Abstract
Chronic kidney disease (CKD) afflicts 15% of adults in the United States, of whom 25% have a family history. Genetic testing is supportive in identifying and possibly confirming diagnoses of CKD, thereby guiding care. Advances in the clinical genetic evaluation include next-generation sequencing with targeted gene panels, whole exome sequencing, and whole genome sequencing. These platforms provide DNA sequence reads with excellent coverage throughout the genome and have identified novel genetic causes of CKD. New pathologic genetic variants identified in previously unrecognized biological pathways have elucidated disease mechanisms underlying CKD etiologies, potentially establishing prognosis and guiding treatment selection. Molecular diagnoses using genetic sequencing can detect rare, potentially treatable mutations, avoid misdiagnoses, guide selection of optimal therapy, and decrease the risk of unnecessary and potentially harmful interventions. Genetic testing has been widely adopted in pediatric nephrology; however, it is less frequently used to date in adult nephrology. Extension of clinical genetic approaches to adult patients may achieve similar benefits in diagnostic refinement and treatment selection. This review aimed to identify clinical CKD phenotypes that may benefit the most from genetic testing, outline the commonly available platforms, and provide examples of successful deployment of these approaches in CKD.
Index Words: Chronic kidney disease, clinical genetics, genetic causes, genetic disorders, genetic testing, next-generation sequencing
Introduction
Chronic kidney disease (CKD) is a major global health problem and is increasing in prevalence.1 In the United States, among the 15% of adults who have CKD, 25% may have a family history.2 By 2040, it is estimated that CKD will have become the fifth-leading cause of death in the United States.3 CKD can be identified by well-established clinical biomarkers, such as serum creatinine levels, cystatin C levels, estimated glomerular filtration rate, and either urinary albumin-creatinine ratio or urinary protein-creatinine ratio4,5 as well as other emerging biomarkers and devices.6,7 However, clinical biomarkers have limited usefulness for determining the cause of CKD or the probability of progression to kidney failure.8, 9, 10, 11
Precision medicine in CKD is aimed at providing an accurate diagnosis, establishing a prognosis, and appropriately individualizing treatment; however, these goals have only been achieved in a few instances despite the high heritability of CKD.8 Presently, approximately 625 genes have been demonstrated to contribute to the development of CKD,12 and it has been suggested that the heritability for estimated glomerular filtration rate and albuminuria may range from 36% to 75% and 16% to 49% of cases, respectively.13 Toward that end, genetic evaluation is an essential component of precision medicine and has the potential to reduce clinical uncertainty of CKD.14 The objective of this study was to review the role of clinical genetics and next-generation sequencing (NGS) in patients with CKD. It is particularly important to bridge the gap in understanding the implications of clinical genetics across the nephrology community, including pediatric and adult nephrologists, because inherited disorders account for varying cases of kidney failure across patient age groups.
Assessing Genetic Contributions to CKD
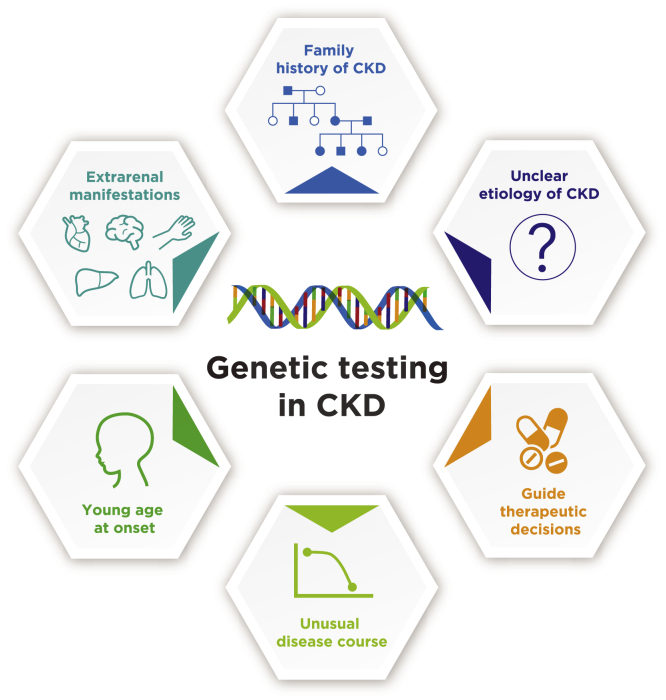
Not all patients with CKD need genetic testing. Some common indications to consider for genetic testing are illustrated in Fig 1. In many patients with CKD, the cause of disease remains unclear despite a thorough evaluation.15 In these instances, genetic testing has the potential to enable formal diagnosis and targeted intervention, as well as reduce the utilization of medications with potential significant side effects.16 Rare single nucleotide variants, small insertion/deletion variants, and copy number variants underlie many Mendelian conditions, and rare kidney diseases can be monogenic. In addition, interactions among multiple genes often determine the clinical phenotypes.17,18 Reaching a definitive diagnosis may require analyzing multiple genes—often from small specimens—inexpensively, quickly, and sensitively. One goal of genetic association studies is to delineate the genetic architecture of complex phenotypes and diseases, such as CKD, to provide new insights into the best approaches to patient management.19
Figure 1.
Indications for genetic testing in CKD. Clinical genetics should be considered in CKD if the etiology is unclear, when a genetic component is clinically suspected (positive family history, early onset, extrarenal manifestations, unusual disease course), or to guide therapy (eg, immunosuppression management, pretransplant evaluation). Abbreviation: CKD, chronic kidney disease.
In addition, genetic testing should be strongly considered when a genetic component is clinically suspected. For example, the presence of a family history of CKD of unclear etiology should prompt genetic evaluation of not only the index case but also other affected family members.10,17,20 Such information can be invaluable for detection of disease in previously undiagnosed relatives, family planning including preimplantation genetic testing, and donor eligibility to evaluate family members as potential kidney donors.21 A genetic component should also be suspected in patients with CKD at young age of onset because nearly all pediatric cases that progress to kidney replacement therapy have genetic underpinnings.22 Patients with CKD and extrarenal manifestations are also at a high risk of a genetic etiology, as exemplified by visual and hearing disturbances in Alport syndrome, retinitis pigmentosa in nephronophthisis, and maturity-onset diabetes of the young (MODY) in the kidney cysts and diabetes syndrome.17,20,23 As a corollary, identification of a genetic cause for CKD will prompt early referrals for early identification and prevention of known or potential extrarenal manifestations.20,21
Even if a clinical diagnosis is strongly suspected, genetic testing can be useful in confirming the diagnosis and reclassifying the disease and in prognostication.10,21 For example, patients with the clinical diagnosis of polycystic kidney disease (PKD) can benefit from the determination of the underlying genetic defect because those with truncating mutations in the PKD1 gene progress to kidney failure, on average, in their 50s, whereas those with nontruncating PKD1 mutations have an average age of kidney failure onset in their 60s, and those with PKD2 mutations exhibit kidney survival until 80 years of age.24 It is important to note, however, the lack of prognostic specificity for individuals with autosomal dominant polycystic kidney disease (ADPKD) despite the population means.25 Establishing a genetic diagnosis can also avoid unnecessary tests and procedures, including in patients with CKD and hematuria with a family history of hematuria. A genetic diagnosis of one of the forms of Alport syndrome may eliminate the need for invasive kidney biopsies in the index case as well as in family members.26
A genetic diagnosis can also be invaluable in personalizing therapeutic decisions. Patients with steroid-resistant nephrotic syndrome due to a genetic etiology will benefit from withholding potentially toxic immunosuppression, which is ineffective, but can be excellent candidates for kidney transplant when they progress to kidney failure because the nephrotic syndrome may not recur in the kidney transplant.20 Establishing the genetic diagnosis also allows for specific therapeutic targeting of the affected pathways, as exemplified by complement blockade in genetic forms of atypical hemolytic uremic syndrome (HUS) and blocking oxalate metabolism in the primary hyperoxalurias.20,27, 28, 29 Increasingly, the confirmation of a genetic diagnosis is becoming essential to enroll patients in pharmaceutical trials, especially in rare diseases such as primary hyperoxalurias and atypical HUS associated with expensive therapies.15
Next-Generation Sequencing (NGS) to Identify Genes in CKD: Recent Studies
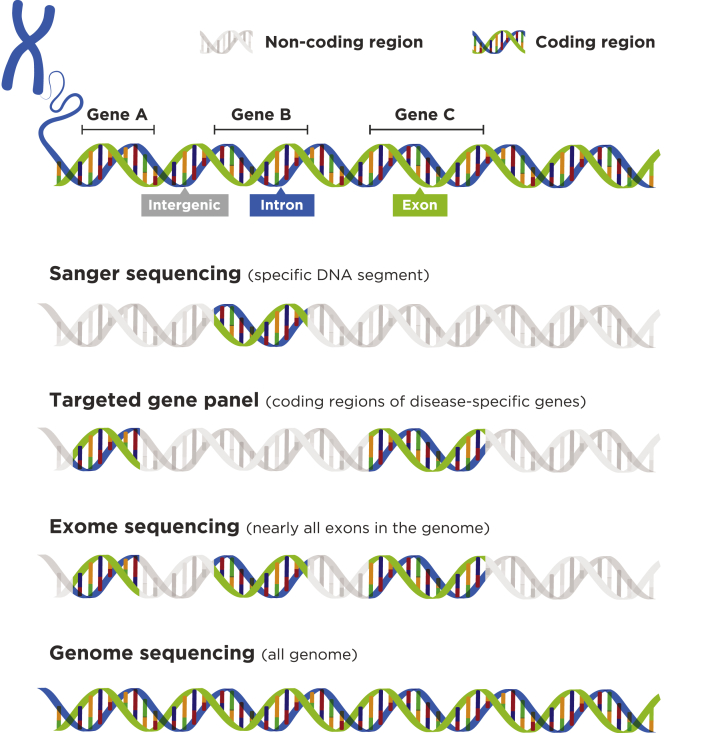
There are currently 3 NGS approaches—targeted panel sequencing, whole exome sequencing (WES), and whole genome sequencing (WGS)—that have been used to improve the diagnosis of hereditary diseases, including CKD (Fig 2).30,31 The relative strengths and weaknesses of these approaches are shown in Table 1.32,33 The emergence of NGS has resulted in a large number of articles on single-gene contributions to kidney disease.34 Several hundred genes have now been identified that may contribute to the development and progression of CKD.35 All NGS approaches listed have contributed to our understanding of specific genes and disease pathways involved in CKD.
