Abstract
Most advanced-stage ovarian cancer patients, including those with epithelial ovarian cancer (EOC), develop recurrent disease and acquisition of resistance to chemotherapy, leading to limited treatment options. Decrease in Let7b miRNA levels in clinical ovarian cancer has been associated with chemoresistance, increased proliferation, invasion, and relapse in EOC. We have established a murine EOC relapsed model by administering paclitaxel (PTX) and stopping therapy to allow for tumor regrowth. Global microRNA profiling in the relapsed tumor showed significant downregulation of Let7b relative to untreated tumors. Here, we report the use of hyaluronic acid (HA)-based nanoparticle formulation that can deliver Let7b miRNA mimic to tumor cells and achieve cellular programming both in vitro and in vivo. We demonstrate that a therapeutic combination of Let7b miRNA and PTX leads to significant improvement in anti-tumor efficacy in the relapsed model of EOC. We further demonstrate that the combination therapy is safe for repeated administration. This novel approach of cellular reprogramming of tumor cells using clinically relevant miRNA mimic in combination with chemotherapy could enable more effective therapeutic outcomes for patients with advanced-stage relapsed EOC.
Keywords: ovarian cancer, IDG-VEGF, relapsed/MDR model, microRNA-Let7b, hyaluronic-acid-based nanoparticles
Graphical abstract

Most patients with advanced-stage epithelial ovarian cancer (EOC) develop recurrent disease and acquisition of resistance to chemotherapy, leading to limited treatment options. In this study we evaluate a novel approach of therapeutic combination of Let7b miRNA and paclitaxel and demonstrate significant improvement in anti-tumor efficacy in the relapsed model of EOC.
Introduction
Ovarian cancer is the most lethal of all gynecological cancers, with an estimated 22,410 cases and 13,770 deaths in the 2021.1 In advanced epithelial ovarian cancer (EOC), after initial treatment with surgery, chemotherapy is given, which involves 3–6 cycles of carboplatin plus paclitaxel (PTX). However, there is recurrence or relapse observed in more than 80% of patients.2 One of the main reasons for such a high rate of relapse in advanced stages of EOC is the acquisition of multidrug resistance (MDR). Acquired resistance to chemotherapy can be due to multiple factors that include (1) molecular alterations in tumor cells, leading to loss of tumor suppressor genes, such as PTEN, RAD51, and BRCA1 or BRCA2 mutations3; (2) decrease in intracellular uptake and increased efflux of drugs like cisplatin and PTX due to upregulation of ATP-binding cassette (ABC) transporters, especially multidrug resistance protein 1 (MDR1)4; and (3) role of the tumor microenvironment; in advanced ovarian cancer, tumor metastasizes to the peritoneal cavity, which leads to buildup of ascitic fluid5. Hence, there is an unmet medical need for development of novel therapies that lead to improvement the efficacy in relapsed ovarian cancer patients.
Preclinical mouse models are important in the discovery and development of novel therapies. The syngeneic ID8,6 Syngeneic ID8 model expressing vascular endothelial growth factor (ID8-VEGF),7 or CRISPR edited ID8 mouse models8 are often used to model the advanced-stage ovarian cancer and interrogate the interaction between tumor cells and various components of the tumor microenvironment (TME), such as immune cells (T cells and macrophages), cytokines, growth factors, and extracellular vesicles (EVs). Although the intraperitoneal (i.p.) ID8 or ID8-VEGF mouse model simulates features of advanced-stage human ovarian cancer, little is known about the molecular events that lead to acquisition of MDR phenomenon post chemotherapy. As a result of this, evaluation of novel therapies a few weeks post tumor implantation would not be able to inform about the efficacy in recurrent disease settings seen in the clinic, where patients with a good response initially have recurrence with acquired MDR.9 A mouse model that recapitulates the recurrence of tumor growth after a few cycles of chemotherapy may better simulate the treatment pathway and the molecular changes at the cellular level that occur in human metastatic disease. Therefore, we hypothesized that the repeat dosing of PTX in the ID8-VEGF model followed by a therapy-free interval would lead to recurrence of/relapse of tumor growth and development of a resistant phenotype with a differential molecular profile compared with an untreated group.
Mature microRNAs (miRNAs) are 19- to 24-nucleotide-long, single-stranded RNAs that regulate gene expression at the post transcriptional level by sequence-specific binding to the 3ʹ untranslated region (3ʹ UTR) of a target mRNA. Several types of human cancers are characterized by (1) upregulation of oncogenic miRNAs that lead to downregulation of tumor suppressor genes and (2) inactivation or deletion of tumor suppressor miRNAs that leads to overexpression of oncogenes, overall leading to increased proliferation, invasion, metastasis, and survival.10 Several miRNA profiling studies in clinical samples have revealed changes in miRNA levels, depending on the stage of the disease and response to therapy, indicating their role in diagnosis and treatment of the ovarian cancer. Members of the Let-7 family (a/b/d/e/f) and miR 200 family (a/b/c) and miR-21 have been found to be de-regulated in advanced-stage patients and have been linked to the chemoresistance and shorter progression-free survival.11 Previous studies that explored systemic delivery of exogenous Let-7b and miRNA-34 have demonstrated targeting of key oncogenes, KRAS, c-MYC, HMGA2, and LIN28, and also prevent progression of human non-small-cell lung cancer (NSCLC) tumor xenografts.12,13 Systemic delivery of naked miRNAs to tumor cells in vivo is highly challenging due to several extracellular and intracellular barriers, including poor permeability due to negative charge and rapid degradation due to nucleases.14 To this end, we have exploited the delivery potential of hyaluronic-acid-poly(ethylene imine) (HA-PEI) conjugate, which has emerged as an ideal delivery system for nucleic acids due to its nonimmunogenic, biodegradable, and noninflammatory properties.15,16 Our previous studies demonstrated successful delivery of small interfering RNA (siRNA) by HA-PEI nanoparticles (NPs) to tumor tissues for anti-cancer therapy.17,18 More recently, previous members of our laboratory have shown that i.p. delivery of miRNAs, such as miR-125b and miR34-a, using HA-PEI NPs has been able to achieve cellular reprograming and improved anti-tumor efficacy in mouse model lung and ovarian cancers.19, 20, 21 In the current study, we have extended the application of HA-PEI NPs for delivery of Let7b miRNA (a tumor suppressor miRNA associated with chemoresistance) to ovarian tumor cells (ID8-VEGF) using an i.p. route of administration to overcome the systemic barriers and improve the delivery of miRNA to the ovarian cancer cells in the peritoneal cavity.
The goal of the current study is to achieve improvement in the anti-tumor efficacy by using combination therapy of HA-PEI-Let7b NPs with PTX in a relapse model of ovarian cancer. To this end, we have developed a relapse model of ovarian cancer (ID8-VEGF) mouse model to recapitulate recurrence of tumor growth after chemotherapy. We then systematically performed gene expression and miRNA profiling in this relapse model of ID-VEGF and identified tumor suppressor miRNA associated with relapse. Reprogramming of cellular miRNA and mRNA levels of ovarian cancer cells was achieved using HA-PEI-Let7b NPs. A combination of HA-PEI-Let7b and PTX has been shown to improve the anti-tumor efficacy in a relapsed model of ovarian cancer, indicating its potential as a treatment option of relapsed ovarian cancer patients.
Results
Molecular profiling of ID8-PTX-MDR/relapsed model of EOC
Development of ID8-PTX relapsed/MDR model of EOC
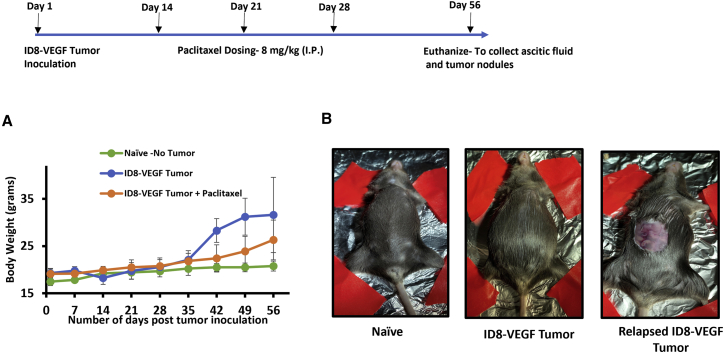
In order to recapitulate the clinical development of relapse in advanced-stage ovarian cancer in the ID8-VEGF tumor model, 2 weeks post tumor inoculation, mice were dosed with 3 doses (8 mg/kg) of PTX and the therapy was stopped to allow for recurrence/relapse of tumor (Figure 1). The tumor growth was monitored by total body weight, which is primarily due to buildup of the ascitic fluid in the abdomen. The control group in which the ID8-VEGF tumor cells were implanted in the peritoneal cavity showed a rapid increase in body weight after 5 weeks post tumor inoculation (Figure 1A), due to the buildup of ascitic fluid in the peritoneal cavity. In contrast, the mice in the ID8-VEGF + PTX treated group showed a delayed growth/relapse of tumor only after 7 weeks post tumor inoculation, indicating an initial response to PTX treatment but the occurrence of relapse when the PTX treatment was stopped. These observations led us to believe that repeat dosing of PTX in a ID8-VEGF model mimics relapse of tumor observed in clinical settings.
