Abstract
Understanding mechanisms underlying the heterogeneity of multipotent stem cells offers invaluable insights into biogenesis and tissue development. Extracellular matrix (ECM) stiffness has been acknowledged as a crucial factor regulating stem cell fate. However, how cells sense stiffness cues and adapt their metabolism activity is still unknown. Here we report the novel role of mitochondrial phosphoenolpyruvate carboxykinase (PCK2) in enhancing osteogenesis in 3D ECM via glycolysis. We experimentally mimicked the physical characteristics of 3D trabeculae network of normal and osteoporotic bone with different microstructure and stiffness, observing that PCK2 promotes osteogenesis in 3D ECM with tunable stiffness in vitro and in vivo. Mechanistically, PCK2 enhances the rate-limiting metabolic enzyme pallet isoform phosphofructokinase (PFKP) in 3D ECM, and further activates AKT/extracellular signal-regulated kinase 1/2 (ERK1/2) cascades, which directly regulates osteogenic differentiation of MSCs. Collectively, our findings implicate an intricate crosstalk between cell mechanics and metabolism, and provide new perspectives for strategies of osteoporosis.
Keywords: Osteogenesis, Osteoporosis, Extracellular matrix, Stiffness, PCK2, Mesenchymal stem cells
Graphical abstract
Highlights
-
•
As the key rate-limiting enzyme of gluconeogenesis, PCK2 manipulates osteogenesis in stiff and soft ECM in vitro and in vivo.
-
•
PCK2 regulates osteogenic capacity of BMMSCs in 3D ECM with different stiffness, via modulating glycolysis and regulating PFKP-AKT/ERK signaling pathways.
Abbreviations
- ALP
alkaline phosphatase
- AFM
atomic force microscopy
- ARS
alizarin red S
- DAPI
4ʹ,6-diamidino-2-phenylindole
- DHAP
dihydroxyacetone phosphate
- α-MEM
minimum essential medium-α;
- ECM
extracellular matrix
- EDTA
ethylene diamine tetra acetic acid
- FBS
fetal bovine serum
- F-1, 6-BP
Fructose 1, 6-bisphosphate
- F-6-P
Fructose 6-phosphate
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GelMA
methacrylate gelatin
- G-6-P
glucose 6-phosphate
- H&E
hematoxylin and eosin
- BMMSCs
bone marrow derived mesenchymal stem cells
- IHC
immunohistochemistry
- NC
negative control
- PEP
phosphoenolpyruvate
- OVX
ovariectomy
- PCK2
mitochondrial phosphoenolpyruvate carboxykinase
- PEPCK
phosphoenolpyruvate carboxykinase
- PFKP
pallet isoform phosphofructokinase
- PFKM
muscle isoform phosphofructokinase
- PFKL
liver isoform phosphofructokinase
- PM
proliferation culture medium
- RUNX2
Runt-related transcription factor 2
- SEM
standard error of mean
- siRNA
small interfering RNA
- vector
scramble control
- YAP
yes-associated protein
1. Introduction
One of the most fundamental life processes during biogenesis and development is the harmonious establishment of organs and tissues; and stem cell heterogeneity plays a conspicuous part in it [1]. Aging and various pathological states are caused by the overcharge of the stem cell lineage commitment [2]. Understanding the mechanisms of how stem cell fate is decided is pivotal for constructing these vital structures like bone and interpreting the etiology of diseases.
Bone marrow-derived mesenchymal stem cells (BMMSCs) reside in a complex mesenchymal niche that surrounded by multiple of biochemical and biophysical cues, which play intricate roles in regulating cell proliferation, morphology, self-renew and multi-differentiation commitment [3,4]. Among these factors, stiffness of the cellular microenvironment is increasingly inclined as a critical regulator in stem cell fate decisions [3,5]. Mounting in vitro evidence has revealed that stiff gels (∼30 kPa) favor osteogenesis, while soft matrix (∼5 kPa) facilitates adipogenesis, both in two-dimensional (2D) and three-dimensional (3D) model [6,7]. Mechanisms mediating mechano-transduction in 2D culture are substantially well learned. However, the knowledge of how 3D microenvironment stiffness decides stem cell fate is still limited. The emerging development of the mechanobiological study has bridged the bioengineering and physical cues, and thus boosting our knowledge of the extracellular matrix (ECM) stiffness on regulation of disease [[8], [9], [10]]. Tissue stiffen is related to cancer and cardiovascular disease, while decreased ECM of tissue also takes place with fibrosis, advanced age, and degenerated diseases, in virtue of accelerated matrix deposition, and cross-linking of existing matrix, etc [11]. The occurrence of osteoporosis is accompanied by the change of three-dimensional (3D) physical microenvironment of bone tissue, including the loose reticular trabeculae network, the decreased stiffness of trabecular bone [12]; in this 3D ECM, osteogenic capacity of MSCs is weakened [13]. Since matrix stiffness is a promising mechanical target to alter stem cell fate, understanding how 3D ECM stiffness regulates the osteogenic differentiation of MSCs [4] has great potential to activate the deteriorated regenerative potential of MSCs in osteoporosis bone, and facilitate the knowledge of osteoporotic etiology, and the new engineering strategies for the treatment of osteoporosis.
The complex microenvironment provides biomechanical cues that elicit intrinsic signaling cascades in cells. Previous studies have shown that various membrane ligands sense matrix hardness and regulate stem cell fate [14]. However, the knowledge of complicated intracellular biochemical reactions in the cytoplasm subsequently coordinating to sense matrix stiffness is still unexplored. It is well-established that stiffness-dependent regulation of stem cell differentiation is mediated by the activation and localization of the nuclear transcription regulator, Yes-associated protein (YAP), which leads to activation of downstream gene signaling for osteogenic differentiation [15]. Other mech-transduction factors modulating the osteogenic commitment of MSCs, via triggering intracellular signaling, include integrin α5/β1, focal adhesion kinase (FAK), mitogen-activated protein kinase (MAPK), and Wnt signaling [16,17]. These finding indicates that biochemical signals could remodel the cytoskeleton via regulating actomyosin contractility, and thus modulating cell functions like growth, differentiation, morphogenesis, autophagy and apoptosis [6]. It is noteworthy that all these physiological processes cost energy. Recently, studies have begun to bring insights into the possible interconnection between ECM properties and glucose metabolism [18,19]. For example, the Phosphoinositide 3-Kinase (PI3K) directly integrates glycolysis and cytoskeletal dynamics via AKT signaling, to regulate cell behaviors [19]. These studies provoke us to explore how stem cells adjust the metabolic activity by sensing variable mechanical cues in 3D ECM with different stiffness.
The key gluconeogenic enzyme, phosphoenolpyruvate carboxykinase (PEPCK) promotes cell metabolic flexibility to resist energy stress, and thus contributing to anabolic metabolism. Cytoplasmic isoform PCK1 and mitochondrial isoform PCK2 are the two isoforms of PEPCK [20]. In different cell lines, PCK2 expression was upregulated under low glucose conditions [[20], [21], [22], [23]]. The cyclic AMP-dependent transcription factor 4 (ATF4) and endoplasmic reticulum stress could elevate the expression of PCK2 [22]. Moreover, through catalyzing oxaloacetate (OAA) from glutamine, PCK2 directly regulates tricarboxylic acid (TCA) cycle function. Our previous work showed that PCK2 promotes MSCs differentiate into osteoblasts in autophagy dependent manner [24]. However, the cues regulating the osteogenesis of MSCs by PCK2 in the complex 3D microenvironment, remain to be poorly understood. A recent study revealed a mechanism by which the key enzyme of glycolysis responds to architectural features of the cytoskeleton in matrix with different stiffness, thus integrating cell metabolism to the mechanical properties of microenvironment [25]. PCK2 has been concurrently demonstrated to mediate gluconeogenesis by replenishing glycolytic pools [[20], [21], [22], [23]], but the interplay of PCK2 and glycolysis in 3D ECM with different stiffness is still unknown.
Here, we report the role of PCK2 in promoting MSCs osteogenesis in 3D ECM via regulating glycolysis (Fig. 1A). By adjusting the crosslinking degree and percentage of methacrylate gelatin (GelMA), the physical 3D network characteristics of trabeculae of normal and osteoporotic bone were experimentally mimicked in vitro. We find that PCK2 could enhance osteogenesis in 3D ECM with tunable stiffness in vitro and in vivo. Mechanistically, PCK2 elevates the glycolysis via rate-limiting metabolic enzyme pallet isoform phosphofructokinase (PFKP) in 3D ECM, and further activates downstream cascades, which directly regulates osteogenic differentiation of MSCs. Together, understanding how PCK2 assists osteogenic differentiation of MSCs responding to the stiffness of 3D ECM provides new approaches for bone regeneration under the condition of osteoporosis, from a novel perspective of glucose metabolism.
Fig. 1.
The physical microenvironment of physiological-mimetic normal bone is different with the pathological-mimetic osteoporotic bone in micro and nano scale. (A) A schematic hypothesis of how PCK2 regulates BMMSCs osteogenesis in 3D ECM with different stiffness. (B) Upper panel: representative SEM images of GelMA with tunable stiffness (GM30, 6% and GM90, 8%). The scale bar is 100 μm. Lower panel: representative SEM images of normal femur and osteoporotic femur from sham and OVX rats (3-month). Scale bar is 100 μm. (C) Surface plot images of sham and OVX trabecular bone by AFM imaging (5 × 5 μm). Scale bar is 1 μm. The region of interest: prominent mineral deposits (red arrows), limited mineral-depleted areas (blue arrows). The collagen fibers and mineral crystals were packed in trabeculae and presented as nodular like surfaces. Bundles combining fibrillar collagen and embedded minerals (yellow arrows) may be observed. Light areas correspond to high zones on the sample surface. (D) The typical mechanics heatmaps of 16 detected positions in trabecular bone of sham and OVX femurs by AFM (n = 5). (E) Quantification of stiffness indicated that the trabecular bone of sham femurs was stiffer than that of OVX femurs (***P < 0.001, by students' t-test). All the data was repeated three times and the represents means ± SEM.
2. Materials and methods
2.1. Establishment of osteoporotic model by ovariectomy and sham operations
The Institutional Animal Care and Use Committee of Peking University (Number: LA2021040) has approved the animal surgical procedures in this study. 8-week-old female Sprague Dawley rats (SD rats, Vital River Corporation, Beijing, China) were randomly assigned to the sham and OVX groups (20 rats per group). Pentobarbital sodium (50 mg/kg, i.p.) was used for general anesthesia by intraperitoneal injections for all the rats. Bilateral OVX or sham operation was performed through standard methods [26].
2.2. Preparation of SEM samples for GelMA and femurs
GelMA was bought from Yongqinquan Intelligent Equipment Co., Ltd., Suzhou, China. According to the product information of GelMA, the photo initiator 0.25% (w/v) for GelMA hydrogel dissolved in deionized water, and then GM30/60/90 was mixed with the photo initiator in appropriate percentage (GM30, 6%; GM60, 6%; GM90, 8%). The bottom of 2 ml centrifuge tubes was cut off, and the GM30/60/90 hydrogel precursor was added to the tubes. Light curing by 405 nm light was maintained for 90s until the hydrogel was completely crosslinked. The cured samples were put into the refrigerator at - 80 °C for freezing for 2 h (note: do not open the door of refrigerator during freezing). The frozen samples were quickly transferred to the vacuum freeze dryer for fully dry (∼18 h). The freeze-dried samples were cut into uniform 1 mm slices with a sharp blade for SEM observation.
After 12 weeks, the rats from sham and OVX groups were sacrificed and the femurs were isolated, respectively. 1–3 mm-thickness transverse sections of fresh femoral head were cut using Low Speed Diamond Wheel Saw (Model 650, SBT) and polished using lapping machine (EM TXP, Leica) prior to SEM tests. After removing organic material and shaking off excess tissue from the trabecular pores of the sample, the samples were put in a 60 °C oven for 8 h on a glass dish to evaporate any residue. After that, we placed samples under vacuum overnight for SEM observation.
