Abstract
With an exponential increase in extracellular vesicle (EV) studies in the past decade, focus has been placed on standardization of experimental design to ensure inter‐study comparisons and validity of conclusions. In the case of in vitro assays, the composition of cell culture media is important to consider for EV studies. In particular, levels of lipoproteins, which are critical components of the interstitial fluid, should be taken into consideration. Results from this study reveal that lipoprotein levels in cell culture medium impact the effects that EVs have on recipient cells. Additionally, evidence of EV binding and fusion to lipoprotein‐like structures in plasma is provided. However, it is unclear whether the impact of lipoproteins in cell culture is due to direct interactions with EVs, indirect effects, or a combination of both mechanisms. Taken together, cell culture studies performed in the absence of physiological levels of lipoproteins are unlikely to reflect interactions that occur between EVs and recipient cells in an in vivo environment.
1. INTRODUCTION
The promising role of extracellular vesicles (EVs) as therapeutic agents, diagnostic tools, and mediators of (patho)physiological processes has led to an exponential increase in EV studies over the past decade. The rapid growth of the field has highlighted the importance of developing guidelines to improve inter‐study reproducibility and validity of conclusions (Théry et al., 2018). This commentary emphasizes the impact of cell culture medium composition, primarily the presence of lipoproteins, on EV studies performed in vitro.
The assessment of EV effects on cultured cells has similar limitations as other in vitro studies. For most cell culture studies, attempts are made to recapitulate key features of an in vivo environment, including temperature, pH, humidity, and the presence of serum components. Serum provides critical biomolecules for cell function and resembles the content of the extracellular fluid in vivo. However, interplay between EVs and recipient cells is often studied in isolation without considering the impact of EV interactions with extracellular components, such as those present in serum. In fact, numerous studies assess cellular uptake and functional effects of EVs in serum‐free media (Delenclos et al., 2017; Jurgielewicz et al., 2020; Schneider et al., 2017) or in media with EV‐depleted serum (Bhat et al., 2018; Boysen et al., 2020; Busatto et al., 2021; Carobolante et al., 2020; Casella et al., 2018; Emam et al., 2018; Gonda et al., 2018; Joshi et al., 2020; Kaur et al., 2014; Komuro et al., 2021; Ross et al., 2021; Tang et al., 2018; Toribio et al., 2019; Wu et al., 2021; Xu et al., 2019; Yue et al., 2017; Zhao et al., 2019; Zhou et al., 2017). In serum‐free media, EVs are devoid of the biomolecular corona that forms in vivo upon exposure to proteins and other components in the blood (Tóth et al., 2021). The biomolecular corona is known to substantially impact the function and uptake of synthetic lipid vesicles (Yang et al., 2021), and may have similar effects on EVs, making it challenging to predict physiological function in serum‐free conditions. In the case of EV‐depleted serum, some non‐EV components may also be reduced in the depletion process. EV‐depleted foetal bovine serum (FBS) is usually obtained by ultracentrifugation or polymer‐based precipitation (Lehrich et al., 2018), which results in the co‐isolation of lipoproteins that overlap in size and density (Busatto et al., 2020; Simonsen, 2017; Sódar et al., 2016; Tian et al., 2020). For example, EV depletion through polymer‐based precipitation (Exo‐FBS, System Biosciences) (Lehrich et al., 2018) results in a 93% reduction in low‐density lipoprotein (LDL)/very‐low‐density lipoprotein (VLDL) cholesterol and a 19% reduction in high‐density lipoprotein (HDL) cholesterol (Figure 1a). Notably, the extent of lipoprotein removal is likely to be highly dependent on the specific protocol used for EV depletion. Circulating lipoproteins are six orders of magnitude more abundant than circulating EVs (Simonsen, 2017), and are also critical components of the interstitial fluid (Sloop et al., 1987). Therefore, assessing the impact of EVs on recipient cells in the absence of physiological levels of lipoproteins is unlikely to be indicative of in vivo effects.
FIGURE 1.
