Abstract
Octocorals are among the most prolific sources of biologically active compounds. A significant part of their specialized metabolites richness is linked to the abundance of their associated microbiota. Consequently, research on the bioprospecting potential of microorganisms associated with these marine invertebrates has gained much interest. Here, we describe the draft genome of Gordonia hongkongensis strain EUFUS-Z928 isolated from the octocoral Eunicea fusca. The genome was assembled de novo from short-read whole-genome sequencing data. Additionally, functional annotation of predicted genes was performed using the RAST tool kit, including genome mining for specialized metabolite biosynthetic gene clusters using the antiSMASH v6.0 tool. The genome sequence data of G. hongkongensis EUFUS-Z928 can provide information for further analysis of the potential biotechnological use of this microorganism and guide the characterization of other related actinobacterial isolates. Likewise, this information increases the analytical capacity for studying the genus Gordonia.
Keywords: Actinobacteria, Marine actinomycete, Rare actinobacteria, Corynebacteriales
Specifications Table
| Subject | Biological sciences |
| Specific subject area | Biotechnology, Microbiology: Bacteriology, Omics: Genomics |
| Type of data | Table Figure Draft genome sequence data |
| How the data were acquired | Whole-genome sequencing using Illumina NovaSeq 6000 platform for short reads |
| Data format | Raw Analyzed |
| Description of data collection | Strain EUFUS-Z928 was isolated from the octocoral Eunicea fusca. High-quality DNA was extracted and sequenced using Illumina NovaSeq 6000 (short reads). Raw paired-end reads were de novo assembled following the Shovill pipeline. The assembly was scaffolded with the MEDUSA algorithm, and annotation was performed using PATRIC web resources. Detection of specialized metabolite biosynthesis gene clusters was conducted with the antiSMASH tool. |
| Data source location | • Institution: Universidad de La Sabana • City/Town/Region: Chía, Cundinamarca • Country: Colombia • GPS coordinates for collected samples: 11°15′02.1″N 74°13′16.0″W |
| Data accessibility | Repository name: OSF Data identification number: R4UZ8 Direct URL to data: https://osf.io/r4uz8/. |
Value of the Data
-
•
The draft genome data of Gordonia hongkongensis strain EUFUS-Z928 provides valuable information for the study of the evolution of the genus Gordonia and its biotechnological potential.
-
•
These data are valuable for environmental and clinical microbiology, bioprospecting, and biotechnology researchers.
-
•
These data can be used for genome mining to discover novel metabolite biosynthesis pathways.
-
•
Given the potential shown by Gordonia species in bioremediation, these data serve to conduct comparative genomics work further and allow a better understanding of the mechanisms involved in bioremediation processes.
1. Data Description
The strain EUFUS-Z928 was isolated from the octocoral Eunicea fusca collected in Santa Marta Bay, Colombia. Table 1 shows the results of the de novo and scaffolded genome assembly of the strain EUFUS-Z928. Scaffolding substantially improved the assembly by reducing the number of contigs by 76.23% and leaving an L50 and L75 of 1 (N50=5,295,384). The scaffolding was performed using as reference the genomes of the closest relatives according to the overall genome relatedness indices (OGRI) results obtained on the de novo assembly (Table S1: https://osf.io/q8xus/).
Table 1.
Characteristics of the de novo assembly and scaffolded genome of strain EUFUS-Z928.
