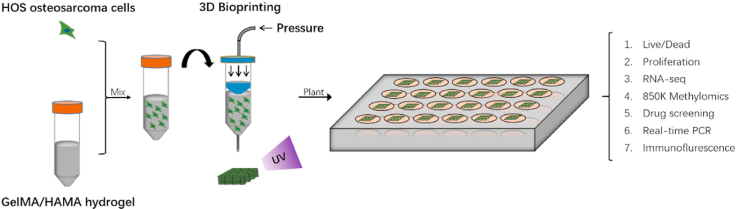
Abstract
Current in vitro models for osteosarcoma investigation and drug screening, including two-dimensional (2D) cell culture and tumour spheroids (i.e. cancer stem-like cells), lack extracellular matrix (ECM). Therefore, results from traditional models may not reflect real pathological processes in genuine osteosarcoma histological structures. Here, we report a three-dimensional (3D) bioprinted osteosarcoma model (3DBPO) that contains osteosarcoma cells and shrouding ECM analogue in a 3D frame. Photo-crosslinkable bioinks composed of gelatine methacrylamide and hyaluronic acid methacrylate mimicked tumour ECM. We performed multi-omics analysis, including transcriptomics and DNA methylomics, to determine differences between the 3DBPO model and traditional models. Compared with 2D models and tumour spheroids, our 3DBPO model showed significant changes in cell cycle, metabolism, adherens junctions, and other pathways associated with epigenetic regulation. The 3DBPO model was more sensitive to therapies targeted to the autophagy pathway. We showed that simulating ECM yielded different osteosarcoma cell metabolic characteristics and drug sensitivity in the 3DBPO model compared with classical models. We suggest 3D printed osteosarcoma models can be used in osteosarcoma fundamental and translational research, which may contribute to novel therapeutic strategy discovery.
Keywords: Bioprinting, Osteosarcoma, In vitro model, Multi-omics, Drug screening, 3D culture
Graphical abstract
The process of bioprinting and further analyses.
Highlights
-
•
3DBPO model behaved better than traditional 2D and CSC models in simulating in vivo osteosarcoma microenvironment.
-
•
3DBPO model showed significant changes in many signaling pathways associated with epigenetic regulation.
-
•
3DBPO model was particularly sensitive to autophagy-related drugs.
1. Introduction
Osteosarcoma is the most common primary bone malignancy. Despite recent therapeutic advances, the use of broad-spectrum tyrosine kinase inhibitor sorafenib [1], VEGFR inhibitor apatinib [2], recombinant human endostatin endostar, etc. [3], osteosarcoma still does harm to children and young adults [4]. The tumour microenvironment (TME) is considered a major contributor to cancer progression and metastasis [5]. A constituent of the TME, the extracellular matrix (ECM), modulates many cellular behaviours, including communication, migration, adhesion, proliferation, and differentiation [6]. As such, it profoundly affects the progression of osteosarcoma. For pre-clinical evaluation, such as drug screening of osteosarcoma, traditional two-dimensional (2D) cell culture and tumour spheroids (i.e. cancer stem-like cells [CSCs]) are widely used [[7], [8], [9]]. However, they are unable to mimic genuine osteosarcoma histological structure because of the lack of ECM [10], while animal models face ethical issues and so-called ‘3R’ principle disadvantages [11,12]. Therefore, novel in vitro osteosarcoma models that properly recapitulate the biologically native interactions between ECM and osteosarcoma cells are urgently needed.
In recent years, 3D bioprinting technologies have made great progress in replicating the delicate architecture of different tissues and mimicking the realistic in vivo microenvironment due to its ability to deposit supporting biomaterials and living cells at the same time in a cell-friendly manner [[13], [14], [15]]. In addition, 3D-bioprinting is an ideal tool applied in organ modelling, multifunctional tissue repairing, and the production of biological sensors [[16], [17], [18], [19], [20], [21]]. Mimicking the spatial structure of osteosarcoma, 3D bioprinting can assist in the assessment of cell behaviour related to ECM and more accurate drug screening.
High-throughput sequencing can provide comprehensive analyses on gene expression patterns of cancer tissues, predict cancer subtypes at the molecular level, and deepen the understanding of related issues [22,23]. Various genetic and epigenetic factors affect gene expression, causing various cancer phenotypes [24]. Multi-omics analysis combining transcriptome and methylome could systematically detect the variation of different pathways, discover biomarkers, and help identify therapeutic targets and inform the medication management. This could more accurately summarise the occurrence and development of the disease than single analysis [25]. However, multi-omics analysis has not previously been applied in osteosarcoma model research.
In this study, using 3D bioprinting, we fabricated an in vitro osteosarcoma model containing osteosarcoma cells and gelatine methacrylamide (GelMA)/hyaluronic acid methacrylate (HAMA) hydrogel. To allow the culture medium to pass through the model and to enable nutrient and waste exchange, we created a grid-like scaffold structure. GelMA/HAMA hydrogel promises good cell viability [14,26] and plays the role of ECM. The gelatine provides basic cell adhesive motifs and hyaluronic acid is an important component of the tumour matrix [27,28]. The 3D bioprinted osteosarcoma (3DBPO) model is more similar to real osteosarcoma tissues than traditional 2D or CSC models owing to its nutrient-exchange structure and the addition of an ECM analogue.
Multi-omics analysis was applied to confirm the variety of pathways and help us identify potential therapeutic targets that have not been found in traditional models. We found that in this system, osteosarcoma cells manifested different proliferation, metabolism, and adherens junctions. Moreover, the 3DBPO models showed high autophagy level and had significant sensitivity to autophagy inhibitor chloroquine (CHQ). Our observations suggest that this 3D osteosarcoma model is a promising platform for fundamental and translational research on osteosarcoma.
2. Materials and methods
2.1. Materials
GelMA, HAMA, and phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) were obtained from StemEasy Biotechnology Company (Jiangsu, China). HOS, 143B and U2-OS (human osteosarcoma cell lines) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cell culture materials including Dulbecco's modified Eagle medium (DMEM) and Dulbecco's modified Eagle medium/F12 (DMEM/F12) were purchased from HyClone Laboratories (ThermoFisher, Waltham, MA USA), Eagle's minimum essential medium (EMEM) was purchased from American type culture collection (ATCC, Manassas, USA), foetal bovine serum (FBS) was purchased from Gibco (ThermoFisher, Waltham, MA USA), and gentamycin sulphate (15 mg/mL solution) was purchase from Sangon Biotech. Cytokines, basic fibroblast growth factor (bFGF), and human epidermal growth factor (hEGF) were purchased from PeproTech. N2 was purchased from ThermoFisher Scientific. Clear round bottom ultra-low attachment microplates (24 wells) were purchased from CORNING. TRIzol reagent was purchased from Invitrogen (Carlsbad, CA, USA). A reverse transcription kit was purchased from Promega (GoScript™ transcription kit, A2791, Mannheim, GE). SYBR Green qPCR Master Mix and cell counting kit-8 (CCK-8) assay were purchased from Bimake (Houston, USA). Live/Dead assays were purchased from KeyGEN BioTECH (Jiangsu, China). Antibodies, including anti-ULK1 (8054), anti-TSC2 (4308), anti-LC3A/B (12741) and anti-rabbit IgG (4409; 4412) were purchased from Cell Signaling Technology (CST, USA). Phalloidin-fluorescein isothiocyanate (phalloidin-FITC, C1033) and 4′,6-diamidino-2-phenylindole (DAPI, C1002) were purchased from Beyotime (Shanghai, China). Fluorescein Isothiocyanate (FITC) was purchased from Yeasen (Shanghai, China). Chemotherapeutic drugs, including cisplatin, doxorubicin, methotrexate, sorafenib, apatinib, erastin, everolimus, and chloroquine, were purchased from Selleck Chemicals (Houston, TX, USA). Osteosarcoma tissue microarrays were purchased from Bioaitech (L714901, Xi'an, China).
2.2. 2D in vitro cell culture
Osteosarcoma cells (HOS, 143B, and U2-OS) were cultured at 37 °C with 5% CO2 in DMEM containing 4.5 g/L glucose supplemented with 10% (v/v) FBS and 1% (v/v) gentamycin sulphate. The cell culture media were changed routinely every 2 days. When the cells grew to nearly 80% density, they were detached from the culture dishes with trypsin, resuspended in cell culture media and plated again.
2.3. Spheroid culture of HOS stem cells (CSC culture)
As described previously [9], HOS, 143B, or U2-OS cells (1 × 104 cells/mL) were seeded in 24-well clear round bottom ultra-low attachment microplates with DMEM/F12 containing N2 (1:100), bFGF (10 ng/mL) and hEGF (10 ng/mL). A total of 100 μL of culture medium was added every 3 days to make up for evaporated liquid, but the media were not changed. The cells were cultured at 37 °C with 5% CO2 for 12 days.
2.4. Bioink preparation and 3D bioprinting
GelMA (5% [w/v]) and LAP (0.5% [w/v]) were dissolved in phosphate-buffered saline (PBS) at 50 °C, and HAMA (1% [w/v]) was added into the solution. After stirring overnight at 37 °C, the solution was filtered through a 0.22-μm filter. To prepare cell-laden hydrogels, HOS, 143B or U2-OS cells were detached from the culture dishes with trypsin, centrifuged at 300 g for 5 min, and resuspended in the GelMA/HAMA solution at a final concentration of 1 × 107 cells/mL avoiding light.