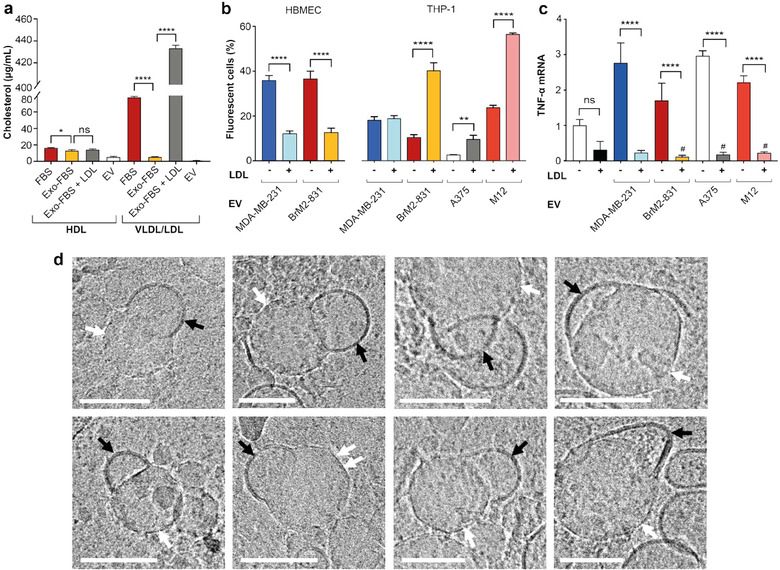
Levels of lipoproteins in foetal bovine serum (FBS) and impact of low‐density lipoprotein on extracellular vesicle (EV) effects. (a) Very‐low‐density lipoprotein (VLDL)/LDL cholesterol and high‐density lipoprotein (HDL) cholesterol in regular FBS, EV‐depleted FBS (Exo‐FBS), Exo‐FBS supplemented with LDL (500‐μg/mL apoB‐100), and MDA‐MB‐231‐derived EVs (1010/mL). b) Association of fluorescently labelled MDA‐MB‐231, BrM2‐831, A375, and M12‐derived EVs (109/mL) with human brain microvascular endothelial cells (HBMECs) and THP‐1 human monocytes. Adapted with permission from Busatto et al. (2020). (c) mRNA levels of tumour necrosis factor α (TNF‐α; normalized to β‐actin) in THP‐1 monocytes exposed to MDA‐MB‐231, BrM2‐831, A375, and M12‐derived EVs (109/mL) and/or LDL (50‐μg/mL apolipoprotein/apoB‐100). (d) Cryogenic transmission electron microscopy images of crude human plasma. EVs (black arrows) are seen fusing/binding to lipoprotein‐like structures (white arrows) that display crystallization in polygonal‐like faceted shapes (example shown with double arrows). Scale bars, 100 nm. Cell studies were performed in Exo‐FBS. Data represent mean ± standard deviation (SD) of three replicates. Statistics by analysis of variance (ANOVA) with the Tukey's multiple comparison post hoc analysis. *p < 0.05; **p < 0.01; ****p < 0.0001. #p < 0.05 compared to untreated cells (without added EVs and LDL). ns, not significant
2. I IMPACT OF LIPOPROTEINS ON EXTRACELLULAR VESICLES
Studies have demonstrated that interactions can occur between EVs and lipoproteins (Busatto et al., 2020; Sódar et al., 2016), and LDL can cause the association between cells and fluorescently labelled EVs to increase, decrease or remain unchanged depending on the source and recipient cell line (Figure 1b). It is important to note that the transfer of lipophilic probes from the EV membrane to LDL cannot be excluded, and may occur to some extent (Simonsen, 2019). Among the (patho)physiological roles of EVs, intercellular communication between cancer cells and immune cells is an important and widely studied area of research (Hou & Chen, 2021). Here, the impact of lipoprotein levels on interactions between cancer cell‐derived EVs and monocytes was assessed. Notably, the presence of physiological levels of LDL dramatically changed cancer cell‐derived EV‐induced effects on recipient monocytes, as illustrated by tumour necrosis factor‐α (TNF‐α) expression (Figure 1c). Specifically, in the absence of physiological levels of LDL, EV exposure led to a substantial increase in TNF‐α, while the addition of both LDL and EVs caused a decrease in the expression of this cytokine compared to untreated cells. Previous studies demonstrated that oxidized LDL can suppress TNF‐α in stimulated macrophages (Hamilton et al., 1990). In this study, the addition of LDL alone caused a decrease in TNF‐α compared to untreated monocytes; however, this change was not statistically significant (Figure 1c). It is likely that the presence of physiological levels of LDL overrides EV‐mediated stimulation of monocytes (TNF‐α increase), which may occur independently of any interactions between these two types of biogenic nanoparticles. Regardless of whether lipoproteins and EVs directly interact, it is evident that conflicting results in terms of monocyte stimulation are obtained with or without LDL supplementation in an EV‐depleted medium.