| Features | de novo Assembled Genome | Scaffolded Genome |
|---|---|---|
| Genome size (bp) | 5,329,221 | 5,333,421 |
| Total number of contigs | 122 | 29 |
| Largest contig (bp) | 599,980 | 5,295,384 |
| N50 (bp) | 252,700 | 5,295,384 |
| N75 (bp) | 105,922 | 5,295,384 |
| L50 | 7 | 1 |
| L75 | 15 | 1 |
| GC (%) | 67.97 | 67.96 |
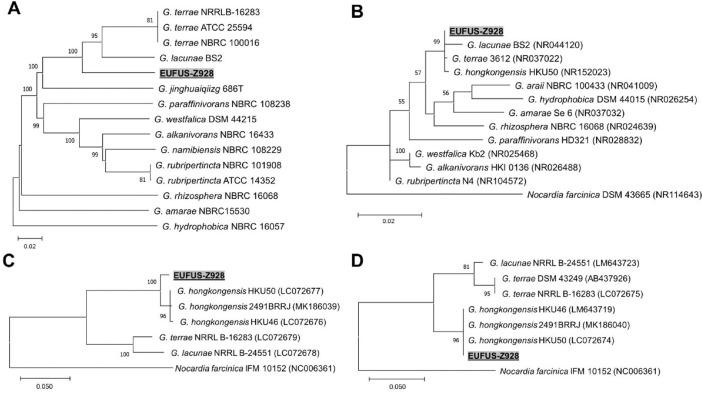
The genome-based classification and identification found the strain EUFUS-Z928 to be closely related to Gordonia terrae and Gordonia lacunae type strains (Table 2, Fig. 1A). Phylogeny analysis with the 16S rRNA gene also found a close relationship with Gordonia hongkongensis (Fig. 1B). Finally, phylogenetic analysis with the sequences of the genes coding for protein translocase subunit SecA1 (secA1) and DNA gyrase subunit B (gyrB) allowed classification of strain EUFUS-Z928 as G. hongkongensis (Fig. 1C and D). It is important to clarify that at the time of the analysis, G. honkongensis genomes were not available in the Type Strain Genome Server (TYGS); therefore, it was impossible to include them in the whole genome-based phylogram.
Table 2.
Overall genome relatedness indices (OGRI) between EUFUS-Z928 and the closely related type strain genomes.
| Strain | dDDHa (d0, in %) |
dDDHa (d4, in %) |
dDDHa (d6, in %) |
G+CΔb (in %) |
ANIbc (%) |
ANImc (%) |
|---|---|---|---|---|---|---|
| G. terrae NRRL B-16283 | 70.80 | 34.50 | 61.50 | 0.15 | 87.68 | 88.83 |
| G. terrae NCTC 10669 | 70.80 | 34.50 | 61.40 | 0.15 | 87.70 | 88.81 |
| G. terrae NBRC 100016 | 70.40 | 34.40 | 61.10 | 0.12 | 87.69 | 88.83 |
| G. lacunae BS2 | 69.40 | 35.50 | 61.00 | 0.12 | 88.09 | 89.19 |
digital DNA–DNA hybridization (DDH): formula d0 (length of all high-scoring segment pairs (HSPs) divided by total genome length), formula d4 (sum of all identities found in HSPs divided by overall HSP length), formula d6 (sum of all identities found in HSPs divided by total genome length).
G+C content difference.
Average nucleotide identity based on BLAST (ANIb) and MUMmer (ANIm).
Fig. 1.
Phylograms of strain EUFUS-Z928 based on (A) whole-genome sequences, (B) 16S rRNA gene sequences, (C) gyrB gene sequences and (D) secA1 gene sequences. The phylogenetic trees were drawn to scale, with branch lengths measured in the number of substitutions per site. The percentage of bootstrap replicates >50% (out of 100 for whole-genome and out of 1000 for single-gene trees) that supported each node are shown. Genome BLAST Distance Phylogeny approach was used for the whole-genome cladogram using the TYGS server. Single-gene phylogenetic trees were inferred by maximum likelihood with the IQ-TREE algorithm.
