Abstract
STUDY QUESTION
Are epididymosomes implicated in protein transfer from the epididymis to spermatozoa?
SUMMARY ANSWER
We characterized the contribution of epididymal secretions to the sperm proteome and demonstrated that sperm acquire epididymal proteins through epididymosomes.
WHAT IS KNOWN ALREADY
Testicular sperm are immature cells unable to fertilize an oocyte. After leaving the testis, sperm transit along the epididymis to acquire motility and fertilizing abilities. It is well known that marked changes in the sperm proteome profile occur during epididymal maturation. Since the sperm is a transcriptional and translational inert cell, previous studies have shown that sperm incorporate proteins, RNA and lipids from extracellular vesicles (EVs), released by epithelial cells lining the male reproductive tract.
STUDY DESIGN, SIZE, DURATION
We examined the contribution of the epididymis to the post-testicular maturation of spermatozoa, via the production of EVs named epididymosomes, released by epididymal epithelial cells. An integrative analysis using both human and mouse data was performed to identify sperm proteins with a potential epididymis-derived origin. Testes and epididymides from adult humans (n = 9) and adult mice (n = 3) were used to experimentally validate the tissue localization of four selected proteins using high-resolution confocal microscopy. Mouse epididymal sperm were co-incubated with carboxyfluorescein succinimidyl ester (CFSE)-labeled epididymosomes (n = 4 mice), and visualized using high-resolution confocal microscopy.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Adult (12-week-old) C57BL/CBAF1 wild-type male mice and adult humans were used for validation purposes. Testes and epididymides from both mice and humans were obtained and processed for immunofluorescence. Mouse epididymal sperm and mouse epididymosomes were obtained from the epididymal cauda segment. Fluorescent epididymosomes were obtained after labeling the epididymal vesicles with CFSE dye followed by epididymosome isolation using a density cushion. Immunofluorescence was performed following co-incubation of sperm with epididymosomes in vitro. High-resolution confocal microscopy and 3D image reconstruction were used to visualize protein localization and sperm-epididymosomes interactions.
MAIN RESULTS AND THE ROLE OF CHANCE
Through in silico analysis, we first identified 25 sperm proteins with a putative epididymal origin that were conserved in both human and mouse spermatozoa. From those, the epididymal origin of four sperm proteins (SLC27A2, EDDM3B, KRT19 and WFDC8) was validated by high-resolution confocal microscopy. SLC27A2, EDDM3B, KRT19 and WFDC8 were all detected in epithelial cells lining the human and mouse epididymis, and absent from human and mouse seminiferous tubules. We found region-specific expression patterns of these proteins throughout the mouse epididymides. In addition, while EDDM3B, KRT19 and WFDC8 were detected in both epididymal principal and clear cells (CCs), SLC27A2 was exclusively expressed in CCs. Finally, we showed that CFSE-fluorescently labeled epididymosomes interact with sperm in vitro and about 12–36% of the epididymosomes contain the targeted sperm proteins with an epididymal origin.
LARGE SCALE DATA
N/A.
LIMITATIONS, REASONS FOR CAUTION
The human and mouse sample size was limited and our results were descriptive. The analyses of epididymal sperm and epididymosomes were solely performed in the mouse model due to the difficulties in obtaining epididymal luminal fluid human samples. Alternatively, human ejaculated sperm and seminal EVs could not be used because ejaculated sperm have already contacted with the fluids secreted by the male accessory sex glands, and seminal EVs contain other EVs in addition to epididymosomes, such as the abundant prostate-derived EVs.
WIDER IMPLICATIONS OF THE FINDINGS
Our findings indicate that epididymosomes are capable of providing spermatozoa with a new set of epididymis-derived proteins that could modulate the sperm proteome and, subsequently, participate in the post-testicular maturation of sperm cells. Additionally, our data provide further evidence of the novel role of epididymal CCs in epididymosome production. Identifying mechanisms by which sperm mature to acquire their fertilization potential would, ultimately, lead to a better understanding of male reproductive health and may help to identify potential therapeutic strategies to improve male infertility.
STUDY FUNDING/COMPETING INTEREST(S)
This work was supported by the Spanish Ministry of Economy and Competitiveness (Ministerio de Economía y Competividad; fondos FEDER ‘una manera de hacer Europa’ PI13/00699 and PI16/00346 to R.O.; and Sara Borrell Postdoctoral Fellowship, Acción Estratégica en Salud, CD17/00109 to J.C.), by National Institutes of Health (grants HD040793 and HD069623 to S.B., grant HD104672-01 to M.A.B.), by the Spanish Ministry of Education, Culture and Sports (Ministerio de Educación, Cultura y Deporte para la Formación de Profesorado Universitario, FPU15/02306 to F.B.), by a Lalor Foundation Fellowship (to F.B. and M.A.B.), by the Government of Catalonia (Generalitat de Catalunya, pla estratègic de recerca i innovació en salut, PERIS 2016-2020, SLT002/16/00337 to M.J.), by Fundació Universitària Agustí Pedro i Pons (to F.B.), and by the American Society for Biochemistry and Molecular Biology (PROLAB Award from ASBMB/IUBMB/PABMB to F.B.). Confocal microscopy and transmission electron microscopy was performed in the Microscopy Core facility of the Massachusetts General Hospital (MGH) Center for Systems Biology/Program in Membrane Biology which receives support from Boston Area Diabetes and Endocrinology Research Center (BADERC) award DK57521 and Center for the Study of Inflammatory Bowel Disease grant DK43351. The Zeiss LSM800 microscope was acquired using an NIH Shared Instrumentation Grant S10-OD-021577-01. The authors have no conflicts of interest to declare.
Keywords: epididymis, epididymosomes, exosomes, extracellular vesicles, protein transfer, sperm proteins, sperm maturation, sperm proteome, male fertility, post-testicular maturation
Introduction
Spermatogenesis is a complex process that takes place in the seminiferous tubules of the testis and involves a highly regulated germ cell differentiation process, giving rise to the male gamete (Oliva and Dixon, 1991; de Kretser et al., 1998; Hecht, 1998; Oliva and Castillo, 2011; Carrell et al., 2016; Gervasi and Visconti, 2017). The resulting testicular spermatozoa, though morphologically differentiated, are still functionally immature cells, unable to move progressively and lacking the ability to fertilize an oocyte (Orgebin-Crist, 1967, 1969; Schoysman and Bedford, 1986; Dacheux and Dacheux, 2014; Jodar et al., 2017). In humans and mice, the motility and the fertilizing potential of spermatozoa are acquired after leaving the testis, when testicular sperm transit through the epididymis, and all along the male reproductive tract. During this journey, the sperm cells are in contact with the fluids secreted by the epididymis, prostate, seminal vesicles and bulbourethral glands, which collectively produce the seminal plasma (Camargo et al., 2018). This complex fluid is crucial for sperm nutrition, maturation and survival, and plays a key role in fertilization by preventing premature capacitation and contributing to the sperm-to-oocyte recognition and interaction (Drabovich et al., 2014; Jodar et al., 2017; Camargo et al., 2018; Samanta et al., 2018; Barrachina et al., 2019). These post-testicular modifications occur in a male gamete that is transcriptionally and translationally inert due to the high DNA compaction of the sperm nuclei and extrusion of the majority of the cytoplasm (Dacheux et al., 2012; Castillo et al., 2015; Jodar et al., 2016, 2017; Barrachina et al., 2018). Therefore, post-testicular functional maturation of the spermatozoa is attributed to the acquisition of components from the seminal plasma (Reilly et al., 2016; Jodar et al., 2017; Zhou et al., 2018; Barrachina et al., 2019; Hernández-Silva and Chirinos, 2019). Previous studies have shown the participation of extracellular vesicles (EVs) released by the different male accessory sex glands in the post-testicular maturation process, which results in substantial changes in the protein, RNA and lipid content of the sperm cell (Saez et al., 2003; Aalberts et al., 2014; Belleannée, 2015; Martin-DeLeon, 2015; Sullivan, 2016; Barceló et al., 2018, 2019; García-Rodríguez et al., 2018).
Recently, our group compiled a catalog of human sperm proteins from published datasets, identifying up to 6871 proteins present in the sperm cell (Castillo et al., 2018). By an integrative analysis using proteomic and transcriptomic data from sperm, testis, epididymis, prostate and seminal vesicles, 165 human sperm proteins were identified as presumably extra-testicularly acquired, with some of them being involved in sperm function and fertilization (Castillo et al., 2018). This finding further highlighted the impact of secretions from the post-testicular male reproductive tract on the sperm proteome and sperm function. Interestingly, about half of these proteins (n = 93) showed an epididymal-origin, emphasizing the importance of epididymal secretions for the post-testicular maturation of the sperm cell.
The human epididymis is a 6–7 m long tubular organ that connects the testis to the vas deferens. After being produced by the testis, sperm cells travel through and are stored in the epididymis for up to 2 weeks. During this period, sperm cells undergo maturation and acquire motility and the ability to fertilize the oocyte (Orgebin-Crist, 1969; Breton et al., 2016; Sullivan et al., 2019). Anatomically, the epididymis is divided into four main regions: initial segments, caput, corpus and cauda epididymides (Turner, 1995; Belleannée et al., 2013a; Breton et al., 2016; Sullivan and Mieusset, 2016), and it is further subdivided into smaller segments that have different functional characteristics, as described elsewhere (Turner et al., 2003; Johnston et al., 2005; Jelinsky et al., 2007; Dacheux et al., 2016). The epididymis consists of a pseudostratified epithelium composed by principal cells (PCs), clear cells (CCs), narrow cells and basal cells (BCs), among others, whose role is essential for the creation of a unique luminal environment for sperm maturation, concentration, protection and storage (Breton et al., 2016; Zhou et al., 2018). BCs cover the base of the epididymis in all epididymal regions, although they were shown to transiently cross the tight-junction barrier to sample the luminal content in specific segments (Shum et al., 2008; Breton et al., 2016; Roy et al., 2016). PCs are the most abundant cell type in the epididymis and participate in the transepithelial transport of water, solutes, ions and protein secretion (Robaire et al., 2006; Da Silva et al., 2007; Park et al., 2017; Zhou et al., 2018; Breton et al., 2019). PCs are traditionally believed to be the main cells producing EVs, called epididymosomes (Sullivan et al., 2007; Caballero et al., 2010; Martin-DeLeon, 2015; Zhou et al., 2018; James et al., 2020), but recent findings indicate that CCs may also release epididymosomes (Battistone et al., 2019b). CCs are proton-secreting cells that express the proton pump V-ATPase in their apical membrane, and are responsible for luminal acidification and endocytosis (Hermo et al., 1988; Brown et al., 1997; Breton et al., 2016, 2019; Zhou et al., 2018).
It is well known that the epididymal lumen contains an impressive population of epididymosomes, which seem to play an important role in the post-testicular maturation of spermatozoa (Yanagimachi et al., 1985; Sullivan et al., 2007; Sullivan and Saez, 2013; Sullivan, 2015; Gervasi and Visconti, 2017). The interaction between epididymosomes and epididymal sperm has also been demonstrated through different approaches, such as the use of biotinylated proteins, fluorescent markers or transgenic mice (Frenette et al., 2002, 2010; Schwarz et al., 2013; Reilly et al., 2016; Nixon et al., 2019; Zhou et al., 2019; Battistone et al., 2019b). In order to decipher the impact of epididymosomes on sperm function, several groups have conducted comprehensive molecular profiling of the epididymosome content in different species, aimed at unraveling the proteomic (Frenette et al., 2006, 2010; Thimon et al., 2008b; Girouard et al., 2011; Nixon et al., 2019) and transcriptomic (Belleannée et al., 2013a,b; Reilly et al., 2016; Sharma et al., 2016, 2018) composition of epididymosomes. While these studies have provided evidence that epididymosomes may be implicated in the transfer of secreted epididymal proteins to sperm (Frenette and Sullivan, 2001; Frenette et al., 2010; D’Amours et al., 2012), to the best of our knowledge, direct protein transfer to spermatozoa through epididymosomes is still poorly characterized.
Therefore, the main aim of this study is to experimentally validate direct sperm acquisition of epididymal proteins through epididymosomes. First, we used protein data from human and mouse sperm to infer proteins with a putative epididymis-derived origin that are potentially transferred to sperm by epididymosomes in both species. Using immunofluorescence analysis, we then experimentally demonstrated the extra-testicular sperm acquisition of some of these inferred proteins through epididymosomes.
