Abstract
Smad ubiquitination regulatory factor 2 (Smurf2) plays various roles in cancer progression. However, the correlation between Smurf2 and clinical outcomes has not been determined in patients diagnosed with colorectal cancer and colorectal liver metastases. We analyzed 66 patients with colorectal cancer who developed liver metastases. Smurf2 expression was assessed using immunohistochemical analysis of primary and metastatic liver tumors. High Smurf2 expression in both primary and metastatic tumors was significantly associated with longer overall survival time and time to surgical failure. Multivariate analyses revealed that low Smurf2 expression in primary tumors was an independent predictor of poor prognosis. In vitro experiments using colon cancer cell lines demonstrated that short interfering RNA knockdown of Smurf2 increased cell migration and tumor sphere formation. Western blot analyses revealed that Smurf2 knockdown increased the protein expression of epithelial cell adhesion molecule (EpCAM). Thus, in summary, high Smurf2 expression in cancer cells was found to be an independent predictor of better prognosis in patients with primary colorectal cancer and consequent liver metastases. The tumor-suppressive role of Smurf2 was found to be associated with cell migration and EpCAM expression; hence, Smurf2 can be considered a positive biomarker of cancer stem cell-like properties.
Subject terms: Cancer, Metastasis
Introduction
Colorectal cancer (CRC) is a leading cause of cancer-related death1. Distant metastasis is a strong factor marking poor prognosis in patients with CRC. Previous studies have demonstrated that a total of 25–30% of CRC patients were also diagnosed with liver metastasis2–4. Therefore, an accurate preoperative diagnosis and an appropriate treatment plan are essential to obtain better prognoses in patients with CRC.
Protein degradation mediated by the ubiquitin/proteasome system (UPS) controls various biological functions and is critical for maintaining homeostasis5. Additionally, the UPS plays important roles in several cancers5,6. Smad ubiquitination regulatory factor 2 (Smurf2) is a member of the homologues to E6-AP carboxyl terminus (HECT) family of E3 ubiquitin ligases. This E3 ligase was initially identified as a regulatory factor of transforming growth factor beta (TGF-β) signal transduction7. Since the identification of Smurf2, its various roles have been explored not only as a regulator of TGF-β but also as a direct regulator of the cell cycle and cancer development8. Although considerable evidence demonstrating the involvement of Smurf2 in cancer biology has been accumulated, the role of Smurf2 remains controversial. A previous study demonstrated the tumor-promoting role of Smurf29,10, whereas other studies have demonstrated the tumor-suppressive role of Smurf211–14. Accordingly, the role of Smurf2 in cancer biology seems to be “context-dependent”. Some studies have explored the tumor-suppressive role of Smurf2 and the corresponding mechanisms in the progression of CRC using a cell line and/or an animal model. Gao et al. demonstrated the tumor-suppressive role of Smurf2 using in vitro and in vivo studies. Additionally, the tumor-suppressive role of Smurf2 was reported to be associated with the degradation of YY1 and downregulation of SENP1/c-myc15. Yu et al. demonstrated that Smurf2 suppressed CRC cell proliferation and tumorigenesis through an interaction with sirtuin 1: Smurf2 depletion leads to sirtuin 1 upregulation and induces the tumor formation and growth of CRC in vitro and in vivo12. Li et al. demonstrated that Smurf2 reduces aerobic glycolysis and cell proliferation by promoting ChREBP ubiquitination and degradation via the proteasome pathway in CRC cells16. Pu et al. demonstrated that Smurf2 inhibited cell growth and metastasis in colon cancer and that Smurf2 regulation was involved in the anticancer effects of schisandrin B in both in vitro and in vivo models17. In contrast to the above studies that demonstrated the various mechanisms of Smurf2 in CRC using cell lines and/or animal models, one study demonstrated that Smurf2 expression was upregulated in CRC specimens and revealed that high Smurf2 expression was associated with impaired overall survival in microsatellite stable CRC, but not in microsatellite instable CRC18. Although this study assessed the association between Smurf2 expression and clinical outcomes, it only assessed primary CRC. No studies have yet comprehensively evaluated the role of Smurf2 in primary CRC and the corresponding liver metastases. Therefore, the aim of the current study was to evaluate the expression of Smurf2 in CRC and the corresponding colorectal liver metastases as well as its correlation with patient clinical outcomes. Furthermore, the molecular mechanisms underlying the clinical results reported herein were explored.
Methods
Patients and human tissue samples
CRC and corresponding liver metastasis tissues were obtained from 66 consecutive patients who underwent surgical resection for primary CRC and liver metastases at Chiba University Hospital (Chiba, Japan) between January 2005 and December 2014. Patients who underwent two-stage hepatectomy and primary tumor resection at other hospitals were excluded. Patients with synchronous liver metastases initially underwent primary tumor resection. As a control group, tissues were obtained from 60 consecutive patients with stage II or III CRC who did not develop distant metastases. These patients underwent surgical resection of the primary CRC between 2012 and 2014. The ethics committee of Chiba University, Graduate School of Medicine (Chiba, Japan) approved the protocol of the present study (approval number: 2405). The study protocol conformed to the provisions of the Declaration of Helsinki. Written informed consent was obtained from each patient before surgery. The clinical samples and their background information were obtained from the same database as in our previous study19.
Indication criteria for surgical resection of metastatic tumors
The indications for resection of colorectal metastases were as follows: (1) curative resection of the primary tumor is possible or has already been performed; (2) curative resection of metastases is possible; (3) preservation of the physiological functions of the remaining tissue is possible (e.g., ≥ 40% of the total liver volume). The criteria are same with our previous study19.
Immunohistochemistry (IHC)
Formalin-fixed paraffin-embedded tissue samples were cut into 4-µm-thick slices and deparaffinized. In the IHC for Smurf2, antigen retrieval was performed by microwaving in citric acid buffer (0.01 mol/L, pH 6.0) for 25 min. Subsequently, endogenous peroxidase activity was blocked using 3% hydrogen peroxide in methanol for 15 min. Non-specific proteins were blocked using 5% bovine serum albumin for 10 min. Following protein blocking, the slides were incubated at 4 °C overnight with the anti-Smurf2 monoclonal antibody (1:50 dilution; cat. no. sc-393848; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Counterstaining was performed with hematoxylin before dehydration, penetration, and mounting. The protocol of IHC is described in our previous study19.