The GelMA/HAMA and cell-laden bioink solutions were loaded into the syringe of an EnvisionTEC 3D-Bioplotter (EnvisionTEC, Germany). Before printing, we designed a layer-by-layer scaffold (10 × 10 × 1.5 mm, 5 layers), with a 0.3 mm thickness for each layer and a 1.3 mm distance between each strip. We set the printing speed at 10 mm/s, the pressure at 2 bar, and the printing temperature at 10 °C during printing. The cell-laden bioink was printed as the scaffold through a 400 μm nozzle (EFD Nordson, Switzerland), and blue light (405 nm, Io = 15 mW cm−2) was then used to crosslink the bioink for 60 s, followed by being planted in normal culture dishes and incubation in DMEM complete medium (containing 4.5 g/L glucose supplemented with 10% (v/v) FBS and 1% (v/v) gentamycin sulphate).
2.5. Rheology analysis
A rheometer with parallel upper and lower plates (Thermo HAAKE MARS 60, 35 mm in diameter) was used to evaluate the rheological properties of the hydrogel prepolymer solution. The amplitude sweep was performed at 10 °C (the temperature during bioprinting) at a fixed frequency of 1 Hz. A temperature sweep assay from 15 to 45 °C was conducted at a rate of 1 °C per minute and a sampling frequency of 1 Hz. The frequency sweep assay was carried out on mixtures between 100 and 0.1 Hz at a fixed temperature of 10 °C, and the storage moduli (G′) and loss moduli (G″) were recorded. Flow sweep assay was performed at a shear rate ranging from 0.1 to 200 s−1 at 10 °C.
2.6. Compression assay
The compression testing of the 3D bioprinted models was performed by an Instron 5697 universal material testing system (Instron, USA). Before testing, all the hydrogel models were cut into cubes 5 mm in length. The compression strength is the applied stress required to break the hydrogel. The compression module is the slope of the initial linear segment of the stress–strain curves.
2.7. Atomic force microscopy (AFM)
Topography and stiffness analyses of the hydrogel were performed using a Bruker 2.7 atomic force microscope (AFM) with an E-piezoelectric scanner (Bruker Dimension Icon, Billerica, MA, USA) set to the PeakForce quantitative nanomechanics mode. The apparent Young's modulus was extrapolated by fitting the force curves with the Derjaguin–Muller–Toporov model. The stiffness of the hydrogel was obtained by calculating the average Young's modulus of three different samples using the measurements of one 10 × 10 μm square per sample. Data analysis was carried out in Nanoscope Analysis software (Bruker).
2.8. Live/dead assay of bioprinted HOS cells
Briefly, the bioprinted hydrogels were rinsed with PBS gently 3 times (1 min each time), followed by immersion in 2 μM calcein acetoxymethyl (Calcein-AM) and 8 μM propidium lodide (PI) for 30 min at 37 °C while avoiding exposure to light. Then, the hydrogels were washed with PBS 3 times (1 min each time). A confocal laser scanning microscope (CLSM; TCS SP8, Leica, Wetzlar, Germany) was used to observe the live (green fluorescence) and dead cells (red fluorescence). The number of live and dead cells was counted by ImageJ.
2.9. RNA isolation and RT-qPCR
The HOS, 143B or U2-OS cells of the 2D culture were washed with PBS 3 times, and Trizol reagent was then added to the microplate to isolate the total RNA. The HOS cells of the spheroid culture were collected in a 50-mL centrifuge tube, centrifuged at 300 g for 5 min, and resuspended in Trizol reagent. Bioprinted hydrogels were washed with PBS 3 times (1 min each time) and then transferred into glass grinders. Subsequently, the 3D constructs were ground to powder with Trizol reagent.
The sample solution was collected in microcentrifuge tubes. The following RNA extraction steps were carried out according to the manufacturer's protocol. Using the GoScript™ Reverse Transcription Kit, 1 μg of total RNA template from each group was reverse transcribed into cDNA. Real-time PCR was performed using a real-time PCR platform (ABI Prism 7500 system, Applied BioSystems, Foster City, CA, USA) with the SYBR Green qPCR Master Mix. Normalised gene expression levels were calculated by the 2−ΔΔct method with normalisation to alpha-tubulin. All the primers used in the real-time PCR are listed in Table S1.
2.10. DNA isolation
The HOS cells of the 2D culture and spheroid culture were collected, centrifuged, and quick-frozen in liquid nitrogen. 3D hydrogels were cut into small pieces, quick-frozen, and ground. The following steps were then carried out based on the procedures for extracting normal cell DNA.
2.11. Omics analysis
2.11.1. RNA-seq analysis
RNA isolation was performed as described above. According to the manufacturer's protocol, 100-bp paired-end libraries were synthesised using a TruSeq™ RNA sample preparation kit (Illumina, USA). An Illumina HiSeq 2500 platform was used to sequence the libraries. Library construction and sequencing were performed at Sinomics Corporation (Shanghai, China). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis and gene set enrichment analysis (GSEA) were performed to identify the differentially expressed mRNAs. Three repetitions per group were used for these analyses.
2.11.2. Methylation array analysis
Illumina Epitect BeadChip 850K HumanMethylation Array (Illumina, Inc., San Diego, CA, United States), which measures the methylation levels at more than 850,000 cytosine positions, was used to assess the methylation of the whole DNA. The DNA was hybridised to Beadchips according to the manufacturers protocol. Subsequently, the signal intensities were analysed by an iScan Microarray Scanner (Illumina). Three repetitions per group were used for this analysis.
2.12. Cell counting kit-8 (CCK-8) assay
2.12.1. 2D models
On day 0, HOS, 143B or U2-OS cells were plated in 96-well plates at a density of 2000 cells per well. On days 1, 4, and 7, cells were incubated with 100 μL of complete phenol red-free medium containing 10% CCK-8 reagent at 37 °C for 2 h. A microplate reader (Tecan, M200pro, Switzerland) was used to measure the absorbance at 450 nm. After each test, the medium was replaced by fresh complete DMEM medium.
2.12.2. CSC models
On days 1, 4, and 7, 10% of the liquid volume per well of CCK-8 reagent was added to each well. After the cells were incubated at 37 °C for 2 h, 100 μL supernatant from each well was transferred into 96-well plates. The measuring methods were the same as mentioned above.
2.12.3. 3D models
On day 1, the hydrogels were placed in 24-well plates and washed with PBS 3 times, followed by incubation in 500 μL of complete phenol red-free medium containing 10% CCK-8 reagent at 37 °C for 2 h. Next, 100 μL supernatant from each well was transferred into 96-well plates. The measuring methods were the same as described above. Subsequently, the hydrogels were plated in new complete DMEM medium. The experiment was repeated on days 4 and 7.
The cell proliferation curve was drawn in GraphPad Prism 7. Five repetitions per group were used for this analysis.
2.13. Quantification of chemotherapeutic drug efficacy
HOS cells of 2D culture were plated in 96-well microplates (1 × 105 cells/mL). The next day, the medium was replaced by medicate medium. The cells were incubated with cisplatin (DDP), doxorubicin (DXR), methotrexate (MTX), erastin (ERA), apatinib (APAT), sorafenib (SORA), everolimus (EVE), or chloroquine (CHQ) at different concentrations (0, 0.01 μM, 0.1 μM, 1 μM, 10 μM, or 100 μM) for 48 h. Thereafter, the CCK-8 assay was used to detect cell viability.
As for the HOS cells of spheroid culture, the drug susceptibility tests were begun on day 7 of culture. The drug concentrations and incubation time were the same as mentioned above. Then, the cell spheres incubated at the same drug concentration were collected after the CCK-8 assay.
On day 7 of culture, 3D hydrogel models were incubated with the drugs at different concentrations (0, 0.01 μM, 0.1 μM, 1 μM, 10 μM, 100 μM, or 500 μM) for 48 h. Then, the CCK-8 assay was used to detect the cell viability.
GraphPad Prism 7 was used to make drug effect curves and calculate IC50.
2.14. Immunofluorescence
Samples were fixed in 4% paraformaldehyde at 37 °C for 20 min, and were then permeabilised by 0.1% Triton for 15 min and blocked with 10% goat serum for 1 h. Next, the samples were incubated with primary antibodies at 4 °C overnight, including anti-ULK1 (1:50), anti-TSC2 (1:100) and anti-LC3A/B (1:100). After rinsing with PBS 3 times, the samples were incubated with anti-rabbit IgG secondary antibody (1:500) at room temperature for 1 h. Cell nuclei were stained using DAPI, and the samples were observed and imaged using CLSM (Leica TCS SP8). The staining procedure for the 2D, CSC, and 3D models were the same.
2.15. Cytoskeleton staining
The HOS cells of the 2D culture were fixed in 4% paraformaldehyde at 37 °C for 20 min, then incubated with phalloidin-FITC for 2 h. Cell nuclei were stained using DAPI, and the cells were observed and imaged using CLSM.
2.16. Drug permeability test
Briefly, after washing with PBS, the three kinds of models were incubated with FITC (10 μg/mL) for 2 h. The models were then observed and imaged using CLSM.
2.17. Survival analysis
The R2: Genomics Analysis and Visualization Platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi) was used to analyse the overall survival probability of patients with osteosarcoma with respect to TSC2, ULK1 and LC3 expression levels. In this study, Mixed Osteosarcoma (Mesenchymal)-Kuijjer-127 dataset comprising 88 patients with survival information who were followed for 240 months was used. The patients were divided into two groups with high and low gene expression and were analysed using Kaplan–Meier survival analysis. P < 0.05 was considered statistically significant. All data in the R2 platform is free and accessible online.