Proteomic profiling of the EV biomolecular corona in blood plasma has demonstrated the presence of apolipoproteins (apo) (Tóth et al., 2021). However, it is unclear whether the EV biomolecular corona encompasses intact lipoproteins or just their protein components. In this study, EVs isolated by tangential flow filtration displayed low levels of HDL cholesterol (6 μg/mL; Figure 1a), which may include cholesterol present in the EV lipid bilayer. Alternatively, EVs may be tightly bound to HDL contaminants. The presence of free HDL in EV samples is unlikely, as tangential flow filtration effectively separates EVs from 14‐nm components (Busatto et al., 2018), which are larger than HDL (5–10 nm) (Busatto et al., 2020). The EVs displayed even lower levels of VLDL/LDL cholesterol (1 μg/mL; Figure 1a). Low levels of VLDL/LDL in Exo‐FBS (Figure 1a) may limit potential associations with EVs, as interactions are likely to be highly concentration‐dependent. It is unknown whether binding and/or fusion occurs between EVs and lipoproteins in an in vivo environment. In this study, cryogenic transmission electron microscopy (cryo‐TEM) images of crude human plasma were obtained, revealing that EVs frequently bind and fuse with lipoprotein‐like structures (at least one occurrence observed in 52% of 1 μm2 image frames) (Figure 1d). The observed lipoprotein‐like structures are within the size range of VLDL and chylomicrons (Busatto et al., 2020), and some display crystallization in polygonal faceted shapes, which are distinct from the smooth curvature of the EV lipid bilayer (Figure 1d). Previous studies have demonstrated that VLDL has a polyhedral shape (Yu et al., 2016), similar to the lipoprotein‐like structures observed in this study (Figure 1d). A crystalline phase with straight facets is a feature that has been observed in particles with lipid hydrocarbon chains, a result of hydrocarbon‐chain freezing (Jacoby et al., 2015), supporting the notion that at least some of the observed structures are likely to be lipoproteins as opposed to protein aggregates.
Given that there are numerous published studies on cell uptake and functional effects of EVs in the absence of physiological levels of lipoproteins, the purpose of these results is to urge the EV community to consider the role of serum components in EV studies. Additionally, the source of serum may impact EV results, as previously demonstrated for synthetic nanoparticles that displayed different interactions with serum components and recipient cells depending on whether human, mouse, rabbit, sheep or bovine serum was added to the cell culture medium (Lee et al., 2020; Müller et al., 2018; Schöttler et al., 2016; Solorio‐Rodríguez et al., 2017; Yang et al., 2021). It is worth noting that bovine and human lipoprotein profiles differ (Kaabia et al., 2018), which should also be considered when using media with FBS to assess EV effects in human cells. In particular, LDL cholesterol is much lower in FBS than in human plasma (upper limit in healthy individuals: 1000‐μg/mL apoB‐100) (Cao et al., 2018) (Figure 1a).
In addition to the impact of EV interactions with lipoproteins, the use of serum‐free or EV/lipoprotein‐reduced media is likely to affect recipient cells. Several studies have shown the EV‐depleted FBS results in reduced cell growth (Angelini et al., 2016; Aswad et al., 2016; Eitan et al., 2015; Lehrich et al., 2018; Liao et al., 2017), similarly to serum starvation, indicating that factors that promote cell proliferation are removed in the isolation process. It has been suggested that the dependence of cells on whole serum could be due to co‐isolated non‐EV components (Liao et al., 2017). Despite the reduction in cell growth, the use of EV/lipoprotein reduced‐serum is strongly recommended when obtaining EVs from cell culture sources to minimize contamination from nanoparticles in serum (Shelke et al., 2014). However, this does not justify the use of serum‐free or EV‐depleted serum for EV uptake and functional studies, unless a specific rationale is provided for removing serum components that are abundant in an in vivo environment.