Immunohistochemical evaluation of Smurf2
Using an inverted microscope (BX40; Olympus Corporation, Tokyo, Japan), the expression levels of Smurf2 were evaluated independently by two investigators accompanied by a pathologist, all of whom were blinded to any clinical information. The intensity of tumor cell staining was scored as follows: 0, negative staining; 1, weak staining; 2, moderate staining; and 3, strong staining. Patients with a score of 0 or 1 were classified as having low expression, and those with scores of 2 or 3 were classified as having high expression. The immunohistochemical evaluations were performed after establishing an inter-observer consensus using samples from preliminary experiments.
Human colon cancer cell lines and culture conditions
The human colon cancer cell line DLD-1 and the human colon cancer lymph node metastasis cell line SW620 were purchased from the American Type Culture Collection (USA). The DLD-1 cell line was cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The SW620 cell line was cultured in Leibovitz’s 15 medium (Gibco) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.).
Western blot analysis
Proteins were extracted from the cultured cells using RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA). Each protein sample was lysed in a buffer (Laemmli Sample Buffer; Bio-Rad Laboratories, Inc., Richmond, CA, USA) containing 5% 2-mercaptoethanol and incubated at 97 °C for 10 min. After measuring the protein concentration of each sample using the Pierce™ BCA Protein Assay kit (Thermo Fisher Scientific, Inc.), 10 µg of protein was separated by electrophoresis on 5-12.5% XV PANTERA Gels (DRC, Tokyo, Japan) and transferred onto a membrane (PerkinElmer, Inc., Waltham, MA, USA). The membranes were blocked in 5% skim milk diluted with 0.1% Tris-buffered saline with Tween-20 at room temperature (15–25 °C) for 60 min. The membranes were then incubated at 4 °C overnight with the following primary antibodies: anti-Smurf2 polyclonal antibody (1:2,000 dilution; cat. no. ab94483; Abcam plc), anti-EpCAM polyclonal antibody (1:1,000 dilution; cat. no. HPA026761; Sigma-Aldrich) and anti-β-actin monoclonal antibody (1:5,000 dilution; cat. no. 5125; Cell Signaling Technology Inc., Beverly, MA, USA). Subsequently, the membranes were incubated in blocking buffer at room temperature (15–25 °C) for 60 min with anti-rabbit IgG horseradish peroxidase secondary antibody (1:2,000 dilution; cat. No. Sc-2301; Santa Cruz Biotechnology). The membranes were then incubated with an enhanced chemiluminescence detection reagent (Chemi-Lumi One Ultra; Nacalai Tesque, Inc., Kyoto, Japan) and developed with an LAS-4000UV mini luminescent image analyzer (Fujifilm, Tokyo, Japan). Band intensities were quantified by densitometric analysis using ImageJ software version 1.51 (National Institutes of Health, Bethesda, MD, USA) and were used to calculate the relative protein level normalized to β-actin. The protocol of western blot is described in our previous study19.
Short interfering RNA (siRNA) transfection
The double-stranded siRNAs used to knock down Smurf2 expression were as follows: siSMURF2, Stealth siRNA (cat. no. HSS127687 and HSS127688; Thermo Fisher Scientific, Inc.). Negative control siRNA (AllStars negative control siRNA; Qiagen) was used as the control for all siRNA experiments. These siRNAs (final concentration, 2 nmol/L) were transfected into DLD-1 and SW620 cells using Lipofectamine™ RNAiMAX transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) as previously described19. These cells were used for subsequent assays, 24 h post-transfection.
Wound-healing assay
A wound-healing assay was conducted to assess cell migration ability using Culture-Insert 2 well in a 35-mm μ-dish (ibidi GmbH, Martinsried, Germany). DLD-1 and SW620 cells transfected with siSmurf2s or siControl were seeded at a density of 3.0 × 104 cells/well and 1.0 × 105 cells/well, respectively, and were cultured for 24 h. Cells were pre-treated with 5 µg/mL mitomycin C at 2 h prior to removal of the Culture-Insert. After removing the Culture-Insert, the wells were filled with the appropriate medium and allowed to heal for 24 h for DLD-1 and for 72 h for SW620. The open wound area was measured under a microscope at 50 × magnification.
Cell proliferation assay
Cell proliferation was examined using CCK-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) as previously described19. The DLD-1 and SW620 cells transfected with siSmurf2s or siControl were seeded at a density of 1,000 cells/well in 96-well plates. CCK-8 (10 μL/well) solution was added to measure cell viability at 0, 24, 48, 72, and 96 h. After 2 h of incubation, the absorbance of each well was measured at 450 nm.
Cell invasion assay
Cell Biolabs CytoSelect™ 24-well cell invasion assay kits (Cell Biolabs, San Diego, CA, USA), utilizing basement membrane-coated inserts, were used as previously described19. Briefly, DLD-1 and SW620 cells transfected with siSmurf2s or siControl were suspended in serum-free medium. Following overnight starvation, the cells were seeded at a density of 1.0 × 105 cells/well in the upper chamber and incubated with the medium in the lower chamber for 48 h. The invasive cells passing through the basement membrane layer were stained, and the absorbance of each well was measured at 560 nm after extraction.
Sphere formation assay
Tumor sphere formation assays were carried out as previously described19 with minor modifications. The DLD‐1 and SW620 cells transfected with siRNA or siControl were seeded in 96‐well ultra‐low attachment plates (Corning Inc., New York, NY, USA) at 15 cells/well and cultured in a sphere medium comprising DMEM‐F12 (1:1) medium (Gibco, Grand Island, NY, USA) containing 20 ng/mL human epidermal growth factor (BD biosciences, San Jose, CA, USA), 20 ng/mL human basic fibroblast growth factor (Invitrogen; Thermo Fisher Scientific, Inc.), 1% B27 Supplement (Invitrogen), 1% N2 Supplement (Invitrogen), 100 μM beta-ME (Bio-Rad Laboratories, Inc., Richmond, CA, USA), 1% Non-essential AA (Invitrogen), and 1% penicillin/streptomycin. After incubation for 7 d at 37 °C, the cells were evaluated and the number of spheres was counted using an inverted microscope (Axio Observer Z1; Carl Zeiss, Oberkochen, Germany). DLD1 with a diameter larger than 50 μm and SW620 with a diameter larger than 25 μm were counted as spheres. The sphere formation rate was calculated as scored sphere number/total plating cells.