Figure 2.
Different approaches to genetic testing. Sanger sequencing is limited to a narrow portion of the genome. In targeted gene panels, only coding portions of a specific set of genes are targeted. Whole exome sequencing captures almost all coding sequences, and whole genome sequencing covers nearly all regions of the genome.
Table 1.
Comparison of Next-Generation DNA Sequencing Strategies32
| Targeted Panels | Whole Exome Sequencing | Whole Genome Sequencing | |
|---|---|---|---|
| Strengths |
|
|
|
| Weaknesses |
|
|
|
Note: This table (modified from original) is used under a Creative Commons CC BY-NC 4.0 License specific to the article published by Elsevier Limited on behalf of the European Society for Medical Oncology: Horak P, Fröhling S, Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016;1:e000094. https://doi.org/10.1136/esmoopen-2016-000094.
Abbreviations: CNV, copy number variant; ncRNA, noncoding RNA; SNV, single nucleotide variant.
Targeted Gene Panels in CKD
Results from the PodoNet Registry, which is focused on steroid-resistant nephrotic syndrome, showed that an NGS-based panel approach that included >30 podocyte-related genes complemented by WES supported the determination of genetic diagnoses in 24% of the patients screened. It also extended the spectrum of genetic diseases presenting with a steroid-resistant nephrotic syndrome phenotype (mutation of the COL4A3-5 and CLCN5 genes), as well as contributing to the discovery of new causative genes (eg, MYOE1, PTPRO).36 NGS techniques have now identified >50 causal genes in hereditary podocytopathies.37
A targeted NGS panel covering 39 genes implicated in focal segmental glomerulosclerosis (FSGS) and steroid-resistant nephrotic syndrome—including COL4A3, COL4A4, or COL4A5—was applied to 81 adults from 76 families (24 families with a history of kidney disease).38 Pathogenic mutations were confirmed in 10 patients (6 with family history) from 9 families. The collagen type IV mutations—evident in 8 patients from 6 families—represented 56% of definitely pathogenic mutations and provided a diagnosis of Alport syndrome in 6 patients. In addition, a histologic diagnosis of thin basement membrane nephropathy, also a manifestation of a COL4A mutation, was established in 2 patients. NGS identified collagen gene mutations in 38% of families with familial FSGS and 3% with sporadic FSGS; >50% of the mutations occurred in COL4A5.38
The value of targeted gene panels has been demonstrated by a kidney disease gene panel that provides a comprehensive and cost-effective approach to the genetic diagnosis of patients with cystic and glomerular inherited kidney diseases. In a study of 421 patients with known or suspected inherited kidney diseases, a 140-gene kidney disease panel for the genetic diagnosis of suspected cystic or glomerular inherited kidney diseases was used for confirmation of disease diagnoses.39 Patients were assigned to a validation cohort (n = 116) with known mutations and a diagnostic cohort (n = 305) with suspected inherited cystic (n = 207) or glomerular (n = 98) diseases.39 Seventy-eight percent of cystic and 62% of glomerular cases in the diagnostic cohort had a genetic cause.39 In patients suspected to have cystic inherited kidney disease, genetic analysis provided diagnosis in 25 of 31 prenatal (81%), 46 of 64 pediatric (72%), and 90 of 112 adult patients (80%).39 In those with suspected glomerular inherited kidney diseases, all 5 patients with congenital-onset glomerular inherited kidney disease (detected in the perinatal period), 26 of 50 pediatric patients (52%), and 30 of 43 adult patients (70%) achieved a molecular diagnosis.39 A high level of sensitivity (99%) was reported in the validation cohort with 134 of 135 previously known mutations accurately detected.39 This panel allowed an etiologic diagnosis in 75% of patients and was particularly useful in those with nonspecific or atypical phenotypes.39 For example, in patients with nonspecific cystic disease of unknown etiology, nearly 70% were shown to have ADPKD by genetic analysis.39
Gribouval et al40 studied 135 patients with adult-onset steroid-resistant nephrotic syndrome and/or FSGS who were highly selected for potentially nonimmune FSGS. An NGS panel that screened for 35 genes was used.40 Pathogenic mutations were found in 30 patients (22.2%), including 16 (11.8%) with mutations in known monogenic SRNS genes and 14 (10.4%) with mutations in APOL1 alleles at high risk. Of the 16 patients with monogenic steroid-resistant nephrotic syndrome mutations, 7 (43.7%) had mutations in COL4A.40 These findings are consistent with prior results indicating that COL4A mutations were the most frequent type of mutation in FSGS.38
Another study on NGS examined variants in a panel of 109 genes associated with FSGS, nephrotic syndrome, congenital anomalies of the kidney and urinary tract, or nephronophthisis. Study participants included probands with a pathologic diagnosis of FSGS and relatives with >500-mg protein excretion per day, comprising 193 individuals from 179 families. Forty-three patients were familial cases belonging to 29 pedigrees and 150 were sporadic. The cohort had 56% men (51% in the familial cases and 57% in the sporadic cases). The mean age of disease onset was 34 years, and 78% of the patients presented with normal kidney function.41 The overall diagnostic rate was 11% (20 of 179 families). This rather low diagnostic rate may still be considered clinically significant because a genetic etiology had not been previously identified in this cohort, with important implications for guiding immunosuppressive therapies as well as planning for a future kidney transplant. Surprisingly, pathogenic variants in COL4A3, COL3A3, or COL4A5 accounted for 55% (11 of 20) of cases attributed to a single gene, with most found in COL4A5. It was concluded that FSGS is commonly the result of COL4A disorders, as well as podocyte and kidney developmental defects.41 Table 2 summarizes key results from the studies discussed in this section.42,43
Table 2.