2.3. Atomic force microscopy (AFM)
AFM measurements were all performed by an AFM (BioScope Resolve, Bruker) with PeakForce QNM mode at room temperature. Cellular elastic modulus was measured by AFM nanoindentation experiments with the ramp mode in fluid. A silicon nitride cantilever (PFQNM-LC-A-CAL, Bruker) was used with pre-calibrated spring constant of 0.091 N/m, tip height of 17 μm, and tip radius of 65 nm. The deflection sensitivity of the cantilever was calibrated by thermal tuning prior to nanoindentation experiments. Force curves of cells were captured with a ramp size of 1.5 μm, a ramp rate of 1 Hz and a relative deflection error threshold of 6 nm. We obtained 16 force mappings per sample, and 30 different points were analyzed for each condition.
1 mm-thickness transverse sections of fresh bone were cut using Low Speed Diamond Wheel Saw (Model 650, SBT) and polished using lapping machine (EM TXP, Leica) prior to AFM tests. The cut sections were attached to glass slides with transparent nail polish. AFM imaging and nanoindentation experiments for femoral head sections were performed in ambient conditions using a silicon cantilever (RTESPA-300, Bruker) with a nominal spring constant of 40 N/m and a tip radius of 8 nm. The probe was calibrated with Sader method before force curve detection. Topology images were scanned with a scan size of 5 × 5 μm2 and a resolution of 512 × 512 pixels. Force curves of femoral head sections were captured with a ramp size of 600 nm, a ramp rate of 1 Hz and a relative deflection error threshold of 2 nm. AFM data were analyzed with Nanoscope Analysis software (version 1.8, Bruker). The elastic modulus of the samples was obtained through Hertzian model assuming the Poisson ratio of the cells (0.5) and bones (0.35).
2.4. Physical properties of GelMA
Mechanical modulus: The photo initiator 0.25% (w/v) for GelMA hydrogel dissolved in deionized water, and then GM30/60/90 was mixed with the photo initiator in appropriate percentage (GM30, 6%; GM60, 6%; GM90, 8%). 200 μL hydrogel precursor was taken out to inject into the mold with diameter of 8 mm (n = 4). After light curing by 405 nm ultraviolet light for 90s, the cylindrical GelMA was put on the hydrogel micro-force tester (EFL-MT-5600, EFL) to measure the mechanical properties with compression testing. The rate of stress relaxation of GelMA gels was recorded and the first 10% slope of the strain on the stress-strain curve was taken as its compression modulus [27].
Fourier-transform infrared (FTIR) spectroscopy test: the FTIR spectroscopy test was performed by ATR mode via potassium bromide pressing, using Bruker VERTEX80v FTIR spectrometer. Before the measurement, the prepared GelMA-30/60/90 were vacuum dried and ground with potassium bromide powder.
Swelling Ratio Test: to test the swelling ratio, the GelMA-30/60/90 (n = 4 for each group) were soaked in 500 μl PBS buffer at 37 °C, and weighed at specific time points (0, 2, 4, 8, 12, 24, 48 and 72 h). Before measuring the weight of samples, the excess surface water was removed. The swelling ratio for each sample was calculated with the equation: Swelling Ratio = (W2-W1)/W1 × 100%, where W2 was the real-time weight at every point, and W1 was the initial weight of the sample.
Degradation test: to measure the enzymatic degradation properties, the prepared GelMA-30/60/90 (n = 4 for each group) were immersed in 500 μl PBS containing 1U ml-1 collagenase type II at 37 °C. At a predefined time, the samples were removed and washed with DI water to eliminate residual salts. After frozen and dried thoroughly, the weight of each sample was measured as W2, and the dry mass initially was W1. The degradation percent was calculated using the following equation: Degradation Percent = (W1-W2)/W1 × 100%.
2.5. Encapsulation of BMMSCs within GelMA
Primary BMMSCs were bought from ScienCell Company (San Diego, CA, USA). All the cell experiments involved in this study were repeated three times using BMMSCs from three different batches. Proliferation medium for BMMSCs included Minimum Essential Medium α (α-MEM, Gibco, Grand Island, NY, USA), 10% fetal bovine serum (Gibco, Grand Island, NY, USA), and 1% penicillin/streptomycin (Gibco, Grand Island, NY, USA). The cells were cultured in a humidified incubator with 5% CO2 at 37 °C. Achieving a final cell concentration of 106/mL, BMMSCs were blended with the GelMA solution prior to cross-linking. After light curing by 405 nm ultraviolet light for 90s, the BMMSCs were uniformly encapsulated into lured GelMA. Then, the proliferation medium was added to the culture plate.
2.6. RNA sequencing and bioinformatics analysis
BMMSCs were transfected with vector and PCK2 overexpression-lentivirus in GM30/60/90 after 7 days in proliferation medium. The whole transcriptome RNA sequencing was performed by Beijing Cnkingbio Biotechnology Co.LTD. Three replicates of each sample were sequenced. The normalized transcript expression level was calculated using the reads per kilobase of transcript per million (RPKM). Hierarchical clustering was performed based on differentially expressed mRNAs using R package (Pheatmap, version:1.0.12). Gene Ontology (GO) analysis was performed to uncover the gene regulatory network based on biological process and molecular function. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed to find out significant pathways from the differential expression genes. The bioinformatics analysis was conducted by Beijing Cnkingbio Biotechnology Co.LTD.
2.7. Lentivirus and si-RNA transfection
Lentiviruses targeting PCK2 (sh-PCK2) and negative control (NC) vectors (NC) for human were purchased from GenePharma Co. (Suzhou, China). Lentiviruses containing PCK2 and the scramble control (vector) for human and rats were purchased from Vector Builder Co. (Guangzhou, China). When cells were fused to 60–70%, lentivirus and 5 mg/ml polybrene (Sigma) was incubated. After 72–96 h, cells were treated with puromycin (1 μg/ml, Sigma–Aldrich) to select the transfection positive cells. The short interfering (si) RNAs targeting PFKP (si-PFKP) and the negative control (si-NC) were purchased from Vector Builder Co. (Guangzhou, China). The transfection agent was Lipofectamine 3000 (Invitrogen, Carlsbad, USA).
2.8. Cell viability test and survival rate
The Calcein-AM/Propidium iodide (PI) Double stain kit (Live/Dead assay kit, 40747ES76, Yeasen biotechnology) was used to determine the cell viability. Briefly, BMMSCs and GelMA mixtures were cultured in ibiTreat μ-plates, nine-well (Ibidi, cat. no. 80826) and crosslinked. Then cells were stained with calcein-AM and PI in assay buffer. The cell samples were incubated at 37 °C for 20 min, washed three times with PBS, and immediately observed under the confocal microscope (Leica TCS-SP8 STED 3X). To measure the survival rate, BMMSCs were cultured in prepared GelMA-30/60/90 (n = 5 for each group). The survival rate for each sample was calculated with the equation: survival rate = W1/(W1+W2) × 100%, where W1 was the live cells, and W2 was the dead cells of the sample.
2.9. Immunofluorescence for cells
Cells were encapsulated in GM30, GM60 and GM90, and seeded in ibiTreat μ-plates, nine-well (Ibidi, cat. no. 80826). Immunofluorescence experiments for cells were conducted as standard protocols. Briefly, cell samples were washed three times with phosphate buffered saline (PBS) to remove the residue medium, fixed with 4% paraformaldehyde for 15min, and permeabilized for 10 min in 0.1% (v/v) Triton X-100 at 4 °C. Then the cell samples were blocked for 15 min QuickBlock™ Blocking Buffer for Immunol Staining (P0260, Beyotime biotechnology) at 4 °C and incubated overnight at 4 °C with the primary antibodies. Then, the cells were washed three times with Immunol Staining Wash Buffer (P0106C, Beyotime biotechnology) and incubated with the secondary antibodies for 1 h at room temperature. The nuclei were stained with DAPI (C0065, Solarbio, Beijing). The cells were washed three times by Immunol Staining Wash Buffer before observation. Images were captured under a confocal microscope (Leica TCS-SP8 STED 3X). The primary antibodies used were as follows: anti-PCK2 (6924S, Cell signaling technology), anti-PFKP (ab119796, Abcam), anti-RUNX2(ab12556, Abcam). The secondary antibodies included Goat anti-mouse IgG(H + L) Alexa Fluor-647 (550047, ZEN Bio), Goat anti-rabbit IgG(H + L) Alexa Fluor-594 (550043, ZEN Bio), Goat anti-rabbit IgG(H + L) Alexa Fluor-488 (550037, ZEN Bio), Actin-Tracker Red 594 (C2205S, Beyotime biotechnology).
2.10. Immunofluorescence and immunohistochemistry for tissues
For femur samples, the preparation process was performed by Beijing Genepool Biotechnology Company Limited. Briefly, tissue samples were fixed for 2 days with 4% paraformaldehyde and dehydrated according to standard protocols. Paraffin embedded sections with 5 mm thick were placed in a 65 °C oven for 2 h, dewaxed to water, and washed with PBS for three times for 5 min each time. The slices were placed in EDTA buffer for microwave repair and boiled for 10min. The sections were blocked with 5% normal goat blocking serum for 20 min at room temperature, and then incubated with the primary antibodies anti-PCK2 (6924S, Cell signaling technology, 1:250) and anti-PFKP (ab119796, Abcam, 1:250) overnight at 4 °C. Washed with PBS for 3 times, 5min each time, the secondary antibodies corresponding to the species of the primary antibody were added to cover the slices in the circle, and incubated at room temperature in dark for 60 min. The nuclei were stained by DAPI for 60 min. After the slices were slightly dried, they were sealed with anti-fluorescence quenching sealing agent. Then the images were scanned and analyzed. Immunohistochemical (IHC) staining was also used for validation of PFKP expression, the antibody was ab119796 (anti-PFKP, Abcam, 1:100, Cambridge, UK).
2.11. ALP staining and quantification of activity
BMMSCs encapsulated in GM30/60/90 were cultured in 48-well plates with the same number. After 7 days of culture in proliferation medium, the cells were washed 3 times with PBS, fixed in 95% cold ethanol, and washed with PBS again. Based on the standard protocol, alkaline phosphatase (ALP) Staining Kit (CWBIO, Beijing, China) was used for ALP staining. The BMMSCs were then gently washed with PBS, and pictures were scanned. For ALP quantification, BMMSCs in encapsulated GelMA were incubated with GelMA hydrogel lysate (EFL-GM-LS-001, EFL) for 2 h. The supernatant was removed, and the cells were in the precipitant. Protein lysate and protease inhibitor were added to the cells. Then, cells collected were centrifuged at 14000×g for 15 min at 4 °C after being lysed with 1% (v/v) Triton X-100 on ice. A BCA protein assay kit (Pierce Thermo Scientific, Waltham, MA, USA) was used for detecting the total protein concentration. The ALP activity was determined by the ALP assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) and calculated as traditional methods.
2.12. qPCR analysis
BMMSCs encapsulated in GM30/60/90 were treated with GelMA hydrogel lysate (EFL-GM-LS-001, EFL). After 2 h, total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and the concentration of RNA was measured by the Nano Drop 8000 spectrophotometer (Pierce Thermo Scientific). Takara kit was used for reverse transcription. SYBR Green Master Mix (Roche Applied Science, Mannheim, Germany) was used to test the mRNA expression of each gene by qPCR, with a 7500 Real-Time PCR Detection System (Applied Biosystems, Foster City, CA, USA). The primer sequences used in the study were listed in Table S1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was listed as the internal control.
2.13. Western blot
Cell pellets were extracted from GelMA as described above. The pellets were lysed in RIPA lysis buffer (medium) with cocktail (Huaxingbochuang) and phosphatase inhibitors (Huaxingbochuang), and the protein concentration was determined using the BCA kit. The Western blot was performed as previously described [24]. The following antibodies were used: anti-GAPDH (ab9485, Abcam), anti-PCK2 (6924S, Cell signaling technology), anti-PFKP (ab119796, Abcam), anti-PFKM (ab154804, Abcam), anti-PFKL (ab154804, Abcam), anti-FAK (ab40794, Abcam), anti-vinculin (13901, Cell signaling technology), anti- Runt-related transcription factor 2 (RUNX2) (ab12556, Abcam), anti-phospho-AKT S473 (4060; Cell Signaling Technology, Boston, MA, USA), anti-AKT (4691, Cell Signaling Technology), anti-phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (4370, Cell Signaling Technology), anti-p44/42 MAPK (Erk1/2) (4695, Cell Signaling Technology).