This commentary aims to shed light on overlooked consequences of the interplay between EVs and other nanosized components in serum. If such interactions are not taken into considerations, results may be less reflective of an in vivo setting. We urge the EV community to consider the effects of lipoproteins on EV interactions with recipient cells in culture.
3. METHODS
Cell lines were obtained and cultured, as previously described (Busatto et al., 2020). For EV isolation, cells were seeded in media with 10% Exo‐FBS (System Biosciences), and conditioned medium was collected after 48 h (90% confluency and over 95% viability). EVs were isolated, purified and concentrated by tangential flow filtration, while nanoparticle tracking analysis, cryo‐TEM, atomic force microscopy and Western blotting were used for EV authentication. Experimental details of EV isolation and characterization have previously been reported (Busatto et al., 2020). A cholesterol assay kit (ab65390; Abcam) was used to measure the levels of LDL/VLDL and HDL in FBS (Gibco, Thermo Fisher Scientific), Exo‐FBS (System Biosciences), Exo‐FBS supplemented with commercial LDL (500‐μg/mL apoB‐100) and MDA‐MB‐231 EVs (1010/mL) according to the manufacturer's instructions. For cell culture studies, EVs (1010/mL) were incubated with commercial LDL (500‐μg/mL apoB‐100) at 37°C for 2 h. Notably, the upper limit of apoB‐100 in plasma of healthy individuals (1000 μg/mL) is twofold higher than the concentration used for EV incubation (Cao et al., 2018). Human brain microvascular endothelial cells (HBMECs) and human monocytes (THP‐1) were then exposed to LDL‐incubated EVs (109/mL) for 3 h (cell association assay) or 24 h (TNF‐α). For flow cytometry experiments, EVs were fluorescently labelled with DiI and excess fluorescent dye was removed by size‐exclusion chromatography (experimental details are provided in Busatto et al. (2020)). For quantitative real‐time polymerase chain reaction (qRT‐PCR), total RNA was extracted with a RNeasy Mini kit (Qiagen) and quantified using NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). RNA was reverse‐transcribed into first‐strand cDNA using the iScript cDNA Synthesis Kit (Bio‐Rad). qRT‐PCR was performed with a 7500 Real‐Time PCR System (Applied Biosystems) using QuantiTect SYBR Green PCR Kit (Qiagen) according to the manufacturer's instructions with primers for TNF‐α (forward: 5′‐CCCCAGGGACCTCTCTCTAA‐3′; reverse: 5′‐TGAGGTACAGGCCCTCTGAT‐3′) and β‐actin (forward: 5′‐CCAACCGCGAGAAGATGA‐3′; reverse: 5′‐ CCAGAGGCGTACAGGGATAG‐3′). Data were analysed using the Bio‐Rad MyiQ Software system and were expressed as relative gene expression using the 2‐ΔΔCt method. The Mayo Clinic Biospecimens Review Group (ID: 17‐010290) provided approval for preclinical research use of human de‐identified, frozen (−20°C), residual plasma (anticoagulant: citrate phosphate dextrose‐adenine‐1). Cryo‐TEM imagining of plasma samples was performed with a FEI (now Thermo Fisher Scientific, Waltham, MA, USA) Talos 200C high‐resolution TEM at −180°C as previously described (Busatto et al., 2020). Briefly, a crude plasma drop (∼3 μL) was placed on a perforated carbon film on a 200 mesh TEM grid in a controlled environment vitrification system. The TEM grid was then plunged into freezing ethane (−183°C). Images were acquired at an acceleration voltage of 200 kV, a Volta phase‐plate was used to enhance image contrast, and recording was performed at low electron exposure (FEI Falcon III direct‐imaging camera).
