed by subjecting them to antigenicity and allergenicity checks. The epitopes with high antigenic potential and non-allergenic status then proceeded to the next step of vaccine design. During the CTL epitope prediction for protein A0A2H0ZE1, epitopes “LSVSGKINY” and “SAISYFARY”, with high antigenicity scores (0.85, 0.84), and a combined score >1 (showing higher binding affinity), were selected. For protein A0A2H0ZXY4, two epitopes, “ETDTNTMKL and SSSIRTSLV” at corresponding positions of 468–476 and 263–271 were selected. Similarly, the remaining five proteins, with accession IDs A0A2H1A5Q4, A0A2H1A279, A0A2H1A7U2, A0A2H0ZDW4 and A0A2H1A5R4, having corresponding positions of 422–430, 142–150, 187–195, 377–385, 224–232, 243–251, 281–289, 102–110, 241–249, 197–205, had high combined scores ranging from 0.9 to 3.3. High antigenicity scores above 2.9 were shortlisted. All T-Cell epitopes selected for each protein were also screened for allergenicity status before inclusion in the candidate vaccine designs, as shown in Table 2 .
Table 2.
Predicted CTL (Cytotoxic T Lymphocytes) epitopes, their position, combined scores, antigenicity, and allergenicity profiling.
| UniProt ID/Putative Vaccine name | CTL Epitopes | Residues Start | Residues End | Combined score (Binding Affinity) | MHC binder | Antigenicity Score | Antigenicity Status | Allergenicity |
|---|---|---|---|---|---|---|---|---|
| A0A2H0ZE14/MEVC-1 | LSVSGKINY SAISYFARY |
137 229 |
145 237 |
1.1 1 |
YES YES |
0.85 0.84 |
Antigen Antigen |
Non Allergens |
| A0A2H0ZXY4/MEVC-2 | ETDTNTMKL SSSIRTSLV |
468 263 |
476 271 |
1.7 1.2 |
YES YES |
2.8 1 |
Antigen Antigen |
Non Allergens |
| A0A2H1A5Q4/MEVC-3 | CSNKEKLSY SVVDMFANY |
422 142 |
430 150 |
2.6 0.9 |
YES YES |
1.5 0.9 |
Antigen Antigen |
Non Allergens |
| A0A2H1A279/MEVC-4 | YSAADDLDY GVDVKKGKY |
187 377 |
195 385 |
3.3 1.9 |
YES YES |
0.7 2 |
Antigen Antigen |
Non Allergens |
| A0A2H1A7U2/MEVC-5 | STITPGWDY VAGYPTEFY |
224 243 |
232 251 |
2.6 1 |
YES YES |
2.1 2.9 |
Antigen Antigen |
Non Allergens |
| A0A2H0ZDW4/MEVC-6 | SSDPNGLIK IVQDVWTKY |
281 102 |
289 110 |
1.7 1 |
YES YES |
1.2 2.9 |
Antigen Antigen |
Non Allergens |
| A0A2H1A5R4/MEVC-7 | EIDPSKDGY NTDSDGRTF |
241 197 |
249 205 |
1.4 1.3 |
YES YES |
1.4 2.3 |
Antigen Antigen |
Non Allergens |
For protein A0A2H0ZE14, specific B cell epitopes were also predicted and two of these epitopes, “CQWEQDYNANDFSIN” and “AFSIGGDLPPYAFSAL”, were selected based on high binding scores (0.9, 0.8) and antigenicity scores (1.9, 0.8). Similarly, for the protein A0A2H0ZXY4, two B cell epitopes, “NGHLTWYVGSDPTLTV” and “IEAETDTNTMKLGLAS”, having start positions (167, 106) and high antigenicity scores (1.8, 2), were selected. Furthermore, for the remaining five proteins, A0A2H1A5Q4, A0A2H1A279, A0A2H1A7U2, A0A2H0ZDW4, and A0A2H1A5R4), high binding scores in the range 0.5–1.4 and high antigenicity scores up to 1.5 were selected. Further analysis was performed for all the studied proteins and corresponding B-cell epitopes were examined for allergenicity status as shown in Table 3 .
Table 3.
Predicted B cell epitopes, their position, scores, antigenicity, and allergenicity profiling.
| UniProt ID/Putative Vaccine name | B-Cell Epitopes | Start position | Binding Score | Antigenicity score | Antigenicity Status | Allergenicity |
|---|---|---|---|---|---|---|
| A0A2H0ZE14/MEVC-1 | CQWEQDYNANDFSINV AFSIGGDLPPYAFSAL |
167 106 |
0.9 0.88 |
1.9 0.8 |
Antigen Antigen |
Non Allergens |
| A0A2H0ZXY4/MEVC-2 | NGHLTWYVGSDPTLTV IEAETDTNTMKLGLAS |
554 465 |
0.92 0.92 |
1.8 2 |
Antigen Antigen |
Non Allergens |
| A0A2H1A5Q4/MEVC-3 | IGDYFACGDDDAKADF ACKRDVKYLADAKTNV |
209 73 |
0.91 0.88 |
0.8 1.5 |
Antigen Antigen |
Non Allergens |
| A0A2H1A279/MEVC-4 | YIKGVDYQPGGSSVTS LVSDYADYTRPVFLSE |
31 238 |
0.86 0.86 |
0.8 1.3 |
Antigen Antigen |
Non Allergens |
| A0A2H1A7U2/MEVC-5 | TITPGWDYTVVSKVNS GHSIYYTNRFKLTDMK |
225 119 |
0.95 0.92 |
0.8 0.5 |
Antigen Antigen |
Non Allergens |
| A0A2H0ZDW4/MEVC-6 | KFLKVKRDRHNPKVQE RRTFLTYNKPSNKVTI |
14 129 |
0.88 0.86 |
0.8 1.3 |
Antigen Antigen |
Non Allergens |
| A0A2H1A5R4/MEVC-7 | DKSVGNWAPYVAGANT DDGELSKPFSDKDYCV |
183 91 |
0.93 0.87 |
0.8 1 |
Antigen Antigen |
Non Allergens |
For potential HTL epitopes selection, SVM-based analysis was performed for the protein A0A2H0ZE1 and two Interferon γ (IFN) inducing HTL epitopes, “AHGQPTMCQWEQDYN” and “KQSSKVGVGLQVETA”, were selected. Similarly, the protein A0A2H0ZXY4 returned two HTL epitopes, “CYVGYLQPRTFLL” and “YYVSYLQPRTFLL”, with high antigenicity scores (0.8–0.5) and potential IFN induction. The analysis for the remaining five studied proteins (A0A2H1A5Q4, A0A2H1A279, A0A2H1A7U2, A0A2H0ZDW4, A0A2H1A5R4) possessing high antigenic potential (ranging 0.5–3.1) helped in the identification of putative IFN inducing HTL epitopes and were classified against each protein as shown in Table 4 . The final shortlisted epitopes (CTL, B cell, and HTL) from Table 2, Table 3, Table 4 were included in further MEVC design against each target protein.