2.18. Immunohistochemistry staining and pathological correlation analysis of osteosarcoma tissue microarrays
The osteosarcoma tissue microarrays collected osteosarcoma tissue samples from 70 patients. For IHC (Immunohistochemistry) staining, osteosarcoma tissue microarrays were incubated with anti-LC3A/B (1:500). IHC staining analysis was then carried out to divide the samples into LC3-high and LC3-low two groups. 70 osteosarcoma tissue samples were finally subjected to pathological correlation analysis.
2.19. Statistical analysis
Unless otherwise mentioned, all data in this study are expressed as mean ± standard deviation (SD) of at least three independent replications. Two-tailed Student's t tests were applied for comparisons between two groups, and one-way analysis of variance with Bonferroni's post hoc analysis was employed for comparisons among three or more groups. A four-parameter logistical model was used to fit the drug effect curves, and the drug concentrations at which cell viability descended to 50% were determined (IC50). Fisher's exact tests or Chi-square tests were applied for pathological correlation analysis. Log-rank tests were used for analysing Kaplan–Meier survival curve. All data analysis was performed using Prism 9 (GraphPad, La Jolla, CA, USA).
3. Results
3.1. Physical properties and biocompatibility of GelMA/HAMA hydrogel
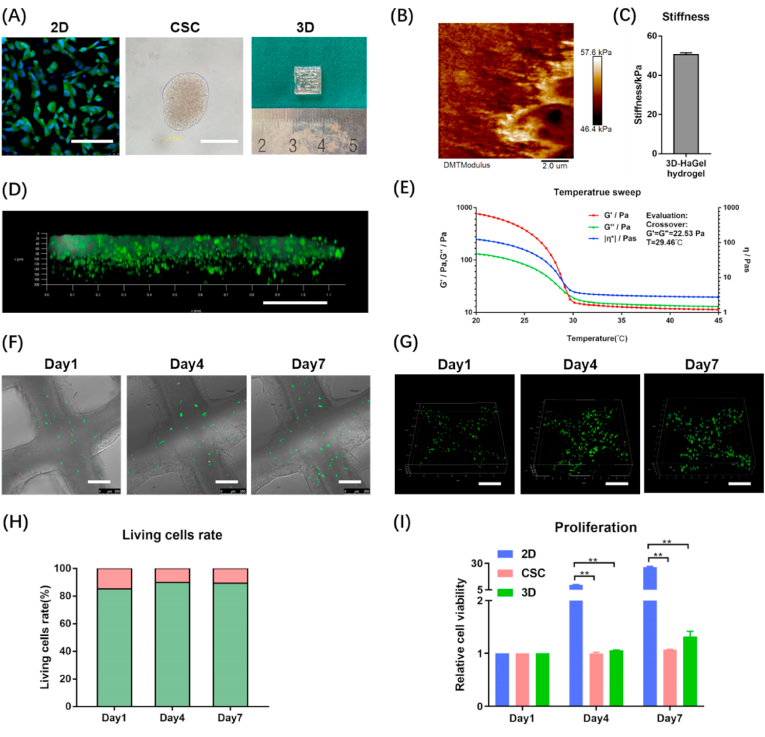
As shown in Fig. 1(A), we created three in vitro osteosarcoma models: 2D cell culture, tumour spheroids (i.e., CSCs), and 3D bioprinted model. In 2D models, HOS were seeded in normal attachment culture dishes. In CSC models, HOS were seeded in ultra-low attachment culture dishes and cultured in medium with bFGF, hEGF, and N2, which would prompt HOS to recover stem cell properties and grow into cell spheroids (nearly 2000 cells per spheroid). The stemness properties of the CSC models were validated by real-time PCR (Fig. S1). Measured by AFM, the value of apparent Young's modulus in 3D bioprinted model was 50.5 ± 1.0 kPa (Fig. 1(B–C)). The compression test showed that the 3D bioprinted model had compressive strength at 52.6 ± 8.2 kPa and compressive modulus at 47.9 ± 10.7 kPa (Fig. S2(A)). As shown in Fig. 1(D), we used FITC to mimic chemotherapy drugs because its molecular weight (389.4) is very close to those of commonly used drugs. FITC could permeate the entire 3D osteosarcoma model within 2 h, confirming that the 3D models have good drug permeability. Then, rheological behaviours of the GelMA/HAMA bioink were determined. We conducted amplitude sweep first and found the critical strain value of the linear viscoelastic region (LVR) is 76.99% (Fig. S2(B)). Temperature sweep analysis revealed that the hydrogel was solid below 29.46 °C and liquid above 29.46 °C (Fig. 1(E)). Flow sweep analysis identified the bioink shear thinning behaviours (Fig. S2(C)). In the oscillatory frequency range of 0.1–100 Hz and at 10 °C, the viscous modulus (G″) was less than elastic modulus (G′) at the whole period (Fig. S2(D)). Based on all the rheological data, the bioink belongs to weak gel and is suitable for 3D bioprinting [28].
Fig. 1.
Physical properties and biocompatibility of GelMA/HAMA hydrogel. (A) The appearance of 2D monolayer cell (Phalloidin-FITC and DAPI staining), tumour spheroids [i.e., cancer stem-like cells (CSC)] and 3D hydrogel models (HOS). Scale bar = 100 μm. (B) The AFM image indicates the apparent Young's modulus of 3D hydrogel in a square of 10 μm*10 μm. (C) The average apparent Young's modulus of 3D hydrogel. (D) Drug permeability of 3D model tested by FITC. Scale bar = 250 μm. Temperature sweep analysis (E) of the bioink used in 3D tumour models. Brightfield confocal photos (F) and 3D reconstruction photos (G) of HOS viability measured by Live/Dead assays on days 1, 4, 7. Scale bar = 250 μm. (H) Living cells rate was quantified by image analysis of the 3D hydrogel models. The green part of column represents living cells, while the red part of column represents dead cells. (I) The CCK-8 assays were carried out on days 1, 4, and 7 to measure the HOS cell proliferation in 2D, CSC and 3D models. The data are expressed as mean ± SD (n = 3). **p < 0.01 indicate statistical significance.
Next, we carried out live/dead assay to examine the viability of HOS cells after bioprinting and took brightfield (Fig. 1(F)) and 3D (Fig. 1(G)) confocal photos. The morphology of the HOS cells growing in the scaffold could be observed. The cells were scattered on day 1 and they proliferated and gathered together until day 7. ImageJ analyses revealed the following living cells rates: 85.17 ± 0.84% on day 1, 89.90 ± 1.88% on day 4, and 89.39 ± 1.91% on day 7 (Fig. 1(H)). Furthermore, CCK-8 assay showed that HOS cells in this hydrogel could proliferate well, and the proliferation rate in 3D was inferior to that in 2D (Fig. 1(I)). Meanwhile, we used other two osteosarcoma cell lines, 143B and U2-OS, to creat the three models, and found that they had similar proliferation characteristics to HOS (Fig. S3(A-B)). To exclude the impact of different media, 3D models were also cultured in CSCs culture medium containing bFGF, hEGF and N2 in another experiment. The results showed no significant differences regardless of whether growth factors were added (Fig. S4(A)). These findings suggest that osteosarcoma cells could survive and proliferate in the GelMA/HAMA matrix after bioprinting, and the hydrogel has excellent biocompatibility.