Materials and methods
In silico prediction of sperm proteins with an epididymal origin
The initial identification of potential sperm proteins with an epididymis origin was based on a previous integrative analysis carried out in our laboratory, which provided a list of 165 human sperm proteins potentially acquired after testicular maturation (from the epididymis, prostate and/or seminal vesicles) (Castillo et al., 2018) (Fig. 1). From those, 93 showed a putative epididymal origin (Fig. 1). This dataset was analyzed herein in order to identify sperm proteins with a potential epididymal origin that were conserved among species, using mice and humans as model systems (Fig. 1). Thus, we assessed whether those 93 human sperm proteins were detected in proteomic data from mouse testis (Paz et al., 2006; Zhu et al., 2006; Huang et al., 2008; Guo et al., 2010; Gan et al., 2013; Qi et al., 2014; Xie et al., 2018; Xu et al., 2018), epididymal sperm (Baker et al., 2008; Dorus et al., 2010; Chauvin et al., 2012; Claydon et al., 2012; Guyonnet et al., 2012; Skerget et al., 2015; Vicens et al., 2017), epididymis (Chaurand et al., 2003; Da Silva et al., 2010; Liu et al., 2015a,b) and epididymosomes (Nixon et al., 2019) (Supplementary Table SI). Specifically, we first selected the human sperm proteins with a putative epididymal origin that were also found in the mouse epididymal sperm proteome, and that were absent in the mouse testis proteome. Subsequently, we further complemented the dataset with information from the mouse epididymis and epididymosome protein reports. However, since the available proteomic data for mouse epididymis and epididymosomes is scarce, this information was not considered as a condition for selecting the potential candidates to be validated herein. The Gene Ontology (GO) Knowledgebase from the GO Consortium (http://www.geneontology.org/) (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) was used to conduct a GO Enrichment analysis on Cellular Components terms (Mi et al., 2019). This tool was powered by the PANTHER v15.0 database (Release date 11 June 2019) and the significance of the enrichment analysis was calculated by a Fisher’s exact test. P-values <0.05 after false discovery rate (FDR) adjustment were considered statistically significant.
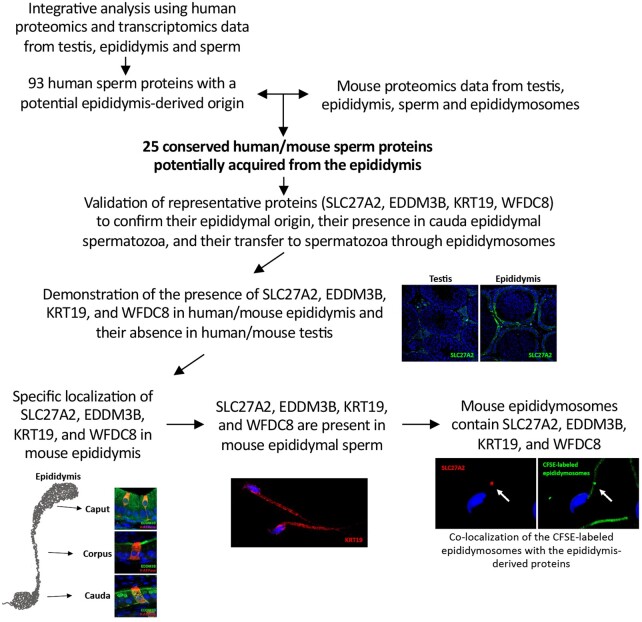
Figure 1.
Overall workflow to identify and validate conserved human/mouse sperm proteins acquired from the epididymis through epididymosomes.
Biological material and ethical approval
Adult (12-week-old) C57BL/CBAF1 wild-type male mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA), and at least three mice were analyzed for each experiment. All experimental procedures were reviewed and approved by the Massachusetts General Hospital Subcommittee on Research Animal Care and performed in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Prior to dissection, animals were anesthetized with pentobarbital (60 mg/kg intraperitoneally) and euthanized by cervical dislocation.
Human epididymis (n = 9) and testis samples (n = 9) were obtained from the Department of Pathology, Hospital Clínic de Barcelona (Barcelona, Spain), and all samples were analyzed for each set of experiments. Epididymis tissues were obtained post-mortem after myocardial infarction (n = 2) or hemorrhagic stroke (n = 1), and from orchidectomy performed on patients with testicular tumors (n = 6). Testicular biopsies were obtained from infertile patients with normal spermatogenesis (n = 9). Patients were 38 ± 5.3 years old (mean ± SEM). Written informed consent was obtained from all subjects in accordance with the ethical standards of the institutional and national research committee, and with the Declaration of Helsinki and the Department of Health and Human Services Belmont Report. All sample storage and processing were approved by the Clinical Research Ethics Committee of the Hospital Clínic de Barcelona (Barcelona, Spain).
Testis and epididymis preparation
Testes and epididymides from adult male mice were removed and fixed for 16 h by immersion in paraformaldehyde (PFA) lysine periodate solution (containing 4% PFA) at 4°C with shaking. After several washes in phosphate buffered saline (PBS), tissues were incubated at 4°C in PBS containing 30% sucrose for 2 days. Subsequently, tissues were embedded in Tissue-Tek OCT compound (Sakura Finetek, Torrance, CA, USA), and mounted and frozen on a cutting block in a Reichert Frigocut microtome. Tissues were cut at 10-μm thickness, and sections were placed onto Fisherbrand Superfrost Plus microscope slides (Fisher Scientific, Pittsburgh, PA, USA).
Testes and epididymides from adult humans were fixed in 4% formol and Sakura molecular fixative (Tissue-Tek Xpress, Sakura, Torrance, CA, USA) for 16 h each. Samples were processed on the Sakura Tissue-Tek Xpress x120 tissue processor (Tissue-Tek Xpress, Sakura), where samples were dehydrated in pre-processing solution (Tissue-Tek Xpress, Sakura) for 30 min, followed by three washes of 30 min in Processing reagent (Tissue-Tek Xpress, Sakura) and paraffin-embedded. Tissues were cut in a Tissue-Tek AutoSection microtome at 2 μm and 4 μm thickness.
Sperm collection
Mouse epididymal fluid containing spermatozoa was obtained from four adult mice as follows. After induction of anesthesia, the epididymides were removed, and the cauda segment was separated from the rest of the tissue. Cauda epididymides were minced (3 cuts) and placed in a physiological solution adjusted to pH 6.6 and containing 50 mM NaCl, 50 mM K-gluconate, 1.2 mM MgSO4, 0.6 mM CaCl2, 4 mM NaOAc, 1 mM Trisodium citrate, 6.4 mM NaH2PO4, 3.6 mM Na2HPO4 (360 mOsm/kg) for 15 min at 37°C. Afterward, the remaining tissue was discarded, and the medium was centrifuged at 400 g for 5 min at room temperature (RT) to separate epididymal spermatozoa from the epididymal fluid. Only mouse cauda sperm was collected since these sperm have already contacted with fluids from all sections of the epididymis: initial segments, caput, corpus and cauda. Sperm cells were immediately used for the co-incubation assay with epididymosomes.
Epididymosomes labeling and isolation
The mouse cauda epididymal fluid from eight epididymides was diluted 1:1 in pH 6.6 solution, centrifuged at 1200 g for 15 min, 3000 g for 15 min and 10 000 g for 30 min at 4°C, and the supernatant was subsequently filtered through a 0.20 μm membrane. Afterward, the supernatant was ultracentrifuged (100 000 g, 70 min, 4°C) and the pellet, which contained epididymosomes and other microvesicles, was resuspended in pH 6.6 solution. Vesicle labeling was conducted by incubation with 7.5 μM carboxyfluorescein succinimidyl ester (CFSE eBioscience™ CFSE, Invitrogen) (Fig. 2A; Condition A: Vesicles-CFSE), following the manufacturer’s instructions. The following negative controls were included: (i) vesicles incubated with anhydrous dimethylsulphoxide as a diluent control, without CFSE (Fig. 2A; Condition B: Vesicles-No CFSE); and (ii) pH 6.6 solution-only incubated with 7.5 μM CFSE (Fig. 2A; Condition C: CFSE) to ensure CFSE does not undergo non-specific aggregation. After 30 min incubation at 37°C in the dark, samples were diluted in pH 6.6 solution and centrifuged at 100 000 g for 70 min at 4°C to remove unbound CFSE. In order to enrich our preparation in epididymosomes, the pelleted vesicles were resuspended in pH 6.6 solution and subjected to a density gradient ultracentrifugation, as previously described (Vojtech et al., 2014). Briefly, the resuspended vesicles were loaded on top of a 30% sucrose/D2O density cushion followed by centrifugation at 100 000 g for 90 min at 4°C. The remaining supernatant was loaded on top of a 25% sucrose/D2O density cushion and centrifuged again at 100 000 g for 14 h at 4°C. Finally, both 25% and 30% sucrose cushions containing epididymosomes were combined, diluted in pH 6.6 solution, and epididymosomes were pelleted after a final ultracentrifugation step (100 000 g for 70 min at 4°C). Through this approach, first labeling vesicles with CFSE dye and, afterward, isolating epididymosomes using a 25–30% sucrose cushion, we ensured that no CFSE dye remained in the medium. The Bradford method (Quick Start™ Bradford Protein Assay; Bio-Rad Laboratories, Hercules, CA, USA) was used for the quantification of the total protein of epididymosomes.
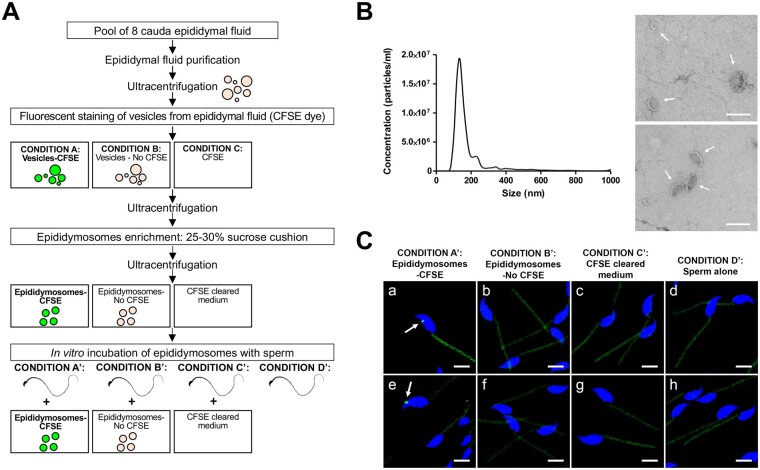
Figure 2.
Characterization of the epididymosomes and assessment of the incubation of CFSE-labeled epididymosomes with mice sperm. (A) Methodological workflow to fluorescently label the epididymosomes with the CFSE dye, followed by their co-incubation with mice sperm (Condition A’), and the negative controls (Condition B’, C’, D’). (B) Morphological characterization of the epididymosomes. The panels show the size distribution and particles concentration determined by nanoparticle tracking analysis (NTA) (left panel) and representative TEM images of isolated epididymosomes (right panel, in arrows). Bars = 200 nm. (C) Confocal microscopy images showing CFSE-labeled epididymosomes (arrows; green) attached to the mouse sperm head (Condition A’ (a, e)). No fluorescent epididymosome-like particles were observed for the negative controls (Condition B’ (b, f), C’ (c, g), D’ (d, h)). Note that autofluorescence in the green channel was present in the sperm midpiece due to a high concentration of mitochondria in this region. Sperm nuclei are labeled with DAPI in blue. Bars = 5 µm. Images show a 3D reconstruction of two colors z-stacked image acquired using the Zeiss LSM-800 confocal microscope. CFSE, carboxyfluorescein succinimidyl ester; DAPI, 4′,6-diamidino-2-phenylindole; TEM, transmission electron microscopy.
Characterization of the epididymosomes by nanoparticle tracking analysis and transmission electron microscopy
Size distribution and concentration of isolated epididymosomes were determined by nanoparticle tracking analysis (NTA) with NanoSight LM10 equipment (Malvern) (Fig. 2B) using the following parameters: camera at 30 frames per second, camera level at 14, temperature at 22°C, and video recording time 60 s. Nanosight NTA Software (version 3.2) analyzed raw data videos by quintuplicate. Vesicle morphology was analyzed using transmission electron microscopy (TEM) (Fig. 2B). Briefly, epididymosomes were fixed with 2% PFA in PBS, transferred onto 200 mesh formvar/carbon-coated nickel grids, and contrast-stained in 2% methylcellulose (tylose)-uranyl acetate solution. Note that vesicles are negatively stained with this method. Examination of preparations was done using a JEOL JEM 1011 transmission electron microscope at 80 kV. Images were collected using an AMT digital imaging system with proprietary image capture software (Advanced Microscopy Techniques, Danvers, MA, USA).