Statistical analysis
The correlations between Smurf2 staining and patient characteristics were evaluated using the χ2 test, Student’s t-test, or Mann–Whitney U-test, as appropriate. Survival rates were calculated using the Kaplan–Meier analyses and assessed using the log-rank test. Survival data were evaluated using the univariate and multivariate Cox proportional regression analyses. When analyzing the correlation between Smurf2 expression in the primary tumor and long-term outcomes, overall survival (OS), time to liver metastases, and time to surgical failure (TSF) were calculated from the date of primary tumor resection. TSF is defined as the interval between the time of initial surgery to the time of the first unresectable recurrence or death20. When analyzing the correlation between Smurf2 expression in liver metastases and the long-term outcome, OS, disease-free survival, and TSF were calculated from the date of initial hepatectomy. The in vitro experiments were performed at least three times independently, and data were analyzed using the Welch’s t-test and multivariate analysis of variance. Statistical significance was set at P < 0.05. Values are expressed as the mean ± standard error of the mean. The above series of statistical analyses were performed using JMP® PRO 13 software (SAS Institute Inc., Cary, NC, USA).
Results
High Smurf2 expression in the primary tumor is associated with better prognosis
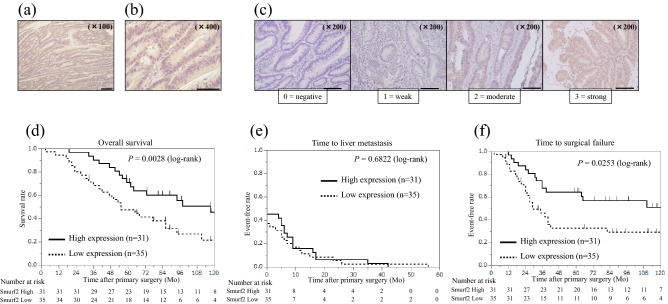
Smurf2 protein expression was examined in primary tumors using IHC. Smurf2 was predominantly expressed in the cytoplasm of cancer cells in primary tumors (Fig. 1a,b). Furthermore, Smurf2 expression was evaluated based on the scoring system (Fig. 1c). In primary tumors, high Smurf2 expression was observed in 31 patients (47.0%), whereas low Smurf2 expression was observed in 35 patients (53.0%). Smurf2 expression profiles in primary tumors and clinicopathological features are shown in Table 1. Smurf2 expression was not associated with any clinicopathological features assessed in the present study. The Kaplan–Meier analysis showed that patients with high Smurf2 expression had a significantly better OS time and TSF after primary surgery than that of patients with low Smurf2 expression (P = 0.0028 and 0.0253, respectively; Fig. 1d,f). No significant difference in the time to liver metastasis was found between patients showing high and low Smurf2 expression (Fig. 1e).
Figure 1.
Smurf2 expression in primary CRC and the long-term outcome based on the Smurf2 expression. Immunohistochemistry analysis for Smad ubiquitination regulatory factor 2 (Smurf 2) expression in (a, b) primary colorectal cancer (CRC). Scale bar = 200 µm (a) and 100 µm (b). (c) Representative images of each score evaluating Smurf2 expression in primary CRC. Scale bar = 200 µm (c). The Kaplan–Meier analysis for (d) overall survival, (e) time to liver metastasis, and (f) time to surgical failure based on the Smad ubiquitination regulatory factor 2 (Smurf 2) expression in primary CRC.
Table 1.
Clinicopathologic features of patients diagnosed with colorectal cancer showing high and low Smad ubiquitination regulatory factor 2 (Smurf2) expression.
| Smurf2 expression | P value | ||
|---|---|---|---|
| High (n = 31) | Low (n = 35) | ||
|
Age at primary surgery (years), Median (range) |
69 (50–82) | 65 (46–81) | 0.0952 |
| Gender, Male/Female | 21/10 | 23/12 | 1.0000 |
| Site of tumor, Colon/Rectum | 19/12 | 21/14 | 1.0000 |
| Site of tumor, Right/Left/Rectum | 6/13/12 | 13/8/14 | 0.1577 |
| Degree of differentiation, tub, pap/por, muc | 30/1 | 32/3 | 0.6161 |
| Ly, 0–1/2–3 | 27/4 | 28/7 | 0.5207 |
| V, 0–1/2–3 | 16/15 | 15/20 | 0.6217 |
| T stage (8th edition), 1/2/3/4 | 1/0/17/13 | 0/2/17/16 | 0.5423 |
| Lymph node metastasis, + / − | 17/14 | 22/13 | 0.6177 |
| CEA, median (range) | 8 (1.4–5610) | 6.9 (0.7–5700) | 0.8786 |
| CA19-9, median (range) | 12 (0.1–702) | 33.3 (0–16,200) | 0.1561 |
| KRAS mutation, Wild/Mutant | 9/4 | 9/7 | 0.7021 |
| Adjuvant chemotherapy, + / − | 5/26 | 5/30 | 1.0000 |
| Interval to liver metastasis (months), mean ± SD | 6.0 ± 1.9 | 5.1 ± 1.8 | 0.7820 |
| Timing of metastasis, Synchronous/metachronous | 17/14 | 22/13 | 0.6177 |
| Number of liver metastatic tumors, mean ± SD | 2.9 ± 0.5 | 3.0 ± 0.5 | 0.8808 |
| Number of liver metastatic tumors, Solitary/Multiple | 10/21 | 13/22 | 0.7973 |
| Size of liver metastatic tumors (cm), mean ± SD | 3.2 ± 0.4 | 4.3 ± 0.4 | 0.0877 |
| Site of liver metastasis, Unilateral/Bilateral | 17/14 | 20/15 | 1.0000 |
| Hepatectomy, Minor/Major | 28/3 | 28/7 | 0.3138 |
| H factor, H1/H2/H3 | 24/7/0 | 19/15/1 | 0.0914 |
| Extrahepatic metastases, + / − | 6/25 | 6/29 | 1.0000 |
CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19–9; KRAS, Kirsten rat sarcoma viral oncogene.