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
This work is partially supported by the Mayo Clinic Center for Regenerative Medicine in Florida (JW), the Mayo Clinic Center for Biomedical Discovery (JW), the National Institute of Allergy and Infectious Diseases, National Institutes of Health under award numbers R21AI152318 (JW), the American Heart Association under award number 20TPA35490415 (JW), Programma Operativo Nazionale Ricerca e Innovazione, Italian Ministry of Education, University and Research, Italy, under award number DOT13A8025 (DI, JW) and the China Scholarship Council (YY). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Sara Busatto, Yubo Yang and Dalila Iannotta contributed equally to this study.
Contributor Information
Sara Busatto, Email: sara.busatto@childrens.harvard.edu.
Joy Wolfram, Email: j.wolfram@uq.edu.au.
REFERENCES
- Angelini, F. , Ionta, V. , Rossi, F. , Miraldi, F. , Messina, E. , & Giacomello, A. (2016). Foetal bovine serum‐derived exosomes affect yield and phenotype of human cardiac progenitor cell culture. Bioimpacts, 6, 15–24. 10.15171/bi.2016.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad, H. , Jalabert, A. , & Rome, S. (2016). Depleting extracellular vesicles from fetal bovine serum alters proliferation and differentiation of skeletal muscle cells in vitro. BMC Biotechnology [Electronic Resource], 16, 32. 10.1186/s12896-016-0262-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, A. , Sharma, A. , & Bharti, A. C. (2018). Upstream Hedgehog signaling components are exported in exosomes of cervical cancer cell lines. Nanomedicine (Lond), 13, 2127–2138. 10.2217/nnm-2018-0143 [DOI] [PubMed] [Google Scholar]
- Boysen, A. T. , Whitehead, B. , Stensballe, A. , Carnerup, A. , Nylander, T. , & Nejsum, P. (2020). Fluorescent labeling of helminth extracellular vesicles using an in vivo whole organism approach. Biomedicines, 8, 213. 10.3390/biomedicines8070213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto, S. , Iannotta, D. , Walker, S. A. , Di Marzio, L. , & Wolfram, J. (2021). A simple and quick method for loading proteins in extracellular vesicles. Pharmaceuticals (Basel), 14, 356. 10.3390/ph14040356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto, S. , Vilanilam, G. , Ticer, T. , Lin, W.‐L. , Dickson, D. , Shapiro, S. , Bergese, P. , & Wolfram, J. (2018). Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cells, 7, 273. 10.3390/cells7120273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto, S. , Walker, S. A. , Grayson, W. , Pham, A. , Tian, M. , Nesto, N. , Barklund, J. , & Wolfram, J. (2020). Lipoprotein‐based drug delivery. Advanced Drug Delivery Reviews, 159, 377–390. 10.1016/j.addr.2020.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto, S. , Yang, Y. , Walker, S. A. , Davidovich, I. , Lin, W.‐H. , Lewis‐Tuffin, L. , Anastasiadis, P. Z. , Sarkaria, J. , Talmon, Y. , Wurtz, G. , & Wolfram, J. (2020). Brain metastases‐derived extracellular vesicles induce binding and aggregation of low‐density lipoprotein. Journal of Nanobiotechnology, 18, 162. 10.1186/s12951-020-00722-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. , Steffen, B. T. , Guan, W. , Remaley, A. T. , Mcconnell, J. P. , Palamalai, V. , & Tsai, M. Y. (2018). A comparison of three apolipoprotein B methods and their associations with incident coronary heart disease risk over a 12‐year follow‐up period: The Multi‐Ethnic Study of Atherosclerosis. Journal of Clinical Lipidology, 12, 300–304. 10.1016/j.jacl.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carobolante, G. , Mantaj, J. , Ferrari, E. , & Vllasaliu, D. (2020). Cow milk and intestinal epithelial cell‐derived extracellular vesicles as systems for enhancing oral drug delivery. Pharmaceutics, 12, 226. 