Co-incubation of sperm with CFSE-labeled epididymosomes
CFSE-labeled epididymosomes isolated as described above were resuspended in pH 6.6 solution supplemented with 1 mM ZnCl2, and incubated with cauda epididymal sperm for 3 h at 37°C (Fig. 2A; Condition A’: Epididymosomes-CFSE), based on previously reported methodologies (Frenette et al., 2002; Reilly et al., 2016). Negative controls included: (i) sperm incubated with epididymosomes not labeled with CFSE (Fig. 2A; Condition B’: Epididymosomes-No CFSE); (ii) sperm incubated with medium without CFSE, in which the CFSE dye from initial Condition C has been properly removed from the medium through sequential ultracentrifugations (Fig. 2A; Condition C’: CFSE cleared medium); and (iii) sperm alone (Fig. 2A; Condition D’: Sperm alone). After 3 h-incubation, sperm cells were washed three times (400 g, 5 min) in pH 6.6 solution to remove any unbound CFSE-labeled epididymosomes and fixed in 1% PFA. Afterwards, the efficacy of both the isolation and the labeling of epididymosomes, and the interaction with mouse spermatozoa were assessed by confocal microscopy (Fig. 2C).
Immunofluorescence
Immunofluorescence was performed as previously described (Battistone et al., 2019a). For the human paraffin-embedded sections, the process included a xylene deparaffinization (2 × 100% xylene), followed by washes in ethanol (2 × 100% EtOH), and tissue rehydration through graded series of ethanol (90%, 70%, 50%, 30%). Afterward, all samples (including human/mouse testes and epididymides, and mouse sperm alone or incubated with epididymosomes) were hydrated in PBS and, for antigen retrieval, samples were treated with PBS containing 1% SDS and 0.1% Triton for 4 min. The anti-WFDC8 and anti-EDDM3B antibodies required an additional antigen retrieval step, and slides were microwaved for 1 min in 1 mM EDTA, 10 mM Tris (pH 9.0). Slides were blocked in 1% bovine serum albumin, and incubated with the primary antibody at 4°C for 16 h, and with the corresponding secondary antibody for 1 h at RT. The primary antibodies used were: affinity-purified rabbit polyclonal antibody against keratin type I cytoskeletal 19 (KRT19; 0.01 µg/µl; #10712-1-AP, Proteintech, Rosemont, IL, USA), affinity-purified rabbit polyclonal antibody against the very long-chain acyl-CoA synthetase (SLC27A2/FATP2; 2 µg/ml; #14048-1-AP, Proteintech, Rosemont, IL, USA), affinity-purified rabbit polyclonal antibody against the epididymal secretory protein E3-beta (EDDM3B; 0.02 µg/µl; #ARP53756_P050, Aviva Systems Biology, San Diego, CA, USA), and protein G purified rabbit polyclonal antibody against the WAP four-disulfide core domain protein 8 (WFDC8; 0.08 µg/µl; #CSB-PA818226LA01HU, Cusabio Technology LLC, Houston, TX, USA). Tissue co-labeling with a chicken polyclonal antibody against the V-ATPase B1 subunit (0.3 µg/ml; made and purified in the Breton lab: (Pǎunescu et al., 2010)) was used for epididymal CCs identification, as previously described (Castro et al., 2017; Park et al., 2017). The secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA, USA) used were: goat anti-rabbit IgG conjugated to Alexa Fluor 488 (30 μg/ml; #111-545-144), donkey anti-rabbit IgG conjugated to Cy3 (7.5 μg/ml; #711-165-152) and donkey anti-chicken IgY conjugated to Cy3 (7.5 μg/ml; #703-165-155). All antibodies were diluted in DAKO medium (Dako, Carpinteria, CA, USA). For the negative controls, the incubations were performed with the secondary antibody only. Slides were mounted using SlowFade™ Diamond Antifade Mountant (Invitrogen, Eugene, OR, USA) with 4′,6-diamidino-2-phenylindole (2 μg/ml; Vector Laboratories, Burlingame, CA, USA). Images were captured using an LSM800 confocal microscope (Zeiss Laboratories, Thornwood, NY, USA). Z-stack images were processed with Volocity software for 3D reconstruction.
Quantification analysis of the epididymosomes
Following IF, a quantification analysis of the epididymosomes was performed using an LSM800 confocal microscope (Zeiss Laboratories, Thornwood, NY, USA). Specifically, the presence of each of the four epididymis-derived proteins (SLC27A2, EDDM3B, KRT19 and WFDC8) in CFSE-labeled epididymosomes was evaluated and quantified from multiple confocal images (22–52 confocal images/protein). At least 150 CFSE-labeled epididymosomes (or clusters of epididymosomes) containing or not containing a given protein were quantified for each protein tested. Because EVs could form clusters and the observed fluorescent signals could represent more than one epididymosome, our quantification analysis might be over- or under- estimating the real amount of epididymosomes with a given epididymis-derived protein.
Results
In silico prediction of sperm proteins with an epididymal origin in humans and mice
The previously published list of 93 human sperm proteins with a putative epididymal acquisition by Castillo et al. (2018) was further complemented with additional proteomics data of mouse testis, sperm, epididymis and epididymosomes (Chaurand et al., 2003; Paz et al., 2006; Zhu et al., 2006; Baker et al., 2008; Huang et al., 2008; Da Silva et al., 2010; Dorus et al., 2010; Guo et al., 2010; Chauvin et al., 2012; Claydon et al., 2012; Guyonnet et al., 2012; Gan et al., 2013; Qi et al., 2014; Skerget et al., 2015; Liu et al., 2015a,b; Vicens et al., 2017; Xie et al., 2018; Xu et al., 2018; Nixon et al., 2019) (Supplementary Table SI). Through this broadened insilico analysis, we identified a subset of 25 sperm proteins potentially acquired from the epididymis in both species (Table I, Supplementary Table SI, Fig. 1). Interestingly, a notable enrichment of the GO Cellular Components ‘extracellular exosome’ (GO: 0070062) and ‘apicolateral plasma membrane’ (GO: 0016327) was observed in this data set (P-value after FDR adjustment < 0.01).
Table I.
List of sperm proteins with a putative epididymis-derived origin in both human and mouse species.
| Human sperm proteins with putative extra-testicular acquisition (from Castillo et al., 2018) |
Mouse proteomics data |
|||||
|---|---|---|---|---|---|---|
| Predicted organ of origin | Accession number | Gene name | Testis | Epididymal sperm | Epididymis | Epididymosomes |
| Epididymis | Q9UKQ2 | ADAM28 | – | × | × | × |
| Epididymis | Q9H2U9 | ADAM7 | – | × | × | × |
| Epididymis | Q8IUA0 | WFDC8 | – | × | × | – |
| Epididymis | Q5GAN3 | RNASE13 | – | × | × | – |
| Epididymis | P30613 | PKLR | – | × | – | × |
| Epididymis | P56851 | EDDM3B | – | × | – | – |
| Epididymis | Q7RTZ1 | OVCH2 | – | × | – | – |
| Epididymis | Q08648 | SPAG11B | – | × | – | – |
| Epididymis | Q075Z2 | BSPH1 | – | × | – | – |
| Epididymis | Q3B820 | FAM161A | – | × | – | – |
| Epididymis/seminal vesicles | O14975 | SLC27A2 | – | × | × | – |
| Epididymis/seminal vesicles | P22748 | CA4 | – | × | – | × |
| Epididymis/seminal vesicles/prostate | P05787 | KRT8 | – | × | × | × |
| Epididymis/seminal vesicles/prostate | P05783 | KRT18 | – | × | × | × |
| Epididymis/seminal vesicles/ prostate | P08727 | KRT19 | – | × | × | × |
| Epididymis/seminal vesicles/prostate | P12830 | CDH1 | – | × | × | × |
| Epididymis/seminal vesicles/prostate | P09668 | CTSH | – | × | × | – |
| Epididymis/seminal vesicles/prostate | E7ET76 | GGT1 | – | × | × | × |
| Epididymis/seminal vesicles/prostate | Q8IW92 | GLB1L2 | – | × | × | – |
| Epididymis/seminal vesicles/prostate | P02788 | LTF | – | × | – | × |
| Epididymis/seminal vesicles/prostate | O95994 | AGR2 | – | × | × | – |
| Epididymis/seminal vesicles/prostate | O14493 | CLDN4 | – | × | – | – |
| Epididymis/seminal vesicles/prostate | Q96BQ1 | FAM3D | – | × | – | – |
| Epididymis/seminal vesicles/prostate | Q9Y584 | TIMM22 | – | × | – | – |
| Epididymis/seminal vesicles/prostate | P06280 | GLA | – | × | – | – |
The subset of human sperm proteins with a predicted acquisition after testicular maturation published by Castillo et al. (2018) was complemented by integrating published mouse proteomic datasets (see an extended version in Supplementary Table SI). Proteins whose epididymis-derived origin has been experimentally validated in this study are indicated in bold.
×, Protein detection, –, Protein absence.
Localization of the inferred epididymis-derived sperm proteins
To validate the results derived from the insilico prediction, we selected four out of the 25 sperm proteins identified above to determine their tissue localization. The selection criteria used to select protein candidates included human sperm proteins with a putative epididymal origin that were also identified in the mouse epididymal sperm proteome, and that were absent in the mouse testis proteome. Due to the low availability of proteomic data for mouse epididymis and epididymosomes, the absence of our protein candidates in these proteomic datasets was not considered an exclusion criterion. The selected proteins were the very long-chain acyl-CoA synthetase (SLC27A2/FATP2), the epididymal secretory protein E3-beta (EDDM3B), the keratin type I cytoskeletal 19 (KRT19) and the WAP four-disulfide core domain protein 8 (WFDC8). Specifically, our protein candidates included a sperm protein already identified in mouse epididymis and epididymosomes proteomes (KRT19), two sperm proteins already identified in mouse epididymis proteome but not in mouse epididymosomes proteome (WFDC8 and SLC27A2), and a sperm protein not identified yet in mouse epididymis and epididymosomes proteomes (EDDM3B) (Table I).
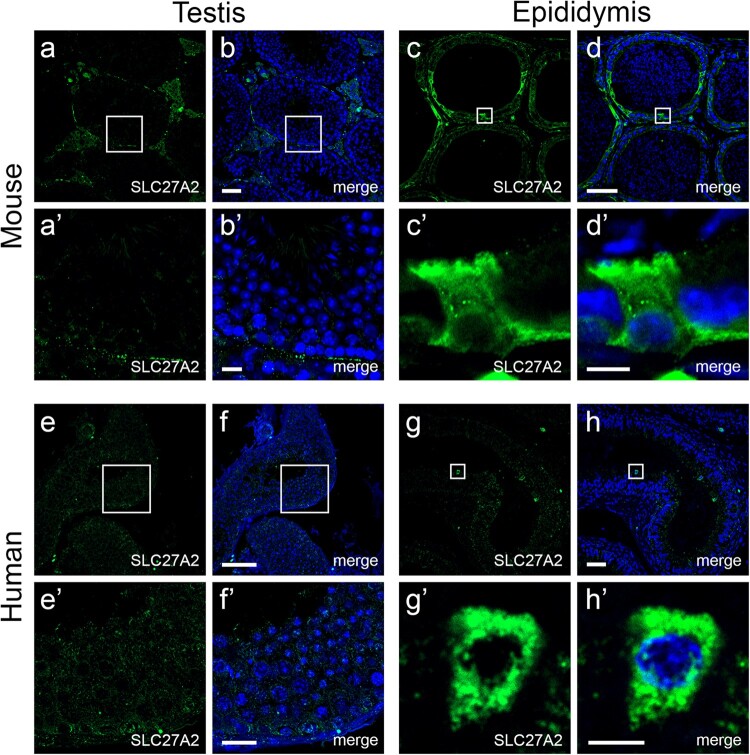
Immunofluorescence analyses through high-resolution confocal microscopy showed the presence of all the selected proteins (SLC27A2, EDDM3B, KRT19 and WFDC8) in mouse and human epididymal epithelial cells, and their absence in mouse and human seminiferous tubules (Fig. 3, Supplementary Fig. S1 and Table SII). Note that although KRT19 and WFDC8 were detected in the connective tissue surrounding the seminiferous tubules (Leydig or interstitial cells), no labeling was observed in testicular germ cells (Supplementary Fig. S1).