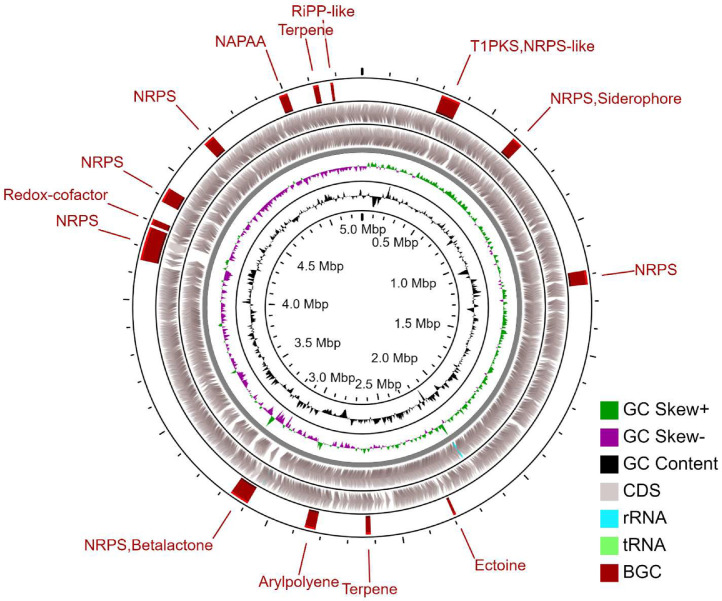
A total of 5042 genes were annotated in the genome of G. honkongensis EUFUS-Z928 (the complete annotation data can be found in Table S2: https://osf.io/2ra3k/). Of these, 4987 corresponded to coding sequences (CDS), most of them (62.44%) with a functional assignment (Table 3). Additionally, analysis with the antiSMASH v6.0 tool identified 14 biosynthetic gene clusters (BGCs) (Table 3 and Fig. 2), among which NRPS and Terpene had more than 1 cluster (i.e., 4 and 2, respectively).
Table 3.
Annotation results of the G. hongkongensis EUFUS-Z928 genome.
| Feature | Values |
|---|---|
| tRNAa | 47 |
| rRNAa | 8 |
| CDSa | 4987 |
| Hypothetical proteins | 1873 |
| Proteins with functional assignments | 3114 |
| Proteins with GO assignmentsa | 991 |
| Proteins with Subsystem assignmentsa | 1640 |
| BGCb | 14 |
| Arylpolyene | 1 |
| Ectoine | 1 |
| NAPAA | 1 |
| NRPS | 4 |
| NRPS, Betalactone | 1 |
| NRPS, Siderophore | 1 |
| Redox-cofactor | 1 |
| RiPP-like | 1 |
| T1PKS,NRPS-like | 1 |
| Terpene | 2 |
According to the RAST tool kit using the PATRIC service center.
According to the antiSMASH v6.0 tool.
Fig. 2.
Circular genome view of G. hongkongensis EUFUS-Z928. The inner ring shows the length of the genome. The following two rings show the GC content and GC skew, respectively. The gray rings correspond to the CDSs annotated by the RAST tool kit in each DNA direction. The outer ring indicates the BGCs annotated by antiSMASH v6.0.
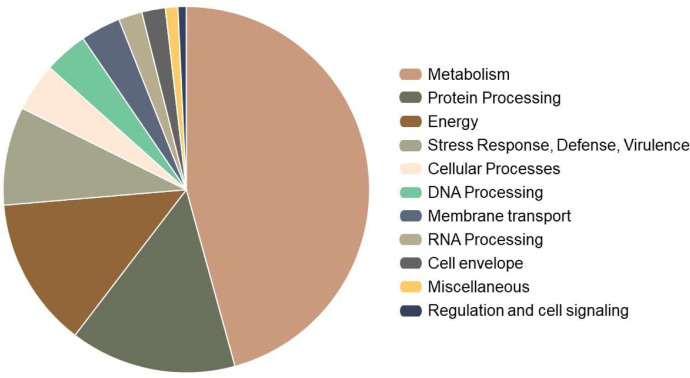
Regarding proteins with assignments to subsystems, as shown in Fig. 3, Metabolism (45.61%), Protein Processing (14.76%), Energy (13.23%), and Stress Response, Defense, Virulence (8.72%) were the subsystems with the highest assignments. In the latter, genes related to antibiotic resistance (n = 43), arsenic resistance (n = 5), as well as genes related to protection against oxidative stress such as mycothiol (n = 10) and protection from reactive oxygen species (n = 3) stand out. Complete information on the 1640 genes assigned to subsystems is shown in Table S3 (https://osf.io/6j5zs/).