3.2. Osteosarcoma cells in 3D culture show unique gene expression compared to those in traditional in vitro models
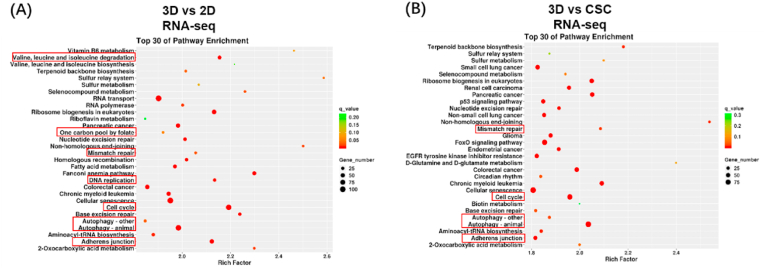
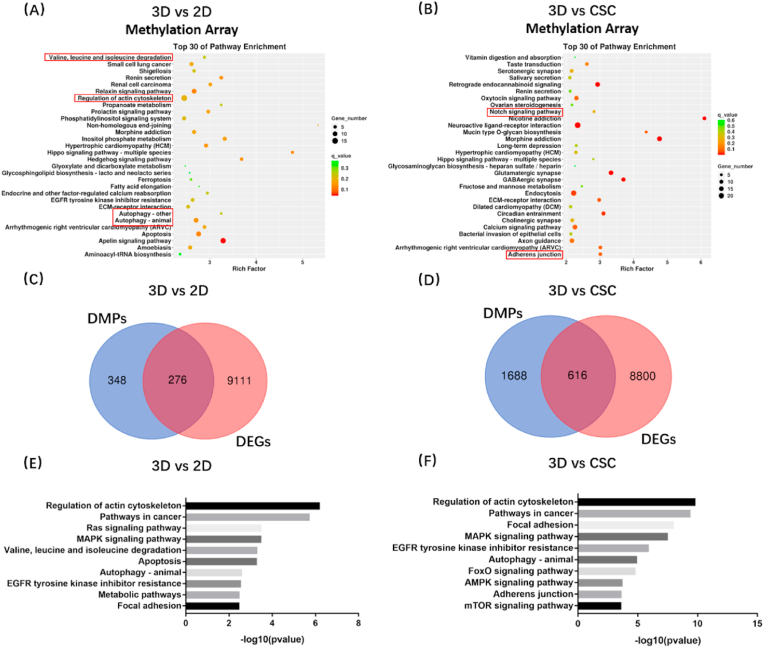
To investigate the difference in the transcriptome among the three in vitro osteosarcoma models, we extracted RNA samples of HOS cells from 3D hydrogels, cell spheroids, and 2D monolayer cells and then assessed genome-wide gene expression by transcriptomic RNA sequencing (RNA-seq). KEGG enrichment analysis among 2D, CSC, and 3D models was conducted; the top 30 enrichment pathways are listed in a scatter diagram (Fig. 2(A-B)). We analysed the results of 3D vs 2D, 3D vs CSC, and CSC vs 2D groups and found that some enrichment pathways were significantly changed, notably DNA replication, valine leucine and isoleucine degradation, and adherens junction pathways. Furthermore, GSEA showed the pathways could be roughly divided into three categories. Cell cycle-related pathways (e.g. DNA replication, mismatch repair, cell cycle, and one carbon pool by folate), cell metabolism-related pathways (e.g. glycosaminoglycan biosynthesis chondroitin sulphate, other glycan degradation and valine leucine and isoleucine degradation, and one carbon pool by folate), and cellular community-related pathway (e.g. adherens junction) (Fig. S5). GSEA revealed that the culture methods significantly affected proliferation, metabolism, and adherens junction of osteosarcoma cells. Obviously, compared to 2D models, both the 3D and CSC osteosarcoma models showed weaker proliferation, and 3D models had less adherens junction-related genes expression. The overview of KEGG enrichment metabolic pathways further showed that the metabolic level of 3D cultured osteosarcoma cells significantly changed compared to that of traditional models, including 2D monolayer and CSC (Fig. S6(A-B)). In addition, we examined the association among 2D, CSC, and 3D osteosarcoma models (Fig. S7). Based on the three models’ Pearson correlation coefficient, we found that CSC models were similar to 2D models (average Pearson correlation coefficient = 0.87), while 3D models were a little different from 2D (average Pearson correlation coefficient = 0.75) and CSC models (average Pearson correlation coefficient = 0.73). These results suggest that the 3DBPO models are distinguished from traditional models at the transcriptomic level, mainly manifesting in the differences in cell cycle, metabolism, adherens junctions, and other pathways.
Fig. 2.
Transcriptomic data showed that the gene expression of 3D culture HOS cells is different from 2D and CSC models. (A–B) KEGG enrichment analysis of differentially expressed genes. The pathways of interest have been marked by red frames. The differential expression pathways could be roughly divided into three categories: cell cycle-related pathways, cell metabolism-related pathways and cellular community-related pathways.
3.3. Osteosarcoma cells in the 3D model show weak proliferation, weak adherens junction, weak anabolism, and strong catabolism
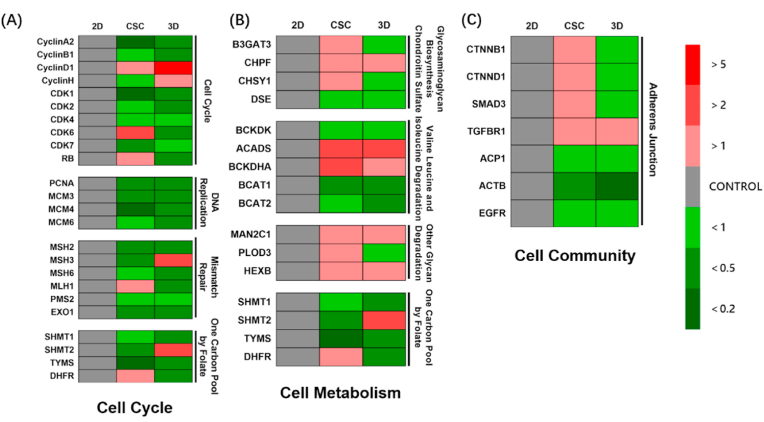
We conducted qPCR for specific genes to further verify the change of each selected enrichment pathway. First, the genes of cell cycle-related pathways were detected, including cyclin, cyclin-dependent kinase, mini-chromosome maintenance, MutL-homolog, serine hydroxymethyltransferase (SHMT) families, and other genes (Fig. 3(A)). Most cell cycle-related genes were down-regulated in 3D osteosarcoma models compared to 2D models, indicating weaker proliferation, while CSC and 3D cultures behaved similarly.
Fig. 3.
Specific gene expression of HOS osteosarcoma cells for 2D monolayer, CSC and 3D bioprinted models. (A) Partial gene expression of cell cycle related pathways, including cell cycle, DNA replication, mismatch repair and one carbon pool by folate. (B) Partial gene expression of cell metabolism related pathways, including glycosaminoglycan biosynthesis chondroitin sulfate, valine leucine and isoleucine degradation, other glycan degradation and one carbon pool by folate. (C) Partial genes expression related to adherens junction. Red or green blocks represent up-regulation or down-regulation of gene expression compared to 2D control group. Colour depth represents fold change. All data are from n = 4 independent experiments.
Next, we detected the genes of metabolism-related pathways (Fig. 3(B)). It was found that the genes of glycosaminoglycan biosynthesis chondroitin sulphate pathway and adherens junction-related genes were down-regulated in the 3D model (Fig. 3(C)). In contrast, the genes of the glycosaminoglycan biosynthesis chondroitin sulphate pathway were upregulated in the CSC model. More degradation of branched-chain amino acids (BCAAs) was identified in 3D and CSC models than in the 2D model. Other glycan degradation pathways increased, whereas one carbon pool by folate decreased in both 3D and CSC. Our findings suggest that the 3D osteosarcoma model shows weak anabolism and strong catabolism, while CSC is almost the same as the 3D osteosarcoma model, except for glycosaminoglycan biosynthesis.
Finally, we measured the gene expressions of the adherens junction pathway (Fig. 3(C)). Osteosarcoma cells in 3D exhibited weaker adherens junction than those in 2D, as demonstrated in ovarian adenocarcinoma [29]. The cells in CSC showed moderate gene expression of adherens junction in 2D and 3D. Together, these qPCR results confirm weaker proliferation, adherens junction, and anabolism of osteosarcoma cells but stronger catabolism in 3D culture than in 2D culture.
3.4. Osteosarcoma cells in 3D culture show different epigenetic regulation compared to those in traditional in vitro models
We next carried out DNA methylomics analysis (Illumina Epitect BeadChip 850K HumanMethylation Array) to determine epigenetic changes in 3D and conventional models. Similarly, the KEGG analyses of differential methylation positions (DMPs) were conducted among 2D, CSC, and 3D models (Fig. 4(A-B)). Valine, leucine, and isoleucine degradation; autophagy; and adherens junction pathways were also found in the top 30 enrichment pathways, suggesting that these pathways were changed significantly in 3D culture compared to 2D or CSC and 3D culture modulates gene expression of these pathways through epigenetic regulation.
Fig. 4.
3D bioprinted osteosarcoma models (HOS) showed differential methylation positions in KEGG enrichment analysis compared with 2D (A) and CSC models (B). The pathways of interest have been marked by red frames. Conjoint analysis of transcriptomics and DNA methylomics between 3D bioprinted osteosarcoma models and traditional models (C–F). Venn diagram analysis identified overlapping target gene between differential methylation positions (DMPs) and differentially expressed genes (DEGs), which found 276 genes in 3D vs 2D groups (C) and 616 genes in 3D vs CSC groups (D). KEGG enrichment analysis of overlapping genes in 3D vs 2D groups (E) and 3D vs CSC groups (F).
To identify the pathways changed in both transcriptomics and DNA methylomics, we performed multi-omics combined analysis in 3DBPO models and traditional models. In 3D vs 2D groups, there were 348 DMPs and 9111 differentially expressed genes (DEGs); among them, 276 genes belong to both DMPs and DEGs (Fig. 4(C)). In 3D vs CSC groups, there were 1688 DMPs and 8800 DEGs; among them, 616 genes belong to both DMPs and DEGs (Fig. 4(D)). Subsequently, we conducted KEGG enrichment analysis again for the genes that had been crossed (Fig. 4(E-F)). The pathways had significant expression and methylation differences between 3D models and 2D or CSC models. Valine, leucine, and isoleucine degradation; metabolic pathways; regulation of actin cytoskeleton; and focal adhesion were related to obviously changed pathways observed previously, including cell proliferation, metabolism, and cellular community. Moreover, multi-omics combined analysis revealed that autophagy-related genes were enriched in two groups, suggesting increased autophagy levels. The results confirmed that expression differences of these pathways are mediated by methylation regulation, while other pathways, including proliferation-related pathways, may be mediated by alternative mechanisms.