Figure 1.
Development of the ID8-PTX-MDR/relapse model of ovarian cancer
Time line showing tumor development and relapse. ID8-VEGF cells were inoculated in female C57BL/6 mice, followed by three doses of paclitaxel treatment (8 mg/kg i.p.) on day 14, 21, and 28 (as indicated by the arrows), and therapy was stopped for development of relapse and naive mice ID8-VEGF ovarian cancer cell line. (A) Graph showing tumor growth (ascitic fluid buildup) for the control group (n = 8) after day 35 and relapse of tumor growth in the PTX groups (n = 8) after day 49. (B) Representative images of mice from the naive (no abdominal swelling) group; control group, showing abdominal swelling due to buildup of ascitic fluid; and PTX group, showing ascites in the peritoneal cavity.
Repeat dosing of PTX induces changes in mRNA levels of resistance-associated genes in an EOC model
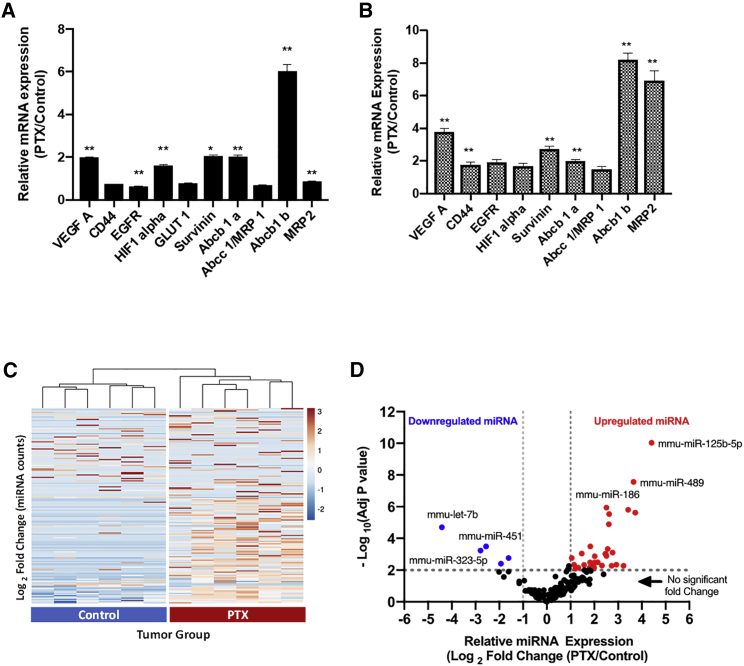
To further understand if the repeat dosing of PTX has altered the molecular profile of the tumor cells to a resistant phenotype, we have chosen a panel of oncogenes that are involved in pathways related to hallmarks of cancer (VEGF, EGFR, CD44, survivin, etc.) and genes that are upregulated involved in MDR cancer (ABC transporters and related genes). qPCR-based relative quantification of mRNA expression levels of tumor cells from the PTX group (normalized to the control group) (Figure 2A) showed 2- to 6-fold upregulation of VEGF-A, survivin, and MDR genes (ABCB1a and ABCB1b) and statistical significance (p < 0.01). Similarly, mRNA levels of these genes in the metastatic tumor nodules from the diaphragm showed 2- to 8-fold upregulation (Figure 2B). Overall, based on these results, we believe that repeat dosing of PTX led to the changes in mRNA levels of key oncogenes and MDR-associated genes in both the tumor cells in the ascitic fluid as well as the tumor nodules recapitulating the molecular changes in human disease.
Figure 2.
Molecular profiling in tumor cells and nodules from ID8-PTX-dosed ovarian cancer mouse model
qPCR data showing upregulation of gene expression for MDR and other genes normalized to β-actin. (A) ID8 cells sorted from ascitic fluid of tumor bearing mice ID8-VEGF control and ID8-VEGF-PTX-dosed animals (n = 6). (B) Tumor nodules derived from diaphragm of ID8-VEGF control and ID8-VEGF-PTX-dosed animals. (C) Heatmap with unsupervised hierarchical clustering (columns) dendrograms show clustering of control and PTX groups-based miRNA expression levels (rows) using NanoString analysis. Scale showing fold changes in miRNA expression. (D) Differentially expressed miRNA in ID8 tumor cells derived from ascitic fluid using NanoString analysis of tumor cells derived from control and PTX-treated ID8-VEGF tumor bearing mice (n = 6). p values obtained (A and B) with Student’s t test: ∗∗p < 0.001 or ∗∗∗∗p < 0.0001, compared with the control group; bars represent fold changes; adjusted p values was obtained using Bonferroni correction method. Upregulated miRNA was defined as log2 fold >1 (fold change >2) and p < 0.01 and downregulated miRNA was defined log2 fold change <−1 (fold change <−2) and p < 0.01.
Differential expression of clinically relevant miRNA upon tumor relapse
Several miRNA profiling studies in clinical samples of ovarian cancer have revealed their changes in miRNA levels, depending on the stage of the disease and response to therapy, indicating their role in diagnosis and treatment of the ovarian cancer. So, we sought to profile the miRNA changes in both the PTX/relapse group as well as the control group to understand the differentially expressed miRNA using NanoString assay. To quantify the miRNA expressions in the control and relapse groups a panel of 700 miRNA probes (Nanostring nCounter mouse miRNA Panel v3) relevant for ovarian cancer was used for the assay, and the expression levels, for a total of 173 miRNAs, whose counts were greater than the assay normalization controls, were used for differential analysis. For a high-level overview of data, miRNA expression levels (counts) are plotted as heatmap with unsupervised hierarchical clustering (Figure 2C). Overall, the heatmap shows distinct pattern with higher number of miRNAs upregulated (52) in the PTX group than the control group. Unsupervised hierarchical clustering reveals that miRNA levels of individual PTX samples is similar (correlated) to each other and different from control samples. This is visually represented by the common nodes joining the samples which have closest distance or pairing together based on miRNA expression level. Overall, this indicates distinct molecular profile of the PTX group (relapse) compared with the control group.
To identify differentially expressed miRNAs that are associated with chemoresistance, we have employed the following criteria:
-
1.
miRNAs that are differentially expressed based on the NanoString data—criteria for differential expression were defined as upregulated miRNA if log2 fold change > 2 and p < 0.01, the criteria for downregulated miRNA if log2 fold change < −2 and p < 0.01. These criteria were chosen based on clinical miRNA profiling data in ovarian cancer (The Cancer Genome Atlas [TCGA] database).23
-
2.
Clinically relevant miRNAs and evidence—miRNAs that are associated with shorter progression-free survival and chemoresistant/relapse and their expression levels from TCGA database and other clinical data studies
For visualization of miRNA that fits the criteria 1, listed above, data from NanoString assay were plotted in the form of volcano plots (Figure 2D). Differentially expressed miRNA, i.e., downregulated miRNA in the PTX group, are data points in blue, and data points in red are upregulated in the PTX group. The list of upregulated and downregulated miRNA is provided in Tables S1 and S2. Next, we wanted to understand if these differentially expressed miRNAs are clinically relevant and if they show similar trends in sample sets from chemoresistant ovarian cancer patients. We relied on public databases, such as TCGA, for ovarian cancer and other clinical datasets reported in literature. Overall, based on the miRNA profiling studies from our in vivo model and available clinical data, we were able to filter a few miRNAs that are clinically relevant and associated with chemoresistance, and these are summarized in Tables 1 and 2.
Table 1.
Downregulated miRNA from NanoString data that are clinically relevant
Table 2.