In the univariate analysis, the primary tumor site (colon vs. rectum, right-sided colon vs. left-sided colon), lymph node metastasis, surgical margin, H factor, and Smurf2 expression were correlated with OS. Among these, rectal cancer, positive lymph node metastasis, surgical margin (R1 and R2), and low Smurf2 expression were identified as independent poor prognosis factors (P = 0.0019, 0.0265, 0.0410, and 0.0370, respectively; Cox proportional hazards model; Table 2) in multivariate analyses. These data suggest that the higher expression of Smurf2 in primary tumors is associated with better prognosis.
Table 2.
Prognostic factors for overall survival (OS) in colorectal cancer.
| Prognostic factors | n | 5-year OS rate (%) | Univariate P value |
Multivariate HR (95% CI) |
P value |
|---|---|---|---|---|---|
| Age at primary surgery, < 65/ ≥ 65 | 44/22 | 54.6/60.8 | 0.7742 | ||
| Sex, Male/Female | 44/22 | 53.8/68.2 | 0.7217 | ||
| Site of primary tumor, Colon/Rectum | 40/26 | 74.7/34.6 | 0.0022 | 0.30 (0.129–0.647) | 0.0019 |
| Site of primary tumor, Right/Left | 19/47 | 78.6/51.1 | 0.0486 | 0.93 (0.339–2.541) | 0.8864 |
| CEA < 5/ ≥ 5 | 18/48 | 65.5/56.3 | 0.1631 | ||
| CA19-9 < 37/ ≥ 37 | 43/23 | 64.5/47.8 | 0.2476 | ||
| Degree of differentiation, tub, pap/por, muc | 62/4 | 57.6/75.0 | 0.9649 | ||
| ly, 0–1/2–3 | 55/11 | 56.4/71.6 | 0.9228 | ||
| v, 0–1/2–3 | 31/35 | 71.0/47.5 | 0.1033 | ||
| T stage (8th edition), 1–3/4 | 37/29 | 64.9/50.6 | 0.2924 | ||
| Lymph node metastasis, − / + | 27/39 | 69.7/51.3 | 0.0112 | 0.43 (0.195–0.909) | 0.0265 |
| KRAS mutation, Wild/Mutant | 11/18 | 55.6/45.5 | 0.6377 | ||
| Adjuvant chemotherapy after primary surgery, − / + | 56/10 | 60.2/50.0 | 0.3630 | ||
|
Timing of liver metastasis, Synchronous/metachronous |
27/39 | 53.9/65.8 | 0.2591 | ||
| Metastasis other than liver, − / + | 54/12 | 62.5/41.7 | 0.1181 | ||
| Number of liver metastasis, Solitary/Multiple | 23/43 | 69.1/53.5 | 0.1929 | ||
| Size of largest tumor (cm), < 5/ ≥ 5 | 53/13 | 65.6/46.2 | 0.3057 | ||
| Hepatectomy, Minor/Major | 56/10 | 62.6/40.0 | 0.0687 | ||
| Surgical margin, R0/R1,R2 | 35/31 | 67.9/48.4 | 0.0287 | 0.49 (0.241–0.972) | 0.0410 |
| H factor, H1/H2-H3 | 43/23 | 71.6/34.8 | 0.0171 | 0.70 (0.327–1.548) | 0.3713 |
|
Expression of Smurf 2 in primary tumor, High/Low |
31/35 | 71.0/47.6 | 0.0265 | 0.47 (0.232–0.957) | 0.0370 |
CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19–9; KRAS, Kirsten rat sarcoma viral oncogene.
High Smurf2 expression in liver metastases is associated with better prognosis
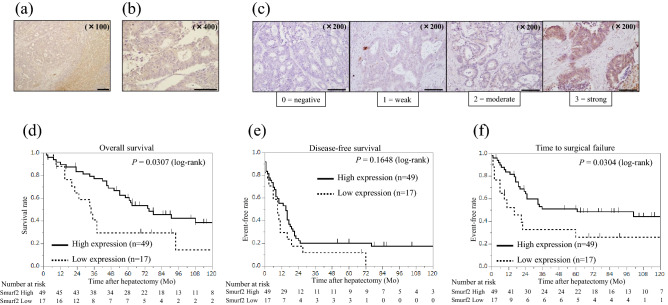
Smurf2 protein expression was examined in metastatic tumors in the liver using IHC. Smurf2 was predominantly expressed in the cytoplasm of cancer cells in liver metastases (Fig. 2a,b ). Further, Smurf2 expression was evaluated based on the scoring system (Fig. 2c). Clinicopathological features were compared between high and low levels of Smurf2 expression in liver metastases. As shown in Table 3, high Smurf2 expression was observed in 49 patients (74.2%), whereas low Smurf2 expression was observed in 17 patients (25.8%). Smurf2 expression was not associated with any clinicopathological features assessed in the present study. The Kaplan–Meier analysis showed that patients with high Smurf2 expression in liver metastases had significantly better OS time and TSF after hepatectomy than that of patients with low Smurf2 expression (P = 0.0307 and 0.0304, respectively; Fig. 2d,f). However, no significant difference in the disease-free survival (DFS) time after hepatectomy was found between patients showing high and low Smurf2 expression (Fig. 2e). These data suggest that higher Smurf2 expression in liver metastases is associated with better prognosis.
Figure 2.
Smurf2 expression in colorectal liver metastases and the long-term outcome based on Smurf2 expression. Immunohistochemistry analysis for Smad ubiquitination regulatory factor 2 (Smurf 2) expression in (a, b) colorectal liver metastases. Scale bar = 200 µm (a) and 100 µm (b). (c) Representative images of each score evaluating Smurf2 expression in colorectal liver metastases. Scale bar = 200 µm (c). The Kaplan–Meier analysis for (d) overall survival, (e) disease-free survival, and (f) time to surgical failure based on the Smad ubiquitination regulatory factor 2 (Smurf 2) expression in metastatic liver tumors.