Results With Targeted Gene Panels in CKD
| Study | Participants | Key Results |
|---|---|---|
| Rasouly et al 201912 | A convenience sample of exome sequence data from 7,974 self-declared healthy adults | Evaluation of the prevalence of candidate pathogenic variants in 625 genes correlated with Mendelian kidney and genitourinary disorders. In total, 23.3% had a candidate pathogenic variant, most of which were due to previously reported variants with high allele frequencies. |
| Trautmann et al 201836 | Registry including >2,000 pediatric patients with steroid-resistant nephrotic syndrome | NGS-based gene panel screening with >30 podocyte-related genes complemented by WES achieved genetic diagnoses in 24% of the patients screened, broadened the spectrum of genetic disease entities presenting with steroid-resistant nephrotic syndrome phenotype (COL4A3-5, CLCN5), and added to the discovery of new disease causative genes (MYOE1, PTPRO). |
| Bullich et al 201839 | 421 patients with known or suspected inherited kidney diseases | 78% of cystic and 62% of glomerular cases in the diagnostic cohort had a genetic cause. |
| Gast et al 201638 |
81 adults from 76 families (24 families with a history of kidney disease) | NGS identified collagen gene mutations in 38% of families with familial FSGS, and 3% with sporadic FSGS; >50% of the mutations occurred in COL4A5. |
| Gribouval et al 201840 | 135 patients with adult-onset steroid-resistant nephrotic syndrome and/or FSGS | Pathogenic mutations were found in 30 patients (22%), including 16 (12%) with mutations in known monogenic SRNS genes and 14 (10%) with APOL1 alleles at high risk. Of the 16 patients with monogenic steroid-resistant nephrotic syndrome mutations, 7 (44%) had mutations in COL4A. |
| Yao et al 201941 |
Probands with a pathologic diagnosis of FSGS and relatives with >500-mg protein excretion/d (total of 193 individuals from 179 families) | Pathogenic variants in COL4A3, COL4A4, or COL4A5 accounted for 55% (11 of 20) of cases associated with a single gene; most were found in COL4A5. |
| Papazachariou et al 201742 | 24 families with members having familial microscopic hematuria assessed for the presence of one of a genetically heterogeneous group of conditions, including the collagen IV nephropathies, heritable complement C3/CFHR5 nephropathy, and glomerulopathy with fibronectin deposits | In 17 of the 24 families (71%), 15 pathogenic mutations in COL4A3, COL4A4, or COL4A5—9 of them novel—were identified. |
| Mantovani et al 202043 | 212 patients with suspected ADPKD | Causative variants were detected in 61.3% of index patients and others of uncertain clinical significance in 12.5%. Most (88%) variants were in PKD1 and 12% were in PKD2. Overall, 80 of 158 variants (50.6%) were not reported in prior studies, indicating wide allelic heterogeneity. |
Abbreviations: ADPKD, autosomal dominant polycystic kidney disease; APOL1, apolipoprotein L1; CKD, chronic kidney disease; CLCN5, chloride voltage-gated channel 5; COL4A, collagen type IV α-chain; FSGS, focal segmental glomerulosclerosis; MYOE1, myosin 1E; NGS, next-generation sequencing; PKD, polycystic kidney disease; PTPRO, protein tyrosine phosphatase receptor type O; WES, whole exome sequencing.
Whole Exome Sequencing (WES) in CKD
Whereas targeted sequencing is often restricted to select predetermined gene panels, the WES approach allows for interrogation of the entire exome, thereby identifying previously unknown variants.10 WES in CKD has been instrumental in identifying mutations in heterogeneous CKD cohorts and in known clinical groups (eg, nephronophthisis and medullary cystic kidney disease), as shown in Table 3.44, 45, 46, 47, 48, 49
Table 3.
Results With Whole Exome Sequencing in Patients With CKD
| Study | Participants | Key Results |
|---|---|---|
| Groopman et al 201944 | 3,315 patients with CKD | Genetic causes were identified in 307, or nearly 10%, of the 3,315 patients, and 30% of patients with a genetic finding were discovered to have abnormalities in the COL4A genes. Among the patients discovered to have COL4A mutations, 62% had been misdiagnosed before the study or had not been diagnosed with Alport syndrome or TBMN. |
| Lata et al 201845 | 92 adults with CKD of unknown cause, familial nephropathy, or hypertension | WES provided a diagnosis in 24% of patients, which included 9 probands with CKD of unknown cause and 13 distinct genetic disorders. Loss-of-function mutations were identified in PARN in 2 probands with tubulointerstitial fibrosis. |
| Wang et al 201846 | 9 MODY probands with biopsy-proven DKD and their families | Evaluation of gene function, protein-protein interactions, and phenotypic differences between probands and parents demonstrated that COL4A3 variants were involved in the progression of DKD. |
| Cameron-Christie et al 201947 | 3,150 patients with broad clinical subcategories of CKD and a control cohort of 9,563 healthy, unrelated individuals | The genes PKD1, PKD2, and COL4A5 reached significance across the entire cohort with CKD, whereas COL4A3 and COL4A4 were highly ranked across multiple analysis models. Signals in COL4A3, COL4A4, and COL4A5 emerged in the combined cohort, suggesting that rare variations in these genes might contribute to many clinical forms of nephropathy. |
| Yamamoto et al 201748 | Single family with medullary cystic kidney disease | WES demonstrated a novel frameshift mutation before the variable number of tandem repeat region in the MUC1 gene. |
| Tang et al 201949 | 2 patients with infantile nephronophthisis, and 3 patients had juvenile nephronophthisis | The 2 patients with infantile nephronophthisis had NPHP3 mutations, with one carrying compound heterozygous mutations (c.1358A>G, c.2369A>G) and the other simultaneously carrying a c.1174C>T IVS26-3A>G cleavage site mutation from the father and a nonsense mutation (p.392R>X, 939) from the mother. Two patients with juvenile nephronophthisis had frameshift mutations (c.1583 to 1596: deletion) and homozygous point mutations (7 c.640G>T) of the NPHP1 gene. The fifth patient had a complex heterozygous mutation of the NPHP2 gene (c.2686G>A, c.1943A>G). |
Abbreviations: CKD, chronic kidney disease; COL4A, collagen type IV α-chain; DKD, diabetic kidney disease; MODY, maturity-onset diabetes of the young; MUC1, mucin 1; NPHP, nephrocystin; PARN, poly(A)-specific ribonuclease; PKD, polycystic kidney disease; TBMN, thin basement membrane nephropathy; WES, whole exome sequencing.
A pilot study assessed the diagnostic utility of WES in a selected referral population of 92 adults with CKD of unknown cause, familial nephropathy, or hypertension.45 WES provided a diagnosis encompassing 13 distinct genetic disorders in 24% of these patients, 9 of whom were probands with an unknown cause of CKD. Loss-of-function mutations were identified in poly(A)-specific ribonuclease in 2 probands with tubulointerstitial fibrosis. Poly(A)-specific ribonuclease mutations have been previously detected in a syndrome that includes lung, bone marrow, and liver fibrosis; these findings implicate poly(A)-specific ribonuclease mutations in kidney fibrosis.45 The genetic results influenced treatment in most patients and included targeted surveillance, familial screening to aid donor selection for transplant, and changes in medication.45 A small-scale study used WES to identify genetic factors associated with diabetic kidney disease. Nine MODY probands with biopsy-proven diabetic kidney disease were included in this study. WES was used to determine suspected MODY probands and their families.46 Individuals from 9 families had a diagnosis of HNF1B-MODY, CEL-MODY, PAX4-MODY, and WFS1-MODY; 196 selected variants of 25 genes associated with increased susceptibility to diabetic kidney disease were identified. Evaluation of gene function, protein-protein interactions, and phenotypic differences between probands and parents demonstrated that COL4A3 variants were involved in the progression of diabetic kidney disease.46
In a 2019 study, Groopman et al44 isolated DNA from 3,315 patients with CKD of any cause and performed proband-only WES on the samples obtained. Genetic causes were identified in 307, or nearly 10%, of the 3,315 patients, and 30% of patients with a genetic finding were discovered to have abnormalities in the COL4A genes.44 Among the patients discovered to have COL4A mutations, 62% had been misdiagnosed with clinical diagnoses such as FSGS, hypertensive nephropathy, or unspecified glomerulopathy before the study or had not been diagnosed with Alport syndrome or thin basement membrane nephropathy.44 These results illustrate the potential for genetic findings to provide new clinical insight, with implications for medical management.