Upregulated miRNA from NanoString data that are clinically relevant
Overall, based on the results of gene expression profiling and miRNA profiling, we believe that we have developed a relapse model of ovarian cancer that has a distinct molecular profile and is associated with PTX resistance. miRNA analysis revealed that Let7b miRNA was found to be the most significantly downregulated in the PTX group and several clinical literature reports have validated the role of the Let-7 family as tumor suppressor miRNA in ovarian cancer patients. Combining these two data points, we hypothesized that transfection of Let-7b miRNA mimic using HA-PEI NPs would reprogram the cellular miRNA levels of ID8 tumor cells, downregulate key oncogenic targets of Let7b, and lead to improvement in potency of PTX in the resistant ID8-VEGF cell line.
Combination of HA-PEI-Let7b mimic and PTX improves the efficacy in ID8-VEGF cells in vitro
In vitro evaluation of combination of Let7b with PTX yields shows improvement in IC50 of PTX in ID8-VEGF-resistant cells
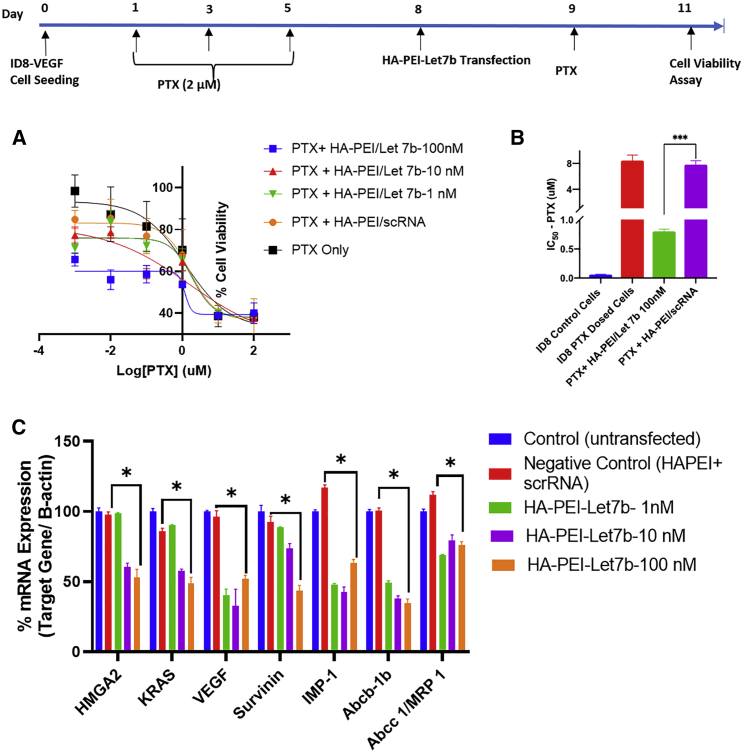
Naked miRNA mimics have poor transfection efficiency because they suffer from delivery challenges in both in vitro and in vivo. HA-PEI is a platform-based NP delivery system that has been developed by previous laboratory members for the delivery of a various sizes of nucleic acids, such as siRNA, plasmid DNA, and miRNA. The goal of these in vitro studies was to evaluate if Let7b (nM) delivered using HA-PEI NPs (HA-PEI-Let7b) in combination with PTX could improve the half-maximal inhibitory concentration (IC50) of PTX in ID8-VEGF cells. ID8-VEGF cells were pre-dosed with PTX (2 μM) to develop resistance, as shown in the time line (Figure 3), and the % cell viability was evaluated after when ID8-PTX-dosed cells are treated with PTX in combination HA-PEI-Let7b NPs, with Let7b concentrations ranging from 0.1 to 100 nM (Figure 3A). These results suggested that the combination of PTX + HA-PEI-Let-7b-100 nM improves the potency of PTX at 0.1, 0.01, and 0.001 μM doses compared with PTX + HA-PEI-scRNA (negative control). Further, the % cell viability data was used to obtain the IC50 value of combination PTX + HA-PEI-Let7b 100 nM and other controls (Figure 2C). As expected, we observed an increase in IC50 value of ID8-PTX-dosed cells (8.1 μM) compared with ID8-control (0.059 μM) by ∼140-fold (Figure 3B), indicating increased resistance to PTX therapy. Interestingly, the combination of PTX + HA-PEI-Let7b-100 nM brings down the IC50 (0.62 μM) by 13-fold. Overall, these data indicate the potential of HA-PEI-Let7b to improve the efficacy of PTX even cells, which are refractory to PTX treatment.
Figure 3.
In vitro evaluation of combination therapy of HA-PEI-let7b and PTX for assessing improvement in potency of PTX and reprogramming of target mRNA levels
ID8-VEGF were dosed with PTX for development of resistance (day 1, 3, and 5) and allowed to recover, followed by HA-PEI let7b nanoparticle dosing and MTS assay readout. (A) Graph % cell viability of various combinations of Let7b (0.1–100 nM) with PTX (0.001–100 μM) measured at 72 h time point. scRNA and untreated control were used for calculating relative cell viability.
(B) Graph showing the IC50 values calculated from the nonlinear curve fitting of % cell viability (y axis) and log10 PTX concentration (μM) on x axis. All conditions were performed with n = 4 wells in a 96-well plate format with 1,500 cells/well.
(C) Evaluation of knockdown of Let7b target mRNA in ID8-control cells 48 h post HA-PEI-Let-7b transfection at 1, 10, and 100 nM Let7b concentrations using qPCR. Scrambled RNA control (scRNA) was used as negative control. Untransfected controls were for normalizing the relative gene expression. B-actin was used as a housekeeping gene. All experiments were performed with n = 3 biological replicates and n = 3 technical/assay replicates. Student's t test was used for statistical significance (∗p < 0.05, ∗∗∗p < 0.001).
Downregulation of mRNA levels of validated let7b targets using HA-PEI-Let7b in ID8-VEGF cells
Let7b is a tumor suppressor miRNA, which mediates its anti-tumor efficacy by downregulation of several target oncogenic signaling pathways, such as HMGA2 KRAS, survivin, VEGF-A, etc.22 In addition to these genes is known to decrease the levels of MDR-related genes, such as IMP-1 and ABCB11.23 So, we wanted to evaluate the improvement in potency achieved in PTX is due to mRNA repression of the multiple target genes at various doses of Let7b mimic, using qPCR. Transfection experiments performed in ID8-VEGF cells using HA-PEI-Let7b NPs at Let7b doses of 1, 10, and 100 nM show % mRNA levels of various genes at 48 h post transfection normalized to the control (untransfected control) using qPCR (Figure 3C). Overall, 30%–70% downregulation of target mRNA was observed when the cells were transfected with HA-PEI -Let-7b at a 100 nM dose. For genes HMGA2, KRAS, and survivin, which are direct targets of Let7b, we do see a dose-dependent effect of Let7b for 100 and 1 nM doses. For other genes, such as VEGF and MDR genes, which are not direct targets, we do not observe a dose-dependent effect, and this may be due to an imperfect complementarity of Let7b sequence to these particular target genes. Finally, the downregulation of the above genes, which are involved in various hallmarks of cancer, such as evading growth suppressors, inducing angiogenesis, evading apoptosis, and enhanced drug efflux transporters, can lead to the inhibition of cell proliferation, which was observed in the previous experiments.