3.5. 3D bioprinted models are much more sensitive to chemotherapeutics related to autophagy pathway
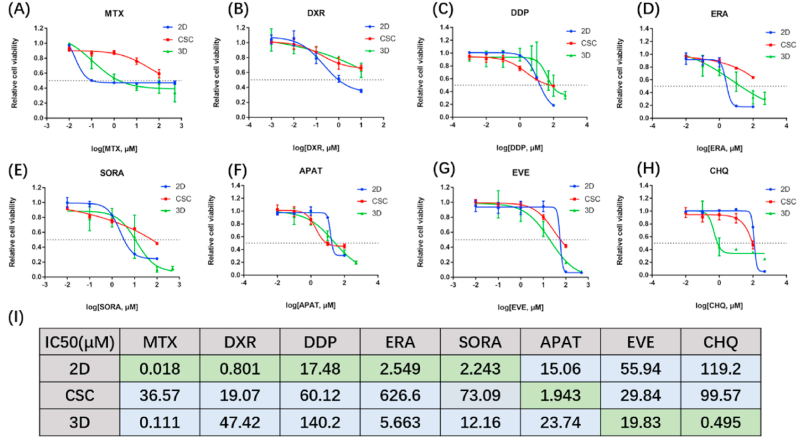
For chemotherapeutic drug screening studies, we firstly tested drug permeability of three models by FITC (Fig. 1(D), Fig. S8). We found that FITC could completely permeate both 2D and 3D models in 2 h, while CSC models were only permeated around the perimeter, demonstrating that drug permeability is better in 2D and 3D models than in the CSC model.
Methotrexate (MTX) is a non-selective inhibitor of dihydrofolate reductase (DHFR), doxorubicin (DXR) is an inhibitor of DNA topoisomerase II, and cisplatin (DDP) is an inorganic platinum complex that can inhibit DNA synthesis by forming DNA adducts in tumour cells. The three drugs all interfere DNA synthesis and thus inhibit tumour proliferation. Recent findings confirm the MAP (methotrexate, doxorubicin, and cisplatin) regimen as the gold standard for osteosarcoma patients [30]. Therefore, we tested the drug sensitivity of the three chemotherapeutic drugs in our models. As shown in Fig. 5(A-C, I), 2D osteosarcoma models showed better drug sensitivity than 3D models, probably due to the different proliferation rate (Fig. 1(I)). The faster osteosarcomas proliferate, the more sensitive they are to drugs that inhibit proliferation.
Fig. 5.
Chemotherapeutic drugs cytotoxicity evaluation for three cell culture models (HOS). (A–H) Drug effect curves of eight clinical chemotherapeutics for osteosarcoma, including MTX (A), DXR (B), DDP (C), ERA (D), SORA (E), APAT (F), EVE (G) and CHQ (H). 2D models showed best sensitivity to MTX, DXR, DDP, ERA and SORA than the other two models. CSC models showed best sensitivity to APAT than the other two models. 3D models showed best sensitivity to EVE and CHQ than the other two models. Data are represented as mean ± SD (n = 3). (I) Corresponding IC50 calculated from the above eight drug effect curves respectively. MTX = methotrexate, DXR = doxorubicin, DDP = cisplatin, ERA = erastin, SORA = sorafenib, APAT = apatinib, EVE = everolimus, CHQ = chloroquine.
Next, we focused on some novel targeted therapies. Erastin (ERA) is an activator of ferroptosis. Ferroptosis together with other chemotherapeutic drugs plays a synergistic role in boosting the growth suppression of hypoxic osteosarcoma in vivo [31,32]. Sorafenib (SORA), a tyrosine kinase inhibitor, was previously reported to inhibit growth, angiogenesis, and metastatic ability of osteosarcoma pre-clinical models via the ERK1/2, MCL-1, and ERM pathways [1]. Apatinib (APAT), an inhibitor of VEGFR, is used for relapsed and unresectable osteosarcoma after the failure of first-line or second-line chemotherapy in a clinical trial (NCT02711007) [2], and contributes to the clinical treatment of doxorubicin [33]. By calculating the IC50, it was found that the efficacy of these three targeted drugs was similar in 2D and 3D models (Fig. 5(D-F, I)).
Everolimus (EVE) is an mTOR inhibitor targeting TSC2 and activates autophagy, while chloroquine (CHQ) is an inhibitor of autophagy. Generally speaking, autophagy removes damaged organelles, such as mitochondria, and is a self-protection mechanism of cells. However, since excessive autophagy has the potential to induce cell death, autophagy inducers have also been considered potential cancer treatments [34]. The analyses of the IC50 of EVE and CHQ showed that autophagy-related drugs were more effective in 3D osteosarcoma models than in 2D models (Fig. 5(G-I)). Moreover, 3D models were much more sensitive to autophagy inhibitor CHQ than to autophagy activator EVE. In addition, the pathways that are associated with the gold standard drugs or novel potential targeted drugs are all detected in the multi-omics analysis mentioned above, indicating that multi-omics analysis can be used for drug screening.
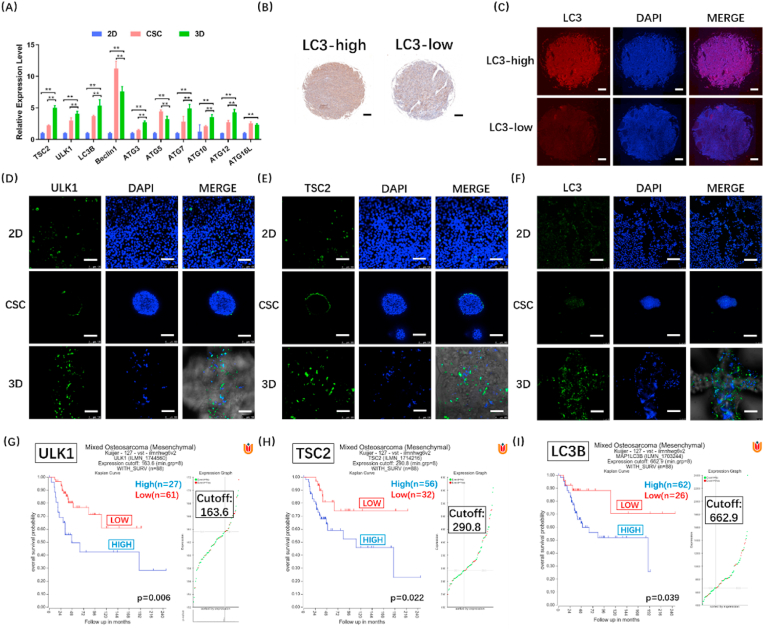
3.6. 3D-bioprinted osteosarcoma models manifest strong autophagy
Autophagy has been demonstrated to be the self-protection mechanism of osteosarcoma, and autophagy inhibitor CHQ shows more significant curative effect in 3D models than in 2D models. Therefore, we further analysed the autophagy level in in vitro models after excluding the influence of growth factors (Fig. S4(B)). RT-qPCR results showed higher expression of autophagy-related genes in 3D models (HOS) compared to 2D and CSC models (Fig. 6(A)). Similar results were found in 143B and U2-OS cells (Fig. S9(A-B)). LC3 is the core protein of autophagy. We used osteosarcoma tissue microarray to determine LC3 expression in clinical samples. By analysing the LC3 positive cells density in the samples of different stages, we found that LC3 expression level was significantly increased in the osteosarcoma of stage IVB (p < 0.05) (Fig. S10). According to the results of immunohistochemical analysis, the samples were then divided into two groups: high LC3 expression (LC3-high) and low LC3 expression (LC3-low) (Fig. 6(B–C)). Pathological correlation analysis also revealed that LC3 expression significantly correlated with the tumour stages (p < 0.05), suggesting high LC3 expression could increase the malignancy of osteosarcoma. The statistical results and clinical data were listed in Table S2. In Fig. 6(D-F), a panel of images obtained from immunofluorescence using anti-ULK1, anti-TSC2 and anti-LC3 antibodies in three models is shown. ULK1 is an activating kinase and a marker of autophagy, and TSC2 suppresses the activation of mTORC1, thus inducing autophagy. Strong ULK1, TSC2 and LC3 expression levels were observed in 3D models, revealing more autophagy in 3D models than in 2D and CSC models. Next, we used a database in R2 platform, genome-wide gene expression analysis of high-grade osteosarcoma, to compare the autophagy levels between 3D models and in vivo study (Fig. 6(G-I)). We found that autophagy was obviously related to survival, and the stronger the autophagy, the shorter the lifetime. The autophagy level was distinctly high in 3D models, but not in 2D and CSC models. Similar to in vivo findings, autophagy had a great influence on survival in 3D models, suggesting that the effect of autophagy on osteosarcoma is better reproduced in 3D models. Furthermore, we found that osteosarcoma in 3D models was particularly sensitive to CHQ, and the stronger the autophagy, the shorter the lifetime; the findings suggest the important role of autophagy in osteosarcoma. In conclusion, these findings confirm that our 3DBPO models exhibited autophagy levels closer to those in vivo than did conventional in vitro 2D and CSC models. Furthermore, the results obtained using our 3DBPO models are also consistent with the clinical drug screening results.
Fig. 6.
The verification of autophagy level in vitro (HOS) and in vivo. Autophagy-related genes expression were determined by RT-qPCR and compared between 3D models and 2D or CSC models (A). The data are expressed as mean ± SD (n = 4). **p < 0.01 indicate statistical significance respectively. Representative immunohistochemistry (B) and immunofluorescence (C) photos of LC3-high and LC3-low osteosarcoma biopsies. Scale bar = 200 μm. Immunofluorescence staining of autophagy markers in 2D, CSC and 3D osteosarcoma models. ULK1 (D), TSC2 (E) and LC3 (F) are shown in green, while nuclei are shown in blue. Scale bar = 100 μm. Kaplan-Meier plot of the overall survival for 88 patients according to ULK1 (G), TSC2 (H) and LC3B (I) expression defined by cut-offs of 163.6 (G), 290.8 (H) and 662.9(I).