Cameron-Christie et al47 evaluated the potential of WES for detection of causal genes for nephropathy in a large cohort of 3,150 patients with broad clinical subcategories of CKD and a control cohort of 9,563 healthy, unrelated individuals. They applied a collapsing analysis to evaluate the contribution of rare variants to CKD in the case samples.47 The genes PKD1, PKD2, and COL4A5 reached significance across the entire cohort with CKD, whereas COL4A3 and COL4A4 were highly ranked across multiple analysis models. Signals in COL4A3, COL4A4, and COL4A5 emerged in the combined cohort, suggesting that rare variations in these genes may contribute to many clinical forms of nephropathy.47
Whole Genome Sequencing (WGS) in CKD
The WGS approach detects DNA variations outside the exons that can lead to genetic disorders, namely in the introns and noncoding regions. Such variations are not detected by WES, which focuses only on the exons. WGS detects complex trait variants, including copy number variants and variants in genes situated in intronic and other noncoding segments of the genome.10,31,50
WGS results from 2 large databases were used to measure the frequency of high-confidence mutations presumed to be causative in ADPKD and autosomal dominant polycystic liver disease and to estimate lifetime ADPKD prevalence using strict criteria for defining rare variants in genes involved in ADPKD (PKD1, PKD2), autosomal dominant polycystic liver disease (PRKCSH, SEC63, GANAB, ALG8, SEC61B, LRP5), and potential cystic disease modifiers.51 Determination of high-confidence pathogenic mutations in WGS resulted in a lower boundary for lifetime prevalence of ADPKD of 9.3 cases per 10,000 sequenced. Genes potentially relevant as cyst modifiers and truncating mutations in autosomal dominant polycystic liver disease genes were identified in 103.9 and 20.2 per 10,000 sequenced, respectively. A greater-than-expected frequency of loss-of-function mutations in autosomal dominant polycystic liver disease genes also suggested the possibility of unrecognized cases and incomplete penetrance.51
WGS has also been used to detect genetic differences in a cohort of Finnish siblings with type 1 diabetes who were discordant for the presence or absence of diabetic nephropathy. Control patients had diabetes for 15-37 years without any complications. Results indicated that when clustered at the gene level, diabetic nephropathy-associated variants were enriched in a network representing proteins fundamental for podocyte function, including protein kinases and protein tyrosine kinase 2.52 Several new genes were found to be strongly associated with diabetic nephropathy, including PRKCE, PTK1, PRKCI, ABTB1, and ALOX5. The significant association with protein kinase genes is especially intriguing, strongly implicating the protein kinase C family in the pathogenesis of diabetic nephropathy and suggesting protein kinase C inhibitors as attractive novel therapies.52 Studies using WGS are summarized in Table 4.53,54
Table 4.
Results With Whole Genome Sequencing in Patients With CKD
| Study | Participants | Key Results |
|---|---|---|
| Lanktree et al 201851 | Results from 2 large databases were used to measure the frequency of high-confidence mutations presumed to be causative in ADPKD and ADPLD | Genes potentially relevant as cyst modifiers and truncating mutations in ADPLD genes were identified in 103.9 and 20.2 per 10,000 sequenced, respectively. A greater-than-expected frequency of loss-of-function mutations in ADPLD genes also suggested the possibility of unrecognized cases and incomplete penetrance. |
| Guo et al 202052 | Finnish siblings with type 1 diabetes who were discordant for the presence or absence of diabetic nephropathy | Diabetic nephropathy-associated variants were enriched in a network representing proteins essential for podocyte function when clustered at the gene level and include protein kinases and protein tyrosine kinase 2. |
| Larrue et al 202053 | 2 patients with nephronophthisis | WGS identified 2 putative disease-causing intronic mutations in the NPHP3 gene, including one deep intronic variant. Both affected splicing, resulting in a truncated nephrocystin-3 protein. |
| Levine et al 202054 | 146 patients with primary membranoproliferative GN and 6,442 individuals without kidney disease (controls) | A significant common variant locus was identified at 6p21.32 (rs35406322) overlapping the HLA locus. |
Abbreviations: ADPKD, autosomal dominant polycystic kidney disease; ADPLD, autosomal dominant polycystic liver disease; CKD, chronic kidney disease; GN, glomerulonephropathy; HLA, human leukocyte antigen; NPHP3, nephrocystin 3; WGS, whole genome sequencing.
Clinical Relevance of Genetic Testing in CKD
To translate genetic findings into improved patient care, longitudinal studies of large cohorts of individuals with genetic diagnoses are required. Long-term follow-up is needed for patients who have participated in NGS studies to examine the role of genetic information on clinical management, health care utilization, and outcomes.55 There are a few published examples of genetic information guiding treatment strategy, and further studies are needed; however, Table 5 provides examples of the clinical utility of a genetic diagnosis of kidney disease.56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73 As noted earlier, because a genetic cause is implicated in nearly all children who receive kidney replacement therapy and in approximately 10% of adults (including 964 of 3,315 patients [29%] aged ≥65 years in the study by Groopman et al44), it can be offered to appropriate patients of varying age groups.22
Table 5.
Clinical Impact of Genetic Testing in Kidney Diseases
| Indication | Genetic Finding | Genetic Diagnosis | Clinical Impact | References |
|---|---|---|---|---|
| Steroid-resistant nephrotic syndrome | Homozygous Fin-major mutation in NPHS1 | Nephrotic syndrome type 1 (OMIM #256300) | Increased risk of posttransplant disease recurrence | 57, 58 |
| COQ2 mutation | CoQ10 deficiency 1 (OMIM #607426) | CoQ10 supplementation can attenuate proteinuria and extrarenal complications such as encephalopathy | 59, 60 | |
| COL4A3 or COL4A4 missense mutation | Alport syndrome (OMIM #104200; #203780) or TBMD (OMIM #141200) |
|
61-64 | |
| Cystic renal dysplasia | 17q12 deletion | Renal cysts and diabetes syndrome (OMIM #137920) | Multisystem work-up for associated extrarenal complications, including testing for diabetes, exocrine pancreatic insufficiency, hepatic function, neurologic anomalies, and/or neurocognitive impairment | 65-67 |
| Nephrolithiasis | APRT mutation | APRT deficiency (OMIM #614723) | Xanthine dehydrogenase inhibition to prevent crystalline nephropathy and allograft loss | 68, 69 |
| Episodic hypertension | SDHD mutation | Hereditary paraganglioma-pheochromocytoma syndrome (OMIM #168000) |
|
70, 71 |
| Failure to thrive, hepatomegaly, and hyperuricemia | G6PC mutation | Glycogen storage disease Ia (OMIM #232200) |
|
72, 73 |
Note: Reproduced from Groopman et al56 with permission of Springer Nature.
Abbreviations: APRT, adenine phosphoribosyltransferase; COL4A, collagen type IV α-chain; CoQ10, coenzyme Q10; COQ2, coenzyme Q2, polyprenyltransferase; G6PC, glucose-6-phosphatase catalytic subunit; NPHS1, nephrin; SDHD, succinate dehydrogenase complex subunit D; TBMD, thin basement membrane disease.