2.14. Metabolic analysis
The bioenergetic profile of BMMSCs transfected with vector-, PCK2 overexpression-, NC-, PCK2 knockdown-lentivirus in GM30/60/90 after 3 days in proliferation medium was assessed using the Seahorse XF96 Extracellular Flux Analyzers (Agilent) that provides extracellular acidification rate (ECAR), an index of glycolysis function. To avoid edge effect to unequal cell numbers and growth rates, all tests were performed with cells of 1.2 × 104 per well. The next day, the medium was changed to XF base DMEM PH7.4 (Seahorse Bioscience) supplemented with 3 mM oligomycin, glucose (10 mM), 2-DG (50 mM) for ECAR. Metabolomic profile of BMMSCs transfected with vector-, PCK2 overexpression-lentivirus in GM30/60/90 after 3 days in proliferation medium were performed by Lipidall Technologies Company Limited, using Ultra-high Performance Liquid Chromatography (UPLC) and Tandem quadrupole time-of fligh (QTOF).
2.15. In vivo bone defect implantation
Twelve weeks after surgery, BMMSCs transfected with NC, PCK2 and sh-PCK2 lentivirus were 3D cultured in GM30, GM60 and GM90 for 7 days with proliferation medium [28,29]. Rats were divided randomly into 18 groups (n = 5): (1) sham rats were implanted with GM30/NC-transfected BMMSCs (sham/GM30/NC); (2) sham rats were implanted with GM30/PCK2-transfected BMMSCs (sham/GM30/PCK2); (3) sham rats were implanted with GM30/sh-PCK2-transfected BMMSCs (sham/GM30/sh-PCK2); (4) sham rats were implanted with GM60/NC-transfected BMMSCs (sham/GM60/NC); (5) sham rats were implanted with GM60/PCK2-transfected BMMSCs (sham/GM60/PCK2), (6) sham rats were implanted with GM60/sh-PCK2-transfected BMMSCs (sham/GM60/sh-PCK2); (7) sham rats were implanted with GM90/NC-transfected BMMSCs (sham/GM90/NC); (8) sham rats were implanted with GM90/PCK2-transfected BMMSCs (sham/GM90/PCK2), (9) sham rats were implanted with GM90/sh-PCK2-transfected BMMSCs (sham/GM90/sh-PCK2), (10) OVX rats were implanted with GM30/NC-transfected BMMSCs (OVX/GM30/NC); (11) OVX rats were implanted with GM30/PCK2-transfected BMMSCs (OVX/GM30/PCK2); (12) OVX rats were implanted with GM30/sh-PCK2-transfected BMMSCs (OVX/GM30/sh-PCK2); (13) OVX rats were implanted with GM60/NC-transfected BMMSCs (OVX/GM60/NC); (14) OVX rats were implanted with GM60/PCK2-transfected BMMSCs (OVX/GM60/PCK2); (15) OVX rats were implanted with GM60/sh-PCK2-transfected BMMSCs (OVX/GM60/sh-PCK2); (16) OVX rats were implanted with GM90/NC-transfected BMMSCs (OVX/GM90/NC); (17) OVX rats were implanted with GM90/PCK2-transfected BMMSCs (OVX/GM90/PCK2); (18) OVX rats were implanted with GM90/sh-PCK2-transfected BMMSCs (OVX/GM90/sh-PCK2).
First, 1% pentobarbital sodium was used to anesthetize via an intraperitoneal injection. After incision and blunt dissection of skin to expose the distal femur, a cylindrical hole with depth, 3.5 mm and diameter, 2 mm was vertically drilled through the cortical bone in each distal femur. The process was cooled with saline water. The approximate distance between the circle defect and distal femur growth plate was 3–5 mm. The BMMSCs/GM complex was implanted into the bone defect. The muscular fascia, subcutaneous tissue, and skin were then sutured around the holder in sequence. Rats were asphyxiated by excessive inhalation of CO2 after 8 weeks.
2.16. Micro-computed tomography and bone histomorphometry analysis
The femurs of rats were isolated and then fixed with 10% formalin for 24 h. After that, micro-CT images were obtained at a resolution of 8.82 μm, with a tube voltage of 60 kV, a tube current of 500 μA, and an exposure time of 1500 ms. Multimodal 3D visualization software (Inveon Research Workplace; Siemens, Munich, Germany) was used for three-dimensional (3D) reconstructions. The parameters calculated in the region of interest (operation area) included Bone mineral density (BMD) and bone volume/total volume (BV/TV). After fixed in 10% formalin for 24 h and decalcification for 1 month, the femurs were then dehydrated, embedded with resin, and sliced for H&E staining and Masson staining.
2.17. Isolation of primary BMMSCs from rat femurs
The femurs of SD rats from sham and OVX group (rBMMSCs) were extracted by traditional flushing method. After centrifugation for 5 min at 1200 rpm, the cells were collected and cultured in fresh α-MEM mixed with 20% FBS and 1% penicillin/streptomycin. Non-adherent cells were removed at 48 h and attached cells were maintained until they reached confluency of 90%∼100% [30]. Then, the BMMSCs isolated from rats were cultured in α-MEM mixed with 10% FBS and 1% penicillin/streptomycin.
2.18. Cell surface phenotype detection
Flow cytometry was performed to detect surface marker of rBMMSCs. The rBMMSCs were collected in passage four and were resuspended in 100 μl flow cytometry staining buffer (Cyagen Biosciences, Inc., Guangzhou, China) at a concentration of 1 × 106 cells/ml and incubated for 30 min at 4 °C. The specific primary antibodies included positive surface markers anti-CD44, anti-CD90 and anti-CD73, negative surface markers anti-CD34, anti-CD29 and anti-CD45, which were obtained from the Cyagen Mesenchymal Stem Cell Characterization kit (cat. no. RAXMX-09011; Cyagen Bioscience Inc.) After incubation, the samples were washed twice in 1 ml flow cytometry staining buffer, then centrifuged at 250×g for 5 min. Then, the supernatant was removed. After incubation at 4 °C with secondary antibodies of PE for 30 min in the dark, the samples were washed twice with flow cytometry staining buffer. Then the samples were centrifuged at 250×g for 5 min and resuspended in 200 μl of flow cytometry staining buffer for cytometric analysis. PE-labeled cells with these were analyzed by flow cytometry (FACSCalibur, Becton Dickinson Biosciences, Franklin Lakes, NJ, USA) and the FlowJo software (Tree Star, San Carlos, CA, USA).
2.19. Statistical analysis
Results were demonstrated as mean ± SEM. Analysis between two paired samples was performed using a two-tailed unpaired Student's t-test by GraphPad Prism Software (La Jolla, CA). Analysis between more than two sample groups was conducted using a one-way unstacked ANOVA using GraphPad Prism Software (La Jolla, CA). P values less than 0.05 were considered statistically significant. Statistical significance was defined as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, or #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001.
3. Results and discussion
3.1. Micro-nano mechanical properties of 3D ECM model in vitro and in vivo
Recently, ECM stiffness is emerging as an attracting mechanical clue that impels the progression of disease via regulating cellular behaviors [17]. Tissue stiffen is related to cancer and cardiovascular disease, while decreased ECM of tissue also takes place with fibrosis, advanced age, and degenerated diseases, in virtue of accelerated matrix deposition, and cross-linking of existing matrix, etc [11]. Osteoporosis is a degenerated skeletal disorder characterized by a deterioration in bone tissue and strength, with a subsequently increase of fracture risk, which brought variations of microarchitecture, such as mineral-matrix ratio and collagen cross-linking, and thus triggering changes of physical microenvironment mechanics [31]. Trabecular bone properties have drawn much attention in the treatment and etiology of osteoporosis, since it provides structural support and flexibility and fills the proximal and distal ends of all bones. The lower quality of trabeculae network from osteoporotic bone may bring about fracture susceptibility [12]. BMMSCs residing in bone marrow are the promising reservoirs for bone regeneration and repair, which also receive biochemical stimuli from native stem cell niche of bone [32]. Therefore, understanding how physical properties of in vivo niche regulates BMMSCs osteogenesis is of significance. For this purpose, the femoral neck samples of sham operated rats (sham) and ovariectomized rats (OVX) were used as in vivo model, for normal and osteoporotic bone, respectively. Meanwhile, a biomaterial model was fabricated whose matrix stiffness could be modulated, contributing to simulating an experimental model with different 3D ECM stiffness for in vitro study. The stiffness of GelMA with different percentages (6, 8 and 10%) and different methacrylation levels of gel (GM30, GM60, GM90) is elevated by increased methacrylation degree and gelatin concentration, from 1 kPa to 30 kPa (Fig. S1). Previous studies have reported that osteogenic commitment occurring mainly at ECM stiffness of 11–30 kPa and adipogenic lineage primarily in 2.5–5 kPa ECM stiffness [6,7]. Therefore, the effects of GelMA with stiffness of 6 kPa (GM30 6%), 10 kPa (GM60 6%) and 25 kPa (GM90 8%) were explored in this study. Besides, the FTIR, the antiswelling, mechanical modulus and biodegradable properties of GM30, GM60 and GM90 were shown in Fig. S1A-D, which were consistent with previous study [33].
We focused on assessing mechanical properties of trabecular bone at the level of single trabeculae, through high-resolution images of its stiffness performance. Nano-scale tools such as atomic force microscopes (AFM) have recently been practical for assessing substrate mechanical properties not only of tissues, but also single cell on cellular and subcellular scales [3]. To learn the trabeculae of normal and osteoporotic bone from micro-nano aspect, we analyzed the structure of trabeculae by electron microscope and measured its mechanical property by AFM imaging and nanoindentation (Fig. 1B–D, Fig. S2A). As showed in Fig. 1B, in the normal bone structure, trabeculae are closely arranged in a 3D reticular network; while in the osteoporotic bone, the collagen cross-links are deteriorated, and the number of trabeculae was obviously decreased, and breakage between trabeculae is increased. A similar internal porous network with trabeculae was observed in GelMA as indicated in Fig. 1B and Fig. S2B. For GM30, the pore was large and loose, and the wall is thin; while in GM90, it exhibited a dense network and the pore is small, and the wall is relatively thick; the physical properties of GM60 was between GM30 and GM90. Therefore, it could be postulated that GelMA with different ECM density could experimentally mimic the physical characteristics of trabeculae network of normal and osteoporotic bone in vitro.
AFM analysis of the trabecular bone was shown in Fig. 1C and D. AFM imaging demonstrated that more characteristic zones of mineral deposits (red arrow) and collagen fibers (yellow arrow), while less typical zones of mineral depleted areas (blue arrow), were observed in sham group, compared with OVX group (Fig. 1C). By AFM nanoindentation, we detected 16 positions of each sample (n = 5) at the surface of trabeculae, and draw the typical mechanics heatmaps (Fig. 1D). The stiffness of trabeculae from sham femur was 5.8 GPa, quantitatively twice that of trabeculae from OVX femur (Fig. 1E). The results were consistent with previous studies which observed a decrease in elastic modulus of trabecular bone from OVX rats [34] and decreased bone formation in osteoporotic human cancellous bone [35]. These findings suggested that remarkable differences of stiffness of trabeculae is one of the main changes brought by osteoporosis, provoking us to explore the role of 3D ECM stiffness to regulate osteogenic differentiation of BMMSCs.