10.3390/pharmaceutics12030226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella, G. , Colombo, F. , Finardi, A. , Descamps, H. , Ill‐Raga, G. , Spinelli, A. , Podini, P. , Bastoni, M. , Martino, G. , Muzio, L. , & Furlan, R. (2018). Extracellular vesicles containing IL‐4 modulate neuroinflammation in a mouse model of multiple sclerosis. Molecular Therapy, 26, 2107–2118. 10.1016/j.ymthe.2018.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delenclos, M. , Trendafilova, T. , Mahesh, D. , Baine, A. M. , Moussaud, S. , Yan, I. K. , Patel, T. , & Mclean, P. J. (2017). Investigation of endocytic pathways for the internalization of exosome‐associated oligomeric alpha‐synuclein. Frontiers in Neuroscience, 11, 172. 10.3389/fnins.2017.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitan, E. , Zhang, S. , Witwer, K. W. , & Mattson, M. P. (2015). Extracellular vesicle‐depleted fetal bovine and human sera have reduced capacity to support cell growth. Journal of Extracellular Vesicles, 4, 26373. 10.3402/jev.v4.26373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emam, S. E. , Ando, H. , Abu Lila, A. S. , Shimizu, T. , Ukawa, M. , Okuhira, K. , Ishima, Y. , Mahdy, M. A. , Ghazy, F.‐.E. S. , & Ishida, T. (2018). A novel strategy to increase the yield of exosomes (extracellular vesicles) for an expansion of basic research. Biological & Pharmaceutical Bulletin, 41, 733–742. 10.1248/bpb.b17-00919 [DOI] [PubMed] [Google Scholar]
- Gonda, A. , Kabagwira, J. , Senthil, G. N. , Ferguson Bennit, H. R. , Neidigh, J. W. , Khan, S. , & Wall, N. R. (2018). Exosomal survivin facilitates vesicle internalization. Oncotarget, 9, 34919–34934. 10.18632/oncotarget.26182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, T. A. , Ma, G. P. , & Chisolm, G. M. (1990). Oxidized low density lipoprotein suppresses the expression of tumor necrosis factor‐alpha mRNA in stimulated murine peritoneal macrophages. Journal of Immunology, 144, 2343–2350 [PubMed] [Google Scholar]
- Hou, P.‐P. , & Chen, H.‐Z. (2021). Extracellular vesicles in the tumor immune microenvironment. Cancer Letters, 516, 48–56. 10.1016/j.canlet.2021.05.032 [DOI] [PubMed] [Google Scholar]
- Jacoby, G. , Cohen, K. , Barkan, K. , Talmon, Y. , Peer, D. , & Beck, R. (2015). Metastability in lipid based particles exhibits temporally deterministic and controllable behavior. Science Reports, 5, 9481. 10.1038/srep09481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, B. S. , De Beer, M. A. , Giepmans, B. N. G. , & Zuhorn, I. S. (2020). Endocytosis of extracellular vesicles and release of their cargo from endosomes. ACS Nano, 14, 4444–4455. 10.1021/acsnano.9b10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgielewicz, B. J. , Yao, Y. , & Stice, S. L. (2020). Kinetics and specificity of HEK293T extracellular vesicle uptake using imaging flow cytometry. Nanoscale Research Letters, 15, 170. 10.1186/s11671-020-03399-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaabia, Z. , Poirier, J. , Moughaizel, M. , Aguesse, A. , Billon‐Crossouard, S. , Fall, F. , Durand, M. , Dagher, E. , Krempf, M. , & Croyal, M. (2018). Plasma lipidomic analysis reveals strong similarities between lipid fingerprints in human, hamster and mouse compared to other animal species. Science Reports, 8, 15893. 10.1038/s41598-018-34329-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Singh, S. P. , Elkahloun, A. G. , Wu, W. , Abu‐Asab, M. S. , & Roberts, D. D. (2014). CD47‐dependent immunomodulatory and angiogenic activities of extracellular vesicles produced by T cells. Matrix Biology, 37, 49–59. 10.1016/j.matbio.2014.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro, H. , Kawai‐Harada, Y. , Aminova, S. , Pascual, N. , Malik, A. , Contag, C. H. , & Harada, M. (2021). Engineering extracellular vesicles to target pancreatic tissue in vivo. Nanotheranostics, 5, 378–390. 10.7150/ntno.54879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. Y. , Son, J. G. , Moon, J. H. , Joh, S. , & Lee, T. G. (2020). Comparative study on formation of protein coronas under three different serum origins. Biointerphases, 15, 061002. 10.1116/6.0000396 [DOI] [PubMed] [Google Scholar]