Prognosis
Age-adjusted total kidney volume represented by the Mayo imaging classification estimates an individual’s unique rate of kidney growth and offers an individualized approach to prognostication in ADPKD.74 Genetic assessment of patients with ADPKD also demonstrated that most with PKD1 protein-truncating in-frame indels or mutations were at elevated risk of CKD progression and were likely to benefit from new treatments.75 One example may include the use of medications, such as tolvaptan, a selective vasopressin V2 receptor antagonist that delays the increase in kidney cyst volume and thereby prevents CKD progression as well as pain due to cysts.76,77 Another example is bardoxolone methyl, a novel nuclear erythroid 2–related factor activator under clinical development that improves mitochondrial function and has an anti-inflammatory action in ADPKD.78 Conversely, some patients with PKD2 or nontruncating PKD1 mutations are unlikely to reach kidney failure by 60 years of age, and such patients can be reassured that conventional treatment (such as inhibition of the renin-angiotensin-aldosterone system) and control of proteinuria and hypertension may be sufficient.75
Diagnosis
Gene panel testing has the potential to provide a genetic diagnosis in a substantial number of patients with suspected Alport syndrome or steroid-resistant nephrotic syndrome, and it should be undertaken early during care. Testing using NGS should have the capacity to detect copy number variants for genes associated with these conditions.79 Clinical genetic testing has the potential to stratify patients and support clinical decision making regarding their care.79 Such patients may benefit from more detailed WGS to better identify pathogenetic factors for therapeutic targeting (such as COQ2 defects that code for proteins in the coenzyme Q10 pathway, as mentioned above).79 Identification of a pathogenic variant may also result in modification or elimination of immunosuppressive treatments and augment rational planning for immunosuppression after kidney transplant.20,79
Thrombotic microangiopathy, which includes atypical HUS, thrombotic thrombocytopenic purpura, and Shiga toxin–associated HUS, requires prompt diagnosis because of differing pathophysiology and importance of early treatment for thrombotic thrombocytopenic purpura and atypical HUS.80 In atypical HUS, a rare variant of thrombotic microangiopathy, several studies have demonstrated heterozygous pathogenic activating mutations in the genes encoding the alternative pathway components C3 and factor B and loss-of-function mutations in the genes encoding the regulators FH (including CFH/CFHR fusions), FI, and CD46.81 Because the genetic mutations are predisposing versus causative with incomplete penetrance, additional disease risk modifiers and environmental triggers (eg, pregnancy, infection) may be contributory.81 Eculizumab is a recombinant humanized monoclonal antibody that can be tailored to the patient once a diagnosis of atypical HUS has been confirmed.28
Variants of Uncertain Significance
Genetic testing may result in the identification of variants of uncertain significance, which represent variants with insufficient evidence to determine the role in disease. The American College of Medical Genetics and Genomics guidelines note that such variants should not be used in clinical decision making82; however, some institutional policies require reporting of variants of uncertain significance to patients.83 One study found that the diagnostic yield of 250 exome sequencing samples increased from 24.8% to 46.8% after manual review of available data just 5 years later.84 Retrospective identification of clinically relevant (or irrelevant) variants, therefore, requires continual review of emerging data, which may be augmented by semiautomated processes.84
Genetic Testing and Kidney Biopsy
In addition to clarifying the diagnosis in CKD of unknown etiology, genetic testing may obviate the need for kidney biopsy in some patients. In a 2021 study of 204 patients with suspected hereditary CKD, WES identified a molecular diagnosis in 80 (39%) patients, including 10 (13%) for whom genetic testing negated the need for a kidney biopsy.85 In a retrospective analysis of 273 patients who underwent kidney transplantation that evaluated a genetics-first approach in improving diagnosis of kidney failure in patients younger than 50 years, targeted WES resulted in a genetic diagnosis in 21%-51% of patients. Among those who received a kidney biopsy for diagnosis of CKD before study enrollment, biopsy results would have provided no additional diagnostic value for 43% of patients if genetic testing had been performed first.86
Genetic Counseling
Pre- and posttest counseling is recommended for all patients who undergo genetic testing, regardless of the modality used.83 Pretest counseling provides patients and their families with an understanding of the opportunities and limitations of genetic testing and ensures that informed consent is obtained.83,87 Posttest counseling should be provided by a licensed genetic counselor and offers guidance for patients on the implications of their testing results for disease risk and management, psychosocial implications, and recommendations for family screening.83,87 Guidelines are available from the American College of Medical Genetics and Genomics to support discussions on the interpretation of sequence variants based on supporting evidence for the role in disease pathogenicity.82
Genetic Testing Services
Genetic testing for kidney diseases is available through various companies including, but not limited to, Invitae Corporation, Natera, Inc, and Centogene US, LLC.88, 89, 90 Invitae offers a Progressive Renal Disease Panel that analyzes 195 genes for kidney disorders such as Alport syndrome, FSGS, and nephrotic syndrome. Natera, through Renasight, tests 385 genes associated with CKD to assess a patient’s genetic cause for kidney disease or whether there is an increased hereditary risk due to family history. Centogene provides genetic testing for >300 different genes associated with inherited kidney-related diseases. There may also be clinical and academic studies enrolling patients that provide genetic testing; otherwise, patients are recommended to inquire with their local genetic counselors or the National Society of Genetic Counselors for more information.91,92
Other Clinical Considerations
A genetic diagnosis is important but can be accompanied by challenges such as physician education, sequence reanalysis, and return on results.56 Additionally, there may be clinical challenges for patients with CKD of unclear etiology whose genetic test determines a diagnosis that is currently without treatment or ongoing clinical trials. In these instances, providing continued clinical follow-up, communication about relevant ongoing research, and directing the patient and family to genetic counseling and advocacy support groups can add important value to the patient’s care.
Case Study Vignette
Unlike other kidney diseases, genetic testing is rarely performed in patients with ADPKD.93 The importance of genetic testing in PKD was reported by Cornec-Le Gall et al94 in a 7-generation pedigree, which yielded a diagnosis of 2 different cystic disorders. Targeted NGS of 65 candidate genes in a patient with an ADPKD-like phenotype without a familial PKD2 mutation yielded a COL4A1 mutation and a diagnosis of hereditary angiopathy with nephropathy, aneurysms, and muscle cramps syndrome. This case demonstrates the utility of targeted NGS in differential diagnoses and identifying disease variability. An accurate diagnosis of ADPKD will be important to optimize patient care and management.
Conclusion
Precision medicine has the potential to considerably improve care for patients with CKD. Accurate diagnosis, prognosis—and to a lesser extent—decisions on treatment have already been advanced by genetic assessment of patients with CKD, most notably with NGS, including targeted gene panels, WES, and WGS. These now widely available and relatively low-cost approaches have shown novel genetic variants in previously unrecognized biological pathways, highlighting disease mechanisms with a potential role in CKD etiology, morbidity, and mortality. Combining data obtained from the NGS approaches described in this review with clinical phenotypic results from large and diverse cohorts will help define genotype-phenotype relationships for different types of CKD and support individualized approaches to treatment.
Nephrologists can benefit from understanding the advantages of NGS and genotype-phenotype relationships in patients with CKD and the factors inducing variability in each disease and the same causative mutation. With decreasing costs of high-throughput sequencing, new discoveries are likely, and genetic testing could become a commonly available and affordable resource for routine clinical care. The optimal application of genomics in the prevention and treatment of CKD requires the adoption of this mode of investigation by practicing nephrologists across the health care spectrum. Whereas the benefits of genetic assessment in patients with kidney disease have been recognized by nephrologists who manage pediatric patients,14 the advantages of precision medicine including genetic evaluation with NGS should be extended to adult patients with CKD to pinpoint causes of disease and rationally guide therapy.15
Article Information
Authors’ Full Names and Academic Degrees
Prasad Devarajan, MD, Glenn M. Chertow, MD, MPH, Katalin Susztak, MD, PhD, Adeera Levin, MD, Rajiv Agarwal, MD, Peter Stenvinkel, MD, PhD, Arlene B. Chapman, MD, and Bradley A. Warady, MD
Support
The article was edited for language and grammar, and support was provided for generating the figures by Fallon Medica LLC. This service was paid for by Reata Pharmaceuticals. Neither Fallon Medica LLC nor Reata Pharmaceuticals were involved in writing content or development of the article’s scope.