3.2. PCK2 is involved in 3D ECM stiffness mediated osteogenesis
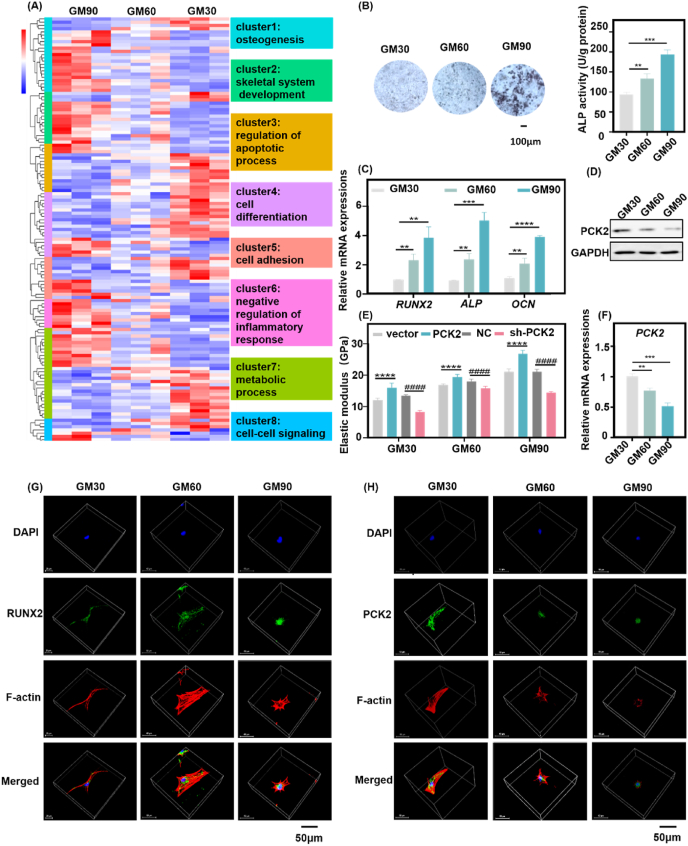
In consistent with the previous study [32], BMMSCs in GelMA exhibited a good viability and were distributed uniformly throughout the hydrogels (Figs. S3A–C). The cell survival ratios in different GelMA after 3, 7 or 14 days were shown (Fig. S3D). The live/dead assay at day 3, day 7 and day 14 demonstrated that almost all of the cells were alive (green). The survival ratio was more than 90%, both in GM30/60/90 at day 3, day 7 and day 14. Therefore, GelMA hydrogel was selected as an appropriate 3D cell culture system to encapsulate cells because it exhibits excellent biological performance. To assess the relationship between 3D ECM stiffness and BMMSCs osteogenesis, we performed RNA-seq on BMMSCs 3D cultured (after 40 h) in GelMA with stiffness of 6 kPa (GM30, 6%), 10 kPa (GM60, 6%) and 25 kPa (GM90, 8%) to broadly map the transcriptome-wide changes responding with 3D ECM stiffness. Because at 40 h, BMMSCs had not exhibited significant cell proliferation [6]. Hierarchical gene clustering and pathway analysis showed that osteogenesis, positive regulation of osteoblastic differentiation, and skeletal system development were remarkably different between the cells cultured in GelMA with different stiffness (Fig. 2A, clusters 1,2, Fig. S4A). Alkaline phosphatase (ALP) staining and activity, qRT-PCR analysis, Western blots and images of confocal laser scanning microscopy (CLSM) showed that the osteogenic differentiation of BMMSCs was increased with the elevation of 3D ECM stiffness (Fig. 2B–D, F, G). When sensing the stiffness cue from ECM, the integrin-and-cadherin-mediated adhesions would couple the ECM to the actin cytoskeleton [36]. Mechanical feedbacks intricately coupled by the activity of integrin-mediated adhesions the focal adhesion kinase (FAK) and vinculin signaling pathways to reorganize the cytoskeleton [25]. The immunofluorescence staining demonstrated that BMMSCs in soft 3D ECM displayed more stretched actin bundles and large cell volume, while actin cytoskeleton of BMMSCs in stiffen 3D ECM were relatively compressed and the cell volume was small. Meanwhile, the immunostaining intensity of RUNX2 is elevated with the increase of 3D ECM (Fig. 2G). These results confirmed that the 3D ECM stiffness has great impact on the osteogenesis of BMMSCs, consistent with previous studies investigating the role of stiffness on MSCs [6,7].
Fig. 2.
PCK2 is involved in the regulation of osteogenesis responding to 3D ECM stiffness. A) Hierarchical gene clustering of RNA-seq showed that various genes related to corresponding biological processes were different between MSCs cultured in 3D ECM with three different stiffness (GM30, 6%, GM60 6% and GM90, 8%). K-means clusters are indicated by colored bars. B) ALP staining and activity indicated that osteogenic differentiation of MSCs was the highest in GM90, followed by GM60, while GM30 induced the lowest osteogenesis (**P < 0.01, ***P < 0.001, by students' t-test). Scale bar is 100 μm. C) The qPCR analysis showed that osteogenic-related genes were increased significantly by high stiffness of 3D ECM (**P < 0.01, ***P < 0.001, ****P < 0.0001, by students' t-test). D) Western blot showed that the protein expression of PCK2 was decreased with the increased 3D ECM stiffness. E) Overexpression of PCK2 dramatically enhanced the cellular stiffness while depletion of PCK2 decreased the cellular stiffness in 3D ECM (****P < 0.0001, compared to vector cells, ####P < 0.0001, compared to NC cells, by students' t-test, sh-PCK2: PCK2-knckdown cells). F) The qPCR analysis demonstrated that the mRNA expression of PCK2 was decreased with the increased 3D ECM stiffness (**P < 0.01, ***P < 0.001, by students' t-test). G) Immunofluorescence showed that 3D ECM stiffening increased RUNX2 intensity. Scale bar is 50 μm. H) Immunofluorescence staining showed that BMMSCs in GM30 displayed the highest intensity of PCK2, followed by those in GM60, while there was barely any PCK2 activation in the cells cultured in GM90. Scale bar is 50 μm. All the data represents three times of repeat and are means ± SEM.
Our transcriptome-wide RNA-seq results highlighted the important role of 3D ECM stiffness on regulating pluripotency of stem cell such as KLF4, BMP2, BMP4 and Wnt5b (Fig. S4A). Pathway enrichment for gene analysis on the differentially expressed genes from each comparison showed significant enrichment for processes between GelMA with different stiffness, including signaling pathway of regulating stem cell multipotency, human diseases (inflammatory bowel diseases), environmental information processing (cytokine-cytokine receptor interaction), tyrosine metabolism, and FoxO signaling (Figs. S4B and C). Another study has reported that the RNA-seq on MSCs cultured in alginate hydrogels with moduli from 3 kPa to 30 kPa captured the 1637 differentially expressed (DE) genes. Among them, genes related to bone formation were upregulated with increased stiffness, which further confirmed our results. Some mechanosensitive genes, such as Sdc3, Wnt4, Colla1 and I11rl1 were upregulated both in 18 kPa and 30 kPa, while MMP9 and FGFR3 and Chd4 (an epigenetic modifier) was downregulated in 18 kPa and upregulated in 30 kPa [6]. Our results also showed that there were 764 DE genes in GM60, compared to GM30 (Fig. S4D), 977 DE genes in GM90 compared to GM30 (Fig. S4E), 594 genes in GM90 compared to GM60 (Fig. S4F). Some osteogenic related genes, such as BMP4, SIRT1 and WNT10B were upregulated both in GM60 and GM90, while MMP9 and COL10A1 (chondrogenic related) was downregulated in GM90 (Fig. S4G). Together, our work combined with previous studies stressed the importance of stiffness on the role of regulating stem cell fate.
Notably, gene clustering also revealed that a number of genes regulating cell metabolism, apoptosis, cell differentiation and adhesion transcripts (Fig. 2A, clusters 3–5,7) and related signaling pathways (Figs. S4A and B) were different between cells in stiff ECM and cells in soft ECM. As a key enzyme for gluconeogenesis, PCK2 was reported to promote osteogenic differentiation of MSCs in our previous work [24]. It provokes us to further explore the role of PCK2 on reversing the damaged osteogenic differentiation of BMMSCs in the soft 3D microenvironment. The mRNA and protein expression of PCK2 was decreased with the impaired stiffness of 3D ECM (Fig. 2D, F). Immunofluorescence staining showed that BMMSCs in GM30 displayed the highest intensity of PCK2, followed by those in GM60, while there was barely PCK2 activation in the cells cultured in GM90 (Fig. 2H). The possible reason may be as follows. Compared with the soft ECM, the osteogenic differentiation BMMSCs is stronger in the stiff ECM. The relatively low expression of PCK2 in the stiff ECM may be preventable for the excessive osteogenesis of cells and maintain the balance of cell differentiation. With the increasing demand for osteogenic differentiation (such as repair of bone defects), the energy demand is higher. Our previous study has shown that PCK2 could regulate autophagy and promote osteogenesis when energy is deficient [24]. Therefore, when cells face higher energy demand, autophagy or other energy metabolism pathways may be activated through PCK2 to meet deeper requirements of osteogenesis. Nevertheless, with the increase of autophagy, PCK2 might be degraded in turn to prevent excessive differentiation and maintain the homeostasis of differentiation and energy supply. However, the mechanism in-depth remains to be further explored.
Elevating cytoskeletal prestress increases cellular stiffness and enhance cell stiffening, enables long-range cytoplasmic mechano-transduction via integrins, and triggers rapid gene expression, promotes stem cell differentiation, whereas lowering cytoskeletal prestress maintains embryonic stem cell pluripotency [37]. Previous study has reported that change of matrix stiffness (from stiff to soft) could trigger proteasomal degradation of the rate-limiting metabolic enzyme phosphofructokinase (PFK) via disassembly of stress fibers, and thus downregulating glycolysis [25]. Our results demonstrated that PCK2 could respond to stiffness of 3D ECM, regulating osteogenic differentiation of BMMSCs, therefore, we wonder whether PCK2 exerts an effect on cellular stiffness in 3D ECM with different stiffness. The PCK2 overexpression and knockdown BMMSCs stable cell lines were established as described in our previous study [24]. Through AFM, we detected 16 positions of each cell and found prominent discrepancies among these four kinds of cells in different ECM. The stiffness of cells was elevated with the increase of ECM stiffness. Specifically, overexpression of PCK2 promoted the stiffness of BMMSCs, while knockdown of PCK2 suppressed the rigidness of BMMSCs in 3D ECM (Fig. 2E). Therefore, these data suggests that BMMSCs recognize stiffness of 3D ECM through myriads of complex biochemical signaling pathways to regulate a series of life processes, including osteogenesis, skeletal development and metabolism. Specifically, PCK2 plays a key role in responding to the stiffness but the effect of PCK2 on osteogenesis in 3D ECM is still unknown.
3.3. PCK2 promotes osteogenesis responding to the stiffness of 3D ECM
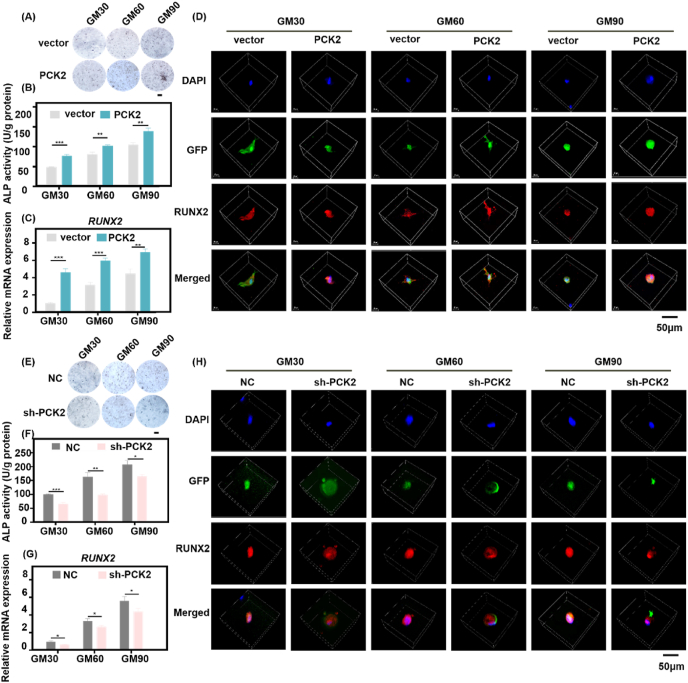
Sensing the physical characteristics of the ECM exerts an impact on cell metabolism partly by modulating the expression of enzymes responding to mechano-transduced cytoskeletal dynamics [5]. It has attracted increasing interest that stiffness plays an important role on metabolic cues in determining stem cell fate [6]. Gluconeogenesis is an important anabolic metabolism to provide energy when cells meeting nutrition-shortage, and PEPCK is the rate-limiting enzyme catalyzes OAA to PEP. As the mitochondrial isoform of PEPCK, PCK2 has been reported to play a pivotal role in monitoring glucose homeostasis and TCA cycle activity [21]. To gain deeper insight, we explored the specific roles of PCK2 on the regulation of osteogenesis in 3D ECM. Our previous work revealed the novel effect of PCK2 promoting osteogenic differentiation of MSCs [24]. We also found that PCK2 could reverse osteoporosis phenotype by maintaining the function of BMMSCs in osteoporotic mice (unpublished data), which gave us a clue that PCK2 may be a core target for regulating the BMMSCs osteogenesis and reversing the degenerative osteogenic differentiation ability of BMMSCs in the soft 3D ECM. To further make clear the role of PCK2 for its effect on osteogenesis of BMMSCs in 3D ECM, gain-and-loss function was used. PCK2 overexpressing-BMMSCs and PCK2 knockdown-BMMSCs were embedded in GM30 (6%), GM60 (6%) and GM90 (8%). After 7 days, the ALP staining and activity were tested. There is more ALP production in GM90 group, compared with GM60 and GM30. The ALP activity of PCK2-overexpression cells was more than the vector group, while ALP activity of PCK2-knockdown cells was lower than that of negative control group (NC), both in the three kinds of ECM with different stiffness (Fig. 3A, B, E, F). The RUNX2 and ALP mRNA expression was significantly upregulated in the BMMSCs cultured in the GM90 ECM, and overexpression of PCK2 intensified this promotion effect while depletion of PCK2 downregulated the enhancement (Fig. 3C, G, Figs. S5A and B). RUNX2 is a significant transcription factor which translocated into the nucleus of MSCs, and thus upregulating the expression of osteogenic related genes to enhance the osteogenic differentiation [5]. Immunofluorescence staining also showed that the highest intensity of RUNX2 in BMMSCs with GM90, followed by BMMSCs with GM60, while there was barely RUNX2 activation in GM30. While BMMSCs in soft ECM displayed highly stretched morphologies, BMMSCs in stiff ECM exhibited a circular and smaller shape. In PCK2-expressing BMMSCs, the intensity of RUNX2 was upregulated while in PCK2-depletion BMMSCs the fluorescent expression of RUNX2 was downregulated. This trend was similar both in the three different physical microenvironments with gradient ascending stiffness (Fig. 3D, H). These results confirm that the osteogenic capacity of BMMSCs is raised in stiffer 3D ECM than soft microenvironment. Furthermore, PCK2 could respond to the stiffness and promote the osteogenic ability of BMMSCs in 3D ECM.