Table 4.
Predicted HTL (Helper T Lymphocytes) epitopes, their position, scores, IFN status, antigenicity and allergenicity profiling.
| UniProt ID/Putative Vaccine name | HTL-Epitopes | Method | IFN | Antigenicity score | Antigenicity Status | Allergenicity |
|---|---|---|---|---|---|---|
| A0A2H0ZE14/MEVC-1 | AHGQPTMCQWEQDYN KQSSKVGVGLQVETA |
SVM SVM |
Positive Positive |
0.81 2.6 |
Antigen Antigen |
Non Allergens |
| A0A2H0ZXY4/MEVC-2 | SRNSNSSFTSSDQGE ENKDGEKWKLVFSDE |
SVM SVM |
Positive Positive |
0.8 0.5 |
Antigen Antigen |
Non Allergens |
| A0A2H1A5Q4/MEVC-3 | GSGSGSSGSGSSGSS PTTNTVTGSPITVGT |
SVM SVM |
Positive Positive |
3.1 2 |
Antigen Antigen |
Non Allergens |
| A0A2H1A279/MEVC-4 | TSQNKDPLSDPDICA DPLSDPDICARDIAL |
SVM SVM |
Positive Positive |
0.9 1.1 |
Antigen Antigen |
Non Allergens |
| A0A2H1A7U2/MEVC-5 | KPATGESFSASGGSA SRVRQATLSAQKRRR |
SVM SVM |
Positive Positive |
1.8 0.5 |
Antigen Antigen |
Non Allergens |
| A0A2H0ZDW4/MEVC-6 | LRGDFDVSYTEADNS PSSDPNGLIKSTVGR |
SVM SVM |
Positive Positive |
0.5 0.9 |
Antigen Antigen |
Non Allergens |
| A0A2H1A5R4/MEVC-7 | NDDGELSKPFSDKDY SGGSTGKSLGASYCV |
SVM SVM |
Positive Positive |
0.8 3.8 |
Antigen Antigen |
Non Allergens |
3.3. Putative vaccine designing
Identification of antigenic epitopes through proteome-wide screening is vital in designing multi-epitope-based vaccines during immunoinformatic approaches. The mapped CTL, B-cell, and HTL epitopes for the selected seven essential proteins of C. auris were further subjected to in silico vaccine design. The shortlisted epitopes (Table 2, Table 3, Table 4) were used in the full-length MEVC designs and joined together using different linkers (i.e., EAAK, AAY, GPGPG, and KK) against each target protein as shown in Table 5 . This also included MEVC design based on the whole proteome, including epitopes from all the seven target proteins.
Table 5.
Showing the target protein-specific and proteome-wide designed MEVCs, their length, antigenicity and allergenicity profiling.