4. Discussion
Current pre-clinical cancer models, especially the widely employed 2D models, show limited efficacy in reproducing tumour complexity [35,36]. 3D-bioprinting is a revolutionary technology and an ideal method to replicate the meticulous structure of tissues and the interrelationship between tumour cells and the ECM. However, to date, only a few studies have employed 3D-bioprinting in cancer model research [13,35,37], and few in vitro models have been confirmed by multi-omics analysis to manifest the full range of gene expression. In addition, the similarities and differences between these models and traditional models remain unclear. Notably, 3D-bioprinting has not been applied to in vitro osteosarcoma models. The main goal of this study was to fabricate an in vitro osteosarcoma cell/ECM model that could serve as a platform for fundamental and translational research and contribute to novel therapeutic strategy discovery. We used the multi-omics analysis methods to characterise the models at multiple levels and found that inhibiting autophagy was an attractive strategy for the treatment of osteosarcoma, a finding not demonstrated by traditional models. Our present findings demonstrate the feasibility of 3D printed osteosarcoma models and suggest that the combination of this model with multi-omics analysis might help in the discovery of therapeutic targets.
Previous studies confirmed that 5% GelMA/1% HAMA hydrogel has good printability and could be printed at a suitable resolution, with the structure maintained after printing [14,26]. GelMA retains the RGD and MMP degradation peptide sequences of gelatine, thus maintaining the ability to support cell adhesion, proliferation, and differentiation [28]. Hyaluronic acid (HA), a biopolymer with extraordinary biophysical properties [38], is recognised as a major ECM component in different tissues and exists in abundance in bone ECM [27,[38], [39], [40]]. The addition of HAMA increases the printability of bioinks. Hence, GelMA and HAMA are high-performing bioinks and suitable analogues of osteosarcoma ECM. Appropriate UV exposure intensity for this hydrogel was found to be 15 mW cm−2 [14]. Thus, crosslinking of GelMA/HAMA could be performed with retention of cell viability. According to AFM measurements, the value of apparent Young's modulus in our 3D bioprinted model was 50.5 ± 1.0 kPa (Fig. 1(B–C)), which is equivalent to the previous reported optimum matrix stiffness (50 kPa) for osteosarcoma cell [41]. Summarily, we fabricated a bioactive 3D osteosarcoma hydrogel scaffold, which could accurately model the interaction between osteosarcoma cells and ECM.
The HOS cell line (harbouring a TP53 mutation) used in the present study has been widely applied in osteosarcoma phenotypic research [42,43]. In this study, there were two control groups, 2D monolayer culture model and tumour spheroids (i.e., CSCs). In the 2D model, HOS cells were planted in normal culture dishes, and their phenotype would be affected by the stiffness of culture dishes and the lack of ECM. Different from the 2D model, the CSC model consists of the spontaneously aggregated tumour spheroid by HOS cells induced by a variety of cytokines and nonadherent surface. Osteosarcoma cells restore the self-renewal ability and microenvironmental features of in vivo human tumour tissues during aggregation [44]. Meanwhile, the CSC model has been used for more than 40 years in cancer research as an intermediate between 2D monolayer cultures and in vivo studies [7]. Although the CSC model is better able to simulate in vivo microenvironment than the 2D model, it shows lower throughput [45] and bad permeability [44] compared with the 2D model. The 2D and CSC are classical in vitro models and have been used to evaluate the bioprinted model. In the research, the 2D and 3D bioprinted models were both cultured in DMEM complete medium, while the CSC models were culture in DMEM/F12 medium supplemented with growth factors. According to the ATCC's protocol, HOS cell line could be also cultured in EMEM complete medium. To exclude the effect of different medium, we investigated the expression of some metabolic and autophagy marker genes when the 3D models (HOS) were cultured in DMEM, DMEM/F12 or EMEM complete medium (Fig. S11). The results showed that, autophagy levels were not influenced by different medium. The expression of only a few metabolic genes was affected, and the difference was small. Especially in the comparison between DMEM and DMEM/F12, only the expression difference of CHPF and DHFR was significant. In conclusion, different medium would not cause significant difference of metabolism and autophagy levels.
In transcriptomics analysis, the different expression pathways are divided into three categories, i.e. cell cycle-related, metabolism-related, and cell community-related pathways. Mismatch repair occurs after DNA replication and helps maintain DNA replication stability and control gene mutation. Folate is the carbon source of DNA. Since pathways of DNA replication, mismatch repair, cell cycle and one carbon pool by folate are all involved in the process of the cell cycle, we classified them into cell cycle-related pathways (Fig. 3(A)). Moreover, since pathways of glycosaminoglycan biosynthesis chondroitin sulphate, other glycan degradation, valine leucine and isoleucine degradation, and one carbon pool by folate are involved in glycan metabolism, BCAAs metabolism, and folate metabolism, we classified them into cell metabolism-related pathways (Fig. 3(B)). Adherens junction is involved in cell movement and cell-cell and cell-ECM signal transduction and belongs to cell community-related pathways (Fig. 3(C)). In this study, the genes selected to be verified by qPCR were significantly regulated in RNA-seq or characteristic genes of each pathway. Consistent with the CCK-8 assay (Fig. 1(I)), we found that expression levels of almost all of the genes of cell cycle-related pathways were lower in the CSC and 3D models than in 2D models. This finding is similar to that in a previous study on clear cell renal cell carcinoma [46]. As for metabolism-related pathways, 3D models showed weak anabolism and strong catabolism, and the CSC model was almost the same as the 3D model, except for glycosaminoglycan biosynthesis, confirming that spatial structure has a great influence on cell metabolism. Consistently, Rodríguez-Enríquez S et al. obtained the same results in tumour spheroids [47]. Glycosaminoglycan (GAG) is responsible for mediating the processes of cell attachment and growth factor signalling [48]. The 3D model showed down-regulated GAG synthesis, consistent with its weak cell proliferation and adherens junction. A reasonable explanation is that inhibition of GAG synthesis disrupts osteosarcoma cell proliferation, consistent with the disruption of communication between the ECM and the actin cytoskeleton [48]. However, the CSC model showed weak cell proliferation but upregulated GAG synthesis, probably owing to its stem cell-like characteristic. BCAAs are the source of energy and promote tumour development and progression. BCKDH and ACADS are the promoters of BCAA catabolism, while BCKDK, BCAT1, and BCAT2 are the suppressors of BCAA catabolism. CSC and 3D models showed more degradation of BCAAs than 2D models; the finding also explains the weak proliferation in CSC and 3D models. In the pathway of one carbon pool by folate, SHMT, TYMS, and DHFR participate in the nuclear de novo dTMP synthesis pathway [49]; thus, CSC and 3D models might show weak dTMP anabolism. Indeed, we found that osteosarcoma cells in 3D printing models exhibited weaker adherens junction than those in 2D or CSC cultures. Notably, some studies suggest that adhesion protein expression is positively correlated with the malignancy degree of sarcoma [50], while the opposite perspective prevails in epithelial tumours [[51], [52], [53]].
DNA methylomics analysis can investigate gene expression difference in DNA level and explain transcriptomics variety upstream. Generally, methylation of the promoter region will lead to down-regulation of the gene. We combined the DEGs of transcriptomics and DMPs of methylomics, determined the intersection, and performed KEGG enrichment analysis (Fig. 4(C–F)). We found that autophagy was enriched based on both transcriptomics and methylomics using KEGG analyses but not GSEA (Fig. S5). The reason might be the algorithm of GSEA, which distinguished the genes changed in opposite trends. In addition, EGFR, MAPK and AMPK signalling pathway may be potential therapeutic targets.
Autophagy is generally recognised as a self-protection mechanism of cells because of its ability to remove damaged organelles and proteins [[54], [55], [56]]. Meanwhile, it promotes chemotherapy resistance and tumour survival [57]. However, excessive autophagy would induce cell death [34]. Therefore, both activators and inhibitors of autophagy are available in osteosarcoma treatment. In the present study, we found that CHQ, an inhibitor of autophagy, was more effective than EVE, an activator of autophagy (Fig. 5(G-H)). The result may relate to EVE behaved well in pre-clinical research [58], while a phase 2 clinical trial showed that 45% of patients were free from progression at 6 months, which is less than the prespecified threshold of a phase 3 trial [59]. Through analysis (drug screening, osteosarcoma tissue arrays, immunofluorescence, and delving into databases), we verified that autophagy promotes osteosarcoma development, and 3D bioprinted models could accurately mimic the autophagy level in vivo (Fig. 5, Fig. 6). Indeed, the current therapy for osteosarcoma usually combines CHQ with other chemotherapy drugs, such as rapamycin [60], cisplatin [61], PD-L1 inhibitor [62], and paclitaxel [56], and its feasibility can be demonstrated using our 3DBPO models.
This study had some limitations. Firstly, the major findings from the in vitro 3DBPO model need to be verified furtherly in animal model. Besides, the broad conjoint analysis used in this study did not distinguish between the genes of upregulated and downregulated methylation. An improved analysis could perhaps be afforded if upregulated methylation positions are combined with downregulated expression genes. In addition, because the microenvironment of osteosarcoma involves other elements, such as endothelial cells, immune cells and stromal cells, the current 3DBPO models could not fully replicate TME. Nevertheless, this study highlights research directions that should be investigated in the future. We plan to coculture osteosarcoma cells with other cells, add other bone matrix components into our system, and build more complicated structures, such as bio-mimetic blood vessels and osteosarcoma organoids.