Fig. 3.
Overview of the assignments to functional subsystems of the G. hongkongensis EUFUS-Z928 genome according to the PATRIC annotation service.
According to the List of Prokaryotic names with Standing in Nomenclature, 47 species of the genus Gordonia have been reported so far (https://lpsn.dsmz.de/genus/gordonia; consulted on 04/02/2022). Although several strains of Gordonia are opportunistic pathogens, their potential for bioremediation of polluted environments [1] makes them a valuable biological resource in several research areas. The whole-genome sequence and functional annotation data of G. hongkongensis EUFUS-Z928 provides valuable information to facilitate the design and execution of more in-depth studies such as comparative genomics and genome mining.
2. Experimental Design, Materials and Methods
2.1. Strain isolation and DNA extraction
Strain EUFUS-Z928 was isolated from a sample of the octocoral Eunicea fusca (collected by diving at Punta de Betín, 11°15′02.1″N 74°13′16.0″W, Santa Marta, Magdalena, Colombia). The isolation was carried out using a modified Zobell medium (1.25 g of yeast extract, 3.75 g of peptone, 18 g of NaCl, 2 g of MgCl2, 0.525 g of KCl, 0.075 g of CaCl2 and 15 g of agar dissolved in enough distilled water to make 1 l of solution) supplemented with nalidixic acid (50 μg/mL). Genomic DNA extraction was performed using the Quick-DNA Fungal/Bacterial Microprep kit (Zymo Research Corporation, Irvine, CA, USA) following the manufacturer's instructions. The quality of the extracted DNA was verified by agarose gel electrophoresis and quantified using Qubit 1X dsDNA High Sensitivity kit (Invitrogen, Life Technologies, CA, USA).
2.2. Whole genome sequencing, assembly and annotation
Whole-genome sequencing was performed by Macrogen Inc. (Korea) using Illumina paired-end sequencing technology. Short read (151 bp) libraries were prepared using TruSeq Nano DNA Library Prep kit (Part # 15041110, Rev. D, Illumina, Inc., San Diego, CA, USA) and sequencing on Illumina NovaSeq 6000 platform. The raw sequence reads were quality filtering, trimming and de novo assembled applying the Shovill pipeline v1.1.0 (with default parameters) [2], employing SPAdes as the assembler tool [3]. Contigs shorter than 200 bp were removed. To check the quality of the de novo assembly, the genome completeness was analyzed by the BUSCO tool [4] (it reached 99.8%), and the ContEst16S algorithm [5] did not identify contamination in the assembled genome. Genome sequencing and assembly data are available from NCBI BioProject with accession PRJNA798903.
Genome scaffolding was performed using the MeDuSa web server [6] with the reference genomes Gordonia sp. SGD-V-85 (RefSeq assembly accession: GCF_001456905) and Gordonia terrae (RefSeq assembly accession: GCF_901542405). These genomes were selected considering the results of the OGRI with the de novo assembled genome. The de novo assembled and scaffolded genomes were compared using the QUAST web server [7]. Genome annotation was done according to the RAST tool kit using the PATRIC service center [8]. To detect and characterize the content of specialized metabolite BGCs, we annotated the genome using the antiSMASH v6.0 tool [9]. A graphical circle map was generated on the CGView server to visualize the annotation results [10].
2.3. Phylogeny analysis
The analysis was conducted on both a genome-wide and single gene basis; including 16S ribosomal RNA gene (well established for phylogenetic analysis of bacteria), secA1, and gyrB genes, which have also been used for Gordonia phylogenetic analysis with more discriminatory power to identification at the species level [11]. The OGRI were calculated using the TYGS [12] and JSpeciesW [13] web servers. The sequences of 16S rRNA, secA1, and gyrB genes were retrieved from our annotated genome G. hongkongensis EUFUS-Z928. Phylogenetic trees for single gene analysis were estimated based on the maximum likelihood method using the IQ-Tree tool [14] (bootstrap values were calculated from 1000 replicates). Phylograms were generated using MEGA v11.0.10 [15]. Whole-genome phylogeny analyses were inferred using the Genome BLAST Distance approach in the TYGS server.