Intraperitoneal delivery of Let7b miRNA in ID8-VEGF tumor model
Restoration of Let7b levels in tumor nodules and downregulation of oncogenes using HA-PEI-let7b nanoparticles
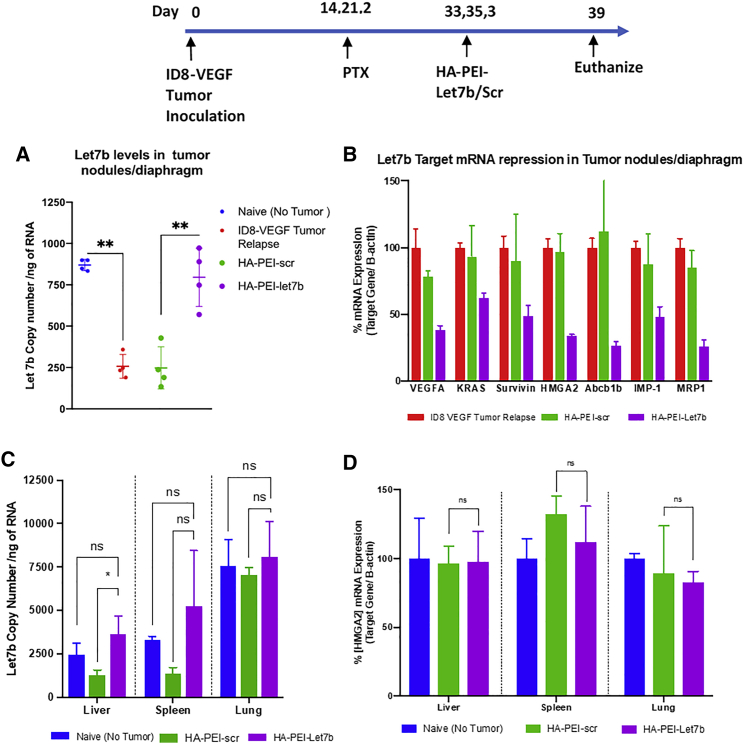
Based on the miRNA profiling data, Let7b miRNA levels are downregulated in relapsed ovarian cancer in vivo. We wanted to evaluate if administering Let7b mimic exogenously using HA-PEI-Let7b NPs via the i.p. route would restore the levels of endogenous levels of Let7b in ID8-VEGF tumor inoculated mice pre-dosed with PTX (Figure 4).To this end, we have quantified Let7b miRNA levels in the diaphragm/tumor nodules using qPCR (Figure 4A). The results show that Let7b levels are downregulated in resistant mice by 3.5 fold (∼250 copies/ng of RNA) compared with Let7b levels in naive mice (∼850 copies/ng of RNA) and levels of Let7b miRNA are restored to almost baseline (∼750 copies/ng of RNA) in mice treated with 3 doses of HA-PEI-Let7b NPs. We further wanted to understand if the delivered Let7b miRNA is able to downregulate the target genes, such as HMGA2, KRAS, survivin, VEGFA, etc., in the tumor nodules. Relative quantification of mRNA levels of tumor nodules extracted from ID8-VEGF tumor bearing mice dosed with Let7b using qPCR shows a downregulation/repression of all the 8 oncogenic mRNAs to ∼30%–70% (Figure 4B) compared with the control and HA-PEI-scr (HA-PEI nanoparticles encapsulted with scrambled RNA) - Negative control for HA-PEI-Let7b Nanoparticles. Further magnitude of changes in these oncogenic and MDR-related genes that we observe in vivo agrees with the data that we saw in vitro. Overall, based on these results, we believe that the restoration of Let7b levels in ID8-VEGF-resistant mice using HA-PEI-let7b NPs leads to downregulation of key oncogenes, which can further improve the efficacy in combination with PTX.
Figure 4.
Restoration of Let7b levels in tumor nodules and downregulation of oncogenes using HA-PEI-let7b nanoparticles
Time line showing tumor development in the ID8-VEGF mouse model and dosing schedule for HA-PEI-Let7b. (A) Quantification of absolute copies of Let7b in tumor nodules 48 h post 3 doses Let7b dosing (i.p.) (n = 4 per group). (B) Relative quantification of Let-7b mediated target gene expression in changes 48 h post 3 doses of HA-PEI-Let7b/scr (n = 4 per group). (C) Biodistribution of Let7b in liver, spleen, and lung 48 h post 3 doses of HA-PEI-Let7b dosing. (D) Relative quantification of HMGA2 mRNA levels in liver, spleen, and lung tissues 48 h post 3 doses of HA-PEI-Let7b. Statistical significance was evaluated using Student’s t test by comparing ID8 tumor relapse control or naive (no tumor) with HA-PEI-Let-7b groups (∗p < 0.05,∗∗p < 0.01, ns = not statistically significant).
Intraperitoneal route of administration of Let7b miRNA leads to minimal off-target effects in non-tumor tissues
Having established that the i.p. administration of HA-PEI-Let7b NPs was able to restore the levels of Let7b in tumor nodules, we wanted to understand the exposure of Let7b in the tumor bearing + HA-PEI-let7b treated group compared with the naive (no tumor) and the tumor bearing + HA-PEI-scr (negative control). To this end, we have quantified the levels of Let7b miRNA using qPCR in organs, such as liver, spleen, and lung (Figure 4C). Because we have administered the HA-PEI-Let7b NPs via the i.p. route, it is not surprising to observe elevated levels of Let7b 3 doses 48 h in peritoneal organs, such as liver and spleen of the tumor bearing + HA-PEI-Let7b treated group compared with the tumor bearing + HA-PEI-scr. Interestingly, we do not observe an increase in the Let7b levels in lung tissue of the HA-PEI-Let7b group compared with HA-PEI-scr, indicating that the particles may be localized to the peritoneal organs. However, these Let7b levels are not statistically significant compared with the naive (no tumor) untreated group. Additionally, we have evaluated if these elevated levels can lead to off-target effects in these organs by quantifying the mRNA levels of HMGA2 gene, which is the primary target for Let7b miRNA (Figure 4D). To our surprise, the elevated let7b levels do not lead to a significant change in the relative mRNA levels of HMGA2 in all the three organs, i.e. liver, spleen, and lung, and of the HA-PEI-Let7b compared with the HA-PEI-scr group. These results support our strategy of combination therapy of Let7b for targeting tumor nodules using the i.p. route with minimal off-target effects.
Improved efficacy of combination of HA-PEI-let7b and PTX in the relapsed model of EOC
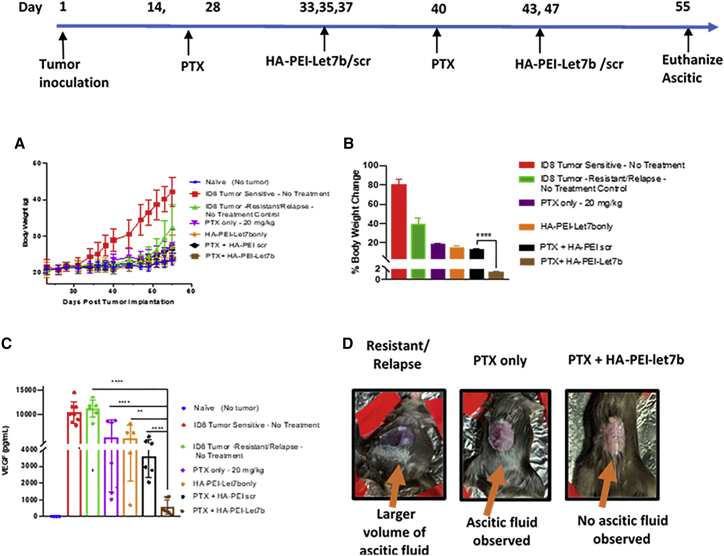
Encouraged by the results obtained from in vitro studies and the delivery potential of HA-PEI-Let7b NPs to restore the levels of Let7b in tumor tissues, we wanted to evaluate the combination therapy (HA-PEI-Let7b + PTX) for improvement in anti-tumor efficacy in vivo using the ID8-VEGF relapse model. The doses of Let7b (1 mg/kg) and PTX (20 mg/kg) were chosen based on previous studies performed by our laboratory in the ID8-VEGF model using HA-PEI NPs.12,19,24 The time line for tumor development, dosing frequency for monotherapy (PTX alone or HA-PEI Let7b alone), or combination therapy PTX HA-PEI-Let7b is shown in Figure 5. Unlike other solid tumors, in the ID8 model of ovarian cancer, tumor proliferates in the peritoneal cavity and it is difficult to measure the exact size of tumor externally. In the ID8-VEGF model of ovarian cancer, as a result of VEGF secretion, physiological changes occur in the walls of the peritoneal cavity, leading to buildup of malignant ascitic fluid in the abdomen, which in turn leads to increase in body weight. Hence, we chose to measure body weight changes as a measure of increase in buildup of malignant ascites fluid (Figures 5A and 5B). For the ID8-VEGF tumor-sensitive group, 4–5 weeks post tumor growth inoculation, we observed an increase in body weight (∼80% compared with the naive group) by day 55, as shown in (Figures 5A and 5B). For the ID8 tumor-resistant/relapse group, which was treated with 3 doses of PTX, body weight changes happen approximately after day 49 (7 weeks), indicating a relapse, and continues to grow until day 55 (∼40% body weight compared with naive). For all monotherapy groups (PTX only or Let7b only) and PTX + HA-PEI-scr, body weight changes observed are less than ∼20%, indicating the inhibition of ascitic fluid formation due to therapy. However, for the combination PTX + HA-PEI-Let7b, the body weight changes are less than 2% (Figure 5B), and the magnitude of reduction in body weight is statistically significant compared with the PTX + HA-PEI-scr group. Figure 5D shows representative images of the ID8 tumor bearing mice, showing abdominal bloating in control and reduction of the same in the PTX and PTX + HA-PEI-Let7b groups on day 55.