- Lehrich, B. , Liang, Y. , Khosravi, P. , Federoff, H. , & Fiandaca, M. (2018). Fetal bovine serum‐derived extracellular vesicles persist within vesicle‐depleted culture media. International Journal of Molecular Sciences, 19, 3538. 10.3390/ijms19113538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Z. , Muth, D. C. , Eitan, E. , Travers, M. , Learman, L. N. , Lehrmann, E. , & Witwer, K. W. (2017). Serum extracellular vesicle depletion processes affect release and infectivity of HIV‐1 in culture. Science Reports, 7, 2558. 10.1038/s41598-017-02908-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, L. K. , Simon, J. , Rosenauer, C. , Mailänder, V. , Morsbach, S. , & Landfester, K. (2018). The transferability from animal models to humans: Challenges regarding aggregation and protein corona formation of nanoparticles. Biomacromolecules, 19, 374–385. 10.1021/acs.biomac.7b01472 [DOI] [PubMed] [Google Scholar]
- Ross, M. , Atalla, H. , Karrow, N. , & Mallard, B. A. (2021). The bioactivity of colostrum and milk exosomes of high, average, and low immune responder cows on human intestinal epithelial cells. Journal of Dairy Science, 104, 2499–2510. 10.3168/jds.2020-18405 [DOI] [PubMed] [Google Scholar]
- Schneider, D. J. , Speth, J. M. , Penke, L. R. , Wettlaufer, S. H. , Swanson, J. A. , & Peters‐Golden, M. (2017). Mechanisms and modulation of microvesicle uptake in a model of alveolar cell communication. Journal of Biological Chemistry, 292, 20897–20910. 10.1074/jbc.M117.792416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler, S. , Klein, K. , Landfester, K. , & Mailänder, V. (2016). Protein source and choice of anticoagulant decisively affect nanoparticle protein corona and cellular uptake. Nanoscale, 8, 5526–5536. 10.1039/c5nr08196c [DOI] [PubMed] [Google Scholar]
- Shelke, G. V. , Lässer, C. , Gho, Y. S. , & Lötvall, J. (2014). Importance of exosome depletion protocols to eliminate functional and RNA‐containing extracellular vesicles from fetal bovine serum. Journal of Extracellular Vesicles, 3, 24783. 10.3402/jev.v3.24783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen, J. B. (2017). What are we looking at? Extracellular vesicles, lipoproteins, or both? Circulation Research, 121, 920–922. 10.1161/CIRCRESAHA.117.311767 [DOI] [PubMed] [Google Scholar]
- Simonsen, J. B. (2019). Pitfalls associated with lipophilic fluorophore staining of extracellular vesicles for uptake studies. Journal of Extracellular Vesicles, 8, 1582237. 10.1080/20013078.2019.1582237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloop, C. H. , Dory, L. , & Roheim, P. S. (1987). Interstitial fluid lipoproteins. Journal of Lipid Research, 28, 225–237 [PubMed] [Google Scholar]
- Sódar, B. W. , Kittel, Á. , Pálóczi, K. , Vukman, K. V. , Osteikoetxea, X. , Szabó‐Taylor, K. , Németh, A. , Sperlágh, B. , Baranyai, T. , Giricz, Z. , Wiener, Z. , Turiák, L. , Drahos, L. , Pállinger, É. , Vékey, K. , Ferdinandy, P. , Falus, A. , & Buzás, E. I. (2016). Low‐density lipoprotein mimics blood plasma‐derived exosomes and microvesicles during isolation and detection. Science Reports, 6, 24316. 10.1038/srep24316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solorio‐Rodríguez, A. , Escamilla‐Rivera, V. , Uribe‐Ramírez, M. , Chagolla, A. , Winkler, R. , García‐Cuellar, C. M. , & De Vizcaya‐Ruiz, A. (2017). A comparison of the human and mouse protein corona profiles of functionalized SiO2 nanocarriers. Nanoscale, 9, 13651–13660. 10.1039/c7nr04685e [DOI] [PubMed] [Google Scholar]