Table 3.
Clinicopathologic features of patients diagnosed with colorectal liver metastases showing high and low Smad ubiquitination regulatory factor 2 (Smurf2) expression.
| Smurf2 expression | P value | ||
|---|---|---|---|
| High (n = 49) | Low (n = 17) | ||
|
Age at primary surgery (years), Median (range) |
68 (46–84) | 69 (50–79) | 0.9356 |
| Gender, Male/Female | 32/17 | 12/5 | 0.7726 |
| Site of tumor, Colon/Rectum | 31/18 | 9/8 | 0.5668 |
| Site of tumor, Right/Left/Rectum | 15/16/18 | 4/5/8 | 0.7397 |
|
Degree of differentiation, tub, pap/por, muc |
47/2 | 15/2 | 0.2714 |
| Ly, 0–1/2–3 | 40 / 9 | 15 / 2 | 0.7145 |
| V, 0–1/2–3 | 24/25 | 7/10 | 0.7786 |
| T stage (8th edition), 1/2/3/4 | 0/1/25/23 | 1/1/9/6 | 0.2342 |
| Lymph node metastasis, + / − | 29/20 | 10/7 | 1.0000 |
|
CEA before hepatectomy, median (range) |
50.7 ± 170.0 | 947.2 ± 288.6 | 0.0094 |
| CA19-9, median (range) | 129.9 ± 497.7 | 1825.8 ± 845.0 | 0.0886 |
| KRAS mutation, Wild/Mutant | 9/4 | 9/7 | 0.7021 |
| Adjuvant chemotherapy, + / − | 36/13 | 13/4 | 1.0000 |
| Interval to liver metastasis (months), mean ± SD | 5.6 ± 1.5 | 5.3 ± 2.6 | 0.9106 |
|
Timing of metastasis, Synchronous/metachronous |
29/20 | 10/7 | 1.0000 |
| Number of liver metastatic tumors, mean ± SD | 3.0 ± 0.4 | 2.6 ± 0.7 | 0.6250 |
| Number of liver metastatic tumors, Solitary/Multiple | 17/32 | 6/11 | 1.0000 |
| Size of liver metastatic tumors (cm), mean ± SD | 3.6 ± 0.4 | 4.1 ± 0.6 | 0.4674 |
|
Site of liver metastasis, Unilateral/Bilateral |
17/14 | 20/15 | 1.0000 |
| Hepatectomy, Minor/Major | 45/4 | 11/6 | 0.0143 |
| H factor, H1/H2/H3 | 33/15/1 | 10/7/0 | 0.6689 |
| Extrahepatic metastases, + / − | 9/40 | 3/14 | 1.0000 |
| Resection margin, R0/R1/R2 | 27/14/8 | 8/6/3 | 0.8372 |
| Adjuvant chemotherapy after hepatectomy, + / − | 36/13 | 13/4 | 1.0000 |
| Recurrence after hepatectomy (all organs), + / − | 38/11 | 16/1 | 0.1632 |
|
Intrahepatic recurrence after hepatectomy, + / − Number of tumors, mean ± SD |
26/23 2.4 ± 0.5 |
10/7 3.8 ± 0.8 |
0.78100.1185 |
| Recurrence in multiple organs, + / − | 6/43 | 15/2 | 1.0000 |
| Repeat resection (all organs), + / − | 24/14 | 6/10 | 0.1332 |
| Repeat hepatectomy, + / − | 13/13 | 2/8 | 0.1422 |
CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19–9; KRAS, Kirsten rat sarcoma viral oncogene.
Differential expression of Smurf2 in patients with liver metastases and patients with stage II or III CRC
When comparing Smurf2 expression in patients with liver metastases and patients with stage II or III CRC (i.e., without any distant metastases), high Smurf2 expression was observed in 47.0% of patients with liver metastases and 73.3% of patients with stage II or III CRC. Smurf2 expression was significantly higher in patients with stage II or III disease than in patients with liver metastases (P = 0.0035) (Supplementary Fig. 1). These data suggest that expression of Smurf2 in primary tumors is associated with CRC progression.
Acceleration of tumor cell migration through Smurf2 knockdown
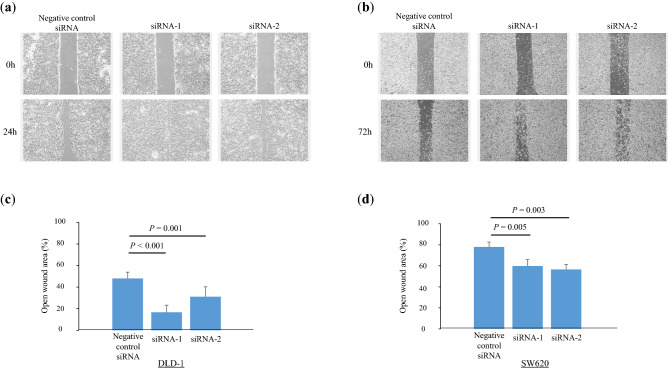
The clinical data indicated that Smurf2 might play a tumor-suppressive role in CRC; therefore, in vitro experiments were performed to elucidate the molecular mechanisms by which Smurf2 regulates the behavior of CRC cells. The human colon cancer cell line DLD-1 and the human colon cancer lymph node metastasis cell line SW620 were used for these experiments. To assess the effects of Smurf2 on cell migration, we performed a wound-healing assay following the knockdown of Smurf2 using siRNAs. The wound-healing assay revealed that the knockdown of Smurf2 significantly prompted wound healing (i.e., increased cell migration) (DLD-1 siRNA1 P < 0.001, siRNA2 P = 0.001, SW620 siRNA1 P = 0.005, siRNA2 P = 0.003) (Fig. 3). These data suggest that the knockdown of Smurf2 expression accelerates tumor cell migration.
Figure 3.