Figure 3.
Localization of the epididymis-derived protein SLC27A2 in mouse and human testes and epididymides. Confocal microscopy images showing the absence of SLC27A2 protein in germ cells from seminiferous tubules of mouse (a, b) and human (e, f) testes, and the presence of SLC27A2 protein (green) in mouse (c, d) and human (g, h) epididymal epithelial cells. Nuclei are labeled with DAPI in blue. The bottom panels (’) show a higher magnification representation of the areas delineated by the boxes in the upper panels. Bars = 50 µm for upper panels, Bars = 10 µm for lower panels (b’, f’), Bars = 5 µm for lower panels (d’, h’). See Supplementary Fig. S1 for the localization of EDDM3B, KRT19, WFDC8 proteins. DAPI, 4′,6-diamidino-2-phenylindole.
Cellular localization of the epididymis-derived proteins SLC27A2, EDDM3B, KRT19 and WFDC8 in different segments of the epididymis
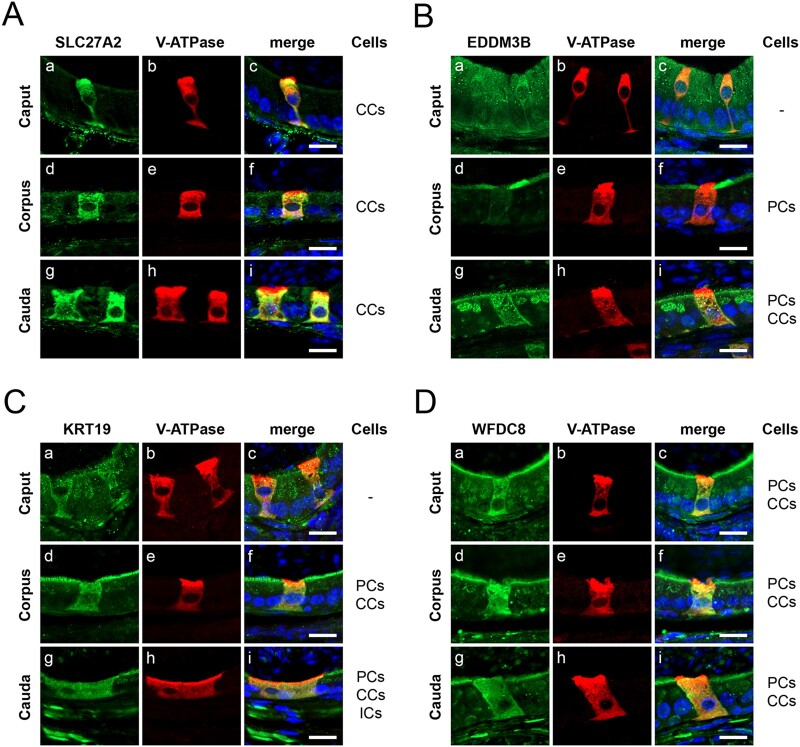
Immunofluorescence analyses of the four selected proteins in the different regions of the epididymis (caput, corpus and cauda) demonstrated their segment-dependent cellular expression in the mouse epididymis (Fig. 4, Supplementary Table SII). SLC27A2 was expressed in epididymal CCs (V-ATPase B1 subunit positive) localized along the entire duct, comprising caput, corpus and cauda epididymides (Fig. 4A). EDDM3B expression was restricted to corpus and cauda epididymides and displayed a different cellular expression pattern (Fig. 4B). In the corpus, EDDM3B was only expressed on the apical membrane of PCs (V-ATPase B1 subunit negative) while, in the cauda, EDDM3B was present in CCs and in the apical membrane and Golgi-like organelles of PCs (Fig. 4B). Likewise, KRT19 was not expressed in the caput epididymis, but it was detected in both CCs and the apical region of PCs in corpus and cauda epididymides (Fig. 4C). KRT19 was also detected in interstitial cells of the cauda epididymis (Fig. 4C). Finally, WFDC8 displayed high expression in CCs and the apical membrane of PCs in all epididymal segments (Fig. 4D).
Figure 4.
Specific localization of the epididymis-derived proteins SLC27A2, EDDM3B, KRT19 and WFDC8 in the mouse epididymis. Double immunolabeling of the epididymis-derived proteins (green) and V-ATPase B1 subunit (red) as a marker for epididymal clear cells (CCs), in the caput, corpus and cauda regions of mouse epididymis. (A) SLC27A2 (green) was specifically detected in CCs (red) on the overall epididymis segments. (B) EDDM3B (green) was detected in the apical portion of principal cells (PCs) located in the corpus; EDDM3B was also detected in CCs (red), and in the apical portion and Golgi-like organelles of PCs in the cauda. (C) KRT19 (green) was present in the apical portion of PCs and in CCs (red) in both corpus and cauda, and in interstitial cells in cauda. (D) WFDC8 (green) was identified in CCs (red) and in the apical portion of PCs all along the epididymal segments. Nuclei are labeled with DAPI in blue. Bars = 10 µm. DAPI, 4′,6-diamidino-2-phenylindole.
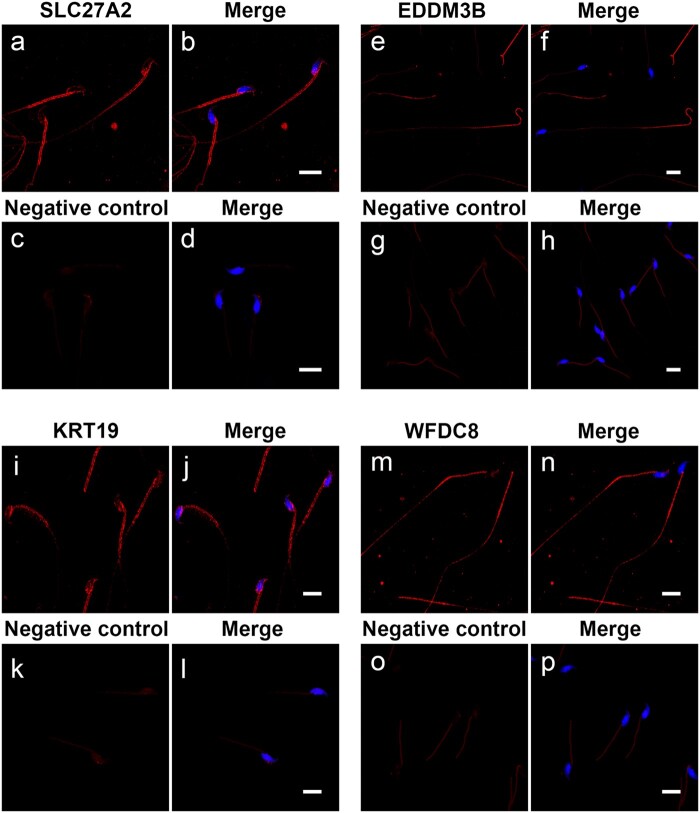
The epididymis-derived proteins SLC27A2, EDDM3B, KRT19 and WFDC8 are present in epididymal sperm
Immunofluorescence analyses showed that SLC27A2, EDDM3B, KRT19 and WFDC8 proteins are present in sperm isolated from the mouse cauda epididymides, suggesting their acquisition during epididymal transit (Fig. 5, Supplementary Table SII). Interestingly, the four selected epididymis-derived proteins displayed a differential localization in mouse spermatozoa: SLC27A2 and KRT19 were detected within the post-acrosomal sheath of the sperm head and in the sperm midpiece; EDDM3B was present in the posterior part of the sperm flagellum; and WFDC8 was predominantly detected in the sperm midpiece, with a far less intense signal in the sperm head and along the sperm flagellum (Fig. 5). This analysis was only performed in mouse epididymal sperm since human ejaculated sperm have already been in contact with all the fluids secreted along the male reproductive tract.
Figure 5.
Presence of the epididymis-derived proteins SLC27A2, EDDM3B, KRT19 and WFDC8 in mouse sperm. Mouse spermatozoa were isolated from cauda epididymis and immunolabeled for SLC27A2, EDDM3B, KRT19 and WFDC8. The four epididymis-derived proteins (red) were detected in cauda sperm, but displayed a different localization pattern: SLC27A2 (a, b) and KRT19 (i, j) proteins were located in the sperm midpiece and post-acrosomal region; EDDM3B (e, f) was present in the posterior part of the sperm tail; and WFDC8 (m, n) was mainly present in sperm midpiece, with a faint signal in the sperm head and tail. No fluorescence signal was observed in each respective negative control (c–d, g–h, k–l, o–p). Sperm nuclei are labeled with DAPI in blue. Representative confocal microscopy images are presented as maximum intensity projections. Bars = 10 µm. DAPI, 4′,6-diamidino-2-phenylindole.
Epididymosomes contain the epididymis-derived proteins SLC27A2, EDDM3B, KRT19 and WFDC8
Epididymosomes and other vesicles from mouse epididymal fluid from the cauda epididymides were firstly labeled with the fluorescent dye CFSE, and epididymosomes were isolated by ultracentrifugation in 25% and 30% sucrose cushions (Fig. 2A). TEM analysis showed the presence of round or cup-shaped vesicles enclosed by a phospholipid bilayer (Fig. 2B; right panel). The size distribution and concentration of the epididymosomes were determined by NTA, showing that the epididymosomes had a diameter of 181.9 ± 3.6 nm (mean ± SEM) (Fig. 2B; left panel). A second smaller population of epididymosomes of higher size indicates the potential occurrence of some vesicle clusters. Both TEM and NTA techniques confirmed the presence of epididymosome-like vesicles with spherical morphology and within the normal range of EVs.
After co-incubation of CFSE-labeled epididymosomes with mouse sperm, fluorescent epididymosomes attached to spermatozoa were detected (Fig. 2C; Condition A’). Negative controls discarded any unspecific labeling (Fig. 2C; Conditions B’ and D’) as well the presence of CFSE fluorescent compounds or aggregates (Fig. 2C; Condition C’). These data confirmed the ability of epididymosomes to interact with mouse sperm after an invitro incubation.
We next investigated whether the attached epididymosomes contained the epididymis-derived proteins SLC27A2, EDDM3B, KRT19 and WFDC8. As shown in Fig. 6, positive CFSE-labeled epididymosomes attached to mouse cauda spermatozoa contain the four selected epididymal proteins (Supplementary Table SII), reinforcing the hypothesis that epididymosomes transfer proteins to spermatozoa. Note that due to the high expression of these proteins in epididymosomes, the intensity of the laser was decreased to capture the images and the epididymis-derived proteins are no longer observed across the sperm as was shown in Figs 5 and 6. Nonetheless, the four epididymis-derived proteins are indeed detected in mouse sperm, in addition to epididymosomes, when displaying the same laser parameters used for confocal microscopy images shown in Fig. 5 and Supplementary Fig. S2. The quantification of fluorescent EVs, which represent one or more CFSE-labeled epididymosomes, showed that 36.4% (60 of 165) contain SLC27A2, 20% (30 of 150) contain EDDM3B, 12.2% (19 of 155) contain KRT19 and 20.9% (32 of 153) contain WFDC8.
Figure 6.