| UniProt IDs/Vaccine Name | Protein Specific and Proteome-wide MEVC Constructs | Number of Amino Acids | Antigenicity score | Antigenicity Status | Allergenicity |
|---|---|---|---|---|---|
| A0A2H0ZE14/MEVC-1 | MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKPEAAKLSVSGKINYAAYSAISYFARYGPGPGAHGQPTMCQWEQDYNGPGPGKQSSKVGVGLQVETAKKCQWEQDYNANDFSINVKKAFSIGGDLPPYAFSAL | 165 | 1 | Antigen | Non-Allergen |
| A0A2H0ZXY4/MEVC-2 | MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKPEAAKETDTNTMKLAAYSSSIRTSLVGPGPGSRNSNSSFTSSDQGEGPGPGENKDGEKWKLVFSDEKKNGHLTWYVGSDPTLTVKKIEAETDTNTMKLGLAS | 165 | 0.96 | Antigen | Non-Allergen |
| A0A2H1A5Q4/MEVC-3 | MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKPEAAKCSNKEKLSYAAYSVVDMFANYGPGPGGSGSGSSGSGSSGSSGPGPGPTTNTVTGSPITVGTKKIGDYFACGDDDAKADFKKACKRDVKYLADAKTNV | 165 | 0.98 | Antigen | Non-Allergen |
| A0A2H1A279/MEVC-4 | MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKPEAAKYSAADDLDYAAYGVDVKKGKYGPGPGTSQNKDPLSDPDICAGPGPGDPLSDPDICARDIALKKYIKGVDYQPGGSSVTSKKLVSDYADYTRPVFLSE | 165 | 1 | Antigen | Non-Allergen |
| A0A2H1A7U2/MEVC-5 | MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKPEAAKSTITPGWDYAAYVAGYPTEFYGPGPGKPATGESFSASGGSAGPGPGSRVRQATLSAQKRRRKKTITPGWDYTVVSKVNSKKGHSIYYTNRFKLTDMK | 165 | 1 | Antigen | Non-Allergen |
| A0A2H0ZDW4/MEVC-6 | MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKPEAAKSSDPNGLIKAAYIVQDVWTKYGPGPGLRGDFDVSYTEADNSGPGPGPSSDPNGLIKSTVGRKKKFLKVKRDRHNPKVQEKKRRTFLTYNKPSNKVTI | 165 | 0.72 | Antigen | Non-Allergen |
| A0A2H1A5R4/MEVC-7 | MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKPEAAKEIDPSKDGYAAYNTDSDGRTFGPGPGNDDGELSKPFSDKDYGPGPGSGGSTGKSLGASYCVKKDKSVGNWAPYVAGANTKKDDGELSKPFSDKDYCV | 165 | 1.1 | Antigen | Non-Allergen |
| MEVC-WP | MRVLYLLFSFLFIFLMPLPGVFGGIGDPVTCLKSGAICHPVFCPRRYKQIGTCGLPGTKCCKKPEAAKLSVSGKINYAAYSAISYFARYAAYETDTNTMKLAAYSSSIRTSLVAAYCSNKEKLSYAAYSVVDMFANYAAYYSAADDLDYAAYVAGYPTEFYAAYSSDPNGLIKAAYIVQDVWTKYAAYEIDPSKDGYAAYNTDSDGRTFGPGPGAHGQPTMCQWEQDYNGPGPGSRNSNSSFTSSDQGEGPGPGGSGSGSSGSGSSGSSGPGPGTSQNKDPLSDPDICAGPGPGKPATGESFSASGGSAGPGPGLRGDFDVSYTEADNSGPGPGNDDGELSKPFSDKDYKKCQWEQDYNANDFSINVKKNGHLTWYVGSDPTLTVKKIGDYFACGDDDAKADFKKYIKGVDYQPGGSSVTSKKTITPGWDYTVVSKVNSKKKFLKVKRDRHNPKVQEKKDKSVGNWAPYVAGANT | 475 | 1.09 | Antigen | Non-Allergen |
These linkers are vital to enabling epitope display, maintaining epitope separation, and preventing folding, to ensure an effective immune response. Moreover, the addition of an adjuvant enhances the immunogenic potential of MEVC peptide vaccine designs [39]. All the designed MEVC constructs were therefore subjected to the addition of a non-toxic adjuvant called human beta defensin-2 (hBD-2) following an EAAAK linker. The hBD-2 possesses the property of self-production with expression levels that enable it to induce a robust immune response against the attached antigen [55,56].
3.4. Protein specific vaccine designs
For each of the seven proteins, MEVCs 165 amino acids in length were designed and processed for calculation of antigenic and allergenic potential to confirm its experimental feasibility. Of the final putative vaccine constructs (MEVC-1, MEVC-2, MEVC-3, MEVC-4, MEVC-5, MEVC-6, MEVC-7) for the individual proteins A0A2H0ZE1, A0A2H0ZXY4, A0A2H1A5Q4, A0A2H1A279, A0A2H1A7U2, A0A2H0ZDW4, A0A2H1A5R4, the lowest antigenicity score was 0.72 and the highest 1.1.
3.5. Whole proteome (WP) based vaccine design
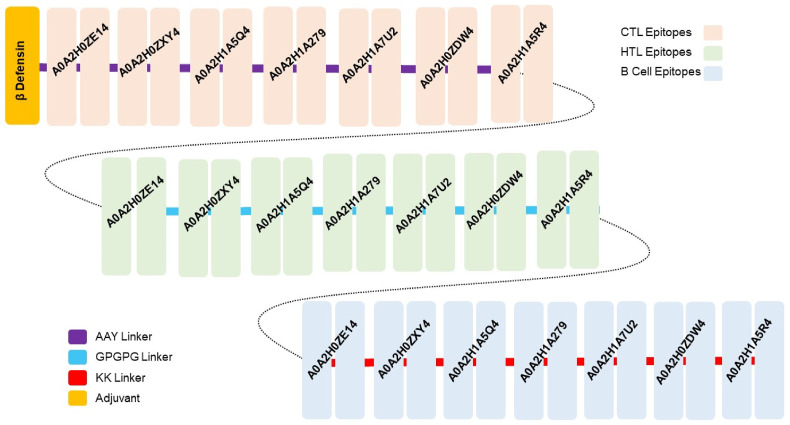
The whole proteome-based final MEVC (MEVC-WP) was constructed with 14 CTL, 14 HTL, and 14 B-Cell epitopes linked together. The length of the final construct was 475 amino acids with a high antigenicity score of >1, demonstrating high experimental feasibility. The general schematic diagram of the putative vaccine construct with included epitopes from different proteins and suitable added linkers is presented in Fig. 2 , where each mapped vaccine and its components are presented in different colors.
Fig. 2.
The topographical organization of the MEVC-WP, including the adjuvant position, CTL HTL, B cell epitopes, and their respective linkers shown in different colors. The Figure also shows each component epitope utilized in protein-specific candidate vaccine designs (MEVCs,1–7).
3.6. Physiochemical properties
The putative vaccine constructs were then analyzed through the ProtParam server to verify several physiochemical parameters vital to confirming the stability of the vaccine designs. These parameters included calculation of theoretical pI (7.4–9.9), molecular weight (17–50 kd), and other parameters determining the feasibility and structural stability of the vaccine for further experimental designs. Moreover, the demonstrated half-life, aliphatic index (thermos-stability), and GRAVY (Grand average of hydropathicity) were also explored. These results reflected the hydrophilic and stable nature of the designed MEVCs. The different physiochemical properties for protein-specific and proteome-wide putative vaccines are given in Table 6 .