Figure 5.
Improved efficacy of combination of HA-PEI-Let7b and PTX in the ID8-PTX-MDR/relapse model of ovarian cancer
Timeline for ID8-VEGF relapse development using repeat dosing of PTX (3 doses, 8 mg/kg, i.p.) on day 14, 21, and 28, followed by dosing regimen for combination therapy (PTX, 20 mg/kg, and HA-PEI-Let7b, 1 mg/kg, i.p.) (A) Graph showing tumor growth using body weight change on y axis as a measure of ascitic fluid buildup post tumor inoculation for various control and treatment groups. (B) Graph showing % body weight change normalized to naive (no tumor) on the y axis group on day 55 (euthanasia) for various control and treatment groups (n = 8 per, n = 4 for naive). (C) Measurement of anti-tumor efficacy of combination therapy using VEGF ELISA.VEGF levels (pg/mL) measured from supernatants of ascitic fluid on day 55 (n = 6–8 per group). (D) Representative images of ID8-VEGF-resistant tumor bearing mice on day 55 post tumor inoculation. Untreated control group (resistant/relapse) showing ascitic fluid buildup leading to abdominal bloating, PTX group only with ascites in the peritoneal cavity, and PTX + HA-PEI-Let7b group with no ascites formation (p values for A, B, and C: ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, and∗∗p < 0.01 were obtained from Student’s t test by comparing PTX + HA-PEI- Let7b with other groups)
It has been demonstrated that high levels of VEGF contribute to i.p. dissemination of ovarian cancer, leading to the subsequent growth of i.p. tumors and formation of malignant ascites.25 Hence, we have chosen to quantify VEGF levels in ascitic fluid as biomarker for tumor growth and to evaluate the impact of combination therapy as a measure of anti-tumor efficacy (Figure 5C). As expected, both the control groups (sensitive and resistant) show very high levels of VEGF (mean ∼11,000 pg/mL), but when monotherapy was given (PTX, 20 mg/kg, or HA-PEI-Let7b, 1 mg/kg) or negative control for combination therapy (PTX + HA-PEI-scRNA), a 2.2-fold reduction in VEGF levels (mean ∼5000 pg/mL) is observed, which is still high compared with the naive or no treatment group (mean ∼10 pg/mL). Interestingly the combination therapy of PTX + HA-PEI-Let7b (5 doses) shows an almost 20-fold reduction in VEGF levels (∼500 pg/mL). Although these results are encouraging, there is room for improvement in reduction, which can be achieved by increasing the number of doses of HA-PEI-Let7b. Unfortunately, this was not possible in the current study because of two reasons A. ascitic fluid was observed in in PTX + HA-PEI-scr for the fifth dose (day 47) and B. the control groups (sensitive and resistant/relapse) had to be euthanized due to high volume (>10 mL) of ascitic fluid buildup by day 55, as per the rules of our animal protocol.
Discussion
In this article, we have evaluated the effect of combination therapy of a clinically relevant miRNA (Let7b miRNA) delivered using HA-PEI NPs and PTX to address the unmet medical need of relapse of tumor growth in advanced-stage ovarian cancer. ID8-VEGF is a syngeneic, immunocompetent, and orthotopic mouse model that has been developed to simulate several clinically relevant characteristics of human disease, such as metastasis of tumor to peritoneal cavity (i.p. space), buildup of malignant ascites fluid, and molecular features of elevated levels of VEGF in ascites.7,26 In our first in vivo study, we have extended this model to recapitulate the relapse of tumor using a repeat dosing approach, where cycles of chemotherapy (weekly doses of PTX) were given and therapy was stopped to evaluate if the tumor relapses. We then systematically performed molecular profiling using qPCR to confirm upregulation of mRNA levels of oncogenes, such as VEGF-A; survivin; and MDR genes, such as ABCB1a and ABCB1b, in the relapse/PTX treated group compared with the tumor bearing untreated/control group. The panel of genes was chosen based on the gene expression profiling studies performed on resistant clinical ovarian cancer. Interestingly, the magnitude of these changes is a similar order of magnitude to what has been observed in cases of clinical samples using data from clinical literature.4 Although a few other genes (EGFR, CD44, and HIF1-alpha) were also evaluated, the expression levels were not elevated compared with the control group, and this could be because the control group is also tumor bearing, which could have already elevated levels of these genes in the tumor cells. It is worth noting that the changes in the genes were seen not only in the tumor cells from the ascitic fluid but also in the metastatic tumor nodules of the PTX treated/relapse group.
In order to understand the miRNAs associated with the relapse of tumor in our animal model, we performed miRNA profiling of the tumor cells derived from the ascitic fluid using NanoString assay. We were able confirm a number of validated downregulated miRNAs, such as tumor suppressor Let7b, miR 451, and miR 148a, and similarly upregulated miRNAs, such as miR 551b ,miR 205, miR 125 b, etc., were also observed to have similar trends in clinical studies.27,28 Although there were a number of other miRNAs that were differentially expressed from our data, we have prioritized clinical relevance and miRNAs with highest magnitude of fold changes and statistical significance as key criteria to further filter down miRNA for combination therapy. We then wanted to exploit such information obtained the miRNA profiling analysis in order to prevent relapse of tumor by supplementing miRNA that are downregulated and are associated with chemoresistance and poor prognosis. In the current study, based on the NanoString assay, Let7b miRNA levels were found to be the most significantly downregulated in the relapse group. Based on the clinical evidence in EOC patients, decreased expression of Let7b correlates with aggressive, high-grade tumors and poor prognosis; accordingly, high Let7b levels are associated with better prognosis and prolonged patient survival.29 Functional studies have shown that in EOC, let-7 downregulates BRCA1, RAD51, PARP, and IGF1, resulting in increased sensitivity to chemotherapy, and longer progression-free survival and overall survival.30 Let-7 induction of chemosensitivity seen in vitro and in vivo is due to inhibition of LIN28A/B, STAT3, E2F1, IMP1, and chemoresistance genes MDR1, ABCG2, and MMP9.31,32
Taking together (1) the role of Let7b as tumor suppressor miRNA, (2) its ability to target multiple oncogenes, and (3) most importantly, its role in resistance development, we believe that strategy of tumor delivery of Let7b would be result in cellular reprogramming of miRNA, leading to prevention of relapse of cancer in resistant ovarian cancer. To achieve tumor delivery of miRNA, we have chosen HA-PEI as delivery vehicle. HA-PEI is a modular and versatile delivery system, which has the capability to deliver a wide variety of nucleic acids, such as siRNA, miRNA, and plasmid DNA, by modifying the ratio of the polymer (HA-PEI) to the nucleic acid. Previous studies conducted by our laboratory have demonstrated that HA-PEI/HA-PEG containing siRNA NPs can achieve efficient gene silencing by targeting genes MDR1 and survivin-resistant ovarian and lung cancer mouse models, respectively.24,33 More recently, multiple studies have demonstrated that exogenous delivery of miRNA 125b, and miRNA 34a using HA-PEI NPs in combination with PTX has shown improved anti-tumor efficacy in aggressive mouse of models of ovarian and lung cancers.19,21 Let7b mimics have been recently explored as a therapeutic strategy to suppress tumor growth using different delivery approaches, such as encapsulation of Let7b mimics using neutral lipid emulsion (NLE) in combination with miR34 and lipid conjugated Let7b in previous studies of mouse models of NSCLC (lung cancer).12,34 However, in aggressive cases of ovarian cancer, we believe that Let7b alone would not be enough and hence have decided to combine with PTX as a combination therapy. Results from in vitro studies in ID8-VEGF cells have shown that intracellular delivery Let7b was able to significantly (10-fold) decrease the IC50 of ID8-VEGF cells that were pre-dosed with PTX. We were also able to show downregulation of validated targets of Let7b, such as HMGA2, VEGF-A, KRAS, etc., which have been shown to improve the sensitivity of ID8-VEGF cells to PTX. To successfully translate this approach in vivo, miRNA mimic needs to be delivered to the tumor nodules, and, to this end, we were able to show that 3 doses of HA-PEI-Let7b NPs are able to restore Let7b miRNA levels in tumor nodules compared with naive (no tumor) mice. In the same study, we were able to demonstrate that the increased levels of Let7b were also able to downregulate mRNA levels of above-mentioned oncogenes in vivo.