Fig. 3.
PCK2 enhances osteogenesis in response to the stiffness of 3D ECM. A-C) PCK2 overexpression increased the A) ALP staining B) and activity and C) RUNX2 mRNA expression in 3D ECM (**P < 0.01, ***P < 0.001, by students' t-test). Scale bar is 100 μm. D) Immunostaining image showed that PCK2 overexpression significantly enhanced the immunostaining intensity of RUNX2 in GM30/60/90. Scale bar is 50 μm. E-G PCK2 knockdown decreased the E) ALP staining and F) activity and G) RUNX2 mRNA expression in 3D ECM (*P < 0.05, **P < 0.01, ***P < 0.001, by students' t-test). Scale bar is 100 μm. H) Immunostaining image showed that depletion of PCK2 dramatically decreased the immunostaining intensity of RUNX2 in 3D ECM. Scale bar is 50 μm. All the results were repeated three times and represents means ± SEM.
3.4. Glycolysis is mechanically modulated by PCK2
Since PCK2 is a critical enzyme in glucose metabolism, we decided to investigate the interconnection between cell mechanics and metabolism. Cellular mechano-transduction relies on adhesion receptors, such as integrins, intrinsic transducing signals that mechanically integrated the cytoskeleton to the ECM, and the cytoskeletal subsequently remodels, which has impact on the topological distribution and function of metabolic organelles, cargoes, and enzyme [38]. Glycolysis and oxidative phosphorylation cooperate in harmony to maintain the metabolic demands in normal cells. When the energy demand increase, cells often display enhanced aerobic glycolysis to meet the metabolic challenge [39]. Previous study has revealed that increased glycolysis is associated with osteoblast differentiation via Wnt signaling [40]. Moreover, inhibiting Wnt3a-induced glycolytic enzymes impaired osteoblast differentiation in vitro, while deletion of Lrp5 in mice reduced glycolytic enzymes in bone, resulting to a decrease in postnatal bone mass [41]. However, whether glycolysis was the deciding factor for osteogenesis in 3D ECM is uncertain. So, we examined how the osteogenic activities change by inhibiting the cells’ glycolysis. It has been reported that treatment of 2-DG (5 mM) for 12 h could block the glycolysis in vitro [42,43]. After 7 days, the ALP staining showed that the 2-DG treatment suppressed the osteogenesis of BMMSCs in GM30, GM60 and GM90 (Fig. S6A), while the mRNA expression of RUNX2 and ALP was consistent with the trend (Figs. S6B and C). These results demonstrated the important role of glycolysis in regulating osteogenic differentiation in 3D microenvironment with different stiffness.
Recent studies have gradually to discover possible interactions between mechanical cues such as stiffness and cell metabolic activities, and cell function [18,25,38]. Nonetheless, a direct relationship between mechanical clues from the 3D microenvironment and cellular intrinsic metabolic responses is poorly understood. From the data above, we could infer that PCK2 is a positive regulator of osteogenesis responding to stiffness of 3D ECM. But the underlying mechanism remains to be investigated. As a key enzyme of glucose metabolism, we further explored how PCK2 promotes osteogenesis after sensing the cues of stiffness from 3D ECM. In order to identify the metabolic pathways involved in the regulation of osteogenesis of BMMSCs by PCK2, an untargeted large-scale metabolomic analysis of PCK2-expressing BMMSCs following their implantation in GM30, GM60 and GM90 was performed. A panel of metabolites (mainly from the TCA cycle, fatty, and amino acid, glycolysis, purine metabolism, and the pentose phosphate pathway) were profiled, and then we constructed a heatmap according to hierarchical clustering analysis (Fig. S7A). Plots of differential metabolites showed that compared to BMMSCs cultured in GM30, gluconic acid, Glycine, xanthosine, pantothenic acid, l-glutamine were upregulated in BMMSCs cultured in GM90 (Fig. 4A). Metabolite Set Enrichment Analysis (MSEA) demonstrated that the metabolite sets enriched by differential metabolites between overexpression of PCK2 and vector group in GM90 included Pentose phosphate pathway, glycolysis and gluconeogenesis. The enrichment fold change was more than 3 times, and P value was less than 0.05 (Fig. 4B). The metabolite sets enriched by differential metabolites between overexpression of PCK2 and vector group in GM30/GM60 were shown in Figs. S7B and C. Indeed, amino acids (including l-Arginine, l-Glutamine, l-Leucine, Gluconic acid and l-Tyrosine) were significantly increased in PCK2 overexpression group of GM30, compared to vector group (Fig. S7D). The expression of glutamine and sarcosine were upregulated in PCK2 overexpression group of GM60, compared to vector group (Fig. S7E). However, PCK2 overexpression upregulated the succinyladenosine and downregulated nucleosides (including uridine 3′-monophosphate) in GM90 (Fig. S7F). For fatty acids, these differences were mainly not significant between PCK2 overexpression and vector group in GM30/60/90 (Fig. S7A). It has been reported that glutamine could promote osteogenic differentiation of BMMSCs via providing ATP through TCA cycle to meet energetic and synthetic demands [44]. A study revealed that glutamine-dependent mTOR activation could promote glycolysis, which deeply linked glutamine and glycolysis [45]. However, the intrinsic mechanism about how BMMSCs use glucose, fatty acids, and amino acids in 3D ECM with different stiffness remains unclear, especially lacking in vivo confirmation. In subsequent studies, genetic mice with conditional modification of encoding metabolic genes will provide key tools in this effort. Metabolomic profiling revealed that glucose-6-phosphate accumulated, coupled with the depletion of downstream intermediates (Fructose 1,6-bisphosphate, Fructose 6-phosphate, etc.) strongly demonstrating a block in the upper state of glycolysis when the MSCs transferring to soft 3D ECM from stiff 3D ECM (Fig. 4C). Besides of the physical property (stiffness), PCK2 also induced metabolic remodeling of BMMSCs in 3D ECM. These modifications may be ascribed to the regulation of PCK2 on the glucose metabolism but it needed to be further explored. PCK2 overexpression inhibited the accumulation of glucose-6-phosphate, whereas increased the downstream intermediates of glycolysis (Fig. 4C). The concentration of fructose 1, 6-bisphosphate and fructose 6-phosphate also confirmed this trend (Fig. 4D). These results demonstrated that PCK2 could regulate glycolysis in sensing the stiffness of 3D ECM.
Fig. 4.
PCK2 regulates glycolytic function by responding to 3D ECM stiffness. (A) Differentially expressed top metabolites between GM30/GM90. (B) Metabolite sets enrichment overview in PCK2 overexpression and vector group in GM90. -m, -metabolism, PPP, pentose phosphate pathway. The P value was less than 0.05. (C) Relative abundance of glucose-derived metabolites of PCK2 overexpression and vector BMMSCs in stiff and soft 3D ECM in a heat map (n = 4 independent cultures), red, accumulation; purple, depletion. G-6-P, glucose 6-phosphate; DHAP, dihydroxyacetone phosphate, F-1, 6-BP, Fructose 1, 6-bisphosphate, F-6-P, Fructose 6-phosphate. (D) Overexpression of PCK2 upregulated the concentration of F-1, 6 BP and enhanced the accumulation of G-6-P in GM30/60/90. G-6-P, glucose 6-phosphate, F-1, 6-BP, Fructose 1, 6-bisphosphate. (E) ECAR profile showed glycolytic function in PCK2-overexpression and vector-transfected cells in GM30, GM60 and GM90. Vertical lines indicate the time of addition of glucose (10 mmol/l), oligomycin (3 μmol/l), and 2-DG (50 mmol/l). (F-H) Quantification of F) basal ECAR, G) glycolysis and H) glycolysis capacity from (E). n = 15 for each group (*P < 0.05, ***P < 0.001, ****P < 0.0001, by students' t-test). (I) ECAR profile showed glycolytic flux in PCK2-knockdown and NC-transfected BMMSCs in GM30, GM60 and GM90. Vertical lines indicate the time of addition of glucose (10 mmol/l), oligomycin (3 μmol/l), and 2-DG (50 mmol/l). (J-L) Quantification of (J) basal ECAR, (K) glycolysis and (L) glycolysis capacity from (I). n = 15 for each group (****P < 0.0001, by students' t-test). All the results were repeated three times and represents means ± SEM.
Then we further measured the glycolytic function after gain-and-loss of PCK2 in 3D ECM with different rigidness directly by measuring the extracellular acidification rate (ECAR). As shown by Seahorse Flux analysis in Fig. 4G and H, the glycolytic rate and capacity of BMMSCs in the ECM with the highest stiff (GM90) was higher than cells in GM60 and GM30. Moreover, PCK2 overexpression in 3D ECM significantly upregulated glycolysis and also increased maximal glycolytic capacity (Fig. 4E–H). Correspondingly, depletion of PCK2 remarkably downregulated the basal ECAR, glycolysis and the glycolytic capacity (Fig. 4I-L). These findings suggested that glycolysis is positively regulated by the 3D ECM stiffness, and PCK2 promotes the glycolytic function in sensing the cues of stiffness from microenvironment. We next performed related pathway analysis, co-localization, and loss-and-gain-of function to further investigate the mechanism that PCK2 regulated glycolysis sensing the stiffness cue.