Table 6.
Predicted physiochemical properties of the designed MEVCs.
| Uniprot IDs/Vaccine name | GC-Content | CAI-Value | Molecular weight | Theoretical pI | Neg charged residues (Asp + Glu): | Positively charged residues (Arg + Lys) | Half-life | Total number of atoms | Aliphatic index | Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|---|---|---|---|
| A0A2H0ZE1/MEVC-1 | 52.32 | 1 | 17845.71 | 9.11 | 9 | 17 | Mammals >30 h Yeast >20 h E.coli >10 h |
2494 | 70.97 | −0.072 |
| A0A2H0ZXY4/MEVC-2 | 51.11 | 1 | 17759.41 | 8.89 | 15 | 20 | Mammals >30 h Yeast >20 h E.coli >10 h |
2489 | 67.94 | −0.293 |
| A0A2H1A5Q4/MEVC-3 | 51.51 | 1 | 17101.72 | 9.14 | 11 | 20 | Mammals >30 h Yeast >20 h E.coli >10 h |
2386 | 62.67 | −0.098 |
| A0A2H1A279/MEVC-4 | 53.73 | 1 | 17653.41 | 8.36 | 17 | 20 | Mammals >30 h Yeast >20 h E.coli >10 h |
2478 | 76.24 | −0.146 |
| A0A2H1A7U2/MEVC-5 | 53.53 | 0.98 | 17870.82 | 9.9 | 7 | 24 | Mammals >30 h Yeast >20 h E.coli >10 h |
2520 | 62.67 | −0.236 |
| A0A2H0ZDW4/MEVC-6 | 51.51 | 0.97 | 18136.25 | 9.87 | 11 | 28 | Mammals >30 h Yeast >20 h E.coli >10 h |
4193 | 72.61 | −0.366 |
| A0A2H1A5R4/MEVC-7 | 52.12 | 1 | 17483.89 | 8.36 | 18 | 21 | Mammals >30 h Yeast >20 h E.coli >10 h |
4193 | 57.33 | −0.395 |
| MEVC-WP | 51.43 | 0.98 | 50504.93 | 7.4 | 48 | 49 | Mammals >30 h Yeast >20 h E.coli >10 h |
6932 | 52.65 | −0.547 |
3.7. Structural modelling of the designed MEVC
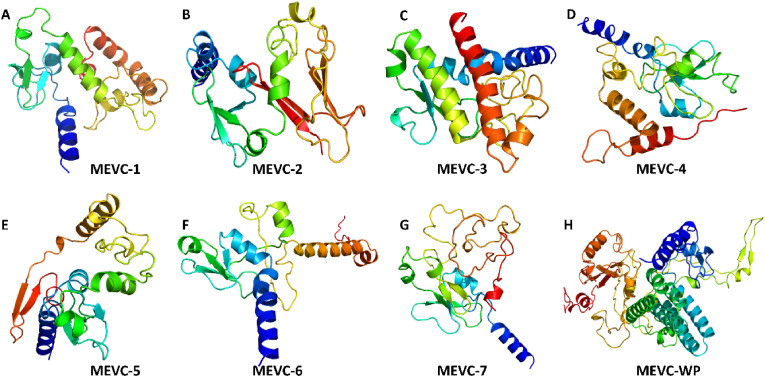
The widely used Robetta server generates highly efficacious structural models through parsing of potent domains and by utilizing different strategies including comparative modeling and de novo sequence-based structural prediction. Based on the confidence levels of available matching protein structures, templates are used to produce a comparative model. In contrast, the de novo Robetta fragment insertion method is used for proteins with no available matching structures. 3D structural modeling for each of the constructed MEVCs was performed with the utilities of the Robetta server. All generated models were visually evaluated using PyMOL software and the refined best models, as shown in Fig. 3 , were included for further analysis.
Fig. 3.
Structures of the final multi-epitopes subunit vaccine (MEVC) models. (A–H) represents the final multi-epitopes subunit vaccine structures designed for each target.