Wound-healing assay in colorectal cancer cells. (a, b) Representative images of wound healing in (a) human colon cancer cell line DLD-1 cells and (b) human colon cancer lymph node metastasis cell line SW620 cells treated with siControl and siSmurf2. (c, d) Open wound area was measured under a microscope at 50 × magnification in (c) human colon cancer cell line DLD-1 cells and (d) human colon cancer lymph node metastasis cell line SW620 cells.
Knockdown of Smurf2 expression did not alter tumor cell proliferation
To assess the effects of Smurf2 on tumor cell proliferation in vitro, we performed CCK-8 assays following the knockdown of Smurf2 using siRNAs. The CCK-8 assay revealed that the knockdown of Smurf2 did not alter the proliferation of the DLD-1 and SW620 cells (Supplementary Fig. 2). These data suggest that the knockdown of Smurf2 expression does not alter tumor cell proliferation.
Knockdown of Smurf2 expression did not alter invasiveness of tumor cells
Cell invasiveness is an important property in the metastatic cascade of cancer; therefore, the effect of Smurf2 on cell invasiveness was assessed. Cell invasion assays revealed that the knockdown of Smurf2 did not alter the invasiveness of the DLD-1 and SW620 cells (Supplementary Fig. 3). These data suggest that the knockdown of Smurf2 expression does not alter the invasiveness of tumor cells.
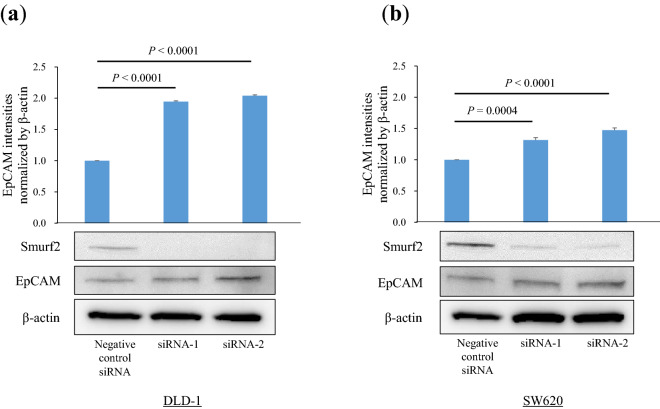
Knockdown of Smurf2 expression increased the EpCAM expression in colon cancer cells
Western blot analyses were performed to evaluate the effects of Smurf2 expression on the protein expression levels of EpCAM. Western blot analyses revealed that Smurf2 knockdown significantly increased EpCAM protein expression in the DLD-1 and SW620 cells (DLD-1 siRNA1 P < 0.0001, siRNA2 P < 0.0001, SW620 siRNA1 P = 0.0004, siRNA2 P < 0.0001) (Fig. 4). These data suggest that the knockdown of Smurf2 expression increased the EpCAM expression in colon cancer cells.
Figure 4.
Western blot analyses for epithelial cell adhesion molecule (EpCAM) protein expression. Western blot analysis was performed to evaluate EpCAM protein expression in (a) human colon cancer cell line DLD-1 cells and (b) human colon cancer lymph node metastasis cell line SW620 cells treated with siControl and siSmurf2.
Knockdown of Smurf2 expression increased sphere formation
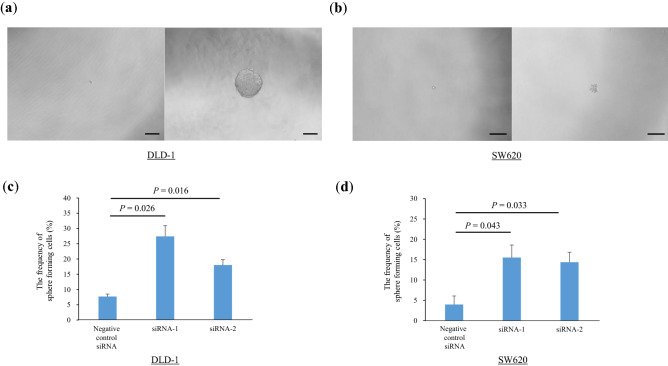
To assess the effect of Smurf2 on the cancer stem cell-like properties, we performed a sphere formation assay. The sphere formation rate was significantly higher in siSmurf2 transfected cells than in the negative control siRNA-transfected cells (DLD-1 siRNA1 P = 0.026, siRNA2 P = 0.016, SW620 siRNA1 P = 0.043, siRNA2 P = 0.033) (Fig. 5). These data suggest that the knockdown of Smurf2 expression promotes sphere formation.
Figure 5.
Sphere formation assay in colorectal cancer cells. (a, b) Representative pictures of sphere formation in DLD-1 (a) and SW620 (b) cells treated with siControl and siSmurf2. (c, d) The sphere formation rate were evaluated in DLD-1 (c) and SW620 (d) cells treated with siControl and siSmurf2.
Discussion
To the best of our knowledge, the present study is the first to demonstrate the prognostic impact of Smurf2 expression on clinical outcomes in patients with CRC and liver metastasis. Our results show that high Smurf2 expression in both primary CRC tumors and corresponding liver metastases was significantly associated with a better prognosis in patients with CRC and liver metastases. A novel finding in the present study is that Smurf2 modulates EpCAM expression. Using in vitro experiments, we demonstrated that the knockdown of Smurf2 expression accelerated cell migration, promoted sphere formation, and increased the expression of EpCAM, a cancer stem cell marker. These results indicate that Smurf2 acts as a negative regulator in CRC development and the metastatic cascade. These underlying mechanisms might be the cause of the early and unresectable recurrence of CRC, which ultimately leads to shorter survival times in patients with low Smurf2 expression.