- Tang, X. , Lu, H. , Dooner, M. , Chapman, S. , Quesenberry, P. J. , & Ramratnam, B. (2018). Exosomal Tat protein activates latent HIV‐1 in primary, resting CD4+ T lymphocytes. JCI Insight, 3. 10.1172/jci.insight.95676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M. , Ticer, T. , Wang, Q. , Walker, S. , Pham, A. , Suh, A. , Busatto, S. , Davidovich, I. , Al‐Kharboosh, R. , Lewis‐Tuffin, L. , Ji, B. , Quinones‐Hinojosa, A. , Talmon, Y. , Shapiro, S. , Rückert, F. , & Wolfram, J. (2020). Adipose‐derived biogenic nanoparticles for suppression of inflammation. Small, 16, 1904064. 10.1002/smll.201904064 [DOI] [PubMed] [Google Scholar]
- Toribio, V. , Morales, S. , López‐Martín, S. , Cardeñes, B. , Cabañas, C. , & Yáñez‐Mó, M. (2019).Development of a quantitative method to measure EV uptake. Science Reports, 9, 10522. 10.1038/s41598-019-47023-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth, E. Á. , Turiák, L. , Visnovitz, T. , Cserép, C. , Mázló, A. , Sódar, B. W. , Försönits, A. I. , Petővári, G. , Sebestyén, A. , Komlósi, Z. , Drahos, L. , Kittel, Á. , Nagy, G. , Bácsi, A. , Dénes, Á. , Gho, Y. S. , Szabó‐Taylor, K. É. , & Buzás, E. I. (2021).Formation of a protein corona on the surface of extracellular vesicles in blood plasma. Journal of Extracellular Vesicles, 10, e12140. 10.1002/jev2.12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, D. , Chang, X. , Tian, J. , Kang, L. , Wu, Y. , Liu, J. , Wu, X. , Huang, Y. , Gao, B. , Wang, H. , Qiu, G. , & Wu, Z. (2021). Bone mesenchymal stem cells stimulation by magnetic nanoparticles and a static magnetic field: Release of exosomal miR‐1260a improves osteogenesis and angiogenesis. Journal of Nanobiotechnology, 19, 209. 10.1186/s12951-021-00958-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z.‐H. , Miao, Z.‐W. , Jiang, Q.‐Z. , Gan, D.‐X. , Wei, X.‐G. , Xue, X.‐Z. , Li, J.‐Q. , Zheng, F. , Qin, X.‐X. , Fang, W.‐G. , Chen, Y.‐H. , & Li, B. (2019). Brain microvascular endothelial cell exosome‐mediated S100A16 up‐regulation confers small‐cell lung cancer cell survival in brain. FASEB Journal, 33, 1742–1757. 10.1096/fj.201800428R [DOI] [PubMed] [Google Scholar]
- Yang, K. , Reker‐Smit, C. , Stuart, M. C. A. , & Salvati, A. (2021). Effects of protein source on liposome uptake by cells: Corona composition and impact of the excess free proteins. Advanced Healthcare Materials, 10, 2100370. 10.1002/adhm.202100370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Kuang, Y.‐L. , Lei, D. , Zhai, X. , Zhang, M. , Krauss, R. M. , & Ren, G. (2016). Polyhedral 3D structure of human plasma very low density lipoproteins by individual particle cryo‐electron tomography1. Journal of Lipid Research, 57, 1879–1888. 10.1194/jlr.M070375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Y. , Garikipati, V. N. S. , Verma, S. K. , Goukassian, D. A. , & Kishore, R. (2017). Interleukin‐10 deficiency impairs reparative properties of bone marrow‐derived endothelial progenitor cell exosomes. Tissue Engineering Part A, 23, 1241–1250. 10.1089/ten.TEA.2017.0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z. , Mcgill, J. , Gamero‐Kubota, P. , & He, M. (2019). Microfluidic on‐demand engineering of exosomes towards cancer immunotherapy. Lab on A Chip, 19, 1877–1886. 10.1039/c8lc01279b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Zhang, W. , Yao, Q. , Zhang, H. , Dong, G. , Zhang, M. , Liu, Y. , Chen, J.‐K. , & Dong, Z. (2017). Exosome production and its regulation of EGFR during wound healing in renal tubular cells. American Journal of Physiology. Renal Physiology, 312, F963–F970. 10.1152/ajprenal.00078.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]