3.8. Interaction of MEVCs with the immune receptor (TLR-2)
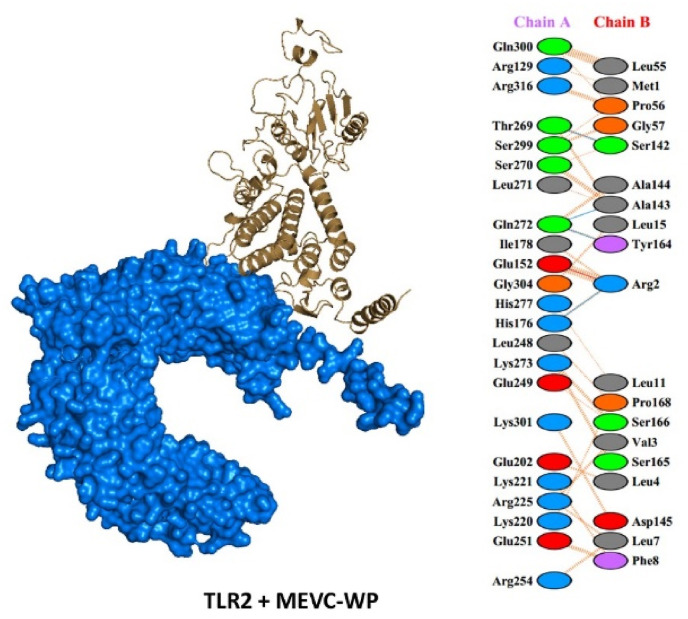
The HAWKDOCK server was employed to evaluate vaccine-TLR-2 interaction. The top ten docking models of vaccine-TLR-2 were generated by the server, ranked based on docking complex score. The docking scores acquired for each of the docking complexes are given in Table 7 . For MEVC1 the docking score was −5832.94 kcal/mol, with three hydrogen bonds and 1 salt bridge reported. Similarly, the MEVC4 also formed three hydrogen bonds and 1 salt bridge with a docking score −4261.51 kcal/mol. Unlike MEVC1 and MEVC4 the total number of hydrogen bonds were higher in MEVC2, with six hydrogen bonds and 1 salt bridge, and a docking score of −5260.80 kcal/mol. MEVC5 and MEVC7 formed 5 hydrogen bonds, with MEVC5 also having 2 salt bridges. The docking score for MEVC5 and MEVC7 were reported to be −5263.43 and −4600.89 kcal/mol. On the other hand, MEVC6 formed a single hydrogen bond and two salt bridges with a docking score of −3449.97 kcal/mol, however, the non-bonded contacts were higher in this complex. For MEVCPW the docking score was −4935.33 kcal/mol with four hydrogen bonds and 1 salt bridge reported. The results decisively suggest that the designed vaccine candidates robustly interact with the immune receptor. The best complex (lowest energy score) for each of the designed vaccine candidates were subjected to interaction analysis by utilizing PDBsum, a web tool for the graphical representation of macromolecular complexes. The docking complexes, hydrogen bonds, salt bridges, and non-bonding contacts between putative vaccines and TLR-2 are shown in Fig. 4 and Fig. S1 (A-G).
Table 7.
Docking Scores of MEVCs with TLR-2. The table also tabulate the total number of salt-bridges, hydrogen bonds and non-bonded contacts in each putative vaccine-TLR2 complex.
| Complex Name | Salt bridges | Hydrogen Bonds | Non-bonded contact | Docking Score |
|---|---|---|---|---|
| TLR2-MEVC1 | 1 | 3 | 162 | −5832.94 |
| TLR2-MEVC2 | 1 | 6 | 169 | −5260.80 |
| TLR2-MEVC3 | 0 | 2 | 103 | −4597.81 |
| TLR2-MEVC4 | 1 | 3 | 113 | −4261.51 |
| TLR2-MEVC5 | 2 | 5 | 161 | −5263.43 |
| TLR2-MEVC6 | 2 | 1 | 195 | −3449.97 |
| TLR2-MEVC7 | 0 | 5 | 119 | −4600.89 |
| TLR2-MEVCPW | 1 | 4 | 125 | −4935.33 |
Fig. 4.
A representative image of TLR-2 (Alpha Fold-O60603-F1) and vaccine (MEVC-WP) docked complex. It demonstrates the surface view of TLR-2 (receptor) shown in Marine blue, while the multi-epitope subunit vaccine (MEVC-WP) is represented in the cartoon view by brown color.
3.9. Codon optimization and in-silico cloning
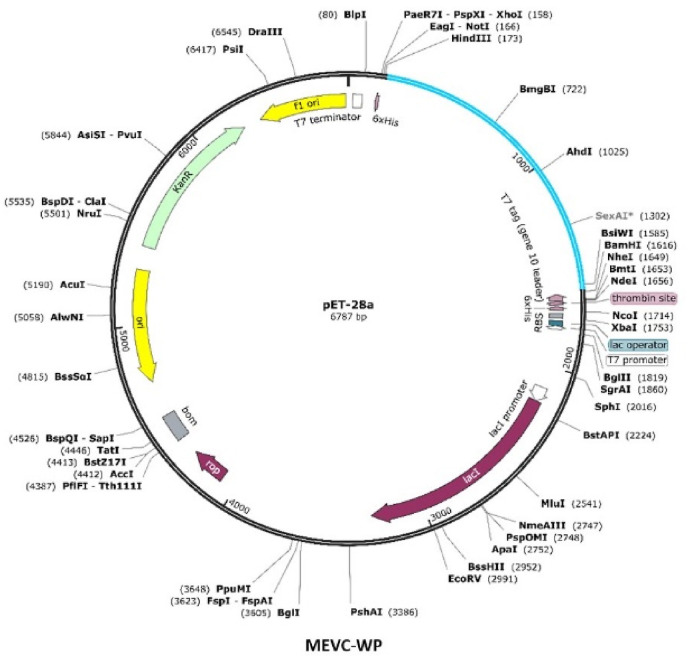
All constructed vaccine sequences were improved by utilizing the Jcat server for optimal expression in E. coli strain K-12. For each of the vaccine constructs, the codons were optimized, and reverse translated to obtain the corresponding nucleotides. The codon adaptation index (CAI) values were also calculated through the Jcat server for each of the vaccine constructs. The higher calculated CAI scores, in the range 0.98–1, and optimal percentage of GC contents (51–53%) ( Table 6 ) indicated that the vaccine protein is expressed at a high level in E. coli. The 5′ and 3’ ends restriction enzymes selected for cloning of the gene sequence of each vaccine into pET28a (+) were XhoI and EcoRI. The software package Snapgene was utilized for the insertion of putative vaccine sequences into the pET28a (+) plasmids to obtain the cloning designs (Fig. 5 ).
Fig. 5.
The in-silico plasmid maps showing pet28avector with inserted vaccine (gene) sequences with XhoI and ECOR1 as enzyme restriction sites at the N and C terminal respectively. It represents the cloned sequence of a vaccine designed against the whole proteome named as MEVC-WP (Sky Blue Color).