EpCAM is a type-1 transmembrane glycoprotein that was initially considered a tumor antigen in colorectal carcinoma. Since its first description in 197921, several studies have demonstrated the prognostic value of EpCAM in several cancers22–27. EpCAM is overexpressed in various cancers and the corresponding metastatic lesions28. Although the prognostic impact of EpCAM differs depending on the type of cancer, Seeber et al. demonstrated that high EpCAM expression in colon cancer cells was associated with aggressive tumor biology, causing multiple recurrences in the early phase following surgery in patients with CRC26. Our clinical data demonstrates that the prognosis was significantly worse in patients with low Smurf2 expression in both primary tumors and metastatic tumors compared to the prognosis of patients with high Smurf2 expression. Although our data may be too limited to draw a definitive conclusion, it is plausible that a decrease in Smurf2 expression could cause high EpCAM expression and consequently result in poor prognosis in patients with low Smurf2 expression. Additionally, our clinical data demonstrates that the TSF was significantly shorter in patients with low Smurf2 expression in both primary tumors and metastatic tumors. When considering the treatment strategy for CRC liver metastasis (CRLM), repeat resection for distant metastases, including liver and lung metastases, has been demonstrated to yield a survival outcome comparable to that of the initial hepatectomy29–31. Therefore, conventional recurrence-free survival is not necessarily associated with OS. However, TSF has been reported to be well correlated with OS in patients with CRLM20. Given these findings, low Smurf2 expression could be a cause of early and unresectable recurrence (i.e., shorter TSF), probably owing to the stem cell-like properties of cancer cells, and could be a good predictor of poor prognosis in patients. The mechanism(s) underlying shorter TSF in patients with low Smurf2 expression may include the involvement of Smurf2 in the stem cell-like properties of cancer cells. EpCAM expression has been associated with the stem cell-like properties of cancer cells22. Yamashita et al. demonstrated that there are distinctive EpCAM-positive cancer cell subpopulations in hepatocellular carcinoma, which have malignant potential for self-renewal, de-dedifferentiation, tumor initiation, invasiveness, and establishment of distant metastases32,33. Furthermore, positive EpCAM expression has been significantly associated with the development of distant metastases and shorter disease-free interval in patients with breast cancer34. Meanwhile, the sphere formation ability is also an important indicator of the stem cell-like properties of cancer cells19. Supporting the EpCAM expression results, Smurf2 knockdown was associated with the increase in the sphere formation rate, indicating that a decrease in Smurf2 expression increased the stem cell-like properties of cancer cells. Collectively, these data suggest that Smurf2 was associated with stem cell-like properties of colon cancer cells. These factors might contribute to the early and unresectable recurrence of CRC. The detailed molecular mechanisms by which Smurf2 modulates EpCAM expression and sphere-formation ability remain unclear. These findings need to be explored in future studies to expand the clinical implications of Smurf2.
Smurf2 expression was significantly lower in CRC with liver metastases than in stage II or III CRC. As shown in this study, low Smurf2 expression was associated with accelerated cell migration and an increase in stem cell-like properties and CTCs, which indicates the aggressive behavior of cancer cells. Therefore, Smurf2 might be an important modulator in CRC progression from primary tumor development to metastatic cascade and even recurrence following initial hepatectomy.
Our in vitro data demonstrate that Smurf2 knockdown significantly accelerated cancer cell migration, but not cell proliferation or invasion; the reasons for the latter remain unclear. Although previous studies have indicated that Smurf2 plays some roles in cell proliferation and invasion, reports of whether Smurf2 acts as a tumor-suppressor or tumor-promoter are varied9,12–14,18. Differences in the experimental models, cell line, or cell culture conditions might drive the variations in these results. Nonetheless, our in vitro results concerning cell proliferation were in line with our clinical results, demonstrating that Smurf2 expression was not significantly associated with tumor number or tumor size. Regarding the discrepancy in the association of Smurf2 with cell migration and with cell invasion, it may be considered that Smurf2 is not a potent modulator of cell motility. We speculate that these results may be consistent with the clinical results indicating that Smurf2 was not associated with DFS. In summary, the contribution of Smurf2 towards cell proliferation and invasion remains unknown and requires clarification in future studies.
The present study has several limitations. First, data were collected retrospectively from the database of a single institution. Therefore, the sample size was small, and the patients’ backgrounds were heterogeneous. Consequently, several prognostic factors were not randomized in the analyses of long-term outcomes. Second, the effect of perioperative chemotherapy was not assessed, because various regimens were used during the study period and the indication for perioperative chemotherapy was decided by physician’s preference in some cases. Therefore, it was difficult to evaluate the effect of systemic chemotherapy accurately. Third, we examined the effects of Smurf2 using loss-of-function experiments. However, gain-of-function experiments and in vivo experiments should be performed to verify our data and elucidate the role of Smurf2 in CRC progression more accurately.
In conclusion, high Smurf2 expression in cancer cells is an independent predictor of better prognosis in patients with primary CRC and corresponding liver metastases. The tumor-suppressive role of Smurf2 was found to be associated with cell migration and EpCAM expression, which is associated with stem cell-like properties of cancer cells. Further studies are warranted to verify our clinical data in a larger cohort and to explore the detailed molecular mechanisms underlying the role of Smurf2, which could ultimately lead to the development of therapeutic targets.
Supplementary Information
Acknowledgements
This research was supported by the Japan Society for the Promotion of Science KAKENHI (Grant No. JP1809164 to N. Sakai).