We then focused on evaluation of combination therapy of HA-PEI-Let7b and PTX on anti-tumor efficacy in the relapse model of ID-VEGF ovarian cancer. The therapeutic strategy was to supplement the downregulated Let7b exogenously using dosing HA-PEI-Let7b NPs (between day 28 and 49) via the i.p. route of administration. We chose to use quantitate VEGF levels as biomarker for efficacy, because VEGF levels in ascitic fluid are correlated with advanced tumor stages and elevated VEGF ascites levels were negatively correlated with patient survival.35,36 The results indicate that the combination therapy (HA-PEI-Let7b + PTX) was successful in not only reducing the ascitic fluid buildup but also reducing the levels of VEGF (∼500 pg/mL) by almost 10-fold compared with monotherapy (PTX or HA-PEI-Let7b). While the combination therapy did not completely reduce the VEGF levels to what is usually observed in benign peritoneal fluid in humans (median,184.5 pg/mL), these levels were certainly lower than the range of VEGF levels (2575–4650 pg/mL) from ascites of clinical ovarian cancer. We believe that there is still room for improvement by increasing the number of doses of HA-PEI-Let7b (1mg/kg) beyond to maintain lower levels of VEGF. Additionally, combining the delivery of other miRNAs, such as miR 451 or miR 148a, which has shown to be downregulated from profiling studies, may be able to improve the efficacy by targeting genes, such as CARF and c-Met. However, this requires extensive evaluation both in vitro and in vivo to make sure the combinations of miRNA are safe and do not lead off-target effects.
Materials and methods
ID8-VEGF cell culture
ID8-VEGF ovarian cancer cells were obtained as a gift from Dr. Michael Goldberg (Dana-Farber Cancer Institute). ID8-VEGF cells were cultured in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS), 5% penicillin streptomycin/S, 1% L-glutamine, and 0.5% sodium pyruvate in 5% CO2 at 37°C. All the reagents were obtained from Thermo Fisher Scientific (Carlsbad, CA).
Synthesis of HA-PEI and formulation of HA-PEI-Let7b nanoparticles
HA-PEI-Let7b NPs were synthesized, as described previously.37 Briefly, 100 mg sodium hyaluronate, 20–40 kDa (Lifecore Biomedical, Chaska, MN), was modified by a coupling reaction with 15 mg of branched poly(ethyleneimine) (bPEI ), 10 kDa (Polysciences, Warrington, PA), using 50 mg of N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, Thermo Fisher Scientific, Rockford, IL) and 50 mg of sulfo-N-hydroxysuccinimide (NHS). For formation of Let7b, miRNA mimic (Thermo Fisher Scientific, Carlsbad, CA) was dissolved in nuclease-free water and added to the HA-PEI polymer (1:27 weight ratio) to form HA-PEI-Let7b NPs.
Development of the relapsed model of ID8-VEGF syngeneic orthotopic model of EOC
All mice used in the below studies were purchased from Charles River Laboratories (Wilmington, MA). Female C57BL/6 mice (8–10 weeks old) were inoculated with 4 million ID8-VEGF cells through i.p. injection. For the development of the relapse model, tumor inoculated mice were dosed with 8 mg/kg of PTX on days 14, 21, and 28 post tumor inoculations, and the treatment was stopped. PTX (Sigma-Aldrich, St. Louis, MO) was solubilized in 50/50 (v/v) Cremophor EL/Ethanol (EMD Serono, Billerica, MA) at a concentration of 6 mg/mL and diluted in PBS for dosing at 8 mg/kg. Relapse of tumor was monitored by total body weight and the buildup of the ascitic fluid in the abdomen for the ID8-VEGF tumor bearing untreated, PTX treated, and naive groups.
Molecular profiling of relapsed/MDR model of EOC
In order to evaluate if the effects of repeat dosing of PTX altered the molecular profile of mRNA and miRNA of tumor cells, animals from both control and PTX (n = 6) were sacrificed 8 weeks post tumor inoculation. An i.p. lavage was performed using ice-cold 1× PBS in animals that did not develop tumor and, for animals that showed abdominal bloating, ascitic fluid was drained using a 5 mL syringe loaded with a 25G needle. Tumor nodules in the i.p. space and on the diaphragm were also collected. Ascitic fluid samples were subjected to red blood cell (RBC) lysis and stained with CD45 antibody (BD Biosciences, San Jose, CA), followed by cell sorting equipment (BD FACSAria Fusion, BD Biosciences), and 1 million cells per sample were collected using CD45 negative and GFP positive as gates for sorting cells. The collected cells for each were split into two equal parts (0.5 million) for mRNA expression using qPCR and miRNA expression profiling using NanoString assay.
Tumor MDR gene expression analysis using qPCR
Total RNA was isolated from the sorted cells using the mirVANA RNA isolation kit (Thermo Fisher Scientific, Carlsbad, CA), using the vendor’s protocol. Isolated total RNA was quantified using UV absorbance at 260 nm (NanoDrop 2000, Thermo Fisher Scientific, Wilmington, DE). TaqMan-based gene expression assays (qPCR) were to evaluate the mRNA expression of VEGF, EGFR, CD44, survivin, etc., and genes involved in MDR (ABC transporters and related genes). Briefly, 1 μg of RNA was converted cDNA using reverse transcription (High Capacity Reverse Transcription kit, Thermo Fisher Scientific, Carlsbad, CA) followed by qPCR reaction (TaqMan gene expression master mix) using Light Cycler 480 (Roche, Branford, CT). B-actin was used a housekeeping gene. Ct values obtained for each sample were converted to relative expression, and fold changes (normalized to the untreated/control group) were obtained using the ΔΔCt method; p values were obtained using Student’s t test (GraphPad Prism, San Diego, CA). All reagents for performing qPCR were obtained from Thermo Fisher Scientific (Carlsbad, CA). TaqMan primers (assay ID) were VEGFA (Mm00437306_m1), KRAS (Mm00517492_m1), survivin (Mm00599749_m1), HMGA2 (Mm04183367_g1), Abcb1a (Mm00440761_m1), Abcb1b (Mm00440736_m1), IMP1 (Mm00501602_m1), MRP1 (Mm00456156_m1), and B-actin (Mm00607939_s1).
Global miRNA expression analysis using NanoString
We used NanoString nCounter platform (NanoString Technologies, Seattle, WA) to evaluate miRNA expression with the nCounter mouse miRNA Panel v. 3 (NanoString Technologies). The panel included unique oligonucleotide tags for over 800 highly curated human miRNAs (from miRbase v. 21) and five housekeeping genes for normalization of expression between samples. Each sample was analyzed by using 100 ng of total RNA for processing and consecutive hybridization (21 h at 65°C) to the probe pairs, consisting of a reporter probe, which carries the signal on its 5′ end, and a capture probe, which holds biotin on its 3′ end. After hybridization, sample cleanup and counting were performed, according to the manufacturer’s instructions. Total RNA was extracted using RNAzol (Molecular Research Center), according to the manufacturer’s instructions. RNA quality was assessed using Agilent TapeStation 4200 (Agilent, San Jose, CA). All RNA samples had an RNA integrity number (RIN) above 7. Raw counts were analyzed using NanoString nSolver software, v. 4.0. We calculated geometric means of negative ligation controls plus two SDs for all samples. All count values below this threshold were excluded from normalization. After that, all the normalization steps were performed, according to manufacturers' instructions. Only miRNAs that were expressed in 75% or more of samples in at least one group were further analyzed. We then selected miRNAs that showed statistical significance with p < .05. Differentially expressed miRNAs in each group were detected by one-way ANOVA using Partek Genomics Suite v. 7, and false discovery rate (FDR) was calculated for multiple comparisons using the q-value, FDR ≤0.1. The significance level for all analyses was set at p < .05.
In vitro evaluation of Let7b delivery with HA-PEI nanoparticles and transfection in ID8-VEGF cells
HA-PEI-Let7b NPs were prepared by complexing HA-PEI conjugate with Let7b mimics at various doses (0.1–100 nM). For evaluation of combination therapy (HA-PEI-Let7b + PTX), ID8 -VEGF cells were plated in a 96-well plate and, for development of resistance, ID8-VEGF cells were treated with a 2 uM dose of PTX every other day in a T75 flask and subsequently plated in 96 wells (1,500 cells/well), 12 h before transfection. Let7b transfection was performed at 24 h, followed by PTX dosing in various combinations. Scrambled miRNA (Thermo Fisher Scientific, Carlsbad, CA) was used as a negative control; Let7b alone and PTX alone for each dose were also used to assess the effect of combination PTX + Let7b; % cell viability for each well was evaluated using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega, Madison, WI), per vendor protocol. Different treatment conditions normalized to untreated cells were performed using MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay 48 h post PTX treatment.