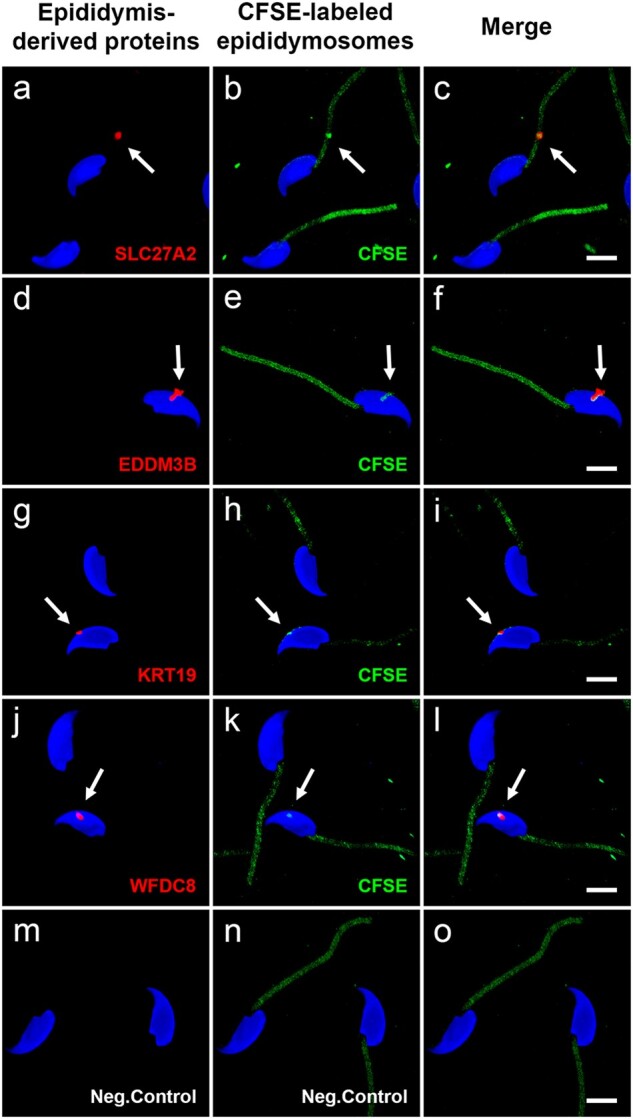
Mouse epididymosomes incubated with cauda sperm in vitro contain the epididymis-derived proteins SLC27A2, EDDM3B, KRT19 and WFDC8. Mouse epididymosomes contain the epididymis-derived proteins SLC27A2, EDDM3B, KRT19 and WFDC8. Several CFSE-labeled epididymosomes (green) attached to mouse cauda spermatozoa are positive (red, arrows) for SLC27A2 (a, b, c), EDDM3B (d, e, f), KRT19 (g, h, i) and WFDC8 (j, k, l) proteins. Due to the high expression of the epididymis-derived proteins in epididymosomes, the intensity of the red laser was decreased to capture the images (see Supplementary Fig. S2). Negative control images (m, n, o) show the incubation of cauda sperm with CFSE cleared medium (Condition C’; no presence of epididymosomes), in order to discard the presence of CFSE fluorescent compounds or aggregates. Negative control shows autofluorescence in sperm midpiece due to high NADH concentration in sperm midpiece mitochondria. Sperm nuclei are labeled with DAPI in blue. Bars = 5 µm. Images show a 3D reconstruction of three colors z-stacked image acquired using the Zeiss LSM-800 confocal microscope. CFSE, carboxyfluorescein succinimidyl ester; DAPI, 4′,6-diamidino-2-phenylindole.
Discussion
While testicular spermatozoa are morphologically differentiated cells, they lack progressive motility and the ability to fertilize an oocyte. The sperm cell undergoes post-testicular maturation through contact with different secretions from the post-testicular male reproductive tract (epididymis, prostate and seminal vesicles) (Jodar et al., 2017; Camargo et al., 2018; Barrachina et al., 2019). Our group recently revealed a subset of potentially extra-testicularly acquired human sperm proteins (Castillo et al., 2018). Here, we extend our previous human insilico analysis by exploring the conservation of this dataset in the mouse model and focusing on sperm maturation acquired during epididymal transit. Consequently, we report for the first time herein a subset of 25 sperm proteins with an epididymal origin that is conserved in both human and mouse species. Through high-resolution confocal microscopy, we confirm the epididymal origin of four selected proteins acquired by sperm and demonstrate that these proteins are transferred to the maturing sperm through epididymosomes.
The functional maturation of ejaculated spermatozoa is achieved in a post-testicular context, mainly driven via interaction of the maturing sperm with epididymal secretions and epididymosomes (Sullivan et al., 2007; Sullivan, 2015, 2016; Breton et al., 2016; Zhou et al., 2018; James et al., 2020). These sperm proteins with an epididymis origin include: (i) proteins involved in sperm maturation and function, such as ADAM7 (Cornwall and Hsia, 1997; Oh et al., 2005; Han et al., 2011; Cho, 2012; Choi et al., 2015), ADAM28 (Howard et al., 2000, 2001; Cho, 2012), EDDM3B (Amaral et al., 2014; Légaré et al., 2017), SPAG11B (Hamil et al., 2000; Zhou et al., 2004; Radhakrishnan et al., 2009; Dorin and Barratt, 2014; Narmadha and Yenugu, 2015), GGT1 (Garrido et al., 2009; Lee and Foo, 2014), LTF (Piomboni et al., 2008; Belleannée et al., 2011; Zumoffen et al., 2015; Nowicka-Bauer et al., 2018; Hernández-Silva and Chirinos, 2019), GLB1L2 (Samanta et al., 2019) and BSPH1 (Plante et al., 2012, 2014, 2016; Plante and Manjunath, 2015); (ii) proteins involved in fertilization, including ADAM7 (Kim et al., 2006; Choi et al., 2015) and LTF (Zumoffen et al., 2015; Hernández-Silva and Chirinos, 2019); (iii) proteins that provide antioxidant protection to sperm, such as GGT1 (Walker et al., 2006) and LTF (Guyonnet et al., 2011; Nowicka-Bauer et al., 2018); (iv) proteins involved in male reproductive tract immunity, such as WFDC8 (Thimon et al., 2008a; Rajesh et al., 2011), SPAG11B (Radhakrishnan et al., 2009; Fei et al., 2012; Dorin and Barratt, 2014; Narmadha and Yenugu, 2015), LTF (Piomboni et al., 2008; Guyonnet et al., 2011; Nowicka-Bauer et al., 2018) and FAM3D (Zhu et al., 2002; Peng et al., 2016); and (v) a protein that could be relevant for energy production and/or lipid synthesis for membranes, SLC27A2 (Caimari et al., 2010; Black et al., 2016).
In addition, LTF and FAM161A might be required for proper early embryogenesis, since their abundance seems to be reduced in sperm samples from unexplained infertile male patients with poor blastocyst development after an IVF cycle using an oocyte donor (McReynolds et al., 2014). However, this hypothesis is in contrast to the generation of viable embryos in vitro using testicular spermatozoa (Dozortsev et al., 2006; Kang et al., 2018). Further functional studies are, thus, required to clarify the potential role of extra-testicularly acquired sperm proteins in early embryogenesis. Interestingly, structural integrity components were also identified within the list of epididymis-derived sperm proteins, such as some members of the keratin gene family (KRT8, KRT18 and KRT19). Of note, in addition to their structural role, keratins also participate in organelle and protein transport, differentiation and proliferation (Magin et al., 2007; Skerget et al., 2015).
In the present study, we provide evidence that the sperm proteins SLC27A2, EDDM3B, KRT19 and WFDC8, are present in epithelial cells lining both the human and mouse epididymis. Specifically, KRT19 was detected in PCs and CCs, and SLC27A2 was detected in CCs. However, the expression patterns of EDDM3B and WFDC8 were species-dependent, with both proteins being detected in PCs and CCs in the mouse epididymis, but only in CCs in the human epididymis. SLC27A2 and WFDC8 were detected in all epididymal segments, whereas EDDM3B and KRT9 were absent from the caput. In addition, while EDDM3B was present in PCs and CCs in the cauda, it was detected only in PCs in the corpus. Thus, the expression of these proteins appears to be under the control of local stimuli within the epididymis. The expression of some of these proteins in PCs was expected because of the well-known participation of these cells in protein biosynthesis and secretion. Our observation that they are all expressed in CCs is in agreement with a recent study indicating that these cells are also involved in the production of epididymosomes via apocrine secretion (Battistone et al., 2019b). However, because CCs have a high endocytic rate (Hermo and Robaire, 2002; Zhou et al, 2018; Breton et al., 2019), it remains possible that they could internalize proteins excreted by PCs. This might be the case for EDDM3B, KRT19 and WFDC8, which are also expressed by PCs in the mouse epididymis, but not for SLC27A2, which is expressed exclusively in CCs. In addition, the transcripts for EDDM3B, KRT19, WFDC8 and SLC27A2 have all been recently identified in CCs (Battistone et al., 2019b). Our results thus further support the novel role of CCs in the transfer of proteins to sperm during epididymal transit.
The strategy followed in the present study, combining the co-incubation of sperm with CFSE-labeled epididymosomes together with targeted fluorescent protein labeling, confirmed the presence of epididymosomes that interact with sperm and contain the epididymis-derived proteins SLC27A2, KRT19, EDDM3B and WFDC8. In addition, our quantification analysis revealed that a significant percentage of epididymosomes (12–36%) contain these epididymis-derived proteins. However, we cannot entirely exclude the possibility that some epididymosomes form clusters, as shown in the NTA analysis, which would then over- or under-estimate the actual percentage of epididymosomes containing each target protein. Despite this limitation, our results provide a proof-of-concept that epididymosomes participate in protein transfer from the epididymis to sperm during the post-testicular maturation process. In fact, there is growing evidence supporting the potential trafficking of RNAs and proteins from the epididymis to sperm through epididymosomes (Krapf et al., 2012). Sharma et al. (2016, 2018) previously demonstrated the epididymis-to-sperm transfer of small RNAs through mouse epididymosomes by incubating them with epididymal or testicular sperm, whereas Reilly et al. (2016) showed that mouse epididymosomes can directly interact with sperm and deliver a selective set of miRNAs. Also, the proteins P25b, ELSPBP1 and GBB2 were proposed as being transferred to sperm by epididymosomes in bovine species. P25b is a protein whose concentration increases in cauda spermatozoa, and it is also present in epididymosomes (Frenette and Sullivan, 2001). The co-incubation of caput sperm with cauda epididymal fluid resulted in an increased sperm concentration of P25b, indirectly suggesting that the epididymosomes might be responsible for the acquisition of this protein by sperm (Frenette and Sullivan, 2001). Besides, an elegant strategy co-incubating biotinylated cauda epididymosomes with caput sperm revealed that two biotinylated proteins, namely ELSPBP1 and GBB2, were transferred to caput sperm from epididymosomes, as identified by using a mass spectrometry analysis of biotinylated proteins (Frenette et al., 2010). Interestingly, flow cytometry assay revealed that the epididymosome-mediated transfer of ELSPBP1 to epididymal sperm only occurred in dead spermatozoa, suggesting that the transfer of ELSPBP1 may be a tag for the recognition of dead sperm and, therefore, may serve as a quality control mechanism for defective sperm in the epididymis (D’Amours et al., 2012).
In the current study, we provide a visual demonstration of the direct interaction between spermatozoa and epididymosomes that contain proteins known to be transferred to the sperm cells during their epididymal maturation. Of note, we found that CFSE-labeled epididymosomes were found to be interacting with different regions of spermatozoa. Similarly, Schwarz et al. (2013) showed that biotin-labeled epididymosomes transfer proteins to bovine sperm, to the post-acrosomal region, but also to the neck, mid-piece and principal pieces. On the contrary, other studies using biotinylated epididymosomes showed that the epididymosome-sperm interaction was predominantly restricted to the post-acrosomal domain of the sperm head (Nixon et al., 2019; Zhou et al., 2019). Another study has also previously shown that the sperm head and mid-piece of the flagellum were the predominant sites for epididymosome-sperm interaction after co-incubating CFSE-labeled epididymosomes with caput sperm (Reilly et al., 2016). While we were able to observe fluorescently CFSE-labeled epididymosomes interacting with sperm, Reilly et al. (2016) found a more disperse staining pattern of the CFSE-labeled epididymosomes after a co-incubation period with sperm. Differences in the methodology used in our study compared to the study by Reilly and collaborators could explain this discrepancy: (i) while the source of sperm and epididymosomes was caput in the Reilly study, we used cauda epididymis; and (ii) the steps to label and isolate epididymosomes were also distinct. Reilly et al. (2016) isolated epididymosomes using a discontinuous OptiPrep gradient and, afterward, the epididymosomes were labeled with CFSE and were subjected to final ultracentrifugation at 100 000 g. In our case, we first labeled the vesicles present in the epididymal cauda fluid and, to fully ensure that no CFSE dye remained in the medium, we then isolated the epididymosomes using a 25–30% sucrose cushion which was followed by another ultracentrifugation at 100 000 g. Indeed, several reports showed CFSE-labeled EVs interacting or being taken up by cells (Tian et al., 2014; Morales-Kastresana et al., 2017; Domínguez Rubio et al., 2020), consistent with our results. Furthermore, Vilanova-Perez et al. (2020) recently revealed individual fluorescent EVs interacting with mammalian sperm invitro using total internal reflection fluorescence microscopy imaging, and no specific EVs-sperm fusion site was observed. Battistone et al. (2019b) using high-resolution confocal microscopy, observed EGFP+ epididymosomes in the lumen of the epididymis in vivo that were attached to the head, midpiece and tail of the sperm. In addition, Pǎunescu et al. (2014) found several epididymosome-like membranous vesicles on the surface of the cytoplasmic droplet located in the sperm tail using high-resolution helium ion microscopy. Further studies are now required to decipher whether there is a specific union site for epididymosomes-sperm, and to shed light on the mechanisms by which the epididymosomes are able to interact and fuse with the sperm membrane.