Financial Disclosure
Dr Devarajan has participated as a consultant/advisor for Reata Pharmaceuticals, Inc, Dicerna Pharmaceuticals, Inc, Alnylam Pharmaceuticals Inc, Natera, Inc, BioPorto Diagnostics, Inc, and UpToDate, Inc, and has received research support from the National Institutes of Health (NIH) (P50DK096418). Dr Chertow has participated on the Board of Directors for Satellite Healthcare, Inc, and has participated as a consultant/advisor for Akebia Therapeutics, Inc, Amgen, Inc, Ardelyx, Inc, AstraZeneca, Baxter Healthcare, CloudCath, Cricket Health, DiaMedica Therapeutics, Inc, DURECT Corporation, DxNow, Inc, Gilead Sciences, Inc, Miromatrix Medical, Inc, Outset Medical, Inc, Reata Pharmaceuticals, Inc, Sanifit, and Vertex Pharmaceuticals, Inc. Dr Susztak has served on the advisory board for Jnana Therapeutics; has participated as a consultant for Bayer AG, AstraZeneca plc, Maze Therapeutics, and Jnana Therapeutics; and has received research support from Gilead Sciences, Inc, GlaxoSmithKline plc, Regeneron Pharmaceuticals, Inc, Boehringer Ingelheim International GmbH, Novartis AG, Calico LLC, Merck & Co, Inc, and Bayer AG. Dr Levin has served on the advisory committee for Reata Pharmaceuticals, Inc, and has received grants from Otsuka America Pharmaceutical, Inc, AstraZeneca plc, and Boehringer Ingelheim International GmbH. Dr Agarwal has participated as a consultant and has served on the advisory committees for Relypsa, Inc, AbbVie, Inc, Amgen, Inc, AstraZeneca plc, Bayer AG, Boehringer Ingelheim International GmbH, Celgene Corp, a Bristol-Myers Squibb Company, Daiichi Sankyo Company, Ltd, Eli Lilly and Co, Gilead Sciences, Inc, GlaxoSmithKline plc, Johnson & Johnson Services, Inc, Merck & Co, Inc, Novartis International AG, Sandoz International GmbH, ZS Pharma, Inc, Akebia Therapeutics, Inc, Takeda Pharmaceutical Co Ltd, Sanofi SA, Reata Pharmaceuticals, Inc, Ironwood Pharmaceuticals, Inc, Otsuka America Pharmaceutical, Inc, Opko Health, Inc, and Bird Rock Bio, Inc, and has received grants from the NIH and Veterans Affairs. Dr Stenvinkel has served on scientific advisory boards for Reata Pharmaceuticals, Inc, AstraZeneca plc, and Baxter Healthcare. Dr Chapman has participated as a consultant/advisor for Otsuka America Pharmaceutical, Inc, Reata Pharmaceuticals, Inc, Sanofi SA, and UpToDate, Inc. Dr Warady has served on the advisory committee for Reata Pharmaceuticals, Inc, as well as the medical advisory committee for the Alport Syndrome Foundation; has participated as a consultant for Bayer AG, Akebia Therapeutics, Inc, Relypsa, Inc, Amgen Inc, FibroGen, Inc, and UpToDate, Inc; and has received research support from the NIH and Baxter Healthcare.
Peer Review
Received September 21, 2021. Evaluated by 4 external peer reviewers, with direct editorial input from the Editor-in-Chief. Accepted in revised form December 26, 2021.
Footnotes
Complete author and article information provided before references.
References
- 1.GBD Chronic Kidney Disease Collaboration Global, regional, and national burden of chronic kidney disease, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020;395:709–733. doi: 10.1016/S0140-6736(20)30045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClellan W.M., Satko S.G., Gladstone E., Krisher J.O., Narva A.S., Freedman B.I. Individuals with a family history of ESRD are a high-risk population for CKD: implications for targeted surveillance and intervention activities. Am J Kidney Dis. 2009;53:S100–S106. doi: 10.1053/j.ajkd.2008.07.059. [DOI] [PubMed] [Google Scholar]
- 3.Foreman K.J., Marquez N., Dolgert A., et al. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet. 2018;392:2052–2090. doi: 10.1016/S0140-6736(18)31694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y.N., Ma S.X., Chen Y.Y., et al. Chronic kidney disease: biomarker diagnosis to therapeutic targets. Clin Chim Acta. 2019;499:54–63. doi: 10.1016/j.cca.2019.08.030. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Giacoman S., Madero M. Biomarkers in chronic kidney disease, from kidney function to kidney damage. World J Nephrol. 2015;4:57–73. doi: 10.5527/wjn.v4.i1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng G., Liu D. Klotho: a promising biomarker closely related to kidney transplant. Exp Clin Transplant. 2018;16:253–258. doi: 10.6002/ect.2017.0329. [DOI] [PubMed] [Google Scholar]
- 7.Celec P., Vlková B., Lauková L., Bábíčková J., Boor P. Cell-free DNA: the role in pathophysiology and as a biomarker in kidney diseases. Expert Rev Mol Med. 2018;20:e1. doi: 10.1017/erm.2017.12. [DOI] [PubMed] [Google Scholar]
- 8.Cañadas-Garre M., Anderson K., Cappa R., et al. Genetic susceptibility to chronic kidney disease—some more pieces for the heritability puzzle. Front Genet. 2019;10:453. doi: 10.3389/fgene.2019.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gluck C., Ko Y.A., Susztak K. Precision medicine approaches to diabetic kidney disease: tissue as an issue. Curr Diab Rep. 2017;17:30. doi: 10.1007/s11892-017-0854-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan A., Eijgelsheim M., Vogt L., Knoers N.V.A.M., de Borst M.H. Diagnostic yield of next-generation sequencing in patients with chronic kidney disease of unknown etiology. Front Genet. 2019;10:1264. doi: 10.3389/fgene.2019.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fiorentino M., Bolignano D., Tesar V., et al. Renal biopsy in patients with diabetes: a pooled meta-analysis of 48 studies. Nephrol Dial Transplant. 2017;32:97–110. doi: 10.1093/ndt/gfw070. [DOI] [PubMed] [Google Scholar]
- 12.Rasouly H.M., Groopman E.E., Heyman-Kantor R., et al. The burden of candidate pathogenic variants for kidney and genitourinary disorders emerging from exome sequencing. Ann Intern Med. 2019;170:11–21. doi: 10.7326/M18-1241. [DOI] [PubMed] [Google Scholar]
- 13.Piras D., Zoledziewska M., Cucca F., Pani A. Genome-wide analysis studies and chronic kidney disease. Kidney Dis (Basel) 2017;3:106–110. doi: 10.1159/000481886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris P.C. The time for next-generation molecular genetic diagnostics in nephrology is now! Kidney Int. 2018;94:237–239. doi: 10.1016/j.kint.2018.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cocchi E., Nestor J.G., Gharavi A.G. Clinical genetic screening in adult patients with kidney disease. Clin J Am Soc Nephrol. 2020;15:1497–1510. doi: 10.2215/CJN.15141219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hays T., Groopman E.E., Gharavi A.G. Genetic testing for kidney disease of unknown etiology. Kidney Int. 2020;98:590–600. doi: 10.1016/j.kint.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connaughton D.M., Kennedy C., Shril S., et al. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95:914–928. doi: 10.1016/j.kint.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Posey J.E. Genome sequencing and implications for rare disorders. Orphanet J Rare Dis. 2019;14:153. doi: 10.1186/s13023-019-1127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe W.L., Jr., Reddy T.E. Genomic approaches for understanding the genetics of complex disease. Genome Res. 2015;25:1432–1441. doi: 10.1101/gr.190603.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann N., Braun D.A., Amann K., et al. Whole-exome sequencing enables a precision medicine approach for kidney transplant recipients. J Am Soc Nephrol. 2019;30:201–215. doi: 10.1681/ASN.2018060575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayasinghe K., Quinlan C., Stark Z., et al. Renal genetics in Australia: kidney medicine in the genomic age. Nephrology (Carlton) 2019;24:279–286. doi: 10.1111/nep.13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devuyst O., Knoers N.V.A.M., Remuzzi G., Schaefer F. Board of the Working Group for Inherited Kidney Diseases of the European Renal Association and European Dialysis and Transplant Association. Rare inherited kidney diseases: challenges, opportunities, and perspectives. Lancet. 2014;383:1844–1859. doi: 10.1016/S0140-6736(14)60659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armstrong M.E., Thomas C.P. Diagnosis of monogenic chronic kidney diseases. Curr Opin Nephrol Hypertens. 2019;28:183–194. doi: 10.1097/MNH.0000000000000486. [DOI] [PubMed] [Google Scholar]