3.5. PCK2 enhances glycolysis through regulation of PFKP
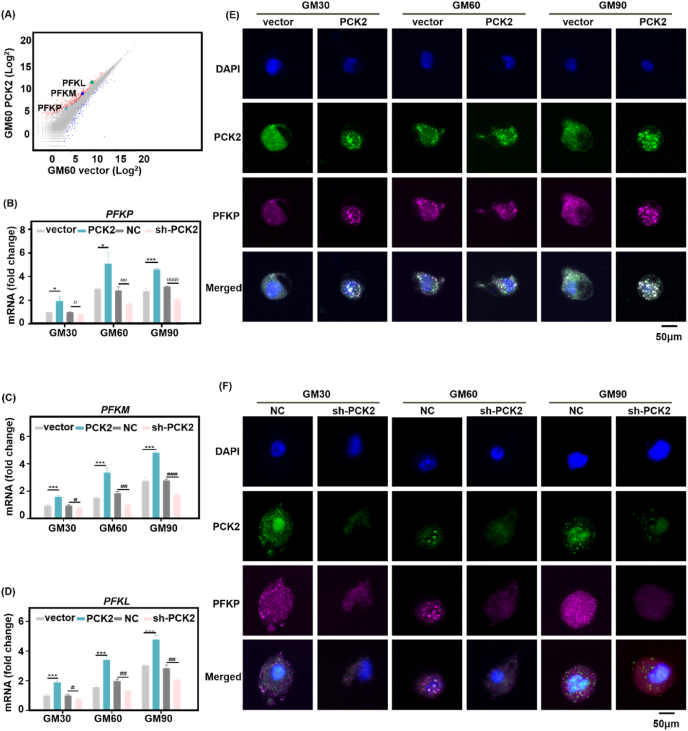
The expression of key enzymes could be the rate-limiting factor of glycolysis. The diagram of glycolytic pathway was shown in Fig. S8A. Through RNA-seq by assessing the gene and pathway changes, we found that the gene expression of all three metabolic enzyme phosphofructokinases (PFK) were up regulated in PCK2-expressing BMMSCs, this trend was similar both in the soft and stiff 3D ECM (Fig. 5A, Figs. S8B and C). The three isoforms are the platelet (PFKP), liver (PFKL) and muscle isoforms (PFKM) [46,47]. All the three PFK isoforms exhibited a systematic increased trend in expression on stiff microenvironment (Fig. 5B–D), compared to the soft ECM, which is consistent with other study [25]. The qPCR analysis showed that the mRNA expression of PFKP, PFKM, PFKL was upregulated in PCK2 overexpression BMMSCs in GM30, GM60 and GM90, while their expression was downregulated in PCK2 knockdown cells (Fig. 5B–D).
Fig. 5.
PCK2 promotes glycolysis via regulating PFKP in vitro. (A) Scatter plot (A), the color of wathet represents PFKP, the color of navy blue represents PFKM, the color of green represents PFKLshowed that the expression of the three isoforms of PFK was upregulated in PCK2 overexpression group in GM60, compared to vector group. (B-D) qPCR analysis showed that expression of PFKP, PFKM, PFKL in the PCK2 overexpression group was upregulated, compared with vector group in 3D ECM with tunable stiffness (*P < 0.05, ***P < 0.001, compared to vector cells, #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001, compared to NC group, by students' t-test). (E, F) Representative images of immunohistochemical staining of PCK2 (green), PFKP (purplish red), and nucleus (blue) of PCK2-overexpression-, PCK2-knockdown-, vector- and NC-transfected BMMSCs cultured in 3D ECM with different stiffness. Scale bar is 50 μm. All the results were repeated three times and represents means ± SEM.
The most commonly expressed PFK isoform in human tissues and cells is the platelet isoform of PFK (PFKP) [48], whose expression increased in clear cell renal cell carcinoma [49], non-small cell lung cancer [25], and acute myelogenous leukemia [50], contributes to glycolysis, to maintain the progression of these diseases. A study has reported that, a target mutant cell line exhibits higher glucose production and lower glucose uptake. ChIP-seq, ATAC-seq, RNA-seq, and metabolomics data showed that the DNA binding affinity and sites of PFKP to transcription factor-7-like 2 (TCF7L2, associated with increased risk of type 2 diabetes) increased, resulting to the increase of PCK1 [51]. But the relationship between PFKP and PCK2 is poorly understood. Therefore, we focused on this isoform to further explore how PCK2 mechanically regulate osteogenesis and the relationship between PFKP. The distribution of PFKP and PCK2 was assessed by immunofluorescence staining. As shown in Fig. 5E and F, the fluorescence strength of PFKP was strongest in GM90, followed by GM60, and the fluorescence intensity of PFKP was the weakest in GM30. The result was consistent with the previous study that the expression of PFKP was decreased when the cells transferred from stiff to soft substrate [25]. Moreover, the co-localization staining intensity and spots of PFKP and PCK2 were raised with the increase of ECM stiffness. The merged immune-staining dots were grown in number and intensity in PCK2-overexpression cells, while the co-localization of PFKP and PCK2 was weaker in PCK2-knockdown cells, which gave the clue that PFKP actually acted downstream to PCK2.
To further evaluate the stiffness-dependent regulation of PCK2 on MSCs osteogenic differentiation in vivo, and further explore the interaction of PFKP and PCK2, we implanted the BMMSCs-GelMA complex into the femur of rats. The sham and OVX rats (3-month after OVX surgery) were divided into 9 group: PCK2 overexpression, PCK2 knockdown, and negative control BMMSCs with GM30, GM60 and GM90, respectively. The rats were sacrificed after 8 weeks. Microcomputed tomography (micro-CT) images showed that sham-operated femurs exhibited normal trabeculae structure and more new bone-like tissue in operation area, generally compared to OVX group (Fig. 6A, D, Fig. S10B). As shown in H&E staining, PCK2 overexpression resulted in much more uniform, acidophilic osteoid tissue, while PCK2 depletion brought about less osteoid tissues, compared to NC group, both in sham and OVX rats (Figs. S9A and B, Fig. S10A). Meanwhile, in sham and OVX group, much more even interweaved extracellular matrix with collagen fiber accumulated in PCK2-overexpression group, while less collagen was observed in PCK2-knockdown group, displayed in Masson's trichrome staining. And more bone-like tissue formation was observed in GM90 group, compared to GM60 and GM30 group (Figs. S9A and B, Fig. S10A). Consistently, the quantitative measurement of micro-CT also suggested that the bone mineral density (BMD) and the percentage of BV to TV in the PCK2 overexpression group was more than the control group, while these parameters were remarkably less in PCK2-depletion group. The trend was similar in sham and OVX group (Fig. 6B, C, E, F). Furthermore, IHC staining showed that PFKP exhibited less expression in PCK2 depletion group compared to NC group, but PCK2-overexpression resulted in more expression of PFKP, both in sham and OVX rats. (Figs. S9C and D). We also observed that PCK2 overexpression increased co-localization of PCK2 and PFKP in the operation sites, and knockdown of PCK2 decreased this trend both in GM30, GM60 and GM90, which confirmed the in vitro results. The tendency of sham and OVX group was consistent (Fig. 6A, D, Fig. S10B). Together, our in vitro and in vivo data suggests that there is a colocalization of PCK2 and PFKP in BMMSCs, indicating that their expression pattern is intercorrelated, which proves that PFKP actually acted downstream to PCK2. However, how PCK2 regulates PFKP in responding to the stiffness of 3D ECM remains to be further explored.
Fig. 6.
PCK2 is mechanically modulated to promote osteogenesis via regulation of PFKP in vivo. (A) Upper panel: representative μCT images of bone regeneration in sham rat femur defects 8 weeks after PCK2-overexpression-, PCK2-knockdown-, NC-transfected BMMSC/GelMA implantation, showing the most abundant new bone formation in the PCK2 overexpression/GM90 of sham group. Scale bars: 1 mm. The middle-lower panel: The tracking of PCK2-PFKP colocalization in the bone defect of rat femur 8 week after PCK2-overexpression-, PCK2-knockdown-, NC-transfected BMMSC/GelMA implantation, showing that PCK2 overexpression/sham induced significantly more PCK2 and PFKP protein expression and more colocalization spots than and NC/sham group, followed by PCK2 knockdown/sham group, both in GM90 and GM30. Scale bars: 1000 μm (middle panel) and 100 μm (lower panel). (B, C) Quantitative analysis of the (B) bone mineral density (BMD) and (C) percentage of BV to TV of newly formed bone tissue (****P < 0.0001, versus vector group, by students' t-test). D) OVX (3-month after OVX surgery) group showed the similar trend with (A), and the general intensity of PCK2 and PFKP colocalization was decreased than sham group. Scale bars are the same with (A). (E, F) Quantitative analysis of the (E) bone mineral density (BMD) and (F) percentage of BV to TV of newly formed bone tissue (****P < 0.0001, versus vector group, by students' t-test). All the results were repeated three times and represents means ± SEM.
3.6. PCK2 regulates PFKP-mediated osteogenesis via ERK1/2 and AKT phosphorylation signaling axis responding to stiffness cues
Based on the results above, it is clear that as a key rate-limiting enzyme for glucose metabolism, PCK2 promotes osteogenesis in 3D ECM via colocalization of PFKP in vitro and in vivo. But what is the underlying molecular mechanism by which PCK2 enhanced osteogenic commitment of MSCs in 3D ECM? Then we investigated the underlying molecular signaling pathway that PCK2 enhances 3D ECM stiffness-mediated osteogenesis via PFKP. Stem cell lineage commitment is modulated by the complicated network of intricate signaling pathways regulated by biochemical and biomechanical stimuli from ECM, including AKT/ERK1/2, YAP/TAZ, and RhoA/ROCK pathway [52]. A study has reported that oxidized ATM promotes glycolysis via upregulating PFKP, and the accumulation of cytoplasmic citrate triggers AKT/ERK1/2 signaling to enhance progression of disease [53]. The involvement of AKT/ERK1/2 signaling pathway in ECM-mediated stimulation of osteogenesis was investigated this decade [52]. For example, collagen I was reported to increase the activation of ERK1/2 and AKT to promote osteogenic differentiation of hMSCs [54]. Moreover, ERK1/2 signaling pathway is well known as the upstream of RUNX2, and the activation of RUNX2 occurs via the ERK1/2 pathway to promote osteogenesis [55]. To further investigate the molecular pathway through which PCK2 regulates 3D ECM stiffness-induced osteogenic differentiation, Western blot analysis was conducted to assess changes of AKT/ERK1/2 in PCK2-expressing and PCK2-knockdown cells with different 3D ECM. We detected a remarkable increase in AKT and ERK1/2 phosphorylation in PCK2 overexpression cells, both in GM30, GM60 and GM90. However, knockdown of PCK2 significantly downregulated phosphorylation of AKT/ERK1/2 in the three 3D ECM with gradient stiffness (Fig. 7 A, B, Figs. S11A and B). Concurrently, we noticed that an accumulation in FPKP, PFKM and PFKL was observed with the increase of 3D ECM rigidness, and PCK2 overexpression upregulated the expression of these three isoforms of PFK, while depletion of PCK2 downregulated this effect (Fig. 7 A, B, Figs. S11A and B).
Fig. 7.
PCK2 regulates osteogenic differentiation via stiffness transduced PFKP-AKT/ERK-dependent glycolysis. (A, B) Protein expression of p-AKT and p-ERK1/2 was upregulated by (A) PCK2 overexpression, and (B) their expressions were downregulated by depletion of PCK2 in GM30, GM60 and GM90. Moreover, (A) overexpression of PCK2 also elevated the protein expression of PFKP, PFKM and PFKL, (B) whereas PCK2 knockdown suppressed their expressions. (C-F) Inhibition of PFKP suppressed the (C) ALP staining and (D) activity of PCK2 in 3D ECM. Scale bar is 100 μm. Moreover, (E, F) the transfection of si-PFKP blocked the elevation of osteogenic-related genes by PCK2 overexpression, both in GM30, GM60 and GM90 (**P < 0.01, ***P < 0.001, ****P < 0.0001 versus vector BMMSCs, #P < 0.05, ##P < 0.01, ###P < 0.001, versus PCK2-expressing BMMSCs, by students' t-test). (G, H) BMMSCs isolated from sham and OVX rats (3-month after surgery) were transfected with vector and PCK2 overexpression lentivirus. In 3D ECM, PCK2 overexpression promoted the damaged osteogenic capacity of OVX BMMSCs, confirmed by (G) ALP staining and (H) activity, and the mRNA expressions of (I) Runx2 and (J) Alp (**P < 0.01, ***P < 0.001, versus sham rBMMSCs, #P < 0.05, ##P < 0.01, ###P < 0.001, ####P < 0.0001, versus OVX rBMMSCs, by students' t-test). Scale bar is 100 μm. All the results were repeated three times and represents means ± SEM.