Downregulation of mRNA levels of validated Let7b targets using HA-PEI-Let7b nanoparticles in ID8-VEGF cells
The experiment steps involved plating of ID8 control cells at a seeding density of 50,000 cells/well in 6-well plates followed by transfection of Let-7b at various doses (1–200 nM) and scRNA along non-transfected control with n = 3 biological replicates; 48 h post transfection, dosing media were removed and cells were detached using trypsin and pelleted, followed by RNA isolation using the mirVANA total RNA isolation kit (the RNA thus obtained was reverse transcribed and gene expression of specific genes, VEGFA, KRAS, survivin, HMGA2, Abcb1b [MDR 1], IMP-1, MRP1, and B-actin, was amplified, detected, and quantified by RT-qPCR using B-actin as a housekeeping gene).
In vivo delivery and distribution of Let7b in tumor and peritoneal organs upon intraperitoneal administration with HA-PEI nanoparticles
Tumor development and HA-PEI NPs were prepared, as described previously; 48 h post 3 doses of HA-PEI-Let7b (1 mg/kg) or HA-PEI-scRNA (negative control miRNA), all the mice were euthanized using a CO2 chamber. Ascitic fluid/i.p. lavage was collected and stored at −80°C for further processing. Tumor nodules were observed on the diaphragm for the resistant/relapse control group, and, for the other groups, diaphragm tissue was collected. Total RNA was isolated from all mice using a mirVANA miRNA isolation kit. mRNA levels of target genes were quantified using TaqMan gene expression assays (RT-qPCR), as described earlier. B-actin was used as endogenous control, and % mRNA expression levels were obtained by normalizing the respective mRNA expression levels observed in the resistant control group. The mRNA target gene expression panel for Let7b mRNA targets was the same as previously described. For quantifying the absolute levels of Let7b (copies/nanograms of total RNA), TaqMan Let7b miRNA assay (assay ID 002619) was employed, according to vendor protocol.
Therapeutic efficacy of combination Let7b and PTX in vivo in a relapsed/MDR model of EOC
To evaluate the efficacy of combination therapy, an orthotopic and syngeneic relapse/resistant model of ovarian cancer was developed in C57BL/6 mice, as mentioned in the previous section. Both naive (no tumor inoculation) and ID8-VEGF-sensitive groups (no PTX treatment at day 14, 21, and 28) were also included; 5 doses of HA-PEI-Let7b (1 mg/kg) NPs were administered i.p. for the combination therapy (PTX + HA-PEI-Let7b) as well as Let7b only in the ID8-VEGF relapse groups. Similarly, PTX (20 mg/kg) was administered i.p. on day 40 for the combination (PTX + HA-PEI-Let7b) and monotherapy (PTX) groups alone for ID8-VEGF relapse groups of mice. PTX + HA-PEI-Scr (1 mg/kg) was used a negative control. Different control and treatment groups were euthanized between days 53 and 55, using a CO2 chamber. An i.p. lavage was performed using ice-cold 1× PBS in animals that did not develop tumor and, for animals that showed abdominal bloating, ascitic fluid was drained using a 5 mL syringe loaded with 25G needle. Following i.p. lavage, peritoneal cavity was opened, and tumor nodules in i.p. space, diaphragm, and other organs, such as liver, lung, and spleen, were collected and frozen at −80C until further processing. VEGF levels (pg/mL) from ascitic fluid isolated from all the groups were quantified using Mouse VEGF Quantikine ELISA kit (R&D Systems, Devens, MA), per the vendor’s protocol.
Statistical analyses and reporting
Statistical significance for various datasets (except NanoString) was evaluated by Student’s t test using GraphPad Prism (GraphPad, San Diego, CA).
Acknowledgments
Financial support for this work was provided by the United States National Cancer Institute of the National Institute of Health through grants R21-CA179652 and R56-CA198492 and the Northeastern University-Dana Farber Cancer Center Joint Program on Cancer Drug Development.
Author contributions
S.K.G.: conceptualization, method development, carrying out the experiments, data analysis, interpretation, writing – original draft, re-writing, and editing; M.R.: carrying out the experiments, data analysis, and writing; A.S.: carrying out the experiments, data analysis, and writing; M.S.T.: conceptualization, data analysis, interpretation, and manuscript editing; M.M.A.: conceptualization, supervision, data analysis, interpretation, and manuscript editing.
Declaration of interests
The authors declare that they have no affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent licensing arrangements) or non-financial interest (such as personal or professional relationships, affiliations, knowledge, or beliefs) in the subject matter or materials discussed in this manuscript.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.03.005.
Supplemental information
References
- 1.Siegel R.L., Miller K.D., Jemal A., Jemal A. Cancer statistics, 2017. CA Cancer J. Clin. 2021;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N., Noone A.M., Krapcho M., Miller D., Bishop K., Kosary C.L., Yu M., Ruhl J., Tatalovich Z., Mariotto A., et al. National Cancer Institute; 2017. SEER Cancer Statistics Review, 1975-2014. [Google Scholar]
- 3.Patch A.M., Christie E.L., Etemadmoghadam D., Garsed D.W., George J., Fereday S., Nones K., Cowin P., Alsop K., Bailey P.J., et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature. 2015;521:489–494. doi: 10.1038/nature14410. [DOI] [PubMed] [Google Scholar]
- 4.Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18:452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed N., Stenvers K.L. Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front Oncol. 2013;3:256. doi: 10.3389/fonc.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roby K.F., Taylor C.C., Sweetwood J.P., Cheng Y., Pace J.L., Tawfik O., Persons D.L., Smith P.G., Terranova P.F. Development of a syngeneic mouse model for events related to ovarian cancer. Carcinogenesis. 2000;21:585–591. doi: 10.1093/carcin/21.4.585. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Yang N., Garcia J.R., Mohamed A., Benencia F., Rubin S.C., Allman D., Coukos G. Generation of a syngeneic mouse model to study the effects of vascular endothelial growth factor in ovarian carcinoma. Am. J. Pathol. 2002;161:2295–2309. doi: 10.1016/s0002-9440(10)64505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walton J.B., Farquharson M., Mason S., Port J., Kruspig B., Dowson S., Stevenson D., Murphy D., Matzuk M., Kim J., et al. CRISPR/Cas9-derived models of ovarian high grade serous carcinoma targeting Brca1, Pten and Nf1, and correlation with platinum sensitivity. Sci. Rep. 2017;7:16827. doi: 10.1038/s41598-017-17119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennessy B.T., Coleman R.L., Markman M. Ovarian cancer. Lancet. 2009;374:1371–1382. doi: 10.1016/S0140-6736(09)61338-6. [DOI] [PubMed] [Google Scholar]