The current paradigm is that epididymosomes are mainly released by PCs via apocrine secretion (Hermo and Jacks, 2002; Rejraji et al., 2006; Sullivan, 2016; Zhou et al., 2018). However, in our study, we observed that the epididymis-derived sperm protein SLC27A2 is exclusively expressed by epididymal CCs, and is present in epididymosomes that are interacting with spermatozoa, suggesting that CCs may also have the ability to release epididymosomes. This is in accordance with a recently published article suggesting that CCs are also releasing epididymosomes, using a transgenic mouse model that specifically expresses EGFP+ in CCs (Battistone et al., 2019b). Furthermore, a proteomic analysis of mouse epididymosomes identified some proteins, such as ATP6V0A4 (a4) and ATP6V1G1 (G1) (Nixon et al., 2019), which are specifically expressed in CCs (Da Silva et al., 2010; Battistone et al., 2019b). Altogether, our results provide additional evidence supporting the novel role of CCs in sperm protein transfer through epididymosomes and, therefore, their potential participation in sperm maturation.
Epididymosome interaction with maturing epididymal sperm is of growing interest, and advances are being made to characterize the real contribution of epididymosomes on the reproductive function. The data obtained in the current study provides additional evidence that epididymosomes harbor an epididymis-derived protein cargo that can be delivered to the spermatozoa, thereby providing the inert transcriptionally and translationally sperm cell with new proteins and, consequently, modulating the sperm proteome. However, it is surprising to note that three of the four proteins (SLC27A2, EDDM3B and WFDC8) shown here to be specifically expressed in epididymis and present in epididymosomes are not listed in the mouse epididymosome proteome profile recently published by Nixon et al. (2019). This fact reflects the current need for further studies using additional approaches to complete the proteome characterization of epididymosomes, which will further clarify the role of the epididymis in sperm function. Furthermore, because of the limitation of collecting human epididymal fluid samples, it is not possible to provide evidence about whether human epididymosomes carry the epididymis-derived proteins. Nonetheless, the fact that three of the four sperm epididymis-derived proteins (WFDC8, EDDM3B, and KRT19) are present in human seminal plasma (Jodar et al., 2017) points to the possibility that human epididymosomes also contain these epididymis-derived proteins, a finding that would provide meaningful information on the role of epididymosomes in human male reproduction. Future studies will be required to determine whether the other proteins identified in our insilico analysis may also be provided to spermatozoa via epididymosomes. Understanding the role played by the epididymis in sperm function through epididymosomes will contribute to improving our current knowledge of male fertility and to identifying altered extra-testicular molecular targets that could directly impact male reproductive health.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Supplementary Material
Acknowledgements
The authors thank all the participants who took part in this study, as well as all medical staff and technicians who helped in obtaining and/or processing the human samples. The authors would also like to thank Diane E. Capen and Dr Dennis Brown for their help and expertise in the sample preparation and epididymosomes characterization by TEM analysis.
Authors’ roles
F.B., M.J. and R.O. were involved in the study design and conceptualization. F.B. and M.A.B performed the experiments. F.B., M.A.B., J.C., C.M., M.J., S.B. and R.O. were involved in data interpretation. C.M. collected human samples and evaluated and interpreted clinical data. F.B., J.C. and M.J. drafted the original manuscript. All authors contributed to the writing of the manuscript, made critical comments and approved the final version.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness (Ministerio de Economía y Competividad; fondos FEDER ‘una manera de hacer Europa’ PI13/00699 and PI16/00346 to R.O.; and Sara Borrell Postdoctoral Fellowship, Acción Estratégica en Salud, CD17/00109 to J.C.), by National Institutes of Health (grants HD040793 and HD069623 to S.B., grant HD104672-01 to M.A.B.), by the Spanish Ministry of Education, Culture and Sports (Ministerio de Educación, Cultura y Deporte para la Formación de Profesorado Universitario, FPU15/02306 to F.B.), by a Lalor Foundation Fellowship (to F.B. and M.A.B.), by the Government of Catalonia (Generalitat de Catalunya, pla estratègic de recerca i innovació en salut, PERIS 2016-2020, SLT002/16/00337 to M.J.), by Fundació Universitària Agustí Pedro i Pons (to F.B.), and by the American Society for Biochemistry and Molecular Biology (PROLAB Award from ASBMB/IUBMB/PABMB to F.B.). Confocal microscopy and transmission electron microscopy was performed in the Microscopy Core facility of the Massachusetts General Hospital (MGH) Center for Systems Biology/Program in Membrane Biology which receives support from Boston Area Diabetes and Endocrinology Research Center (BADERC) award DK57521 and Center for the Study of Inflammatory Bowel Disease grant DK43351. The Zeiss LSM800 microscope was acquired using an NIH Shared Instrumentation Grant S10-OD-021577-01.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Aalberts M, Stout TAE, Stoorvogel W.. Prostasomes: extracellular vesicles from the prostate. Reproduction 2014;147:R1–R14. [DOI] [PubMed] [Google Scholar]
- Amaral A, Paiva C, Attardo Parrinello C, Estanyol JM, Ballescà JL, Ramalho-Santos J, Oliva R.. Identification of proteins involved in human sperm motility using high-throughput differential proteomics. J Proteome Res 2014;13:5670–5684. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MA, Hetherington L, Reeves GM, Aitken RJ.. The mouse sperm proteome characterized via IPG strip prefractionation and LC-MS/MS identification. Proteomics 2008;8:1720–1730. [DOI] [PubMed] [Google Scholar]
- Barceló M, Castells M, Bassas L, Vigués F, Larriba S.. Semen miRNAs contained in exosomes as non-invasive biomarkers for prostate cancer diagnosis. Sci Rep 2019;9:13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló M, Mata A, Bassas L, Larriba S.. Exosomal microRNAs in seminal plasma are markers of the origin of azoospermia and can predict the presence of sperm in testicular tissue. Hum Reprod 2018;33:1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrachina F, Jodar M, Delgado-Dueñas D, Soler-Ventura A, Estanyol JM, Mallofré C, Ballescà JL, Oliva R.. Stable-protein pair analysis as a novel strategy to identify proteomic signatures: application to seminal plasma from infertile patients. Mol Cell Proteomics 2019;18:S77–S90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrachina F, Soler-Ventura A, Oliva R, Jodar M.. Sperm nucleoproteins (histones and protamines). In: A Zini, A Agarwal (eds). A Clinician’s Guide to Sperm DNA and Chromatin Damage. Cham, Switzerland: Springer International Publishing AG, 2018, 31–51. [Google Scholar]
- Battistone MA, Merkulova M, Park Y-J, Peralta MA, Gombar F, Brown D, Breton S.. Unravelling purinergic regulation in the epididymis: activation of V-ATPase-dependent acidification by luminal ATP and adenosine. J Physiol 2019a;597:1957–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistone MA, Spallanzani RG, Mendelsohn AC, Capen D, Nair AV, Brown D, Breton S.. Novel role of proton-secreting epithelial cells in sperm maturation and mucosal immunity. J Cell Sci 2019b;133:jcs.233239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleannée C. Extracellular microRNAs from the epididymis as potential mediators of cell-to-cell communication. Asian J Androl 2015;17:730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belleannée C, Calvo É, Caballero J, Sullivan R.. Epididymosomes convey different repertoires of microRNAs throughout the bovine epididymis. Biol Reprod 2013a;89:30. [DOI] [PubMed] [Google Scholar]
- Belleannée C, Labas V, Teixeira-Gomes A-P, Gatti JL, Dacheux J-L, Dacheux F.. Identification of luminal and secreted proteins in bull epididymis. J Proteomics 2011;74:59–78. [DOI] [PubMed] [Google Scholar]
- Belleannée C, Légaré C, Calvo E, Thimon V, Sullivan R.. microRNA signature is altered in both human epididymis and seminal microvesicles following vasectomy. Hum Reprod 2013b;28:1455–1467. [DOI] [PubMed] [Google Scholar]
- Black PN, Ahowesso C, Montefusco D, Saini N, DiRusso CC.. Fatty acid transport proteins: targeting FATP2 as a gatekeeper involved in the transport of exogenous fatty acids. Medchemcomm 2016;7:612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Nair AV, Battistone MA.. Epithelial dynamics in the epididymis: role in the maturation, protection, and storage of spermatozoa. Andrology 2019;7:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Ruan Y, Park Y-J, Kim B.. Regulation of epithelial function, differentiation, and remodeling in the epididymis. Asian J Androl 2016;18:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D, Smith PJ, Breton S.. Role of V-ATPase-rich cells in acidification of the male reproductive tract. J Exp Biol 1997;200:257–262. [DOI] [PubMed] [Google Scholar]
- Caballero J, Frenette G, Sullivan R.. Post testicular sperm maturational changes in the bull: important role of the epididymosomes and prostasomes. Vet Med Int 2010;2011:757194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caimari A, Oliver P, Rodenburg W, Keijer J, Palou A.. Slc27a2 expression in peripheral blood mononuclear cells as a molecular marker for overweight development. Int J Obes (Lond) 2010;34:831–839. [DOI] [PubMed] [Google Scholar]
- Camargo M, Intasqui P, Bertolla RP.. Understanding the seminal plasma proteome and its role in male fertility. Basic Clin Androl 2018;28:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell DT, Aston KI, Oliva R, Emery BR, de Jonge CJ.. The “omics” of human male infertility: integrating big data in a systems biology approach. Cell Tissue Res 2016;363:295–312. [DOI] [PubMed] [Google Scholar]
- Castillo J, Estanyol JM, Ballescà JL, Oliva R.. Human sperm chromatin epigenetic potential: genomics, proteomics, and male infertility. Asian J Androl 2015;17:601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo J, Jodar M, Oliva R.. The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum Reprod Update 2018;24:535–555. [DOI] [PubMed] [Google Scholar]
- Castro MM, Kim B, Hill E, Fialho MCQ, Puga LCHP, Freitas MB, Breton S, Machado-Neves M.. The expression patterns of aquaporin 9, vacuolar H+-ATPase, and cytokeratin 5 in the epididymis of the common vampire bat. Histochem Cell Biol 2017;147:39–48. [DOI] [PubMed] [Google Scholar]
- Chaurand P, Fouchécourt S, DaGue BB, Xu BJ, Reyzer ML, Orgebin-Crist M-C, Caprioli RM.. Profiling and imaging proteins in the mouse epididymis by imaging mass spectrometry. Proteomics 2003;3:2221–2239. [DOI] [PubMed] [Google Scholar]
- Chauvin T, Xie F, Liu T, Nicora CD, Yang F, Camp DG, Smith RD, Roberts KP.. A systematic analysis of a deep mouse epididymal sperm proteome. Biol Reprod 2012;87:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C. Testicular and epididymal ADAMs: expression and function during fertilization. Nat Rev Urol 2012;9:550–560. [DOI] [PubMed] [Google Scholar]
- Choi H, Han C, Jin S, Kwon JT, Kim J, Jeong J, Kim J, Ham S, Jeon S, Yoo YJ. et al. Reduced fertility and altered epididymal and sperm integrity in mice lacking ADAM7. Biol Reprod 2015;93:70. [DOI] [PubMed] [Google Scholar]
- Claydon AJ, Ramm SA, Pennington A, Hurst JL, Stockley P, Beynon R.. Heterogenous turnover of sperm and seminal vesicle proteins in the mouse revealed by dynamic metabolic labeling. Mol Cell Proteomics 2012;11:M111.014993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornwall GA, Hsia N.. ADAM7, a member of the ADAM (a disintegrin and metalloprotease) gene family is specifically expressed in the mouse anterior pituitary and epididymis. Endocrinology 1997;138:4262–4272. [DOI] [PubMed] [Google Scholar]