5. Conclusions
In this study, 3D osteosarcoma models containing osteosarcoma cells and a shrouding ECM analogue were bioprinted successfully. Using the 3D models, we revealed the importance of autophagy in osteosarcoma. Multi-omics analysis demonstrated differential effects of chemotherapy drugs among different models. Moreover, we found that CHQ, an inhibitor of autophagy, worked well in 3D models. The findings suggest that 3D printed osteosarcoma models can be applied for fundamental and translational osteosarcoma research, and may contribute to novel therapeutic strategy discovery.
CRediT authorship contribution statement
Yixuan Lin: Conceptualization, Data curation, Formal analysis, Validation, Writing – original draft, Writing – review & editing. Yiqi Yang: Formal analysis, Validation, Methodology, Supervision, Writing – review & editing. Kai Yuan: Investigation, Validation, Methodology. Shengbing Yang: Investigation, Methodology. Shuhong Zhang: Methodology, Validation, Formal analysis. Hanjun Li: Conceptualization, Formal analysis, Methodology, Resources, Writing – review & editing. Tingting Tang: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Shanghai Science and Technology Development Fund (18DZ2291200), the National Natural Science Foundation of China (92068205 and 81802679), and China Postdoctoral Science Foundation (2018M632136 and 2019T120348) to HL. The authors thank Shanghai Graphic Design Information Co., Ltd. (Shanghai, China) for providing technical support and services for the 3D plotter (Envisiontec). The authors thank shiyanjia lab for the support of rheology, compression and AFM analysis.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.03.029.
Contributor Information
Hanjun Li, Email: hanklee@sjtu.edu.cn.
Tingting Tang, Email: ttt@sjtu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Pignochino Y., et al. Sorafenib blocks tumour growth, angiogenesis and metastatic potential in preclinical models of osteosarcoma through a mechanism potentially involving the inhibition of ERK1/2, MCL-1 and ezrin pathways. Mol. Cancer. 2009;8:118. doi: 10.1186/1476-4598-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y., Liu R., Wang W., Wang C., Zhang N., Shao X., He Q., Ying M. Advances in targeted therapy for osteosarcoma based on molecular classification. Pharmacol. Res. 2021;169:105684. doi: 10.1016/j.phrs.2021.105684. [DOI] [PubMed] [Google Scholar]
- 3.Xu M., Xu C.-X., Bi W.-Z., Song Z.-G., Jia J.-P., Chai W., Zhang L.-H., Wang Y. Effects of endostar combined multidrug chemotherapy in osteosarcoma. Bone. 2013;57:111–115. doi: 10.1016/j.bone.2013.07.035. [DOI] [PubMed] [Google Scholar]
- 4.Griend RV 1996 osteosarcoma and its variants Orthop. Clin. N. Am. 27 575-581. [PubMed]
- 5.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui J., Dean D., Hornicek F.J., Chen Z., Duan Z. The role of extracelluar matrix in osteosarcoma progression and metastasis. J. Exp. Clin. Cancer Res. 2020;39:178. doi: 10.1186/s13046-020-01685-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier C.D., Wirtz E.C., Knafler G.J., Morris W.Z., Getty P.J., Greenfield E.M. Micrometastatic drug screening platform shows heterogeneous response to MAP chemotherapy in osteosarcoma cell lines. Clin. Orthop. Relat. Res. 2018;476:1400–1411. doi: 10.1007/s11999.0000000000000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L., Geng H., Mei C., Chen L. Zoledronic acid enhanced the antitumor effect of cisplatin on orthotopic osteosarcoma by ROS-PI3K/AKT signaling and attenuated osteolysis. Oxid. Med. Cell. Longev. 2021;2021:6661534. doi: 10.1155/2021/6661534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H., Han X., Yang S., Wang Y., Dong Y., Tang T. FOXP1 drives osteosarcoma development by repressing P21 and RB transcription downstream of P53. Oncogene. 2021;40:2785–2802. doi: 10.1038/s41388-021-01742-4. [DOI] [PubMed] [Google Scholar]
- 10.Cui X., Hartanto Y., Zhang H. Advances in multicellular spheroids formation. J. R. Soc. Interface. 2017;14:20160877. doi: 10.1098/rsif.2016.0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knight A. Animal experiments scrutinised: systematic reviews demonstrate poor human clinical and toxicological utility. ALTEX. 2007;24:320–325. doi: 10.14573/altex.2007.4.320. [DOI] [PubMed] [Google Scholar]
- 12.Lewis D.I. Animal experimentation: implementation and application of the 3Rs. Emerg. Top. Life Sci. 2019;3:675–679. doi: 10.1042/ETLS20190061. [DOI] [PubMed] [Google Scholar]
- 13.Heinrich M.A., Bansal R., Lammers T., Zhang Y.S., Michel Schiffelers R., Prakash J. vol. 31. 2019. p. 1806590. (3D‐Bioprinted Mini‐Brain: A Glioblastoma Model to Study Cellular Interactions and Therapeutics Advanced Materials). [DOI] [PubMed] [Google Scholar]
- 14.Yang Y., Wang M., Yang S., Lin Y., Zhou Q., Li H., Tang T. Bioprinting of an osteocyte network for biomimetic mineralization. Biofabrication. 2020;12 doi: 10.1088/1758-5090/aba1d0. [DOI] [PubMed] [Google Scholar]
- 15.Wang M., Li H., Yang Y., Yuan K., Zhou F., Liu H., Zhou Q., Yang S., Tang T. A 3D-bioprinted scaffold with doxycycline-controlled BMP2-expressing cells for inducing bone regeneration and inhibiting bacterial infection. Bioact. Mater. 2021;6:1318–1329. doi: 10.1016/j.bioactmat.2020.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman F.E., Burdis R., Kelly D.J. Printing new bones: from print-and-implant devices to bioprinted bone organ precursors. Trends Mol. Med. 2021;27:700–711. doi: 10.1016/j.molmed.2021.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Deng C., Yang J., He H., Ma Z., Wang W., Zhang Y., Li T., He C., Wang J. 3D bio-printed biphasic scaffolds with dual modification of silk fibroin for the integrated repair of osteochondral defects. Biomater. Sci. 2021 doi: 10.1039/d1bm00535a. [DOI] [PubMed] [Google Scholar]
- 18.Haring A.P., Jiang S., Barron C., Thompson E.G., Sontheimer H., He J.-Q., Jia X., Johnson B.N. 3D bioprinting using hollow multifunctional fiber impedimetric sensors. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab94d0. [DOI] [PubMed] [Google Scholar]
- 19.Qiao H., Tang T. Engineering 3D approaches to model the dynamic microenvironments of cancer bone metastasis. Bone Res. 2018;6:3. doi: 10.1038/s41413-018-0008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Chen H., Long X., Xu T. Three-dimensional-engineered bioprinted in vitro human neural stem cell self-assembling culture model constructs of Alzheimer's disease. Bioact. Mater. 2022;11:192–205. doi: 10.1016/j.bioactmat.2021.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie F., et al. Three-dimensional bio-printing of primary human hepatocellular carcinoma for personalized medicine. Biomaterials. 2021;265:120416. doi: 10.1016/j.biomaterials.2020.120416. [DOI] [PubMed] [Google Scholar]
- 22.Warren A., Chen Y., Jones A., Shibue T., Hahn W.C., Boehm J.S., Vazquez F., Tsherniak A., McFarland J.M. Global computational alignment of tumor and cell line transcriptional profiles. Nat. Commun. 2021;12:22. doi: 10.1038/s41467-020-20294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicot C. RNA-seq reveals novel CircRNAs involved in breast cancer progression and patient therapy response. Mol. Cancer. 2020;19:76. doi: 10.1186/s12943-020-01198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S., et al. Multi-omics analysis of primary cell culture models reveals genetic and epigenetic basis of intratumoral phenotypic diversity. Dev. Reprod. Biol. 2019;17:576–589. doi: 10.1016/j.gpb.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seal D.B., Das V., Goswami S., De R.K. Estimating gene expression from DNA methylation and copy number variation: a deep learning regression model for multi-omics integration. Genomics. 2020;112:2833–2841. doi: 10.1016/j.ygeno.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 26.Lam T., Dehne T., Krüger J.P., Hondke S., Endres M., Thomas A., Lauster R., Sittinger M., Kloke L. Photopolymerizable gelatin and hyaluronic acid for stereolithographic 3D bioprinting of tissue-engineered cartilage. J. Biomed. Mater. Res. Part B. 2019;107:2649–2657. doi: 10.1002/jbm.b.34354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chanmee T., Ontong P., Itano N. vol. 375. 2016. pp. 20–30. (Hyaluronan: A Modulator of the Tumor Microenvironment Cancer Lett). [DOI] [PubMed] [Google Scholar]
- 28.Fan Y., Yue Z., Lucarelli E., Wallace G.G. Hybrid printing using cellulose nanocrystals reinforced GelMA/HAMA hydrogels for improved structural integration. Adv. Healthcare Mater. 2020;9:2001410. doi: 10.1002/adhm.202001410. [DOI] [PubMed] [Google Scholar]
- 29.Kutova OM, Sencha LM, Pospelov AD, Dobrynina OE, Brilkina AA, Cherkasova EI, Balalaeva IV 2020 comparative analysis of cell–cell contact abundance in ovarian carcinoma cells cultured in two- and three-dimensional in vitro models Biology 9 446. [DOI] [PMC free article] [PubMed]
- 30.Meazza C, Asaftei SD 2021 State-of-the-art, approved therapeutics for the pharmacological management of osteosarcoma Expet Opin. Pharmacother. 1–12 10.1080/14656566.2021.1936499. [DOI] [PubMed]
- 31.Liu Q., Wang K. The induction of ferroptosis by impairing STAT3/Nrf2/GPx4 signaling enhances the sensitivity of osteosarcoma cells to cisplatin. Cell Biol. Int. 2019;43:1245–1256. doi: 10.1002/cbin.11121. [DOI] [PubMed] [Google Scholar]
- 32.Fu J., Li T., Yang Y., Jiang L., Wang W., Fu L., Zhu Y., Hao Y. Activatable nanomedicine for overcoming hypoxia-induced resistance to chemotherapy and inhibiting tumor growth by inducing collaborative apoptosis and ferroptosis in solid tumors. Biomaterials. 2021;268:120537. doi: 10.1016/j.biomaterials.2020.120537. [DOI] [PubMed] [Google Scholar]
- 33.Tian Z.C., Wang J.Q., Ge H. Apatinib ameliorates doxorubicin-induced migration and cancer stemness of osteosarcoma cells by inhibiting Sox2 via STAT3 signalling. J. Orthopaed. Transl. 2020;22:132–141. doi: 10.1016/j.jot.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Farrill JS, Gordon N. Autophagy in Osteosarcoma Date 2014. Cham: Springer International Publishing 10.1007/978-3-319-04843-7_8.