Author contributions
N.S. and N.S. designed the study and acquired data. N.S., N.S., K.F., T.T., S.K., and S.T. analyzed and interpreted the data. N.S. drafted the manuscript. G.O., H.M., H.M., and M.O. performed the critical revision of the manuscript. All authors reviewed and approved the manuscript for publication.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nami Sato and Nozomu Sakai.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-09390-8.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Manfredi S, et al. Epidemiology and management of liver metastases from colorectal cancer. Ann. Surg. 2006;244:254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackl C, et al. Treatment of colorectal liver metastases in Germany: A ten-year population-based analysis of 5772 cases of primary colorectal adenocarcinoma. BMC Cancer. 2014;14:810. doi: 10.1186/1471-2407-14-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engstrand J, Nilsson H, Stromberg C, Jonas E, Freedman J. Colorectal cancer liver metastases—a population-based study on incidence, management and survival. BMC Cancer. 2018;18:78. doi: 10.1186/s12885-017-3925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 6.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: Multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J. Biol. Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 8.Osmundson EC, Ray D, Moore FE, Kiyokawa H. Smurf2 as a novel mitotic regulator: From the spindle assembly checkpoint to tumorigenesis. Cell Div. 2009;4:14. doi: 10.1186/1747-1028-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuchi M, et al. High-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Res. 2002;62:7162–7165. [PubMed] [Google Scholar]
- 10.David D, Jagadeeshan S, Hariharan R, Nair AS, Pillai RM. Smurf2 E3 ubiquitin ligase modulates proliferation and invasiveness of breast cancer cells in a CNKSR2 dependent manner. Cell Div. 2014;9:2. doi: 10.1186/1747-1028-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukunaga E, et al. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. J. Biol. Chem. 2008;283:35660–35667. doi: 10.1074/jbc.M710496200. [DOI] [PubMed] [Google Scholar]
- 12.Yu L, et al. Ubiquitination-mediated degradation of SIRT1 by SMURF2 suppresses CRC cell proliferation and tumorigenesis. Oncogene. 2020;39:4450–4464. doi: 10.1038/s41388-020-1298-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang WL, Zhang JH, Wu XZ, Yan T, Lv W. miR-15b promotes epithelial-mesenchymal transition by inhibiting SMURF2 in pancreatic cancer. Int. J. Oncol. 2015;47:1043–1053. doi: 10.3892/ijo.2015.3076. [DOI] [PubMed] [Google Scholar]
- 14.Chandhoke AS, et al. The ubiquitin ligase Smurf2 suppresses TGFbeta-induced epithelial-mesenchymal transition in a sumoylation-regulated manner. Cell Death Differ. 2016;23:876–888. doi: 10.1038/cdd.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Q, Wang S, Zhang Z. E3 ubiquitin ligase SMURF2 prevents colorectal cancer by reducing the stability of the YY1 protein and inhibiting the SENP1/c-myc axis. Gene Ther. 2021 doi: 10.1038/s41434-021-00289-z. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, et al. The ubiquitination ligase SMURF2 reduces aerobic glycolysis and colorectal cancer cell proliferation by promoting ChREBP ubiquitination and degradation. J. Biol. Chem. 2019;294:14745–14756. doi: 10.1074/jbc.RA119.007508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pu Z, et al. Schisandrin B Attenuates colitis-associated colorectal cancer through SIRT1 linked SMURF2 signaling. Am. J. Chin. Med. 2021 doi: 10.1142/S0192415X21500841. [DOI] [PubMed] [Google Scholar]
- 18.Klupp F, et al. E3 ubiquitin ligase Smurf2: A prognostic factor in microsatellite stable colorectal cancer. Cancer Manag. Res. 2019;11:1795–1803. doi: 10.2147/CMAR.S178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takagi Y, et al. High expression of Kruppel-like factor 5 is associated with poor prognosis in patients with colorectal cancer. Cancer Sci. 2020;111:2078–2092. doi: 10.1111/cas.14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oba M, et al. Discrepancy between recurrence-free survival and overall survival in patients with resectable colorectal liver metastases: A potential surrogate endpoint for time to surgical failure. Ann. Surg. Oncol. 2014;21:1817–1824. doi: 10.1245/s10434-014-3504-1. [DOI] [PubMed] [Google Scholar]
- 21.Herlyn M, Steplewski Z, Herlyn D, Koprowski H. Colorectal carcinoma-specific antigen: Detection by means of monoclonal antibodies. Proc. Natl. Acad. Sci. USA. 1979;76:1438–1442. doi: 10.1073/pnas.76.3.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lugli A, et al. Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br. J. Cancer. 2010;103:382–390. doi: 10.1038/sj.bjc.6605762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varga M, et al. Overexpression of epithelial cell adhesion molecule antigen in gallbladder carcinoma is an independent marker for poor survival. Clin. Cancer Res. 2004;10:3131–3136. doi: 10.1158/1078-0432.ccr-03-0528. [DOI] [PubMed] [Google Scholar]
- 24.Spizzo G, et al. High Ep-CAM expression is associated with poor prognosis in node-positive breast cancer. Breast Cancer Res. Treat. 2004;86:207–213. doi: 10.1023/B:BREA.0000036787.59816.01. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, et al. Clinicopathologic, molecular, and prognostic implications of the loss of EPCAM expression in colorectal carcinoma. Oncotarget. 2016;7:13372–13387. doi: 10.18632/oncotarget.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seeber A, et al. Predominant expression of truncated EpCAM is associated with a more aggressive phenotype and predicts poor overall survival in colorectal cancer. Int. J. Cancer. 2016;139:657–663. doi: 10.1002/ijc.30099. [DOI] [PubMed] [Google Scholar]
- 27.Goossens-Beumer IJ, et al. Clinical prognostic value of combined analysis of Aldh1, Survivin, and EpCAM expression in colorectal cancer. Br. J. Cancer. 2014;110:2935–2944. doi: 10.1038/bjc.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gires O, Pan M, Schinke H, Canis M, Baeuerle PA. Expression and function of epithelial cell adhesion molecule EpCAM: Where are we after 40 years? Cancer Metastasis Rev. 2020;39:969–987. doi: 10.1007/s10555-020-09898-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oba M, et al. Survival benefit of repeat resection of successive recurrences after the initial hepatic resection for colorectal liver metastases. Surgery. 2016;159:632–640. doi: 10.1016/j.surg.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Hishida T, et al. Does Repeated Lung Resection Provide Long-Term Survival for Recurrent Pulmonary Metastases of Colorectal Cancer? Results of a Retrospective Japanese Multicenter Study. Ann. Thorac. Surg. 2017;103:399–405. doi: 10.1016/j.athoracsur.2016.08.084. [DOI] [PubMed] [Google Scholar]
- 31.Bellier J, et al. Repeated resections of hepatic and pulmonary metastases from colorectal cancer provide long-term survival. World J. Surg. 2018;42:1171–1179. doi: 10.1007/s00268-017-4265-3. [DOI] [PubMed] [Google Scholar]
- 32.Yamashita T, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamashita T, et al. Discrete nature of EpCAM+ and CD90+ cancer stem cells in human hepatocellular carcinoma. Hepatology. 2013;57:1484–1497. doi: 10.1002/hep.26168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agboola AJ, et al. EpCAM expression is an indicator of recurrence in basal-like breast cancer. Breast Cancer Res. Treat. 2012;133:575–582. doi: 10.1007/s10549-011-1813-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.