- 24.Cornec-Le Gall E., Audrézet M.P., Chen J.M., et al. Type of PKD1 mutation influences renal outcome in ADPKD. J Am Soc Nephrol. 2013;24:1006–1013. doi: 10.1681/ASN.2012070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heyer C.M., Sundsbak J.L., Abebe K.Z., et al. Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27:2872–2884. doi: 10.1681/ASN.2015050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adam J., Connor T.M., Wood K., et al. Genetic testing can resolve diagnostic confusion in Alport syndrome. Clin Kidney J. 2014;7:197–200. doi: 10.1093/ckj/sft144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ariceta G., Dixon B.P., Kim S.H., et al. The long-acting C5 inhibitor, ravulizumab, is effective and safe in pediatric patients with atypical hemolytic uremic syndrome naïve to complement inhibitor treatment. Kidney Int. 2021;100:225–237. doi: 10.1016/j.kint.2020.10.046. [DOI] [PubMed] [Google Scholar]
- 28.Legendre C.M., Licht C., Muus P., et al. Terminal complement inhibitor eculizumab in atypical hemolytic-uremic syndrome. N Engl J Med. 2013;368:2169–2181. doi: 10.1056/NEJMoa1208981. [DOI] [PubMed] [Google Scholar]
- 29.Garrelfs S.F., Frishberg Y., Hulton S.A., et al. Lumasiran, an RNAi therapeutic for primary hyperoxaluria type 1. N Engl J Med. 2021;384:1216–1226. doi: 10.1056/NEJMoa2021712. [DOI] [PubMed] [Google Scholar]
- 30.United States National Library of Medicine; Medline Plus What are whole exome sequencing and whole genome sequencing? https://medlineplus.gov/genetics/understanding/testing/sequencing/
- 31.Sun Y., Ruivenkamp C.A., Hoffer M.J., et al. Next-generation diagnostics: gene panel, exome, or whole genome? Hum Mutat. 2015;36:648–655. doi: 10.1002/humu.22783. [DOI] [PubMed] [Google Scholar]
- 32.Horak P., Fröhling S., Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016;1 doi: 10.1136/esmoopen-2016-000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hehir-Kwa J.Y., Pfundt R., Veltman J.A. Exome sequencing and whole genome sequencing for the detection of copy number variation. Expert Rev Mol Diagn. 2015;15:1023–1032. doi: 10.1586/14737159.2015.1053467. [DOI] [PubMed] [Google Scholar]
- 34.Arora V., Anand K., Chander Verma I. Genetic testing in pediatric kidney disease. Indian J Pediatr. 2020;87:706–715. doi: 10.1007/s12098-020-03198-y. [DOI] [PubMed] [Google Scholar]
- 35.Pollak M.R., Friedman D.J. The genetic architecture of kidney disease. Clin J Am Soc Nephrol. 2020;15:268–275. doi: 10.2215/CJN.09340819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trautmann A., Lipska-Ziętkiewicz B.S., Schaefer F. Exploring the clinical and genetic spectrum of steroid resistant nephrotic syndrome: the PodoNet Registry. Front Pediatr. 2018;6:200. doi: 10.3389/fped.2018.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopp J.B., Anders H.J., Susztak K., et al. Podocytopathies. Nat Rev Dis Primers. 2020;6:68. doi: 10.1038/s41572-020-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gast C., Pengelly R.J., Lyon M., et al. Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2016;31:961–970. doi: 10.1093/ndt/gfv325. [DOI] [PubMed] [Google Scholar]
- 39.Bullich G., Domingo-Gallego A., Vargas I., et al. A kidney-disease gene panel allows a comprehensive genetic diagnosis of cystic and glomerular inherited kidney diseases. Kidney Int. 2018;94:363–371. doi: 10.1016/j.kint.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 40.Gribouval O., Boyer O., Hummel A., et al. Identification of genetic causes for sporadic steroid-resistant nephrotic syndrome in adults. Kidney Int. 2018;94:1013–1022. doi: 10.1016/j.kint.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 41.Yao T., Udwan K., John R., et al. Integration of genetic testing and pathology for the diagnosis of adults with FSGS. Clin J Am Soc Nephrol. 2019;14:213–223. doi: 10.2215/CJN.08750718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papazachariou L., Papagregoriou G., Hadjipanagi D., et al. Frequent COL4 mutations in familial microhematuria accompanied by later-onset Alport nephropathy due to focal segmental glomerulosclerosis. Clin Genet. 2017;92:517–527. doi: 10.1111/cge.13077. [DOI] [PubMed] [Google Scholar]
- 43.Mantovani V., Bin S., Graziano C., et al. Gene panel analysis in a large cohort of patients with autosomal dominant polycystic kidney disease allows the identification of 80 potentially causative novel variants and the characterization of a complex genetic architecture in a subset of families. Front Genet. 2020;11:464. doi: 10.3389/fgene.2020.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groopman E.E., Marasa M., Cameron-Christie S., et al. Diagnostic utility of exome sequencing for kidney disease. N Engl J Med. 2019;380:142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lata S., Marasa M., Li Y., et al. Whole-exome sequencing in adults with chronic kidney disease: a pilot study. Ann Intern Med. 2018;168:100–109. doi: 10.7326/M17-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Zhang J., Zhao Y., et al. COL4A3 gene variants and diabetic kidney disease in MODY. Clin J Am Soc Nephrol. 2018;13:1162–1171. doi: 10.2215/CJN.09100817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cameron-Christie S., Wolock C.J., Groopman E., et al. Exome-based rare-variant analyses in CKD. J Am Soc Nephrol. 2019;30:1109–1122. doi: 10.1681/ASN.2018090909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto S., Kaimori J.Y., Yoshimura T., et al. Analysis of an ADTKD family with a novel frameshift mutation in MUC1 reveals characteristic features of mutant MUC1 protein. Nephrol Dial Transplant. 2017;32:2010–2017. doi: 10.1093/ndt/gfx083. [DOI] [PubMed] [Google Scholar]
- 49.Tang X., Xu H., Shen Q., et al. Gene mutation and clinical analysis of nephronophthisis diagnosed using whole exome sequencing: experience from China. Clin Nephrol. 2019;92:89–94. doi: 10.5414/CN109571. [DOI] [PubMed] [Google Scholar]
- 50.Zhao E.Y., Jones M., Jones S.J.M. Whole-genome sequencing in cancer. Cold Spring Harb Perspect Med. 2019;9:a034579. doi: 10.1101/cshperspect.a034579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanktree M.B., Haghighi A., Guiard E., et al. Prevalence estimates of polycystic kidney and liver disease by population sequencing. J Am Soc Nephrol. 2018;29:2593–2600. doi: 10.1681/ASN.2018050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo J., Rackham O.J.L., Sandholm N., et al. Whole-genome sequencing of Finnish type 1 diabetic siblings discordant for kidney disease reveals DNA variants associated with diabetic nephropathy. J Am Soc Nephrol. 2020;31:309–323. doi: 10.1681/ASN.2019030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larrue R., Chamley P., Bardyn T., et al. Diagnostic utility of whole-genome sequencing for nephronophthisis. NPJ Genom Med. 2020;5:38. doi: 10.1038/s41525-020-00147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levine A.P., Chan M.M.Y., Sadeghi-Alavijeh O., et al. Large-scale whole-genome sequencing reveals the genetic architecture of primary membranoproliferative GN and C3 glomerulopathy. J Am Soc Nephrol. 2020;31:365–373. doi: 10.1681/ASN.2019040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Groopman E.E., Gharavi A.G. Expanding opportunities and emerging challenges: broadening the scope of genetic testing in nephrology. Kidney Int. 2019;95:743–746. doi: 10.1016/j.kint.2018.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Groopman E.E., Rasouly H.M., Gharavi A.G. Genomic medicine for kidney disease. Nat Rev Nephrol. 2018;14:83–104. doi: 10.1038/nrneph.2017.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holmberg C., Jalanko H. Congenital nephrotic syndrome and recurrence of proteinuria after renal transplantation. Pediatr Nephrol. 2014;29:2309–2317. doi: 10.1007/s00467-014-2781-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patrakka J., Ruotsalainen V., Reponen P., et al. Recurrence of nephrotic syndrome in kidney grafts of patients with congenital nephrotic syndrome of the Finnish type: role of nephrin. Transplantation. 2002;73:394–403. doi: 10.1097/00007890-200202150-00013. [DOI] [PubMed] [Google Scholar]
- 59.Ashraf S., Gee H.Y., Woerner S., et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123:5179–5189. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heeringa S.F., Chernin G., Chaki M., et al. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J Clin Invest. 2011;121:2013–2024. doi: 10.1172/JCI45693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Savige J., Gregory M., Gross O., Kashtan C., Ding J., Flinter F. Expert guidelines for the management of Alport syndrome and thin basement membrane nephropathy. J Am Soc Nephrol. 2013;24:364–375. doi: 10.1681/ASN.2012020148. [DOI] [PubMed] [Google Scholar]
- 62.Kashtan C.E. Alport syndrome. In: Adam MP, Ardinger HH, Pagon RA, et al, eds. GeneReviews [Internet]. August 28, 2001. Updated February 21, 2019. https://www.ncbi.nlm.nih.gov/books/NBK1207/ University of Washington, Seattle; 1993-2021.