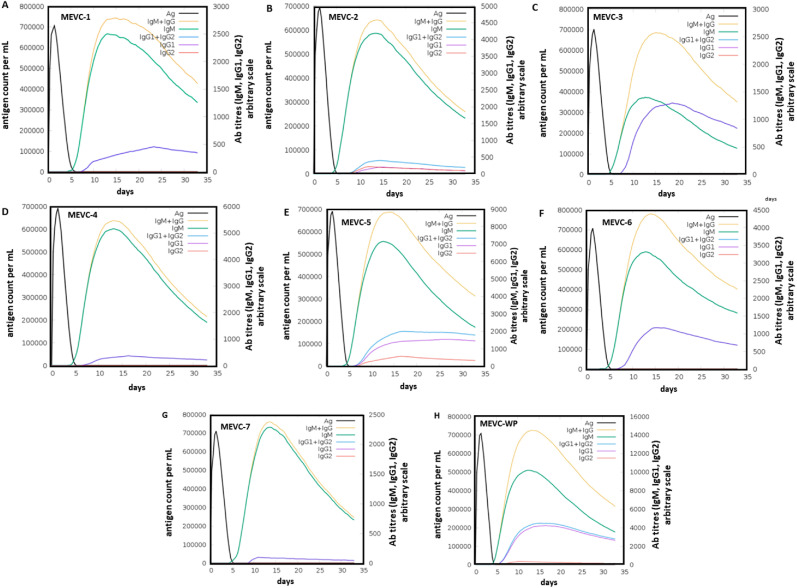
3.10. Immune simulation of the proposed vaccine and immune response
To evaluate the potential antigen-based induction of the human immune response, each of the designed vaccines was examined as an injected antigen with in silico immune simulation. As shown in (Fig. 6 A) the injected antigen MEVC1 (A0A2H0ZE14), after achieving the highest antigen counts at day 2, was slowly neutralized until day 5. Afterward, a strong antibody (IgM + IgG) response was observed with the highest achieved titers of >700,000 au/ml between days 10–15. Similar high antigen counts (day 2) were found for the designed vaccines MEVC2, MEVC3, MEVC4, MEVC5, MEVC6, MEVC7, and MEVCWP, following the same pattern of neutralization (day 5) for all the designed vaccines (Fig. 6). However, variation in the antibody titers was significant between 10 and 15 days post-injection for each of the designed MEVCs. The highest titers of antibodies (IgM + IgG) for MEVC2 were observed at about 600,000 au/ml (Fig. 6B). For MEVC3, a similar higher ratio 700,000 au/ml of antibody (IgM + IgG) titers was observed (Fig. 6C). For MEVC4, the antibody titers were about 600,000 au/ml (Fig. 6D). Similarly, for MEVC5, MEVC6 and MEVC7, the observed antibody titers were ≥700,000 au/ml (Fig. 6E-G). Additionally, the MEVC designed against the whole proteome of C. auris, MEVCWP, also showed about 700,000 au/ml for combined (IgM + UgG) antibody titers (Fig. 6H). This was followed by IgM-specific antibody titers of about 500,000 au/ml. These results showed great immunogenic potential for each designed vaccine, all of which triggered a robust immune response. Consequently, the designed vaccine candidates may trigger the production of protective immunity against the human pathogen C. auris in further in vivo and in vitro models.
Fig. 6.
The immune simulation graphs showing plotted antigen count/ml/day against Ab titers for each of the antigenic vaccine (MEVC) designs. (A–G) Represents the immune simulation graph for vaccines designed against the seven individual MEVC (MEVC-1, MEVC-2, MEVC-3, MEVC-4, MEVC-5, MEVC-6 and MEVC-7), (H) Represents the immune simulation graph for the vaccine designed against the whole proteome named as MEVC-WP.
3.11. Limitations of the study
This study was based on in silico analysis to shortlist putative epitopes which need validation in experimental vaccine designs. Similarly, the physiochemical properties of each vaccine candidate were explored but also need further verification in the wet lab. The important step involving the expression and purification of the protein needs advanced testing using recombinant DNA technologies. Further evaluations are needed to justify the clinical manifestations associated with the vaccine designs against Candida auris.
4. Conclusion
In conclusion, a subtractive proteomics approach has been utilized to shortlist seven protein targets for putative vaccine design against Candida auris. Different epitopes for CTL, B-cell, and HTL have been explored for each of the shortlisted target proteins. Additionally, a whole proteome-based vaccine has been designed which includes each target protein-specific vaccine design. The constructed MEVCs have been modeled and evaluated for potential immunization against C. auris. The present study provides new and valuable epitope candidates and prompts future vaccine development against this pathogen. The suggested highly antigenic MEVCs in this study have paved a way for experimental processing of vaccines against Candida auris, however, despite showing immune reinforcement potential, the candidate vaccines need further validation through experimental processing to confirm their efficacy and safety.
Declaration of competing interest
Authors declare there is no declaration of interest.