To extendedly examine the potential role of PCK2/PFKP signaling in regulating osteogenic differentiation in 3D physical microenvironment, si-RNA was used to inhibit PFKP expression, and the knockdown efficiency was measured by the qRT-PCR (Fig. S11C). Analysis of ALP staining/activity (Fig. 7C and D) and the mRNA expression of RUNX2 and ALP (Fig. 7E and F) suggested that depletion of PFKP in PCK2-expressing cells remarkably suppressed the upregulated osteogenic differentiation effect of PCK2-expressing cells, both in the three ECM with different stiffness. To further explore the regulatory effect of PCK2 on osteogenesis via PFKP-AKT/ERK pathway, LY294002 (50 μM) [54] was used to block the activation of ERK and AKT in the PCK2-expressing cells in GM30/GM60/GM90. The results displayed that elevated osteogenic differentiation of PCK2-overexpression cells in 3D ECM was significantly suppressed by the inhibition of ERK and AKT (Figs. S11D–F). We also extracted the primary BMMSCs from sham and OVX femurs, and identified their phenotype (Figs. S12A–F). The BMMSCs isolated from OVX was transfected with PCK2 overexpression lentivirus. The deteriorated osteogenic differentiation of BMMSCs from OVX was upregulated by the overexpression of PCK2, confirmed by analysis of ALP activity and relative mRNA expressions of Runx2 and Alp (Fig. 7G–J). Therefore, we could infer that PCK2 has the potential to reverse the decreased osteogenic capacity of BMMSCs in soft ECM. Together, these results gave further evidence that PCK2 responded to stiffness signal from 3D ECM, and thus facilitating osteogenesis via PFKP. It could be deduced that AKT/ERK1/2 induced PFKP-mediated-glycolysis might be in part responsible for the stiffening-regulated osteogenesis.
Cues from tissue microenvironment are crucial for deciding stem cell fate in vivo under physiological and pathological circumstances. The fate of MSCs could be decided by coordinate interaction between MSCs and their microenvironment [3]. The biophysical and biochemical extracellular cues from microenvironments, such as the stiffness of the ECM, maintain and modulate the osteogenic differentiation capacity of MSCs [56]. Here, our study shed some light on this process. Specifically, as shown in Fig. 1A, osteoporosis brings about loose organization of trabeculae network, with decreased trabeculae number and increased gap in osteoporotic femurs, accompanied by the changes of physical niche of mechanical properties including ECM stiffness. The osteogenic differentiation of BMMSCs with tailorable stiffness that experimentally mimicked normal (stiff) and osteoporosis (soft) 3D ECM was measured in the present study. As the key enzyme of gluconeogenesis, PCK2 could sense the stiffness of 3D ECM, and thus elevating glycolysis through PFKP.
Cellular mechano-signaling depends on adhesion receptors, such as integrins, FAK and vinculin, to interact signals that mechanically entrain the cytoskeleton to the ECM, and thus influencing the topological distribution of metabolic organelles, cargoes, and enzymes [57]. The PI3K pathway has been reported to modulate cytoskeletal functions and glucose metabolism. Full activation of glycolysis by PI3K requires both AKT activation and Rac-dependent actin (RAC1, an important member of RHO-family GTPases) remodeling to regulate the glycolytic enzyme aldolase, and thus promoting glycolysis [19]. The contractile from ECM could activate focal adhesion kinase (FAK) and extracellular signal-regulated kinase (ERK) 1/2 signaling, which could further activate SMAD and promote the nuclear localization of RUNX2 to promote osteogenesis [2,58]. Another study has reported that PFKP could upregulated intercellular citrate accumulation, and activate AKT/ERK/MMP2/9 signaling cascade [53]. Our data showed that the phosphorylation of AKT and ERK1/2 was positively correlated with stiffness-mediated-osteogenesis by gain-and-loss-of function (Fig. 7A, B, Figs. S11A, B, D-F). In addition, suppression of PFKP inhibited the osteogenic capacity of PCK2-expressing cells in 3D ECM with different stiffness (Fig. 7C–F). Moreover, PCK2 could elevate the damaged osteogenic differentiation of BMMSCs from OVX rats, both in the stiff and soft microenvironment (Fig. 7G–J). Overall, these results demonstrated that PCK2 could sense the stiffness of 3D ECM to promote osteogenesis via regulating PFKP-AKT/ERK1/2-mediated glycolysis.
However, there are still some limitations in this study. For example, endogenous cells around ECM scaffolds also play important role in mediating glycolysis and osteogenesis functions in vivo. Therefore, whether the surface mechanical modulus of GelMA gels indeed regulate the fates of the recruited cells in vivo remains unexplored.
4. Conclusion
In summary, our study elucidates the unraveling role of PCK2 in promoting osteogenesis in 3D ECM with tunable stiffness. The underlying mechanism is through regulating PFKP-mediated glycolysis. These findings highlight the critical importance of structural mechanisms of aging and metabolic disease, such as osteoporosis. The revealed key molecular signals mediating stiffness-dependent osteogenesis will be conducive to the dynamics of stem cell heterogeneity and metabolic adaption occurring upon physiological and pathological condition. Our findings will also provide a more precise and efficient metabolic target strategy for regulating stem cell fate to treat osteogenesis. Moreover, the reciprocal intricate crosstalk between ECM stiffness and metabolism will offer better understanding of the etiology and strategies of degenerative bone diseases such as osteoporosis from the novel metabolic perspective.
CRediT authorship contribution statement
Zheng Li: Conceptualization, Methodology, Validation, Investigation, Formal analysis, Writing – original draft, Writing – review & editing, Funding acquisition. Muxin Yue: Methodology, Data curation, Software, Formal analysis. Xuenan Liu: Methodology, Data curation, Software, Formal analysis. Yunsong Liu: Methodology, Data curation. Longwei Lv: Methodology, Data curation. Ping Zhang: Conceptualization, Writing – review & editing, Funding acquisition. Yongsheng Zhou: Conceptualization, Writing – review & editing, Supervision, Funding acquisition, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by National Natural Science Foundation of China (81870742, 81970911); Beijing Natural Science Foundation (7202233); Key Project of the National Natural Science Foundation of China (81930026); Project funded by China Postdoctoral Science Foundation (2020TQ0020, 2021M700280); Research Foundation of Peking University School and Hospital of Stomatology (PKUSS20210102). We thank Dr. Siying Qin at the National Center for Protein Science at Peking University for technical assistance in AFM operation and data analysis, Dr. Yiqun Liu and Pengyuan Dong at the National Center for Protein Science and Core Facilities of Life Sciences in Peking University for technical assistance in SEM sample preparation, operation, and Xiaorui Hao at Electron Microscopy Laboratory at Peking University for bone sample preparation for AFM measurements. The image acquisition of confocal microscope was supported by Biological Imaging and Analysis Laboratory, Medical and Health Analytical Center, Peking University, especially Jing Wu. We appreciate Rong Guo of Beijing Cnkingbio Biotechnology Co.LTD for the bioinformatics assistance.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.03.036.
Contributor Information
Ping Zhang, Email: zhangping332@hsc.pku.edu.cn.
Yongsheng Zhou, Email: kqzhouysh@hsc.pku.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cossu G., Birchall M., Brown T., De Coppi P., Culme-Seymour E., Gibbon S., Hitchcock J., Mason C., Montgomery J., Morris S., Muntoni F., Napier D., Owji N., Prasad A., Round J., Saprai P., Stilgoe J., Thrasher A., Wilson J. Lancet Commission: stem cells and regenerative medicine. Lancet. 2018;391:883–910. doi: 10.1016/S0140-6736(17)31366-1. [DOI] [PubMed] [Google Scholar]
- 2.Wei Y., Jiang S.J., Si M.T., Zhang X.H., Liu J., Wang Z., Cao C., Huang J., Huang H., Chen L.L., Wang S., Feng C., Deng X.L., Jiang L. Chirality controls mesenchymal stem cell lineage diversification through mechanoresponses. Adv. Mater. 2019;31 doi: 10.1002/adma.201900582. [DOI] [PubMed] [Google Scholar]
- 3.Smith L.R., Cho S., Discher D.E. Stem cell differentiation is regulated by extracellular matrix mechanics. Physiology. 2018;33:16–25. doi: 10.1152/physiol.00026.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy W.L., McDevitt T.C., Engler A.J. Materials as stem cell regulators. Nat. Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang C., Tibbitt M.W., Basta L., Anseth K.S. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darnell M., Gu L., Mooney D. RNA-seq reveals diverse effects of substrate stiffness on mesenchymal stem cells. Biomaterials. 2018;181:182–188. doi: 10.1016/j.biomaterials.2018.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huebsch N., Arany P.R., Mao A.S., Shvartsman D., Ali O.A., Bencherif S.A., Rivera-Feliciano J., Mooney D.J. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat. Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monnier V.M., Mustata G.T., Biemel K.L., Reihl O., Lederer M.O., Zhenyu D., Sell D.R. Cross-linking of the extracellular matrix by the maillard reaction in aging and diabetes: an update on "a puzzle nearing resolution. Ann. N. Y. Acad. Sci. 2005;1043:533–544. doi: 10.1196/annals.1333.061. [DOI] [PubMed] [Google Scholar]
- 9.Handorf A.M., Zhou Y., Halanski M.A., Li W.J. Tissue stiffness dictates development, homeostasis, and disease progression. Organogenesis. 2015;11:1–15. doi: 10.1080/15476278.2015.1019687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashiro Y., Yanagisawa H. The molecular mechanism of mechanotransduction in vascular homeostasis and disease. Clin. Sci. (Lond.) 2020;134:2399–2418. doi: 10.1042/CS20190488. [DOI] [PubMed] [Google Scholar]
- 11.Lampi M.C., Reinhart-King C.A. Targeting extracellular matrix stiffness to attenuate disease: from molecular mechanisms to clinical trials. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aao0475. [DOI] [PubMed] [Google Scholar]
- 12.Toledano M., Toledano-Osorio M., Guerado E., Caso E., Osorio E., Osorio R. Assessing bone quality through mechanical properties in postmenopausal trabecular bone. Injury. 2018;49(Suppl 2):S3–S10. doi: 10.1016/j.injury.2018.07.035. [DOI] [PubMed] [Google Scholar]
- 13.Chavassieux P., Chapurlat R., Portero-Muzy N., Roux J.P., Garcia P., Brown J.P., Libanati C., Boyce R.W., Wang A., Grauer A. Bone-Forming and antiresorptive effects of romosozumab in postmenopausal women with osteoporosis: bone histomorphometry and microcomputed tomography analysis after 2 and 12 Months of treatment. J. Bone Miner. Res. 2019;34:1597–1608. doi: 10.1002/jbmr.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y., Du S., Quan S., Jiang F., Yang C., Li J. Effects of biophysical cues of 3D hydrogels on mesenchymal stem cells differentiation. J. Cell. Physiol. 2021;236:2268–2275. doi: 10.1002/jcp.30042. [DOI] [PubMed] [Google Scholar]