- 10.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 11.Mezzanzanica D., Bagnoli M., De Cecco L., Valeri B., Canevari S. Role of microRNAs in ovarian cancer pathogenesis and potential clinical implications. Int. J. Biochem. Cell Biol. 2010;42:1262–1272. doi: 10.1016/j.biocel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Kasinski A.L., Kelnar K., Stahlhut C., Orellana E., Zhao J., Shimer E., Dysart S., Chen X., Bader A.G., Slack F.J. A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene. 2015;34:3547–3555. doi: 10.1038/onc.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stahlhut C., Slack F.J. Combinatorial action of MicroRNAs let-7 and miR-34 effectively synergizes with erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle. 2015;14:2171–2180. doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bumcrot D., Manoharan M., Koteliansky V., Sah D.W. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganesh S., Iyer A.K., Gattacceca F., Morrissey D.V., Amiji M.M. In vivo biodistribution of siRNA and cisplatin administered using CD44-targeted hyaluronic acid nanoparticles. J. Control Release. 2013;172:699–706. doi: 10.1016/j.jconrel.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran T.H., Krishnan S., Amiji M.M. MicroRNA-223 induced repolarization of peritoneal Macrophages using CD44 targeting hyaluronic acid nanoparticles for anti-inflammatory effects. PLoS One. 2016;11:e0152024. doi: 10.1371/journal.pone.0152024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Iyer A.K., Singh A., Milane L., Choy E., Hornicek F.J., Amiji M.M., Duan Z. Cluster of differentiation 44 targeted hyaluronic acid based nanoparticles for MDR1 siRNA delivery to overcome drug resistance in ovarian cancer. Pharm. Res. 2015;32:2097–2109. doi: 10.1007/s11095-014-1602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesh S., Iyer A.K., Morrissey D.V., Amiji M.M. Hyaluronic acid based self-assembling nanosystems for CD44 target mediated siRNA delivery to solid tumors. Biomaterials. 2013;34:3489–3502. doi: 10.1016/j.biomaterials.2013.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parayath N.N., Gandham S.K., Leslie F., Amiji M.M. Improved anti-tumor efficacy of paclitaxel in combination with MicroRNA-125b-based tumor-associated macrophage repolarization in epithelial ovarian cancer. Cancer Lett. 2019;461:1–9. doi: 10.1016/j.canlet.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parayath N.N., Parikh A., Amiji M.M. Repolarization of tumor-associated Macrophages in a genetically engineered nonsmall cell lung cancer model by intraperitoneal administration of hyaluronic acid-based nanoparticles encapsulating MicroRNA-125b. Nano Lett. 2018;18:3571–3579. doi: 10.1021/acs.nanolett.8b00689. [DOI] [PubMed] [Google Scholar]
- 21.Trivedi M., Singh A., Talekar M., Pawar G., Shah P., Amiji M. MicroRNA-34a encapsulated in hyaluronic acid nanoparticles induces epigenetic changes with altered mitochondrial bioenergetics and apoptosis in non-small-cell lung cancer cells. Sci. Rep. 2017;7:3636. doi: 10.1038/s41598-017-02816-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chirshev E., Oberg K.C., Ioffe Y.J., Unternaehrer J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl Med. 2019;8:24. doi: 10.1186/s40169-019-0240-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling H., Fabbri M., Calin G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganesh S., Iyer A.K., Weiler J., Morrissey D.V., Amiji M.M. Combination of siRNA-directed gene silencing with cisplatin reverses drug resistance in human non-small cell lung cancer. Mol. Ther. Nucleic Acids. 2013;2:e110. doi: 10.1038/mtna.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy J.A., Masse E.M., Herzberg K.T., Meyers M.S., Yeo K.T., Yeo T.K., Sioussat T.M., Dvorak H.F. Pathogenesis of ascites tumor growth: vascular permeability factor, vascular hyperpermeability, and ascites fluid accumulation. Cancer Res. 1995;55:360–368. [PubMed] [Google Scholar]
- 26.Janát-Amsbury M.M., Yockman J.W., Anderson M.L., Kieback D.G., Kim S.W. Comparison of ID8 MOSE and VEGF-modified ID8 cell lines in an immunocompetent animal model for human ovarian cancer. Anticancer Res. 2006;26:2785–2789. [PubMed] [Google Scholar]
- 27.Weidle U.H., Birzele F., Kollmorgen G., Nopora A. Potential microRNA-related targets for therapeutic intervention with ovarian cancer metastasis. Cancer Genomics Proteomics. 2018;15:1–15. doi: 10.21873/cgp.20061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaluvally-Raghavan P., Jeong K.J., Pradeep S., Silva A.M., Yu S., Liu W., Moss T., Rodriguez-Aguayo C., Zhang D., Ram P., et al. Direct upregulation of STAT3 by MicroRNA-551b-3p deregulates growth and metastasis of ovarian cancer. Cell Rep. 2016;15:1493–1504. doi: 10.1016/j.celrep.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shell S., Park S.M., Radjabi A.R., Schickel R., Kistner E.O., Jewell D.A. Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. U S A. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao M., Cai J., Cai L., Jia J., Xie L., Zhu Y., Huang B., Jin D., Wang Z. Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double strand break repair. J. Ovarian Res. 2017;10:24. doi: 10.1186/s13048-017-0321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyerinas B., Park S.M., Murmann A.E., Gwin K., Montag A.G., Zillhardt M., Hua Y.J., Lengyel E., Peter M.E. Let-7 modulates acquired resistance of ovarian cancer to Taxanes via IMP-1-mediated stabilization of multidrug resistance 1. Int. J. Cancer. 2012;130:1787–1797. doi: 10.1002/ijc.26190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyerinas B., Park S.M., Shomron N., Hedegaard M.M., Vinther J., Andersen J.S., Feig C., Xu J., Burge C.B., Peter M.E. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 33.Yang X., Iyer A.K., Singh A., Choy E., Hornicek F.J., Amiji M.M., Duan Z. MDR1 siRNA loaded hyaluronic acid-based CD44 targeted nanoparticle systems circumvent paclitaxel resistance in ovarian cancer. Sci. Rep. 2015;5:8509. doi: 10.1038/srep08509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Segal M., Biscans A., Gilles M.E., Anastasiadou E., De Luca R., Lim J., Khvorova A., Slack F.J. Hydrophobically modified let-7b miRNA enhances biodistribution to NSCLC and downregulates HMGA2 in vivo. Mol. Ther. Nucleic Acids. 2020;19:267–277. doi: 10.1016/j.omtn.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudlowski C., Pickart A.K., Fuhljahn C., Friepoertner T., Schlehe B., Biesterfeld S., Schroeder W. Prognostic significance of vascular endothelial growth factor expression in ovarian cancer patients: a long-term follow-up. Int. J. Gynecol. Cancer. 2006;16:183–189. doi: 10.1111/j.1525-1438.2006.00307.x. [DOI] [PubMed] [Google Scholar]
- 36.Dalal V., Kumar R., Kumar S., Sharma A., Kumar L., Sharma J.B., Roy K.K., Singh N., Vanamail P. Biomarker potential of IL-6 and VEGF-A in ascitic fluid of epithelial ovarian cancer patients. Clin. Chim. Acta. 2018;482:27–32. doi: 10.1016/j.cca.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Parayath N.N., Amiji M.M. Preparation of hyaluronic acid-based nanoparticles for macrophage-targeted MicroRNA delivery and transfection. Methods Mol. Biol. 2020;2118:99–110. doi: 10.1007/978-1-0716-0319-2_7. [DOI] [PubMed] [Google Scholar]
- 38.Gao Y., Meng H., Liu S., Hu J., Zhang Y., Jiao T., Liu Y., Ou J., Wang D., Yao L., et al. LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum. Mol. Genet. 2015;24:841–852. doi: 10.1093/hmg/ddu502. [DOI] [PubMed] [Google Scholar]
- 39.Kovalchuk O., Filkowski J., Meservy J., Ilnytskyy Y., Tryndyak V.P., Chekhun V.F., Pogribny I.P. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 40.Ling S., Ruiqin M., Guohong Z., Ying W. Expression and prognostic significance of microRNA-451 in human epithelial ovarian cancer. Eur. J. Gynaecol. Oncol. 2015;36:463–468. [PubMed] [Google Scholar]
- 41.Gong L., Wang C., Gao Y., Wang J. Decreased expression of microRNA-148a predicts poor prognosis in ovarian cancer and associates with tumor growth and metastasis. Biomed. Pharmacother. 2016;83:58–63. doi: 10.1016/j.biopha.2016.05.049. [DOI] [PubMed] [Google Scholar]
- 42.Wang W., Dong J., Wang M., Yao S., Tian X., Cui X., Fu S., Zhang S. miR-148a-3p suppresses epithelial ovarian cancer progression primarily by targeting c-Met. Oncol. Lett. 2018;15:6131–6136. doi: 10.3892/ol.2018.8110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Sun Y.M., Lin K.Y., Chen Y.Q. Diverse functions of miR-125 family in different cell contexts. J. Hematol. Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chong G.O., Jeon H.S., Han H.S., Son J.W., Lee Y.H., Hong D.G., Lee Y.S., Cho Y.L. Differential MicroRNA expression profiles in primary and recurrent epithelial ovarian cancer. Anticancer Res. 2015;35:2611–2617. [PubMed] [Google Scholar]
- 45.Niu K., Shen W., Zhang Y., Zhao Y., Lu Y. MiR-205 promotes motility of ovarian cancer cells via targeting ZEB1. Gene. 2015;574:330–336. doi: 10.1016/j.gene.2015.08.017. [DOI] [PubMed] [Google Scholar]
- 46.Chu P., Liang A., Jiang A., Zong L. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol. Lett. 2018;15:7571–7578. doi: 10.3892/ol.2018.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.