- D’Amours O, Frenette G, Bordeleau L-J, Allard N, Leclerc P, Blondin P, Sullivan R.. Epididymosomes transfer epididymal sperm binding protein 1 (ELSPBP1) to dead spermatozoa during epididymal transit in bovine. Biol Reprod 2012;87:94. [DOI] [PubMed] [Google Scholar]
- Da Silva N, Pisitkun T, Belleannée C, Miller LR, Nelson R, Knepper MA, Brown D, Breton S.. Proteomic analysis of V-ATPase-rich cells harvested from the kidney and epididymis by fluorescence-activated cell sorting. Am J Physiol Cell Physiol 2010;298:C1326–C1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva N, Shum WWC, Breton S.. Regulation of vacuolar proton pumping ATPase-dependent luminal acidification in the epididymis. Asian J Androl 2007;9:476–482. [DOI] [PubMed] [Google Scholar]
- Dacheux J-L, Belleannée C, Guyonnet B, Labas V, Teixeira-Gomes A-P, Ecroyd H, Druart X, Gatti J-L, Dacheux F.. The contribution of proteomics to understanding epididymal maturation of mammalian spermatozoa. Syst Biol Reprod Med 2012;58:197–210. [DOI] [PubMed] [Google Scholar]
- Dacheux J-L, Dacheux F.. New insights into epididymal function in relation to sperm maturation. Reproduction 2014;147:R27–R42. [DOI] [PubMed] [Google Scholar]
- Dacheux J-L, Dacheux F, Druart X.. Epididymal protein markers and fertility. Anim Reprod Sci 2016;169:76–87. [DOI] [PubMed] [Google Scholar]
- de Kretser DM, Loveland KL, Meinhardt A, Simorangkir D, Wreford N.. Spermatogenesis. Hum Reprod 1998;13:1–8. [DOI] [PubMed] [Google Scholar]
- Domínguez Rubio AP, Martínez J, Palavecino M, Fuentes F, Sánchez López CM, Marcilla A, Pérez OE, Piuri M.. Transcytosis of Bacillus subtilis extracellular vesicles through an in vitro intestinal epithelial cell model. Sci Rep 2020;10:3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorin JR, Barratt CLR.. Importance of b-defensins in sperm function. Mol Hum Reprod 2014;20:821–826. [DOI] [PubMed] [Google Scholar]
- Dorus S, Wasbrough ER, Busby J, Wilkin EC, Karr TL.. Sperm proteomics reveals intensified selection on mouse sperm membrane and acrosome genes. Mol Biol Evol 2010;27:1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozortsev D, Neme R, Diamond MP, Abdelmassih S, Abdelmassih V, Oliveira F, Abdelmassih R.. Embryos generated using testicular spermatozoa have higher developmental potential than those obtained using epididymal spermatozoa in men with obstructive azoospermia. Fertil Steril 2006;86:606–611. [DOI] [PubMed] [Google Scholar]
- Drabovich AP, Saraon P, Jarvi K, Diamandis EP.. Seminal plasma as a diagnostic fluid for male reproductive system disorders. Nat Rev Urol 2014;11:278–288. [DOI] [PubMed] [Google Scholar]
- Fei Z, Hu S, Xiao L, Zhou J, Diao H, Yu H, Fang S, Wang Y, Wan Y, Wang W. et al. mBin1b transgenic mice show enhanced resistance to epididymal infection by bacteria challenge. Genes Immun 2012;13:445–451. [DOI] [PubMed] [Google Scholar]
- Frenette G, Girouard J, D’Amours O, Allard N, Tessier L, Sullivan R.. Characterization of two distinct populations of epididymosomes collected in the intraluminal compartment of the bovine cauda epididymis. Biol Reprod 2010;83:473–480. [DOI] [PubMed] [Google Scholar]
- Frenette G, Girouard J, Sullivan R.. Comparison between epididymosomes collected in the intraluminal compartment of the bovine caput and cauda epididymidis. Biol Reprod 2006;75:885–890. [DOI] [PubMed] [Google Scholar]
- Frenette G, Lessard C, Sullivan R.. Selected proteins of “prostasome-like particles” from epididymal cauda fluid are transferred to epididymal caput spermatozoa in bull. Biol Reprod 2002;67:308–313. [DOI] [PubMed] [Google Scholar]
- Frenette G, Sullivan R.. Prostasome-like particles are involved in the transfer of P25b from the bovine epididymal fluid to the sperm surface. Mol Reprod Dev 2001;59:115–121. [DOI] [PubMed] [Google Scholar]
- Gan H, Cai T, Lin X, Wu Y, Wang X, Yang F, Han C.. Integrative proteomic and transcriptomic analyses reveal multiple post-transcriptional regulatory mechanisms of mouse spermatogenesis. Mol Cell Proteomics 2013;12:1144–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Rodríguez A, de la Casa M, Peinado H, Gosálvez J, Roy R.. Human prostasomes from normozoospermic and non-normozoospermic men show a differential protein expression pattern. Andrology 2018;6:585–596. [DOI] [PubMed] [Google Scholar]
- Garrido N, Martínez-Conejero JA, Jauregui J, Horcajadas JA, Simón C, Remohí J, Meseguer M.. Microarray analysis in sperm from fertile and infertile men without basic sperm analysis abnormalities reveals a significantly different transcriptome. Fertil Steril 2009;91:1307–1310. [DOI] [PubMed] [Google Scholar]
- Gervasi MG, Visconti PE.. Molecular changes and signaling events occurring in spermatozoa during epididymal maturation. Andrology 2017;5:204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girouard J, Frenette G, Sullivan R.. Comparative proteome and lipid profiles of bovine epididymosomes collected in the intraluminal compartment of the caput and cauda epididymidis. Int J Androl 2011;34:e475–e486. [DOI] [PubMed] [Google Scholar]
- Guo X, Shen J, Xia Z, Zhang R, Zhang P, Zhao C, Xing J, Chen L, Chen W, Lin M. et al. Proteomic analysis of proteins involved in spermiogenesis in mouse. J Proteome Res 2010;9:1246–1256. [DOI] [PubMed] [Google Scholar]
- Guyonnet B, Dacheux F, Dacheux J-L, Gatti J-L.. The epididymal transcriptome and proteome provide some insights into new epididymal regulations. J Androl 2011;32:651–664. [DOI] [PubMed] [Google Scholar]
- Guyonnet B, Zabet-Moghaddam M, SanFrancisco S, Cornwall GA.. Isolation and proteomic characterization of the mouse sperm acrosomal matrix. Mol Cell Proteomics 2012;11:758–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamil KG, Sivashanmugam P, Richardson RT, Grossman G, Ruben SM, Mohler JL, Petrusz P, O’Rand MG, French FS, Hall SH.. HE2beta and HE2gamma, new members of an epididymis-specific family of androgen-regulated proteins in the human. Endocrinology 2000;141:1245–1253. [DOI] [PubMed] [Google Scholar]
- Han C, Park I, Lee B, Jin S, Choi H, Kwon JT, Kwon Y, Kim DH, Park ZY, Cho C.. Identification of heat shock protein 5, calnexin and integral membrane protein 2B as Adam7-interacting membrane proteins in mouse sperm. J Cell Physiol 2011;226:1186–1195. [DOI] [PubMed] [Google Scholar]
- Hecht NB. Molecular mechanisms of male germ cell differentiation. BioEssays 1998;20:555–561. [DOI] [PubMed] [Google Scholar]
- Hermo L, Dworkin J, Oko R.. Role of epithelial clear cells of the rat epididymis in the disposal of the contents of cytoplasmic droplets detached from spermatozoa. Am J Anat 1988;183:107–124. [DOI] [PubMed] [Google Scholar]
- Hermo L, Jacks D.. Nature’s ingenuity: bypassing the classical secretory route via apocrine secretion. Mol Reprod Dev 2002;63:394–410. [DOI] [PubMed] [Google Scholar]
- Hermo L, Robaire B.. Epididymal cell types and their functions. In: Robaire B, Hinton BT (eds). The Epididymis: From Molecules to Clinical Practice. Boston, USA: Springer, 2002, 81–102. [Google Scholar]
- Hernández-Silva G, Chirinos M.. Proteins from male and female reproductive tracts involved in sperm function regulation. Zygote 2019;27:5–16. [DOI] [PubMed] [Google Scholar]
- Howard L, Maciewicz RA, Blobel CP.. Cloning and characterization of ADAM28: evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem J 2000;348:21–27. [PMC free article] [PubMed] [Google Scholar]
- Howard L, Zheng Y, Horrocks M, Maciewicz RA, Blobel C.. Catalytic activity of ADAM28. FEBS Lett 2001;498:82–86. [DOI] [PubMed] [Google Scholar]
- Huang X-Y, Guo X-J, Shen J, Wang Y-F, Chen L, Xie J, Wang N-L, Wang F-Q, Zhao C, Huo R. et al. Construction of a proteome profile and functional analysis of the proteins involved in the initiation of mouse spermatogenesis. J Proteome Res 2008;7:3435–3446. [DOI] [PubMed] [Google Scholar]
- James ER, Carrell DT, Aston KI, Jenkins TG, Yeste M, Salas-Huetos A.. The role of the epididymis and the contribution of epididymosomes to mammalian reproduction. IJMS 2020;21:5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Turner TT, Bang HJ, Finger JN, Solarz MK, Wilson E, Brown EL, Kopf GS, Johnston DS.. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod 2007;76:561–570. [DOI] [PubMed] [Google Scholar]
- Jodar M, Sendler E, Krawetz SA.. The protein and transcript profiles of human semen. Cell Tissue Res 2016;363:85–96. [DOI] [PubMed] [Google Scholar]
- Jodar M, Soler-Ventura A, Oliva R; Molecular Biology of Reproduction and Development Research Group. Semen proteomics and male infertility. J Proteomics 2017;162:125–134. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, DiCandeloro P, Wilson E, Kopf GS, Turner TT.. mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod 2005;73:404–413. [DOI] [PubMed] [Google Scholar]
- Kang Y-N, Hsiao Y-W, Chen C-Y, Wu C-C.. Testicular sperm is superior to ejaculated sperm for ICSI in cryptozoospermia: an update systematic review and meta-analysis. Sci Rep 2018;8:7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Oh J, Woo J-M, Choi E, Im SH, Yoo YJ, Kim DH, Nishimura H, Cho C.. Expression and relationship of male reproductive ADAMs in mouse. Biol Reprod 2006;74:744–750. [DOI] [PubMed] [Google Scholar]
- Krapf DC, Ruan Y, Wertheimer EV, Battistone MA, Pawlak JB, Sanjay A, Pilder SH, Cuasnicu P, Breton S, Visconti PE.. cSrc is necessary for epididymal development and is incorporated into sperm during epididymal transit. Dev Biol 2012;369:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LK, Foo KY.. Recent insights on the significance of transcriptomic and metabolomic analysis of male factor infertility. Clin Biochem 2014;47:973–982. [DOI] [PubMed] [Google Scholar]
- Légaré C, Akintayo A, Blondin P, Calvo E, Sullivan R.. Impact of male fertility status on the transcriptome of the bovine epididymis. Mol Hum Reprod 2017;23:355–369. [DOI] [PubMed] [Google Scholar]
- Liu X, Liu F-J, Jin S-H, Shen X-F, Wang Y-W.. In-depth Proteomic mapping of mouse (Mus musculus) epididymal constructive basis for sperm maturation. Proteome Sci 2015a;13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang W, Liu F.. New insight into the castrated mouse epididymis based on comparative proteomics. Reprod Fertil Dev 2015b;27:551–556. [DOI] [PubMed] [Google Scholar]
- Magin TM, Vijayaraj P, Leube RE.. Structural and regulatory functions of keratins. Exp Cell Res 2007;313:2021–2032. [DOI] [PubMed] [Google Scholar]
- Martin-DeLeon P. Epididymosomes: transfer of fertility-modulating proteins to the sperm surface. Asian J Androl 2015;17:720–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McReynolds S, Dzieciatkowska M, Stevens J, Hansen KC, Schoolcraft WB, Katz-Jaffe MG.. Toward the identification of a subset of unexplained infertility: a sperm proteomic approach. Fertil Steril 2014;102:692–699. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD.. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 2019;47:D419–D426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Kastresana A, Telford B, Musich TA, McKinnon K, Clayborne C, Braig Z, Rosner A, Demberg T, Watson DC, Karpova TS. et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci Rep 2017;7:1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narmadha G, Yenugu S.. Molecular modeling of the human sperm associated antigen 11 B (SPAG11B) proteins. Syst Biol Reprod Med 2015;61:78–88. [DOI] [PubMed] [Google Scholar]
- Nixon B, De Iuliis GN, Hart HM, Zhou W, Mathe A, Bernstein I, Anderson AL, Stanger SJ, Skerrett-Byrne DA, Jamaluddin MFB. et al. Proteomic profiling of mouse epididymosomes reveals their contributions to post-testicular sperm maturation. Mol Cell Proteomics 2019;18:S91–S108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka-Bauer K, Lepczynski A, Ozgo M, Kamieniczna M, Fraczek M, Stanski L, Olszewska M, Malcher A, Skrzypczak W, Kurpisz MK.. Sperm mitochondrial dysfunction and oxidative stress as possible reasons for isolated asthenozoospermia. J Physiol Pharmacol 2018;69:403–417. [DOI] [PubMed] [Google Scholar]