Ethics Statements
The samples used by this research were of Colombian origin, and they were obtained according to Amendment No. 5 to ARG Master Agreement No. 117 of 26 May 2015, granted by the Ministry of Environment and Sustainable Development, Colombia.
CRediT authorship contribution statement
Jeysson Sánchez-Suárez: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Visualization. Luis Díaz: Project administration, Funding acquisition, Supervision, Writing – review & editing. Javier Melo-Bolivar: Software, Validation, Formal analysis, Data curation. Luisa Villamil: Project administration, Funding acquisition, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Prof. Sven Zea for his help in the octocoral sampling, Prof. Juan Armando Sánchez for octocoral identification, Prof. Javier Gómez, and the staff of Marine Bioprospecting Laboratory –LabBIM– at “INVEMAR” for the logistic support. We are also grateful to Jorge Rodríguez for his assistance with laboratory procedures.
Funding
This work was supported by Minciencias (Ministerio de Ciencia, Tecnología e Innovación –Colombia– [project code 123080864187, contract 80740-168-2019]); and by Universidad de La Sabana (General Research Directorate, project ING-175-2016).
Data Availability
References
- 1.Sowani H., Kulkarni M., Zinjarde S. Harnessing the catabolic versatility of Gordonia species for detoxifying pollutants. Biotechnol. Adv. 2019;37:382–402. doi: 10.1016/j.biotechadv.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 2.T. Seemann, R. Edwards, A. Goncalves da Silva, K. Kiil, Shovill-assemble bacterial isolate genomes from Illumina paired-end reads, (2020). https://github.com/tseemann/shovill. (Accessed January 3, 2022).
- 3.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 5.Lee I., Chalita M., Ha S.M., Na S.I., Yoon S.H., Chun J. ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017;67:2053–2057. doi: 10.1099/ijsem.0.001872. [DOI] [PubMed] [Google Scholar]
- 6.Bosi E., Donati B., Galardini M., Brunetti S., Sagot M.F., Lió P., Crescenzi P., Fani R., Fondi M. MeDuSa: a multi-draft based scaffolder. Bioinformatics. 2015;31:2443–2451. doi: 10.1093/bioinformatics/btv171. [DOI] [PubMed] [Google Scholar]
- 7.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis J.J., Wattam A.R., Aziz R.K., Brettin T., Butler R., Butler R.M., Chlenski P., Conrad N., Dickerman A., Dietrich E.M., Gabbard J.L., Gerdes S., Guard A., Kenyon R.W., Machi D., Mao C., Murphy-Olson D., Nguyen M., Nordberg E.K., Olsen G.J., Olson R.D., Overbeek J.C., Overbeek R., Parrello B., Pusch G.D., Shukla M., Thomas C., VanOeffelen M., Vonstein V., Warren A.S., Xia F., Xie D., Yoo H., Stevens R. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blin K., Shaw S., Kloosterman A.M., Charlop-Powers Z., van Wezel G.P., Medema M.H., Weber T. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49:W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stothard P., Grant J.R., Van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Brief. Bioinform. 2019;20:1576–1582. doi: 10.1093/bib/bbx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang Y., Takeda K., Yazawa K., Mikami Y. Phylogenetic studies of Gordonia species based on gyrB and secA1 gene analyses. Mycopathologia. 2009;167:95–105. doi: 10.1007/s11046-008-9151-y. [DOI] [PubMed] [Google Scholar]
- 12.Meier-Kolthoff J.P., Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter M., Rosselló-Móra R., Oliver Glöckner F., Peplies J. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics. 2016;32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trifinopoulos J., Nguyen L.T., von Haeseler A., Minh B.Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.