- 15.Chang L., Azzolin L., Di Biagio D., Zanconato F., Battilana G., Lucon Xiccato R., Aragona M., Giulitti S., Panciera T., Gandin A., Sigismondo G., Krijgsveld J., Fassan M., Brusatin G., Cordenonsi M., Piccolo S. The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature. 2018;563:265–269. doi: 10.1038/s41586-018-0658-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun M., Chi G., Xu J., Tan Y., Xu J., Lv S., Xu Z., Xia Y., Li L., Li Y. Extracellular matrix stiffness controls osteogenic differentiation of mesenchymal stem cells mediated by integrin α5. Stem Cell Res. Ther. 2018;9:52. doi: 10.1186/s13287-018-0798-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stanton A.E., Tong X., Yang F. Extracellular matrix type modulates mechanotransduction of stem cells. Acta Biomater. 2019;96:310–320. doi: 10.1016/j.actbio.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan W.J., Mullen P.J., Schmid E.W., Flores A., Momcilovic M., Sharpley M.S., Jelinek D., Whiteley A.E., Maxwell M.B., Wilde B.R., Banerjee U., Coller H.A., Shackelford D.B., Braas D., Ayer D.E., de Aguiar Vallim T.Q., Lowry W.E., Christofk H.R. Extracellular matrix remodeling regulates glucose metabolism through TXNIP destabilization. Cell. 2018;175:117–132. doi: 10.1016/j.cell.2018.08.017. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu H., Juvekar A., Lyssiotis C.A., Lien E.C., Albeck J.G., Oh D., Varma G., Hung Y.P., Ullas S., Lauring J., Seth P., Lundquist M.R., Tolan D.R., Grant A.K., Needleman D.J., Asara J.M., Cantley L.C., Wulf G.M. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell. 2016;164:433–446. doi: 10.1016/j.cell.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montal E.D., Dewi R., Bhalla K., Ou L., Hwang B.J., Ropell A.E., Gordon C., Liu W.J., DeBerardinis R.J., Sudderth J., Twaddel W., Boros L.G., Shroyer K.R., Duraisamy S., Drapkin R., Powers R.S., Rohde J.M., Boxer M.B., Wong K.K., Girnun G.D. PEPCK coordinates the regulation of central carbon metabolism to promote cancer cell growth. Mol. Cell. 2015;60:571–583. doi: 10.1016/j.molcel.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leithner K., Hrzenjak A., Trötzmüller M., Moustafa T., Köfeler H.C., Wohlkoenig C., Stacher E., Lindenmann J., Harris A.L., Olschewski A., Olschewski H. PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene. 2015;34:1044–1050. doi: 10.1038/onc.2014.47. [DOI] [PubMed] [Google Scholar]
- 22.Méndez-Lucas A., Hyroššová P., Novellasdemunt L., Viñals F., Perales J.C. Mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) is a pro-survival, endoplasmic reticulum (ER) stress response gene involved in tumor cell adaptation to nutrient availability. J. Biol. Chem. 2014;289:22090–22102. doi: 10.1074/jbc.M114.566927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leithner K., Triebl A., Trötzmüller M., Hinteregger B., Leko P., Wieser B.I., Grasmann G., Bertsch A.L., Züllig T., Stacher E., Valli A., Prassl R., Olschewski A., Harris A.L., Köfeler H.C., Olschewski H., Hrzenjak A. The glycerol backbone of phospholipids derives from noncarbohydrate precursors in starved lung cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6225–6230. doi: 10.1073/pnas.1719871115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z., Liu X., Zhu Y., Du Y., Liu X., Lv L., Zhang X., Liu Y., Zhang P., Zhou Y. Mitochondrial phosphoenolpyruvate carboxykinase regulates osteogenic differentiation by modulating AMPK/ULK1-Dependent autophagy. Stem Cell. 2019;37:1542–1555. doi: 10.1002/stem.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J.S., Burckhardt C.J., Lazcano R., Solis L.M., Isogai T., Li L., Chen C.S., Gao B., Minna J.D., Bachoo R., DeBerardinis R.J., Danuser G. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature. 2020;578:621–626. doi: 10.1038/s41586-020-1998-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saleh N., Nassef N.A., Shawky M.K., Elshishiny M.I., Saleh H.A. Novel approach for pathogenesis of osteoporosis in ovariectomized rats as a model of postmenopausal osteoporosis. Exp. Gerontol. 2020;137 doi: 10.1016/j.exger.2020.110935. [DOI] [PubMed] [Google Scholar]
- 27.G Y.R., Mei F., Huang Y., Ma S.Q., Wei Y., Zhang X.H., Xu Mingming, He Y., H B.C., C L.L., Deng X.L. vol. 7. 2021. pp. 364–376. (Matrix Stiffness Modulates Tip Cell Formation through the P-PXN-Rac1-YAP Signaling axis). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regmi S., Pathak S., Kim J.O., Yong C.S., Jeong J.H. Mesenchymal stem cell therapy for the treatment of inflammatory diseases: challenges, opportunities, and future perspectives. Eur. J. Cell Biol. 2019;98 doi: 10.1016/j.ejcb.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen M.K., Jeon O., Dang P.N., Huynh C.T., Varghai D., Riazi H., McMillan A., Herberg S., Alsberg E. RNA interfering molecule delivery from in situ forming biodegradable hydrogels for enhancement of bone formation in rat calvarial bone defects. Acta Biomater. 2018;75:105–114. doi: 10.1016/j.actbio.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M., Liu X.M., Li Z., Du Y.G., Liu X.J., Lv L.W., Zhang X., Liu Y.S., Zhang P., Zhou Y.S. Asymmetrical methyltransferase PRMT3 regulates human mesenchymal stem cell osteogenesis via miR-3648. Cell Death Dis. 2019;10:581. doi: 10.1038/s41419-019-1815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai L., Xiong X., Kong X., Xie J. The role of the lysyl oxidases in tissue repair and remodeling: a concise review. Tissue Eng. Regen. Med. 2017;14:15–30. doi: 10.1007/s13770-016-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou G., Liu W., Cui L., Wang X., Liu T., Cao Y. Repair of porcine articular osteochondral defects in non-weightbearing areas with autologous bone marrow stromal cells. Tissue Eng. 2006;12:3209–3221. doi: 10.1089/ten.2006.12.3209. [DOI] [PubMed] [Google Scholar]
- 33.Xiao W., He J., Nichol J.W., Wang L., Hutson C.B., Wang B., Du Y., Fan H., Khademhosseini A. Synthesis and characterization of photocrosslinkable gelatin and silk fibroin interpenetrating polymer network hydrogels. Acta Biomater. 2011;7:2384–2393. doi: 10.1016/j.actbio.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hengsberger S., Ammann P., Legros B., Rizzoli R., Zysset P. Intrinsic bone tissue properties in adult rat vertebrae: modulation by dietary protein. Bone. 2005;36:134–141. doi: 10.1016/j.bone.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Sudhaker Rao D., Ajdelsztajn L., Ciarelli T.E., Lavernia E.J., Fyhrie D.P. Human iliac crest cancellous bone elastic modulus and hardness differ with bone formation rate per bone surface but not by existence of prevalent vertebral fracture. J. Biomed. Mater. Res. B Appl. Biomater. 2008;85:68–77. doi: 10.1002/jbm.b.30918. [DOI] [PubMed] [Google Scholar]
- 36.Leckband D.E., le Duc Q., Wang N., de Rooij J. Mechanotransduction at cadherin-mediated adhesions. Curr. Opin. Cell Biol. 2011;23:523–530. doi: 10.1016/j.ceb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Chowdhury F., Huang B., Wang N. Cytoskeletal prestress: the cellular hallmark in mechanobiology and mechanomedicine. Cytoskeleton (Hoboken) 2021;78:249–276. doi: 10.1002/cm.21658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tharp K.M., Higuchi-Sanabria R., Timblin G.A., Ford B., Garzon-Coral C., Schneider C., Muncie J.M., Stashko C., Daniele J.R., Moore A.S., Frankino P.A., Homentcovschi S., Manoli S.S., Shao H., Richards A.L., Chen K.H., Hoeve J.T., Ku G.M., Hellerstein M., Nomura D.K., Saijo K., Gestwicki J., Dunn A.R., Krogan N.J., Swaney D.L., Dillin A., Weaver V.M. Adhesion-mediated mechanosignaling forces mitohormesis. Cell Metabol. 2021;33:1322–1341. doi: 10.1016/j.cmet.2021.04.017. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg's contributions to current concepts of cancer metabolism. Nat. Rev. Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 40.Chen H., Ji X., Lee W.C., Shi Y., Li B., Abel E.D., Jiang D., Huang W., Long F. Increased glycolysis mediates Wnt7b-induced bone formation. Faseb. J. 2019;33:7810–7821. doi: 10.1096/fj.201900201RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esen E., Long F. Aerobic glycolysis in osteoblasts. Curr. Osteoporos. Rep. 2014;12:433–438. doi: 10.1007/s11914-014-0235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong W.J., Yang H.H., Guan X.X., Xiong J.B., Sun C.C., Zhang C.Y., Luo X.Q., Zhang Y.F., Zhang J., Duan J.X., Zhou Y., Guan C.X. Inhibition of glycolysis alleviates lipopolysaccharide-induced acute lung injury in a mouse model. J. Cell. Physiol. 2019;234:4641–4654. doi: 10.1002/jcp.27261. [DOI] [PubMed] [Google Scholar]
- 43.Moreno-Fernandez M.E., Giles D.A., Oates J.R., Chan C.C., Damen M., Doll J.R., Stankiewicz T.E., Chen X., Chetal K., Karns R., Weirauch M.T., Romick-Rosendale L., Xanthakos S.A., Sheridan R., Szabo S., Shah A.S., Helmrath M.A., Inge T.H., Deshmukh H., Salomonis N., Divanovic S. PKM2-dependent metabolic skewing of hepatic Th17 cells regulates pathogenesis of non-alcoholic fatty liver disease. Cell Metabol. 2021;33:1187–1204. doi: 10.1016/j.cmet.2021.04.018. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu Y., Newman H., Shen L., Sharma D., Hu G., Mirando A.J., Zhang H., Knudsen E., Zhang G.F., Hilton M.J., Karner C.M. Glutamine metabolism regulates proliferation and lineage allocation in skeletal stem cells. Cell Metabol. 2019;29:966–978. doi: 10.1016/j.cmet.2019.01.016. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng M., Xiong G., Cao Z., Yang G., Zheng S., Qiu J., You L., Zheng L., Zhang T., Zhao Y. LAT2 regulates glutamine-dependent mTOR activation to promote glycolysis and chemoresistance in pancreatic cancer. J. Exp. Clin. Cancer Res. 2018;37:274. doi: 10.1186/s13046-018-0947-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanner L.B., Goglia A.G., Wei M.H., Sehgal T., Parsons L.R., Park J.O., White E., Toettcher J.E., Rabinowitz J.D. Four key steps control glycolytic flux in mammalian cells. Cell Syst. 2018;7 doi: 10.1016/j.cels.2018.06.003. 49-62.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ros S., Schulze A. Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metabol. 2013;1:8. doi: 10.1186/2049-3002-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 49.Chatterjee P., Gupta V.B., Fagan A.M., Jasielec M.S., Xiong C., Sohrabi H.R., Dhaliwal S., Taddei K., Bourgeat P., Brown B.M., Benzinger T., Bateman R.J., Morris J.C., Martins R.N. Decreased platelet APP isoform ratios in autosomal dominant Alzheimer's disease: baseline data from a DIAN cohort subset. Curr. Alzheimer Res. 2015;12:157–164. doi: 10.2174/1567205012666150204125732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y., Xu Q., Ji D., Wei Y., Chen H., Li T., Wan B., Yuan L., Huang R., Chen G. Inhibition of pentose phosphate pathway suppresses acute myelogenous leukemia. Tumour Biol. 2016;37:6027–6034. doi: 10.1007/s13277-015-4428-5. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X., Ye P., Huang H., Wang B., Dong F., Ling Q. TCF7L2 rs290487 C allele aberrantly enhances hepatic gluconeogenesis through allele-specific changes in transcription and chromatin binding. Aging (Albany NY) 2020;12:13365–13387. doi: 10.18632/aging.103442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saidova A.A., Vorobjev I.A. Lineage commitment, signaling pathways, and the cytoskeleton systems in mesenchymal stem cells. Tissue Eng. B Rev. 2020;26:13–25. doi: 10.1089/ten.TEB.2019.0250. [DOI] [PubMed] [Google Scholar]
- 53.Peng M., Yang D., Hou Y., Liu S., Zhao M., Qin Y., Chen R., Teng Y., Liu M. Intracellular citrate accumulation by oxidized ATM-mediated metabolism reprogramming via PFKP and CS enhances hypoxic breast cancer cell invasion and metastasis. Cell Death Dis. 2019;10:228. doi: 10.1038/s41419-019-1475-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Tsai K.S., Kao S.Y., Wang C.Y., Wang Y.J., Wang J.P., Hung S.C. Type I collagen promotes proliferation and osteogenesis of human mesenchymal stem cells via activation of ERK and Akt pathways. J. Biomed. Mater. Res. 2010;94:673–682. doi: 10.1002/jbm.a.32693. [DOI] [PubMed] [Google Scholar]
- 55.Lee H.P., Stowers R., Chaudhuri O. Volume expansion and TRPV4 activation regulate stem cell fate in three-dimensional microenvironments. Nat. Commun. 2019;10:529. doi: 10.1038/s41467-019-08465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lutolf M.P., Gilbert P.M., Blau H.M. Designing materials to direct stem-cell fate. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Papalazarou V., Zhang T., Paul N.R., Juin A., Cantini M., Maddocks O.D.K., Salmeron-Sanchez M., Machesky L.M. The creatine-phosphagen system is mechanoresponsive in pancreatic adenocarcinoma and fuels invasion and metastasis. Nat. Metabol. 2020;2:62–80. doi: 10.1038/s42255-019-0159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang C., Liu X., Zhao K., Zhu Y., Hu B., Zhou Y., Wang M., Wu Y., Zhang C., Xu J., Ning Y., Zou D. miRNA-21 promotes osteogenesis via the PTEN/PI3K/Akt/HIF-1α pathway and enhances bone regeneration in critical size defects. Stem Cell Res. Ther. 2019;10:65. doi: 10.1186/s13287-019-1168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.