- Oh J, Woo J-M, Choi E, Kim T, Cho B-N, Park ZY, Kim YC, Kim DH, Cho C.. Molecular, biochemical, and cellular characterization of epididymal ADAMs, ADAM7 and ADAM28. Biochem Biophys Res Commun 2005;331:1374–1383. [DOI] [PubMed] [Google Scholar]
- Oliva R, Castillo J.. Sperm nucleoproteins. In: Zini A, Agarwal A (eds). Sperm Chromatin. New York, USA: Springer, 2011, 45–60. [Google Scholar]
- Oliva R, Dixon GH.. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Prog Nucleic Acid Res Mol Biol 1991;40:25–94. [DOI] [PubMed] [Google Scholar]
- Orgebin-Crist M-C. Studies on the function of the epididymis. Biol Reprod 1969;1:155–175. [DOI] [PubMed] [Google Scholar]
- Orgebin-Crist MC. Sperm maturation in rabbit epididymis. Nature 1967;216:816–818. [DOI] [PubMed] [Google Scholar]
- Park Y-J, Battistone MA, Kim B, Breton S.. Relative contribution of clear cells and principal cells to luminal pH in the mouse epididymis. Biol Reprod 2017;96:366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pǎunescu TG, Ljubojevic M, Russo LM, Winter C, McLaughlin MM, Wagner CA, Breton S, Brown D.. cAMP stimulates apical V-ATPase accumulation, microvillar elongation, and proton extrusion in kidney collecting duct A-intercalated cells. Am J Physiol Renal Physiol 2010;298:F643–F654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pǎunescu TG, Shum WWC, Huynh C, Lechner L, Goetze B, Brown D, Breton S.. High-resolution helium ion microscopy of epididymal epithelial cells and their interaction with spermatozoa. Mol Hum Reprod 2014;20:929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz M, Morín M, Del Mazo J.. Proteome profile changes during mouse testis development. Comp Biochem Physiol D Genomics Proteomics 2006;1:404–415. [DOI] [PubMed] [Google Scholar]
- Peng X, Xu E, Liang W, Pei X, Chen D, Zheng D, Zhang Y, Zheng C, Wang P, She S. et al. Identification of FAM3D as a new endogenous chemotaxis agonist for the formyl peptide receptors. J Cell Sci 2016;129:1831–1842. [DOI] [PubMed] [Google Scholar]
- Piomboni P, Gambera L, Serafini F, Campanella G, Morgante G, de Leo V.. Sperm quality improvement after natural anti-oxidant treatment of asthenoteratospermic men with leukocytospermia. Asian J Androl 2008;10:201–206. [DOI] [PubMed] [Google Scholar]
- Plante G, Manjunath P.. Epididymal Binder of SPerm genes and proteins: what do we know a decade later? Andrology 2015;3:817–824. [DOI] [PubMed] [Google Scholar]
- Plante G, Prud'homme B, Fan J, Lafleur M, Manjunath P.. Evolution and function of mammalian binder of sperm proteins. Cell Tissue Res 2016;363:105–127. [DOI] [PubMed] [Google Scholar]
- Plante G, Thérien I, Lachance C, Leclerc P, Fan J, Manjunath P.. Implication of the human Binder of SPerm Homolog 1 (BSPH1) protein in capacitation. Mol Hum Reprod 2014;20:409–421. [DOI] [PubMed] [Google Scholar]
- Plante G, Thérien I, Manjunath P.. Characterization of recombinant murine binder of sperm protein homolog 1 and its role in capacitation. Biol Reprod 2012;87:20, 1–11. [DOI] [PubMed] [Google Scholar]
- Qi L, Liu Z, Wang J, Cui Y, Guo Y, Zhou T, Zhou Z, Guo X, Xue Y, Sha J.. Systematic analysis of the phosphoproteome and kinase-substrate networks in the mouse testis. Mol Cell Proteomics 2014;13:3626–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan Y, Hamil KG, Tan J-A, Grossman G, Petrusz P, Hall SH, French FS.. Novel partners of SPAG11B isoform D in the human male reproductive tract. Biol Reprod 2009;81:647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajesh A, Madhubabu G, Yenugu S.. Identification and characterization of Wfdc gene expression in the male reproductive tract of the rat. Mol Reprod Dev 2011;78:633–641. [DOI] [PubMed] [Google Scholar]
- Reilly JN, McLaughlin EA, Stanger SJ, Anderson AL, Hutcheon K, Church K, Mihalas BP, Tyagi S, Holt JE, Eamens AL. et al. Characterisation of mouse epididymosomes reveals a complex profile of microRNAs and a potential mechanism for modification of the sperm epigenome. Sci Rep 2016;6:31794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejraji H, Sion B, Prensier G, Carreras M, Motta C, Frenoux J-M, Vericel E, Grizard G, Vernet P, Drevet JR.. Lipid remodeling of murine epididymosomes and spermatozoa during epididymal maturation. Biol Reprod 2006;74:1104–1113. [DOI] [PubMed] [Google Scholar]
- Robaire B, Hinton BT, Orgebin-Crist M-C. The Epididymis. In: Neill JD (ed). Knobil and Neill's Physiology of Reproduction. St. Louis, USA: Elsevier Academic Press, 2006, 1071–1148. [Google Scholar]
- Roy J, Kim B, Hill E, Visconti P, Krapf D, Vinegoni C, Weissleder R, Brown D, Breton S.. Tyrosine kinase-mediated axial motility of basal cells revealed by intravital imaging. Nat Commun 2016;7:10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez F, Frenette G, Sullivan R.. Epididymosomes and prostasomes: their roles in posttesticular maturation of the sperm cells. J Androl 2003;24:149–154. [DOI] [PubMed] [Google Scholar]
- Samanta L, Parida R, Dias TR, Agarwal A.. The enigmatic seminal plasma: a proteomics insight from ejaculation to fertilization. Reprod Biol Endocrinol 2018;16:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta L, Sharma R, Cui Z, Agarwal A.. Proteomic analysis reveals dysregulated cell signaling in ejaculated spermatozoa from infertile men. Asian J Androl 2019;21:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoysman RJ, Bedford JM.. The role of the human epididymis in sperm maturation and sperm storage as reflected in the consequences of epididymovasostomy. Fertil Steril 1986;46:293–299. [DOI] [PubMed] [Google Scholar]
- Schwarz A, Wennemuth G, Post H, Brandenburger T, Aumüller G, Wilhelm B.. Vesicular transfer of membrane components to bovine epididymal spermatozoa. Cell Tissue Res 2013;353:549–561. [DOI] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016;351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, Rando OJ.. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev Cell 2018;46:481–494.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum WWC, Da Silva N, McKee M, Smith PJS, Brown D, Breton S.. Transepithelial projections from basal cells are luminal sensors in pseudostratified epithelia. Cell 2008;135:1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerget S, Rosenow MA, Petritis K, Karr TL.. Sperm proteome maturation in the mouse epididymis. PLoS One 2015;10:e0140650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R. Epididymosomes: a heterogeneous population of microvesicles with multiple functions in sperm maturation and storage. Asian J Androl 2015;17:726–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan R. Epididymosomes Role of extracellular microvesicles in sperm maturation. Front Biosci (Schol Ed) 2016;8:106–114. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Frenette G, Girouard J.. Epididymosomes are involved in the acquisition of new sperm proteins during epididymal transit. Asian J Androl 2007;9:483–491. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Légaré C, Lamontagne-Proulx J, Breton S, Soulet D.. Revisiting structure/functions of the human epididymis. Andrology 2019;7:748–757. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Mieusset R.. The human epididymis: its function in sperm maturation. Hum Reprod Update 2016;22:574–587. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Saez F.. Epididymosomes, prostasomes, and liposomes: their roles in mammalian male reproductive physiology. Reproduction 2013;146:R21–35. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimon V, Calvo E, Koukoui O, Légaré C, Sullivan R.. Effects of vasectomy on gene expression profiling along the human epididymis. Biol Reprod 2008a;79:262–273. [DOI] [PubMed] [Google Scholar]
- Thimon V, Frenette G, Saez F, Thabet M, Sullivan R.. Protein composition of human epididymosomes collected during surgical vasectomy reversal: a proteomic and genomic approach. Hum Reprod 2008b;23:1698–1707. [DOI] [PubMed] [Google Scholar]
- Tian T, Zhu Y-L, Zhou Y-Y, Liang G-F, Wang Y-Y, Hu F-H, Xiao Z-D.. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem 2014;289:22258–22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TT. On the epididymis and its role in the development of the fertile ejaculate. J Androl 1995;16:292–298. [PubMed] [Google Scholar]
- Turner TT, Bomgardner D, Jacobs JP, Nguyen QAT.. Association of segmentation of the epididymal interstitium with segmented tubule function in rats and mice. Reproduction 2003;125:871–878. [DOI] [PubMed] [Google Scholar]
- Vicens A, Borziak K, Karr TL, Roldan ERS, Dorus S.. Comparative sperm proteomics in mouse species with divergent mating systems. Mol Biol Evol 2017;34:1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilanova-Perez T, Jones C, Balint S, Dragovic R, Dustin ML, Yeste M, Coward K.. Exosomes derived from HEK293T cells interact in an efficient and noninvasive manner with mammalian sperm in vitro. Nanomedicine (Lond) 2020;15:1965–1980. [DOI] [PubMed] [Google Scholar]
- Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M. et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 2014;42:7290–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MJ, Rylett CM, Keen JN, Audsley N, Sajid M, Shirras AD, Isaac RE.. Proteomic identification of Drosophila melanogaster male accessory gland proteins, including a pro-cathepsin and a soluble gamma-glutamyl transpeptidase. Proteome Sci 2006;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Shen H, Zhang H, Yan J, Liu Y, Yao F, Wang X, Cheng Z, Tang T-S, Guo C.. Quantitative proteomics analysis reveals alterations of lysine acetylation in mouse testis in response to heat shock and X-ray exposure. Biochim Biophys Acta Proteins Proteom 2018;1866:464–472. [DOI] [PubMed] [Google Scholar]
- Xu B, Zhang Y, Zhan S, Wang X, Zhang H, Meng X, Ge W.. Proteomic profiling of brain and testis reveals the diverse changes in ribosomal proteins in fmr1 knockout mice. Neuroscience 2018;371:469–483. [DOI] [PubMed] [Google Scholar]
- Yanagimachi R, Kamiguchi Y, Mikamo K, Suzuki F, Yanagimachi H.. Maturation of spermatozoa in the epididymis of the Chinese hamster. Am J Anat 1985;172:317–330. [DOI] [PubMed] [Google Scholar]
- Zhou CX, Zhang Y-L, Xiao L, Zheng M, Leung KM, Chan MY, Lo PS, Tsang LL, Wong HY, Ho LS. et al. An epididymis-specific beta-defensin is important for the initiation of sperm maturation. Nat Cell Biol 2004;6:458–464. [DOI] [PubMed] [Google Scholar]
- Zhou W, De Iuliis GN, Dun MD, Nixon B.. Characteristics of the epididymal luminal environment responsible for sperm maturation and storage. Front Endocrinol (Lausanne) 2018;9:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Stanger SJ, Anderson AL, Bernstein IR, De Iuliis GN, McCluskey A, McLaughlin EA, Dun MD, Nixon B.. Mechanisms of tethering and cargo transfer during epididymosome-sperm interactions. BMC Biol 2019;17:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y-F, Cui Y-G, Guo X-J, Wang L, Bi Y, Hu Y-Q, Zhao X, Liu Q, Huo R, Lin M. et al. Proteomic analysis of effect of hyperthermia on spermatogenesis in adult male mice. J Proteome Res 2006;5:2217–2225. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Xu G, Patel A, McLaughlin MM, Silverman C, Knecht K, Sweitzer S, Li X, McDonnell P, Mirabile R. et al. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics 2002;80:144–150. [DOI] [PubMed] [Google Scholar]
- Zumoffen CM, Massa E, Caille AM, Munuce MJ, Ghersevich SA.. Effects of lactoferrin, a protein present in the female reproductive tract, on parameters of human sperm capacitation and gamete interaction. Andrology 2015;3:1068–1075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.