- 63.Bekheirnia M.R., Reed B., Gregory M.C., et al. Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol. 2010;21:876–883. doi: 10.1681/ASN.2009070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kidney Disease Improving Global Outcomes Glomerulonephritis Work Group KDIGO clinical practice guideline for glomerulonephritis. Kidney Int Suppl. 2012;2:139–274. [Google Scholar]
- 65.Verhave J.C., Bech A.P., Wetzels J.F.M., Nijenhuis T. Hepatocyte nuclear factor 1β-associated kidney disease: more than renal cysts and diabetes. J Am Soc Nephrol. 2016;27:345–353. doi: 10.1681/ASN.2015050544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clissold R.L., Hamilton A.J., Hattersley A.T., Ellard S., Bingham C. HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum. Nat Rev Nephrol. 2015;11:102–112. doi: 10.1038/nrneph.2014.232. [DOI] [PubMed] [Google Scholar]
- 67.Mitchel M.W., Moreno-De-Luca D., Myers S.M., et al. 17q12 recurrent deletion syndrome. In: Adam MP, Ardinger HH, Pagon RA, et al, eds. GeneReviews [Internet]. December 8, 2016. Updated October 15, 2020. https://www.ncbi.nlm.nih.gov/books/NBK401562/ University of Washington, Seattle; 1993-2021. [PubMed]
- 68.Bollée G., Harambat J., Bensman A., Knebelmann B., Daudon M., Ceballos-Picot I. Adenine phosphoribosyltransferase deficiency. Clin J Am Soc Nephrol. 2012;7:1521–1527. doi: 10.2215/CJN.02320312. [DOI] [PubMed] [Google Scholar]
- 69.Runolfsdottir H.L., Palsson R., Agustsdottir I.M., Indridason O.S., Edvardsson V.O. Kidney disease in adenine phosphoribosyltransferase deficiency. Am J Kidney Dis. 2016;67:431–438. doi: 10.1053/j.ajkd.2015.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Else T., Greenberg S., Fishbein L. Hereditary paraganglioma-pheochromocytoma syndromes. In: Adam MP, Ardinger HH, Pagon RA, et al, eds. GeneReviews [Internet]. May 21, 2008. Updated October 4, 2018. https://www.ncbi.nlm.nih.gov/books/NBK1548/ University of Washington, Seattle; 1993-2021.
- 71.Rednam S.P., Erez A., Druker H., et al. Von Hippel–Lindau and hereditary pheochromocytoma/paraganglioma syndromes: clinical features, genetics, and surveillance recommendations in childhood. Clin Cancer Res. 2017;23:e68–e75. doi: 10.1158/1078-0432.CCR-17-0547. [DOI] [PubMed] [Google Scholar]
- 72.Bali D.S., El-Gharbawy A., Austin S., Pendyal S., Kishnani P.S. Glycogen storage disease type I. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews [Internet]. April 19, 2006. Updated October 14, 2021. https://www.ncbi.nlm.nih.gov/books/NBK1312/ University of Washington, Seattle; 1993-2021. [PubMed]
- 73.Froissart R., Piraud M., Boudjemline A.M., et al. Glucose-6-phosphatase deficiency. Orphanet J Rare Dis. 2011;6:27. doi: 10.1186/1750-1172-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chebib F.T., Torres V.E. Assessing risk of rapid progression in autosomal dominant polycystic kidney disease and special considerations for disease-modifying therapy. Am J Kidney Dis. 2021;78:282–292. doi: 10.1053/j.ajkd.2020.12.020. [DOI] [PubMed] [Google Scholar]
- 75.Lanktree M.B., Iliuta I.A., Haghighi A., Song X., Pei Y. Evolving role of genetic testing for the clinical management of autosomal dominant polycystic kidney disease. Nephrol Dial Transplant. 2019;34:1453–1460. doi: 10.1093/ndt/gfy261. [DOI] [PubMed] [Google Scholar]
- 76.Erickson K.F., Chertow G.M., Goldhaber-Fiebert J.D. Cost-effectiveness of tolvaptan in autosomal dominant polycystic kidney disease. Ann Intern Med. 2013;159:382–389. doi: 10.7326/0003-4819-159-6-201309170-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Torres V.E., Chapman A.B., Devuyst O., et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med. 2012;367:2407–2418. doi: 10.1056/NEJMoa1205511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pergola P., Appel G., Awad A., et al. Initial results from a phase 2 trial of the safety and efficacy of bardoxolone methyl in patients with autosomal dominant polycystic kidney disease and IgA nephropathy. Nephrol Dial Transplant. 2018;33:i635. [Google Scholar]
- 79.Sen E.S., Dean P., Yarram-Smith L., et al. Clinical genetic testing using a custom-designed steroid-resistant nephrotic syndrome gene panel: analysis and recommendations. J Med Genet. 2017;54:795–804. doi: 10.1136/jmedgenet-2017-104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raina R., Krishnappa V., Blaha T., et al. Atypical hemolytic-uremic syndrome: an update on pathophysiology, diagnosis, and treatment. Ther Apher Dial. 2019;23:4–21. doi: 10.1111/1744-9987.12763. [DOI] [PubMed] [Google Scholar]
- 81.Brocklebank V., Wood K.M., Kavanagh D. Thrombotic microangiopathy and the kidney. Clin J Am Soc Nephrol. 2018;13:300–317. doi: 10.2215/CJN.00620117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Richards S., Aziz N., Bale S., et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knoers N., Antignac C., Bergmann C., et al. ERA-EDTA Working Group for Inherited Kidney Diseases (WGIKD); the Molecular Diagnostics Taskforce of the European Rare Kidney Disease Reference Network (ERKNet). Genetic testing in the diagnosis of chronic kidney disease: recommendations for clinical practice. Nephrol Dial Transplant. 2022;37:239–254. doi: 10.1093/ndt/gfab218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu P., Meng L., Normand E.A., et al. Reanalysis of clinical exome sequencing data. N Engl J Med. 2019;380:2478–2480. doi: 10.1056/NEJMc1812033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jayasinghe K., Stark Z., Kerr P.G., et al. Clinical impact of genomic testing in patients with suspected monogenic kidney disease. Genet Med. 2021;23:183–191. doi: 10.1038/s41436-020-00963-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Snoek R., van Jaarsveld R.H., Nguyen T.Q., et al. Genetics-first approach improves diagnostics of ESKD patients <50 years old. Nephrol Dial Transplant. 2022;37:349–357. doi: 10.1093/ndt/gfaa363. [DOI] [PubMed] [Google Scholar]
- 87.Pinto E Vairo F., Kemppainen J.L., Lieske J.C., Harris P.C., Hogan M.C. Establishing a nephrology genetic clinic. Kidney Int. 2021;100:254–259. doi: 10.1016/j.kint.2021.05.008. [DOI] [PubMed] [Google Scholar]
- 88.Invitae Corporation Invitae progressive renal disease panel. https://www.invitae.com/en/physician/tests/75000/
- 89.Natera Inc. Renasight kidney gene panel. https://www.natera.com/organ-health/renasight-genetic-testing/
- 90.Centogene N.V. Nephrology. https://www.centogene.com/diagnostics/ngspanels/nephrology.html
- 91.NephCure Kidney International Inc. Avenues for genetic testing. https://nephcure.org/livingwithkidneydisease/treatment-options/avenues-for-genetic-testing/
- 92.National Society of Genetic Counselors Find a genetic counselor. https://findageneticcounselor.nsgc.org/?reload=timezone
- 93.Cornec-Le Gall E., Torres V.E., Harris P.C. Genetic complexity of autosomal dominant polycystic kidney and liver diseases. J Am Soc Nephrol. 2018;29:13–23. doi: 10.1681/ASN.2017050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cornec-Le Gall E., Chebib F.T., Madsen C.D., et al. HALT Progression of Polycystic Kidney Disease Group Investigators. The value of genetic testing in polycystic kidney diseases illustrated by a family with PKD2 and COL4A1 mutations. Am J Kidney Dis. 2018;72:302–308. doi: 10.1053/j.ajkd.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]