Acknowledgements
Dong-Qing Wei is supported by grants from the Key Research Area Grant 2016YFA0501703 of the Ministry of Science and Technology of China, the National Science Foundation of China (Grant No. 32070662, 61832019, 32030063), the Science and Technology Commission of Shanghai Municipality (Grant No.: 19430750600), as well as SJTU JiRLMDS Joint Research Fund and Joint Research Funds for Medical and Engineering and Scientific Research at Shanghai Jiao Tong University (YG2021ZD02). The computations were partially performed at the PengCheng Lab. and the Center for High-Performance Computing, Shanghai Jiao Tong University.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105462.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Brown G., Denning D., Gow N., Levitz S., Netea M., White T. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Du H., Bing J., Hu T., Ennis C.L., Nobile C.J., Huang G. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schelenz S., Hagen F., Rhodes J.L., Abdolrasouli A., Chowdhary A., Hall A., Ryan L., Shackleton J., Trimlett R., Meis J.F. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control. 2016;5:1–7. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow N.A., Gade L., Tsay S.V., Forsberg K., Greenko J.A., Southwick K.L., Barrett P.M., Kerins J.L., Lockhart S.R., Chiller T.M. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect. Dis. 2018;18:1377–1384. doi: 10.1016/S1473-3099(18)30597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borman A.M., Szekely A., Johnson E.M. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med. Mycol. 2017;55:563–567. doi: 10.1093/mmy/myw147. [DOI] [PubMed] [Google Scholar]
- 6.Rhodes J., Fisher M.C. Global epidemiology of emerging Candida auris. Curr. Opin. Microbiol. 2019;52:84–89. doi: 10.1016/j.mib.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Abdolrasouli A., Armstrong‐James D., Ryan L., Schelenz S. In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris. Mycoses. 2017;60:758–763. doi: 10.1111/myc.12699. [DOI] [PubMed] [Google Scholar]
- 8.Welsh R.M., Bentz M.L., Shams A., Houston H., Lyons A., Rose L.J., Litvintseva A.P. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J. Clin. Microbiol. 2017;55:2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plachouras D., Lötsch F., Kohlenberg A., Monnet D.L. Candida auris: epidemiological situation, laboratory capacity and preparedness in the European Union and European Economic Area. Euro Surveill. 2020;25:2000240. doi: 10.2807/1560-7917.ES.2020.25.12.2000240. January 2018 to May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee W.G., Shin J.H., Uh Y., Kang M.G., Kim S.H., Park K.H., Jang H.-C. First three reported cases of nosocomial fungemia caused by Candida auris. J. Clin. Microbiol. 2011;49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhary A., Voss A., Meis J. Multidrug-resistant Candida auris:‘new kid on the block’in hospital-associated infections? J. Hosp. Infect. 2016;94:209–212. doi: 10.1016/j.jhin.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhary A., Sharma C., Duggal S., Agarwal K., Prakash A., Singh P.K., Jain S., Kathuria S., Randhawa H.S., Hagen F. New clonal strain of Candida auris, Delhi, India: new clonal strain of Candida auris, Delhi, India. Emerg. Infect. Dis. 2013;19:1670. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magobo R.E., Corcoran C., Seetharam S., Govender N.P. Candida auris–associated candidemia, South Africa. Emerg. Infect. Dis. 2014;20:1250. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emara M., Ahmad S., Khan Z., Joseph L., Al-Obaid I.m., Purohit P., Bafna R. 2015. Candida Auris Candidemia in Kuwait; p. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvo B., Melo A.S., Perozo-Mena A., Hernandez M., Francisco E.C., Hagen F., Meis J.F., Colombo A.L. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J. Infect. 2016;73:369–374. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart S.R., Etienne K.A., Vallabhaneni S., Farooqi J., Chowdhary A., Govender N.P., Colombo A.L., Calvo B., Cuomo C.A., Desjardins C.A. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017;64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudramurthy S.M., Chakrabarti A., Paul R.A., Sood P., Kaur H., Capoor M.R., Kindo A.J., Marak R.S., Arora A., Sardana R. Candida auris candidaemia in Indian ICUs: analysis of risk factors. J. Antimicrob. Chemother. 2017;72:1794–1801. doi: 10.1093/jac/dkx034. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhary A., Sharma A. The lurking scourge of multidrug resistant Candida auris in times of COVID-19 pandemic. Journal of Global Antimicrobial Resistance. 2020;22:175. doi: 10.1016/j.jgar.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang S., Tanji H., Yin K., Zhang S., Sakaniwa K., Huang J., Yang Y., Li J., Ohto U., Shimizu T. Rationally designed small-molecule inhibitors targeting an unconventional pocket on the TLR8 protein–protein interface. J. Med. Chem. 2020;63:4117–4132. doi: 10.1021/acs.jmedchem.9b02128. [DOI] [PubMed] [Google Scholar]
- 20.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 21.Manohar P., Loh B., Nachimuthu R., Hua X., Welburn S.C., Leptihn S. Secondary bacterial infections in patients with viral pneumonia. Front. Med. 2020;7:420. doi: 10.3389/fmed.2020.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossato L., Negrão F.J., Simionatto S. Could the COVID-19 pandemic aggravate antimicrobial resistance? Am. J. Infect. Control. 2020;48:1129–1130. doi: 10.1016/j.ajic.2020.06.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muñoz J.F., Gade L., Chow N.A., Loparev V.N., Juieng P., Berkow E.L., Farrer R.A., Litvintseva A.P., Cuomo C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chatterjee S., Alampalli S.V., Nageshan R.K., Chettiar S.T., Joshi S., Tatu U.S. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genom. 2015;16:1–16. doi: 10.1186/s12864-015-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.T. Khan, M. Abdullah, T.F. Toor, F.N. Almajhdi, M. Suleman, A. Iqbal, L. Ali, A. Khan, Y. Waheed, D.-Q. Wei, Evaluation of the whole proteome of achromobacter xylosoxidans to identify vaccine targets for mRNA and peptides-based vaccine designing against the emerging, leukemia (AML), 8 9. [DOI] [PMC free article] [PubMed]