- 35.Herreros-Pomares A., et al. 3D printing novel in vitro cancer cell culture model systems for lung cancer stem cell study. Mater. Sci. Eng. C. 2021;122:111914. doi: 10.1016/j.msec.2021.111914. [DOI] [PubMed] [Google Scholar]
- 36.Huerta-Reyes M., Aguilar-Rojas A. Three-dimensional models to study breast cancer (Review) Int. J. Oncol. 2021;58:331–343. doi: 10.3892/ijo.2021.5176. [DOI] [PubMed] [Google Scholar]
- 37.Chen H., Cheng Y., Wang X., Wang J., Shi X., Li X., Tan W., Tan Z. 3D printed in vitro tumor tissue model of colorectal cancer. Theranostics. 2020;10:12127–12143. doi: 10.7150/thno.52450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toole B.P., Wight T.N., Tammi M.I. Hyaluronan-cell interactions in cancer and vascular disease. J. Biol. Chem. 2002;277:4593–4596. doi: 10.1074/jbc.R100039200. [DOI] [PubMed] [Google Scholar]
- 39.Zou L., Zou X., Chen L., Li H., Mygind T., Kassem M., Bünger C. Effect of hyaluronan on osteogenic differentiation of porcine bone marrow stromal cells in vitro. J. Orthop. Res. 2008;26:713–720. doi: 10.1002/jor.20539. [DOI] [PubMed] [Google Scholar]
- 40.Nikitovic D., Tzardi M., Berdiaki A., Tsatsakis A., Tzanakakis G.N. Cancer microenvironment and inflammation: role of hyaluronan. Front. Immunol. 2015;6 doi: 10.3389/fimmu.2015.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jabbari E., Sarvestani S., Daneshian L., Moeinzadeh SJPo. vol. 10. 2015. (Optimum 3D Matrix Stiffness for Maintenance of Cancer Stem Cells Is Dependent on Tissue Origin of Cancer Cells). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W., Li X., Meng F.B., Wang Z.X., Zhao R.T., Yang C.Y. Effects of the long non-coding RNA HOST2 on the proliferation, migration, invasion and apoptosis of human osteosarcoma cells. Cell. Physiol. Biochem. 2017;43:320–330. doi: 10.1159/000480412. [DOI] [PubMed] [Google Scholar]
- 43.Lan H., Hong W., Fan P., Qian D., Zhu J., Bai B. Quercetin inhibits cell migration and invasion in human osteosarcoma cells. Cell. Physiol. Biochem. 2017;43:553–567. doi: 10.1159/000480528. [DOI] [PubMed] [Google Scholar]
- 44.Hirschhaeuser F., Menne H., Dittfeld C., West J., Mueller-Klieser W., Kunz-Schughart L.A. Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 2010;148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 45.Yu D., et al. Identification of synergistic, clinically achievable, combination therapies for osteosarcoma. Sci. Rep. 2015;5 16991 doi: 10.1038/srep16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbott A., Bond K., Chiba T., Sims-Lucas S., Oxburgh L., Coburn J.M. Development of a mechanically matched silk scaffolded 3D clear cell renal cell carcinoma model. Mater. Sci. Eng. C. 2021;126:112141. doi: 10.1016/j.msec.2021.112141. [DOI] [PubMed] [Google Scholar]
- 47.Rodríguez-Enríquez S., Gallardo-Pérez J.C., Avilés-Salas A., Marín-Hernández A., Carreño-Fuentes L., Maldonado-Lagunas V., Moreno-Sánchez R. Energy metabolism transition in multi-cellular human tumor spheroids. J. Cell. Physiol. 2008;216:189–197. doi: 10.1002/jcp.21392. [DOI] [PubMed] [Google Scholar]
- 48.Kumarasuriyar A, Lee I, Nurcombe V, Cool SM 2009 De-sulfation of MG-63 cell glycosaminoglycans delays in vitro osteogenesis, up-regulates cholesterol synthesis and disrupts cell cycle and the actin cytoskeleton J. Cell. Physiol. 219 572-583 10.1002/jcp.21700. [DOI] [PubMed]
- 49.Kamynina E., Lachenauer E.R., DiRisio A.C., Liebenthal R.P., Field M.S., Stover P.J. vol. 114. 2017. Arsenic trioxide targets MTHFD1 and SUMO-dependent nuclear de novo thymidylate biosynthesis; pp. E2319–E2326. (Proceedings of the National Academy of Sciences). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan L.Y., et al. Desmoglein 2 promotes vasculogenic mimicry in melanoma and is associated with poor clinical outcome. Oncotarget. 2016;7:46492–46508. doi: 10.18632/oncotarget.10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janiszewska M., Primi M.C., Izard T. Cell adhesion in cancer: beyond the migration of single cells. J. Biol. Chem. 2020;295:2495–2505. doi: 10.1074/jbc.rev119.007759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brennan D., Mahoney M.G. Increased expression of Dsg2 in malignant skin carcinomas. Cell Adhes. Migrat. 2009;3:148–154. doi: 10.4161/cam.3.2.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin S., et al. DSG2 expression is correlated with poor prognosis and promotes early-stage cervical cancer. Cancer Cell Int. 2020;20:206. doi: 10.1186/s12935-020-01292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marchandet L., Lallier M., Charrier C., Baud'Huin M., Ory B., Lamoureux F. vol. 13. 2021. p. 683. (Mechanisms of Resistance to Conventional Therapies for Osteosarcoma Cancers (Basel)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Degenhardt K., et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim H.J., Lee S.G., Kim Y.-J., Park J.-E., Lee K.Y., Yoo Y.H., Kim J.-M. Cytoprotective role of autophagy during paclitaxel-induced apoptosis in Saos-2 osteosarcoma cells. Int. J. Oncol. 2013;42:1985–1992. doi: 10.3892/ijo.2013.1884. [DOI] [PubMed] [Google Scholar]
- 57.Maiuri M.C., Zalckvar E., Kimchi A., Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 58.Pignochino Y., et al. The combination of sorafenib and everolimus abrogates mTORC1 and mTORC2 upregulation in osteosarcoma preclinical models. Clin. Cancer Res. 2013;19:2117–2131. doi: 10.1158/1078-0432.CCR-12-2293. [DOI] [PubMed] [Google Scholar]
- 59.Grignani G., et al. Sorafenib and everolimus for patients with unresectable high-grade osteosarcoma progressing after standard treatment: a non-randomised phase 2 clinical trial. Lancet Oncol. 2015;16:98–107. doi: 10.1016/S1470-2045(14)71136-2. [DOI] [PubMed] [Google Scholar]
- 60.Ishibashi Y., Nakamura O., Yamagami Y., Nishimura H., Fukuoka N., Yamamoto T. Chloroquine enhances rapamycin-induced apoptosis in MG63 cells. Anticancer Res. 2019;39:649. doi: 10.21873/anticanres.13159. [DOI] [PubMed] [Google Scholar]
- 61.Shen C., Wang W., Tao L., Liu B., Yang Z., Tao H. Chloroquine blocks the autophagic process in cisplatin-resistant osteosarcoma cells by regulating the expression of p62/SQSTM1. Int. J. Mol. Med. 2013;32:448–456. doi: 10.3892/ijmm.2013.1399. [DOI] [PubMed] [Google Scholar]
- 62.Liu Z., Wang H., Hu C., Wu C., Wang J., Hu F., Fu Y., Wen J., Zhang W. Targeting autophagy enhances atezolizumab-induced mitochondria-related apoptosis in osteosarcoma. Cell Death Dis. 2021;12:164. doi: 10.1038/s41419-021-03449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.