- 26.Zeb A., Ali S.S., Azad A.K., Safdar M., Anwar Z., Suleman M., Nizam-Uddin N., Khan A., Wei D.-Q. Genome-wide screening of vaccine targets prioritization and reverse vaccinology aided design of peptides vaccine to enforce humoral immune response against Campylobacter jejuni. Comput. Biol. Med. 2021;133:104412. doi: 10.1016/j.compbiomed.2021.104412. [DOI] [PubMed] [Google Scholar]
- 27.UniProt The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gul H., Ali S.S., Saleem S., Khan S., Khan J., Wadood A., Rehman A.U., Ullah Z., Ali S., Khan H. Subtractive proteomics and immunoinformatics approaches to explore Bartonella bacilliformis proteome (virulence factors) to design B and T cell multi-epitope subunit vaccine. Infect. Genet. Evol. 2020;85:104551. doi: 10.1016/j.meegid.2020.104551. [DOI] [PubMed] [Google Scholar]
- 29.Khan S., Ali S.S., Zaheer I., Saleem S., Ziaullah, Zaman N., Iqbal A., Suleman M., Wadood A., Rehman A.U. Proteome-wide mapping and reverse vaccinology-based B and T cell multi-epitope subunit vaccine designing for immune response reinforcement against Porphyromonas gingivalis. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1819423. [DOI] [PubMed] [Google Scholar]
- 30.Solanki V., Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 2018;8:1–19. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R., Ou H.Y., Zhang C.T. DEG: a database of essential genes. Nucleic Acids Res. 2004;32:D271–D272. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu C.S., Chen Y.C., Lu C.H., Hwang J.K. Prediction of protein subcellular localization. Proteins: Structure, Function, and Bioinformatics. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 33.Nagpal G., Usmani S.S., Raghava G.P. A web resource for designing subunit vaccine against major pathogenic species of bacteria. Front. Immunol. 2018;9:2280. doi: 10.3389/fimmu.2018.02280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:1–7. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. 2005. Protein Identification and Analysis Tools on the ExPASy Server, the Proteomics Protocols Handbook; pp. 571–607. [Google Scholar]
- 36.Stranzl T., Larsen M.V., Lundegaard C., Nielsen M. NetCTLpan: pan-specific MHC class I pathway epitope predictions. Immunogenetics. 2010;62:357–368. doi: 10.1007/s00251-010-0441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan T., Khan A., Nasir S.N., Ahmad S., Ali S.S., Wei D.-Q. CytomegaloVirusDb: multi-Omics knowledge database for Cytomegaloviruses. Comput. Biol. Med. 2021:104563. doi: 10.1016/j.compbiomed.2021.104563. [DOI] [PubMed] [Google Scholar]
- 38.Khan T., Khan A., Wei D.-Q. MMV-db: vaccinomics and RNA-based therapeutics database for infectious hemorrhagic fever-causing mammarenaviruses. Database. 2021:2021. doi: 10.1093/database/baab063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan A., Khan S., Saleem S., Nizam-Uddin N., Mohammad A., Khan T., Ahmad S., Arshad M., Ali S.S., Suleman M. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput. Biol. Med. 2021;133:104420. doi: 10.1016/j.compbiomed.2021.104420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma N., Patiyal S., Dhall A., Pande A., Arora C., Raghava G.P. AlgPred 2.0: an improved method for predicting allergenic proteins and mapping of IgE epitopes. Briefings Bioinf. 2021;22:bbaa294. doi: 10.1093/bib/bbaa294. [DOI] [PubMed] [Google Scholar]
- 41.Saha S., Raghava G.P.S. Prediction of continuous B‐cell epitopes in an antigen using recurrent neural network. Proteins: Structure, Function, and Bioinformatics. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 42.Ponomarenko J., Bui H.-H., Li W., Fusseder N., Bourne P.E., Sette A., Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dhanda S.K., Vir P., Raghava G.P. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct. 2013;8:1–15. doi: 10.1186/1745-6150-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arai R., Ueda H., Kitayama A., Kamiya N., Nagamune T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 2001;14:529–532. doi: 10.1093/protein/14.8.529. [DOI] [PubMed] [Google Scholar]
- 45.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weng G., Wang E., Wang Z., Liu H., Zhu F., Li D., Hou T. HawkDock: a web server to predict and analyze the protein–protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. 2019;47:W322–W330. doi: 10.1093/nar/gkz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A. Highly accurate protein structure prediction with AlphaFold. Nature. 2021:1. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castiglione F., Bernaschi M., C-immsim . Proceedings of the Sixteenth International Symposium on Mathematical Theory of Networks and Systems (MTNS2004) 2004. Playing with the immune response. [Google Scholar]
- 49.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33:W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khatoon N., Pandey R.K., Prajapati V.K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-08842-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng B., Suleman M., Zafar Z., Ali S.S., Nasir S.N., Hussain Z., Waseem M., Khan A., Hassan F., Wang Y. Towards an ensemble vaccine against the pegivirus using computational modelling approaches and its validation through in silico cloning and immune simulation. Vaccines. 2021;9:818. doi: 10.3390/vaccines9080818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khan A., Khan T., Nasir S.N., Ali S.S., Suleman M., Rizwan M., Waseem M., Ali S., Zhao X., Wei D.-Q. BC-TFdb: a database of transcription factor drivers in breast cancer. Database. 2021:2021. doi: 10.1093/database/baab018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium U. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tani K., Murphy W.J., Chertov O., Salcedo R., Koh C.Y., Utsunomiya I., Funakoshi S., Asai O., Herrmann S.H., Wang J.M. Defensins act as potent adjuvants that promote cellular and humoral immune responses in mice to a lymphoma idiotype and carrier antigens. International immunology. 2000;12:691–700. doi: 10.1093/intimm/12.5.691. [DOI] [PubMed] [Google Scholar]
- 56.Ferris L.K., Mburu Y.K., Mathers A.R., Fluharty E.R., Larregina A.T., Ferris R.L., Falo L.D., Jr. Human beta-defensin 3 induces maturation of human Langerhans cell–like dendritic cells: an antimicrobial peptide that functions as an endogenous adjuvant. J. Invest. Dermatol. 2013;133:460–468. doi: 10.1038/jid.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.